Abstract
Background
Unbiased estimates of penetrance are challenging but critically important to make informed choices about strategies for risk management through increased surveillance and risk-reducing interventions.
Methods
We studied the penetrance and clinical outcomes of 7 breast cancer susceptibility genes (BRCA1, BRCA2, TP53, CHEK2, ATM, PALB2, and PTEN) in almost 13 458 participants unselected for personal or family history of breast cancer. We identified 242 female participants with pathogenic or likely pathogenic variants in 1 of the 7 genes for penetrance analyses, and 147 women did not previously know their genetic results.
Results
Out of the 147 women, 32 women were diagnosed with breast cancer at an average age of 52.8 years. Estimated penetrance by age 60 years ranged from 17.8% to 43.8%, depending on the gene. In clinical-impact analysis, 42.3% (95% confidence interval = 31.3% to 53.3%) of women had taken actions related to their genetic results, and 2 new breast cancer cases were identified within the first 12 months after genetic results disclosure.
Conclusions
Our study provides population-based penetrance estimates for the understudied genes CHEK2, ATM, and PALB2 and highlights the importance of using unselected populations for penetrance studies. It also demonstrates the potential clinical impact of genetic testing to improve health care through early diagnosis and preventative screening.
Multiple studies of cancer penetrance for breast cancer susceptibility genes have been conducted since the identification of BRCA1 and BRCA2 (BRCA1/2) in 1994 and 1995, respectively (1,2). Early penetrance studies were often enriched for participants with strong family histories of breast cancer (3,4). Some studies have highlighted that penetrance estimates are dependent on the method of ascertainment, and disease risks of familial cases are higher than risk estimates derived from the general population (5-10). Using breast cancer patients and their families, penetrance of BRCA1/2 by age 70 years was estimated as high as 65%-85% and 70%-84%, respectively (3,11–13). In contrast, a population-based study estimated penetrance of 52% (16%) for BRCA1 and 32% (SD = 17%) for BRCA2 by age 70 years (14). The difference highlights the importance of unbiased penetrance estimate, although the latter is more challenging to recruit a sufficiently large number of individuals with pathogenic variants in the breast cancer susceptibility genes. Unbiased estimates of penetrance are critically important to accurately estimate risk over the life course and make informed choices about strategies for risk management through increased surveillance and risk-reducing interventions including prophylactic surgery. This can only be done if population-based genetic testing is deployed.
A few studies have estimated the penetrance of BRCA1/2 pathogenic variants in the general population (14–16). Population-based penetrance estimates are not available for the other breast cancer susceptibility genes with lower frequency of pathogenic variants (eg, TP53 and PTEN), more recently identified genes (eg, PALB2), and those that likely have more moderate penetrance (eg, ATM and CHEK2). Increasingly, there is consideration of population-based genomic health screening for adults for conditions for which surveillance is effective, including breast cancer (17). Our study objective is to provide less biased estimates of breast cancer penetrance in women for the commonly assessed breast cancer susceptibility genes (18) in clinical genetic testing.
Methods
Study Cohort and Sequencing Panel
In phase III of the Electronic Medical Records and Genomics (eMERGE) network, 13 458 female participants were enrolled at 10 clinical sites and had sequencing for a panel of 109 genes (19). The focus of this analysis is 7 breast cancer susceptibility genes included on the eMERGE III panel (BRCA1, BRCA2, PALB2, PTEN, TP53, ATM, and CHEK2). All 7 genes have shown strong or moderate association with breast cancer in previous studies (9,10). Variants identified through sequencing were classified according to the American College of Medical Genetics and Genomics and Association for Molecular Pathology guidelines (20,21) with ClinGen sequence variant interpretation working group modifications for codes PM2, PM3, BA1, PP5/BP6, PS2/PM6, and PVS1, which can be found on ClinGen’s website (https://clinicalgenome.org/working-groups/sequence-variant-interpretation/). Variant classification and genetic reports were provided by Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories (19). Pathogenic or likely pathogenic (P/LP) variants were confirmed by Sanger sequencing (19). We focused on female participants in this study as the risk for breast cancer in men with P/LP variants is statistically significantly lower than that for women. Putative somatic variants (variant allele fraction in blood samples <0.3) were excluded from downstream analysis. Participants who previously knew their genetic results may be more likely to seek genetic answers after a diagnosis of cancer, so they may be more likely to have a higher risk for breast cancer or have undergone risk reduction or enhanced surveillance actions after they received their genetic results. Therefore, only women with P/LP variants who were unaware of the genetic risk were included in the penetrance and clinical impact analysis. For clinical impact analysis, the sample was further restricted to those participants who consented to return of results (RoR) (22) and did not have breast cancer before RoR. We assessed clinical impact at 12 months post-RoR. All 10 clinical sites obtained consent from participants under institutional review board–approved protocols (23).
Identification of Breast Cancer Diagnosis and Post-RoR Risk Management
The electronic health records (EHR) of participants with breast cancer susceptibility P/LP variants were manually queried at 12 months post-RoR for any history of incident breast cancer and the date recorded in the EHR to ascertain if the event occurred prior to or after RoR. We measured the post-RoR performance of the National Comprehensive Cancer Network (24,25) guideline-recommended risk management including breast cancer surveillance and prevention procedures such as breast magnetic resonance imaging (MRI), breast ultrasound, mammograms, breast cancer risk-reducing medication, and prophylactic mastectomy and oophorectomy. We also recorded breast biopsies as a diagnostic test. Records were queried for prior patient knowledge of the identified breast cancer susceptibility P/LP variants. EHR extraction was completed at each clinical site and entered into a central REDCap database (26,27).
Statistical Analysis
We used Kaplan-Meier method (28) to estimate the age-specific penetrance of breast cancer in the women with P/LP variants in each breast cancer susceptibility gene. The participants were censored at their current age or age of prophylactic mastectomy. We reestimated penetrance whenever an event (breast cancer diagnosis or censor) occurred in the curve of penetrance of breast cancer. We also estimated breast cancer penetrance by decade from 30 to 70 years. Confidence intervals (CI) were calculated using Greenwood formula (29). We used 2-sided binomial test to estimate P value and set significance level as .05. The analysis was done in R version 3.6.3.
Results
Clinical Characteristics
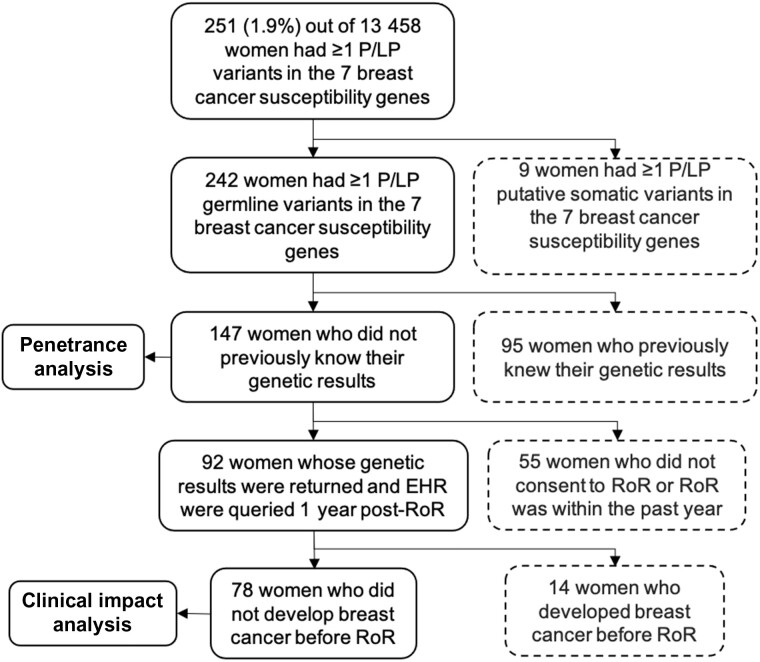
Of the 13 458 eMERGE III female individuals who were sequenced (Supplementary Table 1, available online), we identified 242 women with at least 1 P/LP variant in 1 of the 7 breast cancer susceptibility genes. A flowchart of inclusion criteria for participants is shown in Figure 1. We retained 147 women who had germline P/LP variants and did not know their genetic results for the penetrance analysis. The clinical characteristics of this female cohort are shown in Table 1 and Supplementary Figure 1 (available online). The majority (73.5%) were of European and non-Latina ancestry, followed by 15.6% African American, 8.8% Latina, and 2.7% East Asian by self-reported ancestry. The average age was 55.1 (SD = 18) years. One woman had 2 P/LP variants including 1 frameshift variant in BRCA2 and 1 missense variant in CHEK2. By the date of last chart review, 32 (21.8%, 95% CI = 15.1% to 28.5%) women had developed breast cancer, and 2 of them were diagnosed post-RoR. The average age of breast cancer diagnosis was 52.8 years (95% CI = 30.8 to 74.8), and 4 (2.7%, 95% CI = 0.8% to 5.4%) women had a prophylactic mastectomy or oophorectomy after eMERGE RoR. Although not statistically significant (P = .11), by age 50 years, a higher proportion of African American (7 out of 13, 53.8%) and Latina (5 out of 11, 45.5%) individuals developed breast cancer compared with European-ancestry individuals (20 out of 72, 27.8%), using the binomial test.
Figure 1.
Study cohorts for penetrance analysis and clinical impact analysis. Participants in the dashed box were excluded. EHR = electronic health record; P/LP = pathogenic or likely pathogenic; RoR = return of results.
Table 1.
Clinical characteristics of 147 women with germline pathogenic or likely pathogenic variants in the 7 breast cancer (BC) susceptibility genesa
| Characteristic | All 7 BC susceptibility genes | ATM | BRCA1 | BRCA2 | CHEK2 | PALB2 | PTEN | TP53 |
|---|---|---|---|---|---|---|---|---|
| No. of women | 147 | 21 | 17 | 39 | 48 | 15 | 3 | 5 |
| Mean age (SD), y | 55 (18) | 63 (13) | 43 (19) | 50 (17) | 59 (16) | 61 (19) | 32 (22) | 51 (25) |
| Race/ethnicity, No. (%) | ||||||||
| Europe | 108 (73.5) | 10 (47.6) | 15 (88.2) | 30 (76.9) | 40 (83.3) | 9 (60.0) | 2 (66.7) | 2 (40.0) |
| African American | 23 (15.6) | 6 (28.6) | 2 (11.8) | 5 (12.8) | 2 (4.2) | 5 (33.3) | 1 (33.3) | 2 (40.0) |
| Latina | 13 (8.8) | 4 (19.0) | 0 (0.0) | 2 (5.1) | 5 (10.4) | 1 (6.7) | 0 (0.0) | 1 (20.0) |
| East Asian | 4 (2.7) | 1 (4.8) | 0 (0.0) | 2 (5.1) | 1 (2.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Mean age of diagnosis (SD), y | 53 (11) | 54 (8) | 45 (4) | 52 (16) | 56 (12) | 55 (6) | NA | 41 (5) |
| BC, No. (%) | 32 (21.8) | 6 (28.6) | 3 (17.6) | 6 (15.4) | 11 (22.9) | 4 (26.7) | 0 (0.0) | 2 (40.0) |
| BC after testing, No. (%) | 2 (1.4) | 1 (4.8) | 0 (0.0) | 0 (0.0) | 1 (2.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Prophylactic mastectomy, No. (%) | 3 (2.0) | 0 (0.0) | 1 (5.9) | 1 (2.6) | 1 (2.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| RoR, No. (%)b | 78 (53.1) | 14 (66.7) | 8 (47.1) | 21 (53.8) | 31 (64.6) | 4 (26.7) | 1 (33.3) | 0 (0.0) |
Breast cancer status was ascertained by the last chart review. Genes were sorted alphabetically. One woman with 2 P/LP variants in BRCA2 and CHEK2 was included in both penetrance and clinical-impact analysis. Percentage was calculated for each gene.
Return of results (RoR) row shows the number of women who received their genetic results and did not have breast cancer before the RoR.
Variant Characteristics
The 147 women had 100 unique P/LP variants in the 7 breast cancer susceptibility genes. Fourteen variants were recurrent in this cohort (Supplementary Table 2, available online), the most frequent of which were CHEK2: c.470T>C (n = 26) and CHEK2: c.1100delC (n = 7)—2 European founder variants. The most commonly reported variant type in this cohort was putative loss-of-function (LoF) including frameshift, stop-gain, or splice for BRCA2, BRCA1, ATM, and PALB2. CHEK2 had both missense and putative LoF variants beside 2 deletions. Compared with 90% of 85 putative LoF variants that were annotated in ClinVar with at least 2 stars in review status (assertion criteria provided, multiple submitters, and no conflicts), fewer (62% of 13) missense variants were well documented in ClinVar. The numbers of women with P/LP variants and the numbers of unique P/LP variants for each gene are shown in the Supplementary Table 3 (available online). For ATM, BRCA1, BRCA2, and PALB2, more than 80% of their P/LP variants were annotated in ClinVar with 2 or 3 stars in review status. However, out of 13 P/LP CHEK2 variants in 48 women, only 7 (54%) of the variants in 7 (15%) women have been well annotated in ClinVar.
Prevalence and Penetrance
Among 147 women with at least 1 P/LP variant in the 7 breast cancer susceptibility genes, 56 (0.38%, 95% CI = 0.30% to 0.46%) unselected individuals had BRCA1/2 P/LP variants. This prevalence of 0.38% is consistent with the 0.2%-0.7% prevalence of BRCA1/2 P/LP variants reported in previous studies (9,15,30,31). The frequency of BRCA1/2 P/LP variants differs across race and ethnicity: 41.7% in the 108 European-ancestry individuals, 30.4% in the 23 African Americans, 15.4% in the 13 Latina, and 50.0% in the 4 Asians. CHEK2 P/LP variants are also common and were present in 48 (0.33%, 95% CI = 0.25% to 0.40%) female individuals without previous genetic results. Most (68.8%) of the 48 individuals had 1 of 2 common CHEK2 variants c.470T>C (n = 26) or c.1100delC (n = 7). The prevalence of individuals with the 2 CHEK2 variants ranges from 0.0% to 4.9% in European populations (Supplementary Table 4, available online). This implies CHEK2 P/LP variants might be more common than BRCA1/2 P/LP variants in certain European populations. ATM and PALB2 P/LP variants accounted for 14.3% and 10.2%, respectively, of those 147 women.
Shown in Table 2, BRCA1 and TP53 had high breast cancer penetrance and included early-onset breast cancer diagnosed before age 50 years. BRCA2, ATM, PALB2, and CHEK2 were more commonly associated with later-onset breast cancer, for which the penetrance estimates ranged from 19% to 31% by age 60 years. Even the moderate-penetrance genes conferred a measurably higher risk of breast cancer than average-risk women (32). We compared the penetrance estimated using the 147 women and penetrance reported from previous studies in Figure 1. Supplementary Table 5 (available online) summarizes published penetrance studies for the 7 breast cancer susceptibility genes (14,33–37). We also calculated the penetrance estimates using all 242 women including those who previously knew their genetic results, shown in Supplementary Figure 2 (available online). As expected, the penetrance estimated, including women with prior knowledge of a breast cancer susceptibility, is higher than the penetrance estimated using unselected women.
Table 2.
Penetrance (95% confidence interval) by decades of 6 breast cancer susceptibility genes
| Gene | <30 y | <40 y | <50 y | <60 y | <70 y |
|---|---|---|---|---|---|
| ATM | 0.0 (0.0 to 0.0) | 0.0 (0.0 to 0.0) | 10.3 (8.7 to 11.8) | 25.5 (18.9 to 32.1) | 31.2 (21.8 to 40.7) |
| BRCA1 | 0.0 (0.0 to 0.0) | 0.0 (0.0 to 0.0) | 33.3 (17.9 to 48.7) | 33.3 (17.9 to 48.7)a | 33.3 (17.9 to 48.7)a |
| BRCA2 | 0.0 (0.0 to 0.0) | 3.3 (3.1 to 3.6) | 11.1 (9.6 to 12.5) | 19.8 (15.9 to 23.6) | 19.8 (15.9 to 23.6) |
| CHEK2 | 0.0 (0.0 to 0.0) | 2.4 (2.3 to 2.5) | 7.2 (6.6 to 7.8) | 17.8 (15.2 to 20.4) | 23.6 (19.4 to 27.8) |
| PALB2 | 0.0 (0.0 to 0.0) | 0.0 (0.0 to 0.0) | 8.3 (6.9 to 9.8) | 23.0 (16.2 to 29.8) | 29.4 (19.4 to 39.4) |
| TP53 | 0.0 (0.0 to 0.0)a | 25.0 (10.9 to 39.1)a | 43.8 (8.7 to 78.8)a | 43.8 (8.7 to 78.8)a | 43.8 (8.7 to 78.8)a |
| General populationb | — | 0.6 | 2.1 | 4.5 | 7.8 |
When the number of uncensored women who had P/LP variants is below 5 (sample size is too small), the penetrance estimate is not accurate.
Cumulative risk of breast cancer in populations was derived from incidence rate of breast cancer in 10 years from the current age. The incidence rate was obtained from cancer statistics review 1975-2017 sponsored by the National Cancer Institute Surveillance, Epidemiology, and End Results program (32).
Twenty-one women had ATM P/LP variants, and 18 of them carried putative LoF variants (Supplementary Table 3, available online). Six women developed breast cancer at an average age of 54.2 years (Table 1). Three ATM variants were recurrent. Two women with the same missense variant ATM: c.7271T>G, and 1 woman with a different missense variant ATM: c.6095G>A developed breast cancer at a young average age of 48.7 years (data not shown). Our estimated penetrance increased from 10.3% (95% CI = 8.7% to 11.8%) by age 50 years to 25.5% (95% CI = 18.9% to 32.1%) by age 60 years (Table 2).
We identified 17 women with BRCA1 P/LP variants, and 3 of them had developed breast cancer at an average age of 45.0 years (Table 1). The penetrance was 33.3% (95% CI = 17.9% to 48.7%) by age 50 years (Table 2).
There were 39 women with BRCA2 P/LP variants. The number is twice that for BRCA1, a ratio consistent with previous findings (30,38). All reported P/LP BRCA2 variants in the eMERGE cohort were putative LoF. Of the 39 women, 6 had a history of breast cancer at an average age of 51.7 years (Table 1). The penetrance increased steadily from age 40 to 60 years with a cumulative risk of 19.8% (95% CI = 15.9% to 23.6%) by 60 years (Table 2).
CHEK2 P/LP variants were identified in 48 women in the eMERGE cohort. Twenty-six and 7 women carried the missense variant c.470T>C or frameshift variant c.1100delC, respectively (data not shown). The missense variant is a moderate-risk variant associated with a 1.5-fold increase in breast cancer risk (39–41). The variant c.1100delC was previously reported to be associated with breast cancer with an odds ratio above 2 (4,10,42,43). Although CHEK2 has a relatively low penetrance before age 50 years, the cumulative risk was 23.6% (95% CI = 19.4% to 27.8%) by age 70 years (Table 2).
Out of 15 women with PALB2 P/LP variants, 4 had breast cancer at an average age of 50.6 years (Table 1). All P/LP PALB2 variants were putative LoF. The penetrance increased the most between age 50 and 60 years from 8.3% (95% CI = 6.9% to 9.8%) to 23.0% (95% CI = 16.2% to 29.8%) (Table 2).
We identified 3 women with P/LP variants in PTEN. None of the women had breast cancer with the current ages of 15, 24, and 57 years. The penetrance of PTEN was not further analyzed because of the limited sample size.
Of the 5 women with TP53 P/LP variants, 3 of them had missense variants. Two women with missense variants developed breast cancer at the ages of 37.1 and 44.4 years. Although the sample size is small, breast cancer penetrance for TP53 was estimated at 43.8% (95% CI = 8.7% to 78.8%) by age 50 years (Table 2).
Clinical Impact
We limited the analysis of clinical impact after RoR to the 78 women not previously aware of the genetic results and without a breast cancer diagnosis before RoR (data not shown). Based on the chart review at 12 months post-RoR, 26 women had mammograms, 11 had breast MRI, and 5 had breast ultrasound. Three women had breast biopsies that led to the diagnosis of a new breast cancer in 2 women. From the perspective of risk reduction, 3 women had prophylactic bilateral mastectomies and 1 also had a prophylactic oophorectomy, and another woman had only a prophylactic oophorectomy. One woman started tamoxifen to reduce breast cancer risk. Overall, 33 (42.3%, 95% CI = 31.3% to 53.3%) women took 1 or more clinical actions, not including the breast biopsies, and 9 (11.5%, 95% CI = 4.4% to 18.7%) had at least 2 breast cancer surveillance procedures after RoR.
Discussion
Our study enrollment criteria were broad, and the 147 women used in the penetrance analysis were among approximately 13 000 adult female participants unselected for personal or family history of breast cancer. Therefore, our penetrance estimates apply to a general adult female population.
Across the 7 studied genes, BRCA1 and TP53 variants were associated with high penetrance, and variants in BRCA2, ATM, PALB2, and CHEK2 had more moderate penetrance. Our findings suggest that CHEK2 P/LP variants could be as prevalent as BRCA1/2 in certain populations such as the Finnish and Polish with CHEK2 founder variants.
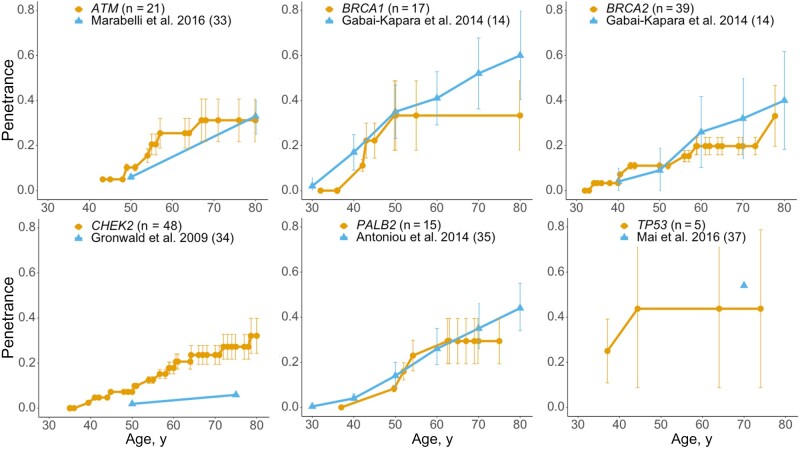
Comparing penetrance estimates from the eMERGE cohort with the previous studies (Figure 2), the penetrance by age is comparable for all genes, although there are some statistically significant differences at certain ages. For example, our estimated penetrance for ATM by age 50 years was 10.3% (95% CI = 8.7% to 11.8%), which is statistically significantly higher than 6.0% (95% CI = 4.6% to 7.4%) reported by Marabelli et al. (33), however, the absolute difference is small. The penetrance for CHEK2 in our study is higher than that reported by Gronwald et al. (34), but their study did not provide confidence intervals. Our CHEK2 estimates are similar to those estimated in a large recent study (9). These results, based on small sample sizes, should be validated when larger numbers of participants are available.
Figure 2.
Breast cancer penetrance (orange) of 6 breast cancer susceptibility genes compared with penetrance (blue) in the literature. See Supplementary Table 5 (available online) for a summary of the recent penetrance studies that were used to compare with these population-based penetrance estimates for the 6 breast cancer genes. The sample size of the Electronic Medical Records and Genomics phase III penetrance cohort is annotated in parentheses after the gene. Error bars indicate 95% confidence intervals. The 95% confidence interval of penetrance for CHEK2 and TP53 are not available from the literature. The genes were sorted alphabetically.
We highlight the importance of estimating penetrance by comparing penetrance estimates before and after removing women who knew their genetic results prior to enrollment in eMERGE. Penetrance of BRCA1/2 increased dramatically when women who knew their genetic result were included (Supplementary Figure 2, available online), suggesting use of selected populations inflates penetrance estimates. Our strategy of excluding women with previous genetic test results is less biased than previous methods using women selected for family history. However, women who were previously tested are more likely to have a family history of cancer, so our penetrance estimates may be potentially deflated by removing more women with family history of cancer compared with a general population.
Routine genetic screening could lead to improvement in long-term clinical outcomes with tailored health surveillance. Among 242 women with P/LP variants in the 7 genes, 147 (61%) of them were not previously aware of their genetic results. This research supports the feasibility of identification of women at increased risk of breast cancer on a population level.
The 7 genes we studied are included in the National Comprehensive Cancer Network–suggested breast cancer gene panel for cancer risk management. Among 74 women who first learned their genetic results through eMERGE, 42.3% (95% CI = 31.3% to 53.3%) had taken actions related to their genetic results within 12 months, although the uptake of mammograms (33.3%), breast MRI (14.1%), and mastectomy (3.8%) was lower than that found in a study of women with BRCA1/2 P/LP variants in which uptake of the same actions was 45.8%, 32.2%, and 3.5%, respectively, within the first year post-RoR (30). With this small sample size of 74 women, 2 new breast cancer cases were diagnosed within 12 months after RoR, demonstrating the potential impact of returning this information.
Although eMERGE III had more than 13 000 female participants sequenced, the number of women with P/LP variants in breast cancer susceptibility genes is small compared with studies selected for family history. Unbiased penetrance estimates using larger sample sizes are still needed and could impact breast cancer surveillance for women with P/LP variants in understudied genes such as PALB2 and ATM. This study only recruited living participants, so those deceased individuals with P/LP variants are not represented in our penetrance estimates.
The genes we studied are commonly tested clinically when assessing breast cancer risk (18). Other genes that are less commonly associated with breast cancer but are included with comprehensive clinical breast cancer genetic assessments including CDH1, STK11, NBN, and NF1 are not included in the eMERGE III gene panel because of the low prevalence and unclear penetrance. Future studies with much larger sample sizes are needed to estimate penetrance for genes with low population prevalence.
A large portion of women did not consent to receive their genetic results, were already aware of their results prior to participation in eMERGE III, or had developed breast cancer before RoR, limiting the sample size for the clinical outcome analysis. We only have access to data from the EHR at the eMERGE sites, therefore, clinical actions performed outside of the eMERGE sites were not assessed. One-year of follow-up may be insufficient time to assess impact, but we were limited by the project period. There was variability across eMERGE sites in how results were returned (22) that could have resulted in differences in participant understanding and actions taken. We did not assess lifestyle modifications adopted by participants to reduce cancer risk such as maintaining a healthy body weight, performing moderate physical activity, and reducing alcohol consumption.
Our study highlights the challenges of unbiased recruitment for sufficiently large numbers of people to estimate cancer risk as we consider population-based genetic testing and risk stratification. We demonstrate that genetic screening for breast cancer susceptibility genes has the potential to diagnose cancers at earlier more treatable stages with enhanced surveillance and to perform risk reducing surgeries such as prophylactic mastectomies and oophorectomies.
Funding
This work was supported by the NHGRI through the following grants: U01HG8657 (Group Health Cooperative/University of Washington); U01HG8685 (Brigham and Women’s Hospital); U01HG8672 (Vanderbilt University Medical Center); U01HG8666 (Cincinnati Children’s Hospital Medical Center); U01HG6379 (Mayo Clinic); U01HG8679 (Geisinger Clinic); U01HG8680 (Columbia University Health Sciences); U01HG8684 (Children’s Hospital of Philadelphia); U01HG8673 (Northwestern University); U01HG8701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG8676 (Partners Healthcare/Broad Institute); and U01HG8664 (Baylor College of Medicine).
Notes
Role of the funder: The funder, National Human Genome Research Institute (NHGRI), had a role in study design, collection, analysis, and interpretation of the data, writing the manuscript, and decision to submit the manuscript for publication. The program director for eMERGE at NHGRI, Rongling Li, reviewed and revised this manuscript and approved the submission of the manuscript.
Disclosures: AM Murad is a paid consultant for Concert Genetics. The other authors have no conflicts of interest to disclose.
Author contributions: Conceptualization: WKC, KDC; project administration: AF; data generation: MLGH, AHB, MSW, MES, CH, LJR, JFP, GLW, AMM, GPJ, ASG, EAR, IBS, DRC, EBL, KAL, NBH, JLW, SH, JW, NS, CW, KDC, WKC; data curation: XF, JW, NS, WKC, MLGH, AHB, MSW, MES, CH, LJR, JFP, GLW, AMM, GPJ, ASG, EAR, IBS, DRC, EBL, KAL, NBH, JLW, SH; formal analysis and visualization: XF; writing original draft: XF, JW, WKC, KDC; supervision: WKC, KDC, YS. All authors reviewed the manuscript and approved the submission of this manuscript.
Acknowledgements: The authors acknowledge the participating patients and families who generously contributed to the eMERGE III data. We thank the participating sites and coordinating centers for the participant recruitment and follow-up data collection and data curation.
Data Availability
All data are publicly available in the dbGaP repository under phs001616.v1.p2. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001616.v2.p2.
Supplementary Material
References
- 1. Narod SA, Foulkes WD.. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4(9):665–676. [DOI] [PubMed] [Google Scholar]
- 2. Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789–792. [DOI] [PubMed] [Google Scholar]
- 3. Ford D, Easton DF, Stratton M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62(3):676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meijers-Heijboer H, van den Ouweland A, Klijn J, et al.; for the CHEK2-Breast Cancer Consortium. Low-penetrance susceptibility to breast cancer due to CHEK2()1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55–59. [DOI] [PubMed] [Google Scholar]
- 5. Evans DG, Shenton A, Woodward E, et al. Penetrance estimates for BRCA1 and BRCA2 based on genetic testing in a clinical cancer genetics service setting: risks of breast/ovarian cancer quoted should reflect the cancer burden in the family. BMC Cancer. 2008;8(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Minikel EV, Zerr I, Collins SJ, et al. Ascertainment bias causes false signal of anticipation in genetic prion disease. Am J Hum Genet. 2014;95(4):371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner H, Jackson L.. Evidence for penetrance in patients without a family history of disease: a systematic review. Eur J Hum Genet. 2020;28(5):539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antoniou A, Pharoah PDP, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu C, Hart SN, Gnanaolivu R, et al. A population-based study of genes previously implicated in breast cancer. N Engl J Med. 2021;384(5):440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Breast Cancer Association Consortium. Breast cancer risk genes—association analysis in more than 113,000 women. N Engl J Med. 2021;384:428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuchenbaecker KB, Hopper JL, Barnes DR. et al. ; and the BRCA1 and BRCA2 Cohort Consortium. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402–2416. [DOI] [PubMed] [Google Scholar]
- 12. Antoniou AC, Durocher F, Smith P. et al. ; for the INHERIT BRCAs program members. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2006;8(1):R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Easton DF, Ford D, Bishop DT.. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet. 1995;56(1):265–271. [PMC free article] [PubMed] [Google Scholar]
- 14. Gabai-Kapara E, Lahad A, Kaufman B, et al. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc Natl Acad Sci USA. 2014;111(39):14205–14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abul-Husn NS, Soper ER, Odgis JA, et al. ; for the Regeneron Genetics Center. Exome sequencing reveals a high prevalence of BRCA1 and BRCA2 founder variants in a diverse population-based biobank. Genome Med. 2019;12(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grzymski JJ, Elhanan G, Morales Rosado JA, et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat Med. 2020;26(8):1235–1239. [DOI] [PubMed] [Google Scholar]
- 17. Buchanan AH, Lester Kirchner H, Schwartz MLB, et al. Clinical outcomes of a genomic screening program for actionable genetic conditions. Genet Med. 2020;22(11):1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. National Comprehensive Cancer Network. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic (version 1.2021); 2020. [DOI] [PubMed]
- 19. eMERGE Consortium. Harmonizing clinical sequencing and interpretation for the eMERGE III network. Am J Hum Genet. 2019;105(3):588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards S, Aziz N, Bale S, et al. ; for the ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Riggs ER, Andersen EF, Cherry AM, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22(2): 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiesner GL, et al. Returning results in the genomic era: initial experiences of the eMERGE network. J Pers Med. 2020;10(2):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fossey R, Kochan D, Winkler E, et al. Ethical considerations related to return of results from genomic medicine projects: the eMERGE Network (phase III) experience. J Pers Med. 2018;8(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. National Comprehensive Cancer Network. Breast cancer risk reduction (version 1.2020); 2020. https://www.nccn.org/professionals/physician_gls/pdf/breast_risk.pdf. Accessed August 1, 2020.
- 25.National Comprehensive Cancer Network. Breast cancer screening and diagnosis (version 1.2019); 2019. https://www.nccn.org/professionals/physician_gls/pdf/breast-screening.pdf. Accessed August 1, 2020.
- 26. Harris PA, Taylor R, Minor BL, et al.; for the REDCap Consortium. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaplan EL, Meier P.. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282): 457–481. [Google Scholar]
- 29. Greenwood M. A report on the natural duration of cancer. In: Ministry of Health Reports on Public Health and Medical Subjects. London: H.M.S.O; 1926:1–26. [Google Scholar]
- 30. Hao J, et al. Healthcare utilization and costs after receiving a positive BRCA1/2 result from a genomic screening program. J Pers Med. 2020;10(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lippi G, Mattiuzzi C, Montagnana M.. BRCA population screening for predicting breast cancer: For or against? Ann Transl Med. 2017;5(13):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. SEER Cancer Statistics Review. Table 4.17 cancer of the female breast (invasive). 1975-2017. https://seer.cancer.gov/csr/1975_2017/browse_csr.php?sectionSEL=4&pageSEL=sect_04_table.17. Accessed August 1, 2020.
- 33. Marabelli M, Cheng SC, Parmigiani G.. Penetrance of ATM gene mutations in breast cancer: a meta-analysis of different measures of risk. Genet Epidemiol. 2016;40(5):425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gronwald J, Cybulski C, Piesiak W, et al. Cancer risks in first-degree relatives of CHEK2 mutation carriers: effects of mutation type and cancer site in proband. Br J Cancer. 2009;100(9):1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(6):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han M-R, Zheng W, Cai Q, et al. Evaluating genetic variants associated with breast cancer risk in high and moderate-penetrance genes in Asians. Carcinogenesis. 2017;38(5):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mai PL, Best AF, Peters JA, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122(23):3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maxwell KN, Domchek SM, Nathanson KL, et al. Population frequency of germline BRCA1/2 mutations. J Clin Oncol. 2016;34(34):4183–4185. [DOI] [PubMed] [Google Scholar]
- 39. Kilpivaara O, Vahteristo P, Falck J, et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer. 2004;111(4):543–547. [DOI] [PubMed] [Google Scholar]
- 40. Han FF, Guo CL, Liu LH.. The effect of CHEK2 variant I157T on cancer susceptibility: evidence from a meta-analysis. DNA Cell Biol. 2013;32(6):329–335. [DOI] [PubMed] [Google Scholar]
- 41. Liu C, Wang Y, Wang Q-S, et al. The CHEK2 I157T variant and breast cancer susceptibility: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2012;13(4):1355–1360. [DOI] [PubMed] [Google Scholar]
- 42. Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71(2):432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. CHEK2 Breast Cancer Case-Control Consortium. CHEK21100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74(6):1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available in the dbGaP repository under phs001616.v1.p2. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001616.v2.p2.