Abstract
The genus Escherichia comprises five species and at least five lineages currently not assigned to any species, termed ‘ Escherichia cryptic clades’. We isolated an Escherichia strain from an international traveller and resolved the complete DNA sequence of the chromosome and an IncI multidrug resistance plasmid using Illumina and Nanopore whole-genome sequencing (WGS). Strain OPT1704T can be differentiated from existing Escherichia species using biochemical (VITEK2) and genomic tests [average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH)]. Phylogenetic analysis based on alignment of 16S rRNA sequences and 682 concatenated core genes showed similar results. Our analysis further revealed that strain OPT1704T falls within Escherichia cryptic clade IV and is closely related to cryptic clade III. Combining our analyses with publicly available WGS data of cryptic clades III and IV from Enterobase confirmed the close relationship between clades III and IV (>96 % interclade ANI), warranting assignment of both clades to the same novel species. We propose Escherichia ruysiae sp. nov. as a novel species, encompassing Escherichia cryptic clades III and IV (type strain OPT1704T=NCCB 100732T=NCTC 14359T).
Keywords: bacterial taxonomy, Enterobacteriaceae, Escherichia
Introduction
Within the genus Escherichia , five species are recognized: Escherichia coli [1], Escherichia hermannii [2], Escherichia fergusonii [3], Escherichia albertii [4] and, most recently, Escherichia marmotae [5]. It has been proposed to reassign E. hermannii to Atlantibacter hermannii [6], but this reassignment is not yet formally approved. Several other species were previously reassigned from the genus Escherichia to other genera, such as E. vulneris (now Pseudescherichia vulneris [7]), E. blattae (now Shimwellia blattae [8]) and E. adecarboxylata (now Leclercia adecarboxylata [9]). All five current Escherichia species have been associated with the potential to cause animal and/or human disease [2, 10–13]. Several Escherichia strains cannot be assigned to any of the five existing species [14]. Based on analysis of genomic data, these strains cluster into several groups, which were termed ‘ Escherichia cryptic clades’, numbered I through VI [14, 15]. Recently, cryptic clade V was formally recognized as a separate species ( E. marmotae ), leaving at least five cryptic clades that have not been delineated at the species level [5]. Here we report the novel species, Escherichia ruysiae sp. nov., isolated from faecal material of an international traveller. Escherichia ruysiae sp. nov. encompasses the closely related Escherichia cryptic clades III and IV.
Isolation and ecology
We discovered a cryptic clade IV strain in our collection, previously identified as extended-spectrum β-lactamase (ESBL)-producing E. coli as part of the combat study, which investigated the acquisition of ESBL-producing Enterobacteriaceae (ESBL-E) during international travel [16]. This isolate, OPT1704T, was further characterized in detail.
The strain was isolated from a human faecal sample provided immediately after an individual’s return from a 1 month journey to several Asian countries. No ESBL-E were detected in a faecal sample collected immediately before departure, suggesting that the ESBL gene, and possibly strain OPT1704T, were acquired during travel. The traveller reported diarrhoea during travel but no antibiotic usage. No ESBL-E were isolated in follow-up faecal samples, suggesting loss of the OPT1704T strain or the ESBL gene within 1 month after return from travel.
Genome features
The whole-genome sequence of strain OPT1704T was determined using a combination of the Illumina HiSeq and Oxford Nanopore Technologies (ONT) sequencing platforms. Strain OPT1704T was grown in liquid lysogeny broth (LB) at 37 °C. DNA for Illumina sequencing was extracted using the Qiagen Blood and Tissue kit (cat. no. 69 506, Qiagen) and the sequencing library was prepared using the Illumina Nextera XT DNA Library Preparation kit (cat. no. FC-131–1096, Illumina), both according to manufacturer’s instructions. DNA for ONT sequencing was extracted using the Qiagen MagAttract HMW DNA extraction kit (cat. no. 67 563, Qiagen) and the sequencing library was prepared using the native barcoding and ligation sequencing kits (cat. nos. EXP-NBD114 and SQK-LSK109, respectively, ONT) according to manufacturer’s instructions. The Illumina sequencing run yielded a total of 6.3×106 paired-end reads, with a mean read length of 151 bp. Default parameters were used in bioinformatic analyses unless noted otherwise. Illumina reads were filtered using fastp (using flag ‘--disable-length-filtering’, version 0.19.5 [17]) and downsampled using seqtk (version 1.3-r106, https://github.com/lh3/seqtk) to provide a theoretical coverage depth of 100× with the assumption that strain OPT1704T has a genome size of approximately 5×106 bp. The ONT sequencing run yielded a total of 2.5×104 reads, with a mean read length of 9078 bp before filtering. ONT reads were filtered on length and on read identity using Filtlong (version 0.2.0, https://github.com/rrwick/Filtlong) with Illumina reads as a reference, leaving 1.5×104 reads with a mean length of 12 580 bp. This provided a theoretical coverage depth of ~38× of ONT reads. The combined assembly using Unicycler (version 0.4.6 [18]) of Illumina and Nanopore reads resulted in a completely assembled genome, consisting of one circular chromosome (4 651 588 bp) and one circular plasmid (116 086 bp). The G+C content of the complete strain OPT1704T genome was 50.6 mol%.
Putative resistance and virulence genes were predicted from the complete genome using ABRicate (https://github.com/tseemann/abricate) with the CARD [19] and VFDB [20] databases. Strain OPT1704T harbours six resistance genes on its IncI plasmid, associated with reduced susceptibility to fluoroquinolones (qnrS1), aminoglycosides (aph [6]-Id and aph(3′′)-Ib), cephalosporins (blaCTX-M-14), trimethoprim (dfrA14) and sulphonamides (sul2), corresponding with its reduced susceptibility to fluoroquinolones (norfloxacin, MIC: 2 mg l−1 and ciprofloxacin, MIC: 0.5 mg l−1), cephalosporins (cefuroxime, MIC: >32 mg l−1 and cefotaxime, MIC: 4 mg l−1) and trimethoprim–sulfamethoxazole (MIC: >8 mg l−1), assessed using VITEK2 (bioMérieux). However, strain OPT1704T was susceptible to tobramycin (MIC: ≤1 mg l−1) and gentamicin (MIC: ≤1 mg l−1) despite the presence of aminoglycoside resistance genes aph(6)-Id and aph(3′′)-Ib. The aph(6)-Id gene encodes an aminoglycoside-modifying enzyme that mediates resistance against streptomycin [21]. The aph(3′′)-Ib gene encodes an aminoglycoside-modifying enzyme mediating resistance against tobramycin and gentamicin [22] but the aph(3′′)-Ib variant identified in strain OPT1704T possesses a Glu18Lys mutation which maps to the catalytic phosphorylase kinase domain (assessed with InterPro [23]). This could potentially inhibit enzymatic function, explaining the observed susceptibility to gentamicin and tobramycin, based on clinical breakpoints [24]. Furthermore, several putative virulence genes were predicted from the genome sequence associated with siderophore function (chuX, entS, fepABD), fimbriae (fimBCDGI), a type II secretion system (gspGHI) and capsular polysaccharide biosynthesis (kpsD). These predicted virulence genes, when present in Escherichia coli , are not typically associated with a specific clinical syndrome such as diarrhoeal disease.
Physiology and chemotaxonomy
Strain OPT1704T formed circular, grey-white colonies on a Columbia sheep (COS) blood agar plate when incubated overnight at 37 °C. No haemolysis was observed. Individual cells were observed using TEM and were rod-shaped and on average 0.7×1.9 µm in size (Fig. S1, available in the online version of this article). Bacteria were fixed with McDowell fixative (4 % v/v PFA and 1 % v/v GA in 0.1 M phosphate buffer) with 1.5 % lysine acetate (Merck) for 4 h and postfixed with 1 % osmium tetraoxide (Electron Microscopy Sciences) for 1 h. Afterwards, the bacteria were dehydrated using an ethanol series and embedded in Epon 812 (Ladd Research). Copper grids covered with formvar were used to collect 60–70 nm sections made using a Leica EM FC6 ultramicrotome. Sections were stained with uranyl acetate (Merck) and lead citrate (Laurylab). Electron micrographs were collected using an FEI Tecnai T12 Biotwin electron microscope operated at 120 kV and equipped with an EMSIS Xarosa camera. Subsequently, we tested motility using the hanging-drop method, oxidase presence using an oxidase strip (cat. no. 40 560, Sigma Aldrich) and catalase presence using H2O2 [25]. The strain was shown to be Gram-stain-negative, non-motile, oxidase-negative and catalase-positive. The strain was capable to grow in the absence of oxygen. On COS blood plates, it showed growth in the temperature range of 20–42 °C. The strain was also able to grow in NaCl concentrations ranging from 0–6 % w/v in lysogeny broth overnight at 37 ˚C, but not at NaCl concentrations from 7–10 % w/v (1 % steps). MALDI-TOF (Bruker) and VITEK2 (BioMérieux) systems both identified strain OPT1704T as E. coli with high confidence scores (score >2 for MALDI-TOF and ‘Excellent identification’ for VITEK2, see Supplemental information for MALDI-TOF spectrum). Comparison of the output of the VITEK2 biochemical test with published biochemical reactions of other Escherichia species revealed that E. ruysiae sp. nov. OPT1704T is distinct from other Escherichia species based on a combination of biochemical markers (Table 1) [2–5, 26].
Table 1.
Comparison of biochemical markers which differentiate E. ruysiae sp. nov. from other Escherichia species
Data for E. albertii, E. coli, E. fergusonii and E. marmotae summarised from literature [2–5, 26]. + and – indicate that ≥85 % of tested strains is positive or negative for that biochemical marker, respectively.
|
ONPG |
+ |
+ |
+ |
+ |
+ |
− |
|
Lysine decarboxylase |
− |
+ |
+ |
+ |
− |
+ |
|
Ornithine decarboxylase |
+ |
+ |
+* |
+ |
+ |
− |
|
Fermentation of: |
||||||
|
Adonitol |
− |
− |
− |
+ |
− |
− |
|
d-Xylose |
− |
− |
+ |
+ |
+ |
+ |
|
Cellobiose |
− |
− |
− |
+ |
+ |
− |
|
d-Sorbitol |
+ |
− |
+ |
− |
− |
+ |
*50–85 % of E. coli possess this biochemical property.
16s rRNA gene and whole-genome phylogeny
Next, we calculated 16S rRNA sequence similarities, average nucleotide identity (ANI) values and digital DNA–DNA hybridization (dDDH) values between strain OPT1704T and type strains of the other Escherichia species, representative genomes of the other three Escherichia cryptic clades and S. enterica serovar Typhimurium (Table 2). Representative genomes for the Escherichia cryptic clades I, II, III and VI were selected from Enterobase [27], using the genomes with the highest contiguity. Clades VII and VIII in Enterobase only consisted of a single strain and were not used in further analyses. We used three separate tools to calculate ANI (fastANI [28], OrthoANIu [29] and ANI calculator from Enveomics [30]). Multiple ANI calculation algorithms were employed to increase confidence in the genomic species delineation, as different ANI algorithms can output different ANI values. In this study, calculated ANI values were similar across ANI calculation algorithms. We also included calculation of the dDDH values between strains using the DSMZ Genome-to-Genome Distance Calculator [31]. The output of formula 2 of the DSMZ Genome-to-Genome Distance Calculator was used, as recommended by the authors of the tool. 16S rRNA genes were extracted from whole genomes using barrnap (version 0.9, https://github.com/tseemann/barrnap) and single nucleotide polymorphisms (SNPs) between strains were counted using snp-dists (version 0.6, https://github.com/tseemann/snp-dists). Extracted 16S rRNA gene segments were 1538 bp long for all strains and were manually aligned and checked. The alignment is provided in the supplementary material.
Table 2.
Comparison of strain OPT1704T 16S rRNA and whole-genome sequence with type strains of E. albertii , E. coli , E. fergusonii , E. marmotae , representative genomes of Escherichia cryptic clades I, II, III and VI and S. enterica serovar Typhimurium
In bold are the values that warrant assignment of strain OPT1704T to a novel species (<98.7 % 16S rRNA sequence similarity, <95–96 % ANI, <70 % dDDH). ANI, average nucleotide identity; dDDH, digital DNA–DNA hybridization.
|
E. ruysiae sp. nov. OPT1704T |
|||||
|---|---|---|---|---|---|
|
16S rRNA sequence similarity (%) |
ANI (%, fastANI) |
ANI (%, OrthoANIu) |
ANI (%, ANI calculator Enveomics) |
dDDH (%) |
|
|
NBRC 107761T |
98.6 |
90.0 |
90.0 |
89.2 |
39.8 |
|
ATCC 11775T |
98.7 |
92.8 |
92.4 |
92.0 |
48.3 |
|
ATCC 35469T |
98.9 |
89.4 |
88.2 |
89.7 |
36.7 |
|
E. hermanni NCTC 12129T |
98.0 |
80.2 |
77.7 |
80.1 |
21.4 |
|
HT073016T |
98.9 |
92.2 |
92.2 |
91.4 |
47.1 |
|
Typhimurium LT2T |
97.5 |
82.1 |
80.7 |
81.8 |
24.0 |
|
Escherichia cryptic clade I 89–3506 |
99.0 |
92.5 |
92.1 |
91.8 |
47.8 |
|
Escherichia cryptic clade II MOD1-EC7253 |
99.2 |
92.0 |
91.7 |
91.0 |
45.5 |
|
Escherichia cryptic clade III E4694 |
99.7 |
96.6 |
96.5 |
96.3 |
70.8 |
|
Escherichia cryptic clade VI UHCL_3L |
98.4 |
91.6 |
91.7 |
91.3 |
45.9 |
Strain OPT1704T showed 98.7–98.9 % 16S rRNA sequence similarity to E. coli ATCC 11775T, E. fergusonii ATCC 35469T and E. marmotae HT073016T, which would not warrant assignment to a novel species based on the current threshold for species delineation (less than 98.7 % sequence similarity [32]). However, the threshold for species delineation on the basis of 16S rRNA sequence has changed often and thresholds of up to 99 % sequence similarity have been proposed previously [33]. In contrast, ANI analysis and dDDH did support assignment of strain OPT1704T to a novel species, together with the representative strain of Escherichia cryptic clade III (Table 2). The analyses also suggested that strain OPT1704T falls within the genus Escherichia . This novel species, encompassing both Escherichia cryptic clades III and IV, was assigned E. ruysiae sp. nov. with strain OPT1704T as the proposed type strain.
Assigning a novel species to a particular genus is challenging and currently no clear guidelines exist. Several approaches have been proposed, such as phylogenomics [32] or counting shared genes [34]. Our phylogenomic analyses show OPT1704T clusters closely with other Escherichia species, and clusters further away from E. hermannii and S. enterica . This clustering pattern is well supported by the bootstrapping analysis, for both the alignment 16S rRNA genes and 682 concatenated core genes. Another commonly used approach is calculating the percentage of conserved proteins (POCP) [34]. Strain OPT1704T had the highest POCP with organisms in the genus Escherichia , clearly above the ‘universal’ cutoff of 50 % shared proteins. This cutoff seems to be inadequate for Enterobacterales: Salmonella enterica and Escherichia hermannii showed a POCP of more than 60 % with all true Escherichia strains (Table S1). As the original POCP approach was developed using only 17 genera and has not been verified for Enterobacterales, the high rate of genetic exchange in this order [35] may necessitate an alternative POCP cutoff to the previously proposed cutoff of 50 %.
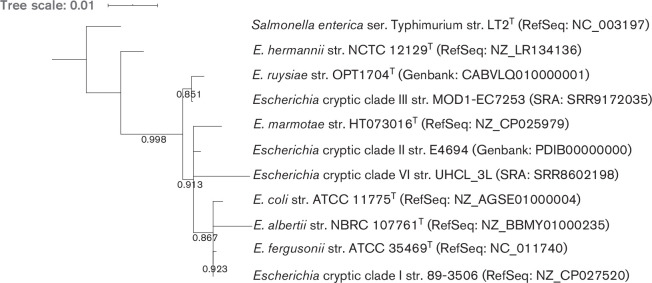
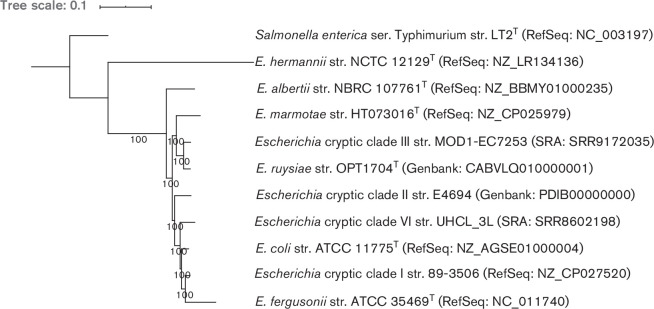
To gain a better understanding of the genus Escherichia , we produced two phylogenies, based on 16S rRNA sequence (Fig. 1) and on an alignment of 682 core genes (Fig. 2). In short, rRNA genes were predicted from whole genomes using barrnap (version 0.9, https://github.com/tseemann/barrnap) and a tree was generated using FastTree (version 2.1.10 [36]). For the core gene alignment, genomes were first annotated with Prokka (version 1.14.0 [37]) and a core gene alignment was produced using Roary (version 3.12.0 [38]) and mafft (version 7.307 [39]). The phylogeny was inferred using a generalised time reversible model using base frequencies from the SNP alignment and free rate heterogeneity (GTR+F+R4 model) in IQ-tree (version 1.6.6 [40]), as advised by ModelFinder [41]. Phylogenies were rooted on the Salmonella enterica serovar Typhimurium strain LT2T genome. Both phylogenies showed that strain OPT1704T clusters closely with the strain MOD1-EC7259 from Escherichia cryptic clade III, and away from the current Escherichia species. Comparing the number of SNPs extracted from the core genome of the strains included in Table 2 showed the same results, as the two strains with the smallest number of SNPs were strains OPT1704T (clade IV) and MOD1-EC7259 (clade III, Table S2).
Fig. 1.
16S phylogeny of Escherichia ruysiae sp. nov. OPT1704T with type strains of other Escherichia species, other Escherichia cryptic clades and Salmonella enterica serovar Typhimurium as the outgroup. Numbers indicate bootstraps on a scale of 0 to 1. Phylogeny available at https://itol.embl.de/tree/14511722611226771596704407.
Fig. 2.
Phylogeny based on 682 concatenated core genes including Escherichia ruysiae OPT1704T with type strains of other Escherichia species, other Escherichia cryptic clades and Salmonella enterica serovar Typhimurium as outgroup. Numbers indicate bootstraps on a scale of 0 to 100. Phylogeny available at https://itol.embl.de/tree/14511722611160731596708541.
Chun et al. [32] proposed that strains with >95–96 % genome-wide ANI between each other should be assigned to the same species. If cryptic clades III and clade IV would share >95–96 % ANI, this would mean both clades should be assigned to the same novel species, E. ruysiae sp. nov. To assess this for a larger number of strains than the type strains presented in Table 2, we downloaded all 65 available WGS from clade III and clade IV strains from Enterobase and compared ANI between all genomes using fastANI (version 1.1 [28]). This analysis revealed that within 33 clade III genomes, the median ANI is 98.6 % (range: 97.7–99.9 %), while within 32 clade IV genomes, the median ANI is 98.9 % (range: 98.6–99.9 %; Table S3). Between clade III and clade IV genomes, the median ANI is 96.5 % (range: 96.1–96.8 %). This suggests clades III and IV should be assigned to the same novel species, E. ruysiae sp. nov. Subsequently, based on ANI analysis we selected 10 representative clade IV genomes and 10 representative clade III genomes to be analysed using the TYGS platform [42]. The TYGS platform employs dDDH estimation and 16S and core genome phylogenetics to define species and subspecies within a given set of genomes, with an upload limit of 20 user-provided genomes. The TYGS analysis also indicated that clade III and clade IV should be assigned to a single species, but could be delineated into two separate subspecies (table S4). Currently, no IJSEM guidelines exist for the delineation of subspecies based on genomic data. However, E. ruysiae sp. nov. could potentially be delineated further into two subspecies (representing the current clades III and IV, respectively) in the future, after a type strain for cryptic clade III has been identified. In the meantime, we propose to term clades III and IV genomic lineages of E. ruysiae sp. nov.
Finally, we annotated the genomes of the strains provided in Table 2 using the EggNOG database [43] and extracted categories of cluster of orthologous genes (COG categories). Strain OPT1704T did not encode a different profile of COG categories compared to other Escherichia type strains (Table S5). Possibly, a gene ontology analysis which includes more genomes from all species might elucidate the different functional profiles, but this is out of the scope of the current study.
Based on phenotypic and genotypic data presented above, the niche for E. ruysiae sp. nov. cannot be exactly defined yet. Although OPT1704T was isolated from human faeces, earlier studies have indicated that strains belonging to E. ruysiae sp. nov. do not adhere well to human-derived cell lines [44]. This finding is highlighted by the fact that we could not detect OPT1704T anymore using ESBL microarray a month after we first detected it, although this might also be caused by loss of the ESBL gene. In conclusion, it seems that the human gut is not the primary niche for E. ruysiae sp. nov.
Description of Escherichia ruysiae sp. nov.
Escherichia ruysiae (ruy′si.ae N.L. gen. n. ruysiae named after Anna Charlotte Ruys, Professor of Microbiology at the University of Amsterdam from 1940 to 1969).
Cells are Gram-stain-negative, facultatively anaerobic, non-sporulating, non-motile rods with a size of approximately 1×2 µm. Colonies are circular, convex, grey-white and semi-transparent when grown overnight at 37 °C on COS agar plates. The species is catalase-positive and oxidase-negative and grows at temperatures between 20 and 42 °C and NaCl concentrations between 0 and 6 % w/v. In the VITEK2 GN biochemical test set it yields positive results for β-galactosidase, d-glucose, maltose, d-mannitol, d-mannose, d-sorbitol, trehalose, sucrose, d-tagatose, γ-glutamyl-transferase, fermentation glucose, tyrosine arylamidase, succinate alkalinization, α-galactosidase, ornithine decarboxylase, courmarate, β-glucoronidase, 0/129 resistance (Comp. Vibrio.) and Ellman and negative for Ala-Phe-Pro-Arylamidase, adonitol, l-pyrrolydonyl-arylamidase, l-arabitol, cellobiose, H2S production, β-N-acetyl glucosaminidase, glutamyl arylamidase Pna, β-glucosidase, β-xylosidase, β-alanine arylamidase Pna, l-proline arylamidase, lipase, palatinose, urease, citrate (sodium), malonate, 5-keto-d-gluconate, l-lactate alkalinization, α-glucosidase, β-N-acetyl-galactosaminidase, phosphatase, glycine arylamidase, lysine decarboxylase, l-histidine assimilation, Glu-Gly-Arg-arylamidase, l-malate assimilation and l-lactate assimilation (Table S6).
The type strain, OPT1704T (=NCCB 100732T=NCTC 14359T), was isolated from faecal material of an international traveller returning from Asia.
The 16S rRNA sequence is deposited in ENA under accession LR745848. Raw Illumina and Nanopore whole-genome sequencing data, as well as the complete genome assembly are deposited under project PRJEB34275.
Supplementary Data
Funding information
The COMBAT study was funded by Netherlands Organization for Health, Research and Development (ZonMw; 50-51700-98-120) and EU-H2020 programme (COMPARE, 643476).
Acknowledgements
The authors would like to thank Rob Weijts and Patricia Brinke for their help in phenotypic characterization of type strain OPT1704T of Escherichia ruysiae sp. nov., and Arie van der Ende, Thomas Roodsant and Kees van der Ark for the helpful discussions. We thank SURFsara (www.surfsara.nl) for the support in using the Lisa Compute Cluster.
The members of the COMBAT consortium, in alphabetical order: Maris S. Arcilla, Martin C.J. Bootsma, Perry J. van Genderen, Abraham Goorhuis, Martin Grobusch, Jarne M. van Hattem, Menno D. de Jong, Damian C. Melles, Nicky Molhoek, Astrid M.L. Oude Lashof, John Penders, Constance Schultsz, Ellen E. Stobberingh, Henri A. Verbrugh.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The COMBAT study was approved by the Medical Research Ethics Committee, Maastricht University Medical Centre (METC 12-4-093). All participants provided written informed consent.
Footnotes
Abbreviations: ANI, average nucleotide identity; COS, Columbia agar+sheep blood; dDDH, digital DNA–DNA hybridization; ESBL, extended-spectrum β-lactam; ONT, Oxford Nanopore Technologies; POCP, percentage of conserved proteins; SNP, single nucleotide polymorphism; WGS, whole-genome sequencing.
One supplementary figure and six supplementary tables are available with the online version of this article.
References
- 1.Castellani A, Chambers AJ. Manual of tropical medicine. William Wood. 1919 [Google Scholar]
- 2.Brenner DJ, Davis BR, Steigerwalt AG, Riddle CF, McWhorter AC, et al. Atypical biogroups of Escherichia coli found in clinical specimens and description of Escherichia hermannii sp. nov. J Clin Microbiol. 1982;15:703–713. doi: 10.1128/JCM.15.4.703-713.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer JJ, Fanning GR, Davis BR, O'Hara CM, Riddle C, et al. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol. 1985;21:77–81. doi: 10.1128/JCM.21.1.77-81.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huys G, Cnockaert M, Janda JM, Swings J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int J Syst Evol Microbiol. 2003;53:807–810. doi: 10.1099/ijs.0.02475-0. [DOI] [PubMed] [Google Scholar]
- 5.Liu S, Jin D, Lan R, Wang Y, Meng Q, et al. Escherichia marmotae sp. nov., isolated from faeces of Marmota himalayana. Int J Syst Evol Microbiol. 2015;65:2130–2134. doi: 10.1099/ijs.0.000228. [DOI] [PubMed] [Google Scholar]
- 6.Hata H, Natori T, Mizuno T, Kanazawa I, Eldesouky I, et al. Phylogenetics of family Enterobacteriaceae and proposal to reclassify Escherichia hermannii and Salmonella subterranea as Atlantibacter hermannii and Atlantibacter subterranea gen. nov., comb. nov. Microbiol Immunol. 2016;60:303–311. doi: 10.1111/1348-0421.12374. [DOI] [PubMed] [Google Scholar]
- 7.Alnajar S, Gupta RS. Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infection, Genetics and Evolution. 2017;54:108–127. doi: 10.1016/j.meegid.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Priest FG, Barker M. Gram-Negative bacteria associated with brewery yeasts: reclassification of Obesumbacterium proteus biogroup 2 as Shimwellia pseudoproteus gen. nov., sp. nov., and transfer of Escherichia blattae to Shimwellia blattae comb. nov. Int J Syst Evol Microbiol. 2010;60:828–833. doi: 10.1099/ijs.0.013458-0. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K, Sakazaki R, Kosako Y, Yoshizaki E. Leclercia adecarboxylata gen. nov., comb. nov., formerly known as Escherichia adecarboxylata . Curr Microbiol. 1986;13:179–184. doi: 10.1007/BF01568943. [DOI] [Google Scholar]
- 10.Liu S, Feng J, Pu J, Xu X, Lu S, et al. Genomic and molecular characterisation of Escherichia marmotae from wild rodents in Qinghai-Tibet plateau as a potential pathogen. Sci Rep. 2019;9:1–9. doi: 10.1038/s41598-019-46831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ooka T, Seto K, Kawano K, Kobayashi H, Etoh Y, et al. Clinical Significance of Escherichia albertii . Emerg Infect Dis. 2012;18:488–492. doi: 10.3201/eid1803.111401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo T, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 2003;5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 13.Savini V, Catavitello C, Talia M, Manna A, Pompetti F, et al. Multidrug-Resistant Escherichia fergusonii: a case of acute cystitis. J Clin Microbiol. 2008;46:1551–1552. doi: 10.1128/JCM.01210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walk ST. The "Cryptic" Escherichia . EcoSal Plus. 2015;6 doi: 10.1128/ecosalplus.ESP-0002-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangiredla J, Mammel MK, Barnaba TJ, Tartera C, Gebru ST, et al. Draft genome sequences of Escherichia albertii, Escherichia fergusonii, and strains belonging to six cryptic lineages of Escherichia spp. Genome Announc. 2018;6:e00271–18. doi: 10.1128/genomeA.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, et al. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (combat study): a prospective, multicentre cohort study. Lancet Infect Dis. 2017;17:78–85. doi: 10.1016/S1473-3099(16)30319-X. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2004;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundin GW, Bender CL. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1993;59:1018–1024. doi: 10.1128/AEM.59.4.1018-1024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ojdana D, Sieńko A, Sacha P, Majewski P, Wieczorek P, et al. Genetic basis of enzymatic resistance of E. coli to aminoglycosides. Adv Med Sci. 2018;63:9–13. doi: 10.1016/j.advms.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters Version 9. 2019.
- 25.Leber AL. Clinical Microbiology Procedures Handbook. John Wiley & Sons; 2020. [Google Scholar]
- 26.Abbott SL, O'Connor J, Robin T, Zimmer BL, Janda JM. Biochemical properties of a newly described Escherichia species, Escherichia albertii . J Clin Microbiol. 2003;41:4852–4854. doi: 10.1128/JCM.41.10.4852-4854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z, Alikhan N-F, Mohamed K, Fan Y, Achtman M, et al. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-R LM, Konstantinidis KT. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016;4:e1900v1 [Google Scholar]
- 31.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 33.Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 34.Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, et al. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, et al. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat Commun. 2020;11:3602. doi: 10.1038/s41467-020-17278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 38.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vignaroli C, Di Sante L, Magi G, Luna GM, Di Cesare A, et al. Adhesion of marine cryptic Escherichia isolates to human intestinal epithelial cells. Isme J. 2015;9:508–515. doi: 10.1038/ismej.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.