Significance
Caloric restriction (CR) increases life and health span, but our understanding of the evolutionary basis of the effect remains unclear. For many years, the disposable soma hypothesis provided the main evolutionary explanation, suggesting animals under restriction divert resources away from reproduction toward somatic maintenance. This idea was recently challenged by the “clean cupboards” hypothesis, suggesting life extension under CR is a coincidental by-product of energy conservation. This paper tests between these ideas by looking for common signatures in the metabolomic response across multiple tissues to graded CR levels in mice. There was no evidence for widespread increases in somatic protection but instead largely idiosyncratic responses in individual tissues. The common pathways identified appeared more consistent with the clean cupboards hypothesis.
Keywords: calorie restriction, aging, evolution, life span, metabolomics
Abstract
The evolutionary context of why caloric restriction (CR) activates physiological mechanisms that slow the process of aging remains unclear. The main goal of this analysis was to identify, using metabolomics, the common pathways that are modulated across multiple tissues (brown adipose tissue, liver, plasma, and brain) to evaluate two alternative evolutionary models: the “disposable soma” and “clean cupboards” ideas. Across the four tissues, we identified more than 10,000 different metabolic features. CR altered the metabolome in a graded fashion. More restriction led to more changes. Most changes, however, were tissue specific, and in some cases, metabolites changed in opposite directions in different tissues. Only 38 common metabolic features responded to restriction in the same way across all four tissues. Fifty percent of the common altered metabolites were carboxylic acids and derivatives, as well as lipids and lipid-like molecules. The top five modulated canonical pathways were l-carnitine biosynthesis, NAD (nicotinamide adenine dinucleotide) biosynthesis from 2-amino-3-carboxymuconate semialdehyde, S-methyl-5′-thioadenosine degradation II, NAD biosynthesis II (from tryptophan), and transfer RNA (tRNA) charging. Although some pathways were modulated in common across tissues, none of these reflected somatic protection, and each tissue invoked its own idiosyncratic modulation of pathways to cope with the reduction in incoming energy. Consequently, this study provides greater support for the clean cupboards hypothesis than the disposable soma interpretation.
Calorie restriction (CR) is the most robust and repeatable approach for slowing the rate of aging, leading to extensions of both health and life span (1, 2). First discovered over 100 y ago (3), its impacts are conserved across a broad range of taxa from yeast to nonhuman primates (4). Nevertheless, despite this long history and decades of work, the molecular mechanisms that underpin its effects remain unclear and disputed (5). For the past 10 y, our group has been exploring the impacts of graded levels of CR on the phenotypic responses of mice. The details of this project can be found on the Open Science Framework (https://osf.io/9yath/). This body of work was motivated by analyses that showed the impact of restriction on life span in rodents is linearly related to the level of restriction, up to at least 65% restriction (6, 7). Hence, we reasoned that whatever mechanisms underlie the life span effect, they must also increase in a linear fashion with the level of restriction over this range (8). A second aspect of this design was the observation that mice under restriction are generally given their food once per day. They then consume it over a relatively short period, after which they starve until the next ration of food is delivered. This is consequently a form of “time-restricted feeding” (TRF), which has been shown to have life span impacts independent of calorie intake. Indeed, it has been suggested that the impact of CR may be mostly due to this TRF effect. Consequently, we included in our experiments a control group that had ad libitum (AL) access to food for only 12 h per day (8).
A major critique of studies of CR is that most designs involve animals simply being given less food relative to their baseline intake. This achieves the aim of restricting calories but also means the animals receive less of all of the main macronutrients (protein, fat, and carbohydrates). Studies in Drosophila have suggested that the impact of CR may be due to restriction of protein not calories (9). Similar conclusions were reached in studies of mice (10). However, these conclusions are at odds with several CR studies in rats and mice where the level of protein was compensated in the diets so that as calories fell, the level of protein intake did not, and the life span impact of CR in these cases was not affected (11). In other words, CR without protein restriction produced the same life span benefits. Acknowledging this debate, in our experiments with graded CR, we also included a second series of diets where mice were exposed to a fixed level of calories but increasing protein restriction to match that on the CR diets. This allowed us to ascertain which of the responses to CR might be caused by protein restriction alone.
Thus far, our work on these mice has detailed the phenotypic responses to graded restriction in terms of the impacts on body composition (12), circulating hormones, glucose homeostasis and oxidative stress (13), body temperature and torpor (8), physical activity and behavior (13–15), cellular senescence in the colon (4), alterations in basal metabolic rate (16), gene expression profiles by RNA-seq in the hypothalamus (17, 18), liver (19), and white adipose tissue (20), and metabolomic responses in the liver (21), plasma (22), brown adipose tissue (BAT) (23), and brain (cerebellum) (24). This has led to an unprecedented overall picture of the responses to graded levels of CR in a single group of animals (Dataset S1).
Under restriction, animals experience an immediate imbalance between their incoming energy supply and energy expenditure. They sense this via changes in peripheral hormone levels, including leptin and insulin. Detected in the hypothalamus, these changes set off a cascade of neuropeptide responses that coordinate how the animals react. At the morphological level, this includes withdrawing fat from the fat stores to fuel the energy shortfall, growing the alimentary tract, and thereby increasing the energy extracted from the ingested food. The animals also reduce the sizes of their lean tissues, which reduces their energy demands and brings their intake and expenditure back into line. Metabolic responses (addressed in the current paper in detail) create further energy savings and a reduction in body temperature, including in the higher levels of restriction periods of time spent in torpor. Total physical activity did not change, but the patterning of this activity was radically altered. Mice under greater levels of restriction showed increasingly intense levels of activity in the run-up to food delivery each day (food anticipation activity). Understanding this pattern of changes has inspired a hypothesis regarding the underlying evolutionary mechanism regarding the CR effect (25).
Traditionally, to explain the evolutionary significance of the CR impact on life span, researchers have turned to the disposable soma hypothesis (DSH). According to the DSH, life span extension comes about because of a reallocation of energy away from reproduction toward somatic maintenance. It is assumed that this reallocation extends survival until the period of caloric deficit is over, and reproduction can resume, and thereby maximizes fitness. This idea was first put forward by Holliday (26) and Masoro and Austad (27). Later, Shanley and Kirkwood (28) examined the suggestion in a more quantitative manner and showed that it could work, provided certain conditions held (reproductive overhead or an adverse effect of famine on juvenile survival). In our hypothesis (25), we suggested that the only goal of an animal under restriction is to conserve energy so that its energy demands fall to meet its energy supply. An animal not doing this will exhaust its reserves and rapidly die. This interpretation suggests there is no long-term strategic goal vis-à-vis reallocation of energy use between reproduction and survival. Rather the longer-term life span benefits come about as an unintended by-product of the primary goal of making an energy balance. This idea was called the “clean cupboards” hypothesis (25) and is so named from an analogous situation of someone locked in their home receiving daily food supplies that suddenly become inadequate to meet demands. In that circumstance the person would eat all the food in their kitchen cupboards. Coincidental then to making an energy balance, they would get a nice set of clean cupboards, but the point is this would just be an epiphenomenon of making a balance. There was no strategic goal to clean the cupboards. The clean cupboards are like the life span impact of CR. One benefit of this model is that it also allows us to understand why CR also has a range of detrimental impacts, for example on ability to fight off parasitic infections (29, 30) and impaired wound healing (31).
In the current paper, we bring together data from the previous metabolomics work we have completed in mice under graded CR (21–24) to perform an integrated multitissue evaluation of the responses to CR. The main goal was to identify which pathways are modulated in common across all the tissues, with a view to potentially evaluate the disposable soma interpretation of CR, versus the clean cupboards idea. Specifically, we predicted that if the DSH idea were correct, we might expect a coordinated metabolic response across all four tissues to enhance tissue protection. This could, for example, be based on reducing free-radical damage, which has historically been a popular idea for why organisms age and die (6, 32), but could involve any aging-related process. In contrast, the clean cupboards idea also predicts some coordinated responses centered on energy conservation but would also potentially involve many idiosyncratic tissue-specific responses based around conserving energy in any available pathway, and hence a largely uncoordinated response across the individual tissues that could in some cases be harmful, as long as they still saved energy.
Methods
The methodology with respect to metabolomics analysis has been published previously for each tissue: liver (21), plasma (22), BAT (23), and brain (cerebellum) (24).
Experimental Design.
The principal characteristics of the study are described below. Full details of the overall design and rationale have been reported elsewhere (12). The 49 20-wk-old male mice were submitted a baseline period of 14-d monitoring, prior to introducing the restricted or AL diets. The mice were individually housed and randomly allocated to six treatment groups: 12AL, 24AL, and 10%, 20%, 30%, and 40% CR. All mice (except for the 24AL group) were exposed to the same 12-h feeding regime whereby food was only available during the period of darkness, and any remaining food was removed at lights on (06:30 h). The 24AL mice had food AL 24 h per day. The animals were fed a high carbohydrate open-source diet (D12450B: Research Diets) that contained 20% protein, 70% carbohydrate, and 10% fat (by energy). The exact restriction was determined based on food intake of each individual mouse over a 2-wk baseline period. Mice were fed CR (or AL) diets for 12 wk, before being killed at 32 wk of age. The animals were culled between 14:00 and 18:00 h by a CO2 overdose. All samples were stored at –80 °C prior to analysis. Our dataset consisted of metabolomics data from technical triplicates of individual tissue samples for all four tissues across the six different feeding groups.
Animals.
The C57BL/6J male mice were purchased from Charles River. All procedures were reviewed and approved by the University of Aberdeen Welfare and Ethical Review Board and under Home office project license (PPL 60/4366 held by J.R.S.), following ethical approval of the protocols by the local ethical review committee. Additional information about the procedures and measures conducted on these mice can be found in the first article of this series (12).
Spectrophotometric and Chromatographic Techniques.
BAT samples were analyzed using hydrophilic interaction liquid chromatography to detect metabolites in both positive and negative ionization modes. This was carried out on a Dionex UltiMate 3000 RSLC system (Thermo Fisher Scientific) using a ZIC-pHILIC (zwitterionic ion chromatography–hydrophilic interaction chromatography) column. Brain samples were analyzed using hydrophilic interaction liquid chromatography (HILIC), which was carried out on a Dionex UltiMate 3000 RSLC system (Thermo Fisher Scientific) using a ZIC-pHILIC column. For the mass spectrometry (MS) analysis, a Thermo Orbitrap Exactive (Thermo Fisher Scientific) was operated in polarity switching mode. Liver samples were analyzed with liquid chromatography–MS using an Orbitrap Exactive mass spectrometer at the Glasgow Polyomics facility. The ZIC-pHILIC platform detects mainly polar molecules, and nonpolar lipids can be identified in the wash-through. Plasma samples were analyzed with dual chromatography–Fourier transform mass spectrometry using electrospray ionization in the positive mode. To enhance metabolite detection, we used dual chromatography, utilizing both anion exchange and reverse-phase (C18) columns to separate samples. A switching valve allowed data to be collected alternately between the AE and C18 column. Also, a Thermo LTQ-FT mass spectrometer (linear ion trap with a Fourier transform ion cyclotron resonance MS detector) (Thermo Fisher) was set to collect data between 85 and 850 m/z (mass–charge ratio).
MS Data Processing.
BAT.
A total of 3,155 m/z (mass-to-charge ratio) features were detected in the negative ionization mode and 2,754 m/z features in the positive ionization mode. Datasets were combined, and duplicates removed, which reduced the total number of features to 5,860. After filtering, there were 2,876 m/z features present.
Brain.
We extracted m/z, retention times (RTs), and intensities for each sample for 2,754 peaks in the positive ionization mode and 3,155 in the negative ionization mode. After filtering, there were 3,259 m/z features present.
Liver.
A total of 6,550 features across all samples, including adducts, fragments, and metabolites, were detected. We filtered these features to unique metabolite chromatographic peaks, which included 505 identified and 381 unidentified metabolites. Among 886 metabolite features, 1.9% of the metabolite intensities were unclassified due to either low metabolite concentration, poor ionization, machinery limitations, including detection sensitivity and ion suppression or true lack of presence.
Plasma.
A total of 8,043 m/z features were found in the overall profile, 27% of which were detected in both the AE and C18 modes. The majority of the m/z features were unique to the AE analysis. The C18 analysis added 1,622 m/z features that were not detected by the AE separation. The m/z values were filtered based on their replication across individuals and signal-to-noise ratio. Post filtering, a total of 2,954 m/z features were present, 1,702 from AE and 1,252 from C18.
Metabolite Identification.
The package xMSannotator 1.3.1 was used to identify metabolites from the Human Metabolome Database (HMDB), Kyoto Encyclopedia of Genes and Genomes (KEGG), and LipidMaps databases. Unknown metabolites were identified using a clustering algorithm that uses m/z values, retention times, intensities, and potential adducts, and xMSannotator provides a confidence score. Metabolites with a confidence score of less than 2 were excluded from further analysis. Additionally, metabolites with multiple matches were filtered based on the difference between their theoretical and actual monoisotopic mass, and the metabolite ID with the smallest difference was kept, and others were removed.
We identified and classified the metabolite features detected in the four and three out of four tissues in the match-features analysis, using the ID from either KEGG or HMDB (putative analysis). Here, we faced a common issue in untargeted metabolomics—some features were not name identified by the database, and therefore, they were removed from the analysis.
Metabolomic Preprocessing.
Three steps were followed; first, metabolites were normalized using a log2 transformation. Second, only metabolites with a signal-to-noise ratio (SNRi = mean/sample SD) ≥ 15 were kept for analysis. Third, metabolites that were missing from 15% or more of all samples were removed. In the case of the BAT and liver missing values, we imputed using a random forest regression approach using the missForest package for metabolites that had peaks that were significant in at least one group. Furthermore, in the liver, the raw peak intensities for each sample were normalized by centering on the median for each metabolite in each sample using the Metabolomics package, then log2 transformed.
Statistical Modeling of Differential Metabolite Expression.
To detect significantly differentially expressed (SDE) metabolites between treatment groups, an empirical Bayes moderated linear model was fitted to each metabolite in all tissues. The empirical Bayes approach shrinks the estimated sample variances by borrowing information from across metabolites. Comparisons across metabolite fold changes were made between each level of CR (10%, 20%, 30%, and 40%) and 24AL relative to 12AL. P values for each comparison were adjusted using the Benjamini–Hochberg procedure using a false-discovery rate (FDR) of 20%. The package Devium for the orthogonal partial least-square discriminate analysis (O-PLS-DA) analysis was used to complete the validation steps and retrieve metabolite loadings.
Analysis of Covariance.
All the metabolite intensities were log2 transformed prior to one-way ANCOVA analysis. Levene’s test and normality checks were carried out, and the assumptions met. Regarding outliers, we used the interquartile range rule and only removed extreme values in each metabolite. A total of 38 metabolite-matches were present in all four matrices and 323 in three of four tissues. ANCOVA was used to evaluate the behavior of all the detected features (identified and unidentified features) in the four or three of four tissues (factor tissue [four or three levels] BAT, brain, liver, and plasma), while controlling for type of dietary treatment (covariate [six levels] 24AL, 12AL, 10CR, 20CR, 30CR, and 40CR), as well as their interactions (dietary treatment*tissue). The analysis was checked using SPSS Statistics, version 26, software. Furthermore, to assess the biological pathways across tissues, a regression equation analysis for the fit regression model was performed using Minitab software, version 19.
Biological Pathway Analysis.
To determine how multiple metabolites were linked in biological pathways, whether in the single tissues or across them, we performed two pathway analyses in the Ingenuity Pathway Analysis (IPA) program. In the first analysis, we investigated which pathways were correlated to the level of restriction; therefore, we entered unadjusted P values and coefficients from each comparison (values across metabolite fold changes of each level of CR [10%, 20%, 30%, and 40%] and 24AL relative to 12AL) alongside IDs from either KEGG or HMDB. Furthermore, we used mummichog software in this analysis (21–24).
To investigate similar pathways in all four tissues, we made a second analysis using IPA. We entered unadjusted P values from the ANCOVA analysis and slope value from each fit regression equation by match-group, alongside the ID of the representative metabolite from either KEGG or HMDB. The core analysis was only carried out with the metabolites that had a significant dietary treatment effect, a tissue effect, but no tissue by treatment interaction (pattern 2), in an attempt to define pathways that may be associated with common modulation of physiology across all four tissues following dietary treatment (see Results for further details of the alternative patterns resulting from ANCOVA analysis).
Results
CR Alters the Metabolome in a Graded Fashion.
As the CR level increased, the number of SDE metabolites relative to 12AL also increased in all four tissues. The numbers of metabolites that were increased relative to 12AL in each tissue was similar to the numbers of metabolites that decreased (P ≤ 0.05) (Fig. 1). However, most detected metabolites (>85%) in most tissues were unchanged relative to the restriction level. To determine pathways that changed between the 12AL group and the CR levels in each tissue, we performed pathway enrichment analyses in IPA. Fifty pathways (3 in the liver, 7 in the brain, 14 in the BAT, and 26 in the plasma) with multiple metabolites were significantly correlated to the restriction level (Dataset S2). From this analysis, we noted that these pathways are mainly associated with energy metabolism, fatty acid pathways, changes across several amino acids, nucleotide metabolism, a shift from glycolysis to lipolysis, and increased antioxidant defense. Plasma and BAT were more responsive to CR than the liver and brain since they had more significantly modulated pathways with more metabolites within each pathway.
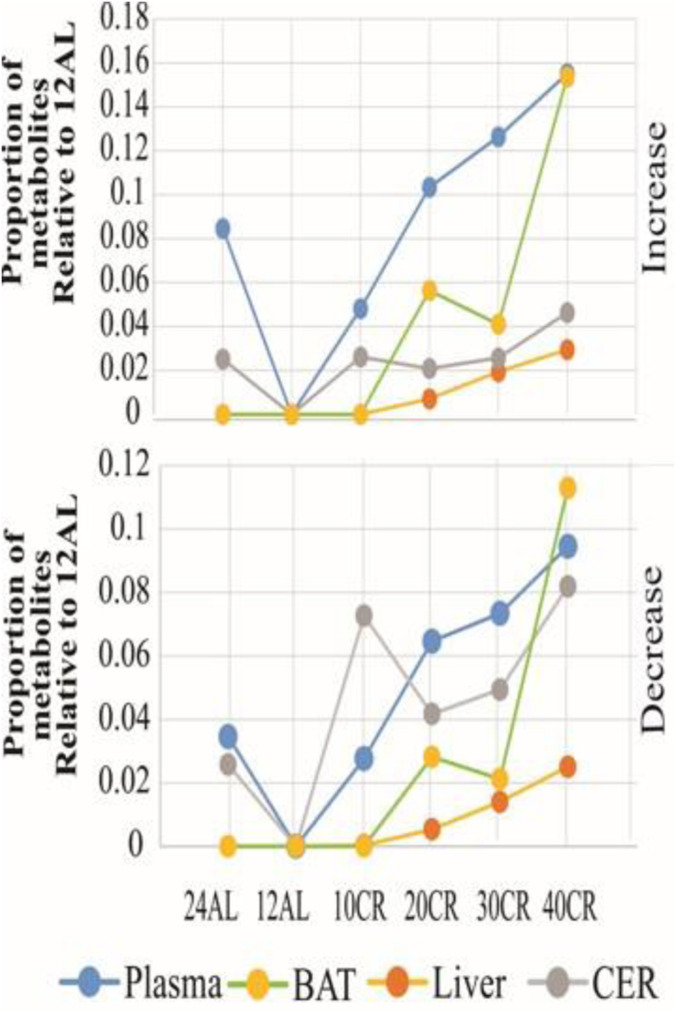
Fig. 1.
Numbers of significantly differentially expressed (SDE) metabolites in response to graded CR in the liver, BAT, brain, and plasma relative to the number of metabolites in mice exposed to the 12AL treatment. The top plot shows the number of increased and the other plot the number of decreased metabolites.
From the O-PLS-DA analysis, we were able to demonstrate significant discrimination between feeding groups. We performed the O-PLS-DA analysis of each tissue on all identified and unidentified features (Fig. 2). The models indicated that dietary treatment explained 40% (BAT) to 60% (plasma) of the features’ variance. Loadings from the O-PLS-DA model determined features that contributed to the separation of groups (Fig. 2). We showed that the metabolome of all four tissues changed significantly in response to the graded CR treatment in a graded fashion from the SDE and O-PLS-DA model analysis.
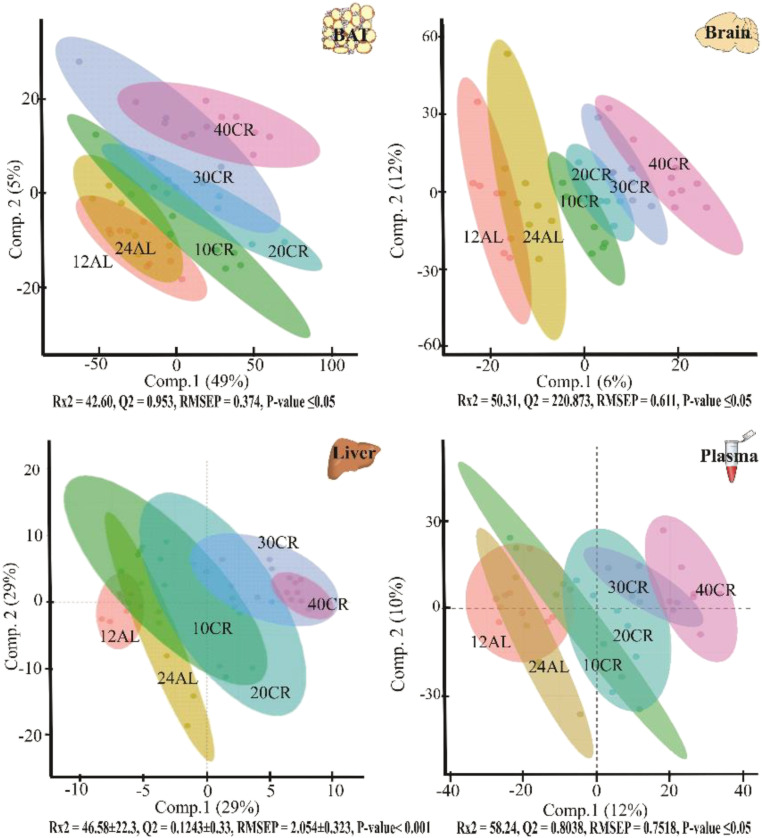
Fig. 2.
Orthogonal partial least-square discriminate analysis (OPLS-DA) demonstrates the differentiation effect of each diet group (12-h ad libitum fed: 12AL; 24-h ad libitum fed: 24AL; 10–40% calorie-restricted: 10CR, 20CR, 30CR, 40CR) on the filtered and normalized metabolites extracted from each tissue. The OPLS-DA plot showed significant separation among samples in the first two principal components (comp.1 and comp.2) based on the model quality parameters that are under each plot.
Key Modulated Pathways in Common across Tissues.
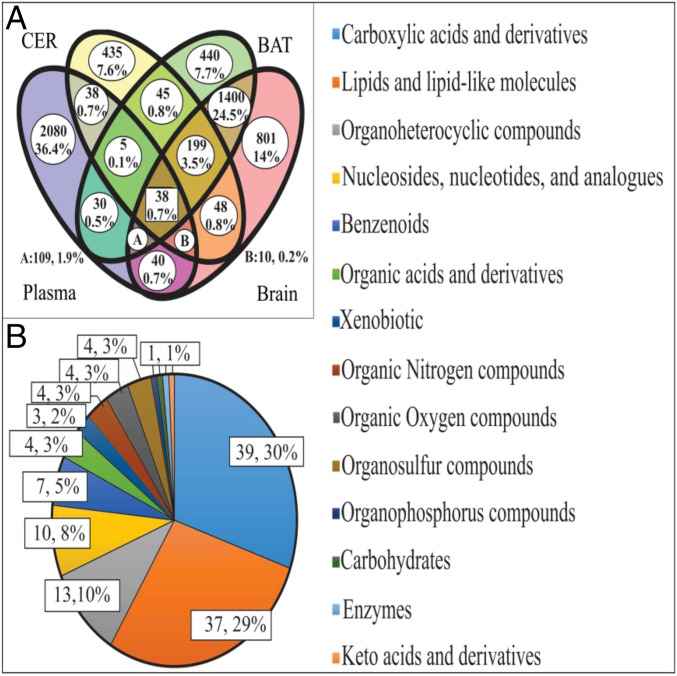
We found multiple matches of m/z features from the dataset from each tissue. Thirty-eight features were shared across all four tissues; 199 features were shared by liver, brain, and BAT; 109 features shared by plasma, BAT, and brain; 10 features shared by plasma, liver, and brain; and 5 features shared by plasma, liver, and BAT (Fig. 3A). Of the 361 m/z features shared by three out of four tissues, we were able to putatively assign IDs to 129 metabolites (35.7% of the total features) (Dataset S3). From ANCOVA analysis, we detected five patterns according to the P values from the test of between-subjects effects (Dataset S3). Pattern 1 was a significant difference between tissues, but no significant diet or diet by tissue interaction. Pattern 2 included significant effects of both tissue and diet but no interaction. Pattern 3 included a significant tissue effect, no overall diet impact, but a significant diet by tissue interaction. Pattern 4 involved significant tissue, diet, and interaction effects. Pattern five indicated no significant effects of tissue, diet, or interaction. Based on the above analysis, 24 metabolites showed responses with patterns 2, 3, and 4 across all four tissues. Of these 17 metabolites showed significant tissue by diet interaction effects. That is, while the metabolites changed in relation to the dietary treatment, the pattern of change was significantly different between the different tissues. Only seven metabolites (29%) showed a significant impact of dietary treatment consistent across the four tissues (dietary effect with no significant diet by tissue interaction: pattern 2). These were l-proline, benzamidine, l-glutamine, urate, 5-methylthioadenosine, sphinganine, and sphingosine.
Fig. 3.
(A) Venn diagram of all detected metabolites in the four different matrices using only two decimals. (B) Categories of metabolites in the second, third, and fourth patterns. Fourteen classes according to the information in the HMDB, KEGG, and PubChem were found.
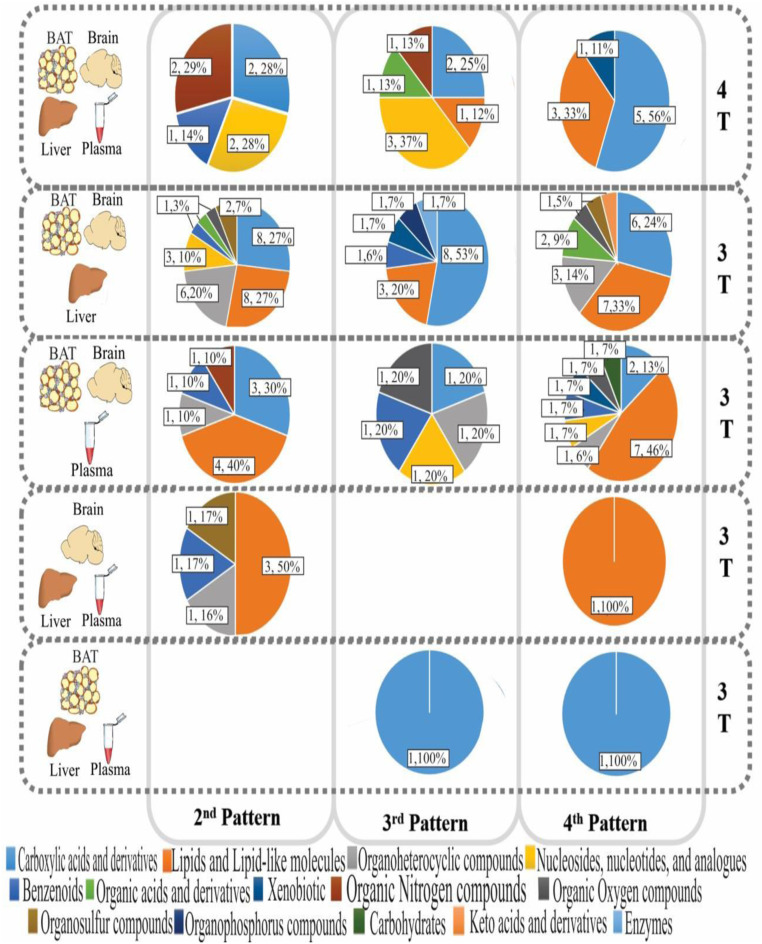
When considering metabolites detected in three out of four tissues, there were 30 metabolites showing pattern 2 in the BAT, brain, and liver; 11 in the BAT, brain, and plasma; 5 in the brain, liver, and plasma; but none in the BAT, liver, and plasma. Hence in total, we identified 52 metabolites showing pattern 2 (diet effect without a diet by tissue interaction) in either all four tissues or three out of four tissues. As with metabolites found in all four tissues, the majority (56%) of identified metabolites that were responsive to dietary treatment did not respond in the same way across tissues. That is, 37 metabolites in BAT, brain, and liver, 20 in BAT, brain, and plasma, 1 in brain, liver, and plasma, and 2 in BAT, liver, and plasma showed a significant diet by tissue interaction.
Using the HMDB, KEGG, and PubChem databases (Figs. 3B and 4 and Dataset S3), we identified 14 chemical classes of metabolite that were responsive to diet. Carboxylic acids and derivatives (amino acids were the main metabolites detected in this category, 79.5%) and lipids and lipid-like molecules (fatty acid esters were the majority molecules in this category, 27.0%) were the two main categories identified that, together, consisted of over 50% of the metabolites that were responsive to CR level.
Fig. 4.
Pie graphs show the metabolite categories and their respective percent and number of metabolites by tissue group and pattern.
Based on our previous rationale, metabolites showing pattern 2 (significant level of restriction and tissue effects, but no interaction) may be the most important metabolites associated with CR’s life span effects, because they would show elevated activation with increasing restriction in a consistent way across tissues. Parameters from regression models linking these 52 metabolites to the level of restriction are presented in Dataset S4. l-Proline was the only metabolite with a negative slope (b = −0.0512) in relation to the CR level. That is, in all tissues, l-proline levels declined as the level of restriction increased. The other six metabolites in all four tissues showed a positive relationship, and hence increased as the level of restriction increased in all tissues: benzamidine (b = 0.0389), l-glutamine (b = 0.02776), urate (b = 0.0272), 5-methylthioadenosine (b = 0.0295), sphingosine (b = 0.0343), and sphinganine (b = 0.0343). In the metabolites found in three of four tissues, N-butyryl-l-homoserine lactone (b = −0.01763; in BAT, brain, and liver), cyclo (l-leucyl-l-phenylalanyl);(3S,6S)-3-benzyl-6-(2-methylpropyl)piperazine-2,5-dione (b = −0.0334; in BAT, brain, and plasma), and deethylatrazine (b = −0.0275, in brain, liver, and plasma) were the only metabolites that declined as the level of restriction increased; the remainder showed positive slopes.
These 52 metabolites were used for IPA pathway analysis. Of the 52 metabolites, only 33 were successfully mapped (Dataset S5). Our analysis of overlapping canonical pathways identified 114 pathways (Dataset S6). The most significantly altered pathways were l-carnitine biosynthesis (4/10 molecules, −logP = 5.49), NAD (nicotinamide adenine dinucleotide) biosynthesis from 2-amino-3-carboxymuconate (3/10 molecules, −logP = 3.76), semialdehyde, S-methyl-5′-this adenosine degradation II (2/4 molecules, −logP = 3.08), NAD biosynthesis II (from tryptophan) (3/19 molecules, −logP = 2.88), and tRNA charging (4/43 molecules, −logP = 2.84). The l-carnitine biosynthesis pathway showed only metabolites up-regulated and detected in BAT, brain, and liver (γ-butyrobetaine, glycine, N6,N6-trimethyl-l-lysine, and succinic acid). The pathways NAD biosynthesis from 2-amino-3-carboxymuconate, NAD biosynthesis II (from tryptophan), and tRNA charging pathway shared molecules (AMP detected in BAT, brain, and liver, and l-glutamine that was detected in all four tissues). These three pathways were related to tryptophan metabolism, which may be connected with the cofactors’ biosynthesis. l-Proline (down-regulated in all four tissues in relation to CR level) and glycine (up-regulated in BAT, brain, and liver) were only detected in the tRNA charging pathway. In the S-methyl-5′-thioadenosine degradation II pathway, 5′-methylthioadenosine and adenine were up-regulated. Besides, these two metabolites are also linked in the spermine biosynthesis pathway, which, in turn, also has connections with the biosynthesis of cofactors and amino acid metabolism pathways (methionine metabolism).
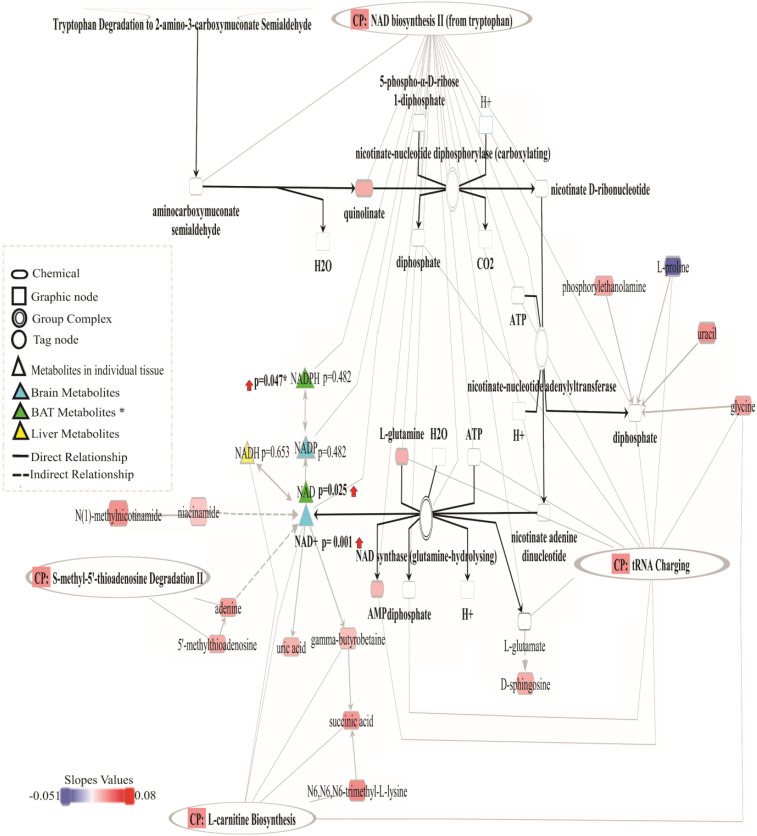
To summarize our results, we show the NAD biosynthesis from the 2-amino-3-carboxymuconate pathway in Fig. 5. It also includes metabolites from other pathways that have been shown to relate to biosynthesis and utilization of NAD.
Fig. 5.
NAD biosynthesis from 2-amino-3-carboxymuconate semialdehyde pathway together with some metabolites from the other four canonical pathways. The second pattern metabolites (except niacinamide of the fourth pattern) were colored based on the correlation coefficient. Brighter red means more up-regulation, and blue, more colorful, more down-regulation. Metabolites shown in triangle symbols were detected in individual tissue, except NADPH, caught in BAT and CER. One-way ANOVA P values are shown next to the symbol (P value with an asterisk indicates the BAT result), and red arrows means a positive increase in the CR groups compared to AL. White metabolites indicate those not identified. Abbreviations: AMP, adenosine monophosphate; ATP, adenosine triphosphate; CP, canonical pathways; CO2, carbon dioxide; H+, hydron; NAD, nicotinamide adenine dinucleotide; NAD+, nicotinamide adenine dinucleotide, oxidized form; NADH, nicotinamide adenine dinucleotide, its reduced form; NADP, nicotinamide adenine dinucleotide phosphate; and NADPH, reduced nicotinamide adenine dinucleotide phosphate.
Discussion
Why animals live longer on a restricted energy budget remains unclear and disputed (1, 5, 25, 33, 34), even though there has been much progress in understanding CR’s nutritional and molecular consequences in recent years (9, 35–37). Although research on several model organisms have shown the importance of mTOR (mechanistic target of rapamycin), insulin/IGF1-like signaling, and sirtuins as nutrient-signaling pathways that potentially mediate the effects of CR (35, 36), these studies do not answer the central issue of why activation of such systems in response to nutrient depletion might have evolved (25). Also, it is relevant to ask, if organisms have the ability to activate these longevity-related pathways, why are they not always switched on? If these pathways were active, they could allow the animals to live longer and healthier lives, decreasing the incidence of age-related diseases (such as cardiovascular disease, neurodegenerative disorders, cancer, and diabetes, and reduced frailty) (34, 38). The classical answer to this conundrum is provided by the DSH (26, 28). This suggests that the longevity assurance (or somatic maintenance) pathways are energetically costly to keep active. Hence, animals have to make a choice in how to use available resources: for somatic protection, or for reproduction. In normal circumstances, they spend some energy on reproduction ensuring survival of their genes, but at the cost of protecting their soma, leading to senescence and death. When energy supply is reduced, however, it is suggested they divert all the lower overall resources into elevated somatic maintenance, switching off reproduction, and thereby ensuring longer survival until the food supply increases sufficiently to resume reproduction. However, there are several problems with this interpretation. For example, it struggles to explain why the life span benefits increase as restriction increases and cannot explain why some aspects of somatic protection are reduced under restriction. These problems are resolved by a recent hypothesis (the clean cupboards hypothesis), which suggests that under restriction all animals are attempting to do is make an immediate energy balance and any life span effects are just an unintended by-product. With this in mind, we studied the effects of CR on metabolomic responses of four tissues, and evaluated how consistent such responses were with the classical DSH theory (26, 28), or with the clean cupboards idea (25).
Across the four tissues, we identified more than 10,000 different metabolic features. Most of these were not responsive to the CR treatment. Hence, while more metabolites were changed as the restriction level increased, even at the 40% CR level, only 25% of metabolites were altered relative to the 12AL group. Moreover, these changes included up- and down-regulated levels in approximately equal measure. Using O-PLS-DA analysis, we showed that for each tissue, it was possible to distinguish the dietary groups. Hence, for each tissue, the individuals under any particular level of restriction were reacting consistently to the restriction. That is, the animals under 40% restriction were all broadly responding in the same way. The liver of one animal under 40% restriction responded similarly to the liver of another animal under 40% restriction. However, importantly, most of the changes we observed were idiosyncratic to the particular tissue. So, while the liver of animal 1 might be responding similarly to the liver of animal 2, these livers responded very differently from the brains in the same two animals. Moreover, the changes included modulation of many tissue-specific pathways. Nevertheless, most of these pathway changes were consistent with the animals utilizing metabolites to fuel their energy metabolism. For example, they included increased levels of TCA cycle intermediaries, increased glycolysis, elevated β-oxidation of fatty acids, and amino acid degradation. These changes are consistent with the clean cupboards interpretation of the metabolic impacts of CR in that the animals appeared to be utilizing metabolites to fuel their metabolism but doing so differently in different tissues.
Only 52 metabolites showed significant changes in the liver, BAT, brain, and plasma that were consistent across the tissues where they were detected (i.e., showed no tissue by diet treatment interaction). IPA mapped only 35 of these metabolites, pointing to just five common pathways that were changed consistently among tissues. In the first place, the pathway showing the most changes was the l-carnitine biosynthesis, where γ-butyrobetaine, glycine, N6,N6,N6-trimethyl-l-lysine, and succinic acid were all increased significantly with respect to the level of restriction. Carnitine is synthesized in vivo from l-lysine and l-methionine, mostly in liver, kidney, and brain (39). Carnitines are involved in transport of fatty acids across the mitochondrial membrane, facilitating β-oxidation, and are critical for maintaining normal mitochondrial function (40). It has been previously suggested that carnitine homeostasis is compromised in several rodent models of metabolic dysregulation. A high-fat diet exacerbated age-related carnitine loss, as levels fell in multiple tissues (skeletal muscle, liver, and kidney), although plasma concentrations were unchanged (41). In rats’ liver, carnitine concentrations increased under CR, probably mediated by activation of PPARα and up-regulation of organic transporter cation (42). Furthermore, β‐oxidation of fatty acids is the primary energy production mechanism under CR and carnitine is essential for that process (40). Regarding the brain, carnitine and its relative esters (such as palmitoyl-l-carnitine and acetyl-carnitine) may be redistributed to the brain during fasting; the brain may use them for energy production or, possibly, for the delivery of acetyl groups (43). In BAT, carnitine is a crucial compound for maintaining the mitochondria’s shape in this tissue (44). This function is linked closely to the fatty acid oxidation required for thermogenesis and the expression of uncoupling protein-1. There is evidence that an excessive accumulation of lipid droplets in BAT in carnitine-deficient mice has been caused by a decreased utilization of long-chain fatty acids. Nevertheless, lipid droplets dramatically decrease in size, and histopathological and ultramicroscopic features are wholly recovered by l-carnitine treatment. Significant increases were also observed in the body temperature and carnitine concentrations of these mice (44). We found that mice showed a progressive decline in mean daily body temperature under CR (8). The possible rise of carnitine levels in BAT might be generated due to body temperature changes that we saw in our mice. The linear relationship between average temperature and the energy restriction level supports the idea that temperature changes are an integral aspect of the life span effect. In short, carnitine biosynthesis indicates elevated fatty acid oxidation since it is essential for the transfer of long-chain fatty acids across the inner mitochondrial membrane for subsequent β-oxidation. Bruss et al. (45) observed that in mice, the increase of fatty acid oxidation was almost entirely accounted for by the rise in the endogenous fatty acid synthesis under CR. We have shown previously that graded CR levels lead to reductions in body mass and altered body composition related to the utilization of both body fat and structural tissue (12). Hence, up-regulation of this pathway shown here is consistent with the idea that animals prioritize fat utilization in their metabolism. Alternatively, this up-regulation of molecules in the carnitine biosynthesis pathway could be a consequence of body temperature changes.
NAD biosynthesis from 2-amino-3-carboxymuconate semialdehyde and NAD biosynthesis II (from tryptophan) were two other pathways identified as significantly up-regulated in relation to CR in a similar manner across all tissues. According to our IPA results, quinolinic acid, l-glutamine, and adenosine monophosphate (AMP) were up-regulated by CR in these pathways. These molecules were up-regulated mainly in BAT, brain, and liver. Of the three metabolites that IPA classified, only quinolinic acid is a precursor for NAD. Glutamine and AMP are found in the NAD pathway but are not specific to it. Quinolinic acid is in the kynurenine pathway of tryptophan catabolism which, together with the enzyme quinolinate phosphoribosyltransferase (QPRT), catalyzes the formation of nicotinic acid mononucleotide (NMN) from quinolinic acid and 5-phosphoribosyl-1-pyrophosphate, fueling NAD synthesis (46). In isolation, changes in quinolinate could either mean that CR-treated animals had increased activity in this pathway generating NAD or that they were responding to lowered levels of niacin in the diet by up-regulating the synthesis of quinolinate from tryptophan. Previously, we performed transcriptomic analyses of the hypothalamus (18), liver (19), and epididymal adipose tissue (20) of these mice. Hence we only had simultaneous transcriptomic and metabolomics data for the liver. The liver transcriptomics data showed that levels of QPRT were down-regulated in relation to the level of restriction [F(1,41) = 3.984; P, 0.006]. This would suggest NAD levels in the liver were likely not elevated due to de novo synthesis via quinolinic acid. Given the interpretation that NAD synthesis was potentially elevated, we manually searched the spectra for metabolites that would match other components of the NAD synthesis pathway (specifically NAD, nicotinic acid adenine dinucleotide [NAAD], nicotinic acid mononucleotide [NAMN], and NAD+ phosphorylated [NADP]) but had failed to enter the list of significantly modulated metabolites because they did not meet the very strict criteria of showing a linear increase (or decrease) with parallel responses across all tissues (pattern 2). We found that NAD and its oxidized form (NAD+) were present in the BAT and brain samples, respectively, showing a significant difference between AL and CR groups. Conversely, CR had no significant influence on the reduced form (nicotinamide adenine dinucleotide + hydrogen, NADH) in the liver. Therefore, we can assume that CR showed a graded effect on the NAD levels in BAT and CER; however, the evidence for an effect in the liver was less certain.
Nonetheless, the up-regulation of nicotinamide phosphoribosyltransferase and nicotinamide nucleotide transhydrogenase (two genes related to the salvage pathway of NAD) detected in the liver transcriptomic analysis (19) suggest that an increase in the NAD levels in the liver might not come exclusively from the de novo pathway. Moreover, we detected two NAD precursors, niacinamide (NAM) and 1-methyl nicotinamide (meNAM), which were not included in the IPA analysis but were up-regulated due to restriction in all tissues. There were also unidentified BAT and brain features with m/z ratios similar to NAAD, NADP, and NAMN but were not specifically identified by xMSAannotator. No compounds with a close m/z to NAAD and NADP were found in liver and plasma, but plasma features were found with m/z like NAMN. These results might support the idea that elevated NAD was due to the reduced dietary niacin and high activity in the tryptophan and/or nicotinamide pathways. Previous investigations have linked the kynurenine pathway activation to aging and treatments that slow aging, including CR (47, 48).
Importantly, an interplay exists between aging, nutritional challenge, and circadian metabolism. Sato et al. (49) observed higher levels of multiple NAD metabolites in aged mice liver under CR and that these mice maintained tighter circadian gene expression over their life span. Furthermore, they observed that circadian reprogramming of physiological homeostasis due to aging or CR was tissue specific (49). Therefore, this provides support to clean cupboard theory and further emphasizes the centrality of NAD to energy generation and resource allocation. Mitchell et al. (50) showed that chronic treatment with nicotinamide was associated with health improvements and lower inflammation in the absence of life span extension in high-fat diet-fed mice. Thus, based on the available literature and our research data, whether these metabolites would undermine or amplify the life span’s effect under CR remained in question. However, the possible health effects related to regulating the activity of many proteins related to energy metabolism by boosting NAD in each tissue could explain our results (48, 49). There is evidence that the synthesis of NAD by the kynurenine pathway may be enhanced when the immune system responds to challenges, indicating its prosurvival value. However, the precise nature of the benefits that the kynurenine pathway confers on fitness is a matter of debate (46).
A fourth modulated pathway common to all four tissues was tRNA charging. Transfer RNAs are best known for their role as adaptors during the translation of the genetic code and essential players in protein synthesis, linking the genetic code with the amino acid sequence of proteins (51). Signaling pathways that sense amino acid abundance are integral to tissue homeostasis and cellular defense (48). Furthermore, the amino acid levels regulate both protein synthesis and autophagic proteolysis (52). Here, we observed that glycine and l-glutamine were up-regulated across all the tissues, which could mean an increase in protein synthesis and autophagy under CR. However, it could also mean a possible adverse effect since a rise in amino acids may negatively alter proteolysis. For example, increases in glutamine inhibited proteolysis in the liver, possibly due to a lysosomotropic effect caused by ammonia released from glutamine (53). This possible negative effect emphasizes that CR might not only generate a range of positive outcomes but also has negative consequences. This dual effect is expected from the clean cupboards hypothesis (25), but not the DSH.
Another molecule that was up-regulated in this pathway was AMP. This molecule contributes to AMP-activated protein kinase (AMPK) activation, an enzyme and key nutrient sensor with the ability to regulate whole-body metabolism since it maximizes ATP generation by promoting catabolic pathways inhibiting anabolic processes that consume ATP (54). During CR, AMPK becomes activated in diverse tissues, including the heart, liver, skeletal muscle, and hypothalamus (55, 56). Furthermore, AMPK phosphorylates acetyl-CoA carboxylase to inhibit fatty acid synthesis. This inhibition of fatty acid synthesis leads to the activation of fatty acid β-oxidation (except in neurons that lack these enzymes) to conserve energy by preventing a futile cycle of fatty acid synthesis and breakdown. Also, the inhibition of fatty acid synthesis preserves nicotinamide adenine dinucleotide phosphate levels and contributes to the increased levels of redox defenses potentially associated with CR’s longevity benefits (55). However, in some cases, CR has failed to activate AMPK (5). In mice under a 35% CR, despite demonstrating the expected metabolic characteristics (lower body weight, plasma leptin, and liver glycogen content), neither heart, skeletal muscle, nor liver showed any evidence of increased activity of either the α1- or α2-isoform of AMPK (57). By contrast, in rats, a CR of 30% causes down-regulation of AMPK, particularly in the liver (58). These opposite results might reflect differences in the CR protocols, which vary in CR’s duration and level.
l-Proline was the only molecule that was down-regulated in the tRNA charging pathway in all four tissues. The depletion of amino acids (essential, nonessential, or conditionally essential) has been associated with an increase in the concentration of uncharged tRNA species activating GCN2 (general amino acid control nonderepressible 2). In mammals, GCN2 can be rapidly activated in the brain and liver upon ingestion of a meal lacking essential amino acids. However, GCN2 can also be triggered by the nondietary depletion of amino acids (for example, arginine). Furthermore, GCN2 has a connection with eIF2α (eukaryotic initiation factor 2α) kinase and the master growth-regulating kinase mTOR (mechanistic target of rapamycin), two key mediators of energy expenditure, among many other functions (48, 52). Recently, a mammalian cell analysis discovered that mitochondrial NADPH is essential to enable proline biosynthesis and that proline level deficiency triggered a strong response in the GCN2–eIF2α–activating transcription factor 4 pathway, which senses depletion of amino acids and the uncharged tRNAs (59). Hence, the reduced proline levels observed here may be connected to the activation of these transduction mechanisms involved in sensing intracellular amino acids. However, this could also mean a response to control the energy expenditure under CR. Another possible explanation for this reduction of l-proline in this pathway could be based on halofuginone’s results (HF; a CR mimetic drug) (60). This prolyl-tRNA synthetase inhibitor activates the amino acid starvation response by mimicking proline deprivation. Treatment with HF was linked to suppression of a subset of TNF-α–stimulated responses, mirroring HF’s suppression of proinflammatory functions in mature TH17 (CD4+ T helper 17) memory cells and its suppression of TGF-β–stimulated fibrotic tissue remodeling. Hence, CR-mediated reductions in inflammation might be partly linked to this l-proline reduction (60). In previous work, we found that circulating TNF-α was reduced with CR (61), and transcriptomic analysis indicated that the transcription factor NF-κB was inhibited at all CR levels, reflecting a reduced state of inflammation (18).
Finally, we only found a single degradation pathway, S-methyl-5′-thioadenosine degradation II, in the top five canonical pathways. In this pathway, the thioether nucleoside, 5′-methylthioadenosine, was significant. This molecule is a product of transpropylamine reactions that lead to spermidine and spermine synthesis. These polyamines are ubiquitous in mammalian cells (62). 5′-Methylthioadenosine was up-regulated in the four tissues analyzed, potentially reflecting increased spermidine and spermine levels in tissue. Spermidine homeostasis involves nutritional uptake, intestinal synthesis by gut microbiota, endogenous biosynthesis, degradation, and active transporter systems between compartments. Spermidine supplementation and CR both result in histone hypo-acetylation with chromatin silencing either by sirtuin-mediated activation of histone deacetylases (CR) or by inhibition of the histone acyl transferase (spermidine) (63). In addition, in many mammalian tissues, 5′-methylthioadenosine phosphorylase (MTAP) is the main enzyme that divides 5′-methylthioadenosine into 5-methylthioribose-1-phosphate and adenine. Adenine was another metabolite that we detected up-regulated by CR. Adenine in this pathway is converted to adenosine and after into AMP. This process creates a path for the metabolic salvaging of the purine portion of ATP used for the synthesis of S-adenosylmethionine, which is recycled for use in 5′-methylthioadenosine synthesis (64). Once again, these changes are consistent with CR modulating pathways to improve metabolic efficiency.
Limitations.
As with any metabolomic analysis, there were some limitations to our study. One of these limitations we faced was the poor overlap of the m/z feature among tissues; this can probably be attributed to using different analytical platforms and integrating datasets of multiomics that measure fundamentally different biomolecules. Besides, nowadays, by untargeted metabolomics, it is only possible to putatively identify a subset of the total m/z values detected, which can introduce bias into metabolite and pathway analysis (65). Despite the limitations, we were able to analyze 6.4% (361 metabolites features) of the metabolites detected in four or three tissues grouping them according to a representative m/z value. Moreover, our metabolic set enrichment analysis was limited by the metabolic coverage used to create the pathways and likely represented only a fraction of the total metabolic pathways altered by CR. Even when we identified compounds, they were not always incorporated into the IPA program’s pathways because the compounds were not in their knowledge database.
With respect to the actual experimental work, another limitation was the short duration of the imposed restriction. Short-term interventions may not fully capture the changes that underpin the lifelong impacts of CR on life span. Thus, it remains unclear whether long-term CR exposure would alter the cross-tissue metabolome the same way. It is possible that the short-term changes we detected here provide more support for the clean cupboards hypothesis, but that longer term, the changes might be more consistent with the DSH. A final limitation of this work was that it was based on a single strain of inbred mice (C57BL/6J) known to be a positive responder to CR. Mouse strains do vary in their responses to CR (66), and hence it is currently unclear whether the responses we detected strain-specific or more generally applicable.
Conclusion
Individual mice, and the tissues within those mice, responded consistently to the imposition of CR. However, there were very few consistent metabolic responses across all tissues. The overall impression was that each tissue invoked its own idiosyncratic modulation of pathways to cope with the reduced flow of incoming energy. In one tissue, it might be elevated β-oxidation of fatty acids, in another amino acid degradation, and in another glycolysis—different responses with a common goal of fueling metabolism. Nevertheless, a small number of pathways did show consistent changes in all tissues. These included l-carnitine and NAD biosynthesis and tRNA charging. There was no strong evidence to support the idea from the DSH that resources were being diverted away from reproduction toward somatic maintenance since no consistent pathways linked to somatic maintenance appeared to be up-regulated. In contrast, the pattern of response was much more consistent with the mice up-regulating metabolic pathways that allowed them to cope with reduced energy intake—the clean cupboards hypothesis.
Supplementary Material
Acknowledgments
Our work on graded calorie restriction has been funded by the United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC) (BB/G009953/1, BB/P009875/1, and BB/J020029/1) and the K. C. Wong Foundation. J.R.S. was supported by a President’s International Fellowship Initiative (PIFI) professorial fellowship from the Chinese Academy of Sciences (CAS), and a Royal Society Wolfson merit award. L.A.G.-F. was supported by a PIFI postdoctoral fellowship from CAS. C.L.G. was supported by a BBSRC EastBio doctoral training partnership award (1438803). D.E.L.P. was supported in part by NIH Grant AGO49494. D.L. was supported by Office of Naval Research Grant N000141512377. We thank Dean Jones, Quinlyn Soltow, and Karl Burgess for technical assistance with the original metabolomics analysis, and the Biological Services Unit staff for caring for the animals.
Footnotes
The authors declare no competing interest.
See Profile, e2111255118, in vol. 118, issue 31.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101977118/-/DCSupplemental.
Data Availability
References
- 1.Speakman J. R., Mitchell S. E., Caloric restriction. Mol. Aspects Med. 32, 159–221 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Weindruch R., Walford R. L., Retardation of Aging and Disease by Dietary Restriction (C. C. Thomas, 1988). [Google Scholar]
- 3.Osborne T. B., Mendel L. B., Ferry E. L., Wakeman A. J., The resumption of growth after long continued failure to grow. J. Biol. Chem. 23, 439–454 (1915). [Google Scholar]
- 4.Fontana L., et al., The effects of graded caloric restriction: XII. Comparison of mouse to human impact on cellular senescence in the colon. Aging Cell 17, e12746 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwangbo D. S., Lee H. Y., Abozaid L. S., Min K. J., Mechanisms of lifespan regulation by calorie restriction and intermittent fasting in model organisms. Nutrients 12, 1194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merry B. J., Oxidative stress and mitochondrial function with aging—the effects of calorie restriction. Aging Cell 3, 7–12 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Speakman J. R., Hambly C., Starving for life: What animal studies can and cannot tell us about the use of caloric restriction to prolong human lifespan. J. Nutr. 137, 1078–1086 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Mitchell S. E., et al., The effects of graded levels of calorie restriction: III. Impact of short term calorie and protein restriction on mean daily body temperature and torpor use in the C57BL/6 mouse. Oncotarget 6, 18314–18337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piper M. D. W., Mair W., Partridge L., Counting the calories: The role of specific nutrients in extension of life span by food restriction. J. Gerontol. A Biol. Sci. Med. Sci. 60, 549–555 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Solon-Biet S. M., et al., Defining the nutritional and metabolic context of FGF21 using the geometric framework. Cell Metab. 24, 555–565 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Speakman J. R., Mitchell S. E., Mazidi M., Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp. Gerontol. 86, 28–38 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Mitchell S. E., et al., The effects of graded levels of calorie restriction: I. Impact of short term calorie and protein restriction on body composition in the C57BL/6 mouse. Oncotarget 6, 15902–15930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lusseau D., et al., The effects of graded levels of calorie restriction: IV. Non-linear change in behavioural phenotype of mice in response to short-term calorie restriction. Sci. Rep. 5, 13198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell S. E., et al., The effects of graded levels of calorie restriction: V. Impact of short term calorie and protein restriction on physical activity in the C57BL/6 mouse. Oncotarget 7, 19147–19170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshun S., et al., The effects of graded levels of calorie restriction XV: Phase space attractors reveal distinct behavioral phenotypes.J. Gerontol. A Biol. Sci. Med. Sci. 75, 858–866 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Mitchell S. E., et al., The effects of graded levels of calorie restriction: VIII. Impact of short term calorie and protein restriction on basal metabolic rate in the C57BL/6 mouse. Oncotarget 8, 17453–17474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derous D., et al., The effects of graded levels of calorie restriction: VII. Topological rearrangement of hypothalamic aging networks. Aging (Albany NY) 8, 917–932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derous D., et al., The effects of graded levels of calorie restriction: VI. Impact of short-term graded calorie restriction on transcriptomic responses of the hypothalamic hunger and circadian signaling pathways. Aging (Albany NY) 8, 642–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derous D., et al., The effects of graded levels of calorie restriction: XI. Evaluation of the main hypotheses underpinning the life extension effects of CR using the hepatic transcriptome. Aging (Albany NY) 9, 1770–1824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derous D., et al., The effects of graded levels of calorie restriction: X. Transcriptomic responses of epididymal adipose tissue. J. Gerontol. A Biol. Sci. Med. Sci. 73, 279–288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green C. L., et al., The effects of graded levels of calorie restriction: IX. Global metabolomic screen reveals modulation of carnitines, sphingolipids and bile acids in the liver of C57BL/6 mice. Aging Cell 16, 529–540 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green C. L., et al., The effects of graded levels of calorie restriction: XIII. Global metabolomics screen reveals graded changes in circulating amino acids, vitamins, and bile acids in the plasma of C57BL/6 mice. J. Gerontol. A Biol. Sci. Med. Sci. 74, 16–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green C., et al., Effects of graded levels of calorie restriction: XIV. Global metabolomics screen reveals brown adipose tissue changes in amino acids, catecholamines, and antioxidants after short-term restriction in C57BL/6 mice. J. Gerontol. A. Biol. Sci. Med. Sci. 75, 218–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green C., et al., The effects of graded levels of calorie restriction: XVI. Metabolomic changes in the cerebellum indicate activation of hypothalamocerebellar connections driven by hunger responses. J. Gerontol. A. Biol. Sci. Med. Sci. 76, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speakman J. R., Why does caloric restriction increase life and healthspan? The “clean cupboards” hypothesis. Natl. Sci. Rev. 7, 1153–1156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holliday R., Food, reproduction and longevity: Is the extended lifespan of calorie-restricted animals an evolutionary adaptation? BioEssays 10, 125–127 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Masoro E. J., Austad S. N., The evolution of the antiaging action of dietary restriction: A hypothesis. J. Gerontol. A Biol. Sci. Med. Sci. 51, B387–B391 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Shanley D. P., Kirkwood T. B. L., Calorie restriction and aging: A life-history analysis. Evolution 54, 740–750 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Kristan D. M., Calorie restriction and susceptibility to intact pathogens. Age (Dordr.) 30, 147–156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardner E. M., Caloric restriction decreases survival of aged mice in response to primary influenza infection. J. Gerontol. A Biol. Sci. Med. Sci. 60, 688–694 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Hunt N. D., et al., Effect of calorie restriction and refeeding on skin wound healing in the rat. Age (Dordr.) 34, 1453–1458 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-Lluch G., Navas P., Calorie restriction as an intervention in ageing. J. Physiol. 594, 2043–2060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken A. W., Adams G., Hartshorne L., Tatar M., Simons M. J. P., The hidden costs of dietary restriction: Implications for its evolutionary and mechanistic origins. Sci. Adv. 6, eaay3047 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamming D. W., Anderson R. M., “Metabolic effects of caloric restriction” in eLS (Wiley, 2014). 10.1002/9780470015902.a0021316.pub2 (Accessed December 23, 2020). [DOI]
- 35.Gems D., Partridge L., Genetics of longevity in model organisms: Debates and paradigm shifts. Annu. Rev. Physiol. 75, 621–644 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Kapahi P., Kaeberlein M., Hansen M., Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res. Rev. 39, 3–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moger-Reischer R. Z., Snider E. V., McKenzie K. L., Lennon J. T., Low costs of adaptation to dietary restriction. Biol. Lett. 16, 20200008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu D., et al., Calorie-restriction-induced insulin sensitivity is mediated by adipose mTORC2 and not required for lifespan extension. Cell Rep. 29, 236–248.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcovina S. M., et al., Translating the basic knowledge of mitochondrial functions to metabolic therapy: Role of l-carnitine. Transl. Res. 161, 73–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mezhnina V., et al., CR reprograms acetyl-CoA metabolism and induces long-chain acyl-CoA dehydrogenase and CrAT expression. Aging Cell 19, e13266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noland R. C., et al., Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J. Biol. Chem. 284, 22840–22852 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luci S., Hirche F., Eder K., Fasting and caloric restriction increases mRNA concentrations of novel organic cation transporter-2 and carnitine concentrations in rat tissues. Ann. Nutr. Metab. 52, 58–67 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Jones L. L., McDonald D. A., Borum P. R., Acylcarnitines: Role in brain. Prog. Lipid Res. 49, 61–75 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Ozaki K., Sano T., Tsuji N., Matsuura T., Narama I., Carnitine is necessary to maintain the phenotype and function of brown adipose tissue. Lab. Invest. 91, 704–710 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Bruss M. D., Khambatta C. F., Ruby M. A., Aggarwal I., Hellerstein M. K., Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am. J. Physiol. Endocrinol. Metab. 298, E108–E116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moffett J. R., et al., Quinolinate as a marker for kynurenine metabolite formation and the unresolved question of NAD+ synthesis during inflammation and infection. Front. Immunol. 11, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jové M., et al., Caloric restriction reveals a metabolomic and lipidomic signature in liver of male mice. Aging Cell 13, 828–837 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green C. L., Lamming D. W., Regulation of metabolic health by essential dietary amino acids. Mech. Ageing Dev. 177, 186–200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato S., et al., Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170, 664–677.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell S. J., et al., Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 27, 667–676.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raina M., Ibba M., tRNAs as regulators of biological processes. Front. Genet. 5, 171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallinetti J., Harputlugil E., Mitchell J. R., Amino acid sensing in dietary-restriction-mediated longevity: Roles of signal-transducing kinases GCN2 and TOR. Biochem. J. 449, 1–10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kadowaki M., Kanazawa T., Amino acids as regulators of proteolysis. J. Nutr. 133, 2052S–2056S (2003). [DOI] [PubMed] [Google Scholar]
- 54.Balyan R., Gautam N., Gascoigne N. R. J., The ups and downs of metabolism during the lifespan of a t cell. Int. J. Mol. Sci. 21, 1–22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bradshaw P. C., Cytoplasmic and mitochondrial NADPH-coupled redox systems in the regulation of aging. Nutrients 11, 504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cantó C., Auwerx J., Calorie restriction: Is AMPK a key sensor and effector? Physiology (Bethesda) 26, 214–224 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez A. A., et al., Metabolic adaptations to fasting and chronic caloric restriction in heart, muscle, and liver do not include changes in AMPK activity. Am. J. Physiol. Endocrinol. Metab. 287, E1032–E1037 (2004). [DOI] [PubMed] [Google Scholar]
- 58.To K., et al., Down-regulation of AMP-activated protein kinase by calorie restriction in rat liver. Exp. Gerontol. 42, 1063–1071 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Tran D. H., et al., Mitochondrial NADP+ is essential for proline biosynthesis during cell growth. Nat. Metab. 3, 571–585 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim Y., et al., Aminoacyl-tRNA synthetase inhibition activates a pathway that branches from the canonical amino acid response in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 117, 8900–8911 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell S. E., et al., The effects of graded levels of calorie restriction: II. Impact of short term calorie and protein restriction on circulating hormone levels, glucose homeostasis and oxidative stress in male C57BL/6 mice. Oncotarget 6, 23213–23237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamatani N., Kubota M., Willis E. H., Frincke L. A., Carson D. A., 5′-Methylthioadenosine is the major source of adenine in human cells. Adv. Exp. Med. Biol. 165 (Pt. B), 83–88 (1984). [DOI] [PubMed] [Google Scholar]
- 63.Kiechl S., et al., Higher spermidine intake is linked to lower mortality: A prospective population-based study. Am. J. Clin. Nutr. 108, 371–380 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Li Y., Wang Y., Wu P., 5′-Methylthioadenosine and cancer: Old molecules, new understanding. J. Cancer 10, 927–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson C. H., Ivanisevic J., Siuzdak G., Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17, 451–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao C. Y., Rikke B. A., Johnson T. E., Diaz V., Nelson J. F., Genetic variation in the murine lifespan response to dietary restriction: From life extension to life shortening. Aging Cell 9, 92–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.