Significance
Obesity-associated metaflammation is an emerging disease, in which proinflammatory macrophages play a decisive role. Here, we identified the soluble mannose receptor (sMR) as a regulator of metaflammation and demonstrated increased levels of sMR in obese mice and humans. Additionally, we identified that increased sMR directly binds to CD45 on macrophages, inhibiting its activity and inducing inflammatory macrophage activation via the Akt/NF-κB pathway. Consequently, increased sMR levels induced inflammatory activation of metabolic tissue macrophages and aggravation of whole-body metabolic homeostasis. These data identify the MR as a regulator of macrophage activation and offer perspectives for therapeutic approaches for the treatment of metaflammation.
Keywords: immunometabolism, mannose receptor, macrophage, obesity, metaflammation
Abstract
Proinflammatory activation of macrophages in metabolic tissues is critically important in the induction of obesity-induced metaflammation. Here, we demonstrate that the soluble mannose receptor (sMR) plays a direct functional role in both macrophage activation and metaflammation. We show that sMR binds CD45 on macrophages and inhibits its phosphatase activity, leading to an Src/Akt/NF-κB–mediated cellular reprogramming toward an inflammatory phenotype both in vitro and in vivo. Remarkably, increased serum sMR levels were observed in obese mice and humans and directly correlated with body weight. Importantly, enhanced sMR levels increase serum proinflammatory cytokines, activate tissue macrophages, and promote insulin resistance. Altogether, our results reveal sMR as regulator of proinflammatory macrophage activation, which could constitute a therapeutic target for metaflammation and other hyperinflammatory diseases.
Metaflammation defines a chronic inflammatory state in response to prolonged excessive nutrient intake and is characterized by low-grade inflammation of metabolic tissues (1). Macrophage reprogramming toward an inflammatory phenotype plays a critical role in obesity-induced metaflammation (2, 3). In lean individuals, macrophages in metabolic tissues maintain tissue homeostasis and insulin sensitivity, potentially through secreting anti-inflammatory cytokines, for example, TGF-β and IL-10 (1). In metaflammation, however, macrophages in adipose tissue and liver are activated through proinflammatory factors in their microenvironment, such as high levels of saturated free fatty acids (FA) and IFN-γ. Consequently, these macrophages produce high amounts of tumor necrosis factor (TNF), which directly inhibits canonical insulin signaling (4), leading to ectopic fat deposition in the liver and in skeletal muscles (5). Additionally, activation of Kupffer cells (KCs), the liver-resident macrophages, promotes recruitment and activation of inflammatory monocytes, which contribute to hepatic insulin resistance and steatosis (6–8).
The MR (also termed CD206) is a type I transmembrane protein belonging to the C-type lectin family, which is mainly expressed by subpopulations of macrophages, dendritic cells, and endothelial cells (9, 10). The MR consists of a cysteine-rich region, a fibronectin type II domain, eight C-type lectin-like domains (CTLDs), a transmembrane region, and a short cytosolic tail. Due to its high affinity for glycosylated antigens, the MR plays an important role in antigen uptake and presentation (11, 12). In addition to its functions as a transmembrane protein, the extracellular part of the MR can be shed by metalloproteases and released into the extracellular space (13, 14). Hence, soluble MR (sMR) can be detected in murine and human serum, and its level was found to be increased in patients with a variety of inflammatory diseases (15–20), correlating with severity of disease and even mortality. However, a physiological role of the sMR has not been studied yet, and it remains unclear whether the sMR can actively trigger inflammation.
Here, we report that sMR enhances macrophage proinflammatory activation, both in vitro and in vivo, and promotes metaflammation. We demonstrate that the sMR directly interacts with CD45 on the surface of macrophages and inhibits its phosphatase activity, leading to Src/Akt/NF-κB–mediated cellular reprogramming toward an inflammatory phenotype. Additionally, we found enhanced sMR serum levels in obese mice and humans and show that sMR-induced activation of macrophages triggers metaflammation in vivo.
Results
Soluble MR Enhances Proinflammatory Cytokine Secretion by Macrophages.
To investigate whether the MR is involved in the proinflammatory activation of macrophages, we first stimulated bone marrow–derived macrophages from wild-type or MR-deficient mice with LPS. We found increased secretion of the proinflammatory cytokines TNF, IL-6, IL-1β, and IL-12 in MR-expressing wild-type macrophages (Fig. 1A). Because the MR itself lacks intracellular signaling motifs and hence no MR-mediated signaling has been described so far, we hypothesized that the sMR, resulting from the shedding of the MR extracellular region (SI Appendix, Fig. S1A), might play a role in macrophage activation through direct interaction with macrophage surface proteins. To investigate this hypothesis, we generated a fusion protein consisting of the Fc region of human IgG1 and the extracellular region of the MR (encompassing the cysteine-rich region, the fibronectin region, and CTLD1-2) (FcMR) (21). We showed that treatment of MR-deficient macrophages with FcMR also enhanced proinflammatory cytokine secretion after LPS stimulation compared to isotype control-treated cells (Fig. 1B). We observed similar results when treating MR-deficient macrophages with commercially available recombinant MR protein, consisting of the complete extracellular region of the protein (Fig. 1C), suggesting that binding of sMR to the macrophage surface might indeed be responsible for the observed effects. To definitively prove that the sMR causes the observed increase in cytokine production, we purified sMR from the supernatant of MR-expressing macrophages (SI Appendix, Fig. S1B) and showed that its administration to MR-deficient macrophages increased the secretion of TNF after LPS stimulation (SI Appendix, Fig. S1C). Similar results were obtained from FcMR-treated primary macrophages isolated from murine liver, spleen, or peritoneal cavity (Fig. 1D) and in human monocyte-derived macrophages (moMϕ) after addition of recombinant human MR (Fig. 1E) or after small interfering RNA (siRNA)-mediated down-regulation of the MR (Fig. 1F). Interestingly, sMR also promoted a shift in cellular energy metabolism toward increased glycolysis in both murine and human macrophages (SI Appendix, Fig. S1 D and E), a bioenergetic hallmark of proinflammatory activation in macrophages (22, 23). Taken together, these data demonstrate that the sMR enhances proinflammatory activation of both murine and human macrophages.
Fig. 1.
sMR induces proinflammatory cytokine secretion by macrophages. (A) Secretion of TNF, IL-6, IL-1β, and IL-12 by LPS-treated WT or MR-deficient (MR−/−) macrophages. (B) TNF, IL-6, and IL-1β secretion by LPS-treated MR-deficient macrophages after incubation with FcMR. (C) Secretion of TNF by LPS-stimulated MR-deficient macrophages after addition of 0.3 µg/mL recombinant murine MR (rMR). (D) Primary murine macrophages were isolated from liver, spleen, or peritoneal cavity (PEC) of WT mice by magnetic separation of F4/80+ cells. Secretion of TNF after LPS treatment and stimulation with FcMR were determined by ELISA. (E) Secretion of TNF by LPS-treated human monocyte-derived macrophages (moMФ) after stimulation with 0.3 µg/mL recombinant human MR (rhMR). (F) Secretion of TNF, IL-6, and IL-1β by LPS-stimulated human moMФ after siRNA-mediated down-regulation of the MR. All graphs are depicted as mean ± SEM; for all experiments, n ≥ 3. *P < 0.05.
sMR Induces a Proinflammatory Phenotype in Macrophages.
To further dissect the effect of the sMR on macrophages, we treated MR-deficient macrophages with FcMR for 4, 12, or 24 h and performed RNA sequencing (RNA-seq) analysis (Fig. 2A). Principal component analysis revealed clear transcriptomic distinction of the samples in all analyzed conditions (Fig. 2B). A heatmap of the 1,366 differentially expressed (DE) genes between FcMR treatment and control presented the substantial changes in gene expression due to the FcMR treatment over time with overlapping and unique gene sets (Fig. 2C and SI Appendix, Fig. S2A). Gene ontology enrichment analysis based on these shared and specific DE gene sets up-regulated upon FcMR treatment clearly confirmed inflammatory activation of macrophages (SI Appendix, Fig. S2B). The most significantly up-regulated genes in response to FcMR treatment further emphasized the strong and dynamic proinflammatory activation of macrophages, with well-known immunological key mediators such as TNF, IL-6, IL-1β, and IL-12 (Fig. 2D and SI Appendix, Fig. S2C). To classify the response elicited by sMR within the broad spectrum of macrophage activation phenotypes, we performed an enrichment analysis using macrophage activation signatures derived from our previous study comprising macrophages treated with 28 different immunological stimuli (24) and the gene sets of FcMR-mediated up-regulated genes per time point. This analysis revealed a striking similarity of FcMR-induced expression patterns to macrophage signatures associated with a chronic inflammatory phenotype, as induced by TNF, PGE2, and P3C (TPP) in our previous stimulation study (Fig. 2E and SI Appendix, Fig. S2D), further substantiating that the sMR reprograms macrophages toward a proinflammatory phenotype.
Fig. 2.
RNA-seq analysis identifies a proinflammatory phenotype in MR-treated macrophages. (A) Schematic overview of the bioinformatics RNA-seq analysis strategy. (B) Principal component analysis based on variance-stabilized counts of all 23,250 present genes. (C) Heatmap of hierarchically clustered, normalized, and z-scaled expression values of the union of 1,366 DE genes between FcMR-treated and control samples. (D) Normalized and z-scaled expression values of the union of the top 25 DE genes of each time point significantly up-regulated in at least two consecutive time points ranked according to their FcMR versus control samples visualized in a heatmap. (E) Dot plot of gene set enrichment analysis results of 49 predefined stimulus-specific macrophage expression signatures comprising 28 different stimuli on the FcMR-specific DE genes for each time point. TPP: TNF, PGE2, and Pam3Cys; PA: palmitic acid.
sMR Activates Macrophages by Binding and Inhibiting CD45.
Next, we investigated the molecular mechanisms regulating sMR-induced macrophage reprogramming and searched for binding partners of the MR on the macrophage surface. To this end, we isolated cell lysates from macrophages that previously underwent surface biotinylation and performed immunoprecipitation using FcMR. Western blot analysis using streptavidin allowed us to monitor cell surface proteins interacting with sMR, including a clear band at the molecular weight of the phosphatase CD45 (between 180 and 220 kDa, depending on the splice variant) (SI Appendix, Fig. S3A), a known binding partner of the MR (25). Indeed, coimmunoprecipitation experiments revealed a physical interaction between the MR and CD45 on macrophages (Fig. 3 A and B).
Fig. 3.
MR inhibits phosphatase activity of CD45 on macrophages. (A) Macrophage cell lysates were immune precipitated using a CD45-specific antibody and stained for FcMR binding by far Western blot. (B) Macrophage lysates were immune precipitated with FcMR or isotype control and stained for CD45 by Western blot. (C) CD45 was precipitated from lysates of FcMR- or isotype control-treated macrophages and incubated with 4-NPP in the presence or absence of the CD45 inhibitor SF1670. Graph depicts CD45-mediated dephosphorylation of 4-NPP measured by colorimetry. Samples precipitated without CD45 antibody were used as controls. (D) CD45 was precipitated from lysates of FcMR- or isotype control-treated macrophages and incubated with the CD45 substrate TATEGQ-pY-QPQ. Graph depicts the phosphorylation status of TATEGQ-pY-QPQ. Samples precipitated without CD45 antibody were used as controls. (E) Secretion of TNF, IL-6, IL-1β, and IL-12 by LPS-stimulated macrophages after siRNA-mediated down-regulation of CD45. (F) Influence of FcMR on secretion of TNF, IL-6, IL-1β, and IL-12 by LPS-stimulated macrophages after siRNA-mediated down-regulation of CD45. All graphs are depicted as mean ± SEM; for all experiments, n ≥ 3. *P < 0.05.
CD45 can be expressed as different isoforms, depending on alternative splicing of its three exons A, B, and C. To identify the CD45 isoform interacting with sMR, we assessed their respective expression using isoform-specific antibodies. Analysis by Western blot and flow cytometry clearly showed the absence of exons A, B, and C in bone marrow–derived macrophages (SI Appendix, Fig. S3B), pointing out that these cells only express the CD45RO isoform. This is in accordance with our RNA-seq data, which showed a specific read coverage of all exons of Cd45 except for exons A, B, and C (SI Appendix, Fig. S3C). Additionally, we showed that primary macrophages from spleen, white adipose tissue (WAT), liver, and the peritoneal cavity also expressed the CD45RO isoform (SI Appendix, Fig. S3D), which is in agreement with previous literature (26). Accordingly, we confirmed the direct interaction of FcMR with CD45RO from primary splenic macrophages by far Western blot (SI Appendix, Fig. S3E).
Since little is known about CD45 phosphatase activity in macrophages, we next investigated whether CD45 is active in these cells. Therefore, we immunoprecipitated CD45 from macrophage lysates and added 4-nitrophenyl phosphate (pNPP), from which dephosphorylation by CD45 can be quantified using colorimetry. We monitored a clear phosphatase activity, which was blocked by a CD45-specific inhibitor (SI Appendix, Fig. S3F), demonstrating the presence of active CD45 in macrophages. Next, we tested the effect of sMR on CD45 phosphatase activity. To this end, we immunoprecipitated CD45 from lysates of FcMR-treated macrophage and showed that dephosphorylation of pNPP was reduced when compared to isotype control-treated cells (Fig. 3C), indicating that the MR inhibited CD45 phosphatase activity. In a second approach, we assessed the dephosphorylation of a synthetic peptide containing pY505 of Lck, a specific substrate of CD45. We showed that preincubation of macrophages with FcMR increased pY505 phosphorylation (Fig. 3D), confirming the inhibitory effect of the MR on CD45 phosphatase activity.
To investigate whether sMR-mediated inhibition of CD45 phosphatase activity plays a role in macrophage activation, we down-regulated CD45 expression using siRNA (SI Appendix, Fig. S3G). Similar to inhibition of CD45 by sMR, CD45 down-regulation resulted in increased expression of TNF, IL-6, IL-1β, and IL-12 after stimulation with LPS (Fig. 3E). Importantly, addition of FcMR after down-regulating CD45 had no further effect on cytokine secretion (Fig. 3F), demonstrating that the activating effect of the MR on macrophages was indeed due to its inhibition of CD45.
sMR-Mediated Inhibition of CD45 Activates an Src/Akt/NF-κB Signaling Cascade in Macrophages.
We next investigated how sMR-mediated inhibition of CD45 results in macrophage reprogramming toward a proinflammatory phenotype. First, we screened for overrepresented transcription factor (TF) binding motifs in the nonprotein coding regions of FcMR-specific up-regulated DE genes. Network visualization of enriched TF binding motifs and their potential target DE genes clearly exposed NF-κB as the dominating transcriptional regulator of differential gene expression across all three time points (Fig. 4A). In fact, from 351 known NF-κB target genes, 118 genes (34%) were significantly DE after FcMR treatment on at least one time point (SI Appendix, Fig. S4 A and B and Dataset S1). Moreover, from all 351 NF-κB target genes, only 269 genes displayed a clear expression (BaseMean expression value ≥ 10) in macrophages, of which 117 genes (43%) were increased by FcMR. This proportion even increased up to 49% for all target genes with BaseMean expression value ≥ 1,000 (61 out of 124 target genes), suggesting a clear activation of NF-κB by the sMR. Indeed, macrophage treatment with FcMR significantly down-regulated IκBα (Fig. 4B), an inhibitor of NF-κB, which disables its nuclear translocation retaining NF-κB in the cytosol. Accordingly, enhanced nuclear translocation of both NF-κB subunits p65 and p50 (Fig. 4C) and increased recruitment of p65 toward the TNF promotor (SI Appendix, Fig. S4D) were observed after treatment with FcMR.
Fig. 4.
MR reprograms macrophages to a proinflammatory phenotype via Src/Akt/NF-κB signaling. (A) Network visualization of significantly enriched (q-value < 0.1) TF binding motifs (blue) of the MSigDB motif gene set and their potential targets (colored in red according to their FC) among the up-regulated DE genes after 4, 12, or 24 h of FcMR treatment. (B) MR-deficient macrophages were treated with FcMR or isotype control. Total IκB was determined by Western blot. (C) MR-deficient macrophages were treated with FcMR or isotype control. p65 and p50 were monitored in the cytosolic and nuclear fraction by Western blot. (D) MR-deficient macrophages were treated with FcMR or isotype control. Src and phosphorylated Src (pSrc) were determined by Western blot. (E) MR-deficient macrophages were treated with 3 μM PP2, 1 μM KX2-391, or 1 μM A419529 and stimulated with LPS. TNF secretion was monitored by ELISA. (F) FcMR or isotype control-treated MR-deficient macrophages were incubated with PP2, KX2-391, or A419259 and stimulated with LPS. Secretion of TNF was determined by ELISA. (G) MR-deficient macrophages were treated with FcMR or isotype control for 30 min. Akt and phosphorylated Akt (pAkt) were determined by Western blot. (H) MR-deficient macrophages were treated with 5 μM MK-2206 and stimulated with LPS. TNF, IL-6, and IL-1β secretion was monitored by ELISA. (I) FcMR or isotype control-treated MR-deficient macrophages were incubated with MK-2206 and stimulated with LPS. Secretion of TNF, IL-6, and IL-1β was determined by ELISA. All graphs are depicted as mean ± SEM; for all experiments, n ≥ 3. *P < 0.05.
Subsequently, we aimed at identifying the signaling cascade leading from FcMR-mediated inhibition of CD45 to activation of NF-κB. Since CD45 can lead to the activation of Src kinases (27), Src in turn can activate Akt (28), and both Src and Akt have been associated with NF-κB activation (29–32), we investigated whether FcMR-mediated inhibition of CD45 resulted in NF-κB activation through signaling via Src and Akt. Indeed, FcMR treatment increased phosphorylation and hence activation of Src (Fig. 4D). Furthermore, blocking Src using three different chemical inhibitors (PP2, KX2-391, and A419259) markedly decreased TNF secretion (Fig. 4E). Of note, the effect of FcMR on TNF secretion was abolished in the presence of these Src inhibitors (Fig. 4F), demonstrating that FcMR-induced macrophage activation depends on Src signaling. Similarly, FcMR treatment clearly increased phosphorylation of Akt (Fig. 4G), and addition of an Akt-specific inhibitor decreased LPS-induced secretion of TNF, IL-6, and IL-1β (Fig. 4H). Also here, the stimulatory effect of FcMR on cytokine secretion was abolished by Akt inhibition (Fig. 4I), showing an important role for Akt signaling in FcMR-enhanced TNF secretion. Accordingly, inhibition of Akt prevented FcMR-induced translocation of NF-κB into the nucleus (SI Appendix, Fig. S4D).
Taken together, these data demonstrate that sMR-mediated inhibition of CD45 results in activation of an Src/Akt signaling pathway leading to nuclear translocation of NF-κB and macrophage reprogramming toward an inflammatory phenotype.
Serum sMR Is Up-Regulated in Obesity and Promotes High-Fat Diet–Induced Metabolic Dysfunctions and Hepatic Steatosis.
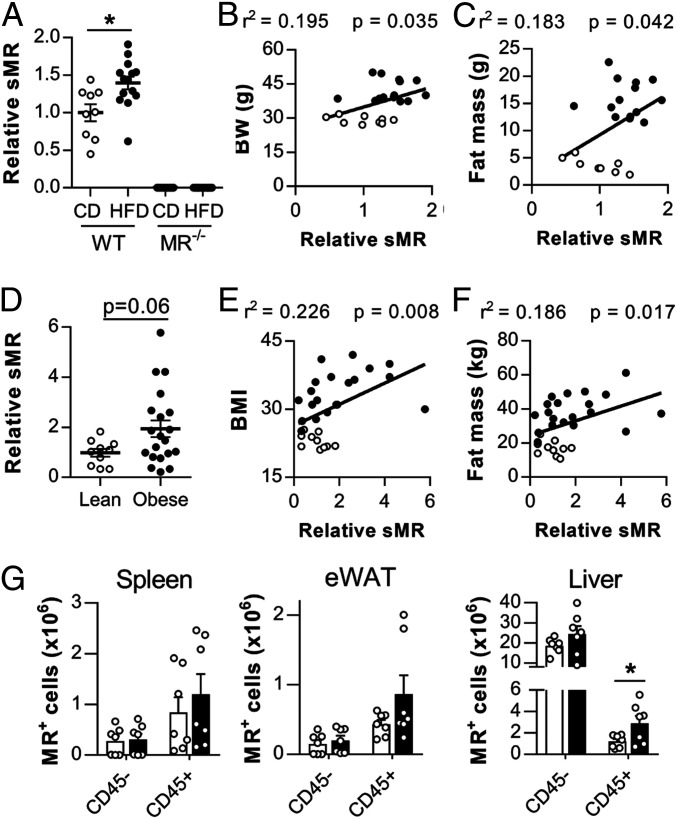
Next, we monitored whether the inflammatory effect of the MR on macrophages regulates inflammatory processes in vivo using a murine model of obesity-induced metaflammation. We first investigated whether high-fat diet (HFD) feeding resulted in changes in serum sMR levels (SI Appendix, Fig. S5A), and we demonstrated significantly increased sMR concentrations in the serum of HFD-fed obese mice, as compared to lean control diet (CD)-fed mice (Fig. 5A). Additionally, serum sMR levels positively correlated with body weight and fat mass of the mice (Fig. 5 B and C). In humans, serum sMR levels were also increased in obese individuals when compared to lean subjects (Fig. 5D) and correlated positively with body mass index and fat mass (Fig. 5 E and F), indicating a direct correlation between serum sMR levels and obesity in both humans and mice.
Fig. 5.
sMR is up-regulated in obesity. (A) sMR levels in the serum of wild-type or MR-deficient mice after HFD or CD feeding. (B and C) Correlation of sMR serum levels and body weight (B) or fat mass (C) of all mice depicted in A. (D) sMR levels in the serum of lean and obese humans. (E and F) Correlation between sMR serum levels and body mass index (E) or fat mass (F). (G) Wild-type mice were fed an HFD or CD for 18 wk. MR-expressing cells were quantified in different organs. Results are expressed as means ± SEM; n = 7 mice per group for G. *P < 0.05.
Subsequently, we analyzed changes in MR-expressing cells as putative source for increased sMR serum levels in HFD-fed mice. In spleen and WAT of both CD- and HFD-fed mice, nearly all MR-expressing cells were CD45+, whereas in liver, CD45− cells also expressed the MR. These latter cells were identified as CD31+CD146+ liver sinusoidal endothelial cells (LSECs), which were indeed previously reported to express the MR (9). Importantly, whereas no differences in MR expression could be detected in CD45− cells, a clear increase in MR+ cells was observed in CD45+ hematopoietic cells in spleen, liver, and WAT of HFD-fed obese mice compared to CD-fed mice (Fig. 5G). Of note, CD45+MR+ cells in all three organs were mainly identified as CD64+F4/80+ macrophages (SI Appendix, Fig. S5 B and C). Taken together, this demonstrates that obesity increased MR-expressing macrophages in spleen, liver, and WAT.
To test whether increased sMR levels regulate macrophage-mediated inflammatory diseases in vivo, we then analyzed the development of obesity-induced metaflammation in MR-deficient mice (Fig. 6A). Whereas no differences in body weights were found between wild-type and MR-deficient mice on CD, MR-deficient mice gained slightly less weight on HFD (Fig. 6B). This effect was not due to a decrease in caloric intake, as HFD-fed MR−/− mice rather displayed a mild increase in food consumption when compared to WT mice (SI Appendix, Fig. S6A). Furthermore, the HFD-induced reductions in locomotor activity, energy expenditure, and carbohydrate oxidation observed in WT obese mice were found to be partly reverted in MR−/− mice (SI Appendix, Fig. S6 B–E). Analysis of body composition showed that the lower body weight in HFD-fed MR-deficient mice resulted exclusively from a reduction in fat mass, without affecting lean mass (Fig. 6C). Accordingly, the weights of epididymal, mesenteric, and subcutaneous (inguinal) WAT, as well as brown adipose tissue (BAT), were lower in HFD-fed MR-deficient mice (SI Appendix, Fig. S7 A and B). This decrease in WAT mass in HFD-fed MR−/− mice appears to be due to reduced adipocyte hyperplasia rather than hypertrophy (SI Appendix, Fig. S8 A–E). Of note, no significant differences between genotypes were observed in gene expression of proteins involved in adipocyte differentiation and FA metabolism in epididymal white adipose tissue (eWAT) (SI Appendix, Fig. S8F) nor in beiging or thermogenic markers in inguinal white adipose tissue (iWAT) and BAT, respectively (SI Appendix, Fig. S8 G and H). Liver weight was also markedly lower in HFD-fed MR-deficient mice as compared to wild-type controls (Fig. 6D), suggesting a reduction in hepatic steatosis. Indeed, MR-deficient mice were completely protected against HFD-induced hepatic steatosis (Fig. 6 F and G). Accordingly, hepatic triglycerides, total cholesterol and phospholipids contents (Fig. 6H, SI Appendix, Fig. S7F), and hepatic gene expression of lipid droplet-associated proteins (SI Appendix, Fig. S7H) were markedly lower in HFD-fed MR−/− mice. Furthermore, the expression of various genes encoding proteins involved in FA transport and storage were significantly decreased in HFD-fed MR−/− mice, whereas no changes in expression of genes implicated in FA oxidation were observed (SI Appendix, Fig. S7I). Circulating alanine aminotransaminase (ALAT) levels were also markedly decreased in HFD-fed MR-deficient mice (SI Appendix, Fig. S7G).
Fig. 6.
MR regulates WAT and liver macrophage activation, hepatic steatosis, and metabolic homeostasis after HFD feeding. (A) Wild-type (WT) and MR-deficient (MR−/−) mice were fed an HFD or CD for 18 wk. (B) Body weight of mice on diet for 18 wk. (C) Lean and fat mass after 18 wk on diet determined by MRI. (D) Liver weight after 18 wk on diet. (E) Homeostasis Model Assessment of Insulin Resistance index (HOMA-IR) based on blood glucose and fasting insulin. (F and G) Hematoxylin and eosin (H&E) staining (F) and quantification (G) of hepatic steatosis. (H) Levels of liver triglycerides. (I) Intraperitoneal insulin tolerance test. Blood glucose levels were measured at the indicated time points, and the area under the curve (AUC) of the glucose excursion curve was calculated as a surrogate measure for whole-body insulin resistance. (J) Intraperitoneal glucose tolerance test. The AUC of the glucose excursion curve was calculated as a surrogate measure for whole-body glucose intolerance. (K–M) Numbers of total monocytes (K), macrophages (L), and CD11c+ macrophages (M) per gram eWAT determined by flow cytometry. (N) Correlation between eWAT CD11c+ macrophages and whole-body insulin resistance, assessed by the AUC of the glucose excursion curve. (O–R) Numbers of total monocytes (O), KCs (P), and CD11c+ KCs (Q) per gram liver determined by flow cytometry. (R) Correlation between CD11c+ KCs and whole-body insulin resistance. Results are expressed as means ± SEM; n = 5 to 15 mice per group.*P < 0.05.
We next assessed metabolic consequences of MR deficiency. Although no differences in metabolic parameters were observed between genotypes in CD-fed mice, HFD-fed MR-deficient mice displayed lower fasting plasma insulin levels than wild-type mice, whereas fasting glucose levels were unchanged (SI Appendix, Fig. S7 C and D). The calculated Homeostasis Model Assessment of Insulin Resistance index was significantly reduced in HFD-fed MR-deficient mice (Fig. 6E), suggesting that insulin resistance is less severe in these mice. In line with this, whole-body insulin sensitivity (Fig. 6I) and glucose tolerance (Fig. 6J) were higher, despite similar insulin levels (SI Appendix, Fig. S7E), in HFD-fed MR-deficient mice compared to wild-type mice. Importantly, the alleviated hepatic steatosis and whole-body metabolic homeostasis were still observed when HFD-fed MR-deficient mice were weight paired to their wild-type counterparts (SI Appendix, Fig. S11 A–D), indicating that MR deficiency protects against HFD-induced metabolic dysfunctions independently of body weight changes. Altogether, these data indicate that the MR might contribute to obesity-induced metabolic dysfunctions.
MR Promotes Inflammatory Macrophage Accumulation in eWAT and Liver during Obesity.
Since our in vitro data demonstrate that the MR reprograms macrophages toward an inflammatory phenotype, we next investigated whether the observed metabolic changes in MR-deficient mice might be caused by reduced proinflammatory macrophage activation in metabolic tissues.
As previously reported, HFD significantly increased obesity-associated proinflammatory CD11c+ adipose tissue macrophages (ATMs) in eWAT of wild-type mice (33), whereas total ATMs and monocytes were not affected (Fig. 6 K–M and SI Appendix, Fig. S9). Remarkably, while no significant differences in total ATMs and monocyte numbers were observed between genotypes, inflammatory CD11c+ ATM numbers were found to be significantly higher in HFD-fed MR-expressing wild-type mice as compared to MR-deficient mice (Fig. 6 K–M) and strongly correlated with whole-body insulin resistance (Fig. 6N).
In the liver, HFD significantly increased proinflammatory CD11c+ KCs and monocytes in wild-type mice, while total KCs were not affected (Fig. 6 O–Q). Similar to what was observed in eWAT, inflammatory CD11c+ KCs, but also monocytes, were more abundant in liver of MR-expressing wild-type mice as compared to MR-deficient mice, of which the CD11c+ KCs again correlated strongly with insulin resistance (Fig. 6R), while total KCs were not affected (Fig. 6 O–Q). This was associated with higher expression of genes involved in proinflammatory macrophage activation in liver and WAT of MR-expressing wild-type mice (SI Appendix, Fig. S10 B and D). Importantly, these differences in proinflammatory macrophage abundances in metabolic tissues were still present when wild-type and MR-deficient mice were weight paired (SI Appendix, Fig. S11 E and F), indicating that also the regulation of obesity-induced proinflammatory macrophages by the MR is independent of changes in body weight. By contrast, MR deficiency neither affected other myeloid and lymphocyte subsets nor T cell–associated cytokines gene expression in eWAT or liver from HFD-fed mice (SI Appendix, Fig. S10).
Taken together, these data provide first indications that the MR might contribute to obesity-induced metaflammation.
sMR Treatment Induces Proinflammatory Cytokines, Metabolic Dysfunctions, and Increased Proinflammatory Macrophages.
To univocally determine the role of the MR in metaflammation, we investigated whether in vivo administration of sMR in lean mice is able to induce macrophage activation and metabolic dysfunctions by intraperitoneally injecting FcMR or isotype control every 3 d for 4 wk (Fig. 7A). We first monitored circulating cytokine levels in response to a single intraperitoneal injection of FcMR in CD-fed mice. In accordance with our in vitro experiments, even a single injection of FcMR acutely increased serum levels of TNF and IL-6 and the chemokine MCP-1/CCL2 compared to isotype control-treated mice (Fig. 7B).
Fig. 7.
Increased MR levels regulate whole-body metabolism and promote inflammation. (A) CD-fed WT mice were injected i.p. with 4.82 μmoles/mouse FcMR or isotype control for 4 wk. (B) Serum cytokine concentrations were determined by cytometric bead array (CBA) at the indicated timepoints post–first injection. (C and D) Graphs depicting body weight over time (C) and overall change in body weight (D). (E and F) Intraperitoneal insulin tolerance test after FcMR treatment of CD-fed mice. Graph in F shows changes in circulating glucose levels at 15 min postinsulin i.p. (G) The number of F4/80+ magnetic cell separation (MACS)-sorted eWAT macrophages was determined after 4 wk of treatment. (H) Expression of inflammatory genes in eWAT was monitored by qPCR. (I) Cytokine secretion by F4/80+ macrophages from eWAT was determined by CBA after stimulation with 100 ng/mL LPS. BW: body weight. Results are expressed as means ± SEM; n = 3 to 5 mice per group. *P < 0.05.
After 4 wk of treatment, we monitored a mild increase in body weight in FcMR-treated mice compared to control mice (Fig. 7 C and D). In addition, insulin sensitivity, as measured by an acute drop in blood glucose levels following insulin intraperitoneal (i.p.) injection, was reduced in FcMR-treated mice compared to control mice (Fig. 7 E and F), confirming the detrimental effect of the sMR on whole-body metabolic homeostasis. Importantly, the effect of FcMR on HFD-induced insulin resistance was even more pronounced when mice were fed an HFD concomitantly with FcMR treatment for 4 wk, underlining the inflammatory effect of the sMR in mice fed an HFD (SI Appendix, Fig. S12).
FcMR treatment increased macrophage numbers in eWAT of lean mice (Fig. 7G). Moreover, gene expression of Il1b, Tnf, Il6, and Ccl2 was increased in eWAT of FcMR-treated lean mice (Fig. 7H). Accordingly, macrophages isolated from these mice showed increased secretion of most of these cytokines upon stimulation with LPS (Fig. 7I), demonstrating that, in lean mice, increased serum sMR levels induce the secretion of proinflammatory cytokines, induce whole-body insulin resistance, and promote macrophage activation in metabolic tissues in vivo.
Discussion
The MR as a member of the C-type lectin family has mainly been described as an endocytic receptor recognizing glycosylated antigens and mediating antigen processing and presentation (34, 35). However, the extracellular region of the MR can be shed by metalloproteases and released as a soluble protein in the extracellular space. Consequently, the sMR is detectable in murine and human serum (13, 14), and recent studies reported an increase of serum sMR levels in a variety of inflammatory diseases and serum sMR levels directly correlated with severity of disease and mortality (15–20). Here, we demonstrated that, in addition to a mere phenotypic correlation, the sMR plays a direct, functional role in macrophage activation, driving reprogramming toward a proinflammatory phenotype. By interacting with and inactivating CD45, the sMR reprograms macrophages via activation of Src/Akt signaling and nuclear translocation of NF-κB. In vivo, sMR levels were increased in obese mice and humans as compared to lean controls, and we found that MR deficiency reduced adipose tissue and liver proinflammatory macrophages and protected against obesity-induced metabolic dysfunctions. Consistently, treatment of lean mice with sMR acutely increased serum proinflammatory cytokines and induced both tissue macrophage activation and systemic insulin resistance.
Although wild type (WT) and MR−/− mice on CD were phenotypically similar in term of body composition and whole-body metabolic homeostasis, MR−/− mice on HFD appeared to be slightly resistant to HFD-induced body weight gain and displayed markedly improved insulin sensitivity and glucose tolerance when compared to WT mice. However, it is important to underline that body weight pairing of obese mice from the two genotypes clearly indicated that the lower body weight observed in HFD-fed MR−/− mice only marginally contributed to their lower hepatic steatosis, improved whole-body metabolic homeostasis, and phenotypic changes in AT and liver macrophages. In addition, we found that sMR injections promoted inflammation but only induced mild insulin resistance in lean WT mice. Although significant, this effect was modest and may be explained by several factors, including the timing of administration and dosage, but also by the fact that lean mice are highly insulin sensitive. Of note, injecting sMR at the start of HFD feeding accelerated the development of insulin resistance, which supports a role for sMR in promoting metaflammation.
In this study, we found increased MR+ cells in spleen, liver, and eWAT upon HFD feeding, which were almost exclusively macrophages. Since cell surface MR can be shed and released as a soluble form, obesity-induced changes in tissue homeostasis may increase MR expression and sMR release by macrophages, creating increased local and systemic sMR levels to promote macrophage-mediated inflammation and metabolic dysfunctions. An important factor in this process could be the ligand-inducible TF PPAR-γ, which is activated, among others, by free FAs. Indeed, single-cell RNA sequencing analysis of adipose tissue immune cells revealed that PPAR signaling is among the up-regulated pathways in obesity-induced lipid-associated macrophages in both mice and humans (36). Since Mrc1 (encoding the MR) is also a direct target gene of PPAR-γ (37), obesity-induced activation of PPAR-γ in macrophages may lead to enhanced transcription of the MR, eventually resulting in increased sMR levels. One of the limitations of our study was the use of constitutive whole-body MR−/− mice instead of a myeloid cell-specific knockout model, which is currently not available. As such, the main cellular source of circulating proinflammatory sMR remains to be confirmed, although it is conceivable that a significant part of the increased circulating sMR during obesity may also be derived from nonmyeloid MR-expressing cells, such as LSECs.
Our data demonstrate that macrophage activation by the MR was due to MR-mediated inhibition of CD45, which in turn leads to activation of Src and Akt, and nuclear translocation of NF-κB. CD45 has been postulated to inhibit Src kinases (27) and Akt (28), a known regulator of NF-κB (30, 31, 38). Here, we show that CD45-mediated dephosphorylation of Src induces Akt-mediated nuclear translocation of NF-κB and that the sMR uses this signaling pathway to induce macrophage reprogramming toward an inflammatory phenotype. MR-mediated macrophage activation depended on Akt signaling, as its inhibition abrogated MR-mediated nuclear translocation of NF-κB and ensuing TNF and IL-6 secretion by macrophages. Additionally, Akt has been postulated as a regulator that can fine-tune NF-κB–mediated responses through regulating efficient binding of p65 to specific target promoters (32). Of note, these authors demonstrated that NF-κB–mediated expression of TNF was particularly sensitive to Akt signaling, which is in accordance with the Akt-dependency of TNF expression after MR-induced activation of macrophages described here.
The immunometabolic phenotype of obese MR-deficient mice resembles that of mice deficient for MGL/CLEC10A, another member of the C-type lectin family. Indeed, these mice displayed reduced hepatic steatosis, insulin resistance, and glucose intolerance upon HFD when compared to wild-type mice, a feature that was associated with lower AT proinflammatory macrophages (39). In another context, MGL/CLEC10A was also shown to bind and inactivate CD45 (40), offering the possibility that MGL/CLEC10A can directly induce macrophage reprogramming by inhibition of CD45, similar to the MR. Future studies will have to reveal whether MGL/CLEC10A indeed plays a direct role in macrophage activation, whether its expression is also increased in HFD-induced obesity, and whether this may be mediated by a soluble form of MGL/CLEC10A.
In summary, we demonstrate that a soluble form of the MR reprograms macrophages toward an inflammatory phenotype by interacting with CD45 on the surface of these macrophages. MR-mediated inhibition of CD45 activated Src and Akt kinases, leading to nuclear translocation of NF-κB and induction of a transcriptional program that ultimately results in enhanced inflammatory cytokine production. Furthermore, sMR levels in serum of obese mice and humans are increased, strongly correlating with body weight and adiposity, a feature that needs however to be confirmed in female mice and larger population studies. Accordingly, MR deficiency resulted in fewer adipose tissue and liver proinflammatory macrophages and protection against hepatic steatosis and metaflammation, whereas increased MR levels induced elevated serum proinflammatory cytokines, macrophage activation, and metabolic dysfunctions. Altogether, our results identify sMR as a regulator of proinflammatory macrophage activation and could contribute to the development of new therapeutic strategies for metaflammation and other hyperinflammatory diseases. Targeting MR-mediated activation of macrophages using antibodies, nanobodies, aptamers, or small molecules that could prevent MR interacting with macrophage CD45RO might open new possibilities for therapeutics aimed at dampening (meta)inflammation.
Materials and Methods
Antibodies and Reagents.
A detailed list of antibodies and reagents is included in SI Appendix, Supplementary Methods.
Generation of Bone Marrow–Derived Macrophages.
Bone marrow was flushed from the femurs and tibias of mice and cultured for 7 d in medium containing 2.5% supernatant of a Granulocyte Macrophage Colony-Stimulating Factor (GM-CSF)–producing cell line (total concentration 150 ng/mL).
Generation and Purification of FcMR.
FcMR proteins (encompassing the CR region, the FN II domain, and CTLD1-2 fused to the Fc region of hIgG1) and isotype controls (Fc region of hIgG1) were generated as described previously (21). For all in vitro experiments, FcMR and isotype controls were used in a concentration of 10 µg/mL.
Monitoring Secretion of TNF, IL-6, IL-1β, and IL-12.
Macrophages were incubated with 10 µg/mL FcMR or isotype control, 300 ng/mL recombinant MR (2535-MM-050, R&D Systems), 30 ng/mL purified sMR, 3 µM PP2, 1 µM KX2-391, 1 µM A419259, or 5 µM MK-2206. After 2 h, LPS was added in the given concentrations. Unless indicated differently, supernatants were collected at 3 h (TNF) or 18 h (IL-6, IL-12p70) post-LPS stimulation. For secretion of IL-1β, cells were incubated with LPS for 3 h and with 10 mM nigericin for another 1 h. Secreted cytokine levels were measured by enzyme-linked immunosorbent assay (ELISA). Levels of TNF, IL-6, IL-1β, and MCP-1 in the circulation at 2 h post-FcMR, isotype control injection, or in culture supernatant of LPS-treated eWAT macrophages were measured using the cytometric bead array kits (BD Biosciences), per manufacturer’s recommendations.
Mice and Diet.
All animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Institute for Laboratory Animal Research and have received approval from the university Ethical Review Boards (DEC No. 12199; Leiden University Medical Center, Leiden, The Netherlands). To reduce variation due to sex hormone cycles on whole-body metabolism, male mice were used for all in vivo experiments. MR−/− mice were generated on C57BL/6J background, regularly backcrossed to C57BL/6J, and compared to age-matched C57BL/6J wild-type mice from the same mouse facility. To minimize eventual effects of genotype-dependent microbiota differences on metabolic and immunological outcomes, the beddings of WT and MR−/− mice were frequently mixed before randomization. All mice were housed in a temperature-controlled room with a 12-h light–dark cycle. Throughout the experiment, food and tap water were available ad libitum. The 8- to 10-wk-old male mice were randomized according to total body weight, lean and fat mass, and fasting plasma glucose, insulin, TC and TG levels, after which they were fed an HFD (45% energy derived from fat, D12451, Research Diets) or a CD (10% energy derived from fat, D12450B, Research Diets) for 18 wk. An a priori power calculation was done. Analysis was performed blinded to the conditions. For in vivo FcMR treatment, C57BL/6J wild-type littermate mice were randomized as above. Subsequently, mice were biweekly intraperitoneally injected with 50 µg FcMR or 6.75 µg isotype control, to yield the same administered dose of hIgG1, for 4 wk while either on CD or concomitant with the start of HFD feeding.
Quantification and Statistical Analysis.
All data are presented as mean ± SEM (SEM). Statistical analysis was performed using GraphPad Prism 8.0 (GraphPad Software) with unpaired t test, one-way or two-way ANOVA, followed by Fisher’s post hoc test. Differences between groups were considered statistically significant at P < 0.05. Outliers were identified according to the two-SD method. Single data points represent mean values of distinct independent experiments (in vitro) or individual mice (in vivo).
Supplementary Material
Acknowledgments
This work was funded by Deutsche Forschungsgemeinschaft (German Research Foundation) SFB1454 (Project 432325352) and under Germany’s Excellence Strategy EXC2151 (Project 390873048) (to S.B.), a European Federation for the Study of Diabetes (EFSD)/Lilly Research Grant Fellowship from the European Federation for the Study of Diabetes (to B.G.), Dutch Research Council (NWO) Graduate School Program Grant 022.006.010 (to H.J.P.v.d.Z.), and the Dutch Organization for Scientific Research (ZonMW TOP Grant 91214131 to B.G. and M.Y.). We thank Frank Otto and Arifa Ozir-Fazalalikhan for their invaluable technical help.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103304118/-/DCSupplemental.
Data Availability
The complete RNA-seq analysis including code and count data can be found under jsschrepping/Embgenbroich_2020 at https://github.com/schultzelab. Additionally, the unprocessed RNA-seq data are available online in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov) under accession number GSE145369.
Change History
August 6, 2021: The author line has been updated.
References
- 1.Brestoff J. R., Artis D., Immune regulation of metabolic homeostasis in health and disease. Cell 161, 146–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotamisligil G. S., Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Lackey D. E., Olefsky J. M., Regulation of metabolism by the innate immune system. Nat. Rev. Endocrinol. 12, 15–28 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil G. S., Murray D. L., Choy L. N., Spiegelman B. M., Tumor necrosis factor alpha inhibits signaling from the insulin receptor. Proc. Natl. Acad. Sci. U.S.A. 91, 4854–4858 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shulman G. I., Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N. Engl. J. Med. 371, 2237–2238 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Neuschwander-Tetri B. A., Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology 52, 774–788 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Lanthier N., et al., Kupffer cell activation is a causal factor for hepatic insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G107–G116 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Morinaga H., et al., Characterization of distinct subpopulations of hepatic macrophages in HFD/obese mice. Diabetes 64, 1120–1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Pomares L., The mannose receptor. J. Leukoc. Biol. 92, 1177–1186 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K., Donovan M. J., Rogers R. A., Ezekowitz R. A., Distribution of murine mannose receptor expression from early embryogenesis through to adulthood. Cell Tissue Res. 292, 311–323 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Burgdorf S., Kautz A., Böhnert V., Knolle P. A., Kurts C., Distinct pathways of antigen uptake and intracellular routing in CD4 and CD8 T cell activation. Science 316, 612–616 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Burgdorf S., Lukacs-Kornek V., Kurts C., The mannose receptor mediates uptake of soluble but not of cell-associated antigen for cross-presentation. J. Immunol. 176, 6770–6776 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Jordens R., Thompson A., Amons R., Koning F., Human dendritic cells shed a functional, soluble form of the mannose receptor. Int. Immunol. 11, 1775–1780 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Martínez-Pomares L., et al., A functional soluble form of the murine mannose receptor is produced by macrophages in vitro and is present in mouse serum. J. Biol. Chem. 273, 23376–23380 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Saha B., et al., Biomarkers of macrophage activation and immune danger signals predict clinical outcomes in alcoholic hepatitis. Hepatology 70, 1134–1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki Y., et al., Macrophage mannose receptor, CD206, predict prognosis in patients with pulmonary tuberculosis. Sci. Rep. 8, 13129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rødgaard-Hansen S., et al., Increased concentrations of the soluble mannose receptor in serum from patients with pneumococcal bacteraemia, and prediction of survival. Infect. Dis. (Lond.) 47, 203–208 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Ding D., Song Y., Yao Y., Zhang S., Preoperative serum macrophage activated biomarkers soluble mannose receptor (sMR) and soluble haemoglobin scavenger receptor (sCD163), as novel markers for the diagnosis and prognosis of gastric cancer. Oncol. Lett. 14, 2982–2990 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loonen A. J. M., et al., Soluble mannose receptor levels in blood correlate to disease severity in patients with community-acquired pneumonia. Immunol. Lett. 206, 28–32 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Andersen E. S., et al. Macrophage-related serum biomarkers soluble CD163 (sCD163) and soluble mannose receptor (sMR) to differentiate mild liver fibrosis from cirrhosis in patients with chronic hepatitis C: A pilot study. Eur. J. Clin. Microbiol. Infect. Dis. 33, 117–122 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Schuette V., et al., Mannose receptor induces T-cell tolerance via inhibition of CD45 and up-regulation of CTLA-4. Proc. Natl. Acad. Sci. U.S.A. 113, 10649–10654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viola A., Munari F., Sánchez-Rodríguez R., Scolaro T., Castegna A., The metabolic signature of macrophage responses. Front. Immunol. 10, 1462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Bossche J., O’Neill L. A., Menon D., Macrophage immunometabolism: Where are we (going)? Trends Immunol. 38, 395–406 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Xue J., et al., Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martínez-Pomares L., et al., Cell-specific glycoforms of sialoadhesin and CD45 are counter-receptors for the cysteine-rich domain of the mannose receptor. J. Biol. Chem. 274, 35211–35218 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Pilling D., Fan T., Huang D., Kaul B., Gomer R. H., Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PLoS One 4, e7475 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shrivastava P., Katagiri T., Ogimoto M., Mizuno K., Yakura H., Dynamic regulation of Src-family kinases by CD45 in B cells. Blood 103, 1425–1432 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Chen J., The Src/PI3K/Akt signal pathway may play a key role in decreased drug efficacy in obesity-associated cancer. J. Cell. Biochem. 110, 279–280 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Bai D., Ueno L., Vogt P. K., Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int. J. Cancer 125, 2863–2870 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie X., et al., Connexin43 mediates NF-κB signalling activation induced by high glucose in GMCs: Involvement of c-Src. Cell Commun. Signal. 11, 38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abu-Amer Y., et al., Tumor necrosis factor-alpha activation of nuclear transcription factor-kappaB in marrow macrophages is mediated by c-Src tyrosine phosphorylation of Ikappa Balpha. J. Biol. Chem. 273, 29417–29423 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Cheng J., Phong B., Wilson D. C., Hirsch R., Kane L. P., Akt fine-tunes NF-κB-dependent gene expression during T cell activation. J. Biol. Chem. 286, 36076–36085 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lumeng C. N., Bodzin J. L., Saltiel A. R., Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117, 175–184 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauen J., et al., Enhanced cross-presentation and improved CD8+ T cell responses after mannosylation of synthetic long peptides in mice. PLoS One 9, e103755 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreer C., et al., N-glycosylation converts non-glycoproteins into mannose receptor ligands and reveals antigen-specific T cell responses in vivo. Oncotarget 8, 6857–6872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaitin D. A., et al., Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178, 686–698.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klotz L., et al., Increased antigen cross-presentation but impaired cross-priming after activation of peroxisome proliferator-activated receptor gamma is mediated by up-regulation of B7H1. J. Immunol. 183, 129–136 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Lluis J. M., Buricchi F., Chiarugi P., Morales A., Fernandez-Checa J. C., Dual role of mitochondrial reactive oxygen species in hypoxia signaling: Activation of nuclear factor-kappaB via c-SRC and oxidant-dependent cell death. Cancer Res. 67, 7368–7377 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Westcott D. J., et al., MGL1 promotes adipose tissue inflammation and insulin resistance by regulating 7/4hi monocytes in obesity. J. Exp. Med. 206, 3143–3156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Vliet S. J., Gringhuis S. I., Geijtenbeek T. B. H., van Kooyk Y., Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 7, 1200–1208 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete RNA-seq analysis including code and count data can be found under jsschrepping/Embgenbroich_2020 at https://github.com/schultzelab. Additionally, the unprocessed RNA-seq data are available online in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov) under accession number GSE145369.