Significance
GHB is a natural brain metabolite of GABA, previously reported to be neuroprotective. However, the high-affinity binding site for GHB has remained elusive for almost 40 y. We here unveil CaMKIIα, a highly important neuronal kinase, as the long-sought-after GHB high-affinity target. Via a specific interaction within the central hub domain of CaMKIIα, GHB analogs act to stabilize the hub oligomer complex. This interaction potentially explains pronounced neuroprotective effects of GHB analogs in cultured neurons exposed to a chemical insult and in mice exposed to ischemia. The postischemic treatment effects of GHB analogs underline these compounds as selective and high-affinity potential drug candidates and CaMKIIα as a relevant pharmacological target for stroke therapy.
Keywords: photoaffinity labeling, x-ray crystallography, HOCPCA, excitotoxicity, photothrombotic stroke
Abstract
Ca2+/calmodulin-dependent protein kinase II alpha subunit (CaMKIIα) is a key neuronal signaling protein and an emerging drug target. The central hub domain regulates the activity of CaMKIIα by organizing the holoenzyme complex into functional oligomers, yet pharmacological modulation of the hub domain has never been demonstrated. Here, using a combination of photoaffinity labeling and chemical proteomics, we show that compounds related to the natural substance γ-hydroxybutyrate (GHB) bind selectively to CaMKIIα. By means of a 2.2-Å x-ray crystal structure of ligand-bound CaMKIIα hub, we reveal the molecular details of the binding site deep within the hub. Furthermore, we show that binding of GHB and related analogs to this site promotes concentration-dependent increases in hub thermal stability believed to alter holoenzyme functionality. Selectively under states of pathological CaMKIIα activation, hub ligands provide a significant and sustained neuroprotection, which is both time and dose dependent. This is demonstrated in neurons exposed to excitotoxicity and in a mouse model of cerebral ischemia with the selective GHB analog, HOCPCA (3-hydroxycyclopent-1-enecarboxylic acid). Together, our results indicate a hitherto unknown mechanism for neuroprotection by a highly specific and unforeseen interaction between the CaMKIIα hub domain and small molecule brain-penetrant GHB analogs. This establishes GHB analogs as powerful tools for investigating CaMKII neuropharmacology in general and as potential therapeutic compounds for cerebral ischemia in particular.
The calcium/calmodulin-dependent protein kinase II alpha subunit (CaMKIIα) is a central mediator of synaptic plasticity and responds to minute fluctuations in calcium (Ca2+) (1). The CaMKIIα holoenzyme is a large protein assembly of 12 to 14 subunits, each consisting of a kinase domain flexibly linked to the central hub domain. The hub domain is conserved through evolution (2). It organizes the holoenzyme into oligomeric structures (3, 4), yet displays remarkable dynamics (5). This correlates well with an emerging functional importance in activation-triggered destabilization and release of vertical dimers that may enable spreading of activity (6, 7). Furthermore, the hub domain has been reported to interact directly with the kinase domains (8–10) to confer allosteric control of kinase activity (9). The importance of preserving hub integrity is further evident from a human patient with a mutation in the hub (p.His477Tyr) causing defective oligomerization and severe neurodevelopmental defects (11). Thus far, pharmacological modulation of the hub domain has never been demonstrated but would constitute an attractive approach to regulate overall kinase function in cases of CaMKIIα aberrant activity.
Functionally, CaMKIIα is activated in a highly cooperative manner, initiated by increases in intracellular Ca2+, Ca2+/CaM binding, and autophosphorylation at residue Thr286 in the regulatory segment (12). This is then accompanied by translocation of CaMKIIα to the postsynaptic density (PSD) (13). In cases of excessive stimuli, such as ischemic brain injury or glutamate-mediated excitotoxicity, Thr286 autophosphorylation permits Ca2+/CaM-independent autonomous activity which can persist for hours (14–16) and cause cell death (17).
The natural brain substance γ-hydroxybutyrate (GHB) is a metabolite of γ-aminobutyric acid (GABA) which has been reported to be neuroprotective in mammals (18–20). GHB binds with high affinity to an until-now unknown specific binding protein highly expressed in forebrain regions (21). This site is distinct from GABAB receptors also known to bind GHB, albeit with low affinity (22). We here reveal CaMKIIα as the long-sought-after specific GHB high-affinity binding site (SI Appendix, Fig. S1). Moreover, we show that the highly selective and brain-penetrant GHB analog 3-hydroxycyclopent-1-enecarboxylic acid (HOCPCA) (23) confers significant neuroprotection in pathological states of CaMKII activation, such as after an ischemic injury. This is plausibly explained by the pronounced effect of GHB analogs on CaMKIIα hub stabilization upon binding and consequently functional regulation of the holoenzyme, although a causal link remains to be fully proven.
Results
CaMKIIα Is the Specific High-Affinity Target for GHB.
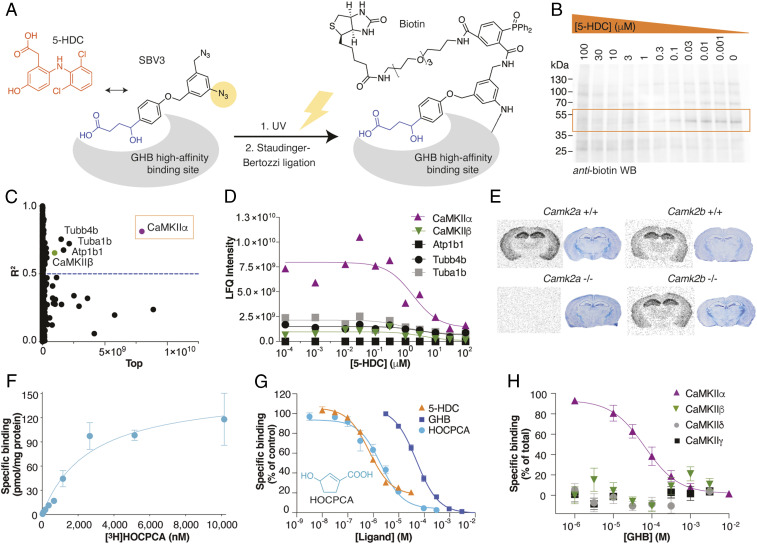
To enable unbiased identification of the elusive GHB high-affinity binding site, we designed a photolabile diazide–labeled GHB analog, 4-(4-((3-azido-5-(azidomethyl)benzyl)oxy)phenyl)-4-hydroxybutanoate (SBV3), based on previous studies on GHB substituted in the 4-position with biaromatic moieties (24, 25). SBV3 bears two orthogonal azides: a photolabile aromatic azide for linking and an aliphatic azide that is not photolabile but is employed in a bioorthogonal click reaction (26) (Fig. 1A). We found that this compound possesses high affinity for the binding site present in cortical homogenate (KI 66 nM; SI Appendix, Fig. S2). Both the Huisgen click reaction and the Bertozzi–Staudinger ligation were employed for biotin labeling of the photolinked target protein under various conditions, with the latter giving the best result (SI Appendix, Fig. S2 A–C). To enable differentiation of endogenously biotinylated proteins from the biotin-labeled GHB target, SBV3 was incubated with rat hippocampal membranes in competition with 5-hydroxydiclofenac (5-HDC), a derivative of diclofenac (27) and incidentally, the most high-affinity GHB ligand reported to date (KI 35 nM; SI Appendix, Fig. S2). Hereby, we could obtain concentration-dependent binding profiles (Fig. 1B and SI Appendix, Fig. S2). Streptavidin affinity purification followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis identified 1,184 proteins that were analyzed for concentration-dependent profiles. CaMKIIα was identified as the top candidate (R2 = 0.81; abundance 7.97 × 109) (Fig. 1C), clearly superior with respect to abundance and goodness of the curve fit to the next-in-line candidates, four of which are in fact known CaMKIIα interactors (28) (Fig. 1D; summarized in SI Appendix, Table S1). Furthermore, the identified molecular mass (55 kDa), the brain regional expression profile (forebrain abundance), and ontogenesis match very well with both CaMKIIα and our previously reported specific GHB binding site (29–31).
Fig. 1.
Identification of CaMKIIα as the specific GHB high-affinity target. (A–D) Target identification using a combination of photoaffinity labeling (PAL), affinity purification, and chemical quantitative proteomics. (A) Bioorthogonal approach using the photolabile diazide–labeled GHB analog SBV3 (GHB moiety in blue), in competition with 5-HDC (orange) for PAL followed by biotin-ligation. (B) Representative anti-biotin Western blot after PAL and concentration-dependent competition with 5-HDC (see also SI Appendix, Fig. S2). (C) Identification of CaMKIIα from LC-MS/MS data as the best hit from nonlinear regression analysis for all proteins and (D) concentration-dependent competition of the best hits (proteins were quantified using the label-free quantification [LFQ] algorithm) (see also Dataset S1). (E) Target validation by [3H]HOCPCA autoradiography using brain slices from Camk2a and Camk2b+/+ and -/- mice (cresyl violet staining for tissue visualization) (see also SI Appendix, Fig. S3). (F–H) Target validation by [3H]HOCPCA binding to whole-cell homogenate from HEK293T cells transfected with CaMKIIα. (F) [3H]HOCPCA saturation binding to CaMKIIα (n = 5); shown is one representative curve (means ± SD). (G) [3H]HOCPCA competition binding to CaMKIIα in the presence of GHB (n = 3), HOCPCA (n = 5), and 5-HDC (n = 3), pooled data (means ± SEM) (see also SI Appendix, Table S1). (H) Subtype-selective specific [3H]HOCPCA binding for CaMKIIα cf CaMKIIβ. Data are pooled (n = 3) for each subtype and depicted as specific binding (% of total) (see also SI Appendix, Fig. S4A).
Validation of CaMKIIα as the High-Affinity GHB Target.
To validate CaMKIIα as the specific GHB target, we carried out a number of experiments in native tissues, cortical homogenate, and recombinant CaMKIIα utilizing selective GHB analogs. These included the cyclic GHB analog HOCPCA that displays no affinity for low-affinity GABAB receptors (23), the commercially available NCS-382, and their derived radioligands (32–34). By means of in vitro autoradiography, we show that [3H]HOCPCA (Fig. 1E), [3H]NCS-382, and [3H]GHB (SI Appendix, Fig. S3) display intact binding to brain tissue of Camk2a+/+, Camk2b+/+, and Camk2b-/- mice but complete lack of binding to slices from Camk2a-/- mice. This striking CaMKII alpha subtype selectivity was further corroborated by the absence of binding of the photoligand SBV3 to homogenate from Camk2a-/- mice (SI Appendix, Fig. S3). Additionally, to validate binding in an overexpressed system, we prepared whole-cell homogenates from CaMKIIα–transfected HEK293T cells and demonstrate a saturable [3H]HOCPCA binding profile (Fig. 1F; summarized in SI Appendix, Table S2), as well as concentration-dependent inhibition by GHB, HOCPCA, and 5-HDC in the expected relative rank order (Fig. 1G; summarized in SI Appendix, Table S3). Finally, no [3H]HOCPCA binding was observed to CaMKIIβ/γ/δ subtypes heterologously expressed in HEK293T cells (Fig. 1H and SI Appendix, Fig. S4). To our knowledge, such CaMKIIα subtype selectivity is completely unforeseen.
Structural Evidence for a Hub Domain Ligand Binding Site.
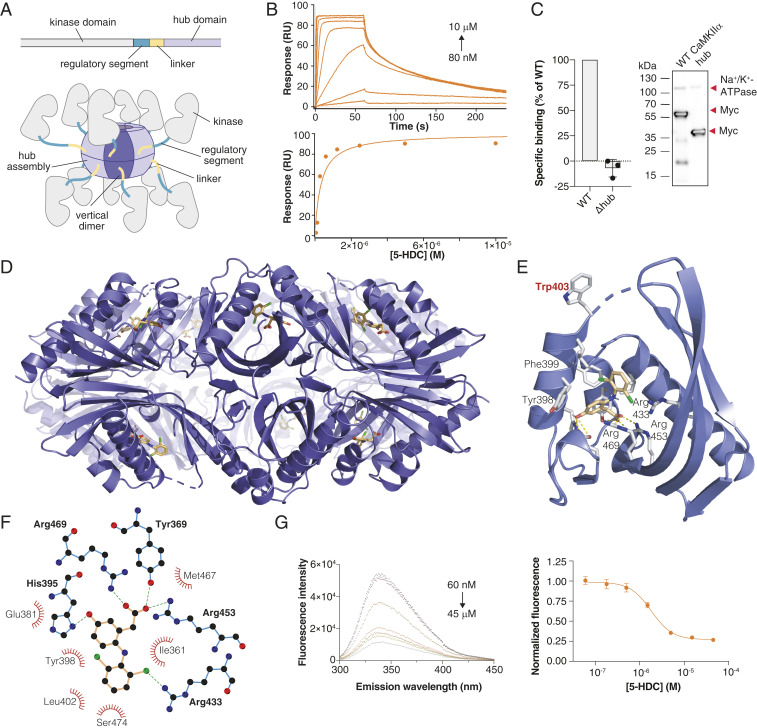
Potential ligand binding sites in CaMKIIα include the kinase domain, the regulatory segment, and the hub domain (Fig. 2A). From previous crystal structures (4), we hypothesized that a deep cavity in the hub that contains several positively charged arginine (Arg) residues is the binding pocket for GHB analogs. By surface plasmon resonance, we detected binding to the isolated CaMKIIα hub by GHB analogs, the strongest interaction being with 5-HDC (KD 0.30 µM) (Fig. 2B and SI Appendix, Fig. S4 B–F). Further evidence for selective binding to the hub was obtained using HEK293T cells transiently expressing a CaMKIIα hub deletion mutant (Δhub, 35 kDa) that showed complete absence of [3H]HOCPCA binding (Fig. 2C). Final proof of the binding location was obtained from cocrystallization with the human hub domain. Using a previously reported stabilized form of the hub (6x Hub) (2), we obtained an x-ray crystal structure of the tetradecameric CaMKIIα hub oligomer bound to 5-HDC (2.2-Å resolution) (Fig. 2 D–F, further structural data are provided in SI Appendix, Fig. S5 and Table S4). The structure highlights direct interactions of 5-HDC with the predicted Arg residues 433, 453, 469, and His395 (Fig. 2F) (4). The structure also reveals a distinct conformational shift in Trp403 located at the edge of the binding pocket upon 5-HDC binding (Fig. 2E). This “Trp flip” was further consolidated using an in-house–developed intrinsic tryptophan fluorescence assay for the isolated hub domain protein (Fig. 2G, modeled in SI Appendix, Fig. S6). Convincingly, in this assay, 5-HDC exhibited a concentration-dependent quenching of the fluorescence in both the 6x Hub mutant and the wild-type (WT) hub protein (half maximal inhibitory concentration [IC50] values of 1.81 and 1.47 µM, respectively), which was, however, not seen for HOCPCA (SI Appendix, Fig. S6). This likely reflects the smaller size of HOCPCA, allowing for the simultaneous occupancy of HOCPCA and Trp403 inside the binding cavity, also supported by modeling (SI Appendix, Fig. S6C).
Fig. 2.
GHB analogs bind the CaMKIIα hub domain. (A) Schematic of a single CaMKIIα subunit composed of a kinase domain (gray), regulatory segment (green), linker (yellow), and hub domain (lilac). Twelve to 14 hub domains oligomerize into the holoenzyme, shown here in an activated form. (B) Concentration-dependent binding of 5-HDC to immobilized CaMKIIα 6x Hub measured by surface plasmon resonance (Top); Langmuir-binding isotherm (Bottom), representative data (see also SI Appendix, Fig. S4 B–F). (C) Absence of [3H]HOCPCA binding to the CaMKIIα mutant lacking the hub domain (Δhub) with representative Western blot showing expected sizes. Na+/K+-ATPase was used as a loading control; red arrows indicate the relevant bands. (D) X-ray crystal structure of 5-HDC bound to the CaMKIIα 6x Hub (14-mer). (E) Close-up view of a single hub subunit showing the key molecular interactions with displacement (flip) of Trp403 with ligand bound highlighted. (F) Ball and stick model of key binding residues (bold), nearby residues, and hydrogen bonds in green-dashed lines (for electron densities, reference SI Appendix, Fig. S5). (G) Quenching of intrinsic tryptophan fluorescence caused by Trp403 flip (6× Hub) with increasing concentrations of 5-HDC (Left) and resulting inhibition curve (Right) (n = 8), pooled data (means ± SEM) (see also SI Appendix, Fig. S6 A–C).
GHB Analogs Stabilize CaMKIIα Hub Oligomer Formation.
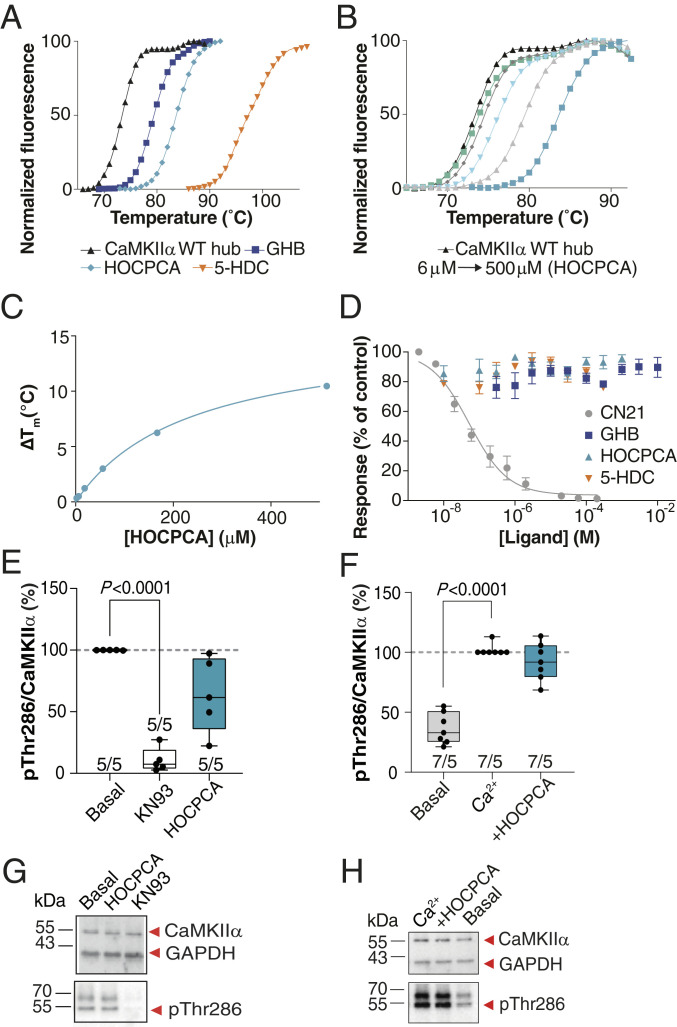
Intrigued by GHB analogs binding in a specific hub cavity and the functional importance of hub oligomerization for normal CaMKIIα function (6, 7, 35), we investigated a potential effect of the compounds on hub dynamics. Employing a thermal shift assay of the purified CaMKIIα WT hub protein using differential scanning fluorometry, we observed pronounced effects on protein denaturation. In the presence of saturating concentrations of each of the three compounds GHB, HOCPCA, and 5-HDC, extensive concentration-dependent stabilization of the hub domain was observed, resulting in respective maximum Tm increases of 13, 15, and 29 °C (Fig. 3 A–C and SI Appendix, Fig. S6 D and E). As shown, the thermal shift curves were concentration-dependent with potencies corresponding with the relative affinity rank order of the compounds obtained in radioligand binding assays on cortical homogenate: 5-HDC > HOCPCA > GHB (SI Appendix, Table S3). However, whereas these data confirm the binding site to the hub and a marked effect on hub stability, they offer no direct interpretation of downstream functional effects resulting from this.
Fig. 3.
GHB analogs stabilize the hub but fail to affect CaMKII enzymatic activity under basal, nonpathological conditions. (A) Right-shifted thermal shift assay melting curves of CaMKIIα WT hub upon binding of GHB, HOCPCA and 5-HDC, (B) HOCPCA concentration dependence, and (C) saturation isotherm; representative data (see also SI Appendix, Fig. S5 D and E). (D) GHB analogs do not affect syntide-II phosphorylation by CaMKIIα, CN21 as positive control, pooled data (n = 3, means ± SEM). (E and F) CaMKIIα Thr286 autophoshorylation quantified by Western blot in cultured cortical neurons (days in vitro [DIV] 18 to 20). No effect of HOCPCA under (E) basal and (F) Ca2+-stimulated conditions. Shown is quantification of mean band intensities of Ca2+-stimulated pThr286 levels normalized to total CaMKIIα expression with 50 to 100 μM Ca2+ alone or together with HOCPCA (3 mM) for 1 h. (G and H) Respective representative Western blots. GAPDH was used as loading control. (E and F) Number in bar diagrams indicates number of experiments/individual cultures. Box plots (boxes, 25 to 75%; whiskers, minimum and maximum; lines, median) (one-way ANOVA, post hoc Dunnett’s test).
Lack of Effects of GHB Ligands on Basal and Stimulated CaMKIIα Activity.
To explore a potential functional relevance of the hub binding site on CaMKII activity, we probed the ability of compounds (GHB, HOCPCA, and 5-HDC; CN21 peptide as control) to inhibit phosphorylation of a commonly used peptide substrate (syntide) in the ADP-Glo assay employing recombinant CaMKIIα and a fixed concentration of CaM. Whereas we observed the expected concentration-dependent inhibition of CaMKII activation by syntide by CN21, no effect was seen of the three GHB analogs applied over a wide concentration span (Fig. 3D).
In order to study the effects of the compounds on the enzymatic activity of CaMKII in a more physiological setting, we also assessed Thr286 autophosphorylation levels in mouse primary cortical neurons, optimized to ensure a native high expression of CaMKIIα and ability to bind [3H]HOCPCA (SI Appendix, Fig. S7). In these studies, as well as in subsequent in vivo studies, HOCPCA was selected as model compound due to its high selectivity and affinity for CaMKIIα (Fig. 1E), as well as its known cellular and brain permeability (32, 36). Following compound stimulation for 1 h, pThr286 levels were measured by Western blotting. Whereas KN93, a reported CaM inhibitor (37), showed significant inhibition of Thr286 autophosphorylation already at basal conditions, HOCPCA (3 mM) did not affect Thr286 autophosphorylation during basal nor Ca2+-stimulated conditions (Fig. 3 E–H). In line with this overall lack of effect on CaMKIIα enzymatic activity, bath application of HOCPCA did not inhibit classical long-term potentiation (LTP) induced in mouse hippocampal slices when given as a single dose (SI Appendix, Fig. S8). These findings underline that binding to the hub does not per se affect the enzymatic function of CaMKIIα under naïve nonpathological conditions.
CaMKIIα Hub Ligands Are Cellular Protectants.
Previous studies have reported GHB to be neuroprotective after ischemia (18–20) by yet-unknown mechanisms. Given our findings that GHB ligands strongly impair CaMKIIα hub dynamics, we hypothesized that an effect might depend on the activation state of the holoenzyme. In order to investigate this, we designed a number of experiments to examine the neuroprotective effect of HOCPCA in neurons subjected to excitotoxicity. Under such circumstances, intracellular Ca2+ levels rise, resulting in CaMKIIα autonomy (16, 17) and translocation to the PSD, where it colocalizes with the NMDA-type glutamate receptor subunit GluN2B (13). As tat-CN21 is known to be neuroprotective after this type of insult to neurons and inhibit GluN2B colocalization (38), we used this compound for comparison.
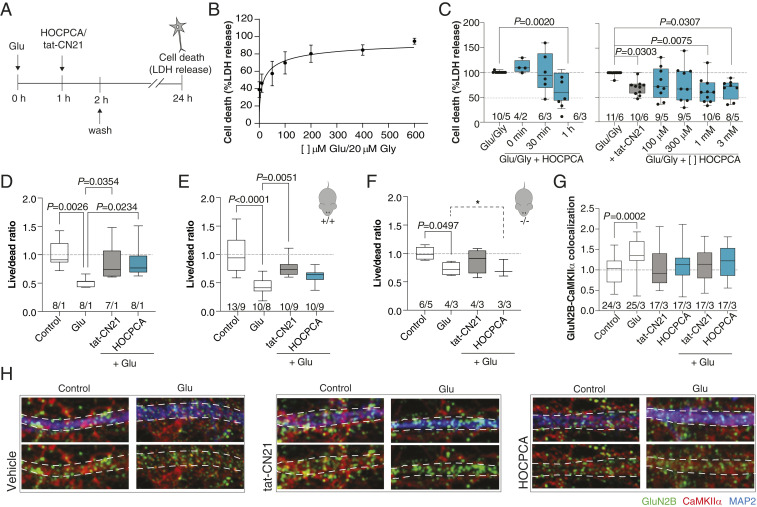
In C57/B6-derived cortical neurons, excitotoxity was induced by a high concentration of l-glutamate (Glu), as described by others (39), and cell viability quantified by lactate dehydrogenase activity (Fig. 4A). Using this protocol, we observed a strong susceptibility to cell death by a brief Glu stimulus (Fig. 4B), which was CaMKII dependent since tat-CN21 significantly reduced cell death (Fig. 4C). Application of HOCPCA (1 mM) immediately or 30 min after stimulation did not have any effect on cell survival (Fig. 4 C, Left). However, application of HOCPCA 1 h after the noxious Glu stimulus, significantly reduced cell death which was found to be concentration dependent (Fig. 4 C, Right). Similar findings were obtained with live/dead staining on primary hippocampal cultures from FvB mice, as also here application of HOCPCA (1 mM) 1 h poststimulation resulted in a significantly increased cell live/dead ratio (Fig. 4D).
Fig. 4.
Neuroprotective effects of HOCPCA in cultured neurons. (A and C) Excitotoxicity experiments in cultured cortical neurons (DIV 16 to 18). (A and B) Timeline and Glu–Gly response curve for testing neuroprotective effects of compounds by lactate dehydrogenase (LDH) release (see also SI Appendix, Fig. S7). Maximum cell death was obtained with 100 μM Glu/20 μM Gly and above (data normalized to maximum cell death), pooled data from five different cultures (means ± SEM). (C) Time- (Left) and concentration-dependent effects of HOCPCA (0.1 to 3 mM) (Right) on cell survival at 24 h in cultured cortical neurons stimulated with 100 to 200/20 µM Glu/Gly for 1 h (tat-CN21 as control). Cell death was normalized to maximum cell death as measured by LDH release (one-way ANOVA, post hoc Dunnett’s test). (D and H) Excitotoxicity experiments in cultured hippocampal neurons isolated from either FvB or C57/B6 mice. (D) HOCPCA (1 mM) improves cell survival when applied 1 h after a Glu-excitotoxic insult (400 μM Glu) in pooled FvB neurons (DIV 16 to 17) (tat-CN21 as control). Live–dead cell ratio was measured by calcein AM and ethidium homodimer-1 staining and normalized to control condition (one-way ANOVA, post hoc Dunnett’s test). (E and F) Cell survival effects in neurons (DIV 16 to 17) isolated from Camk2a+/+ and -/- littermates (C57/B6), showing consistent Glu-induced cell death. The effect of HOCPCA observed in +/+ neurons is partial, yet completely absent in -/- neurons (95% CI, test for equality; dotted line and marked by *). (G) Quantification of GluN2B–CaMKIIα colocalization in hippocampal neurons (DIV 14 to 19) exposed to Glu (400 μM) for 2 min and immediately fixed. (H) Representative immunostained images. The dotted lines indicate the outlines of the dendrite. Colocalization of CaMKIIα (red) with the GluN2B-positive dots (green) was measured within the dotted lines. Note the punctuated CaMKIIα pattern in the dendrite upon stimulation in the vehicle condition, which is absent with HOCPCA (2 mM) and tat-CN21. (C and G) Number in bar diagrams indicates number of experiments/individual cultures. Box plots (boxes, 25 to 75%; whiskers, minimum and maximum; lines, median) (one-way ANOVA, post hoc Dunnett’s test). Number in bar diagrams indicates number of experiments/individual cultures. Box plots (boxes, 25 to 75%; whiskers, minimum and maximum; lines, median) (one-way ANOVA, post hoc Dunnett’s test).
To address target engagement, this experimental setup was repeated in neurons isolated from Camk2a+/+ and Camk2a-/- mouse littermates. These experiments are unavoidably endowed with more variability, as individual embryos were used for neuron isolation to permit littermate matching by genotyping. In Camk2a+/+ neurons, we observed a significant Glu response (56% mean cell death), which was significantly improved in the tat-CN21 condition. HOCPCA application showed a trend, though not significant, toward improved live/dead ratio (18% mean recovery compared with the Glu-only condition). (Fig. 4 E and F). Interestingly, in Camk2a-/- neurons, we observed a milder but still significant Glu response. In these neurons, neither tat-CN21 nor HOCPCA improved cell survival. As HOCPCA presented with a very high P value (>0.9999) in the test for statistical difference between control and HOCPCA conditions, we also tested the meaningful hypothesis that Glu and HOCPCA treatments are equivalent in Camk2a-/- neurons (40). In this test, HOCPCA was found to be equal to Glu with at least 95% confidence (90% CI [−0.202–0.198] live/dead ratio) whereas CN21 was not (90% CI [−0.093–0.381].
Finally, to visualize the direct effect of the compound on CaMKII under these conditions, we examined the GluN2B–CaMKIIα colocalization in hippocampal dendritic spines after Glu stimulation in presence or absence of the compound. As a control, tat-CN21 was taken along again, which blocked the increased GluN2B–CaMKIIα colocalization as expected (38, 41). Strikingly, application of HOCPCA blocked the increase in GluN2B–CaMKIIα colocalization, similar to tat-CN21 (Fig. 4 G and H). These data show that HOCPCA behaves similar to the CaMKII-specific inhibitor tat-CN21 in neurons despite having a different binding domain on CaMKII, thereby adding to the mounting evidence that the observed neuroprotective effect of HOCPCA is mediated via CaMKIIα.
CaMKIIα Hub Ligands Afford Lasting Neuroprotection In Vivo.
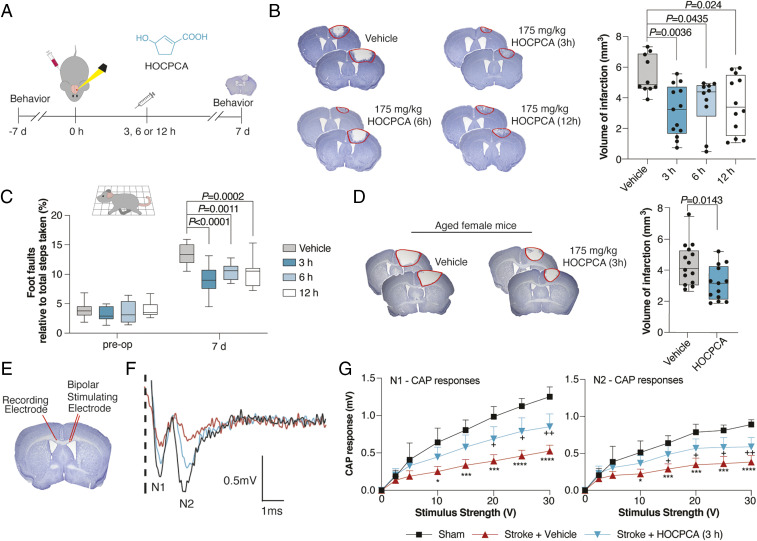
For in vivo validation of the HOCPCA effect, we turned to a model of ischemic stroke, employing a noninvasive, well-validated photothrombotic model of focal ischemic injury in C57/B6 mice, which allows for evaluation of infarct sizes and motor performance (42). Similar to other models of stroke, we confirmed a CaMKIIα–relevant pathogenesis (16), as evidenced by an elevation in pThr286 levels 3 to 12 h postinjury (SI Appendix, Fig. S9). This guided the timing of compound treatment for the in vivo studies (Fig. 5A). Given the reported neuroprotective action of GHB in vivo (18–20), we hypothesized that this effect is mediated at least in part by interaction with the CaMKIIα hub (KI 3.0 µM; SI Appendix, Table S3). However, as GHB is also known to bind to GABAB receptors, and this mediates hypothermia and sedation (22) albeit with much lower affinity (KI 230 µM) (23), we also considered that a GABAB receptor–mediated effect could contribute to the neuroprotective action of GHB. The obvious way to discern this is to use HOCPCA as this compound exhibits submicromolar affinity for the CaMKIIα hub domain (KI 0.13 µM) and, most importantly, has been reported to lack affinity to native GABAB receptors at 1 mM (23). To further underscore the absence of GABAB effects of HOCPCA in vivo, we compared the effects of GHB and HOCPCA on body temperature in C57/B6 mice. Whereas GHB (275 mg/kg) by intraperitoneal (i.p.) injection produced the expected GABAB-mediated strong hypothermia, a high dose of HOCPCA (175 mg/kg) produced no hypothermia (SI Appendix, Fig. S10A).
Fig. 5.
Neuroprotective effects of HOCPCA in vivo. (A) Timeline for testing of HOCPCA by systemic administration in mice. (B) Effect on infarct size after treatment with a single dose of HOCPCA at 3, 6, or 12 h postphotothrombosis in young male mice (one-way ANOVA, post hoc Dunnett’s test), representative cresyl violet staining with doses and n’s indicated (Left) and quantification of stroke volumes (Right). (C) Functional recovery of young male mice assessed in grid-walking task (preoperative, preoperation; two-way ANOVA (time, treatment), post hoc Dunnett’s test) (see also SI Appendix, Fig. S10). (D) Treatment of aged female mice (20 to 24 mo) with HOCPCA (175 mg/kg) (two-tailed Student’s t test), representation as for B. (E–G) Axonal function assessed by electrophysiological recording of compound action potentials (CAPs) 14 d poststroke. Young male mice were treated with a single dose of HOCPCA (175 mg/kg) at 3 h poststroke (blue) cf vehicle (red) and sham (black). (F) Representative recording of CAPs showing negative peak for myelinated (N1) and unmyelinated axons (N2). (G) Amplitudes of CAP peaks for N1 (Right) and N2 (Left). Means ± SD, two-way ANOVA (stimulus strength, treatment), post hoc Tukey’s test, +P < 0.05, ++P < 0.01, sham compared with stroke + HOCPCA, *P < 0.05, ***P < 0.001, ****P < 0.0001 sham compared with stroke + vehicle. Box plots (boxes, 25 to 75%; whiskers, minimum and maximum; lines, median).
Turning now to the stroke model, we initially tested both GHB (275 mg/kg) and HOCPCA (17.5 and 175 mg/kg) at an early time point (30 min poststroke). In both cases, infarct volumes were significantly reduced with the high dose of each compound (SI Appendix, Fig. S10B). Compellingly, when HOCPCA (175 mg/kg) was administered later, at 3, 6, and 12 h postphotothrombotic stroke (Fig. 5A), it resulted in a more-pronounced reduction in infarct volume by ∼40 to 50% measured 7 d poststroke (Fig. 5B). This was accompanied by improvements in motor coordination measured in paw placement on the grid-walking task (Fig. 5C) and in forelimb asymmetry in the cylinder test (SI Appendix, Fig. S10C).
To point to a preclinical relevance, significant treatment effects of single doses of HOCPCA were confirmed in both aged female mice (175 mg/kg; Fig. 5D) and with a lower dose in young males (90 mg/kg) (SI Appendix, Fig. S10 D–F). To provide target engagement by HOCPCA in these effects, we again tried to make use of the Camk2a-/- mice. As seen in other models of stroke (43), our photothrombotic stroke model likewise produced much larger infarcts in Camk2a-/- compared with Camk2a+/+ littermates (SI Appendix, Fig. S10G). Unfortunately, these larger infarcts were accompanied by marked hemorrhagic transformation (SI Appendix, Fig. S10H), hampering a subsequent treatment study.
Finally, to investigate potentially sustained functional effects of HOCPCA (175 mg/kg) after a single, acute treatment at 3 h poststroke, we assessed the interhemispheric transfer and integration of sensorimotor and cognitive information via axons through the corpus callosum 14 d poststroke (44) by recording of compound action potentials (CAPs). This showed an overall significant increase in CAPs with HOCPCA treatment (∼50 to 60% of control) (Fig. 5 F and G). The impairments in axon function and reversal after a single treatment with HOCPCA were further supported by a reversal of the stroke-induced decrease in transport of the neuroanatomical tracer biotinylated dextran amine by HOCPCA (SI Appendix, Fig. S10 I and J).
Discussion
The role of the CaMKIIα hub as a determinant for holoenzyme assembly and structural integrity is well-established (4, 7, 9), yet its functional importance has been less appreciated. This study identifies a binding site in the CaMKIIα hub domain for GHB-related small molecules that appears to be the long-sought-after GHB high-affinity site in the mammalian brain. Although this evolutionarily conserved and abundant forebrain high-affinity binding site was reported already in 1982 (45), it has never been unambiguously annotated (30, 31, 46). Compared with our previous proteomics study (30), in which a large number of potential hits were identified, the current study identifies only one main target: CaMKIIα. This milestone finding was achieved by an inventive approach combining a bifunctional photoligand, allowing for the introduction of a biotin tag for affinity purification, and the competition of the photoaffinity labeling reaction to permit quantitative proteomics. The finding opens for further understanding the role of GHB as a natural GABA metabolite in the mammalian brain.
The CaMKIIα hub structure presented here uniquely has a bound ligand. The central Arg residues 453 and 469 in the pocket make important interactions with the carboxylic acid moiety, as previously suggested (2, 4). The fact that the cocrystal with 5-HDC is a tetradecamer shows how this oligomeric state can coexist with ligand bound. The Trp403 displacement upon binding illustrates a hub conformation with potential functional relevance potentially involved in stabilizing an oligomeric state of the protein. Interestingly, this residue was recently reported as a part of a loop mediating kinase docking in CaMKIIα lacking a linker region (9), suggesting that a conformational change in this part of the hub may also influence allosteric control of kinase activity (9); although under the standard conditions tested here, no effect on substrate phosphorylation or Thr286 autophosphorylation was observed. As Trp403 is unique to CaMKIIα and not in CaMKIIβ/γ/δ subtypes, it may also be an important molecular determinant for CaMKIIα selectivity and/or dynamics.
Our structural and molecular data suggest a prototypical action of small-molecule hub ligands, such as GHB analogs, in providing pharmacological compensation of CaMKIIα overactivity during abnormal activation states of CaMKIIα activity such as excitotoxicity or destabilization. The effect on hub stabilization suggests that compounds can alter hub oligomeric state, but a full understanding of the functional consequences of this remain to be fully investigated. It is envisaged that to detect such effects, assays must be tailormade to pick up modulation via the hub. Here, GHB ligands will serve as important tools.
Using the highly selective CaMKIIα ligand HOCPCA, we provide pharmacological insight into this role of the enigmatic GHB high-affinity site ligands as cellular protectants. The specific interaction with CaMKIIα is obtained from various in vitro binding studies and is further abrogated by target-specific functional effects in neurons: Genetic deletion of CaMKIIα abrogates the effect of HOCPCA in Camk2a-/- neurons, although Glu responses are generally quite low. Furthermore, a significant reduction in PSD translocation of CaMKIIα is observed upon HOCPCA treatment, inferring a CaMKIIα–specific effect of this ligand. The peptide CN21 shares the functional effects with HOCPCA but is not selective for CaMKIIα, which may explain its slightly different efficacy. After all, CN21 is not a hub ligand. The in vitro findings indicating a late-onset effect are further corroborated by extensive neuroprotection observed in vivo in mice when HOCPCA is administered 3 to 12 h poststroke. This points to a clinically relevant time window for patient treatment where brain tissue can be salvaged and where there is currently no other treatment. Given that the effect of HOCPCA on brain connectivity is still apparent 14 d after a single treatment at 3 h poststroke further indicates that compound administration at the right time postinjury can provide lasting effects. It underscores CaMKIIα as a clinically relevant target in stroke therapy. Given the limited treatment options and general poor prognosis for functional recovery after cerebral ischemia (47), these findings offer a mechanism with an extended time window for therapeutic rescue of neuronal tissue at risk. Since HOCPCA does not appear to inhibit LTP, analogs of this type are also promising in terms of producing drug candidates with fewer adverse effects given its unique hub site-of-action.
Overall, our elucidations present a principle for regulating CaMKIIα function via modulation of the hub domain and suggests CaMKIIα hub ligands for ameliorating several states of CaMKII dysregulation such as cerebral ischemia, conditions involving excitotoxicity, and protein destabilization. Whereas the GABAB receptor has long been the only pharmacologically validated GHB target in vivo (22), we here extend this to include CaMKIIα. This prompts considerations of CaMKIIα as contributing factor to the neuroprotective actions of GHB but also in clinical conditions like alcoholism and narcolepsy where GHB is employed (48) and plausibly in relation to the endogenous role of GHB. The structural and molecular insight into the CaMKIIα hub provided here, and the identification of a unique binding site for small molecules, permits ways for tailoring of future therapeutically relevant drug candidates targeting this very important brain kinase.
Materials and Methods
Materials and procedures are described in full in SI Appendix, Supplementary Information Text.
Supplementary Material
Acknowledgments
We thank members of all laboratories for their support. We appreciate insightful comments from Ype Elgersma and Howard Schulman and expert advice from Bernhard Küster and Mike Gibson. A special thanks to Kresten Lindorff-Larsen and Nils Ole Dalby for invaluable input. We thank Mehrnoush A. Jolfei for help with the LTP experiments, Ales Marek for providing [3H]HOCPCA radioligand, and Anders S. Kristensen for providing access to the Safire2 plate reader. For x-ray crystallography data, we thank James Holton and George Meigs at the Advanced Light Source Beamline 8.3.1 for assistance with data collection. Beamline 8.3.1 at the Advanced Light Source is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR-15-328599, the NIH (Grants R01 GM124149 and P30 GM124169), Plexxikon Inc., and the Integrated Diffraction Analysis Technologies program of the US Department of Energy Office of Biological and Environmental Research. The Advanced Light Source (Berkeley, CA) is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under Contract No. DE-AC02-05CH11231, Office of Basic Energy Sciences. This work was supported by the Lundbeck Foundation (Grants R83-2011-8000 to P.W., R77-2011-A6415 to B.F., R139-2012-12270 to P.W., R190-2014-3710 to A.B., R192-2015-666 to U.L., R303-2018-3162 to M.A.S., and R277-2018-260 to P.W.), the Novo Nordisk Foundation (Grants NNF17OC0028664 to P.W., NNF14CC0001 to J.V.O., and NNF19SA0057841 to B.R.K.), the Independent Research Fund Denmark (Grants 8020-00156B to P.W. and 1333-00161B to A.B.K.), a Drug Research Academy and Lundbeck Foundation pre-graduate scholar stipend in Pharmaceutical Neuroscience (to A.S.G.L. and S.J.G.), the Netherlands Organization for Scientific Research (Grant NWO-VIDI 016.Vidi.188.014 to G.M.v.W.), a Royal Society of New Zealand Project Grant, Brain Research New Zealand (to A.N.C.), and the Howard Hughes Medical Institute (J.K.).
Footnotes
Competing interest statement: The University of Copenhagen and Otago Innovation Ltd. have licensed the patent rights for GHB derivatives and their uses (WO/2019/149329) to Ceremedy Ltd., of which B.F., B.R.K., and P.W. are cofounders.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108079118/-/DCSupplemental.
Data Availability
All raw mass spectrometry proteomics data from this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (49), with the dataset identifier PXD019679 (50). The atomic coordinates have been deposited to the Protein Data Bank with the identifier 7REC (51). All other data are available in the main text or the supplementary information.
References
- 1.De Koninck P., Schulman H., Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science 279, 227–230 (1998). [DOI] [PubMed] [Google Scholar]
- 2.McSpadden E. D., et al., Variation in assembly stoichiometry in non-metazoan homologs of the hub domain of Ca2+ /calmodulin-dependent protein kinase II. Protein Sci. 28, 1071–1082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg O. S., et al., Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J. 273, 682–694 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Hoelz A., Nairn A. C., Kuriyan J., Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol. Cell 11, 1241–1251 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg O. S., Deindl S., Sung R. J., Nairn A. C., Kuriyan J., Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell 123, 849–860 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Stratton M., et al., Activation-triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity. eLife 3, e01610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya M., et al., Molecular mechanism of activation-triggered subunit exchange in Ca2+/calmodulin-dependent protein kinase II. eLife 5, e13405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao L. H., et al., A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin- dependent kinase II holoenzyme. Cell 146, 732–745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sloutsky R., et al., Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation. Sci. Signal. 13, eaaz0240 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres-Ocampo A. P., et al., Characterization of CaMKIIα holoenzyme stability. Protein Sci. 29, 1524–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chia P. H., et al., A homozygous loss-of-function CAMK2A mutation causes growth delay, frequent seizures and severe intellectual disability. eLife 7, e32451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharyya M., Karandur D., Kuriyan J., Structural insights into the regulation of Ca2+/calmodulin-dependent protein kinase II (CaMKII). Cold Spring Harb. Perspect. Biol. 12, a035147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayer K. U., De Koninck P., Leonard A. S., Hell J. W., Schulman H., Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature 411, 801–805 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Skelding K. A., Spratt N. J., Fluechter L., Dickson P. W., Rostas J. A., αCaMKII is differentially regulated in brain regions that exhibit differing sensitivities to ischemia and excitotoxicity. J. Cereb. Blood Flow Metab. 32, 2181–2192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed M. E., et al., Beneficial effects of a CaMKIIα inhibitor TatCN21 peptide in global cerebral ischemia. J. Mol. Neurosci. 61, 42–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng G., et al., Autonomous CaMKII activity as a drug target for histological and functional neuroprotection after resuscitation from cardiac arrest. Cell Rep. 18, 1109–1117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coultrap S. J., Vest R. S., Ashpole N. M., Hudmon A., Bayer K. U., CaMKII in cerebral ischemia. Acta Pharmacol. Sin. 32, 861–872 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ottani A., et al., Effect of γ-hydroxybutyrate in two rat models of focal cerebral damage. Brain Res. 986, 181–190 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Sadasivan S., Maher T. J., Quang L. S., Gamma-hydroxybutyrate (GHB), gamma-butyrolactone (GBL), and 1,4-butanediol (1,4-BD) reduce the volume of cerebral infarction in rodent transient middle cerebral artery occlusion. Ann. N. Y. Acad. Sci. 1074, 537–544 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Wendt G., et al., Gamma-hydroxybutyrate, acting through an anti-apoptotic mechanism, protects native and amyloid-precursor-protein-transfected neuroblastoma cells against oxidative stress-induced death. Neuroscience 263, 203–215 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Klein A. B., et al., Autoradiographic imaging and quantification of the high-affinity GHB binding sites in rodent brain using 3H-HOCPCA. Neurochem. Int. 100, 138–145 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Kaupmann K., et al., Specific γ-hydroxybutyrate-binding sites but loss of pharmacological effects of γ-hydroxybutyrate in GABAB(1)-deficient mice. Eur. J. Neurosci. 18, 2722–2730 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Wellendorph P., et al., Novel cyclic γ-hydroxybutyrate (GHB) analogs with high affinity and stereoselectivity of binding to GHB sites in rat brain. J. Pharmacol. Exp. Ther. 315, 346–351 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Høg S., et al., Novel high-affinity and selective biaromatic 4-substituted γ-hydroxybutyric acid (GHB) analogues as GHB ligands: Design, synthesis, and binding studies. J. Med. Chem. 51, 8088–8095 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Sabbatini P., et al., Design, synthesis and in vitro pharmacology of new radiolabelled GHB analogues including photolabile analogues with irreversible binding to the high-affinity GHB binding sites. J. Med. Chem. 53, 6506–6510 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Hosoya T., et al., Novel bifunctional probe for radioisotope-free photoaffinity labeling: Compact structure comprised of photospecific ligand ligation and detectable tag anchoring units. Org. Biomol. Chem. 2, 637–641 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Wellendorph P., Høg S., Skonberg C., Bräuner-Osborne H., Phenylacetic acids and the structurally related non-steroidal anti-inflammatory drug diclofenac bind to specific γ-hydroxybutyric acid sites in rat brain. Fundam. Clin. Pharmacol. 23, 207–213 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Baucum A. J. II, Shonesy B. C., Rose K. L., Colbran R. J., Quantitative proteomics analysis of CaMKII phosphorylation and the CaMKII interactome in the mouse forebrain. ACS Chem. Neurosci. 6, 615–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellendorph P., et al., Novel radioiodinated γ-hydroxybutyric acid analogues for radiolabeling and photolinking of high-affinity γ-hydroxybutyric acid binding sites. J. Pharmacol. Exp. Ther. 335, 458–464 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Absalom N., et al., α4βδ GABAA receptors are high-affinity targets for γ-hydroxybutyric acid (GHB). Proc. Natl. Acad. Sci. U.S.A. 109, 13404–13409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bay T., Eghorn L. F., Klein A. B., Wellendorph P., GHB receptor targets in the CNS: Focus on high-affinity binding sites. Biochem. Pharmacol. 87, 220–228 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Vogensen S. B., et al., New synthesis and tritium labeling of a selective ligand for studying high-affinity γ-hydroxybutyrate (GHB) binding sites. J. Med. Chem. 56, 8201–8205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehta A. K., Muschaweck N. M., Maeda D. Y., Coop A., Ticku M. K., Binding characteristics of the γ-hydroxybutyric acid receptor antagonist [3H](2E)-(5-hydroxy-5,7,8,9-tetrahydro-6H-benzo[a][7]annulen-6-ylidene) ethanoic acid in the rat brain. J. Pharmacol. Exp. Ther. 299, 1148–1153 (2001). [PubMed] [Google Scholar]
- 34.Griem-Krey N., Klein A. B., Herth M., Wellendorph P., Autoradiography as a simple and powerful method for visualization and characterization of pharmacological targets. J. Vis. Exp. 145, 10.3791/58879 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Karandur D., et al., Breakage of the oligomeric CaMKII hub by the regulatory segment of the kinase. eLife 9, e57784 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiesen L., et al., In vitro and in vivo evidence for active brain uptake of the GHB analog HOCPCA by the monocarboxylate transporter subtype 1. J. Pharmacol. Exp. Ther. 354, 166–174 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Wong M. H., et al., The KN-93 molecule inhibits calcium/calmodulin-dependent protein kinase II (CaMKII) activity by binding to Ca2+/CaM. J. Mol. Biol. 431, 1440–1459 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Buonarati O. R., et al., CaMKII versus DAPK1 binding to GluN2B in ischemic neuronal cell death after resuscitation from cardiac arrest. Cell Rep. 30, 1–8.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashpole N. M., Hudmon A., Excitotoxic neuroprotection and vulnerability with CaMKII inhibition. Mol. Cell. Neurosci. 46, 720–730 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Schuirmann D. J., A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. J. Pharmacokinet. Biopharm. 15, 657–680 (1987). [DOI] [PubMed] [Google Scholar]
- 41.Vest R. S., Davies K. D., O’Leary H., Port J. D., Bayer K. U., Dual mechanism of a natural CaMKII inhibitor. Mol. Biol. Cell 18, 5024–5033 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarkson A. N., Huang B. S., Macisaac S. E., Mody I., Carmichael S. T., Reducing excessive GABA-mediated tonic inhibition promotes functional recovery after stroke. Nature 468, 305–309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waxham M. N., Grotta J. C., Silva A. J., Strong R., Aronowski J., Ischemia-induced neuronal damage: A role for calcium/calmodulin-dependent protein kinase II. J. Cereb. Blood Flow Metab. 16, 1–6 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Singer W., Development and plasticity of cortical processing architectures. Science 270, 758–764 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Benavides J., et al., High affinity binding sites for γ-hydroxybutyric acid in rat brain. Life Sci. 30, 953–961 (1982). [DOI] [PubMed] [Google Scholar]
- 46.Andriamampandry C., et al., Cloning and functional characterization of a γ-hydroxybutyrate receptor identified in the human brain. FASEB J. 21, 885–895 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Feigin V. L., Norrving B., Mensah G. A., Global burden of stroke. Circ. Res. 120, 439–448 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Busardò F. P., Kyriakou C., Napoletano S., Marinelli E., Zaami S., Clinical applications of sodium oxybate (GHB): From narcolepsy to alcohol withdrawal syndrome. Eur. Rev. Med. Pharmacol. Sci. 19, 4654–4663 (2015). [PubMed] [Google Scholar]
- 49.Vizcaíno J. A., et al., ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leurs U., Olsen J. V., GHB analogs confer neuroprotection through specific interaction with the CaMKIIα hub domain. Proteomics IDEntifications Database repository (PRIDE). http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD019679. Deposited 6 September 2020.
- 51.McSpadden E. D., Chi C. C., Gee C. L., Kuriyan J., Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Il464His, and Phe467Met mutant human CaMKII alpha hub bound to 5-HDC. Protein Data Bank. 10.2210/pdb7REC/pdb. Deposited 7 July 2020. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw mass spectrometry proteomics data from this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (49), with the dataset identifier PXD019679 (50). The atomic coordinates have been deposited to the Protein Data Bank with the identifier 7REC (51). All other data are available in the main text or the supplementary information.