Abstract
Nitroreductase as a potential biomarker for aggressive tumors has received extensive attention. In this work, a novel NIR fluorescent probe for nitroreductase detection was synthesized. The probe Py-SiRh-NTR displayed excellent sensitivity and selectivity. Most importantly, the confocal fluorescence imaging demonstrated that HepG-2 cells treated with Py-SiRh-NTR under hypoxic conditions showed obvious enhanced fluorescence, which means that the NTR was overexpressed under hypoxic conditions. Moreover, the probe showed great promise that could help us to study related anticancer mechanisms research.
Keywords: nitroreductase, NIR, hypoxic, cell imaging
1. Introduction
Nitroreductase (NTR), as a family of evolution-related proteins, is involved in the reduction of nitrogen-containing compounds [1,2]. NTR has received extensive attention as a potential biomarker for aggressive tumors in recent years [3]. Moreover, NTR can be overexpressed in hypoxic tumors [4]. Hypoxia is a feature of tumor tissue, whereby the median oxygen (O2) concentration in some solid tumors is ∼4% and may even decrease to 0% locally [5,6,7]. In clinical applications, the formation of hypoxic tumors with abnormal microvessels can limit the perfusion of cytotoxic chemotherapeutic drugs, therein decreasing their curative effects compared with that of normoxic tumors [5,6,7]. Therefore, estimating the tumor hypoxia degree is of great importance in predicting anticancer efficacies [7]. There is no doubt that developing an effective method to monitor NTR activity is not only useful for clinical diagnoses and anticancer mechanisms research, but also for new anticancer drug evaluation and tumor therapy.
Various methods have been developed for the detection and identification of NTR. The fluorescent imaging has attracted much attention as a powerful technique in monitoring biological analytes and processes in living systems [8]. Especially, the near-infrared (NIR) bioprobes are advantageous because of the obvious advantages of NIR light including minimum photodamage to biological samples, a high signal-to-noise ratio, and deep tissue penetration [9,10]. The detection of NTR activities within hypoxic tumor cells and tissues using fluorescent methods has been well described recently [11]. However, most of these NTR fluorescent probes need ultraviolet light as the excitation light which could cause photodamage to biological samples. Most importantly, only a few fluorescent probes have the ability to target the activity of NTR in specific subcellular organelle [12,13,14,15,16,17,18,19,20,21,22,23,24,25].
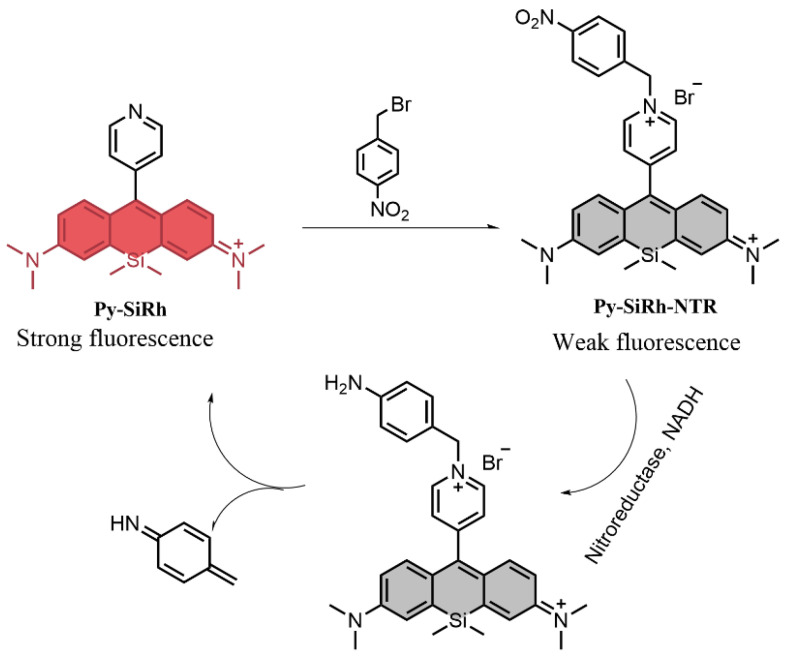
Herein, in this work, we constructed such a novel NIR fluorescent probe to detect nitroreductase (Py-SiRh-NTR; Scheme 1). Our design strategy could be finished via three-step synthesis to obtain the benzyl pyridinium salt to construct Py-SiRh-NTR. Further, we hypothesized that the benzyl pyridinium salt group could be triggered by nitroreductase, followed by the linker self-immolation to form Py-SiRh with strong near-infrared fluorescence emission.
Scheme 1.
The structure of the probe and sensing mechanism [10].
2. Results and Discussion
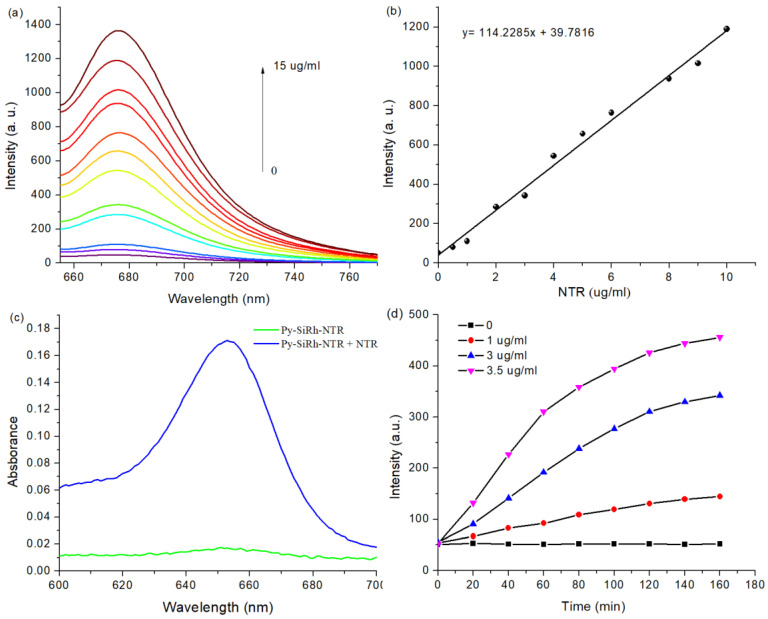
Firstly, we tested the response of Py-SiRh-NTR to NTR. The probe was incubated in 10 mM PBS buffer solution at 37 °C, containing 200 µM NADH. The fluorescence spectra were depicted in Figure 1a; the fluorescent probe Py-SiRh-NTR showed a very weak fluorescence at 680 nm. After the addition of NTR, the solution showed an obvious fluorescence enhancement, and the fluorescence intensity reached the maximum after 180 min. The titration experiments indicated that a higher NTR concentration induced the significant fluorescence enhancement (28-fold). Especially, the fluorescence intensity has an excellent linear relationship with the concentrations of NTR in 0−10 μg/mL (Figure 1b). The detection limit was calculated to be as low as 0.07 μg/mL. The results indicated that the sensitivity of Py-SiRh-NTR is comparable to the published NTR probes. Moreover, the solution of Py-SiRh-NTR has no absorption at about 655 nm, but after the addition of NTR, the absorption at 655 nm increased distinctly (Figure 1c). The reaction product with NTR was verified by high resolution mass spectrometry. A peak at m/z = 386.2045 assigned to Py-SiRh was observed, that verified the proposed response mechanism. Thus, we could assuredly say that the proposed response mechanism was well demonstrated.
Figure 1.
(a) Fluorescence spectra changes of Py-SiRh-NTR (10 μM) treated with the increasing concentrations of NTR in the presence of 200 μM NADH for 180 min at 37 °C in PBS λex = 645 nm. (b) The linear correlation between the fluorescence intensity of Py-SiRh-NTR and the concentration of NTR. (c) The absorption spectra of Py-SiRh-NTR (10 μM) in the absence of NTR and in the presence of NTR. (d) Time-dependent fluorescent response spectra of Py-SiRh-NTR (10 μM) in the presence of varied concentrations of NTR (from bottom to top): 0, 1.0, 3.0, 3.5 μg/mL.
Then, the time-dependent fluorescence response of Py-SiRh-NTR was examined (Figure S1, Supplementary Material). All of the fluorescence kinetic curves are shown in Figure 1d. It was apparent to see that the fluorescence intensity of Py-SiRh-NTR depended on the concentration of NTR. In other words, a higher concentration of NTR could induce the faster fluorescence intensity, enhanced. The time taken to reach the reaction equilibrium of Py-SiRh-NTR was tested for 180 min. Furthermore, the kinetic parameters for this NTR-catalyzed reaction were also verified. A Lineweaver–Burke plot of 1/V (V is the initial reaction rate) versus the reciprocal of the probe Py-SiRh-NTR concentration is shown in Figure S2 [26]. According to the Michaelis–Menten equation [26,27], the Michaelis constant (Km) and maximum of the initial reaction rate (Vmax) were examined to be 52.2 μM and 0.074 μM/S, respectively.
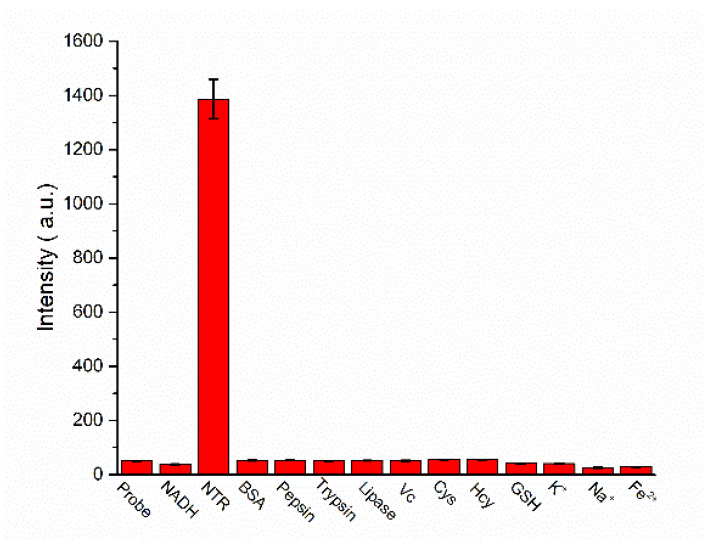
Subsequently, the selectivity of the probe was tested for NTR over other biologically relevant species, such as trypsin, lipase, bull serum albumin (BSA), pepsin, amino acids (cysteine, homocysteine, glutathione) and some inorganic salts (K+, Na+, Fe2+). The results were shown in Figure 2, and it was apparent to see that only the NTR could make an obvious fluorescence enhancement compared to the controls. It was demonstrated that the Py-SiRh-NTR is highly selective for NTR over other typical physiological species, which may be necessary for the bio-imaging in tangled living cells. All of the results of the experiments clearly demonstrated that this NIR fluorescent probe can be an excellent tool to detect the NTR in living cells.
Figure 2.
Fluorescence response of Py-SiRh-NTR toward various biologically relevant species. NTR (15 μg/mL), BSA, ABM, Trypsin, Lipase (20 μg/mL) and other biologically relevant species (100 μM).
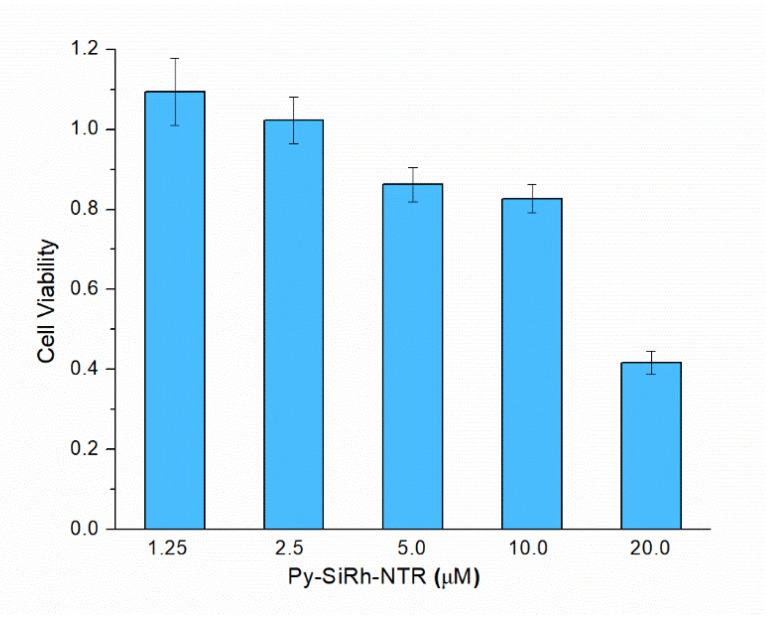
Encouraged by the fluorescent experiments, we attempted to examine whether the Py-SiRh-NTR could perform well in living cells. Before the bioimaging, a standard MTS assay was carried out to test the biocompatibility. The results revealed that the viabilities of HepG-2 cells were not noticeably affected by incubation with different concentrations of Py-SiRh-NTR (1.0–10.0 μM) for 24 h (Figure 3). Thus, the low cytotoxicity of the probe in the measured concentration range is well demonstrated. Therefore, in the subsequent bio-imaging experiments, 2 μM Py-SiRh-NTR was thus used.
Figure 3.
Cytotoxicity of Py-SiRh-NTR in cultured HepG-2 cells. Cell viability was measured by MTS assays, and the results were reported as percentage relative to untreated cells (mean ± SD).
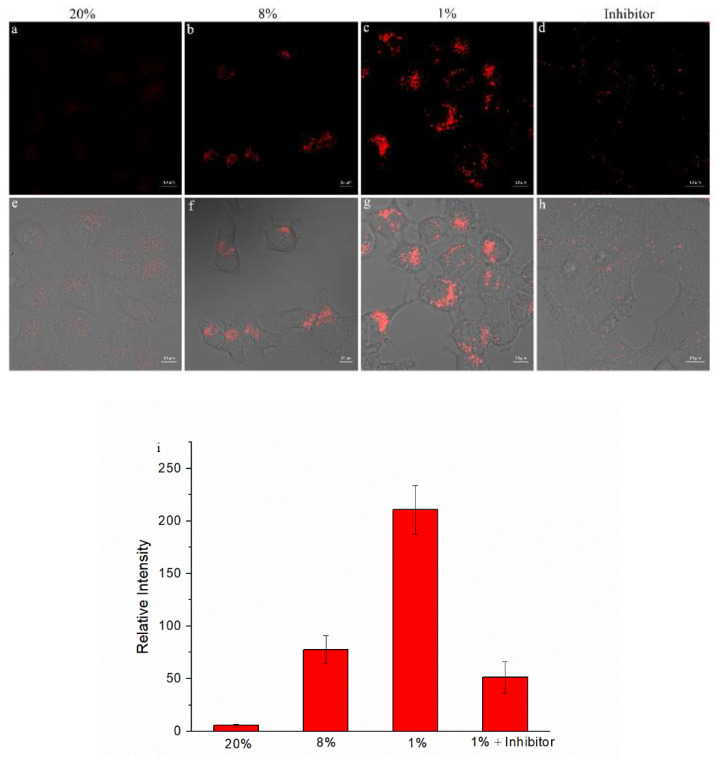
Then, we used the confocal fluorescence microscopy on HepG-2 cells to test the imaging ability of Py-SiRh-NTR for the intracellular NTR monitoring, as we know that the tumor cells could overexpress NTR under hypoxic conditions. Therefore, in this experiment, the HepG-2 cells were incubated with 2 μM Py-SiRh-NTR and treated under normoxic conditions (20% O2) and hypoxic conditions (1% O2) for 4 h to compare the concentration of NTR under different oxygen conditions. The results are shown in Figure 4; HepG-2 cells incubated under normoxic conditions showed a very weak near-infrared fluorescence that indicated the low concentration of NTR. On the contrary, HepG-2 cells displayed increased near-infrared fluorescence intensity when treated under hypoxic conditions, indicating that the HepG-2 cells could overexpress NTR in hypoxic conditions. Most importantly, when the HepG-2 cells were pretreated with the dicoumarin, a well-known NTR inhibitor under hypoxic conditions for 4 h [28], no obvious near-infrared fluorescence signal was detected. This verified that the near-infrared fluorescence signal was exactly from the production of the Py-SiRh-NTR with NTR in HepG-2 cells under hypoxic conditions. The results showed that the Py-SiRh-NTR could monitor the activity of nitroreductase in living cells and evaluate the degree of tumor hypoxia.
Figure 4.
Confocal fluorescence imaging of HepG2 cells incubated with Py-SiRh-NTR (2 μM) under different oxygen concentration conditions. (a–d) The near-IR channel fluorescence image (λex = 633 nm). (e–h) The merged image of near-IR channel fluorescence image and bright filed imaging. (i) Average fluorescence intensity from images of (a–d).
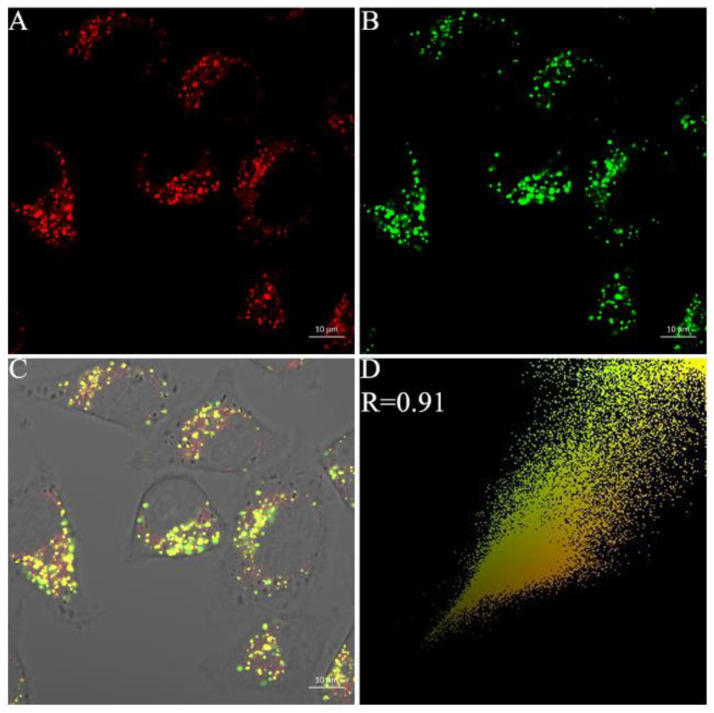
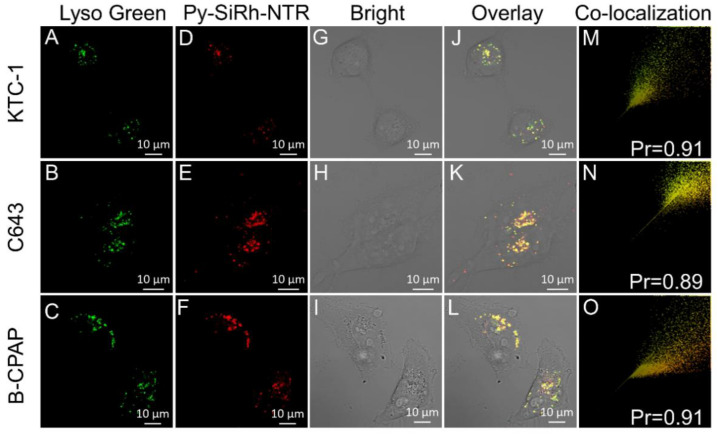
We also examined the lysosome-targeting ability of the probe after reaction with NTR. HepG-2 cells were treated with Py-SiRh-NTR under the hypoxic condition of 1% O2 for 4 h, according to our confocal fluorescence microscopy results, and then incubated with LysoTracker green (0.5 μM, 15 min). As shown in Figure 5, the excellent overlapping images of the NIR fluorescence channel and green fluorescence channel displayed a high Pearson coefficient, which indicated that Py-SiRh-NTR could specifically localize in lysosomes after the probe finished the reaction with the NTR under the hypoxic condition. Finally, we also tested the co-localization ability with KTC-1, C643 and B-CPAP cells, and achieved a good Pearson coefficient (Figure 6).
Figure 5.
Co-localization of Py-SiRh-NTR and LysoTracker green in HepG2 cells. The cells were incubated under the hypoxiccondition of 1% O2 with Py-SiRh-NTR (2 μM), and treated with LysoTracker green (2 μM). (A) The fluorescence from the Py-SiRh-NTR. (B) The fluorescence from the LysoTracker green. (C) Merged image of images (A,B). (D) Intensity correlation plot between Py-SiRh-NTR and LysoTracker green.
Figure 6.
Co-localization of Py-SiRh-NTR and LysoTracker green in KTC-1, C643 and B-CPAP cells. The cells were incubated with Py-SiRh-NTR (2 μM), and treated with LysoTracker green (200 nM). (A–C) The fluorescence from the LysoTracker green; (D–F) The fluorescence from the Py-SiRh-NTR; (G–I) Brightfield images of cells. (J–L) Merged image of images LysoTracker green and Py-SiRh-NTR. (M–O) Intensity correlation plot between Py-SiRh-NTR and LysoTracker green.
3. Conclusions
In summary, we have designed the Py-SiRh-NTR as a brand new NIR fluorescent probe for highly selective detection of nitroreductase and hypoxic-tumor-cell imaging. The probe shows good solubility and is highly sensitive to NTR, which could be used to discern the degree of hypoxia tumor cells. To be specific, the Py-SiRh-NTR provides an outstanding fluorescent diagnostic tool which can be used to study the NTR activity-related pathological process of cancers. The bright potential diagnostic application of Py-SiRh-NTR is apparent.
Supplementary Materials
The following are available online. The detail synthesis of Py-SiRh-NTR, 1HNMR, 13NMR and HRMS characterization of the probe Py-SiRh-NTR, as well as fluorescence responding to NTR (Figure S1) and Lineweaver-Burk plot for the enzyme-catalyzed reaction (Figure S2), as well as the normalized absorption spectra and fluorescence emission spectra of Py-SiRh (Figure S3) are shown in Supplementary materials.
Author Contributions
Conceptualization, Z.L. and K.L.; methodology, F.L.; formal analysis, H.Z.; investigation, Y.X.; data curation, H.Z.; writing—original draft preparation, K.L.; writing—review and editing, Y.X.; supervision, Z.L.; project administration, K.L.; funding acquisition, Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Support Program for Science and Technology Department of Sichuan Province (2020YJ0237) and the 1·3·5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC18025).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data present in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.de Oliveira I.M., Henriques J.A.P., Bonatto D. In silico identificationg of a new group of specific bacterial and fungal nitroreductases-like proteins. Biochem. Biophys. Res. Commun. 2007;355:919–925. doi: 10.1016/j.bbrc.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 2.Hecht H.J., Erdmann H., Park H.J., Sprinzl M., Schmid R.D. Crystal structure of NADH oxidase from Thermus thermophilus. Nat. Struct. Biol. 1995;2:1109–1114. doi: 10.1038/nsb1295-1109. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J., Chai X., He X.P., Kim H.J., Yoon J., Tian H. Fluorogenic probes for disease-relevant enzymes. Chem. Soc. Rev. 2019;48:683–722. doi: 10.1039/C7CS00907K. [DOI] [PubMed] [Google Scholar]
- 4.Race P.R., Lovering A.L., Green R.M., Ossor A., White S.A., Searle P.F., Wrighton C.J., Hyde E.I. Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme. J. Biol. Chem. 2005;280:13256–13264. doi: 10.1074/jbc.M409652200. [DOI] [PubMed] [Google Scholar]
- 5.Caballero A., Lazaro J.J., Ramos J.L., Esteve-Nunez A. PnrA, a new nitroreductase-family enzyme in the TNT-degrading strain Pseudomonas putida JRL11. Environ. Microbiol. 2005;7:1211–1219. doi: 10.1111/j.1462-2920.2005.00801.x. [DOI] [PubMed] [Google Scholar]
- 6.Celik A., Yetis G. An unusually cold active nitroreductase for prodrug activations. Bioorg. Med. Chem. 2012;20:3540–3550. doi: 10.1016/j.bmc.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Komatsu H., Harada H., Tanabe K., Hiraoka M., Nishimoto S.-I. Indolequinone-rhodol conjugate as a fluorescent probe for hypoxic cells: Enzymatic activation and fluorescence properties. MedChemComm. 2010;1:50–53. doi: 10.1039/c0md00024h. [DOI] [Google Scholar]
- 8.Germain R.N., Robey E.A., Cahalan M.D. A decade of imaging cellular motility and interaction dynamics in the immune system. Science. 2012;336:1676–1681. doi: 10.1126/science.1221063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan C., Shi L., Guo Z., Zhu W. Molecularly near-infrared fluorescent theranostics for in vivo tracking tumor-specific chemotherapy. Chin. Chem. Lett. 2019;30:1849–1855. doi: 10.1016/j.cclet.2019.08.038. [DOI] [Google Scholar]
- 10.Zhang H., Li K., Li L.L., Yu K.K., Yu X.-Q. Pyridine-Si-xanthene: A novel near-infrared fluorescent platform for biological imaging. Chin. Chem. Lett. 2019;30:1063–1066. doi: 10.1016/j.cclet.2019.03.017. [DOI] [Google Scholar]
- 11.Liu Y., Teng L., Chen L., Ma H., Liu H.W., Zhang X.B. Engineering of a near-infrared fluorescent probe for real-time simultaneous visualization of intracellular hypoxia and induced mitophagy. Chem. Sci. 2018;9:5347–5353. doi: 10.1039/C8SC01684D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z.R., Tang Y., Xu A., Lin W. A new fluorescent probe with a large turn-on signal for imaging nitroreductasein tumor cells and tissues by two-photon microscopy. Biosens. Bioelectron. 2017;89:853–858. doi: 10.1016/j.bios.2016.09.107. [DOI] [PubMed] [Google Scholar]
- 13.Li Y., Sun Y., Li J., Su Q., Yuan W., Li F. Ultrasensitive near-infrared fluorescence-enhanced probe for in vivo nitroreductase imaging. J. Am. Chem. Soc. 2015;137:6407–6416. doi: 10.1021/jacs.5b04097. [DOI] [PubMed] [Google Scholar]
- 14.Xu S., Wang Q., Zhang Q., Zhang L., Zuo L., Jiang J.D., Hu H.Y. Real time detection of ESKAPE pathogens by a nitroreductase-triggered fluorescence turn-on probe. Chem. Commun. 2017;53:11177–11180. doi: 10.1039/C7CC07050K. [DOI] [PubMed] [Google Scholar]
- 15.Luo S., Zou R., Wu J., Landry M.P. A probe for the detection of hypoxic cancer cells. ACS Sens. 2017;2:1139–1145. doi: 10.1021/acssensors.7b00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J., Liu H., Hu X., Li J., Tang W.H. Efficient two-photon fluorescent probe for nitroreductase detection and hypoxia imaging in tumor cells and tissues. Anal. Chem. 2015;87:11832–11839. doi: 10.1021/acs.analchem.5b03336. [DOI] [PubMed] [Google Scholar]
- 17.Zhu K., Qina T., Zhao C., Luo Z., Wang L. A novel fluorescent turn-on probe for highly selective detection of nitroreductase in tumor cells. Sens. Actuators B Chem. 2018;276:397–403. doi: 10.1016/j.snb.2018.08.134. [DOI] [Google Scholar]
- 18.Liu Y., Liu W., Li H., Yan W., Yang X., Zhang J. Two-photon fluorescent probe for detection of nitroreductase and hypoxia-specific microenvironment of cancer stem cell. Anal. Chim. Acta. 2018;1024:177–186. doi: 10.1016/j.aca.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Gebremedhin K., Li Y., Yao Q., Xiao M., Peng X.J. Development of a red-light emission hypoxia-sensitive two-photon fluorescent probe for in vivo nitroreductase imaging. J. Mater. Chem. B. 2019;7:408–414. doi: 10.1039/C8TB02635A. [DOI] [PubMed] [Google Scholar]
- 20.Evans S.M., Kim K., Moore C.E., Uddin M.I., Capozzi M.E. Molecular probes for imaging of hypoxia in the retina. Bioconjug. Chem. 2014;25:2030–2037. doi: 10.1021/bc500400z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu G., Tang Y., Ma Y., Xu A., Lin W.Y. A new aggregation-induced emission fluorescent probe for rapid detection of nitroreductase and its application in living cells. Spectrochim. Acta A. 2018;188:197–201. doi: 10.1016/j.saa.2017.06.065. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J., Shi W., Li L., Gong Q., Ma H.M. A Lysosome-Targeting Fluorescence Off-On Probe for Imaging of Nitroreductase and Hypoxia in Live Cells. Chem. Asian J. 2016;11:2719–2724. doi: 10.1002/asia.201600012. [DOI] [PubMed] [Google Scholar]
- 23.Wong R.H., Kwong T., Yau K.H., Au-Yeung H.Y. Real time detection of live microbes using a highly sensitive bioluminescent nitroreductase probe. Chem. Commun. 2015;51:4440–4442. doi: 10.1039/C4CC10345A. [DOI] [PubMed] [Google Scholar]
- 24.Ao X., Bright S.A., Taylor N.C., Elmes R.B. 2-Nitroimidazole based fluorescent probes for nitroreductase; monitoring reductive stress in cellulo. Org. Biomol. Chem. 2017;15:6104–6108. doi: 10.1039/C7OB01406F. [DOI] [PubMed] [Google Scholar]
- 25.Tanabe K., Hirata N., Harada H., Hiraoka M., Nishimoto S. Emission under Hypoxia: One-Electron Reduction and Fluorescence Characteristics of an Indolequinone-Coumarin Conjugate. ChemBioChem. 2008;9:426–432. doi: 10.1002/cbic.200700458. [DOI] [PubMed] [Google Scholar]
- 26.Li H., Lei W., Wu J., Li S., Zhou G., Liu D., Yang X., Wang S., Li Z., Zhang J. An upconverting nanotheranostic agent activated by hypoxia combined with NIR irradiation for selective hypoxia imaging and tumour therapy. J. Mater. Chem. B. 2018;6:2747–2757. doi: 10.1039/C8TB00637G. [DOI] [PubMed] [Google Scholar]
- 27.Napp J., Behnke T., Fischer L., Würth C., Wottawa M., Katschinski D.M., Alves F., Resh-Genger U., Schäferling M. Targeted Luminescent Near-Infrared Polymer-Nanoprobes for In Vivo Imaging of Tumor Hypoxia. Anal. Chem. 2011;83:9039–9046. doi: 10.1021/ac201870b. [DOI] [PubMed] [Google Scholar]
- 28.Chevalier A., Zhang Y., Khdour O.M., Kaye J.B., Hecht S.M. Mitochondrial Nitroreductase Activity Enables Selective Imaging and Therapeutic Targeting. J. Am. Chem. Soc. 2016;138:12009–12018. doi: 10.1021/jacs.6b06229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data present in this study are available on request from the corresponding author.