Abstract
Transcription factors (TFs) harboring broad-complex, tramtrack, and bric-a-brac (BTB) domains play important roles in development and disease. These BTB domains are thought to recruit transcriptional modulators to target DNA regions. However, a systematic molecular understanding of the mechanism of action of this TF family is lacking. Here, we identify the zinc finger BTB-TF Zbtb2 from a genetic screen for regulators of exit from pluripotency and demonstrate that its absence perturbs embryonic stem cell differentiation and the gene expression dynamics underlying peri-implantation development. We show that ZBTB2 binds the chromatin remodeler Ep400 to mediate downstream transcription. Independently, the BTB domain directly interacts with nucleosome remodeling and deacetylase and histone chaperone histone regulator A. Nucleosome remodeling and deacetylase recruitment is a common feature of BTB TFs, and based on phylogenetic analysis, we propose that this is a conserved evolutionary property. Binding to UBN2, in contrast, is specific to ZBTB2 and requires a C-terminal extension of the BTB domain. Taken together, this study identifies a BTB-domain TF that recruits chromatin modifiers and a histone chaperone during a developmental cell state transition and defines unique and shared molecular functions of the BTB-domain TF family.
Keywords: epigenetics, transcription factor, chromatin remodeling, embryonic stem cell, histone chaperone, ZBTB2, HiRA, NuRD, Ep400
Abbreviations: 2C, 2-cell embryo; 2i, two inhibitors; 2iLIF, N2B27 containing LIF, CHIR, and PD0325901; AP-MS, affinity purification–MS; bFGF, basic FGF; BTB, broad-complex, tramtrack, and bric-a-brac; BTB-link, extended BTB domain; CHIR, CHIR99021; ChIP-seq, chromatin immunoprecipitation sequencing; DOX, doxycycline; EpiLC, epiblast-like cell; FDR, false discovery rate; FGF, fibroblast growth factor; GSK3, glycogen synthetase kinase 3; HiRA, histone regulator A; i, inhibitor; KD, knock-down; LFQ, label-free quantification; LIF, leukemia inhibitory factor; MEK, mitogen-activated protein kinase kinase; mESC, mouse embryonic stem cell; N2B27, chemically defined base medium; NuRD, nucleosome remodeling and deacetylase; PBST, PBS–0.1% Tween-20; qPCR, quantitative PCR; serum-LIF, fetal calf serum–containing medium supplemented with LIF; TF, transcription factor; Y2H, yeast-2-hybrid; Znf, zinc-finger domain
Transcription factors (TFs) are key determinants of gene expression and, therefore, play a major role in development and disease (1). TFs interpret the regulatory code of the genome by binding to DNA and regulating transcription (1). Although DNA-binding is well characterized (2), the mechanisms by which TFs modulate transcription are not completely understood. Many TFs present a modular protein architecture containing DNA-binding domains and domains that interact with transcriptional activators or repressors (1). Zinc-finger (Znf) domains are the most common family of DNA-binding domains and are often found in combination with Kruppel-associated box or broad-complex, tramtrack, and bric-a-brac (BTB) domains (3). Kruppel-associated box domains recruit KAP1 (4) and therefore mediate transcriptional repression. In contrast, there is no comprehensive understanding of the transcriptional role of BTB domains.
There are three groups of TFs containing BTB domains (BTB TFs): the Zbtb, the Bach, and the Nacc families. They are defined by their DNA-binding domains: Znf for Zbtb, bZIP for Bach, and BEN for Nacc (5). In human and mouse, the Bach and the Nacc families contain only two members each, and the Zbtb family comprises 49 members. Several of them are critical regulators of fate allocation and differentiation across many organs and systems (6). A striking example is hematopoiesis, in which BTB TFs direct the differentiation of several lineages (7). The BTB domains are invariably found at the N terminus of BTB TFs and the DNA-binding domains at the C terminus, separated by a long nonconserved linker region (7). Although BTB domains function in homo- and hetero-dimerization and protein–protein interactions, the BTB domains found in TFs and CUL3 ubiquitin ligases constitute separate families (5). Mechanistic studies of ZBTB factors have focused on the proto-oncogenes Bcl6 (Zbtb27) and Lrf (Zbtb7a) and on the tumor suppressor Plzf (Zbtb16) and found that they act as transcriptional repressors via the recruitment of complexes such as nuclear receptor corepressor/silencing mediator of retinoic acid and thyroid hormone receptor (NCOR/SMRT), BCL6 corepressor (BCOR), SIN3 transcription regulator family member A/B (SIN3A/B), and nucleosome remodeling and deacetylase (NuRD) (7).
Mouse embryonic stem cells (mESCs) are a developmentally relevant cell type that captures the pluripotent state of the preimplantation mouse epiblast and that recapitulates developmental progression upon release into differentiation in vitro (8). Furthermore, a large number of genomic datasets have been derived from mESCs, making them an ideal model for mechanistic studies of transcription. mESCs are maintained by provision of the cytokine leukemia inhibitory factor (LIF) in a fetal calf serum–containing medium (Serum-LIF) (9) or of inhibitors (i) of glycogen synthetase kinase 3 (GSK3) and mitogen-activated protein kinase kinase (MEK) in a chemically defined medium (N2B27) (10). LIF, GSK3i, and MEKi stabilize a pluripotency TF network centered on OCT4, SOX2, and NANOG (8). mESCs grown in the presence of the two inhibitors (2i) are called naïve and display higher and more homogeneous expression levels of pluripotency TFs than Serum-LIF–grown cells (11). Naïve and Serum-LIF cell states are interconvertible (12), whereas naïve cells can efficiently differentiate into postimplantation epiblast-like cells (EpiLCs) (13). In both naïve and Serum-LIF conditions, mESCs transition through a 2-cell embryo (2C)-like state that is thought to reflect properties of cleavage-stage embryos and that is characterized by expanded developmental potency (14), increased histone mobility (15), and a specific gene expression signature that includes the upregulation of endogenous retroviruses (16). While the TF DUX directly binds and activates the promoters of 2C-like genes (17), it is less clear how other chromatin regulators, such as PRC1.6, Ep400 (18), or CAF-1 (19), mechanistically regulate 2C-like genes.
The NuRD complex is composed of HDAC1/2, CHD4, GATAD2A/B, RBBP4/7, MTA1/2/3, and MBD2/3 (20). MBD2 and MBD3 are mutually exclusive subunits and define the two functionally distinct MBD2–NuRD and MBD3–NuRD complexes (21). Mbd3 has been subject to several genetic studies in mESCs, showing that NuRD drives mESC differentiation (22, 23, 24) and fine-tunes gene expression by modulating chromatin accessibility (25).
The histone regulator A (HiRA) complex acts as the H3.3 histone chaperone at euchromatic loci (26). HiRA has been proposed to be recruited by naked DNA and to have a nucleosome gap–filling function (27). It is composed of the subunits HIRA, CABIN1, and UBN1/2 (28). It has been suggested that UBN1 and UBN2 are part of two independent, but functionally indistinguishable, complexes UBN1-HiRA and UBN2-HiRA (29). Euchromatic H3.3 is found in H2A.Z-containing nucleosomes (30) that are incorporated into chromatin by the chromatin remodeling complex Ep400 (31), yet if and how Ep400 interacts with H3.3 chaperones is unclear. HiRA is required for exit from the naïve pluripotency during differentiation (32), while the Ep400 complex is essential for the maintenance of mESCs (33).
Here, we identify the BTB-TF Zbtb2 in a genetic screen for regulators of exit from mESC pluripotency and report a detailed mechanistic analysis of its function, showing that ZBTB2 recruits ZNF639, MBD3-NuRD, UBN2-HiRA, and the Ep400 complex. Transcriptome analysis reveals that ZBTB2 interactors form two functionally distinct modules, one encompassing ZNF639, NuRD, and HiRA and the other corresponding to the Ep400 complex. We show that NuRD and HiRA associate with ZBTB2 via the subunits GATAD2A/B and UBN2, respectively. We systematically test these interactions across all BTB TFs in yeast-2-hybrid (Y2H) screens. We find that ZBTB2 harbors an extension of the BTB domain that mediates a unique interaction with UBN2. GATAD2A/B interaction is instead a common feature of BTB TFs and shared across several BTB-TF phylogenetic branches, making NuRD recruitment a candidate ancestral feature of TF-associated BTB domains. Our study therefore reveals unique and shared molecular functions of the BTB domain TF family.
Results
A sensitized genetic screen identifies Zbtb2 as regulator of the exit from pluripotency
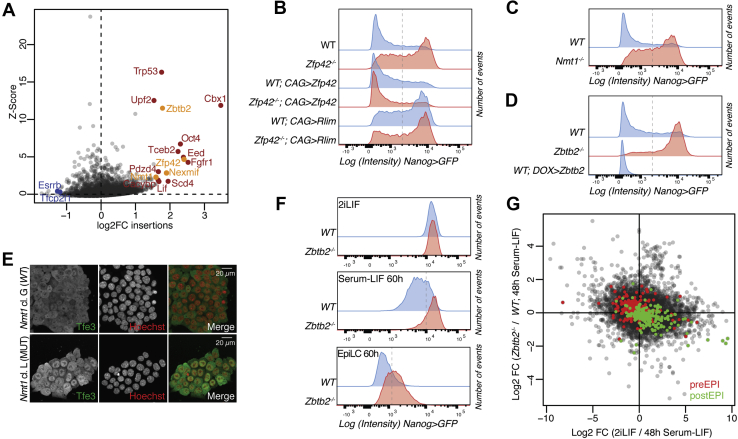
We performed a sensitized genetic screen for maintenance of pluripotency in the presence of the GSK3i CHIR99021 (CHIR) (34), which is unable to block mESC differentiation in the absence of LIF or the MEKi PD0325901 (35). As the role of CHIR in mESC maintenance is well characterized (36, 37), this medium formulation should increase sensitivity for other less-understood pathways. We mutagenized haploid mESCs (38) harboring an Oct4>GFP-puro reporter with retroviruses carrying a splicing acceptor site for insertional mutagenesis and Oct4 enhancer elements for overexpression (39), leading to both loss and gain of function alleles. We cultured the mutagenized library in N2B27 + CHIR + Puromycin to select for undifferentiated cells and harvested the cells after 16 to 23 days. We mapped the insertions by high-throughput inverse PCR and determined insertion enrichment compared with the starting library for every gene (Fig. 1A, Table S1). Confirming the specificity of our setup, Fgfr1 and Lif were among the highest scoring hits. FGFR1 is the main fibroblast growth factor (FGF) receptor in mESCs and acts upstream of MEK activation (40), and chemical inhibition of FGFR signaling has been shown to substitute for MEKi in mESC maintenance (10). Lif, evidently a gain of function hit, is also able to sustain pluripotency in combination with CHIR (34). Oct4 is a technical false-positive hit, as it can drive the expression of the Oct4>GFP-puro reporter irrespectively of the cell state. Confidence in our analysis was also bolstered by the identification of Esrrb and Tfcp2l1 among the insertion-depleted genes, as these are transcriptional mediators of CHIR activity (8, 36, 41). Some of the highest scoring screen hits, such as Cbx1 (42), Eed (43, 44), Trp53 (45), and Upf2 (46), have previously been implicated in the exit from pluripotency. We therefore decided to validate the hits Zbtb2, Zfp42, Nexmif, and Nmt1.
Figure 1.
Screen results and validation.A, Z-scores and enrichment fold-changes of retroviral insertions within the gene bodies of indicated genes after selection in N2B27 + CHIR + Puromycin. In blue are pluripotent network TFs. Red labels previously described differentiation driver genes and orange novel candidates. B–D and F, Nanog>GFP intensities after switching from 2iLIF to N2B27 + CHIR + bFGF for 3 days (B–D) or as indicated in panel F of indicated genotypes. Dashed lines indicate the thresholds for quantifications presented in Fig. S1, D and F. E, TFE3 immunofluorescence in Nmt1−/− and WT mESCs in 2iLIF. Nuclei were counterstained with Hoechst. G, scatterplot of log2 fold-changes (FC) in gene expression of indicated contrasts. Green labels post-implantation epiblast (postEPI)-specific and red pre-implantation epiblast (preEPI)-specific genes (56). 2iLIF, N2B27 containing LIF, CHIR, and PD0325901; bFGF, basic FGF; CHIR, CHIR99021; mESCs, mouse embryonic stem cells; N2B27, chemically defined medium.
The Nexmif gene lies upstream of the Rlim transcription start site, and RLIM is a known E3 ubiquitin ligase for ZFP42 (47), so we reasoned that the Nexmif insertion enrichment would lead to Rlim overexpression. We therefore generated CRISPR KO clones (Table S2) for Zfp42 (Fig. S1A), Nmt1 (Fig. S1B), and Zbtb2 (Fig. S1C), and Rlim overexpressing cells using naïve TNG-A mESCs, a conventional diploid cell line harboring a Nanog>GFP reporter (48). Upon exposure to N2B27 + CHIR, concomitant with the addition of recombinant basic FGF (bFGF) to increase the stringency of the assay, all three mutants and the Rlim-overexpressing cells showed delayed downregulation of the Nanog reporter when compared with WT cells (Fig. 1, B–D, and Fig. S1D), indicating that they are bona fide loss-of-function screen hits. Overexpression of Rlim in the Zfp42 mutant did not modify the phenotype of the Zfp42 single mutant (Fig. 1B and Fig. S1D), supporting the epistatic relationship between Rlim and Zfp42. While Zfp42 overexpression did not affect differentiation (Fig. 1B and Fig. S1D), strong Zbtb2 constitutive overexpression using a CAG promoter caused cell death (not shown). We therefore tested the effect of moderate Zbtb2 overexpression, achieved by a doxycycline (DOX)-inducible promoter, and observed accelerated differentiation upon Zbtb2 induction (Fig. 1D and Fig. S1D).
Nmt1 encodes an N-myristoyltransferase (49), and myristoylation is required for the function of FRS2, an essential component for FGFR1 signaling (50). We therefore speculated that loss of Nmt1 would inhibit mESC differentiation by dampening mitogen-activated protein kinase signaling. However, ERK phosphorylation upon exposure to bFGF was unperturbed in Nmt1−/− cells (Fig. S1E). We therefore turned our attention to another myristoylated protein, LAMTOR1 (51), which is required for TFE3 nuclear exclusion (52) and, in turn, for the exit from pluripotency (53). Immunofluorescence staining revealed abnormal nuclear localization of TFE3 in Nmt1−/− mESCs (Fig. 1E), suggesting that ectopically active TFE3 mediates the differentiation delay in the absence of Nmt1.
Although Zbtb2 had already been suggested to play a role in the differentiation of Serum-LIF mESCs (54), we decided to focus our efforts on this factor in the hope of gaining insights that are broadly applicable to BTB TFs. We first sought to better characterize Zbtb2’s role in the exit from the naïve state and determine when the earliest developmental defect would arise. We first tested differentiation of N2B27 containing LIF, CHIR, and PD0325901 (2iLIF) cells using the EpiLC differentiation protocol, which faithfully mimics the preimplantation to postimplantation epiblast transition (13), and observed a delay in Nanog reporter downregulation in Zbtb2−/− cells although Nanog levels were unchanged in steady-state 2iLIF cultures (Fig. 1F and Fig. S1F). We then turned to differentiation in Serum-LIF, which establishes a developmentally advanced pluripotent state (55). Even in this assay, Nanog>GFP downregulation was delayed in Zbtb2−/− cells (Fig. 1F and Fig. S1F). To determine transcriptome-wide changes, we performed RNA-Seq of WT and Zbtb2−/− cells in 2iLIF and during differentiation. We found that gene expression changes that accompany the 2iLIF to Serum-LIF transition in WT cells were dampened in Zbtb2−/− cells exposed to Serum-LIF for 48 h (h) (Pearson correlation coefficient R = −0.19, Fig. 1G, Table S3). When we specifically focused on genes regulated during embryonic development (56), we found that Zbtb2−/− cells in Serum-LIF, when compared with WT controls, were impaired in upregulating genes that are expressed in the postimplantation epiblast and in downregulating genes that are predominantly transcribed in the preimplantation epiblast (Fig. 1G). This coherent deregulation of developmental genes was specific to the Serum-LIF transition and undetectable in steady-state 2iLIF cells (Fig. S1G). In summary, loss of Zbtb2 delays and Zbtb2 overexpression increases mESC differentiation, demonstrating an instructive role of ZBTB2 in cell-state transitions.
An extended BTB domain binds UBN2 and GATAD2B; NuRD interaction is stabilized by ZNF639
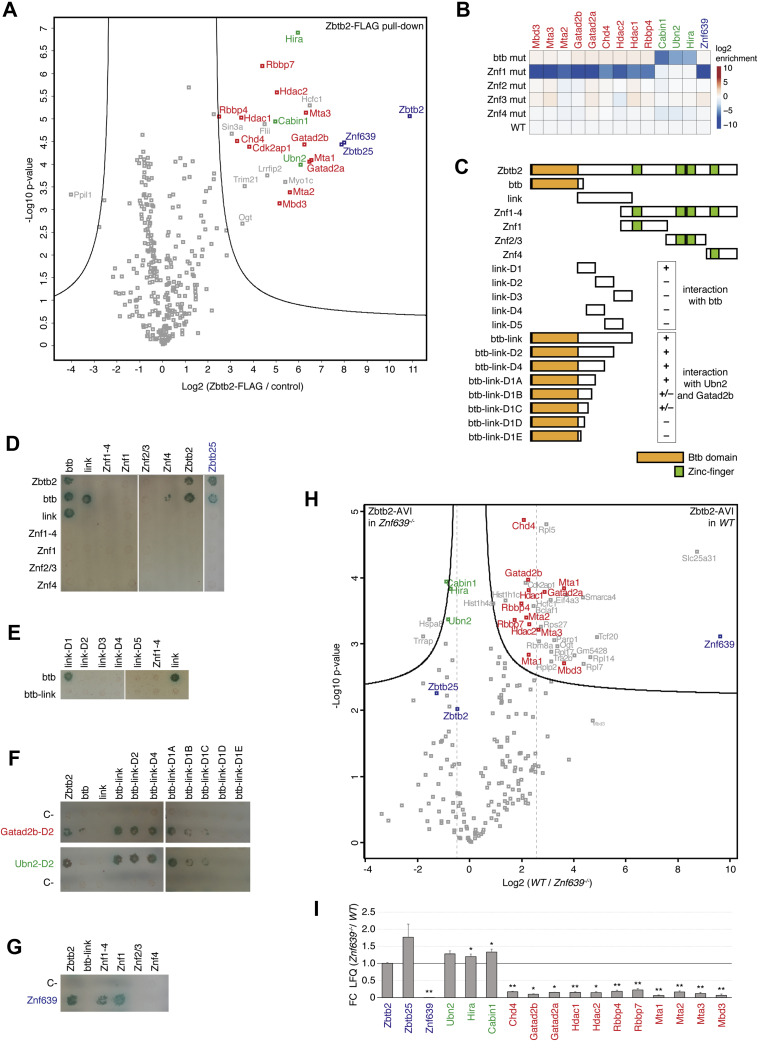
To understand the mechanisms by which Zbtb2 exerts its function, we set out to perform affinity purification–MS (AP-MS) of ZBTB2 in mESCs. Owing to the absence of antibodies detecting the endogenous protein, we introduced a 3xFLAG into the Zbtb2 locus by genome editing, producing a ZBTB2–3xFLAG fusion protein under the control of endogenous regulatory elements (Fig. S2A and Table S2). Upon anti-FLAG pull-down in the presence of nuclease to avoid DNA- and RNA-bridged interactions, we identified ZBTB25 and ZNF639, previously reported to bind to ZBTB2 (54), and all subunits of the NuRD and of the HiRA complexes as specific ZBTB2 interactors (Fig. 2A). We did not detect MBD2 or UBN1 peptides, demonstrating copurification of MBD3–NuRD and UBN2–HiRA complexes, specifically. Next, we decided to determine the role of individual ZBTB2 domains by purifying ZBTB2–AVI fusion proteins with mutations in the BTB and Znf domains (Table S4). These were expressed in a Zbtb2−/− Zbtb25−/− background (Fig. S2C) to prevent indirect interactions by bait dimerization with endogenous ZBTB2 or ZBTB25, and under the control of DOX. Although quantitative PCR (qPCR) revealed a ∼6-fold overexpression (Fig. S2D), the interactomes of endogenous ZBTB2–3xFLAG and overexpressed WT ZBTB2–AVI are indistinguishable (Table S5). Furthermore, DOX induction of untagged Zbtb2 and Zbtb2-AVI similarly induced differentiation (Fig. S2B), confirming functionality of the Zbtb2–AVI constructs. Mutation of the BTB domain, however, caused loss of the interaction with UBN2–HiRA and of Znf1 abolished the interaction with ZNF639 and NuRD, whereas mutation of the other Znfs did not significantly alter the interactome (Fig. 2B and Table S5).
Figure 2.
An extended BTB domain binds UBN2 and GATAD2B; NuRD interaction is stabilized by ZNF639.A, volcano plot of protein enrichments in anti-FLAG AP-MS from mESCs expressing endogenous ZBTB2-3xFLAG compared with parental untargeted mESCs. ZBTB2 and partner TFs are indicated in blue, NuRD subunits in red, and HiRA subunits in green. B, bait-normalized log2 fold-enrichment of ZNF639, HiRA subunits, and NuRD subunits in AP-MS from mESCs overexpressing ZBTB2-AVI alleles with indicated mutations compared with overexpressing WT ZBTB2-AVI. All baits (Table S4) were induced with DOX in Zbtb2−/−Zbtb25−/− mESCs. C, diagram of ZBTB2 constructs used for Y2H analysis; plus and minus indicate positive and negative interactions as in panels E and F. D–G, colony growth on SD-Ade/-His/-Trp/-Leu/+X-alpha-Gal/+Aureobasidin A (QDOXA) plates of strains expressing indicated proteins. Bait constructs are vertical (D and E) and horizontal (F and G), and prey constructs are horizontal (D and E) and vertical (F and G). H, volcano plot of protein enrichments in AP-MS of overexpressed ZBTB2-AVI in WT compared with Znf639−/− mESCs. Color code as in Figure 2A. Dashed lines highlight the difference in NuRD subunit abundance relative to the bait ZBTB2. I, bait-normalized interaction changes as in panel H. ∗p-value < 0.01 and ∗∗p-value < 0.001. AP-MS, affinity purification–MS; BTB, broad-complex, tramtrack, and bric-a-brac; DOX, doxycycline; HiRA, histone regulator A; mESCs, mouse embryonic stem cells; NuRD, nucleosome remodeling and deacetylase; TFs, transcription factors; Y2H, yeast-2-hybrid.
Loss of interaction upon domain mutation does not imply direct physical binding, as it could be due to an indirect functional dependency or bridging factor. We, therefore, turned to Y2H assays. First, we addressed dimerizing properties of ZBTB2. We detected homodimerization of ZBTB2’s BTB domain and heterodimerization with ZBTB25 (Fig. 2, C and D). To our surprise, we also found a strong interaction of ZBTB2’s linker region with ZBTB2’s BTB domain but not with the full-length ZBTB2 protein (Fig. 2, C and D). The linker is 136 amino acids in length and predicted to be unstructured. We generated linker region deletions and found that the segment immediately adjacent to the BTB domain (link-D1) mediates interaction with the BTB domain (Fig. 2, C and E), suggesting the existence of an extended BTB domain structure. We therefore used this extended BTB domain (BTB-link) in further assays. Next, we tested the direct interaction of ZBTB2 with the HiRA complex subunits HIRA, CABIN1, and UBN2. We found that UBN2, but not HIRA or CABIN1, binds full-length ZBTB2 and the BTB-link domain, but neither the isolated BTB domain or linker region (Fig. S2E). Deletion analysis of UBN2 identified a minimal interacting region of 68 amino acids that is outside of annotated domains (Fig. S2, F and G). The BTB-link domain therefore mediates homodimerization and heterodimerization with ZBTB2 and ZBTB25, respectively, and interaction with UBN2.
We then tested binding to the NuRD subunits RBBP4, MBD3, MTA2, MTA3, HDAC1, GATAD2A, and GATAD2B. Based on our AP-MS results (Fig. 2B), we expected that Znf1 would mediate such an interaction, yet the only direct interactions we identified were between GATAD2A and ZBTB2, and between GATAD2B and both ZBTB2 and the BTB-link domain (Fig. S2, H and I). No interactions were found with Znf1, the BTB domain, or the linker region. Additional Y2H assays revealed that the C-terminal half of GATAD2B, which includes its Gata-type Znfs, mediates interaction with the BTB-link domain (Fig. S2, J and K).
As the direct interactions with both UBN2 and GATAD2B were mediated by the BTB-link domain, but not the BTB domain or linker alone, we sought to determine the minimal BTB domain extension required for either interaction. Through serial truncations, we found that a 44-amino acid extension was necessary for both (Fig. 2, C and F). This, together with the direct interaction between the conserved BTB domain and this 44-amino acid fragment (Fig. 2, C and E), strongly suggests a functionally essential structural extension of ZBTB2’s BTB domain.
Further Y2H assays identified a direct interaction between ZNF639 and ZBTB2’s Znf1 domain (Fig. 2G), as expected by the AP-MS data (Fig. 2B). Znf1 was required for the interaction with NuRD in AP-MS (Fig. 2B) but did not mediate any direct interaction with NuRD subunits (Fig. S2, H and I). Therefore, we wondered whether ZNF639 is required to stabilize or enhance the interaction between NuRD and ZBTB2, which is mediated by the extended BTB domain. To test this hypothesis, we performed AP-MS of ZBTB2-AVI in wt and Znf639−/− mESCs (Fig. S2L and Table S2) and found an ∼8-fold reduction in NuRD interaction upon loss of Znf639 (Fig. 2, H and I).
In summary, our AP-MS and Y2H experiments show that the BTB domain of ZBTB2 mediates homodimerization and heterodimerization with ZBTB25, an extended BTB domain recruits the UBN2–HiRA complex through the UBN2 subunit, and the interaction with MBD3–NuRD is direct via BTB-link binding GATAD2A/B, but also requires Znf1 recruiting ZNF639.
Recruitment of HiRA is a unique property of ZBTB2, while GATAD2A/B interaction is a conserved feature of TF-associated BTB domains
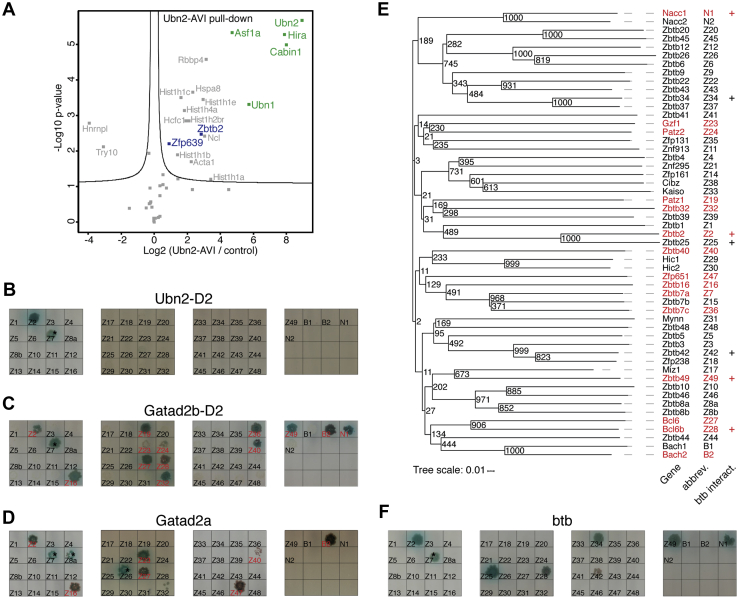
Interaction of a BTB TF with the HiRA complex has not been reported before, whereas the interaction with NuRD has been previously shown for ZBTB7A (57). We therefore wondered if HiRA and NuRD recruitment would be conserved across BTB TFs. We first attempted to identify additional HiRA-interacting BTB domain proteins by performing AP-MS of DOX-induced UBN1-AVI and UBN2-AVI in mESCs. Although Ubn1-AVI and Ubn2-AVI transcript levels were ∼4.5-fold and ∼2-fold higher than the respective endogenous mRNAs (Fig. S3A), no additional HiRA-interacting BTB domain proteins were identified. In fact, the only TFs we recovered in the UBN2-AVI pull-down (Fig. 3A) were ZBTB2 and its partner ZNF639 but not ZBTB25, and in the UBN1-AVI pull-down (Fig. S3B) ZBTB2 and MEF2D, a known direct interactor of CABIN1 (58).
Figure 3.
Recruitment of HiRA is a unique property of ZBTB2, while GATAD2A/B interaction is a conserved feature of TF-associated BTB domains.A, volcano plot of protein enrichments in AP-MS of mESCs overexpressing UBN2-AVI compared with control BirA-only–expressing mESCs. ZBTB2 and partner TFs are indicated in blue and HiRA subunits in green. B–D and F, colony growth on QDOXA plates of strains expressing extended BTB domains of Zbtb (Z#), Bach (B#), and Nacc (N#) bait constructs (labeling as abbreviated in panel E), and Ubn2-D2 (B), Gatad2b-D2 (C), Gatad2a (D), and ZBTB2 BTB domain (F) prey constructs. GATAD2A/B interactors are indicated in red. Asterisks mark autoactivating bait constructs (Fig. S3, C and D). E, phylogenetic tree of TF-associated BTB domains with bootstrap values. GATAD2A/B interactors, based on panel C and D and (57) are indicated in red. The plus indicates BTB domains dimerizing with the BTB domain of ZBTB2 (btb interact.), based on panel F. AP-MS, affinity purification–MS; BTB, broad-complex, tramtrack, and bric-a-brac; HiRA, histone regulator A; mESCs, mouse embryonic stem cells; TFs, transcription factors.
As not all BTB TFs are expressed in mESCs, we turned to Y2H to systematically assay ability to bind to UBN2 or GATAD2A/B. We performed Y2H screens of the BTB domains of the 49 Zbtb factors, Nacc1/2, and Bach1/2, extending the conserved BTB domains by at least 60 amino acids, in case other BTB domains would possess extended structures similar to Zbtb2. Strikingly, only ZBTB2’s BTB-link domain interacted with UBN2 (Fig. 3B and Fig. S3C), corroborating the UBN1/2 AP-MS data to support that HiRA recruitment by ZBTB2 is unique. Surprisingly, we identified BTB domains of 14 BTB TFs to interact with GATAD2A or GATAD2B (Fig. 3, C and D and Fig. S3, C and D). We were not able to confirm binding to ZBTB7A’s BTB domain (57) because of autoactivation in the Y2H assays (Fig. S3, C and D). Taken together, at least 15 of 54 BTB TFs bind to NuRD subunits, suggesting that GATAD2A/B interaction is a common function of TF-associated BTB domains.
We wondered whether the GATAD2A/B-interacting domains would be phylogenetically related. Phylogenies based on the complete sequence of Zbtb TFs do not reflect the similarities within the BTB domains (59) because of the influence of the Znf domains. We therefore constructed a phylogenetic tree based on BTB domain sequences (Fig. 3, E and Table S6). Although confidence for the evolutionary older branches was low, GATAD2A/B-interacting BTB domains do not form a clade but are scattered throughout the tree, including the Bach and Nacc clades. This shows that GATAD2A/B recruitment is an ancestral property of TF-associated BTB domains.
To test for heterodimerization with ZBTB2 and if this correlates with binding to GATAD2A/B, we repeated the Y2H BTB domain family screen using ZBTB2’s BTB domain as bait (Fig. 3F and Fig. S3C). This identified six BTB domains heterodimerizing with ZBTB2, of which 3 also bound to GATAD2A/B. Specificity for heterodimerization and GATAD2A/B binding are therefore separate properties of the BTB domain.
Although ZBTB25’s BTB domain heterodimerizes with ZBTB2, it did not bind UBN2 or GATAD2A/B (Fig. 3, B–F). We therefore hypothesized that binding to ZBTB25 is not relevant to ZBTB2’s role in cell fate transition. To test this, we characterized the phenotype of Zbtb25−/− and Zbtb2−/− Zbtb25−/− mESCs (Fig. S3E) in the 2iLIF to Serum-LIF transition and compared it with Zbtb2−/− cells. In fact, loss of Zbtb25 did not delay Nanog>GFP downregulation or modify the phenotype of Zbtb2 single mutants (Fig. S3, F and G).
In summary, we tested the conservation of UBN2 and GATAD2A/B interaction by AP-MS and Y2H. While UBN2 recruitment appears to be unique to ZBTB2, GATAD2A/B interaction is a common and ancestral feature of BTB TFs.
ZBTB2 interacts with the Ep400 complex in a HiRA-independent manner
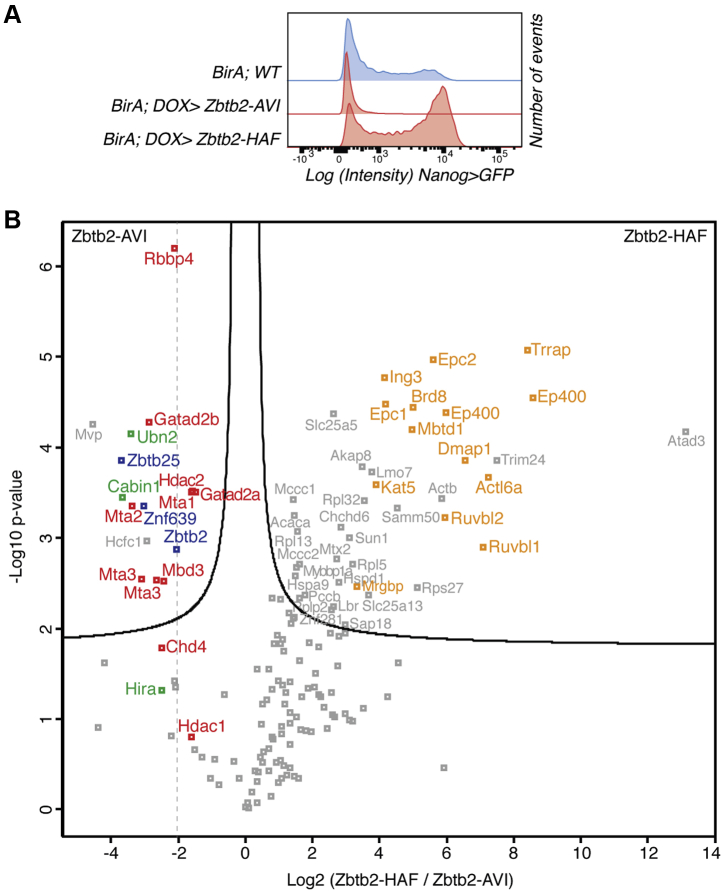
While evaluating the functionality of tagged Zbtb2 constructs for AP-MS, we found that overexpression of ZBTB2 fused with an extended C-terminal 3xHA-AVI-3xFLAG-tag (HAF-tag) caused a delay in Nanog>GFP reporter downregulation (Fig. 4A). This phenotype is opposite to ZBTB2-AVI overexpression but similar to loss of Zbtb2, suggesting that ZBTB2-HAF acts dominant negative. To understand the underlying molecular defect, we compared the interactomes of the ZBTB2-AVI and ZBTB2-HAF fusion proteins. To our surprise, we found that the entire Ep400 complex copurified with the dominant-negative ZBTB2–HAF, while none of the other interactors was lost (Fig. 4B). This prompted us to look more carefully at the ZBTB2–AVI AP-MS data and consistently found Ep400 subunit peptides across independent experiments (Table S5). ZBTB2 directly binds the Ep400 complex subunit KAT5 in Y2H assays (60), suggesting that ZBTB2–HAF stabilizes a physiological, but transient or weak interaction.
Figure 4.
A dominant negative Zbtb2 construct stabilizes the interaction with the Ep400 complex.A, Nanog>GFP intensities after switching from 2iLIF to N2B27 +CHIR +bFGF in the presence of DOX of indicated genotypes. B, volcano plot of protein enrichments in AP-MS of mESCs overexpressing ZBTB2-HAF compared with those overexpressing ZBTB2-AVI. ZBTB2 and partner TFs are indicated in blue, NuRD subunits in red, HiRA subunits in green, and Ep400 subunits in orange. AP-MS, affinity purification–MS; bFGF, basic FGF; CHIR, CHIR99021; HiRA, histone regulator A; N2B27, chemically defined medium; NuRD, nucleosome remodeling and deacetylase; TFs, transcription factors.
Ep400 incorporates H2A.Z/H3.3 histones into chromatin (31). We therefore hypothesized that association of Ep400 with ZBTB2 could be mediated by the HiRA complex, which is an H3.3 chaperone (28). To test this, we performed AP-MS of ZBTB2-HAF in wt and in Ubn2−/− mESCs (Fig. S4A). As expected, lack of UBN2 caused loss of the HiRA interaction, but the association with Ep400 was not affected (Fig. S4B). Interestingly, we noted a substantial increase of ZBTB25 in ZBTB2 AP-MS in Ubn2−/− cells (Fig. S4B). UBN2 and ZBTB25 may therefore compete for interaction with ZBTB2. Together with the inability of ZBTB25 to interact with UBN2 (Fig. 3B), this supports the idea that ZBTB25 is a negative regulator of HiRA recruitment by ZBTB2. We therefore conclude that ZBTB2 interacts weakly or transiently with Ep400 and independent of HiRA cobinding.
Ep400 and Znf639/NuRD/HiRA constitute independent Zbtb2 functional modules
To address the functional role of ZBTB2’s protein interactions, we set out to compare the loss-of-function phenotype of Zbtb2 with that of its binding partners. Depletion of the Ep400 complex subunits Ep400 and Kat5 causes loss of mESC self-renewal (33), while KO of Mbd3 (24) or Hira (32) induces resistance to exit from the mESC state during differentiation. However, these phenotypes may arise from pleiotropic, Zbtb2-unrelated roles. As the ZBTB2–HiRA interaction is dependent on UBN2, and the ZBTB2–NuRD interaction is stabilized by ZNF639, we reasoned that analysis of Ubn2−/− and Znf639−/− cells might reveal Zbtb2-specific functions. Compared with Zbtb2 mutants, Znf639−/− and Ubn2−/− mESCs showed moderate delays in Nanog>GFP downregulation upon Serum-LIF transition (Fig. 5, A and B). Although weak, these phenotypes are consistent with Znf639 and Ubn2, and by extension NuRD and HiRA, cooperating with Zbtb2 in cell fate transitions.
Figure 5.
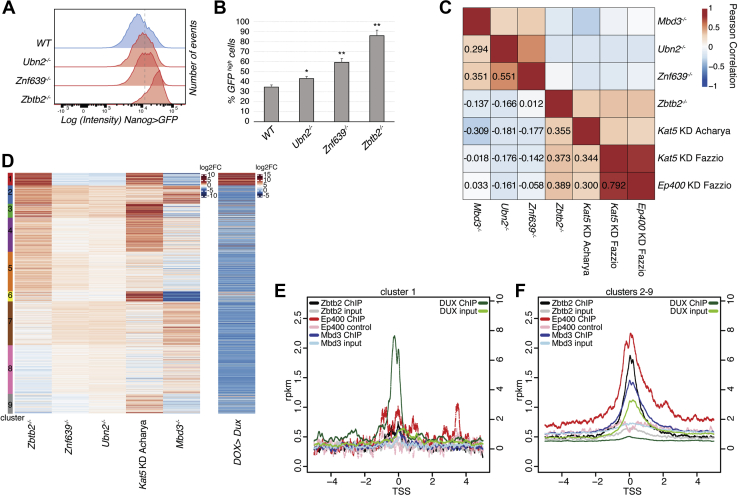
Ep400 and ZNF639/NuRD/HiRA constitute independent functional modules.A and B, Nanog>GFP intensities after switching from 2iLIF to Serum-LIF for 3 days of indicated genotypes (A). Dashed line indicates the threshold for quantification of GFP-high cells as the average and SD of biological triplicates (B). ∗p-values < 0.01 and ∗∗p-values < 0.001 compared with the WT control. C, pairwise Pearson correlations of differential gene expression relative to respective control cells for Ubn2−/−, Znf639−/−, and Zbtb2−/− mutant cells after 48 h in Serum-LIF, and for Mbd3−/− (22), Kat5 KD (33, 61), and Ep400 KD (33) in Serum-LIF. Only genes deregulated in Zbtb2−/− cells were considered (1420 genes). D, k-means clustering of differential gene expression as in Figure 2C and upon Dux overexpression (17). E and F, ZBTB2 (54), EP400 (62), and MBD3 (25) (left scale), and DUX (17) (right scale) ChIP-seq reads per kilobase of transcript per million mapped reads (rpkm) centered on transcriptional start sites (TSSs) of cluster 1 (E) and cluster 2 to 9 genes (F), according to panel D. HiRA, histone regulator A; KD, knock-down.; NuRD, nucleosome remodeling and deacetylase; Serum-LIF, fetal calf serum–containing medium supplemented with LIF. ChIP-seq, chromatin immunoprecipitation sequencing.
To investigate these functional relationships further, we performed RNA-Seq of Zbtb2−/−, Znf639−/−, and Ubn2−/− mESCs after 48 h in Serum-LIF. Consistent with Nanog reporter phenotypes, preimplantation epiblast-enriched transcripts were upregulated and postimplantation epiblast-specific genes were downregulated in Znf639 and Ubn2 mutant cells in Serum-LIF, although to a lesser extent than observed in Zbtb2 KO cells (Fig. S5A). To describe Zbtb2’s transcriptional role, we focused on genes significantly changing upon loss of Zbtb2 and included published transcriptome data of Mbd3−/− (22), Ep400, and Kat5 knock-down (KD) (33, 61) mESCs in Serum-LIF (Table S3). We found that transcriptional alterations in Zbtb2−/− cells correlated with those upon KD of Ep400 (R = 0.39) and Kat5 (R = 0.37 and 0.36), showing a mechanistic relationship between Zbtb2 and the Ep400 complex (Fig. 5C). Changes in Mbd3 mutants, in contrast, correlated with those in Znf639 KO cells (R = 0.36), corroborating the strong reduction of NuRD binding to ZBTB2 in Znf639 mutants. Furthermore, alterations in Mbd3, Znf639, and Ubn2 mutants correlated reciprocally, pointing toward a functional ZNF639/NuRD/HiRA unit (Fig. 5C). Strikingly, there was no general correlation between Zbtb2/Ep400-dependent and Znf639/Mbd3/Ubn2-dependent gene expression, raising the hypothesis that Ep400 and ZNF639/NuRD/HiRA cooperate with ZBTB2 within functionally independent modules.
For more detailed insight, we performed k-means clustering of differential gene expression (Fig. 5D). Cluster 1 genes were upregulated in Zbtb2−/− and Kat5 KD cells and downregulated in Znf639−/−, Mbd3−/−, and Ubn2−/− cells. Cluster 1 contains the strongest changing genes in Zbtb2−/− cells, and closer inspection revealed them to be enriched for 2C-like genes, as corroborated by the correlation with DUX-induced genes (Fig. 5D) (17). Such opposite functions are consistent with independent complexes competing for limiting ZBTB2 amounts, where loss of the ZNF639/NuRD/HiRA complex would lead to an increase in Ep400/ZBTB2-dependent 2C-like gene repression.
Taken together, our findings show that Zbtb2, Znf639, and Ubn2 loss of function causes a developmental delay that is reflected in the deregulation of preimplantation and postimplantation epiblast-specific genes. However, a systematic analysis of ZBTB2 target genes revealed that gene expression changes upon loss of Ep400 and Kat5 correlate best with Zbtb2 mutants, while alterations upon loss of Znf639, Ubn2, and Mbd3 correlate reciprocally, but not with Zbtb2 mutants. This suggests the existence of two functionally distinguishable ZBTB2 modules, one associated with the Ep400 complex, causing most Zbtb2-dependent gene expression changes, and the other with ZNF639/NuRD/HiRA. Opposite roles in regulating 2C-like gene expression suggest that these modules act antagonistically within one protein complex or independently in competing biochemical complexes.
ZBTB2 inhibits 2C-like gene transcription indirectly but binds and represses its own promoter
The Ep400 complex represses expression of 2C-like genes without binding to their regulatory DNA sequences (18). We therefore wondered if ZBTB2 is similarly depleted at 2C-like genes. As ZBTB2 mostly binds to promoters (54), we analyzed ZBTB2, EP400, KAT5 (62), MBD3, and CHD4 (25) occupancy at promoters of genes belonging to cluster 1 (2C-like genes) or clusters 2 to 9 (Fig. 5, E and F and Fig. S5, B and C). We found that these factors are depleted at cluster 1 promoters, as opposed to DUX which is highly enriched, as expected (17). Therefore, 2C-like gene repression by ZBTB2/Ep400 is either indirect through the regulation of other genes or mediated through DNA-binding–independent mechanisms.
Nevertheless, ZBTB2 is enriched at promoters, where it colocalizes with EP400, KAT5, MBD3, and CHD4 (Fig. S5D). Therefore, we wondered what the activity of ZBTB2 on bound genes is. As one of the most prominent ZBTB2 peaks in ZBTB2 chromatin immunoprecipitation sequencing (ChIP-seq) lays on the promoter of Zbtb2 itself (Fig. S5E), we hypothesized that Zbtb2 transcription provides a readout for ZBTB2 activity. Indeed, Zbtb2 transcripts were upregulated in Zbtb2 mutants in which the gene structure was not affected (Figs. S1C and S5, E and F). Conversely, overexpression of ZBTB2 from a plasmid that contained silent coding sequence mutations and that allowed us to distinguish ectopic and endogenous transcripts downregulated endogenous Zbtb2 mRNA levels (Fig. S5F). ZBTB2 therefore inhibits its own expression, likely in a direct negative feedback loop. Notably, Kat5 KD by siRNA transfection, similar to Zbtb2 mutants, induced Zbtb2 transcription (Fig. S5G).This is consistent with joint roles of ZBTB2 and Ep400 in gene regulation, at least of Zbtb2.
In conclusion, ZBTB2 represses transcription either via promoter binding, as is the case for autoregulation, or via indirect mechanisms, as is the case for 2C-like genes.
Discussion
In this study, we exploited a genetic screen in mESCs to mechanistically and functionally characterize the BTB-TF ZBTB2, gaining insights that are broadly applicable to BTB TFs.
We made use of a sensitized setup and took advantage of a loss-of-function and gain-of-function haploid mESC library, identifying previously described and novel regulators of exit from pluripotency (Fig. 1A). We validated the role of Zfp42/Rlim, Nmt1, and Zbtb2 in an independent cell line and using an unrelated reporter (Fig. 1, B–D), confirming their role in mESC differentiation. Previous studies of Zfp42 (63, 64) led to the prevailing idea that Zfp42 is dispensable for pluripotency or development, to the point that Zfp42−/− cells have been used in genetic screens for the exit from pluripotency (32, 52, 65). Our data also support the relationship between Rlim and Zfp42 (47) and call for further investigation of the function of this genetic axis in pluripotency. Nmt1 is the major N-myristoyltransferase in mESCs and is essential in early mouse development (49). As some of its substrates are known (51), we tested two possible modes of action, via FRS2 (50) and mitogen-activated protein kinase signaling, or via LAMTOR1 and TFE3 localization (52). While we were not able to detect changes in ERK phosphorylation (Fig. S1E), we noticed a clear increase in nuclear TFE3 (Fig. 1E), which is compatible with a TFE3-dependent differentiation delay.
With the aim of gaining fundamental insights into transcriptional regulation of cell state transition, we focused our work on the BTB-TF Zbtb2. A previous report showed morphological delay upon LIF removal in Zbtb2−/− cells (54) but did not characterize this phenotype further. We found that Zbtb2−/− naïve mESCs are defective in differentiating into the EpiLC and Serum-LIF cell states (Fig. 1F). Upon transitioning to Serum-LIF, in particular, Zbtb2−/− cells fail to timely upregulate postimplantation epiblast genes and downregulate preimplantation epiblast genes (Fig. 1G). To understand the underlying molecular mechanism, we performed a thorough biochemical analysis of ZBTB2-containing protein complexes and showed (1) that ZBTB2’s BTB domain mediates homodimerization, heterodimerization with ZBTB25, interaction with UBN2-HiRA via UBN2, and interaction with MBD3-NuRD via GATAD2A/B, (2) that ZBTB2 first Znf interacts with ZNF639 and that this interaction is required to establish or stabilize binding of ZBTB2 to NuRD, and (3) that ZBTB2 interacts weakly or transiently with the Ep400 complex. Ep400 incorporates H3.3-containing histones into chromatin (31), for which HiRA is a chaperone (28). Nevertheless, we show that Ep400 recruitment to ZBTB2 is independent of HiRA (Fig. S4B). Although we identified binding to Ep400 using a dominant-negative ZBTB2 construct (ZBTB2-HAF), we propose that this is a physiologically relevant interaction because (1) low levels of Ep400 subunits are detectable in AP-MS of the functional ZBTB2-AVI construct (Table S5), (2) transcriptional changes upon loss of Zbtb2 and Ep400/Kat5 broadly correlate (Fig. 5, C and D), (3) ZBTB2 and KAT5 repress transcription of the direct target Zbtb2 (Fig. S5, F and G), and (4) a direct interaction between ZBTB2 and the Ep400 subunit KAT5 was identified by high-throughput Y2H screening (60). Further studies are required to provide definitive evidence for the ZBTB2–Ep400 interaction and explore its physiological significance.
These molecular insights have important implications for our understanding of histone variant deposition. The prevailing models for HiRA recruitment hypothesize a transcription-coupled mechanism (66) or intrinsic affinity for nucleosome-free DNA (27). As ZBTB2 is able to recruit HiRA and localize at promoters, we propose that HiRA recruitment at transcription start sites can be mediated by ZBTB2. The HiRA complex binds H3.3 (28), yet there is no known HiRA-associated chromatin remodeler for H3.3 deposition, such as ATRX for the H3.3 chaperone DAXX (67). Our work provides the first physical link between HiRA and a chromatin remodeler. The hypothesis that HiRA might be coupled to Ep400 is further supported by Ep400’s preference for H2A.Z/H3.3 nucleosome deposition (31).
Although BTB domain-containing TFs are acknowledged to play biologically important and disease-relevant roles (6), a general understanding of their evolution and molecular function is lacking. Building on our characterization of ZBTB2’s interactome, we systematically assayed the biochemical properties of BTB TFs. Using Y2H, we found that UBN2 recruitment is a unique feature of ZBTB2. GATAD2A/B binding, instead, is shared by at least 15 BTB TFs and is therefore the most common feature of BTB TFs reported to date. Instances of this interaction are present in the Nacc family, in the Bach family, and in many apparently unrelated branches of the Zbtb family (Fig. 3E), suggesting that GATAD2A/B binding is an ancestral property of BTB TFs. Intriguingly, the Human Reference Protein Interactome Mapping Project (60) identified the BTB-TFs ZBTB1, ZBTB2, ZBTB8A, ZBTB14, and BACH2 to directly interact with KAT5, suggesting that Ep400 recruitment might also be a conserved property of BTB TFs. The dimerization specificity of BTB domains is incompletely understood and has been proposed to be enforced by a quality control mechanism (68). In contrast, our Y2H data for ZBTB2 heterodimerization show that dimerization specificity is a BTB domain–intrinsic property and unrelated to interaction with other partners. For example, ZBTB25’s BTB domain heterodimerizes with ZBTB2’s BTB domain (Fig. 3F) but does not interact with GATAD2A/B or UBN2, whereas ZBTB2’s BTB domain does so (Fig. 3, B–D). This raised the possibility that ZBTB25 might modulate ZBTB2’s avidity for HiRA and NuRD, which was confirmed, in the case of HiRA, by the competition between UBN2 and ZBTB25 for ZBTB2 binding (Fig. S4B). Nevertheless, this regulatory mechanism does not seem to play an important role, as the lack of Zbtb25 does not affect differentiation in either WT or Zbtb2−/− cells (Fig. S3, F and G).
The linker region between the BTB domain and the first Znf of ZBTB proteins is not conserved (5) and, although it can mediate protein interactions, is usually considered to work as a flexible unstructured linker (7). We found that the first 44 amino acids of ZBTB2’s linker interact with the conserved BTB domain, but not with the extended BTB domain (Fig. 2E), and that extension by these 44 amino acids is necessary for the BTB domain to bind UBN2 and GATAD2B (Fig. 2F). We interpret this as evidence for a structured extension of the BTB domain of ZBTB2. As ZBTB7A does not require a BTB domain extension to interact with GATAD2B (57), we hypothesize that this is a unique feature of ZBTB2 that evolved with its ability to bind UBN2.
To assess the function of the characterized ZBTB2 interactions, we generated Znf639−/− and Ubn2−/− mESCs and analyzed their differentiation phenotypes and transcriptional alterations. Znf639−/− and Ubn2−/− mESCs show a delay in Nanog>GFP downregulation (Fig. 5, A and B) that, although weaker than in Zbtb2 mutants, is consistent with the reported phenotypes of Mbd3−/− (24) and Hira−/− cells (32). At the transcriptional level, Mbd3, Ubn2, and Znf639 mutants correlate reciprocally (Fig. 5C), behaving as a functional unit, which is consistent with the role of ZNF639 in stabilizing NuRD interaction (Fig. 2H). Although we do not know whether ZNF639, UBN2, and NuRD simultaneously bind ZBTB2, this suggests that they work synergistically, rather than regulating independent genes. Znf639/NuRD/HiRA module mutants show no transcriptional correlation with Zbtb2−/− cells, suggesting that ZBTB2 directs gene regulation predominantly through another interactor, such as the Ep400 complex. In fact, Zbtb2-specific expression changes correlate with those upon Ep400 and Kat5 depletion (Fig. 5C), pointing to the existence of two separate ZBTB2 effector modules, one associated with Ep400 and the other with ZNF639/NuRD/HiRA. The phenotypic convergence of Zbtb2, Znf639, Mbd3, and Ubn2/Hira on promoting exit from naïve pluripotency might be due to coherent regulation of preimplantation- and postimplantation-specific genes (Fig. S5A). The role of Ep400 in driving mESC differentiation remains to be determined because Ep400 and Kat5 are essential for ESC self-renewal (33).
ZBTB2 and the Ep400 complex (18) repress 2C-like genes, whereas ZNF639, MBD3, and UBN2 promote their transcription (Fig. 5D). Nevertheless, all these factors are depleted at the promoters of 2C-like genes (Fig. 5, E and F and Fig. S5, B and C), demonstrating that this regulation is indirect. The underlying mechanism remains to be determined. Because the H3.3 histone chaperone DAXX/ATRX directly inhibits 2C-like gene expression (69, 70), this mechanism may involve ZBTB2 modulating H3.3 dynamics through Ep400 and HiRA. A similar mechanism may contribute to the regulation of 2C-like genes by the canonical H3 chaperone CAF1 (19). However, ZBTB2 can also repress transcription in a sequence-specific and direct manner, which we demonstrate by taking advantage of a prominent ZBTB2 ChIP-seq peak at the Zbtb2 promoter and showing that ZBTB2 regulates its own transcription (Fig. S5F). KAT5 acts similar to ZBTB2 in Zbtb2 autoregulation (Fig. S5G), providing functional support for ZBTB2 and Ep400 acting together in gene regulation.
In summary, this study presents a detailed biochemical and transcriptional analysis of ZBTB2 and identifies how chromatin modifiers and histone chaperones are recruited by this TF during cell state transition. We use these molecular insights to systematically analyze BTB TFs and to propose a comprehensive concept for their evolution and function. This work will serve as a resource for the study of BTB TFs and inspire further systematic approaches to this important family of TFs.
Experimental procedures
Genetic screen
175∗106 haploid mESCs B1A4 Oct4>GFP-Puro (38) were transduced with each of the retroviruses reFlipROSAβgeo(Cre)∗0, reFlipROSAβgeo(Cre)∗+1, reFlipROSAβgeo(Cre)∗+2, and rsFlipROSAβgeo∗+2 (39), selected with Neomycin, and frozen. After thawing, cells were allowed to recover in Serum-LIF (GMEM (Sigma), 10% fetal bovine serum (Sigma), 1 mM sodium pyruvate (Gibco), 2 mM L-glutamine (Gibco), 0.1 mM nonessential amino acids (Gibco), 0.1 mM 2-mercaptoethanol (Sigma), and 1000 U/ml mLIF (Chao laboratory, Basel)) for 24 h and then transferred on day 0 to N2B27 (Dulbecco's modified Eagle's medium [DMEM]/F12 medium (Life Technologies) and Neurobasal medium (Gibco) 1:1, N2 supplement 1/200 (homemade), B-27 serum-free supplement 1/100 (Gibco), 2 mM L-glutamine (Gibco), 0.1 mM 2-mercaptoethanol (Sigma)) + 3 μM CHIR (Steward laboratory, Dresden). Fresh medium was provided every 2 days, and the cells were replated every 4 days until day 13, when the medium was changed to N2B27 + 3 μM CHIR + 1 μg/ml Puromycin (Gibco). Half of the cells were harvested on day 16, then again on day 20, and finally all on day 23. Cell pellets were digested overnight at 56 °C in 10 mM Tris HCl, pH 7.5, 10 mM EDTA, 10 mM NaCl, 0.5% SDS, and 1 mg/ml proteinase K (Macherey-Nagel), and then with 0.1 mg/ml RNase A (QIAGEN) for 2 h at 37 °C. DNA was ethanol-precipitated, washed twice with 70% ethanol and resuspended in water. Five microgram of DNA was digested with DpnII or MseI (NEB) for 5 h at 37 °C, purified with the PCR cleanup kit (QIAGEN), ligated in 250 μl with 3 μl of T4 ligase (NEB) at 16 °C for 36 h, and purified with the PCR cleanup kit (QIAGEN). Half of the eluate was redigested with NheI and the other half with PvuII (NEB), and the reactions were pooled and purified by the PCR cleanup kit (QIAGEN) eluted in 50 μl. Ten microliters of the eluate was used as a template for PCR with KOD (TaKaRa) with an extension time of 1 min and 10 s and an annealing temperature of 58 °C, for 35 cycles, with primers aatgatacggcgaccaccgagatctacacGCCAGTCCTCCGATTGA and caagcagaagacggcatacgagatBBBBBBAGTTCCTATTCCGAAGTTCCTATTCTCTA (B = barcode). PCRs were purified with the PCR cleanup kit (QIAGEN) and subjected to next-generation sequencing with sequencing primer TGATTGACTACCCGTCAGCGGGGGTCTTTCA and indexing primer TATACTTTCTAG+A+GAATAGGAACTTCGGAATA+G+GAACT (+N = LNA modification).
Sequence reads were processed to remove adaptors (Msel TTAA and Dpnll GATC) and then mapped to the mouse reference genome (mm9 only chromosomes 1–19, X, Y, and M) using Bowtie version 1.0.0 with parameters -v 3 -m 1 —best —strata. Output SAM files were sorted and converted to BAM files using SAMtools version 0.1.19-44428cd. Genomic tracks were generated using genomeCoverageBed from bedtools, version 2.25.0, using only the first base pair position of each read according to the strand. The number of insertions per gene (using only insertions supported by more than one read) normalized to 10,000 reads per library was generated using the ENSEMBL gtf annotation Mus_musculus.NCBIM37.67. Most of this analysis was processed using the unix command awk. The R version 3.3.2 was used to compute a mean log2 fold change (using a pseudocount of 0.5) contrasting each group to the corresponding control and a z-score within each library (Table S1).
Cell culture
TNG-A mESCs (48) were cultured in 2iLIF (N2B27, 1 μM PD0325901 (Steward laboratory), 3 μM CHIR, and 1000 U/ml mLIF) on gelatin-coated tissue culture plates. For medium switch experiments, cells were washed with PBS, detached with Accutase (Sigma), centrifuged for 3 min at 300g in DMEM/F12–0.1% BSA, resuspended in the new medium, counted, and replated. In experiments with DOX-inducible constructs, 1 μg/ml DOX (Sigma) was added to the final medium. Cells were transitioned to N2B27 + 3 μM CHIR + 12 ng/ml bFGF (Smith laboratory) on gelatin-coated plates at a density of 2500/cm2, or to EpiLC medium (N2B27 base, 20 ng/ml activin A, 12 ng/ml bFGF (Smith laboratory), and 1% KSR (Life Technologies)) on fibronectin-coated plates at a density of 25,000/cm2, or to Serum-LIF on gelatin-coated plates at a density of 2500/cm2. At the moment of analysis, cells were washed with PBS, resuspended with trypsin (Life Technologies), centrifuged for 3 min at 300g in DMEM/F12–0.1% BSA, resuspended in DMEM/F12–0.1% BSA, and flowed on an LSRII SORP Analyzer (Becton Dickinson). Percentage of GFP-high cells was quantified with BD FACSDiva 8.0.1, and flow profiles were made with FlowJo (FlowJo, LLC). For Kat5 KD, 105 WT naïve TNG-A mESCs were transfected with 20 pmol Kat5 siRNA (FlexiTube GeneSolution, GS81601, QIAGEN) or negative control siRNA (AllStars Negative Control, 1027281, QIAGEN) and 4-μl Lipofectamine RNAi-MAX (Life Technologies). The next day, the medium was changed to fresh 2iLIF, and the cells were harvested 3 days after transfection.
Mutant cell lines and overexpression constructs
A TNG-A clone stably expressing Cas9 (TbC1) was derived by transfection of PB-LR5.1-EF1a-bsdr2ACas9 (derived from pPB-LR5.1-EF1a-puro2ACas9, gift of Kosuke Yusa, Wellcome Trust Sanger Institute) and PBase (PiggyBac Transposase, (53)). To generate KO cell lines, TbC1 cells were reverse-transfected with 400 ng of U6>sgRNA plasmids (George Church, plasmid #41824, Addgene) according to Table S2 and 3-μl Lipofectamine 2000 (Life Technologies). Single cells were sorted in 96-well plates in 2iLIF and expanded. To generate Zbtb2-3xFLAG knock-in cell lines, E14 mESCs were transfected with 400 ng of U6>sgRNA plasmid (George Church, plasmid #41824, Addgene), 800-ng hCas9 plasmid (George Church, plasmid # 41815, Addgene), and 800 ng of homologous recombination template plasmid (Fig. S2A and Table S2) with 3 μl Lipofectamine 2000. After 7 days, GFP-positive cells were purified and, after 2 days, transfected with a Cre recombinase-expressing plasmid (gift of Hitoshi Niwa). Single GFP-negative cells were purified after 4 days into 96-well plates and expanded. For constitutive or inducible expression, the cDNA of the gene of interest was cloned in pPB-CAG-DEST-pgk-hph (CAG>) (53) or pPB-TRE-DEST-rTA-HSV-neo (DOX>) (52), respectively, and 1 μg plasmid was reverse-transfected together with 1 μg PBase and 3 μl Lipofectamine 2000 in TbC1 cells. The next day, the fresh medium with the appropriate selection was added and the cells were analyzed after at least 1 week of selection.
Immunofluorescence
Cells were plated on a laminin-coated 96-well glass plate (Greiner Bio-One), fixed with PBS–4% PFA for 20 min, washed twice with PBS, permeabilized with PBS–0.1% Triton X-100 for 10 min at room temperature (RT), incubated in the blocking solution (3% donkey serum (Sigma), 1% BSA in PBS–0.1% Tween-20 (PBST)) for 1 h at RT, incubated with anti-Tfe3 antibody (RRID:AB_1857931, Cat# HPA023881, Sigma) 1/1000 in the blocking solution overnight at 4 °C, washed three times with PBST, stained for 2 h at RT with the secondary antibody donkey anti-rabbit IgG—Alexa Fluor 555 (Life Technologies) 1/500 in PBST, counterstained with PBST-Hoechst33342 1/5000 (Life Technologies), washed twice with PBS, and imaged at an LSM-710 scanning head confocal microscope (Zeiss). Images were exclusively cropped with no further manipulation.
Western blot
WT and Nmt1−/− mESCs grown in 2iLIF were washed twice with PBS and incubated for 30 min or 6 h in N2B27 + 12 ng/ml bFGF, put on ice, harvested, and lysed in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA) with fresh cOmplete Protease Inhibitor Tablet (Roche) and Phosphatase Inhibitor Tablet (Roche). Ten microgram of cell lysate was resolved on a 10% SDS-PAGE and wet-blotted on nitrocellulose. Separate membranes were probed with Erk1/2 antibody (#9102, Cell Signaling Technology) or Phospho-Erk1/2 antibody (#9101, Cell Signaling Technology) 1/1000 in PBST–5% BSA.
AP-MS
For FLAG pull-down of endogenous ZBTB2-3xFLAG, 107 E14 Zbtb2-FLAG or WT E14 (negative control) mESCs were switched from 2iLIF to Serum-LIF culture conditions and harvested after 48 h as described below. For the Streptavidin pull-downs of mESCs overexpressing AVI-tagged baits, 2∗105 WT or mutant naïve TNG-A cells grown in 2iLIF were transfected with 1 μg pgk>BirA plasmid and 1 μg DOX>prey plasmid or no plasmid as negative control with 3-μl Lipofectamine 2000. The next day, the fresh 2iLIF medium with hygromycin (selection for the pgk>BirA plasmid) was applied. Cells were grown in a selective medium for 3 to 4 days, and then, the medium was changed to Serum-LIF + 1 μg/ml DOX. After 48 h, 107 cells were harvested with trypsin, washed in PBS–0.1%BSA and PBS, and nuclei were extracted in 10 mM Tris HCl, pH 7.5, 10 mM KCl, 1 mM DTT, and 0.5% IGEPAL, with cOmplete protease inhibitor for 20 min on ice. Nuclei were lysed by rotation for 1 h at 4 °C in 20 mM Tris HCl 7.5, 100 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 10% glycerol, 0.5% Triton X-100, cOmplete protease inhibitor, phosphatase inhibitor, and 250 U/ml benzonase. Lysates were clarified by centrifugation for 5 min at 12,000g at 4 °C; 5 μl of protein-G Dynabeads conjugated with 2 μg of FLAG-M2 Ab or 10 μl of M280 Streptavidin-Dynabeads was added and incubated at 4 °C rotating for 4 h. Beads were then washed three times with 20 mM Tris HCl, pH 7.5, 150 mM NaCl, with 0.5% IGEPAL and twice without IGEPAL. Beads were digested with Lys-C at RT for 4 h and then with trypsin overnight at 37 °C.
The generated peptides were acidified with TFA to a final concentration of 0.8% and analyzed by capillary LC-MS/MS with an EASY-nLC 1000 using the two-column setup (Thermo Scientific). The peptides were loaded with 0.1% formic acid and 2% acetonitrile in water onto a peptide trap (Acclaim PepMap 100, 75 μm × 2 cm, C18, 3 μm, 100 Å) at a constant pressure of 800 bar. Peptides were separated, at a flow rate of 150 nl/min with a linear gradient of 2 to 6% buffer B in buffer A in 3 min followed by a linear increase from 6 to 22% in 40 min, 22 to 28% in 9 min, 28 to 36% in 8 min, and 36 to 80% in 1 min, and the column was finally washed for 14 min at 80% B (buffer A: 0.1% formic acid, buffer B: 0.1% formic acid in acetonitrile) on a 50 μm × 15 cm ES801 C18, 2 μm, 100 Å column (Thermo Scientific) mounted on a DPV ion source (New Objective) connected to a Orbitrap Fusion (Thermo Scientific). The data were acquired using 120,000 resolution for the peptide measurements in the Orbitrap and a top T (3s) method with HCD fragmentation for each precursor and fragment measurement in the ion trap according the recommendation of the manufacturer (Thermo Scientific). Protein identification and relative quantification of the proteins was done with MaxQuant version 1.5.3.8 using Andromeda as search engine (71) and label-free quantification (LFQ, (72)) as described (73). The mouse subset of the UniProt database from January 2017, containing 59,066 proteins, was used as reference. The protein and peptide false discovery rate (FDR) was set to 0.01. MaxQuant version 1.5.3.8. used additionally a decoy database of the same size and a contaminant database with 245 known contaminant proteins. Strict trypsin cleavage was used allowing also cleavages N-terminal of proline, and a maximum of two missed cleavages were allowed. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine and N-terminal protein acetylation were set as dynamic modifications. The peptide tolerance for the first search was 20 ppm and for the main search 4.5 ppm, and the mass tolerance for the fragment ions was 0.5 Da. The minimum length of the peptides was set to six amino acids and the maximum peptide mass to 8000 Da. MaxQuant results were loaded into Scaffold_4.10.0 (https://www.proteomesoftware.com/).
The LFQ values were analyzed with Perseus v.1.6.2.2 as follows: entries identified only by site or reverse and potential contaminants were removed, values were Log2-transformed, entries identified in less than two replicates in any group were removed, and missing values were imputed based on the normal distribution of each replicate with a 0.25-fold width and a downshift of 1.8-fold. Volcano plots are based on two-sided t test, and threshold curves on an S0 = 0.1 and FDR = 0.0054 for overexpressed ZBTB2 baits, S0 = 2 and FDR =0.01 for the endogenous ZBTB2-3xFLAG bait, and S0 = 0.1 and FDR = 0.02 for UBN1/2 baits.
For the heat map representation in Figure 2B, missing LFQ values in experimental triplicates of ZBTB2 mutant AP-MS were imputed with a 1.8-fold downshift and a 0.25-fold distribution width of the actual distribution of detected proteins using the fitdistr function from the Cran package MASS (https://cran.r-project.org/web/packages/MASS/index.html), as described (74). To correct for varying Zbtb2 amounts in different APs, protein enrichments in each AP were normalized to the respective Zbtb2 bait. To compare interaction strengths, the enrichment of each interactor was normalized to its enrichment in WT Zbtb2 purifications.
Y2H
Y2H assays were performed with the plasmids and strains from the Matchmaker Gold Yeast Two-Hybrid System (Takara Bio) according to manufacturer’s protocol, with the following plasmid modifications. For N-terminal AD-fusions, pGADT7 was digested with NdeI and BamHI and religated with the oligo TAGTGGTGGAACAAAAATGGGCCCGAATTCCCGGGATCGATTAACTGAGTAG, to create ApaI and ClaI sites for In-Fusion cloning (Takara Bio). For C-terminal -AD and -DBD fusions, TTTAAACTATTTGGGCCCATTTTTGTTCCACCACTATAAGCTTGGAGTTGATTGTATGCTTGG and either AAATGGGCCCAAATAGTTTAAACCGCGGTGGATCTGGTGGAATGGATAAAGCGGAATTAATTCCCGAG or AAATGGGCCCAAATAGTTTAAACCGCGGTGGATCTGGTGGAATGAAGCTACTGTCTTCTATCGAACAAGC were used for site-directed mutagenesis of pGADT7 and pGBKT7, respectively, to create a new MCS with ApaI and SacII sites for In-Fusion cloning (Takara Bio). pGBKT7-C and pGADT7-C were digested with NdeI and BamHI and religated with the oligo TATGCCAGCTGCTAAAAGAGTTAAATTGGATTAG to create a new c-Myc nuclear localization sequence. Briefly, pGBKT7 and pGADT7 plasmids were transformed into the yeast strains Y2HGold and Y187, respectively. Several colonies were picked and grown overnight in SD-Trp or SD-Leu for pGBKT7 or pGADT7 plasmids, respectively. When the cultures reached an A600 ∼0.5, they were mated overnight in YPD medium and grown on an SC-Trp/-Leu plate for 2 to 3 days, and plate replicas were made on an SD-Ade/-His/-Trp/-Leu/+X-alpha-Gal/+Aureobasidin A (QDOXA) plates. Pictures of the plates were taken after 2 to 4 days, and images were exclusively cropped, with no further processing.
Phylogeny
Btb domain sequences were retrieved with the ‘Architecture analysis’ tool of SMART (smart.embl.de; query: “BTB AND ZnF_C2H2” in “Mus musculus”) or individually with the ‘Sequence analysis’ tool. The phylogenetic tree was calculated, based on a multiple sequence alignment generated with T-Coffee (https://www.ebi.ac.uk/Tools/msa/tcoffee/), using the neighbor-joining clustering method provided by the ClustalW2 package (http://www.clustal.org/clustal2/) running 1000 iterations. The tree was visualized with iTOL (https://itol.embl.de/).
Gene expression analysis
RNA-Seq reads and published data (see table below) were aligned to the mouse GRCm38/mm10 genome using qAlign from the Bioconductor package QuasR (75), with default parameters except for aligner=”Rhisat2” and splicedAlignment=TRUE. For aligning RNA-Seq reads for Dux overexpression (17), paired=”fr” was additionally used. Alignments were quantified for known UCSC genes obtained from the TxDb.Mmusculus.UCSC.mm10.knownGene package using qCount. Microarray data (33) were analyzed and normalized using the Bioconductor package limma (76). In this dataset, Tip60 KD replicate 3 is an outlier, and Ep400 KD replicates 1 and 3 do not show Ep400 transcript reduction and were therefore excluded from the analysis.
Differential gene expression (Table S3) was determined using edgeR (77). In Figure 1G, Figs. S1G, and S5A, preEPI is the combination of the EPI and ICM + EPI markers, and postEPI the PE gene set (56). Pearson correlation coefficients (Fig. 5C) were calculated using R’s cor function. For heat map visualization (Fig. 5D), significantly deregulated genes in Zbtb2 mutants were considered (Table S3), which are the genes that showed an absolute log2 expression FC of greater than 1 with an FDR of less than 0.005 between WT and Zbtb2 KO cells in at least one of the conditions: 2iLIF, 24-h EpiLC, 48-h EpiLC, 24-h Serum-LIF, 48-h Serum-LIF. All contrast shown in Figure 5D were used for clustering.
ChIP-seq analysis
Published datasets (see table below) were aligned to the mouse GRCm38/mm10 genome using qAlign and profiled using qProfile from the Bioconductor package QuasR. 137435 ATAC peaks (78) were called using Macs2 (79), of which 22,826 were in promoters (±1 kb of annotated transcriptional start sites). For heat map visualization (Fig. S5D), ChIP-seq signals were profiled in these promoters and ChIP enrichment calculated over respective inputs (Zbtb2, H3K4me3, Chd4, Mbd3, Dux) or controls (Kat5, Ep400). For metaplots (Fig. 5, E and F and Fig. S5, B and C), ChIP-seq signals were profiled in promoter regions of cluster genes that were extracted from the TxDb.Mmusculus.UCSC.mm10.knownGene Bioconductor package (https://bioconductor.org/packages/release/data/annotation/html/TxDb.Mmusculus.UCSC.mm10.knownGene.html).
qPCR
RNA was extracted with the RNeasy Mini Kit (QIAGEN), and 1 μg of it was reverse-transcribed with SuperScript III Reverse Transcriptase (Life Technologies). qPCR was performed with the TaqMan Fast Universal PCR Master Mix (Thermo Fisher) and the TaqMan probes GAPDH (4352339E), Zbtb2 (Mm01605943_g1), Kat5 (Mm01231512_m1), Ubn1 (Mm01343954_m1), and Ubn2 (Mm00723981_m1). The DOX>Zbtb2 plasmid that cannot be detected by Mm01605943_g1 was produced by fusion PCR of the endogenous cDNA and the following gBlock (Integrated DNA Technologies): ATGGATTTGACCAACCATGGACTTATTCTACTGCAGCAGTTAAACGCTCAGCGAGAGTTTGGTTTCCTGTGTGACTGCACGGTTGCAATCGGCGATGTGTATTTTAAAGCCCATAAGAGTGTGTTGGCAAGTTTTAGTAACTATTTCAAAATGCTTTTCGTGCACCAAACATCAGAGTGTGTGAGATTAAAACCAACAGATATCCAACCAGATATCTTTTCATACTTATTGCATTTAATGTATACCGGGAAGATGGCCCCACAGCTCATCGACCCTGTGAGGCTAGAGCAAGGGATCAAATTCCTGCACGCATACCCCCTCATCCAGGAAGCCAGCCTTGCCAGCCAAGGCAGCTTTTCCCATCCCGAGCAAGTCTTCCCTCTGGCCTCATCCTTGTACGGCATTCAGATTGCAGACCATCAGCTGAGACAAGCCACCAAGATGAATTTAGGGCCTGAGAAACTTGGACGGGAGCCTAGGCCACAGGCATCCAGGATGA. The construct was validated by plasmid qPCR with Mm01605943_g1.
Statistical tests
p-values presented in Figures 2I and 5B and Figs. S1D, S2D, S3G, and S5, F and G were calculated using two-tailed equal variance t-tests.
Data availability
The genomic data generated in this study have been deposited at ArrayExpress (E-MTAB-9796, E-MTAB-9797, E-MTAB-9798). The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD025520 and PXD025466.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Hans-Rudolf Hotz (Friedrich Miescher Institute [FMI]) for help with the phylogenetic analysis; Hubertus Kohler (FMI) for cell sorting; Vytautas Iesmantavicius and Jan Seebacher (FMI) for help with computational analysis of mass spectrometry data and helpful discussions; Verena Waehle and Melanie Rittirsch (FMI) for technical assistance; Min Jia and Jeff Chao (FMI) for providing LIF; and Austin Smith (University of Exeter) for providing TNG-A cells.
Author contributions
D. O. and J. B. conceptualization; D. O., A. F. B., D. H., and J. B. formal analysis; D. O. and J. B. supervision; D. O. and J. B. funding acquisition; D. O. validation; D. O., S. P., D. H., and S. A. S. investigation; D. O. visualization; D. O., D. H., S. A. S., and U. E. methodology; D. O. writing–original draft; D. O. project administration; D. O. and J. B. writing–review and editing; U. E. resources.
Funding and additional information
D. O. was supported by the European Molecular Biology Organization (EMBO) (ALTF 1632-2014) and Marie Curie Actions (LTFCOFUND2013, GA-2013-609409). Research in the laboratory of J. B. is supported by the Novartis Research Foundation.
Edited by John Denu
Contributor Information
Daniel Olivieri, Email: daniel.olivieri@fmi.ch.
Joerg Betschinger, Email: joerg.betschinger@fmi.ch.
Supporting information
References
- 1.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K., Zheng H., Goity A., van Bakel H., Lozano J.-C., Galli M. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins T., Stone J.R., Williams A.J. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helleboid P., Heusel M., Duc J., Piot C., Thorball C.W., Coluccio A., Pontis J., Imbeault M., Turelli P., Aebersold R., Trono D. The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification. EMBO J. 2019;38 doi: 10.15252/embj.2018101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stogios P.J., Downs G.S., Jauhal J.J., Nandra S.K., Privé G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevrier S., Corcoran L.M. BTB-ZF transcription factors, a growing family of regulators of early and late B-cell development. Immunol. Cell Biol. 2014;92:481–488. doi: 10.1038/icb.2014.20. [DOI] [PubMed] [Google Scholar]
- 7.Maeda T. Regulation of hematopoietic development by ZBTB transcription factors. Int. J. Hematol. 2016;104:310–323. doi: 10.1007/s12185-016-2035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- 9.Smith A.G., Heath J.K., Donaldson D.D., Wong G.G., Moreau J., Stahl M., Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 10.Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva J., Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galonska C., Ziller M.J., Karnik R., Meissner A. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015;17:462–470. doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 14.Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bošković A., Eid A., Pontabry J., Ishiuchi T., Spiegelhalter C., Ram E.V.S.R., Meshorer E., Torres-Padilla M.-E. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014;28:1042–1047. doi: 10.1101/gad.238881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macfarlan T.S., Gifford W.D., Agarwal S., Driscoll S., Lettieri K., Wang J., Andrews S.E., Franco L., Rosenfeld M.G., Ren B., Pfaff S.L. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.-W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L., Nix D.A., Peterson C.M., Tapscott S.J., Carrell D.T., Cairns B.R. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Terrones D., Gaume X., Ishiuchi T., Weiss A., Kopp A., Kruse K., Penning A., Vaquerizas J.M., Brino L., Torres-Padilla M.-E. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat. Genet. 2018;50:106–119. doi: 10.1038/s41588-017-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishiuchi T., Enriquez-Gasca R., Mizutani E., Bošković A., Ziegler-Birling C., Rodriguez-Terrones D., Wakayama T., Vaquerizas J.M., Torres-Padilla M.-E. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- 20.Xue Y., Wong J., Moreno G.T., Young M.K., Côté J., Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 21.Guezennec X.L., Vermeulen M., Brinkman A.B., Hoeijmakers W.A.M., Cohen A., Lasonder E., Stunnenberg H.G. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds N., Latos P., Hynes-Allen A., Loos R., Leaford D., O’Shaughnessy A., Mosaku O., Signolet J., Brennecke P., Kalkan T., Costello I., Humphreys P., Mansfield W., Nakagawa K., Strouboulis J. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaji K., Nichols J., Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 24.Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 25.Bornelöv S., Reynolds N., Xenophontos M., Gharbi S., Johnstone E., Floyd R., Ralser M., Signolet J., Loos R., Dietmann S., Bertone P., Hendrich B. The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell. 2018;71:56–72.e4. doi: 10.1016/j.molcel.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg A.D., Banaszynski L.A., Noh K.-M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R.C., Miller J.C., Lee Y.-L. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D.C., Pchelintsev N.A., Adams P.D., Jansen L.E.T., Almouzni G. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 29.Xiong C., Wen Z., Yu J., Chen J., Liu C.-P., Zhang X., Chen P., Xu R.-M., Li G. UBN1/2 of HIRA complex is responsible for recognition and deposition of H3.3 at cis-regulatory elements of genes in mouse ES cells. BMC Biol. 2018;16:110. doi: 10.1186/s12915-018-0573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin C., Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradhan S.K., Su T., Yen L., Jacquet K., Huang C., Côté J., Kurdistani S.K., Carey M.F. EP400 deposits H3.3 into promoters and enhancers during gene activation. Mol. Cell. 2016;61:27–38. doi: 10.1016/j.molcel.2015.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leeb M., Dietmann S., Paramor M., Niwa H., Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14:385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fazzio T.G., Huff J.T., Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 35.Wray J., Kalkan T., Smith A.G. The ground state of pluripotency. Biochem. Soc. Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 36.Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Göttgens B., Niwa H., Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elling U., Taubenschmid J., Wirnsberger G., O’Malley R., Demers S.-P., Vanhaelen Q., Shukalyuk A.I., Schmauss G., Schramek D., Schnuetgen F., von Melchner H., Ecker J.R., Stanford W.L., Zuber J., Stark A. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnütgen F., Hansen J., De-Zolt S., Horn C., Lutz M., Floss T., Wurst W., Noppinger P.R., von Melchner H. Enhanced gene trapping in mouse embryonic stem cells. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molotkov A., Mazot P., Brewer J.R., Cinalli R.M., Soriano P. Distinct requirements for FGFR1 and FGFR2 in primitive endoderm development and exit from pluripotency. Dev. Cell. 2017;41:511–526.e4. doi: 10.1016/j.devcel.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu D., Ye S., Ruiz B., Zhou X., Liu D., Zhang Q., Ying Q.-L. Klf2 and Tfcp2l1, two Wnt/β-catenin targets, act synergistically to induce and maintain naive pluripotency. Stem Cell Rep. 2015;5:314–322. doi: 10.1016/j.stemcr.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattout A., Aaronson Y., Sailaja B.S., Ram E.V.R., Harikumar A., Mallm J.-P., Sim K.H., Nissim-Rafinia M., Supper E., Singh P.B., Sze S.K., Gasser S.M., Rippe K., Meshorer E. Heterochromatin protein 1β (HP1β) has distinct functions and distinct nuclear distribution in pluripotent versus differentiated cells. Genome Biol. 2015;16:213. doi: 10.1186/s13059-015-0760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leeb M., Pasini D., Novatchkova M., Jaritz M., Helin K., Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tee W.-W., Shen S.S., Oksuz O., Narendra V., Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin T., Chao C., Saito S., Mazur S.J., Murphy M.E., Appella E., Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 46.Li T., Shi Y., Wang P., Guachalla L.M., Sun B., Joerss T., Chen Y., Groth M., Krueger A., Platzer M., Yang Y., Rudolph K.L., Wang Z. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015;34:1630–1647. doi: 10.15252/embj.201489947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gontan C., Achame E.M., Demmers J., Barakat T.S., Rentmeester E., van IJcken W., Grootegoed J.A., Gribnau J. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- 48.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 49.Yang S.H., Shrivastav A., Kosinski C., Sharma R.K., Chen M.-H., Berthiaume L.G., Peters L.L., Chuang P.-T., Young S.G., Bergo M.O. N-myristoyltransferase 1 is essential in early mouse development. J. Biol. Chem. 2005;280:18990–18995. doi: 10.1074/jbc.M412917200. [DOI] [PubMed] [Google Scholar]
- 50.Kouhara H., Hadari Y.R., Spivak-Kroizman T., Schilling J., Bar-Sagi D., Lax I., Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 51.Thinon E., Serwa R.A., Broncel M., Brannigan J.A., Brassat U., Wright M.H., Heal W.P., Wilkinson A.J., Mann D.J., Tate E.W. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat. Commun. 2014;5:4919. doi: 10.1038/ncomms5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villegas F., Lehalle D., Mayer D., Rittirsch M., Stadler M.B., Zinner M., Olivieri D., Vabres P., Duplomb-Jego L., Bont E.S.J.M.D., Duffourd Y., Duijkers F., Avila M., Geneviève D., Houcinat N. Lysosomal signaling licenses embryonic stem cell differentiation via inactivation of Tfe3. Cell Stem Cell. 2019;24:257–270.e8. doi: 10.1016/j.stem.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 53.Betschinger J., Nichols J., Dietmann S., Corrin P.D., Paddison P.J., Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karemaker I.D., Vermeulen M. ZBTB2 reads unmethylated CpG island promoters and regulates embryonic stem cell differentiation. EMBO Rep. 2018;19 doi: 10.15252/embr.201744993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalkan T., Smith A. Mapping the route from naive pluripotency to lineage specification. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130540. doi: 10.1098/rstb.2013.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boroviak T., Loos R., Lombard P., Okahara J., Behr R., Sasaki E., Nichols J., Smith A., Bertone P. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Masuda T., Wang X., Maeda M., Canver M.C., Sher F., Funnell A.P.W., Fisher C., Suciu M., Martyn G.E., Norton L.J., Zhu C., Kurita R., Nakamura Y., Xu J., Higgs D.R. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351:285–289. doi: 10.1126/science.aad3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Youn H.-D., Sun L., Prywes R., Liu J.O. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286:790–793. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 59.Siggs O., Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–3369. doi: 10.4161/cc.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luck K., Kim D.-K., Lambourne L., Spirohn K., Begg B.E., Bian W., Brignall R., Cafarelli T., Campos-Laborie F.J., Charloteaux B., Choi D., Coté A.G., Daley M., Deimling S., Desbuleux A. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Acharya D., Hainer S.J., Yoon Y., Wang F., Bach I., Rivera-Pérez J.A., Fazzio T.G. KAT-independent gene regulation by Tip60 promotes ESC self-renewal but not pluripotency. Cell Rep. 2017;19:671–679. doi: 10.1016/j.celrep.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen P.B., Chen H.V., Acharya D., Rando O.J., Fazzio T.G. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015;22:999–1007. doi: 10.1038/nsmb.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masui S., Ohtsuka S., Yagi R., Takahashi K., Ko M.S., Niwa H. Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells. BMC Dev. Biol. 2008;8:45. doi: 10.1186/1471-213X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scotland K.B., Chen S., Sylvester R., Gudas L.J. Analysis of Rex1 (zfp42) function in embryonic stem cell differentiation. Dev. Dyn. 2009;238:1863–1877. doi: 10.1002/dvdy.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang S.-H., Kalkan T., Morrisroe C., Smith A., Sharrocks A.D. A genome-wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarai N., Nimura K., Tamura T., Kanno T., Patel M.C., Heightman T.D., Ura K., Ozato K. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J. 2013;32:2392–2406. doi: 10.1038/emboj.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grover P., Asa J.S., Campos E.I. H3–H4 histone chaperone pathways. Annu. Rev. Genet. 2018;52:1–22. doi: 10.1146/annurev-genet-120417-031547. [DOI] [PubMed] [Google Scholar]
- 68.Mena E.L., Kjolby R.A.S., Saxton R.A., Werner A., Lew B.G., Boyle J.M., Harland R., Rape M. Dimerization quality control ensures neuronal development and survival. Science. 2018;362 doi: 10.1126/science.aap8236. [DOI] [PubMed] [Google Scholar]
- 69.Sadic D., Schmidt K., Groh S., Kondofersky I., Ellwart J., Fuchs C., Theis F.J., Schotta G. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 2015;16:836–850. doi: 10.15252/embr.201439937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elsässer S.J., Noh K.-M., Diaz N., Allis C.D., Banaszynski L.A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 72.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hubner N.C., Bird A.W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 75.Gaidatzis D., Lerch A., Hahne F., Stadler M.B. QuasR: Quantification and annotation of short reads in R. Bioinformatics. 2015;31:1130–1132. doi: 10.1093/bioinformatics/btu781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olivieri D., Castelli E., Kawamura Y.K., Papasaikas P., Lukonin I., Rittirsch M., Hess D., Smallwood S.A., Stadler M.B., Peters A.H.F.M., Betschinger J. Cooperation between HDAC3 and DAX1 mediates lineage restriction of embryonic stem cells. EMBO J. 2021;40 doi: 10.15252/embj.2020106818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data generated in this study have been deposited at ArrayExpress (E-MTAB-9796, E-MTAB-9797, E-MTAB-9798). The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD025520 and PXD025466.