Abstract
Background & Aims
Epithelial cells form a monolayer at mucosal surface that functions as a highly selective barrier. Lysophosphatidic acid (LPA) is a bioactive lipid that elicits a broad range of biological effects via cognate G protein-coupled receptors. LPA receptor 5 (LPA5) is highly expressed in intestinal epithelial cells, but its role in the intestine is not well-known. Here we determined the role of LPA5 in regulation of intestinal epithelial barrier.
Methods
Epithelial barrier integrity was determined in mice with intestinal epithelial cell (IEC)-specific LPA5 deletion, Lpar5ΔIEC. LPA was orally administered to mice, and intestinal permeability was measured. Dextran sulfate sodium (DSS) was used to induce colitis. Human colonic epithelial cell lines were used to determine the LPA5-mediated signaling pathways that regulate epithelial barrier.
Results
We observed increased epithelial permeability in Lpar5ΔIEC mice with reduced claudin-4 expression. Oral administration of LPA decreased intestinal permeability in wild-type mice, but the effect was greatly mitigated in Lpar5ΔIEC mice. Serum lipopolysaccharide level and bacterial loads in the intestine and liver were elevated in Lpar5ΔIEC mice. Lpar5ΔIEC mice developed more severe colitis induced with DSS. LPA5 transcriptionally regulated claudin-4, and this regulation was dependent on transactivation of the epidermal growth factor receptor, which induced localization of Rac1 at the cell membrane. LPA induced the translocation of Stat3 to the cell membrane and promoted the interaction between Rac1 and Stat3. Inhibition of Stat3 ablated LPA-mediated regulation of claudin-4.
Conclusions
This study identifies LPA5 as a regulator of the intestinal barrier. LPA5 promotes claudin-4 expression in IECs through activation of Rac1 and Stat3.
Keywords: LPA, Barrier, LPA5, Rac1, Stat3
Abbreviations used in this paper: AJ, adherens junction; ATX, autotaxin; DSS, dextran sulfate sodium; EGFR, epidermal growth factor receptor; FD-4, fluorescein isothiocyanate-labeled 4 kDa dextran; GEF, guanine nucleotide exchange factor; GI, gastrointestinal; GPCR, G protein-coupled receptor; IBD, inflammatory bowel disease; IEC, intestinal epithelial cell; IF, immunofluorescence; JAMs, junctional adhesion molecules; LPA, lysophosphatidic acid; LPA5, LPA receptor 5; LPS, lipopolysaccharide; NHE3, Na+/H+ exchanger 3; PBS, phosphate-buffered saline; ROCK, RhoA-associated kinase; RT-PCR, reverse transcriptase polymerase chain reaction; SD, standard deviation; shRNA, short hairpin RNA; Stat, signal transducers and activators of transcription; TER, transepithelial electrical resistance; TJ, tight junction
Graphical abstract
Summary.
Bioactive lipid lysophosphatidic acid (LPA) exerts multiple effects, but our understanding of its role in the intestine is limited. Using LPA5 receptor-deficient mice, we explore the effect of LPA on intestinal permeability and describe a new function of LPA5.
Intestinal lumen is lined with epithelial cells forming a barrier that prevents unimpeded movement of solutes and fluid and protects the host from the luminal milieu. Epithelial cells are joined together by a highly organized apical junctional complex, which includes tight junction (TJ) and adherens junction (AJ). TJs are made up of a number of proteins, including transmembrane claudins, occludin, and junctional adhesion molecules (JAMs).1 The AJ consists of cadherin adhesion receptors and cytoplasmic proteins associated with them, including catenins and actin filaments.2 A breach of the intestinal epithelial barrier can elicit a broad range of diseases, including inflammatory bowel disease (IBD), celiac disease, and colorectal cancer.3
Although lipids are the major constituents of cell membranes, lipids can also function as a mediator of intercellular and extracellular processes.4 Among the naturally occurring lipids, lysophosphatidic acid (LPA) has been linked to various pathologic conditions such as cancer, fibrosis, inflammation, and atherosclerosis. LPA signals through 6 distinct G protein-coupled receptors (GPCRs), termed LPA1–LPA6 (encoded by Lpar1-Lpar6 genes in rodents). The expression level of each LPA receptor varies widely among different tissues and cell types.5,6 Mouse gastrointestinal (GI) tract expresses at least 5 distinct LPA receptors at various levels, although the locations of each LPA receptor are not known.7 Bioactive LPA in serum and plasma is mainly produced by autotaxin (ATX), a secreted lysophospholipase D that converts lysophosphatidylcholine to LPA.8 Elevated level of ATX is associated with intestinal inflammation, and inhibition of ATX is shown to reduce inflammation in the gut and restore intestinal epithelial cell (IEC) differentiation.9, 10, 11, 12 Orally administered LPA increases tumor incidence in ApcMin/+ mice by stimulating cancer cell proliferation.7,13 However, the ability of LPA to modulate cell proliferation and migration is equally critical for the maintenance of the epithelial barrier in the gut. We have shown recently that LPA maintains intestinal epithelial integrity by facilitating wound closure via LPA1-dependent stimulation of IECs.14 Lpar1-/- mice develop more severe colitis, an effect associated with reduced epithelial mucosa restoration and barrier defect.14,15 The expression level of LPA5, also known as GPR92 or GPR93, is relatively high in the GI tract where Lpar5 mRNA is detected in the epithelial cells and lymphocytes.16,17 LPA5 differs from LPA1 or LPA2 in that its amino acid sequence shares high homology with the purinergic family of GPCR.16 In addition, LPA5 has a unique preference for an LPA species with an ether linkage, and it can also be activated by the dietary protein hydrolysate and peptone.18,19 Lpar5 is detected in sensory nerves of the mouse enteric nerve system where it is activated by mesenteric lymphatic fluid, suggesting its role as a nutrient sensor.20 In general, LPA stimulates cell migration, but a recent study suggested that LPA5 is an anti-migration receptor.21
We have shown previously that LPA5 regulates the brush border Na+/H+ exchanger NHE3 (Slc9A3), which is a major Na+ transporter in the intestine.22, 23, 24 Oral administration of LPA attenuates intestinal water loss, a hallmark of diarrhea, in an experimental model of diarrhea via activation of NHE3.22,25 Despite its abundant expression in the GI tract, the role of LPA5 in IECs is not known beyond the regulation of NHE3. In this study, we investigated the role of LPA5 in regulation of intestinal epithelial barrier function using mice lacking Lpar5 in IECs. Our study demonstrated that loss of LPA5 resulted in epithelial barrier defect, and we have delineated critical signaling pathways that underlie LPA5-dependent regulation of intestinal epithelial barrier.
Results
Dysregulation of Intestinal Epithelial Barrier in the Absence of LPA5
We have reported recently that mice lacking Lpar5 in IECs, Lpar5ΔIEC, do not display a gross morphologic change in the intestine, and the basal intestinal functions assessed by stool frequency and fecal water content are unchanged.25 However, the comparison of bowel movement does not reveal a subtle change that may affect the intestinal epithelium. Because Lpar5 expression is high in the intestine,16,19,26 we aimed to study whether Lpar5 loss in IECs alters epithelial integrity by examining the epithelial barrier function. We compared intestinal permeability between Lpar5f/f and Lpar5ΔIEC mice by oral administration of fluorescein isothiocyanate-labeled 4 kDa dextran (FD-4). Measuring the fluorescence levels in the serum revealed that FD-4 flux across the intestinal mucosa was significantly elevated in Lpar5ΔIEC mice compared with control Lpar5f/f mice (Figure 1A), indicating a more permeable intestinal epithelium in the absence of LPA5. To determine whether this increase in epithelial permeability is associated with a change in TJ protein expression, we determined mRNA expression levels of several TJ proteins in the mouse colon. Quantification of mRNA levels by quantitative reverse transcription polymerase chain reaction (RT-PCR) showed that claudin-4 (Cldn4) mRNA abundance was reduced in Lpar5ΔIEC mice compared with Lpar5f/f mice, but ZO-1 (TJPN1), occludin (Ocln), JAM-A (F11R), claudin-1 (Cldn1), claudin-2 (Cldn2), and claudin-7 (Cldn7) transcript levels were not statistically different between them (Figure 1B). Western blotting of mucosal lysates from the small intestine and colon confirmed decreased claudin-4 expression in Lpar5ΔIEC mice versus Lpar5f/f mice (Figure 1C). The expression level of E-cadherin, the primary epithelial AJ protein, was not altered by Lpar5 loss. Immunofluorescence (IF) confocal microscopic analysis showed that the membrane expression of claudin-4 of epithelial cells in the small intestine and colon of Lpar5ΔIEC mice was down-regulated (Figure 1D).
Figure 1.
Lpar5 loss dysregulates epithelial barrier in the mouse intestine. (A) FD-4 was orally administered to Lpar5f/f and Lpar5ΔIEC mice, and FD-4 levels in the serum were determined 4 hours later. ∗∗P < .01. n = 5. (B) The mRNA levels of TJ proteins in colonic mucosa of Lpar5f/f and Lpar5ΔIEC mice were determined by quantitative RT-PCR. The mRNA expression level of each protein was normalized to β-actin mRNA expression level. ∗∗P < .01. n = 5. Data are expressed as mean ± SD. (C) Representative Western blots of junctional proteins in the small intestine (left) and distal colon (right) are shown. Molecular weight (MW) of each protein is shown in parentheses. Comparable results were observed in 3 independent experiments. (D) Confocal IF images for claudin-4 (green), F-actin (red), and nuclei (blue: DAPI) are shown. Representative images from 3 Lpar5f/f and 4 Lpar5ΔIEC mice are shown. Scale bar = 50 μm.
LPA-Mediated Regulation of Epithelial Permeability In Vitro and In Vivo Is Dependent on LPA5
To establish a cause-effect relationship between LPA5 and epithelial barrier function, we used 2 IEC lines, SK-CO15 and Caco-2bbe cells. LPA5 expression in these cell lines was assessed by determining Lpar5 mRNA levels by RT-PCR. As in our previous study,27 both lines expressed Lpar1 and Lpar2. Lpar5 mRNA expression was low in SK-CO15 cells or undetectable in Caco-2bbe cells by RT-PCR (Figure 2A). Knockdown of LPA5 in SK-CO15 cells using 2 different short hairpin (sh) RNA (shRNA) specific for LPA5, shLPA5, resulted in >60% decrease in Lpar5 mRNA levels (Figure 2B). To correlate LPA5 expression and epithelial barrier function, we determined transepithelial electrical resistance (TER) in SK-CO15 cells stably expressing HA-LPA5 or shLPA5. HA-LPA5 expression increased TER by more than 75%, whereas knockdown using shRNA resulted in decrease (18.2% by shLPA5-777 and 25.6% by shLPA5-361) in TER (Figure 2C). Consistently, stable expression of HA-LPA5 in Caco-2bbe cells increased TER by 38% (Figure 2D). We next sought to determine whether activation of LPA5 dynamically regulates TER. Cells grown on Transwell inserts were cultured in medium supplemented with 0.5% fetal bovine serum overnight. Cells were then treated with LPA or a carrier (0.1% bovine serum albumin in phosphate-buffered saline [PBS]), and TER was measured hourly for 4 hours. In control SK-CO15/pCDH cells, LPA resulted in a statistically significant increase in TER (Figure 2E). The effect was markedly elevated in cells expressing HA-LPA5 (Figure 2F), whereas knockdown of Lpar5 completely ablated the effect (Figure 2G). We postulated that if LPA indeed regulates epithelial barrier, orally administered LPA should strengthen the epithelial barrier function in the mouse intestine. To test this idea, we gavaged LPA or carrier to Lpar5f/f and Lpar5ΔIEC mice for 3 days, followed by oral administration of FD-4 to determine intestinal permeability. LPA administration led to a significant decrease in serum FD-4 concentration in Lpar5f/f mice compared with carrier-treated controls (Figure 2H), suggesting that LPA enhances intestinal barrier function in vivo. The difference in serum FD-4 levels between LPA and carrier treatment was significantly reduced in Lpar5ΔIEC mice. However, a smaller and yet statistically significant difference in FD-4 levels in Lpar5ΔIEC mice suggests that another LPA receptor, in addition to LPA5, may contribute to LPA-dependent intestinal epithelial barrier function.
Figure 2.
LPA decreases epithelial permeability in vitro and in vivo. (A) Lpar1, Lpar2, and Lpar5 mRNA in SK-CO15 cells and Caco-2bbe cells were amplified by PCR. Representative image from 3 independent experiments is shown. (B) Lpar5 mRNA expression levels in SK-CO15 cells expressing HA-LPA5 or shLPA5 were determined. Data (mean ± SD) are presented as fold change over control cells. n = 3. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 compared with pCDH. (C) TER across monolayers of SK-CO15 cells expressing HA-LPA5 or shLPA5 was determined. n = 6. ∗∗P < .01 compared with pCDH. (D) TER was determined in Caco-2bbe cells expressing pCDH or HA-LPA5. n = 3. ∗∗P < .01. SK-CO15 cells transfected with (E) pCDH, (F) HA-LPA5, or (G) shLPA5 were grown on Transwell inserts. Cells were treated with LPA or carrier (0.01% bovine serum albumin in PBS), and TER was determined hourly for 4 hours. Data are expressed as mean ± SD. ∗∗P < .01, ∗∗∗P < .001 versus control. n = 3. (H) LPA was orally administered for 3 days (45 nmol/L/day), and FD-4 was delivered orally 4 hours before collecting serum. Data are expressed as mean ± SD. n = 4. ∗P < .05. ∗∗P <.01.
Increased Bacterial Translocation in Lpar5ΔIEC Mice
Increased epithelial permeability allows infiltration of microorganisms into the intestinal mucosal and peripheral organs. We first determined serum lipopolysaccharide (LPS) levels in Lpar5f/f and Lpar5ΔIEC mice. Serum LPS concentration in Lpar5ΔIEC mice was elevated by more than 7-fold versus Lpar5f/f mice (Figure 3A). These results were corroborated by increased bacterial loads in Lpar5ΔIEC mice. The liver, ileum, and proximal and distal colon of mice were homogenized, and serial dilutions of the homogenates were plated on Agar overnight. Bacterial counts were markedly greater in Lpar5f/f mouse tissues (Figure 3B). In particular, there was more than 20-fold difference between Lpar5ΔIEC and Lpar5f/f livers, demonstrating increased dissemination of bacteria to peripheral organs in Lpar5ΔIEC mice.
Figure 3.
Serum LPS and bacterial loads are elevated in Lpar5ΔIECmice. (A) Serum LPS levels were determined in Lpar5f/f and Lpar5ΔIEC mice. n = 5. (B) Bacterial loads in liver, ileum, proximal colon (P Colon), and distal colon (D Colon) of Lpar5f/f and Lpar5ΔIEC mice were determined. n = 4 for each group. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 versus Lpar5f/f. Data are expressed as mean ± SD.
Increased Susceptibility to Dextran Sulfate Sodium–Induced Colitis in the Absence of LPA5
Increasing evidence suggests epithelial barrier dysfunction as a significant contributing factor to the pathogenesis of IBD.28,29 To evaluate whether the dysregulation of the epithelial barrier by Lpar5 loss is sufficient to alter intestinal inflammatory disease processes, we used the dextran sulfate sodium (DSS)-induced colitis model. Age and gender matched Lpar5f/f and Lpar5ΔIEC mice were given 2% DSS in drinking water for 5 days, followed by a 6-day period of recovery with normal water. During the DSS administration, Lpar5ΔIEC mice developed more severe colitis compared with control mice as evidenced by greater effects on body weight loss, occult blood, and diarrhea (Figure 4A–D). On the last day of the experiment, the disease state in Lpar5f/f mice was close to the baseline, whereas colitis was still active in Lpar5ΔIEC mice. Determining the expression levels of several proinflammatory cytokines, including tumor necrosis factor-α, interferon-γ, interleukin 6, and interleukin 1β, further corroborated that DSS-induced colitis in Lpar5ΔIEC mouse colon was exacerbated (Figure 4E). Consistently, histologic analysis revealed increased immune cell infiltration and mucosal damage in Lpar5ΔIEC mice than in Lpar5f/f mice (Figure 4F and G).
Figure 4.
Lpar5 loss increases the severity of colitis induced by DSS.Lpar5f/f and Lpar5ΔIEC mice were given 2% DSS for 5 days, followed by 6 days of recovery. Each day the mice were weighed (A), and their stools were collected to determine the Hemoccult scores (B) and diarrhea scores (C). (D) Mean disease activity indices are shown. n = 8. ∗∗P < .01, ∗∗∗P < .001 versus Lpar5f/f. (E) Expression levels of tumor necrosis factor-α, interferon-γ, interleukin 6, and interleukin 1β mRNA in mouse colon isolated on day 1 (before DSS), day 5 (end of DSS), and day 11 (end of recovery) were determined and normalized to β-actin mRNA levels. Data are expressed as mean ± SD. ∗P < .05. ∗∗P < .01. (F) Representative images of Swiss roll mounts of whole mouse colon collected from Lpar5f/f and Lpar5ΔIEC mice treated with 2% DSS for 5 days are shown. Scale bars = 200 μm. (G) Histologic damage index scores from Swiss roll mounts of colon after DSS treatment. n = 3. ∗P < .05.
LPA5 Regulates Claudin-4 Expression
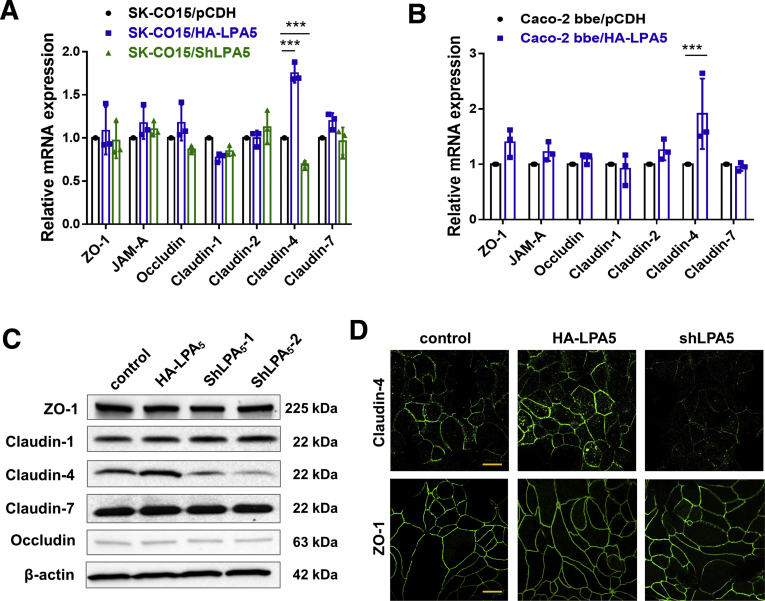
In Lpar5ΔIEC mouse intestine, we observed a significant decrease in claudin-4 expression (Figure 1). To confirm this observation, we determined the mRNA levels of several TJ proteins in SK-CO15 cells expressing HA-LPA5 or shLPA5. In line with the findings in the mouse intestine, there was a LPA5-dependent change in claudin-4 mRNA expression. Specifically, HA-LPA5 expression or Lpar5 knockdown altered claudin-4 mRNA expression without significantly altering mRNA expression levels of ZO-1, JAM-A, occludin, and claudin-1, -2, and -7 (Figure 5A). The LPA5-dependent modulation of claudin-4 expression was confirmed in Caco-2bbe cells expressing HA-LPA5 (Figure 5B). The change in claudin-4 mRNA expression in SK-CO15 cells was mirrored by a similar change in claudin-4 protein expression (Figure 5C). IF confocal microscopy on SK-CO15 cells showed that HA-LPA5 expression increased claudin-4 IF signal levels at the plasma membrane between juxtaposed cells, whereas shLPA5 attenuated the IF signal (Figure 5D). Although the effects of shLPA5 on TER and claudin-4 expression assured the role of LPA5 in epithelial barrier regulation in SK-CO15 cells, we could not detect LPA5 protein expression using anti-LPA5 antibodies, both commercial and homemade, which suggested that LPA5 expression in SK-CO15 cells must be relatively low. For this reason, we chose to use SK-CO15 cells that were stably transfected with HA-LPA5 for the following experiments.
Figure 5.
Claudin-4 expression is altered by LPA5in IECs. (A) The mRNA expression levels of several junctional proteins were determined in SK-CO15 cells transfected with pCDH, HA-LPA5, or shLPA5. mRNA levels are presented relative to β-actin mRNA level. n = 3. ∗∗∗P < .001. (B) Transcript levels of junctional proteins in Caco-2bbe cells expressing HA-LPA5 or control pCDH were determined and presented as a ratio relative to β-actin mRNA expression. n = 3. ∗∗∗P < .001. (C) Expression of several junctional proteins in SK-CO15 cells was determined by Western blotting. Representative immunoblots are shown. n = 4. (D) Representative confocal IF images showing cellular expression patterns of claudin-4 and ZO-1 in SK-CO15 cells. Scale bars = 10 μm.
Decreased claudin-4 mRNA expression by Lpar5 loss in the mouse intestine implied that LPA5 regulates claudin-4 at the level of transcription. To substantiate this notion, we determined claudin-4 mRNA and protein expression in cells treated with LPA. LPA increased claudin-4 mRNA in SK-CO15 cells, and the effects were elevated in SK-CO15/HA-LPA5 cells (Figure 6A, upper). As expected, similar changes in claudin-4 protein expression were observed in these cells (Figure 6A, lower). In Caco-2bbe cells lacking endogenous expressed LPA5, claudin-4 expression was not affected by LPA, but LPA had a robust effect on claudin-4 mRNA and protein expression in Caco-2bbe/HA-LPA5 cells (Figure 6B).
Figure 6.
LPA transcriptionally regulates claudin-4. SK-CO15 (A) and Caco-2bbe cells (B) expressing pCDH or HA-LPA5 were treated with LPA for the indicated duration (h). Claudin-4 mRNA (upper; mean ± SD) and protein levels (lower) were determined. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 versus 0 hours. Representative Western blots are shown. n = 3.
LPA5-Dependent Regulation of Claudin-4 Is Mediated via Transactivation of Epidermal Growth Factor Receptor
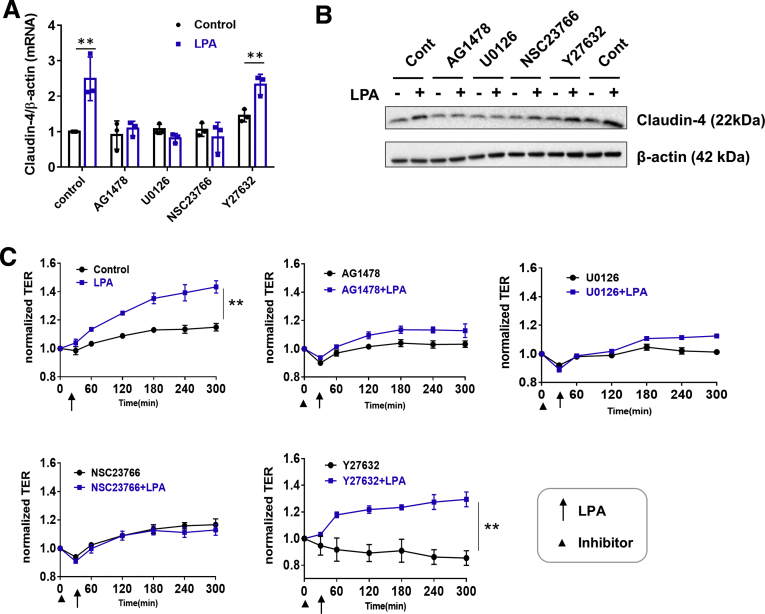
The activation of NHE3 by LPA5 involves transactivation of the epidermal growth factor receptor (EGFR), which results in the activation of RhoA/RhoA-associated kinase (ROCK) and MEK-ERK pathways.23,24 To explore whether these signaling pathways regulate LPA-dependent claudin-4 transcription, SK-CO15/HA-LPA5 cells were treated with LPA in the presence of EGFR inhibitor AG1478, MEK1/2 inhibitor U0126, Rac1 inhibitor NSC23766, or ROCK inhibitor Y27632. The increase in claudin-4 mRNA expression by LPA was blocked in cells pretreated with AG1479, U0126, or NSC23766, but not with Y27632 (Figure 7A). The blockade in LPA-dependent claudin-4 mRNA by these inhibitors resulted in similar changes to claudin-4 protein expression (Figure 7B). To demonstrate that augmented epithelial barrier function by LPA is a consequence of increased claudin-4 expression, we determined TER under the same conditions that blocked LPA-mediated claudin-4 transcriptional regulation. Increased TER in response to LPA was mitigated by inhibition of EGFR, MEK, or Rac1, but not by inhibition of ROCK (Figure 7C), demonstrating a correlation between claudin-4 expression and epithelial barrier function.
Figure 7.
LPA5mediated claudin-4 induction is dependent on Rac1. SK-CO15 cells expressing HA-LPA5, pretreated with AG1478 (1 μmol/L), U0126 (10 μmol/L), NSC23766 (100 μmol/L), or Y27632 (5 μmol/L), were treated with LPA for 2 hours. Claudin-4 mRNA (A) and protein (B) expression were determined. Data are expressed as mean ± SD. n = 3. ∗∗P < .01. (C) SK-CO15/HA-LPA5 cells on Transwells, pretreated with an inhibitor for 30 minutes, were treated with LPA or carrier (0.1 % bovine serum albumin in PBS) from the apical side of the cells. TER was measured hourly. Representative changes in TERs from 3 or more independent experiments are shown. Data are expressed as mean ± SD. ∗∗P < .01.
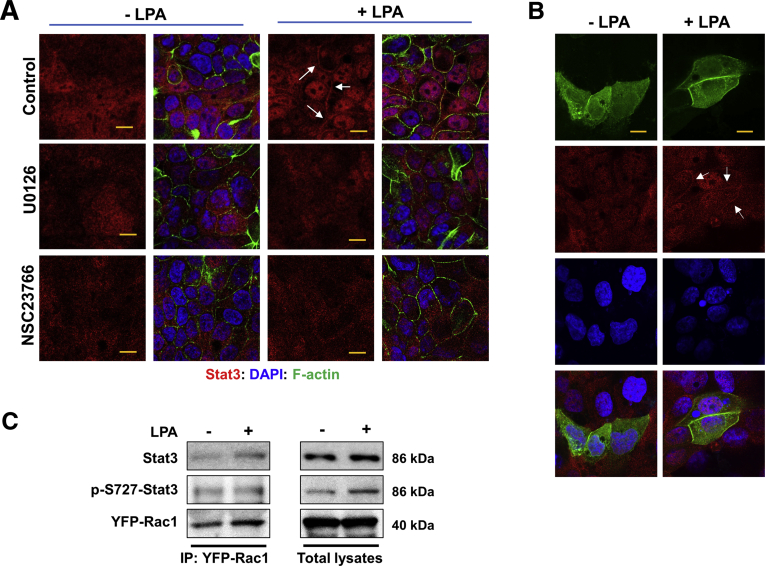
Because the MEK-ERK pathway and Rac1 can control one another,30 we next determined the spatial relationship between ERK and Rac1 activation by LPA5. First, cells pretreated for 30 minutes with NSC23766 were activated with LPA, and phosphorylation of ERK was determined. Rac1 inhibition did not affect LPA-mediated phosphorylation of ERK (Figure 8A), suggesting that Rac1 is not an upstream regulator of ERK in this context. On the other hand, LPA-induced Rac1 activity was ablated by U0126 (Figure 8B). The latter results indicated that the LPA5-EGFR-MEK-ERK cascade modulates Rac1 activity. To confirm the role of the MEK-ERK pathway on Rac1, we transiently expressed YFP-Rac1 in SK-CO15/HA-LPA5 cells and determined cellular distribution of YFP-Rac1. In the resting cells, YFP-Rac1 fluorescence signal was diffusely distributed in the cytoplasm, although some distinct fluorescent punta were also visible (Figure 8C, Control - upper lane). LPA treatment resulted in the appearance of YFP-Rac1 on the cell membrane (Figure 8C, LPA - upper lane). This cell membrane targeting of YFP-Rac1 was blocked by the inhibition of Rac1, EGFR, or MEK (Figure 8C, lower 3 lanes). These results suggest that the LPA5-EGFR-ERK signaling causes the movement of Rac1 to the cell junction. However, Rac1 is not a transcription factor, and it is unlikely that Rac1 directly modulates claudin-4 transcription.
Figure 8.
LPA5activates Rac1 activity via an ERK-dependent mechanism. (A) Phosphorylation of ERK by LPA was determined in presence or absence of NSC23766. (B) Rac1 activity was determined as described in Materials and Methods. n = 3. Data are expressed as mean ± SD. ∗∗P < .01. (C) Representative confocal IF images of SK-CO15/HA-LPA5 cells transiently transfected with YFP-Rac1 are shown. Cells were treated with carrier (left) or LPA (right) for 10 minutes in presence or absence of NSC23766, AG1478, or U0126. YFP-Rac1 (green), F-actin (red), and nuclei (blue) are shown. Scale bar = 10 μm.
LPA-Induced Regulation of Claudin-4 Is Dependent on Rac1 and Stat3
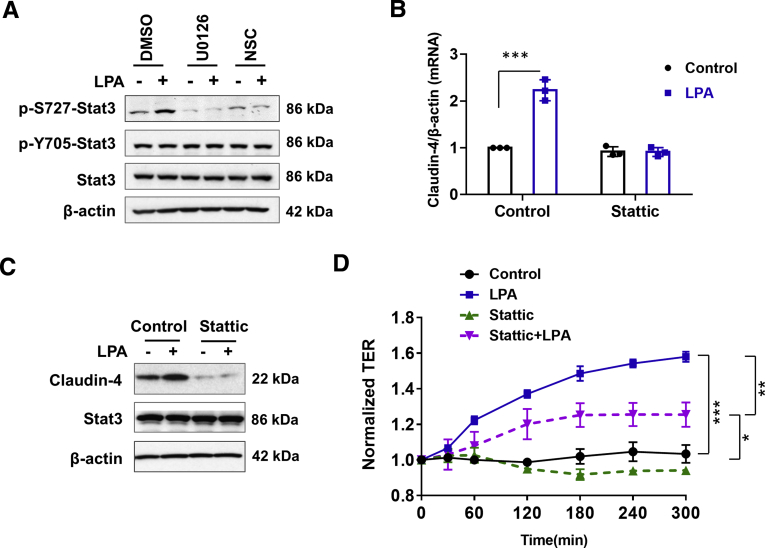
Rac1 was initially discovered for its ability to stimulate the polymerization of actin filaments, but Rac1 has been shown to have distinct roles in the regulation of gene transcription.31 Among the transcription factors that are regulated by Rac1, the signal transducers and activators of transcription (STATs) are known to regulate cell permeability, and Rac1 can bind and activate Stat3.32,33 Because phosphorylation and translocation of STAT to the nucleus are regulated by various growth factors including LPA, we determined whether LPA5 regulates Stat3 by determining phosphorylation of Stat3.32 LPA increased phosphorylation at S727 (p-S727-Stat3). On the other hand, the phosphorylation level at Y705 (p-Y705) was relatively high under basal conditions, and it was not significantly altered by LPA (Figure 9A). LPA-induced p-S727 was inhibited by U0126 and NSC23766 (Figure 9A), indicating that the MEK-ERK-Rac1 signaling is necessary for Stat3 activation. Moreover, inhibition of Stat3 with Stattic, a small molecule inhibitor of Stat3, ablated up-regulation of claudin-4 mRNA and protein by LPA (Figure 9B and C). Stattic also inhibited LPA-induced increase in TER (Figure 9D), indicating that LPA5 regulates epithelial barrier function by Stat3-dependent induction of claudin-4.
Figure 9.
LPA5phosphorylates Stat3. (A) p-S727 and p-Y705 of Stat3 were determined in cells treated with LPA in presence or absence of U0126 or NSC23766. Effect of Stat3 inhibitor Stattic on claudin-4 mRNA (B) and protein (C) was determined. Mean ± SD. n = 3. ∗∗∗P < .001 versus control. (D) TER in SK-CO15/HA-LPA5 cells was determined in presence or absence of Stattic. Data are expressed as mean ± SD. ∗P < .05. ∗∗P < .01. ∗∗∗P < .001.
IF confocal microcopy of SK-CO15/HA-LPA5 cells showed that LPA induced translocation of Stat3 from the cytoplasm to the nucleus (Figure 10A, upper lane). Surprisingly, in addition to its nuclear expression, Stat3 was visible along the edge of juxtaposed cells. The localization of Stat3 at the nucleus and cell membrane was blocked by U0126 and NSC23766 (Figure 10A, middle and lower lanes), further suggesting that Rac1 activity is necessary for the activation of Stat3 by LPA. To determine whether LPA regulates the interaction between Rac1 and Stat3, Stat3 and YFP-Rac1 cellular localization was analyzed. In cells under basal conditions, both Stat3 and YFP-Rac1 were diffusely distributed in the cytoplasm (Figure 10B, left column). LPA induced the appearance of Stat3 in the nucleus and on the cell membrane, where it co-localized with YFP-Rac1 (Figure 10B, right column). However, the induction of nuclear and membranous appearance of Stat3 was rapid, and we could not discern which event occurred first. Nonetheless, the presence of Stat3 at the cell membrane suggested that LPA induces the interaction between Rac1 and Stat3, in line with a previous study that activated Rac1 forms a complex with Stat3 in COS-1 cells.33 To confirm their interaction, we performed co-immunoprecipitation of YFP-Rac1 and Stat3. Co-immunoprecipitation of Stat3 with YFP-Rac1 under basal condition suggested that Rac1 and Stat3 interact without LPA treatment (Figure 10C). The interaction of YFP-Rac1 with Stat3 under basal conditions was not surprising because transfected YFP-Rac1 partially retained its activity without LPA treatment (Figure 8B). Importantly, LPA stimulated co-immunoprecipitation of Stat3 with YFP-Rac1 (Figure 10C), indicating that LPA promotes the Rac1-Stat3 interaction. However, we were surprised that we could not observe increased co-immunoprecipitation of p-S727-Stat3 with YFP-Rac1 in response to LPA. These results suggest that LPA induces the interaction of Rac1 with Stat3, and this interaction precedes phosphorylation of Stat3 at S727.
Figure 10.
LPA causes translocation of Stat3 to cell membrane and nuclei. (A) Representative confocal IF images of Stat3 (red) in cells treated with LPA are shown. Cells were treated with LPA or carrier in presence or absence of U126 or NSC23766. Scale bar = 10 μm. (B) SK-CO15/HA-LPA5 cells transiently expressing YFP-Rac1 were treated with LPA to determine cellular localization of Stat3 (red) and YFP-Rac1 (green). Arrows indicate membrane localization of Stat3. Scale bar = 5 μm. Results from 3 independent experiments are shown. (C) YFP-Rac1 was immunoprecipitated from lysate of SK-CO15/HA-LPA5/YFP-Rac1 cells, followed by immunoblotting using anti-Stat3 or anti-p-S727-Stat3 antibody. Representative blots from 3 independent experiments are shown. Right panels show Stat3 and p-S727-Stat3 in cell lysate.
Discussion
Our previous studies have demonstrated the role of LPA5 in the regulation of NHE3, which plays a major role in sodium and fluid absorption in the intestine.22, 23, 24 However, Lpar5 is highly expressed in mature enterocytes, and cellular signaling by LPA5 is expected to have a broad range of effects based on general functions of other LPA receptors. In the current study, we show that Lpar5 loss in IECs dysregulates epithelial barrier, resulting in increased paracellular permeability in the mouse intestine. Lpar5ΔIEC mice had increased serum FD-4 level compared with Lpar5f/f mice, suggesting increased leakiness of the intestinal epithelial monolayer in Lpar5ΔIEC mice. In addition, we found elevated serum LPS level and increased bacterial translocation into the intestinal mucosa and the liver of Lpar5ΔIEC mice. These results suggest that loss of Lpar5 compromises innate immunity by disrupting the intestinal epithelial barrier function.
IECs express multiple forms of claudin that have different expression patterns within the GI tract.34 Claudin-4, which was the first claudin shown to regulate the TJ permeability in cultured cells, is highly expressed in the villus epithelial cells in the intestine of rats and mice.35, 36, 37 Claudin-1 and claudin-2 expression is more restricted to the crypt base in the small intestine and colon.36 Claudin-7 is roughly evenly distributed along the crypt-villus axis, but its expression is greater on the basolateral surface of the epithelial cell than on the apical membrane.37,38 We observed that Lpar5 loss specifically led to decreased claudin-4 expression in both mouse intestine and colonic epithelial cell lines. In contrast, claudin-1, claudin-2, or claudin-7 expression was not altered by Lpar5 loss. It is noteworthy that the current findings differ from our previous study of LPA1 where Lpar1 loss in mice resulted in decreased expression of claudin-2, -4, and -7.15 Cellular distribution of LPA5 or other LPA receptors in native intestinal tissue is not known, but HA-LPA5 expressed in Caco-2bbe cells preferentially partitioned to the apical surface over the basolateral surface.23 This differs from LPA1, which localizes to both apical and basolateral membranes.14 Therefore, the differences in the expression patterns of LPA5 versus LPA1 within the intestinal epithelium and along the crypt-villus axis are likely to influence the expression of a specific claudin.
LPA and precursors of LPA, such as phosphatidic acid, are present in a variety of food products.39,40 Orally administered LPA has been linked to biological effects, including wound healing, electrolyte balance, and cancer in the GI tract.14,22,39,41,42 A recent study showed that LPA can mitigate villus blunting and restore apical transporter expression in the small intestine of myosin 5B-deficient mice, a rodent model of microvillus inclusion disease.43 Our current study demonstrated that orally delivered LPA can stimulate the epithelial barrier function in vivo, and LPA5 plays a major role in manifesting this effect. Interestingly, we found that LPA was able to decrease FD-4 flux in Lpar5ΔIEC mice, albeit by a lesser extent compared with control mice. We postulate that the residual effect on LPA-dependent epithelial permeability is mediated by LPA1. Lpar1-deficient mice have delayed wound healing and decreased epithelial barrier function.14,15 Because LPA1 is expressed in the intestine of Lpar5ΔIEC mice, it is likely that LPA1 mediates LPA-induced epithelial permeability in these mice.
Intestinal epithelial barrier dysfunction is a major cause of the development and progression of IBD, and the association between epithelial dysfunction and DSS-induced colitis has been demonstrated.44, 45, 46 Administration of 2% DSS, which is in a low range used in similar experiments, exacerbated colitis in Lpar5ΔIEC mice. In particular, weight loss and diarrhea were much greater in Lpar5ΔIEC mice. Diarrhea is one of the most common symptoms of IBD, and NHE3 has been shown to be down-regulated in patients with IBD.47,48 A positive correlation between reduced NHE3 expression and increased severity of colitis has been demonstrated in rodent models of colitis.49,50 Because LPA5 regulates NHE3, it is possible that Lpar5 loss reduces NHE3 expression or function that may have increased diarrhea in DSS-treated Lpar5ΔIEC mice. A recent study of NHE3 regulation in Lpar5ΔIEC mice showed that basal NHE3 mRNA expression was decreased in Lpar5-deficient IECs, although no significant change in NHE3 activity was observed.25 On the other hand, LPA-mediated activation of NHE3 or fluid absorption was completely absent in these mice.25 It is not yet known whether the lack of LPA-mediated NHE3 regulation is sufficient to cause more severe diarrhea in the setting of IBD or infectious diseases. Because dysregulation of NHE3 is a frequent cause of diarrhea, the lack of NHE3 stimulation by LPA and dietary peptone may attenuate NHE3-dependent fluid absorption and hence increases fluid loss in the gut, resulting in more severe diarrhea. However, this possibility remains to be experimentally evaluated.
The majority of extracellular LPA is generated by ATX, which is elevated in inflamed mucosa of human and mouse intestine. Excess amounts of LPA generated within the damaged tissues can trigger proinflammatory and pro-angiogenic responses by immune cells, fibroblasts, or cancer cells. Therefore, inhibition of ATX mitigates inflammation, as demonstrated in rodent models of IBD.9, 10, 11, 12 Because of the potential benefit of ATX inhibition to treat chronic inflammation, the current findings of LPA5-dependent regulation of intestinal epithelial barrier and the protection from DSS-induced colitis by Lpar5 loss in IECs appear paradoxical. In the intestine, ATX is expressed by endothelial venules and B cells,9,11 and it seems unlikely that ATX-derived LPA can access LPA5 at the apical side of the epithelium. Hence, we speculate that orally administered LPA primarily acts on apically expressed LPA receptors such as LPA1 and LPA5 to maintain epithelial barrier and fluid homeostasis in the GI tract.
The transactivation of EGFR by various GPCR agonists, including LPA, thrombin, and endothelin-1, was first reported by the Ullrich group.51 An important aspect of EGFR transactivation is that it provides a mechanism that enables GPCR agonists to active the MEK-ERK signaling pathway. As expected, LPA5 regulates the MEK-ERK pathway via EGFR transactivation.23 EGFR is generally thought to be located in the basolateral membrane of polarized epithelial cells, but others have demonstrated apically located EGFR.52,53 Previous studies have shown the presence of EGFR on both apical and basolateral sides of monolayers formed from primary canine oxyntic epithelial cells, but apical EGFR and not basolateral EGFR regulates epithelial barrier to gastric acid.54,55 In Caco-2bbe cells, EGFR is expressed on both apical and basolateral membranes, but the regulation of NHE3 by LPA5 is observed only when apical EGFR is stimulated.23 Although we did not specifically compare the role of apical vs basolateral EGFR in claudin-4 regulation, we infer from our previous study that apically located EGFR mediates claudin-4 regulation by LPA.
The Rho GTPase family constitutes important effectors of LPA receptors. LPA causes actin cytoskeletal rearrangement through activation of RhoA, which results in barrier dysfunction in endothelial cells and migration of cancer cells.56,57 We have shown recently that LPA mediates mouse colonic epithelial YAMC cell migration through a Rac1-dependent mechanism.14 On the other hand, LPA inhibits RhoA in YAMC cells, and RhoA inactivation results in spreading and migration of fibroblasts.14,58 In Caco-2bbe cells, LPA5 regulates RhoA independent of the MEK-ERK pathway.23,24 On the contrary, we found in the current study that LPA-induced stimulation of Rac1 activity was mediated via the MEK-ERK pathway. The latter observation is in line with previous studies demonstrating the upstream regulatory role of the MEK-ERK signaling in Rac activation.59,60 NSC23766, which specifically inhibits Rac1 activation through interfering its binding to a Rac1-specific guanine nucleotide exchange factor (GEF),61 blocked targeting of YFP-Rac1 to cell-cell junction by LPA. It is not known which GEF is responsible for Rac1 activation by LPA5. LPA is known to regulate the Rac-specific GEF Tiam1, which regulates Rac1 activity via its recruitment to epithelial junction.62, 63, 64 A future study is needed to determine whether LPA5 regulates Tiam1 and whether Tiam1 activates Rac1 in the context of epithelial barrier regulation.
LPA often mediates its proinflammatory or oncogenic effects through transcriptional regulation of Myc, Stat, nuclear factor kappa B, and β-catenin.65, 66, 67, 68 We found that inhibition of Stat3 by Stattic attenuated LPA-mediated claudin-4 transcriptional activation, demonstrating that Stat3 is a transcriptional factor regulating claudin-4 expression. Stat3 is mostly latent in the cytoplasm until activated through receptor-mediated phosphorylation.69 Y705 phosphorylation is generally regarded as a dominant actuator of Stat3 signaling, but Y705 was phosphorylated under basal conditions, and LPA did not significantly alter p-Y705 in SK-CO15 cells. Interestingly, cell-cell adhesion can induce p-Y705-Stat3, and it is thought to confer differentiated epithelial morphology of cancer cells.70,71 Therefore, it is possible that the polarized epithelial morphology of SK-CO15 and Caco-2bb cells maintains phosphorylation at Y705. Instead of phosphorylating Stat3 at Y705, LPA increased phosphorylation at S727. Phosphorylation at S727 (p-S727) has been documented to be associated with Stat3 activation,72,73 although others have shown a negative role of p-S727 on Stat3.74,75
It has been shown previously that activated Rac1 complexes with Stat3.33 In addition, Stat3 activation by Ca2+-dependent cell-cell adhesion has been suggested.70 Our findings that LPA promoted co-immunoprecipitation of Rac1 and Stat3 and their co-localization at the cell membrane are in line with these earlier studies. However, we were not able to detect increased Rac1-p-S727-Stat3 interaction or the presence of p-S727-Stat3 at the cell junction. Although this failure could have been due to differences in antibodies used against total Stat3 versus p-S727-Stat3, it also suggests that membrane-targeted Rac1 recruits Stat3 and not p-S727-Stat3.
One potential concern in the current study is the use of pharmacologic inhibitors despite their broad usage to target their respective proteins. Like all pharmacologic inhibitors, some of the inhibitors used here show off-target effects.76 On the other hand, other studies have confirmed the specificity of these inhibitors by comparing them with gene knockdown.77, 78, 79 Likewise, we have previously demonstrated the identical effect of AG1478 and EGFR knockdown on LPA5-mediated ERK activation in Caco-2bbe cells.23 Another potential limitation of the current study is the use of intestinal cell lines SK-CO15 and Caco-2bbe for the mechanistic insight into LPA5 function. Although these cell lines are used widely as model IECs22,80,81 and decreased claudin-4 expression by Lpar5 loss was recapitulated in these cell lines, it is unlikely that these cells have kept all the features of native colonic epithelial cells. With these limitations in mind, we suggest that claudin-4 regulation by LPA5 is a 3-step process. First, LPA-LPA5 signaling initiates activation of EGFR-MEK-ERK, which stimulates the translocation of Rac1 to the cell membrane. Second, Rac1 recruits and interacts with Stat3 at the cell membrane where Stat3 is phosphorylated at S727. Third, activated Stat3 rapidly dissociates from Rac1 and translocates to the nucleus to transcribe claudin-4.
In summary, our study demonstrates that LPA enhances the epithelial barrier function via LPA5. The absence of LPA5 results in increased epithelial permeability, which allows bacterial translocation across the intestinal luminal membrane. LPA5 regulates the epithelial barrier by transcriptionally modulating claudin-4 via a mechanism requiring Rac1 and Stat3. Together with our previous studies on LPA1 facilitating epithelial wound healing in the intestine, the present study suggests a potential benefit of using LPA to enhance intestinal epithelial barrier integrity.
Materials and Methods
Cell Culture and Plasmids
SK-CO15 and Caco-2bbe human colonic epithelial cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 50 U/mL penicillin, 50 μg/mL streptomycin, and 1 mmol/L sodium pyruvate.82,83 Lentiviral plasmid (pLKO.1-puro) containing shRNA targeting LPA5 (shLPA5) and control lentivirus containing non-targeting scrambled sequences (shCon) were obtained from Sigma-Aldrich (St Louis, MO). The silence of Lpar5 was confirmed by RT-PCR. Cells that were transfected with pCDH harboring N-terminal hemagglutinin (HA)-tagged human LPA5, HA-LPA5, were maintained in growth medium supplemented with 2 μg/ml puromycin for SK-CO15 cells and 8 μg/mL for Caco-2bbe cells as previously described.23 YFP-Rac1 was a gift from Joel Swanson (Addgene plasmid #11391, Watertown, MA). Cells were grown on plastic dishes or Transwell inserts (Corning, Tewksbury, MA) for 7 days after confluence before all assays.
Chemicals and Antibodies
LPA (18:1; 1-oleoyl-2-hydroxy-sn-glycero-3-phosphate) was purchased from Avanti Polar Lipids (Alabaster, AL) and prepared according to the manufacturer’s instructions. For in vitro study, LPA was used at the final concentration of 10 μmol/L in PBS containing 0.1% bovine serum albumin, unless otherwise specified. An equal volume of PBS containing 0.1% bovine serum albumin was added as a control. All chemicals, including FD-4, AG1478, U0126, NSC23766, and Y27632, were obtained from Sigma-Aldrich or EMD Millipore (Billerica, MA). The following commercial antibodies were used: rabbit anti-claudin-1, rabbit anti-claudin-4, rabbit anti-occludin, and rabbit anti-ZO-1 (Thermo Fisher Scientific, Waltham, MA); goat anti-E-cadherin and mouse anti-Rac1 (R&D Systems, Minneapolis, MN); mouse anti-β-actin (Sigma-Aldrich); and rabbit anti-Stat3 and rabbit anti-p-Stat3 (Cell Signaling, Danvers, MA).
Animals
Generation of Lpar5f/f mice and Lpar5ΔIEC (Lpar5f/f;Vil-cre) mice was previously reported.25 Littermates were used in all experiments. Genotypes were determined by PCR using the following primers: Vil-Cre forward, CAA GCC TGG CTC GAC GGC C and reverse, CGC GAA CAT CTT CAG GTT CT; loxP forward, CCA GGC AGA GAG AGG AAG TG and reverse, TGG CCT CAG AAG ATT TGC TC. Experiments with animals were performed under approval by the Institutional Animal Care and Use Committees of the Atlanta Veterans Administration Medical Center and Emory University (Atlanta, GA) and in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals.
Intestinal Permeability In Vivo
Intestinal permeability was determined using FD-4 as a mucosal tracer flux marker as described previously.15 Briefly, FD-4 in PBS (100 mg/mL; Sigma-Aldrich) was administered by gastric gavage at a final dose of 60 mg/100 g body weight. Four hours later, mice were euthanized, and blood samples were collected by cardiac puncture. Blood was centrifuged at 5000 rpm for 10 minutes, and serum was diluted in an equal volume of PBS. Fluorescence intensity in the serum was measured at an excitation of 485 nm and an emission of 525 nm (485/528 nm) using a Synergy 2 (BioTek, Winooski, VT) plate reader. Concentrations of FD-4 were tabulated against a standard curve.
Oral Administration of LPA
Male mice of 9 weeks of age were pretreated either with LPA (150 μL of 300 μmol/L stock) or PBS by gavage daily for 3 days. On day 3, mice were given oral administration of FD-4, and intestinal permeability was determined.
Measurement of Serum LPS
LPS in the serum was determined by using a Pierce Chromogenic Endotoxin Quant kit according to the manufacturer’s instruction (Thermo Fisher Scientific).
Measurement of Bacteria Loads
Mice were euthanized by using isoflurane and cervical dislocation. The liver was perfused by injecting 10 mL PBS into the hepatic portal vein. The perfused liver was removed and stored on ice. The ileum and colon were removed. Sections of the liver, ileum, proximal colon, and distal colon were excised and weighted. After homogenizing, serial dilutions of the homogenates were plated on Agar plates, and plates were incubated at 37°C for 24–36 hours.
DSS-Induced Colitis
Eight- to 12-week-old male mice (n = 16 per strain) were permitted free access to 2% DSS (w/v; mol wt, 36,000–50,000; Affymetrix, Inc, Santa Clara, CA) in drinking water for 5 days to induce acute colitis. Half of the mice were euthanized at the end of DSS treatment, and the remaining half were given normal water for the next 6 days to recover. The body weight of each mouse was measured and recorded daily. Assessment of stool consistency and the presence of occult blood by a guaiac test (Hemoccult Sensa; Beckman Coulter, Fullerton, CA) were determined daily for each mouse. The disease activities (diarrhea, occult blood, and weight loss) were quantified as we described previously.14 Mice were euthanized by using isoflurane and cervical dislocation on day 5 or 11. Colon was removed, flushed with chilled (4°C) Ca2+ and Mg2+ free PBS, and fixed in RNAlater (Thermo Fisher Scientific) for RNA isolation. Whole colon tissues were fixed in 10% buffered formalin overnight for histologic analysis. Paraffin-embedded sections were stained with H&E for microscopic assessment of colitis.
Histology
For each animal, a histologic examination was performed on 3 samples of the distal colon. Histologic parameters were quantified in a blinded manner (MW and YH) using the scores previously published.84 Three independent parameters used were severity of inflammation (0–3: none, slight, moderate, severe); depth of injury (0–3: none, mucosal, mucosal and submucosal, transmural); and crypt damage (0–4: none, basal one-third damaged, basal two-thirds damaged, only surface epithelium intact, entire crypt and epithelium lost). The score of each parameter was multiplied by a factor reflecting the percentage of tissue involvement (×1: 0%–25%, ×2: 26%–50%, ×3: 51%–75%, ×4: 76%–100%), and these values were summed to obtain a total score.
In Vitro Permeability Measurement
SK-CO15 cells and Caco-2 cells were grown on Transwell inserts with 0.4-μm pore size (Corning). TER of a monolayer was measured using an epithelial V-Ω meter (World Precision Instruments, Inc, Sarasota, FL). Resistance of cells on filter was calculated by subtracting the resistance of the membrane plus medium from the resistance of the membrane plus medium plus cells. Each experiment was measured in triplicate, and the average value was taken. This value was then multiplied by the area of the Transwell membrane (0.9 cm2) to obtain a final value in Ω × cm2.
Western Immunoblot and Immunoprecipitation
Lysate from mouse intestinal tissues or cultured cells was prepared, and Western blotting was performed as previously described.14 For immunoprecipitation of YFP-Rac1, SK-CO15 cells were lysed in cold lysis buffer (20 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 5 mmol/L glycerophosphate, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L Na2EDTA, 1 mmol/L EGTA, 1 mmol/L Na3VO4,1 mmol/L NaF, 10 mmol/L leupeptin, 1% Triton X-100, protease inhibitors mixture, and 2.5 mmol/L N-ethylmaleimide). Protein concentration was determined by bicinchoninic acid assay (Sigma-Aldrich). Equal amounts of cell lysates (typically 500 μg) were incubated overnight with anti-green fluorescent protein antibody. The immunocomplex was purified by incubating with protein G-Sepharose beads for 1 hour, followed by 2 washes in lysis buffer and 1 wash in PBS. Immunocomplexes were eluted from the beads in 2× Laemmli buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotted with anti-Stat3 or anti-pS727-Stat3 antibody.
Quantitative RT-PCR
Total RNA was extracted from colon mucosal scrapes or cultured cells using the RNeasy Mini kit (Qiagen, Hilden, Germany). One μg of total RNA was used for cDNA synthesis using a First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instruction. Quantitative PCR was performed with iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) on a Mastercycler Realplex (Eppendorf, Hamburg, Germany). Expression levels determined in triplicate per sample were normalized to β-actin. PCR primer sequences are listed in Table 1.
Table 1.
List of PCR Primers
| Genes | Primer sequence, 5′-3′ |
|
|---|---|---|
| Forward | Reverse | |
| hβ-actin | GGACTTCGAGCAAGAGATGG | AGCACTGTGTTGGCGTACAG |
| hClaudin-1 | CGATGAGGTGCAGAAGATGA | CCAGTGAAGAGAGCCTGACC |
| hClaudin-2 | TTGTGACAGCAGTTGGCTTC | TCATGCCCACCACAGAGATA |
| hClaudin-4 | AAGTGACAGGGTGTGGTGGT | TACCCGGAACAGAGGAGATG |
| hClaudin-7 | AATTTTCATCGTGGCAGGTC | GGACAGGAACAGGAGAGCAG |
| hJAM-A | AGCCATCAGTCAAGGGTCAC | GAGGCGGAGGTTACAGTGAG |
| hOccludin | TATGGAGGAAGTGGCTTTGG | ATGCCCAGGATAGCACTCAC |
| hZO-1 | CAGCAACTTTCAGACCACCA | GTGCAGTTTCACTTGGCAGA |
| hLPA5R | ATCTTCCTGCTGTGCTTCGT | CCCTCGGCGCTAAAGTAGTA |
| mβ-actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| mClaudin-1 | AAGGGTGTTGCCATTGACAT | TGCAACATAGGCAGGACAAG |
| mClaudin-2 | AAGGTGCTGCTGAGGGTAGA | TTGAGCATTCAAAGCACAGG |
| mClaudin-4 | TGGAACCCTTCCGTTGATTA | CACTGGGCTGCTTCTAGGTC |
| mClaudin-7 | CTGGTGTTGGGCTTCTTAGC | TGATGACCAATCCAGGAACA |
| mJAM-A | AGCCAGATCACAGCTCCCTA | GACAGAGGAGGGGACACTGA |
| mOccludin | AGTGGGTCAGGGAATATCCA | TCAGCAGCAGCCATGTAACTC |
| mZO-1 | GTCCCTGTGAGTCCTTCAGC | GAAGGGCTCCTTGTGGGATA |
| mTNF-α | TCGTAGCAAACCACCAAGTG | AGATAGCAAATCGGGTGACG |
| mIFN-γ | TCTCCAGAAACCCTCACTGGT | TCAGCGGATTCATCTGCTTCG |
| mIL-1β | TGAAATGCCACCTTTTGACA | GTAGCTGCCACAGGCTTCTCC |
| mIL-6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| mLPA5R | GCTCCAGTGCCCTGACTATC | GGGAAGTGACAGGGTGAAGA |
Rac1 GTPase Activity Assay
GTP-bound Rac1 was determined using a G-LISA Activation Assay Kit according to the manufacturer’s instructions (Cytoskeleton, Denver, CO).
Confocal IF
After being flushed with cold PBS, the medial small intestine and colon segments were incubated overnight in 30% sucrose in PBS for cryoprotection. Six-micron cryostat sections were prepared and stored at –80°C until needed. The frozen sections were fixed with ice-cold 100% ethanol and acetone at the ratio of 1:1 for 10 minutes at –20°C. The cell monolayers on Transwell filters were washed 3 times with ice-cold PBS, followed by fixation with 4% paraformaldehyde in PBS for 10 minutes at room temperature. For claudin-4 staining, cells were fixed with 100% ice cold ethanol for 20 minutes at –20°C. After fixation, tissues and cells were permeabilized with 0.2% Triton X-100 for 10 minutes, blocked in PBS containing 5% normal goat serum for 30 minutes, and incubated for 1 hour with a specific primary antibody at room temperature. The monolayers were rinsed 5 times with PBS for 5 minutes and incubated with fluorescence tag-labeled secondary antibodies for 30 minutes at room temperature. After five 5-minute washes with PBS, specimens were mounted with ProLong Glass Antifade Reagent (Invitrogen) and observed under a Nikon A1R HD confocal microscope (Nikon Instruments Inc, New York, NY) coupled to a Plan Apo λ 60x Oil lens.
Statistical Analysis
Statistics were performed using independent samples, two-tailed unpaired Student t test or analysis of variance, followed by Tukey post hoc test using Prism 6 software (GraphPad Software, La Jolla, CA). Results are presented as mean ± standard deviation (SD). A value of P <.05 was considered significant.
CRediT Authorship Contributions
Mo Wang (Conceptualization: Supporting; Formal analysis: Lead; Investigation: Lead; Methodology: Equal; Project administration: Supporting; Validation: Lead; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Peijian He (Conceptualization: Supporting; Investigation: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Yiran Han (Investigation: Supporting; Writing – review & editing: Supporting)
Lei Dong (Funding acquisition: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Chris Yun, PhD (Conceptualization: Lead; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by grants from the National Institutes of Health (R01DK116799) and the Veterans Administration Merit Award (I01BX004459). MW was supported by the State Scholarship Fund of the China Scholarship Council. Confocal microscopic analyses were supported in part by the Integrated Cellular Imaging Shared Resources of Winship Cancer Institute of Emory University and NIH/NCI under award P30CA138292.
Contributor Information
Lei Dong, Email: dong556@126.com.
C. Chris Yun, Email: ccyun@emory.edu.
References
- 1.Tsukita S., Furuse M., Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 2.Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol. 2014;15:397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 3.Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 4.Chiurchiu V., Leuti A., Maccarrone M. Bioactive lipids and chronic inflammation: managing the fire within. Frontiers in Immunology. 2018;9:38. doi: 10.3389/fimmu.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi J.W., Herr D.R., Noguchi K., Yung Y.C., Lee C.W., Mutoh T., Lin M.E., Teo S.T., Park K.E., Mosley A.N., Chun J. LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol. 2010;50:157–186. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 6.Houben A.J., Moolenaar W.H. Autotaxin and LPA receptor signaling in cancer. Cancer Metastasis Rev. 2011;30:557–565. doi: 10.1007/s10555-011-9319-7. [DOI] [PubMed] [Google Scholar]
- 7.Lin S., Lee S.J., Shim H., Chun J., Yun C.C. The absence of LPA receptor 2 reduces the tumorigenesis by ApcMin mutation in the intestine. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1128–G1138. doi: 10.1152/ajpgi.00321.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Meeteren L.A., Ruurs P., Stortelers C., Bouwman P., van Rooijen M.A., Pradere J.P., Pettit T.R., Wakelam M.J., Saulnier-Blache J.S., Mummery C.L., Moolenaar W.H., Jonkers J. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol Cell Biol. 2006;26:5015–5022. doi: 10.1128/MCB.02419-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanda H., Newton R., Klein R., Morita Y., Gunn M.D., Rosen S.D. Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nat Immunol. 2008;9:415–423. doi: 10.1038/ni1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hozumi H., Hokari R., Kurihara C., Narimatsu K., Sato H., Sato S., Ueda T., Higashiyama M., Okada Y., Watanabe C., Komoto S., Tomita K., Kawaguchi A., Nagao S., Miura S. Involvement of autotaxin/lysophospholipase D expression in intestinal vessels in aggravation of intestinal damage through lymphocyte migration. Lab Invest. 2013;93:508–519. doi: 10.1038/labinvest.2013.45. [DOI] [PubMed] [Google Scholar]
- 11.Lin S., Haque A., Raeman R., Guo L., He P., Denning T.L., El-Rayes B., Moolenaar W.H., Yun C.C. Autotaxin determines colitis severity in mice and is secreted by B cells in the colon. FASEB J. 2019;33:3623–3635. doi: 10.1096/fj.201801415RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He P., Haque A., Lin S., Cominelli F., Yun C.C. Inhibition of autotaxin alleviates inflammation and increases the expression of sodium-dependent glucose cotransporter 1 and Na(+)/H(+) exchanger 3 in SAMP1/Fc mice. Am J Physiol Gastrointest Liver Physiol. 2018;315:G762–G771. doi: 10.1152/ajpgi.00215.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S., Wang D., Iyer S., Ghaleb A.M., Shim H., Yang V.W., Chun J., Yun C.C. The absence of LPA2 attenuates tumor formation in an experimental model of colitis-associated cancer. Gastroenterology. 2009;136:1711–1720. doi: 10.1053/j.gastro.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S.J., Leoni G., Neumann P.A., Chun J., Nusrat A., Yun C.C. Distinct phospholipase C-beta isozymes mediate lysophosphatidic acid receptor 1 effects on intestinal epithelial homeostasis and wound closure. Mol Cell Biol. 2013;33:2016–2028. doi: 10.1128/MCB.00038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S., Han Y., Jenkin K., Lee S.J., Sasaki M., Klapproth J.M., He P., Yun C.C. Lysophosphatidic acid receptor 1 is important for intestinal epithelial barrier function and susceptibility to colitis. Am J. Pathol. 2018;188:353–366. doi: 10.1016/j.ajpath.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C.W., Rivera R., Gardell S., Dubin A.E., Chun J. GPR92 as a new G12/13- and Gq-coupled lysophosphatidic acid receptor that increases cAMP, LPA5. J Biol Chem. 2006;281:23589–23597. doi: 10.1074/jbc.M603670200. [DOI] [PubMed] [Google Scholar]
- 17.Choi S., Lee M., Shiu A.L., Yo S.J., Aponte G.W. Identification of a protein hydrolysate responsive G protein-coupled receptor in enterocytes. Am J Physiol Gastrointest Liver Physiol. 2007;292:G98–G112. doi: 10.1152/ajpgi.00295.2006. [DOI] [PubMed] [Google Scholar]
- 18.Williams J.R., Khandoga A.L., Goyal P., Fells J.I., Perygin D.H., Siess W., Parrill A.L., Tigyi G., Fujiwara Y. Unique ligand selectivity of the GPR92/LPA5 lysophosphatidate receptor indicates role in human platelet activation. J Biol Chem. 2009;284:17304–17319. doi: 10.1074/jbc.M109.003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S., Lee M., Shiu A.L., Yo S.J., Hallden G., Aponte G.W. GPR93 activation by protein hydrolysate induces CCK transcription and secretion in STC-1 cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1366–G1375. doi: 10.1152/ajpgi.00516.2006. [DOI] [PubMed] [Google Scholar]
- 20.Poole D.P., Lee M., Tso P., Bunnett N.W., Yo S.J., Lieu T., Shiu A., Wang J.C., Nomura D.K., Aponte G.W. Feeding-dependent activation of enteric cells and sensory neurons by lymphatic fluid: evidence for a neurolymphocrine system. Am J Physiol Gastrointest Liver Physiol. 2014;306:G686–G698. doi: 10.1152/ajpgi.00433.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jongsma M., Matas-Rico E., Rzadkowski A., Jalink K., Moolenaar W.H. LPA is a chemorepellent for B16 melanoma cells: action through the cAMP-elevating LPA5 receptor. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S., Yeruva S., He P., Singh A.K., Zhang H., Chen M., Lamprecht G., de Jonge H.R., Tse M., Donowitz M., Hogema B.M., Chun J., Seidler U., Yun C.C. Lysophosphatidic acid stimulates the intestinal brush border Na+/H+ exchanger 3 and fluid absorption via LPA5 and NHERF2. Gastroenterology. 2010;138:649–658. doi: 10.1053/j.gastro.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoo B.K., He P., Lee S.J., Yun C.C. Lysophosphatidic acid 5 receptor induces activation of Na(+)/H(+) exchanger 3 via apical epidermal growth factor receptor in intestinal epithelial cells. Am J Physiol Cell Physiol. 2011;301:C1008–C1016. doi: 10.1152/ajpcell.00231.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.No Y.R., He P., Yoo B.K., Yun C.C. Regulation of NHE3 by lysophosphatidic acid is mediated by phosphorylation of NHE3 by RSK2. Am J Physiol Cell Physiol. 2015;309:C14–C21. doi: 10.1152/ajpcell.00067.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkin K.A., He P., Yun C.C. Expression of lysophosphatidic acid receptor 5 is necessary for the regulation of intestinal Na(+)/H(+) exchanger 3 by lysophosphatidic acid in vivo. Am J Physiol Gastrointest Liver Physiol. 2018;315:G433–G442. doi: 10.1152/ajpgi.00130.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W.S., Rozenblatt-Rosen O., Shi H.N., Yilmaz O., Xavier R.J., Regev A. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yun C.C., Sun H., Wang D., Rusovici R., Castleberry A., Hall R.A., Shim H. LPA2 receptor mediates mitogenic signals in human colon cancer cells. Am J Physiol. 2005;289:C2–C11. doi: 10.1152/ajpcell.00610.2004. [DOI] [PubMed] [Google Scholar]
- 28.Antoni L., Nuding S., Wehkamp J., Stange E.F. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2014;20:1165–1179. doi: 10.3748/wjg.v20.i5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang J., Leong R.W., Wasinger V.C., Ip M., Yang M., Phan T.G. Impaired intestinal permeability contributes to ongoing bowel symptoms in patients with inflammatory bowel disease and mucosal healing. Gastroenterology. 2017;153:723–731 e721. doi: 10.1053/j.gastro.2017.05.056. [DOI] [PubMed] [Google Scholar]
- 30.Vial E., Sahai E., Marshall C.J. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 2003;4:67–79. doi: 10.1016/s1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 31.Benitah S.A., Valeron P.F., van Aelst L., Marshall C.J., Lacal J.C. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim Biophys Acta. 2004;1705:121–132. doi: 10.1016/j.bbcan.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Leve F., Peres-Moreira R.J., Binato R., Abdelhay E., Morgado-Diaz J.A. LPA induces colon cancer cell proliferation through a cooperation between the ROCK and STAT-3 pathways. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon A.R., Vikis H.G., Stewart S., Fanburg B.L., Cochran B.H., Guan K.L. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- 34.Luissint A.C., Parkos C.A., Nusrat A. Inflammation and the intestinal barrier: leukocyte-epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology. 2016;151:616–632. doi: 10.1053/j.gastro.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Itallie C., Rahner C., Anderson J.M. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J Clin Invest. 2001;107:1319–1327. doi: 10.1172/JCI12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahner C., Mitic L.L., Anderson J.M. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology. 2001;120:411–422. doi: 10.1053/gast.2001.21736. [DOI] [PubMed] [Google Scholar]
- 37.Lili L.N., Farkas A.E., Gerner-Smidt C., Overgaard C.E., Moreno C.S., Parkos C.A., Capaldo C.T., Nusrat A. Claudin-based barrier differentiation in the colonic epithelial crypt niche involves Hopx/Klf4 and Tcf7l2/Hnf4-alpha cascades. Tissue Barriers. 2016;4 doi: 10.1080/21688370.2016.1214038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita H., Chiba H., Yokozaki H., Sakai N., Sugimoto K., Wada T., Kojima T., Yamashita T., Sawada N. Differential expression and subcellular localization of claudin-7, -8, -12, -13, and -15 along the mouse intestine. J Histochem Cytochem. 2006;54:933–944. doi: 10.1369/jhc.6A6944.2006. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka T., Horiuchi G., Matsuoka M., Hirano K., Tokumura A., Koike T., Satouchi K. Formation of lysophosphatidic acid, a wound-healing lipid, during digestion of cabbage leaves. Biosci Biotechnol Biochem. 2009;73:1293–1300. doi: 10.1271/bbb.80813. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T., Kassai A., Ohmoto M., Morito K., Kashiwada Y., Takaishi Y., Urikura M., Morishige J., Satouchi K., Tokumura A. Quantification of phosphatidic acid in foodstuffs using a thin-layer-chromatography-imaging technique. J Agric Food Chem. 2012;60:4156–4161. doi: 10.1021/jf300147y. [DOI] [PubMed] [Google Scholar]
- 41.Li C., Dandridge K.S., Di A., Marrs K.L., Harris E.L., Roy K., Jackson J.S., Makarova N.V., Fujiwara Y., Farrar P.L., Nelson D.J., Tigyi G.J., Naren A.P. Lysophosphatidic acid inhibits cholera toxin-induced secretory diarrhea through CFTR-dependent protein interactions. J Exp Med. 2005;202:975–986. doi: 10.1084/jem.20050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutsumi T., Inoue M., Okamoto Y., Ishihara A., Tokumura A. Daily intake of high-fat diet with lysophosphatidic acid-rich soybean phospholipids augments colon tumorigenesis in Kyoto Apc Delta rats. Dig Dis Sci. 2017;62:669–677. doi: 10.1007/s10620-016-4434-5. [DOI] [PubMed] [Google Scholar]
- 43.Kaji I., Roland J.T., Watanabe M., Engevik A.C., Goldstein A.E., Hodges C.A., Goldenring J.R. Lysophosphatidic acid increases maturation of brush borders and SGLT1 activity in MYO5B-deficient mice, a model of microvillus inclusion disease. Gastroenterology. 2020;159:1390–1405 e1320. doi: 10.1053/j.gastro.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martini E., Krug S.M., Siegmund B., Neurath M.F., Becker C. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4:33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khounlotham M., Kim W., Peatman E., Nava P., Medina-Contreras O., Addis C., Koch S., Fournier B., Nusrat A., Denning T.L., Parkos C.A. Compromised intestinal epithelial barrier induces adaptive immune compensation that protects from colitis. Immunity. 2012;37:563–573. doi: 10.1016/j.immuni.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naydenov N.G., Feygin A., Wang D., Kuemmerle J.F., Harris G., Conti M.A., Adelstein R.S., Ivanov A.I. Nonmuscle myosin IIA regulates intestinal epithelial barrier in vivo and plays a protective role during experimental colitis. Sci Rep. 2016;6:24161. doi: 10.1038/srep24161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan S., Alex P., Dassopoulos T., Zachos N.C., Iacobuzio-Donahue C., Donowitz M., Brant S.R., Cuffari C., Harris M.L., Datta L.W., Conklin L., Chen Y., Li X. Downregulation of sodium transporters and NHERF proteins in IBD patients and mouse colitis models: potential contributors to IBD-associated diarrhea. Inflamm Bowel Dis. 2009;15:261–274. doi: 10.1002/ibd.20743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janecke A.R., Heinz-Erian P., Muller T. Congenital sodium diarrhea: a form of intractable diarrhea, with a link to inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2016;63:170–176. doi: 10.1097/MPG.0000000000001139. [DOI] [PubMed] [Google Scholar]
- 49.Larmonier C.B., Laubitz D., Thurston R.D., Bucknam A.L., Hill F.M., Midura-Kiela M., Ramalingam R., Kiela P.R., Ghishan F.K. NHE3 modulates the severity of colitis in IL-10-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2011;300:G998–G1009. doi: 10.1152/ajpgi.00073.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janecke A.R., Heinz-Erian P., Muller T. Reduced NHE3 activity results in congenital diarrhea and can predispose to inflammatory bowel disease. Am J Physiol Regul Integr Comp Physiol. 2017;312:R311. doi: 10.1152/ajpregu.00545.2016. [DOI] [PubMed] [Google Scholar]
- 51.Daub H., Weiss F.U., Wallasch C., Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 52.Gonnella P.A., Siminoski K., Murphy R.A., Neutra M.R. Transepithelial transport of epidermal growth factor by absorptive cells of suckling rat ileum. J Clin Invest. 1987;80:22–32. doi: 10.1172/JCI113051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuwada S.K., Lund K.A., Li X.F., Cliften P., Amsler K., Opresko L.K., Wiley H.S. Differential signaling and regulation of apical vs basolateral EGFR in polarized epithelial cells. Am J Physiol. 1998;275:C1419–C1428. doi: 10.1152/ajpcell.1998.275.6.C1419. [DOI] [PubMed] [Google Scholar]
- 54.Chen M.C., Goliger J., Bunnett N., Soll A.H. Apical and basolateral EGF receptors regulate gastric mucosal paracellular permeability. Am J Physiol Gastrointest Liver Physiol. 2001;280:G264–G272. doi: 10.1152/ajpgi.2001.280.2.G264. [DOI] [PubMed] [Google Scholar]
- 55.Chen M.C., Solomon T.E., Kui R., Soll A.H. Apical EGF receptors regulate epithelial barrier to gastric acid: endogenous TGF-alpha is an essential facilitator. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1098–G1106. doi: 10.1152/ajpgi.00507.2001. [DOI] [PubMed] [Google Scholar]
- 56.Amerongen G.P.V., Vermeer M.A., van Hinsbergh V.W.M. Role of RhoA and Rho kinase in lysophosphatidic acid-induced endothelial barrier dysfunction. Arterioscl Throm Vas. 2000;20:E127–E133. doi: 10.1161/01.atv.20.12.e127. [DOI] [PubMed] [Google Scholar]
- 57.Murray D., Horgan G., MacMathuna P., Doran P. NET1-mediated RhoA activation facilitates lysophosphatidic acid-induced cell migration and invasion in gastric cancer. Br J Cancer. 2008;99:1322–1329. doi: 10.1038/sj.bjc.6604688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arthur W.T., Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell. 2001;12:2711–2720. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tushir J.S., D’Souza-Schorey C. ARF6-dependent activation of ERK and Rac1 modulates epithelial tubule development. EMBO J. 2007;26:1806–1819. doi: 10.1038/sj.emboj.7601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin S., Buel G.R., Nagiec M.J., Han M.J., Roux P.P., Blenis J., Yoon S.O. ERK2 regulates epithelial-to-mesenchymal plasticity through DOCK10-dependent Rac1/FoxO1 activation. Proc Natl Acad Sci U S A. 2019;116:2967–2976. doi: 10.1073/pnas.1811923116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao Y., Dickerson J.B., Guo F., Zheng J., Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hordijk P.L., ten Klooster J.P., van der Kammen R.A., Michiels F., Oomen L.C., Collard J.G. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 63.van Leeuwen F.N., Olivo C., Grivell S., Giepmans B.N.G., Collard J.G., Moolenaar W.H. Rac activation by lysophosphatidic acid LPA1 receptors through the guanine nucleotide exchange factor Tiam1. J Biol Chem. 2003;278:400–406. doi: 10.1074/jbc.M210151200. [DOI] [PubMed] [Google Scholar]
- 64.Guillemot L., Paschoud S., Jond L., Foglia A., Citi S. Paracingulin regulates the activity of Rac1 and RhoA GTPases by recruiting Tiam1 and GEF-H1 to epithelial junctions. Mol Biol Cell. 2008;19:4442–4453. doi: 10.1091/mbc.E08-06-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H., Bialkowska A., Rusovici R., Chanchevalap S., Shim H., Katz J.P., Yang V.W., Yun C.C. Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Kruppel-like factor 5. J Biol Chem. 2007;282:15541–15549. doi: 10.1074/jbc.M700702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee S.J., Ritter S.L., Zhang H., Shim H., Hall R.A., Yun C.C. MAGI-3 competes with NHERF-2 to negatively regulate LPA2 receptor signaling in colon cancer cells. Gastroenterology. 2011;140:924–934. doi: 10.1053/j.gastro.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stortelers C., Kerkhoven R., Moolenaar W. Multiple actions of lysophosphatidic acid on fibroblasts revealed by transcriptional profiling. BMC Genomics. 2008;9:387. doi: 10.1186/1471-2164-9-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plastira I., Bernhart E., Goeritzer M., DeVaney T., Reicher H., Hammer A., Lohberger B., Wintersperger A., Zucol B., Graier W.F., Kratky D., Malle E., Sattler W. Lysophosphatidic acid via LPA-receptor 5/protein kinase D-dependent pathways induces a motile and pro-inflammatory microglial phenotype. J Neuroinflammation. 2017;14:253. doi: 10.1186/s12974-017-1024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shah M., Patel K., Mukhopadhyay S., Xu F., Guo G., Sehgal P.B. Membrane-associated STAT3 and PY-STAT3 in the cytoplasm. J Biol Chem. 2006;281:7302–7308. doi: 10.1074/jbc.M508527200. [DOI] [PubMed] [Google Scholar]
- 70.Onishi A., Chen Q., Humtsoe J.O., Kramer R.H. STAT3 signaling is induced by intercellular adhesion in squamous cell carcinoma cells. Exp Cell Res. 2008;314:377–386. doi: 10.1016/j.yexcr.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.D’Amico S., Shi J., Martin B.L., Crawford H.C., Petrenko O., Reich N.C. STAT3 is a master regulator of epithelial identity and KRAS-driven tumorigenesis. Genes Dev. 2018;32:1175–1187. doi: 10.1101/gad.311852.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li C., Iness A., Yoon J., Grider J.R., Murthy K.S., Kellum J.M., Kuemmerle J.F. Noncanonical STAT3 activation regulates excess TGF-β1 and collagen I expression in muscle of stricturing Crohn’s disease. J Immunol. 2015;194:3422–3431. doi: 10.4049/jimmunol.1401779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qin H.R., Kim H.J., Kim J.Y., Hurt E.M., Klarmann G.J., Kawasaki B.T., Duhagon Serrat M.A., Farrar W.L. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68:7736–7741. doi: 10.1158/0008-5472.CAN-08-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung J., Uchida E., Grammer T.C., Blenis J. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol. 1997;17:6508–6516. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mandal T., Bhowmik A., Chatterjee A., Chatterjee U., Chatterjee S., Ghosh M.K. Reduced phosphorylation of Stat3 at Ser-727 mediated by casein kinase 2: protein phosphatase 2A enhances Stat3 Tyr-705 induced tumorigenic potential of glioma cells. Cell Signal. 2014;26:1725–1734. doi: 10.1016/j.cellsig.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 76.Davies S.P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borikova A.L., Dibble C.F., Sciaky N., Welch C.M., Abell A.N., Bencharit S., Johnson G.L. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J Biol Chem. 2010;285:11760–11764. doi: 10.1074/jbc.C109.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akunuru S., James Zhai Q., Zheng Y. Non-small cell lung cancer stem/progenitor cells are enriched in multiple distinct phenotypic subpopulations and exhibit plasticity. Cell Death Dis. 2012;3:e352. doi: 10.1038/cddis.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan Y., Zhou F., Zhang R., Claret F.X. Stat3 inhibitor Stattic exhibits potent antitumor activity and induces chemo- and radio-sensitivity in nasopharyngeal carcinoma. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Birkl D., Quiros M., Garcia-Hernandez V., Zhou D.W., Brazil J.C., Hilgarth R., Keeney J., Yulis M., Bruewer M., Garcia A.J., O’Leary M.N., Parkos C.A., Nusrat A. TNFalpha promotes mucosal wound repair through enhanced platelet activating factor receptor signaling in the epithelium. Mucosal Immunology. 2019 doi: 10.1038/s41385-019-0150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monteiro A.C., Sumagin R., Rankin C.R., Leoni G., Mina M.J., Reiter D.M., Stehle T., Dermody T.S., Schaefer S.A., Hall R.A., Nusrat A., Parkos C.A. JAM-A associates with ZO-2, afadin, and PDZ-GEF1 to activate Rap2c and regulate epithelial barrier function. Mol Biol Cell. 2013;24:2849–2860. doi: 10.1091/mbc.E13-06-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lisanti M.P., Caras I.W., Davitz M.A., Rodriguez-Boulan E. A glycophospholipid membrane anchor acts as an apical targeting signal in polarized epithelial cells. J Cell Biol. 1989;109:2145–2156. doi: 10.1083/jcb.109.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoo B.K., Yanda M.K., No Y.R., Yun C.C. Human intestinal epithelial cell line SK-CO15 is a new model system to study Na(+)/H(+) exchanger 3. Am J Physiol Gastrointest Liver Physiol. 2012;303:G180–G188. doi: 10.1152/ajpgi.00069.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krieglstein C.F., Cerwinka W.H., Sprague A.G., Laroux F.S., Grisham M.B., Koteliansky V.E., Senninger N., Granger D.N., de Fougerolles A.R. Collagen-binding integrin alpha1beta1 regulates intestinal inflammation in experimental colitis. J Clin Invest. 2002;110:1773–1782. doi: 10.1172/JCI200215256. [DOI] [PMC free article] [PubMed] [Google Scholar]