Abstract
Viruses use internal Ribosome Entry Sites (IRES) to hijack host ribosomes and promote cap-independent translation. While well-studied in bulk, the dynamics of IRES-mediated translation remain unexplored at the single-molecule level. Here, we developed a bicistronic biosensor encoding distinct repeat epitopes in two open reading frames (ORFs), one translated from the 5’-cap, the other from the Encephalomyocarditis Virus IRES. When combined with a pair of complementary probes that bind the epitopes co-translationally, the biosensor lights up in different colors depending on which ORF is translated. Using the sensor together with single-molecule tracking and computational modeling, we measured the kinetics of cap-dependent versus IRES-mediated translation in living human cells. We show that bursts of IRES translation are shorter and rarer than bursts of cap translation, although the situation reverses upon stress. Collectively our data support a model for translational regulation primarily driven by transitions between translationally active and inactive RNA states.
INTRODUCTION
While most eukaryotic mRNAs are translated in the canonical cap-dependent manner1, some eukaryotic messages and many viral RNAs use an internal ribosome entry site (IRES) to recruit ribosomes and initiate translation in a cap-independent manner. For example, important eukaryotic genes, such as VEGF, P53, MYC, and the HOX locus, all encode IRES elements to recruit ribosomes in conditions where canonical cap-dependent translation is repressed.2–5 Similarly, viruses have evolved IRES sequences to hijack host ribosomes during infection.6,7 Viral IRES sequences can be categorized into several distinct classes, each containing unique structural RNA motifs that attract different subsets of host translation factors,8,9 although most viral IRES sequences require fewer factors than the cap to support translation.7,8,10–12 Viruses exploit this during infection by triggering cellular stress to globally repress cap-dependent translation and free up host ribosomes. The end result is a large pool of host initiation factors and ribosomal subunits that are free to bind and initiate at viral IRES sequences at the peril of the host cell.13–22
Most previous experimental analyses of IRES-mediated translation use bicistronic transcripts encoding an IRES between two reporter proteins, 23,24 and quantify IRES activity by the ratio of upstream and downstream reporter expression. 9,25,26 Although bicistronic reporters contain IRES sequences out of context, their inherent one-to-one cap:IRES stoichiometry ensures that both cap and IRES experience nearly identical subcellular environments, making it possible to fairly compare cap-dependent and IRES-mediated translation. Another advantage is they allow a precise dissection of the IRES element itself, independent of other compounding factors.
So far, bicistronic reporters have been beneficial for deducing the relative IRES activity in cells hours or days after transfection.27 However, these studies have lacked the spatiotemporal resolution needed to visualize the sites of IRES translation and quantify translation initiation and elongation kinetics in real-time. This has made it difficult to assess the heterogeneity of IRES-mediated translation among individual RNA or within specific subcellular environments. Methods to study IRES-mediated translation with higher spatiotemporal resolution are needed to precisely understand how IRES-mediated translation differs from cap-dependent translation.
Here, we develop a real-time biosensor to quantify IRES-mediated translation dynamics with single-molecule resolution in living cells. We engineered complementary repeat epitopes into a bicistronic reporter, such that cap- versus IRES- translation could be monitored in two colors simultaneously from a single RNA using Nascent Chain Tracking (NCT).28 The resulting biosensor captures the dynamics of cap versus IRES translation with resolutions of tens-of-nanometers in space and sub-seconds in time. Application to the IRES from the Encephalomyocarditis Virus (EMCV) uncovers the spatial organization and dynamics of single IRES-mediated translation sites compared to those driven by the cap under normal and stressful conditions in living U2OS cells. Given the ubiquity of non-canonical translation initiation, we anticipate our biosensor will find broad application to understand the mechanisms of viral and eukaryotic IRES-mediated translation in diverse cellular conditions.
RESULTS
A multicolor biosensor to compare cap and IRES translation at the single-molecule level in living cells
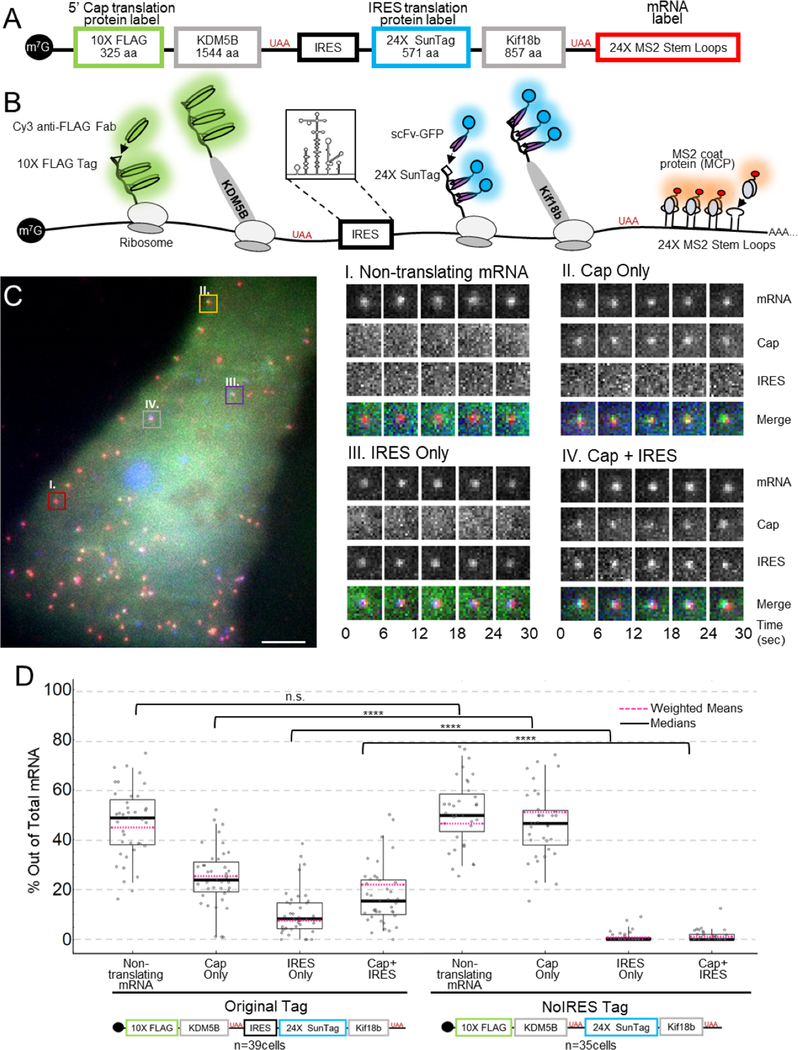
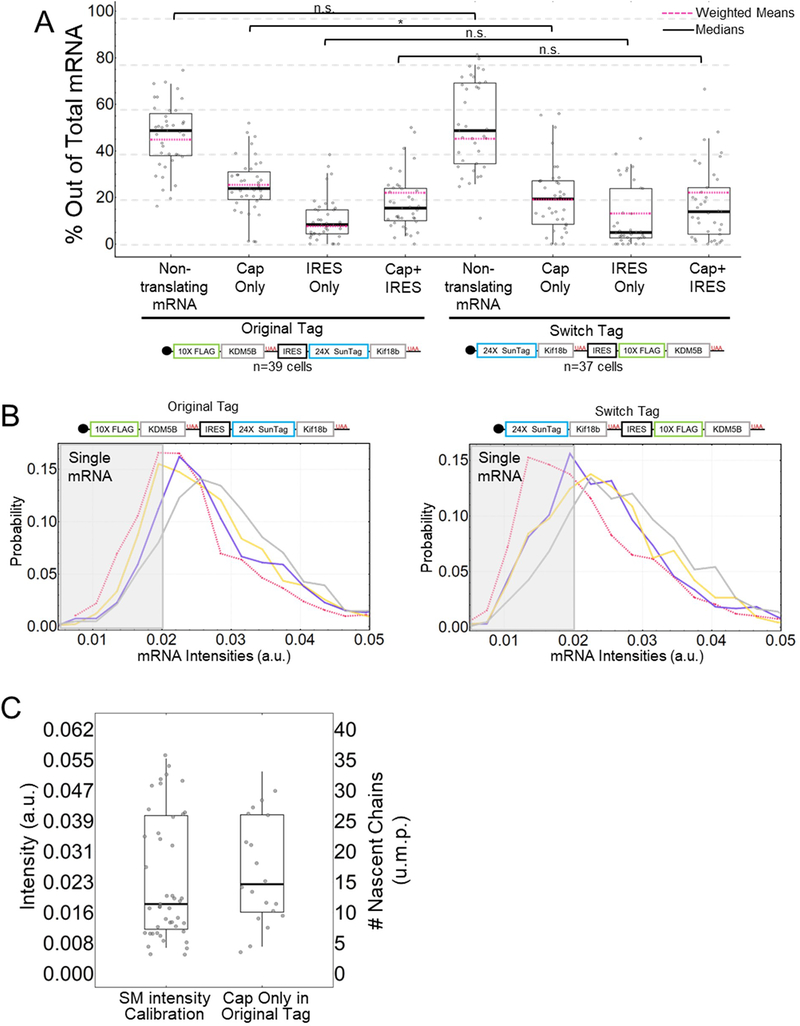
To fairly compare IRES and cap translation, we constructed a bicistronic NCT biosensor that is bound by different probes depending on the manner of translation initiation (Fig. 1a–b). Encoded under cap-dependent translation is a lysine demethylase KDM5B, N-terminally fused to a spaghetti monster tag (SM) encoding 10×FLAG epitopes. The FLAG SM tag is bound by fluorescently conjugated fragments of anti-FLAG antibodies (Fab), allowing visualization of cap translation soon after the first FLAG epitope emerges from the ribosome.28,29 Encoded under IRES-mediated translation is a Kinesin-like protein Kif18b, N-terminally tagged with 24×SunTag epitopes30 that are quickly bound by single chain variable fragments (scFv) fused to GFP as the SunTag epitope emerges from the ribosome.28,30 In addition, the biosensor contains 24×MS2 stem loops in the 3’ untranslated region (UTR) that are bound by Halo-tagged MS2 coat proteins (MCP) (Fig. 1a–b).29
Figure 1:
A multicolor biosensor to compare Cap and IRES translation at the single-molecule level in living cells. (A) Overview of the construct. (B) Schematic of the system. Cap-dependent protein reporter (green) is labeled by anti-FLAG Fab conjugated to Cy3 that binds the 10× FLAG peptide epitopes in the N-terminus. IRES-mediated protein reporter (blue) is labeled by a GCN4 scFv fused to a GFP that binds the 24× SunTag peptide epitopes. RNA (red) is marked by MCP-Halo labeled with JF646 that binds to repeated MS2 stem loops in the 3’ UTR. (C) Left. Representative cell imaged 6 hours after plasmid and probe loading. Different colored boxes within the cell illustrate different types of translation spots seen within a single cell. Right. Examples of co-moving spots. I – non-translating mRNA (red). II – single mRNA translating Cap Only (yellow). III – single mRNA translating IRES Only (purple). IV – single mRNA translating in a Cap and IRES manner (gray). (D) Quantification of species percentages out of total mRNA for both the Original Tag and the noIRES control. Each point represents the percent of that species in a cell. The p-values are based on a two-tailed Mann-Whitney test: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. The thick black line indicates the median and the dashed red line represents the weighted (by mRNA/cell) mean, the box indicates the 25–75% range, and the whiskers indicate the 5–95% range. Source data for panel d (Original Tag) is available online.
As a first application, we inserted the IRES element from EMCV into our biosensor. EMCV is a small single-stranded RNA virus that causes many mammalian diseases and which is widely used in research settings to coexpress distinct gene products from a single transcript.32,33.31 The EMCV IRES sequence is 553 nucleotides in length and contains a methionine start codon for the preferred open reading frame (ORF) (Fig. 1b).32,33 Previous assays have shown the EMCV IRES recruits ribosomes without the need of a 5’ cap or many canonical translation initiation factors.32,33 However, little is known about when, where, and how EMCV IRES-mediated translation occurs in living cells at the single-molecule level.
To visualize these aspects of EMCV IRES translation, we bead-loaded34 DNA encoding our biosensor, along with purified anti-FLAG Cy3-Fab, anti-SunTag GFP-scFv, and HaloTag-MCP into U2OS cells 3–6 hours before imaging. With this combination of probes, translation sites exhibit protein labeled by Fab or scFv co-moving with mRNA labeled by MCP. In addition to non-translating mRNA (Fig. 1c panel I), we identified translation sites labeled by just Fab (Fig. 1c panel II), just scFv (Fig. 1c panel III), and both Fab and scFv (Fig. 1c panel IV), indicating Cap-only translation, IRES-only translation, and Cap+IRES translation, respectively (Supplementary Movie 1).
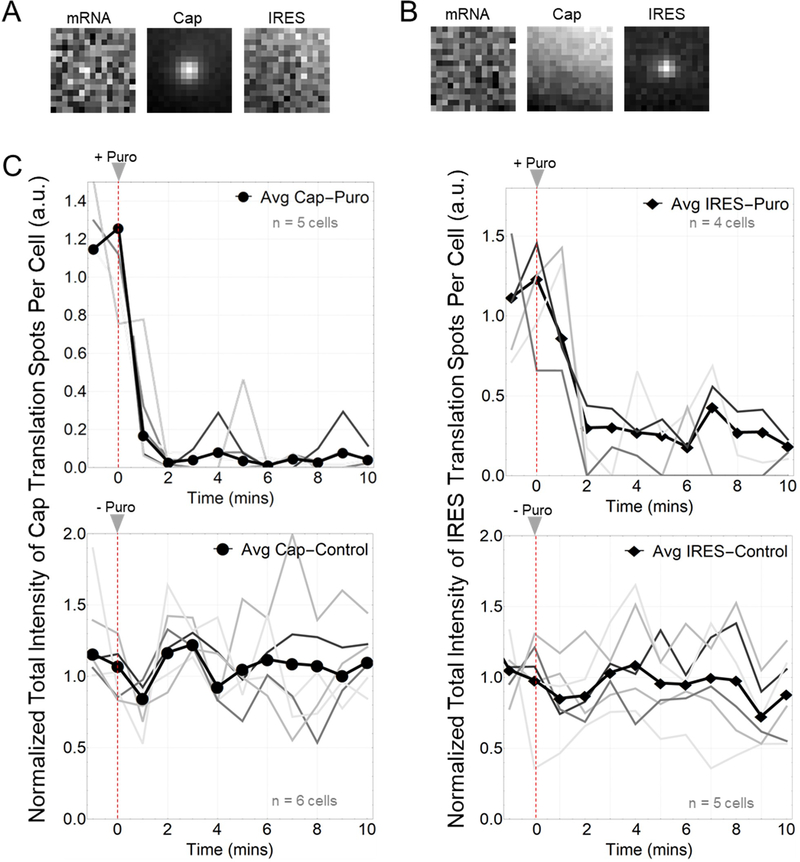
We performed two control studies to confirm that spots were active translation sites. First, to rule out fluorescence bleed-through from the protein channels to the mRNA channel, we repeated experiments without labeling mRNA. Regardless of the intensity of the translation signal, no bleed-through was observed in the mRNA channel (Extended Data Fig. 1a–b). All other bleed-through possibilities were ruled out by direct observations of distinct populations of non-translating mRNA, IRES-only translation sites, and Cap-only translation sites. Second, to show that the translation sites were active, we treated cells with 50 ug/mL puromycin, an elongation inhibitor that releases nascent chains from ribosomes,35 and we confirmed a rapid disappearance of all Fab or scFv translation signals within translation sites (Extended Data Fig. 1c).
To better quantify the heterogeneity of translation, we took 2.5 minute movies (25 frames x 13 z-planes per volume x 3 colors = 975 images per movie) of 39 cells from eight experiments. Movies were acquired using HILO illumination to maximize signal-to-noise.36 In total, we observed 3748 mRNA, of which 1784 were being translated. 24% of mRNA were translated in a Cap-only manner, 8% in an IRES-only manner, and 15% in a cap- and IRES-manner simultaneously (Cap+IRES) (Fig. 1d, left). As a control, we removed the IRES element from our biosensor, and we observed almost complete reduction in anti-SunTag GFP-scFv signals (Fig. 1d, right), ruling out ribosomal run-through from cap to IRES. All else equal, these data demonstrate that the IRES element alone can capture and initiate host ribosomes, but not as efficiently as the cap in a bicistronic context. Given the large fraction of transcripts we observed being translated in both a cap- and IRES-manner, these data also demonstrate that a single bicistronic transcript can be translated simultaneously in two open reading frames.
The EMCV IRES can be translated throughout the entire cell cytoplasm
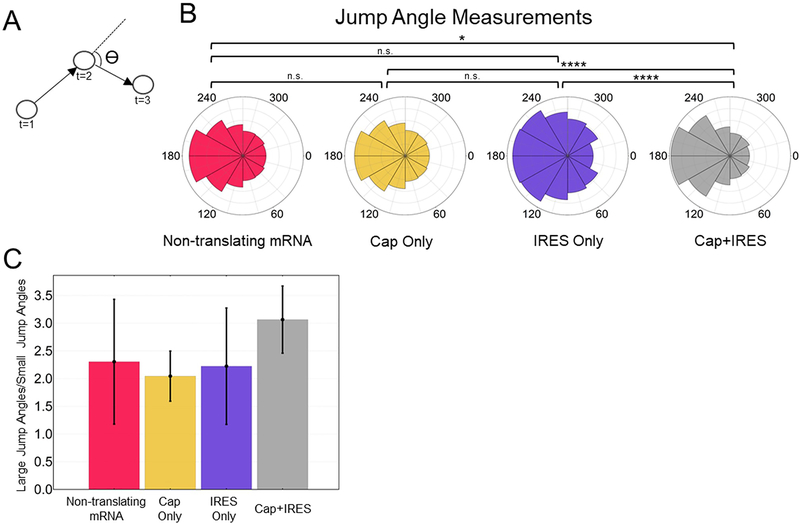
We wondered why there were fewer EMCV IRES translation spots compared to cap. One possibility is the IRES sequence itself requires a specific environment. For example, EMCV IRES-mediated translation is known to occur around replication complexes that localize to specific regions of cells31. While this compartmentalization is likely mediated by many factors, the IRES sequence itself could play an active role in the process or require proper localization for translation. To test this, we quantified the propensity for IRES-mediated translation sites to compartmentalize in four ways. We first measured (1) the tendency of IRES-mediated translation sites to cluster and (2) their average distance to the nuclear periphery. This revealed the EMCV IRES has no preference for clustering (Extended Data Fig. 2a) and only a minor preference for translation in the perinuclear region (Extended Data Fig. 2b). We next tracked individual translation sites (Supplementary Movie 2) to quantify (3) their mobilities and (4) their degrees of confinement. From the 3771 mRNA we tracked, we calculated their cumulative distributions of diffusion coefficients and average mean squared displacements (Extended Data Fig. 2c–d). In both analyses, IRES-only translation sites were statistically indistinguishable from cap-only translation sites. Likewise, the confinement of IRES-only and cap-only translation sites, as quantified by a preference for 180 degree jumps over 0 degree jumps,37 were also indistinguishable (Extended Data Fig. 2e–f). According to all four metrics, IRES-mediated translation sites localize and move within cells similarly to cap-dependent translation sites. Collectively, these data suggest the EMCV IRES does not require a specialized environment for translation.
IRES and cap translation sites have a similar size and ribosomal organization
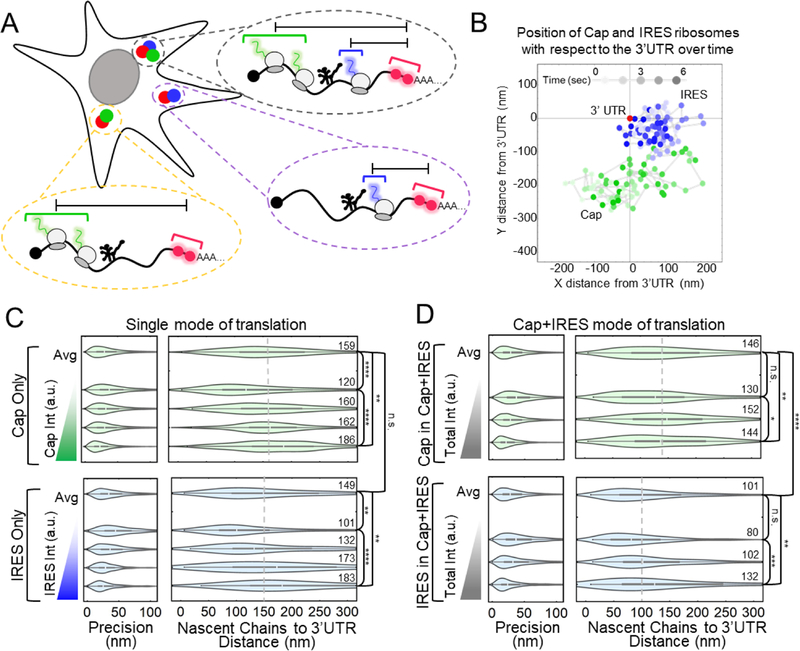
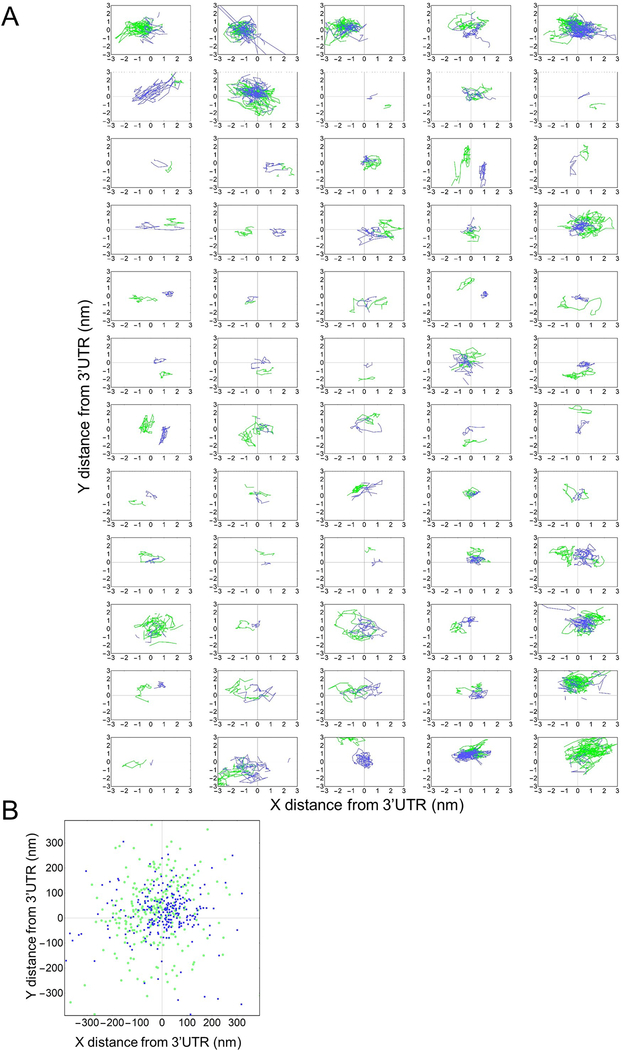
Since we could not distinguish IRES-mediated translation sites based on their subcellular localization or mobility, we wondered if there were other features that could explain the lower probability of observing IRES-mediated translation sites. In particular, we wondered if there were differences in the size of single IRES translation sites compared to single cap translation sites. Recently, the Zenklusen38 and Parker39 laboratories used smFISH to show that cap translating ribosomes tend to stretch out translation sites, i.e., heavily translated mRNA cover a greater volume within the cell, in contrast to models of mRNA looping.40 Because IRES-mediated translation sites do not require looping factors, they could have a different ribosomal organization than cap translation sites.31 To test this, we took advantage of the long ORFs in our biosensor. The 1D distance from the center of the cap ORF and IRES ORF to the center of the 3’UTR marked by the MS2 signal is 8.5 kb and 3 kb, respectively (Fig. 1a). Assuming cap and IRES ribosomes initiate stochastically, their average positions provide an approximation for the center of each ORF. This allowed us to measure the distance between the centers of the IRES ORF, cap ORF and 3’UTR, all within single translation sites (Fig. 2a–b, Extended Data Fig. 3).
Figure 2:
IRES and Cap translation sites stretch out as ribosomes load. (A) Schematic showing how the measurements from the 3’UTR to the Cap and IRES nascent chains were conducted within single cells. (B) Graph showing IRES and Cap nascent chain positions relative to 3’UTR through time of a representative Cap+IRES translation spot. X and Y distances displayed in nanometers (nm) and time (sec) is represented as a gradient in spot color. 75 frames were imaged at a rate of 77 msec per frame. (C) Distributions of the distance between Cap or IRES nascent chains and the 3’UTR in Cap Only (top, n=793) and IRES Only (bottom, n=213) translation sites. In each box, the average of all translation sites is shown on top (Avg), and equal-sized subsets sorted by their total nascent chain signal intensity below. Precision is estimated from the distribution of distances between two consecutive timepoints, e.g. any two connected points in B. The median distance is reported to the right of each distribution. (D) Distributions of the distance between Cap (top, n=296) or IRES (bottom, n=259) nascent chains and the 3’UTR in Cap+IRES translation sites. For the box and whisker plots, the white lines indicate the medians, the boxes indicate the 25–75% range, and the whiskers indicate the 5–95% range. The p-values are based on a two-tailed Mann-Whitney test: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Source data for panels c and d are available online.
We began with Cap-only and IRES-only translation sites, to see if they differ in size. In contrast to this, the median distance between cap-dependent ribosomes and the 3’UTR in Cap-only sites was 159 nm, a value statistically indistinguishable from the 149 nm median distance we measured between IRES-mediated ribosomes and the 3’UTR in IRES-only translation sites (Fig. 2c). This similarity suggests that when the IRES ORF is not being translated (as in Cap-only sites), it is compact. Furthermore, by ranking translation sites by their total intensity (i.e., total ribosomal content or degree of translation), we found that as the brightness of the Cap-only or IRES-only signals increased, the distance between those signals and the MS2 signal marking the 3’ UTR also increased (Fig. 2c). These data corroborate in living cells what the Zenklusen38 and Parker39 labs observed in fixed cells; namely, that translating ribosomes tend to stretch out mRNA. However, the similarity in the size of Cap-only and IRES-only translation sites suggests these features alone cannot account for the relatively low probability of IRES-mediated translation.
For completeness, we examined the size of Cap+IRES translation sites (Fig. 2d). In these sites, the median distance between cap-dependent ribosomes and the 3’UTR was 146 nm, similar to the 159 nm distance we measured in Cap-only translation sites (p-value = 0.42). In contrast, the median distance between IRES-mediated ribosomes and the 3’UTR was just 101 nm, significantly less than the 149 nm distance we measured in IRES-only translation sites (p-value = 6E-6). These data indicate the upstream cap-dependent ribosomes restrict the freedom of downstream IRES-mediated ribosomes, causing them to spread out less. Here it makes sense that IRES-mediated ribosomes tend to be closer to the 3’UTR than cap-dependent ribosomes because the IRES itself is considerably 3’ of the cap. Finally, we again confirmed that as the brightness of translation signals increased, the distances between these signals and the MS2 signal marking the 3’ UTR also increased (Fig. 2d).
The EMCV IRES recruits and initiates 2–3 times fewer ribosomes than the cap
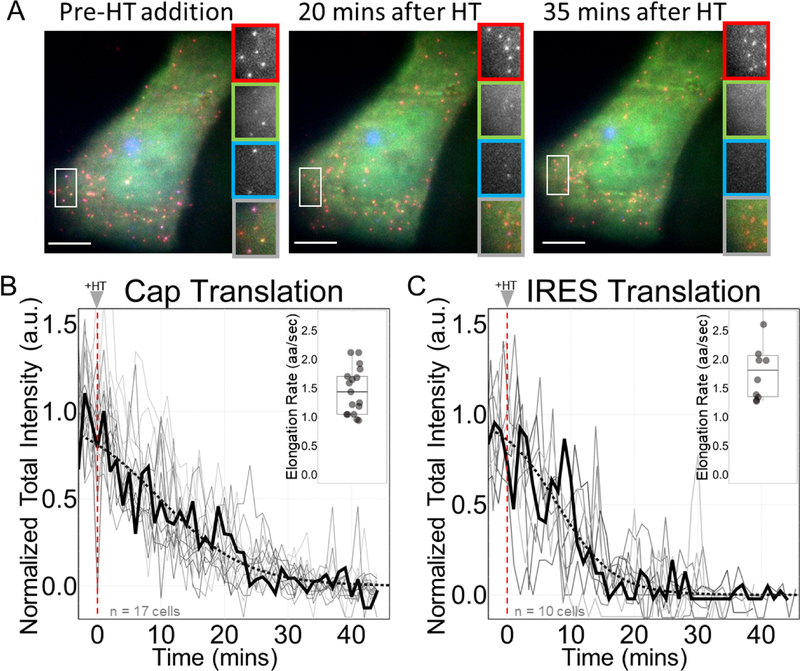
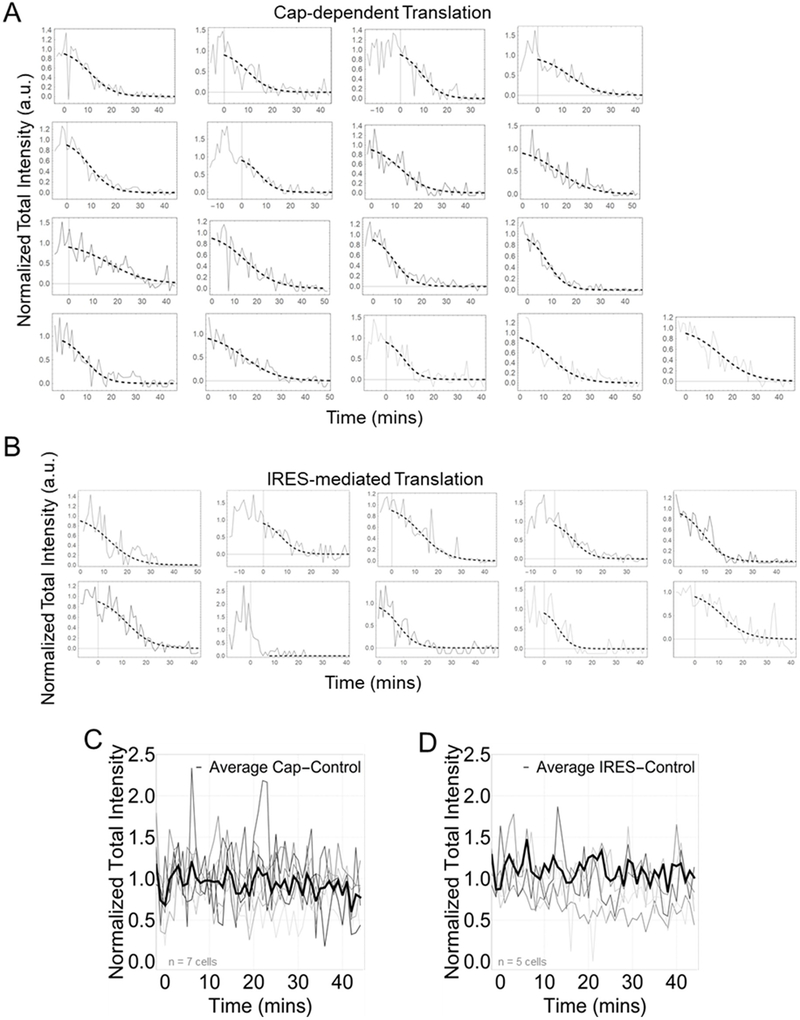
Having measured the prevalence, subcellular distribution, and spatial organization of single EMCV IRES translation sites, we next focused on their dynamics. For this, we inhibited translation initiation using Harringtonine41 so all elongating ribosomes runoff each ORF and translation fluorescence signals decay (Fig. 3 and Extended Data Fig. 4).42 Fits to the decays yielded similar run-off completion times of 45 ± 6 min for cap-dependent ribosomes versus 43 ± 17 min for IRES-mediated ribosomes (corresponding to relatively43 slow average elongation rates of 1.44 ± 0.40 aa/sec and 1.81 ± 2.39 aa/sec, respectively). Since these run-off times are similar, we conclude that elongation is not responsible for the low probability of observing IRES translation.
Figure 3:
Elongation is not a rate-limiting step lowering the efficiency of IRES-mediated translation. (A) Cells before harringtonine (HT) addition, 20 minutes after HT addition, and 35 minutes after HT addition with crops of the mRNA channel (red), Cap channel (green), IRES channel (blue), and merge (gray). Scale bars represent 10μm. (B-C) Normalized total intensity of nascent chains in Cap-dependent (left) and IRES-mediated translation sites (right). Each gray line represents a single cell treated with HT. The black line shows a representative cell. The dotted black line is a phenomenological fit of the representative cell. The inset shows the estimated elongation rates of each cell in amino acids per second (aa/sec). The thick black line indicates the median. The box indicate the 25%−75% range, and the whiskers indicate the 5%−95% range. All cells were imaged for 45 minutes with a 1-minute interval between each capture. Harringtonine was added after 5 captures marked by the red dotted line at time point 0. Intensity values are in arbitrary units (a.u.). n=17 cells for Cap Translation. n=10 cells for IRES Translation. Source data for panels b and c are available online.
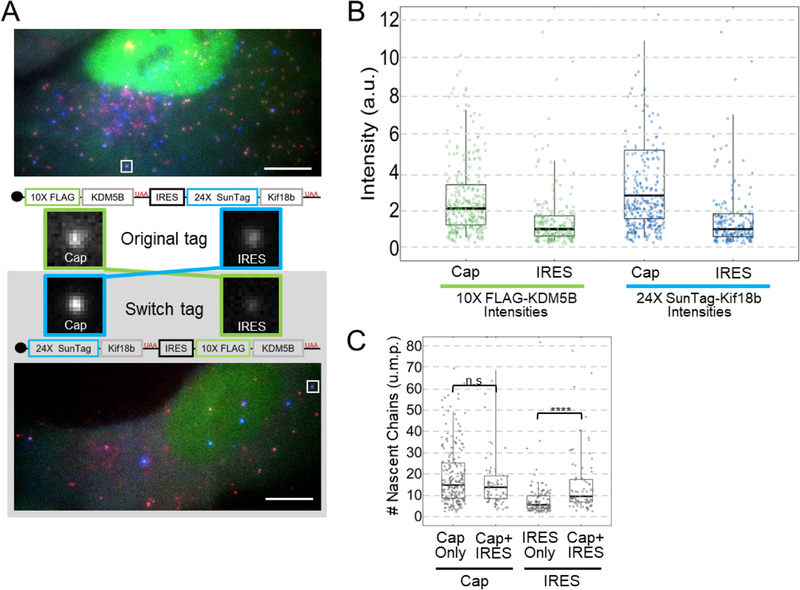
We next turned our attention to ribosome recruitment and initiation. To accurately measure these, we needed to compare the intensities of nascent chain signals within translation sites. A direct comparison was not possible because the cap-dependent and IRES-mediated nascent chains differ in sequence and number of tags, and are labeled by complementary fluorophores and probes with different binding kinetics and quantum yields. To enable a fairer comparison, we developed a “Switch Tag” in which the reporters were swapped (Fig. 4a). This allowed us to compare the same reporter under the control of both the cap (e.g., in the Original Tag) and the IRES (in the Switch Tag). In this way, we could ensure differences in the intensity of translation sites would reflect differences in ribosome recruitment or initiation dynamics rather than differences in fluorophore or probe detection kinetics or codon biases within epitope tags.
Figure 4:
The Cap recruits and initiates 2–3 times more ribosomes than the IRES. (A) One representative cell expressing the Original Tag (from n=39 cells) (top) or the Switch Tag (from n=37 cells) (bottom) with a Cap+IRES translation spot highlighted by the white square. Scale bars are 10μm. Crops of the representative sites are shown in the middle. The construct schematic with the corresponding crop illustrates how the intensity comparisons between Cap and IRES were conducted. (B) Box and Whisker plots showing the intensity comparisons between Cap and IRES. Left graphs shows intensity comparisons of 10× FLAG-KDM5B nascent chain signals from Cap in the Original Tag (n=302 spots) and IRES in the Switch Tag (n=167 spots). Right graphs shows intensity comparisons of 24× SunTag-Kif18b from Cap in the Switch Tag (n=262 spots) and IRES in the Original Tag (n=201 spots). Intensity measurements are in arbitrary units (a.u.). (C) Intensities of Cap in Cap Only (Original Tag) translation sites were compared to Cap in Cap+IRES (Original Tag), IRES in IRES Only (Switch Tag) and IRES in Cap+IRES (Switch Tag) to obtain numbers of ribosomes in units of mature protein (u.m.p.) on all types of translating species. Cap Only sites (n=226 spots) have a median of 14.6 ribosomes, Cap+IRES sites (n=76 spots) have a median of 13.6 Cap-dependent ribosomes and 9.4 IRES-mediated ribosomes. IRES Only sites (n=121 spots) have a median of 5.4 ribosomes. The p-values are based on a two-tailed Mann-Whitney test: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. For the box and whisker plots, the thick black lines indicate the medians, the boxes indicate the 25–75% range, and the whiskers indicate the 5–95% range. Source data are available online.
Reassuringly, when the Switch Tag was expressed in cells, it yielded nearly the same percentages of each type of translation site as the Original Tag (Extended Data Fig. 5a), demonstrating the 10×FLAG and 24×SunTag reporters do not interfere with translation dynamics and have similar detection efficiencies. A direct comparison of the intensity of translation sites encoding 10×FLAG-KDM5B initiated in a cap-dependent manner (from the Original Tag) versus an IRES-mediated manner (from the Switch Tag) gave a median intensity ratio of 2.1 ± 0.1 (Fig. 4b). Similarly, a direct comparison of the intensity of translation sites encoding 24×SunTag-Kif18b initiated in a cap-dependent manner (from the Switch Tag) versus an IRES-mediated manner (from the Original Tag) gave a median intensity ratio of 2.8 ± 0.2 (Fig. 4b, Extended Data Fig. 5b).27 The similarity of the ratios we measured indicate the presence of two to three times fewer ribosomes in IRES-mediated translation sites than cap-dependent translation sites.
To obtain absolute ribosome occupancies, we developed a calibration construct that forms translation sites with a known number of ribosomes (further details in the Methods) (Extended Data Fig. 5c). Comparing the intensity of these translation sites to 10×FLAG-KDM5B translation sites in the Original and Switch Tags (Fig. 4c) revealed that Cap+IRES translation sites have a median of 13.6 cap-dependent ribosomes and a median of 9.4 IRES-mediated ribosomes, while Cap-only translation sites have a median of 14.6 ribosomes (p value = 0.196), and IRES-only translation sites have a median of just 5.4 ribosomes (p value = 5.83E-8). Thus, cap-dependent translation sites have more ribosomes than IRES-mediated translation sites, consistent with the higher percentage of mRNA translated in a cap-dependent versus IRES-mediated manner. These data demonstrate that under normal physiological conditions IRES-mediated translation of the biosensor is less productive than cap-dependent translation, both at the population and single-molecule levels.
Computational modeling reveals ribosomal recruitment limits IRES translation
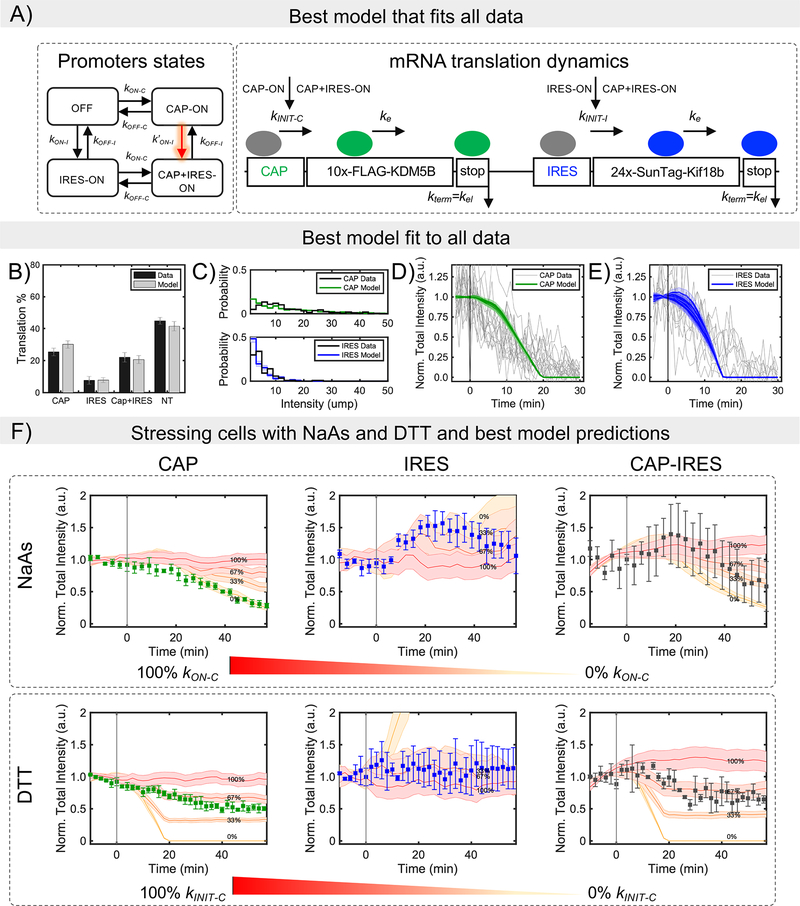
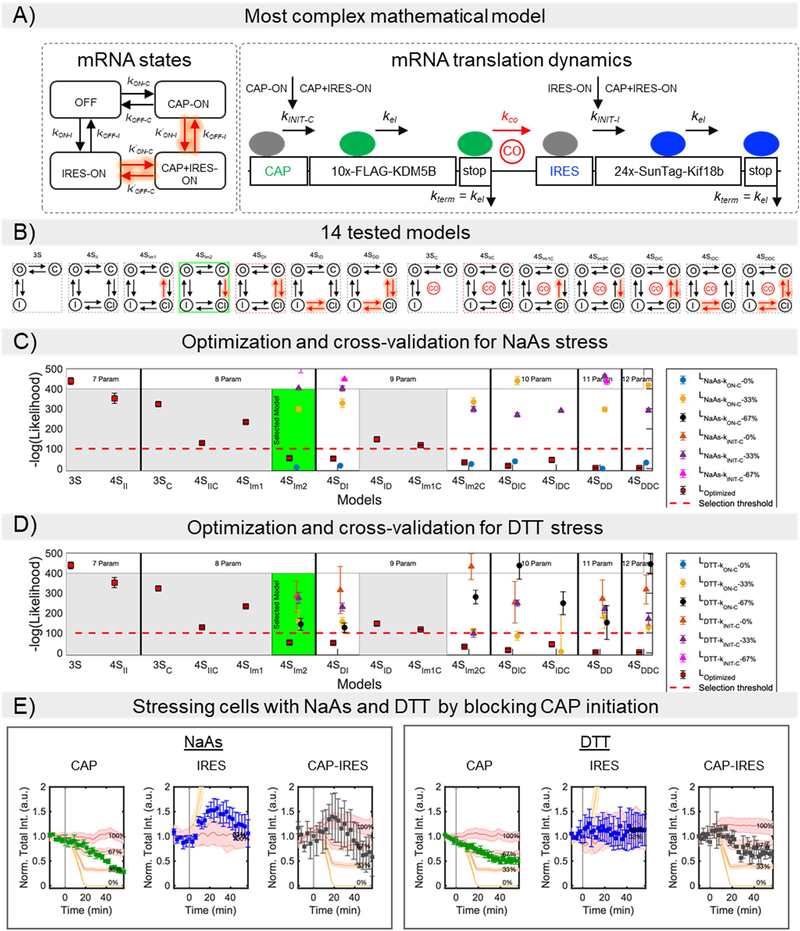
According to our experiments, the relatively low probability of IRES-mediated translation is due to rate-limiting steps that precede elongation, presumably either ribosome recruitment or initiation. To distinguish these possibilities, we developed a set of models with varying levels of complexity. All models consider the kinetics of individual ribosomes translating along an mRNA, with stochastic initiation and codon-dependent elongation proportional to the prevalence of the associated tRNA in the human genome.43 Models differ in the number of states an mRNA can transition between: Three-state models include an inactive mRNA state (OFF), an active mRNA state that allows cap translation (Cap-ON), and an active mRNA that allows IRES translation (IRES-ON). Four-state models include an additional active mRNA state (Cap+IRES-ON) that allows both cap and IRES-translation (Fig. 5a).
Figure 5:
Modeling bicistronic translation of the multicolor biosensor. (A) The mathematical model considers four mutually exclusive RNA states: non-translating (OFF), Cap-dependent (CAP-ON), IRES-mediated (IRES-ON), and both Cap and IRES (CAP+IRES-ON), in which initiation can take place from the Cap or IRES as indicated. Elongation and termination processes continue independent of RNA state. To capture interdependence between Cap and IRES states, multiple hypotheses were tested, and the best model was selected after parameter optimization and model reduction (Figure S6C–D). The selected model considers 4 promoter states, in which the IRES activation rate depends on the Cap-state (model 4SIm2). (B) Mean values and SD for the model and mean values and SEM for experimental data for the fraction of Cap-dependent (Cap), IRES-mediated (IRES), both Cap-dependent and IRES-mediated (Cap+IRES), and non-translating spots (NT). The prevalence of translation events are shown as the percentage of total RNA. For simulation n = 4000 trajectories were used, SD was calculated using bootstrapping (X50 with sampling of 300 trajectories). For experimental data, n=39 cells. (C) Experimental and model intensity distributions for Cap-dependent and IRES-mediated translation. Distributions consider only those spots that have intensities greater than or equal to one unit of mature protein (u.m.p.). (D) Decrease in intensity after Harringtonine application for Cap-mediated translation spots and (E) IRES-mediated translation spots. To denote variability, 10 independent model simulations are plotted in D and E. (F) Experimental data and simulated predictions for translation inhibition by the chemical stressors NaAs and DTT. Chemical stressors were simulated by reducing the Cap activation rates at the RNA state level (i.e. blocking kON-C) for NaAs and by reducing Cap-initiation (i.e. blocking kINIT-C) rates for DTT. Experimental data are represented by the square symbols. Errors bars in the experimental data are the mean and SEM and for the simulations mean and SD. NaAs: n=32 cells. DTT: n=28 cells. Simulations: n = 4,000 trajectories. The values given in the figure represents the percentage of inhibition. Cap, Cap-only spots; IRES, IRES-only spots; and Cap-IRES, Cap translation intensity in spots with both Cap and IRES intensities. Source data for panels b-f are available online.
The stochastic dynamics for all models were simulated over large ranges of potential parameters and automated searches were conducted to identify combinations of mechanisms and parameters that maximize the likelihood of all data, including the fraction of translating spots (Fig. 1), Harringtonine run-off kinetics (Fig. 3), and the translation site intensity distributions (Fig. 4; also see Computational Methods and Fig. 5b–e). In total, we considered 14 unique models with between 7 and 12 free parameters, some of which included interdependence between cap- and IRES- translation, either in the form of enhanced transition rates between states or via reinitiation of ribosomes from cap to IRES (Equations 1 and 2; See Computational Methods and Extended Data Fig. 6a–b). The simplest model that reproduces all data has eight parameters (Fig. 5a and Table 1): (1) a baseline elongation rate of = 1.7 aa/sec, agreeing with our earlier estimate and consistent with previously measured rates,44,45 (2) an initiation rate kINIT-C ~ 1/21 sec−1 for cap-dependent translation; (3) an initiation rate kINIT-I~ 1/20 sec−1 for IRES-mediated translation; (4) cap activation bursts with refractory periods (1/kON-C) of 34.5 min and (5) durations of (1/kOFF-C) of 8.3 min, leading to the synthesis of kINIT-C /kOFF-C = 24 nascent proteins on average per cap burst; (6) In the absence of cap, the model predicts that typical bursts of IRES translation would have a refractory period (1/kON-I) of 91.3 min and (7) a duration of (1/kOFF-I) of 2.5 min, leading to the synthesis of 7.5 nascent proteins on average per IRES burst. According to these fitted parameters, the efficiency of IRES translation is not limited by initiation (since kINIT-I ~ kINIT-C ~ 1/20 sec−1), but rather the IRES spends less time in a translationally active state that can recruit ribosomes. In addition to the above seven parameters (which can be reduced to six by setting the cap and IRES initiation rates equal), one additional parameter was required to fit the data: an enhancement in IRES activation when cap translation is on (i.e. k’ON-I > kON-I). Specifically, in the presence of cap, the IRES refractory period is reduced from 91.3 min to 11 min, leading to a 6.9 fold increase in IRES translation. This enhancement was required to capture the large percentage of Cap+IRES translation sites (which is greater than one would predict if cap and IRES translation were independent) (Fig. 1d, left) and the larger number of IRES-mediated ribosomes in Cap+IRES translation sites compared to IRES-only translation sites (Fig. 4c). These data and the best-fit model therefore provide evidence that translation of an upstream ORF can positively impact translation of a non-overlapping downstream ORF.
Table 1. Estimated parameter values for the final selected model.
4 promoter states and promoter activation for IRES influenced by Cap.
| Parameter | Description | Value |
|---|---|---|
| kINIT–C | Initiation rate for Cap | 4.8×10−2± 4.6×10−2 sec−1 |
| kINIT–I | Initiation rate for IRES | 4.9×10−2± 5.3×10−2 sec−1 |
| *ke | Average elongation rate | 1.7 ± 0.18 aa/sec |
| kON–C | Transition rate from Soff to SCAP or SIRES to SCAP–IRES | 4.8×10−4± 9.7×10−5 sec−1 |
| kOFF-C | Transition rate from SCAP to Soff. | 2×10−3± 1.8×10−3 sec−1 |
| kON–I | Transition rate from Soff to SIRES. | 1.8×10−4± 5.5×10−5 sec−1 |
| kOFF–I | Transition rate from SIRES to Soff. | 6.6×10−3± 2.7×10−3 sec−1 |
| k′ ON–I | Transition rate from SCAP to SCAP–IRES. | 1.5×10−3±1.0×10−3 sec−1 |
*ke is the elongation rate calculate as the average elongation of all codons.
Predicting cap and IRES translation in response to specific cellular stresses
It is well known that viral infections cause increased levels of cellular stress. During stress, cap-dependent translation decreases, while viral RNAs continue to be translated, in part due to IRES-mediated ribosomal recruitment to viral RNAs. Extensive studies have shown that both viral and some endogenous IRES sequences remain translationally active during cellular stress.13,15,16
To visualize the impact of stress on IRES-mediated translation at the single-molecule level, we exposed cells expressing our biosensor to NaAs46, which induces oxidative stress, and DTT47, which induces ER stress. Because our reporter is bicistronic, the cap and IRES experience the same microenvironment during stress, thereby providing an internal control to fairly compare the impact of stress on cap versus IRES translation. Upon exposure to both types of stress, the intensity of Cap-only translation sites decreased (Fig. 5f, left). In contrast, the intensity of IRES-only translation sites remained steady or increased (Fig. 5f, middle), while Cap+IRES translation sites displayed an intermediate response (Fig. 5f, right). These data demonstrate EMCV IRES translation is robust to cellular stress compared to cap translation, as is necessary for efficient viral translation in cells during infection.
We next used our best-fit model to predict the stress-response. We hypothesized stress decreases cap-dependent translation in one of two ways (Supplementary Note 1). Either translation is inhibited at the level of ribosome recruitment by blocking mRNA activation (by decreasing kON-C ) (Fig. 5f), or translation is inhibited at the level of initiation (by decreasing kINIT-C ) (Extended Data Fig. 6e). We tested each mechanism assuming 33%, 67% and 100% reductions to the corresponding rate (Extended Data Fig. 6c–d). The best fit models suggest that oxidative and ER stress affect cap-translation via slightly different mechanisms. NaAs stress is best predicted by blocking 100% of cap activation (log-likelihood of 187 ± 4 vs 582 ± 2 for the model in which cap-initiation is blocked), while DTT stress is best predicted by blocking ~33% of cap-initiation (log-likelihood of 599 ± 9 vs 1026 ± 52 for the model in which cap activation is blocked).
DISCUSSION
Since their discovery in 199848, IRES sequences have been studied intensely to elucidate their mechanisms to recruit ribosomes in a cap-independent manner1. However, limited spatiotemporal resolution had prevented the real-time analysis of IRES-mediated translation. To confront this problem we created a unique bicistronic biosensor to quantify when, where, and to what degree the EMCV IRES is translated at the single-molecule level in living cells. According to our data, the EMCV IRES is an excellent mimic of the cap, capable of being translated in similar subcellular environments, moving with similar kinetics, and sharing a similar underlying ribosomal organization upon translation. Furthermore, our computational fits reveal that just like the cap, the EMCV IRES can initiate ribosomes at a rate of ~1 ribosome every 20 seconds. However, despite these similarities, we find the IRES only recruits one ribosome for every two or three recruited to the cap under normal conditions. This inefficiency arises because the IRES spends less time than the cap in a translationally active state capable of recruiting ribosomes. Cellular stress reverses the situation, with the EMCV IRES continuing unperturbed, but the cap now spending more time in an inactive state. Our collective data therefore support a model in which the transition from a translationally inactive to active RNA state (capable of recruiting ribosomes) is the main factor controlling the balance between cap and IRES translation.
A hallmark of IRES-mediated translation is its dependence on a subset of host translation factors.32 Due to these lax requirements, one would think that translation of the IRES could occur in special microenvironments that, for example, are enriched or depleted in specific translation factors. For example, EMCV IRES translation may occur around replication complexes.31 Using our biosensor, we find little evidence this process is facilitated by the IRES sequence alone. Instead, the EMCV IRES appears to be translated all throughout the cell cytoplasm in a manner that does not involve clustering and does not alter translation site mobility. This lack of distinction means the EMCV IRES does not require a specialized environment for translation. Furthermore, it demonstrates additional factors are needed to localize EMCV IRES translation during viral infection.31 In the future it will be interesting to determine which factors are required for localization by creating biosensors that place the EMCV IRES in more natural monocistronic viral contexts.
According to our results, both the EMCV IRES and cap transition between translationally active and inactive states. Moreover, these transitions appear to be modulated to control translational output similar to how promoter activation and deactivation control transcriptional bursts.44 This general principle of regulation could be the result of sharing a common subset of translation factors. According to our best-fit model, bursts of IRES translation are both shorter in duration (2.5 min for IRES versus 8.3 min for cap) and separated by longer inactive refractory periods (91.3 min for IRES versus 34.5 min for cap) than bursts of cap translation. Given the complex structure of the EMCV IRES,9 which presumably undergoes dynamic changes, our results suggest the IRES has trouble adopting and maintaining a conformation that can recruit ribosomes and support translation. In contrast, the cap relies on a larger set of factors, including the cap-binding protein eIF4E and scaffolding protein eIF4G. Presumably these additional factors work together to better maintain a conformation that is attractive to ribosomes and more amenable to translation.
An interesting observation with our biosensor was that cap translation enhances IRES translation, but not the other way around. This make sense given the subset of factors the IRES requires compared to the cap. When cap translation is on, all factors necessary for IRES translation are present at high concentrations. This would enhance the probability the IRES gets translated. In contrast, when the IRES is on, not all factors required for cap translation are available, including eIF4E and eIF4G. Without these factors, cap-dependent translation is not enhanced.
The molecular mechanisms that govern the enhancement of IRES-mediated translation in the presence of cap-dependent translation in our biosensor remain unclear. One possibility given our live-cell confirmation that ribosomes stretch out translation sites is that the stretching alters the structure of the IRES. This could impact the IRES in a number of ways. The IRES could be stabilized (increased kOFF), its folding could become faster (increased kON), or ribosomes coming off the cap could reinitiate at the IRES (the addition of kCO). According to our simulations, all of these improve the fits to our data, but faster folding alone was sufficient to improve the fit to near optimum values (see Extended Data Fig. 6c). Thus, our data suggest the stretching out of actively translated transcripts may help the IRES transition into a translationally active state.
Despite the lower overall translation efficiency of the EMCV IRES compared to the cap under normal conditions, the upside of relying on a subset of factors is IRES-mediated translation can persist and surpass cap-dependent translation during stress, a situation viruses have evolved in their ongoing arms race with eukaryotic cells. We see that in NaAs stress, IRES-mediated translation remains strong, presumably because this stress targets eIF4E,20 which is not required for IRES translation. Though IRES translation also remained strong compared to cap in DTT stress, the effect was smaller than with NaAs, presumably because DTT stress impacts a different set of translation factors than NaAs. In the future, it will be interesting to investigate which factors play the biggest roles and also which IRES sequences are most robust.
Our technology to monitor IRES-mediated translation in living cells provides a new perspective on non-canonical translation that will complement technologies like ribosome profiling45 and in vitro single-molecule assays.49 Our biosensor extends the breadth of NCT28, which until now had been exclusively used to investigate cap-dependent translation.28,30,47,50 By incorporating IRES sequences in a bicistronic context, we demonstrated how two distinct modes of translation can be fairly compared at the single-molecule level in living U2OS cells. This approach can be extended to study a variety of viral and eukaryotic IRES sequence in diverse cell types. This will help uncover the true breadth of IRES translation in vivo, but there are two important caveats to keep in mind. First, viral IRES sequences are not usually in a bicistronic context. Although there are exceptions, such as the IRESs of the Dicistroviridae,51 placing an IRES 3’ of another ORF can alter its translation (as we saw in the enhanced IRES translation within Cap+IRES translation sites). Second, IRES sequences (including EMCV) are usually part of uncapped viral RNA that never experience the cell nucleus. Capped bicistronic reporters could pick up nuclear factors that alter IRES translation. In the future, these caveats can be addressed by extending our technology to more natural IRES constructs, perhaps by loading or transfecting monocistronic RNA containing both a 5’ IRES and repeat epitopes into cells or by infecting cells with engineered virions. Ultimately, as single-molecule experiments and computational analyses improve, we anticipate that integrated biosensors and stochastic models like those introduced here will provide new insights into not only how viral IRES sequences recruit ribosomes, but also how eukaryotic IRES elements function during development and cellular survival situations.
METHODS
Plasmid construction
The Original Tag (SM-KDM5B-EMCV-SunTag-Kif18b-MS2) contains a spaghetti monster (SM) with 10× FLAG epitopes, a SunTag with 24× SunTag epitopes, and an MS2 repeat with 24× MS2 stem loops. The coding region of the SunTag and Kif18b was obtained by polymerase chain reaction (PCR) of a pCMV-SunTag-Kif18b-PP7 template (Addgene #128606), using the following primers: 5’-GCC GAA AGG TTT AAA CGC TAG CTC TGG AGG AGA AGA ACT TTT GAG CAA GAA T-3’; 5’-AGT AAC AGT CCG CCT AGG TCC TTA TCG GAC ACC TTG GT-3’. The PCR product contained arms of homology to the acceptor plasmid (SM-KDM5B-MS2; Addgene #81084). The acceptor plasmid was cut with Nhel (New England BioLabs) between the end of KDM5B and the MS2 stem loops. The PCR product and cut acceptor plasmid were assembled via Gibson Assembly (homemade mixture). The resulting plasmid was SM-KDM5B-Nhe1-SunTag-Kif18b-MS2, which was also used as the NolRES construct. The EMCV IRES sequence was amplified by PCR from EMCV_IRES_pcDNA4TO_H2B_SunTag24x_v1 (Addgene #246719) using the following primers: 5’- CCG AAA GGT TTA AAC GCT AGC ACG TTA CTG GCC GAA −3’; 5’- TTC TTC TCC TCC AGA GCT AGC TAT TAT CAT CGT GTT TTT CAA AGG AAA −3’. The PCR product contained arms of homology to the acceptor plasmid (SM-KDM5B-Nhe1-SunTag-Kif18b-MS2). The acceptor plasmid was cut with Nhel (New England BioLabs) between the end of KDM5B and beginning of SunTag. The PCR product and cut acceptor plasmid were assembled via Gibson Assembly. The start codon for SunTag-Kif18b is within the EMCV IRES sequence.
For the construction of the Switch Tag (SunTag-Kif18b-EMCV-SM-KDM5B-MS2), the coding region of the SunTag and Kif18b was obtained by PCR of a pCMV-SunTag-Kif18b-PP7 template (Addgene #128606), using the following primers: 5’-TCG CTG TGA TCG TCA CTT GGC GGA CAC CAT GGA AGA ACT TTT GAG CAA GAAT-3’; 5’- CGT CCT TGT AGT CCA TGG TGG CGG CGC GCC GTC TTA GAT ATC GGA CAC CTTG-3’. The PCR product contained arms of homology to the acceptor plasmid (SM-KDM5B-MS2 Addgene #81084). The acceptor plasmid was cut with Notl (New England BioLabs) at the beginning of SM. The PCR product and cut plasmid were assembled via Gibson Assembly. The resulting plasmid was SunTag-Kif18b-Nhe1-SM-KDM5B-MS2. The EMCV IRES sequence was amplified by PCR from EMCV_IRES_pcDNA4TO_H2B_SunTag24×_v1 (Addgene #246719) using the following primers: 5’- CCA AGG TGT CCG ATA TCT AAG ACG GCG TTA CTG GCC GAA GCC GCT -’3; 5’-CCT TGT AGT CCA TGG TGG CGG CGC ATA TTA TCA TCG TGT TTT TCA AAG GAA AAC CAC-3’. The PCR product contained arms of homology to the acceptor plasmid (SunTag-Kif18b-AscI-SM-KDM5B-MS2). The acceptor plasmid was cut with AscI (New England BioLabs) between the end of Kif18b and beginning of SM. The PCR product and cut acceptor plasmid were assembled via Gibson Assembly. The start codon for SM-KDM5B is within the EMCV IRES sequence.
anti-FLAG Fab generation and dye-conjugation
Pierce mouse IgG1 preparation kit (Thermo Scientific) was used to generate Fab according to the manufacturer’s instruction. Briefly, immobilized ficin in the presence of 25 mM cysteine was used to digest FLAG antibodies (Wako, 012–22384 Anti DYKDDDDK mouse IgG2b monoclonal) to create Fab. Fab were separated from the Fc region using NAb Protein A column. After elution, Fab were concentrated to 1 mg/ml and conjugated to Cy3. Cy3 N-hydroxysuccinimide ester (Invitrogen) was dissolved in DMSO and stored at −20°C. 100 μg of Fab were diluted into 100 μl of 100 mM NaHCO3 (pH 8.5). 1.33 μl of Cy3 was added to this solution and incubated with end-over-end rotation for 1–2 hours at room temperature. The conjugated Fab were then eluted from a PBS pre-equilibrated PD-mini G-25 desalting column (GE Healthcare) that removed unconjugated dye. Conjugated Fabs were then concentrated using an Ultrafree 0.5 filter (10k-cut off; Millipore) to 1 mg/ml. The Fab:dye ratio was calculated using the absorbance at 280 and 550 nm, and using the extinction coefficient of Fab with the dye correction factor at 280 nm provided by the manufacturers (0.08 for Cy3). The degree of labeling was calculated using the following formula:
MCP and scFv-GFP purification
His-tagged MCP/scFv-GFP was purified with Ni-NTA-agarose (Qiagen) following the manufacturer’s instructions with minor modifications. Briefly, the bacteria were lysed in a PBS-based buffer containing a complete set of protease inhibitors (Roche), binding to the Ni-NTA resin was carried out in the presence of 10 mM imidazole. After washing with 20 and 50 mM imidazole in PBS, the protein was eluted with 300 mM imidazole in PBS, and directly used for experiments. The rest was dialyzed against a HEPES-based buffer (10% glycerol, 25 mM HEPES pH 7.9, 12.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 0.01 % NP-40 detergent, and 1 mM DTT) and stored at −80 °C after snap-freezing by liquid nitrogen.
Cell culture, transfection, and beadloading
U2OS cells were grown in DMEM (Thermo Scientific) supplemented with 10% (v/v) FBS, 1 mM L-glutamine and 1% (v/v) Penicillin-streptomycin (DMEM+). One to two days prior to experiments, cells were plated into a 35 mm MatTek chamber at approximately 70–80% confluency. Two to four hours prior to experiments, cells were put in OPTI-MEM (Thermo Scientific) supplemented with 10% (v/v) FBS (OPTI-MEM+). Cells were then bead-loaded with fluorescently labeled Fab, GFP-fused scFv, MCP-HaloTag protein and purified DNA of interest. Briefly, 100 μg/ml of fluorescently labeled Fab, 100 μg/ml of purified GFP-fused scFv, 33 μg/ml of purified MCP HaloTag protein, and 750ng of DNA of interest were prepared in a total volume of 4μl of 1×PBS. After removing OPTI-MEM and FBS, the 4μL solution was pipetted to the top of the cells. Then, ~106 μm glass beads (Sigma Aldrich) were sprinkled evenly over the cells. The chamber was tapped firmly 12 times on the bench, and OPTI-MEM+ was added back to the cells. Two hours after bead-loading, cells were washed twice with phenol-red-free DMEM+ such that all beads were removed. 200 nM of JF646-HaloTag ligand was next added (1μL of 200nM to 1mL of phenol-red-free DMEM+). After 20 minutes of incubation at 37 ° C, the cells were washed twice with phenol-red-free DMEM+ to remove excess ligand. 2 mL of phenol-red-free DMEM+ was added back to the cells. Translation experiments were conducted immediately after washing. U2OS cells were purchased from ATCC and were authenticated by STR profiling by ATCC and morphological assessments. We also confirmed that all cell lines tested negative for mycoplasma contamination.
Single molecule tracking microscopy
To track single-molecule mRNA translation events, we used a custom-built widefield fluorescence microscope with a highly inclined illumination scheme.28,36 Briefly, the excitation beams, 488, 561 and 637 nm solid-state lasers (Vortran), were coupled and focused on the back focal plane of the objective (60X, NA 1.49 oil immersion objective, Olympus). The emission signals were split by an imaging grade, ultra-flat dichroic mirror (T660lpxr, Chroma) and detected by two aligned EM-CCD cameras (iXon Ultra 888, Andor) by focusing with 300 mm tube lenses (this lens combination produces 100X images with 130 nm/pixel). Live cells were placed into an incubation chamber (Okolab) at 37 °C and 5% CO2 on a piezoelectric stage (PZU-2150, Applied Scientific Instrumentation). The focus was maintained with the CRISP Autofocus System (CRISP-890, Applied Scientific Instrumentation). Image acquisition was performed using open source Micro-Manager.52 With this setting, one camera detected far-red emission signals while the other detected either red or green emission signals.
Far-red signals were excited with the 637 nm laser with a 731/137 nm emission filter (FF01–731/137/25, Semrock). Red and green signals were separated by the combination of the excitation lasers and the emission filters installed in a filter wheel (HS-625 HSFW TTL, Finger Lakes Instrumentation); namely, the 561 nm laser and 593/46 nm emission filter (FF01–593/46–25, Semrock) were used for Cy3 imaging, and the 488 nm laser and 510/ 42 nm emission filter (FF01–510/42–25, Semrock) were used for sfGFP or A488 imaging. The lasers, filter wheel, cameras, and the piezoelectric stage were synchronized by an Arduino Mega board (Arduino). The exposure time of the cameras was selected as 53.64 msec throughout the experiments. The readout time for the cameras from the combination of imaging size, readout mode, and the vertical shift speed was 23.36 msec, resulting in an imaging rate of 13 Hz (77 msec per image). The excitation laser lines were digitally synched to ensure they only illuminated cells when the camera was exposing to avoid excessive photobleaching. To capture the entire volume of the cytoplasm of U2OS cells, 13 z stacks with a step size of 500 nm (6 μm in total) were acquired using the piezoelectric stage. Because one image of Cy3 was captured on one camera and one image of sfGFP/A488 + JF646 was captured on the other camera in the same stack of the cell, the z-position within the cell changed every two images. The position of the filter wheel was changed during the camera readout time. This resulted in a total cellular imaging rate of 0.5 Hz (2 s per volume for 3-colors). Note that all colors described in the text and that are shown in the figures are based on the color of the excitation laser: RNA in red (JF646) and protein in green (Cy3) or blue (sfGFP).
Cell imaging conditions with no drugs added for all constructs
Cells beadloaded with SM-KDM5B-EMCV-SunTag-Kif18b-MS2 (Original Tag), SunTag-Kif18b-EMCV-SM-KDM5B-MS2 (Switch Tag), or SM-KDM5B-SunTag-Kif18b-MS2 (NolRES Tag), Cy3 labeled anti-FLAG Fab, Halo-MCP protein (labeled with JF646-HaloTag ligand), and anti-SunTag scFv-GFP were imaged with a 6 second interval between every 13 captures (one entire cell volume) for 25–50 total time-points. Laser powers for all images were: 15mW for 637nm, 9mW for 488nm, and 5mW for 561nm with an ND10 neutral density filter at the beam expander.
Particle tracking
Collected images were first pre-processed with Fiji.53 Briefly, the 3D images were projected to 2D images by a maximum intensity projection and background subtracted. Post-processed images were then analyzed by a custom-written Mathematica (Wolfram Research) routine to detect and track particles in the RNA channel (red color). Specifically, particles were emphasized with a band-pass filter such that the positions could be detected using the built-in Mathematics routine ComponentMeasurements “IntensityCentroid.” Detected particles were linked through time by allowing a maximum displacement of 5 pixels between consecutive frames. Particle tracks lasting at least 5 frames were selected. To properly account for the offset between the two cameras, a geometric transformation function (see method below) was applied to the coordinates of the center of mRNAs. For each frame of each track, 15×15 (pixels × pixels) crops centered on the registered mRNA coordinate were made and averaged through time. Using Mathematics’s bandpass filter and ComponentMeasurements described above, the time-averaged crops corresponding to each track were categorized based on the presence of detectable signals in the green and blue nascent chain channels: Red − mRNA not translating, Red + Green = Yellow − mRNA translating in Cap Only for Original Tag or IRES Only for Switch Tag, Red + Blue = Purple − mRNA translating in IRES Only for Original Tag and Cap Only for the Switch Tag, Red + Green + Blue = White − mRNA translating in both cap and IRES manner. Once the spots were categorized in this automated fashion, all spots were again hand-checked to minimize error.
Finally, the original 2D maximum intensity projected images corresponding to each hand-checked track were fit to find their precise coordinates and intensities (using the built-in Mathematica routine NonlinearModelFit) to a 2D Gaussians of the following form:
where IBG is the background intensity, I the particle peak intensity, (σx, σy) the spread of the particle, and (x0, y0) the particle location. From these data, the intensity, position through time, and number of spots over time in each track were quantified for downstream analysis.
Fast imaging for mean square displacement analysis
For fast particle tracking to accurately quantify the mean square displacements, single planes of cells loaded with the Original Tag construct, Cy3 anti-FLAG Fab, anti-GCN4 scFv-GFP, and Halo-MCP were imaged with an imaging rate of 77 msec.
Geometric transformation function
The offset between the two cameras was registered using the built-in Mathematica routine FindGeometricTransform. To find the transformation function that best aligned the fitted positions, 100 nm diameter Tetraspeck beads evenly spread out across the image field-of-view were imaged on the same day experiments were taken. Only the fitted particle positions were registered to avoid introducing any distortion into images. Therefore, a slight offset can be observed between the red and the green or blue particles even though they are within a diffraction limited spot according to our registration.
Calibrating translation site intensity
We wanted to quantify the units of mature protein (i.e. number of nascent chains or active ribosomes) at a translation site using its intensity signal. For this purpose, we imaged two calibration constructs.28 The two calibration constructs were equal in length, one containing the spaghetti monster 10×FLAG tag (SM-BetaActin) which contains 10 repeats of the FLAG epitope, the other containing just a single FLAG epitope (1×FLAG-filler-BetaActin). Note the spaghetti monster 10×FLAG tag (SM) is the same tag utilized in the Original Tag (SM-KDM5B-IRES-ST-Kif18b) and the Switch Tag (ST-Kif18b-IRES-SM-KDM5B). The 1×FLAG-filler-BetaActin tag was used to measure the number of ribosomes translating in a translation site. With the 1×FLAG-filler-BetaActin calibration construct, each nascent chain in a translation site contains just one FLAG epitope labeled by a single Fab conjugated (on average) to a single Cy3 fluorophore. By imaging this 1×FLAG construct at high laser powers such that individual translation sites and single Cy3 fluorophores (confirmed by single-step photobleaching, see below) can both be visualized, the ratio of total Cy3 signal in translation sites to single Cy3 fluorophore intensity signals approximates the number of nascent chains (or ribosomes) per translation site. To quantify this, we imaged cells beadloaded with 1×FLAG-filler-BetaActin and Cy3 anti-FLAG Fab in a single optical plane at high laser powers (50 mW for 561nm and 15mW for 637nm laser). A short movie was acquired, after which cells were continually illuminated to photobleach them to the point at which single probe fluorescence could easily be detected by single-step photobleaching. At this point, a second short 250-frame movie was acquired. The intensity of polysomes (verified by the presence of an RNA signal intensity) from the first frame of the first movie was then measured (as described in the ‘Particle tracking’ section above) and compared to the plateau intensity of a single Cy3 just prior to single-step photobleaching. The ratio of polysome to Cy3 intensities gives us an estimate for the number of ribosomes on a single RNA. By averaging all measured RNA together, we obtain a mean value for the ribosomal occupancy of a single, translating, 1×FLAG-filler-BetaActin mRNA. From these measurements, 11.4 ± 2.0 ribosomes were estimated to be translating the 1×FLAG-filler-BetaActin calibration construct.
Since the 1×FLAG-filler-BetaActin calibration construct and the SM-BetaActin calibration construct (with 10×FLAG) are the same length with the same promoters and 3’ and 5’ UTRs, their translation sites should contain roughly the same number of ribosomes (11.4 ± 2.0). With a known number of ribosomes on SM-BetaActin, we wished quantify the number of ribosomes on the cap ORF in the Original Tag (SM-KDM5B-IRES-ST-Kif18b) and the IRES ORF in the Switch Tag (ST-Kif18b-IRES-SM-Kif18b). To do this, we imaged cells in two different chambers. In one chamber, we beadloaded cells with anti-FLAG Fab (Cy3) and SM-BetaActin. In the second chamber, we beadloaded cells with anti-FLAG (Cy3) and the Original Tag (SM-KDM5B-IRES-ST-Kif18b). Both chambers were imaged on the same day with the same imaging conditions (50 mW for 561nm and 15mW for 637nm laser). By taking the intensity ratio of SM-BetaActin containing 11.4 ± 2.0 ribosomes and Cap Only translation sites in the Original Tag (SM-KDM5B-IRES-ST-Kif18b), we measured the ribosomal occupancy of the cap ORF to be 14.6 ± 5.6 ribosomes. Taking the intensity ratio of these translation sites and all other SM translation sites in the Original Tag (Cap in Cap+IRES translation sites) and Switch Tag (ST-Kif18b-IRES-SM-KDM5B) (IRES in IRES Only and Cap+IRES translation sites) gave the number of ribosomes translating in all possible translation sites, as shown in Fig. 4c.
Ribosome run-off experiments using Harringtonine treatment and elongation estimates
To measure average elongation rates, cells beadloaded with the Original Tag (SM-KDM5B-EMCV-SunTag-Kif18b-MS2), Cy3 labeled anti-FLAG Fab, Halo-MCP protein (labeled with JF646-HaloTag ligand), and anti-SunTag scFv-GFP were imaged with a 60 second interval between every 13 frames (one entire cell volume) for 50 total time-points. Laser powers were the same as previously described for general imaging. After acquiring 5 time-points of pre-treated images, cells were treated with a final concentration of 3 pg/mL of Harringtonine (Cayman Chemical). After treatment, cells were imaged for the remaining 45 time-points as described. As a photobleaching control, cells were imaged at the exact same imaging conditions described previously however no drug was added.
To generate ribosomal run-off curves, images were analyzed with the particle tracker as previously described. In each frame of each cell image, nascent chain signal intensities from all translation sites were totaled resulting in an intensity decay curve over time for each individual cell. Each decay curve was normalized to the average value from the first five frames (preceding drug addition). Each individual curve was fit to the following phenomenological equation:
where the linear part of the fit (the slope at ax=b) provides a good estimate of the elongation rates in amino acids over time in seconds.42 From this, elongation rates for each cell were calculated.
Puromycin treatment
Cells beadloaded with the Original Tag (SM-KDM5B-EMCV-SunTag-Kif18b-MS2) or the Switch Tag (SunTag-Kif18b-EMCV-SM-KDM5B-MS2), Cy3 labeled anti-FLAG Fab, Halo-MCP protein (labeled with JF646-HaloTag ligand), and scFv-GFP were imaged with a 60 second interval between every 13 frames (one entire cell volume) for 15 total time-points. After acquiring 5 time-points of pre-treated images, cells were treated with a final concentration of 50μg/mL of puromycin. After treatment, cells were imaged for the remaining 10 time-points as described previously. As a photo-bleaching control, cells were imaged at the exact same imaging conditions described previously with no drug added. Three biological replicates were taken.
Sodium Arsenite (NaAs) and Dithiothreitol (DTT) treatment
Cells beadloaded with the Original Tag (SM-KDM5B-EMCV-SunTag-Kif18b-MS2) or Switch Tag (SunTag-Kif18b-EMCV-SM-KDM5B-MS2), Cy3 labeled anti-FLAG Fab, Halo-MCP protein (labeled with JF646-HaloTag ligand), and anti-GCN4 scFv-GFP were imaged with a 180 second interval for NaAs and 120 for DTT between every 13 frames (one entire cell volume) for 35 total time-points. After acquiring 5 time-points of pre-treated images, cells were treated with a final concentration of 0.5 mM of NaAs or 0.75 mM of DTT. After treatment, cells were imaged for the remaining time-points. As a photo-bleaching control, cells were imaged at the exact same imaging conditions described previously with no drug added. Four biological replicates were taken.
Statistics and Reproducibility
Fig 1c: Representative image from n=39 independent biological replicates (cells).
Fig 1d: n=39 independent biological replicates (cells) for the Original Tag and n=35 independent biological replicates (cells) for the NolRES Tag.
Fig 2b: Representative track out of 296 tracked Cap+IRES translation sites
Fig 2c top: Avg of 296 tracked Cap+IRES translation sites from 11 independent biological replicates (cells). High intensity group has 99 tracked mRNA, the medium intensity group has 99 tracked mRNA, and the low intensity group has 98 tracked mRNA.
Fig 2c bottom: Avg of 259 tracked Cap+IRES translation sites from 11 cells. High intensity group has 87 tracked mRNA, the medium intensity group has 86 tracked mRNA, and the low intensity group has 86 tracked mRNA.
Fig 2d Top: Avg of 793 tracked Cap Only translation sites from 11 independent biological replicates (cells). High intensity group has 199 tracked mRNA, the medium-high intensity group has 198 tracked mRNA, the medium-low intensity group has 198, and the low intensity group has 198 tracked mRNA.
Fig 2d Bottom: Avg of 213 tracked Cap Only translation sites from 11 cells. High intensity group has 53 tracked mRNA, the medium-high intensity group has 54 tracked mRNA, the medium-low intensity group has 53, and the low intensity group has 53 tracked mRNA.
Fig 3a: Representative cell from the 17 independent biological replicates (cells)
Fig 3b: n=17 independent biological replicates (cells).
Fig 3c: n=10 independent biological replicates (cells).
Fig 4a top: Representative cell of Original Tap out of 39 independent biological replicates (cells)
Fig 4a bottom: Representative cell of Switch Tag out of 37 independent biological replicates (cells)
Fig 4b from left to right: n= 302 spots out of 39 independent biological replicates (cells), n=167 spots out of 37 independent biological replicates (cells), n=262 spots out of 37 independent biological replicates (cells), n=201 spots out of 39 independent biological replicates (cells).
Fig 4c from left to right: 226 spots out of 39 independent biological replicates (cells), n=76 spots out of 39 independent biological replicates (cells), n=121 spots out of 37 independent biological replicates (cells), n=76 spots out of 37 independent biological replicates (cells).
Fig 5a: 14 unique 4 state models were considering with between 7 and 12 free parameters. The best model had 8 free parameters.
Fig 5b. Experimental data represent the fraction of translation spots weighted by the number of experimentally measured spots per cell. n=39 independent biological replicates (cells). Simulations n = 4,000 trajectories. Simulated mean and SD values were calculated using bootstrapping (X50 with sampling of 300 trajectories).
Fig 5c. Experimental distributions were calculated form a total of n = 226 spots for Cap and n = 121 for IRES. Simulations n = 4,000 trajectories. Simulated mean and SD values were calculated using bootstrapping (X50 with sampling of 1000 trajectories).
Fig 5d–e: For experimental Cap and IRES intensities n=10 and n =17 independent biological replicates (cells), respectively. Simulations n = 4,000 trajectories. Lines represent the mean and shadow bars represent the SD, and 10 independent repetitions are shown.
Fig 5f: NaAs: n=32 independent biological replicates (cells). DTT: n=28 independent biological replicates (cells). Simulations: n = 4,000 trajectories. Lines represent the mean and shadow bars represent the SD.
Extended Data Fig. 3a: Representative data set out of n=296 translation spots from n=11 independent biological replicates (cells).
Extended Data Fig. 3b: Each point represents the median distance from the 3’UTR for either Cap in green of IRES in blue within n=296 Cap+IRES translation sites.
Extended Data Fig. 6c–d: Relative log-likelihood values for the optimization process are calculated according to Eq. 23 (Supplementary Information). For the NaAs and DTT cross-validation experiments log-likelihood values are calculated according to Eq. 26 (Supplementary Information). The log-likelihood reported are relative to the minimum value from all models. A selection threshold (dashed red line) was defined by a log-likelihood of 100 worse than the most complex and best fitting model. Relative log-likelihood values over 500 are not plotted. Symbols represent and error bars represent mean values and SD of 3 independent optimization runs.
Extended Data Fig. 6e: NaAs: n=32 independent biological replicates (cells). DTT: n=28 independent biological replicates (cells). Model was simulated for 4,000 trajectories with a burn-in period of 10,000 seconds. Lines represent the mean and shadow bars represent the SD.
REPORTING SUMMARY
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
CODE AVAILABILITY
All codes and required data are available at: https://github.com/MunskyGroup/Koch_Aguilera_etal_2020.git
DATA AVAILABILITY STATEMENT
Source Data are available with the paper online.
Raw images can be found at: https://doi.org/10.6084/m9.figshare.12751853.v1
Extended Data
Extended Data Fig. 1. Controls for bleedthrough and active translation.
(A-B) Five frame average of a Cap Only and IRES Only translation spots. mRNA marker dye, JF646, was not added to these cells. Cells were imaged for 3-minutes with a 6-second interval between each capture.
(C) Top graphs show normalized total intensity over time for Cap-dependent (left) and IRES-mediated translation spots (right), after addition of puromycin. Gray lines indicate individual cells. Thick dark line indicates the average total intensity of all cells. Red dashed lines indicate time at which puromycin was added. Cap-dependent: n=5 cells. IRES-mediated: n= 5 cells. Bottom graphs show normalized total intensity of Cap-dependent (left) and IRES-mediated (right) translation spots without the addition of puromycin. Cap-dependent: n=6 cells. IRES-mediated: n= 5 cells. All cells (control and drug treated) were imaged for 10-minutes with a 1-minute interval. Error bars represent S.E.M.
Extended Data Fig. 2. IRES and cap translation site localization and mobility.
(A) Quantification of translating and non-translating mRNA distances in micrometers (μm) to nearest-neighbor translation spot within single cells. Each point represents the average distance per cell. n=39 cells.
(B) Quantification of distance in μm from the nucleus of translating and non-translating mRNA. Each point represents the average distance from the edge of the nucleus per cell. n=39 cells.
(C) Representative cell out of n=11 cells imaged with fast imaging conditions. An example mRNA is highlighted with a white circle and a track through time of that mRNA is graphed below.
(D) Cumulative distribution function plot of non-translating mRNA (red), IRES Only (purple), Cap Only (yellow), and Cap+IRES (gray) species based on their diffusion coefficients (μm2 /sec). Inset shows the Mean Square Displacements (MSD) of the different species over time in seconds. n=3771 total tracked mRNA (translating and non-translating), n=11 cells.
(E) Schematic showing how the jump angles are measured. Error bars represent the standard error of the mean (S.E.M).
(F) Circular plots of the jump angle distributions for non-translating mRNA, Cap Only, IRES Only and Cap+IRES translation sites.. For the box and whisker plots, the thick black lines indicate the medians, the boxes indicate the 25%−75% range, and the whiskers indicate the 5%−95% range. The p-values are based on a two-tailed Mann-Whitney test: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Extended Data Fig. 3. Measuring distances between Cap and IRES nascent chains in Cap+IRES translation spots.
(A) Representative data set of measured distances of Cap (light green) and IRES (blue) nascent chains to 3’UTR through time in single Cap+IRES translation tracks.
(B) Median distances of Cap and IRES nascent chains to 3’UTR of each Cap+IRES track. Distances are measured in nanometers (nm). 3’ UTR coordinates were fixed at (0,0) for all analyses. n= 296 translation spots.
Extended Data Fig. 4. Ribosomal run-off curves from single cells after addition of Harringtonine.
(A) Harringtonine-induced ribosomal run-off curves from single cells. Each curve shows the decay in nascent chain signal intensity from all Cap-dependent and
(B) IRES-mediated translation sites within a single cell post-Harringtonine. Run-off curves were phenomenologically fit to a Tanh function to align curves in time for averaging in Figure 4. The slope of fitted curves at a normalized intensity value of 0.5 was used to estimate the elongation rate.
(C) Cap-dependent (n=7 cells) and (D) IRES-mediated (n=5 cells) translation controls in which no drugs were added. Each gray line shows the total nascent chain signal intensity from all translation sites in an individual cell. The thick black line is a representative cell. Intensity in arbitrary units (a.u.). All cells were imaged for 45 minutes with a 1-minute interval between each capture.
Extended Data Fig. 5. Original Tag comparison to Switch Tag, single mRNA selection, and polysome intensity calibrations.
(A) Quantification of the percentages of each type of translation sites for the Original Tag (left, n=39 cells) and the Switch Tag (right, n=37 cells). Each point represents a single-cell measurement.
(B) Probability histograms showing distributions of mRNA intensities of non-translating mRNA (Red), Cap Only (Yellow), IRES Only (Purple), and Cap+IRES (Gray) translation sites for the Original Tag and the Switch Tag. The gray boxes represent the mRNA intensity threshold used to eliminate multiple mRNAs. Intensities in arbitrary units (a.u.).
(C) Translation site calibration measurements. The intensities of Cap in Cap Only translation sites (n=20spots) in the Original Tag were compared to a 10×Flag calibration system (n=47spots) with a known number of ribosomes. These comparisons lead to a calculated number of 14.6 ribosomes in Cap Only translation sites using the Original Tag. For the box and whisker plots, the thick black lines indicate the medians (A and C), and the dashed red line indicate the weighted means (A) the boxes indicate the 25%−75% range, and the whiskers indicate the 5%−95% range. The p-values are based on a two-tailed Mann-Whitney test: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Extended Data Fig. 6. Model of the bicistronic gene construct.
(A) The most complete mathematical model considers four mutually exclusive RNA states: non-translating (OFF), Cap-dependent (CAP-ON), IRES-mediated (IRES-ON), and both Cap and IRES (CAP+IRES-ON). All transition rate values between RNA states are free-independent parameters. A cross-over mechanism (CO symbol in the figure), by which a ribosome that completes the translation of the Cap-dependent protein could immediately re-initiate translation of the IRES-mediated protein, is represented by the reaction parameter kCO.
(B) Comparison of 14 different sub-models. The sub-models test different hypotheses, including variations of the number of mRNA states (3 or 4 states), dependency on Cap and IRES switching states, and/or the existence of the cross-over mechanism. A complete description is given in the Supplementary Information (S.I.).
(C) Cross-validation is used to compare two possible mechanisms of translation inhibition under NaAs stress. The first mechanism mimics the inhibition of the Cap activation rates at the promoter level (LNaAs- state-cap; i.e., block of kON-C and k’ON-C). The second mechanism considers blocking ribosomal initiation for Cap (LNaAs-INIT-CAP; i.e., block of kINIT-c).
(D) Optimization process and cross-validation for the DTT stress. The same inhibitory mechanisms described in C are tested for DTT stress.
Relative Log-likelihood values for the optimization process are calculated according to (S.I.) Eq. 23 and Eq. 26 for the NaAs and DTT cross-validation experiments, respectively. A selection threshold (dashed red line) was defined by a log-likelihood of 100 worse than the most complex and best fitting model. Models above the selection threshold were discarded (gray background), and their cross-validation log-likelihood values are not shown. The best model shown (green background) was chosen as the model with fewest free parameters below the selection threshold. A complete description is given in the Statistics and Reproducibility section.
(E) Model simulations for the best-fit model 4SIm2 under NaAs and DTT stresses. The figure shows the effect of blocking ribosomal initiation and activation for Cap.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Luke Lavis for kindly providing JF646 labeled HaloTag ligand and Dr. Hataichanok (Mam) Scherman for purifying Halo-MCP and GFP-scFv (anti-SunTag). We thank Dr. Lachlan Whitehead and collaborators for the Imagej Zoom plugin. We thank all members of the Stasevich and Munsky labs for their support and helpful discussions. BM and LA were supported by a grant from the W.M. Keck Foundation and by the NIH (grant no. 5R35GM124747). TJS, AK, and TM were supported by the NIH (grant no. R35GM119728).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Jackson RJ, Hellen CUT & Pestova TV The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stéphanie Bornes et al. Translational Induction of VEGF Internal Ribosome Entry Site Elements During the Early Response to Ischemic Stress. Circ. Res. 100, 305–308 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Xue S et al. RNA regulons in Hox 5’ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA & Willis AE C-Myc 5’ untranslated region contains an internal ribosome entry segment. Oncogene 16, 423–428 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Ray PS, Grover R & Das S Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 7, 404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Firth AE & Brierley I Non-canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doudna JA & Sarnow P Translation Initiation by Viral Internal Ribosome Entry Sites. 25 (2007). [Google Scholar]
- 8.Komar AA, Mazumder B & Merrick WC A new framework for understanding IRES-mediated translation. Gene 502, 75–86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieft JS Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 33, 274–283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filbin ME & Kieft JS Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 19, 267–276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baird SD Searching for IRES. RNA 12, 1755–1785 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Miragall O, Quinto S. L. de & Martinez-Salas E Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 139, 172–182 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Gendron K, Ferbeyre G, Heveker N & Brakier-Gingras L The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 39, 902–912 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson P, Yang D, Zhang H & Hemida M Viral Replication Strategies: Manipulation of ER Stress Response Pathways and Promotion of IRES-Dependent Translation. (2013). [Google Scholar]
- 15.Holcik M & Sonenberg N Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Hanson PJ et al. IRES-Dependent Translational Control during Virus-Induced Endoplasmic Reticulum Stress and Apoptosis. Front. Microbiol. 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hetz C The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Hooper PL, Hightower LE & Hooper PL Loss of stress response as a consequence of viral infection: implications for disease and therapy. Cell Stress Chaperones 17, 647–655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez J, Yaman I, Sarnow P, Snider MD & Hatzoglou M Regulation of Internal Ribosomal Entry Site-mediated Translation by Phosphorylation of the Translation Initiation Factor eIF2a. J. Biol. Chem. 277, 19198–19205 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Sonenberg N & Hinnebusch AG Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson SR & Sarnow P Regulation of host cell translation by viruses and effects on cell function. Curr. Opin. Microbiol. 3, 366–370 (2000). [DOI] [PubMed] [Google Scholar]
- 22.J. P et al. Viral Replication Strategies: Manipulation of ER Stress Response Pathways and Promotion of IRES-Dependent Translation. in Viral Replication (ed. Rosas-Acosta G) (InTech, 2013). doi: 10.5772/56072. [DOI] [Google Scholar]
- 23.Weingarten-gabbay S & Segal E Toward a systematic understanding of translational regulatory elements in human and viruses. 6286, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plank TDM, Whitehurst JT & Kieft JS Cell type specificity and structural determinants of IRES activity from the 5’ leaders of different HIV-1 transcripts. Nucleic Acids Res. 41,6698–6714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies MV & Kaufman RJ The Sequence Context of the Initiation Codon in the Encephalomyocarditis Virus Leader Modulates Efficiency of Internal Translation Initiation. J VIROL 66, 9 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gale M, Tan S-L & Katze MG Translational Control of Viral Gene Expression in Eukaryotes. Microbiol. Mol. Biol. Rev. 64, 239–280 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Quinto SL, Lafuente E & Martínez-Salas E IRES interaction with translation initiation factors: Functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7, 1213–1226 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morisaki T et al. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Viswanathan S et al. High-performance probes for light and electron microscopy. Nat. Methods 12, 568–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan X, Hoek TA, Vale RD & Tanenbaum ME Dynamics of Translation of Single mRNA Molecules In Vivo. Cell 165, 976–989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carocci M & Bakkali-Kassimi L The encephalomyocarditis virus. Virulence 3, 351–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamond N, Deforges J, Ulryck N & Sargueil B 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs. Nucleic Acids Res. 42, 10373–10384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bochkov YA & Palmenberg AC Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. BioTechniques 41, 283–292 (2006). [DOI] [PubMed] [Google Scholar]
- 34.McNeil PL & Warder E Glass beads load macromolecules into living cells. J. Cell Sci. 88, 669–678 (1987). [DOI] [PubMed] [Google Scholar]
- 35.Azzam ME & Algranati ID Mechanism of Puromycin Action: Fate of Ribosomes after Release of Nascent Protein Chains from Polysomes. Proc. Natl. Acad. Sci. U. S. A. 70, 3866–3869 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokunaga M, Imamoto N & Sakata-Sogawa K Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Cattoglio C et al. Determining cellular CTCF and cohesin abundances to constrain 3D genome models. eLife 8, e40164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adivarahan S et al. Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol. Cell 72, 727–738.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khong A & Parker R mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol. 217, 4124–4140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svitkin YV et al. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J. 28, 58–68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma AK et al. A chemical kinetic basis for measuring translation initiation and elongation rates from ribosome profiling data. PLOS Comput. Biol. 15, e1007070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morisaki T & Stasevich TJ Quantifying Single mRNA Translation Kinetics in Living Cells. Cold Spring Harb. Perspect. Biol. 10, a032078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aguilera LU et al. Computational design and interpretation of single-RNA translation experiments. PLOS Comput. Biol. 15, e1007425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munsky B, Neuert G & Oudenaarden A van. Using Gene Expression Noise to Understand Gene Regulation. Science 336, 183–187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingolia NT, Hussmann JA & Weissman JS Ribosome Profiling: Global Views of Translation. Cold Spring Harb. Perspect. Biol. 5 (2019) doi: 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz-Ramos R, Lopez-Carrillo L, Rios-Perez AD, De Vizcaya-Ruiz A & Cebrian ME Sodium arsenite induces ROS generation, DNA oxidative damage, HO-1 and c-Myc proteins, NF-κB activation and cell proliferation in human breast cancer MCF-7 cells. Mutat. Res. Toxicol. Environ. Mutagen. 674, 109–115 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Wang C, Han B, Zhou R & Zhuang X Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell 165, 990–1001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelletier J & Sonenberg N Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325 (1988). [DOI] [PubMed] [Google Scholar]
- 49.Marshall RA, Aitken CE, Dorywalska M & Puglisi JD Translation at the Single-Molecule Level. Annu. Rev. Biochem. 77, 177–203 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Wu B, Eliscovich C, Yoon YJ & Singer RH Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanamori Y & Nakashima N A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7, 266–274 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-only REFERENCES
- 52.Edelstein AD et al. Advanced methods of microscope control using pManager software. J. Biol. Methods 1, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source Data are available with the paper online.
Raw images can be found at: https://doi.org/10.6084/m9.figshare.12751853.v1