Abstract
Stress is ubiquitous to life and can irreparably damage essential biomolecules and organelles in cells. To survive, organisms must sense and adapt to stressful conditions. One highly conserved adaptive stress response is through the posttranslational modification of proteins by the small ubiquitin-like modifier (SUMO). Here, we examine the effects of acute ethanol stress on protein sumoylation in the budding yeast Saccharomyces cerevisiae. We found that cells exhibit a transient sumoylation response after acute exposure to ≤7.5% vol/vol ethanol. By contrast, the sumoylation response becomes chronic at 10% ethanol exposure. Mass spectrometry analyses identified 18 proteins that are sumoylated after acute ethanol exposure, with 15 known to associate with chromatin. Upon further analysis, we found that the chromatin structural proteins Smc5 and Smc6 undergo ethanol-induced sumoylation that depends on the activity of the E3 SUMO ligase Mms21. Using cell-cycle arrest assays, we observed that Smc5 and Smc6 ethanol-induced sumoylation occurs during G1 and G2/M phases but not S phase. Acute ethanol exposure also resulted in the formation of Rad52 foci at levels comparable to Rad52 foci formation after exposure to the DNA alkylating agent methyl methanesulfonate (MMS). MMS exposure is known to induce the intra-S-phase DNA damage checkpoint via Rad53 phosphorylation, but ethanol exposure did not induce Rad53 phosphorylation. Ethanol abrogated the effect of MMS on Rad53 phosphorylation when added simultaneously. From these studies, we propose that acute ethanol exposure induces a change in chromatin leading to sumoylation of specific chromatin structural proteins.
INTRODUCTION
Stress is an inevitable consequence of life. Every organism experiences this unwelcome and detrimental phenomenon. At the cellular level, stress is often caused by alterations in intra- or extracellular environments. Prolonged exposure of cells to stress conditions such as oxidation, temperature shifts, hypoxia, osmolarity alterations, genotoxic events, and a multitude of others can lead to damage of DNA, RNA, proteins, and other macromolecules. Consequently, the ability to sense and adapt to changing extracellular conditions is integral to cell survival. An effective response to exogenous stressors is elicited through activation of cellular stress response pathways that alter gene expression and/or protein interactions or activity in a coordinated effort to reestablish and maintain cellular homeostasis (Galluzzi et al., 2018). The inability to respond quickly and adapt to stress can lead to cell death, and failure to adapt to prolonged stress conditions underlies many human pathologies such as heart disease, neurodegeneration, and cancer (Fulda et al., 2010).
Posttranslational modifications (PTMs) play a key role in aiding cell survival during stress conditions. One PTM found to increase during a number of stress conditions is the small ubiquitin-like modifier (SUMO). Similar to protein ubiquitination, protein sumoylation utilizes an enzymatic cascade that leads to attachment of SUMO molecules to target substrates (Johnson et al., 1997; Okuma et al., 1999). SUMO modifications can alter protein localization, protein–protein interactions, and aid in protein stability and solubility (Geiss-Friedlander and Melchior, 2007). While global protein sumoylation is known to increase across a broad array of stresses, the majority of the target proteins and the kinetics by which they are sumoylated are distinct (Tempé et al., 2008; Guo and Henley, 2014; Lewicki et al., 2015). Although studies have reported the involvement of protein sumoylation in cellular stress responses and various targets (Zhou et al., 2004; Miller et al., 2013; Oeser et al., 2016), the function and regulation of specific protein sumoylation events during distinct stresses still remains poorly understood.
To understand better the key targets and functions of protein sumoylation during stress conditions, we have been recently studying the proteins that become sumoylated during acute ethanol exposure in the budding yeast Saccharomyces cerevisiae. The utilization of different yeasts for the purpose of ethanol production has been exploited for centuries (Mohd Azhar et al., 2017; Parapouli et al., 2020). Due to the industrial importance of ethanol production, a considerable amount of research has examined the differences in ethanol tolerance among laboratory and industrial yeast strains (Lewis et al., 2010; Steensels and Verstrepen, 2014). While yeast cells can tolerate relatively high concentrations of ethanol, this does not prevent them from experiencing cellular stress during acute and chronic exposure to ethanol. Chronic exposure to high concentrations of ethanol has been shown to alter membrane fluidity and lipid composition, increase reactive oxygen species (ROS) production through decoupling oxidative phosphorylation in the mitochondria, and cause protein misfolding (Auesukaree, 2017). Despite these investigations, there is minimal understanding of the molecular determinants and cellular processes that contribute to ethanol tolerance. In this study, we explore the effects of acute ethanol stress on protein sumoylation.
RESULTS
Global sumoylation kinetics in yeast depend on ethanol concentration
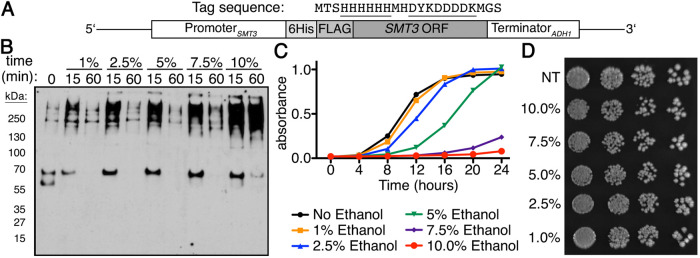
We previously examined global sumoylation response patterns over time in S. cerevisiae to various stressors that included ethanol (10% vol/vol; Oeser et al., 2016). In that study, acute exposure to high ethanol stress resulted in a steady accumulation of SUMO conjugates over a 60-min time course. While we were interested in sumoylation induced by 10% vol/vol ethanol, we also wanted to determine whether the sumoylation patterns observed remained unchanged at ethanol concentrations lower than 10% (vol/vol), or whether the sumoylation effect was only observed at ethanol concentrations that limit yeast growth. Utilizing a yeast strain where the endogenous SUMO gene, SMT3, was tagged with a His6-FLAG sequence at its 5′ end (Figure 1A), we examined sumoylation during ethanol stress at the following concentrations (vol/vol): 1%, 2.5%, 5%, 7.5%, and 10%. At concentrations lower than 10%, we found that the ethanol sumoylation response was transient with a pronounced increase in SUMO conjugates at 15 min that returned to basal levels by 60 min (Figure 1B), indicating that at lower ethanol concentrations yeast can mount an adaptive response that mitigates the need for chronic sumoylation.
FIGURE 1:
Global sumoylation kinetics are dependent on ethanol concentration. (A) Schematic for His6-FLAG-SMT3 located at the SMT3 locus and expressed from the endogenous SMT3 promoter. His6-FLAG sequence shown, sequences underlined. (B) Comparison of global sumoylation changes that occur during cellular exposure of ethanol (1%, 2.5%, 5%, 7.5%, and 10% vol/vol) over 60 min. Changes in sumoylation patterns were examined by Western analysis using an anti-FLAG antibody. (C) Quantitative measure of growth rates in liquid culture generated by Bioscreen C automated growth curve analysis. Cells were grown in triplicate at 30°C in rich media with 0%, 1%, 2.5%, 5%, 7.5%, or 10% ethanol for 24 h with continuous shaking. Absorbance at 600 nm was measured every 30 min and average absorbance (at 600 nm) was plotted vs. time. Error bars show SD for triplicate samples. (D) His6-FLAG-SMT3 cells were grown to midlog phase in rich liquid media, then treated for 60 min in rich media with 0%, 1%, 2.5%, 5%, 7.5%, or 10% ethanol before being 10-fold serially diluted onto rich media plates lacking ethanol and incubated at 30°C for 3 d.
To determine what levels of ethanol affect yeast cell growth, we queried the effect of ethanol concentration on cell growth when cells were chronically exposed to each of these concentrations. To do this, we performed liquid culture growth assays over a period of 24 h. Cells in 1% ethanol exhibited growth identical to cells with no ethanol treatment, while cells in 2.5% and 5% ethanol were delayed before entering into exponential growth. Cells in 7.5% and 10% ethanol, however, did not achieve exponential growth during the 24-h period (Figure 1C). To be certain that acute exposure to ethanol did not impact overall cell growth, we performed spot dilution tests on media lacking ethanol after the cells were exposed to the same concentrations of ethanol as listed above for 1 h in liquid culture. We did not observe any growth deficiency between untreated and ethanol-treated cells (Figure 1D), indicating that acute ethanol exposure does not impact cell growth as observed with chronic exposure (Figure 1C).
A chromatin structure maintenance complex is sumoylated during acute ethanol exposure
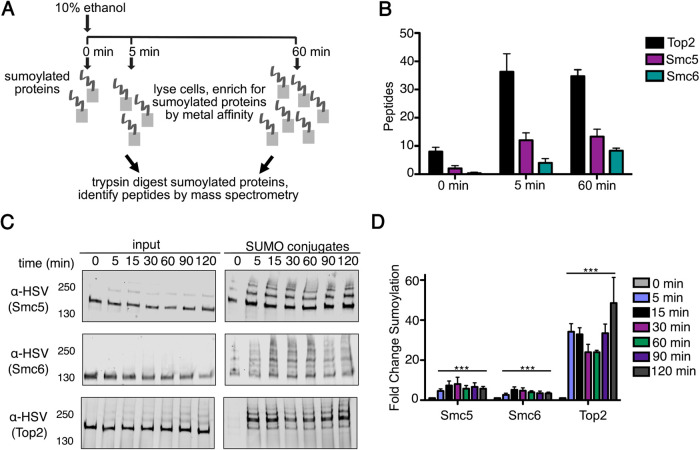
To identify proteins sumoylated during acute ethanol stress, we utilized a label-free mass spectrometry (MS) approach as previously described (Oeser et al., 2016). Although we found that sumoylation is transient at lower ethanol concentrations, we chose to query sumoylation at 10% ethanol to maximize the potential proteins that could be identified at early and late time points. Similar to our previous studies (Oeser et al., 2016), we used metal affinity chromatography to enrich for sumoylated species from His6-FLAG-SMT3 cell lysates derived from cultures before ethanol treatment and after ethanol treatment, in this case 5 and 60 min post ethanol exposure (Figure 2A). Sumoylated proteins were subsequently isolated by gel purification and subject to trypsin digestion before MS analysis. Total peptide counts were examined to determine which proteins showed increases at 5 min or 60 min of 10% ethanol exposure.
FIGURE 2:
The Smc5-Smc6 chromatin complex is sumoylated during acute ethanol exposure. (A) Mass spectrometry strategy to identify proteins sumoylated during acute ethanol exposure. (B) Total peptide counts identified for the top three proteins (Top2, Smc5, and Smc6) at 0, 5, and 60 min of 10% ethanol stress. (C) Cells expressing His6-FLAG-SMT3 and either C-terminally 3xHSV epitope tagged Smc5, Smc6, or Top2 from their endogenous promoters were subject to acute ethanol (10% vol/vol) over a 120-min time course. Cell lysates (input) and purified sumoylated proteins (SUMO conjugates) were examined via Western analysis using an anti-HSV antibody to detect Smc5, Smc6, and Top2. (D) Fold change sumoylation of Smc5, Smc6, and Top2 in 10% ethanol. Each value represents the mean, and SEM values are indicated as bars (n = 3), two-way ANOVA with Bonferroni post hoc test was used to compare untreated zero time point vs. subsequent time points in the presence of ethanol; significant differences (p < 0.0001) are indicated (***).
Proteins were classified as being sumoylated in response to ethanol stress if the summed peptide counts from the 5-min and 60-min replicates exceeded the 0-min peptide counts by threefold or greater and were statistically significant (p ≤ 0.1) in their changes (Oeser et al., 2016; Supplemental Table 1). From the MS analysis, we found 18 proteins that appear to be sumoylated during acute ethanol exposure (Table 1), with 15 out of 18 identified as chromatin-binding and 7 out of 18 identified as chromatin-binding transcription factors by a gene ontology (GO) analysis (Table 2: Gcr1, Tec1, Hap1, Ste12, Cst6, Met4, and Upc2). Of the 18 proteins identified, the top three proteins were the chromatin structural proteins Top2, Smc5, and Smc6 (Figure 2B). We were most intrigued by Smc5 and Smc6 because they form a highly conserved complex with a known role in DNA replication and repair (Gill, 2004; Tsuyama et al., 2006; Duan et al., 2009b; Irmisch et al., 2009; Gallego-Paez et al., 2014; Menolfi et al., 2015).
TABLE 1:
Proteins sumoylated during acute ethanol exposure.
| Accession | Gene | Total Peptides | Description | ||
|---|---|---|---|---|---|
| 0 min | 5 min | 60 min | |||
| YNL088W | TOP2 | 24 | 109 | 104 | Topoisomerase II |
| YOL034W | SMC5 | 6 | 36 | 40 | Role in DNA replication and repair; in complex with Smc6 |
| YLR383W | SMC6 | 1 | 12 | 25 | Role in DNA replication and repair; in complex with Smc6 |
| YPL075W | GCR1 | 0 | 9 | 16 | Transcription activator |
| YOR038C | HIR2 | 1 | 3 | 13 | Regulation of histone gene transcription |
| YBR083W | TEC1 | 0 | 3 | 10 | Transcription factor, filamentation genes |
| YGR071C | ENV11 | 1 | 2 | 9 | Vacuolar function |
| YBL097W | BRN1 | 2 | 3 | 9 | Subunit of condensin complex |
| YLR256W | HAP1 | 1 | 21 | 9 | Zinc finger transcription factor |
| YDR485C | VPS72 | 1 | 6 | 8 | Htz1p-binding component of the SWR1 complex |
| YIL126W | STH1 | 1 | 4 | 8 | ATPase component of RSC chromatin remodeling complex |
| YHR084W | STE12 | 0 | 3 | 6 | Transcription factor activated by MAPK |
| YBR081C | SPT7 | 2 | 2 | 6 | Subunit of SAGA complex |
| YIL036W | CST6 | 0 | 17 | 4 | Basic leucine-zipper (bZIP) transcription factor |
| YJL092W | SRS2 | 0 | 4 | 1 | DNA helicase and DNA-dependent ATPase |
| YKL054C | DEF1 | 0 | 4 | 1 | RNAPII degradation factor |
| YNL103W | MET4 | 0 | 10 | 0 | Leucine-zipper transcriptional activator |
| YDR213W | UPC2 | 0 | 7 | 0 | Sterol regulatory element-binding protein |
TABLE 2:
Gene ontology analysis.
| GO terms from the molecular function ontology | |||
|---|---|---|---|
| GO term (GO ID) | Genes annotated | GO term usage | Genome frequency |
| DNA binding (GO:0003677) | TEC1, UPC2, ENV11, STE12, CST6, STH1, SRS2, DEF1, HAP1, SMC6, MET4, TOP2, SMC5, HIR2, GCR1 | 15 of 18 genes, 83.33% | 602 of 6411 annotated genes, 9.39% |
| DNA-binding transcription factor activity (GO:0003700) | TEC1, UPC2, STE12, CST6, HAP1, MET4, GCR1 | 7 of 18 genes, 38.89% | 180 of 6411 annotated genes, 2.81% |
We confirmed Smc5, Smc6, and Top2 were sumoylated during ethanol stress by enriching for sumoylated proteins from His6-FLAG-SMT3 cell lysates. To do this, we created C-terminally 3xHSV tagged versions of Smc5, Smc6, and Top2 that were expressed from their endogenous promoters. By Western analysis, each protein demonstrated multiple higher molecular weight SUMO conjugates after 10% (vol/vol) ethanol stress over a 2-h period (Figure 2C). We chose to verify Smc5, Smc6, and Top2 sumoylation over a 2-h time course to confirm that sumoylation remained stable in 10% ethanol. We next quantified the ethanol-induced sumoylation of Smc5, Smc6, and Top2 by measuring the intensity of the entire sumoylation ladder for each protein at each time point of ethanol exposure. Although this does not provide absolute numbers as it would for single bands, it is useful for a comparison of ethanol treatment with no treatment. Using this method, we found the ethanol-induced increase in sumoylation was statistically significant in all cases relative to no treatment (Figure 2D). Even though we confirmed that ethanol stress induced sumoylation of our top three hits (Smc5, Smc6, and Top2), we specifically chose to pursue Smc5 and Smc6 ethanol-induced sumoylation for these studies because their sumoylation during acute ethanol stress has not yet been reported.
As an additional analysis, we expressed 3xHSV-Smc5 in a yeast strain in which all lysine residues of the SMT3 gene itself have been mutated to arginine residues (Esteras et al., 2017), thus allowing us to determine whether the ethanol-induced sumoylation of Smc5 is due to multiple monosumoylation events or a chain of polysumoylation. In the case of Smc5, there was a predominant single sumoylation band pattern when expressed in the mutated SMT3 strain (Supplemental Figure 1). However, we did observe a slightly higher molecular weight sumoylated species that indicates there might be additional monosumoylation sites if SUMO chain formation is eliminated. While identification of all Smc5 and Smc6 sumoylation sites is of interest for future studies, here we chose to examine the broader features of ethanol-induced Smc5 and Smc6 sumoylation.
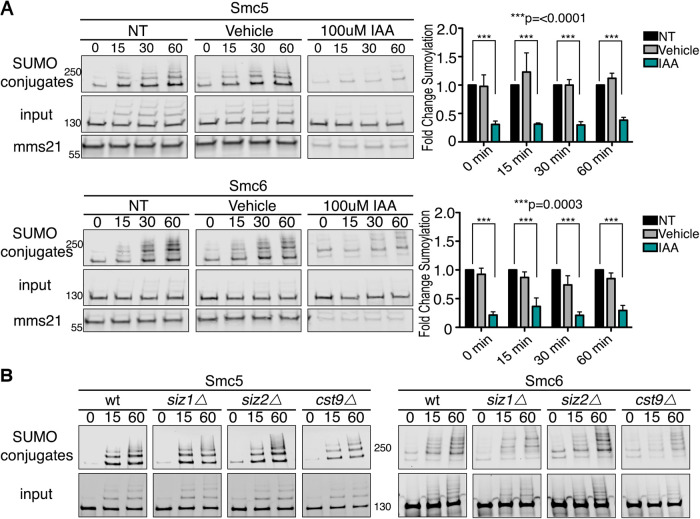
The E3 SUMO ligase Mms21 is necessary for the sumoylation of Smc5 and Smc6 during acute ethanol stress
After identifying Smc5 and Smc6 as sumoylation targets during acute ethanol stress, we wanted to determine the enzymes required for their ethanol-induced sumoylation. Unlike the ubiquitination system, which has more than 100 E3 ubiquitin ligases in yeast (De Bie and Ciechanover, 2011), the sumoylation system only has four known E3 SUMO ligases: Siz1, Siz2, Cst9, and Mms21 (Hay, 2001; Gill, 2004). It has been previously reported that the E3 for Smc5/6 is Mms21 during DNA damage (Duan et al., 2009a; Bermúdez-López et al., 2015; Liang et al., 2018), but it was unclear if this was the case during acute ethanol exposure. SIZ1, SIZ2, and CST9 are not essential genes and can be deleted. MMS21 is essential and we therefore used an auxin-inducible degron (AID) depletion strategy (Nishimura et al., 2009; Havens et al., 2012), wherein Mms21 was fused to an auxin degron and its depletion was induced by addition of auxin to the media. We found that, after Mms21 depletion by addition of auxin for 90 min, Smc5 and Smc6 sumoylation induced by acute ethanol exposure decreased approximately 75% during the 60-min time course (Figure 3A), consistent with the similar reduction in Mms21 levels. We also investigated whether the nonessential E3’s were involved in sumoylation induced by acute ethanol exposure. We found that complete loss of Siz1, Siz2, or Cst9 did not significantly reduce Smc5 and Smc6 sumoylation during acute ethanol stress (Figure 3B). Although we conclude that Mms21 is the likely E3 SUMO ligase for ethanol-induced sumoylation of Smc5 and Smc6 because it is a known interactor with the Smc5-Smc6 complex (Bermúdez-López et al., 2015; Horigome et al., 2016; Kim et al., 2016; Peng et al., 2018), there remains the possibility that the nonessential E3 SUMO ligases (Siz1, Siz2, and Cst9) share some redundancy such that individual deletions might not have a pronounced effect on Smc5 and Smc6 ethanol-induced sumoylation.
FIGURE 3:
The E3 SUMO ligase Mms21 promotes sumoylation of Smc5 and Smc6 during ethanol stress. (A) Cells expressing His6-FLAG-SMT3, MMS21-3HSV-AID, and C-terminally 3xHSV tagged Smc5 or Smc6 from their endogenous promoters were grown to early log phase, then treated with NT, vehicle, or auxin (IAA) for 90 min before exposure to acute ethanol for 60 min. Cell lysates (input) and purified sumoylated proteins (SUMO conjugates) were examined via Western analysis with an anti-HSV antibody to detect Mms21, Smc5, and Smc6. Inputs and Mms21 samples were run on the same gel and blot while SUMO conjugates were run separately on the same gel and blot. Blot images were cropped for optimal visualization. Fold changes in sumoylation were calculated utilizing values from Image Studio Lite software. Values were normalized to inputs and NT samples set to a value of 1.0. Error bars show SEM for triplicate samples. NT samples compared with IAA were significantly different in Smc5 and Smc6 with ***, p = <0.0001 and ***, p = 0.0003, respectively. (B) Wild-type (WT), siz1Δ, siz2Δ, or cst9Δ cells expressing His6-FLAG-SMT3 and either 3xHSV Smc5 or Smc6 were subjected to 60-min of acute ethanol (10% vol/vol) exposure. Cell lysates (input) and metal affinity purified sumoylated proteins (SUMO conjugates) were examined by Western analysis with an anti-HSV antibody. As in A, inputs and SUMO conjugates were run separately, but each on the same gel and blot. Blot images were only cropped for optimal visualization.
Ethanol-induced Smc5 and Smc6 sumoylation occurs in G1 and G2/M phases but is reduced in S phase
Sumoylation of proteins is essential for progression through the cell cycle and SUMO conjugation to its targets dynamically changes during different cell-cycle stages under normal conditions (Talamillo et al., 2020). A common feature of yeast cells is that exposure to stress often affects progression through the cell cycle (Jorgensen and Tyers, 2004). For example, yeast cells have been shown to halt in G1 after exposure to stress (Qu et al., 2019). This pause is generally thought to allow cells the time to resolve or adapt to the stress before proceeding through the cell cycle (Qu et al., 2019).
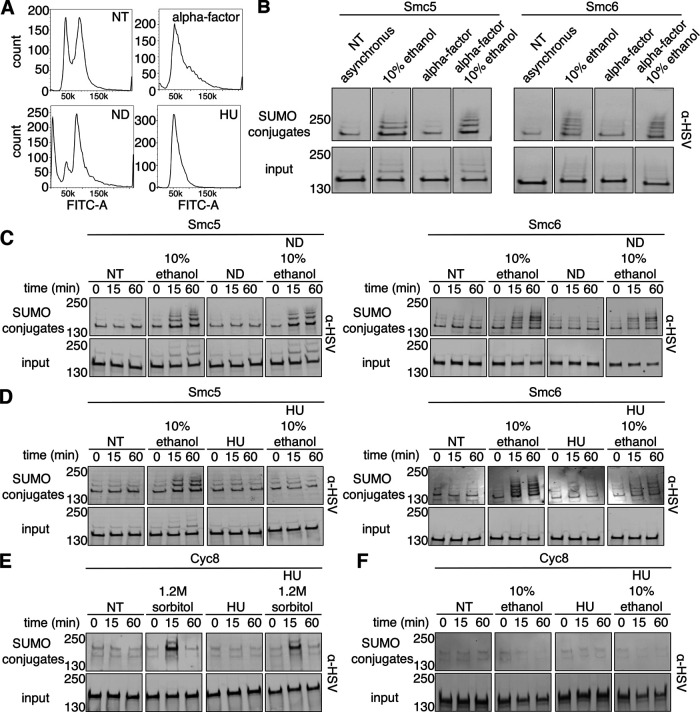
Chromatin undergoes regulated changes during the cell cycle (Ma et al., 2015; Antonin and Neumann, 2016). Because the Smc5-Smc6 complex is chromatin-associated and its chromatin localization is altered specifically during S phase (Jeppsson et al., 2014), we were interested in determining the stages of the cell cycle where Smc5 and Smc6 undergo ethanol-dependent sumoylation. We arrested cells in G1 using α-factor, early S phase using hydroxyurea (HU), and G2/M using nocodazole (ND). Cells arrested in G1 were generally observed to be either large and unbudded or with shmoo morphology (Supplemental Figure 2). Cells arrested in S phase were generally observed to be large with small buds. G2/M-arrested cells were generally observed as dumbbells. We next examined DNA content of cells in asynchronous culture after arrest at each stage in the cell cycle by staining cells with SYTOX green followed by examination using flow cytometry (Haase and Reed, 2002). Untreated cells exhibited 1C and 2C DNA content profiles consistent with asynchronous growth (Figure 4A). Cells arrested in G1 with α-factor had a 1C DNA content with a slight shoulder indicating a small subset of cells in early S phase. Our strains contain an intact BAR1 gene, which encodes a protease that cleaves and inactivates α-factor (Ciejek and Thorner, 1979), therefore some minor escape from G1 arrest was expected. Cells arrested in early S phase with HU had a 1C DNA content peak as has been previously described (Paillé et al., 2019). This peak is indicative of early S-phase arrest in asynchronous culture because hydroxyurea depletes dNTPs required for DNA synthesis in early S phase. Cells arrested in G2/M with ND had a 2C DNA content peak as anticipated.
FIGURE 4:
Ethanol exposure leads to Smc5 and Smc6 sumoylation in G1 and G2/M phases that is reduced in S phase. (A) Fluorescence histograms of asynchronous His6-FLAG-SMT3 cultures stained with SYTOX green: no treatment (NT), G1 arrest with α-factor, G2/M arrest with nocodazole (ND), and S arrest with hydroxyurea (HU). (B) Asynchronous cells expressing His6-FLAG-SMT3 and either C-terminally 3xHSV-tagged Smc5 or Smc6 from their endogenous promoters were arrested in G1 with α-factor for 90 min before treatment with 10% ethanol for 60 min. Cell lysates (input) and purified sumoylated proteins (SUMO conjugates) were examined by Western analysis with an anti-HSV antibody. (C, D) Similar experiments to B where asynchronous cells were arrested in G2/M with nocodazole and S phase with HU, respectively, for 90 min before exposure to 10% ethanol for a 60-min time course. (E) Asynchronous cells expressing His6-FLAG-SMT3 and 3xHSV Cyc8 from its endogenous promoter were arrested in S phase with HU for 90 min before 60 min of hyperosmotic (1.2 M sorbitol) stress. Cell lysates (input) and purified sumoylated proteins (SUMO conjugates) were examined by Western analysis using an anti-HSV antibody. (F) Similar experiment to E where asynchronous cells were arrested in S phase with HU for 90 min before 60 min of 10% ethanol stress. Inputs were run on one gel and blot and SUMO conjugates were run on one gel and blot with the exception of B where inputs and SUMO conjugates were run on the same gel and blot. Images were cropped for optimal visualization.
After confirming the appropriate arrest at each cell-cycle stage by microscopy (Supplemental Figure 2) and flow cytometry (Figure 4A), we examined whether Smc5 and Smc6 were sumoylated during each stage. We found that arrest in G1 allowed the same ethanol-induced sumoylation as asynchronous cells (Figure 4B). We also found that G2/M arrest allowed Smc5 and Smc6 ethanol-induced sumoylation similar to asynchronous cells (Figure 4C). By contrast, cells arrested in early S phase showed considerably reduced ethanol-induced sumoylation of Smc5 and Smc6 (Figure 4D). To determine that HU addition did not inhibit global sumoylation in the cell, we also examined sumoylation of the transcriptional corepressor Cyc8 during HU treatment and hyperosmotic stress (1.2 M sorbitol), which is a condition we previously found Cyc8 to be sumoylated (Oeser et al., 2016). Hyperosmotic stress-induced Cyc8 sumoylation still occurred after early S-phase arrest (Figure 4E), thereby ruling out an inhibitory effect of HU addition on global sumoylation. While we previously showed that Cyc8 is not sumoylated during acute ethanol stress (Oeser et al., 2016), we wanted to be certain that this was also the case after early S-phase arrest. We found that Cyc8 is not sumoylated after acute ethanol exposure during early S-phase arrest (Figure 4F). Altogether, Smc5 and Smc6 undergo ethanol-induced sumoylation during α-factor and ND arrests in G1 and G2/M phases, respectively, but do not during HU arrest in early S phase.
Smc5 and Smc6 sumoylation patterns during other stress conditions
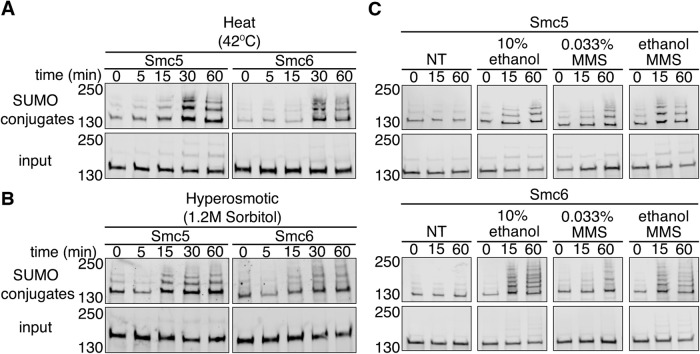
We previously reported that global sumoylation patterns and kinetics differ between distinct stress conditions (Oeser et al., 2016). To gain better insight into Smc5 and Smc6 sumoylation, we examined their sumoylation patterns during heat stress (42°C), hyperosmotic stress (1.2 M sorbitol), and DNA damage (exposure to methyl methanesulfonate [MMS]). After exposing cells to 42°C heat shock over a time course of 60 min, we found that Smc5 and Smc6 SUMO conjugates accumulated at a slower rate compared with exposure to ethanol (Figure 5A), though the pattern of sumoylation banding was similar to ethanol. While we did not identify Smc5 or Smc6 in our prior analysis of hyperosmotic stress (Oeser et al., 2016), we also subjected cells to hyperosmotic stress (1.2 M sorbitol) over 60 min to be thorough. The results were more complicated than ethanol and heat stress. We found that there was a rapid decrease in Smc5 and Smc6 sumoylation after 5 min before a rapid increase at 15 min that remained stable for the duration of the time course (Figure 5B). The decrease in Smc5 and Smc6 sumoylation is consistent with what we previously observed during hyperosmotic stress; proteins are desumoylated to provide a free pool of SUMO that is readily available for Tup1 and Cyc8 sumoylation (Oeser et al., 2016). There was an increase in Smc5 and Smc6 sumoylation following the initial decrease during hyperosmotic stress; we think that might be a return to basal sumoylation seen before hyperosmotic stress as the cells adapt to the stress and Tup1-Cyc8 are desumoylated, thus increasing the free SUMO pool (Oeser et al., 2016). However, it is possible that Smc5 and Smc6 are increasingly sumoylated during later time exposure of cells to hyperosmotic stress. We note it is difficult to assess what the indirect rebound effects on global sumoylation may be as cellular SUMO pools are restored during adaptation to hyperosmotic stress versus what the direct regulated events may be on protein sumoylation in later stages of adaption.
FIGURE 5:
Smc5 and Smc6 sumoylation patterns during other stress conditions. (A, B) Cells expressing His6-FLAG-SMT3 and C-terminally 3xHSV epitope tagged Smc5 or Smc6 from their endogenous promoters were exposed to either heat (42°C) or hyperosmotic (1.2 M sorbitol) stress over a 60-min time course. Cell lysates (input) and metal affinity purified sumoylated proteins (SUMO conjugates) were examined by Western analysis using an anti-HSV antibody. (C) Similar experiment to A but cells were subjected to either no treatment, 10% ethanol, 0.033% MMS, or 10% ethanol and 0.033% MMS over 60 min.
It has been reported that Smc5 and Smc6 are sumoylated after exposure to levels of MMS that induce DNA damage (Cremona et al., 2012; Chung and Zhao, 2015; Sarangi et al., 2015; Zapatka et al., 2019). We wanted to verify that Smc5 and Smc6 were sumoylated during an acute time course of MMS exposure, and whether the addition of ethanol with MMS treatment had any increased effect. We predicted that if both had the same effect, that addition of ethanol would not change Smc5 or Smc6 sumoylation beyond MMS treatment. We exposed cells to 0.033% MMS and found that Smc5 and Smc6 were sumoylated after addition of the DNA-damaging agent (Figure 5C). However, addition of ethanol with MMS increased Smc5 and Smc6 sumoylation when compared with MMS treatment alone but was generally equivalent to that of ethanol treatment alone (Figure 5C). We conclude that ethanol treatment may have a stronger dominant effect on Smc5 and Smc6 sumoylation in an asynchronous culture when compared with acute MMS treatment due to the stages of the cells in the cell cycle. We postulate the more pronounced effect of ethanol in an asynchronous culture might be due to the likelihood that maximal MMS-induced sumoylation of Smc5 and Smc6 occurs during S phase. This would be analogous to the MMS-induced sumoylation of the RecQ helicase Sgs1, which is limited to S phase (Bermúdez-López et al., 2016). Ethanol-induced sumoylation of Smc5 and Smc6 occurs during G1 and G2/M (Figure 4). G1 and G2/M are the predominant cell-cycle phases in an asynchronous culture (Figure 4A), so the results of the experiment in Figure 5C are consistent with a dominant effect of ethanol on Smc5 and Smc6 sumoylation during asynchronous growth when compared with MMS treatment alone
Acute ethanol stress induces Rad52 foci formation but not Rad53 phosphorylation
Chronic, long-term treatment of yeast cells with ethanol has been reported to increase DNA mutation rates through error-prone DNA polymerases being recruited to replication forks delayed in their progression (Voordeckers et al., 2020). Thus, we wanted to explore whether acute ethanol exposure led to any visible measures of chromatin structural alterations. It was not obvious how to measure in vivo global chromatin structural changes during an acute response of 60 min or less. We looked for examples in the literature but could not find any publications to our knowledge that queried in vivo chromatin changes in yeast under acute conditions of stress. Therefore, we decided to first examine in vivo global chromatin structure using chromatin staining and histone H2B localization by fluorescence microscopy (Figure 6A). We did not observe any obvious general differences over the 60-min time course of acute ethanol treatment.
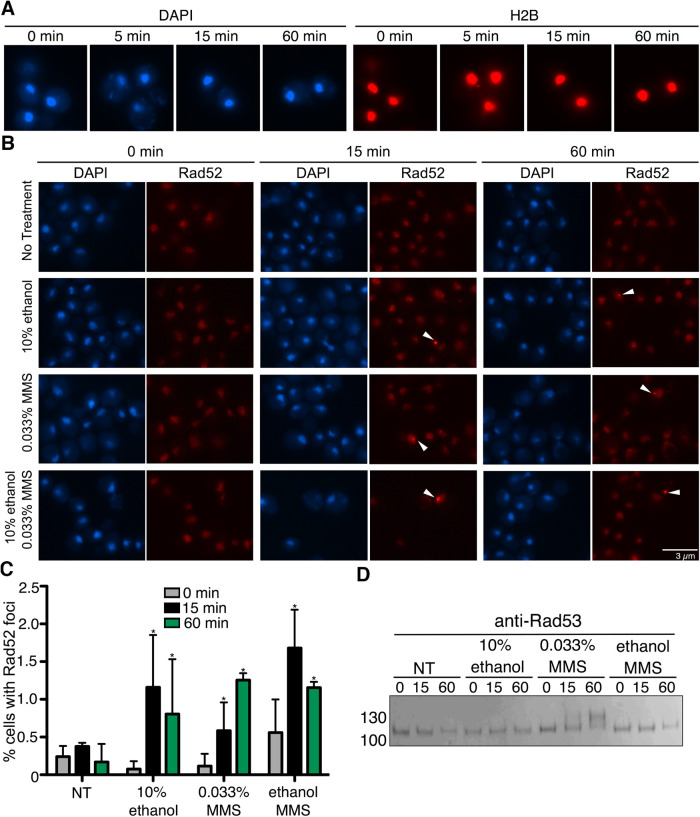
FIGURE 6:
Ethanol causes formation of Rad52 foci but not Rad53 phosphorylation. (A) H2B-mCherry cells were exposed to acute ethanol over a 60-min time course. Cells fixed at indicated time points and imaged by fluorescent microscopy. (B) Rad52-tdTomato cells were exposed to acute ethanol (10%), 0.033% MMS, or combined ethanol and MMS over a 60-min time course. Cells fixed at indicated time points and imaged by fluorescent microscopy. Five fields of cells for each condition with ≥40 cells/field were counted for the presence of nuclear foci (C). (D) Cells were exposed to acute ethanol (10%), 0.033% MMS, or combined ethanol and MMS over a 60-min time course. Rad53 phosphorylation was observed by Western analysis using an anti-Rad53 antibody.
We next examined more specific measures of chromatin changes. In vivo ways to examine chromatin changes resulting from damaging agents like MMS, which is an alkylating agent that methylates guanine and adenine moieties (Beranek, 1990), have been developed through observing foci formed by the homologous recombination protein Rad52 fused to tdTomato (Estrem and Moore, 2019), and phosphorylation of the intra-S-phase checkpoint kinase Rad53 (Sanchez et al., 1996). Though many studies examining Rad52 foci are usually conducted after extensive DNA damage caused by irradiation (Smith et al., 2019) and/or during specific phases of the cell cycle (Barlow and Rothstein, 2009; Smith et al., 2019), we chose to remain with the acute ethanol exposure conditions in an asynchronous culture that we used for the studies in the prior figures, especially because asynchronous cultures predominantly contain cells in G1 and G2/M phases where ethanol-induced sumoylation is most prominent (Figure 4).
For these studies, we examined cells that were either untreated, treated with 0.033% MMS or 10% ethanol alone, or 0.033% MMS plus 10% ethanol together. Compared to no treatment, 15-min treatment of cells with either 10% ethanol or 0.033% MMS caused the formation of Rad52 foci in about 1% of cells in an asynchronous cell culture (Figure 6, B and C), and this is consistent with that observed in prior studies examining MMS treatment in asynchronous cultures (Barlow and Rothstein, 2009; Smith et al., 2019). There was no increase in Rad52 foci frequency in cells treated with both 10% ethanol and 0.033% MMS relative to cells treated with either stress individually (Figure 6, B and C). By this measure, ethanol and MMS have a similar effect on Rad52 foci formation. Interestingly, acute ethanol treatment did not induce phosphorylation of Rad53, whereas MMS did cause Rad53 phosphorylation (Figure 6D), indicating that acute ethanol exposure does not trigger the intra-S-phase checkpoint like MMS exposure (Barlow and Rothstein, 2010). Importantly, addition of ethanol and MMS together did not trigger Rad53 phosphorylation, suggesting that ethanol exposure effects are epistatic to MMS exposure effects. Altogether, the collective data suggests that acute ethanol stress impacts chromatin, but the effects have some differences compared with the DNA-damaging agent MMS.
DISCUSSION
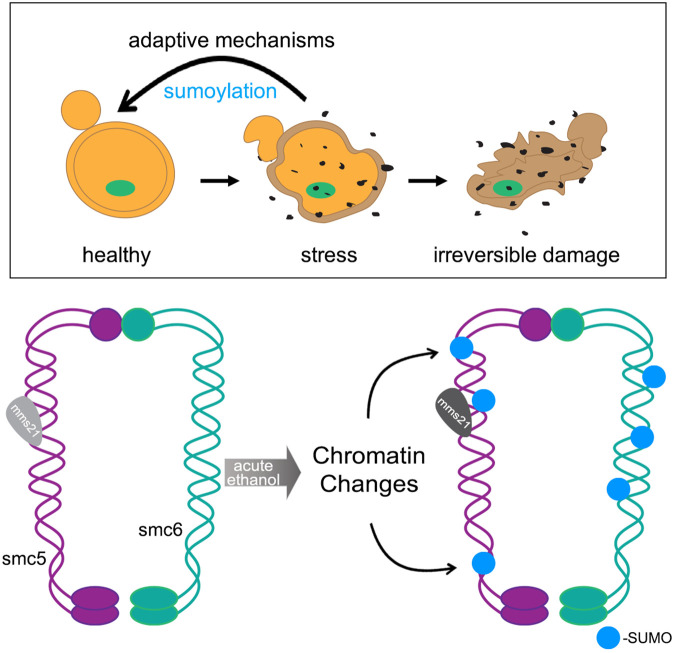
Adaptation to stress is essential for cellular survival, and the cell utilizes distinct multifaceted approaches to reestablish homeostasis during particular stress conditions. In the absence of stress adaptation, prolonged cellular stress can lead to the irreversible damage of cellular components that can ultimately impact cell viability (Figure 7). In the case of ethanol, chronic exposure to high concentrations of ethanol leads to alterations in membrane fluidity and lipid composition, increased production of ROS through altering oxidative phosphorylation in the mitochondria, and causes protein denaturing and misfolding (Kato et al., 2011, 2019; Auesukaree, 2017). In this study, we chose to examine proteins that become sumoylated during acute ethanol exposure. Of the 18 proteins we identified in a mass spectrometric analysis that become increasingly sumoylated during acute ethanol stress, we found that 15 of the 18 proteins are known to be chromatin-associated proteins (Table 2). This is not unexpected as protein sumoylation is known to regulate multiple chromatin-associated processes including the DNA damage checkpoint (Wu et al., 2014; Munk et al., 2017), regulation of chromosome structure (Tanaka et al., 1999; Nacerddine et al., 2005), and chromosome movement (Seeber and Gasser, 2017; Zhao, 2018).
FIGURE 7:
Model for cellular stress and Smc5/6 sumoylation.
Although not the main focus of this study, we think it is important to comment on the scope of proteins identified in the MS analysis. Of the 18 proteins discovered, 7 are known to function as transcription factors (Tables 1 and 2: Gcr1, Tec1, Hap1, Ste12, Cst6, Met4, and Upc2). The particular functions of these transcription factors reflect the ways in which ethanol alters cellular physiology. The cellular effects of ethanol exposure include alterations in membrane fluidity and lipid composition (Tóth et al., 2014), changes in glucose and amino acid uptake (Yang et al., 2012), a reduction in the activities of glycolytic enzymes (Tóth et al., 2014), and disruption of membrane integrity (Stanley et al., 2010). In terms of membrane fluidity, Upc2 is a transcription factor that undergoes regulated cleavage from the ER membrane to activate sterol biosynthesis genes (Joshua and Höfken, 2017), and it is known that ethanol exposure leads to the increased synthesis and presence of unsaturated fatty acids and ergosterol in the membrane (Henderson and Block, 2014). For changes in glucose and amino acid uptake, Gcr1 regulates genes involved in glycolysis (Hossain et al., 2016), and Met4 regulates genes in the sulfur amino acid biosynthesis pathway (McIsaac et al., 2012). Tec1 and Ste12 are involved in regulating filamentous and invasive growth pathways (Mayhew and Mitra, 2014). Cst6 is also known to regulate stress and carbon utilization pathways (Pohlers et al., 2017), and deletions are sensitive to ethanol stress (Liu et al., 2016). Altogether, it is notable that the cellular processes affected by ethanol exposure are regulated by transcription factors that are sumoylated during acute ethanol stress. Alteration of transcription factor activity in these pathways is consistent with yeast cells mounting an adaptative response to manage the cellular dysfunction that occurs with exposure to ethanol. Further studies are needed to verify the ethanol-induced sumoylation of these transcription factors and to understand the transcriptional responses that might occur through their ethanol-induced sumoylation and subsequent alteration of their function.
In addition to transcription factors, major proteins that showed increased sumoylation upon ethanol exposure were Smc5, Smc6, and Top2, which are known to form a highly conserved chromatin structure complex (Aragón, 2018). The Smc5-Smc6 complex is one of four highly conserved structural maintenance of chromosomes complexes found in eukaryotes and is best known for its role in DNA repair and overall genome stability (Aragón, 2018). It has been described that Smc5 and Smc6 sumoylation occurs as a regulatory consequence of stalled replication forks and has a functional role in modulating replication-associated repair and error-free DNA bypass via the Mph1 helicase (Zapatka et al., 2019). The Smc5-Smc6 complex also has been shown to interact with the E3 SUMO ligase Mms21 to promote the sumoylation of the STR helicase complex that acts in the removal of recombination intermediates (Bonner et al., 2016); however, we did not find members of the complex Sgs1, Top3, or Rmi1 to undergo increased sumoylation in our MS analysis (Supplemental Table 1). We did find that Smc5 and Smc6 are sumoylated during arrest of cells in the G1 and G2/M phases of the cell cycle after acute ethanol exposure (Figure 4), but we did not observe Smc5 and Smc6 sumoylation during early S-phase arrest induced by addition of HU. We believe this indicates that there are specific windows when ethanol-induced Smc5 and Smc6 sumoylation occurs. Considering that Smc5 and Smc6 have nearly identical chromatin localizations during G1 and G2/M phases but different ones during S phase (Jeppsson et al., 2014), it may be that chromatin context is important for Smc5 and Smc6 sumoylation during ethanol stress.
We also observed an increase in Smc5 and Smc6 sumoylation after exposure of cells to either ethanol or the DNA alkylating agent MMS (Figure 5C); however, ethanol-induced sumoylation had a predominant effect over MMS-induced sumoylation. It is possible that acute ethanol stress-induced Smc5-Smc6 sumoylation may operate through a different mechanism than MMS stress. We found that both ethanol and MMS exposure induced Rad52 foci to a similar extent (Figure 6, B and C), leading us to conclude that both might lead to Smc5 and Smc6 sumoylation through a chromatin structural change, though the nature of the change might be different. In fact, ethanol exposure did not trigger the intra-S-phase checkpoint as seen by the phosphorylation of Rad53, whereas MMS treatment did lead to Rad53 phosphorylation (Figure 6D). Currently, the function of ethanol-induced Smc5 and Smc6 sumoylation is not clear. Future experiments will be needed to determine whether Smc5 and Smc6 sumoylation is due to DNA damage, altered chromatin structure, or a response to protein misfolding that could lead to both DNA damage and/or chromatin structural loss. Consistent with this idea, we note that heat shock also induced Smc5 and Smc6 sumoylation, and heat shock can also lead to similar changes in protein/chromatin structure as well as DNA damage (Niskanen and Palvimo, 2017).
Overall, we conclude from the data presented here that there are two responses the cell elicits during acute ethanol exposure through sumoylation: one is to protect chromatin structure and the other is to mount an adaptive response through altered gene transcription. We previously found that sumoylation modulates a transient phase separation in the Tup1-Cyc8 transcriptional corepressor complex (Oeser et al., 2016), indicating a chromatin-modifying activity for sumoylation during hyperosmotic stress. From a transcriptional perspective, we know from our previous work (Oeser et al., 2016; Nadel et al., 2019) and other studies (Zhou et al., 2004; Stielow et al., 2008; Zhao, 2018) that genes involved either directly in transcription or its modulation have been reported to be sumoylated. How stress-induced sumoylation affects protein activity, localization, or stability remains an open question in the field, and it may be dictated by the magnitude and duration of the stress. Our studies indicate that sumoylation targets chromatin-associated proteins during ethanol stress adaptation, and support the idea that transient sumoylation is a common regulatory phenomenon during stress conditions.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strains and plasmids
Yeast strains and plasmids used in this study are listed in Table 3. Standard yeast genetic methods were used for this study (Guthrie, 1991). All gene deletions were verified by colony PCR and phenotypic analyses when available.
TABLE 3:
Yeast strains.
| Strain | Genotype | Reference |
|---|---|---|
| RGY 5266 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX | Oeser et al., 2016 |
| RGY 5824 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, cyc8Δ::NatMX, Cyc8-3HSV::LEU2 | Oeser et al., 2016 |
| RGY 6005 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, Smc6-3HSV::KanMX | This study |
| RGY 6014 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, Smc5-3HSV::KanMX | This study |
| RGY 6339 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, MMS21-3HSV-IAA1.T10::KanMX, AFB2::LEU2, Smc5-3HSV::URA3 | This study |
| RGY 6340 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, MMS21-3HSV-IAA1.T10::KanMX, AFB2::LEU2, Smc6-3HSV::URA3 | This study |
| RGY 6346 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, siz2Δ::KanMX, Smc5-3HSV::URA3 | This study |
| RGY 6347 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, cst9Δ::KanMX, Smc6-3HSV::URA3 | This study |
| RGY 6361 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, siz1Δ::KanMX, Smc5-3HSV::URA3 | This study |
| RGY 6362 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, siz1Δ::KanMX, Smc6-3HSV::URA3 | This study |
| RGY 6363 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, siz2Δ::KanMX, Smc6-3HSV::URA3 | This study |
| RGY 6344 | met15Δ0, his3Δ1, ura3Δ0, leu2Δ0, 6His-FLAG-SMT3::HIS3MX, cst9Δ::KanMX, Smc5-3HSV::URA3 | This study |
| yJM2468 | ura3-52, lys2-801, leu2-Δ1, his3-Δ200, trp1-Δ63, RAD52-tdTomato::HIS3MX | Estrem and Moore, 2019 |
Growth and stress conditions
Cells were grown to a density of ∼1.5 × 107 cells/ml at 30°C in yeast extract peptone dextrose (YPD) media before stress induction. All 0 time point samples were collected before stress induction. For ethanol stress, volume per volume amounts were added to cultures for a final concentration of 10% vol/vol ethanol. For hyperosmotic stress, equal volumes of culture and YPD + 2.4 M sorbitol were combined for a final concentration of 1.2 M sorbitol. For heat stress, cells were pelleted and resuspended in YPD media warmed to 42°C and placed in a shaking platform 42°C incubator.
Sumoylated protein purification
Aliquots (50 ml) of cells were collected at designated time points after stress and flash-frozen in liquid nitrogen. Harvested cells were lysed by vortexing with acid washed glass beads at 4°C in 1 ml denaturing lysis buffer (8 M urea, 50 mM Tris, pH 8.0, 0.05% SDS with 2 mM phenylmethylsulfonyl fluoride and 20 mM NEM [N-ethylmaleimide]). Aliquots representing 5% of the input were set aside. Cell lysates were incubated with TALON resin (Novagen) overnight at 4°C. Resin was washed three times with wash buffer (8 M urea, 50 mM Tris, pH 8.0, 200 mM NaCl, 0.05% SDS). Sumoylated proteins were eluted from beads with loading buffer (8 M urea, 10 mM MOPS, 10 mM EDTA, 1% SDS, 0.01% bromophenol blue, pH 6.8) and incubated at 65°C for 10 min.
Western analysis
Total cell lysates and purified sumoylated proteins were resolved by SDS–PAGE using 4–20% gradient gels (BioRad). Western analyses were performed with mouse anti-FLAG (1:2500; Sigma), mouse anti-HSV (1:2500; Novagen), rabbit anti-HSV (1:2000; Abcam), and rabbit anti-Rad53 (1:2000; Abcam).
MS analyses
Sumoylated proteins from cells exposed to 0, 5, or 60 min of 10% (vol/vol) ethanol stress were enriched by metal affinity chromatography as described above. Samples were run 1 cm into a 4–20% SDS–PAGE gel and bands were excised. Proteins in gel slices were digested with trypsin and digestion products desalted and dried by vacuum centrifugation. Dried peptide mixtures were resuspended in 7 µl of 0.1% formic acid. Five microliters was analyzed using a LTQ OrbiTrap mass spectrometer (Thermo Scientific). Complete MS methods were performed as previously described (Richardson et al., 2012).
The protein database search algorithm X!Tandem (Craig and Beavis, 2004) was used to identify peptides from the Saccharomyces Genome Database (http://www.yeastgenome.org). Peptide false discovery rates were measured using Peptide Prophet (Keller et al., 2002). Identified peptides were filtered using Peptide Prophet scores of ≥0.55 (∼10% false discover rate). The entire dataset is in Supplemental Table S1. The significance of the changes in peptide counts between 0, 5, and 60 min of ethanol stress was determined by a two-tailed, homoscedastic Student’s t test. Data was filtered by a p ≤ 0.1 and a ± threefold change in summed peptide counts. Final filtered data is in Table 1.
Auxin-degron depletion experiments
Cells were grown to a density of ∼0.86 × 107 cells/ml at 30°C in yeast complete media before addition of either NT (no treatment), vehicle (1:1000, 95% EtOH), or 100 μM 3-indoleacetic acid (IAA; Sigma). Cells were then incubated for 90 min at 30°C and before stress induction. Ethanol (10% [vol/vol]) was then added to cultures for an additional hour with 50 ml aliquots of cells collected at designated time points flash-frozen in liquid nitrogen before proceeding with sumoylated protein purification.
Cell-cycle analysis
Asynchronous cells were diluted back to a density of ∼0.39 × 107 cells/ml in YPD or YPD+1% dimethyl sulfoxide (nocodazole) media before cell-cycle halt and stress induction. For G1 arrest, 50 μg/ml α-factor (Sigma) was added immediately to cultures and incubated at 30°C for 90 min. For G2/M arrest, cells were incubated at 30°C for 2 h, then treated with 0.05 mg/ml nocodazole (Sigma) for an additional hour. For S arrest, 100 mM hydroxyurea (Sigma) was added immediately to cultures and incubated at 30°C for 90 min. All cell arrests were verified by pelleting a 200 µl aliquot of culture and examining under a phase contrast microscope (Nikon). Approximately 75–90% of cells per field were observed to be arrested at a given phase (Supplemental Figure 2). For G1 arrest cells were observed to be either large and unbudded or with shmoo morphology. Cells arrested in S phase were observed to be large with small buds. While G2/M-arrested cells were observed as dumbbells.
Flow cytometry analysis
Yeast cell-cycle analyses by flow cytometry were conducted similar to those previously described (Richardson et al., 2012). Cells were grown in 5-ml cultures to a density of ∼0.8 × 107 and exposed to the following conditions: no treatment, α-factor, nocodazole, or hydroxyurea and arrested as described above. After arrest, cells were harvested by centrifugation, fixed in 70% ethanol, washed with 50 mM sodium citrate containing 0.25 mg/ml RNase A, incubated at 95°C for 15 min, and then 37°C for 2 h. Fixed cells were resuspended in 50 mM sodium citrate, stained with 2µM SYTOX green (Invitrogen), and sonicated for 1 s at an amplitude of 10% immediately before analysis by flow cytometry. Cell-cycle analyses were performed with FlowJo software.
Fluorescence microscopy
Aliquots of cells at each time point after ethanol stress were removed, fixed in 4% paraformaldehyde solution for 15 min at room temperature, then washed with 1X phosphate-buffered saline.
Cells were imaged on a Nikon Eclipse 90i with a 100× objective, filters for GFP (HC HiSN Zero Shift filter set with excitation wavelength [450–490 nm], a dichroic mirror [495 nm], and emission filter [500–550 nm]), tdTomato (HC HiSN Zero Shift filter set with excitation wavelength [530–560 nm], a dichroic mirror [570 nm], and emission filter [590–650 nm]), or DAPI (HC HiSN Zero Shift filter set with excitation wavelength [325–375 nm], a dichroic mirror [400 nm], and emission filter [435–485 nm]), and a Photometrics Cool Snap HQ2 cooled CCD camera with NIS-Elements acquisition software.
Image processing
All blots were scanned using a Licor Odyssey CLx and ImageStudio Lite. All images were processed with a MacBook Pro or iMac computer (Apple) using Photoshop (Adobe).
Rigor and reproducibility
All biochemical and microbiological assays were performed in triplicate. For fluorescence microscopy, three separate researchers quantified foci formation. Statistics used were paired Student’s t tests (Figures 3 and 6) and two-way ANOVA with Bonferroni post hoc test (Figure 2).
Supplementary Material
Acknowledgments
We thank Jeffrey Moore (University of Colorado Anschutz Medical Campus) for the Rad52-tdTomato strain. We thank Luis Aragón (MRC Clinical Sciences Centre, Imperial College, London) for the yeast strain CCG9474 that allowed for identification of SUMO chains. We thank Matt Kaeberlein’s lab (University of Washington) for experimental assistance with the growth curves. We thank Xiaolan Zhao (Sloan Kettering Institute) and Sue Biggins (Fred Hutchinson Cancer Research Center) for critical advice. This work was supported by a National Institutes of Health/ National Institute of General Medical Sciences (NIH/NGIMS) training grant no. T32GM-007270 (A.I.B.), a Howard Hughes Medical Institute Gilliam fellowship GT10825 (A.I.B.), a NIH/NIGMS grant no. R01AG-031136 (R.G.G.), NIH/NIGMS grants no. R01GM-114112 and no. R35GM-136234 (R.G.G.).
Abbreviations used:
- AID
auxin inducible degron
- GFP
green fluorescent protein
- HU
hydroxyurea
- MMS
methyl methanesulfonate
- ND
nocodazole
- NEM
N-ethylmaleimide
- NT
no treatment
- PTM
posttranslational modification
- ROS
reactive oxygen species
- SMC
structural maintenance of chromosomes
- SUMO
small ubiquitin like modifier.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E20-11-0715) on March 31, 2021.
REFERENCES
- Antonin W, Neumann H (2016). Chromosome condensation and decondensation during mitosis. Curr Opin Cell Biol 40, 15–22. [DOI] [PubMed] [Google Scholar]
- Aragón L (2018). The Smc5/6 Complex: new and old functions of the enigmatic long-distance relative. Annu Rev Genet 52, 89–107. [DOI] [PubMed] [Google Scholar]
- Auesukaree C (2017). Molecular mechanisms of the yeast adaptive response and tolerance to stresses encountered during ethanol fermentation. J Biosci Bioeng 124, 133–142. [DOI] [PubMed] [Google Scholar]
- Barlow JH, Rothstein R (2009). Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J 28, 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Rothstein R (2010). Timing is everything: cell cycle control of Rad52. Cell Div 5, 29–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek DT (1990). Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res, Fundam Mol Mech Mutagen 231, 11–30. [DOI] [PubMed] [Google Scholar]
- Bermúdez-López M, Pociño-Merino I, Sánchez H, Bueno A, Guasch C, Almedawar S, Bru-Virgili S, Garí E, Wyman C, Reverter D, et al. (2015). ATPase-dependent control of the Mms21 SUMO ligase during DNA repair. PLoS Biol 13, e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez-López M, Villoria MT, Esteras M, Jarmuz A, Torres-Rosell J, Clemente-Blanco A, Aragon L (2016). Sgs1’s roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev 30, 1339–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JN, Choi K, Xue X, Torres NP, Szakal B, Wei L, Wan B, Arter M, Matos J, Sung P, et al. (2016). Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep 16, 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung I, Zhao X (2015). DNA break-induced sumoylation is enabled by collaboration between a sumo ligase and the ssDNA-binding complex RPA. Genes Dev 29, 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciejek E, Thorner J (1979). Recovery of S. cerevisiae a cells from G1 arrest by α factor pheromone requires endopeptidase action. Cell 18, 623–635. [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2004). TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467. [DOI] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Zhao X (2012). Sumoylation and the DNA damage response. Biomolecules 2, 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie P, Ciechanover A (2011). Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ 18, 1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H (2009a). Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell 35, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Yang Y, Chen YH, Arenz J, Rangi GK, Zhao X, Ye H (2009b). Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5-6 subcomplex and the hinge regions of Smc5 and Smc6. J Biol Chem 284, 8507–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteras M, Liu IC, Snijders AP, Jarmuz A, Aragon L (2017). Identification of SUMO conjugation sites in the budding yeast proteome. Microb Cell 4, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem C, Moore JK (2019). Astral microtubule forces alter nuclear organization and inhibit DNA repair in budding yeast. Mol Biol Cell 30, 2000–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Gorman AM, Hori O, Samali A (2010). Cellular stress responses: cell survival and cell death. Int J Cell Biol 2010, 214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Paez LM, Tanaka H, Bando M, Takahashi M, Nozaki N, Nakato R, Shirahige K, Hirota T (2014). Smc5/6-mediated regulation of replication progression contributes to chromosome assembly during mitosis in human cells. Mol Biol Cell 25, 302–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Yamazaki T, Kroemer G (2018). Linking cellular stress responses to systemic homeostasis. Nat Rev Mol Cell Biol 19, 731–745. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007). Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8, 947–956. [DOI] [PubMed] [Google Scholar]
- Gill G (2004). SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18, 2046–2059. [DOI] [PubMed] [Google Scholar]
- Guo C, Henley JM (2014). Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life 66, 71–77. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991). Guide to yeast genetics and molecular biology. Methods Enzymol 194, 1–863. [PubMed] [Google Scholar]
- Haase SB, Reed SI (2002). Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1, 117–121. [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL (2012). A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT (2001). Protein modification by SUMO. Trends Biochem Sci 26, 332–333. [DOI] [PubMed] [Google Scholar]
- Henderson CM, Block DE (2014). Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomycescerevisiae. Appl Environ Microbiol 80, 2966–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C, Bustard DE, Marcomini I, Delgoshaie N, Tsai-pflugfelder M, Cobb JA, Gasser SM (2016). PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev 30, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Claggett JM, Edwards SR, Shi A, Pennebaker SL, Cheng MY, Hasty J, Johnson TL (2016). Posttranscriptional regulation of Gcr1 expression and activity is crucial for metabolic adjustment in response to glucose availability. Mol Cell 62, 346–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch A, Ampatzidou E, Mizuno K, O’Connell MJ, Murray JM (2009). Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J 28, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson K, Carlborg KK, Nakato R, Berta DG, Lilienthal I, Kanno T, Lindqvist A, Brink MC, Dantuma NP, Katou Y, et al. (2014). The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet 10, e1004680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G (1997). The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J 16, 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M (2004). How cells coordinate growth and division. Curr Biol 14, 1014–1027. [DOI] [PubMed] [Google Scholar]
- Joshua IM, Höfken T (2017). From lipid homeostasis to differentiation: old and new functions of the zinc cluster proteins Ecm22, Upc2, Sut1 and Sut2. Int J Mol Sci 18, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Yamamoto Y, Izawa S (2011). Severe ethanol stress induces assembly of stress granules in Saccharomyces cerevisiae. Yeast 28, 339–347. [DOI] [PubMed] [Google Scholar]
- Kato S, Yoshida M, Izawa S (2019). Btn2 is involved in the clearance of denatured proteins caused by severe ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 19, 1–8. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Kim DH, Harris B, Wang F, Seidel C, McCroskey S, Gerton JL (2016). Mms21 SUMO ligase activity promotes nucleolar function in Saccharomyces cerevisiae. Genetics 204, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicki MC, Srikumar T, Johnson E, Raught B (2015). The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J Proteomics 118, 39–48. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP (2010). Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 186, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Li BZ, Tan AP, Kolodner RD, Putnam CD, Zhou H (2018). SUMO E3 ligase Mms21 prevents spontaneous DNA damage induced genome rearrangements. PLoS Genet 14, e1007250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Bergenholm D, Nielsena J (2016). Genome-wide mapping of binding sites reveals multiple biological functions of the transcription factor Cst6p in Saccharomyces cerevisiae. MBio 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kanakousaki K, Buttitta L (2015). How the cell cycle impacts chromatin architecture and influences cell fate. Front Genet 5, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew D, Mitra RD (2014). Transcription factor regulation and chromosome dynamics during pseudohyphal growth. Mol Biol Cell 25, 2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Petti AA, Bussemaker HJ, Botstein D (2012). Perturbation-based analysis and modeling of combinatorial regulation in the yeast sulfur assimilation pathway. Mol Biol Cell 23, 2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menolfi D, Delamarre A, Lengronne A, Pasero P, Branzei D (2015). Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance. Mol Cell 60, 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD (2013). Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in arabidopsis. Mol Cell Proteomics 12, 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Azhar SH, Abdulla R, Jambo SA, Marbawi H, Gansau JA, Mohd Faik AA, Rodrigues KF (2017). Yeasts in sustainable bioethanol production: a review. Biochem Biophys Reports 10, 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk S, Sigurðsson JO, Xiao Z, Batth TS, Franciosa G, von Stechow L, Lopez-Contreras AJ, Vertegaal ACO, Olsen JV (2017). Proteomics reveals global regulation of protein SUMOylation by ATM and ATR kinases during replication stress. Cell Rep 21, 546–558. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A (2005). The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9, 769–779. [DOI] [PubMed] [Google Scholar]
- Nadel CM, Mackie TD, Gardner RG (2019). Osmolyte accumulation regulates the SUMOylation and inclusion dynamics of the prionogenic Cyc8-Tup1 transcription corepressor. PLoS Genet 15, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6, 917–922. [DOI] [PubMed] [Google Scholar]
- Niskanen EA, Palvimo JJ (2017). Chromatin SUMOylation in heat stress: to protect, pause and organise?: SUMO stress response on chromatin. BioEssays 39, 1–10. [DOI] [PubMed] [Google Scholar]
- Oeser ML, Amen T, Nadel CM, Bradley AI, Reed BJ, Jones RD, Gopalan J, Kaganovich D, Gardner RG (2016). Dynamic Ssumoylation of a conserved transcription corepressor prevents persistent inclusion formation during hyperosmotic stress. PLoS Genet 12, e1005809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H (1999). In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun 254, 693–698. [DOI] [PubMed] [Google Scholar]
- Paillé A, Charton R, Muguet A, Griesenbeck J, Smerdon MJ, Conconi A (2019). Analyses of rRNA gene chromatin in cell cycle arrested Saccharomycescerevisiae cells. Data Br 25, 104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parapouli M, Vasileiadis A, Afendra AS, Hatziloukas E (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol 6, 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng XP, Lim S, Li S, Marjavaara L, Chabes A, Zhao X (2018). Acute Smc5/6 depletion reveals its primary role in rDNA replication by restraining recombination at fork pausing sites. PLOS Genet 14, e1007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlers S, Martin R, Hellwig D, Kniemeyer O, Saluz HP, Van Dijck P, Ernst JF, Brakhage A, Kurzai O (2017). Lipid signaling via Pkh1/2 regulates fungal CO2 sensing through the kinase Sch9. MBio 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Jiang J, Liu X, Wei P, Yang X, Tang C (2019). Cell cycle inhibitor Whi5 records environmental information to coordinate growth and division in yeast. Cell Rep 29, 987–994.e5. [DOI] [PubMed] [Google Scholar]
- Richardson LA, Reed BJ, Charette JM, Freed EF, Fredrickson EK, Locke MN, Baserga SJ, Gardner RG (2012). A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep 2, 372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ (1996). Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357–360. [DOI] [PubMed] [Google Scholar]
- Sarangi P, Steinacher R, Altmannova V, Fu Q, Paull TT, Krejci L, Whitby MC, Zhao X (2015). Sumoylation influences DNA break repair partly by increasing the solubility of a conserved end resection protein. PLoS Genet 11, e1004899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Gasser SM (2017). Chromatin organization and dynamics in double-strand break repair. Curr Opin Genet Dev 43, 9–16. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Bryant EE, Joseph FJ, Rothstein R (2019). DNA damage triggers increased mobility of chromosomes in G1-phase cells. Mol Biol Cell 30, 2620–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA (2010). The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol 109, 13–24. [DOI] [PubMed] [Google Scholar]
- Steensels J, Verstrepen KJ (2014). Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu Rev Microbiol 68, 61–80. [DOI] [PubMed] [Google Scholar]
- Stielow B, Sapetschnig A, Krüger I, Kunert N, Brehm A, Boutros M, Suske G (2008). Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol Cell 29, 742–754. [DOI] [PubMed] [Google Scholar]
- Talamillo A, Barroso-Gomila O, Giordano I, Ajuria L, Grillo M, Mayor U, Barrio R (2020). The role of SUMOylation during development. Biochem Soc Trans 48, 463–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y (1999). Characterization of a fission yeast SUMO-1 homologue, Pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol 19, 8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempé D, Piechaczyk M, Bossis G (2008). SUMO under stress. Biochem Soc Trans, 36, 874–878. [DOI] [PubMed] [Google Scholar]
- Tóth ME, Vígh L, Sántha M (2014). Alcohol stress, membranes, and chaperones. Cell Stress Chaperones 19, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama T, Inou K, Seki M, Seki T, Kumata Y, Kobayashi T, Kimura K, Hanaoka F, Enomoto T, Tada S (2006). Chromatin loading of Smc5/6 is induced by DNA replication but not by DNA double-strand breaks. Biochem Biophys Res Commun 351, 935–939. [DOI] [PubMed] [Google Scholar]
- Voordeckers K, Colding C, Grasso L, Pardo B, Hoes L, Kominek J, Gielens K, Dekoster K, Gordon J, Van der Zande E, et al. (2020). Ethanol exposure increases mutation rate through error-prone polymerases. Nat Commun 11, https://0.1038/s41467-020-17447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Ouyang J, Mori E, Nguyen HD, Maréchal A, Hallet A, Chen DJ, Zou L (2014). SUMOylation of ATRIP potentiates DNA damage signaling by boosting multiple protein interactions in the ATR pathway. Genes Dev 28, 1472–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KM, Lee NR, Woo JM, Choi W, Zimmermann M, Blank LM, Park JB (2012). Ethanol reduces mitochondrial membrane integrity and thereby impacts carbon metabolism of Saccharomyces cerevisiae. FEMS Yeast Res 12, 675–684. [DOI] [PubMed] [Google Scholar]
- Zapatka M, Pociño-Merino I, Heluani-Gahete H, Bermúdez-López M, Tarres M, Ibars E, Solé-Soler R, Gutiérrez-Escribano P, Apostolova S, Casas C, et al. (2019). Sumoylation of Smc5 promotes error-free bypass at damaged replication forks. Cell Rep 29, 3160–3172. [DOI] [PubMed] [Google Scholar]
- Zhao X (2018). SUMO-mediated regulation of nuclear functions and signaling processes. Mol Cell 71, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H (2004). Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279, 32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.