ABSTRACT
Topoisomerase I (Topo I) of Escherichia coli, encoded by topA, acts to relax negative supercoils in DNA. Topo I deficiency results in hypernegative supercoiling, formation of transcription-associated RNA-DNA hybrids (R-loops), and DnaA- and oriC-independent constitutive stable DNA replication (cSDR), but some uncertainty persists as to whether topA is essential for viability in E. coli and related enterobacteria. Here, we show that several topA alleles, including ΔtopA, confer lethality in derivatives of wild-type E. coli strain MG1655. Viability in the absence of Topo I was restored with two perturbations, neither of which reversed the hypernegative supercoiling phenotype: (i) in a reduced-genome strain (MDS42) or (ii) by an RNA polymerase (RNAP) mutation, rpoB*35, that has been reported to alleviate the deleterious consequences of RNAP backtracking and transcription-replication conflicts. Four phenotypes related to cSDR were identified for topA mutants: (i) one of the topA alleles rescued ΔdnaA lethality; (ii) in dnaA+ derivatives, Topo I deficiency generated a characteristic copy number peak in the terminus region of the chromosome; (iii) topA was synthetically lethal with rnhA (encoding RNase HI, whose deficiency also confers cSDR); and (iv) topA rnhA synthetic lethality was itself rescued by ΔdnaA. We propose that the terminal lethal consequence of hypernegative DNA supercoiling in E. coli topA mutants is RNAP backtracking during transcription elongation and associated R-loop formation, which in turn leads to transcription-replication conflicts and to cSDR.
IMPORTANCE In all life forms, double-helical DNA exists in a topologically supercoiled state. The enzymes DNA gyrase and topoisomerase I act, respectively, to introduce and to relax negative DNA supercoils in Escherichia coli. That gyrase deficiency leads to bacterial death is well established, but the essentiality of topoisomerase I for viability has been less certain. This study confirms that topoisomerase I is essential for E. coli viability and suggests that in its absence, aberrant chromosomal DNA replication and excessive transcription-replication conflicts occur that are responsible for lethality.
KEYWORDS: topoisomerase I, R loops, constitutive stable DNA replication, transcription-replication conflict, DNA supercoiling
INTRODUCTION
DNA in all cells is negatively supercoiled, and in bacteria such as Escherichia coli, two enzymes, gyrase and topoisomerase I (Topo I), ordinarily act oppositely to maintain the homeostasis of DNA superhelical density (reviewed in references 1 to 4). DNA gyrase is a hetero-tetrameric enzyme (comprised of two subunits each of GyrA and GyrB proteins) that is ATP dependent and introduces negative supercoils, whereas Topo I (encoded by topA) is an 865-amino-acid monomer that catalyzes relaxation of supercoiled DNA in an energy-independent reaction. An important role for Topo I is in the dissipation of negative supercoils, in accord with the twin-domain supercoiling model (5, 6), that are generated behind RNA polymerase (RNAP) in the transcription elongation complex (TEC).
That gyrase deficiency leads to bacterial death is well established. On the other hand, the essentiality of Topo I for viability, in either E. coli or closely related bacteria such as Salmonella enterica and Shigella flexneri, is somewhat less certain (7–13). One difficulty has been that topA mutants rapidly accumulate suppressors which are often in the genes encoding the gyrase subunits (8–10, 13–17), and consistent with their opposing actions, gyrase and Topo I mutations can, in combination, partially cancel one another’s sickness or inviability (18, 19). Growth of E. coli topA mutants is also improved upon overexpression or amplification of genes encoding the topoisomerase III (13, 18, 20) or IV (10, 16, 19).
Topo I deficiency is associated with an increased prevalence of R-loops (RNA-DNA hybrids) in the cells, which has been attributed to reannealing of the 5′ end of nascent RNA into hypernegatively supercoiled DNA behind the TEC under these conditions (21–23; for reviews, see references 4, 24, and 25). Overexpression of RNase HI (encoded by rnhA), which degrades RNA in RNA-DNA hybrids, can alleviate some of the phenotypes of topA mutants (18, 22, 23, 26–28), and conversely, topA rnhA mutants exhibit exacerbated sickness (13, 26, 29). In principle, R-loops can exert toxicity by acting as roadblocks to subsequent transcription (30, 31) and to replication (32–34); a third mechanism for toxicity is by serving as sites for initiation of aberrant chromosomal replication, as further outlined below. That R-loop formation is modulated by DNA supercoiling has been shown also in the CRISPR-Cas9 system (35) and in eukaryotic cells (36–38).
Recent evidence indicates that transcription-replication conflicts can themselves lead to increased formation of R-loops in the genome following RNAP backtracking at the sites of conflict (39–42; for reviews, see references 43 to 45). It has also been suggested that extended RNAP backtracking could be associated with R-loop formation from the 3′ end of the nascent RNA (40, 46).
R-loops are physiological initiators of ColE1 plasmid replication (47), but in addition, their excessive occurrence (as in rnhA mutants) can lead to pathological initiation of chromosomal DNA replication in both bacteria (reviewed in references 48 to 50) and eukaryotes (51). Such aberrant replication in bacteria is referred to as constitutive stable DNA replication (cSDR) since, unlike ordinary chromosomal replication, which is initiated at oriC with the aid of the unstable protein DnaA, it continues long after inhibition of protein synthesis in the cells. cSDR can be identified biochemically as persistent DNA synthesis following addition of translational inhibitors such as chloramphenicol or spectinomycin.
cSDR can also be identified genetically as rescue of lethality associated with loss of DnaA function, which is a more stringent test of cSDR, since it demonstrates the capability to duplicate the entire chromosome in the absence of oriC-initiated replication (48, 49). During its progression around the bacterial chromosome, such aberrant replication would be expected also to encounter (i) increased head-on conflicts with TECs on heavily transcribed genes (especially the rrn operons) that have evolved to be codirectional with oriC-initiated replisome progression and (ii) increased arrest at Ter sites flanking the terminus region which are bound by the Tus protein (52, 53). The occurrence of cSDR in rnhA mutants has been established through both biochemical and genetic assays (48). The protein DksA, which participates in avoidance or resolution of transcription-replication conflicts (54, 55), is also required for viability of rnhA dnaA mutants (56).
In recent work, Drolet’s group showed by biochemical assays that cSDR occurs in Topo I-deficient cells (28). Ogawa and coworkers and Kaguni and coworkers also showed that specificity for replication initiation from oriC in vitro requires both RNase HI and Topo I (57, 58).
In this study, we examined several topA insertion and deletion alleles for both their viability and their ability to rescue ΔdnaA lethality in E. coli. Our results indicate that topA-null alleles are lethal in the wild-type strain MG1655 but that they are viable in MDS42, which is an engineered derivative lacking all prophages and transposable elements (59). The null mutants of MG1655 were viable with rpoB*35, which encodes an RNAP variant that has been reported to alleviate the deleterious effects of transcription-replication conflicts (40, 52, 60–65). Both in MDS42 and with rpoB*35, the viable Topo I-deficient derivatives continued to exhibit increased negative supercoiling. One topA allele could also rescue ΔdnaA lethality, providing genetic confirmation of cSDR in Topo I-deficient strains. We propose that bacterial lethality in the absence of Topo I is caused by RNAP backtracking during transcription elongation and associated R-loop formation, which in turn lead to transcription-replication conflicts and to cSDR.
RESULTS
Description of topA insertion and deletion mutations and the assay to test for their viability.
Three pairs of topA mutations were constructed on the E. coli chromosome by the recombineering approach of Datsenko and Wanner (66), each pair being composed of an FRT (FLP recombination target)-flanked Kanr element and the corresponding derivative with Flp recombinase-mediated site-specific excision of the Kanr element to leave behind a “scar” of 27 in-frame codons; these are designated below by the suffixes “::Kan” and “::FRT,” respectively.
The three pairs of mutations represent the following (Fig. 1A): (i) deletion of all but the first codon and the last six codons (860 to 865) of the topA open reading frame (ΔtopA), that is, similar to the various gene knockouts of the Keio collection (67); (ii) insertion beyond codon 480 in topA (topA-Ins480), this position being chosen because an earlier study had shown that a Tetr insertion allele at this site was viable and associated with increased frequency of transposon precise excisions (11); and (iii) deletion from codon 480 to codon 860 in topA (topA-480Δ).
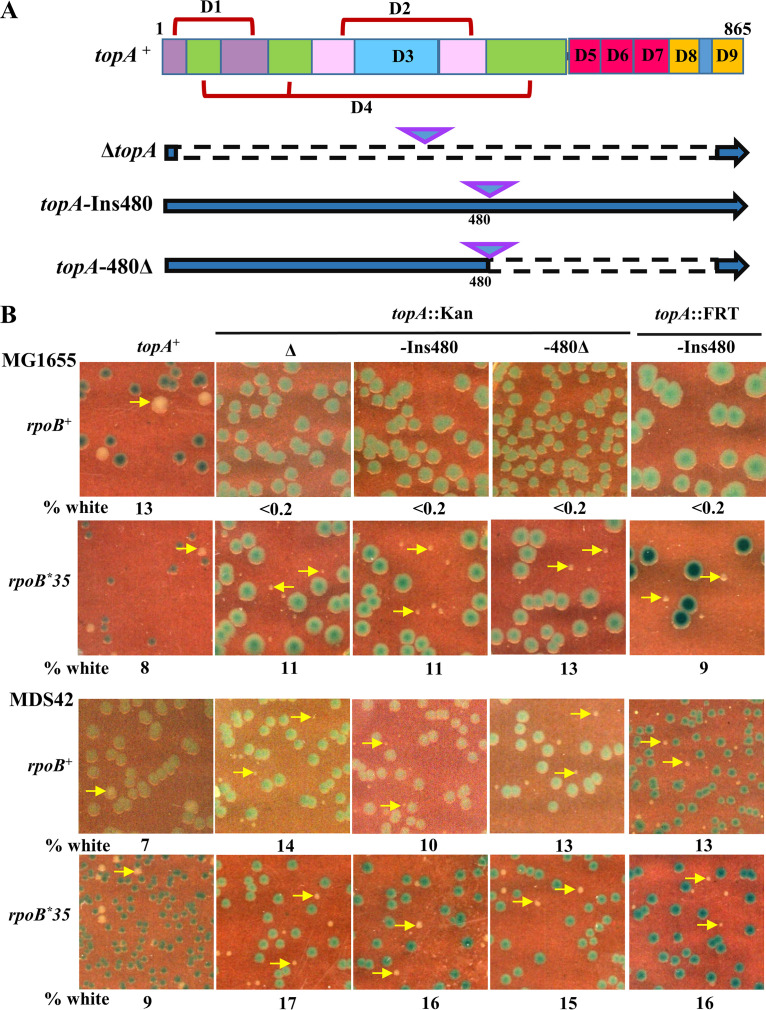
FIG 1.
(A) Representations of topA+ ORF delineating the encoded protein's domains D1 to D9 (adapted from references 87 and 88) and of the constructed topA alleles (three pairs) wherein the interrupted line segments represent deletions and each inverted triangle represents the pair comprising either Kanr insertion (::Kan allele) or its FRT derivative (::FRT allele). Domains D1, D2, and D4 of Topo I are each composed of two or more polypeptide segments that are split in the linear representation but are in proximity to one another in the three-dimensional protein structure. (B) Blue-white screening assay, on LB medium, of MG1655 and MDS42 strain derivatives with the topA+ shelter plasmid pHYD2390 and the different topA alleles as indicated at the top of each column; the rpoB allele status is indicated to the left of each row. Examples of white colonies are marked by yellow arrows. From left to right, strains used for the panels were pHYD2390 derivatives, as follows: row 1, GJ13519, GJ15603, GJ15604, GJ15688, and GJ16921; row 2, GJ16703, GJ16813, GJ16814, GJ16815, and GJ16854; row 3, GJ12134, GJ16816, GJ16817, GJ16818, and GJ18977; and row 4, GJ16819, GJ16820, GJ16821, GJ16822, and GJ17777.
All the topA mutations were constructed and maintained in derivatives that were also ΔlacZ on the chromosome and carried a shelter plasmid derivative of a single-copy IncW replicon encoding trimethoprim (Tp) resistance (68) with topA+ and lacZ+ genes. Since this plasmid’s segregation into daughter cells during cell division is not tightly controlled, around 10% of cells in a population are plasmid free. Only provided that these latter cells are viable, they grow as white colonies on Tp-free medium supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), whereas the plasmid-bearing cells grow as blue colonies on these plates. The appearance of white colonies which can be subsequently purified, therefore, is a demonstration of viability in the absence of the topA+ shelter plasmid, and we employed similar blue-white screening assays earlier for tests of viability with other essential genes, such as rho, nusG, dnaA, and rne (69–71).
Viability of topA mutations in MDS42 and MG1655 rpoB*35.
With the blue-white assay described above, we found that none of the six topA alleles is viable in MG1655 (Fig. 1B). These observations are consistent with those of Stockum et al. (13), who also employed a similar approach to conclude that topA is essential in E. coli.
By the same blue-white assay, we could show that all the topA mutations are viable in strain MDS42 and in rpoB*35 derivatives of both MG1655 and MDS42, on both LB and defined media (Fig. 1B; also, see Fig. S1A in the supplemental material); the growth of white colonies of the MDS42 topA derivatives was improved in the presence of rpoB*35, on both media (Fig. S1B). MDS42 is a derivative of MG1655 with 14% of its genome (comprising all prophages and transposable elements) deleted (59), while rpoB*35 is a mutation in RNAP that has been reported to render the enzyme less prone to backtracking or arrest and more accommodative of conflicts with replication (40, 52, 60–65).
In microscopy experiments (Fig. S2), cell size and morphology were unchanged with the rpoB*35 mutation alone in both MG1655 and MDS42. Cells of the MDS42 topA mutant were filamented, and the filamentation was to a large extent suppressed in the topA rpoB*35 derivative. The topA rpoB*35 derivative of MG1655 was also moderately filamented.
Growth rate experiments in liquid cultures did not yield reliable data because of extended lag times and accumulation of suppressors in the topA derivatives. Suppressor accumulation has also been documented earlier for topA mutants by other workers (8, 13). Based on the observation that the “white” topA mutant clones in the blue-white screening assay grow to colonies of around 108 cells in 48 h, we estimated a doubling time of around 100 min.
For reasons that are explained in Discussion, we tested whether suppression of topA lethality by rpoB*35 in MG1655 is abolished in the absence of the UvrD DNA helicase in the cells. In the blue-white assay, viable colonies of MG1655 rpoB*35 topA were obtained even in a ΔuvrD background, indicating that the suppression is UvrD independent (Fig. S3A).
Rescue of topA lethality in MDS42 or by rpoB*35 is not due to reversal of hypernegative supercoiling.
Lethality caused by loss of Topo I is associated with greatly elevated levels of negative supercoiling in vivo, and at least some suppressors of inviability, such as mutations in gyrA or gyrB and overexpression or amplification of genes encoding topoisomerases III or IV, also confer reversal of the hypernegative supercoiling phenotype (14–16, 18, 19; for reviews, see references 4 and 24). To examine whether the viability of topA null mutants in MDS42 and with rpoB*35 is correlated with reversal of hypernegative supercoiling, we determined their supercoiling status, by chloroquine-agarose gel electrophoresis (21, 72) of a reporter plasmid pACYC184 (73) in preparations made from the different strain derivatives.
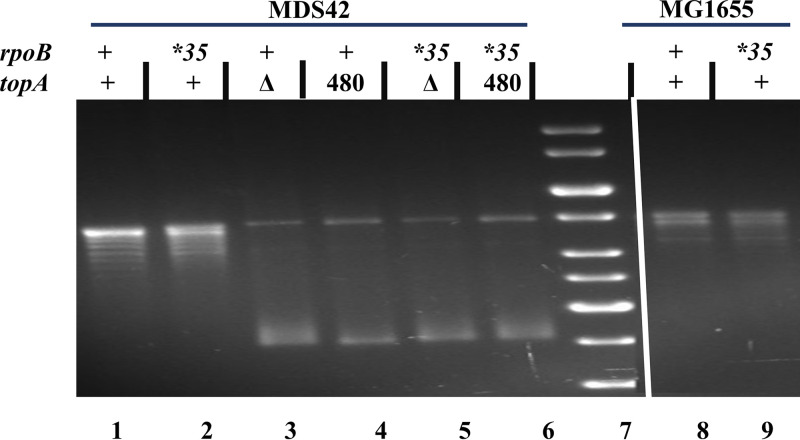
The results indicate that (i) in MDS42, both topA mutations tested confer increased supercoiling (Fig. 2, compare lanes 3 to 6 with lanes 1 to 2); (ii) rpoB*35 does not alter supercoiling, in both the topA+ (compare lanes 1 and 2, or compare lanes 8 and 9) and topA derivatives (compare lanes 3 and 4, or compare lanes 5 and 6); and (iii) supercoiling levels are not different between the strain backgrounds of MG1655 and MDS42 for both rpoB+ (compare lanes 1 and 8) and the rpoB*35 mutant (compare lanes 2 and 9). We conclude that when lethality conferred by loss of Topo I is suppressed either by genome size reduction in MDS42 or rpoB*35 or by both perturbations together, there is no concomitant reduction in the hypernegative supercoiling status of DNA in these mutants.
FIG 2.
Supercoiling status of reporter plasmid pACYC184 in topA+ and topA derivatives, as determined by chloroquine-agarose gel electrophoresis. Genotypes at topA and rpoB loci are indicated on top of each lane; for topA, Δ and 480 refer to ΔtopA::FRT and topA-Ins480::FRT, respectively. Strains were pACYC184 derivatives: lane 1, GJ12134; lane 2, GJ16819; lane 3, 18976; lane 4, GJ18977; lane 5, GJ17776; lane 6, GJ17777; lane 8, GJ18601; lane 9, GJ18910. Lane 7, DNA size standards.
topA lethality in MG1655 is not rescued by ectopic expression of the R-loop helicase UvsW.
Ectopic expression of the phage T4-encoded R-loop helicase UvsW (74) was previously shown to rescue lethality associated with increased R-loop prevalence in several different E. coli mutants. The latter include strains with combined deficiency of RNases HI and HII (69) or of RNase HI and RecG (75), as well as those with deletions of genes rho or nusG involved in factor-dependent transcription termination (69).
Since Topo I deficiency phenotypes are also associated with increased occurrence of intracellular R-loops and are partially suppressed by RNase HI overexpression (18, 21–23, 26–28), we employed the blue-white assays to examine whether UvsW expression (from a Ptac-UvsW chromosomal construct, induced with IPTG [isopropyl-β-d-thiogalactopyranoside]) could rescue MG1655 topA lethality; an MG1655 Δrho derivative (whose lethality is known to be rescued by UvsW) was chosen as a control. The results indicate that UvsW expression does not confer viability to the MG1655 topA derivative, whereas it could do so to the Δrho mutant (Fig. S3B). UvsW expression was associated with impaired growth of the topA+ blue colonies; this growth impairment was exemplified both by a marked decrease in plating efficiency and by occurrence of blue haloes around the colonies, suggestive of cell lysis. That UvsW expression is toxic to wild-type E. coli has been reported earlier (69).
Rescue of ΔdnaA lethality by Topo I deficiency.
As mentioned above, Topo I deficiency was earlier shown by a biochemical assay to confer cSDR, but whether it can rescue lethality associated with loss of DnaA function (that is, the genetic assay for cSDR) has not been determined. We adapted the blue-white assay to test whether any of the topA mutations can rescue lethality associated with loss of DnaA function, by constructing a Tpr lacZ+ shelter plasmid that carried both topA+ and dnaA+. Three different dnaA alleles were used in these experiments: ΔdnaA::Kan (70), which is a Keio-style insertion-deletion that has all but the first codon and the last six codons of the 468-codon-long dnaA ORF removed; its FRT derivative, ΔdnaA::FRT (70); and dnaA177 (76), whose DNA sequence determination revealed that it carries both a missense mutation in codon 267 (resulting in a Thr-to-Ilv substitution) and an amber nonsense mutation in codon 450. The strains also carried Δtus and rpoB*35 mutations, which facilitate cSDR-directed chromosome duplication by overcoming the problems posed, respectively, by the Ter sites and by excessive head-on transcription-replication conflicts (52, 70, 77, 78).
Of the six topA mutations tested that had been shown above to be lethal in MG1655, one (topA-Ins480::FRT) was able to rescue lethality of ΔdnaA::FRT and of dnaA177 at 30°C on both minimal and LB media (Fig. 3 and Fig. S4A), while the others yielded no viable white colonies (Fig. 3A). Even with topA-Ins480::FRT, there was no rescue imposed by DnaA deficiency at 37°C or 42°C (Fig. 3B; see also row 5 in each of the panels of Fig. S5), nor were viable colonies recovered with the ΔdnaA::Kan allele (Fig. 3A). On the other hand, ΔdnaA::Kan lethality was rescued by other cSDR-provoking mutations such as rnhA and dam (data not shown).
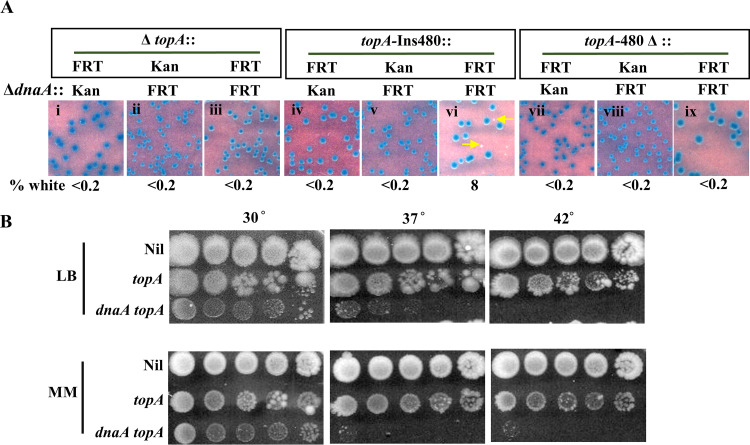
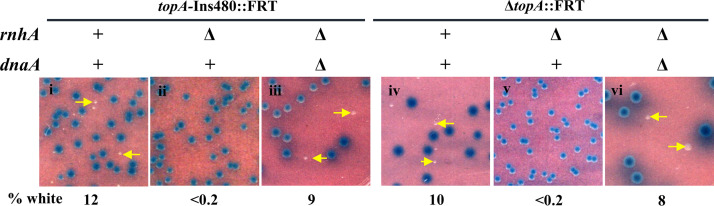
FIG 3.
Suppression of ΔdnaA lethality by topA. (A) Blue-white screening assay at 30°C on glucose-minimal medium A with topA+ dnaA+ shelter plasmid pHYD2390 in MG1655 ΔdnaA Δtus rpoB*35 derivatives carrying different topA alleles; the nature of the ΔdnaA allele (::Kan or ::FRT) and the topA allele is shown at the top of each panel. Examples of white colonies are marked by arrows. Strains employed for the different panels were pHYD2390 derivatives: i, GJ17786; ii, GJ17790; iii, GJ18940; iv, GJ17787; v, GJ17791; vi, GJ18941; vii, GJ17788; viii, GJ17792; and ix, GJ18942. (B) Serial dilution spotting on LB and glucose-minimal medium A (MM) at the indicated temperatures of isogenic topA dnaA derivatives of MG1655 Δtus rpoB*35, as follows: Nil, topA+ dnaA+ (GJ17784/pHYD2390); topA, topA-Ins480::FRT dnaA+ (GJ17784); and topA dnaA, topA-Ins480::FRT ΔdnaA::FRT (that is, the white colony from panel vi of Fig. 4A [GJ18941]); note that the topA+ ΔdnaA derivative of this strain is inviable.
Two distinct and interesting interpretations are suggested from these data: (i) unlike the other five topA alleles, topA-Ins480::FRT might possess a low level of DNA relaxation activity (since it encodes a full-length polypeptide with just a 27-amino-acid linker inserted between residues 480 and 481 of Topo I) which is not sufficient for viability per se in MG1655 but nonetheless is necessary for viability in the derivatives whose sole source of chromosome duplication is cSDR and (ii) expression of the essential dnaN gene immediately downstream of, and in the same operon as, dnaA is achieved from a fortuitous outward reading promoter in the Kanr element of the ΔdnaA::Kan allele, but this promoter is rendered inactive under Topo I-deficient conditions.
Notwithstanding these unusual features, our data clearly establish that cSDR in a Topo I-deficient derivative can act to rescue the lethality associated with total absence of DnaA in the cells. This viability is contingent on absence of the Tus protein (Fig. S4B). On the other hand, the DinG helicase, which has been shown to be needed for ΔdnaA viability of RecG- or Dam-deficient cells (70), was not required in the Topo I-deficient strain, nor did its absence impede viability of the topA mutant in the dnaA+ derivative (Fig. S4C).
Copy number analysis of different chromosomal regions in topA mutants.
In an exponentially growing population of bacterial cells, DnaA-initiated replication imposes a bidirectional gradient of copy number for different regions of the circular chromosome, with the peak near oriC and trough in the terminus region (53). If a dnaA+ strain also suffers a perturbation that activates cSDR (such as deficiency of RNase HI, RecG, Dam, or multiple DNA exonucleases), the DNA copy number pattern is characterized by superposition of a “midterminus peak” on the bidirectional gradient described above (52, 70, 77–82). Previously, we proposed that the midterminus peak represents a population aggregate of replication forks progressing from stochastically firing cSDR origins that are widely distributed across the genome (53, 70), although other groups have suggested that it represents a discrete origin of replication (52, 77–79), or occurrence of overreplication when oppositely directed forks converge at the terminus (80, 81).
Brochu and coworkers have shown earlier that Topo I-deficient mutants exhibit the midterminus peak (19), but their strains also carried additional genetic changes, such as a gyrB(Ts) mutation and amplification of the genes encoding subunits of topoisomerase IV. For our DNA copy number analysis studies, we used dnaA+ strains of the MDS42 background without or with rpoB*35 and additionally with the topA-Ins480::FRT mutation (that is, the allele associated with rescue of ΔdnaA inviability).
The whole-genome sequence reads obtained from each of the strains were aligned to the MDS42 reference sequence, and normalized read counts for the different chromosomal regions were determined. No suppressor mutation in any of the candidate genes was identified in the topA mutants, while the presence of the topA mutation itself and of the CAC-to-CAA codon change (His-to-Gln substitution) associated with the rpoB*35 allele (60) was confirmed in each of the relevant strains.
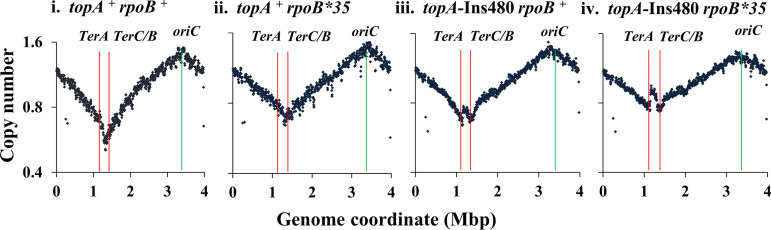
The parental (topA+ rpoB+) strain exhibited the expected bidirectional copy number gradient from oriC to Ter (Fig. 4, panel i), which was also largely preserved in its rpoB*35 derivative (Fig. 4, panel ii). The topA mutant derivatives of both these strains showed distinct midterminus peaks superimposed on the oriC-to-Ter gradient (Fig. 4, panels iii and iv, respectively).
FIG 4.
Copy number analysis by deep sequencing in topA mutant derivatives of MDS42. Relative copy numbers are plotted as semi-log graphs for overlapping 10-kb intervals across the genome (the relevant genotype of each strain is indicated at the top of each panel); positions of oriC, TerA, and TerC/B are marked, and the gap at around 0.3 Mbp in each of the plots corresponds to the argF-lac deletion present in the strains. Strains used for the different panels are as follows: i, GJ12134; ii, GJ16819; iii, GJ18977; and iv, GJ17777.
These results therefore confirm that a topA mutation capable of conferring ΔdnaA viability is associated with a midterminus peak of DNA copy number in dnaA+ derivatives.
Mutual suppression between lethal mutations: loss of DnaA suppresses topA-rnhA synthetic lethality.
Deficiency of either Topo I or RNase HI is associated with cSDR, and Stockum et al. (13) as well as Drolet and coworkers (26, 29) previously reported lethality or aggravated sickness in the double-deficient strains. We too found in this study that introduction of the rnhA mutation into otherwise viable topA derivatives (that is, in the MG1655-derived strain with rpoB*35 and Δtus mutations) confers synthetic lethality; the two topA alleles tested were the FRT derivatives of topA-Ins480 and ΔtopA (Fig. 5, compare panels i and ii for the former and iv and v for the latter). The lethalities were rescued in the presence of ΔdnaA (Fig. 5, panels iii and vi, respectively), indicating that two otherwise lethal mutant combinations (topA rnhA and dnaA) could mutually suppress one another. Robust viability of the triple mutants was observed on both rich and defined media at 30°C and 37°C but less so at 42°C (Fig. S5).
FIG 5.
Synthetic topA rnhA lethality, suppressed by ΔdnaA. Blue-white screening assay at 30°C on glucose minimal medium A with the topA+ dnaA+ shelter plasmid pHYD2390 in MG1655 Δtus rpoB*35 derivatives carrying different alleles of topA, rnhA, and dnaA as indicated at the top of each panel. Strains employed for the different panels were pHYD2390 derivatives: i, GJ17784; ii, GJ19609; iii, GJ18951; iv, GJ17783; v, GJ19608; and vi, GJ18983.
We performed PCR experiments to confirm that the chromosomal topA locus in each of the viable triple-mutant topA rnhA dnaA derivatives was indeed disrupted (and had not, for example, become topA+ by gene conversion from the wild-type allele on the shelter plasmid). Two primer pairs were used simultaneously to distinguish between the topA+, ΔtopA::FRT, and topA-Ins480::FRT alleles, which yielded amplicons of 500, 328, and 581 bp, respectively (Fig. S6A and B). The results established that the signatures for the topA-Ins480 and ΔtopA mutations were present, and that the one for topA+ was absent, in the triple-mutant strains (Fig. S6B, lanes 5 and 7, respectively).
As discussed below, these results suggest that it is excessive chromosomal replication which kills cells doubly defective for Topo I and RNase HI.
DISCUSSION
The enzymes Topo I and DNA gyrase act to maintain the homeostatic balance of DNA negative supercoiling in E. coli. Topo I relaxes negative supercoils, especially those occurring behind RNAP during transcription elongation, and thus prevents the nascent transcript from reannealing with the template DNA strand to form R-loops.
In this study, we confirmed that Topo I-deficient E. coli mutants are inviable and furthermore have identified two novel means by which the lethality can be independently and additively suppressed: (i) by deletion of the nonessential 14% of the genome comprising prophages and transposable elements, as in strain MDS42, and (ii) by the rpoB*35 mutation, encoding an altered RNAP, which has been variously described (not mutually exclusive) to mimic the transcriptional effects of ppGpp (60), to reduce RNAP backtracking (40), and to mitigate the effects of transcription-replication conflicts by destabilizing the TEC (40, 52, 62, 63, 65). Neither of the suppressors acts by reversing hypernegative supercoiling in the topA mutants. We have also shown that Topo I deficiency, in the presence of additional rpoB*35 and Δtus mutations, can rescue ΔdnaA lethality, thereby providing genetic confirmation for occurrence of cSDR in the Topo I-deficient derivatives.
rpoB*35 and RNAP backtracking.
As mentioned above, several workers have provided evidence that the rpoB*35-encoded substitution in RNAP destabilizes the TEC in vitro (40, 62) and protects against transcription-replication conflicts in vivo (65), including during cSDR (52, 70, 77, 78). Trautinger and Lloyd (61) reported that rpoB*35 suppresses the Ts phenotype of greA greB double mutants and the UV sensitivity of an mfd mutant, which they interpret as evidence that it may function by preventing backtracking, thus facilitating dissociation of stalled TECs. Likewise, rpoB*35 also suppresses rep uvrD lethality, which has been ascribed to direct reduction of replicative barriers posed by TECs under these conditions (63).
On the other hand, there is one report from the Nudler group that RpoB*35-substituted RNAP exhibits increased backtracking in vitro in the presence of UvrD (64). This property of RpoB*35-RNAP appears to be strictly UvrD dependent, and the same group has shown in other studies (40) that relative to wild-type RNAP, the RpoB*35 enzyme is resistant to pausing and backtracking.
It is therefore reasonable to conclude that the RpoB*35 enzyme is in general more resistant than wild-type RNAP to arrest and backtracking during transcription elongation, except perhaps in the specific context when a high concentration of UvrD dimers occurs following DNA damage. Our finding in this study, that the suppression of topA lethality by rpoB*35 is UvrD independent (Fig. S3A), is noteworthy in this context.
Mechanism of lethality in Topo I-deficient strains.
The fact that rpoB*35 restores growth to MG1655 in the absence of Topo I without affecting the hypernegative supercoiling status of the mutants suggests that it is the downstream consequences of increased negative supercoiling, namely, RNAP backtracking and impairment of TEC progression leading to transcription-replication conflicts, which are responsible for topA lethality. Pathological R-loop formation is also expected to be an important feature at the arrested TECs, but whether it precedes or follows RNAP backtracking remains to be determined. In the topA mutant, rpoB*35 would also relieve the sickness during cSDR engendered by transcription-replication conflicts especially at the rrn operons.
To explain topA viability in MDS42, we propose that the regions of the genome that are deleted in this strain (comprising prophages and transposable elements) are preferentially enriched for sites of R-loop formation, TEC arrest and transcription-replication conflict. Loss of the proteins Rho and NusG, which are involved in factor-dependent transcription termination and reportedly in R-loop avoidance (31, 69, 83–85), is also better tolerated in MDS42 than in MG1655, and especially so in the presence of rpoB*35 (65, 69, 86).
Finally, why does ectopic expression of the R-loop helicase UvsW not rescue topA lethality, although it can rescue the lethalities associated with loss of RNase H enzymes, RecG, Rho, or NusG (69, 75)? One possibility is that R-loop formation in Topo I-deficient strains is a consequence, and not a cause, of RNAP backtracking and arrest, so that R-loop removal per se would not mitigate the primary problem. An alternative possibility is that Topo I itself is required to relax the negative supercoils arising from UvsW’s helicase action on R-loops, and hence that UvsW is unable to act efficiently to unwind R-loops specifically in the topA mutants. The fact that RNase HI overexpression can suppress topA sickness phenotypes (26, 27) lends support to the second model.
Topo I deficiency and cSDR.
Martel and coworkers provided biochemical evidence for cSDR in Topo I-deficient cultures (28), which is presumably initiated from R-loops in these cells; our data establish that such cSDR is sufficient to sustain viability in the absence of DnaA, in derivatives carrying Δtus and rpoB*35 mutations. The latter two mutations are expected to facilitate the completion of replication of the circular chromosome by forks initiated from a site(s) other than oriC (52, 70, 77, 78). Our data also support the earlier suggestion (70) that incubation at 30°C is more permissive than that at 37°C or 42°C for rescue by cSDR of ΔdnaA lethality.
Of the six different topA mutations that were inviable in MG1655-derived strains, it was only the topA-Ins480::FRT allele that could confer viability to the ΔdnaA derivatives. As explained above, this mutation generates a modified version of Topo I in which a 27-amino-acid linker is inserted between residues 480 and 481 of the polypeptide. From the Topo I monomer crystal structure (87, 88), it is expected that the linker is situated at or near the junction between residues that make up domain D2 and those that make up domain D4; it is therefore possible that the linker allows the (albeit inefficient) folding of the polypeptide to yield a correct tertiary structure. The residual Topo I activity of this protein might be needed for proper chromosome segregation after cSDR in the ΔdnaA mutants (20).
oriC-initiated replication contributes to topA-rnhA synthetic lethality.
We have shown that although the ΔtopA and ΔrnhA combination is synthetically lethal, the ΔtopA ΔrnhA ΔdnaA mutant is viable. Thus, oriC-initiated replication is a contributor to topA rnhA toxicity, which suggests that it is excessive replication (sum of that from oriC and cSDR, the latter contributed additively by both rnhA and topA) which confers toxicity. These results are in agreement with those from Usongo and coworkers (20, 29), who reported earlier that mutations affecting either replication from oriC or replication restart functions can alleviate the sickness of cells deficient for both Topo I and RNase HI activities.
MATERIALS AND METHODS
Growth media, bacterial strains, and plasmids.
The rich and defined growth media were, respectively, LB and minimal A with 0.2% glucose (89), and unless otherwise indicated, the growth temperature was 37°C. Supplementation with X-Gal and the antibiotics ampicillin, kanamycin (Kan), tetracycline (Tet), chloramphenicol (Cm), and trimethoprim (Tp) were at the concentrations described earlier (90). Isopropyl-β-d-thiogalactoside (IPTG) was added at the indicated concentrations. E. coli strains used are listed in Table S1 in the supplemental material.
Plasmids described earlier include pMU575 (Tpr, single-copy-number vector with lacZ+) (68); pACYC184 (Tetr Cmr, p15A replicon) (73); pHYD2388 (70) and pHYD2411 (69) (Tpr, pMU575 derivatives with, respectively, dnaA+ and rho+); and pKD13, pKD46 and pCP20, described by Datsenko and Wanner (66), for recombineering experiments and Flp recombinase-catalyzed excision between a pair of FRT sites. The construction of two derivatives of plasmid pMU575 is described in the supplemental material: pHYD2382, carrying topA+, and pHYD2390, carrying topA+ dnaA+.
Blue-white screening assays.
To determine lethality or viability of strains with chromosomal topA or dnaA mutations, derivatives carrying the shelter plasmid pHYD2382 (topA+) or pHYD2390 (topA+ dnaA+) were grown overnight in Tp-supplemented medium, subcultured into medium without Tp for growth to mid-exponential phase, and plated at suitable dilutions on X-Gal plates without Tp. The percent white colonies was determined (minimum of 500 colonies counted), and representative images were captured.
Plasmid supercoiling assays.
Strains carrying plasmid pACYC184 were grown in LB to mid-exponential phase, and plasmid preparations were made with the aid of a commercial kit. Plasmid supercoiling status in each of the preparations was determined essentially as described elsewhere (72), following electrophoresis on 1% agarose gels with 5 μg/ml chloroquine at 3 V/cm for 17 h.
Copy number analysis by deep sequencing.
Copy number determinations of the various genomic regions were performed essentially as described (70). Genomic DNA was extracted by the phenol-chloroform method from cultures grown in LB to mid-exponential phase, and paired- and single-end deep sequencing was performed on Illumina platforms to achieve around 100-fold coverage for each preparation. Sequence reads were aligned to the MDS42 reference genome (accession number AP012306.1), and copy numbers were then determined by a moving-average method after normalization of the base read count for each region to the aggregate of aligned base read counts for that culture.
Other methods.
Procedures were as described for P1 transduction (91) and for recombinant DNA manipulations, PCR, and transformation (92). Different chromosomal topA mutations were generated by recombineering (66) as described in the supplemental material. For microscopy experiments, cells from cultures grown in LB to mid-exponential phase were immobilized on 1% agarose pads and visualized by differential interference contrast imaging with the aid of a Zeiss Axio Imager Z2 microscope.
Data availability.
Genome sequence data from this work have been submitted under accession number PRJNA670792 and are available for public access at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA670792.
ACKNOWLEDGMENTS
We thank Jillella Mallikarjun for sequencing the dnaA177 allele, Aswin Seshasayee and T. V. Reshma for help with deep sequencing, V. Balaji for assistance with microscopy, and the COE team members for advice and discussions.
This work was supported by Government of India funds from (i) DBT Centre of Excellence (COE) project for Microbial Biology – Phase 2, (ii) SERB project CRG/2018/000348, and (iii) DBT project BT/PR34340/BRB/10/1815/2019. J.G. was the recipient of the J C Bose fellowship and INSA Senior Scientist award.
We declare that there are no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
J. Gowrishankar, Email: shankar@iisermohali.ac.in.
Ann M. Stock, Rutgers University-Robert Wood Johnson Medical School
REFERENCES
- 1.Vos SM, Tretter EM, Schmidt BH, Berger JM. 2011. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol 12:827–841. 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen SH, Chan NL, Hsieh TS. 2013. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170. 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 3.Bush NG, Evans-Roberts K, Maxwell A. 2015. DNA topoisomerases. EcoSal Plus 6:ESP-0010-2014. 10.1128/ecosalplus.ESP-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brochu J, Breton EV, Drolet M. 2020. Supercoiling, R-loops, replication and the functions of bacterial type 1A topoisomerases. Genes 11:249. 10.3390/genes11030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu LF, Wang JC. 1987. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A 84:7024–7027. 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorman CJ. 2019. DNA supercoiling and transcription in bacteria: a two-way street. BMC Mol Cell Biol 20:26. 10.1186/s12860-019-0211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sternglanz R, DiNardo S, Voelkel KA, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang JC. 1981. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci U S A 78:2747–2751. 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson SM, Higgins CF, Lilley DM. 1984. The genetic control of DNA supercoiling in Salmonella typhimurium. EMBO J 3:1745–1752. 10.1002/j.1460-2075.1984.tb02041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni Bhriain N, Dorman CJ. 1993. Isolation and characterization of a topA mutant of Shigella flexneri. Mol Microbiol 7:351–358. 10.1111/j.1365-2958.1993.tb01127.x. [DOI] [PubMed] [Google Scholar]
- 10.McNairn E, Ni Bhriain N, Dorman CJ. 1995. Overexpression of the Shigella flexneri genes coding for DNA topoisomerase IV compensates for loss of DNA topoisomerase I: effect on virulence gene expression. Mol Microbiol 15:507–517. 10.1111/j.1365-2958.1995.tb02264.x. [DOI] [PubMed] [Google Scholar]
- 11.Reddy M, Gowrishankar J. 1997. Identification and characterization of ssb and uup mutants with increased frequency of precise excision of transposon Tn10 derivatives: nucleotide sequence of uup in Escherichia coli. J Bacteriol 179:2892–2899. 10.1128/jb.179.9.2892-2899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stupina VA, Wang JC. 2005. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J Biol Chem 280:355–360. 10.1074/jbc.M411924200. [DOI] [PubMed] [Google Scholar]
- 13.Stockum A, Lloyd RG, Rudolph CJ. 2012. On the viability of Escherichia coli cells lacking DNA topoisomerase I. BMC Microbiol 12:26. 10.1186/1471-2180-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiNardo S, Voelkel KA, Sternglanz R, Reynolds AE, Wright A. 1982. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 31:43–51. 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 15.Pruss GJ, Manes SH, Drlica K. 1982. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell 31:35–42. 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 16.Raji A, Zabel DJ, Laufer CS, Depew RE. 1985. Genetic analysis of mutations that compensate for loss of Escherichia coli DNA topoisomerase I. J Bacteriol 162:1173–1179. 10.1128/jb.162.3.1173-1179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q, Pongpech P, DiGate RJ. 2001. Type I topoisomerase activity is required for proper chromosomal segregation in Escherichia coli. Proc Natl Acad Sci U S A 98:9766–9771. 10.1073/pnas.171579898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broccoli S, Phoenix P, Drolet M. 2000. Isolation of the topB gene encoding DNA topoisomerase III as a multicopy suppressor of topA null mutations in Escherichia coli. Mol Microbiol 35:58–68. 10.1046/j.1365-2958.2000.01671.x. [DOI] [PubMed] [Google Scholar]
- 19.Brochu J, Vlachos-Breton E, Sutherland S, Martel M, Drolet M. 2018. Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli. PLoS Genet 14:e1007668. 10.1371/journal.pgen.1007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Usongo V, Drolet M. 2014. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet 10:e1004543. 10.1371/journal.pgen.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drolet M, Bi X, Liu LF. 1994. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem 269:2068–2074. 10.1016/S0021-9258(17)42136-3. [DOI] [PubMed] [Google Scholar]
- 22.Masse E, Phoenix P, Drolet M. 1997. DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J Biol Chem 272:12816–12823. 10.1074/jbc.272.19.12816. [DOI] [PubMed] [Google Scholar]
- 23.Masse E, Drolet M. 1999. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem 274:16659–16664. 10.1074/jbc.274.23.16659. [DOI] [PubMed] [Google Scholar]
- 24.Drolet M. 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59:723–730. 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 25.Belotserkovskii BP, Tornaletti S, D'Souza AD, Hanawalt PC. 2018. R-loop generation during transcription: formation, processing and cellular outcomes. DNA Repair (Amst) 71:69–81. 10.1016/j.dnarep.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A 92:3526–3530. 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masse E, Drolet M. 1999. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J Mol Biol 294:321–332. 10.1006/jmbi.1999.3264. [DOI] [PubMed] [Google Scholar]
- 28.Martel M, Balleydier A, Sauriol A, Drolet M. 2015. Constitutive stable DNA replication in Escherichia coli cells lacking type 1A topoisomerase activity. DNA Repair (Amst) 35:37–47. 10.1016/j.dnarep.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Usongo V, Martel M, Balleydier A, Drolet M. 2016. Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity. DNA Repair (Amst) 40:1–17. 10.1016/j.dnarep.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Hraiky C, Raymond MA, Drolet M. 2000. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem 275:11257–11263. 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 31.Raghunathan N, Kapshikar RM, Leela JK, Mallikarjun J, Bouloc P, Gowrishankar J. 2018. Genome-wide relationship between R-loop formation and antisense transcription in Escherichia coli. Nucleic Acids Res 46:3400–3411. 10.1093/nar/gky118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25:2041–2056. 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gómez-González B, Aguilera A. 2019. Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev 33:1008–1026. 10.1101/gad.324517.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Muse T, Aguilera A. 2019. R loops: from physiological to pathological roles. Cell 179:604–618. 10.1016/j.cell.2019.08.055. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov IE, Wright AV, Cofsky JC, Aris KDP, Doudna JA, Bryant Z. 2020. Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc Natl Acad Sci U S A 117:5853–5860. 10.1073/pnas.1913445117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Hage A, French SL, Beyer AL, Tollervey D. 2010. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24:1546–1558. 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, Pommier Y, Tazi J, Coquelle A, Pasero P. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11:1315–1324. 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang T, Wallis M, Petrovic V, Challis J, Kalitsis P, Hudson DF. 2019. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol 9:190222. 10.1098/rsob.190222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boubakri H, de Septenville AL, Viguera E, Michel B. 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29:145–157. 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. 2011. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146:533–543. 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, Woodward JJ, Dreifus JE, Merrikh H. 2017. Replication-transcription conflicts generate R-loops that orchestrate bacterial ctress survival and pathogenesis. Cell 170:787–799. 10.1016/j.cell.2017.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. 2017. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170:774–786. 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nudler E. 2012. RNA polymerase backtracking in gene regulation and genome instability. Cell 149:1438–1445. 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lang KS, Merrikh H. 2018. The clash of macromolecular titans: replication-transcription conflicts in bacteria. Annu Rev Microbiol 72:71–88. 10.1146/annurev-micro-090817-062514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuzminov A. 2018. When DNA topology turns deadly—RNA polymerases dig in their R-loops to stand their ground: new positive and negative (super)twists in the replication-transcription conflict. Trends Genet 34:111–120. 10.1016/j.tig.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zatreanu D, Han Z, Mitter R, Tumini E, Williams H, Gregersen L, Dirac-Svejstrup AB, Roma S, Stewart A, Aguilera A, Svejstrup JQ. 2019. Elongation factor TFIIS prevents transcription stress and R-loop accumulation to maintain genome stability. Mol Cell 76:57–69. 10.1016/j.molcel.2019.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh T, Tomizawa J. 1980. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A 77:2450–2454. 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kogoma T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61:212–238. 10.1128/.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drolet M, Brochu J. 2019. R-loop-dependent replication and genomic instability in bacteria. DNA Repair (Amst) 84:102693. 10.1016/j.dnarep.2019.102693. [DOI] [PubMed] [Google Scholar]
- 50.Sinha AK, Possoz C, Leach DRF. 2020. The roles of bacterial DNA double-strand break repair proteins in chromosomal DNA replication. FEMS Microbiol Rev 44:351–368. 10.1093/femsre/fuaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuckey R, Garcia-Rodriguez N, Aguilera A, Wellinger RE. 2015. Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proc Natl Acad Sci U S A 112:5779–5784. 10.1073/pnas.1501769112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG. 2013. Avoiding chromosome pathology when replication forks collide. Nature 500:608–611. 10.1038/nature12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gowrishankar J. 2015. End of the beginning: elongation and termination features of alternative modes of chromosomal replication initiation in bacteria. PLoS Genet 11:e1004909. 10.1371/journal.pgen.1004909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. 2010. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141:595–605. 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Mooney RA, Grass JA, Sivaramakrishnan P, Herman C, Landick R, Wang JD. 2014. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell 53:766–778. 10.1016/j.molcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myka KK, Kusters K, Washburn R, Gottesman ME. 2019. DksA-RNA polymerase interactions support new origin formation and DNA repair in Escherichia coli. Mol Microbiol 111:1382–1397. 10.1111/mmi.14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogawa T, Pickett GG, Kogoma T, Kornberg A. 1984. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A 81:1040–1044. 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaguni JM, Kornberg A. 1984. Topoisomerase I confers specificity in enzymatic replication of the Escherichia coli chromosomal origin. J Biol Chem 259:8578–8583. 10.1016/S0021-9258(17)39769-7. [DOI] [PubMed] [Google Scholar]
- 59.Pósfai G, Plunkett G, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046. 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 60.McGlynn P, Lloyd RG. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35–45. 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 61.Trautinger BW, Lloyd RG. 2002. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J 21:6944–6953. 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. 2005. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell 19:247–258. 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 63.Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS, Lloyd RG, McGlynn P. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell 36:654–666. 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamarthapu V, Epshtein V, Benjamin B, Proshkin S, Mironov A, Cashel M, Nudler E. 2016. ppGpp couples transcription to DNA repair in E. coli. Science 352:993–996. 10.1126/science.aad6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Washburn RS, Gottesman ME. 2011. Transcription termination maintains chromosome integrity. Proc Natl Acad Sci U S A 108:792–797. 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrews AE, Lawley B, Pittard AJ. 1991. Mutational analysis of repression and activation of the tyrP gene in Escherichia coli. J Bacteriol 173:5068–5078. 10.1128/jb.173.16.5068-5078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leela JK, Syeda AH, Anupama K, Gowrishankar J. 2013. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A 110:258–263. 10.1073/pnas.1213123110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raghunathan N, Goswami S, Leela JK, Pandiyan A, Gowrishankar J. 2019. A new role for Escherichia coli Dam DNA methylase in prevention of aberrant chromosomal replication. Nucleic Acids Res 47:5698–5711. 10.1093/nar/gkz242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ali N, Gowrishankar J. 2020. Cross-subunit catalysis and a new phenomenon of recessive resurrection in Escherichia coli RNase E. Nucleic Acids Res 48:847–861. 10.1093/nar/gkz1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dattananda CS, Rajkumari K, Gowrishankar J. 1991. Multiple mechanisms contribute to osmotic inducibility of proU operon expression in Escherichia coli: demonstration of two osmoresponsive promoters and of a negative regulatory element within the first structural gene. J Bacteriol 173:7481–7490. 10.1128/JB.173.23.7481-7490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156. 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dudas KC, Kreuzer KN. 2001. UvsW Protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol Cell Biol 21:2706–2715. 10.1128/MCB.21.8.2706-2715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carles-Kinch K, George JW, Kreuzer KN. 1997. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J 16:4142–4151. 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wechsler JA, Gross JD. 1971. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet 113:273–284. 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- 77.Dimude JU, Stockum A, Midgley-Smith SL, Upton AL, Foster HA, Khan A, Saunders NJ, Retkute R, Rudolph CJ. 2015. The consequences of replicating in the wrong orientation: bacterial chromosome duplication without an active replication origin. mBio 6:e01294-15. 10.1128/mBio.01294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Midgley-Smith SL, Dimude JU, Rudolph CJ. 2019. A role for 3’ exonucleases at the final stages of chromosome duplication in Escherichia coli. Nucleic Acids Res 47:1847–1860. 10.1093/nar/gky1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maduike NZ, Tehranchi AK, Wang JD, Kreuzer KN. 2014. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol Microbiol 91:39–56. 10.1111/mmi.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wendel BM, Courcelle CT, Courcelle J. 2014. Completion of DNA replication in Escherichia coli. Proc Natl Acad Sci U S A 111:16454–16459. 10.1073/pnas.1415025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wendel BM, Cole JM, Courcelle CT, Courcelle J. 2018. SbcC-SbcD and ExoI process convergent forks to complete chromosome replication. Proc Natl Acad Sci U S A 115:349–354. 10.1073/pnas.1715960114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Azeroglu B, Mawer JS, Cockram CA, White MA, Hasan AM, Filatenkova M, Leach DR. 2016. RecG directs DNA synthesis during double-strand break repair. PLoS Genet 12:e1005799. 10.1371/journal.pgen.1005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harinarayanan R, Gowrishankar J. 2003. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol 332:31–46. 10.1016/s0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- 84.Gowrishankar J, Harinarayanan R. 2004. Why is transcription coupled to translation in bacteria? Mol Microbiol 54:598–603. 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 85.Gowrishankar J, Leela JK, Anupama K. 2013. R-loops in bacterial transcription: their causes and consequences. Transcription 4:153–157. 10.4161/trns.25101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. 2008. Termination factor Rho and its cofactors NusA and NusG silence foeign DNA in E. coli. Science 320:935–938. 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan K, Zhou Q, Cheng B, Zhang Z, Joachimiak A, Tse-Dinh YC. 2015. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res 43:11031–11046. 10.1093/nar/gkv1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cao N, Tan K, Zuo X, Annamalai T, Tse-Dinh Y-C. 2020. Mechanistic insights from structure of Mycobacterium smegmatis topoisomerase I with ssDNA bound to both N- and C-terminal domains. Nucleic Acids Res 48:4448–4462. 10.1093/nar/gkaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 90.Anupama K, Leela JK, Gowrishankar J. 2011. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol 82:1330–1348. 10.1111/j.1365-2958.2011.07895.x. [DOI] [PubMed] [Google Scholar]
- 91.Gowrishankar J. 1985. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J Bacteriol 164:434–445. 10.1128/jb.164.1.434-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Table S1, and Fig. S1 to S6. Download JB.00195-21-s0001.pdf, PDF file, 1.0 MB (1MB, pdf)
Data Availability Statement
Genome sequence data from this work have been submitted under accession number PRJNA670792 and are available for public access at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA670792.