Abstract
Histone post-translational modifications are essential for the regulation of gene expression in eukaryotes. Gcn5 (KAT2A) is a histone acetyltransferase that catalyzes the post-translational modification at multiple positions of histone H3 through the transfer of acetyl groups to the free amino group of lysine residues. Gcn5 catalyzes histone acetylation in the context of a HAT module containing the Ada2, Ada3 and Sgf29 subunits of the parent megadalton SAGA transcriptional coactivator complex. Biochemical and structural studies have elucidated mechanisms for Gcn5’s acetyl- and other acyltransferase activities on histone substrates, for histone H3 phosphorylation and histone H3 methylation crosstalks with histone H3 acetylation, and for how Ada2 increases Gcn5’s histone acetyltransferase activity. Other studies have identified Ada2 isoforms in SAGA-related complexes and characterized variant Gcn5 HAT modules containing these Ada2 isoforms. In this review, we highlight biochemical and structural studies of Gcn5 and its functional interactions with Ada2, Ada3 and Sgf29.
Keywords: Gene regulation, Histone modification, Transcriptional regulation, Epigenetic modification, Histone crosstalks, Protein isoforms
1. Introduction
The discovery that the yeast Gcn5 transcriptional adaptor [1] is a histone acetyltransferase was a watershed moment in eukaryotic gene regulation because it provided a direct link between histone modifications and transcriptional regulation [2]. Previous studies had already established that Gcn5 did not act alone, but was associated with the Ada2 and Ada3 transcriptional adaptor proteins [3-6]. The subsequent fractionation of native Gcn5-containing complexes from yeast revealed that Gcn5, together with Ada2 and Ada3, were associated in the SAGA (Spt-Ada-Gcn5-Acetyltransferase) megadalton complex and a smaller ADA complex [7]. Whereas the isolated Gcn5 protein could only acetylate naked histones in vitro, the SAGA and ADA complexes possessed the ability to acetylate the more physiological substrate of nucleosomes in which histones package DNA into disc-like units.
Ada2, Ada3 and Gcn5 form a stable protein complex, first shown by in vitro cotranslation and then by in vivo reconstitution in E. coli [5,6,8]. The Ada2/Ada3/Gcn5 complex is necessary and sufficient for nucleosomal HAT activity and thus constitutes the minimal Gcn5 complex for nucleosomal histone acetylation. The SAGA subunit protein Sgf29 binds to the Ada2/Ada3/Gcn5 complex and completes the SAGA HAT module.
This review focuses on Gcn5’s histone acetyltransferase activity in the context of the Ada2/Ada3/Gcn5/Sgf29 module, with a particular emphasis on mechanistic and structural findings.
2. Gcn5
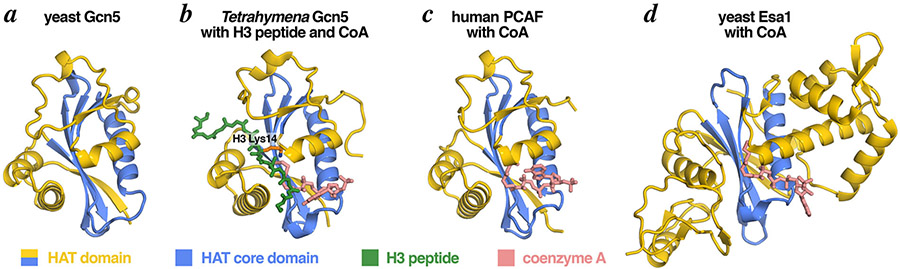
Gcn5 (KAT2A) is the catalytic subunit of the HAT module in SAGA (Fig. 1). Gcn5’s HAT domain (Fig. 2) has an α/β architecture with a central sheet of six β-strands surrounded by five α-helices [9] (Fig. 3a). The globular fold is conserved across species from yeast Gcn5 to Tetrahymena Gcn5 [10] and humans Gcn5 and PCAF (Fig. 3b, c) [11,12]. The HAT core domain is formed by four antiparallel β-strands (β2-β5) which lie over an amphipathic α-helix (α3). This HAT core domain is shared with other CoA-dependent HATs such as yeast Esa1 (Fig. 3d).
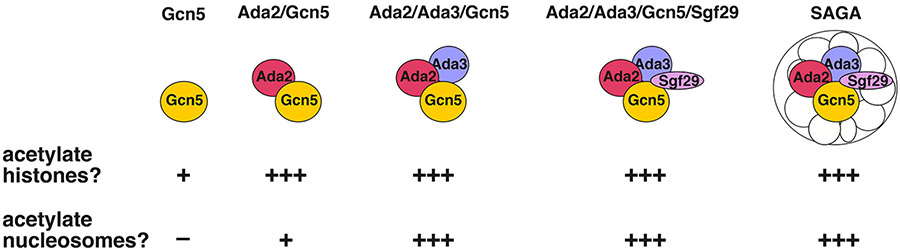
Figure 1: Acetylation efficiency of different Gcn5-containing complexes.
The Gcn5 catalytic subunit possesses weak histone acetyltransferase activity on its own. Ada2 increases HAT activity on histones, and Ada3 is needed for robust nucleosome acetyltransferase activity. Sgf29 mediates processive acetylation and completes the SAGA HAT module. Comparison of HAT activity based on Balasubramanian et al, 2002 [8] and Ringel et al, 2015 [46].
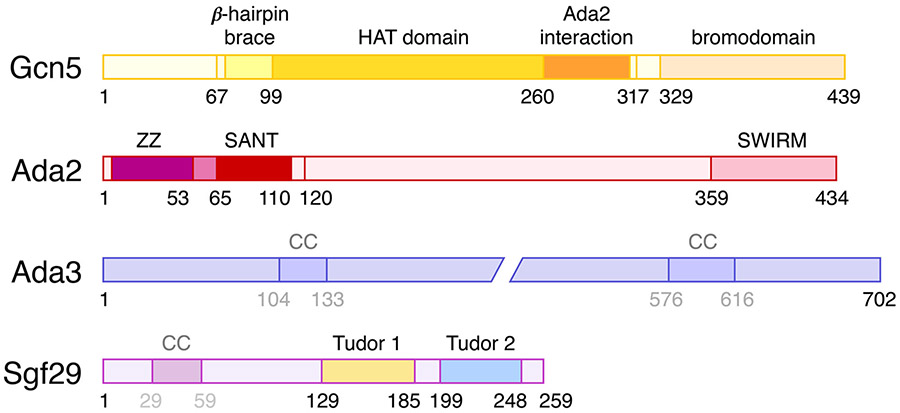
Figure 2: Domain organization of the SAGA HAT module subunits.
The known domains and motifs in Gcn5, Ada2, Ada3 and Sgf29 are shown. In addition, predicted coiled-coil regions (CC) are shown for Ada3 and Sgf29. Little is currently known about the structure of Ada3.
Figure 3: The conserved structure of the Gcn5 HAT domains.
The Gcn5 HAT domain contains a central HAT core domain (blue) and structurally more divergent N- and C-terminal segments (yellow). The HAT core domain is structurally conserved among orthologous Gcn5 enzymes (yeast in panel a, Tetrahymena in panel b), closely related HAT enzymes like PCAF (panel c) and more distantly related HAT enzymes like Esa1 (panel d). The crystal structure of the Tetrahymena ternary complex (panel b) shows Gcn5 bound to its two substrates, CoA (a nonenzymatic surrogate for acetyl-CoA) and an H3 histone peptide. Molecular graphics prepared in PyMOL [66] using PDB coordinates 1YGH, 1PU9, 1CM0, 1MJA.
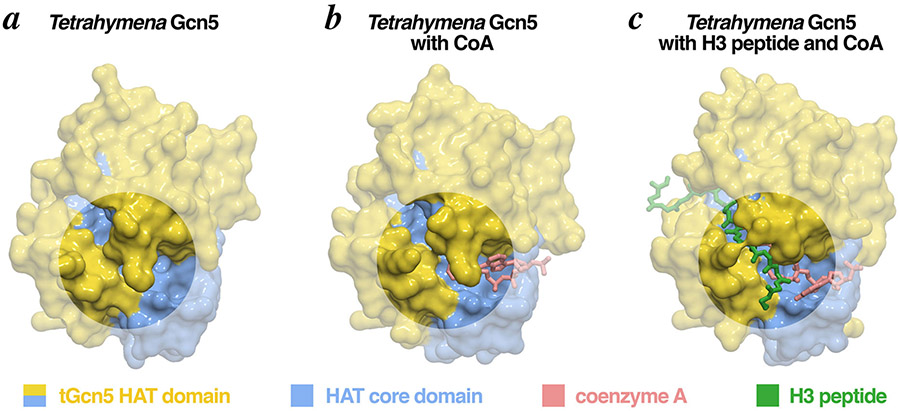
Acetylation by Gcn5 requires two substrates, a histone substrate and an acetyl-CoA cosubstrate. Detailed kinetic studies establish that Gcn5 follows ordered bi-bi kinetics, with acetyl-CoA binding first to the HAT domain before the histone peptide substrate [13]. The crystal structure of the ternary complex of the Tetrahymena Gcn5 HAT domain bound to CoA and histone H3 peptide together with additional structures of the Tetrahymena Gcn5 HAT domain on its own (apo form) and in a binary complex bound to Coenzyme A (CoA) provided the structural explanation why [10]. The structures show the HAT domain contains two pronounced clefts in an L shape. Acetyl-CoA resides in the smaller cleft while the histone H3 peptide occupies the larger cleft (Fig. 3b). In the apo form, the CoA end of the peptide binding cleft of Gcn5 is obstructed, blocking the H3 peptide from binding (Fig. 4a, b). Binding of the coenzyme A moiety causes a modest opening of the peptide binding cleft, a conformational change which is then apparently used by the histone peptide to access the peptide binding cleft (Fig. 4c). These structures also provided important details for how the catalytic Tetrahymena Glu122 (equivalent to yeast Glu173) acts as a general base to increase the nucleophilicity of the substrate Lys14 α-amino group for attack on acetyl-CoA [10,14]. Gcn5’s catalytic mechanism is discussed further in the Albaugh and Denu contribution in this special issue.
Figure 4: Surface representations of the Gcn5 HAT domain in complex with its substrates.
The Gcn5 HAT domain contains two pronounced clefts which accommodate the histone peptide and acetyl-CoA substrates. Binding of acetyl-CoA to the smaller cleft causes a conformational change that opens the larger, peptide binding cleft, allowing the H3 peptide to bind (circles highlight the conformational changes). Molecular graphics prepared in PyMOL using PDB coordinates 1QST, 1QSR and 1PU9.
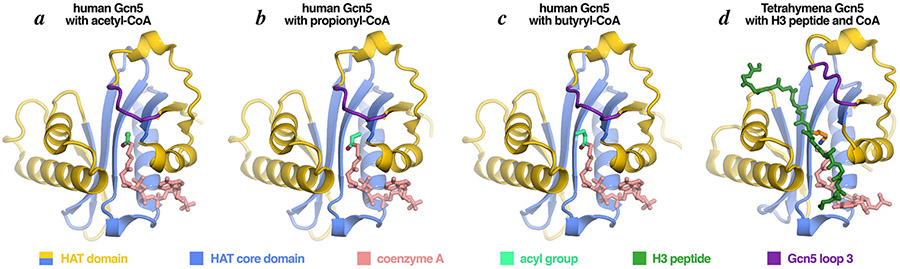
Much of the investigations into Gcn5’s enzymatic activity has focused on its acetyltransferase activity because of the strong association between gene activity and histone acetylation. However, Gcn5 is not limited to modifying lysine side chains with an acetyl group. The Gcn5 HAT domain has been shown to acylate histone lysine side chains with a surprisingly large range of acyl groups (Fig. 5). Human Gcn5 can propionylate and butyrylate histone H3 on Lys14 in vitro and in vivo and both the H3K14pr and H4K14bu marks are associated with transcriptionally active chromatin [15]. Structural studies had shown that acetyl-CoA, propionyl-CoA and butyryl-CoA all can bind to Gcn5 but the bound butyryl-CoA was not positioned for catalysis and the butyryl group would occupy the same space as the substrate target lysine (Fig. 6) [16]. Significant conformational changes occur upon the subsequent binding of the substrate peptide, as observed in the ternary Tetrahymena Gcn5/CoA/histone peptide structures, where the Gcn5 HAT domain loop 3 is pushed away to accommodate histone peptide binding (Fig. 6d).
Figure 5: Acyl groups used by Gcn5.
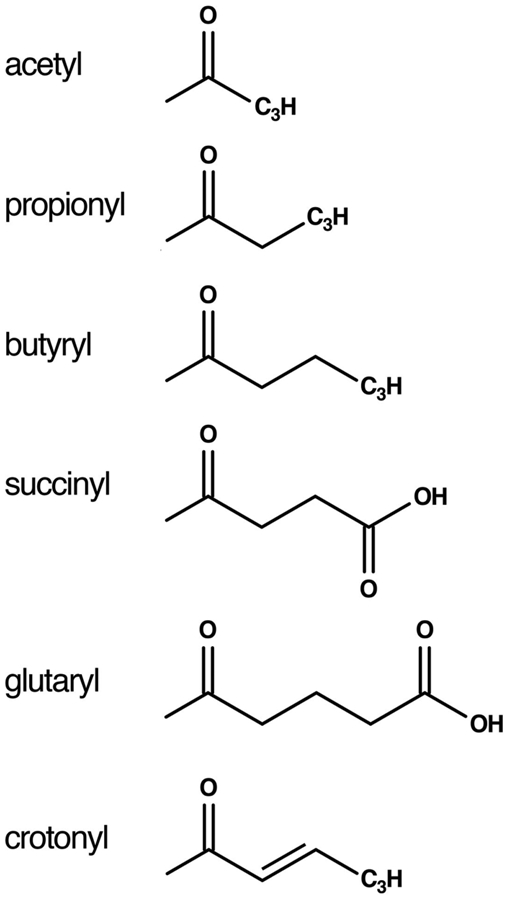
Chemical structures of acyl groups in acyl-CoA cosubstrates used by Gcn5 to acylate lysine residues.
Figure 6: Crystal structures of Gcn5 HAT domains bound to different acyl-CoA cosubstrates and conformational changes associated with substrate binding.
The Gcn5 HAT domain can bind to acetyl-CoA and the bulkier propionyl- and butyryl-CoA with modest structural changes (panels a, b, c). Subsequent binding of the histone peptide and catalysis appears to require significant conformational changes in Gcn5 loop 3 (panel d). Loop 3 also moves to make way for the bulky succinyl acyl group of succinyl CoA (panel d) which interacts with Tyr465 in the loop. Molecular graphics prepared in PyMOL using PDB coordinates 1Z4R, 5H84, 5H86, 1PU9.
In a separate study, human Gcn5 was reported to act as a histone H3 succinyltransferase. However, it has also been reported that the protein bovine serum albumin can be succinylated non-enzymatically by succinyl-CoA and that succcinyl-CoA is signficantly more reactive in non-enzymatic acylation than acetyl-CoA, propionyl-CoA or butyryl-CoA [17-19]. As such, it is not entirely clear whether the observed histone H3 succinylation was actually catalyzed by Gcn5 or if it occurred non-enzymatically. Human Gcn5 has also been reported to glutarylate H4K91 but its glutaryltransferase activity was not confirmed in vitro using purified components [20] and like succinyl-CoA, glutaryl-CoA can non-enzymatically acylate proteins. It is worth noting that Gcn5-catalyzed acylation is not limited to saturated fatty acids: the yeast Ada2/Ada3/Gcn5 complex crotonylates nucleosome substrates in vitro and Gcn5-dependent crotonylation regulates transcription of selected genes in yeast [21].
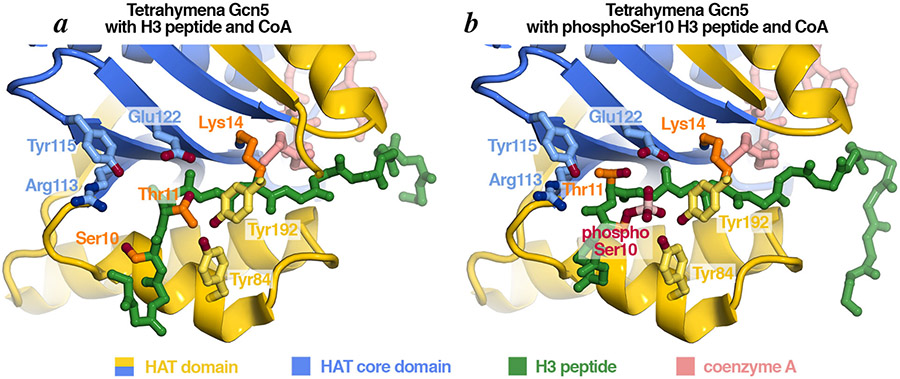
Studies of the Gcn5 HAT domain have also been instrumental in elucidating the first molecular basis for histone modification crosstalks [23]. Phosphorylation of H3 Ser10 increases the efficiency of Gcn5 acetylation of H3 peptides by increasing binding affinity to the H3 peptide substrate. In this case, the crosstalk between histone modification (Ser10 phosphorylation and Lys14 acetylation) occurs in cis in the same peptide region. Crystallographic structures of H3 peptides with or without Ser10 phosphorylation show a large rearrangement for part of the H3 peptide upon Ser10 phosphorylation (Fig. 7). These changes reposition Thr11 for more extensive interactions with the Gcn5 HAT domain, consistent with the increased binding affinity of the Ser10 phosphorylated H3 peptide. This conformational change is limited to the H3 peptide, with no significant changes to the Gcn5 HAT domain or to CoA binding.
Figure 7: Mechanism of phosphorylation-acetylation epigenetic histone modification crosstalk.
Histone H3 S10 phosphorylation induces conformational changes in the H3 peptide that increase its interactions with the catalytic site of Tetrahymena Gcn5 (compare H3 Thr11 positions), increasing the binding affinity for the substrate. Key Gcn5 residues that mediate these interactions are shown. Molecular graphics prepared in PyMOL using PDB coordinates 1PU9 and 1PUA.
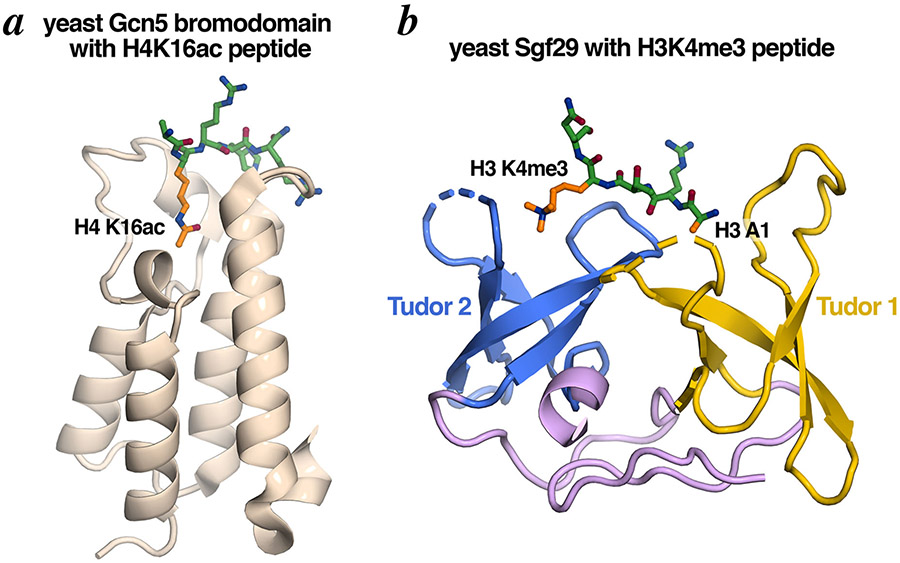
Gcn5 contains a bromodomain at its C-terminus. The bromodomain is a chromatin modification reader of acetylated lysine residues and is found in many transcriptional regulators. The structure of the Gcn5 bromodomain was determined on its own by NMR and in complex with an H4 tail peptide containing acetylated Lys16 by crystallography [24,25]. The structures show a four-helix bundle with a deep hydrophobic pocket which accommodates the acetylated lysine residue (Fig. 8a).
Figure 8: HAT module domains that mediate histone modification crosstalk.
(a) The Gcn5 bromodomain serves as a reader of acetylated H4K16. (b) The Sgf29 tandem Tudor domain recognizes methylated H3K4. Molecular graphics prepared in PyMOL using PDB coordinates 1E6I and 3MP1.
3. Ada2
Recognizable domains in Ada2 include a ZZ zinc finger domain and a SANT domain at the N-terminus and a SWIRM domain at the C-terminus (Fig. 2). Deletion and mutational analysis of the Ada2 ZZ and SANT domains showed that the SANT domain, but not the ZZ domain, is required for interactions with Gcn5 and for normal growth in yeast [6,26]. The SANT (Swi3, Ada2, N-Cor, and TFIIB) domain is an approximately 55 residue α-helical domain with structural homology to the myb DNA-binding domain. Consistent with this, several chromatin remodeling enzymes employ the SANT domain as part of their DNA-binding modules to interact with DNA [27-31]. The SANT domain has also been proposed to act as a histone tail binding motif in chromatin-regulatory enzymes [32]. The interactions between Ada2 and Gcn5 increase Gcn5’s HAT activity on histone substrates in vitro and are necessary for transcriptional activation in vivo [8,26,33]. The Gcn5 HAT domain is not sufficient to interact with Ada2 and at least 20 residues C-terminal to the Gcn5 HAT domain are required for Ada2 interaction.
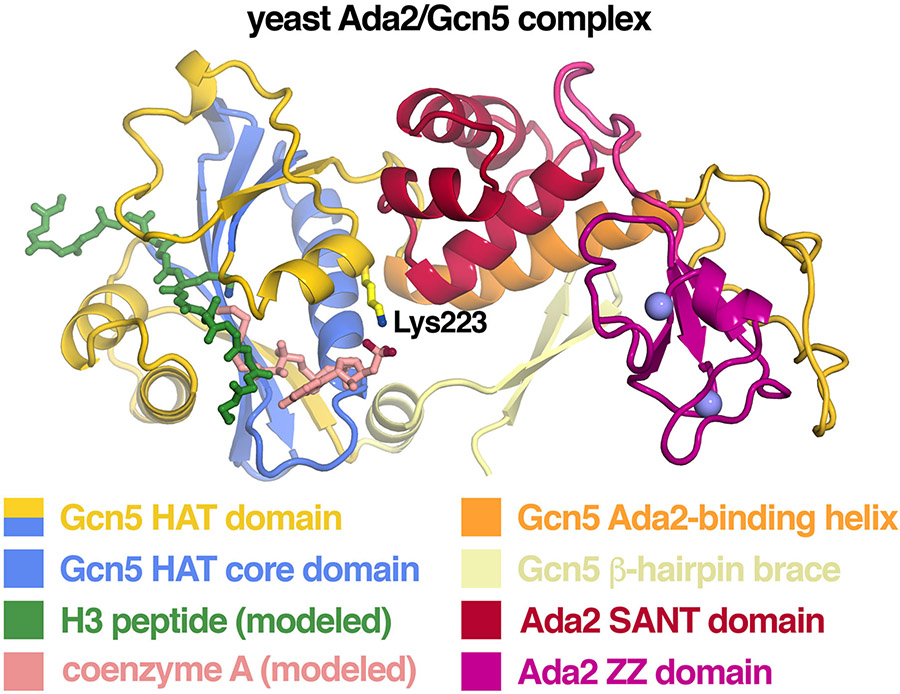
The structural basis for how the Ada2 SANT domain interacts with Gcn5 and thus brings Gcn5 into the SAGA complex was provided by the crystal structure of the yeast Ada2/Gcn5 complex [34]. Earlier attempts to crystallize this complex produced crystals with poor diffraction characteristics. This technical limitation was overcome through the use of a synthetic antibody fragment screened in vitro for binding to the Ada2/Gcn5 complex as a crystallization chaperone. The structure includes the Gcn5 HAT domain with N-terminal and C-terminal extensions bound to the Ada2 ZZ and SANT domains (Fig. 9). The Gcn5 HAT domain is essentially unchanged on its own or complexed with Ada2. The 20 residues immediately C-terminal to the Gcn5 HAT domain (residues 260-280) shown to be critical for interacting with Ada2 form an α-helix that points away from the HAT domain. This Gcn5 α-helix makes extensive interactions with the Ada2 SANT domain on one side and with the Gcn5 β-hairpin brace which precedes the Gcn5 HAT domain.
Figure 9: Structure of the Ada2/Gcn5 complex indicates the Ada2 SANT domain helps Gcn5 bind to acetyl-CoA.
Crystal structure of the yeast Ada2/Gcn5 complex with modeled H3 peptide and CoA based on the Tetrahymena Gcn5/CoA/H3 peptide structure. The Ada2 SANT is unlikely to directly affect histone peptide binding since it is located far away from the peptide binding site. Instead, the SANT domain is positioned by interactions with the Gcn5 β-hairpin brace and the Ada2-binding helix to constrain Gcn5 Lys223 to bind directly to the 3’ phosphate of CoA. The Ada2 zinc binding ZZ domain extends even further away from the Gcn5 HAT domain. Molecular graphics prepared in PyMOL using PDB coordinates 6CW2.
At first glance, the Ada2/Gcn5 complex structure creates a predicament: the Ada2 SANT domain is known to increase Gcn5’s HAT activity on histone peptides, but modeling an H3 peptide and coenzyme A based on the orthologous Tetrahymena Gcn5/H3 peptide/CoA structure positions the Ada2 SANT domain on the opposite side of the Gcn5 HAT domain from the H3 peptide. The Ada2 SANT domain is too far away to make direct contact with the H3 peptide, arguing against the SANT domain acting as a histone tail recruiting module. Instead, it appears that the Ada2 SANT domain activates Gcn5’s HAT activity by recruiting the acetyl-CoA cofactor. This mechanism is consistent with Gcn5’s ordered bi-bi reaction mechanism where acetyl-CoA binds first to Gcn5 before the histone peptide substrate can bind. However, the distance between the SANT domain and the cofactor precludes direct recruitment of the cofactor. Rather, the Ada2/Gcn5 crystal structure with the modeled H3 peptide and CoA cofactor indicates that the Ada2 SANT domain corrals the Gcn5 HAT domain residue Lys223’s side chain to interact with the coenzyme A cofactor. This structural hypothesis was substantiated with isothermal calorimetry binding studies and HAT activity assays which show that Gcn5 Lys223, a residue conserved across species, plays a critical role in acetyl-CoA binding and acetyltransferase activity.
The Ada2/Gcn5 structure shows that the Ada2 ZZ zinc-binding domain interacts with an extended chain of Gcn5(282-312) beyond the Gcn5 α-helix that interacts with the Ada2 SANT domain. This “head module” of the Ada2/Gcn5 complex was observed to rotate with respect to the rest of the structure in two different crystal forms. The physiological significance of this conformational flexibility and in fact, the biological roles of the Ada2 ZZ domain are currently not known. Unlike the Ada2 SANT domain, the ZZ domain is apparently not essential for growth or for activity in an in vivo transcription assay [26]. The ZZ domain in some other chromatin proteins binds to unmodified or acetylated histone H3 tails [35], but it is unclear if the Ada2 ZZ domain possesses a similar activity.
The C-terminus of yeast Ada2 and its Ada2a orthologs contain a SWIRM (Swi3, Rsc8 and Moira) domain, comprised of an α-helical core containing a helix-turn-helix motif [36,37]. The structural similarities to the linker histone proteins prompted experiments which show that the yAda2 SWIRM domain binds to DNA and to dinucleosomes, but not to mononucleosomes [36]. This suggests the possibility that the yAda2 SWIRM domain might bind to linker or extranucleosomal DNA.
4. Ada3
In contrast to the substantial structural characterization of Gcn5 and Ada2, much of Ada3’s function or structure remains unclear. Ada3 forms a ternary complex with Gcn5 and Ada2, apparently through interactions with Ada2 [5,6,8]. Ada3 is necessary for the Ada2/Ada3/Gcn5 HAT module to acetylate nucleosome versus histone or peptide substrate [8] but the mechanism for this is not known. We also lack clear structural information for Ada3, which does not appear to contain recognizable domains or motifs aside from some predicted coiled-coil regions (Fig. 2). Hybrid methods combining chemical cross-linking, mass spectrometry and molecular modeling have been used to produce a structural model for a mammalian Ada2/Ada3/Gcn5/Sgf29 complex but this model needs validation [38]. The recent cryoelectron microscopy structures for the yeast SAGA complex are important and exciting developments, but unfortunately the Ada2/Ada3/Gcn5/Sgf29 HAT module is poorly defined presumably due to conformational diversity [39,40]. Two α-helical regions in this density interact with the SAGA TAF6 subunit and were attributed to Ada3 in the structure from the Schultz and Ben-Shem groups [39]. More details from structures of the SAGA and related complexes are described in the Hemlinger, Papai, Devys and Tora contribution in this issue.
5. Sgf29
Sgf29 (SAGA associated factor of 29 kDa) was identified as a SAGA subunit in a proteomics analysis of the SAGA complex [41]. Sgf29 is not required for the integrity of the SAGA complex but global histone H3 acetylation is decreased in vivo in the absence of Sgf29 [42] even though Sgf29 is not necessary for the SAGA complex to acetylate histones or nucleosomes in vitro [43]. Proteomic and biochemical analyses confirm that Sgf29 is a core subunit of SAGA and ADA family of complexes [44]. The rat ortholog of Sgf29 appears to associate with the Ada2/Ada3/Gcn5 HAT module via coiled-coil interactions with Ada3 [45] and the recombinant S. pombe four protein complex of Ada2, Ada3, Gcn5 and Sgf29 has been expressed and characterized [46] showing that these four proteins are sufficient to form a subcomplex.
Sgf29 exerts its effect on H3 acetylation though histone modification crosstalk with H3 methylation. The C-terminal tandem Tudor domain of human Sgf29 was determined to be necessary and sufficient for binding to H3K4me3, with the second Tudor domain playing a critical role in the binding [47]. Crystal structures of the yeast and human Sgf29 tandem Tudor domains bound to unmodified and methylated H3 peptides show that each Tudor domain forms a typical twisted five-stranded anti-parallel β-barrel Tudor fold with a not so typical face-to-face orientation of the Tudor domains [43] (Fig. 8b). The bound H3K4me3 peptide binds in a negatively charged pocket created by the two Tudor domains with the Tudor 1 domain binding to the first H3 residue (Ala1) and the Tudor 2 domain binding to the di- or trimethylated Lys4 residue. The physical association of a H3K4me3 binding module in Sgf29 with the Ada2/Ada3/Gcn5 histone acetyltransferase module thus provides a mechanism to account for why H3 acetylation is correlated with H3K4 methylation [43,48].
This histone H3 acetylation-histone H3 methylation crosstalk appears to explain why the SAGA complex is particularly active at promoters. Promoters are enriched for H3K4me3, which recruits the SAGA complex through Sgf29 binding to this histone mark at promoters where increased histone acetylation is mediated by the SAGA HAT module. Elegant in vitro studies employing recombinant S. pombe Ada2/Ada3/Gcn5/Sgf29 complexes and recombinant true H3K4me3 nucleosomes indicate that this four subunit HAT module binds to H3K4me3 and stimulates acetylation on the same H3 tail in cis [46]. The presence of H3K4 trimethylation affected the Michaelis constant Km, but not the catalytic rate constant kcat for histone peptide substrates. This indicates that H3K4 methylation improves HAT activity by increasing binding of the Ada2/Ada3/Gcn5/Sgf29 module to the methylated substrate and not by directly influencing the HAT module. The Ada2/Ada3/Gcn5/Sgf29 module was also found to preferentially acetylate H3K4me3 nucleosomes over unmodified nucleosomes when presented with both substrates simultaneously, thus explaining how the SAGA complex can hyperacetylate gene promoters while leaving gene bodies less acetylated.
6. The Ada2/Ada3/Gcn5 and Ada2/Ada3/Gcn5/Sgf29 HAT modules
Gcn5 preferentially acetylates free histone H3 at position K14, but association with subunits of the SAGA or ADA complexes expands this target preference [49]. The minimal complex for nucleosomal acetylation, Ada2/Ada3/Gcn5 was observed to possess the same lysine specificity as the full SAGA complex [8]. Steady-state kinetic studies using histone H3 polypeptide substrate show that once H3K14 has been acetylated, Gcn5 can acetylate H3K9, H3K23, H3K18, H3K27 and H3K36 in decreasing order of preference [50]. A similar but not identical substrate specificity was detected for the Ada2/Ada3/Gcn5 HAT module using the same histone H3 substrate [51]. The Gcn5 bromodomain plays an important role in both cooperativity and selectivity of histone acetylation. Cooperative acetylation of nucleosomes by the Ada2/Ada3/Gcn5 HAT module and by the SAGA complex was determined to be mediated by the Gcn5 bromodomain acetyl-lysine binding activity [52]. This acetyl-lysine binding activity also regulates lysine-site-specificity of the Ada2/Ada3/Gcn5 HAT module, which acetylates H3K14 and H3K23 independent of the Gcn5 bromodomain and H3K9, H3K18, H3K27 and H3K36 dependent on the Gcn5 bromodomain [51]. It appears that the Ada2/Ada3/Gcn5 HAT module binds to acetylated H3K14 via the Gcn5 bromodomain, stimulating acetylation of H3K18. This tethering mechanism of Gcn5 to H3K14ac is reminiscent of Sgf29’s binding to H3K4me3 to achieve hyperacetylation, and both tethering schemes appear to be employed by the SAGA complex [46].
The Ada2/Ada3/Gcn5 HAT module is the minimal SAGA complex that will acetylate nucleosomes, but the nucleosome core particle with 145-147 bp of DNA wrapped around the histone octamer core is apparently not the preferred binding or acetylation substrate of SAGA. Mittal et al were able to perform technically demanding kinetic studies of the yeast SAGA complex to show SAGA bound nucleosomes containing 15 bp of extranucleosomal or linker DNA on one side and 95 bp extranucleosomal DNA on the other side twice as tightly as a 147 bp 601 nucleosome with minimal extranucleosomal DNA [53]. The +95/+15 nucleosome also increased SAGA’s acetylation catalytic rate constant (kcat) by a factor of 4. Although the specific SAGA subunit or domain that mediates this increased binding is not known, the Ada2 SWIRM domain which appears to interact with extranucleosomal DNA is a reasonable candidate [36].
The SAGA complex’s deubiquitylase module contains the yeast Sca7/Sgf73 or the human ataxin-7 subunit. Expansion of the polyglutamine sequence near the amino terminus of the ataxin-7 protein causes the neurodegenerative disease spinocerebellar ataxia type 7 (SCA7). The pathogenic ataxin-7-60Q protein containing 60 glutamines, but not the non-pathogenic ataxin-7-10Q protein containing 10 glutamines, formed a complex with the yeast Ada2/Ada3/Gcn5 complex apparently through direct interactions with Gcn5 [54]. It is worth noting that SAGA’s deubiquitylase and HAT modules appear to be physically close to each other in the cryoelectron microscopy structures of the SAGA complex [39,40]. Ataxin-7-60Q significantly reduced the HAT activity of the Ada2/Ada3/Gcn5 complex on both histone and nucleosome substrates in vitro. These in vitro results were substantiated by studies in yeast cells which show that the ataxin-7-60Q protein, but not the ataxin-7-10Q protein, inhibited H3K9 acetylation at both GAL1 and GAL7 promoters.
7. Ada2 isoforms in the HAT module
In contrast to yeast which contains only one Ada2 isoform, higher eukaryotes contain Ada2a and Ada2b isoforms with 25-50% sequence identity between isoforms in plants, Drosophila and humans [55-58]. The Ada2b isoform is found in the SAGA complex while the Ada2a isoform is found in the ATAC (Ada Two A-containing) complex. Both SAGA and ATAC complexes contain the catalytic core of Ada2a or Ada2b plus Ada3, Gcn5 and Sgf29, and the corresponding human ternary and quaternary complexes of Ada2a/Ada3/Gcn5, Ada2b/Ada3/Gcn5, Ada2a/Ada3/Gcn5/Sgf29 and Ada2b/Ada3/Gcn5/Sgf29 possess HAT activity on histones [59]. On mononucleosome substrates, a recombinant human Ada2a/Ada3/Gcn5 ATAC subcomplex possessed significantly lower H3 HAT activity compared to the equivalent SAGA Ada2b/Ada3/Gcn5 subcomplex [60]. However in a different study, both the human ATAC and SAGA complexes showed similar, weak H3 HAT activity using mononucleosome or polynucleosome substrates [61]. In addition to the Gcn5 HAT subunit, Drosophila and human ATAC complexes contains a second catalytic HAT subunit, Atac2. The Atac2 HAT subunit confers upon ATAC complexes histone H4 HAT activity, at least on histone substrates [61-63]. The C-terminal regions of Ada2a and Ada2b appear to mediate incorporation into Drosophila SAGA versus ATAC complexes [64].
Further complicating matters, Drosophila contains two splice isoforms of Ada2b: Ada2b-PA and Ada2b-PB [65,66]. The shorter Ada2b-PA isoform forms the Chiffon histone acetyltransferase (CHAT) complex with Ada3, Gcn5, Sgf29 and Chiffon [67]. Chiffon is the Drosophila ortholog of the Dbf4 regulatory subunit of the Cdc7 kinase required for chromosomal DNA replication. The longer Ada2b-PB isoform is incorporated into a complex together with Ada3, Gcn5 and Sgf29. This Drosophila ADA complex possesses histone and nucleosomal H3K9 and/or H3K14 acetyltransferase activity [68], and appears to be a metazoan counterpart to the yeast ADA complex which contains Ada2, Ada3, Gcn5, Sgf29 and two yeast ADA complex-specific subunits, Ahc1 and Ahc2 [7,44,69]. The functional role of these smaller Gcn5-containing HAT complexes is not clear, but the absence of the Tra1 subunit which binds to acidic transcriptional activator suggests functions besides activator-dependent transcriptional regulation.
8. Outlook
Much has been learned about the structure and function of Gcn5 on its own and in the context of the Ada2/Ada3/Gcn5/Sgf29 HAT module. We understand the molecular mechanisms for how the Gcn5 HAT domain binds to and acetylates its H3 histone tail substrate, how phosphorylation can affect acetylation in cis and how Ada2 binding increases Gcn5’s HAT activity. Nevertheless, much remains to be discovered. We need to better understand conformational changes in Gcn5 that occur upon acetyl-coenzyme A cofactor and histone peptide substrate binding and that enable non-acetyl acylation of histone substrates. We still need to determine the mechanism of nucleosome acetylation by the Ada2/Ada3/Gcn5/Sgf29 HAT module. This will require elucidating what regions or domains of the HAT module interact with the nucleosome and what surfaces of the nucleosome are engaged by the HAT module. Structural descriptions of the intact HAT module on its own and in complex with the nucleosome would be highly desirable. We will also need to investigate how histone crosstalks mediated by the Gcn5 bromodomain and the Sgf29 Tudor domain occur when the Ada2/Ada3/Gcn5/Sgf29 HAT module interacts with its nucleosome substrate. Given the apparent modularity of the SAGA complex with respect to its acetylation and deubiquitylation enzymatic activities, it is likely that studies of the Ada2/Ada3/Gcn5/Sgf29 HAT module will be highly relevant to understanding the acetylation activity of SAGA and SAGA-like complexes.
Acknowledgements
Jose M. Espinola Lopez was supported by an NIH Eukaryotic Gene Regulation Pre-doctoral training grant T32 GM125592 and with matching funds from Penn State University. This work was also supported by NIGMS R35 grant GM127034 to Song Tan.
References
- [1].Georgakopoulos T, Thireos G, Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription, Embo J. 11 (1992) 4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. , Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation, Cell. 84 (1996) 843–851. doi: 10.1016/S0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- [3].Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L, Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors, Embo J. 13 (1994) 4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Georgakopoulos T, Gounalaki N, Thireos G, Genetic evidence for the interaction of the yeast transcriptional co-activator proteins GCN5 and ADA2, Mol. Gen. Genet 246 (1995) 723–728. doi: 10.1007/BF00290718. [DOI] [PubMed] [Google Scholar]
- [5].Horiuchi J, Silverman N, Marcus GA, Guarente L, ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex, Mol Cell Biol. 15 (1995) 1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Candau R, Berger SL, Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo, J Biol Chem. 271 (1996) 5237–5245. doi:doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- [7].Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, et al. , Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex, Genes Dev. 11 (1997) 1640–1650. doi:doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- [8].Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S, Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation, J Biol Chem. 277 (2002) 7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- [9].Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, et al. , Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator, Proc Natl Acad Sci USA. 96 (1999) 8931–8936. doi:doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, et al. , Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide, Nature. 401 (1999) 93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- [11].Clements A, Rojas JR, Trievel RC, Wang L, Berger SL, Marmorstein R, Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A, EMBO Journal. 18 (1999) 3521–3532. doi: 10.1093/emboj/18.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schuetz A, Bernstein G, Dong A, Antoshenko T, Wu H, Loppnau P, et al. , Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A, Proteins. 68 (2007) 403–407. doi: 10.1002/prot.21407. [DOI] [PubMed] [Google Scholar]
- [13].Tanner KG, Langer MR, Kim Y, Denu JM, Kinetic mechanism of the histone acetyltransferase GCN5 from yeast, J Biol Chem. 275 (2000) 22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- [14].Langer MR, Tanner KG, Denu JM, Mutational analysis of conserved residues in the GCN5 family of histone acetyltransferases, J Biol Chem. 276 (2001) 31321–31331. doi: 10.1074/jbc.M103839200. [DOI] [PubMed] [Google Scholar]
- [15].Kebede AF, Nieborak A, Shahidian LZ, Le Gras S, Richter F, Gómez DA, et al. , Histone propionylation is a mark of active chromatin, Nat Struct Mol Biol. 24 (2017) 1048–1056. doi: 10.1038/nsmb.3490. [DOI] [PubMed] [Google Scholar]
- [16].Ringel AE, Wolberger C, Structural basis for acyl-group discrimination by human Gcn5L2, Acta Crystallogr D Struct Biol. 72 (2016) 841–848. doi: 10.1107/S2059798316007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].James AM, Smith CL, Smith AC, Robinson AJ, Hoogewijs K, Murphy MP, The Causes and Consequences of Nonenzymatic Protein Acylation, Trends Biochem Sci. 43 (2018) 921–932. doi: 10.1016/j.tibs.2018.07.002. [DOI] [PubMed] [Google Scholar]
- [18].Wagner GR, Bhatt DP, O'Connell TM, Thompson JW, Dubois LG, Backos DS, et al. , A Class of Reactive Acyl-CoA Species Reveals the Non-enzymatic Origins of Protein Acylation, Cell Metab. 25 (2017) 823–837.e8. doi: 10.1016/j.cmet.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wagner GR, Hirschey MD, Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases, Molecular Cell. 54 (2014) 5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bao X, Liu Z, Zhang W, Gladysz K, Fung YME, Tian G, et al. , Glutarylation of Histone H4 Lysine 91 Regulates Chromatin Dynamics, Molecular Cell. 76 (2019) 660–675.e9. doi: 10.1016/j.molcel.2019.08.018. [DOI] [PubMed] [Google Scholar]
- [21].Kollenstart L, de Groot AJL, Janssen GMC, Cheng X, Vreeken K, Martino F, et al. , Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription, Journal of Biological Chemistry. 294 (2019) 20122–20134. doi: 10.1074/jbc.RA119.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y, et al. , KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase, Nature. 552 (2017) 273–277. doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clements A, Poux AN, Lo W-S, Pillus L, Berger SL, Marmorstein R, Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase, Molecular Cell. 12 (2003) 461–473. doi:doi: 10.1016/s1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- [24].Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE, Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain, J Mol Biol. 304 (2000) 355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- [25].Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, et al. , The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p, 19 (2000) 6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sterner DE, Wang X, Bloom MH, Simon GM, Berger SL, The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex, J Biol Chem. 277 (2002) 8178–8186. doi: 10.1074/jbc.M108601200. [DOI] [PubMed] [Google Scholar]
- [27].Grüne T, Brzeski J, Eberharter A, Clapier CR, Corona DFV, Becker PB, et al. , Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI, Molecular Cell. 12 (2003) 449–460. doi:doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- [28].Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T, The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains, Embo J. 30 (2011) 2596–2609. doi: 10.1038/emboj.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, et al. , Structure and mechanism of the chromatin remodelling factor ISW1a, 472 (2011) 448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- [30].Farnung L, Vos SM, Wigge C, Cramer P, Nucleosome-Chd1 structure and implications for chromatin remodelling, Nature. 550 (2017) 539–542. doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sundaramoorthy R, Hughes AL, El-Mkami H, Norman DG, Ferreira H, Owen-Hughes T, Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome, Elife. 7 (2018) 977. doi: 10.7554/eLife.35720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL, Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes, Molecular Cell. 10 (2002) 935–942. doi:doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- [33].Candau R, Zhou JX, Allis CD, Berger SL, Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo, EMBO Journal. 16 (1997) 555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sun J, Paduch M, Kim S-A, Kramer RM, Barrios AF, Lu V, et al. , Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2, Proc Natl Acad Sci USA. 23 (2018) 201805343. doi: 10.1073/pnas.1805343115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Y, Mi W, Xue Y, Shi X, Kutateladze TG, The ZZ domain as a new epigenetic reader and a degradation signal sensor, Crit. Rev. Biochem. Mol. Biol 54 (2019) 1–10. doi: 10.1080/10409238.2018.1564730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou M-M, Structure and chromosomal DNA binding of the SWIRM domain, Nat Struct Mol Biol. 12 (2005) 1078–1085. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
- [37].Yoneyama M, Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, et al. , Structural and functional differences of SWIRM domain subtypes, J Mol Biol. 369 (2007) 222–238. doi: 10.1016/j.jmb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- [38].Nguyen-Huynh N-T, Sharov G, Potel C, Fichter P, Trowitzsch S, Berger I, et al. , Chemical cross-linking and mass spectrometry to determine the subunit interaction network in a recombinant human SAGA HAT subcomplex, Protein Sci. 24 (2015) 1232–1246. doi: 10.1002/pro.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pápai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, Ben-Shem A, Structure of SAGA and mechanism of TBP deposition on gene promoters, Nature. 577 (2020) 711–716. doi: 10.1038/s41586-020-1944-2. [DOI] [PubMed] [Google Scholar]
- [40].Wang H, Dienemann C, Stützer A, Urlaub H, Cheung ACM, Cramer P, Structure of the transcription coactivator SAGA, Nature. 577 (2020) 717–720. doi: 10.1038/s41586-020-1933-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA, Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry, Mol Cell Biol. 22 (2002) 4723–4738. doi: 10.1128/mcb.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shukla A, Chaurasia P, Bhaumik SR, Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability, Cell Mol Life Sci. 66 (2009) 1419–1433. doi: 10.1007/s00018-008-8605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, et al. , Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation, Embo J. 30 (2011) 2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, et al. , Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes, Mol. Syst. Biol 7 (2011) 503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kurabe N, Katagiri K, Komiya Y, Ito R, Sugiyama A, Kawasaki Y, et al. , Deregulated expression of a novel component of TFTC/STAGA histone acetyltransferase complexes, rat SGF29, in hepatocellular carcinoma: possible implication for the oncogenic potential of c-Myc, Oncogene. 26 (2007) 5626–5634. doi: 10.1038/sj.onc.1210349. [DOI] [PubMed] [Google Scholar]
- [46].Ringel AE, Cieniewicz AM, Taverna SD, Wolberger C, Nucleosome competition reveals processive acetylation by the SAGA HAT module, Proc Natl Acad Sci USA. 112 (2015) E5461–70. doi: 10.1073/pnas.1508449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, et al. , Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers, Cell. 142 (2010) 967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- [48].Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL, Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail, J Biol Chem. 282 (2007) 27923–27934. doi: 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- [49].Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL, Expanded lysine acetylation specificity of Gcn5 in native complexes, J Biol Chem. 274 (1999) 5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- [50].Kuo Y-M, Andrews AJ, Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3, PLoS ONE. 8 (2013) e54896. doi: 10.1371/journal.pone.0054896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cieniewicz AM, Moreland L, Ringel AE, Mackintosh SG, Raman A, Gilbert TM, et al. , The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3, Mol. Cell Proteomics 13 (2014) 2896–2910. doi: 10.1074/mcp.M114.038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Li S, Shogren-Knaak MA, The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes, J Biol Chem. 284 (2009) 9411–9417. doi: 10.1074/jbc.M809617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mittal C, Culbertson SJ, Shogren-Knaak MA, Distinct requirements of linker DNA and transcriptional activators in promoting SAGA-mediated nucleosome acetylation, Journal of Biological Chemistry. 293 (2018) 13736–13749. doi: 10.1074/jbc.RA118.004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Burke TL, Miller JL, Grant PA, Direct inhibition of Gcn5 protein catalytic activity by polyglutamine-expanded ataxin-7, Journal of Biological Chemistry. 288 (2013) 34266–34275. doi: 10.1074/jbc.M113.487538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF, Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression, Nucleic Acids Res. 29 (2001) 1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kusch T, Guelman S, Abmayr SM, Workman JL, Two Drosophila Ada2 homologues function in different multiprotein complexes, Mol Cell Biol. 23 (2003) 3305–3319. doi: 10.1128/mcb.23.9.3305-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Muratoglu S, Georgieva S, Pápai G, Scheer E, Enünlü I, Komonyi O, et al. , Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes, Mol Cell Biol. 23 (2003) 306–321. doi:doi: 10.1128/mcb.23.1.306-321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Barlev NA, Emelyanov AV, Castagnino P, Zegerman P, Bannister AJ, Sepulveda MA, et al. , A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription, Mol Cell Biol. 23 (2003) 6944–6957. doi: 10.1128/mcb.23.19.6944-6957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Riss A, Scheer E, Joint M, Trowitzsch S, Berger I, Tora L, Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5, Journal of Biological Chemistry. 290 (2015) 28997–29009. doi: 10.1074/jbc.M115.668533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gamper AM, Kim J, Roeder RG, The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis, Mol Cell Biol. 29 (2009) 266–280. doi: 10.1128/MCB.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, et al. , The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes, Cell Mol Life Sci. 67 (2010) 611–628. doi: 10.1007/s00018-009-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Suganuma T, Gutiérrez JL, Li B, Florens L, Swanson SK, Washburn MP, et al. , ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding, Nat Struct Mol Biol. 15 (2008) 364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- [63].Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, et al. , The double-histone-acetyltransferase complex ATAC is essential for mammalian development, Mol Cell Biol. 29 (2009) 1176–1188. doi: 10.1128/MCB.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Vamos EE, Boros IM, The C-terminal domains of ADA2 proteins determine selective incorporation into GCN5-containing complexes that target histone H3 or H4 for acetylation, FEBS Lett. 586 (2012) 3279–3286. doi: 10.1016/j.febslet.2012.06.051. [DOI] [PubMed] [Google Scholar]
- [65].Qi D, Larsson J, Mannervik M, Drosophila Ada2b is required for viability and normal histone H3 acetylation, Mol Cell Biol. 24 (2004) 8080–8089. doi: 10.1128/MCB.24.18.8080-8089.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, et al. , The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions, Mol Cell Biol. 25 (2005) 8215–8227. doi: 10.1128/MCB.25.18.8215-8227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Torres-Zelada EF, Stephenson RE, Alpsoy A, Anderson BD, Swanson SK, Florens L, et al. , The Drosophila Dbf4 ortholog Chiffon forms a complex with Gcn5 that is necessary for histone acetylation and viability, J Cell Sci. 132 (2019) jcs214072. doi: 10.1242/jcs.214072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Soffers JHM, Li X, Saraf A, Seidel CW, Florens L, Washburn MP, et al. , Characterization of a metazoan ADA acetyltransferase complex, Nucleic Acids Res. 47 (2019) 3383–3394. doi: 10.1093/nar/gkz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, Berger SL, et al. , The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae, Mol Cell Biol. 19 (1999) 6621–6631. doi:doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Schrödinger, LLC, The PyMOL Molecular Graphics System, Version 1.8, (2015).