Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an infectious disease that has spread worldwide. Current treatments are limited in both availability and efficacy, such that improving our understanding of the factors that facilitate infection is urgently needed to more effectively treat infected individuals and to curb the pandemic. We and others have previously demonstrated the significance of interactions between the SARS-CoV-2 spike protein, integrin α5β1, and human ACE2 to facilitate viral entry into host cells in vitro. We previously found that inhibition of integrin α5β1 by the clinically validated small peptide ATN-161 inhibits these spike protein interactions and cell infection in vitro. In continuation with our previous findings, here we have further evaluated the therapeutic potential of ATN-161 on SARS-CoV-2 infection in k18-hACE2 transgenic (SARS-CoV-2 susceptible) mice in vivo. We discovered that treatment with single or repeated intravenous doses of ATN-161 (1 mg/kg) within 48 h after intranasal inoculation with SARS-CoV-2 lead to a reduction of lung viral load, viral immunofluorescence, and improved lung histology in a majority of mice 72 h post-infection. Furthermore, ATN-161 reduced SARS-CoV-2-induced increased expression of lung integrin α5 and αv (an α5-related integrin that has also been implicated in SARS-CoV-2 interactions) as well as the C–X–C motif chemokine ligand 10 (Cxcl10), further supporting the potential involvement of these integrins, and the anti-inflammatory potential of ATN-161, respectively, in SARS-CoV-2 infection. To the best of our knowledge, this is the first study demonstrating the potential therapeutic efficacy of targeting integrin α5β1 in SARS-CoV-2 infection in vivo and supports the development of ATN-161 as a novel SARS-CoV-2 therapy.
Keywords: ATN-161, Cxcl10, hACE2, Integrins, SARS-CoV-2
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the agent causative of coronavirus disease 2019 (COVID-19), is a highly transmissible respiratory pathogen in which an estimated 14% of all patients will develop serious conditions, with a subsequent mortality rate of 1.4–3.4% [1], [2], [3]. Several non-pharmacological interventions have been implemented to slow down the spread of SARS-CoV-2 [4], [5], [6], [7]. However, to date there is no universally agreed direct therapy available to treat COVID-19. It is important to study effective therapeutic targets focusing on disrupting aspects of the viral replication process, including SARS-CoV-2 host cell entry [8], which uses its spike proteins to bind to the angiotensin-converting enzyme 2 (hACE2) receptor on targeted cells to facilitate its entry and replication [9]. The virus infects the cells after the proteolytic cleavage of the spike protein by the transmembrane serine protease 2 (TMPRSS2) or Cathepsin B or L or FURIN is required for spike protein priming and virus infection [10].
Integrins are family of cell adhesion receptors that may play an important role in SARS-CoV-2 host cell entry and infection due to the spike protein containing an integrin binding motif arginine-glycine-aspartate (RGD) sequence [11], [12], [13], [14]. Integrins are composed of non-covalently linked α and β subunits that recognize and bind to extracellular matrix (ECM) proteins and mediate cell survival, proliferation, differentiation, and migration [15], [16], [17], [18]. Integrin dimers are expressed in most cells, including endothelial cells, epithelial cells in the respiratory tract [19], and are known to be involved in the infectious etiology of other viruses; such as human cytomegalovirus [20], Epstein–Barr virus [21], rotavirus [22], Kaposi's sarcoma-associated virus (HHV-8) [23] and Ebola [24]. Importantly, the β1 family of integrins are closely associated with ACE2 [25]. A recent study reviewed novel mutation (K403R) in the spike protein that does not exist in other strains of the coronavirus, creating an RGD motif, which could be recognized by integrins [2]. Therefore, the new RGD motif in SARS-CoV-2 could increase the binding potency of ACE2-positive target cells in association with β1 integrins as well as potentially facilitating infection of ACE2-negative cells [2]. This could help explain the faster and aggressive spread of virus as compared to SARS-CoV-1, which belongs to the same family of beta coronaviruses [26]. Targeting therapies to disrupt the spike-binding event may also inhibit viral replication by halting host cell entry and offers a promising approach to treat COVID-19 [8].
Our laboratory has previously shown that the SARS-CoV-2 spike protein is capable of binding to α5β1 integrin in cell-free ELISA assays [27], an observation which has since been repeated in epithelial cell-containing systems in vitro [28]. We further demonstrated that the small pentapeptide α5β1 integrin inhibitor ATN-161 could inhibit this binding and also decrease SARS-CoV-2 infection and cytopathy in cultured vero-E6 cells [27]. ATN-161 has several properties that make it potentially attractive as a novel SARS-CoV-2 therapy; it is safe and well-tolerated with no dose-limiting toxicity in phase I cancer clinical trials [29], has demonstrated in-vivo efficacy in mice against a different beta coronavirus, porcine hemagglutinating encephalomyelitis virus (PHEV) [30] as well as other disease/injury models [31], [32], and can also bind to and inhibit integrin αvβ3 which is similar to α5β1 integrin that has also been implicated in SARS-CoV-2 infection [33], [34]. In the present study we examined the therapeutic potential of ATN-161 in a human ACE2 receptor-expressing K18 mouse model of SARS-CoV-2 infection.
2. Methods
2.1. Mice and ethics statement
Male heterozygous K18-hACE c57BL/6 J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J, 10-weeks-old) were obtained from The Jackson Laboratory. Mice were housed in the animal facility at Tulane University School of Medicine. The Institutional Animal Care and Use Committee of Tulane University reviewed and approved all procedures for sample handling, inactivation, and removal from a BSL3 containment (permit number 3430 (#5)).
2.2. SARS-CoV-2 infection
Mice were inoculated with either saline or SARS-CoV-2 via intranasal administration by the ABSL3-trained staff with a dose of 2 × 105 TCID50/mouse to induce viral infection in these animals [35], [36]. The infected mice were observed daily to record body weight and clinical signs of illness (e.g., fur ruffling, less activity). After 3 days post infection (dpi), the mice were euthanized by CO2 asphyxiation followed by cervical dislocation and lungs were harvested for histology, immunofluorescence and qRT-PCR analysis.
2.3. Treatment interventions
Mice received ATN-161 (1 mg/kg, Medkoo Biosciences, Morrisville, NC, USA) via retro orbital i.v. injections. The subjects were divided into 6 groups; two control groups were non-infected: 1. saline treated control (saline, n = 3) and 2. ATN-161 treated mice (ATN-161, n = 3). These mice resided in the BSL1 portion of the Tulane Department of Comparative Medicine animal housing facility. Four SARS-CoV-2 infected groups received either saline or ATN-161 prepared fresh daily (3. SARS-CoV-2 + saline (n = 5), 4. SARS-CoV-2 + ATN-161 once at 2 h (n = 5), 5. SARS-CoV-2 + ATN-161 daily (2, 24, and 48 h, (n = 5)) and 6. SARS-CoV-2 + ATN-161 once at 48 h (n = 5) treatment administration following intra nasal viral inoculation with SARS-CoV-2.
2.4. Viral copy number determination
Tissues were weighed and homogenized in Trizol Lysis Reagent (Invitrogen). RNA was extracted from lung homogenates according to the instructions of the RNA extraction kit manufacturer (RNeasy Plus Mini Kit; Qiagen) post phase separation using Trizol reagent. Total RNA (Five microliters) was added in duplicate to a 0.2-ml standard 96-well optical microtiter plate (Cat. No.N8010560; Thermo Fisher). qRT-PCR reaction was set by using TaqPath 1-Step Master Mix (Cat. No. A28527; Thermo Fisher) with a primers and a FAM-labeled probe targeting the N1 amplicon of N gene (2019-nCoV RUO Kit, Cat. No. 10006713; IDT-DNA) of SARS-CoV-2 (https://www.ncbi.nlm.nih.gov/nuccore; accession number MN908947), following the manufacturer's instructions. Viral load was calculated by the linear regression function by Cq values acquired from 2019 nCoV qRT-PCR Probe Assays. The viral copy numbers from the lung samples are represented as copies/100 ng of RNA as followed using a published assay [37], [35], [38]. Subgenomic mRNA (sgmRNA) encoding the E gene was quantified using FAM-labeled primers (sgm-N-for: 5′-CGATCTCTTGTAGATCTGTTCTC-3′, sgm-N-Probe:5′-FAMTAACCAGAATGGAGAACGCAGTGGG-TAMRA-3′,sgm-N-reverse:5′-GGTGAACCAAGACGCAGTAT-3′), following the manufacturer's instructions. Subgenomic N viral copy number was calculated by standard Cq values. The viral copy numbers from the lung samples are represented as copies/100 ng of RNA followed using a published assay [38], [35].
2.5. Gene expression
The lung tissue was homogenized in Trizol Lysis Reagent (Invitrogen). RNA was extracted from lung homogenates according to the instructions of the RNA extraction kit manufacturer (RNeasy Plus Mini Kit; Qiagen) post phase separation using Trizol reagent. RNA was converted to cDNA using iScript reverse transcriptase master mix (Bio-Rad). qPCR was carried out with QuantStudio 3 Real-Time PCR Systems (Life Technologies) using TaqMan PCR Master Mix and premixed primers/probe sets (Thermo Fisher Scientific) sets specific for Itgav (Mm00434486_m1), Cxcl10 (Mm00445235_m1), Itga5 (Mm00439797_m1), Itgb3 (Mm00443980_m1), Itgb1 (Mm01253233_m1), ACE2 (Hs01085333_m1), and Hypoxanthine phosphoribosyltransferase, Hprt (Mm01545399_m1, Control gene) (Life Technologies) were used gene expression. Data were analyzed comparing control to SARS-CoV-2 infected mice and are presented as a fold change of control.
2.6. Histology
The harvested whole right lungs (three lobes) were fixed in Z-fix (Anatech Ltd., Battle Creek, MI, USA). Paraffin sections (5 μm in thickness) were used for hematoxylin and eosin and Masson's trichrome stains to identify morphological changes in lungs. Slides were imaged with a digital slide scanner (Zeiss Axio Scan; Zeiss, White Plains, NY). Representative photo micrographs at 20× magnification were acquired from the whole scanned right lung using the Aperio Image Scope software (version 12.3.2.8013, Leica, Buffalo Grove, IL, USA). The disease severity of lung lobe sections collected from different mice in each group, uninfected control/ATN-161 n = 6; SARS-CoV-2-infected (5 mice for vehicle, 5 mice for each 3 ATN-161 groups) were quantified by a pathological score. Whole specimens were examined by using Aperio Image Scope software at 10× in three randomly selected areas to establish a histopathological score in each case. For H&E, pathology score of infiltration of lymphocytes (inflammation), mucus plug, alveolar thickening, thrombosis and hemorrhage (bleeding) were analyzed according to the following criteria: 0- none; 1-uncommon detection in <5% lung fields (200×); 2- detectable in up to 33% of lung fields; 3- detectable in up to 33–66% of lung fields; 4- detectable in >66% of lung fields as described in [39], [40], [41], [42]. Collagen staining was performed using the Masson's trichrome staining (Poly Scientific R&D) method for the detection of collagen positive area, according to the manufacturer's recommendations [40]. Whole specimens areas were captured per lobe; morphometric quantitation of collagen was measured using QuPath v0.2.0 software and the collagen positive area is expressed as square microns.
2.7. SARS-CoV-2 immunofluorescence
Fixed tissue samples were processed via indirect immunofluorescence assays (IFA) for the detection of the SARS-CoV-2 antigen. The slides were deparaffinized in xylenes and rehydrated through an ethanol series, followed by heat-induced antigen retrieval with high pH antigen unmasking EDTA solution. The slides were washed with PBS with 0.1% Triton X-100 and blocked with 10% normal goat serum at room temperature for 1 h. Primary antibody (Polyclonal Anti-SARS Coronavirus (antiserum, Guinea Pig) 1:1000, NR-10361) incubation was achieved at room temperature for 1 h. Slides were washed, and primary antibody detected following 40 min incubation in an appropriate secondary antibody tagged with Alexa Fluor fluorochromes (1:1000) in normal goat serum. After washing in PBS, mounting media with DAPI was used to label the nuclei. Slides were imaged with a digital slide scanner (Zeiss Axio Scan.Z1; Zeiss, White Plains, NY). Fluorescent images were acquired using HALO (Indica Labs, v2.3.2089.70). Threshold and multiplex analyses were performed with HALO for quantitation.
2.8. Liver enzyme assays
Blood samples were collected in BD micro trainer tubes by cardiac puncture, incubated at room temperature for 30 min to coagulate, and then centrifuged for 10 min at 3000 rpm to separate the serum. Serum samples for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) level were assayed by colorimetric determination using Sigma activity assay kits (Sigma Diagnostics, St Louis, MO): AST (MAK055) and ALT (MAK052). Procedures were performed following the manufacturer's instructions.
2.9. Statistics
Statistical tests were performed with GraphPad Prism, 8.4.3 version (GraphPad Software, San Diego, CA). Data are presented as mean ± SEM. Significant differences were designated using omnibus one-way ANOVA and, when significant, followed-up with two-group planned comparisons selected a priori to probe specific hypothesis-driven questions (saline vs SARS-CoV-2 + vehicle; SARS-CoV-2 + vehicle vs each of the various SARS-CoV-2 + ATN-161 treated groups); the Holm-Sidak adjustment was applied to control for multiple comparisons. Non-parametric tests were employed in the event of violations of underlying ANOVA assumptions. Statistical significance was taken at the p < 0.05 level.
3. Results
3.1. ATN-161 impacts the Genomic-N and subgenomic N (Sgm-—N) viral load in SARS-CoV-2 infected K18-hACE2 mice lung tissue
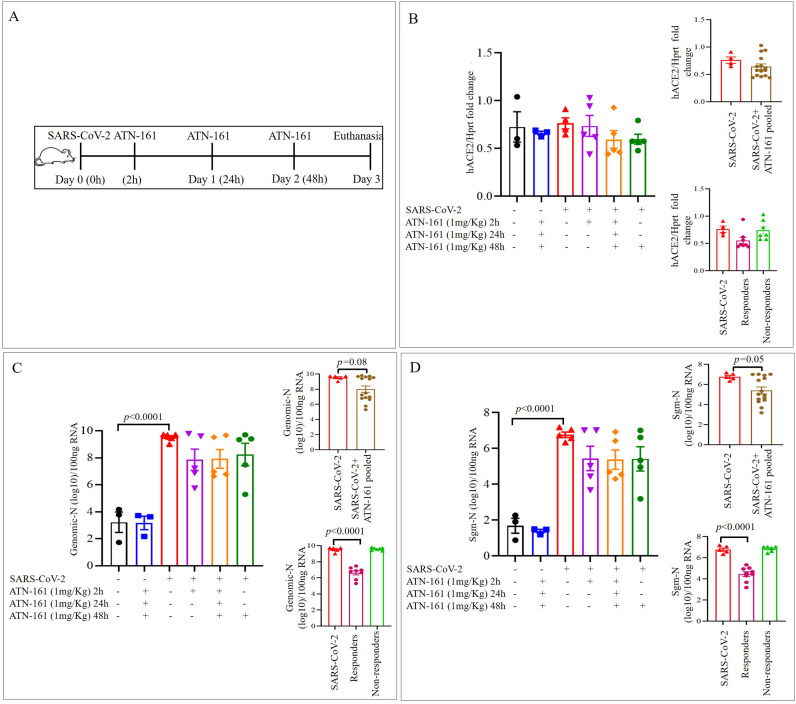
Lung tissue expression of hACE2 and viral loads after 3dpi with SARS-CoV-2 + Saline or ATN-161 administered either once at 2 h or 48 h post-infection or daily, post-infection, were measured, analyzed and compared to Vehicle-treated/uninfected (no SARS-CoV-2 exposure) mice either given Saline or ATN-161 (daily) for 3dpi. (Fig. 1A). For hACE2, the omnibus one-way ANOVA was not significant (Fig. 1B).
Fig. 1.
ATN-161 reduces the Genomic-N and Sub genomic N (Sgm—N) viral load in SARS-CoV-2 infected K18-hACE2 mice lung tissue
Schematic overview of experimental timeline for K18-hACE2 mice (A) 10-week old male K18-hACE2 transgenic mice administered saline (black bar) or ATN-161 (1 mg/kg, blue bar) intravenously (via retro-orbital). Mice were inoculated via the intranasal route with Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (2 × 105 TCID50) + Saline (i.v. by retroorbital rout) (Red bar). ATN-161 (1 mg/Kg) treatment was administered at 3 different time periods, SARS-CoV-2 + ATN-161-2 h post infection (purple bar), SARS-CoV-2 + ATN-161-daily (2 h, 24 h and 48 h) administration (orange bar), and SARS-CoV-2 + ATN-161-48 h administration (green bar) post SARS-CoV-2 intranasal inoculation. Panels B, C, and D top inserts with brown bar represents SARS-CoV-2 + ATN-161 all treatment groups pooled data; Panels B, C, and D bottom insert with pink bar represents responding mice and light green bar represents nonresponding with SARS-CoV-2 + ATN-161 (either 2, daily or 48 h) treatment. 3 days post infection (3 dpi) the mice were euthanized, and RNA isolated from the left lungs by Trizol method for qRT-PCR (B) hACE2 expression in lungs of K18-hACE2 mice (C) viral genomic-N (Total-N) and (D) sub genomic-N mRNA (sgm-—N). Experimental groups are divided into 6 groups. Saline n = 3; ATN-161 n = 3; SARS-CoV-2 (5 mice for vehicle, 5 mice for each 3 ATN-161 groups). Data are presented as mean ± SEM. P values represent saline vs SARS-CoV-2 + vehicle and SARS-CoV-2 + vehicle vs SARS-CoV-2 + ATN-161 treatment groups. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For Genomic-N (Fig. 1C), the omnibus one-way ANOVA was significant [F(5,20) = 12.84, p < 0.0001]. We followed up this significant analysis with two group planned comparisons using the Holm-Sidak correction. We found a significant increase in Genomic-N among SARS-CoV-2 + Saline mice compared to non-infected, Saline-treated mice [t(6) = 5.98, p < 0.0001]. Though we did not detect significant group differences between any other two-group comparison, visual inspection of the graph revealed that there was 1) heterogeneity among the ATN-161 treated groups, suggesting a dichotomy in this population with regards to response to the ATN-161 treatment about the viral load and 2) a general trend towards reduced viral load among all ATN-161 treated groups, regardless of timepoint or number of injections. This indicated to us a potential for our analyses to be underpowered. Therefore, to increase power and reduce variability, we pooled all ATN-161 treated animals into a single group and re-analyzed the data by comparing lung viral load in this group to that of the SARS-CoV-2 + Saline-treated mice. We found a trend towards significance [t(18) = 2.03, p < 0.06] such that ATN-161-treated mice had lower Genomic N viral load in lungs than SARS-CoV-2-infected mice. Then we analyzed the data using the Mann-Whitney U nonparametric test to account for a significant (p < 0.01) violation of the homogeneity of variance assumption (though ANOVA is robust to this type of violation), the effect remained marginal (p = 0.08). We also dichotomized ATN-161-treated animals into ‘responder’ and ‘non-responder’ groups such that mice were considered non-responders if they displayed Genomic N values >2 × 109 (one power of 10 lower than the lowest value observed in the SARS-CoV-2 group), Sgm-N values >1 × 105 (one power of 10 lower than the lowest value observed in the SARS-CoV-2 group), and viral immunohistology staining counts >0.7 (one power of 10 lower than the lowest value observed in the SARS-CoV-2 group). Ns of responders for each ATN-161 treated group were 3, 3, and 2 for ATN-161-2 h, ATN-161-daily, and ATN-161-48 h groups respectively. When we re-analyzed the data by comparing lung viral load in these groups to that of the SARS-CoV-2 + Saline-treated mice, we found a significant omnibus one-way ANOVA [F(2,17) = 35.32, p < 0.0001] such that ATN-161 responders had significantly lower genomic lung viral loads than SARS-CoV-2-infected animals [t(17) = 7.04, p < 0.0001].
For Sgm-N (Fig. 1D), the omnibus one-way ANOVA was significant [F(5,20) = 13.81, p < 0.0001] such that there was a significant increase in Sgm-N among SARS-CoV-2 + Saline mice compared to non-infected, Saline-treated mice [t(6) = 6.13, p < 0.0001]. Analysis of Sgm-N values among pooled ATN-161-treated mice again revealed that ATN-161 reduced Sgm-N viral load [t(18) = 2.23, p < 0.05] that persisted (p = 0.05) when the Mann-Whitney U non-parametric analysis was applied given the significant (p < 0.05) homogeneity of variance in this group. When we re-analyzed the data by comparing lung viral load in the responder/non-responder groups to that of the SARS-CoV-2 + Saline-treated mice, we found a significant omnibus one-way ANOVA [F(2,17) = 39.91, p < 0.0001] such that ATN-161 responders had significantly lower Sgm-N lung viral loads than SARS-CoV-2-infected animals [t(17) = 7.40, p < 0.0001].
3.2. ATN-161 effects on the expression of virus in SARS-CoV-2 infected K18-hACE2 mice lung tissue
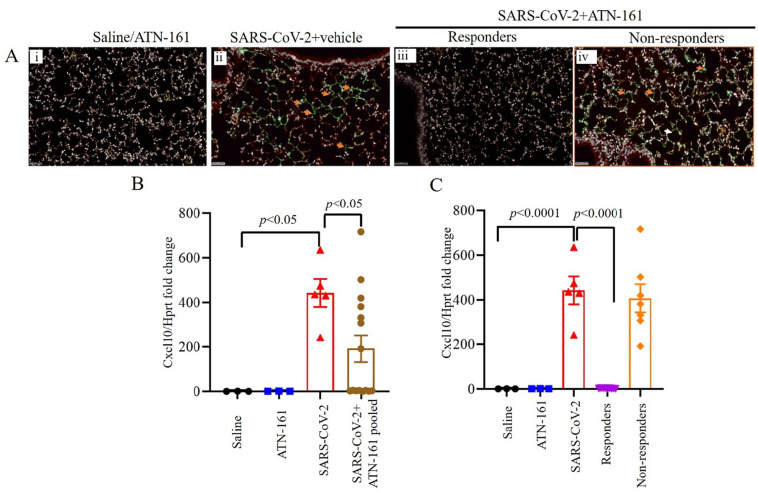
Given the observations in Fig. 1, we continued our analyses comparing non-infected Saline or ATN-161-treated mice with SARS-CoV-2 + Saline and either pooled ATN-161-treated infected mice or ATN-161 responders/non-responders. Immunohistochemistry staining for SARS-CoV-2 positive cells was conducted on all mice. Representative SARS-CoV-2 viral staining images from saline/ATN-161 (i); SARS-CoV-2 + vehicle (ii); Responders (iii) and Non-responders (iv) with SARS-CoV-2 + ATN-161 administration either 2, daily or 48 h administration, post SARS-CoV-2 intranasal inoculation are depicted in Fig. 2A. All K-18 hACE2 mice infected with SARS-CoV-2 + Saline or non-responders from SARS-CoV-2 + ATN-161 administration group had multifocal regions of SARS-CoV-2–positive cells (Fig. 2A ii, iv), whereas treatment with ATN-161, 2 h and daily administered groups show 3 mice are completely negative for SARS-CoV-2 protein and 2 mice are negative (responders) in 48 h ATN-161 group out of 5 mice challenged for SARS-CoV-2.
Fig. 2.
ATN-161 reduces the expression of virus in SARS-CoV-2 infected K18-hACE2 mice lung tissue. Immunohistochemistry staining for SARS-CoV-2 (Ai-iv, orange arrowheads indicates SARS-CoV-2 viral stain (green) positive cells). SARS-CoV-2 viral staining representative image from saline/ATN-161 (i); SARS-CoV-2 + vehicle (ii); Responders (iii) and Non-responders (iv) with SARS-CoV-2 + ATN-161 administration either 2, daily or 48 h administration, post SARS-CoV-2 intranasal inoculation. The tissue analysis at 3 dpi using the protocol described in Fig. 1. All K-18 hACE2 mice infected with SARS-CoV-2 + Saline or non-responders from SARS-CoV-2 + ATN-161 administration group with have multifocal regions of SARS-CoV-2–positive cells (ii, iv), whereas treatment with ATN-161, 2 h and daily administered groups show 3 mice are completely negative for SARS-CoV-2 protein and 2 mice are negative (responders) in 48 h ATN-161 group out of 5 mice challenged for SARS-CoV-2. (B–C) Cxcl10 mRNA expression in lung. Scale bars, 50 μm. Green = SARS-CoV-2; White = nuclei/DAPI; Red = empty/autofluorescence. Data are presented as mean ± SEM. P values represent saline vs SARS-CoV-2 + vehicle and SARS-CoV-2 + vehicle vs SARS-CoV-2 + ATN-161 pooled, SARS-CoV-2 + ATN-161 responders, and SARS-CoV-2 + ATN-161 non-responders. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
One-way ANOVA of C–X–C motif chemokine ligand 10 (Cxcl10) mRNA expression in lung comparing saline or ATN-161 treated non-infected, SARS-CoV-2 + Saline, and SARS-CoV-2 + ATN-161 pooled mice (Fig. 2B) revealed a significant effect [F(3,23) = 4.56, p < 0.05] such that SARS-CoV-2 + Saline mice had significantly higher lung Cxcl10 mRNA expression compared to Saline-treated uninfected mice [t(6) = 3.07, p < 0.05] and pooled ATN-161-treated mice [t(18) = 2.46, p < 0.05]. One-way ANOVA of Cxcl10 mRNA expression in lung comparing saline or ATN-161 treated non-infected, SARS-CoV-2 + Saline, and SARS-CoV-2 + ATN-161 responders and non-responders (Fig. 2C) revealed a significant effect [F(4,21) = 24.38, p < 0.0001], that persisted when the Kruskal-Wallis non-parametric analysis (p < 0.005) was applied given the significant violation of homogeneity of variance (p < 0.05) in this ANOVA analysis. Two-group planned follow-up comparisons revealed that SARS-CoV-2 + Saline mice had significantly higher lung Cxcl10 mRNA expression compared to Saline-treated uninfected mice [t(6) = 5.59, p < 0.0001] and ATN-161-treated responders [t(11) = 7.12, p < 0.0001].
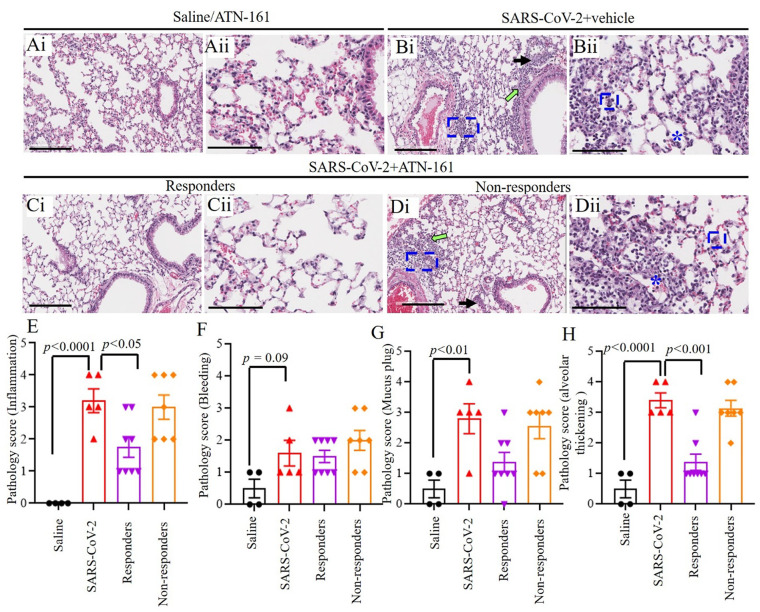
3.3. Histopathological analysis of SARS-CoV-2 infected K18-hACE2 mice lung tissue by hematoxylin and eosin stain
Representative images of hematoxylin and eosin staining of lung sections from saline/ATN-161 (Fig. 3 Ai–Aii); SARS-CoV-2 + vehicle (Fig. 3Bi–ii); Responders (Fig. 3Ci–ii) and non-responders (Fig. 3Di–ii) with SARS-CoV-2 + ATN-161 (either 2 h, daily, or 48 h) administration post-SARS-CoV-2 intranasal inoculation are shown in Fig. 3. Histopathological observations indicated that multifocal lesion, moderate interstitial pneumonia and alveolar septal thickening (Fig. 3Bii, Dii, blue frames), infiltration of lymphocytes (Fig. 3Bi, Di, green arrow), mucus plug and fibrin exudation (Fig. 3Bi, Di, black arrow), bleeding and pulmonary oedema (asterisk) were observed in SARS-CoV-2 + vehicle and SARS-CoV-2 + ATN-161 non responders mice whereas these observations are less or completely absent in SARS-CoV-2 + ATN-161 responders mice. Hematoxylin and eosin staining of lung sections from saline/ATN-161 (Fig. 3Ai–Aii) revealed no histopathological changes in any of the mice. Semi quantitative analysis of histopathological findings like inflammation, bleeding, mucus plug, and alveolar thickening (Fig. 3E–H) demonstrated that the interaction between the virus and pneumocytes, and/or alveolar macrophages triggers alveolar thickening as the major lung feature in severe SARS-CoV-2 infection [43]. These observations were similar to those described in SARS-CoV-2 infection in a mouse model and are consistent with the previously postulated mechanisms underlying pulmonary damage in SARS-CoV study of a lung biopsy of a patient sample [44], [43], [45].
Fig. 3.
Histopathological Analysis of SARS-CoV-2 infected K18-hACE2 mice lung tissue (Ai–Dii) Hematoxylin and eosin staining of lung sections from saline/ATN-161 (Ai–Aii); SARS-CoV-2 + vehicle (Bi–ii); Responders (Ci–ii) and Non-responders (Di–ii) with SARS-CoV-2 + ATN-161(either 2 h, daily, or 48 h) administration post SARS-CoV-2 intranasal inoculation with tissue analysis at 3 dpi using the protocol described in Fig. 1. Histopathological observations indicated that multifocal lesion, moderate interstitial pneumonia and alveolar septal thickening (Bii, Dii, blue frames), infiltration of lymphocytes (Inflammation) (Bi, Di, green arrow), mucus plug and fibrin exudation (Bi, Di, black arrow), bleeding and pulmonary edema (Bii, Dii, asterisk). (E–H) Semi quantitative analysis of histopathological findings is based on subjective scoring as described in methods. Control n = 3; ATN-161 n = 3; SARS-CoV-2 (5 mice for vehicle, 5 mice for each 3 ATN-161 groups). Scale bar: 200 μm (Ai, Bi, Ci, and Di) and 100 μm (Aii, Bii, Cii, and Dii). Data are presented as mean ± SEM. P values represent saline vs SARS-CoV-2 + vehicle and SARS-CoV-2 + vehicle vs SARS-CoV-2 + ATN-161 responders, and SARS-CoV-2 + ATN-161 non-responders. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
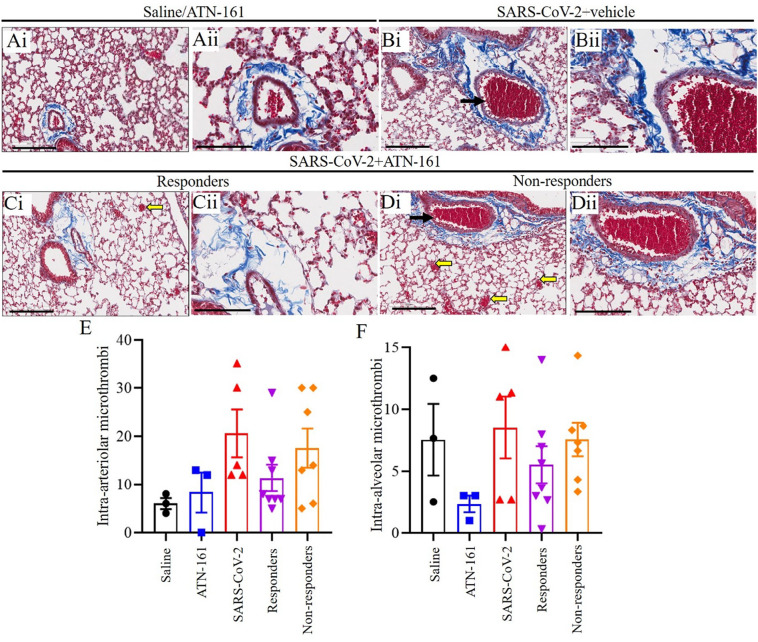
3.4. Histopathological analysis of SARS-CoV-2 infected K18-hACE2 mice lung tissue Masson's Trichrome stain
Representative images of Masson's Trichrome-stained sections (Fig. 4 Ai–Dii) of lung sections from saline/ATN-161 (Fig. 4Ai–Aii); SARS-CoV-2 + vehicle (Fig. 4Bi–ii); Responders (Fig. 4Ci–ii) and Non-responders (Fig. 4Di–ii) with SARS-CoV-2 + ATN-161 (either 2 h, daily, or 48 h) administration post SARS-CoV-2 intranasal inoculation. Histopathological analysis revealed multiple intra-arteriolar microthrombi (black arrows), intra-alveolar microthrombi (yellow arrow) in SARS-CoV-2 + vehicle and SARS-CoV-2 + ATN-161 non responders' mice whereas these observations are less or completely absent in SARS-CoV-2 + ATN-161 responder mice. Masson's Trichrome staining of lung sections from saline/ATN-161 (Fig. 4Ai–Aii) were no histopathological changes in any of the mice. However, quantification of collagen positive area revealed that there were no differences in collagen area between saline non-infected vs SARS-CoV-2 + Saline, SARS-CoV-2 + ATN-161 responders and non-responders. Semi quantitative analysis of histopathological findings of intra-arteriolar microthrombi and, intra-alveolar microthrombi (Fig. 4E–F) demonstrated lung pathology, hemorrhage, and blood clots. The microthrombi in the lung with the hyperinflammatory response in SARS-CoV-2 infected mice lungs occurs predominantly through intussusceptive angiogenesis [46].
Fig. 4.
Histopathological Analysis of SARS-CoV-2 infected K18-hACE2 mice lung tissue. Representative images of Masson's Trichrome-stained sections (Ai–Dii) of lung sections from Saline/ATN-161 (Ai-Aii); SARS-CoV-2 + vehicle (Bi–ii); Responders (Ci–ii) and Non-responders (Di–ii) with SARS-CoV-2 + ATN-161(either 2 h, daily, or 48 h) administration post SARS-CoV-2 intranasal inoculation with tissue analysis at 3 dpi using the protocol described in Fig. 1. Histopathological observations indicated that showing multiple intra-arteriolar microthrombi (black arrows), intra-alveolar microthrombi (yellow arrow). However, the groups did not differ on the extent of fibrosis histopathology (collagen area). Semi quantitative analysis of histopathological findings is based on subjective scoring as described in methods (E–F). Control n = 3; ATN-161 n = 3; SARS-CoV-2 (5 mice for vehicle, 5 mice for each 3 ATN-161 groups). Scale bar: 200 μm (Ai, Bi, Ci, and Di) and 100 μm (Aii, Bii, Cii, and Dii). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
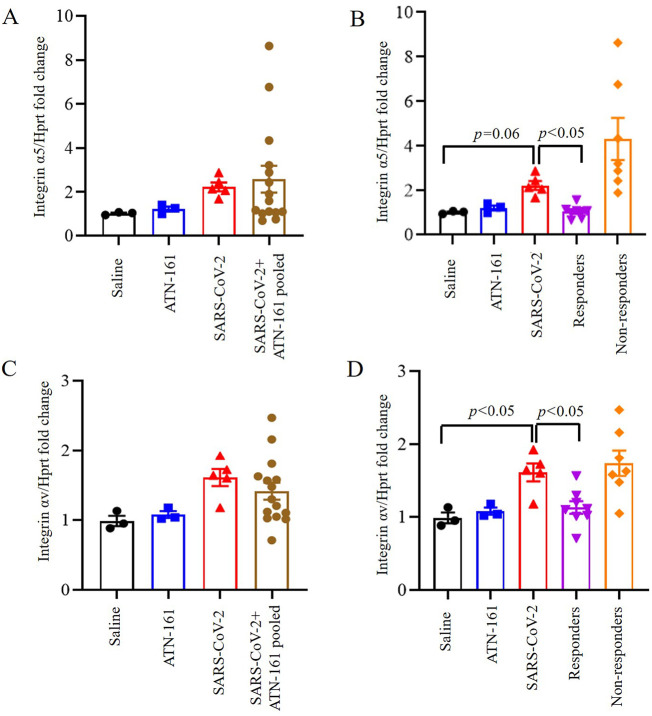
3.5. Induced expression of integrin α5, and integrin αv in the lungs of SARS-CoV-2 infected K18-hACE2 mice and is inhibited by ATN-161 treatment
One-way ANOVA of integrin-α5 expression in lung comparing saline or ATN-treated non-infected, SARS-CoV-2 + Saline, and SARS-CoV-2 + ATN-161 pooled mice was not significant (Fig. 5A). One-way ANOVA of integrin-α5 expression in lung comparing saline or ATN-161 treated non-infected, SARS-CoV-2 + Saline, and SARS-CoV-2 + ATN-161 responders and non-responders (Fig. 5B) revealed a significant effect [F(4,21) = 6.59, p < 0.05], that persisted when the Kruskal-Wallis non-parametric analysis (p < 0.05) was applied given the trend towards a violation of homogeneity of variance in this ANOVA analysis (p = 0.06). Because there was very high variability in the SARS-CoV-2 + ATN-161 non-responders, and we were concerned about the possibility of inflated Type 1 error risk, we conservatively conducted two-group follow-up comparisons using the non-parametric approach (Dunn-corrected). We found that SARS-CoV-2 + Saline mice tended to have higher lung integrin-α5 expression compared to uninfected Saline-treated mice [z = 2.32, p = 0.06].
Fig. 5.
Induced expression of integrin α5, and integrin αv in the lungs of SARS-CoV-2 infected K18-hACE2 mice and is inhibited by ATN-161 treatment. Integrin α5 (A–B), and Integrin αv (C–D) mRNA expression in lung at different time periods of ATN-161 by intravenous injection (i.v) after infection with SARS-CoV-2 (TCID50 = 2 × 105 /0.05 mL) was determined by qRT-PCR. Data are presented as mean ± SEM. P values represent saline vs SARS-CoV-2 + vehicle and SARS-CoV-2 + vehicle vs SARS-CoV-2 + ATN-161 pooled, SARS-CoV-2 + ATN-161 responders, and SARS-CoV-2 + ATN-161 non-responders.
One-way ANOVA of integrin-αv expression in lung comparing saline or ATN-161 treated non-infected, SARS-CoV-2 + Saline, and SARS-CoV-2 + ATN-161 pooled mice was not significant (Fig. 5C). One-way ANOVA of integrin-αv expression in lung comparing saline or ATN-161 treated non-infected, SARS-CoV-2 + Saline, and SARS-CoV-2 + ATN-161 responders and non-responders (Fig. 5D) revealed a significant effect [F(4,21) = 6.04, p < 0.005], such that SARS-CoV-2 + Saline mice had significantly higher lung integrin-αv expression compared to Saline-treated uninfected mice [t(6) = 2.70, p < 0.05] and ATN-161-treated responders [t(11) = 2.67, p < 0.05].
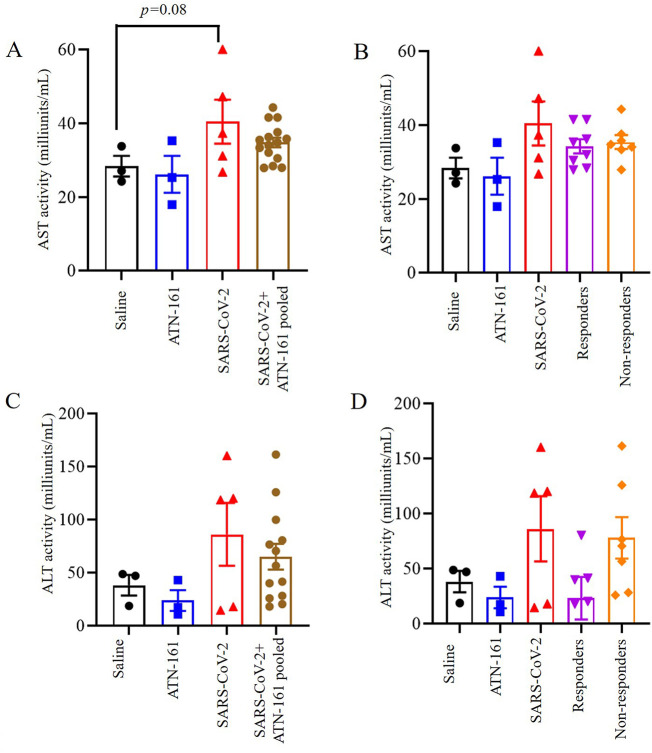
3.6. Effect ATN-161 on hepatic AST and ALT levels in SARS-CoV-2 infected K18-hACE2 mice
Levels of the hepatic transaminase AST trended to increase in the SARS-CoV-2 + vehicle infected group as compared to the saline or ATN-treated non-infected group [t(6) = 2.171, p = 0.08]. One-way ANOVA analysis of AST levels in serum comparing SARS-CoV-2 + Saline, and SARS-CoV-2 + ATN-161 pooled mice, SARS-CoV-2 + ATN-161 responders and non-responders determined a not significant difference between the groups [F(4,21) = 2.04, p > 0.05]. The results are shown in Fig. 6A–B.
Fig. 6.
Effect of ATN-161 on hepatic enzyme levels in the serum of SARS-CoV-2 infected K18-hACE2 mice. (A–B) aspartate aminotransferase (AST); (C–D) Alanine aminotransferase (ALT). Data are presented as mean ± SEM. P values represent saline vs SARS-CoV-2 + vehicle and SARS-CoV-2 + vehicle vs SARS-CoV-2 + ATN-161 pooled, SARS-CoV-2 + ATN-161 responders, and SARS-CoV-2 + ATN-161 non-responders.
Likewise, levels of the hepatic transaminase ALT trended to increase in the SARS-CoV-2 + vehicle infected group as compared to saline or ATN-treated non-infected group [t(6) = 1.468, p = 0.41]. One-way ANOVA analysis of ALT levels in serum demonstrated a downward trend in SARS-CoV-2 + ATN-161 responders as compared to SARS-CoV-2 + Saline. However, SARS-CoV-2 + ATN-161 pooled mice, and non-responders were not different as compared to infected group [F(4,18) = 0.212, p > 0.05]. The results are shown in Fig. 6C–D.
4. Discussion
The coronavirus disease-2019 (COVID-19) caused by SARS-CoV-2 continues to ravage the world. As of March 24, 2021, there were 3,089,162 deaths with a total of 145,759,060 confirmed (COVID-19) cases worldwide [47], [48]. While the development of vaccines and antibody treatments against COVID-19 is promising and globally accepted in the current pandemic [49], the emergence of SARS-CoV-2 viral variants may present major challenges to these therapeutic approaches. Like most viruses, SARS-CoV-2 mutates and continually presents variants. Spike protein mutations are the most common mutation seen in SARS-CoV-2 variants, which facilitate viral entry into the cell and mediate viral propagation by binding to ACE2 receptor. Interestingly, an RDG integrin-binding motif is a novel feature of SARS-CoV-2 spike protein, which is not seen in other coronaviruses [50], [11]. While this feature may have enhanced viral infectivity of SARS-CoV-2, it remains unknown how variant strains of SARS-CoV-2 may affect binding to integrins, with the resultant feature of affecting viral entry and propagation. However, it is worth noting that to the best of our knowledge, none of the currently characterized SARS-CoV-2 variants have directly mutated the RGD motif; This suggests the possibility that the RGD motif is of evolutionary advantage to the virus by supporting its ability to infect hosts.
Integrins are cell surface receptors that may bind to the SARS-CoV-2 spike protein interaction [51], [52], [11], [34]. In our previous study, we demonstrated by ELISA that the SARS-CoV-2 spike protein and ACE2 could bind to α5β1, and that ATN-161 could disrupt both of these interactions [27]. We further demonstrated that ATN-161 could significantly reduce SARS-CoV-2 infection (viral load, cell viability and cytopathy) in Vero (E6) cells in vitro [27]. In this study, we extend the understanding of the role of integrins and explored the therapeutic role of ATN-161 against SARS-CoV-2 infection in vivo in the k18-hACE2 mice model.
K18-hACE2 mice provide a platform for the rigorous screening of candidate drugs before their evaluation in other animal models [53]. This mouse model is widely used to evaluate the pathogenicity of viruses such as SARS-CoV-2 that require or prefer the human form of ACE2 (versus mouse ACE2) to readily infect mice and can be used to study potential therapies [54], [36]. Previous studies provided the evidence that SARS-CoV-2 infection could cause typical interstitial pneumonia and develop respiratory disease in hACE2-expressing mice resembling what is commonly seen in COVID-19 patients [55], [54], [56], [57].
We measured genomic-N (Nucleocapsid (N) protein) and Sgm-N in lungs to assess SARS-CoV-2viral infectivity and replication. Our results showed that SARS-CoV-2–infected hACE2 mice had significantly higher SARS-CoV-2 genomic-N. Similarly, viral sgmRNA copies were detected predominantly in the lung as compared to uninfected saline treated mice. SgmRNA levels of the virus is an adequate surrogate assay for detection of replicating virus (replicating virus separated from the total genome, and form dimers as the virus is replicating its machinery which can continue to produce protein) [35]. Thus, we are confident that our mice were successfully infected with SARS-CoV-2 [36].
We found that ATN-161-treated mice had lower Genomic-N viral load in lungs than saline treated SARS-CoV-2-infected mice. Visual inspection of Genomic-N and sgm-N graph revealed that there was 1) heterogeneity among the ATN-161 treated groups, suggesting a dichotomy in this population with regards to ATN-161 treatment response and viral load, and 2) a general trend towards reduced viral load among all ATN-161 treated groups, regardless of timepoint or number of injections. A limitation of this study is that it employed a limited number of K18 hACE2 mice, a function of the logistical difficulty of doing live virus BSL-3 studies. We also dichotomized ATN-161-treated animals into ‘responder’ and ‘non-responder’ groups such that mice were considered non-responders if they displayed Genomic N values >2 × 109, Sgm-N values >1 × 105 and viral immunohistology staining counts >0.7 values observed in the SARS-CoV-2 group in the SARS-CoV-2 + saline group. ATN-161 responders had significantly lower genomic lung viral loads than SARS-CoV-2-infected animals.
Accordingly, this bimodal distribution of responders may be due to our use of heterozygous (HT) K18-hACE2 mice, as the K18-hACE2 homozygous mouse model completely replaces mACE2 expression with hACE2 under the mAce2 promoter [43]. Thus, it might be possible that our use of HT K18-hACE2 mice in this study produced variable expression patterns of mACE2 vs hACE2, reducing the efficacy of ATN-161 in mice that expressed higher levels of mACE2 relative to hACE2. ATN-161 interference with mACE2-α5β1 interactions would presumably have minimal to no effect on SARS-CoV-2 infection as compared to impacting hACE2-α5β1 interactions. ATN-161 reduced Sgm-N viral load among pooled ATN-161-treated mice. When we re-analyzed the data by comparing lung viral load in the responder/non-responder groups to that of the SARS-CoV-2 + Saline-treated mice, we found that ATN-161 responders had significantly lower Sgm-N lung viral loads than SARS-CoV-2-infected animals. This suggests a lack of statistical power due to low Ns among individual ATN-treated groups; follow-up studies with additional subjects will be conducted to replicate these findings.
All K-18 hACE2 mice infected with SARS-CoV-2 or non-responders from SARS-CoV-2 + ATN-161 administered group had multifocal regions of SARS-CoV-2–positive cells. Our results are further supported by recent findings of lung histological changes in SARS-CoV-2 infected hACE2 mice [36], [43]. Furthermore, we observed that viral immunohistology staining counts were negative in responders for each ATN-161 treated group with n's of 3, 3, and 2 for ATN-161-2 h, ATN-161-daily, and ATN-161-48 h groups, respectively. The infected K18-hACE2 mice did not lose body weight after only 3dpi as expected, but we observed high levels of viral copies and infectious virus in the lungs given as demonstrated by other studies in K18-hACE2 mice [36], [43], [58].
In regards to an inflammatory response to SARS-CoV-2 infection, we observed that Cxcl10 mRNA expression was significantly upregulated in the lungs of SARS-CoV-2 infected mice as has previously been reported [58]. In patients having rapid early viral replication, this is followed by inflammatory responses that contribute to pathology [59]. Post-mortem analysis of human COVID-19 patients showed immune cell accumulation in the lungs [60]. In our study, ATN-161 treatment, either pooled or ATN-161 responders, significantly lowered SARS-CoV-2-induced Cxcl10 levels, a particularly robust result suggesting that ATN-161 has anti-inflammatory properties in the context of SARS-CoV-2 infection. This result is in agreement with our previous studies in experimental ischemic stroke where post-stroke treatment with ATN-161 reduced neuro-inflammation [31].
Morphological changes observed at 3 dpi in infected lungs of K18-hACE2 mice included multifocal lesions, moderate interstitial pneumonia, infiltration of lymphocytes, and fibrin exudation. These were seen in SARS-CoV-2 + vehicle and SARS-CoV-2 + ATN-161 non responders mice but were less or completely absent in SARS-CoV-2 + ATN-161 responder mice as well as absent in uninfected saline/ATN-161 treated mice. These findings are consistent with previous reports from post-mortem examination of patients with COVID-19 and SARS-CoV-2-infected K18-hACE2 mice in other reports [58], [43], [61], [62], [63].
Pulmonary fibrosis, a particularly negative consequence of COVID-19-induced acute respiratory distress syndrome [64], was not prominent in infiltrated areas at 3 dpi in infected lungs of K18-hACE2 mice. However, we observed moderate multiple intra-arteriolar microthrombi, intra-alveolar microthrombi, and large interstitial hemorrhagic areas in SARS-CoV-2 + vehicle and SARS-CoV-2 + ATN-161 non responder mice whereas these observations were less or completely absent in SARS-CoV-2 + ATN-161 responder mice. Severely infected COVID-19 patient autopsy samples showed that microthrombi or immunothrombi is associated with a hyperinflammatory response [64], [65]. Our findings are further supported by recent results in the human immune system humanized mouse model on histological changes in the lungs with SARS-CoV-2 infection [66], [36], [43]. Indeed, we did not observe collagen positive fibrosis with SARS-CoV-2 infection in this K18 mouse model. This possibly may be due to the acute nature of our SARS-CoV-2 infection study and we predict that chronic inflammation would induce fibrosis [67].
Our results suggest a critical role of integrins as an additional receptor to SARS-CoV-2 spike protein cell entry [68], [69]. We observed induced expression of integrin α5 and integrin αv in the lungs of SARS-CoV-2 infected K18-hACE2 mice whereas lung expression of hACE2 levels did not vary in SARS-CoV-2 + Saline or ATN-161 treated mice, suggesting that SARS-CoV-2 infection and/or pathogenesis involves these, and perhaps other integrins, that activate downstream signaling to induce lung pathology [34,70,71]. Indeed, a recent study showed that increased integrin α5β1 and αvβ3 levels in cardiac myocytes, obtained from heart failure patients, correlates with ACE2 expression [73]. This suggests that the concomitant elevation of these integrins and the upregulation of ACE2 in an organ may render it more susceptible to SARS-CoV-2 infection. Hence, as observed in the present study, decreasing the expressions of α5β1 and αVβ3 in the presence of ACE2 may dampen the effect of SARS-CoV-2 infection on lung tissue morphology. Other studies have clearly linked integrin α5β1, and its inhibition, with other viral infections [71], [74]. ATN-161 treatment inhibited porcine hemagglutinating encephalomyelitis virus (PHEV) infection and its increase in the expression of integrin α5β1 in vivo in a mouse model [74]. Similarly, we observed that ATN-161 treatment inhibits SARS-CoV-2-induced integrin α5, and integrin αv in the K18-hACE2 mice lungs among ATN-responders. Importantly, our studies were performed at the dose of ATN-161, 1 mg/kg, that demonstrated efficacy in in vitro and in vivo preclinical studies in blocking angiogenesis, solid tumor growth, and ischemic stroke injury [75], [31], [32]. This dose was also similar to the 0.8 mg/kg dose used in the PHEV anti-viral in vivo studies [74]. However, as we and others have demonstrated U-shaped dose response effects with ATN-161 [31], [27] additional in vivo SARS-CoV-2 dosing studies are necessary and currently underway. However, ATN-161 present several potential advantages as a novel COVID-19 therapy. Unlike other RGD-based peptides, it preferentially binds to and inhibits activated forms of α5β1 in areas of inflammation, hypoxia and angiogenesis [31]. ATN-161 also binds to integrin αvβ3 [31], an additional integrin that has been implicated in SARS-CoV-2 pathogenesis [34], [33], and is safe, well-tolerated in human clinical trials (cancer) with no dose limiting toxicity [29], [76], and can be administered i.v., i.p., and intranasally [32], [77]; The latter may support a more readily accessible means of COVID-19 treatment as well as afford a prophylactic approach which is currently under investigation in our laboratory.
Serum biochemistry indicated a slightly higher level of AST and ALT in SARS-CoV-2-infected mice compared with the saline-treated group that may indicate that there is hepatic damage associated with SARS-CoV-2 infection acutely in this model. This requires future further evaluation, although these results are in agreement with previous studies that showed an increase in both AST and ALT which reflect hepatic function [78], [79], [80], Importantly, ATN-161 treatment did not show an effect on hepatic function in SARS-CoV-2 infected mice. However, there is no difference in AST and ALT levels between non infected saline and ATN-161 treated groups, indicating that ATN-161 treatment by itself did not cause hepatic damage, as expected [81], [76], [80].
5. Conclusion
To the best of our knowledge, this study is the first to demonstrate that integrin blockade can reduce SARS-CoV-2 infection in vivo. Specifically, we demonstrated that post-infection treatment with ATN-161 reduces lung viral load, replication, improves lung histology and reduces lung α5 and αv integrin expression in a majority of SARS-CoV-2 infected k-18 mice. Our results further support the hypothesis that integrins play an important role in SARS-CoV-2 infection as well as support the further investigation of ATN-161, and potentially other anti-integrin agents, as novel therapies for COVID-19.
5.1. Future perspective
Although several studies have predicted the potential for SARS-CoV-2 to bind integrins and thereby infect host cells with or without associated ACE2 interaction, for the first time we present evidence here that inhibition of integrin α5β1 (and αvβ3) in vivo can reduce both SARS-CoV-2 viral load and pathological complications in lung tissue of SARS-CoV-2 in k18-hACE2 mice. Despite the limitations of our study (small sample size, use of heterozygous k18-hACE2 mice), the promise for ATN-161 as a potential therapeutic and prophylaxis after SARS-CoV-2 exposure remains attractive. To date, several small-molecule drug and peptide treatments have been developed for intranasal delivery which reach systemic circulation rapidly, many of which have been formulated to treat diseases which have the ability to alter cognition, such as depression [82]. As the olfactory and trigeminal nerves offer a safe and effective delivery pathway to deliver therapeutic agents to the brain, an intranasal-based therapy to both prevent and treat neurological complications of post-acute COVID-19 syndrome may one day include a formulation of ATN-161 [83], [84].
CRediT authorship contribution statement
Amruta Narayanappa: Experimentation, Analysis, Writing - original draft. Elizabeth B. Engler-Chiurazzi: Analysis, Writing, Editing manuscript. Isabel C. Murray-Brown, Timothy E. Gressett, Ifechukwude J. Biose, Wesley H. Chastain, Jaime B Befeler: Performed literature searches, Histological analysis. Gregory Bix: Conceptualization, Visualization, review, supervision, and funding.
Funding
Tulane University startup funds. to G.J.B and NIH/NIMH K01 MH117343 to E.B.E-C.
Declaration of competing interest
The authors have declared that no conflict of interest exists.
Acknowledgements
We would like to thank Drs. Jay K. Kolls, Xuebin Qin (Tulane University) for their thoughtful comments and advice on this manuscript. Angela Birnbaum, Kristin E. Chandler, Thompson, Carli C, Bavaret Tammy for BSL3 service (Tulane University). Authors are grateful to Naoki Iwanaga, Hemanth Kumar Kandikattu, Smither, Allison R, Brandon J. Beddingfield, Nicholas J. Maness for technical help in experiments. Authors are thankful to Cecily C. Midkiff, Confocal Microscopy and Molecular Pathology Core, Dina Guapp, Histology Core, Delucca, Beatris, Tulane University. The authors appreciate Rebecca Solch, Meshi Paz, Hawkins, Scott V, Department of Neurosurgery at Tulane University, for assistance.
References
- 1.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvacho I., Piesche M. RGD-binding integrins and TGF-beta in SARS-CoV-2 infections - novel targets to treat COVID-19 patients? Clin. Transl. Immunol. 2021;10(3) doi: 10.1002/cti2.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Control CfD . 2020. Prevention. Interim Clinical Guidance for Management of Patients With Confirmed Coronavirus Disease (COVID-19) [Google Scholar]
- 4.Perra N. Non-pharmaceutical interventions during the COVID-19 pandemic: a review. Phys. Rep. 2021 doi: 10.1016/j.physrep.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Group CC-W. Liu Y., Morgenstern C., Kelly J., Lowe R., Jit M. The impact of non-pharmaceutical interventions on SARS-CoV-2 transmission across 130 countries and territories. BMC Med. 2021;19(1):40. doi: 10.1186/s12916-020-01872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spinelli M.A., Glidden D.V., Gennatas E.D., Bielecki M., Beyrer C., Rutherford G., et al. Importance of non-pharmaceutical interventions in lowering the viral inoculum to reduce susceptibility to infection by SARS-CoV-2 and potentially disease severity. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(20)30982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Commentary on Ferguson. Eubank S., Eckstrand I., Lewis B., Venkatramanan S., Marathe M., Barrett C.L. Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. Bull. Math. Biol. 2020;82(4):52. doi: 10.1007/s11538-020-00726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 2020;177 doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan S., Sun H., Bu X., Wan G. New strategy for COVID-19: an evolutionary role for RGD motif in SARS-CoV-2 and potential inhibitors for virus infection. Front. Pharmacol. 2020;11:912. doi: 10.3389/fphar.2020.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tresoldi I., Sangiuolo C.F., Manzari V., Modesti A. SARS-COV-2 and infectivity: possible increase in infectivity associated to integrin motif expression. J. Med. Virol. 2020;92(10):1741–1742. doi: 10.1002/jmv.25831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguirre C., Meca-Lallana V., Barrios-Blandino A., Del Rio B., Vivancos J. Covid-19 in a patient with multiple sclerosis treated with natalizumab: may the blockade of integrins have a protective role? Mult. Scler. Relat. Disord. 2020;44 doi: 10.1016/j.msard.2020.102250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu X., Lu D., Scully M., Kakkar V. The role of integrins in cancer and the development of anti-integrin therapeutic agents for cancer therapy. Perspect. Med. Chem. 2008;2:57–73. [PMC free article] [PubMed] [Google Scholar]
- 16.Amruta N., Rahman A.A., Pinteaux E., Bix G. Neuroinflammation and fibrosis in stroke: the good, the bad and the ugly. J. Neuroimmunol. 2020;346 doi: 10.1016/j.jneuroim.2020.577318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman A.A., Amruta N., Pinteaux E., Bix G.J. Neurogenesis after stroke: a therapeutic perspective. Transl. Stroke Res. 2021;12(1):1–14. doi: 10.1007/s12975-020-00841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandikattu H.K., Venkateshaiah S.U., Kumar S., Mishra A. IL-15 immunotherapy is a viable strategy for COVID-19. Cytokine Growth Factor Rev. 2020;54:24–31. doi: 10.1016/j.cytogfr.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishibashi Y., Relman D.A., Nishikawa A. Invasion of human respiratory epithelial cells by bordetella pertussis: possible role for a filamentous hemagglutinin arg-gly-asp sequence and alpha5beta1 integrin. Microb. Pathog. 2001;30(5):279–288. doi: 10.1006/mpat.2001.0432. [DOI] [PubMed] [Google Scholar]
- 20.Feire A.L., Koss H., Compton T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. U. S. A. 2004;101(43):15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao J., Palefsky J.M., Herrera R., Berline J., Tugizov S.M. The epstein-barr virus BMRF-2 protein facilitates virus attachment to oral epithelial cells. Virology. 2008;370(2):430–442. doi: 10.1016/j.virol.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarate S., Romero P., Espinosa R., Arias C.F., Lopez S. VP7 mediates the interaction of rotaviruses with integrin alphavbeta3 through a novel integrin-binding site. J. Virol. 2004;78(20):10839–10847. doi: 10.1128/JVI.78.20.10839-10847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akula S.M., Pramod N.P., Wang F.Z., Chandran B. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell. 2002;108(3):407–419. doi: 10.1016/s0092-8674(02)00628-1. [DOI] [PubMed] [Google Scholar]
- 24.Gire S.K., Goba A., Andersen K.G., Sealfon R.S., Park D.J., Kanneh L., et al. Genomic surveillance elucidates ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345(6202):1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke N.E., Fisher M.J., Porter K.E., Lambert D.W., Turner A.J. Angiotensin converting enzyme (ACE) and ACE2 bind integrins and ACE2 regulates integrin signalling. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0034747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B., Tian E.K., He B., Tian L., Han R., Wang S., et al. Overview of lethal human coronaviruses. Signal Transduct. Target. Ther. 2020;5(1):89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beddingfield B.J., Iwanaga N., Chapagain P.P., Zheng W., Roy C.J., Hu T.Y., et al. The integrin binding peptide, ATN-161, as a novel therapy for SARS-CoV-2 infection. Basic Translat. Sci. 2021;6(1):1–8. doi: 10.1016/j.jacbts.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park E.J., Myint P.K., Appiah M.G., Darkwah S., Caidengbate S., Ito A., et al. The spike glycoprotein of SARS-CoV-2 binds to beta1 integrins expressed on the surface of lung epithelial cells. Viruses. 2021;13(4) doi: 10.3390/v13040645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cianfrocca M., Kimmel K., Gallo J., Cardoso T., Brown M., Hudes G., et al. Phase 1 trial of the antiangiogenic peptide ATN-161 (Ac-PHSCN-NH 2), a beta integrin antagonist, in patients with solid tumours. Br. J. Cancer. 2006;94(11):1621–1626. doi: 10.1038/sj.bjc.6603171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv X., Li Z., Guan J., Zhang J., Xu B., He W., et al. ATN-161 reduces virus proliferation in PHEV-infected mice by inhibiting the integrin alpha5beta1-FAK signaling pathway. Vet. Microbiol. 2019;233:147–153. doi: 10.1016/j.vetmic.2019.04.029. [DOI] [PubMed] [Google Scholar]
- 31.Doñate F., Parry G.C., Shaked Y., Hensley H., Guan X., Beck I., et al. Pharmacology of the novel antiangiogenic peptide ATN-161 (Ac-PHSCN-NH2): observation of a U-shaped dose-response curve in several preclinical models of angiogenesis and tumor growth. Clin. Cancer Res. 2008;14(7):2137–2144. doi: 10.1158/1078-0432.CCR-07-4530. [DOI] [PubMed] [Google Scholar]
- 32.Edwards D.N., Salmeron K., Lukins D.E., Trout A.L., Fraser J.F., Bix G.J. Integrin alpha5beta1 inhibition by ATN-161 reduces neuroinflammation and is neuroprotective in ischemic stroke. J. Cereb. Blood Flow Metab. 2020;40(8):1695–1708. doi: 10.1177/0271678X19880161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kliche J., Kuss H., Ali M., Ivarsson Y. Cytoplasmic short linear motifs in ACE2 and integrin beta3 link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci. Signal. 2021;14(665) doi: 10.1126/scisignal.abf1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meszaros B., Samano-Sanchez H., Alvarado-Valverde J., Calyseva J., Martinez-Perez E., Alves R., et al. Short linear motif candidates in the cell entry system used by SARS-CoV-2 and their potential therapeutic implications. Sci. Signal. 2021;14(665) doi: 10.1126/scisignal.abd0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han K., Blair R.V., Iwanaga N., Liu F., Russell-Lodrigue K.E., Qin Z., et al. Lung expression of human angiotensin-converting enzyme 2 sensitizes the mouse to SARS-CoV-2 infection. Am. J. Respir. Cell Mol. Biol. 2021;64(1):79–88. doi: 10.1165/rcmb.2020-0354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 37.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwanaga N., Cooper L., Rong L., Beddingfield B., Crabtree J., Tripp R.A. 2020. Novel ACE2-IgG1 Fusions With Improved In Vitro and In Vivo Activity Against SARS-CoV2. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J., Wong L.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589(7843):603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kandikattu H.K., Manohar M., Venkateshaiah S.U., Yadavalli C., Mishra A. Chronic inflammation promotes epithelial-mesenchymal transition-mediated malignant phenotypes and lung injury in experimentally-induced pancreatitis. Life Sci. 2021;119640 doi: 10.1016/j.lfs.2021.119640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kandikattu H.K., Manohar M., Verma A.K., Kumar S., Yadavalli C.S., Venkateshaiah S.U., et al. Macrophages-induced IL-18–mediated eosinophilia promotes characteristics of pancreatic malignancy. Life Sci. Alliance. 2021;4(8) doi: 10.26508/lsa.202000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kandikattu H.K., Venkateshaiah S.U., Verma A.K., Mishra A. Tacrolimus (FK506) treatment protects allergen-, IL-5-and IL-13-induced mucosal eosinophilia. Immunology. 2021;163(2):220–235. doi: 10.1111/imm.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun S.H., Chen Q., Gu H.J., Yang G., Wang Y.X., Huang X.Y., et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28(1):124–133. doi: 10.1016/j.chom.2020.05.020. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen Y.T., Liao F., Hsiao C.H., Kao C.L., Chen Y.C., Wu-Hsieh B.A. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J. Virol. 2006;80(6):2684–2693. doi: 10.1128/JVI.80.6.2684-2693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J.M., Zhou G.D., Sun Y.L., Wang S.S., Yang J.F., Meng E.H., et al. Clinical pathology and pathogenesis of severe acute respiratory syndrome. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2003;17(3):217–221. [PubMed] [Google Scholar]
- 46.Mentzer S.J., Konerding M.A. Intussusceptive angiogenesis: expansion and remodeling of microvascular networks. Angiogenesis. 2014;17(3):499–509. doi: 10.1007/s10456-014-9428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.at:https://www.who.int/emergencies/diseases/novel-coronavirus-2019 ACdC-pWHOA.
- 48.https://coronavirus.jhu.edu/map.html.
- 49.Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. 2021. SARS-CoV-2 B.1.1.7 and B.1.351 Spike Variants Bind Human ACE2 With Increased Affinity. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart P.L., Nemerow G.R. Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 2007;15(11):500–507. doi: 10.1016/j.tim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Montagutelli X., Prot M., Levillayer L., Salazar E.B., Jouvion G., Conquet L. 2021. The B1. 351 and P. 1 Variants Extend SARS-CoV-2 Host Range to Mice. bioRxiv. [Google Scholar]
- 53.Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight. 2020;5(19) doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182(1):50–58. doi: 10.1016/j.cell.2020.05.027. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang C., Huang L., Wang Y., Li X., Ren L., Gu X., et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397(10270):220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yinda C.K., Port J.R., Bushmaker T., Owusu I.O., Avanzato V.A., Fischer R.J. 2020. K18-hACE2 Mice Develop Respiratory Disease Resembling Severe COVID-19. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21(11):1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295(3):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian S., Hu W., Niu L., Liu H., Xu H., Xiao S.Y. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 2020;15(5):700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaller T., Hirschbuhl K., Burkhardt K., Braun G., Trepel M., Markl B., et al. Postmortem examination of patients with COVID-19. JAMA. 2020;323(24):2518–2520. doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox S.E., Akmatbekov A., Harbert J.L., Li G., Quincy Brown J., Vander Heide R.S. Pulmonary and cardiac pathology in African American patients with COVID-19: an autopsy series from New Orleans. Lancet Respir. Med. 2020;8(7):681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brumeanu T.D., Vir P., Shashikumar S., Karim A.F., Kar S., Chung K.K. 2020. A Human-Immune-System Mouse Model for COVID-19 Research (DRAGA Mouse: HLA-A2.HLA-DR4.Rag1KO.IL-2Rgammac KO.NOD) bioRxiv. [DOI] [Google Scholar]
- 67.Leng L., Cao R., Ma J., Mou D., Zhu Y., Li W., et al. Pathological features of COVID-19-associated lung injury: a preliminary proteomics report based on clinical samples. Signal Transduct. Target. Ther. 2020;5(1):240. doi: 10.1038/s41392-020-00355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kliche J., Kuss H., Ali M., Ivarsson Y. Cytoplasmic short linear motifs in ACE2 and integrin ß3 link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci. Signal. 2021;14(665) doi: 10.1126/scisignal.abf1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makowski L., Olson-Sidford W., W-Weisel J. Biological and clinical consequences of integrin binding via a rogue RGD motif in the SARS CoV-2 spike protein. Viruses. 2021;13(2):146. doi: 10.3390/v13020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calver J., Joseph C., John A., Organ L., Fainberg H., Porte J. BMJ Publishing Group Ltd; 2021. S31 The Novel Coronavirus SARS-CoV-2 Binds RGD Integrins and Upregulates avb3 Integrins in Covid-19 Infected Lungs. [Google Scholar]
- 71.Hussein H.A., Walker L.R., Abdel-Raouf U.M., Desouky S.A., Montasser A.K.M., Akula S.M. Beyond RGD: virus interactions with integrins. Arch. Virol. 2015;160(11):2669–2681. doi: 10.1007/s00705-015-2579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bristow M.R., Zisman L.S., Altman N.L., Gilbert E.M., Lowes B.D., Minobe W.A., et al. Dynamic regulation of SARS-cov-2 binding and cell entry mechanisms in remodeled human ventricular myocardium. JACC Basic Transl. Sci. 2020;5(9):871–883. doi: 10.1016/j.jacbts.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lv X., Li Z., Guan J., Hu S., Zhang J., Lan Y., et al. Porcine hemagglutinating encephalomyelitis virus activation of the integrin alpha5beta1-FAK-cofilin pathway causes cytoskeletal rearrangement to promote its invasion of N2a cells. J. Virol. 2019;93(5) doi: 10.1128/JVI.01736-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khalili P., Arakelian A., Chen G., Plunkett M.L., Beck I., Parry G.C., et al. A non–RGD-based integrin binding peptide (ATN-161) blocks breast cancer growth and metastasis in vivo. Mol. Cancer Ther. 2006;5(9):2271–2280. doi: 10.1158/1535-7163.MCT-06-0100. [DOI] [PubMed] [Google Scholar]
- 76.Cianfrocca M.E., Kimmel K.A., Gallo J., Cardoso T., Brown M.M., Hudes G., et al. Phase 1 trial of the antiangiogenic peptide ATN-161 (Ac-PHSCN-NH(2)), a beta integrin antagonist, in patients with solid tumours. Br. J. Cancer. 2006;94(11):1621–1626. doi: 10.1038/sj.bjc.6603171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sundaram A., Chen C., Khalifeh-Soltani A., Atakilit A., Ren X., Qiu W., et al. Targeting integrin alpha5beta1 ameliorates severe airway hyperresponsiveness in experimental asthma. J. Clin. Invest. 2017;127(1):365–374. doi: 10.1172/JCI88555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carvalho J.R., Machado M.V. New insights about albumin and liver disease. Ann. Hepatol. 2018;17(4):547–560. doi: 10.5604/01.3001.0012.0916. [DOI] [PubMed] [Google Scholar]
- 79.Wisniewska H., Skonieczna-Zydecka K., Parczewski M., Niscigorska-Olsen J., Karpinska E., Hornung M., et al. Hepatotropic properties of SARS-CoV-2-preliminary results of cross-sectional observational study from the first wave COVID-19 pandemic. J. Clin. Med. 2021;10(4) doi: 10.3390/jcm10040672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arévalo A., Pagotto R., Pórfido J., Daghero H., Segovia M., Yamasaki K., et al. Ivermectin reduces in vivo coronavirus infection in a mouse experimental model. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-86679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Donate F., Parry G.C., Shaked Y., Hensley H., Guan X., Beck I., et al. Pharmacology of the novel antiangiogenic peptide ATN-161 (Ac-PHSCN-NH2): observation of a U-shaped dose-response curve in several preclinical models of angiogenesis and tumor growth. Clin. Cancer Res. 2008;14(7):2137–2144. doi: 10.1158/1078-0432.CCR-07-4530. [DOI] [PubMed] [Google Scholar]
- 82.Xu J., Tao J., Wang J. Design and application in delivery system of intranasal antidepressants. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.626882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fortuna A., Alves G., Serralheiro A., Sousa J., Falcao A. Intranasal delivery of systemic-acting drugs: small-molecules and biomacromolecules. Eur. J. Pharm. Biopharm. 2014;88(1):8–27. doi: 10.1016/j.ejpb.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 84.Homayun B., Lin X., Choi H.J. Challenges and recent Progress in Oral drug delivery Systems for Biopharmaceuticals. Pharmaceutics. 2019;11(3) doi: 10.3390/pharmaceutics11030129. [DOI] [PMC free article] [PubMed] [Google Scholar]