ABSTRACT
Transport of mRNAs is an important step of gene expression, which brings the genetic message from the DNA in the nucleus to a precise cytoplasmic location in a regulated fashion. Perturbation of this process can lead to pathologies such as developmental and neurological disorders. In this review, we discuss recent advances in the field of mRNA transport made using single molecule fluorescent imaging approaches. We present an overview of these approaches in fixed and live cells and their input in understanding the key steps of mRNA journey: transport across the nucleoplasm, export through the nuclear pores and delivery to its final cytoplasmic location. This review puts a particular emphasis on the coupling of mRNA transport with translation, such as localization-dependent translational regulation and translation-dependent mRNA localization. We also highlight the recently discovered translation factories, and how cellular and viral RNAs can hijack membrane transport systems to travel in the cytoplasm.
KEYWORDS: RNA transport, single molecule imaging, translation, motors, membrane transport, nuclear pores
Introduction
Messenger RNAs are synthesized at specific genomic loci in the nucleoplasm and travel through the nucleus and nuclear pores to produce proteins in the cytoplasm at the right time and place. Some mRNAs require delivery to specific cytoplasmic locations for local translation, and this plays an important role in diverse biological processes such as spatial patterning during embryogenesis, cell fate determination, asymmetric cell division, cell polarity and motility, signalling and neuronal synaptic functions [1]. Perturbations in mRNA localization and its transport can lead to developmental and neurological disorders, as well as cancer [2,3].
Study of RNA transport is a fast-developing field that combines different experimental approaches, among which single molecule imaging techniques hold an important place. They enlighten sub-cellular mRNA distribution and cell-to-cell variations of mRNA expression, which are not easily accessible by ensemble biochemical methods. In this review, we will discuss technological progress in single molecule imaging of mRNA and highlight their contributions to our understanding of mRNA transport from transcription to translation. We will in particular describe how RNAs move through the nucleoplasm, cross the nuclear pores and how they are transported to specific cytoplasmic location to locally produce proteins. This review will also highlight surprising recent results arising from imaging studies that indicate how mRNPs can use membrane transport system to reach their destination, and how nascent proteins can contribute to polysome transport and localization. Several recent reviews cover other aspects of single molecule imaging of RNA metabolism, including transcription, splicing, translation and decay [4–7].
Methods to image single RNAs in fixed cells: single molecule fluorescent in situ hybridization
Single molecule fluorescent in situ hybridization (smFISH) is a widely used RNA imaging technique, which detects RNA in fixed cells with single molecule sensitivity and high efficiency (commonly higher than 80%) [8]. This detection efficiency is achieved through the use of multiple fluorescently labelled oligonucleotide probes directed against the transcript of interest, and the efficiency increases with the probe number. The first smFISH experiments were performed with a set of ten 50-nucleotide long DNA probes labelled with five fluorophores each [8] (Fig. 1A). A variation of this approach, which uses 48 or more short 20-mer oligonucleotides labelled with one fluorophore each, was developed later on [9]. In either method, the single RNA molecules are detected as diffraction-limited spots by fluorescent microscopy. The single molecule sensitivity was demonstrated by measuring the intensity of fluorescence emitted by one probe and by quantifying the amount of probes in the diffraction limited RNA spots, which were shown to correspond to single mRNA molecules [8]. Consequently, these spots can be simply counted to reveal differences in RNA levels between individual cells.
Figure 1.

Single molecule methods of mRNA detection
A – Single molecule FISH (smFISH). 10 probes with 5 fluorophores each are used for mRNA detection with single molecule sensitivity [8]. B – Single molecule inexpensive FISH (smiFISH). The mRNA is detected using non-labelled primary probes, which contain a sequence complementary to target RNA and a read-out sequence identical for all the probes. This read-out sequence hybridizes to a unique secondary probe labelled with 2 fluorophores on its 3ʹ and 5ʹ [13]. C – Sequential smFISH, example of Multiplexed Error Robust FISH (merFISH). RNA targets are first hybridized with encoding probes, containing sequence complementary to target and two read-out sequences. Each RNA target is identified by a combination of uniques read-out sequences in subsequent rounds of hybridization with secondary probes, one secondary probe per round of hybridization. After each round of hybridization with a secondary probe, the signal is detected, registered and the probe is removed. Each RNA is identified with a unique barcode (lower panel, left column), in which a hybridization round functions as a bit. If the RNA gives a signal with a given probe it is assigned 1, in case of no signal it is assigned 0. To increase the sensitivity, sets of 24–100 primary probes are used per mRNA (top panel, left) [16].
To quantify the mRNA numbers in single cells on smFISH images, dedicated analysis softwares are available [9–12]. One of the widely used applications, FISH-quant, enables not only detection and counting of isolated RNA molecules but also quantifies the number of molecules in structures that contain RNA aggregates, for example, transcription sites, nuclear speckles, P-bodies or other foci in the cytoplasm or in the nucleus. FISH-quant performs quantification in three dimensions, taking into account the spatial extension of RNA-containing structures [12]. The latest development of software includes tools for automated cell segmentation [13], but it can also be combined with recent generalist machine learning algorithms, which accurately segment cells and nuclei [14]. FISH-quant provides a fully automated pipeline making it suitable for analysis of high-throughput smFISH experiments and it can be combined with supervised, machine learning, and unsupervised classification methods to examine spatial distribution of RNA in single cells [15].
Several variants of smFISH allowing for multiplexing and cost decrease were recently developed [13,16–19]. They all rely on the use of unlabelled primary probes followed by detection of these probes with a secondary fluorescent oligo, similarly to an indirect immunofluorescence, except that here the interactions depend on base pairing. Using this strategy, a particularly simple and inexpensive smFISH protocol, dubbed smiFISH, was developed [13]. Here, fixed cells are hybridized with a set of multiple non-labelled primary probes (usually 24), which all contain a sequence complementary to a fluorescently labelled secondary probe (Fig. 1B). The same secondary probe is used with different gene-specific primary probes, thus strongly decreasing cost. This method is quite robust, easy to set up, and comes with probe design software [13]. High-throughput versions of such indirect smFISH methods have been achieved by either parallelization [20,21], or multiplexing [16,17]. In the latter case, the use of different fluorophores allows a first level of multiplexing. Such multicolour smFISH can be used to detect several RNA species in the same cell or different parts of the same RNA molecule, for instance, to study RNA conformation in vivo [22–24]. While the number of fluorophores is limited, combinatorial spectral approaches allow to detect up to 32 RNA species at the same time, with each RNA carrying a unique colour code [25].
To further increase multiplexing capabilities, an important breakthrough was the introduction of sequential smFISH methods, MERFISH and seqFISH [16–19]. The most recent development of this approach consists of sequential rounds of hybridization using different secondary probes to reveal distinct populations of primary probes. Between hybridization rounds, the secondary probes of the previous round are removed by photobleaching, chemical treatment or denaturation. After each round, the fluorescent signal is recorded and the signals from different rounds are superposed, allowing to attribute to each RNA molecule a unique code called a barcode [16,17] (Fig. 1C). In barcodes, each round of a secondary probe hybridization functions as a bit, 1 signifies a presence of a signal and 0 – its absence. In this manner, with n sequentially hybridized secondary probes, 2 n-1 RNA species can in principle be coded and distinguished. In practice, some barcodes are not used to make each barcode different from all the others by at least two bits, thus decreasing the number of false identifications arising from decoding errors. Remarkably, MERFISH and seqFISH experiments can be scaled up to near transcriptome level (10,000 RNAs), enabling high-throughput analysis of cell-to-cell variation in gene expression and a determination of the spatial organization of the transcriptome [16,17].
Quantitative smFISH analyses revealed that the numbers of mRNA copies per cell varies stochastically between cells in isogenic populations. It was further shown that these variations can have intrinsic or extrinsic causes and are important for lineage specification during development and for a variety of cellular functions [26–29]. High-throughput smFISH experiments allowed to quantify thousands of mRNAs in the same cell. They showed that the average numbers of mRNA molecules per cell are comprised between 0,1 and 1000 for the majority of mRNAs, but can be lower than 0,1 molecule per cell for few low expressing genes, and go up to more than one thousand molecules for highly expressed genes, such as GAPDH, CYTB, or β-actin after serum induction [8,13,17,19,20]. These numbers are in remarkable agreement with bulk RNA-sequencing data, but smFISH has higher detection efficiency than single cell RNA sequencing [19], making it more suitable for low expressed genes and it outperforms for studies of cell-to-cell variations in mRNA levels, since it can examine thousands of cells in one experiment [17,19]. This enables to elucidate gene-regulatory networks by the analysis of covariations in the expression level of different genes between the cells [16,17,19].
FISH experiments led to a discovery that RNA can adopt a specific localization inside the cell. A pioneering study analysing by FISH more than 2000 transcripts in early Drosophila embryo, demonstrated that 71% of mRNAs exhibited distinct localization patterns, including restriction to specific regions in the embryo, association with subcellular structures, or a particular localization during cell division (Fig. 2A) [30]. High-throughput smFISH examined a distribution of multiple RNAs in mammalian cell lines and tissues. MERFISH experiments on human cell lines showed that among 9050 mRNAs, 16% were enriched in the nucleus and 11% in the endoplasmic reticulum [19]. Three types of mRNA localization were observed in mouse fibroblasts by seqFISH+ analysis of 10,000 genes: nuclear/perinuclear pattern, cytoplasmic and protrusion enriched [17], while Battich and colleagues characterized mitochondria-associated mRNAs in a high-throughput smFISH study [20]. Recently, a dual protein/mRNA screen using smFISH and the corresponding mRNAs encoded proteins localization analysis revealed multiple patterns of mRNA localization, including cell protrusions, cytoplasmic foci, endosomes, Golgi, nuclear envelope, mitotic centrosomes (Fig. 2B) and showed that mRNA localization was often driven by nascent translation [31]. We will discuss in more detail the intimate links between RNA localization, its transport and translation in the following sections.
Figure 2.

Different patterns of mRNA localization revealed by FISH and smFISH
A – FISH on Drosophila embryo (mRNA green/nuclei red). The gene names are indicated on the bottom left and localization patterns above the photos. From left to right: bicoid mRNA displays anterior localization in the embryo; anillin mRNA has perinuclear localization; CG1962 mRNA localizes to microtubules networks and centrosomes during cell division. Reprinted from Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007; 131:174–87. Pages 178, 180, 181. Copyright 2020, with permission from Elsevier. B – smFISH on Hela cells. The genes names are indicated on the bottom left and localization patterns are above the photos. Top – from left to right: KIF5B is localized to the cell protrusions; FLNA is localized to the cell edge; BUB1 – cytoplasmic foci. Orange arrows indicate mentioned locations. Bottom – ASPM mRNA is localized to nuclear envelope and cytoplasmic foci in interphase and to spindle pole during cell division. Left – smFISH, right – nuclei stained with DAPI. Orange arrow indicates interphase cell; pink arrow – mitotic cell. Reprinted from Chouaib R, Safieddine A, Pichon X, Imbert A, Kwon OS, Samacoits A, Traboulsi A-M, Robert M-C, Tsanov N, Coleno E, et al. A Dual Protein-mRNA Localization Screen Reveals Compartmentalized Translation and Widespread Co-translational RNA Targeting. Dev Cell 2020; 54:773–791.e5. Pages 775, 776. Copyright 2020, with permission from Elsevier.
Application of high-throughput smFISH to tissues gives an opportunity to explore qualitative and quantitative aspects of transcriptome in a more physiological context. Indeed, seqFISH+ on mouse brain slices allowed to image simultaneously 10,000 RNAs and to create high-resolution maps of different brain regions shedding light on its organization and cellular interactions [17]. Further improvement of MERFISH, such as coupling with a signal amplification step, enables a dramatic increase of the sensitivity for detection of short and low-abundant transcripts in tissues with high autofluorescence [32].
Methods to image single RNAs in live cells
SmFISH provides information about localization and absolute counts of mRNA molecules in single cells, but it lacks dynamics and temporal resolution. These limits can be overcome by live-cell microscopy approaches. To follow RNA in vivo, fluorescently labelled RNA can be microinjected in the cells [33]. While this is a method of choice for studying small RNAs [34–36], it is well known that transcription-coupled events are important to control the mRNA fate [37–40], and therefore the use of the genetically encoded tagged reporters or fluorescent probes are preferable ways for mRNA imaging. Several such methods currently exist.
The first approach is based on the use of fluorogenic oligonucleotide probes called molecular beacons (MB). MBs form stem-loops that contain a fluorophore and a quencher in a close proximity, leading to reduced fluorescence from unbound beacons, thus minimizing background. Binding of MBs to their RNA target disrupts interaction of the fluorophore with the quencher and increases fluorescence emission (Fig. 3A). The use of these probes was hindered by their relatively low sensitivity, difficulties of their delivery into cells, as well as their degradation. Recent developments allowed to obtain more stable probes, using chemically modified backbones, and opened possibility for RNA detection with single molecule sensitivity in live cells by inserting arrays of MBs binding sequences in the RNA of interest [41].
Figure 3.
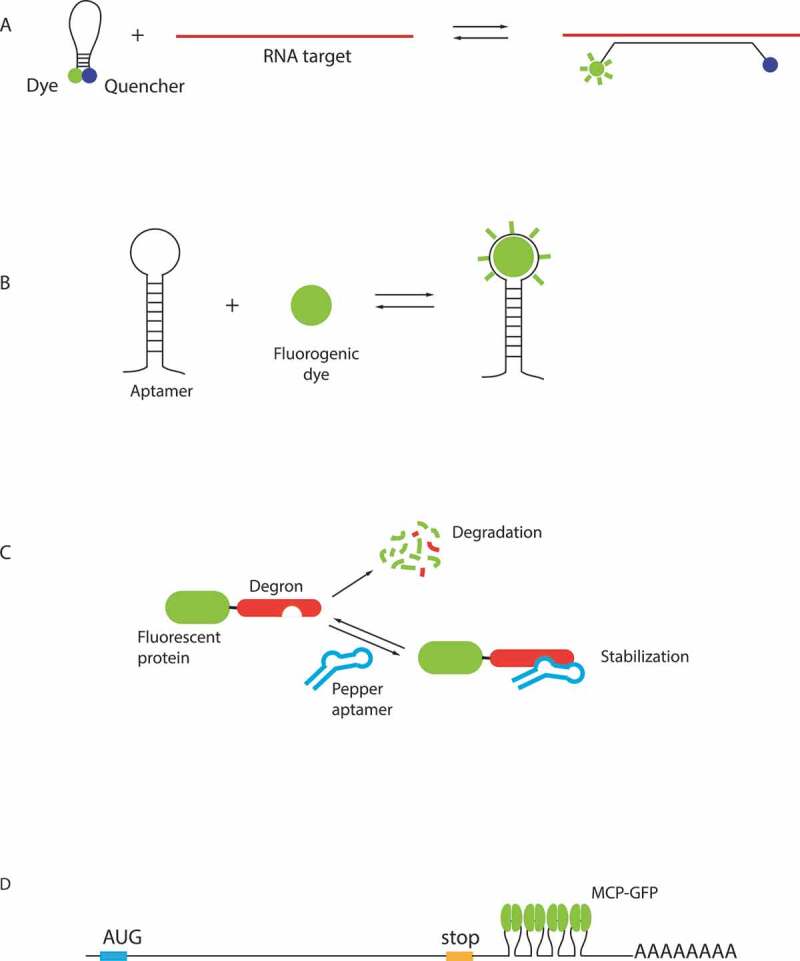
RNA detection in living cells
A – Molecular beacons. Unbound MBs are not fluorescent due to a proximity of a quencher (blue circle) to a fluorophore (red circle). Upon binding to the target RNA, these interactions are disrupted leading to a fluorescence emission. B – Principle of fluorogenic RNA aptamers. RNA aptamers are based on the interaction of a specific RNA structure with a fluorogenic dye (green circle), which becomes fluorescent upon aptamer binding. C – The regulatory RNA aptamer Pepper. Pepper RNA binds a protein destabilization domain fused to a fluorescent protein, leading to its stabilization and allowing the fluorescent signal to be observed [54]. D – MCP-GFP detection of mRNA in live cells. MCP-GFP binds as a dimer to stem-loops from bacteriophage MS2. These stem-loops are usually inserted in 3ʹUTR of the mRNA of interest, and 24 stem-loops enable detection of single-RNA molecules [55,56].
An alternative approach relies on fluorescent RNA aptamers, which are short RNA sequences that fold into specific structures able to bind fluorogenic small molecules (Fig. 3B). A first generation of aptamers developed for RNA live cells imaging includes Spinach, Broccoli, Mango and Corn [42–45]. These aptamers bind small, cell-permeant dyes that activate their fluorescence upon binding, for review see [46,47]. As a proof of principle, several highly expressed RNAs were visualized using these aptamers [45,48]. In case of low-abundant RNAs in yeast, a special denoising algorithm was required to increase signal-to-noise ratio of images [49]. Overall, this first generation of molecules suffered from non-optimal folding and insufficient brightness, preventing to achieve single molecule sensitivity.
A recently developed second generation of aptamers has an improved RNA folding, as well as an increased brightness and higher affinity of the fluorogenic small molecules. Currently, the most promising systems appear to be the Peppers, o-Coral and Mango II, as well as a Broccoli-BI pair that is 10 fold brighter than the original Broccoli-DHFBI [50–53]. Notably, single mRNAs were visualized in live cells using arrays of Broccoli aptamers [53]. An array of 24 Mango II aptamers also enabled a single molecule sensitivity in live and fixed cells and is suitable for super-resolution imaging [51]. Peppers are particularly attractive due to their high brightness exceeding Broccoli and to the availability of fluorescent dyes of different colours from cyan to red, making them suitable for combinatorial labelling. They were used for labelling of mRNA, genomic loci and super-resolution imaging [52]. The Peppers from Yi Yang group [52] should not be confounded with Peppers of the Samie R. Jaffrey group [54], which have the same name but based on a different principal. In this case, traditional fluorescent proteins are fused to a destabilization-regulatory domain, which promotes their degradation, but can be stabilized by binding to an RNA aptamer, helping to decrease a background due to a non-bound protein (Fig. 3C) [54].
As of today, the most sensitive approaches for RNA visualization in live cells are based on RNA-binding proteins that recognize specific RNA sequences or structures (for review see [5]). Several of these RNA-binding proteins come from bacteriophages. The first to be adapted for RNA imaging and still the most widely used today is the bacteriophage MS2 coat protein (MCP) [55]. This is a small protein that binds a 21 nucleotide RNA stem-loop from the MS2 genome with high specificity and sub-nanomolar affinity. To follow RNAs in live cell, an array of MCP-binding motifs is inserted in a gene of interest and co-expressed with the MCP-GFP fusion (Fig. 3). The insertion of multiple MCP-binding motifs leads to fluorescent signal amplification and increases the signal-to-noise ratio. The MCP-GFP methodology was first applied to study mRNA transport in live Saccharomyces cerevisiae cells [55]. This was achieved by inserting six MCP binding stem-loops in the 3ʹ-UTR of ASH1 mRNA that localizes to the bud tip (Fig. 3D). The method was adapted to mammalian cells and it was shown through quantification that 24 MS2 motifs were sufficient to detect single mRNAs with a simple wide-field microscope, which was at that time an important progress in the RNA imaging [56]. The MCP-GFP system has since been successfully implemented to study different steps of RNA metabolism including transcription, splicing, nuclear export, transport in the cytoplasm and degradation [57–66], leading to important discoveries. For example, it allowed to show that transcription is discontinuous and proceeds in bursts in bacteria and eukaryotes, including mammals [59,67–69]. It also enabled to estimate that Polymerase II elongation rates average from 1 to 4 kb/min depending on the organism [58,60,62,67,70–72].
The MCP-GFP also performs well in whole animals, it was applied to study the transport of the endogenous β-actin mRNA in live mouse [69,73]. The transgenic mice expressing MCP-GFP and MS2-tagged RNA were normal, undistinguishable from the wild type animals, indicating that this tagging system did not adversely affect mRNA metabolism. The system was also used in zebrafish and repeatedly in Drosophila embryos to study transcriptional regulation [72,74–81].
Further efforts helped to increase the brightness of single RNA molecules, mainly by increasing the number of MS2 stem-loops. A tag containing 96 MS2 repeats was constructed to visualize transcription in live bacteria [67] and recently, an array of 128 degenerated MS2 stem-loops was engineered and used to visualize HIV-1 transcription in mammalian cells and RNA transport in neurons [70,82]. Here, the brightness of single molecules increased 5 times compared to the original 24 MS2 repeat, helping to image RNAs longer and at higher speed. This high-speed imaging enabled to uncover that HIV-1 and some cellular genes are transcribed by RNA polymerase II convoys, groups of polymerases that move synchronously through the gene [70].
It is important to keep in mind that in some cases MCP binding can change the behaviour of the tagged RNA by interfering with RNA biogenesis. For instance, it was shown in yeast that MCP binding protects short-lived mRNAs in yeast from degradation, leading to RNA truncation and the appearance in the cytoplasm of MCP-protected fragments, which cannot be distinguished from the complete mRNA by live imaging [83,84]. This problem was solved by the use of modified stem loops that are less stably bound by MCP, enabling to restore their normal degradation [85]. Therefore, the MS2-tagged RNAs should be thoroughly controlled by quantifying their abundance and verifying their localization by smFISH, and by comparing them to the native untagged RNAs.
Structurally related to MCP, the coat protein of Pseudomonas aeruginosa bacteriophage PP7 (PCP) was also adapted for visualization of RNA in live cells [86]. The simultaneous labelling with MCP and PCP fused to different fluorophores enables imaging of two RNA species in the same cell, or two parts of the same RNA [62,66,87–89]. Several other RNA-binding proteins were applied for RNA visualization: the human U1A protein of U1snRNP, the bacteriophage λ protein λN and the bacterial protein BglG/SacY [90–92]. The U1A protein is not suitable for RNA visualization in human cells but it was successfully used in S. Cerevisiae and S. Pombe [93,94]. λN and BglG/SacY represent other tags of choice for double or multiple RNA labelling, although they are not as widely used as MCP and PCP.
New approaches to visualize RNA in live cells use Cas9 and Cas13. Catalytically dead versions of these enzymes can be programmed to bind the RNA of interest by short DNA and/or RNA guide sequences [95–97]. At this point, Cas13 shows efficient labelling of different RNAs and has a promising potential for live-cell imaging [97]. Different RNA species can be detected using two orthogonal Cas13 of different origins, and simultaneous labelling of DNA and RNA can be achieved by combining Cas9 and Cas13. This approach opens opportunity for live imaging of endogenous RNA without modifying them with tags [97].
Transport of mRNAs through the nucleoplasm
Transcription, processing, export and localization of mRNAs are tightly linked. Pre-mRNAs are co-transcriptionally capped and, in most cases, are co-transcriptionally spliced [98]. During transcription and after, mRNAs dynamically associate with numerous RNA-binding proteins (RBPs) including splicing, export and 3ʹ-end processing factors, cap-binding complexes, the exon-exon junction complex and poly-A binding proteins [99]. Transcript cleavage and addition of the polyA-tail results in the release of the mRNA from the transcription site [100]. On their way to the cytoplasm, the mRNPs first need to be transported to the nuclear pores and to translocate across them.
MCP labelling in live cells illuminated how mRNPs are transported in the nucleus and its passage through the nuclear pore complex (NPC). By imaging MS2-tagged mRNA reporters in human U2OS cells, it was found that the nuclear phases of gene expression, including mRNA transcription, processing, nucleoplasmic transport and export, take place within 5–40 min, with the exact duration depending on the size of the mRNA and splicing rates [62,63,87,101]. Correctly processed mRNPs are not retained in any particular nuclear domain. These mRNPs are simply released from their transcription sites and freely diffuse throughout the nucleoplasm until they reach nuclear pores. No active, motorized transport of mRNPs has been observed in the nucleus, in contrast to the cytoplasm [64,101,102]. Nucleoplasmic transport is slow and takes minutes compared to nuclear export that occurs in seconds. In the nucleoplasm, mRNPs display sub-diffusive movements and are restricted to chromatin-free spaces. The maintenance of the nuclear organization is strongly dependent on ATP and the energy deprivation provokes a shrinking of the nuclear space, leading to mRNP trapping and reduced diffusion, which is energy-independent on its own [64]. The intra-nuclear diffusion of mRNAs is dependent on their sizes with small molecules diffusing faster than large ones. This effect could be explained by the sieve-like properties of the inter-chromatin space [101].
Transport of mRNAs through the nuclear pores
mRNPs are exported to the cytoplasm through the nuclear pore complexes (NPCs). The NPCs are macromolecular complexes that are structurally conserved between all eukaryotes and consist of nucleoporins (for review see [103,104]). By electron microscopy, the NPCs contain three rings: nuclear, inner and cytoplasmic. On the nuclear side of NPC, eight rod-like filaments form a nuclear basket, while on the cytoplasmic side, eight filaments emanate from the pore. The central channel of the NPC functions as a selective transport gate for macromolecules. It is filled with hydrogel-like fibres consisting of phenylalanine-glycine (FG) repeats from 10 different nucleoporins [103]. These FG-repeats are disordered and flexible, forming a barrier between the nucleus and the cytoplasm. Molecules of up to 40 kDa in size diffuse through the NPC while larger molecules are transported by nuclear transport receptors. These transporters recognize signals on the cargo and transiently interact with the FG repeats, thereby facilitating transport without specific energy requirements. Yet, energy is required to impose transport directionality. mRNPs are transported by transport receptor NXF1/NXT1 (Mex67/Mtr2 in yeast), which binds mRNPs within the nucleus and releases them on the cytoplasmic side of the NPCs. The assembly and disassembly of the transport complexes are accompanied by RNA remodelling, which requires the energy of ATP hydrolysis [100]. The DEAD-box helicases UAP56 (Sub2 in yeast) and DDX19 (Dbp5 in yeast) mediate binding of the mRNP to the NXF1/NXT1 transport receptor and its release in the cytoplasm, respectively [105–107]. Export of mRNPs is tightly linked to quality-control mechanisms. For instance, spliced mRNAs are efficiently exported while intron-containing mRNAs usually accumulate in the nucleus [101,107].
RNPs are large macromolecules and how they cross the nuclear pores remains partly mysterious. MCP labelling allowed to investigate the kinetics of transport through NPCs in vivo. The dimension of the inner channel of the NPC is about 40 nm [103], which is smaller than the resolution of a regular light microscope (~ 200 nm of lateral resolution). To overcome this limitation and to be able to image the passage of mRNPs through the NPC with high precision, new microscopy approaches were developed [61,108]. Super-registration microscopy enables to take simultaneous high-speed movies of two different fluorophores on two precisely aligned cameras. Using this technique, the distances between β-actin mRNPs, labelled with MCP-YFP, and the nuclear pores, labelled with tandem-dimer Tomato, were measured, reaching a sub-diffraction spatial precision of 26 nm and a temporal resolution of 20 ms. Based on these measurements, a 3-step model of nuclear export in mouse cells was proposed: docking (80 ms), transport (5–20 ms), and release (80 ms), with a total export time about 180 ms [61]. The authors found that mRNPs can move bi-directionally within the pores, confirming that unidirectional export requires interaction with cytoplasmic molecules that remodel the mRNP and prevent it from crossing back the pores [61,105,109].
Further increase in temporal and spatial resolution was achieved through single-point edge-excitation sub-diffraction microscopy (SPEED), followed by deconvolution [108]. In this approach, single fluorescently labelled nuclear pore and individual MCP-labelled mRNPs were illuminated with two overlapped laser beams and imaged on a CCD camera with frequency of 500 frames per second, allowing for a temporal resolution of 2 ms and a spatial precision of 8 nm. This approach found the time of NPC passage equal to 12 ms for β-actin and firefly luciferase mRNAs in mouse and human cells [108]. This time of NPC passage differs more than 10 times from previous measurements [61]. The authors explain this discrepancy by a higher temporal resolution of their study, since as soon as they decreased the temporal resolution, only more slowly diffusing molecules were observed. Only 36% of the mRNPs arrived to the cytoplasmic side of the NPCs while others returned back to the nucleus, supporting the idea that quality-control selection of the mRNPs occurs on the nucleoplasmic side and in the central channel of NPCs [108].
Single-molecule imaging revealed that, in both mammalian and yeast cells, the mRNPs were sliding along the nuclear envelope before being transported through the nuclear pores, displaying so-called nuclear periphery scanning [61,110]. Furthermore, instantaneous sampling of 3D nuclear volumes by multi-focus microscopy showed that more than 60% of MCP-labelled β-actin mRNPs in mouse fibroblast nuclei were located within 0.5 μm from the nuclear pores, further supporting the existence of a scanning process during which mRNPs sample nuclear pores before engaging into export [102].
PP7-GFP labelling of mRNAs in live yeast S. cerevisiae combined with genetic approaches helped to uncover the molecular basis of this scanning behaviour. It was found to be dependent on interactions of mRNPs with the nuclear basket proteins Mlp1/2, and mediated by the nuclear poly-A binding protein Nab2 that directly interacts with Mlp1 on one side and the mRNA export receptor Mex67 on the other. Mutations of the nuclear basket proteins Mlp1/2 or Nab2 decreased mRNPs dwell time at the nucleoplasmic side of the pores and led to their release back into the nucleoplasm suggesting a role for nuclear basket in mRNPs remodelling and their quality-control before export [110,111]. Upon arrival to the cytoplasm, mRNPs first docked at the cytoplasmic side of the nuclear pores, probably for a remodelling step after export. The time of export in yeast cells was about 200 ms, similar to measurements obtained by super-registration microscopy in mammalian cells [61,112]. The authors further show that Mex67p is involved in the mechanisms controlling the cytoplasmic release of mRNPs, as well as overall transport directionality [112]. More recent imaging in yeast and mammalian cells indicated that Mex67 and its human homolog NXF1 mainly function at the NPC and not before, and probably associate with mRNPs only at the pore during its crossing [113,114], while the release of mRNPs in the cytoplasm required Mex67/NXF1 and the essential for mRNA export helicase Dbp5 [114].
In the insect Chironimus tentans, an elegant approach was used to observe the endogenous Balbiani Ring mRNPs at a single molecule level by light-sheet microscopy. In this case, mRNPs were tracked using fluorescently-labelled hrp36 protein, a homolog of mammalian hnRNPA1 that binds the pre-mRNA co-transcriptionally, accompanies it during nuclear export and remains associated with polysomes during translation [115]. Upon cytoplasmic injection, fluorescently labelled hrp36 entered the nucleus where it was co-transcriptionally incorporated in Balbiani Ring mRNPs and followed them to the cytoplasm [116,117]. Similar to yeast and mammalian cells, a nuclear periphery scanning was observed with the mRNPs displaying a rate-limiting step at the nuclear basket that may correspond to mRNP remodelling. Only 25% of the particles were successfully exported with an export time ranging between 65 ms and several seconds in agreement with the reported time in yeast and mammalian cells [117]. A single molecule analysis of fluorescently labelled helicase Dbp5, showed a large number of fluorescently labelled Dbp5 molecules at the nuclear envelope, which localized mostly on the cytoplasmic side of the NPC and stayed bound for an average of 55 seconds. The authors proposed that several Dbp5 molecules act in concert for mRNP translocation and release [117].
Transport of mRNPs in the cytoplasm
Once arrived in the cytoplasm, mRNPs are remodelled: they lose some nuclear factors and gains new RBPs that will facilitate their translation and transport to the final cytoplasmic destination [118]. As we previously mentioned, some mRNAs are localized to specific sub-cellular areas, while others are randomly distributed throughout the cytoplasm or non-localized [2]. Note that even non-localized mRNAs should leave the perinuclear area after export to be homogeneously distributed throughout the cytoplasm. Live cell imaging shows that cytoplasmic mRNPs can be subjected to diffusion, anchoring and directed motorized transport through interactions with cellular motors [56,119,120]. These types of movements are stochastic and were observed for non-localized as well as for localized mRNAs. However, the cytoplasmic transport of localized mRNA such as β-actin showed a strong bias towards motorized directed movements, which help to deliver it to the cell periphery where it can get anchored [56]. Motorized directed movement is particularly important in neurons where mRNPs patrol synapses in a highly dynamic fashion resembling a sushi-belt [121]. In neuronal processes, some mRNPs display anterograde transport biases, which are believed to play important roles in RNA sorting to specific distal locations and therefore fundamental for neuronal function [73,82,122].
The trajectories of single RNA molecules can be reconstructed with tracking algorithms [123]. Recently, a software allowing to infer mechanistic characteristics of movement from single trajectories was developed [124]. This approach is based on hidden Markov models and it helps to determine switches between different types of movements: diffusion, transient anchoring and directed motorized transport, depending on cytoplasmic location of mRNPs, thus giving access to the link between mRNA transport and its local functions. Mathematical modelling of trajectories also helps to better characterize the localization mechanism. For instance, recent data in neurons suggest that β-actin mRNPs follow a Levy walk in neuronal processes, i.e. long stochastic transport events followed by local exploration [73]. This may allow the mRNP to efficiently explore the dendritic tree to find relevant synapses.
Molecular mechanisms linking cytoplasmic mRNAs to motors
The transport of mRNA to specific cytoplasmic locations is often mediated by cis-acting localization elements (LEs) that are usually found in the 3ʹ-UTR of the mRNA but can also be embedded in its coding sequence. These RNA signals are bound by RBPs that link the mRNA to transport and anchoring systems through adaptor proteins. Active motorized transport is the most common mechanism of RNA localization. However, other mechanisms such as selective degradation or diffusion coupled to anchoring can be also used, sometimes in combination [125,126]. The LEs display no sequence or structure conservation and can be very heterogeneous in length and structures. Only few of the known LEs display short linear sequence motifs such as the hnRNP A2 responsive element of Myelin Basic Protein mRNA [127], or the bi-partite zipcode of β-actin mRNA [128,129]. The majority of LEs contain diverse secondary structures including stem-loops, bulges, or G-quadruplexes [130]. How such diverse structures are coupled to the cellular transport machineries is not well understood.
One of the best-studied examples is that of ASH1 mRNA, which localizes to the bud tip in yeast cells during mitosis. This mRNA has four LEs that are located in both the 3ʹUTR and the coding sequence, which are bound co-transcriptionally by the RNA-binding protein She2p. In the cytoplasm, the adaptor protein She3p links the mRNA: She2p complex to the myosin motor Myo4, forming the so-called locasome that is transported on actin cables to the bud tip [55,131] (Fig. 4A; for review see [132]). Recently, X-ray crystallography was applied to investigate the structure of one of the four ASH1 LEs, E3, in complex with She2p and She3p proteins [133]. This study shows that two 28 nucleotides stem-loops corresponding to a minimal E3 element are bound by two dimers of She2p. The central region of the E3 stem undergoes structural rearrangements upon this binding and the structure is stabilized by the nuclear Loc1p protein, which is replaced by She3p in the cytoplasm. She3p binds concomitantly the RNA and She2p, helping to achieve a highly specific recognition of the LE, necessary for its transport. She2p and She3p binding require a combination of RNA sequence and structure and it is fulfilled through the action of the globular domain of She2p and the unstructured domain of She3p [133]. It is important to note that the same She-dependent transport system is used in yeast for a variety of mRNAs, which localize to the bud at different phases of the cell cycle [134–136]. Interestingly, ASH1 mRNA transport can involve phase-separated granules, containing multiple mRNA molecules [137,138], a feature that is found in other cases of mRNA transport in different systems such as mouse neurons and developing Xenopus oocytes [73,139]. In contrast, cases of singly transported mRNAs have been described in neuronal dendrites and in Drosophila oocyte [140,141].
Figure 4.

mRNP transport complexes
A – ASH1 mRNA in yeast Saccharomyces cerevisiae. ASH1 is transported on F-actin cables, it is bound by RBP She2p co-transcriptionally, an adaptor She3p links RNA-She2p complex to a motor Myo4D in the cytoplasm [126]. B – RNA-stimulated assembly of transport complex for localized RNAs in Drosophila embryo. RBP Egalitarian (Egl) binds the RNA LE and promotes its interaction with the dimer of Bicaudal-D (BicD). RNA:Egl complex binding to BicD releases its autoinhibitory loop facilitating the interaction with dynein to form an active dynein-dynactin complex for the transport to the minus end of microtubules. Two Egl proteins are associated with one molecule of RNA [144]. C – Transport of septin mRNAs in fungus Ustilago maydis. mRNAs are bound by the RBP Rrm4 together with its partner Grp1 and transported on Rab5a positive early endosomes. The complex mRNA-Rrm4-Grp1, containing also Poly-A binding protein 1 (Pab 1), is tethered on endosomes through the adaptor Upa1. The mRNA is translated during the transport [184]. D – Model of β-catenin mRNA translational regulation. β-catenin mRNA accumulates in cytoplasmic translation factories, where it is translated and the newly-made β-catenin protein is degraded by the destruction complex. Upon WNT signalling activation, the factories are dissolved and the newly synthesized β-catenin migrates to the nucleus to activate transcription [31,192].
In Drosophila embryo, multiple RNAs localize to the minus end of microtubules, both in the oocyte and in polarized epithelial cells of the blastocyst. These mRNAs bear a variety of seemingly unrelated LEs. Nevertheless, they are transported through the association of their LEs with a common adaptor RBP called Egalitarian (Egl), which couples mRNAs to the dynein transport complex through an interaction with Bicaudal-D (BicD) [142]. Structural studies demonstrate that BicD binds both Egl and an endosomal Rab6GTP cargo through the single binding site. While the two cargoes are not co-transported, the use of the same adaptors for motorized transport of RNPs and for endosomal sorting suggest that similar mechanisms could underline these processes [143]. In vitro study using purified proteins to reconstruct RNA transport on microtubules showed that binding of the RNA LE by Egl promotes its interaction with BicD homodimer, which in turn alters the conformation of BicD to promote its assembly with an active dynein-dynactin complex (Fig. 4B). Indeed, an autoinhibitory loop of BicD is released upon binding the RNA:Egl complex and becomes available for interaction with dynein and dynactin. The experiments also suggest that two Egl molecules bind one LEs and that the RNA stabilizes the Egl dimer, which may bind BicD with a higher affinity than monomers. The authors propose that the RNA plays a scaffolding role in the assembly of the transport complex, allowing the regulation of intracellular transport and limiting unproductive transport cycles without RNA [144].
Imaging translation reveals active transport of polysomes as a new mechanism for RNA localization
In line with RNA-driven assembly of transport complexes, it is believed that mRNAs are translationally silenced during their transport and that their translation starts only at the appropriate cytoplasmic location [145]. However, this view is now questioned by new single molecule approaches that provide spatial and temporal information about mRNA translation and its relation with transport.
One of these techniques is based on fluorescence fluctuation spectroscopy (FFS), which monitors fluorescent intensities in a microscopic volume in live cell. The passage of the fluorescently tagged mRNA and ribosomes through this small volume allows to follow their association via a cross-correlation analysis of the fluorescent signals, and to calculate the stoichiometry of the interactions by measuring the brightness of the diffusing macromolecules (Fig. 5A, left). This method can be broadly applied to study dynamics of mRNA interactions with any RBPs within the cell. It was used to demonstrate that association of ribosomes and ZBP1 with β-actin mRNA were mutually exclusive, thus confirming a role of ZBP1 in translation inhibition [146]. Fluorescently labelled mRNA and ribosomes can also be tracked simultaneously using single particle tracking, enabling to create maps of mRNA and ribosome trajectories and to elucidate where and when the mRNA is translated (Fig. 5A, right). Using co-tracking, it was found that translating β-actin mRNAs slowdown their movements and dwell a longer time near focal adhesion sites in fibroblasts [147].
Figure 5.
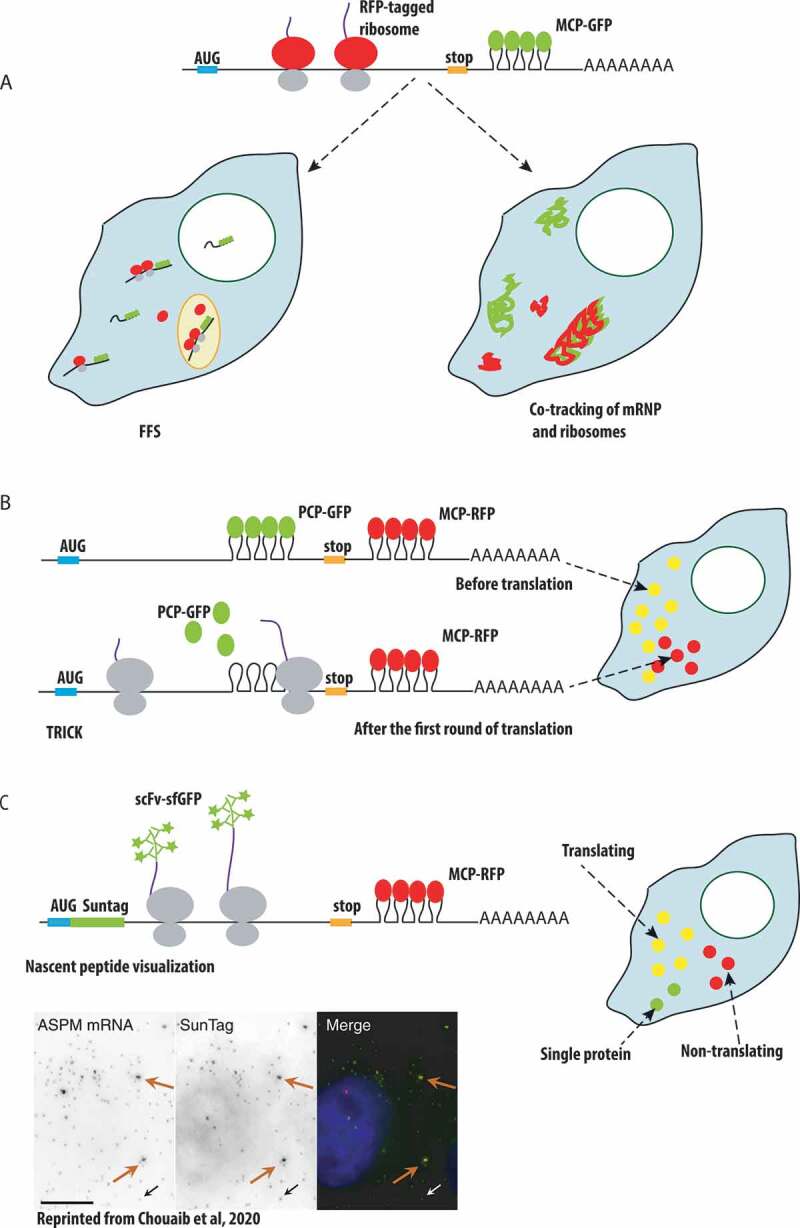
Single-molecule approaches to study mRNA translation in live cells
A – Tracking of mRNAs and ribosomes. RNAs are labelled with MCP-GFP (top, green ovals) and ribosomes with red fluorescent protein (top, red ovals), allowing to follow them by Fluorescence Fluctuation Spectroscopy (FFS) or single-particle tracking. Left: two-photon FFS monitors passages of mRNAs and ribosomes through a microscopic volume in the cell [146]. Right: single particle co-tracking of mRNPs and ribosomes [147]. B – TRICK. The mRNA is labelled with PCP-GFP (left, green ovals) in its coding region and with MCP-RFP (left, red ovals) in its 3ʹ-UTR. Ribosome removes bound PCP-GFP during translation. The non-translated mRNAs are visible in the microscope as yellow dots due to the superposition of green and red colours (right), while after the first round of translation the mRNAs become red dots (right) [88]. C – Visualization of nascent translation. Top left – The mRNA is labelled with MCP-RFP (left, red ovals), the nascent peptide is visualized using single chain antibodies fused to super-folder GFP ScFv-sfGFP (left, green stars), that recognize SunTag, or with anti-Flag antibodies, or Frankenbody in case of protein labelling with repeats of Flag- or HA- tags. Right – Non-translating mRNAs are visible as red dots (right), the single molecules of protein are green dots (right) and actively transcribed mRNA are yellow dots [149]. Bottom – Micrograph of HeLa cells with a SunTagged ASPM allele, showing ASPM mRNA (by smiFISH, left and red), the signal from the SunTag (middle and green); blue, nuclear staining with DAPI. White and black arrows, a single mRNA positive for the SunTag; orange arrow, an mRNA foci positive for the SunTag. Scale bar: 10 mm. Reprinted from Chouaib R, Safieddine A, Pichon X, Imbert A, Kwon OS, Samacoits A, Traboulsi A-M, Robert M-C, Tsanov N, Coleno E, et al. A Dual Protein-mRNA Localization Screen Reveals Compartmentalized Translation and Widespread Co-translational RNA Targeting. Dev Cell 2020; 54:773–791.e5. Page 781, Copyright 2020, with permission from Elsevier.
Another approach to image translation is called Translation RNA Imaging by the Coat-protein Knock-off (TRICK). This method distinguishes the mRNAs that underwent the first round of translation from the mRNAs that have never been translated. Here, double tagging of mRNA with PCP in the coding region and MCP in the 3ʹUTR is used. The passage of the ribosome during translation leads to the PCP removal from the coding region, yielding the mRNAs that have only one colour instead of two (Fig. 5B). Using TRICK, the authors showed that under stress conditions transcripts with 5ʹ-terminal oligopyrimidine (5ʹ-TOP) motif are transcriptionally repressed, and either remain free in the cytoplasm or become sequestered in P-bodies. Upon stress release, the free cytoplasmic mRNAs resumed translation, while the mRNAs localized to P-bodies stayed transcriptionally inactive. TRICK also was applied to the whole Drosophila embryo to confirm that localized Oskar mRNA starts translation only when it reaches the posterior pole of the oocyte [88].
A recent breakthrough in the field was brought by single molecule approaches that directly visualize nascent peptides in live cells. These methods are based on repeated peptide epitopes such as SunTag, FLAG or HA tags (Fig. 5C) [148–150]. The SunTag is a repeated epitope from the yeast Gcn4 protein and it is detected by a recombinant single chain variable fragment antibody fused to sfGFP (scFv-sfGFP), which recognizes the epitope with a very high affinity, in the sub-nanomolar range. The Gcn4 peptide is inserted in multiple copies in the protein of interest, usually 24, such that the signal is amplified and single molecules become visible with a simple wide-field microscope [149]. To visualize translation of single mRNPs, the SunTag is inserted at the N-terminus of the protein of interest. In this case, the peptide epitope becomes bound by the fluorescent single chain antibodies as soon as it emerges from the ribosomes, allowing the visualization of translation at the level of single mRNAs [151–154]. As an alternative to the SunTag, a protein can be tagged with 10 copies of the FLAG tag inserted in the Spaghetti Monster scaffold, and its translation can then be followed using either fluorescently labelled Fab antibody fragments loaded in the cells using beads [148], or a recombinant single chain anti-HA antibody termed Frankenbody. The Frankenbody can be expressed in fusion with different detection modules, including fluorescent proteins and HaloTag, allowing visualization in various colours [150]. Additional pairs of repeated epitope/nanobody also became available for multi-colour read-out of translation [155].
The direct visualization of translation allowed to measure the translation elongation speed, which varies from 5 to 18 amino acids per minute [148,151,153]. Similar to transcription, translation of single mRNAs occurs in a burst-like fashion in Hela cells and neurons [151,153]. The 3ʹUTR seems to play an important role in the spatial regulation of translation in neurons: a reporter mRNA with β-actin 3ʹUTR was translated in proximal dendrites but translationally repressed in distal dendrites, while the 3ʹ-UTR of Arc mRNA increased the number of translating mRNAs in dendrites [152,153]. Interestingly, the onset of translation had little or no effect on mRNA motility in the cytoplasm, and the diffusive movements of polysomes were nearly as rapid as a diffusion of free mRNA molecules non-engaged in translation. The diffusion coefficients of individual polysomes translating the same mRNA showed high heterogeneity, which could probably reflect their interactions with multiple intracellular structures. The type of cytoplasmic transport also varied for different translating mRNAs. For example, POLR2A polysomes displayed only diffusive movements, while Dynein heavy chain polysomes alternated diffusion, anchoring and, surprisingly, motorized transport [151]. This suggests that, in contrast to previous expectations, translation could be instrumental for mRNA transport. In line with this idea, recent evidences indicate that for a series of human mRNAs localizing to centrosomes, nuclear envelope and Golgi apparatus, translation inhibition disrupts RNA localization [31,156,157]. Moreover, in the case of centrosomal mRNAs, reporter experiments and direct polysome imaging with the SunTag demonstrated that the polysomes are subjected to active motorized transport driven by the nascent protein and not by the RNA [21]. This represents a considerable difference from the traditional paradigm of RNA-dependent transport of translationally repressed transcripts. Moreover, both the centrosomal mRNAs and the localization mechanism are conserved in Drosophila [157], revealing a remarkable example of the evolutionary conservation of RNA localization. Finally, some of these centrosomal mRNAs localize to cilia in quiescent cells, and in this case, transport appears to be dependent on both translation and EJC [158], suggesting a concomitant contribution of RNA and protein determinants. Together with previously described case in Drosophila [158,159], it is a second example of the EJC role in mRNA localization.
Interestingly, the mRNA encoding for the Dynein heavy chain appears to be translated in dedicated translation factories, which are small cytoplasmic foci accumulating many copies of the same mRNA [151]. A few other mRNAs were recently found to be translated in similar structures, including β-catenin for which the factories were shown to be involved in the co-translational degradation of the nascent protein (Fig. 4D) [31]. Remarkably, the foci were dissolved upon Wnt signalling activation, which is known to stabilize β-catenin [31]. Therefore, the β-catenin translation factories play a regulatory role by tightly controlling the amount of the key effector of Wnt signalling pathway [31]. Similar regulation can be at place in axonal growth cones, where a coupling of local translation and protein degradation was previously described [160]. The translation factories or analogous foci could also be the sites of the co-translational assembly of protein complexes, a process recently found to occur in yeasts and in mammalian cells [161,162]. Another interesting example of local translational regulation is provided by intestinal epithelium in mouse. In these polarized cells, ribosomes were found to display apical localization leading to a more efficient translation of the apical mRNAs. After food intake, the mRNAs encoding for ribosomal proteins were transported to the apical part of the cells, where they were efficiently translated to produce more ribosomes and sustain more efficient translation associated with nutrients absorption [163]. These findings add another level of complexity in the regulation of localized translation, opening a possibility that in addition to mRNA localization, the translation apparatus can also be localized.
RNA transport on membrane organelles
Endoplasmic reticulum
In different organisms from yeast to mammals, multiple mRNAs encoding membrane and secreted proteins are localized to endoplasmic reticulum (ER). The ER targeting of these mRNAs usually depends on a signal peptide, a short peptide sequence located at the N-terminus of the nascent protein (reviewed in [164]). Although this type of targeting is a predominant way for ER localization, some mRNAs encoding for cytoplasmic proteins were also reported to be localized and translated on ER [165]. Through quantitative imaging in fixed and live cells, recent study demonstrates that a small fraction of cytosolic mRNAs is associated with ER in a translation-dependent manner and it remains on the ER during several rounds of translation. A fraction of the ER-attached mRNAs display a higher ribosome occupancy as compared to their cytoplasmic counterparts, and this could have physiological consequences [166]. Furthermore, ER localization can also occur in a translation-independent manner through RNA encoded signals [167]. For instance, in Xenopus oocyte, Vg1 mRNA is targeted to the ER through its 3ʹUTR localization signal and is then transported on microtubules attached to the ER to reach the vegetal pole [168].
The previously described ASH1 mRNA in yeast was also observed to be co-transported with ER tubular structures by live-cell imaging [169]. However, in this study ASH1 was not expressed from its endogenous promoter leading to changes of its expression during cell cycle. Later studies showed that only the bud-localized mRNAs expressed in S and G2 phases are co-transported with tubular ER to achieve their localization. These mRNAs encode membrane and secreted proteins but their targeting to the ER can be independent from the signal peptide, as in case of the yeast WSC2 mRNA. On the contrary, ASH1 mRNA is expressed during late mitosis and it localizes to the bud tip independently of the ER [135].
Finally, it is important to note that the ER is also used by different RNA viruses during their replication cycle. For instance, infection by flaviviruses modifies the ER and viral RNAs dynamically associate to the reticulum membranes during viral replication [170,171]. Coronaviruses, including SARS-CoV-1, modify ER membranes and create double-membrane organelles, which contain viral RNA and serve as compartments of viral replication and transcription in the cytoplasm of infected cells [172,173]. Similar structure are likely formed by recently appeared SARS-CoV-2 and their investigation will be important for understanding the viral RNAs metabolism.
Endosomes and transport of viral RNAs
RNA transport on endosomes plays a role in the infection cycles of different RNA viruses, including retroviruses. For example, the genomic RNAs of Murine Leukaemia Virus (MLV) travel together with the retroviral proteins Gag and envelope on recycling endosomes inside infected cells, to reach the plasma membrane for budding [57]. Gag specifically recognizes the genomic retroviral RNA and it interacts with endosomal membranes thereby acting as a tether between them. The transport of the HIV-1 genomic RNA is also linked to the endosomal system [174,175]. The endosomal pathway may be used to deliver viral RNA-Gag complexes to the sites of cellular contacts and to facilitate direct cell to cell transmission of HIV-1 [175]. Indeed, HIV-1 infection of primary CD4 + T-cells proceeds through specialized cell-cell contacts called virological synapses, and their formation requires specialized endosomal compartments called secretory lysosomes [176]. In infected macrophages, HIV-1 viruses bud and accumulate in intracellular compartments, which were first classified as late endosomes or multi-vesicular bodies [177–179]. However, later studies have found that they are distinct from canonical endosomes and may be connected to the plasma membrane, although their precise nature remains unclear [180]. Another pathogen, influenza A virus, belonging to negative-strand RNA viruses, transports its viral RNP on Rab11 positive endosomes in a kinesin dependent manner [181–183]. All these examples demonstrate that transport of RNPs on endosomes plays an important role in host–pathogen interactions.
Endosomes and transport of cellular RNAs
Following the rule that viruses use pre-existing cellular mechanisms for their propagation, endosomes also transport cellular RNAs in different organisms. In a pathogenic fungus of corn, Ustilago maydis, localized mRNAs travel on microtubules by hitchhiking on endosomes. U. maydis switches from yeast-like to hyphal (i.e. filamentous) growth during the early phase of corn infection. To precisely target proteins during hyphal growth, numerous mRNAs are bound by the RBP Rrm4 and transported to hyphal tips by Rab5a positive early endosomes (Fig. 4C). The mRNA-Rrm4 complex is tethered to endosomal membranes through an adaptor called Upa1, which binds simultaneously Rrm4 and membrane lipids (reviewed in [132]). Rrm4 functions to transport mRNAs and ribosomes on the same endosomes, helping also to distribute polysomes throughout the hypha [184]. Recently, transcriptome-wide iCLIP has shown that Rrm4 transports on the endosomes more than 2000 transcripts together with its new partner Glycin rich protein 1 (Grp1) (Fig. 4C). The analysis of the RNA-binding sites in this transport complex revealed that Rrm4 and Grp1 bind together and in close proximity to the 3ʹUTR of the majority of mRNAs and this allows for translation concomitantly with transport [185]. This is the case of the important for infection septins, whose mRNAs are bound by Rrm4 and Grp1 in their 3ʹUTRs and are translated and assembled in high order structures on endosomes during their transport [186–188]. Although it is not clear if the nascent protein is involved in the transport process, Rrm4 binds some other mRNAs inside open reading frames, suggesting that these mRNAs could be translationally inhibited, or subjected to translational regulation during their transport [185].
Similar mechanisms could exist in mammalian system, where a novel study found that some RNA-containing granules hijack late endosomes in non-polarized human U2OS cells and primary rat neurons [189]. These granules are tethered on the cytoplasmic side of late endosomes through the protein Annexin 11 (ANXA 11). ANXA11 interacts with RNA through its unstructured N-terminal low complexity region, whose phase separating properties facilitate RNA granule formation. On the other side, ANXA11 binds the lysosomal membranes through its annexin domain in a calcium- and phospholipid-regulated manner. Remarkably, familial cases of the neurodegenerative disease Amyotrophic Lateral Sclerosis (ALS) are associated with mutations in ANXA11. These mutations lead to alterations of neuronal functions through disruption of ANXA11 interactions with late endosomes. This impacts its phase-separation properties important for the formation of RNA granules and perturbs the mRNP transport system that normally brings RNA to distal parts of neurites [189]. This exciting finding provides an interesting parallel with the endosomal mRNA transport system in Ustilago maudis cited above [132]. It also resembles the transport of retroviral RNAs on endosomes mediated by the viral Gag protein [57].
Another discovery provides a link between endosomal trafficking and localized mRNA translation in Retinal Ganglion Cells (RGCs) of Xenopus [190]. In the axons of RGCs, mRNPs and ribosomes were found to be associated with early and late endosomes. Live cell imaging revealed that some mitochondrial proteins were translated on Rab7a positive late endosomes at the sites of long-term contacts with mitochondria, and this local translation was shown to be important for mitochondrial activity. It is plausible that endosomal translation could exert broader functions in axons, since other translating mRNAs were also associated with late endosomes. Importantly, a humans neurological disorder Charcot-Marie-Tooth disease type 2B is associated with Rab7a mutations that disrupt the local translation of mitochondrial proteins. This leads to mitochondrial dysfunction and contributes to axonopathy of sensory and motor neurons [190].
mRNPs association with endosomes seems to be a recurrent theme in mammalian cells. Recently, multiple mRNAs encoding for proteins involved in endocytic and membrane pathways were found to be localized to early endosomes in human cell lines [191]. Among them, mRNA of Early Endosomal Antigen 1(EEA1) is anchored at the endosomes in a translation-dependent manner and its translation might be negatively regulated by RBP CSRP1 at this location [191], suggesting that similarly to Ustilago maudis, the endosomes could be sites of regulated translation in mammals.
Concluding remarks
Studies of mRNA transport by single molecule microscopy revealed essential aspects of the spatial and temporal regulation of gene expression. Spectacular technical advancements enable to address challenging questions to broaden our understanding how mRNA delivers and translates the genetic information. A first open question is the detailed characterization of mRNA transport complexes, which will benefit from the concerted use of single molecule and systematic biochemical approaches for the identification of RBPs, mRNA LEs and motors. A second challenge is to study mRNA transport in its endogenous context and in whole tissues and organisms. These studies are facilitated by the development of CRISPR-Cas9 methods to tag endogenous genes, as well as by Cas13 approaches to directly visualize untagged endogenous mRNAs [95–97]. Finally, the single molecule approaches that visualize nascent translation [151–154] revolutionized the field and opened the possibility to study mRNA transport and translation simultaneously, in response to various stimuli and in different physiological contexts, including in multicellular organisms, where studies are facilitated by rapid progress of imaging technologies such as light sheet microscopy and computer vision. These techniques already provided important insights on the links between transport and translation, and now point us to multiple, exciting future directions.
Acknowledgments
We thank B. Pradet-Balade for critical reading of the manuscript, useful comments and discussion, X. Pichon for helpful feedback on the manuscript. We apologize to all the colleagues whose papers may not have been cited due to the limited space. This work was funded by grants from the Ligue Nationale Contre le Cancer (“équipe labélisée”) and the Agence nationale de la recherche (TRANSFACT).
Funding Statement
This work was supported by the Ligue Contre Le Cancer and by the Agence Nationale de la Recherche (ANR).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Kim SH, Vieira M, Shim JY, et al. Recent progress in single-molecule studies of mRNA localization in vivo. RNA Biol. 2019;16(9):1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bovaird S, Patel D, Padilla J-CA, et al. Biological functions, regulatory mechanisms, and disease relevance of RNA localization pathways. FEBS Lett. 2018;592:2948–2972. [DOI] [PubMed] [Google Scholar]
- [3].Moore S, Järvelin AI, Davis I, et al. Expanding horizons: new roles for non-canonical RNA-binding proteins in cancer. Curr Opin Genet Dev. 2018;48:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Biswas J, Liu Y, Singer RH, et al. Fluorescence imaging methods to investigate translation in single cells. Cold Spring Harb Perspect Biol. 2019;11:a032722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pichon X, Lagha M, Mueller F, et al. Toolbox to image gene expression in single cells: sensitive approaches for demanding challenges. Mol Cell. 2018;71:468–480. [DOI] [PubMed] [Google Scholar]
- [6].Sato H, Das S, Singer RH, et al. Imaging of DNA and RNA in living eukaryotic cells to reveal spatiotemporal dynamics of gene expression. Annu Rev Biochem. 2020;159–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tutucci E, Livingston NM, Singer RH, et al. Imaging mRNA in vivo, from birth to death. Annu Rev Biophys. 2018;47:85–106. [DOI] [PubMed] [Google Scholar]
- [8].Femino AM, Fay FS, Fogarty K, et al. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. [DOI] [PubMed] [Google Scholar]
- [9].Raj A, van den Bogaard P, Rifkin SA, et al. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Femino AM, Fogarty K, Lifshitz LM, et al. Visualization of single molecules of mRNA in situ. Methods Enzymol. 2003;361:245–304. [DOI] [PubMed] [Google Scholar]
- [11].Moffitt JR, Zhuang X.. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol. 2016;572:1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mueller F, Senecal A, Tantale K, et al. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods. 2013;10(4):277–278. [DOI] [PubMed] [Google Scholar]
- [13].Tsanov N, Samacoits A, Chouaib R, et al. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016;44:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stringer C, Michaelos M, Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. bioRxiv. 2020;2020(2):02.931238. [DOI] [PubMed] [Google Scholar]
- [15].Samacoits A, Chouaib R, Safieddine A, et al. A computational framework to study sub-cellular RNA localization. Nat Commun. 2018;9:4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen KH, Boettiger AN, Moffitt JR, et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Eng C-HL, Lawson M, Zhu Q, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 2019;568:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lubeck E, Coskun AF, Zhiyentayev T, et al. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xia C, Fan J, Emanuel G, et al. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci U S A. 2019;116:19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Battich N, Stoeger T, Pelkmans L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods. 2013;10:1127–1133. [DOI] [PubMed] [Google Scholar]
- [21].Safieddine A, Coleno E, Traboulsi A-M, et al. A conserved choreography of mRNAs at centrosomes reveals a localization mechanism involving active polysome transport. bioRxiv. 2020;2020(9):04.282038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Adivarahan S, Livingston N, Nicholson B, et al. Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol Cell. 2018;72(727–738):e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Khong A, Parker R. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J Cell Biol. 2018;217:4124–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].West JA, Mito M, Kurosaka S, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214:817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Methods. 2012;9:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fang M, Xie H, Dougan SK, et al. Stochastic cytokine expression induces mixed T helper cell states. PLoS Biol. 2013;11:e1001618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Itzkovitz S, Lyubimova A, Blat IC, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2011;14:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Raj A, Rifkin SA, Andersen E, et al. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lécuyer E, Yoshida H, Parthasarathy N, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. [DOI] [PubMed] [Google Scholar]
- [31].Chouaib R, Safieddine A, Pichon X, et al. A dual protein-mRNA localization screen reveals compartmentalized translation and widespread co-translational RNA targeting. Dev Cell. 2020;54(773–791):e5. [DOI] [PubMed] [Google Scholar]
- [32].Xia C, Babcock HP, Moffitt JR, et al. Multiplexed detection of RNA using MERFISH and branched DNA amplification. Sci Rep. 2019;9:7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ainger K, Avossa D, Morgan F, et al. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. [DOI] [PubMed] [Google Scholar]
- [35].Pitchiaya S, Androsavich JR, Walter NG. Intracellular single molecule microscopy reveals two kinetically distinct pathways for microRNA assembly. EMBO Rep. 2012;13:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pitchiaya S, Mourao MDA, Jalihal AP, et al. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. Mol Cell. 2019;74(521–533):e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ghosh S, Marchand V, Gáspár I, et al. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol. 2012;19:441–449. [DOI] [PubMed] [Google Scholar]
- [38].Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24:1914–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vera M, Pani B, Griffiths LA, et al. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. eLife. 2014;3:e03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zid BM, O’Shea EK. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014;514:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen M, Yang Y, Krueger CJ, et al. Optimizing molecular beacons for intracellular analysis of RNA. Methods Mol Biol Clifton NJ. 2018;1649:243–257. [DOI] [PubMed] [Google Scholar]
- [42].Dolgosheina EV, Jeng SCY, Panchapakesan SSS, et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol. 2014;9:2412–2420. [DOI] [PubMed] [Google Scholar]
- [43].Filonov GS, Moon JD, Svensen N, et al. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136:16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Song W, Filonov GS, Kim H, et al. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat Chem Biol. 2017;13:1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bouhedda F, Autour A, Ryckelynck M. Light-up RNA aptamers and their cognate fluorogens: from their development to their applications. Int J Mol Sci. 2018;19:44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Trachman RJ, Truong L, Ferré-D’Amaré AR. Structural principles of fluorescent RNA aptamers. Trends Pharmacol Sci. 2017;38:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Strack RL, Disney MD, Jaffrey SR. A superfolding spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods. 2013;10:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Guet D, Burns LT, Maji S, et al. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat Commun. 2015;6:8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bouhedda F, Fam KT, Collot M, et al. A dimerization-based fluorogenic dye-aptamer module for RNA imaging in live cells. Nat Chem Biol. 2020;16:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cawte AD, Unrau PJ, Rueda DS. Live cell imaging of single RNA molecules with fluorogenic Mango II arrays. Nat Commun. 2020;11:1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chen X, Zhang D, Su N, et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat Biotechnol. 2019;37:1287–1293. [DOI] [PubMed] [Google Scholar]
- [53].Li X, Kim H, Litke JL, et al. Fluorophore-promoted RNA folding and photostability enables imaging of single broccoli-tagged mRNAs in live mammalian cells. Angew Chem Int Ed Engl. 2020;59:4511–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wu J, Zaccara S, Khuperkar D, et al. Live imaging of mRNA using RNA-stabilized fluorogenic proteins. Nat Methods. 2019;16:862–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bertrand E, Chartrand P, Schaefer M, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. [DOI] [PubMed] [Google Scholar]
- [56].Fusco D, Accornero N, Lavoie B, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol CB. 2003;13:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Basyuk E, Galli T, Mougel M, et al. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev Cell. 2003;5:161–174. [DOI] [PubMed] [Google Scholar]
- [58].Boireau S, Maiuri P, Basyuk E, et al. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol. 2007;179:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chubb JR, Trcek T, Shenoy SM, et al. Transcriptional pulsing of a developmental gene. Curr Biol CB. 2006;16:1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Darzacq X, Shav-Tal Y, de Turris V, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Grünwald D, Singer RH. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Martin RM, Rino J, Carvalho C, et al. Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. Cell Rep. 2013;4:1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schmidt U, Basyuk E, Robert M-C, et al. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol. 2011;193:819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shav-Tal Y, Darzacq X, Shenoy SM, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Janicki SM, Tsukamoto T, Salghetti SE, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Horvathova I, Voigt F, Kotrys AV, et al. The dynamics of mRNA turnover revealed by single-molecule imaging in single cells. Mol Cell. 2017;68(615–625):e9. [DOI] [PubMed] [Google Scholar]
- [67].Golding I, Paulsson J, Zawilski SM, et al. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. [DOI] [PubMed] [Google Scholar]
- [68].Yunger S, Rosenfeld L, Garini Y, et al. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7:631–633. [DOI] [PubMed] [Google Scholar]
- [69].Lionnet T, Czaplinski K, Darzacq X, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tantale K, Mueller F, Kozulic-Pirher A, et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat Commun. 2016;7:12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Larson DR, Zenklusen D, Wu B, et al. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Garcia HG, Tikhonov M, Lin A, et al. Quantitative imaging of transcription in living drosophila embryos links polymerase activity to patterning. Curr Biol CB. 2013;23:2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Park HY, Lim H, Yoon YJ, et al. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343:422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bothma JP, Garcia HG, Esposito E, et al. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living drosophila embryos. Proc Natl Acad Sci U S A. 2014;111:10598–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Campbell PD, Chao JA, Singer RH, et al. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Dev Camb Engl. 2015;142:1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fernandez C, Lagha M. Lighting up gene activation in living drosophila embryos. Methods Mol Biol Clifton NJ. 2019;2038:63–74. [DOI] [PubMed] [Google Scholar]
- [77].Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in drosophila. Curr Biol CB. 2003;13:1159–1168. [DOI] [PubMed] [Google Scholar]
- [78].Lucas T, Ferraro T, Roelens B, et al. Live imaging of bicoid-dependent transcription in drosophila embryos. Curr Biol CB. 2013;23:2135–2139. [DOI] [PubMed] [Google Scholar]
- [79].Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell. 2006;11:251–262. [DOI] [PubMed] [Google Scholar]
- [80].Zimyanin VL, Belaya K, Pecreaux J, et al. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ferraro T, Esposito E, Mancini L, et al. Transcriptional memory in the drosophila embryo. Curr Biol CB. 2016;26:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bauer KE, Segura I, Gaspar I, et al. Live cell imaging reveals 3ʹ-UTR dependent mRNA sorting to synapses. Nat Commun. 2019;10:3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Garcia JF, Parker R. MS2 coat proteins bound to yeast mRNAs block 5ʹ to 3ʹ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. Rna N Y N. 2015;21:1393–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Heinrich S, Sidler CL, Azzalin CM, et al. Stem-loop RNA labeling can affect nuclear and cytoplasmic mRNA processing. Rna N Y N. 2017;23:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Tutucci E, Vera M, Biswas J, et al. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods. 2018;15:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chao JA, Patskovsky Y, Almo SC, et al. Structural basis for the coevolution of a viral RNA-protein complex. Nat Struct Mol Biol. 2008;15:103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Coulon A, Ferguson ML, de Turris V, et al. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife. 2014;3:e03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Halstead JM, Lionnet T, Wilbertz JH, et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hocine S, Raymond P, Zenklusen D, et al. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods. 2013;10:119–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen J, Nikolaitchik O, Singh J, et al. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U S A. 2009;106:13535–13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Daigle N, Ellenberg J. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat Methods. 2007;4:633–636. [DOI] [PubMed] [Google Scholar]
- [92].Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci. 2000;97:5273–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chung S, Takizawa PA. In vivo visualization of RNA using the U1A-based tagged RNA system. Methods Mol Biol Clifton NJ. 2011;714:221–235. [DOI] [PubMed] [Google Scholar]
- [94].Takeuchi-Andoh T, Ohba S, Shinoda Y, et al. A simplified vector system for visualization of localized RNAs in Schizosaccharomyces pombe. Biosci Biotechnol Biochem. 2016;80:1362–1367. [DOI] [PubMed] [Google Scholar]
- [95].Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Yang L-Z, Wang Y, Li S-Q, et al. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol Cell. 2019;76(981–997):e7. [DOI] [PubMed] [Google Scholar]
- [98].Saldi T, Cortazar MA, Sheridan RM, et al. Coupling of RNA polymerase II transcription elongation with pre-mRNA Splicing. J Mol Biol. 2016;428:2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wende W, Friedhoff P, Sträßer K. Mechanism and regulation of co-transcriptional mRNP assembly and nuclear mRNA export [Internet]. In: Oeffinger M, Zenklusen D, editors. The biology of mRNA: structure and function. Cham: Springer International Publishing; 2019. p. 1–3. DOI: 10.1007/978-3-030-31434-7_1 [DOI] [PubMed] [Google Scholar]
- [100].Stewart M. Polyadenylation and nuclear export of mRNAs. J Biol Chem. 2019;294(9):2977–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Mor A, Suliman S, Ben-Yishay R, et al. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12(6):543–552. [DOI] [PubMed] [Google Scholar]
- [102].Smith CS, Preibisch S, Joseph A, et al. Nuclear accessibility of β-actin mRNA is measured by 3D single-molecule real-time tracking. J Cell Biol. 2015;209(4):609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Knockenhauer KE, Schwartz TU. The nuclear pore complex as a flexible and dynamic gate. Cell. 2016;164(6):1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ashkenazy-Titelman A, Shav-Tal Y, Kehlenbach RH. Into the basket and beyond: the journey of mRNA through the nuclear pore complex. Biochem J. 2020;477:23–44. [DOI] [PubMed] [Google Scholar]
- [105].Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20(4):645–651. [DOI] [PubMed] [Google Scholar]
- [106].Strässer K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413(6856):648–652. [DOI] [PubMed] [Google Scholar]
- [107].Soheilypour M, Mofrad MRK. Quality control of mRNAs at the entry of the nuclear pore: cooperation in a complex molecular system. Nucl Austin Tex. 2018;9:202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ma J, Liu Z, Michelotti N, et al. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat Commun. 2013;4(1):2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lin DH, Correia AR, Cai SW, et al. Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nat Commun. 2018;9(1):2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Saroufim M-A, Bensidoun P, Raymond P, et al. The nuclear basket mediates perinuclear mRNA scanning in budding yeast. J Cell Biol. 2015;211(6):1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Fasken MB, Stewart M, Corbett AH. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J Biol Chem. 2008;283(40):27130–27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Smith C, Lari A, Derrer CP, et al. In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p. J Cell Biol. 2015;211(6):1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Derrer CP, Mancini R, Vallotton P, et al. The RNA export factor Mex67 functions as a mobile nucleoporin. J Cell Biol. 2019;218(12):3967–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ben-Yishay R, Mor A, Shraga A, et al. Imaging within single NPCs reveals NXF1’s role in mRNA export on the cytoplasmic side of the pore. J Cell Biol. 2019;218(9):2962–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Visa N, Alzhanova-Ericsson AT, Sun X, et al. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84(2):253–264. [DOI] [PubMed] [Google Scholar]
- [116].Siebrasse JP, Veith R, Dobay A, et al. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc Natl Acad Sci U S A. 2008;105(51):20291–20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Siebrasse JP, Kaminski T, Kubitscheck U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc Natl Acad Sci U S A. 2012;109(24):9426–9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Khong A, Parker R. The landscape of eukaryotic mRNPs. Rna N Y N. 2020;26(3):229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Katz ZB, Wells AL, Park HY, et al. β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012;26(17):1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Yamagishi M, Ishihama Y, Shirasaki Y, et al. Single-molecule imaging of β-actin mRNAs in the cytoplasm of a living cell. Exp Cell Res. 2009;315(7):1142–1147. [DOI] [PubMed] [Google Scholar]
- [121].Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. Embo J. 2011;30(17):3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Das S, Moon HC, Singer RH, et al. A transgenic mouse for imaging activity-dependent dynamics of endogenous Arc mRNA in live neurons. Sci Adv. 2018;4(6):eaar3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Vallotton P, van Oijen AM, Whitchurch CB, et al. Diatrack particle tracking software: review of applications and performance evaluation. Traffic Cph Den. 2017;18:840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Monnier N, Barry Z, Park HY, et al. Inferring transient particle transport dynamics in live cells. Nat Methods. 2015;12:838–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Chin A, Lécuyer E. RNA localization: making its way to the center stage. Biochim Biophys Acta Gen Subj. 2017;1861:2956–2970. [DOI] [PubMed] [Google Scholar]
- [126].Mofatteh M, Bullock SL. SnapShot: subcellular mRNA localization. Cell. 2017;169(178–178):e1. [DOI] [PubMed] [Google Scholar]
- [127].Munro TP, Magee RJ, Kidd GJ, et al. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem. 1999;274:34389–34395. [DOI] [PubMed] [Google Scholar]
- [128].Chao JA, Patskovsky Y, Patel V, et al. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Patel VL, Mitra S, Harris R, et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012;26:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Pratt CA, Mowry KL. Taking a cellular road-trip: mRNA transport and anchoring. Curr Opin Cell Biol. 2013;25:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol CB. 1999;9:569–578. [DOI] [PubMed] [Google Scholar]
- [132].Niessing D, Jansen R-P, Pohlmann T, et al. mRNA transport in fungal top models. Wiley Interdiscip Rev RNA. 2018;9(1). [DOI] [PubMed] [Google Scholar]
- [133].Edelmann FT, Schlundt A, Heym RG, et al. Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex. Nat Struct Mol Biol. 2017;24:152–161. [DOI] [PubMed] [Google Scholar]
- [134].Aronov S, Gelin-Licht R, Zipor G, et al. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Fundakowski J, Hermesh O, Jansen R-P. Localization of a subset of yeast mRNAs depends on inheritance of endoplasmic reticulum. Traffic Cph Den. 2012;13:1642–1652. [DOI] [PubMed] [Google Scholar]
- [136].Shepard KA, Gerber AP, Jambhekar A, et al. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A. 2003;100:11429–11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Heym RG, Zimmermann D, Edelmann FT, et al. In vitro reconstitution of an mRNA-transport complex reveals mechanisms of assembly and motor activation. J Cell Biol. 2013;203:971–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Lange S, Katayama Y, Schmid M, et al. Simultaneous Transport of Different Localized mRNA Species Revealed by Live-Cell Imaging. Traffic. 2008;9:1256–1267. [DOI] [PubMed] [Google Scholar]
- [139].Neil CR, Jeschonek SP, Cabral SE, et al. L-bodies are novel RNA-protein condensates driving RNA transport in Xenopus oocytes [Internet]. Cell Biol. bioRxiv. 2020. [cited 2020 Oct 13]; Available from: http://biorxiv.org/lookup/doi/10.1101/2020.05.08.084814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Batish M, van den Bogaard P, Kramer FR, et al. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A. 2012;109:4645–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Little SC, Sinsimer KS, Lee JJ, et al. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat Cell Biol. 2015;17:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Dienstbier M, Boehl F, Li X, et al. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Liu Y, Salter HK, Holding AN, et al. Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev. 2013;27:1233–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].McClintock MA, Dix CI, Johnson CM, et al. RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. eLife. 2018;7:e36312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Wu B, Buxbaum AR, Katz ZB, et al. Quantifying protein-mRNA interactions in single live cells. Cell. 2015;162:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Katz ZB, English BP, Lionnet T, et al. Mapping translation “hot-spots” in live cells by tracking single molecules of mRNA and ribosomes. eLife. 2016;5:e10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Morisaki T, Lyon K, DeLuca KF, et al. Real-time quantification of single RNA translation dynamics in living cells. Science. 2016;352:1425–1429. [DOI] [PubMed] [Google Scholar]
- [149].Tanenbaum ME, Gilbert LA, Qi LS, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Zhao N, Kamijo K, Fox PD, et al. A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nat Commun. 2019;10:2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Pichon X, Bastide A, Safieddine A, et al. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J Cell Biol. 2016;214:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wang C, Han B, Zhou R, et al. Real-time imaging of translation on single mRNA transcripts in live cells. Cell. 2016;165:990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Wu B, Eliscovich C, Yoon YJ, et al. Translation dynamics of single mRNAs in live cells and neurons. Science. 2016;352:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Yan X, Hoek TA, Vale RD, et al. Dynamics of translation of single mRNA molecules in vivo. Cell. 2016;165:976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Boersma S, Khuperkar D, Verhagen BMP, et al. Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell. 2019;178(458–472):e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Sepulveda G, Antkowiak M, Brust-Mascher I, et al. Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. eLife. 2018;7:e34959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bergalet J, Patel D, Legendre F, et al. Inter-dependent centrosomal co-localization of the cen and ik2 cis-natural antisense mRNAs in drosophila. Cell Rep. 2020;30(3339–3352):e6. [DOI] [PubMed] [Google Scholar]
- [158].Kwon OS, Mishra RK, Safieddine A, Alasseur Q, Faucourt M, Coleno E, Barbosa I, Bertrand E, Spassky N, Hir HL . Exon Junction Complex dependent mRNA localization is linked to centrosome organization during ciliogenesis. bioRxiv 2020;358960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Palacios IM, Gatfield D, St Johnston D, et al. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. [DOI] [PubMed] [Google Scholar]
- [160].Deglincerti A, Liu Y, Colak D, et al. Coupled local translation and degradation regulate growth cone collapse. Nat Commun. 2015;6:6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Kamenova I, Mukherjee P, Conic S, et al. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat Commun. 2019;10:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Shiber A, Döring K, Friedrich U, et al. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. 2018;561:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Moor AE, Golan M, Massasa EE, et al. Global mRNA polarization regulates translation efficiency in the intestinal epithelium. Science. 2017;357:1299–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Cui XA, Palazzo AF. Localization of mRNAs to the endoplasmic reticulum. Wiley Interdiscip Rev RNA. 2014;5:481–492. [DOI] [PubMed] [Google Scholar]
- [165].Reid DW, Nicchitta CV. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2015;16:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Voigt F, Zhang H, Cui XA, et al. Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep. 2017;21:3740–3753. [DOI] [PubMed] [Google Scholar]
- [167].Pyhtila B, Zheng T, Lager PJ, et al. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. Rna N Y N. 2008;14:445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. [DOI] [PubMed] [Google Scholar]
- [169].Schmid M, Jaedicke A, Du T-G, et al. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr Biol CB. 2006;16:1538–1543. [DOI] [PubMed] [Google Scholar]
- [170].Miorin L, Maiuri P, Hoenninger VM, et al. Spatial and temporal organization of tick-borne encephalitis flavivirus replicated RNA in living cells. Virology. 2008;379:64–77. [DOI] [PubMed] [Google Scholar]
- [171].Miorin L, Romero-Brey I, Maiuri P, et al. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J Virol. 2013;87:6469–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Knoops K, Kikkert M, SHE van den W, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Neuman BW, Angelini MM, Buchmeier MJ. Does form meet function in the coronavirus replicative organelle? Trends Microbiol. 2014;22:642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Lehmann M, Milev MP, Abrahamyan L, et al. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J Biol Chem. 2009;284:14572–14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Molle D, Segura-Morales C, Camus G, et al. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J Biol Chem. 2009;284:19727–19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Jolly C, Welsch S, Michor S, et al. The regulated secretory pathway in CD4(+) T cells contributes to human immunodeficiency virus type-1 cell-to-cell spread at the virological synapse. PLoS Pathog. 2011;7:e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Gaudin R, Berre S, Cunha de Alencar B, et al. Dynamics of HIV-containing compartments in macrophages reveal sequestration of virions and transient surface connections. PloS One. 2013;8:e69450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Jouve M, Sol-Foulon N, Watson S, et al. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe. 2007;2:85–95. [DOI] [PubMed] [Google Scholar]
- [179].Raposo G, Moore M, Innes D, et al. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic Cph Den. 2002;3:718–729. [DOI] [PubMed] [Google Scholar]
- [180].Mariani C, Desdouits M, Favard C, et al. Role of Gag and lipids during HIV-1 assembly in CD4(+) T cells and macrophages. Front Microbiol. 2014;5:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Eisfeld AJ, Kawakami E, Watanabe T, et al. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol. 2011;85:6117–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Ramos-Nascimento A, Kellen B, Ferreira F, et al. KIF13A mediates trafficking of influenza A virus ribonucleoproteins. J Cell Sci. 2017;130:4038–4050. [DOI] [PubMed] [Google Scholar]
- [183].Vale-Costa S, Amorim MJ. Recycling endosomes and viral infection. Viruses. 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Higuchi Y, Ashwin P, Roger Y, et al. Early endosome motility spatially organizes polysome distribution. J Cell Biol. 2014;204:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Olgeiser L, Haag C, Boerner S, et al. The key protein of endosomal mRNP transport Rrm4 binds translational landmark sites of cargo mRNAs. EMBO Rep. 2019;20:e46588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Zander S, Baumann S, Weidtkamp-Peters S, et al. Endosomal assembly and transport of heteromeric septin complexes promote septin cytoskeleton formation. J Cell Sci. 2016;129:2778–2792. [DOI] [PubMed] [Google Scholar]
- [187].Baumann S, König J, Koepke J, et al. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 2014;15:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].König J, Baumann S, Koepke J, et al. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. Embo J. 2009;28:1855–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Liao Y-C, Fernandopulle MS, Wang G, et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell. 2019;179(147–164):e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Cioni J-M, Lin JQ, Holtermann AV, et al. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176(56–72):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Popovic D, Nijenhuis W, Kapitein LC, et al. Co-translational targeting of transcripts to endosomes. bioRxiv. 2020;2020(7):17.208652. [Google Scholar]
- [192].Chin A, Lécuyer E. Translating messages in different neighborhoods. Dev Cell. 2020;54:691–693. [DOI] [PubMed] [Google Scholar]


