Abstract
Background
Previously, we reported that Wnt5a-positive breast cancer can be classified as estrogen receptor (ER)-positive breast cancer; its prognosis is worse than that of Wnt5a-negative breast cancer. This study aimed to investigate the mechanisms underlying the poor prognosis in Wnt5a-positive breast cancer patients.
Methods
In total, 151 consecutive ER-positive breast cancer patients who underwent resection between January 2011 and February 2014 were enrolled. DNA microarray and pathway analyses were conducted using MCF-7 cells stably expressing Wnt5a [MCF-7/Wnt5a (+)]. Based on the outcomes, cell viability/drug sensitivity assays, and mutation analysis were performed using cell cultures and breast cancer tissues. The relationship between Wnt5a and the PI3K–AKT–mTOR signaling pathway was also examined.
Results
The relapse-free survival rate in patients with Wnt5a-positive breast cancer was significantly lower than that in patients with Wnt5a-negative breast cancer (P = 0.047). DNA microarray data suggest that only the cytochrome P450 (CYP) pathway was significantly upregulated in MCF-7/Wnt5a (+) cells (P = 0.0440). Additionally, MCF-7/Wnt5a (+) cells displayed reduced sensitivity to the metabolic substrates of CYP, tamoxifen (P < 0.001), paclitaxel (P < 0.001), and cyclophosphamide (P < 0.001). Of note, PIK3CA mutations were not associated with the expression of Wnt5a in breast cancer tissue and culture cells.
Conclusions
In ER-positive breast cancer, Wnt5a upregulates the CYP metabolic pathway and suppresses tamoxifen, paclitaxel, and cyclophosphamide resistance, all of the three, standard treatment methods for ER-positive breast cancer. Wnt5a is thus potentially involved in the poor prognosis of ER-positive breast cancer independently of the PI3K–AKT–mTOR signaling pathway.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12282-021-01241-0.
Keywords: Wnt5a, Estrogen receptor-positive breast cancer, Cytochrome P450, Pathway analysis, Drug resistance
Introduction
The Wnt pathway is classified into the β-catenin-dependent and -independent pathways [1]. Wnt5a is a typical ligand of the β-catenin-independent pathway, modulating cell polarity and migration via the PCP/CE and Ca2+ pathways [1, 2]. The Wnt5a signaling pathway is involved in the progression of several cancers [1, 3–7]; Wnt5a contributes to the progression of gastric, lung, and prostate cancers [3–5], but serves as a tumor suppressor in thyroid and ovarian cancers [6, 7]. Wnt5a is reportedly overexpressed in approximately 30% of all breast cancer cases, and most Wnt5a-positive breast cancers are estrogen receptor (ER)-positive breast cancers. Of note, the 5-year relapse-free survival (RFS) rate is significantly lower in Wnt5a-positive versus -negative breast cancer [8].
Ovarian cancer cells overexpressing Wnt5a showed low chemosensitivity to paclitaxel and epirubicin [9]. However, in breast cancers, relatively few studies have examined the association between Wnt5a and drug sensitivity. Furthermore, upregulated PI3K–AKT–mTOR signaling is associated with a poor prognosis in ER-positive breast cancer [10–12]. Of note, Wnt5a is reportedly upregulated in ER-positive breast cancers harboring a PIK3CA mutation [13], indicating a close relationship between PI3K signaling and the Wnt5a pathway. Therefore, in the present study, the signaling pathways associated with Wnt5a were investigated to analyze the mechanisms underlying a Wnt5a-mediated drug resistance and poor prognosis.
Materials and methods
Patients and breast cancer tissues (RFS-tracked cases)
Consecutive breast cancer tissues resected between 2011 and 2014 were reviewed as previously described [8]. The follow-up period was extended, and the 8-year RFS was investigated by December 2019 in the present study. To strictly investigate “recurrence of early-stage breast cancer,” the two patients with Stage IV at diagnosis included in our previous study [8], were excluded from this study.
Furthermore, we grouped patients treated with the key drugs, tamoxifen, paclitaxel, cyclophosphamide, epirubicin and 5-fluorouracil, and investigated RFS comparing Wnt5a-positive and -negative patients.
Cell culture
The breast cancer cell lines MCF-7, and MDA-MB-175-VII (RRID: CVCL_0031, and CVCL_1400) were obtained from ATCC (catalog #HTB-22 and #HTB-25, Manassas, VA, USA) and were confirmed not to be listed in the ICLAC Register of Misidentified Cell Lines (version 10). MDA-MB-175-VII is an ER-positive and HER2-negative breast cancer cell line like MCF-7; however, unlike MCF-7, it endogenously expresses Wnt5a [8]. MCF-7 and MDA-MB-175-VII cells were cultured according to the manufacturer’s instructions.
Transfection, and RNA interference
The pPGK-neo/Wnt5a plasmid was transfected into MCF-7 cells using Lipofectamine LTX + PLUS reagent (Life Technologies, Carlsbad, CA, USA). Successfully transfected cells selectively formed colonies in the presence of G418. The colonies were screened for Wnt5a expression through western blotting. Thereafter, certain MCF-7 cells stably expressing Wnt5a [MCF-7/Wnt5a (+)] or not expressing Wnt5a [MCF-7/Wnt5a (−)] were established. Additionally, the siRNA-mediated suppression of Wnt5a in MCF-7 and MDA-MB-175-VII cells was conducted as previously described [8].
Gene microarray analyses
The Oligo DNA microarray analyses were performed using 3D-Gene Human Oligo chip 25 k (Toray Industries, Tokyo, Japan) as previously described [8]. In all three MCF-7/Wnt5a (+) clones, only significantly upregulated (expression ≥ twofold) or downregulated (expression ≤ 1/2) genes were selected. Gene ontology and pathway analyses were performed using the DAVID online tool (Version 6.8, https://david.ncifcrf.gov/). In addition, a heatmap was generated using SHINYHEATMAP.COM (http://shinyheatmap.com/). Among the differentially expressed genes, we searched for those associated with breast cancer/cancer/CYP, as per the PubMed website (https://pubmed.ncbi.nlm.nih.gov/). The genes and interrelationships were then arranged using the application “GeneMania” in Cytoscape (Version 3.8.2, http://cytoscape.org).
Cell viability assay
6 × 103 MCF-7/Wnt5a (+) and MCF-7/Wnt5a (−) cells from ≥ 80% confluent cultures were seeded into a 96-well plate in triplicate. Twenty-four, 48, and 72 h after the administration of 15 µM tamoxifen, 200 nM paclitaxel, 8000 µM cyclophosphamide, 800 nM epirubicin, and 400 µM 5-fluorouracil, 20 µL CellTiter 96® Aqueous One Solution Reagent (Promega, Madison, WI, USA) was added into each well (please see Online Resource 1A). Two hours later, the absorbance was measured according to the manufacture's instruction. Additionally, viability was also measured in the context of MDA-MB-175-VII cells. Briefly, 24 h after seeding (in the same way as for MCF-7 cells) MDA-MB-175-VII were transfected with the Wnt5a-siRNA or negative-siRNA (catalog #4392420 or #4390843, Life Technologies) and viability was measured as above, 24, 48, and 72 h later.
Western blot analysis
For immunoblot analysis, MCF-7 and MDA-MB-175-VII cells were washed with PBS and lysed with lysis buffer containing a Phosphatase Inhibitor Cocktail (Nacalai Tesque Inc., Kyoto, Japan). Proteins were separated via SDS-PAGE and then electro-transferred onto nitrocellulose membranes (Amersham Protran Premium, GE Healthcare, Buckinghamshire, UK). The membranes were probed with various primary and secondary antibodies (Online Resource 1B) and visualized with enhanced chemiluminescence detection reagents (Amersham ECL Select, GE Healthcare, Buckinghamshire, UK). All western blotting experiments were performed in triplicate.
Detection of PIK3CA mutant variants
Among the 151 cases immunoreactive for Wnt5a, PIK3CA mutations were evaluated only in those with a tumor size of ≥ 1 cm in diameter. The QIAamp DNA FFPE Tissue Kit (Qiagen GmbH, Hilden, Germany) was used to extract DNA from formalin-fixed paraffin-embedded (FFPE) tissues. The E542K, E545D/K, and H1047R/L were detected through direct sequencing using the primers listed in Online Resource 1C.
Quantification of Wnt5a mRNA expression
RNA was extracted using the NucleoSpin total RNA FFPE (Takara Bio, Shiga, Japan) from tissues sections sliced from the FFPE block, including the tumor component only. cDNA was synthesized through reverse-transcription using the PrimeScript II High Fidelity RT-PCR Kit (Takara Bio). Wnt5a expression was quantitatively analyzed via real-time PCR using the SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) and the CFX96 real-time PCR detecting system (Bio-Rad). Wnt5a expression was quantified using the ΔCt value. The used primers are listed in Online Resource 1C.
Statistical analysis
Statistical analysis was performed using the EZR [14] and SPSS (Version 20.0, Chicago, IL, USA) software. Welch's t test was used to compare the age, and cell viability between Wnt5a-negative and -positive cells, and Wnt5a-silenced and –non-silenced cells. The clinicopathologic characteristics were analyzed using the Chi-squared test. The significance between RFS curves was analyzed using the generalized Wilcoxon test. The frequency of Wnt5a positivity and the expression levels of Wnt5a mRNA were compared between PIK3CA mutation-negative and -positive cases using the Chi-square test and Welch’s t-test, respectively. P values < 0.05 were considered statistically significant.
Results
Wnt5a-positive breast cancer patients show poor prognosis
A total of 151 patients was enrolled. The median (range) follow-up period was 73.2 (11.7 to 102) months for all patients. The background of the patients is shown in Online Resource 2. The 8-year RFS rate of Wnt5a-positive breast cancer patients was lower than that of Wnt5a-negative patients [(91.9% (95% CI = 85.1–98.7) vs 98.6%, (95% CI = 96.0–100.0), P = 0.047] (Fig. 1). The postoperative treatment regimens used in recurrent patients are listed in Online Resource 3.
Fig. 1.
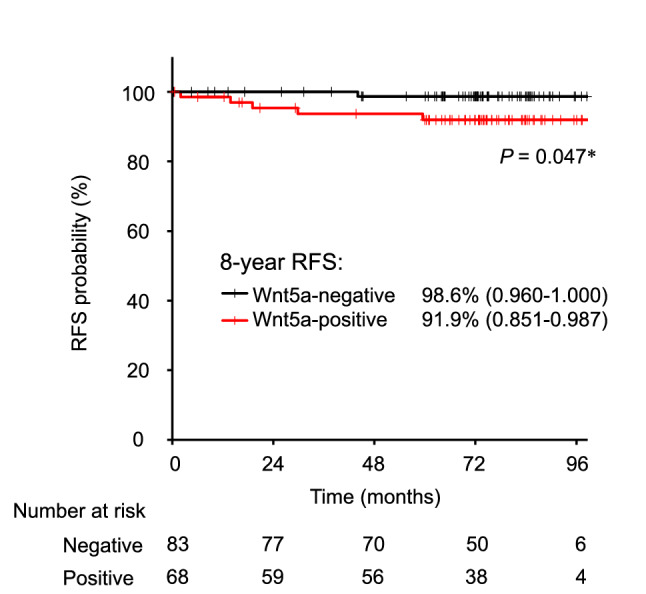
Prognosis of Wnt5a in ER-positive breast cancer patients. Prognosis was estimated via Kaplan–Meier analysis (n = 151); Wnt5a-positive breast cancer patients (n = 68) displayed a lower 8-year RFS probability: *P = 0.047 (Wilcoxon test). RFS relapse-free survival
The CYP pathway is upregulated in Wnt5a-expressing MCF-7 cells
To investigate pathways related to the recurrence of Wnt5a-positive breast cancer, MCF-7 cells stably expressing Wnt5a were established and DNA microarray analyses were performed. A total of 176 candidate genes differentially expressed between MCF-7/Wnt5a (+) and MCF-7/Wnt5a (−) cells, were identified; 76 unique genes were upregulated (expression ≥ twofold) and 100 unique genes were downregulated (expression ≤ 1/twofold; Online Resource 4). The P-value was determined using the DAVID database (Table 1). Interestingly, as per the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, physiological pathways directly or indirectly influencing Wnt5a expression were identified; only the cytochrome P450 (CYP) pathway, involved in drug metabolism [15], was identified among the upregulated genes (P = 0.0440; Table 1). Conversely, certain virus-related pathways were identified among the downregulated genes (Table 1). Interestingly, the JAK-STAT signaling pathway, associated with cancer progression, was attenuated in MCF-7/Wnt5a (+) cells (Table 1). Of note, the upregulated genes were primarily involved in the oxidation–reduction process, response to calcium ions, retinoid metabolic process, response to hypoxia and retinal metabolic process, ATPase binding, and oxidoreductase activity (Table 1). Conversely, most of the downregulated genes include those related to viruses and interferons (Table 1).
Table 1.
Selected gene categories significantly over-represented in Wnt5a-positive breast cancer
| Gene category | Number of genes | % | P-value |
|---|---|---|---|
| Upregulated genes | |||
| KEGG pathway | |||
| Drug metabolism—cytochrome P450 | 3 | 4.2 | 0.044 |
| Chemical carcinogenesis | 3 | 4.2 | 0.059 |
| Gene ontology | |||
| Biological process | |||
| Oxidation–reduction process | 8 | 11 | 0.006 |
| Response to calcium ion | 3 | 4.2 | 0.019 |
| Retinoid metabolic process | 3 | 4.2 | 0.021 |
| Response to hypoxia | 4 | 5.6 | 0.025 |
| Retinal metabolic process | 2 | 2.8 | 0.043 |
| Protein polymerization | 2 | 2.8 | 0.046 |
| Cellular component | |||
| Extracellular exosome | 20 | 28 | 0.003 |
| Early endosome | 5 | 7 | 0.009 |
| Plasma membrane | 23 | 32 | 0.021 |
| Endosome membrane | 4 | 5.6 | 0.027 |
| Molecular function | |||
| ATPase binding | 3 | 4.2 | 0.028 |
| Oxidoreductase activity | 4 | 5.6 | 0.034 |
| Downregulated genes | |||
| KEGG pathway | |||
| Influenza A | 13 | 13.8 | < 0.001 |
| Herpes simplex infection | 13 | 13.8 | < 0.001 |
| Measles | 11 | 11.7 | < 0.001 |
| Hepatitis C | 9 | 9.6 | < 0.001 |
| Hepatitis B | 6 | 6.4 | 0.002 |
| Toxoplasmosis | 5 | 5.3 | 0.004 |
| Epstein–Barr virus infection | 5 | 5.3 | 0.006 |
| RIG-I-like receptor signaling pathway | 4 | 4.3 | 0.008 |
| Chemokine signaling pathway | 5 | 5.3 | 0.024 |
| JAK–STAT signaling pathway | 4 | 4.3 | 0.055 |
| Cytosolic DNA-sensing pathway | 3 | 3.2 | 0.055 |
| Gene ontology | |||
| Biological Process | |||
| Type I interferon signaling pathway | 19 | 20.2 | < 0.001 |
| Defense response to virus | 22 | 23.4 | < 0.001 |
| Response to virus | 19 | 20.2 | < 0.001 |
| Negative regulation of viral genome replication | 13 | 13.8 | < 0.001 |
| Interferon-gamma-mediated signaling pathway | 10 | 10.6 | < 0.001 |
| Response to interferon-alpha | 5 | 5.3 | < 0.001 |
| Innate immune response | 12 | 12.8 | < 0.001 |
| Response to interferon-beta | 4 | 4.3 | < 0.001 |
| Negative regulation of type I interferon production | 4 | 4.3 | < 0.001 |
| Cellular response to interferon-alpha | 3 | 3.2 | < 0.001 |
| Positive regulation of interferon-alpha production | 3 | 3.2 | 0.0015 |
| Positive regulation of interferon-beta production | 3 | 3.2 | 0.0076 |
| Cellular component | |||
| Cytosol | 37 | 39.4 | < 0.001 |
| Cytoplasm | 37 | 39.4 | 0.0073 |
| Plasma membrane | 31 | 33 | 0.0076 |
| Spermatoproteasome complex | 2 | 2.1 | 0.024 |
| Molecular function | |||
| Double-stranded RNA binding | 8 | 8.5 | < 0.001 |
| 2′-5′-oligoadenylate synthetase activity | 4 | 4.3 | < 0.001 |
| NAD + ADP − ribosyltransferase activity | 4 | 4.3 | < 0.001 |
| Single-stranded RNA binding | 4 | 4.3 | 0.001 |
| Nucleotidyltransferase activity | 3 | 3.2 | 0.0067 |
| Helicase activity | 4 | 4.3 | 0.0069 |
| Transferase activity | 4 | 4.3 | 0.0096 |
| CCR5 chemokine receptor binding | 2 | 2.1 | 0.036 |
Relationships between differentially expressed genes, cancer, and CYP
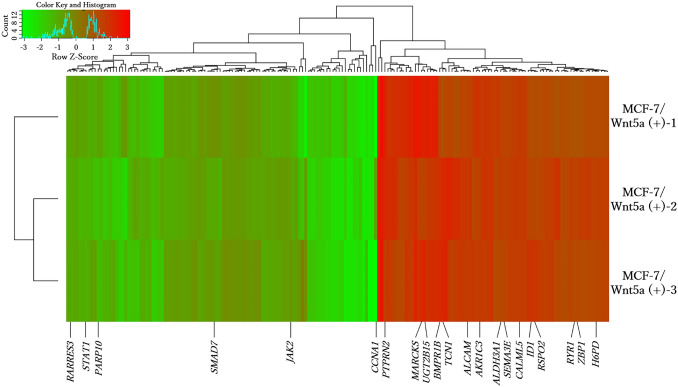
The 176 differentially expressed genes are represented in the form of a heatmap (Fig. 2). Among them, 21 genes are related to breast cancer/cancer/CYP and have been reported to promote cell migration and proliferation, and cancer metastasis. The upregulated genes that correlate positively with the malignancy of ER-positive breast cancer are UGT2B15 [16], AKR1C3 [17], ALCAM [8], and PTPRN2 [18]; the downregulated genes are JAK2, STAT1 [19], CCNA1 [20]. Of note, UGT2B15 and AKR1C3 are genes related to the CYP pathway, and ALCAM and PTPRN2 are genes co-expressed with CCNA1 (Online Resource 5).
Fig. 2.
Gene expression heatmap. Gene expression was analyzed via microarray using three MCF-7/Wnt5a (+) clones (MCF-7/Wnt5a (+) 1 to 3; see Online Resource 4). Gene expression was quantified as the relative ratio based on control cells [MCF-7/Wnt5a (−)]. The heatmap was generated at http://shinyheatmap.com/ after the expression levels were logarithmized
Wnt5a expression decreases the sensitivity to tamoxifen, paclitaxel, and cyclophosphamide
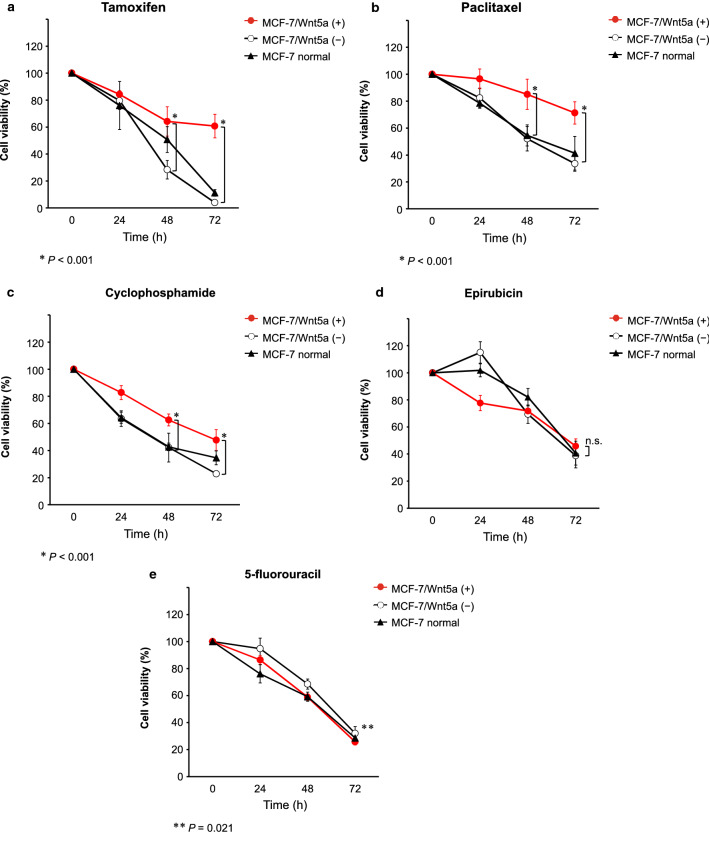
Tamoxifen, paclitaxel and cyclophosphamide, but not epirubicin and 5-fluorouracil, are metabolized by CYP [21–25]. Therefore, the sensitivity to these five drugs, used in the standard treatment of ER-positive breast cancer, was examined. MCF-7/Wnt5a (+) cells exhibited significantly lower sensitivity to tamoxifen, paclitaxel, and cyclophosphamide than MCF-7/Wnt5a (−) cells at both 48 h and 72 h (P < 0.001; Fig. 3a–c). In contrast, no difference was found in resistance to epirubicin (P = 0.12) and MCF-7/Wnt5a (+) cells exhibited higher sensitivity to 5-fluorouracil (P = 0.021; Fig. 3d, e). Importantly, similar trends were observed in the context of MDA-MB-175-VII cells, although without significance in some instances (Online Resource 6).
Fig. 3.
Wnt5a expression diminishes the sensitivity to tamoxifen, paclitaxel, and cyclophosphamide. MCF-7, MCF-7/Wnt5a (+), and MCF-7/Wnt5a (−) cells were exposed to 15 µM tamoxifen (a), 200 nM paclitaxel (b), 8000 µM cyclophosphamide (c), 800 nM epirubicin (d), or 400 µM 5-fluorouracil (e). Results are represented as the mean ± S.D. of 6 measurements. *P < 0.001; **P = 0.021 (Welch’s t-test); n.s. not significant
Additionally, we examined the difference in the RFS between Wnt5a-positive versus -negative breast cancer patients treated with the different above drugs. The RFS probability was lower in the Wnt5a-positive. However, used. However, no significant differences were detected, probably due to the small sample-size (Online Resource 7).
The PI3K signaling pathway is not correlated with Wnt5a expression
Next, the relationship between the expression of Wnt5a and the PI3K and JNK signaling pathways was examined via western blotting in MCF-7/Wnt5a (+) and MCF-7/Wnt5a (−) cells. The expression of phosphorylated JNK, which occurs downstream of the Wnt5a signaling pathway [2], remained unaltered in Wnt5a overexpressing or silenced cells (Fig. 4a). Similarly, there was no difference in the expression of phosphorylated AKT (Fig. 4b).
Fig. 4.
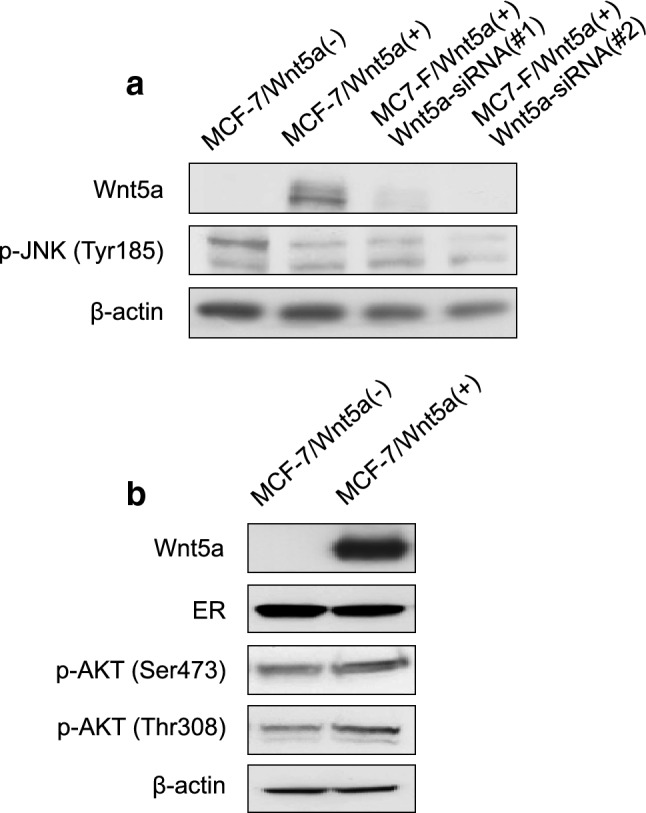
Effect of Wnt5a on the expression of breast cancer-related signaling molecules. The expression of phosphorylated JNK (a) and of phosphorylated AKT (b) was assessed via western blotting. ER estrogen receptor
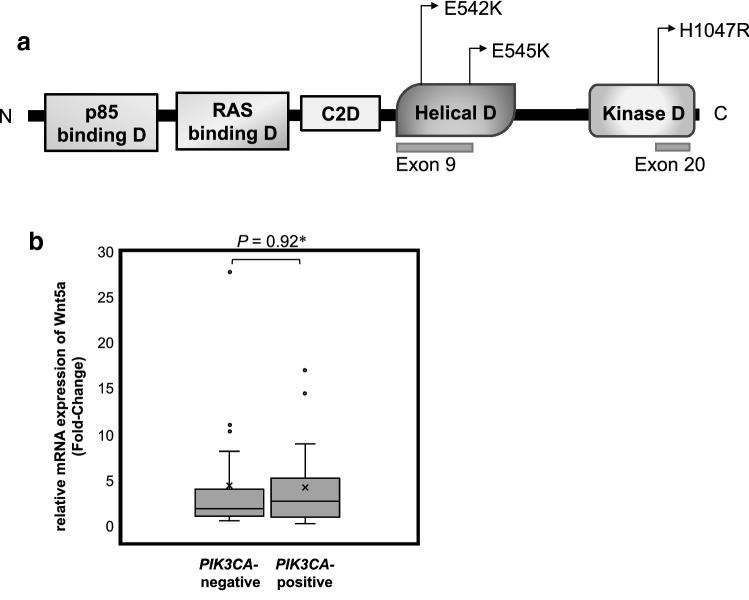
PIK3CA mutations were examined in 40 cases (Table 2) and detected in 19 cases of ER-positive breast cancers (Table 3); three principal mutation sites were identified: E542K, E545K, and H1047R [26] (Fig. 5a). Of note, PIK3CA mutations were observed in 8 and 11 Wnt5a-positive and -negative breast cancer patients, respectively. However, there was no significant difference in the frequency of PIK3CA mutations depending on the expression of Wnt5a (P = 0.73; Table 3). Furthermore, no difference in Wnt5a expression was observed depending on the mutation site (Table 4).
Table 2.
Characteristics of the 40 ER-positive breast cancer patients assessed for the PIK3CA status
| Total (n = 40) | |
|---|---|
| Age (median, range) | 58.5 (87–34) |
| ≦ 50 | 15 |
| > 50 | 25 |
| Tumor size | |
| pT1 ≦ 20 mm | 17 |
| pT2/pT3 > 20 mm | 23 |
| Lymph-node metastasis | |
| Negative | 25 |
| Positive | 15 |
| Progesterone receptor | |
| Negative | 2 |
| Positive | 38 |
| HER2 status | |
| Negative | 37 |
| Positive | 3 |
| Nuclear grade | |
| 1, 2 | 13 |
| 3 | 27 |
| Wnt5a expression (IHC) | |
| Wnt5a-negative | 21 |
| Wnt5a-positive | 19 |
IHC immunohistochemistry, HER2 human epidermal growth factor receptor 2
Table 3.
Wnt5a expression, assessed through immunohistochemistry (IHC), according to the PIK3CA mutation status
| Total (n = 40) | PIK3CA mutation | P-value | |
|---|---|---|---|
| Negative (n = 21) | Positive (n = 19) | ||
| Wnt5a expression (IHC) | |||
| Wnt5a-negative | 10 | 11 | |
| Wnt5a-positive | 11 | 8 | 0.73 |
Fig. 5.
a Occurrence of mutations within exons 9 and 20 of PIK3CA. b Wnt5a mRNA expression according to the PIK3CA mutation status (n = 40). The Wnt5a mRNA expression was examined in PIK3CA mutation-positive and -negative cancers. Statistical significance is highlighted: *P = 0.92 (Welch’s t-test)
Table 4.
PIK3CA mutation sites in ER-positive breast cancers patients (n = 19), as detected through the Sanger method
| Wnt5a expression (IHC) | PIK3CA mutation | P-value | ||
|---|---|---|---|---|
| Exon 9 | Exon 20 | |||
| E542K | E545K | H1047R | ||
| Wnt5a-negative | 0 | 6 | 4 | |
| Wnt5a-positive | 1 | 3 | 5 | 0.50 |
IHC immunohistochemistry
Furthermore, the expression of Wnt5a mRNA. The median (range) expression of Wnt5a mRNA was 1.7 (0.94 to 3.9) in PIK3CA mutation-negative and 2.5 (0.83–5.1) in PIK3CA mutation-positive cases; however, no significant difference was observed between the two groups (P = 0.92; Fig. 5b).
Discussion
The recurrence rate of Wnt5a-positive breast cancer patients is significantly higher than that of Wnt5a-negative breast cancer patients. Therefore, this study investigated the association between the expression of Wnt5a expression and malignancy grade and prognosis. Interestingly, pathway analysis revealed that the CYP metabolic pathway was upregulated after Wnt5a overexpression.
CYP is a key enzyme that oxidizes various substrates and primarily metabolizes drugs in the liver. In our study, CYP upregulation reduced the sensitivity to tamoxifen, paclitaxel, and cyclophosphamide (all metabolized by CYP). Conversely, the sensitivity to epirubicin and 5-fluorouracil (not metabolized by CYP) was not affected. These results suggest that Wnt5a enhances the tamoxifen, paclitaxel, and cyclophosphamide metabolism via CYP, thus decreasing their intracellular concentration. Therefore, the early termination of adjuvant drug therapy for breast cancer may lead to a high recurrence rate of Wnt5a-positive breast cancer. Importantly, as no prior study have shown the association between Wnt5a and CYP in any cancer, these results are novel.
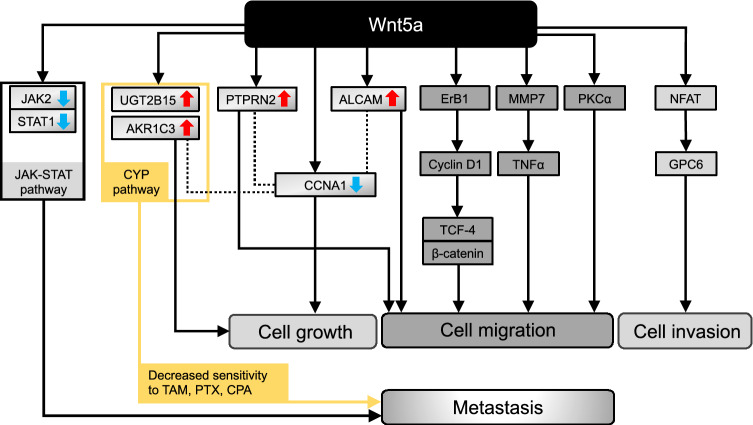
The genes associated in this study with Wnt5a in ER-positive breast cancer are summarized in Fig. 6 together with the previously reported genes [27–30]. Among the upregulated genes, UGT2B15 and AKR1C3 are included in the CYP metabolic pathway as per KEGG pathway analyses, suggesting the association between the CYP and Wnt5a pathways in the context of drug sensitivity [16, 17]. Interestingly, ALCAM and PTPRN2 were also shown to correlate with Wnt5a expression and to promote cell migration [8, 18].
Fig. 6.
The genes associated with Wnt5a in ER-positive breast cancer are summarized. The solid black lines represent the relationships already reported in the literature; the colored solid lines and the dotted lines and yellow line represent the relationships defined in this study. TAM tamoxifen, PTX paclitaxel, CPA cyclophosphamide
Additionally, the downregulation of the JAK-STAT signaling pathway was reported to contribute to breast cancer progression and metastasis [19]. Of note, it has been reported that the downregulation of CCNA1 leads to cell growth in the context of breast cancer [20]. Interestingly, here we determined that CCNA1 is co-expressed with ALCAM, PTPRN2, and AKR1C3, suggesting an association between them.
Based on our previous reports [12, 31, 32] we hypothesized that Wnt5a expression would be associated with the PI3K–AKT–mTOR signaling pathway. However, unexpectedly, no significant correlation was found. These results indicate the involvement of pathways other than PI3K in the recurrence of Wnt5a-positive breast cancer.
Altogether, our results indicate that Wnt5a potentially serves as a drug resistance marker in ER-positive breast cancer. However, further studies, in vivo, are essential to validate our results. Additionally, further studies are required for the development of new drugs targeting drug resistance markers; for instance, the development of a treatment strategy for Wnt5a-positive breast cancer based on anti-Wnt5a antibodies, in line with that developed for Wnt5a-positive gastric cancer [33] should be carried out.
In conclusion, here, we show that Wnt5a upregulates CYP expression and diminishes the sensitivity to key drugs used for treating ER-positive breast cancer, including tamoxifen, paclitaxel, and cyclophosphamide. Wnt5a is potentially involved in the poor prognosis of ER-positive breast cancer independently of the PI3K–AKT–mTOR signaling pathway.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Yusuke Motoi for his assistance with the experiments. We also thank Editage (www.editage.com) for English language editing. This study was funded by a Grant-in-Aid for Young Scientists B (Grant number: 17K16509) and a Grant-in-Aid for Scientific Research C (Grant number: 17K10549) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This study was also supported in part by Nanken-Kyoten, TMDU (2020-A25 to T.K.). The funding sources had no role in study design.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by AA, TK, and MI. The first draft of the manuscript was written by AA and all authors commented on the previous versions of the manuscript. All authors read and approved the final version of the manuscript.
Declarations
Conflict of interest
The authors have no competing interests to declare.
Ethical approval
The study was approved by the institutional review board of Hiroshima University Hospital (No. 926) and was performed in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all participants included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/28/2021
A Correction to this paper has been published: 10.1007/s12282-021-01268-3
References
- 1.Kikuchi A, Yamamoto H, Sato A. Selective activation mechanisms of Wnt signaling pathways. Trends Cell Biol. 2009;19:119–129. doi: 10.1016/j.tcb.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Cao Y, Wang X, Xu C, Gao Z, Zhou H, Wang Y, et al. 4-HPR impairs bladder cancer cell migration and invasion by interfering with the Wnt5a/JNK and Wnt5a/MMP-2 signaling pathways. Oncol Lett. 2016;12:1833–1839. doi: 10.3892/ol.2016.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurayoshi M, Oue N, Yamamoto H, Kishida M, Inoue A, Asahara T, et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and proliferation. Cancer Res. 2006;66:10439–10448. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
- 4.Huang C-L, Liu D, Nakano J, Ishikawa S, Kontan K, Yokomise H, et al. Wnt5a expression is associated with the tumor proliferation and the stromal vascular endothelial growth factor: an expression in non–small-cell lung cancer. J Clin Oncol. 2005;23:8765–8773. doi: 10.1200/JCO.2005.02.2871. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto H, Oue N, Sato A, Hasegawa Y, Yamamoto H, Matsubara A, et al. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene. 2010;29:2036–2046. doi: 10.1038/onc.2009.496. [DOI] [PubMed] [Google Scholar]
- 6.Kremenevskaja N, von Wasielewski R, Rao AS, Schöfl C, Andersson T, Braban G. Wnt-5a has tumor suppressor activity in thyroid carcinoma. Oncogene. 2005;24:2144–2154. doi: 10.1038/sj.onc.1208370. [DOI] [PubMed] [Google Scholar]
- 7.Bitler BG, Nicodemus JP, Li H, Cai Q, Wu H, Hua X, et al. Wnt5a suppresses epithelial ovarian cancer by promoting cellular senescence. Cancer Res. 2011;71:6184–6194. doi: 10.1158/0008-5472.CAN-11-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi Y, Kadoya T, Amioka A, Hanaki H, Sasada S, Masumoto N, et al. Wnt5a-induced cell migration is associated with the aggressiveness of estrogen receptor-positive breast cancer. Oncotarget. 2018;9:20979–20992. doi: 10.18632/oncotarget.24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng C, Zhang X, Yu H, Wu D, Zheng J. Wnt5a as a predictor in poor clinical outcome of patients and a mediator in chemoresistance of ovarian cancer. Int J Gynecol Cancer. 2011;21:280–288. doi: 10.1097/IGC.0b013e31820aaadb. [DOI] [PubMed] [Google Scholar]
- 10.Miller TW, Hennessy BT, González-Angulo AM, Fox EM, Mills GB, Chen H, et al. Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest. 2010;120:2406–2413. doi: 10.1172/JCI41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang SX, Polley E, Lipkowitz S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat Rev. 2016;45:87–96. doi: 10.1016/j.ctrv.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steelman LS, Navolanic P, Chappell WH, Abrams SL, Wong EWT, Martelli AM, et al. Involvement of Akt and mTOR in chemotherapeutic- and hormonal-based drug resistance and response to radiation in breast cancer cells. Cell Cycle. 2011;10:3003–3015. doi: 10.4161/cc.10.17.17119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cizkova M, Cizeron-Clairac G, Vacher S, Susini A, Andrieu C, Lidereau R, et al. Gene expression profiling reveals new aspects of PIK3CA mutation in ER alpha-positive breast cancer: Major implication of the Wnt signaling pathway. PLoS ONE. 2010;5:e15647. doi: 10.1371/journal.pone.0015647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasuda K, Ranade A, Venkataramanan R, Strom S, Chupka J, Ekins S, et al. A comprehensive in vitro and in silico analysis of antibiotics that activate pregnane X receptor and induce CYP3A4 in liver and intestine. Drug Metab Dispos. 2008;36:1689–1697. doi: 10.1124/dmd.108.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 17.Jia Y, Domenico J, Swasey C, Wang M, Gelfand EW, Lucas JJ. Modulated expression of genes encoding estrogen metabolizing enzymes by G1-phase cyclin-dependent kinases 6 and 4 in human breast cancer cells. PLoS ONE. 2004;9:e97448. doi: 10.1371/journal.pone.0097448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengelaub CA, Navrazhina K, Ross JB, Halberg N, Tavazoie SF. PTPRN2 and PLCβ1 promote metastatic breast cancer cell migration through PI(4,5)P2-dependent actin remodeling. EMBO J. 2016;35:62–76. doi: 10.15252/embj.201591973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(169–184):e7. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starlard-Davenport A, Kutanzi K, Tryndyak V, Word B, Lyn-Cook B. Restoration of the methylation status of hypermethylated gene promoters by microRNA-29b in human breast cancer: a novel epigenetic therapeutic approach. J Carcinog. 2013;12:15. doi: 10.4103/1477-3163.115720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coller JK, Krebsfaenger N, Klein K, Endrizzi K, Wolbold R, Lang T, et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol. 2002;54:157–167. doi: 10.1046/j.1365-2125.2002.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonnichsen DS, Liu Q, Schuetz EG, Schuetz JD, Pappo M, Relling MV, et al. Variability in human cytochrome P450 paclitaxel metabolism. J Pharmacol Exp Ther. 1995;275:566–575. [PubMed] [Google Scholar]
- 23.Chang TK, Weber GF, Crespi CL, Waxman DJ. Differential activation of cyclophosphamide and ifosphamide by cytochromes P-450 2B and 3A in human liver microsomes. Cancer Res. 1993;53:5629–5637. [PubMed] [Google Scholar]
- 24.Robert J. Clinical pharmacokinetics of epirubicin. Clin Pharmacokinet. 1994;26:428–438. doi: 10.2165/00003088-199426060-00002. [DOI] [PubMed] [Google Scholar]
- 25.Ichikawa W. Prediction of clinical outcome of fluoropyrimidine-based chemotherapy for gastric cancer patients, in terms of the 5-fluorouracil metabolic pathway. Gastric Cancer. 2006;9:145–155. doi: 10.1007/s10120-006-0373-8. [DOI] [PubMed] [Google Scholar]
- 26.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 27.Kim S, Chun SY, Kwon YS, Nam KS. Crosstalk between Wnt signaling and Phorbol ester-mediated PKC signaling in MCF-7 human breast cancer cells. Biomed Pharmacother. 2016;77:114–119. doi: 10.1016/j.biopha.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Civenni G, Holbro T, Hynes NE. Wnt1 and Wnt5a induce cyclin D1 expression through ErbB1 transactivation in HC11 mammary epithelial cells. EMBO Rep. 2003;4:166–171. doi: 10.1038/sj.embor.embor735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pukrop T, Klemm F, Hagemann T, et al. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc Natl Acad Sci USA. 2006;103:5454–5459. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yiu GK, Kaunisto A, Chin YR, Toker A. NFAT promotes carcinoma invasive migration through glypican-6. Biochem J. 2011;440:157–166. doi: 10.1042/BJ20110530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng L, Chen J, Zhong XR, Luo T, Wang YP, Huang HF, et al. Correlation between activation of PI3K/AKT/mTOR pathway and prognosis of breast cancer in Chinese women. PLoS ONE. 2015;10:e0120511. doi: 10.1371/journal.pone.0120511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanaki H, Yamamoto H, Sakane H, Matsumoto S, Ohdan H, Sato A, et al. An anti-Wnt5a antibody suppresses metastasis of gastric cancer cells in vivo by inhibiting receptor-mediated endocytosis. Mol Cancer Ther. 2012;11:298–307. doi: 10.1158/1535-7163.MCT-11-0682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.