Abstract
The mechanisms linking the function of microbes to host health are becoming better defined but are not yet fully understood. One recently explored mechanism involves microbe-mediated alterations in the host epigenome. Consumption of specific dietary components such as fiber, glucosinolates, polyphenols, and dietary fat has a significant impact on gut microbiota composition and function. Microbial metabolism of these dietary components regulates important epigenetic functions that ultimately influences host health. Diet-mediated alterations in the gut microbiome regulate the substrates available for epigenetic modifications like DNA methylation or histone methylation and/or acetylation. In addition, generation of microbial metabolites such as butyrate inhibits the activity of core epigenetic enzymes like histone deacetylases (HDACs). Reciprocally, the host epigenome also influences gut microbial composition. Thus, complex interactions exist between these three factors. This review comprehensively examines the interplay between diet, gut microbes, and host epigenetics in modulating host health. Specifically, the dietary impact on gut microbiota structure and function that in-turn regulates host epigenetics is evaluated in terms of promoting protection from disease development.
Keywords: Diet, Gut Microbiota, Epigenetics, Butyrate, HDACs, Metabolism
1. Introduction
Gut microbes, either independent of dietary considerations or in combination with nutritional interventions, have been demonstrated to mediate epigenetic modification in host organisms. Epigenetics involves the modification of genes through 1) covalent modifications to histone proteins such as histone acetylation, 2) DNA methylation including methylation to cytosine or adenosine residues, 3) expression of enzymes that regulate DNA methylation, such as DNA methyltransferase or ten-eleven translocation (TET), or histone acetylation including histone deacetylases (HDACs) and histone acyltransferases (HATs) thus altering chromatin structure, and 4) post-transcriptional regulation via noncoding RNAs [1,2]. Histones can undergo post-translational modifications (PTMs) such as methylation, acetylation, ubiquitination, phosphorylation and SUMOylation that contribute to the activation and repression of genes. For example, HAT-mediated acetylation of histone tails typically liberates chromatin allowing for gene transcription, whereas HDAC-mediated deacetylation strengthens the bond between DNA and histones preventing gene transcription [3]. Butyrate, a short chain fatty acid (SCFA) derived from microbial fermentation of non-digestible carbohydrates, is a well-studied example of diet-microbe-host epigenetic interaction that functions as an HDAC inhibitor [4,5]. Other microbe-epigenetic interactions that will be discussed in this review include microbe-mediated DNA methylation, and other examples of histone deacetylation and methylation. Interestingly, host-microbe interactions are reciprocal, meaning that changes in host epigenetics also influence the colonization and composition of the gut microbiota [3,6,7].
The aforementioned epigenetic modifications have been linked to fundamental physiological processes and a number of different diseases including obesity, diabetes, inflammatory bowel diseases (IBD), and cancer [1,2,8,9]. However, to date, the intricacies of these interactions have not been well-defined. The goal of this review is to comprehensively describe the complex interaction between diet, gut microbiota structure and function, and host epigenetics that together influence overall health. We begin by providing a brief overview of the general features of the gut microbiota including their contribution to host physiology, spatial organization, and major phyla of the gastrointestinal (GI) tract. Next, the dietary impact on gut microbiota will be examined, followed by a discussion on how microbes directly influence host epigenetics. Lastly, studies that have collectively examined the interplay between diet, gut microbes, and host epigenetics will be discussed focusing on specific food components including dietary fat, fiber, glucosinolates from cruciferous vegetables, polyphenols as well as the proposed effects of caloric restriction.
2. General Features and Roles of the Gut Microbiota and Intestinal Epigenetics
Human cells are matched in number by the microbiota which exist both on the surface of our bodies and within them [10]. Biomedical researchers have increasingly investigated the relationship between human host and microbe over the past few decades [11]. The microbiota of the GI system harbors great microbial mass and diversity that significantly contributes to physiological processes of the host including digestion and absorption of nutrients, immune development, protection against pathogens, and other important physiological functions [12].
The majority of gut microbes reside in the colon, but also colonize all regions of the gut, albeit at fewer numbers in the small intestine, thus having region-specific distribution [12]. Gut microbes exhibit spatial organization by way of harboring the gut lumen or mucosa [13]. In addition, the gut microbiota shifts in composition over the lifespan, increasing in diversity from birth to about two years of age with the introduction of whole foods and reducing in diversity with aging toward the end of life [14,15].
Concurrent with the establishment of the gut microbiota, host epigenetic processes play a vital role in the development of the gut including differentiation of intestinal stem cells into mature, fully functional cells [16-18]. Up- or downregulation of genes through modifications at DNA enhancers have been found when comparing Lgr5+ intestinal stem cells to the methylomes of their descendants [16,18]. More specifically, a study by Kaaij et al demonstrated enrichment of methylation at histone H3 lysine 4 and acetylation at histone H3 lysine 27 in villus epithelial tissue, and that such loci served as enhancers driving gene expression upon stem cell differentiation [16]. A different study conducted by Kim et al found enrichment of methylation at H3K4me2-related enhancers in secretory and enterocyte cell progenitors compared to Lgr5+ intestinal stem cells [18]. Some have also identified various changes in methylomes between intestinal stem cells and intestinal progenitors at promoters and introns, but other studies have found evidence against changes in histone and DNA methylation transcriptional start sites outside of promoters [16-18]. Alterations in transcriptomes from stem cells to progenitors can be traced back to resulting upregulation of tissue specific genes, like proteins needed for intestinal cell absorption, therefore reinforcing the notion that epigenetic modifications lead to intestinal cell differentiation [19].
Microbe-mediated epigenetic programming begins early in life, during which time the gut microbiota is developing itself. The development of the gut microbiota promotes epigenetic modification of intestinal epithelial cells, as demonstrated by studies using cell lines exposed to probiotics and pathogenic microbes. One such study found that immature fetal intestinal epithelial cells exposed to Lactobacillus acidophilus and Bifidobacterium infantis had significant alterations in DNA methylation at 92 regions of interest (ROIs) compared to untreated cells [6]. These cells also demonstrated DNA methylation changes at 180 ROIs when treated with Klebsiella spp., which is a pathogenic microbe present in infants who are born prematurely [6]. Taken together, epigenetic modifications may be dictated by exposure to a certain microbial milieu beginning at birth. The gut microbiota established in early life is shaped by diet beginning with formula or breast milk and continues throughout adulthood depending on one’s lifestyle and dietary choices.
The most abundant phyla of the GI tract include Actinobacteria, Bacteriodetes, Firmicutes, and Proteobacteria [12,14,20]. While these major phyla are shared across different host species and experimental subjects, a high level of inter-individual variability exists in gut microbiota composition at lower taxonomic levels in regard to their relative abundance and functional capacity. These differences can influence the types of microbial metabolites that are generated when faced with particular substrates as highlighted later in the review [21]. Some researchers postulate that all humans contain a core set of bacteria or bacterial functions necessary for host survival [22]. These functions include nutrient digestion and absorption, bile acid deconjugation, stimulation of the host immune system, metabolism of dietary fiber and generation of short chain fatty acids (SCFAs), and promotion of epithelial integrity [12]. However, gut dysbiosis, the deleterious re-structuring of gut microbiota composition, can disrupt these processes and contribute to the development of multiple pathologies including obesity, cardiovascular disease, type II diabetes, and nonalcoholic fatty liver disease [23]. As such, there has been increasing interest in how the gut microbiota causes such detriments. One explanation is that microbiota influence epigenetic modifications, which can greatly change how genes are expressed in host tissues. Notably, the structure and function of the gut microbiota is influenced by various features of the host diet including the caloric content, macronutrient ratios, and inclusion of bioactive food components.
3. Dietary Impact on the Gut Microbiota
3.1. Dietary influences in early life on the gut microbiota.
Dietary influences on the gut microbiota are speculated to begin in utero [24]. It has been long accepted that the intestines of fetuses are sterile, with microbial growth beginning after the newborn is seeded by bacteria in the environment. This theory has however been challenged by limited studies which have identified microbes from phyla Firmicutes, Tenericutes, Proteobacteria, Bacteriodetes, and Fusobacteria present in human placentas [25]. Additionally, it has been shown that subjects with excess gestational weight displayed an increased abundance of Firmicutes and decreased Proteobacteria along with decreased butyrate metabolism genes in the placenta [26]. Breast-fed versus formula-fed infants also have a markedly different gut microbiota. Healthy infants who were breast-fed for the first 6 months of life demonstrated significant increases in Actinobacteria (Bifidobacterium) and Proteobacteria (Enterobacteriaceae) in their feces from 4 days of age to 120 days [27]. The members of Firmicutes in these same infants also showed significant change, with increases in Veillonella, Lachnospiraceae, Clostridium perfringens, and Lactobacillus but decreases in Staphylococcus and Streptococcus [27]. In contrast, infants who are exclusively formula fed were found to have less abundance in Bifidobacterium than breast fed infants [28]. Longer durations of breast feeding also favored proliferation of Bifidobacterium and Veillonella and decreased abundance of Lachnospiraceae, Ruminococcaceae, and other less prevalent microbes [14]. As solid foods are introduced, the intestinal microbiota in infants continues to shift. Greater microbial richness based on Shannon alpha diversity indexes was seen in 9-month olds with diets higher in protein and fiber and lower in fats [14]. A positive correlation was also found between the amount of “family foods” (defined by this study as foods typically introduced during late infancy) consumed and alpha diversity in their feces [14]. The family foods specifically associated with increased fecal Shannon diversity were porridge, rye bread, vegetable fats, cheese, and dairy milk based on a seven day food record taken by the infants’ parents [14]. Consumption of breast milk had the opposite effect, with these 9-month old infants having a negative correlation between amount of breast milk consumed and alpha diversity [14]. On a macronutrient level, the 9-month old children demonstrated a positive correlation between protein intake and fecal abundance in family Lachnospiraceae, but a negative correlation with Bifidobacteriaceae and protein [14]. Dietary fiber was positively correlated with Pasteurellaceae [14]. This study thus demonstrates that the increase in gut microbial diversity from infancy to childhood is greatly influenced by shifts in diet.
3.2. Dietary impact of plant-based vs. animal-based diets on the gut microbiome.
Differences in gut microbiota composition and function have been found when comparing vegetarian vs omnivorous diets in adults. Long-term vegetarians contain a greater percent composition of Bacteriodes-Prevotella, Bacteriodes thetaiotamicron, and Clostridium clostridioforme but lesser percent of Clostridium coccoides in their feces compared to omnivores [29]. Vegan diets, lacking in all forms of animal products, also promote greater proliferation of Bacteriodaceae than omnivorous diets, but not to the extent of vegetarians based on abundance of OTU counts [30]. A significant increase in alpha diversity in the feces of vegetarians compared to omnivores based on number of OTUs observed and Chao1 analysis was also noted, but interestingly there was no significant difference in diversity between vegans and omnivores. Analysis of nutritional profiles provided by the participants in this study demonstrated a significantly greater caloric intake of carbohydrates by vegetarians compared to omnivores, which may account for the difference in microbial diversity. Although vegan participants also had greater carbohydrate consumption than the omnivores, this comparison did not reach significance. The vegans and vegetarians both had lesser intake of proteins than the omnivores. No difference in lipid consumption by these three groups was found [30].
Altering the amount of fat in the diet also influences gut microbiota composition. In a study comparing sedentary high-fat and low-fat diet fed mice, the high-fat fed mice were found to have an increase in Clostridiaceae and a decrease in Bifidobacteriaceae abundance [31]. In addition, a significant increase in Lachnospiraceae and Ruminococcaceae was reported but significant decreases in Turicibacteraceae and Erysiplotrichaceae abundance. The type of fat consumed also dictates changes observed in the gut microbiota. For example, Huang et al. showed differences in microbial composition in mice fed diets rich in polyunsaturated fatty acids (PUFAs) versus saturated fatty acids from either milk fat- vs lard-based diets compared to a low fat (LF) control diet [32]. Here it was found that all high fat diets promoted an increase in Firmicutes abundance compared to the LF diet. In particular, the lard-based diet elicited an expansion of the phyla Tenericutes and reduction in Proteobacteria compared to the other high fat diets. In a separate study, Devkota et al. (2012) found that a diet high in saturated milk fat promoted the abundance of Proteobacteria compared to a lard-based diet [33].
In addition to changes in microbial composition, high fat diets alter the metagenome and functional capacity of gut microbes. For example, David et al. showed that consumption of an animal-based diet in human participants resulted in the production of bile shifting the structure of the gut microbiome, particularly an increase in bile-tolerant microorganisms such as Bilophila wadsworthia as well as bile salt hydrolase gene expression based on metagenomic analyses [34]. Notably, the outgrowth of this microbe has been linked to inflammatory bowel disease [33]. The shift of structure in the animal-based diet was evident by the decreased the level of Firmicutes (microbes that digest plant polysaccharides) and increased the level Lactococcus lactic, Staphylococcus carnosus, Pediococcus acidilactici, Penicillium sp., and certain fungi [34]. This animal-based diet showed a shift of microbes’ fermentation preference, from carbohydrates to amino acids.
In addition to dietary fat, polyphenols from fruits such as grapes, blueberries, cranberries, and cruciferous vegetables have been demonstrated to significantly alter the gut microbiota and improve metabolic health. For example, mice fed lyophilized table grapes [35] promoted the abundance of microbes such as Akkermansia muciniphila, which has been associated with leanness [36]. Grape-derived polyphenols were also found to decrease high fat (HF)-diet mediated increase in dissimilatory sulfite reductase A (dsrA) gene abundance which is indicative of an outgrowth of sulfidogenic bacteria like B. wadsworthia belonging to the phylum Proteobacteria [37]. Blueberry polyphenols have been found to reduce HF diet-induced weight gain and altered the gut microbiota in mice [38]. In a double-blind crossover placebo-controlled trial, cranberry supplementation in individuals fed an animal-based diet, protected against reductions in SCFA levels and increases in cancer-promoting secondary bile acids lithocholic and deoxycholic acid [39]. Broccoli consumption in humans has been shown to increase the abundance of Firmicutes and decrease the abundance of Bacteroidetes [40]. Glucosinolates found in broccoli are converted into isothiocyanates (ITCs) by bacteria that regulate host epigenetics. Interestingly, ITC production is dependent on the functional capacity of individual microbiomes [21]. Altogether, berry-derived polyphenols and cruciferous vegetables have the capacity to alter the gut microbiota in beneficial ways.
3.3. Caloric restriction impacts the gut microbiota.
Differences in caloric intake also promote shifts in microbial community structure. Obese adolescents who lost over 4.0 kg after 10-weeks of low-calorie diet and exercise had greater abundance of Clostridium coccoides and less abundance of Bacteroides fragilis prior to lifestyle intervention, although it is unclear if diet alone caused the changes seen in this study [41] Additionally, lifelong caloric restriction in mice has been shown to result in changes to the gut microbiota. Mice placed on a 30% caloric restriction showed a decrease in the Firmicutes/Bacteriodetes ratio in their feces from age 62 to 141 weeks, even if the mice were fed high-fat diet versus low-fat diet [42]. These calorie restricted mice showed greater abundance of genera associated with longer-lifespan like Lactobacilli and less abundance in those associated with shorter-lifespan like Lactococcus, Bacteriodales, and Peptostreptococcaceae. Again, these findings were regardless of exercise or dietary fat intake [42]. Even short-term caloric restriction can change the microbiome. For instance, mice on a 2 month long 30% caloric restriction exhibited a significant decrease in relative percent abundance of Firmicutes and an increase in Bacteriodetes, lending to a decrease in the Firmicutes/Bacteriodetes ratio compared to mice with free access to food [43]. Taken together, alterations in dietary composition from macronutrient ratios, type of fat, polyphenolic content as well as the caloric content of the diet can promote structural and functional changes in the gut microbiota.
4. Impact of Gut Microbes on Host Epigenetics
The influence of the gut microbiota on epigenetics has been a new and exciting area of research, as it may explain how microbes can cause disease processes in the GI tract or conversely provide protection from disease. Changes in the concentration of luminal nutrients, such as SCFAs produced by microbiota, result in post-translational alterations to DNA and histones [44]. In general, epigenetic modification allows the body to change gene expression to adapt to various environmental factors without altering the structure or content of said gene [9]. These modifications are reversible; however, they are most often retained even after mitotic or meiotic division. Epigenetic modifications include addition or removal of acetyl, methyl, or crotonyl groups to lysine residues on histones, and methylation of CpG islands on DNA by DNA methyltransferases (DNMTs) [9,45]. Gut microbes have been shown to affect each of these processes resulting in altered physiological function of the host.
4.1. Histone Modifications
Histone crotonylation, which alters chromatin sensitivity to other modifiers, increases in the presence of microbe-derived SCFAs [46]. One study found that colonic epithelial cells with crotonylated histones were more likely to be in S or G2-M phase rather than G1 phase, suggesting it has an effect on the cell cycle [46]. Moreover, crotonylation was significantly decreased in the colons of mice administered antibiotics [46]. In addition to SCFAs, intestinal microbes produce and secrete cofactors for histone acetylation and methylation such as cobalt, iodine, selenium, zinc and other metabolites [47]. Histone acetylation results in gene upregulation, however histone methylation can result in either increased or decreased expression depending on the lysine residue in question [47]. Histone methylation and acetylation has been demonstrated to be dependent on the gut microbiota. For instance, germ-free (GF) mice have significantly less methylation and acetylation at multiple histones compared to conventionally raised (ConvR) mice or conventionalized mice (ConvD, GF mice that received cecal content transfer from conventional mice) [48]. More specifically, acetylation at H4 increased up to 12-fold and methylation on H3 up to 1.5-fold in colonized mice depending on lysine residue, tissue type, and method of colonization [48]. The greater amount and variety of microbes available to produce SCFAs can thereby influence acetylation and methylation and is a possible cause for this observed difference, as colonized mice had significantly higher amounts of acetate, propionate, and butyrate in their cecal contents than did the GF mice [48]. SCFAs produced by microbes during fermentation of carbohydrates have also been shown to inhibit histone deacetylases, further supporting their role in epigenetics [47].
4.2. DNA Methylation
In addition to histone modifications, the gut microbiota also causes epigenetic modification to DNA itself. DNA methylation can have varying results depending on location, with methylation at gene promoters resulting in silencing and methylation at the gene body causing activation [47]. Relative abundance of Firmicutes and Bacteriodetes correlates with methylation of DNA promotors, with increased presence of Firmicutes resulting in the promotion of several obesogenic genes [49]. Although endogenous factors largely control DNA methylation, the presence of pathogenic bacteria can especially disrupt typical methylation and cause disease. A notable example of this is Helobacter pylori infection, which causes aberrant DNA methylation in gastric mucosal cells [8]. More specifically, one study found that of the 276,831 genetic blocks investigated, 16.5% were hypermethylated and 18.9% were hypomethylated in young H. pylori positive humans compared to young healthy humans [8].
A possible result of aberrant DNA methylation is change in miRNA profiles found within gastric cells, which has been implicated in digestive disorders such as atrophic gastritis. For example, Watari et al (2019) found that genes encoding for certain miRNAs were associated with increased risk of gastric cancer in H. pylori positive patients with atrophic gastritis [50]. This study found that patients with active H. pylori infections and gastric cancer had significantly increased methylation at miR-34c in the antrum and corpus of the stomach and at miR-129-2 in the corpus compared to those with active H. Pylori infection without gastric cancer. Patients with gastric cancer and previously eradicated H. Pylori had increased methylation at miR-129-2 in the angulus of the stomach than those with no gastric cancer but previously eradicated H. Pylori. Overall, eradication of H. pylori was found to significantly decrease methylation at miR-124a-3 in non-cancerous mucosal tissue from patients with or without gastric cancer. These results exemplify the benefit of identifying aberrant epigenetic modifications, which could serve as biomarkers for diagnosis and/or targets for disease treatment. One way to combat these deleterious interactions is through dietary modification. For instance, microbe-mediated metabolism of dietary fiber leads to the production of SCFAs that through altering the host epigenome is protective against various disease states (as described in the next section).
5. Impact of Host Epigenetics on the Gut Microbiome
In a reciprocal fashion to gut microbe-mediated changes to the host epigenome, alterations in host epigenetic machinery also dictate the composition of the gut microbiota. For example, Alenghat et al. (2013) demonstrated that mice with HDAC3 deficiency in intestinal epithelial cells (IECs) displayed significantly altered gut microbiota composition including a significant expansion of Proteobacteria and Defferibacteres compared to HDAC3F/F mice [7]. There was also an increase in Firmicutes abundance, but this was not significant. In addition, HDAC3-IEC deficient (HDAC3▵IEC) mice exhibited altered gene expression, histone acetylation, and decreased intestinal barrier function. After these mice were re-derived germ free, they were protected from intestinal barrier dysfunction [7]. Therefore, this study showed both the importance of the gut microbiota in contributing to intestinal barrier health as well as the impact of host epigenetic machinery on gut microbial ecology.
Cortese et al. [6] investigated host-microbe cross-talk in a model of neonatal necrotizing enterocolitis (NEC), an inflammatory bowel disease that affects premature infants. In this study, it was found that prenatal treatment with the glucocorticoid dexamethasone altered the epigenome of the host and the microbiota composition including an increase in Firmicutes and decrease in Bacteroidetes. In addition, IECs were treated with probiotic and pathogenic bacteria, which elicited DNA modifications in over 200 regions [6]. These studies highlight the reciprocal interactions between host epigenome and the gut microbiota [6,7].
6. Diet-Microbe Interactions that Regulate the Host Epigenome
As evidenced by several studies, the host’s epigenome can both affect and be affected by the gut microbiota and that the gut microbiota can be regulated through diet. Introduction of specific nutrients allows for a transient shift of the gut microbiome with reciprocal effects on host health [34]. A number of dietary food components, including dietary fiber, glucosinolates, dietary lipids, and polyphenols, are metabolized by intestinal bacteria and impact host epigenetics, thereby influencing health and disease. The resultant metabolic byproducts and their specific impact on epigenetic machinery are examined below, including a brief discussion on caloric restriction.
6.1. Dietary Fiber and Short Chain Fatty Acids
Research has shown that SCFAs represent a primary energy source for colonocytes [4]. The production of SCFAs is mediated by gut microbes through fermentation of dietary fibers or non-digestible carbohydrates, with levels ranging from 20-140 mmol in the GI tract [51]. SCFAs have been shown to influence many host processes such as energy balance and protection from inflammatory bowel diseases (IBD) and colon cancer. One proposed mechanism of butyrate-mediated protection in colon cancer and IBD is HDAC inhibition. Studies have shown that butyrate-mediated inhibition of HDAC activity led to decreased epithelial cell proliferation and increased cell death [5,51]. Donohoe et al. [5] found that mice supplemented with B. fibrisolvens while being fed a high fiber diet (rich in inulin) were protected from AOM/DSS-induced tumor growth compared to control groups. Due to the Warburg effect, butyrate accumulates in tumor cells thus increasing HDAC inhibition. Consistently, it was found that histone acetylation was increased with high fiber diet and B. fibrisolvens along with increased Caspase 3 activity and Ki-67 assays indicating increased apoptosis and decreased proliferation, respectively. Furthermore, elevated butyrate and histone acetylation was detected in human colorectal adenocarcinoma biopsies [5]. Another study established an association between butyrate-mediated HDAC inhibition and weight loss [52]. First, it was demonstrated that HDAC3 deficiency in IECs (HDAC▵IEC) protected mice from diet-induced obesity. Secondly, it was found that butyrate-mediated weight loss was dependent on HDAC3 as the protective effects of butyrate were lost in the HDAC3▵IEC mice [52]. As previously mentioned Krautkramer et al. (2016) demonstrated that ConvR and ConvD mice had significantly greater histone acetylation compared to GF mice [48]. It was further demonstrated that a western diet consisting of high fat and high sugar decreased SCFA production and decreased histone acetylation in the colon, liver, and white adipose tissue. Introducing SCFAs to GF mice increased histone acetylation and methylation revealing a direct role of microbial metabolites on the host epigenome [48]. These lines of evidence involving the production of butyrate by microbes can influence host epigenetics leading to a significant positive impact on preventing cancer and obesity.
6.2. Cruciferous Vegetables and Isothiocyanates
Isothiocyanates (ITCs) are generated from glucosinolates through myrosinase activity upon chewing and chopping of plants and through bacterial metabolism [53]. Bacteria with thioglucosidase activity are able to convert glucosinolates from cruciferous vegetables, such as turnips, kale, broccoli, cauliflower, and brussels sprouts, into ITCs through hydrolyzation in the colon [54]. Consumption of about 200g of broccoli for 18 days altered the composition of the gut microbes by decreasing the Firmicutes population by 9% and increasing Bacteroidetes abundance by 10% in human subjects [40]. Li et al. (2011) reported that the level of ITCs produced following broccoli consumption is highly dependent on the functional capacity of individual microbiomes as much interindividual variability exists in gut microbiota composition and function in humans [21]. These findings could have important implications for dietary recommendations in oncology, as ITCs have been shown to reduce tumor growth via altering DNA methylation and histone acetylation [51].
One type of ITC that is metabolized by myrosinase is sulforaphane. This molecule has been shown to have protective effects against pancreatic cancer, colon cancer, leukemia and prostate cancer [55-58]. One proposed mechanism is through sulforaphane-mediated inhibition of HDAC activity [56]. Myzak et al demonstrated that sulforaphane inhibited HDAC activity both in vitro and in vivo using colon cancer cells and APCmin mice, respectively [57,58]. Moreover, sulforaphane treatment in the APCmin mice protected against tumor development. Altogether, microbial-mediated production of ITCs represents a strong diet-microbe interaction that has a direct impact on the host epigenome and health.
6.3. Polyphenols
Polyphenols are natural molecular compounds that can be found in vegetables and fruits, as well as cereal, wine, tea, and coffee [59]. Polyphenols have been investigated for having an antimicrobial role [60] and also acting as prebiotic substrates for microbial enzymatic activity thereby generating bioactive byproducts that stimulate host epigenetic responses [59]. For example, phytochemicals curcumin, epigallocatechin gallate (EGCG), and resveratrol result in altered microbiota composition that feeds forward on the host epigenome [61]. First, curcumin was shown to improve colonic cell function through inhibiting HDAC activity [62,63]. McFadden et al. (2015) demonstrated using an AOM/IL10−/− model that curcumin prevented colonic tumor development that was associated with improved microbial diversity [63]. EGCG was reported to reduce high fat diet-induced metabolic derangements through increasing DNA methyltransferase 1 (DNMT1) expression and subsequent CpG hypomethylation in the colon [64]. In addition, it was found in colon cancer cells that EGCG inhibited HDAC activity and reduced CpG hypermethylation [65]. Resveratrol supplementation in men with metabolic syndrome significantly altered community composition of the fecal microbiota including increased the abundance of Akkermansia muciniphila [66]. In addition, a relationship has been reported between elevated sirtuin 1 expression and reduced pro-inflammatory cytokines in resveratrol-treated mice with colitis [67]. However, less information is available regarding a role for resveratrol in regulating host epigenetic responses upon gut microbe transformation.
Flavonoids found in plants and fruits like naringenin and hesperetin have been studied to observe the benefits polyphenols have on the host through microbial interaction in the colon [59,68]. When consumed at the recommended amount of 0.15 – 1.0 grams/day, flavanones have been shown to demonstrate anti-inflammatory and anticancer properties [68]. Narigenin and hesperetin have been shown to inhibit DNMT activity in human squamous carcinoma cells [69]. Collectively, various phytochemicals have been demonstrated to both alter gut microbiota composition and host epigenetic function.
6.4. Dietaiy Fat
Dietary fat, microbe, and host epigenetic interactions have been implicated in the development of obesity as well as the prevention of colon cancer depending on the type of fat delivered. Researchers demonstrated that HDAC3 expression in intestinal epithelial cells is dependent on gut microbes and that HDAC3 mediates the absorption of fat leading to obesity when animals are fed a HF diet as HDAC3-IEC deficient mice had decreased fat absorption [70]. This IEC-specific disruption of HDAC3 was shown to protect the host against obesity, glucose intolerance, and elevated plasma lipids in a murine diet-induced obesity (DIO) models [70]. HDAC3 gene and protein levels were significantly reduced in GF mice, indicating the importance of microbes in regulating HDAC3 levels. Notably, GF mice remain lean on a HF diet supporting the findings that HDAC3 is a mediator of obesity. Additionally, it was found that HDAC3 controls diurnal rhythmicity of genes involved in nutrient transport including the long chain fatty acid transporter Cd36 [70]. These findings are in line with the findings by Donohoe et al. (2014) in which butyrate protected against HF-diet induced obesity through acting as an HDAC inhibitor, as previously described [5]. Deleterious metabolic effects have also been reported for long chain omega 6 polyunsaturated fatty acids (PUFAs) such as linoleic acid that is converted by bacteria including Roseburia, Bifidobacteria, and Lactobacillus into conjugated linoleic acid (CLA) [51]. Daily CLA supplementation has been shown to alter CpG methylation in HF diet fed mice [71]. Similarly, CLA increased CpG methylation in the promoter region of proopiomelanocortin (POMC) in pups resulting in increased food intake and metabolic disorders in adulthood [72].
On the other hand, consumption of specific fatty acids such as omega 3 PUFAs have been shown to have positive impacts on host health including reduced risk of colorectal cancer [73]. For example, it was demonstrated that fish oil supplementation resulted in fewer tumors induced by azoxymethane (AOM) and decreased miRNA expression compared to omega 6 fatty acids [74]. A direct role of microbes in omega 3 fatty acid-mediated protection through epigenetic alterations is unclear, however. Taken together, long chain PUFAs have been reported to directly impact host epigenetics protecting against colon cancer and bacterial metabolic byproducts such as CLA trigger epigenetic alterations, influencing metabolic health.
Other studies have linked high fat diet consumption with epigenetic modification and changes in intestinal cell differentiation, but without investigating if alterations in the gut microbiota were the cause [75,76]. For example, high fat diet induced intestinal stem cell proliferation and regeneration in mice, but reduced differentiation of these cells causing development of shorter villi in the small intestine via upregulation of the transcription factor PPAR-δ [75]. A ketogenic diet characterized by high-fat, low-carbohydrate consumption increased the histone deacetylase inhibitor beta-hydroxybutyrate, which also causes enhanced replication of intestinal stem cells as demonstrated by increased numbers and depth of crypts [76]. This upregulation and resulting increased cell proliferation can therefore cause tumorigenesis, showing a possible link between obesity, high fat diet, and intestinal cancers [75].
6.5. Caloric Restriction
While much evidence exists to establish a connection between caloric restriction and epigenetics and also between caloric restriction and altered gut microbiota composition in improving metabolic health, a direct connection between the three together has not been explored. Fabbiano et al. (2018) demonstrated that short term caloric restriction of 3 days altered the gut microbiota including an increase in the abundance of Verrucomicrobia [77]. Notably, Akkermansia muciniphila, belonging to the phylum Verrucomicrobia, has been reported to promote leanness in a DIO animal model [36]. Subsequent fecal microbiota transplant from calorically-restricted mice improved the metabolic phenotype and browning of subcutaneous inguinal fat including the upregulation of uncoupling protein 1 (Ucp1) compared to control [77]. Indeed, the UCP1 promoter region has been shown to be regulated via DNA methylation and cold exposure was also shown to remodel chromatin facilitating UCP1 gene transcription [78]. A study by Mihaylova et al. revealed that organoids derived from the intestinal crypts of mice fasting for 24 hours were more numerous and propagated better than those from ad libitum fed mice owed to the upregulation of transcription factor PPARδ [79]. This increase in PPARδ resulted in the upregulation of the fatty acid oxidation genes Cpt1a, Pdk4, and Hmgcs2, that presumably contributed to enhanced proliferation seen in the fasting mice. Not only was this effect seen in mice age 10-12 weeks, but also in mice ages 18-22 months, with more organoids forming per crypt used from the old fasting mice than the old ad libitum-fed mice. While all three factors were not collectively examined in the study by Fabbiano et al [77] or Mihaylova et al [79], an interaction between caloric restriction, altered gut microbiota composition and function, and host epigenetic responses related to improved metabolism represents an interesting avenue for future research.
7. Conclusion
Many studies discussed in this review have implicated the relationship between the microbiome, diet, and epigenetics, yet other studies argue against such relationships. For instance, Camp et al demonstrated that microbiota modulate host transcription in various regions of the gut without remodeling chromatin, thus exemplifying the need for more research on this topic [80]. We have only scratched the surface to understand what food components drive changes in the gut microbiome that allows for communication with host epigenetic pathways and the mechanisms involved. What is understood is that these interactions are complex, involve various food components, a number of epigenetic alterations, and reciprocal responses from the host that feedback on the gut microbiota. The epigenetic response to diet is also influenced by the resident microbiota of the host that dictate the composition of metabolic byproducts. Further investigation into diet-microbe-host interactions may lead to the development of therapies to promote human health.
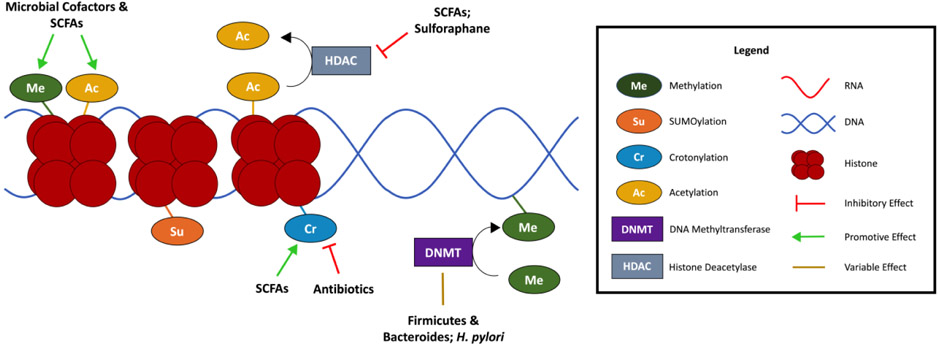
Figure 1. Microbial regulation of epigenetic machinery.
Microbial factors facilitate histone modification such as histone methylation or acetylation. Short chain fatty acids (SCFAs) derived from microbial metabolism of non-digestible carbohydrates directly inhibit histone deacetylases (HDACs). SCFAs also promote histone crotonylation. Microbes impact DNA methyltransferase (DNMT) activity resulting in altered DNA methylation patterns. Altogether, microbe-mediated changes in the epigenome influences host health and disease.
Table 1.
A summary of the effect of the various dietary components or caloric restriction on the gut microbiota.
| Dietary Component/Lifestyle |
Microbiota Change (Phylum, Family, or Genus level) |
References |
|---|---|---|
| Fiber | ↑ Pasteurellaceae ↑ Roseburia, ↑ Eubacterium rectale ↑ Ruminococcus bromii |
[14,34] |
| Protein | ↑ Lachnospiraceae ↓ Bifidobacteriaceae |
[14] |
| Dietary Fat | ↑ Clostridiaceae ↑ Lachnospiraceae ↑ Ruminococcaceae ↓ Bifidobacteriaceae ↓ Turicibacteraceae ↓ Erysiplotrichaceae |
[31,34] |
| Polyphenols & Glucosinolates | ↑ Akkermansia muciniphila ↑ Bacteriodetes ↓ Firmicutes |
[36,37,40] |
| Caloric Restriction | ↑ Bacteroides fragilis ↑ Lactobacilli ↑ Bacteriodetes ↓ Clostridium coccoides ↓ Lactococcus ↓ Bacteriodales ↓ Peptostreptococcaceae ↓ Firmicutes ↓ Firmicutes/Bacteriodetes ratio |
[41,42] |
| Vegetarian Diet | ↑ Bacteriodes-Prevotella ↑ Bacteriodes thetaiotamicron ↑ Clostridium clostridioforme ↓ Clostridium coccoides |
[29] |
| Vegan Diet | ↑ Bacteriodaceae | [29] |
| Animal-Based Diet | ↑ Bilophila wadsworthia ↑ Lactococcus lactic ↑ Staphylococcus carnosus ↑ Pediococcus acidilactici ↑ Penicillium sp. ↓ Firmicutes |
[34] |
| Breast Milk | ↑ Bifidobacterium ↑ Enterobacteriaceae ↑ Veillonella ↑ Lachnospiraceae ↑ Clostridium perfringens ↑ Lactobacillus ↓ Staphylococcus ↓ Streptococcus ↓ Lachnospiraceae ↓ Ruminococcaceae |
[14,27,28] |
Highlights.
Diet has a dramatic impact on gut microbiota composition and function
Microbial metabolism of dietary food components triggers epigenetic changes in the host
Microbial metabolite-mediated epigenetic alterations influence host health and disease
Acknowledgments
Funding: This work was supported by the US Department of Agriculture NIFA, HATCH-NEV00767; the National Institute for General Medical Sciences (NIGMS) of the NIH (P20 GM130459); the National Heart, Lung and Blood Institute of the NIH (R15 HL143496); and the National Science Foundation EPSCOR Track II, (OIA-1826801) to Bradley Ferguson. Funding was also received from Midwestern University for student research projects to Tori Shock and Luis Badang.
Grants, Sponsors, Funding Sources
Midwestern University Internal Funding for Student Research
B.S.F was funded by the US Department of Agriculture NIFA, HATCH-NEV00767; the National Institute for General Medical Sciences (NIGMS) of the NIH, P20 GM130459; the National Heart, Lung and Blood Institute of the NIH, R15 HL143496; and the National Science Foundation EPSCOR Track II, OIA-1826801.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab 2019;29:1028–44. 10.1016/j.cmet.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rosen ED, Kaestner KH, Natarajan R, Patti ME, Sallari R, Sander M, et al. Epigenetics and epigenomics: Implications for diabetes and obesity. Diabetes 2018;67:1923–31. 10.2337/db18-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alenghat T. Epigenomics and the microbiota. Toxicol Pathol 2015;43:101–6. 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Donohoe DR, Garge N, Zhang X, Sun W, Connell TMO, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 2011;13:517–26. 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov 2014;4:1387–97. 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cortese R, Lu L, Yu Y, Ruden D, Claud EC. Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease. Epigenetics 2016;11:205–15. 10.1080/15592294.2016.1155011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alenghat T, Osborne LC, Saenz SA, Kobuley D, Ziegler CGK, Mullican SE, et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature 2013;504:153–7. 10.1038/nature12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yamashita S, Nanjo S, Rehnberg E, Iida N, Takeshima H, Ando T, et al. Distinct DNA methylation targets by aging and chronic inflammation: A pilot study using gastric mucosa infected with Helicobacter pylori. Clin Epigenetics 2019;11:1–14. 10.1186/s13148-019-0789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tabatabaiefar MA, Sajjadi RS, Narrei S. Epigenetics and common non communicable disease. Adv Exp Med Biol 2019;1121:7–20. 10.1007/978-3-030-10616-4_2. [DOI] [PubMed] [Google Scholar]
- [10].Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016;14:1–14. 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huang X, Fan X, Ying J, Chen S. Emerging trends and research foci in gastrointestinal microbiome. J Transl Med 2019;17:1–11. 10.1186/s12967-019-1810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martinez-Guryn K, Leone V, Chang EB. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 2019;26:314–24. 10.1016/j.chom.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: Connecting spatial organization to function. Cell Host Microbe 2017;21:433–42. 10.1016/j.chom.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Laursen MF, Andersen LBB, Michaelsen KF, Mølgaard C, Trolle E, Bahl MI, et al. Infant gut microbiota development Is driven by transition to family foods independent of maternal obesity. MSphere 2016;1:1–16. 10.1128/msphere.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim S, Jazwinski SM. The gut microbiota and healthy aging: A mini-review. Gerontology 2018;64:513–20. 10.1159/000490615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kaaij LTJ, van de Wetering M, Fang F, Decato B, Molaro A, van de Werken HJG, et al. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol 2013;14. 10.1186/gb-2013-14-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kazakevych J, Sayols S, Messner B, Krienke C, Soshnikova N. Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res 2017;45:5770–84. 10.1093/nar/gkx167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim TH, Li F, Ferreiro-Neira I, Ho LL, Luyten A, Nalapareddy K, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 2014;506:511–5. 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Raab JR, Tulasi DY, Wager KE, Morowitz JM, Magness ST, Gracz AD. Quantitative classification of chromatin dynamics reveals regulators of intestinal stem cell differentiation. Dev 2020;147. 10.1242/dev.181966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Riaz Rajoka MS, Shi J, Mehwish HM, Zhu J, Li Q, Shao D, et al. Interaction between diet composition and gut microbiota and its impact on gastrointestinal tract health. Food Sci Hum Wellness 2017;6:121–30. 10.1016/j.fshw.2017.07.003. [DOI] [Google Scholar]
- [21].Li F, Hullar MAJ, Beresford SAA, Lampe JW. Variation of glucoraphanin metabolism in vivo and ex vivo by human gut bacteria. Br J Nutr 2011;106:408–16. 10.1017/S0007114511000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Publ Gr 2015;14:20–32. 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Newsholme P, De Bittencourt PIH. Gut associated bacteria are critical to metabolism, inflammation and health. Curr Opin Clin Nutr Metab Care 2016;19:245–9. 10.1097/MCO.0000000000000293. [DOI] [PubMed] [Google Scholar]
- [24].Chu DM, Meyer KM, Prince AL, Aagaard KM. Impact of maternal nutrition in pregnancy and lactation on offspring gut microbial composition and function. Gut Microbes 2016;7:459–70. 10.1080/19490976.2016.1241357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J, et al. The placenta harbors a unique microbiome. Sci Transl Med 2014;6:1–22. 10.1126/scitranslmed.3008599.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obs Gynecol 2015;212:653. 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Eggesbø M, Moen B, Peddada S, Baird D, Rugtveit J, Midtvedt T, et al. Development of gut microbiota in infants not exposed to medical interventions. APMIS 2012;119:17–35. 10.1111/j.1600-0463.2010.02688.x.Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Harmsen HJM, Wildeboer-Veloo ACM, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 2000;30:61–7. 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- [29].Matijašić BB, Obermajer T, Lipoglavšek L, Grabnar I, Avguštin G, Rogelj I. Association of dietary type with fecal microbiota in vegetarians and omnivores in Slovenia. Eur J Nutr 2014;53:1051–64. 10.1007/s00394-013-0607-6. [DOI] [PubMed] [Google Scholar]
- [30].Losasso C, Eckert EM, Mastrorilli E, Villiger J, Mancin M, Patuzzi I, et al. Assessing the influence of vegan, vegetarian and omnivore oriented westernized dietary styles on human gut microbiota: A cross sectional study. Front Microbiol 2018;9:1–12. 10.3389/fmicb.2018.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One 2014;9. 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. J Parenter Enter Nutr 2013;37:746–54. 10.1177/0148607113486931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Devkota S, Wang Y, Musch MW, Leone V, Fehlner-peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487:104–9. 10.1038/nature1l225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Baldwin J, Collins B, Wolf PG, Martinez K, Shen W, Chuang CC, et al. Table grape consumption reduces adiposity and markers of hepatic lipogenesis and alters gut microbiota in butter fat-fed mice. J Nutr Biochem 2016;27:123–35. 10.1016/j.jnutbio.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci 2013;110:9066–71. 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Collins B, Hoffman J, Martinez K, Grace M, Lila MA, Cockrell C, et al. A polyphenol-rich fraction obtained from table grapes decreases adiposity, insulin resistance and markers of inflammation and impacts gut microbiota in high-fat-fed mice. J Nutr Biochem 2016;31:150–65. 10.1016/j.jnutbio.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jiao X, Wang Y, Lin Y, Lang Y, Li E, Zhang X, et al. Blueberry polyphenols extract as a potential prebiotic with anti-obesity effects on C57BL/6 J mice by modulating the gut microbiota. J Nutr Biochem 2019;64:88–100. [DOI] [PubMed] [Google Scholar]
- [39].Rodríguez-Morató J, Matthan NR, Liu J, de la Torre R, Chen CYO. Cranberries attenuate animal-based diet-induced changes in microbiota composition and functionality: a randomized crossover controlled feeding trial. J Nutr Biochem 2018;62:76–86. 10.1016/j.jnutbio.2018.08.019. [DOI] [PubMed] [Google Scholar]
- [40].Kaczmarek JL, Liu X, Charron CS, Novotny JA, Jeffery EH, Seifried HE, et al. Broccoli consumption affects the human gastrointestinal microbiota. J Nutr Biochem 2019;63:27–34. 10.1016/j.jnutbio.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Santacruz A, Marcos A, Wärnberg J, Martí A, Martin-Matillas M, Campoy C, et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity 2009;17:1906–15. 10.1038/oby.2009.112. [DOI] [PubMed] [Google Scholar]
- [42].Zhang C, Li S, Yang L, Huang P, Li W, Wang S, et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun 2013;4. 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zeng T, Cui H, Tang D, Garside GB, Wang Y, Wu J, et al. Short-term dietary restriction in old mice rejuvenates the aging-induced structural imbalance of gut microbiota. Biogerontology 2019;20:837–48. 10.1007/s10522-019-09830-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015;7:2839–49. 10.3390/nu7042839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schumacher A, Petronis A. Epigenetics of complex diseases: From general theory to laboratory experiments. Curr Top Microbiol Immunol 2006;310:81–115. 10.1007/3-540-31181-5_6. [DOI] [PubMed] [Google Scholar]
- [46].Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun 2018;9:1–15. 10.1038/s41467-017-02651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ye J, Wu W, Li Y, Li L. Influences of the gut microbiota on DNA methylation and histone modification. Dig Dis Sci 2017;62:1155–64. 10.1007/s10620-017-4538-6. [DOI] [PubMed] [Google Scholar]
- [48].Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell 2016;64:982–92. 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kuma H, Kumar H, Fund R, Faiho A, Fundelin K, Fey RE, et al. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. MBio 2014;5:1–4. 10.1128/mBio.02113-14.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Watari J, Ito C, Shimoda T, Tomita T, Oshima T, Fukui H, et al. DNA methylation silencing of microRNA gene methylator in the precancerous background mucosa with and without gastric cancer: Analysis of the effects of H. pylori eradication and long-term aspirin use. Sci Rep 2019;9:1–10. 10.1038/s41598-019-49069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hullar MAJ, Fu BC. Diet, the gut microbiome, and epigenetics. Cancer J (United States) 2014;20:170–5. 10.1097/PPO.0000000000000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Whitt J, Woo V, Lee P, Moncivaiz J, Haberman Y, Denson L, et al. Disruption of epithelial HDAC3 in intestine prevents diet-induced obesity in mice. Gastroenterology 2018;155:501–13. 10.1053/j.gastro.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Getahun SM, Chung FL. Conversion of glucosinolates to isothiocyanates in humans after ingestion of cooked watercress. Cancer Epidemiol Biomarkers Prev 1999;8:447–51. [PubMed] [Google Scholar]
- [54].Romeo L, Iori R, Rollin P, Bramanti P, Mazzon E. Isothiocyanates: An overview of their antimicrobial activity against human infections. Molecules 2018;23:1–18. 10.3390/molecules23030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Vanduchova A, Anzenbacher P, Anzenbacherova E. Isothiocyanate from broccoli, sulforaphane, and its properties. J Med Food 2019;22:121–6. 10.1089/jmf.2018.0024. [DOI] [PubMed] [Google Scholar]
- [56].Myzak MC, Dashwood WM, Orner GA, Ho E, Dashwood RH. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apcmin mice. FASEB J 2006;20:506–8. 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: Inhibition of histone deacetylase. Cancer Res 2004;64:5767–74. 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- [58].Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis 2006;27:811–9. 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem 2013;24:1415–22. 10.1016/j.jnutbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- [60].Duda-Chodak A The inhibitory effect of polyphenols on human gut microbiota. J Physiol Pharmacol 2012;63:497–503. [PubMed] [Google Scholar]
- [61].Evans LW, Athukorala M, Martinez-Guryn K, Ferguson BS. The role of histone acetylation and the microbiome in phytochemical efficacy for cardiovascular diseases. Int J Mol Sci 2020;21:1–18. 10.3390/ijms21114006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dent P, Booth L, Roberts J, Poklepovic A, Hancock J. (Curcumin+sildenafil) enhances the efficacy of 5FU and anti-PD1 therapies in vivo. J Cell Physiol 2020;235:6862–74. [DOI] [PubMed] [Google Scholar]
- [63].Mcfadden RMT, Larmonier CB, Shehab KW, Midura-Kiela M, Ramalingam R, Harrison CA, et al. The role of curcumin in modulating colonic microbiota during colitis and colon cancer prevention. Inflamm Bowel Dis 2015;21:2483–94. 10.1097/MIB.0000000000000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Remely M, Ferk F, Sterneder S, Setayesh T, Roth S, Kepcija T, et al. EGCG prevents high fat diet-induced changes in gut microbiota, decreases of DNA strand breaks, and changes in expression and DNA methylation of Dnmt1 and MLH1 in C57BL/6J male mice. Oxid Med Cell Longev 2017;2017:1–17. 10.1155/2017/3079148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Saldanha SN, Kala R, Tollefsbol TO. Molecular mechanisms for inhibition of colon cancer cells by combined epigenetic-modulating epigallocatechin gallate and sodium butyrate. Exp Cell Res 2014;324:40–53. 10.1016/j.yexcr.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Walker JM, Eckardt P, Aleman JO, Correa J, Liang Y, Iizumi T, et al. The effects of trans-resveratrol on insulin resistance, inflammation, and microbiota in men with the metabolic syndrome: a pilot randomized, placebo controlled clinical trial. J Clin Transl Res 2018;4:122–35. 10.18053/jctres.04.201802.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhang L, Hui XUE, Zhao G, Qiao C, Xiaomei SUN, Pang C, et al. Curcumin and resveratrol suppress dextran sulfate sodium- induced colitis in mice. Mol Med Rep 2019;19:3053–60. 10.3892/mmr.2019.9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Firrman J, Liu L, Argoty GA, Zhang L, Tomasula P, Wang M, et al. Analysis of temporal changes in growth and gene expression for commensal gut microbes in response to the polyphenol naringenin. Microbiol Insights 2018;11:1–12. 10.1177/1178636118775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Busch C, Burkard M, Leischner C, Lauer UM, Frank J, Venturelli S. Epigenetic activities of flavonoids in the prevention and treatment of cancer. Clin Epigenetics 2015;7:1–18. 10.1186/s13148-015-0095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science (80- ) 2019;365:1428–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chaplin A, Palou A, Serra F. Methylation analysis in fatty-acid-related genes reveals their plasticity associated with conjugated linoleic acid and calcium supplementation in adult mice. Eur J Nutr 2017;56:879–91. 10.1007/s00394-015-1135-3. [DOI] [PubMed] [Google Scholar]
- [72].Zhang X, Yang R, Jia Y, Cai D, Zhou B, Qu X, et al. Hypermethylation of Sp1 binding site suppresses hypothalamic POMC in neonates and may contribute to metabolic disorders in adults: Impact of maternal dietary CLAs. Diabetes 2014;63:1475–87. 10.2337/db13-1221. [DOI] [PubMed] [Google Scholar]
- [73].Hall MN, Campos H, Li H, Sesso HD, Stampfer MJ, Willett WC, et al. Blood levels of long-chain polyunsaturated fatty acids, aspirin, and the risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev 2007;16:314–21. 10.1158/1055-9965.EPI-06-0346. [DOI] [PubMed] [Google Scholar]
- [74].Davidson LA, Wang N, Shah MS, Lupton JR, Ivanov I, Chapkin RS. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis 2009;30:2077–84. 10.1093/carcin/bgp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016;531:53–8. 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cheng CW, Biton M, Haber AL, Gunduz N, Eng G, Gaynor LT, et al. Ketone Body Signaling Mediates Intestinal Stem Cell Homeostasis and Adaptation to Diet. Cell 2019;178:1115–1131.e15. 10.1016/j.cell.2019.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Fabbiano S, Suárez-Zamorano N, Chevalier C, Lazarević V, Kieser S, Rigo D, et al. Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell Metab 2018;28:907–921.e7. 10.1016/j.cmet.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shore A, Karamitri A, Kemp P, Speakman JR, Lomax MA. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia 2010;53:1164–73. 10.1007/s00125-010-1701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mihaylova MM, Cheng CW, Cao AQ, Tripathi S, Mana MD, Bauer-Rowe KE, et al. Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell Stem Cell 2018;22:769–778.e4. 10.1016/j.stem.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Camp JG, Frank CL, Lickwar CR, Guturu H, Rube T, Wenger AM, et al. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res 2014;24:1504–16. 10.1101/gr.165845.113. [DOI] [PMC free article] [PubMed] [Google Scholar]