SUMMARY
Sleep is essential for a variety of plastic processes, including learning and memory. However, the consequences of insufficient sleep on circuit connectivity remain poorly understood. To better appreciate the effects of sleep loss on synaptic connectivity across a memory-encoding circuit, we examined changes in the distribution of synaptic markers in the Drosophila Mushroom body (MB). Protein-trap tags for active zone components indicate that recent sleep time is inversely correlated with Bruchpilot (BRP) abundance in the MB lobes; sleep loss elevates BRP while sleep induction reduces BRP across the MB. Overnight sleep deprivation also elevated levels of dSyd-1 and Cacophony, but not other pre-synaptic proteins. Cell-type specific genetic reporters show that MB-intrinsic Kenyon cells (KCs) exhibit increased pre-synaptic BRP throughout the axonal lobes after sleep deprivation; similar increases were not detected in projections from large interneurons or dopaminergic neurons that innervate the MB. These results indicate that pre-synaptic plasticity in KCs is responsible for elevated levels of BRP in the MB lobes of sleep-deprived flies. Because KCs provide synaptic inputs to several classes of post-synaptic partners, we next used a fluorescent reporter for synaptic contacts to test whether each class of KC output connections is scaled uniformly by sleep loss. The KC output synapses that we observed here can be divided into three classes: KCs to MB interneurons, KCs to dopaminergic neurons, and KCs to MB output neurons. No single class showed uniform scaling across each constituent member, indicating that different rules may govern plasticity during sleep loss across cell types.
ETOC Blurb
Weiss and Donlea find that sleep loss increases pre-synaptic Bruchpilot across the Drosophila Mushroom Body (MB) due to plasticity in MB-intrinsic Kenyon cells. Contacts from Kenyon cells to post-synaptic targets show differing changes with sleep loss, indicating that sleep deprivation may differentially alter distinct classes of MB synapses.
INTRODUCTION
In a variety of species, sleep supports the capacity for new learning and is vital for the consolidation of recently formed memories1-6. In Drosophila melanogaster, overnight sleep loss is sufficient to impair acquisition of new associative memories that are encoded in the Mushroom bodies (MBs) 3, and sleep disruptions that follow learning can prevent memory consolidation 2,7. Interestingly, sleep is often elevated or intensified during conditions of heightened synaptic reorganization, including early development 8-10, recovery from neural injury 11,12, and memory consolidation 1,2. Together, these results indicate that sleep may support plastic remodeling in the brain. The consequences of sleep disruptions on synaptic connectivity, however, are not clearly understood. One hypothesis proposes that sleep permits the homeostatic downscaling of synapses throughout the brain, suggesting that sleep loss may impact cognition by saturating synaptic connections across plastic circuits 13-15. This model is supported by several studies that have found an increase in the size or number of synaptic processes after extended waking in both flies and mice 16-19. Additionally, sleep deprivation increases the overall abundance of several synaptic proteins in whole fly brains, suggesting a trend towards synaptic overgrowth during sleep loss 20,21. Conversely, acute sleep induction can result in a net decrease of synaptic protein21 and transcripts22 in fly brain homogenates. Other experiments, however, indicate that sleep deprivation may either weaken or prevent the expansion of synaptic connections in some circuits 9,23-25. These previous studies demonstrate that sleep disruption alters synaptic abundance or size in several neuronal cell types, but it is unclear whether sleep loss uniformly affects all classes of neurons or synapses within a given circuit.
The Drosophila Mushroom Body (MB) provides an ideal structure to characterize how sleep loss may differentially impact distinct types of synaptic contacts within a plastic circuit. The MB is a core associative neuropil that is conserved across arthropod species and is required for the acquisition and encoding of olfactory memories 26-28. In the fruit fly, olfactory information is relayed to the MBs via secondary projection neurons, which synapse onto ~2,200 Kenyon cells (KCs) in each brain hemisphere 29-32. KC axons extend through fasciculated bundles that comprise five distinct MB lobes: α, β, γ, α’, and β' 33. Each axonal lobe of the MBs is divided into compartments that are each innervated by distinct dopaminergic neuron (DAN) types (~21 total) and connect to unique MB output neurons (~22 total types) 29. Associative memories are encoded in the synaptic connections between odor-encoding KCs and valence-encoding MB output neurons (MBONs), with reinforcement signals provided by compartment-specific DANs 34-41. DANs, as a result, encode unconditioned stimuli during learning, and MBONs mediate behavioral output. Additionally, two modulatory interneurons, APL and DPM, project throughout the MB lobes. Both neurons likely receive synaptic inputs from and provide recurrent connections back onto many KCs, but each plays a functionally distinct role in MB functions: APL facilitates sparse coding of odor cues and memory storage 42-44, while DPM supports recurrent activity during memory storage and promotes sleep 45-49. Because of the high degree of interconnectivity within and between cell types, the MB provides an optimal system to test whether sleep loss alters all circuit components to a similar degree, whether one particular connection type may be especially sensitive to sleep loss, or if different constituents may exhibit distinct patterns of reorganization with insufficient sleep. Distinguishing between these models may open opportunities to understand how sleep loss degrades memory encoding in the MB and to develop interventions that maintain plasticity during prolonged waking.
While KCs in the MB γ lobe individually exhibit increased pre-synaptic terminal volume with sleep deprivation 16, the effects of sleep loss on other cell types in the MB have not been systematically examined. Because activity within subpopulations of MB neurons can regulate sleep 34,50-53, understanding the effects of sleep loss on MB connectivity may inform our understanding not only of the cognitive consequences of sleep loss, but also of mechanisms that encode sleep need. Here, we quantify the effects of sleep loss on the abundance of pre-synaptic proteins across neuron types in the MB. We observe a net increase following sleep loss in the abundance of protein-trap reporters for the pre-synaptic proteins Bruchpilot (BRP), dSYD-1, and Cacophony 54-58, but not other synaptic components. Using cell type-specific genetic reporters, we find that the increase in BRP can be localized to MB-intrinsic KCs and not to other neuronal populations in the MB. Because KCs synapse upon many other cell types in the MB, we also tested the effect of sleep loss on the abundance of synaptic contacts between KCs and many of their post-synaptic partners. These experiments find an assortment of responses in synaptic contacts between KCs and different post-synaptic partners, suggesting that sleep deprivation does not uniformly scale all KC output synapses. Instead, sleep loss results in a variety of plastic effects on different classes of KC output synapses, with some increasing their contacts, others weakening their connections, and a final portion remaining unchanged. Our results indicate that different circuit motifs within the MB may be differentially affected by sleep loss, and identify particularly plastic connections that may contribute to impaired memory and increased homeostatic sleep drive following prolonged waking.
RESULTS
BRP abundance in Mushroom Body lobes is inversely related with recent sleep time
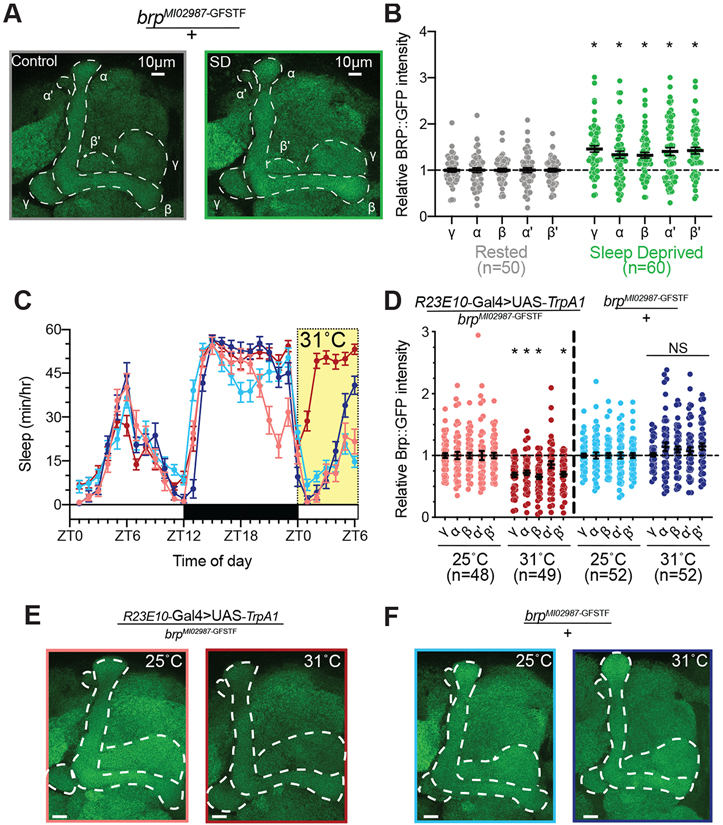
To examine the consequences of sleep loss on MB synaptic connections, we first observed the abundance of GFP-tagged Bruchpilot expressed from a MiMIC protein trap insertion into an intron of the Brp locus 56,57. Brp is a core component of pre-synaptic active zones 55,58, and pre-synaptic Brp protein levels correlate closely with active zone size and release probability 59-61. We mechanically deprived brpMI02987-GFSTF/+ flies of sleep overnight (Figure S1A) then dissected their brains and imaged BRP::GFP fluorescence using confocal microscopy. The BRP::GFP signal increased by ~32-46% in each MB lobe from sleep-deprived brpMI02987-GFSTF/+ flies compared to controls (Figures 1A-B), supporting previous reports of increased pre-synaptic terminal size in MB neurons 16. We also tested the effects of acutely inducing sleep by activating sleep-promoting neurons in a dorsal stratum of the Fan-shaped Body 62. Flies expressing brpMI02987-GFSTF along with the warm-sensitive cation channel TrpA1 under the control of R23E10-Gal4 were heated to 31°C for 6h to acutely increase their sleep from ZT0-6 (Figure 1C) 63,64. Sleep induction reduced BRP::GFP fluorescence in the MB γ, α, β, and β' lobes of experimental flies (R23E10-Gal4>UAS-TrpA1/brpMI02987-GFSTF) compared to siblings that were maintained at 25°C (Figures 1D-E). A 6h exposure to 31°C did not significantly alter either sleep or BRP::GFP fluorescence in brpMI02987-GFSTF/+ controls relative to siblings housed at 25°C (Figures 1D, 1F). Together, these results support the hypothesis that prolonged waking results in a net synaptic expansion in the MBs, and that sleep broadly facilitates the downscaling of these connections.
Figure 1 – Sleep bidirectionally regulates Brp abundance in the mushroom body.
(A) Representative images of endogenous Brp (green) labeled with GFP in brpMI02987-GFSTF/+ flies following 12 hours of rest (left) or 12 hours of overnight sleep deprivation (right). The lobes of the MB are outlined in white.
(B) Quantification of Brp::GFP intensity throughout the MB lobes of brpMI02987-GFSTF/+ flies after 12-hr of overnight sleep loss (green) normalized to rested controls (gray). Two-way ANOVA finds a significant effect of SD (F(1,108)=20.62, p<0.0001, n=50-60 hemispheres/group). Pairwise comparisons using Sidak’s multiple comparisons test found significant increases in Brp::GFP in each MB lobe after sleep deprivation relative to rested siblings (p≤0.002 for each test).
(C) Hourly sleep traces at 25°C (light shading) and 31°C (dark shading) for R23E10-Gal4>UAS-TrpA1/brpMI02987-GFSTF (red) and brpMI02987-GFSTF/+ flies (blue). Thermogenetic activation of dFB neurons in brpMI02987-GFSTF -expressing flies (R23E10-Gal4>UAS-TrpA1/brpMI02987-GFSTF; dark red) increased sleep time compared to siblings that remained at 25°C (light red) and brpMI02987-GFSTF/+ genetic controls that were housed at 25°C (light blue) or shifted to 31°C (dark blue). Flies were temperature shifted from ZT0-6 (yellow shading).
(D) Quantification of Brp::GFP intensity for groups shown in Figure 1C. Sleep induction in R23E10-Gal4>UAS-TrpA1/brpMI02987-GFSTF flies that were shifted to 31°C (dark red) led to a significant decrease in Brp intensity in α, β, β’, and γ lobes compared to siblings that remained at 25°C (light red). Exposure to 31°C did not change Brp::GFP in brpMI02987-GFSTF/+ genetic controls (25°C shown in light blue, 31°C in dark blue). Two-way Repeated Measures ANOVA found a significant group-by-lobe interaction (F(12,784)=3.796, p<0.0001, n=48-52 hemispheres/group).
(E) Representative images of endogenous Brp::GFP labeled in R23E10-Gal4>UAS-TrpA1/brpMI02987-GFSTF flies that were housed at 25°C (left) or given a 6-h exposure at 31°C (right).
(F) Representative images of endogenous Brp::GFP labeled in brpMI02987-GFSTF/+ flies that were housed at 25°C (left) or shifted to 31°C for 6-h (right).
See also Figure S1 for sleep traces from experimental groups shown in Figure 1A-B.
Scale bars depict 10 μm; error bars represent SEM for all panels.
Variable effects of sleep loss on MB abundance of pre-synaptic proteins
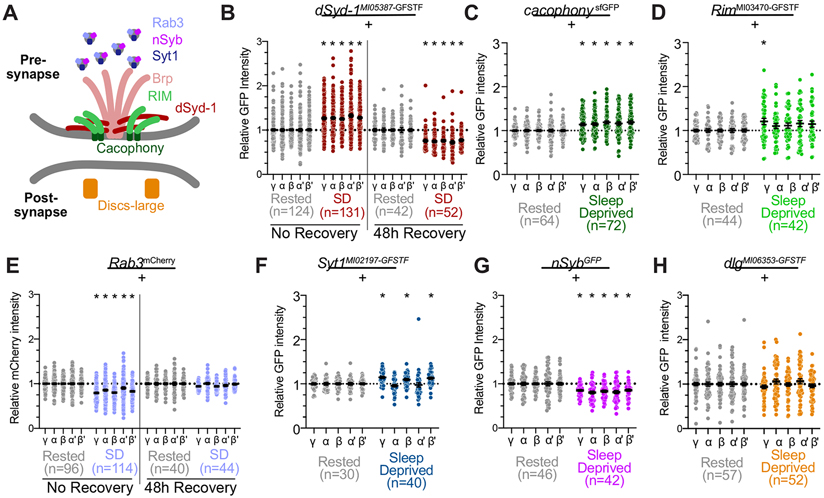
While our above results focus on the effects of sleep loss on BRP levels, previous studies have found evidence for coordinated scaling of several synaptic proteins 20. Other studies, however, suggest that synaptic scaling can change the abundance of some, but not all, pre-synaptic components 21,65. To examine the effects of sleep loss on additional pre-synaptic machinery, we obtained protein trap reporters for six additional pre-synaptic proteins and measured their abundance in the MB after overnight sleep loss (See schematic in Figure 2A; sleep traces and representative images shown in Figure S1B-O). First, we observed the effects of sleep loss on dSyd-1, an active zone component that interacts with BRP and is required for the organization of electron dense T-bars 54. Fluorescence of dSyd-1MI05387-GFSTF 56,57, like BrpMI02987-GFSTF, increased after sleep deprivation by ~25-32% across each MB lobe (Figure 2B). We next measured the effects of sleep loss on Cacophony (Cac), the primary voltage-gated calcium channel at the pre-synapse 66 Like reporters for BRP and dSyd-1, abundance of a GFP-tagged reporter for Cac, CacsfGFP 67, showed a significant increase in MB signal after sleep deprivation (Figure 2C). Overnight sleep loss, however, did not broadly increase the abundance of a protein-trap reporter for Rab3 interacting molecule, RimMI03470-GFSTF (Figure 2D), which influences synaptic accumulation of Cac 68. We also quantified the effects of sleep loss on Rab3, which exhibits vesicle-like staining at the fly active zone and shares the protein domains that are required for vesicle localization with its mammalian homolog 69-71. MBs from sleep deprived flies exhibited a significantly reduced amount of Rab3mCherry 70 in each MB lobe (Figure 2E). Similar quantifications of reporters for two proteins that localize to synaptic vesicles, Syt1MI02197-GFSTF 56,57 and nSybGFP 72. found that sleep loss elevated Syt1MI02197-GFSTF 56,57 abundance locally in the γ, β, and β’ lobes (Figure 2F), while fluorescent intensity of nSybGFP was decreased in each MB lobe of sleep deprived flies (Figure 2G). While the diversity of post-synaptic receptors expressed in MB neurons complicates analysis of post-synaptic plasticity, abundance of the primarily post-synaptic DlgMI06353-GFSTF was not changed by sleep deprivation (Figure 2G). 48h of undisturbed recovery after overnight sleep deprivation restored abundance of Rab3mCherry back to control levels (Figure 2E, right), while dSyd-1MI05387-GFSTF in previously sleep-deprived flies was reduced below the levels observed in undisturbed controls (Figure 2B, right). Together, these results indicate that sleep loss does not uniformly increase the amount of all pre-synaptic proteins across the MB. Instead, the MBs of sleep-deprived flies show broad increases across all lobes in BRP, dSyd-1, and Cac which physically interact 54, and only local, if any, increases in other synaptic components, including Rim, Rab3, Syt1, and nSyb.
Figure 2 – Pre-synaptic proteins show variable responses to sleep loss in the MB lobes.
(A) Schematic illustration of pre-synaptic active zone, including core protein components observed in these studies. Brp localizes in the electron-dense T-bar, where it physically interacts with Syd-1. Both contribute to the recruitment of other pre-synaptic proteins to the AZ. RIM is necessary for proper localization of the Ca2+ channel Cacophony in the pre-synaptic plasma membrane. Rab3 regulates priming of vesicles and organization of AZ proteins. Syt-1 is a Ca2+ sensor located on synaptic vesicles. Nsyb is localized to synaptic vesicles and mediates vesicle fusion. Dlg is a scaffolding protein that is primarily located at the postsynaptic density.
(B) Abundance of dSyd-1::GFP throughout the MB after overnight sleep deprivation (red) compared to rested controls (grey) when flies were dissected either immediately following sleep deprivation (left) or allowed 48h of ad libitum recovery sleep before dissection (right); dSyd-1::GFP intensity in all groups is normalized to rested controls. Two-way ANOVA finds a significant effect of SD (F(3,345)=43.12, p<0.0001, n=42-131 hemispheres/group).
(C) Quantification of Cac::sfGFP intensity in MB axonal lobes following overnight sleep deprivation (green) normalized to rested controls (grey). Two-way ANOVA finds a significant effect of SD (F(1,134)=18.51, p<0.0001, n=64-72 hemispheres/group).
(D) Quantification of Rim::GFP in the MB lobes of sleep deprived RimMI03470-GFSTF flies (green) and rested controls (grey). Two-way ANOVA finds a significant effect of SD (F(1,84)=4.871, p=0.03, n=42-44 hemispheres/group), post-hoc comparisons using Sidak’s multiple comparisons test finds a significant increase in Rim::GFP abundance in the γ lobes (p=0.048), but not in α (p=0.54), β (p=0.38), α’ (p=0.23), or β’ (p=0.27).
(E) Fluorescent intensity of endogenous Rab3::mCherry in the MB lobes of sleep deprived Rab3mCherry/+ (light blue) compared to rested siblings (grey). Data from brains dissected immediately following sleep-deprivation shown on left; right depicts quantification of brains dissected after 48h of recovery from overnight sleep deprivation. Two-way ANOVA finds a significant lobe x group interaction (F(12,1160)=4.472, p<0.0001, n=42-131 hemispheres/group).
(F) Quantification of Syt1::GFP intensity throughout the MB after overnight sleep deprivation (dark blue) compared to rested controls (grey). Two-way ANOVA finds a significant lobe-by-SD interaction (F(4,272)=7.94, p<0.0001, n=30-40 hemipsheres/group); post-hoc comparisons using Sidak’s multiple comparisons test find a significant increase of Syt1::GFP in the γ (p=0.0005), β (p=0.029), and β’ lobes (p=0.0023). No significant change was detected in the α or α’ lobes (p=0.7827 and 0.9937, respectively, by Sidak’s multiple comparisons tests).
(G) Abundance of nSyb::GFP in MB lobes of rested (grey) and sleep-deprived flies (magenta). Two-way repeated measures ANOVA finds a significant effect of sleep deprivation on nSyb::GFP abundance (F(1,86)=19.33, p<0.0001, n=42-46 hemispheres/group).
(H) Dlg::GFP levels in the MB lobes of rested controls (grey) and sleep-deprived siblings (orange). Two-way repeated measures ANOVA finds no significant effect of sleep deprivation (F(1,107)=0.002567, p=0.9597, n=52-57 hemispheres/group).
See also Figure S1 for representative images and sleep traces from each experimental group and genotype.
Scale bars depict 10 μm; error bars represent SEM for all panels.
Pre-synaptic BRP is elevated in KCs but not in other MB cell types
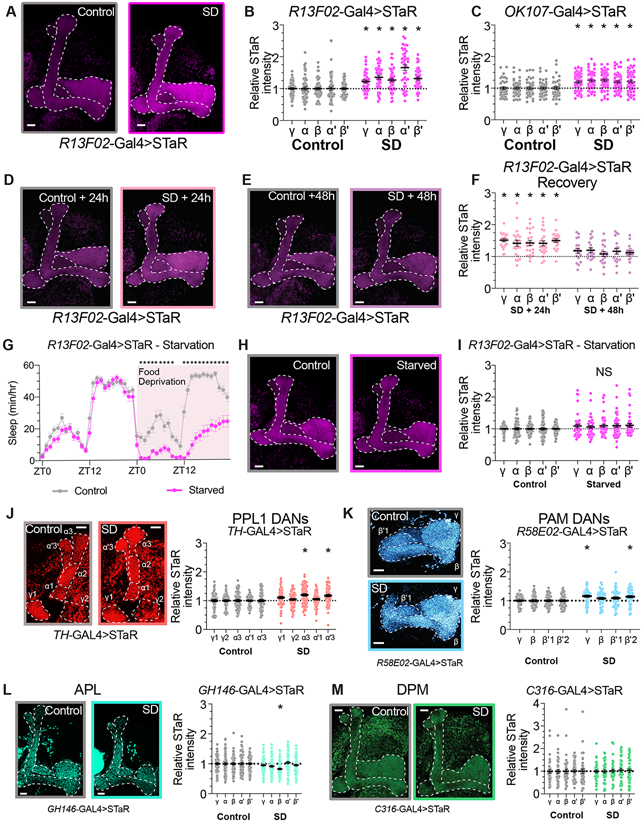
As described above, the abundance of BRP::GFP is significantly elevated across all MB lobes of sleep deprived brpMI02987-GFSTF/+ flies (Figures 1A-B). The use of a protein-trap reporter, however, does not provide information about which cell types within the MB contribute to the increase in BRP abundance that occurs after extended waking. To better identify the specific neuron classes that show elevated BRP levels after sleep loss, we used Synaptic Tagging with Recombination (STaR), a flp-based reporter to specifically label BRP expression in genetically defined classes of neurons 73. Like the protein-trap constructs used above, STaR reports the abundance of a BRP-fusion reporter (BRP::V5) expressed under the control of the endogenous brp promoter. For these studies, we have used a flp-based reporter that fuses smFP_V5 to BRP in genetically targeted neurons 74,75. We began testing the effects of sleep loss on specific classes of MB neurons by labeling BRP in odor-encoding, MB-intrinsic Kenyon cells (KCs) using the genetic driver R13F02-Gal4 63. STaR labelling in KCs using R13F02-Gal4 increased significantly in all KC lobes after overnight sleep loss (Figures 3A-B and S2A). Similar increases in KC expression of BRP were also found using a second broad KC driver, OK107-Gal4 (Figures 3C and S2B) 28. The abundance of smFP_V5-tagged BRP remains elevated in R13F02-positive KCs after 24h of recovery after overnight sleep deprivation, but returns to control levels within approximately 48h of recovery (Figure 3D-F and S2C-D). Because starvation results in sleep loss without the accrual of sleep pressure or learning deficits, we tested whether KC active zones are altered after 24h of starvation 76,77. R13F02-Gal4>STaR flies fed only 2% agar in H2O slept 70±4.73% less than their fed siblings (Figure 3G), but no significant increase in STaR signal was detected in starved flies (Figure 3H-I). These results suggest that the increased BRP abundance that we observe in KCs after sleep loss may be correlated with cognitive impairments and increased sleep drive. To understand whether the increased BRP abundance that we observed is uniformly shared across subpopulations of KCs that innervate different regions of the MB lobes, we used more restricted genetic driver lines to label KCs that project into the α/β, α’/β’, or γ lobes 29. We found that not all KC subsets exhibited similar increases in smFP_V5-tagged BRP; KCs with axons targeted to the α/β core, α/β posterior, and γ dorsal regions showed significant increases in pre-synaptic STaR labelling after sleep deprivation, while there was no effect of sleep disruption on BRP abundance in α/β surface or α’/β’ anterior/posterior KCs (Figure S3). Genetic mosaicism of flp expression using available driver lines prevented consistent measurements of BRP abundance in α’/β’ medial and γ main KC subpopulations. Together, these data indicate that increased BRP abundance within subpopulations of KC neurons contributes significantly to the overall elevation of BRP that we observe in MB lobes following sleep deprivation.
Figure 3 – Increased BRP abundance in Kenyon cell axons after sleep deprivation.
(A) Representative images from R13F02-Gal4>STaR flies after 12 hours of rest (left) or 12 hours of overnight SD (right). Presynapses labelled by STaR (BRP::V5) in magenta.
(B) Quantification of BRP::V5 intensity in rested controls (gray) and after overnight SD (magenta) in KCs labeled by R13F02-Gal4. Two-way ANOVA finds a significant effect of SD (F(1,94)=43.43, p<0.0001, n=42-54 hemispheres/group).
(C) Quantification of BRP::V5 intensity in rested controls (gray) and after overnight SD (magenta) in KCs labeled by OK107-Gal4. Two-way ANOVA finds a significant effect of SD (F(1,94)=19.82, p<0.0001, n=42-54 hemispheres/group).
(D-E) Representative images from R13F02-Gal4>STaR flies following 24- (D) or 48-hours (E) of recovery sleep from overnight SD.
(F) BRP::smFP_V5 intensity quantification for R13F02-Gal4>STaR flies permitted 24- or 48-hours of ad lib recovery sleep following overnight sleep deprivation. Fluorescence intensity is normalized to time-matched rested controls for each SD group. Two-way ANOVA finds a significant effect of group (F(3,82)=21.11, p<0.0001, n=18-24 hemispheres/group). * represents p<0.05 by Sidak’s pairwise comparisons test for SD vs control at the matched timepoint.
(G) Hourly sleep timecourse from R13F02-Gal4>STaR flies that were provided 24h of baseline sleep before either control handling (grey) or food deprivation (magenta). Two-way Repeated Measures ANOVA finds a significant time-by-treatment interaction (F(47,3760)=20.51, p<0.0001, n=39-43 flies/group).
(H) Representative images from R13F02-Gal4>STaR flies after control handling (left) or 24h of food deprivation (right).
(I) Quantification of BRP::smFP_V5 abundance in MB lobes of R13F02-Gal4>STaR flies that have been fed standard fly media (grey) or starved for 24h (magenta). Two-way repeated measures ANOVA finds no significant effect of starvation (F(1,92)=3.229, p=0.0756, n=41-53 hemispheres/group).
(J) Left panel depicts representative images from TH-Gal4>STaR flies after 12 hours of rest (left) or 12 hours of overnight SD (right). Presynapses labelled by STaR (BRP::V5) in red. Right panel shows quantification of BRP::smFP_V5 intensity in rested controls (gray) and after overnight SD (red) in PPL1 dopaminergic neurons labeled by TH-Gal4. Two-way ANOVA finds a significant sleep by MB compartment (F(4,556)=6.184, p<0.0001, n=69-72 hemispheres/group).
(K) Left panel: representative images from R58E02-Gal4>STaR flies labeling BRP in PAM dopaminergic neurons after 12 hours of rest (left) or 12 hours of overnight SD (right). Presynapses labelled by STaR (BRP::V5) in blue. On right, quantification of BRP::smFP_V5 intensity in rested controls (gray) and after overnight SD (blue) in PAM DANs labeled by R58E02-Gal4. Two-way ANOVA finds a significant effect of SD (F(1,104)=7.893, p=0.0059, n=50-56 hemispheres/group).
(L) Left, Representative images from GH146-Gal4>STaR flies after 12 hours of rest (left) or 12 hours of overnight SD (right). Presynapses labelled by STaR (BRP::V5) in green. Right panel shows quantification of BRP::smFP_V5 intensity in rested controls (gray) and after overnight SD (green) in APL labeled by GH146-Gal4. Two-way ANOVA finds a significant lobe by sleep interaction (F(4,480) = 6.672, p<0.0001, n=60-62 hemispheres/group).
(M) On left, representative images from C316-Gal4>STaR flies after 12 hours of rest (left) or 12 hours of overnight SD (right). Presynapses labelled by STaR (BRP::smFP_V5) in green. Right panel depicts quantification of BRP::V5 intensity in rested controls (gray) and after overnight SD (green) in DPM labeled by C316-Gal4. Two-way ANOVA finds no significant effect of SD (F(1,79)=0.04082, p=0.84, n=40-41 hemispheres/group).
See also Figure S2 for sleep traces from experimental groups shown in Figure 3 A-J, and Figure S3 for pre-synaptic BRP quantification from subsets of KC neurons.
Scale bars depict 10 μm; error bars represent SEM for all panels.
Next, we tested whether increased BRP::V5 could also be observed after sleep deprivation (Figure S2E-H) in other cell types that innervate the MB lobes using STaR. Two populations of dopaminergic neurons project into the MB: PPL1 neurons that project into compartments of the γ, α, and α' lobes, which encode punishment, and PAM neurons that terminate in γ, β, and β' lobe zones, which activate in response to rewarding stimuli 29,35,36,39. When we quantified pre-synaptic STaR labelling in PPL1 DANs using TH-Gal4 78, a two-way ANOVA detected a significant effect for sleep deprivation. Pairwise comparisons within compartments of the γ, α, and α' lobes, however, found significant increases only in the α3 and α'3 compartments (Figure 3J). Similarly, we used R58E02-Gal4 to drive STaR expression in PAM DANs projecting into the horizontal lobes of the MB 29,63. While a Two-way ANOVA found a significant effect for sleep deprivation on STaR fluorescence in PAM DANs, pairwise comparisons detect significant increases in labelled Brp only in the γ lobe and β'2 compartment, and not in the β lobe and β'1 compartment (Figure 3K). No increase in BRP::smFP_V5 intensity was detected when we drove STaR in APL (GH146-Gal4) or DPM neurons (C316-Gal4), which are both large interneurons that project broadly across all MB lobes (Figures 3L-M) 44,49. Based on these data, the broad increase in BRP that we observed in MB lobes (Figures 1A-B) after sleep deprivation can be attributed primarily to KCs, not to other MB cell types.
Divergent consequences of sleep loss on KC output synapse classes
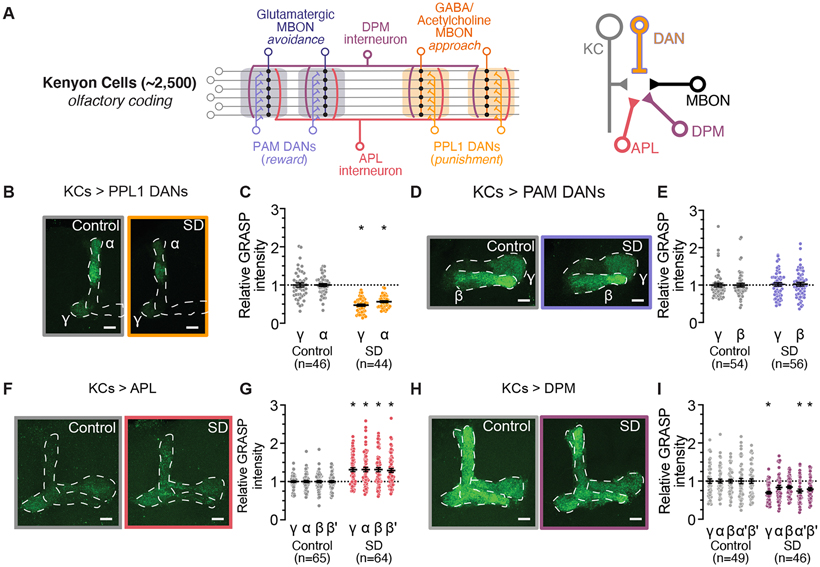
Within the MB lobes, KCs synapse upon several classes of post-synaptic partners, opening the possibility that sleep loss may differentially alter contacts between KCs and each synaptic target. While connections between KCs and MBONs can encode associative memories and odor valence, KCs also provide synaptic inputs to APL and DPM interneurons, and to DANs from both the PAM and PPL1 clusters 79 (See Figure 4A for MB circuit schematic). Each of these synaptic connections contributes to different aspects of olfactory processing and memory encoding 29,34-36,38,43-46. To understand how each type of output synapse from KCs might be influenced by sleep loss, we used GFP Reconstitution across Synaptic Partners (GRASP) 80,81 to observe synaptic contacts between KCs and their various synaptic targets in rested and sleep deprived flies (Sleep traces shown in Figure S5-6). GRASP has previously been used to identify patterns of synaptic contacts in worms 82-84, flies 81,85-88, and mice 89,90 using light microscopy. Here, we expressed an activity-dependent GRASP reporter to label recently active contacts in which KCs release neurotransmitter onto a synaptic partner of interest 81. First, we observed the effects of sleep deprivation on connectivity from KCs to DANs, which is required for memory formation 40. Interestingly, GRASP signal from KCs (MB-LexA) to PPL1 DANs (TH-Gal4) was significantly depressed in the brains of sleep deprived flies (Figures 4B-C) while no effect of sleep loss could be detected on GRASP signal from KCs to PAM DANs (R58E02-Gal4) (Figures 4D-E). Next, we used GRASP to measure synaptic contacts from KCs (MB-LexA) to the APL (GH146-Gal4) and DPM (C316-Gal4) interneurons. KC>APL GRASP signal was increased across the MBs of sleep-deprived flies (Figures 4F-G), while KC>DPM GRASP was significantly decreased in the γ, α', and β' lobes following overnight sleep loss (Figures 4H-I). These results suggest that KC output synapses are not all modulated uniformly during sleep loss, but rather that each KC>interneuron connection may be regulated independently. Interestingly, individual connection types show relatively consistent changes across compartments and lobes of the KC axons, indicating that subpopulations of KCs may share plasticity rules that influence which connections are altered during prolonged waking.
Figure 4 – Effects of SD on synaptic contacts between KCs and DANs, APL & DPM.
(A) Schematic of connectivity between neuronal cell types in the MB. KC axons innervate tiled zones (depicted by shaded regions) that each receive innervation from distinct DANs and provide input to unique MBONs. APL and DPM interneurons receive input from and provide recurrent feedback to KC pre-synapses (Left). KC pre-synapses project onto MBON, DAN, APL, and DPM partners (Right).
(B) Representative images of nsyb GRASP intensity between presynaptic KCs (MB-LexA) and postsynaptic PPL1 DANS (TH-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(C) Quantification of relative KC>PPL1 GRASP intensity after SD (orange) normalized to rested controls (gray). Two-way ANOVA finds a significant effect of SD (F(1,88)=91.81, p<0.0001, n=44-46 hemispheres/group).
(D) Representative images of nsyb GRASP intensity between presynaptic KCs (MB-LexA) and postsynaptic PAM DANs (R58E02-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(E) Quantification of relative KC>PAM GRASP intensity in γ and β lobes after SD (purple), normalized to rested controls (gray). Two-way ANOVA finds no significant effect of SD (F(1,108)=0.09979, p=0.7527, n=54-56 hemispheres/group).
(F) Representative images of nsyb GRASP intensity between presynaptic KCs (MB-LexA) and postsynaptic APL (GH146-Gal4) in the MB lobes of rested controls (left) and in flies subjected to overnight SD (right).
(G) Quantification of relative KC>APL GRASP intensity after SD (red), normalized to rested controls (gray). Two-way ANOVA finds a significant effect of SD (F(1,127)=30.17, p<0.0001, n=64-65 hemispheres/group)
(H) Representative images of nsyb GRASP intensity between presynaptic KCs (MB-LexA) and postsynaptic DPM (C316-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(I) Quantification of relative KC>DPM GRASP intensity after SD (magenta), normalized to rested controls (gray). Two-way ANOVA finds a significant effect of SD (F(1,93)=11.42, p=0.0011, n=46-49 hemispheres/group)
See also Figure S4 for sleep traces from experimental groups shown in Figure 3 B-I.
Scale bars depict 10 μm; error bars represent SEM for all panels.
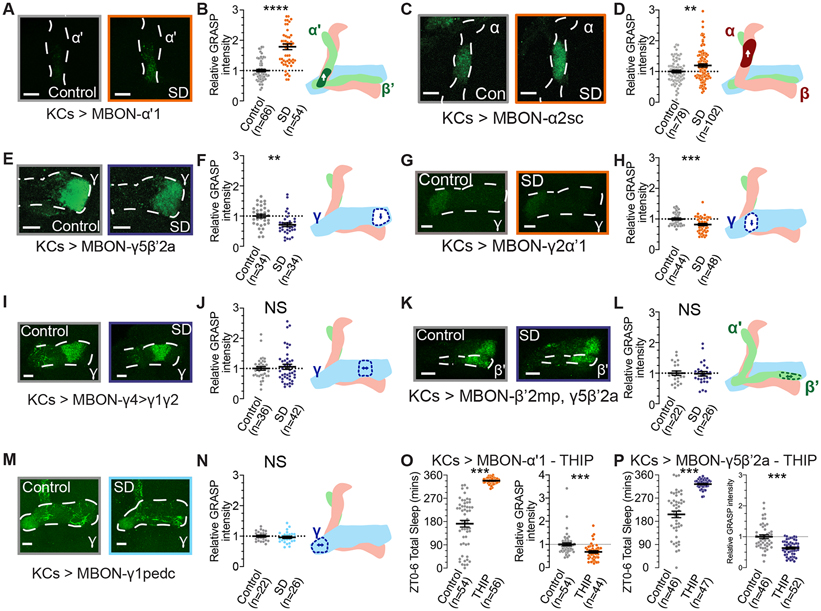
Memories of associative conditioning are encoded within plastic connections between odorcoding KCs and MBONs that innervate individual MB lobe compartments 34,38. To test the effects of sleep loss on KC>MBON connections, we measured GRASP signal to quantify contacts between KCs (MB-LexA) and MBONs in several compartments. As shown in Figure 5, KC>MBON connections varied across different MB compartments. Sleep deprived flies (Sleep data shown in Figure S5) showed consistently elevated GRASP signal from KCs to MBON-α’1 (Figures 5A-B) and from KCs to MBON-α2 (Figures 5C-D), but decreased GRASP between KCs and MBON-γ5β’2a (Figures 5E-F) and from KCs and MBON-γ2a’1 (Figures 5G-H). Other KC to MBON synapses, including those to MBON-γ4>γ1γ2 (Figure 5I-J), MBON-β’2mp,γ5β’2a (Figure 5K-L), and MBON-γ1pedc (Figure 5M-N) are unchanged after overnight sleep deprivation. While discrete MBON subsets produce different neurotransmitters, the neurotransmitter identity of an MBON does not seem to determine pre-synaptic effects of sleep loss (Figure 5; orange groups denote cholinergic MBONs, dark blue shows glutamatergic, and light blue represents GABAergic 29). These results suggest that KC>MBON connections are altered during sleep deprivation in a compartment-by-compartment manner. Further studies will be required to understand the rules that govern the variations in plasticity across compartments, as they are not cleanly predicted by the role of an MBON in encoding valence, or by the neurotransmitters produced by individual MBONs. Connections from KCs to MBON-γ5β’2 and to MBON-γ2a’1, for instance, are both reduced after sleep deprivation, but each expresses a different neurotransmitter and activation of each MBON can result in opposing changes in sleep and behavioral valence 34,53. As shown in our experiments using BrpMI02987-GFSTF in Figure 1, recent sleep history can bidirectionally influence active zone protein abundance in the MB lobes. To test whether acute sleep induction drives changes that are opposite to those observed after sleep deprivation, we pharmacologically increased sleep using the GABA-A receptor agonist THIP and imaged two pairs of KC>MBON connections. In both genotypes, 6h of THIP administration yielded highly significant increases in sleep (Figures 5O-P, left panels). While sleep loss increased KC>MBON- a’1 GRASP and decreased KC>MBON-γ5β’2 GRASP (Figures 5B, F), 6h of sleep-promoting THIP treatment reduced GRASP signal in both KC>MBON connections (Figures 5O-P, right panels). Increased sleep, therefore, may not solely drive synaptic changes that are the converse to those that occur during sleep loss.
Figure 5 – KC>MBON connections exhibit compartment-specific changes with SD.
(A) Representative images of nsyb GRASP intensity between presynaptic KCs (MB-LexA) and postsynaptic MBON-α’1 (MB543B-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(B) Quantification of relative KC>MBON-α’1 GRASP intensity after SD (orange), normalized to rested controls (gray). Two-tailed T-test finds a significant effect of SD (t=8.068, p<0.0001, n=54-66 hemispheres/group).
(C) Representative images of nsyb GRASP intensity between presynaptic KCs (MB-LexA) and postsynaptic MBON-α2sc (R71D08-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(D) Quantification of relative KC>MBON-α2sc GRASP intensity after SD (orange), normalized to rested controls (gray). Two-tailed T-test finds a significant effect of SD (t=2.800, p=0.0057, n=78-102 hemispheres/group).
(E) Representative images of nsyb GRASP intensity in the γ5 compartment between presynaptic KCs (MB-LexA) and postsynaptic MBON-γ5β’2a (R66C08-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(F) Quantification of relative KC>MBON-γ5β’2a GRASP intensity in the γ5 compartment after SD (blue), normalized to rested controls (gray). Two-tailed T-test finds a significant effect of SD (t=3.411, p=0.0011, n=34 hemispheres/group).
(G) Representative images of nsyb GRASP intensity in the γ2 compartment between presynaptic KCs (MB-LexA) and postsynaptic MBON-γ2α’1 (R25D01-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(H) Quantification of relative KC>MBON-γ2α’1 GRASP intensity in the γ2 compartment after SD (orange), normalized to rested controls (gray). Two-tailed T-test finds a significant effect of SD (t=3.793, p=0.0003, n=44-48 hemispheres/group).
(I) Representative images of nsyb GRASP intensity in the γ4 compartment between presynaptic KCs (MB-LexA) and postsynaptic MBON-γ4>γ1γ2 (MB434B-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(J) Quantification of relative KC>MBON-γ4>γ1γ2 GRASP intensity in the γ4 compartment after SD (blue), normalized to rested controls (gray). Two-tailed T-test finds no significant effect of SD (t=0.5245, p=0.6015, n=36-42 hemispheres/group).
(K) Representative images of nsyb GRASP intensity in the β’2 compartment between presynaptic KCs (MB-LexA) and postsynaptic MBON-β’2mp, γ5β’2a (MB011B-Gal4) in rested controls (left) and in flies subjected to overnight SD (right).
(L) Quantification of relative KC>MBON-β’2mp, γ5β’2a GRASP intensity in the β’2 compartment after SD (blue), normalized to rested controls (gray). Two-tailed T-test finds no significant effect of SD (t=0.1928, p=0.8480, n=22-26 hemispheres/group).
(M) Representative images of GRASP labelling from presynaptic KCs (MB-LexA) and postsynaptic MBON-γ1pedc (R12G04-Gal4) in rested controls (left) and flies dissected after overnight sleep loss (right).
(N) Relative quantification of KC>MBON-γ1pedc GRASP intensity in the γ1 compartments of rested (gray) and sleep deprived (light blue) brains. Two-tailed T-test finds no significant effect of SD (t=0.7659, p=0.4476, n=22-26 hemispheres/group).
(O) Left; sleep totals for KC>MBON-α’1 GRASP flies either fed standard fly media (gray) or 0.1 mg/mL THIP (orange). Right; Relative KC>MBON-α’1 GRASP intensity for groups shown in left panel (gray depicts vehicle controls, orange shows 6h treatment with 0.1mg/mL THIP). Two-tailed T-tests find significant effects of THIP treatment on sleep (t=12.95, p<0.0001, n=54-56) and GRASP abundance (t=3.906, p=0.0002, n=44-54 hemispheres/group).
(P) Left; 6h sleep amount for control (gray) and THIP-treated (blue; 0.1mg/mL THIP) KC>MBON-γ5β’2a GRASP flies. Right; Relative intensity of KC>MBON-γ5β’2a GRASP signal in control flies (gray) and flies fed THIP for 6h prior to dissection. Two-tailed T-test finds a significant effect of THIP treatment on sleep (t=10.14, p<0.0001, n=44-47) and on KC>MBON-γ5β’2a GRASP intensity (t=5.492, p<0.0001, n=46-52).
See also Figure S5 for sleep traces from experimental groups in Figure A-L.
Scale bars depict 10 μm; error bars represent SEM for all panels.
DISCUSSION
In this study, we use genetic reporters to quantify the effects of sleep loss on pre-synaptic active zone markers and putative synaptic contacts in the Drosophila MB lobes. We find that abundance of Brp, dSyd-1, and Cacophony broadly increase across all MB lobes after overnight sleep deprivation, and that acutely increasing sleep for six hours is sufficient to reduce Brp levels across the α, β, γ, and β’ lobes. KCs strongly contribute to the increase in Brp across each MB lobe following sleep loss, while pre-synapses of other MB cell types are less sensitive to sleep disruption. Because release of Drosophila neuromodulators likely occurs through a combination of classical neurotransmission and extrasynaptic release 91, our studies do not rule out the possibility that BRP-independent secretion of dopaminergic dense core vesicles might be altered in the MB lobes by sleep loss. The elevated levels of Brp present in KCs of sleep-deprived flies returns to control levels within 48h of ab libitum recovery sleep. While associative learning can recover within only a few hours after sleep deprivation3, our studies indicate that some synaptic consequences of prolonged waking may persist for at least 24h of recovery. These findings parallel those from humans and rodents suggesting that some measures of cognition and neurophysiology recover rapidly after acute sleep loss while others last much longer, even for several days in some cases 92-96. The tractability of Drosophila may provide opportunities for future studies to investigate the processes that mediate recovery from sleep loss and to test whether similar trends in plasticity occur in other neuropil regions across the brain.
Interestingly, sleep deprivation does not seem to increase other active zone components; Rim and Syt1 only show localized changes in some MB lobes, and the primarily post-synaptic marker Dlg shows no significant changes across the MB after sleep loss. Additionally, we find that the abundance of vesicular proteins Rab3 and nSyb decreases across all MB lobes following overnight sleep deprivation. The varying responses between pre-synaptic components may indicate that sleep-deprivation may alter the abundance of some active zone constituents along differing time courses, or that active zone release machinery may be regulated differently than synaptic vesicle pools. The varied responses of each synaptic reporter that we observe suggests that Brp, dSyd-1, and Cac levels may underlie the consequences of sleep loss on MB functioning, but the precise physiological consequences of these changes on KC neurotransmitter release are unclear. Previous work finds that increasing BRP gene copy number drives changes in other active zone proteins that recapitulate protein levels observed in short sleeping mutants, and also increases sleep in a dose-dependent manner 21. It is tempting to speculate that increases in Brp with sleep loss may drive concomitant increases in some core active zone scaffolding components, and compensatory decreases in some proteins regulating synaptic vesicle release. Experiments at the Drosophila larval NMJ indicate that elevated Brp levels increase the rate of spontaneous release and enhance facilitation with pairs of stimuli, while other markers of synapse strength, including the amplitudes of evoked and spontaneous junction potentials, remained unchanged 21. It is unclear whether acute changes in Brp with sleep loss induce the same physiological changes at MB-output synapses, and additional studies will be required to understand how plastic mechanisms that contribute to memory formation might be altered by the pre-synaptic changes described above. Recent work finds that pan-neuronal knockdown of dSyd-1 can reduce sleep and dampen homeostatic rebound, even in flies with elevated BRP 21. Consistent with the idea that dSyd-1 levels may influence sleep pressure, we observed decreased dSyd-1MI05387-GFSTF abundance in previously sleep-deprived flies after 48h of recovery.
While the MB contains several different cell types, pre-synapses in the axons of KCs appear to be uniquely plastic during sleep loss. Our use of an activity-dependent fluorescent GRASP reporter of synaptic contacts observed that sleep loss altered synaptic contacts between KCs and distinct post-synaptic partners in different ways 81. Among these changes, we found that GRASP fluorescence reporting contacts from KCs to PPL1 DANs is strongly decreased after sleep loss, indicating a weakening of the KC>PPL1 DAN contacts. Interestingly, these connections may be vital for recurrent activation within MB compartments during learning and could contribute to prediction error signals 40. While further studies will be required to examine the contribution of these particular connections to learning deficits after sleep loss, human subjects have been reported to exhibit impaired error prediction and affective evaluation in learning tasks following sleep loss 97. Because we observed reduced GRASP signal in KC>PPL1 DAN connections, which mediate aversive reinforcement 39, and not in KC>PAM DAN connections, which influence appetitive reinforcement 36, it is also possible that sleep loss may not equally degrade the encoding of reinforcement signals across all valences or modalities. Recent findings also suggest that not all forms of memory require sleep for consolidation; appetitive olfactory memories can be consolidated without sleep when flies are deprived of food, and sleep-dependent and -independent memory traces in these conditions are stored in separate MB zones 98. We find that the KC>MBON connections that contribute to sleep-dependent memory (KC>γ2α’1) also show an overall decrease in GRASP signal with sleep loss, while those that are vital for sleep-independent memory (MBON-γ1pedc) show no GRASP change after sleep deprivation. These compartment-specific variations in the effects of sleep on both memory and synaptic distribution further indicate that local MB zones may follow distinct plasticity rules under physiological stressors, including sleep loss.
Additionally, GRASP signal from KCs to APL is significantly elevated following sleep loss, suggesting a strengthening of KC>APL connections. KCs and APL form a negative feedback circuit, where KCs activate APL, and APL inhibits KCs; this feedback inhibition maintains sparseness of odor coding and odor specificity of memories 42. It is possible that KCs compensate for increased synaptic abundance accumulated during sleep loss by recruiting inhibition from APL. While further experimentation is needed to examine the role of these connections in the regulation of net synaptic strength during sleep loss, sleep deprivation results in increased cortical excitability in humans and rodents 99,100, and hyperexcitability is often counteracted by increased synaptic inhibition 101,102. Conversely, sleep loss reduces connectivity between KCs and DPM, a second large interneuron that may facilitate recurrent activity in the MB lobes 46,103.
Our results also indicate that KC>MBON synaptic contacts exhibit a variety of changes in response to sleep deprivation. The specific KC>MBON connections that show significantly elevated or reduced GRASP signal here are not clearly assorted based on valence encoding, contribution to specific associative memory assays, or influence on sleep/wake regulation 34,53. Activity in several MB cell types, including α’/β’ KCs, MBON-γ5β’2, MBON-γ2 α’1, DPM, and PAM DANs regulates sleep 34,48,50-53. The observation that KC>MBON-γ5β’2a labelling is reduced with sleep loss complements previous observations of reduced electrical activity in MBON-γ5β’2 following sleep deprivation 53. Other sleep-promoting MB neurons, however, such as DPM48, do not show an overall increase in BRP abundance, suggesting either that other changes in excitability, synaptic drive, or post-synaptic adaptations might drive homeostatic sleep regulation in these cells, or that distinct subsets of connections within the populations that we label here might be sleep-regulatory. The compartment-to-compartment variance in KC>MBON responses to sleep loss also parallels previous findings that plasticity rules can vary between MBONs during heterosynaptic plasticity 37. While our GRASP results suggest diverse changes in putative synaptic contacts with sleep loss, the functional effects of these changes require further study. It is important to note that a significant portion of MB synapses are comprised of connections between either pairs or groups of KCs 79,104. The genetic strategies that we have used in this study have prevented reliable visualization and quantification of these connections. As a result, the effect of sleep loss on KC>KC synapses has not been examined here but may comprise a portion of the increase in KC pre-synaptic abundance that we observe in Figure 3. While our studies identify synaptic classes that exhibit altered GRASP labelling across sleep loss, future studies using super resolution imaging and/or physiology could examine the structural and molecular changes that underlie this plasticity. Connections between neurons in the MB may be also influenced by non-neuronal cell types, including astrocytes. Astrocytic contact with KCs can be reduced by sleep loss 105 and astrocytic calcium levels correlate with sleep pressure 106, which both suggest that astrocytic processes could be positioned to mediate sleep-dependent plasticity in the MB.
The broad conservation of release machinery across active zones within and between cell types has simplified our examination of pre-synaptic plasticity during sleep loss. Assays of both Hebbian and homeostatic plasticity have also identified a variety of post-synaptic adaptations. Interestingly, postsynaptic densities isolated from rodent cortex show significant reorganization of post-synaptic GluR5 receptors, which depends upon the activity of Homer1 107, and sleep-dependent phosphorylation of CaMKII and GluR1 contribute to consolidation of visual cortex plasticity 23. Because MBONs exhibit post-synaptic plasticity during other contexts, including the formation of associative memories 37, sleep deprivation may also alter post-synaptic organization of MBONs or other cell types in the MB. Although the distribution of Dlg is not significantly changed by sleep loss, the rich variety of post-synaptic receptors for acetylcholine, dopamine, GABA, and other signals in the MB requires development of additional reporters to examine these post-synaptic consequences of insufficient sleep in MB neurons. Additionally, while our data outline changes in pre-synaptic protein abundance and pre-synaptic KC contacts that result from sleep loss, the possibility that these synaptic changes may be accompanied by homeostatic compensation in neuronal excitability or firing patterns remains to be tested. Because sleep-deprived flies can recover the capacity to learn after only a brief nap 3, homeostatic adjustments in post-synaptic strength and/or excitability may permit MBs to compensate for pre-synaptic changes that appear to persist for at least 24 hours after sleep deprivation (Figs. 3D-F). Further, recovery sleep or pharmacological sleep enhancement may not simply reverse the effects of sleep loss (Figs 2B, 5O-P) and it is unclear how particular subsets of synaptic proteins or connections may be selected for removal during times of elevated sleep.
The consequences of sleep loss on synaptic connectivity are not clearly understood, but previous work has found net changes in synaptic abundance or size across brain regions 16,17,20,107. We characterize a diverse array of synaptic responses to sleep loss among different cell types within the same circuit. Our findings may suggest that distinct cell types and connections within the MB are governed by heterogeneous plasticity rules during sleep disruption. While previous studies have characterized the synaptic effects of sleep history on individual cell types within plastic circuits, our data provide a more comprehensive understanding of the consequences of sleep loss on MB circuits. While this project outlines the local effects of sleep loss on MB connectivity, it is unclear whether specific neural subsets also drive BRP increases within other neuropil compartments of sleep-deprived brains20. Here, we find an overall increase in the abundance of reporters for some, but not all, pre-synaptic proteins. These pre-synaptic changes are not distributed equally across all cell types; they are most pronounced in MB-intrinsic KCs. Further, output connections from KCs to different classes of synaptic partners show varying patterns of plasticity in MB sub-circuits that contribute to encoding odor valence, comprise recurrent feedback loops, or relay reinforcement signals. Our results indicate that sleep loss may degrade MB-dependent memory by altering several different classes of synapses, but future studies will be required to test the specific roles of changes at individual synapse types and the mechanisms by which prolonged waking reorganizes MB connectivity.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeffrey Donlea (jdonlea@ucla.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets generated during this study. This study did not generate any novel code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly Strains and Environment
Fly stocks were fed standard cornmeal media (per 1L H20: 12g agar, 29g Red Star yeast, 71g cornmeal, 92g molasses, 16mL methyl paraben 10% in EtOH, 10mL propionic acid 50% in H20) at 25°C with 60% relative humidity and entrained to a daily 12hr light, 12hr dark schedule. All flies were reared in environmentally-controlled chambers at 25°C and 60% relative humidity on a 12hr light:12hr dark schedule. BrpMI2978-GFSTF, dSyd-1MI05387-GFSTF, RimMI03470-GFSTF, Syt1MI02197-GFSTF, OK107-Gal4, R13F02-Gal4, R19B03-Gal4, R58E02-Gal4, GH146-Gal4, C316-Gal4, R12G04-Gal4, R25D01-Gal4, R66C08-Gal4, R71D08-Gal4, R23E10-Gal4, UAS-TrpA1, nsyb GRASP effectors (w*; P{w+mC=lexAop-nSyb-spGFP1-10}2, P{w+mC=UAS-CD4-spGFP11}2; MKRS/TM6B) and rab3mCherry were obtained from the Bloomington Drosophila Stock Center, TH-Gal4 was provided by Dr. David Krantz (UCLA), STaR effector flies (w−; 20xUAS-RSR.PEST, 79C23S-RSRT-STOP-RSRT-smGFP_V5-2A-LexA/cyo) were provided by Dr. Orkun Akin (UCLA), and cacsfGFP was a gift from Dr. Kate O’Connor-Giles (Brown University). All MB split-Gal4 fly stocks (MB011B, MB185B, MB371B, MB434B, MB463B, MB543B, MB594B, and MB607B) were generated by the lab of Dr. Gerald Rubin 29,34 and generously provided by the HHMI Janelia Research Campus (http://splitGal4.janelia.org/cgi-bin/splitGal4.cgi).
METHOD DETAILS
Behavior
Sleep was measured as previously described 108. 3-7 day old adult female flies were housed individually in 65mm borosilicate glass tubes (5mm diameter) containing fly food coated with paraffin wax on one end and a foam plug in the other. Locomotor activity was measured using Drosophila Activity Monitors from Trikinetics (Waltham MA, USA) and sleep was analyzed using Visual Basic macros in Microsoft Excel 108 or SCAMP analysis scripts in MATLAB 109. Baseline sleep was monitored in all groups, and sleep deprivation was performed using mechanical stimulation via the SNAP method 108. For starvation experiments, flies either remained on standard fly media for control treatment or were transferred into fresh tubes containing 2% agar in H2O at ZT0, 24h prior to dissection.
Immunohistochemistry and Confocal imaging
Flies were anesthetized on ice, then brains were dissected in PBS and fixed in either 4% paraformaldehyde in PBS for 30 minutes or in 3% glyoxal for 25 minutes (all brains from an individual experiment were treated identically). Following fixation, brains were washed in PBS and PBTX (PBS + 0.3% Triton-x100) and incubated in 3% Normal Goat Serum in PBTX for one hour. For GFP and mCherry immunostaining, brains were incubated in primary antibody overnight followed by secondary antibody for roughly 24 hours. Immunostaining for V5 used a 48-hour incubation period in mouse anti-V5 conjugated with DyLight550 (Bio-Rad). After antibody incubation, brains were washed in PBS and mounted on slides using Vectashield fluorescence mounting medium from Electron Microscopy Services (Burlingame CA, USA). All specimens were imaged on a Zeiss 880 laser scanning confocal microscope using a 40x water immersion objective. Matching imaging settings were used for each brain within individual experiments.
Primary antibodies used: chicken anti-GFP at 1:1000 (Molecular Probes), rabbit anti-DsRed at 1:1000 (Clontech), mouse anti-GFP at 1:100 (Sigma), mouse anti-V5 conjugated with DyLight550 at 1:400 (Bio-Rad).
Secondary antibodies used: goat anti-chicken Alexa 488, goat anti-rabbit Alexa 546, goat antimouse Alexa 488 (Molecular Probes). All secondary antibodies were used at a 1:1000 dilution.
Quantification of mushroom body fluorescent signal intensity used an average intensity projection over 4 z-slices of the lobe of interest, followed by manual outlining of the labelled lobe to measure mean GFP or anti-V5 intensity in Fiji 110.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics
Statistical comparisons were made using t-Tests or One- or Two-Way ANOVAs as appropriate; figure legends describe the statistical tests used for each panel. Where needed, post-hoc pairwise analysis measured the effect of sleep manipulations on each MB lobe using Sidak’s multiple comparisons tests. All statistical comparisons were conducted using GraphPad Prism 8 (San Diego CA, USA). Sample sizes for each experiment are depicted in figure panel or in the appropriate figure legend. All group averages shown in data panels depict mean ± SEM.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental Models: Drosophila Strains | ||
| Canton-S | Gero Miesenböck (University of Oxford) | |
| brp MI02987-GFSTF | Bloomington Drosophila Stock Center | RRID:BDSC_59292 |
| R23E10-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_49032 |
| UAS-TrpA1 (II) | Bloomington Drosophila Stock Center | RRID:BDSC_26263 |
| dSyd-1 MI05387-GFSTF | Bloomington Drosophila Stock Center | RRID:BDSC_59414 |
| cacophony sfGFP | Kate O’Connor-Giles (Brown University) | |
| Rim MI03470-GFSTF | Bloomington Drosophila Stock Center | RRID:BDSC_60200 |
| Syt1 MI02197-GFSTF | Bloomington Drosophila Stock Center | RRID:BDSC_59788 |
| Rab3 mCherry | Bloomington Drosophila Stock Center | RRID:BDSC_81509 |
| nSyb GFP | Troy Littleton (MIT) | |
| dlg MI06353-GFSTF | Bloomington Drosophila Stock Center | RRID:BDSC_59417 |
| STaR effector: w−; 20xUAS-RSR.PEST, 79C23S-RSRT-STOP-RSRT-smGFP_V5-2A-LexA/cyo | Orkun Akin (UCLA) | |
| R13F02-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_48571 |
| OK107-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_854 |
| TH-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_8848 |
| R58E02-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_41347 |
| GH146-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_30026 |
| C316-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_30830 |
| MB594B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68255 |
| MB185B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68267 |
| MB371B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68383 |
| MB463B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68370 |
| MB607B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68256 |
| GRASP Effector: w*; P{w[+mC]=lexAop-nSyb-spGFP1-10}2, P{w[+mC]=UAS-CD4-spGFP11}2; MKRS/TM6B | Bloomington Drosophila Stock Center | RRID:BDSC_64315 |
| MB-LexA | Scott Waddell (University of Oxford) | |
| MB543B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68335 |
| R71D08-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_61645 |
| R66C08-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_49412 |
| R25D01-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_49122 |
| R12G04-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_48523 |
| MB434B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68325 |
| MB011B split-Gal4 | HHMI Janelia Research Campus | RRID:BDSC_68294 |
| Antibodies | ||
| Chicken anti-GFP | ThermoFisher | A10262 RRID:AB_2534023 |
| Anti-chicken Alexa488 | ThermoFisher | A11039 RRID:AB_142924 |
| Mouse anti-V5 DyLight550 | BioRad | MCA1360GA RRID:AB_567249 |
| Mouse anti-GFP | Sigma | G6539–100UL RRID:AB_259941 |
| Anti-mouse Alexa488 | ThermoFisher | A11001 RRID:AB_2534069 |
| Software | ||
| Visual Basic sleep analysis macros | Paul Shaw (Washington University in St. Louis) | |
| Prism 8 | Graphpad (www.graphpad.com) | |
| FIJI | FIJI (Imagej.net/FIJI) | |
| Chemicals | ||
| THIP hydrochloride (4,5,6,7-Tetrahydroisoxazolo[5,4-c]pyridin-3-ol hydrochloride) | Sigma-Aldrich | T101-100MG |
| Phosphate buffered saline tablets | Sigma-Aldrich | P4417-100TAB |
| Triton X-100 | Simga-Aldrich | X100-100ML |
| VECTASHIELD Mounting Medium | Vector Laboratories | H-1000 |
| Glyoxal 3% Fixative | Electron Microscopy Sciences | 16525 |
| Paraformaldehyde 16% Solution | Electron Microscopy Sciences | 15710-S |
Highlights.
Amount of pre-synaptic BRP in Mushroom bodies is inversely related to recent sleep
Increased BRP after sleep loss is restricted specifically to Kenyon cells
Outputs from KCs to different synaptic partners show varied changes with sleep loss
Acknowledgements
We thank all members of the Donlea lab for helpful discussions and feedback during this project, especially Dr. Jaison Omoto and Prabhjit Singh for their technical assistance with histology protocols. Fly stocks were generously provided by Drs. Orkun Akin (UCLA), David Krantz (UCLA), and Kate O’Connor Giles (Brown University), the Bloomington Drosophila Stock Center, and the HHMI Janelia Research Campus. This project was supported by an Early Career Development Award from the Sleep Research Society Foundation to JD, a Career Development Award from the Human Frontiers Science Program to JD (CDA00026-2017-C), NIH grant NS105967 to JD, and a Jessamine K. Hilliard UCLA Neurobiology Graduate Student Grant to JW.
Footnotes
Declaration of Interests
The authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Walker MP, Brakefield T, Morgan A, Hobson JA, and Stickgold R (2002). Practice with sleep makes perfect: sleep-dependent motor skill learning. Neuron 35, 205–211. [DOI] [PubMed] [Google Scholar]
- 2.Ganguly-Fitzgerald I, Donlea JM, and Shaw PJ (2006). Waking experience affects sleep need in Drosophila. Science 313, 1775–1781. [DOI] [PubMed] [Google Scholar]
- 3.Seugnet L, Suzuki Y, Vine L, Gottschalk L, and Shaw PJ (2008). D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol 18, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graves LA, Heller EA, Pack AI, and Abel T (2003). Sleep deprivation selectively impairs memory consolidation for contextual fear conditioning. Learn Memory 10, 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnan HC, Gandour CE, Ramos JL, Wrinkle MC, Sanchez-Pacheco JJ, and Lyons LC (2016). Acute Sleep Deprivation Blocks Short- and Long-Term Operant Memory in Aplysia. Sleep 39, 2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDermott CM, LaHoste GJ, Chen C, Musto A, Bazan NG, and Magee JC (2003). Sleep Deprivation Causes Behavioral, Synaptic, and Membrane Excitability Alterations in Hippocampal Neurons. J Neurosci 23, 9687–9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Yu F, and Guo A (2009). Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep 32, 1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roffwarg HP, Muzio JN, and Dement WC (1966). Ontogenetic development of the human sleep-dream cycle. Science 152, 604–619. [DOI] [PubMed] [Google Scholar]
- 9.Kayser MS, Yue Z, and Sehgal A (2014). A Critical Period of Sleep for Development of Courtship Circuitry and Behavior in Drosophila. Science 344, 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw PJ, Cirelli C, Greenspan RJ, and Tononi G (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. [DOI] [PubMed] [Google Scholar]
- 11.Singh P, and Donlea JM (2020). Bidirectional Regulation of Sleep and Synapse Pruning after Neural Injury. Curr Biology Cb 30, 1063–1076.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanhope BA, Jaggard JB, Gratton M, Brown EB, and Keene AC (2020). Sleep Regulates Glial Plasticity and Expression of the Engulfment Receptor Draper Following Neural Injury. Curr Biology Cb 30, 1092–1101.e3. [DOI] [PubMed] [Google Scholar]
- 13.Tononi G, and Cirelli C (2003). Sleep and synaptic homeostasis: a hypothesis. Brain Res Bull 62, 143–150. [DOI] [PubMed] [Google Scholar]
- 14.Tononi G, and Cirelli C (2014). Sleep and the Price of Plasticity: From Synaptic and Cellular Homeostasis to Memory Consolidation and Integration. Neuron 81, 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tononi G, and Cirelli C (2006). Sleep function and synaptic homeostasis. Sleep Med Rev 10, 49–62. [DOI] [PubMed] [Google Scholar]
- 16.Bushey D, Tononi G, and Cirelli C (2011). Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332, 1576–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vivo L. de, Bellesi M, Marshall W, Bushong EA, Ellisman MH, Tononi G, and Cirelli C (2017). Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science 355, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spano GM, Banningh SW, Marshall W, Vivo L. de, Bellesi M, Loschky SS, Tononi G, and Cirelli C (2019). Sleep Deprivation by Exposure to Novel Objects Increases Synapse Density and Axon-Spine Interface in the Hippocampal CA1 Region of Adolescent Mice. - PubMed - NCBI. Journal of Neuroscience 39, 6613–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gisabella B, Scammell T, Bandaru SS, and Saper CB (2020). Regulation of hippocampal dendritic spines following sleep deprivation. Journal of Comparative Neurology 528, 380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilestro GF, Tononi G, and Cirelli C (2009). Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S, Piao C, Beuschel CB, Götz T, and Sigrist SJ (2020). Presynaptic Active Zone Plasticity Encodes Sleep Need in Drosophila. Curr Biology Cb 30, 1077–1091.e5. [DOI] [PubMed] [Google Scholar]
- 22.Dissel S, Angadi V, Kirszenblat L, Suzuki Y, Donlea J, Klose M, Koch Z, English D, Winsky-Sommerer R, van Swinderen B, et al. (2015). Sleep Restores Behavioral Plasticity to Drosophila Mutants. Curr Biol 25, 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aton SJ, Seibt J, Dumoulin M, Jha SK, Steinmetz N, Coleman T, Naidoo N, and Frank MG (2009). Mechanisms of Sleep-Dependent Consolidation of Cortical Plasticity. Neuron 61, 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durkin J, and Aton SJ (2016). Sleep-Dependent Potentiation in the Visual System Is at Odds with the Synaptic Homeostasis Hypothesis. Sleep 39, 155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aton SJ, Broussard C, Dumoulin M, Seibt J, Watson A, Coleman T, and Frank MG (2013). Visual experience and subsequent sleep induce sequential plastic changes in putative inhibitory and excitatory cortical neurons. Proc National Acad Sci 110, 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zars T, Fischer †M, Schulz R, and Heisenberg M (2000). Localization of a Short-Term Memory in Drosophila. Science 288, 672–675. [DOI] [PubMed] [Google Scholar]
- 27.Erber J, Masuhr Th., and Menzel R (1980). Localization of short-term memory in the brain of the bee, Apis mellifera. Physiol Entomol 5, 343–358. [Google Scholar]
- 28.Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, and O’Kane CJ (1996). Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science 274, 2104–2107. [DOI] [PubMed] [Google Scholar]
- 29.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo T-T, Dionne H, Abbott L, Axel R, Tanimoto H, et al. (2014). The neuronal architecture of the mushroom body provides a logic for associative learning. Elife 3, e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marin EC, Jefferis GSXE, Komiyama T, Zhu H, and Luo L (2002). Representation of the glomerular olfactory map in the Drosophila brain. Cell 109, 243–255. [DOI] [PubMed] [Google Scholar]
- 31.Murthy M, Fiete I, and Laurent G (2008). Testing Odor Response Stereotypy in the Drosophila Mushroom Body. Neuron 59, 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner GC, Bazhenov M, and Laurent G (2008). Olfactory Representations by Drosophila Mushroom Body Neurons. J Neurophysiol 99, 734–746. [DOI] [PubMed] [Google Scholar]
- 33.Crittenden JR, Skoulakis EM, Han K-A, Kalderon D, and Davis RL (1998). Tripartite mushroom body architecture revealed by antigenic markers. Learning & memory (Cold Spring Harbor, N.Y.) 5, 38–51. [PMC free article] [PubMed] [Google Scholar]
- 34.Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais P-Y, Robie AA, Yamagata N, Schnaitmann C, et al. (2014). Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife 3, e04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, and Heisenberg M (2003). Dopamine and Octopamine Differentiate between Aversive and Appetitive Olfactory Memories in Drosophila. J Neurosci 23, 10495–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, and Waddell S (2012). Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492, 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hige T, Aso Y, Modi MN, Rubin GM, and Turner GC (2015). Heterosynaptic Plasticity Underlies Aversive Olfactory Learning in Drosophila. Neuron 88, 985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owald D, Felsenberg J, Talbot CB, Das G, Perisse E, Huetteroth W, and Waddell S (2015). Activity of Defined Mushroom Body Output Neurons Underlies Learned Olfactory Behavior in Drosophila. Neuron 86, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li H, Hirsh J, and Miesenböck G (2009). Writing Memories with Light-Addressable Reinforcement Circuitry. Cell 139, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cervantes-Sandoval I, Phan A, Chakraborty M, and Davis RL (2017). Reciprocal synapses between mushroom body and dopamine neurons form a positive feedback loop required for learning. Elife 6, e23789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry JA, Phan A, and Davis RL (2018). Dopamine Neurons Mediate Learning and Forgetting through Bidirectional Modulation of a Memory Trace. Cell Reports 25, 651–662.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin AC, Bygrave AM, de Calignon A, Lee T, and Miesenböck G (2014). Sparse, decorrelated odor coding in the mushroom body enhances learned odor discrimination. Nat Neurosci 17, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitman JL, Huetteroth W, Burke CJ, Krashes MJ, Lai S-L, Lee T, and Waddell S (2011). A Pair of Inhibitory Neurons Are Required to Sustain Labile Memory in the Drosophila Mushroom Body. Curr Biol 21, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, and Davis RL (2009). The GABAergic anterior paired lateral neuron suppresses and is suppressed by olfactory learning. Nat Neurosci 12, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu D, Keene AC, Srivatsan A, Waddell S, and Davis RL (2005). Drosophila DPM Neurons Form a Delayed and Branch-Specific Memory Trace after Olfactory Classical Conditioning. Cell 123, 945–957. [DOI] [PubMed] [Google Scholar]
- 46.Keene AC, Stratmann M, Keller A, Perrat PN, Vosshall LB, and Waddell S (2004). Diverse Odor-Conditioned Memories Require Uniquely Timed Dorsal Paired Medial Neuron Output. Neuron 44, 521–533. [DOI] [PubMed] [Google Scholar]
- 47.Keene AC, Krashes MJ, Leung B, Bernard JA, and Waddell S (2006). Drosophila Dorsal Paired Medial Neurons Provide a General Mechanism for Memory Consolidation. Curr Biol 16, 1524–1530. [DOI] [PubMed] [Google Scholar]
- 48.Haynes PR, Christmann BL, and Griffith LC (2015). A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 4, R774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, and Quinn WG (2000). The amnesiac Gene Product Is Expressed in Two Neurons in the Drosophila Brain that Are Critical for Memory. Cell 103, 805–813. [DOI] [PubMed] [Google Scholar]
- 50.Joiner WJ, Crocker A, White BH, and Sehgal A (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. [DOI] [PubMed] [Google Scholar]
- 51.Pitman JL, McGill JJ, Keegan KP, and Allada R (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756. [DOI] [PubMed] [Google Scholar]
- 52.Sitaraman D, Aso Y, Rubin GM, and Nitabach MN (2015). Control of Sleep by Dopaminergic Inputs to the Drosophila Mushroom Body. Front Neural Circuit 9, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, and Nitabach MN (2015). Propagation of Homeostatic Sleep Signals by Segregated Synaptic Microcircuits of the Drosophila Mushroom Body. Curr Biol 25, 2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Owald D, Fouquet W, Schmidt M, Wichmann C, Mertel S, Depner H, Christiansen F, Zube C, Quentin C, Körner J, et al. (2010). A Syd-1 homologue regulates pre- and postsynaptic maturation in DrosophilaDSyd-1 regulates synapse maturation. J Cell Biology 188, 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, et al. (2006). Bruchpilot Promotes Active Zone Assembly, Ca2+ Channel Clustering, and Vesicle Release. Science 312, 1051–1054. [DOI] [PubMed] [Google Scholar]
- 56.Nagarkar-Jaiswal S, DeLuca SZ, Lee P-T, Lin W-W, Pan H, Zuo Z, Lv J, Spradling AC, and Bellen HJ (2015). A genetic toolkit for tagging intronic MiMIC containing genes. Elife 4, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagarkar-Jaiswal S, Lee P-T, Campbell ME, Chen K, Anguiano-Zarate S, Gutierrez MC, Busby T, Lin W-W, He Y, Schulze KL, et al. (2015). A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4, 2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle M-C, Schmidt M, Qin G, et al. (2006). Bruchpilot, a Protein with Homology to ELKS/CAST, Is Required for Structural Integrity and Function of Synaptic Active Zones in Drosophila. Neuron 49, 833–844. [DOI] [PubMed] [Google Scholar]
- 59.Akbergenova Y, Cunningham KL, Zhang YV, Weiss S, and Littleton JT (2018). Characterization of developmental and molecular factors underlying release heterogeneity at Drosophila synapses. Elife 7, e38268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hong H, Zhao K, Huang S, Huang S, Yao A, Jiang Y, Sigrist S, Zhao L, and Zhang YQ (2020). Structural remodeling of active zones is associated with synaptic homeostasis. J Neurosci Official J Soc Neurosci 40, 2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matkovic T, Siebert M, Knoche E, Depner H, Mertel S, Owald D, Schmidt M, Thomas U, Sickmann A, Kamin D, et al. (2013). The Bruchpilot cytomatrix determines the size of the readily releasable pool of synaptic vesiclesAZ cytomatrix regulates readily releasable pool. J Cell Biology 202, 667–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, and Shaw PJ (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Reports 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, and Garrity PA (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugie A, Hakeda-Suzuki S, Suzuki E, Silies M, Shimozono M, Möhl C, Suzuki T, and Tavosanis G (2015). Molecular Remodeling of the Presynaptic Active Zone of Drosophila Photoreceptors via Activity-Dependent Feedback. Neuron 86, 711–725. [DOI] [PubMed] [Google Scholar]
- 66.Smith LA, Wang X, Peixoto AA, Neumann EK, Hall LM, and Hall JC (1996). A Drosophila Calcium Channel α1 Subunit Gene Maps to a Genetic Locus Associated with Behavioral and Visual Defects. J Neurosci Official J Soc Neurosci 16, 7868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gratz SJ, Goel P, Bruckner JJ, Hernandez RX, Khateeb K, Macleod GT, Dickman DK, and O’Connor-Giles KM (2019). Endogenous tagging reveals differential regulation of Ca2+ channels at single AZs during presynaptic homeostatic potentiation and depression. Journal of Neuroscience, 3068–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Graf ER, Valakh V, Wright CM, Wu C, Liu Z, Zhang YQ, and DiAntonio A (2012). RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J Neurosci 32, 16586–16596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graf ER, Daniels RW, Burgess RW, Schwarz TL, and DiAntonio A (2009). Rab3 dynamically controls protein composition at active zones. Neuron 64, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams JL, Shearin HK, and Stowers RS (2019). Conditional Synaptic Vesicle Markers for Drosophila. G3 Genes Genomes Genetics 9, g3.200975.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DiAntonio A, Burgess RW, Chin AC, Deitcher DL, Scheller RH, and Schwarz TL (1993). Identification and characterization of Drosophila genes for synaptic vesicle proteins. J Neurosci 13, 4924–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guan Z, Quiñones-Frías MC, Akbergenova Y, and Littleton JT (2020). Drosophila Synaptotagmin 7 negatively regulates synaptic vesicle release and replenishment in a dosage-dependent manner. Elife 9, e55443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Akin O, Nern A, Tsui CYK, Pecot MY, and Zipursky SL (2014). Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Viswanathan S, Williams ME, Bloss EB, Stasevich TJ, Speer CM, Nern A, Pfeiffer BD, Hooks BM, Li W-P, English BP, et al. (2015). High-performance probes for light and electron microscopy. Nat Methods 12, 568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng J, Santiago IJ, Ahn C, Gur B, Tsui CK, Su Z, Xu C, Karakhanyan A, Silies M, and Pecot MY (2018). Drosophila Fezf coordinates laminar-specific connectivity through cell-intrinsic and cell-extrinsic mechanisms. Elife 7, e33962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, and Shaw PJ (2010). The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. Plos Biol 8, e1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keene AC, Duboué ER, McDonald DM, Dus M, Suh GSB, Waddell S, and Blau J (2010). Clock and cycle Limit Starvation-Induced Sleep Loss in Drosophila. Curr Biol 20, 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, and Birman S (2003). Targeted gene expression inDrosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol 54, 618–627. [DOI] [PubMed] [Google Scholar]
- 79.Takemura S, Aso Y, Hige T, Wong A, Lu Z, Xu CS, Rivlin PK, Hess HF, Zhao T, Parag T, et al. (2017). A connectome of a learning and memory center in the adult Drosophila brain. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmann CI (2008). GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. [DOI] [PubMed] [Google Scholar]
- 81.Macpherson LJ, Zaharieva EE, Kearney PJ, Alpert MH, Lin T-Y, Turan Z, Lee C-H, and Gallio M (2015). Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat Commun 6, 10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bayer EA, and Hobert O (2018). Past experience shapes sexually dimorphic neuronal wiring through monoaminergic signalling. Nature 561, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oren-Suissa M, Bayer EA, and Hobert O (2016). Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 533, 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weinberg P, Berkseth M, Zarkower D, and Hobert O (2018). Sexually Dimorphic unc-6/Netrin Expression Controls Sex-Specific Maintenance of Synaptic Connectivity. Curr Biology Cb 28, 623–629.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gordon MD, and Scott K (2009). Motor control in a Drosophila taste circuit. Neuron 61, 373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin T-Y, Luo J, Shinomiya K, Ting C-Y, Lu Z, Meinertzhagen IA, and Lee C-H (2015). Mapping chromatic pathways in the Drosophila visual system. J Comp Neurology 524, 213–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu C, Meng Z, Wiggin TD, Yu J, Reed ML, Guo F, Zhang Y, Rosbash M, and Griffith LC (2019). A Serotonin-Modulated Circuit Controls Sleep Architecture to Regulate Cognitive Function Independent of Total Sleep in Drosophila. Current Biology 29, 3635–3646.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu S, Guo C, Zhao H, Sun M, Chen J, Han C, Peng Q, Qiao H, Peng P, Liu Y, et al. (2019). Drosulfakinin signaling in fruitless circuitry antagonizes P1 neurons to regulate sexual arousal in Drosophila. Nat Commun 10, 4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim J, Zhao T, Petralia RS, Yu Y, Peng H, Myers E, and Magee JC (2011). mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat Methods 9, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi J-H, Sim S-E, Kim J-I, Choi DI, Oh J, Ye S, Lee J, Kim T, Ko H-G, Lim C-S, et al. (2018). Interregional synaptic maps among engram cells underlie memory formation. Sci New York N Y 360, 430–435. [DOI] [PubMed] [Google Scholar]
- 91.Grygoruk A, Chen A, Martin CA, Lawal HO, Fei H, Gutierrez G, Biedermann T, Najibi R, Hadi R, Chouhan AK, et al. (2014). The Redistribution of Drosophila Vesicular Monoamine Transporter Mutants from Synaptic Vesicles to Large Dense-Core Vesicles Impairs Amine-Dependent Behaviors. J Neurosci Official J Soc Neurosci 34, 6924–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saletin JM, Goldstein-Piekarski AN, Greer SM, Stark S, Stark CE, and Walker MP (2016). Human Hippocampal Structure: A Novel Biomarker Predicting Mnemonic Vulnerability to, and Recovery from, Sleep Deprivation. J Neurosci 36, 2355–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu J, Zhou Q, Li J, Chen Y, Shao S, and Xiao Y (2021). Decreased resting-state alpha-band activation and functional connectivity after sleep deprivation. Sci Rep-uk 11, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamazaki EM, Antler CA, Lasek CR, and Goel N (2020). Residual, differential neurobehavioral deficits linger after multiple recovery nights following chronic sleep restriction or acute total sleep deprivation. Sleep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vecsey CG, Baillie GS, Jaganath D, Havekes R, Daniels A, Wimmer M, Huang T, Brown KM, Li X-Y, Descalzi G, et al. (2009). Sleep deprivation impairs cAMP signalling in the hippocampus. Nature 461, 1122–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Havekes R, Park AJ, Tudor JC, Luczak VG, Hansen RT, Ferri SL, Bruinenberg VM, Poplawski SG, Day JP, Aton SJ, et al. (2016). Sleep deprivation causes memory deficits by negatively impacting neuronal connectivity in hippocampal area CA1. eLife 5, e13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spoormaker VI, Schröter MS, Andrade KC, Dresler M, Kiem SA, Goya-Maldonado R, Wetter TC, Holsboer F, Sämann PG, and Czisch M (2011). Effects of rapid eye movement sleep deprivation on fear extinction recall and prediction error signaling. Hum Brain Mapp 33, 2362–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chouhan NS, Griffith LC, Haynes P, and Sehgal A (2020). Availability of food determines the need for sleep in memory consolidation. Nature, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vyazovskiy VV, Olcese U, Lazimy YM, Faraguna U, Esser SK, Williams JC, Cirelli C, and Tononi G (2009). Cortical firing and sleep homeostasis. Neuron 63, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huber R, Mäki H, Rosanova M, Casarotto S, Canali P, Casali AG, Tononi G, and Massimini M (2013). Human Cortical Excitability Increases with Time Awake. Cereb Cortex 23, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xue M, Atallah BV, and Scanziani M (2014). Equalizing excitation–inhibition ratios across visual cortical neurons. Nature 511, 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peng Y-R, Zeng S-Y, Song H-L, Li M-Y, Yamada MK, and Yu X (2010). Postsynaptic Spiking Homeostatically Induces Cell-Autonomous Regulation of Inhibitory Inputs via Retrograde Signaling. J Neurosci 30, 16220–16231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krashes MJ, Keene AC, Leung B, Armstrong JD, and Waddell S (2007). Sequential Use of Mushroom Body Neuron Subsets during Drosophila Odor Memory Processing. Neuron 53, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura S, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, et al. (2020). A connectome and analysis of the adult Drosophila central brain. Elife 9, e57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vanderheyden W, Dongen HPV, Frank M, and Gerstner J (2019). Sleep pressure regulates mushroom body neural-glial interactions in Drosophila. Matters Sel 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Blum ID, Keleş MF, Baz E-S, Han E, Park K, Luu S, Issa H, Brown M, Ho MCW, Tabuchi M, et al. (2020). Astroglial Calcium Signaling Encodes Sleep Need in Drosophila. Curr Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diering GH, Nirujogi RS, Roth RH, Worley PF, Pandey A, and Huganir RL (2017). Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science 355, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shaw PJ, Tononi G, Greenspan RJ, and Robinson DF (2002). Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417, 287–291. [DOI] [PubMed] [Google Scholar]
- 109.Donelson NC, Donelson N, Kim EZ, Slawson JB, Vecsey CG, Huber R, and Griffith LC (2012). High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. Plos One 7, e37250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated during this study. This study did not generate any novel code.