Abstract
The purpose of this study is to study the neuroprotective role of selective serotonin reuptake inhibitor (SSRI), citalopram, against Alzheimer’s disease (AD). Multiple SSRIs, including citalopram, are reported to treat patients with depression, anxiety and AD. However, their protective cellular mechanisms have not been studied completely. In the current study, we investigated the protective role of citalopram against impaired mitochondrial dynamics, defective mitochondrial biogenesis, defective mitophagy and synaptic dysfunction in immortalized mouse primary hippocampal cells (HT22) expressing mutant APP (SWI/IND) mutations. Using quantitative RT-PCR, immunoblotting, biochemical methods and transmission electron microscopy methods, we assessed mutant full-length APP/C-terminal fragments and Aβ levels and mRNA and protein levels of mitochondrial dynamics, biogenesis, mitophagy and synaptic genes in mAPP-HT22 cells and mAPP-HT22 cells treated with citalopram. Increased levels of mRNA levels of mitochondrial fission genes, decreased levels of fusion biogenesis, autophagy, mitophagy and synaptic genes were found in mAPP-HT22 cells relative to WT-HT22 cells. However, mAPP-HT22 cells treated with citalopram compared to mAPP-HT22 cells revealed reduced levels of the mitochondrial fission genes, increased fusion, biogenesis, autophagy, mitophagy and synaptic genes. Our protein data agree with mRNA levels. Transmission electron microscopy revealed significantly increased mitochondrial numbers and reduced mitochondrial length in mAPP-HT22 cells; these were reversed in citalopram-treated mAPP-HT22 cells. Cell survival rates were increased in citalopram-treated mAPP-HT22 relative to citalopram-untreated mAPP-HT22. Further, mAPP and C-terminal fragments werealso reduced in citalopram-treated cells. These findings suggest that citalopram reduces mutant APP and Aβ and mitochondrial toxicities and may have a protective role of mutant APP and Aβ-induced injuries in patients with depression, anxiety and AD.
Introduction
Alzheimer’s disease (AD) is an age-related, multifactorial neurodegenerative disease characterized by memory loss and multiple cognitive changes (1). According to the World Alzheimer’s Report, in 2020 over 50 million people worldwide had AD, and it projects that this number will increase to more than 152 million by 2050. In 2020, the total estimated annual worldwide healthcare cost for persons with AD was $980 billion (2). With the increased human lifespan, AD is a significant health concern in the society (1).
Depression is among the most common neuropsychiatric comorbidities in AD. All nine DSM-IV clinical symptoms of depression, anxiety and apathy may develop in 96% of patients (3). Recent studies suggest that depression comorbidity is a modifiable risk factor in AD among hypertension and diabetes (4). Antidepressant treatment offers intracellular modification that may help to improve neurogenesis, amyloid burden, tau pathology and neuroinflammation (4–16). The COVID19 outbreak elevated anxiety in AD patients in retirement homes and created a new burden to society (17).
Selective serotonin reuptake inhibitors (SSRIs) are commonly prescribed antidepressants to ameliorate depressive symptoms associated with AD (18–23). They target serotonin receptors and transporters thus limiting reabsorption of serotonin (5HT) into presynaptic neurons, resulting in increased serotonin levels in the synaptic cleft, ultimately resulting in an increase in signaling and ligand–receptor binding (24). Increased serotonergic neurotransmission results in anti-depressive and anxiolytic effects (25–27).
Serotonin is a key neurotransmitter that plays a large role in synaptic functions of neurons affected by AD and is also the most predominantly dysregulated neurotransmitter that has been found in AD neurons (28–33). It regulates multiple higher-order physiological processes like neuronal resilience, depression, dementia, energy homeostasis, circadian rhythm, diabetes, neurodegeneration and neurogenesis (5,25–27,29,34–39). Despite its multifaceted impact in the brain, the physiological and molecular mechanism of how serotonin can restore synaptic plasticity in depression and process of neurogenesis and cellular metabolism in dementia is still poorly understood.
Citalopram is an SSRI that is the most prescribed antidepressant in AD patients as it has fewer side effects and has been found to be effective in reducing depression in AD patients (12,22,40). Citalopram is a bicyclic phthalein derivative with antidepressant and anxiolytic activities. Citalopram selectively inhibits the neuronal reuptake of serotonin in presynaptic cells in the central nervous system and enhances the actions of serotonin on its receptors (24,41). SSRIs, including citalopram, are hypothesized to alter the processing of APP and the generation of Aβ and to delay the progression of AD (5,7,10–13). Elevated serotonin signaling was associated with decreased interstitial fluid Aβ peptide and Aβ plaque load (6,8,9).
Besides, SSRI use over 5 years was associated with lower Pittsburgh compound B and Aβ load in cognitively healthy participants (6). SSRIs have also shown to modulate the Aβ peptide(s) soluble species generated and, therefore, can reduce the toxicity associated with oligomeric forms of Aβ (10). Citalopram may also have downstream benefits on phosphorylated Tau (p-Tau) by minimizing the potential for Aβ-tau cross talk (11,14,15). SSRIs may also help by reducing synergistic toxic effects of Aβ and p-Tau, thus reducing the associated synaptic and neuronal dysfunction observed in AD patients (4,11). Treatment of AD patients with citalopram has shown to reduce circulating cytokines and to interact with inflammatory signaling pathways that typically elevated in chronic inflammatory conditions (4,16). All these put together may help reduce cognitive impairment and depression in patients with AD and possibly with other neurodegenerative diseases (10,23,27–29,42,43). However, underlying mechanisms of neuroprotection are not well understood.
Mitochondria, the cellular powerhouses, generate energy to execute higher-order cellular functions such as energy homeostasis, cellular survival, synaptogenesis and neuronal resilience (44,45). Mitochondrial abnormalities, including impaired mitochondrial dynamics, defective biogenesis, defective mitophagy and mitochondrial dysfunction, are largely involved in AD (46–49). Neurotransmitters, neurosteroids and antioxidants maintain and/or increase cellular ATP and respiration rate by increasing mitochondrial biogenesis (50). Aging, neurodegeneration, depression and brain metabolic syndromes have been reported to have serotonin dysfunction, reduce cellular energy and lower cellular respiration (51,47). Serotonin induces cell metabolism by increasing mitochondrial biogenesis and enhancing mitochondrial function (50,52). Citalopram-targeted mitochondrial biogenesis reduces mitochondrial dysfunction and improves cellular resilience (53). Another report also suggests that serotonin enhances mitochondrial movement in hippocampal axons by inhibiting GSK3β activity via Akt pathway (54).
Serotonin’s role in mitochondria in cortical neurons has shown the light on Sirt1, PGC1α-mediated 5HT2A receptor post-synaptic modulation, which increases mitochondrial biogenesis (52,56). Some published studies and preliminary data have demonstrated improved mitochondrial dynamics in a cellular model of serotonin and non-serotonin neuron (55,56). The 5HT2A receptor is also a synaptic marker of serotonin upregulation as serotonin, tryptophan hydroxylase2 and 5HT2R directionally increase simultaneously (19,34,57–60).
Another theory explaining the role of serotonin in mitochondrial function is based on serotonin’s permissive entry to the presynaptic neurons in gut and brain neurons that synthesize serotonin (56–62). Post-translational modification is a result of increase in the histone serotonylation. This leads to an increase in the cellular levels of serotonin, which directly binds to mitochondria via Sirt1-PGC1α in the presence of 5HT2A receptor (52,55). The birth of mitochondria increases mitochondrial mass and improves neuroregeneration by improving adult neurogenesis, synaptogenesis and cell migration (64–65).
Autophagy is often dysregulated in neurodegenerative diseases, and considerable evidence shows that SSRIs promote autophagic processes (66–71). Electron microscopy and protein studies indicate an increased colocalization of autophagosomes and mitochondria and a consequent autophagic clearance of dysfunctional mitochondria (66,68). Therefore, mitophagy could be another important pathway involved in the SSRIs and mitochondria-associated neuroprotective action seen in AD. Consistent with this, the levels of autophagy-related genes ATG4B, ATG5, ATG7 and LC3B were found elevated, along with a parallel increase in mitophagy genes such as Bcl-2 and p62. A simultaneous reduction in the autophagy inhibitor Beclin1 levels was also seen (66,67,70). Hwang et al. demonstrated that SSRI sertraline mechanistically binds to and antagonizes the mitochondrial VDAC1 (voltage dependent anion channel 1) and suppresses tauopathy by promoting the autophagic degradation of microtubule-associated protein tau (MAPT) protein via inducing autophagy (71).
However, SSRI, citalopram is protective in hippocampal neurons that express mutant APP and Aβ in AD is not yet studied. Hippocampus plays an essential role in consolidating information from short-term memory to long-term memory. A short-term memory loss and disorientation are the early symptoms in AD (14–16). Thus, studies on hippocampal tissues are the best source of studying learning and memory functions in both healthy and dementia states. Further, investigation of hippocampal neurons, particularly immortalized neurons that express mutant APP and Aβ, will provide an excellent opportunity to test drug targets of AD.
In the current study, we sought to determine the protective actions of SSRI, citalopram, against mutant APP and Aβ induced: (1) impaired mitochondrial dynamics, (2) defective biogenesis, (3) defective autophagy, (4) defective mitophagy, (5) synaptic damage, (6) cell survival, (7) mutant full-length APP and C-terminal fragments and (8) Aβ levels.
Results
mRNA levels of mitochondrial dynamics and mitochondrial biogenesis genes
Using the reagent TriZol (Invitrogen), we isolated total RNA from all four groups of cells—(1) HTT22 cells, (2) HT22+Citalopram treated, (3) HT22+mutant APP transfected and (4) HT22+mutant APP transfected and Citalopram treated (see Fig. 1). mRNA levels of mitochondrial dynamic genes (fission Drp1 & Fis1 and fusion Mfn1, Mfn2 and Opa1), mitochondrial biogenesis genes (PGC1α, Nrf1, Nrf2 and TFAM), autophagy (LC3A, LC3B, ATG5, Beclin1), mitophagy (PINK1, TERT, BCL2 and BNIP3L) and synaptic genes (PSD95, synaptophysin) were measured by using Sybr-Green chemistry-based quantitative real-time RT-PCR.
Figure 1 .
Flowchart of cells used in the present study for citalopram treatment and experiment conducted.
Mitochondrial dynamics
As shown in Table 1, in mutant APP transfected HT22 cells (refer as mAPP-HT22 from on), mRNA levels of mitochondrial fission genes were significantly increased (Drp1 by 2.1-fold, P < 0.005 and Fis1 by 1.7-fold, P < 0.05) compared with control HT22 cells. In contrast, mRNA expression levels of mitochondrial fusion genes were significantly decreased (Mfn1 by 1.8-fold, P < 0.05; Mfn2 by 2.2-fold, P < 0.05 and Opa1 by 1.9, P < 0.05) in mAPP-HT22 cells relative to control HT22 cells. This indicates the presence of abnormal mitochondrial dynamics in mAPP cells. However, citalopram-treated mAPP-HT22 cells showed reduced Drp1 (2.5-fold decrease, P < 0.005) and Fis1 (2.2-fold, P < 0.005) and increased fusion genes in citalopram-treated mAPP-HT22 relative to citalopram-untreated mAPP-HT22 cells. These observations indicate that citalopram reduced fission activity and enhanced fusion machinery in mAPP cells.
Table 1.
Fold changes of mRNA expression of mitochondrial structural, synaptic, biogenesis, autophagy and mitophagy genes in mutant APP-HT22 cells compared with HT22 cells and also citalopram-treated mutant APP-HT22 cells compared with citalopram-untreated mutant APP-HT22 cells
| Genes | mRNA fold changes | mRNA fold change | |
|---|---|---|---|
| HT22 versus HT22 + mAPP | HT22 + mAPP versus HT22 + mAPP+Cit | ||
| Mitochondrial structural genes | Drp1 | 2.1** | -2.5** |
| Fis1 | 1.7* | -2.2** | |
| Mfn1 | -1.8* | 2.0* | |
| Mfn2 | -2.2* | 2.7* | |
| OPA1 | -1.9 | 2.1* | |
| Synaptic genes | Synaptophysin | -2.2** | 2.7** |
| PSD95 | -2.1** | 2.0* | |
| Serotonin-related genes | SERT | -2.4 | 1.9** |
| TPH 2 | -2.1** | 3.4* | |
| 5HTR1A | -2.0* | 1.8* | |
| 5HTR1B | -2.1 | 2.8 | |
| 5HTR4 | -2.7 | 3.7* | |
| 5HTR6 | 2.1 | -2.1 | |
| Biogenesis genes | Nrf1 | -1.8* | 2.3 |
| PGC1α | -2.1** | 3.1** | |
| Nrf2 | -1.6* | 2.3* | |
| TFAM | -2.5** | 3.1* | |
| Autophagy genes | LC3A | -1.6* | 2.3* |
| LC3B | -1.8* | 2.5* | |
| ATG5 | -2.5 | 2.4* | |
| Beclin1 | -1.5* | 2.2* | |
| Mitophagy genes | TERT | -2.1** | 3.5* |
| PINK1 | -2.4** | 2.0* | |
| BCL2 | -1.5* | 2.1* | |
| BNIP3L | -1.3 | 2.7* | |
| AD genes | MAPT | 2.8* | -2.8 |
| APP | 2.1 | -2.3* | |
| BACE1 | 2.7 | -2.4* | |
| ADAM10 | -2.1* | 3.2* |
*P < 0.05.
**P < 0.005.
Mitochondrial biogenesis
mRNA levels of mitochondrial biogenesis genes were significantly reduced (PGC1a by 2.1-fold, P < 0.005; Nrf1 by 1.8-fold, P < 0.05; Nrf2 by 1.6-fold, P < 0.05 and TFAM by 2.5, P < 0.005) in mAPP-HT22 cells relative to control HT22 cells. However, as shown in Table 1, in citalopram-treated mAPP-HT22 cells, mRNA levels of mitochondrial biogenesis were increased (PGC1a by 3.1-fold, P < 0.005; Nrf1 by 2.3-fold, P < 0.05; Nrf2 by 2.3-fold and TFAM by 3.1-fold, P < 0.005). These observations strongly suggest that citalopram increases mitochondrial biogenesis activity in the presence of mutant APP and amyloid beta in HT22 cells.
Autophagy
In mAPP-HT22 cells relative to control HT22 cells, mRNA levels of autophagy genes were significantly reduced (LC3A by 1.6-fold, P < 0.05; LC3B by 1.8-fold, P < 0.05; ATG5 by 2.5-fold, P < 0.05 and Beclin1 by 1.5-fold, P < 0.05) (Table 1). On the contrary, in citalopram-treated mAPP-HT22 cells relative to citalopram-untreated mAPP-HT22 cells, mRNA levels of autophagy genes were increased (LC3A by 2.3-fold, P < 0.05; LC3B by 2.5-fold, P < 0.05; ATG5 by 2.4-fold, P < 0.05 and Beclin1 by 2.2-fold, P < 0.005). These observations suggest that citalopram increases autophagy activity in AD cells.
Mitophagy
As shown in Table 1 in mAPP-HT22 cells, mRNA levels of mitophagy genes were significantly reduced (PINK1 by 2.4-fold, P < 0.005; TERT by 2.1-fold, P < 0.005, BCL2 by 1.5-fold, P < 0.05 and BNIP3L by 1.3-fold, P < 0.0.5) relative to control HT22 cells, indicating that mutant APP and Aβ reduce mitophagy activities. However, citalopram-treated mAPP-HT22 cells showed opposite effects, meaning mitophagy genes were increased in citalopram-treated mAPP-HT22 cells (PINK1 by 2.0-fold; TERT by 3.5-fold, P < 0.005; BCL2 by 2.1-fold, P < 0.05 and BNIP3L by 2.7-fold, P < 0.0.5) relative to citalopram-untreated mAPP-HT22 cells. These observations indicate that citalopram enhanced mitophagy activity in AD cells.
Synaptic genes
mRNA levels of synaptic genes were significantly reduced (synaptophysin by 2.2-fold, P ≤ 0.05 and PSD95 by 3.1-fold, P < 0.05) in mAPP-HT22 cells relative to control HT22 cells (Table 1). However, in citalopram-treated mAPP-HT22 cells relative to citalopram-untreated mAPP-HT22 cells, mRNA levels of synaptic genes were increased (synaptophysin by 2.7-fold, P < 0.005 and PSD95 by 2.0-fold, P < 0.05) (Table 1). These observations indicate that citalopram increases synaptic activity in AD cells.
Serotonin-related genes
mRNA levels of serotonin-related genes were significantly reduced (SERT by 2.4-fold, P ≤ 0.05; TPH2 by 2.1-fold, P < 0.005; 5HTR1A by 2.0-fold, P < 0.05; 5HTR1B by 2.1-fold; 5HTR4 by 2.7-fold, P < 0.05) in mAPP-HT22 cells relative to control HT22 cells (Table 1). On the contrary, in citalopram-treated mAPP-HT22 cells relative to citalopram-untreated mAPP-HT22 cells, mRNA levels of serotonin-related genes were significantly increased (SERT by 1.9-fold, P ≤ 0.005; TPH2 by 3.4-fold, P < 0.05; 5HTR1A by 1.8-fold, P < 0.05; 5HTR1B by 2.8-fold; 5HTR4 by 3.7-fold, P < 0.05). mRNA levels of 5HTR6 were increased in mAPP-HT22 cells relative to control HT22 cells; however in citalopram-treated mAPP-HT22 cells relative to citalopram-untreated mAPP-HT22 cells, mRNA levels of 5HTR6 were reduced. Overall, these observations suggest that citalopram enhances serotonin levels in disease state.
AD-related genes
To determine the impact of citalopram in AD-related genes, mRNA levels of MAPT, APP, BACE1 and ADAM10 (a-secretase) were measured—(1) in mAPP-HT22 cells relative to control HT22 cells and (2) in mAPP-HT22 cells treated with citalopram relative to citalopram-untreated mAPP-HT22 cells. AD-related genes were upregulated (MAPT by 2.8-fold, P < 0.05; APP by 2.1-fold; BACE1 by 2.7-fold) in mAPP-HT22 cells relative to control HT22 cells (Table 1). However, mRNA levels were significantly reduced (MAPT by 2.8-fold, P < 0.05; APP by 2.3-fold, P < 0.05; BACE1 by 2.4-fold, P < 0.05) in mAPP-HT22 cells treated with citalopram relative to citalopram-untreated mAPP-HT22 cells. These observations indicate that citalopram reduces the toxicities of AD-related genes.
As expected, ADAM10 that represents a-secretase was reduced in mAPP-HT22 cells (ADAM10 by 2.1-fold, P < 0.05) relative to control HT22 cells. ADAM10 levels were significantly increased in citalopram-treated mAPP-HT22 cells (ADAM10 by 3.2-fold, P < 0.05) relative to citalopram-untreated mAPP-HT22 cells.
Citalopram enhances cell viability
To determine the effect of citalopram on cell viability, HT22 cells and HT22 cells transfected with mutant APP cDNA. As shown in Figure 2, cell viability was significantly decreased in mutant APP cells (P = 0.02) relative to control HT22 cells. However, cell viability was increased in citalopram-treated mutant APP cells (P = 0.04) relative to citalopram-untreated mutant APP cells.
Figure 2 .
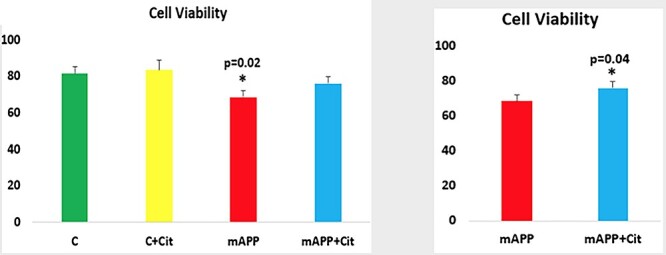
Cell viability assays in HT22 cells and HT22 cells transfected with mutant APP cDNA. Cell viability was significantly decreased in mutant APP cells (P = 0.02) relative to control HT22 cells. However, cell viability was increased in citalopram-treated mutant APP cells (P = 0.04) relative to citalopram-untreated mutant APP cells.
Citalopram suppresses full-length mutant APP
To determine the impact of citalopram on mutant APP and c-terminal fragments of APP, mutant APP cells were treated with citalopram at 20 μM final concentration. As shown in Figure 3, reduced levels of full-length mutant APP were found in citalopram-treated mutant APP cells. Mouse hippocampal cells transfected with mutant APP cDNA and treated with 20 μM citalopram for 48 h. A full-length 110 kDa mAPP protein was found in the transfected cells. Quantitative densitometry analysis of the full-length mAPP in transfected cells shows a significant decrease in the citalopram-treated cells (P = 0.03).
Figure 3 .
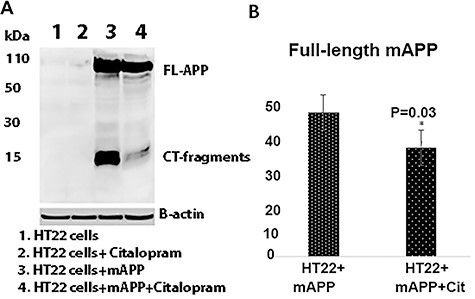
Immunoblotting of mAPP-HT22 cells with 6E10 antibody. (A) A full-length 110 kDa mAPP protein was found in the transfected cells. (B) Quantitative densitometry analysis of the full length mAPP.
Citalopram reduces the levels of Aβ40 and 42
Mutant APP cells were treated with citalopram and assessed Aβ40 and 42 using sandwich ELISA. As shown in Figure 4, mutant APP cells treated with citalopram showed significantly reduced levels of Aβ42 (P = 0.01) relative to citalopram-untreated mutant APP-treated cells. These observations suggest that citalopram affects APP processing and reduces Aβ levels.
Figure 4 .
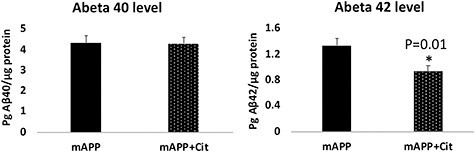
Aβ40 and 42 levels of protein lysates obtained from HT22 cells transfected with mutant APP cDNA and treated with citalopram. (A) Aβ40 did not change much between the treated and untreated groups. (B) On the contrary, a significant reduction in the Aβ42 levels was observed upon citalopram treatment.
Citalopram maintains mitochondrial dynamics
Reduced levels of fission (Drp1—P = 0.01 and Fis1 P = 0.05) and increased levels of fusion proteins (Mfn1—P = 0.003, Mfn2—P = 0.04 and Opa1—P = 0.05) were found in citalopram-treated HT22 cells relative to untreated cells (Fig. 5). Mutant APP cells showed increased fission (Drp1—P = 0.005, Fis1—P = 0.04) and reduced fusion proteins (Mfn1—P = 0.002, Mfn2—P = 0.04 and Opa1—P = 0.05) relative to control HT22 cells; however, relative to mutant APP cells, citalopram-treated mutant APP cells showed reduced fission (Drp1—P = 0.004 and Fis1—P = 0.005) and increased fusion (Mfn1—P = 0.02, Mfn2—P = 0.05 and Opa1—P = 0.04) proteins (Fig. 5). These observations indicate that citalopram reduced Aβ-induced mitochondrial toxicities.
Figure 5 .
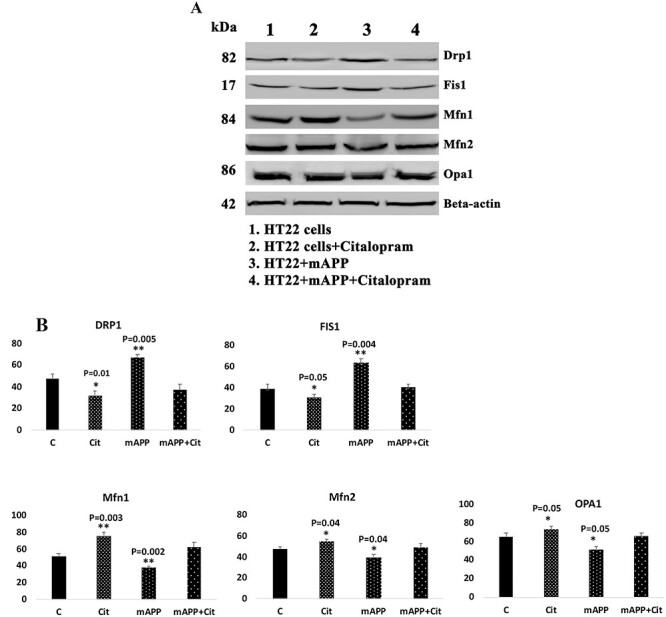
Immunoblotting analysis of mitochondrial dynamics proteins. (A) Representative immunoblots for control and mAPP-HT22 cells with or without citalopram. (B) Quantitative densitometry analysis for mitochondrial dynamics proteins—significantly increased levels of fission proteins Drp1 and Fis1 were observed in cells transfected with mutant APP. Fusion proteins Mfn1, Mfn2 and Opa1 were significantly decreased. On the other hand, citalopram-treated mutant APP showed reduced levels of fission proteins and increased levels of fusion proteins were observed.
Citalopram enhances mitochondrial biogenesis
The protective roles of citalopram were determined against the mutant APP-induced mitochondrial dynamics, biogenesis and synaptic proteins. As shown in Figure 6, increased levels of mitochondrial biogenesis proteins (PGC1α—P = 0.01, NRF1—P = 0.02, NRF2—P = 0.05 and TFAM—P = 0.05) were found in HT22 cells treated with citalopram relative to citalopram-untreated HT22 cells; however, these levels were reduced in mutant APP cells (PGC1α—P = 0.004, NRF1—P = 0.02, NRF2—P = 0.003 and TFAM—P = 0.01) (Fig. 6). Increased levels of PGC1α—P = 0.01, NRF1—P = 0.04, NRF2—P = 0.004 and TFAM—P = 0.03 were found in citalopram-treated mutant APP cells relative to citalopram-untreated APP cells.
Figure 6 .
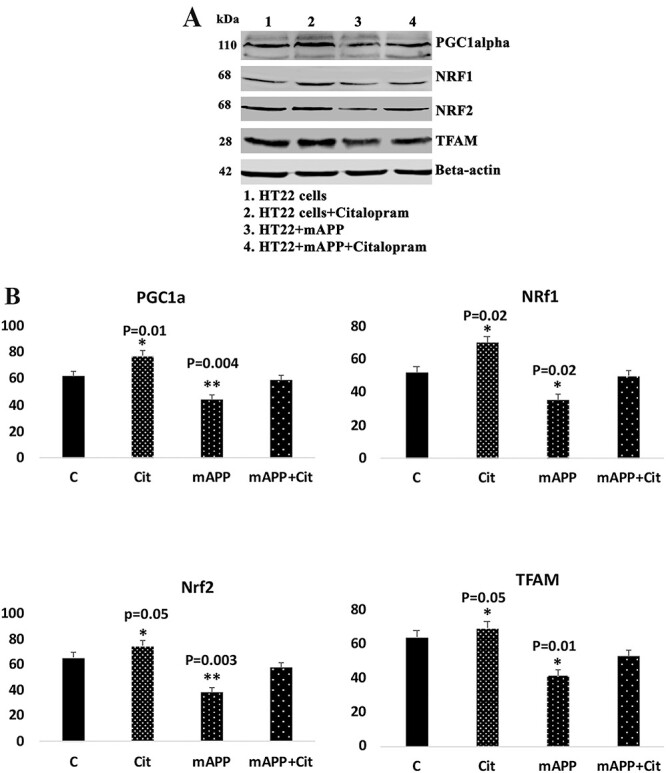
Immunoblotting analysis of mitochondrial biogenesis proteins in HT22 cells and mutant APP cDNA transfected and treated with 20 μm citalopram for 48 h. (A) Representative immunoblots for control HT22 and mAPP-HT22 cells with or without citalopram treatment. (B) Quantitative densitometry analysis showed significant reduction in the levels of PGC1a, NRF1, NRF2 and TFAM upon mAPP cDNA transfection. But levels of all mitochondrial biogenesis proteins increased with citalopram treatment.
Citalopram enhances mitophagy/autophagy
The protective role of citalopram against amyloid beta-induced mitophagy/autophagy was assessed. As shown in Figure 7, in mutant APP cells, reduced levels of autophagy (ATG5—P = 0.002, ATG7—P = 0.01, LC3BI—P = 0.02 and LC3BII—P = 0.02, P62—P = 0.02) and mitophagy (PINK1, P = 0.003, TERT—P = 0.01) proteins were found, and these proteins were increased in citalopram-treated mutant APP cells relative to mutant APP cells, indicating that citalopram reduced defective mitophagy/autophagy in mutant APP cells (Fig. 7).
Figure 7 .
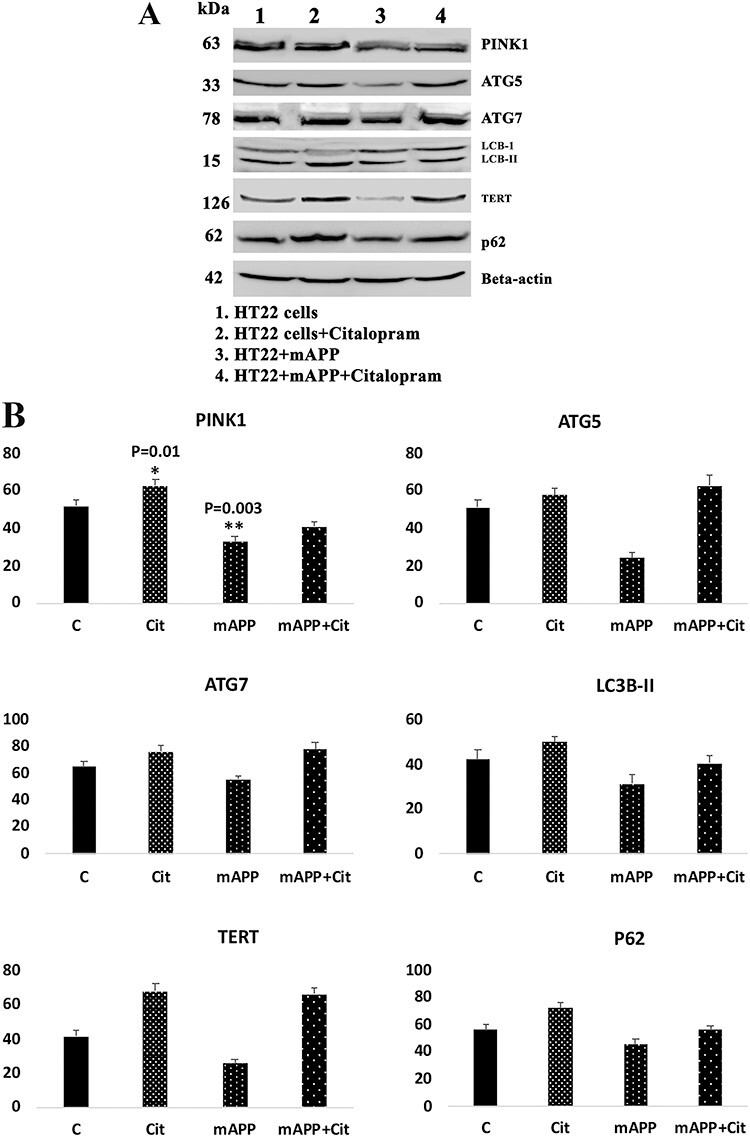
Immunoblotting analysis of mitophagy and autophagy proteins in control HT22 cells and mutant APP cDNA transfected HT22 cells and 20 μm citalopram treated for 48 h. (A) Representative immunoblots for control HT22 cells and mAPP-HT22 cells with or without CIT. (B) Quantitative densitometry analysis of mitophagy and autophagy proteins. Upon mAPP transfection significant reduction was only seen in the levels of PINK1 (P = 0.003), while ATG5, ATG7, TERT, LC3B and P62 reduced though the change was insignificant.
Citalopram enhances synaptic and dendritic activities
As shown in Figure 8, synaptic and dendritic proteins synaptophysin (P = 0.004), PSD95 (P = 0.05) and MAP2 (P < 0.02) were reduced in mutant APP cells; however, these were increased in citalopram-treated mutant APP cells (synaptophysin P = 0.005, PSD95 P = 0.01 and MAP P < 0.01) relative to citalopram-untreated mutant APP cells, indicating that citalopram enhances synaptic activity in the presence of Aβ.
Figure 8 .
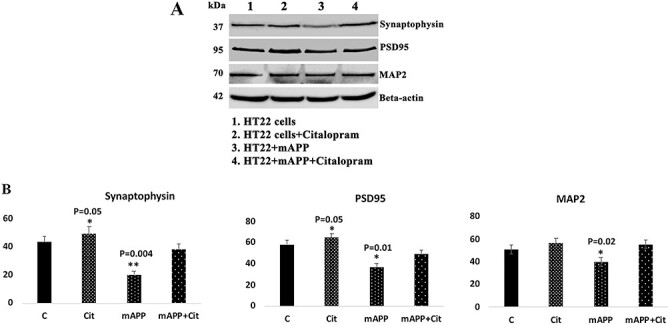
Immunoblotting analysis of synaptic proteins and dendritic protein MAP2. (A) Representative immunoblots for control and mAPP-HT22 cells with or without CIT. (B) Quantitative densitometry analysis of synaptic and MAP2 proteins.
Citalopram reduces mitochondrial number and increases mitochondrial length
Increased number of mitochondria were found in the mutant APP cells (P = 0.03) relative to HT22 cells, suggesting that Aβ fragments hippocampal mitochondria (Fig. 9). Mitochondrial length was decreased in mutant APP (P = 0.01) relative to control cells. On the other hand, decreased number of mitochondria were seen in the citalopram-treated mutant APP cells (P = 0.03) relative to citalopram-untreated mutant APP cells. Mitochondrial length was increased in citalopram-treated mutant APP cells (P = 0.03) relative to citalopram-untreated mutant APP cells (Fig. 9).
Figure 9 .
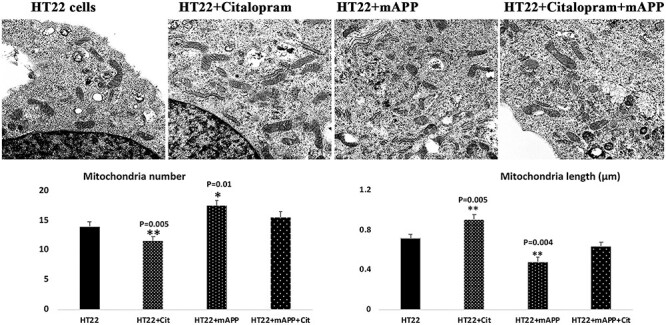
Transmission electron microscopy analysis. Mitochondrial number and length in control HT22 cells and mutant APP cDNA transfected HT22 and treated with 20 μM citalopram for 48 h. (A) Representative transmission electron microscopy images of mitochondria in the treated and untreated cells. (B) Quantitative analysis of mitochondrial number and length in each of the four groups. Significantly increased number of mitochondria was found in HT22 cells transfected with mutant APP relative to untransfected HT22 cells. Mitochondrial length significantly decreased upon mutant APP cDNA transfection. Citalopram treatment decreased the mitochondrial number and increased its length in the control HT22 cells.
Discussion
The objective of our study is to assess the protective effects of SSRI, citalopram, against the toxic effects of mutant APP and Aβ in mouse primary hippocampal (HT22) neurons. The HT22 cells were transfected with mutant APP cDNA that express Swe/Ind mutations and studied mutant APP/Aβ toxic effects. To determine the protective effects of citalopram, mAPP-HT22 cells were treated with citalopram and measured mRNA and protein levels of mitochondrial dynamics, biogenesis, synaptic, dendritic, autophagy/mitophagy genes and assessed cell survival in mAPP-HT22 cells, and untransfected, WT-HT22 cells. In mAPP-HT22 cells relative to HT22 cells, increased levels of mRNA of mitochondrial fission genes; decreased levels of fusion, biogenesis, autophagy, mitophagy and synaptic genes. However, in mAPP-HT22 cells treated with citalopram compared with mAPP-HT22 cells, reduced levels of the mitochondrial fission genes, increased fusion, biogenesis, autophagy, mitophagy and synaptic genes were found. Interestingly, serotonin-related genes were upregulated in citalopram-treated mAPP-HT22 cells relative to citalopram-untreated mAPP-HT22, indicating that citalopram enhances serotonin levels—this is the direct evidence that suggests increased serotonin is responsible for all synaptic and mitochondrial changes. Our protein data agree with mRNA levels. Transmission electron microscopy revealed significantly increased mitochondrial numbers and reduced mitochondrial length in mAPP-HT22 cells; these were reversed in citalopram-treated mAPP-HT22 cells, indicating that citalopram reduces fission activity and enhances fusion machinery of mitochondria. Cell survival rates were increased in citalopram-treated mAPP-HT22 relative to mAPP-HT22 (Fig. 2), indicating that citalopram has ability to reduce mutant APP and/or Aβ and maintains cell survival in a disease state. These observations strongly suggest that citalopram is a potential therapeutic candidate for mutant APP and Aβ-induced injuries in patients with depression, anxiety and AD.
Citalopram reduces mutant APP, C-terminal fragments and Aβ
Our extensive immunoblotting analysis revealed that mutant APP and C-terminal fragments were reduced in citalopram-treated mAPP-HT22 cells relative to mAPP-HT22 cells (Fig. 3). Further, our sandwich analysis of Aβ levels in citalopram-treated mAPP-HT22 cells revealed that significantly reduced Aβ42, but not Aβ40 (Fig. 4), strongly indicates that citalopram affects APP processing, mainly g-secretase in citalopram-treated cells. It is interesting to see significant reduction of mutant full-length APP and C-terminal fragments in citalopram-treated cells and reduced mutant full-length APP makes sense how citalopram impacts amyloid cascade events in disease progression and development, such as cellular homeostasis, mitochondrial biogenesis and synaptic activities in AD cells. Overall, these findings suggest that citalopram reduces mutant APP and Aβ levels. Further research is still needed to understand the precise mechanism of citalopram involvement in APP processing.
Citalopram reversed the impaired mitochondrial dynamics in AD
Current study findings of mAPP and Aβ-induced impaired mitochondrial dynamics concur with earlier studies of hippocampal tissues from 12-month-old APP transgenic mice (72), mouse neuroblastoma cells incubated with Aβ25-35 peptide (73) and APP primary neuronal cultures (74). In addition, impaired mitochondrial dynamics (increased mitochondrial fission and reduced fusion) is extensively reported in diabetes, obesity, hyperglycemia, depression and anxiety conditions. Mitochondrial fission proteins, Drp1 and Fis1, are activated due to (1) abnormal interactions between Drp1 and Aβ that enhance Drp1 GTPase activity (73,75,76) increased free radicals due to hyperglycemia, diabetes/obesity, depression and anxiety conditions that activate Drp1 and Fis1 that enhance Drp1 GTPase activity (44,77). Increased Drp1-GTPase activity fragments mitochondria excessively, leading to impaired mitochondrial dynamics, ultimately defective biogenesis and mitophagy in AD (73,74,76,78). And also, fusion proteins (Mfn1, Mfn2 and Opa1) were significantly reduced in mAPP-HT22 cells, indicating the presence of impaired mitochondrial dynamics in mAPP-HT22 cells.
However, in the current study, citalopram reduced the levels of fission proteins (Drp1 and Fis1) and increased fusion proteins (Mfn1, Mfn2 and Opa1) in mAPP-HT22 cells. As demonstrated, citalopram reduces mAPP/Aβ and Drp1 levels in mAPP-HT22 cells; and these reduced levels of mAPP/Aβ and Drp1 ultimately reduce fission activity and maintain mitochondrial dynamics in AD cells. However, further research still needed to understand the precise role of citalopram in enhancing fusion and reducing fission in AD cells. And further research is still urgently needed to understand how citalopram treatment increases serotonin, particularly synaptic serotonin, and reduces mutant APP/Aβ levels and mutant APP/Aβ-induced mitochondrial free radicals in AD.
It is possible that increased serotonin (due to citalopram treatment) induces/increases a-secretase levels and cleaves Aβ peptide into small pieces and clears from AD cells (79). Thus, increased clearance of Aβ levels and reduced synthesis of Aβ levels (due to increased a-secretase) levels in citalopram-treated AD cells, ultimately reduces Aβ-induced excessive mitochondrial fragmentation. These possibilities need to be studied at the biochemical and molecular levels using postmortem AD brains, AD cell and mouse models.
Defective mitochondrial biogenesis
mRNA and protein levels of mitochondrial biogenesis genes were reduced in mAPP-HT22. These observations indicate that reduced mitochondrial biogenesis is a typical AD feature. Enhancement of mitochondrial biogenesis is an ideal therapeutic approach in AD. Citalopram treatment in multiple clinical trials supports beneficial effects on neurotransmitters and improves AD pathogenesis. This study has shown a mechanism of mitochondrial biogenesis and citalopram treatment, which is a new concept. The result shows that SSRI, citalopram, improves mitochondrial biogenesis, dynamics and by clearing Aβ levels in mAPP-HT22 cells. Clearance improved in neurons expressing amyloid b and SSRI treatment.
Defective mitochondrial biogenesis has been reported in depression, anxiety and AD states. Further, serotonin levels are reported to be low in these conditions. Mechanistically, antidepressants, such as citalopram and others, work by increasing synaptic levels of serotonin and maintain mitochondrial function, which is defective in AD, depression and anxiety conditions. It is possible that serotonin levels were increased in citalopram-treated mAPP-HT22 cells, which results the direct effect of serotonin targeting mitochondria and improving mitochondrial function, thereby improving synaptic mitochondrial function.
Citalopram reversed defective autophagy and mitophagy
In the current study, we found reduced mRNA and protein levels of autophagy ATG5, LC3BI and LC3BII, and mitophagy proteins PINK1 and TERT in hippocampal cells that express mutant APP and Aβ. We also found that mRNA and protein levels of autophagy and mitophagy genes were increased in mAPP-HT22 cells with citalopram treatment. These observations indicate that abnormal accumulation of mutant APP and Aβ affects autophagy, and mitophagy events in hippocampal cells and citalopram reversed APP and Aβ-induced defective autophagy and mitophagy activities in AD.
Further, our observations of defective autophagy and mitophagy in mAPP-HT22 cells strongly agree with mammalian target of rapamycin (mTOR) involvement of mutant APP and Aβ-induced autophagy and mitophagy in AD (80–83). Activation of autophagy and mitophagy is hypothesized as a therapeutic target for AD (79,84,85).
Like other G protein-coupled receptors, citalopram-treated increased serotonin directly activates PI3K/AKT/GSK3/mTOR-mediated cell signaling (86). We have also seen that serotonin directly affects mTOR signaling (unpublished observations, A. Reddy Lab). The processes result in activated autophagy. In multiple neurodegenerative diseases, including AD and MCI, loss of SERT may result in defective autophagy (87). However, as demonstrated in the current study, citalopram treatment mitigates biological pathways involved in AD pathologies via autophagy and mitophagy.
Oxidative stress-induced mitochondrial damage is a common intracellular mechanism that triggers the activation of autophagy to clear damaged mitochondria. Mitophagy is being an integral part of mitochondrial biogenesis, because clearance of mitochondria allows synthesis of new mitochondria.
Citalopram treatment increases synaptic activities
In the current study, we found reduced levels of synaptic proteins, synaptophysin and PSD95, in hippocampal cells that express mutant APP and Aβ, indicating that APP and Aβ affects synaptic proteins. Our observations concur with previous studies (72,88,89). Serotonin levels are low in hippocampal neurons from AD patients and AD mouse models, indicating that reduced serotonin levels affect synaptic functions in AD. Increasing evidence also suggests that spine density is critical for synaptic and cognitive functions in AD patients and AD mice and reduced levels of synaptic and dendritic proteins are undoubtedly responsible for synaptic damage and cognitive functions in AD. However, citalopram treatment increases serotonin in hippocampal neurons, which results in improved synaptic proteins and overall synaptic activity in AD hippocampal neurons. Mutant APP and Aβ expressing cells, when treated with citalopram, reversed the synaptic damage.
Considerable evidence suggests that the cholinergic hypothesis is no more the solo player in the neurotransmitter hypothesis related to AD. Post-transcriptional histone complex modification might play an essential role in the hippocampal neurons by downregulating gene transcription in pathological conditions. On the other hand, neurons synthesize neurotransmitters including serotonin might improve RNA transcription via histone serotonylation. SSRI-induced serotonin transcription in the presynaptic neurons of raphe and gut might improve serotonin transmission in the basal forebrain’s cholinergic neurons. (55,56,61). Further research is still needed to unravel these proposed events in AD.
Citalopram treatment increases serotonin
Increasing evidence suggests that serotonin levels are low in individuals with depression, anxiety, AD and ADRD and other neurological conditions (6,12,16,24,32), indicating that reduced serotonin levels trigger clinical symptoms of depression, anxiety and AD and psychosis conditions. Citalopram treatment is predicted to increase serotonin levels in synaptic clefts and other regions in neurons. Therefore, we measured mRNA levels of several genes-related serotonin in citalopram-treated HT22 cells and citalopram-treated mAPP-HT22 relative citalopram-untreated cells. As expected, mRNA levels of serotonin-related genes were upregulated in both citalopram HT22 cells and citalopram-treated mAPP-HT22 cells (Table 1). These observations strongly suggest that serotonin impacts synaptic, mitochondrial biogenesis, mitochondrial dynamics, autophagy, and mitophagy in AD, depression, anxiety and ADRD conditions.
Mechanistic impact of citalopram in late-onset sporadic AD
SSRI, citalopram, is a prescribed drug for individuals with depression, anxiety, AD and ADRD. However, the mechanistic protective effect of citalopram is largely unknown. It is still unclear, if citalopram works for symptomatic patients, meaning patients with clinical symptoms and disease pathologies, such as mutant APP and Aβ. The prevalence of familial AD is about 1–2% of total AD patients and remaining all late-onset sporadic AD. In familial AD patients, it is expected that mutant APP and Aβ are produced, accumulated over the period of time. In late-onset AD cases, Aβ levels are increased in an age-dependent manner, but not mutant APP levels. Serotonin levels suppressed in both early-onset familial AD and late-onset sporadic AD. The impact of mutant APP on serotonin in both familial and sporadic ADA is unknown and needs to be investigated.
Our study findings demonstrate that both mutant APP, C-terminal fragments and Aβ levels were reduced in citalopram-treated mAPP-HT22 cells. And citalopram-treated mAPP-HT22 cells showed elevated serotonin-related genes and reduced AD-related genes (Table 1), indicating that (1) citalopram treatment rescues/compensates mutant APP and Aβ-induced low serotonin toxicities and (2) the toxicities of AD-related genes in AD. Our study observations are positive and may have therapeutic value for patients with depression, anxiety, AD and ADRD patients.
Although our study findings based on APP mutation and hippocampal neurons (similar to familial AD), the outcome truly implicates late-onset sporadic AD also. However, further research is still needed to understand the role of mutant APP in late-onset sporadic AD. The impact of mutant APP and its relation to citalopram treatment and serotonin in late-onset sporadic AD (most AD condition in real sense) can be addressed using brain tissues from (1) Down syndrome, (2) familial AD with APP, PS1/PS2 mutations and (3) sporadic AD patients treated with citalopram. Overall, well-defined postmortem AD brains with abovementioned brains are needed to answer the mechanistic impact of citalopram in late-onset sporadic AD.
SSRIs, including citalopram, have yielded conflicting data in clinical trials; this may be due to issues with conducting clinical trials, including (1) stage(s) of disease process, (2) dosage level and frequency, (3) DNA polymorphisms in the genomes of patients and (4) gender differences and serotonin levels at the time citalopram treatment. As we all know, a single drug may not be useful for all depression, anxiety, AD and ADRD patients. It is important to measure the levels of blood-based serotonin in AD patients before starting the treatment of citalopram.
In summary, mutant APP and Aβ increase fragmentation, reduce fission, biogenesis, synaptic activity, and mitophagy and autophagy activities in hippocampal cells. Citalopram reduced mutant APP and Aβ-induced mitochondrial deficits, synaptic damage and autophagy/mitophagy defects in hippocampal neurons. Further, the novelty of studying primary hippocampal (HT22) neurons is that these cells can be used for drug screening using high throughput machines and tools. As described in the current study, primary mouse hippocampal neurons after transfection with mutant APP cDNA mimic several features of AD. Unlike transgenic APP mice, primary hippocampal neurons are quick to test drugs such as citalopram and other candidates in order to understand Aβ and mitochondrial/synaptic pathologies and changes in mitophagy and autophagy.
Material and Methods
Chemicals and reagents
HT22 cells were a kind gift from David Schubert, and Dulbecco’s Modified Eagle Medium (DMEM) and Minimum Essential Medium (MEM), penicillin/streptomycin, Trypsin–EDTA and fetal bovine serum were purchased from GIBCO (Gaithersburg, MD USA).
Mutant APP cDNA constructs
We purchased mutant APP SweIND cDNA clone (pCAX-APP Swe/Ind) from Add gene—https://www.addgene.org and further sub-cloned into a mammalian expression vector pRP-Puro-CAG. pRP vector is pUC backbone having CMV promoter and SV40 polyadenylation site with puromycin selection for stable transfection. The sequence output was confirmed with NCBI sequence hAPP [NM_201414.2]*(K595N M596L V642F). Expression of mutant APP Swe/Ind cDNA was verified for APP mutant protein expression. We transfected mutant APP Swe/Ind cDNA into HT22 cells using lipofectamine 3000 for 24 h. Further cells were treated with citalopram (Sigma/Aldrich, CA) 20 μM final concentration for 24 hrs. After 24 h, cells were harvested, and the pellet was used for RNA and protein analysis.
Tissue culture work
The HT22 cells were grown for 3 days in a medium [1:1 mixture of DMEM and OptiMEM, 10% FBS plus penicillin and streptomycin (Invitrogen, Carlsbad, CA)] until the cells are confluent. We performed four independent cell cultures and transfections with mutant APP cDNA treatments for all experiments (n = 4) and treated with citalopram for 24 h.
qRT-PCR analysis
As shown in Figure 1, we used (1) HT22 cells, (2) HT22 cells treated with citalopram, (3) HTT cells transfected with mutant APP cDNA and (4) HT22 cells transfected with mutant APP cDNA + treated with citalopram.
Quantification of mRNA expression of mitochondrial dynamics, mitochondrial biogenesis, autophagy, mitophagy and synaptic genes using real-time RT-PCR. Using the reagent TriZol (Invitrogen, CA), we isolated total RNA from all four groups of cells. Using primer Express Software (Applied Biosystems, CA), we designed the oligonucleotide primers for the housekeeping genes β-actin; GAPDH; mitochondrial biogenesis genes PGC1α, Nrf1, Nrf2 and TFAM; mitochondrial structural genes; fission (Drp1 and Fis1); fusion genes (MFN1, MFN2, Opa1) and autophagy (ATG5 & LC3BI, LC3BII), mitophagy (PINK1 & TERT, BCL2 & BNIPBL) and synaptic and dendritic genes (synaptophysin, PSD95 and MAP2). The primer sequences and amplicon sizes are given in Table 3. We used Sybr-Green-based quantitative real-time RT-PCR. Briefly, 5 μg of DNAse-treated total RNA was used as starting material, to which we added 1 μl of oligo (dT), 1 μl of 10 mm dNTPs, 4 μl of 5× first strand buffer, 2 μl of 0.1 M DTT and 1 μl RNAseout. The reagents RNA, Oligo dT and dNTPs were mixed first, then heated to denature RNA at 65°C for 5 min and briefly chilled on ice until the remaining components were added. The samples were incubated at 42°C for 2 min, and then 1 μl of Superscript III (40 U/μl) was added. The samples were incubated at 42°C for 50 min; the reaction was inactivated by heating at 70°C for 15 min. Based on reverse transcriptase efficiency 100%, the cDNA diluted and used 100 ng/20 μl reaction in triplicate assays using QuantStudio3 (Applied Biosystems). The PCR conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The fluorescent spectra were recorded during the elongation phase of each PCR cycle. A dissociation curve was generated to distinguish nonspecific amplicons. The CT values were calculated with Quant studio and design and a specific setting on the baseline; the amplification plots and CT values were exported from the exponential PCR phase directly into a Microsoft Excel worksheet for further analysis.
Table 3.
Summary of antibody dilutions and conditions used in the immunoblotting analysis of mitochondrial dynamics, mitochondrial biogenesis, synaptic, autophagy and mitophagy proteins in citalopram-treated and -untreated mAPP-HT22 cells and untransfected HT22 cells
| Marker primary antibody—species and dilution | Purchased from company, city and state | Secondary antibody, dilution | Purchased from company, city and state |
|---|---|---|---|
| 6E10 Mouse monoclonal 1:500 | Biolegend, San Diego, CA | Sheep anti-mouse HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Drp1 Rabbit polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Fis1 Rabbit polyclonal 1:500 | Protein Tech Group, Inc, Chicago, IL | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn1 Rabbit polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| Mfn2 Rabbit polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| OPA1 Rabbit polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| SYN Rabbit monoclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| PSD95 Rabbit monoclonal 1:300 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| MAP2 Mouse monoclonal 1:600 | Santa Cruz Biotechnology, Dallas, TX | Sheep anti-mouse HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| PGC1a Rabbit polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| NRF1 Rabbit polyclonal 1:300 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| NRF2 Rabbit polyclonal 1:300 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
| TFAM Rabbit polyclonal 1:300 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare Amersham, Piscataway, NJ |
The mRNA transcript level was normalized against β-actin and the GAPDH at each dilution. In Table 2, a comparison of β-actin, GAPDH and genes of interest shown; relative quantification is performed according to the CT method (Applied Biosystems). Briefly, the comparative CT method involved averaging triplicate samples taken as the CT values for β-actin, GAPDH and genes of interest. β-actin normalization was used in the present study because β-actin CT values were similar for the control of untreated cells and experimental groups. The ΔCT value is obtained by subtracting the average β-actin CT value from the average CT value for interest genes. The ΔCT of WT-HT22 cells was used as the calibrator. The fold change was calculated according to the formula 2− (Δ ΔCT), where ΔΔCT is the difference between ΔCT and the ΔCT calibrator value. Statistical significance was calculated between mRNA expression in HT22 cells between treated and untreated groups.
Table 2.
Summary of qRT-PCR oligonucleotide primers used in measuring mRNA expressions in mitochondrial dynamics and mitochondrial biogenesis, synaptic, autophagy and mitophagy genes in citalopram-treated and -untreated mutant APP HT22 cells
| Gene | DNA sequence (5′-3′) | PCR product size |
|---|---|---|
| Mitochondrial dynamics genes | ||
| Drp1 | Forward primer ATGCCAGCAAGTCCACAGAA | 86 |
| Reverse primer TGTTCTCGGGCAGACAGTTT | ||
| Fis1 | Forward primer CAAAGAGGAACAGCGGGACT | 95 |
| Reverse primer ACAGCCCTCGCACATACTTT | ||
| Mfn1 | Forward primer GCAGACAGCACATGGAGAGA | 83 |
| Reverse primer GATCCGATTCCGAGCTTCCG | ||
| Mfn2 | Forward primer TGCACCGCCATATAGAGGAAG | 78 |
| Reverse primer TCTGCAGTGAACTGGCAATG | ||
| Opa1 | Forward primer ACCTTGCCAGTTTAGCTCCC | 82 |
| Reverse primer TTGGGACCTGCAGTGAAGAA | ||
| Mitochondrial biogenesis genes | ||
| PGC1α | Forward primer GCAGTCGCAACATGCTCAAG | 83 |
| Reverse primer GGGAACCCTTGGGGTCATTT | ||
| Nrf1 | Forward primer AGAAACGGAAACGGCCTCAT | 96 |
| Reverse primer CATCCAACGTGGCTCTGAGT | ||
| Nrf2 | Forward primer ATGGAGCAAGTTTGGCAGGA | 96 |
| Reverse primer GCTGGGAACAGCGGTAGTAT | ||
| TFAM | Forward primer TCCACAGAACAGCTACCCAA | 84 |
| Reverse primer CCACAGGGCTGCAATTTTCC | ||
| Reverse primer AGACGGTTGTTGATTAGGCGT | ||
| Synaptic genes | ||
| Synaptophysin | Forward primer CTGCGTTAAAGGGGGCACTA | 81 |
| Reverse primer ACAGCCACGGTGACAAAGAA | ||
| PSD95 | Forward primer CTTCATCCTTGCTGGGGGTC | 90 |
| Reverse primer TTGCGGAGGTCAACACCATT | ||
| Autophagy genes | ||
| LC3A | Forward primer CCCATCGCTGACATCTATGAAC | 77 |
| Reverse primer AAGGTTTCTTGGGAGGCGTA | ||
| LC3B | Forward primer TCCACTCCCATCTCCGAAGT | 94 |
| Reverse primer TTGCTGTCCCGAATGTCTCC | ||
| ATG5 | Forward primer TCCATCCAAGGATGCGGTTG | 95 |
| Reverse primer TCTGCATTTCGTTGATCACTTGAC | ||
| Beclin1 | Forward primer ACCAGCGGGAGTATAGTGAGT | 98 |
| Reverse primer CAGCTGGATCTGGGCGTAG | ||
| Mitophagy genes | ||
| Pink1 | Forward primer CCATCGGGATCTCAAGTCCG | 70 |
| Reverse primer GATCACTAGCCAGGGACAGC | ||
| TERT | Forward primer GCAAGGTGGTGTCTGCTAGT | 100 |
| Reverse primer AGCTTGCCGTATTTCCCCAA | ||
| BCL2 | Forward primer TCCTTCCAGCCTGAGAGCAA | 73 |
| Reverse primer GCCTGAGAGGAGACGTCCTG | ||
| BNIP3L | Forward primer GCACGTTCCTTCCTCGTCT | 82 |
| Reverse primer GCTCTGTCCCGACTCATGC | ||
| Serotonin-related genes | ||
| TPH2 | Forward primer GATTCAGCGGTGCCAGAAGA | 132 |
| Reverse primer GGAGAACACAACCGCAGTCT | ||
| SERT | Forward primer CAAAACGTCTGGCAAGGTGG | 155 |
| Reverse primer ACACCCCTGTCTCCAAGAGT | ||
| 5HTR1A | Forward primer ACCAGCTTCGGAACATCGTC | 132 |
| Reverse primer CTGTCTCACCGCCCCATTAG | ||
| 5HTR1B | Forward primer TACACGGTCTACTCCACGGT | 121 |
| Reverse primer CGGTCTTGTTGGGTGTCTGT | ||
| 5HTR5 | Forward primer CTCCACGTGGTGTGTCTTCA | 130 |
| Reverse primer GGCATGCTCCTTAGCAGTGA | ||
| 5HTR6 | Forward primer GCATAGCTCAGGCCGTATGT | 115 |
| Reverse primer TCCCGCATGAAGAGGGGATA | ||
| AD-related genes | ||
| APP | Forward primer TTCGCTGACGGAAACCAAGA | 140 |
| Reverse primer CGTCAACAGGCTCGACTTCA | ||
| MAPT | Forward primer TGCCCATGCCAGACCTAAAG | 147 |
| Reverse primer TGTTCCCTAACGAGCCACAC | ||
| BACE1 | Forward primer GGAACCCATCTCGGCATCC | 145 |
| Reverse primer CCCTCAGGTTGTCCACCATC | ||
| ADAM10 | Forward primer ATGGTGTTGCCGACAGTGTT | 150 |
| Reverse primer TTTGGCACGCTGGTGTTTTT | ||
| Housekeeping genes | ||
| B-actin | Forward primer AGAAGCTGTGCTATGTTGCTCTA | 91 |
| Reverse primer TCAGGCAGCTCATAGCTCTTC | ||
| GAPDH | Forward primer TTCCCGTTCAGCTCTGGG | 59 |
| Reverse primer CCCTGCATCCACTGGTGC | ||
Immunoblotting analysis
Immunoblotting analysis was performed using protein lysates prepared HT-22 cells transfected and untransfected mutant APP cDNA using 6E10 antibody that recognizes full-length mutant human APP and Aβ as described in Reddy 2018 (88). We also performed immunoblotting analysis for mitochondrial dynamics, biogenesis, synaptic, autophagy and mitophagy proteins. Details of antibody dilutions are published in Reddy et al. (88). 20 μg protein lysates were resolved on a 4–12% Nu-PAGE gel (Invitrogen). The resolved proteins were transferred to nylon membranes (Novax Inc., San Diego, CA) and were then incubated for 1 h at room temperature with a blocking buffer (5% dry milk dissolved in a TBST buffer). The membranes were incubated overnight with the primary antibodies. The membranes were washed with a TBST buffer three times at 10-min intervals and then incubated for 2 h with appropriate secondary antibody Sheep anti-mouse HRP 1:10000, followed by three additional washes at 10-min intervals. Proteins were detected with chemiluminescence reagents (Pierce Biotechnology, Rockford, IL), and the bands from immunoblots were visualized.
Cell survival assay
Cell-based apoptosis assay was performed using Cellometer Vision CBA Image Cytometry System (Nexcelom Bioscience LLC, Lawrence, MA) with two fluorophore Annexin V-FITC and propidium iodide (PI) staining solution, according to manufacturer’s instructions. Briefly, cells were harvested using trypsin, then spin down at 300 g for 3 min and pellets were washed with 1XPBS; cells were counted using a hematocytometer. Collected 100 000--150 000 cells and cells were resuspended in 40 μl of Annexin V binding buffer. 5 μl each of Annexin V—FITC reagent (green) and PI (red) were added to binding buffer containing cells; gently mix the solution by pipetting up and down 10 times, then incubate for 15 min at RT in the dark; after incubation, add 250 μl of 1XPBS and spin down at 300 g for 3 min, then resuspend the cell pellets in 50 μl of Annexin V binding buffer and then assess the cells apoptosis. Gate purple represents live cells, gate green represents the positive apoptotic cells, gate blue represents the detection of positive necrotic cells and gate red represents debris.
Measurement of soluble Aβ levels
Soluble Aβ levels were conducted using sandwich ELISA as described in Reddy et al. 2017 (90). Briefly, protein lysates were from cell pellets in a Tris-buffered saline (pH 8.0) containing protease inhibitors (20 mg/ml pepstatin A, aprotinin, phosphoramidon and leupeptin; 0.5 mm phenylmethanesulfonyl fluoride and 1 mm ethylene glycol-bis (flaminoethyl ether)-NN tetraacetic acid). Samples were sonicated briefly and centrifuged at 10 000g for 20 min at 4°C. The soluble fraction was used to determine the soluble Aβ by ELISA. For each sample, Aβ1-40 and Aβ1-42 were measured with commercial colorimetric ELISA kits (BioSource International, Camarillo, CA) specific for human. A 96-well plate was used, following the manufacturer’s instructions. Each sample was run in duplicate. Protein concentrations of the homogenates were determined following the BSA method, and Aβ was expressed as pg Aβ/mg protein.
Transmission electron microscopy
Using the TTU Electron Microscopy Core Facility, we acquired images of ultrastructural changes in cells treated and untreated with citalopram. The number and length of mitochondria indicated how our treatment alters synaptic degeneration and regeneration, cells treated with citalopram compared with untreated cells.
Statistical considerations
We performed statistical analyses of mAPP-HT22 cells treated with citalopram compared with untreated cells for mitochondrial count and length, the soluble Aβ40 and Aβ42, mitochondrial proteins (Drp1, Fis1, Mfn1, Mfn2, Opa1, Nrf1, Nrf2, PGC1a, TFAM), mitophagy (PINK1, Tert, Bcl2, BNIP3L), autophagy (LC3B-I, LC3B-II, ATG5, Beclin1) and synaptic and dendritic (synaptophysin, PSD95, MAP2) proteins, using a one-way analysis of variance and t-test statistical analyses. Conflict of Interest statement. None declared.
Funding
Alzheimer’s Association New Investigator Research Grant (2016-NIRG-39787); Center of Excellence for Translational Neuroscience and Therapeutics (PN-CTNT20115-AR); Alzheimer’s Association through a SAGA grant and NIH grant (AG063162 to A.P.R.); NIH grants (R01AG042178, R01AG47812, R01NS105473, AG069333, AG066347 to P.H.R.).
Contributor Information
Arubala P Reddy, Nutritional Sciences Department, Texas Tech University, Lubbock, TX, USA.
Xiangling Yin, Garrison Institute on Aging, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA.
Neha Sawant, Internal Medicine Department, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA.
P Hemachandra Reddy, Internal Medicine Department, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA; Pharmacology & Neuroscience Department, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA; Neurology Department, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA; Speech, Language and Hearing Sciences Departments, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA; Public Health Department, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA.
References
- 1.Selkoe, D.J. (2001) Alzheimer’s disease: genes, proteins, and therapy. Physiol. Rev., 81, 741–766. [DOI] [PubMed] [Google Scholar]
- 2.World Alzheimer Report (2020) Design, Dignity, Dementia: Dementia-Related Design and the Built Environment. https://www.alzint.org/resource/world-alzheimer-report-2020/.
- 3.Novais, F. and Starkstein, S. (2015) Phenomenology of depression in Alzheimer’s disease. J. Alzheimers Dis., 47, 845–855. [DOI] [PubMed] [Google Scholar]
- 4.Dafsari, F.S. and Jessen, F. (2020) Depression—an underrecognized target for prevention of dementia in Alzheimer’s disease. Transl. Psychiatry, 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mdawar, B., Ghossoub, E. and Khoury, R. (2019) Selective serotonin reuptake inhibitors and Alzheimer’s disease. Neural Regen. Res., 15, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirrito, J.R., Disabato, B.M., Restivo, J.L., Verges, J.R., Goebel, W.D., Sathyan, A., Hayreh, D., D'Angelo, G., Benzinger, T., Yoon, H.et al. (2011) Serotonin signaling is associated with lower amyloid-β levels and plaques in transgenic mice and humans. Proc. Natl. Acad. Sci. U. S. A., 108, 14968–14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirrito, J.R., Wallace, C.E., Yan, P., Davis, T.A., Gardiner, W.D., Doherty, B.M., King, D., Yuede, C.M., Lee, J. and Sheline, Y.I. (2020) Effect of escitalopram on Aβ levels and plaque load in an Alzheimer mouse model. Neurology, 95, e2666–e2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheline, Y.I., West, T., Yarasheski, K., Swarm, R., Jasielec, M., Fisher, J.R., Ficker, W.D., Yan, P., Xiong, C., Frederiksen, C.et al. (2014) An antidepressant decreases CSF Aβ production in healthy individuals and in transgenic AD mice. Sci. Transl. Med., 6, 236re4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheline, Y.I., Snider, J.B., Beer, J.C., Seok, D., Fagan, A.M., Suckow, R.F., Lee, J., Waligorska, T., Korecka, M., Aselcioglu, I.et al. (2020) Effect of escitalopram dose and treatment duration on CSF Aβ levels in healthy older adults: a controlled clinical trial. Neurology, 95, e2658–e2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, T.W., 1, Pollock, B.G. and Milgram, N.W. (2007) Potential cognitive enhancing and disease modification effects of SSRIs for Alzheimer’s disease. Neuropsychiatr. Dis. Treat., 3, 627–636. [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson, R.L., Guo, Z., Halagappa, V.M., Pearson, M., Gray, A.J., Matsuoka, Y., Brown, M., Martin, B., Iyun, T., Maudsley, S.et al. (2007) Prophylactic treatment with paroxetine ameliorates behavioral deficits and retards the development of amyloid and tau pathologies in 3xTgAD mice. Exp. Neurol., 205, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsworthy, R.J. and Aldred, S. (2019) Depression in Alzheimer’s disease: an alternative role for selective serotonin reuptake inhibitors? J. Alzheimers Dis., 69, 651–661. [DOI] [PubMed] [Google Scholar]
- 13.Wang, J., Zhang, Y., Xu, H., Zhu, S., Wang, H., He, J., Zhang, H., Guo, H., Kong, J., Huang, Q. and Li, X. (2014) Fluoxetine improves behavioral performance by suppressing the production of soluble β-amyloid in APP/PS1 mice. Curr. Alzheimer Res., 11, 672–680. [DOI] [PubMed] [Google Scholar]
- 14.Ren, Q., Wang, Y., Gong Zhou, Q., Xu and Zhang, Z. (2015) Escitalopram ameliorates Forskolin-induced tau hyperphosphorylation in HEK239/tau441 cells. J. Mol. Neurosci., 56, 500–508. [DOI] [PubMed] [Google Scholar]
- 15.Wang, Y., Ren, Q., Gong, W., Wu, D., Tang, X., Li, X., Wu, F., Bai, F., Xu, L. and Zhang, Z. (2016) Escitalopram attenuates β-amyloid-induced tau hyperphosphorylation in primary hippocampal neurons through the 5-HT1A receptor mediated Akt/GSK-3β pathway. Oncotarget, 7, 13328–13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker, F.R. (2013) A critical review of the mechanism of action for the selective serotonin reuptake inhibitors: do these drugs possess anti-inflammatory properties and how relevant is this in the treatment of depression? Neuropharmacology, 67, 304–317. [DOI] [PubMed] [Google Scholar]
- 17.Haj, M.E., Altintas, E., Chapelet, G., Kapogiannis, D. and Gallouj, K. (2020) High depression and anxiety in people with Alzheimer’s disease living in retirement homes during the COVID-19 crisis. Psychiatry Res., 291, 113294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tune, L.E. (1998) Depression and Alzheimer’s disease. Depress. Anxiety, 8, 91–95. [PubMed] [Google Scholar]
- 19.Bethea, C.L., Reddy, A.P. and Christian, F.L. (2018) How studies of the serotonin system in macaque models of menopause relate to Alzheimer’s disease. J. Alzheimers Dis., 57, 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson, S., Herrmann, N., Rapoport, M.J. and Lanctôt, K.L. (2007) Efficacy and safety of antidepressants for treatment of depression in Alzheimer’s disease: a meta-analysis. Can. J. Psychiatr., 52, 248–255. [DOI] [PubMed] [Google Scholar]
- 21.Claeysen, S., Bockaert, J. and Giannoni, P. (2015) Serotonin: a new hope in Alzheimer’s disease? ACS Chem. Neurosci., 6, 940–943. [DOI] [PubMed] [Google Scholar]
- 22.Felice, D., O'Leary, O.F., Cryan, J.F., Dinan, T.G., Gardier, A.M., Sánchez, C. and David, D.J. (2015) When ageing meets the blues: are current antidepressants effective in depressed aged patients? Neurosci. Biobehav. Rev., 55, 478–497. [DOI] [PubMed] [Google Scholar]
- 23.Cassano, T., Calcagnini, S., Carbone, A., Bukke, V.N., Orkisz, S., Villani, R., Romano, A., Avolio, C. and Gaetani, S. (2019) Pharmacological treatment of depression in Alzheimer’s disease: a challenging task. Front. Pharmacol., 10, 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen, L.J. (1997) Rational drug use in the treatment of depression. Pharmacotherapy, 17, 45–61. [PubMed] [Google Scholar]
- 25.Taylor, C., Fricker, A.D., Devi, L.A. and Gomes, I. (2005) Mechanisms of action of antidepressants: from neurotransmitter systems to signaling pathways. Cell. Signal., 17, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yohn, C.N., Gergues, M.M. and Samuels, B.A. (2017) The role of 5-HT receptors in depression. Mol. Brain, 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Deurwaerdère, P. and Di Giovanni, G. (2020) Serotonin in health and disease. Int. J. Mol. Sci., 21, 3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sepehry, A.A., Lee, P.E., Hsiung, G.Y., Beattie, B.L. and Jacova, C. (2012) Effect of selective serotonin reuptake inhibitors in Alzheimer’s disease with comorbid depression: a meta-analysis of depression and cognitive outcomes. Drugs Aging, 29, 793–806. [DOI] [PubMed] [Google Scholar]
- 29.Meltzer, C.C., Smith, G., DeKosky, S.T., Pollock, B.G., Mathis, C.A., Moore, R.Y., Kupfer, D.J. and Reynolds, C.F., 3rd. (1998) Serotonin in aging, late-life depression, and Alzheimer’s disease: the emerging role of functional imaging. Neuropsychopharmacology, 18, 407–430. [DOI] [PubMed] [Google Scholar]
- 30.Trillo, L., Das, D., Hsieh, W., Medina, B., Moghadam, S., Lin, B., Dang, V., Sanchez, M.M., De Miguel, Z.et al. (2013) Ascending monoaminergic systems alterations in Alzheimer’s disease. Translating basic science into clinical care. Neurosci. Biobehav. Rev., 37, 1363–1379. [DOI] [PubMed] [Google Scholar]
- 31.Strac, D.S., Muck-Seler, D. and Pivac, N. (2015) Neurotransmitter measures in the cerebrospinal fluid of patients with Alzheimer’s disease: a review. Psychiatr. Danub., 27, 14–24. [PubMed] [Google Scholar]
- 32.Kandimalla, R. and Reddy, P.H. (2017) Therapeutics of neurotransmitters in Alzheimer’s disease. J. Alzheimers Dis., 57, 1049–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds, G.P., Mason, S.L., Meldrum, A., De Keczer, S., Parnes, H., Eglen, R.M. and Wong, E.H. (1995) 5-Hydroxytryptamine (5-HT)4 receptors in post mortem human brain tissue: distribution, pharmacology and effects of neurodegenerative diseases. Br. J. Pharmacol., 114, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bethea, C.L. and Reddy, A.P. (2015) Ovarian steroids regulate gene expression related to DNA repair and neurodegenerative diseases in serotonin neurons of macaques. Mol. Psychiatry, 20, 1565–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavares, G.A., Torres, A. and de Souza, J.A. (2020) Early life stress and the onset of obesity: proof of micro RNAs' involvement through modulation of serotonin and dopamine systems’ homeostasis. Front. Physiol., 11, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morin, L.P. (1999) Serotonin and the regulation of mammalian circadian rhythmicity. Ann. Med., 31, 12–33. [DOI] [PubMed] [Google Scholar]
- 37.Zakin, E., Abrams, R. and Simpson, D.M. (2019) Diabetic neuropathy. Semin. Neurol., 39, 560–569. [DOI] [PubMed] [Google Scholar]
- 38.Prabhakar, V., Gupta, D., Kanade, P. and Radhakrishnan, M. (2015) Diabetes-associated depression: the serotonergic system as a novel multifunctional target. Indian. Aust. J. Pharm., 47, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Homberg, J.R., Kolk, S.M. and Schubert, D. (2013) Editorial perspective of the research topic “Deciphering serotonin’s role in neurodevelopment”. Front. Cell. Neurosci., 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, I., Greenblatt, H.K. and Greenblatt, D.J. (2016) Stereo-psychopharmacology: the case of citalopram and escitalopram. Clin. Pharmacol. Drug Dev., 5, 331–335. [DOI] [PubMed] [Google Scholar]
- 41.Parker, N.G. and Brown, C.S. (2000) Citalopram in the treatment of depression. Ann. Pharmacother., 34, 761–771. [DOI] [PubMed] [Google Scholar]
- 42.Brendel, M., Sauerbeck, J., Greven, S., Kotz, S., Scheiwein, F., Blautzik, J., Delker, A., Pogarell, O., Ishii, K., Bartenstein, P.et al. (2018) Alzheimer’s disease neuroimaging initiative. Serotonin selective reuptake inhibitor treatment improves cognition and grey matter atrophy but not amyloid burden during two-year follow-up in mild cognitive impairment and Alzheimer’s disease patients with depressive symptoms. J. Alzheimers Dis., 65, 793–806. [DOI] [PubMed] [Google Scholar]
- 43.Morgese, M.G., Schiavone, S. and Trabace, L. (2017) Emerging role of amyloid beta in stress response: implication for depression and diabetes. Eur. J. Pharmacol., 817, 22–29. [DOI] [PubMed] [Google Scholar]
- 44.Reddy, P.H. (2008) Mitochondrial medicine for aging and neurodegenerative diseases. NeuroMolecular Med., 10, 291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy, P.H. (2009) Role of mitochondria in neurodegenerative diseases: mitochondria as a therapeutic target in Alzheimer’s disease. CNS Spectr., 8, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swerdlow, N.S. and Wilkins, H.M. (2020) Mitophagy and the brain. Int. J. Mol. Sci., 21, 9661. doi: 10.3390/ijms21249661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, W., Zhao, F., Ma, X., Perry, G. and Zhu, X. (2020) Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol. Neurodegener., 15, 30. doi: 10.1186/s13024-020-00376-6PMID: 32471464; PMCID: PMC7257174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng, B., Wang, X., Su, B., Lee, H.G., Casadesus, G., Perry, G. and Zhu, X. (2012) Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J. Neurochem., 120, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, X., Su, B., Lee, H.G., Li, X., Perry, G., Smith, M.A. and Zhu, X. (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J. Neurosci., 29, 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.deOliveira, M.R. (2016) Fluoxetine and the mitochondria: a review of the toxicological aspects. Toxicol. Lett., 258, 185–191. [DOI] [PubMed] [Google Scholar]
- 51.Wong, K.Y., Roy, J., Fung, M.L., Heng, B.C., Zhang, C. and Lim, L.W. (2020) Relationships between mitochondrial dysfunction and neurotransmission failure in Alzheimer’s disease. Aging Dis., 11, 1291–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fanibunda, S.E., Deb, S., Maniyadath, B., Tiwari, P., Ghai, U., Gupta, S., Figueiredo, D., Weisstaub, N., Gingrich, J.A., Vaidya, A.D.B., Kolthur-Seetharam, U. and Vaidya, V.A. (2019) Serotonin regulates mitochondrial biogenesis and function in rodent cortical neurons via the 5-HT2A receptor and SIRT1-PGC-1α axis. Proc. Natl. Acad. Sci. U. S. A., 116, 11028–11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allen, J., Romay-Tallon, R., Brymer, K.J., Caruncho, H.J. and Kalynchuk, L.E. (2018) Mitochondria and mood: mitochondrial dysfunction as a key player in the manifestation of depression. Front. Neurosci., 12, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen, S., Owens, G.C., Crossin, K.L. and Edelman, D.B. (2007) Serotonin stimulates mitochondrial transport in hippocampal neurons. Mol. Cell. Neurosci., 36, 472–483. [DOI] [PubMed] [Google Scholar]
- 55.Farrelly, L.A., Thompson, R.E., Zhao, S., Lepack, A.E., Lyu, Y., Bhanu, N.V., Zhang, B., Loh, Y.E., Ramakrishnan, A., Vadodaria, K.C.et al. (2019) Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature, 567, 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bader, M. (2019) Serotonylation: serotonin signaling and epigenetics. Front. Mol. Neurosci., 12, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhattacharyya, S., Puri, S., Miledi, R. and Panicker, M.M. (2002) Internalization and recycling of 5-HT2A receptors activated by serotonin and protein kinase C-mediated mechanisms. Proc. Natl. Acad. Sci. U. S. A., 99, 14470–14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steiner, J.P., Bachani, M., Wolfson-Stofko, B., Lee, M.H., Wang, T., Li, G., Li, W., Strayer, D., Haughey, N.J. and Nath, A. (2015) Interaction of paroxetine with mitochondrial proteins mediates neuroprotection. Neurotherapeutics, 12, 200–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohira, K., Hagihara, H., Miwa, M., Nakamura, K. and Miyakawa, T. (2019) Fluoxetine-induced dematuration of hippocampal neurons and adult cortical neurogenesis in the common marmoset. Mol. Brain, 12, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song, N.N., Huang, Y., Yu, X., Lang, B., Ding, Y.Q. and Zhang, L. (2017) Divergent roles of central serotonin in adult hippocampal neurogenesis. Front. Cell. Neurosci., 11, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walther, D.J., Stahlberg, S. and Vowinckel, J. (2011) Novel roles for biogenic monoamines: from monoamines in transglutaminase-mediated post-translational protein modification to monoaminylation deregulation diseases. FEBS J., 278, 4740–4755. [DOI] [PubMed] [Google Scholar]
- 62.Anastas, J.N. and Shi, Y. (2019) Histone serotonylation: can the brain have “happy” chromatin? Mol. Cell, 74, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, H., Wang, J., Jiang, J., Stavrovskaya, I.G., Li, M., Li, W., Wu, Q., Zhang, X., Luo, C., Zhou, S.et al. (2014) N-acetyl-serotonin offers neuroprotection through inhibiting mitochondrial death pathways and autophagic activation in experimental models of ischemic injury. J. Neurosci., 34, 2967–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma, J., Gao, Y., Jiang, L., Chao, F.L., Huang, W., Zhou, C.N., Tang, W., Zhang, L., Huang, C.X., Zhang, Y.et al. (2017) Fluoxetine attenuates the impairment of spatial learning ability and prevents neuron loss in middle-aged APPswe/PSEN1dE9 double transgenic Alzheimer’s disease mice. Oncotarget, 8, 27676–27692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin, L., Gao, L.F., Sun, D.S., Wu, H., Wang, Q., Ke, D., Lei, H., Wang, J.Z. and Liu, G.P. (2017) Long-term ameliorative effects of the antidepressant fluoxetine exposure on cognitive deficits in 3 × TgAD mice. Mol. Neurobiol., 54, 4160–4171. [DOI] [PubMed] [Google Scholar]
- 66.Vucicevic, L., Misirkic-Marjanovic, M., Paunovic, V., Kravic-Stevovic, T., Martinovic, T., Ciric, D., Maric, N., Petricevic, S., Harhaji-Trajkovic, L., Bumbasirevic, V.et al. (2014) Autophagy inhibition uncovers the neurotoxic action of the antipsychotic drug olanzapine. Autophagy, 10, 2362–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, Y., Hu, Z., Du, X., Davies, H., Huo, X. and Fang, M. (2017) miR-16 and fluoxetine both reverse autophagic and apoptotic change in chronic unpredictable mild stress model rats. Front. Neurosci., 11, 428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Shu, X., Sun, Y., Sun, X., Zhou, Y., Bian, Y., Shu, Z., Ding, J., Lu, M. and Hu, G. (2019) The effect of fluoxetine on astrocyte autophagy flux and injured mitochondria clearance in a mouse model of depression. Cell Death Dis., 10, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen, V.C., Hsieh, Y.H., Chen, L.J., Hsu, T.C. and Tzang, B.S. (2018) Escitalopram oxalate induces apoptosis in U-87MG cells and autophagy in GBM8401 cells. J. Cell. Mol. Med., 22, 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gassen, N.C. and Rein, T. (2019) Is here a role of autophagy in depression and antidepressant action? Front Psychiatry, 10, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hwang, H.Y., Shim, J.S., Kim, D. and Kwon, H.J. (2020) Antidepressant drug sertraline modulates AMPK-MTOR signaling-mediated autophagy via targeting mitochondrial VDAC1 protein. Autophagy, 9, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manczak, M., Kandimalla, R., Yin, X. and Reddy, P.H. (2018) Hippocampal mutant APP and amyloid beta-induced cognitive decline, dendritic spine loss, defective autophagy, mitophagy and mitochondrial abnormalities in a mouse model of Alzheimer’s disease. Hum. Mol. Genet., 27, 1332–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manczak, M., Mao, P., Calkins, M.J., Cornea, A., Reddy, A.P., Murphy, M.P., Szeto, H.H., Park, B. and Reddy, P.H. (2010) Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer’s disease neurons. J. Alzheimers Dis., 20Suppl 2(Suppl 2), S609–S631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Calkins, M.J., Manczak, M., Mao, P., Shirendeb, U. and Reddy, P.H. (2011) Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum. Mol. Genet., 20, 4515–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manczak, M., Calkins, M.J. and Reddy, P.H. (2011) Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum. Mol. Genet., 20, 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manczak, M. and Reddy, P.H. (2012) Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer’s disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum. Mol. Genet., 21, 2538–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhatti, G.K., Reddy, A.P., Reddy, P.H. and Bhatti, J.S. (2020) Lifestyle modifications and nutritional interventions in aging-associated cognitive decline and Alzheimer’s disease. Front. Aging Neurosci. 2020 Jan 10, 11, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pradeepkiran, J.A., Reddy, A.P., Yin, X., Manczak, M. and Reddy, P.H. (2020) Protective effects of BACE1 inhibitory ligand molecules against amyloid beta-induced synaptic and mitochondrial toxicities in Alzheimer’s disease. Hum. Mol. Genet. 2020 Jan 1, 29, 49–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reddy, P.H. and Beal, M.F. (2008) Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer’s disease. Trends Mol. Med., 14, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xue, X., Wang, L.R., Sato, Y., Jiang, Y., Berg, M., Yang, D.S., Nixon, R.A. and Liang, X.J. (2014) Single-walled carbon nanotubes alleviate autophagic/lysosomal defects in primary glia from a mouse model of Alzheimer’s disease. Nano Lett., 14, 5110–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dunlop, E.A. and Tee, A.R. (2014) mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin. Cell Dev. Biol., 36, 121–129. [DOI] [PubMed] [Google Scholar]
- 82.Cai, Z., Zhou, Y., Liu, Z., Ke, Z. and Zhao, B. (2015) Autophagy dysfunction upregulates beta-amyloid peptides via enhancing the activity of γ-secretase complex. Neuropsychiatr. Dis. Treat., 11, 2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mputhia, Z., Hone, E., Tripathi, T., Sargeant, T., Martins, R. and Bharadwaj, P. (2019) Autophagy modulation as a treatment of amyloid diseases. Molecules, 24, 3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tran, M. and Reddy, P.H. (2020) Defective autophagy and mitophagy in aging and Alzheimer’s disease. Front. Neurosci., (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.John, A. and Reddy, P.H. (2021) Synaptic basis of Alzheimer’s disease: focus on synaptic amyloid beta, P-tau and mitochondria. Ageing Res. Rev., 65, 101208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kitagishi, Y., Kobayashi, M., Kikuta, K. and Matsuda, S. (2012) Roles of PI3K/AKT/GSK3/mTOR pathway in cell signaling of mental illnesses. Depress. Res. Treat., 2012, 752563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith, G.S., Barrett, F.S., Joo, J.H., Nassery, N., Savonenko, A., Sodums, D.J., Marano, C.M., Munro, C.A., Brandt, J., Kraut, M.A.et al. (2017) Molecular imaging of serotonin degeneration in mild cognitive impairment. Neurobiol. Dis., 105, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reddy, P.H., Yin, X., Manczak, M., Kumar, S., Pradeepkiran, J.A., Vijayan, M. and Reddy, A.P. (2018) Mutant APP and amyloid beta-induced defective autophagy, mitophagy, mitochondrial structural and functional changes and synaptic damage in hippocampal neurons from Alzheimer’s disease. Hum. Mol. Genet., 27, 2502–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kandimalla, R., Manczak, M., Yin, X., Wang, R. and Reddy, P.H. (2018) Hippocampal phosphorylated tau induced cognitive decline, dendritic spine loss and mitochondrial abnormalities in a mouse model of Alzheimer’s disease. Hum. Mol. Genet., 27, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reddy, P.H., Manczak, M. and Yin, X. (2017) Mitochondria-division inhibitor 1 protects against amyloid-β induced mitochondrial fragmentation and synaptic damage in Alzheimer’s disease. J. Alzheimers Dis., 58, 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]


