This review focuses on recent advances relating to H2S signaling mechanisms, and highlights interconnections between H2S and H2O2 at the post-translational modification level and with ABA in stomatal movement.
Keywords: Abscisic acid, persulfidation, proteomics, redox modifications, stomatal movement, sulfenylation
Abstract
Hydrogen sulfide (H2S) is a signaling molecule that regulates critical processes and allows plants to adapt to adverse conditions. The molecular mechanism underlying H2S action relies on its chemical reactivity, and the most-well characterized mechanism is persulfidation, which involves the modification of protein thiol groups, resulting in the formation of persulfide groups. This modification causes a change of protein function, altering catalytic activity or intracellular location and inducing important physiological effects. H2S cannot react directly with thiols but instead can react with oxidized cysteine residues; therefore, H2O2 signaling through sulfenylation is required for persulfidation. A comparative study performed in this review reveals 82% identity between sulfenylome and persulfidome. With regard to abscisic acid (ABA) signaling, widespread evidence shows an interconnection between H2S and ABA in the plant response to environmental stress. Proteomic analyses have revealed persulfidation of several proteins involved in the ABA signaling network and have shown that persulfidation is triggered in response to ABA. In guard cells, a complex interaction of H2S and ABA signaling has also been described, and the persulfidation of specific signaling components seems to be the underlying mechanism.
Introduction
Hydrogen sulfide (H2S) is a colorless gas with a characteristic unpleasant odor. In nature, H2S is present in volcanic gas, hot springs, rock salts, and natural gas, as well as in emissions produced as a result of industrial activity. In biological systems, H2S can be considered an ancient molecule since it originates from bacterial anaerobic metabolism. In the absence of oxygen, sulfur-reducing microorganisms use different forms of oxidized sulfur as electron acceptors during the degradation of simple organic matter, producing H2S and CO2 (Offre et al., 2013). H2S is additionally used by sulfur-oxidizing bacteria as an electron donor in anoxygenic photosynthesis to produce oxidized sulfur compounds (Johnston et al., 2009).
H2S has long been considered a toxic molecule dangerous to the environment and complex biological organisms. In mammals, the presence of sulfide in mitochondria causes the inhibition of cytochrome c oxidase of the respiratory chain, as does the presence of carbon monoxide (CO) and nitric oxide (NO) (Cooper and Brown, 2008). However, below a specific concentration threshold, CO, NO, and H2S affect various cellular events and are currently considered to be signaling molecules that function as physiological gasotransmitters (Wang, 2014). In plants, H2S is also recognized to have the same relevance as other signaling molecules, such as NO and hydrogen peroxide (H2O2) (Calderwood and Kopriva, 2014; Aroca et al., 2018; Aroca et al., 2020). All these molecules, including H2S, show toxicity/signaling duality, depending on the concentration threshold.
Although H2S is known to be present in mammalian tissues, its intracellular production and signaling function as a neuromodulator were first established in the late 20th century (Abe and Kimura, 1996). H2S is produced endogenously by cells through different enzymes involved in cysteine metabolism, in both mammals and plants. In plants, H2S is also produced in the photosynthetic sulfate assimilation pathway in the chloroplast (Gotor et al., 2019). Intensive research on H2S has been carried out in recent years both in animals and in plants, and an impressive exponential increase in the number of original publications and reviews has occurred. Consequently, the number of biological functions in which sulfide is known to be involved has rapidly increased. In plants, H2S has been shown to be essential in regulating a wide range of vital processes. H2S improves the tolerance and protection of plants to numerous adverse environmental conditions, and, in this way, it allows plant adaptability and viability, and its beneficial effects play a role in important aspects of development (Zhang et al., 2021b; Zhou et al., 2021b). Sulfide also regulates critical processes, including autophagy and abscisic acid (ABA)-dependent stomatal movement (Gotor et al., 2019; Laureano-Marín et al., 2020; Zhang et al., 2020). In addition, the interplay of H2S with other signaling molecules and phytohormones in multiple physiological processes has been extensively described (Aroca et al., 2020).
Despite the very large number of plant studies that are continuously being conducted, studies on the molecular mechanisms through which sulfide exerts its regulatory effects are still scarce. We believe that this aspect deserves specific attention, and this review highlights the progress obtained in understanding the mechanism of action of sulfide in plant systems. The most recent outcomes on the mechanism of the sulfide control of guard cell ABA signaling are also highlighted.
Hydrogen sulfide action
The reaction mechanism in which H2S participates and exerts regulatory and signaling function is complex, and it is necessary to take into account the complex reactivity of this molecule. H2S encompasses neutral H2S and anionic forms (hydrosulfide, HS–, and sulfide, S2–) with pKa1 and pKa2 values of 6.9 and >12, respectively (Kabil and Banerjee, 2010). Therefore, in aqueous solution, H2S exists in equilibrium with its H+ and HS– anionic forms; these latter are unable to cross organelle membranes. Under physiological pH conditions, two-thirds of H2S exists in the form of HS–. However, the lipid solubility of H2S and its membrane permeability promote the biological distribution of sulfide species within cells (Cuevasanta et al., 2012).
The mechanism of action of H2S is related to the characteristics of acid–base behavior and chemical reactivity with other biochemical molecules, such as low-molecular weight (LMW) thiols, protein thiols, protein metal centers, and biological oxidants. Among these oxidants, the hydroxyl radical (OH·), nitrogen dioxide (NO2·), superoxide radical (O2· –), H2O2, peroxynitrite (ONOOH), and hypochlorite (HOCl) can support H2S oxidation (Li and Lancaster, 2013; Zaffagnini et al., 2019).
Metalloproteins are well-established biochemical targets of H2S that covalently attach to heme porphyrins. Thus, H2S acts as a potent inhibitor of mitochondrial cytochrome c oxidase, inhibiting mitochondrial respiration, releasing cytochrome c during apoptosis, and stimulating procaspase 9 persulfidation (Vitvitsky et al., 2018). H2S can also react quickly and reversibly with other ferric heme proteins such as methemoglobin and leghemoglobin to reduce their iron center and form a complex (Jensen and Fago, 2018; Boubeta et al., 2020). In addition to modifying heme proteins, H2S can also modify Zn-finger proteins, but this leads to persulfidation and rapid thiol oxidation (Lange et al., 2019).
A second mechanism of action of H2S that is well established in mammalian and plant systems is the modification of proteins by the oxidation of cysteine residues to form corresponding persulfides (Filipovic, 2015; Gotor et al., 2019). Protein thiol persulfidation has been widely described for numerous proteins, and it was initially described as S-sulfhydration in mouse liver. Susceptibility of several proteins to modification by sulfide has been determined (Mustafa et al., 2009; Filipovic et al., 2018). In plants, three high-throughput proteomic analyses also revealed the presence of persulfidation in the Arabidopsis proteome, showing >3400 and 5214 proteins susceptible to persulfidation in leaf and root tissue, respectively (Aroca et al., 2015, 2017a; Jurado-Flores et al., 2021). Different studies on this post-translational modification of specific proteins have shown that it results in changes to the function of the proteins, altering their catalytic activity or intracellular location and inducing important physiological effects, ranging from regulation of autophagy, ABA-dependent stomatal closure, ethylene biosynthesis, and root hair growth, to resistance to oxidative stress (Table 1). Persulfides on specific cysteine residues have been described in different Arabidopsis proteins, including abscisic acid-insensitive 4 (ABI4) (Zhou et al., 2021a), cytosolic ascorbate peroxidase 1 (APX1) (Aroca et al., 2015), cytosolic glyceraldehyde-3-phosphate dehydrogenase (GapC1) (Aroca et al., 2017b), actin 2 (ACT2) (Li et al., 2018), l-cysteine desulfhydrase (DES1), respiratory burst oxidase homolog protein D (RBOHD) (Shen et al., 2020), SNF1-related protein kinase SnRK2.6 (Chen et al., 2020), and the autophagic proteins ATG4a and ATG18a (Laureano-Marín et al., 2020; Aroca et al., 2021a). In addition, tomato 1-aminocyclopropane-1-carboxylic acid oxidases 1 and 2 (LeACO1 and LeACO2, respectively) (Jia et al., 2018) and tomato antioxidant enzymes (Li et al., 2020) have also been demonstrated to be persulfidated on specific cysteine residues (Table 1).
Table 1.
Plant proteins persulfidated at specific cysteine residues
| Protein | Accession number | No. of Cys residues | Persulfidated Cys | Effect of the modification | Reference |
|---|---|---|---|---|---|
| Arabidopsis abscisic acid insensitive 4 (ABI4) | At2g40220 | 3 | Cys250 | MAPKKK18 transactivation/increase the MAPK cascade signal in response to ABA | Zhou et al. (2021a) |
| Arabidopsis actin2 (ACT2) | At3g18780 | 4 | Cys287 | Inhibition of actin polymerization/depolymerization of F-actin bundles/inhibition of root hair growth | Li et al. (2018) |
| Arabidopsis cytosolic ascorbate peroxidase1 (APX1) | At1g07890 | 5 | Cys32 | Increase of enzyme activity | Aroca et al. (2015) |
| Arabidopsis autophagy-related protein cysteine protease 4a (ATG4a) | At2g44140 | 12 | Cys170 | Inhibition of proteolytic activity/repression of autophagy | Laureano-Marín et al., (2020) |
| Arabidopsis autophagy-related protein ATG18a | At3g62770 | 8 | Cys103 | Activation of ATG18a binding capacity to specific phospholipids/repression of auotphagy | Aroca et al. (2021a) |
| Arabidopsis l-cysteine desulfhydrase1 (DES1) | At5g28030 | 3 | Cys44 and Cys205 | Increase of enzyme activity/induction of H2S production/ABA-dependent stomatal closure | Shen et al. (2020) |
| Arabidopsis cytosolic glyceraldehyde 3-phosphate dehydrogenase C1 (GapC1) | At3g04120 | 2 | Cys160 | Enhanced nuclear localization | Aroca et al. (2017b) |
| Arabidopsis open stomata1/SNF1-related protein kinase2.6 (OST1/SnRK2.6) | At4g33950 | 6 | Cys131 and Cys137 | Increase of enzyme activity/enhanced interaction with ABA response factor ABF2/ABA-dependent stomatal closure | Chen et al. (2020) |
| Arabidopsis NADPH oxidase respiratory burst oxidase homolog protein D (RBOHD) | At5g47910 | 10 | Cys825 and Cys890 | Increase of enzyme activity/induction of H2O2 production/ABA-dependent stomatal closure | Shen et al. (2020) |
| Tomato 1-aminocyclopropane-1-carboxylic acid oxidases 1 and 2 (ACO1/2) | NP_001234024/NP_001316842 | 4 | Cys60 | Inhibition of enzyme activity/repression of ethylene biosynthesis | Jia et al. (2018) |
| Tomato cytosolic ascorbate peroxidase1 (cAPX1) | NP_001234782.1 | 6 | Cys168 | Increase of enzyme activity/enhanced resistance to oxidative stress | Li et al. (2020) |
| Tomato catalase1 (CAT1) | NP_001234827.1 | 10 | Cys234 | Inhibition of enzyme activity/enhanced resistance to oxidative stress | Li et al. (2020) |
| Tomato peroxidase 5 (POD5) | XP_004235031.1 | 10 | Cys46 and Cys61 | Increase of enzyme activity/enhanced resistance to oxidative stress | Li et al. (2020) |
Shown as the accession number, the number of cysteine residues in the amino acid sequence, the specific persulfidated cysteines, and the effects and biological consequences of the post-translational modification of the proteins.
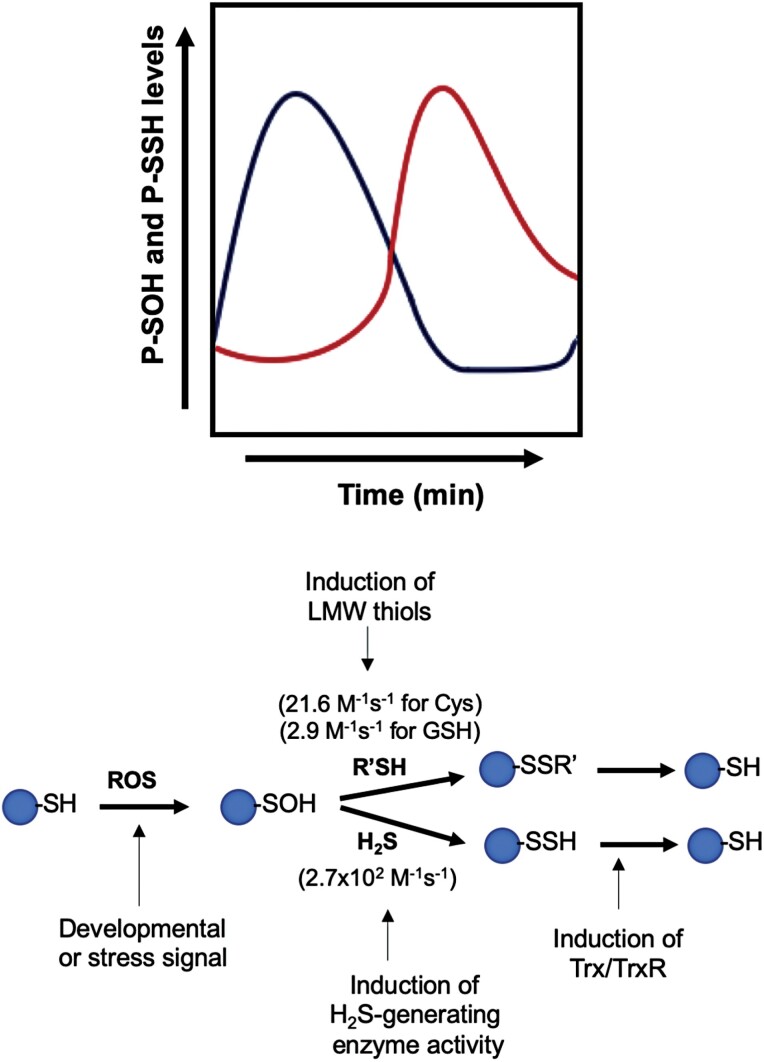
Numerous biochemical and genetic data have established beyond doubt the signaling effect of H2S in cells through persulfidation, with important consequences for numerous physiological and pathological processes in mammals and plants (Yuan et al., 2017; Paul and Snyder, 2018; Aroca et al., 2020). However, the precise mechanism that leads to the modification and the sulfur species that produces the protein persulfide formation is the subject of extensive debate and study. H2S, or its ionic forms, HS– and S2–, cannot react directly with protein thiols and requires the presence of an oxidant. Thus, H2S can react with oxidized cysteine residues as sulfenic acid (R-SOH), but also with protein nitrosothiols (R-SNO) to give protein persulfides, but this latter process is thermodynamically unfavorable (Filipovic et al., 2012). H2S can also chemically react with disulfides (R-S-S-R), but this seems unlikely to occur due to the low level of intracellular H2S and the slow reaction rate (Filipovic, 2015). Therefore, the reaction of H2S with protein sulfenic acid to form protein persulfide seems the most plausible explanation for H2S action. Cysteine residue oxidation represents a way for redox control of protein function and, therefore, H2O2 signaling takes place via the oxidation of cysteine to sulfenic acid, and the direct outcome on proteins is protein sulfenylation (Cuevasanta et al., 2015; Zivanovic et al., 2019; Willems et al., 2020). Although sulfenylated residues (R-SOH) can be reversed to reduced thiol by the action of a diverse set of reducing enzymes, stress conditions can lead to the overoxidation of cysteine residues originating the sulfinic (-SO2H) or sulfonic (-SO3H) motifs that are irreversible (Zaffagnini et al., 2019). However, in the catalytic cycle of peroxiredoxins, it was shown that the sulfinic cysteine can be reduced by sulfiredoxin in the presence of ATP via the formation of a phosphoryl intermediate (Sevilla et al., 2015). Protein sulfenic acid residues can react either with LMW thiols or with H2S (HS− ionic form), but the latter shows a significantly higher rate constant (Fig. 1). In fact, protein sulfenic residues react two orders of magnitude faster with H2S than with glutathione (Cuevasanta et al., 2015). Since H2S reacts with sulfenylated residues to form persulfide, protein persulfidation may play a role in H2O2-based signal transduction by preventing the overoxidation of cysteine residues, resulting in the loss of protein function. Under persistent oxidation stress, persulfidated proteins can react with ROS to form perthiosulfenic acids (-SSOH) and, in the presence of excess oxidant, perthiosulfenic acid can be oxidized to perthiosulfinic (-SSO2H) and perthiosulfonic acid (-SSO3H) (Aroca et al., 2018; Filipovic et al., 2018). These oxidized perthiol residues can be reduced back to thiol by the action of glutathione and thioredoxin systems, as has been demonstrated in mouse liver (Zivanovic et al., 2019; Dóka et al., 2020). The protective effect of persulfidation against overoxidation has also been shown in different cell types, where protein persulfidation increases following a phase-shifted curve after an increase in protein sulfenylation (Zivanovic et al., 2019) (Fig. 1).
Fig. 1.
Schematic representation of the temporal dynamic of protein sulfenylation (P-SOH) and persulfidation (P-SSH) in different cell types (after Zivanovic et al., 2019). After a transient ROS production induced by developmental or stress signals, the levels of sulfenylation in proteins are increased, accompanied by an increase in the activity of sulfide-generating enzymes and/or induction of low molecular weight (LMW) thiols, followed by a transient increase in protein persulfidation reversed by the action of reducing enzymes such as the thioredoxin system (Trx/TrxR). The rate constants for the reaction of R-SOH with LMW thiols and H2S at physiological pH 7.4 are shown (Cuevasanta et al., 2015).
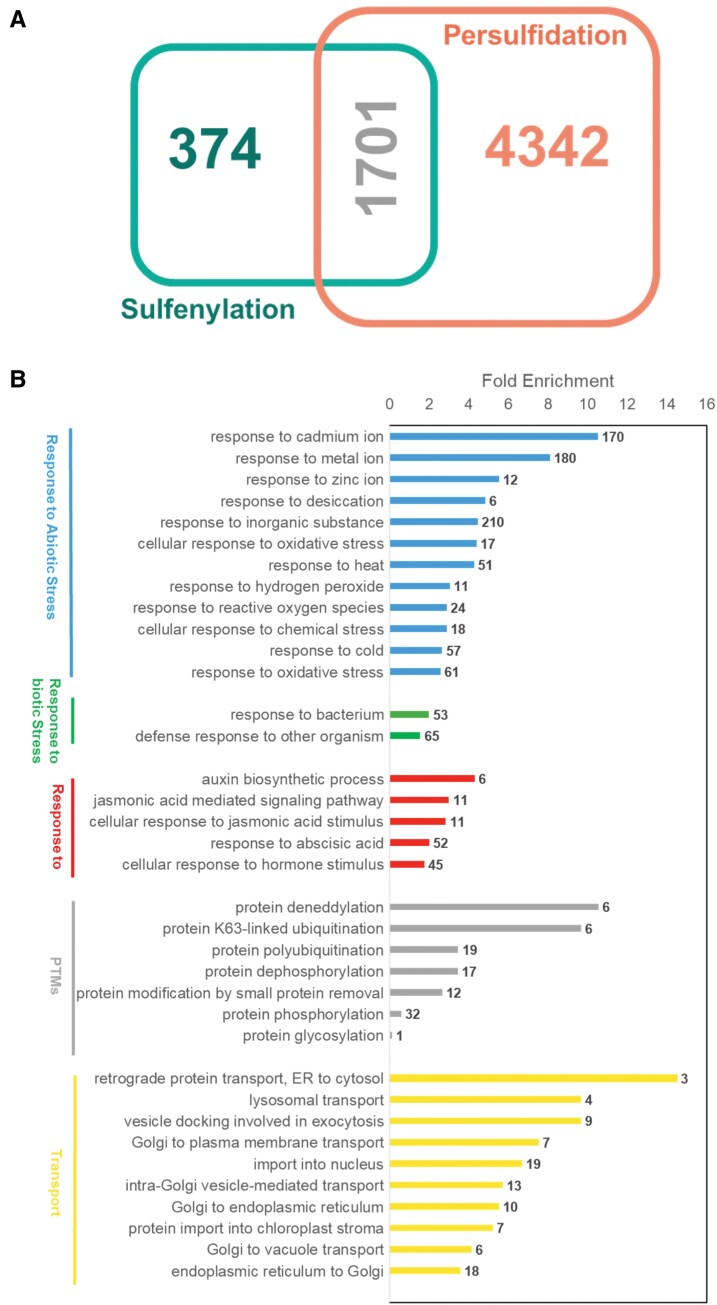
Redox regulation has been shown to be involved in many signaling processes that regulate environmental (biotic and abiotic) stress responses (Alvarez et al., 2012; Hancock, 2019), development (Jia et al., 2015; Deng et al., 2020), or autophagy and cell death (Gotor et al., 2013; Pérez-Pérez et al., 2014; Xie et al., 2014), processes where H2S is also involved. There is a lot of evidence that there is an overlap between ROS and H2S; therefore, together, protein cysteine oxidation and persulfidation may represent a mechanism for the modulation of signaling processes induced by developmental or environmental stress events. In recent years, many proteins have been described as sulfenylation targets in Arabidopsis in several works, revealing >2000 targets for this modification (De Smet et al., 2019; Huang et al., 2019; Wei et al., 2020). In a comparison performed between the sulfenylated and the previously identified persulfidated proteins, >6000 targets (Aroca et al., 2015, 2017a; Laureano-Marín et al., 2020; Jurado-Flores et al., 2021) revealed that 82% of the sulfenylated proteome described in Arabidopsis also undergo persulfidation (Fig. 2A). A total of 1701 proteins are targets for either sulfenylation or persulfidation, a number that must be underestimated taking into account that the Arabidopsis samples were very different in all these proteomic analyses. Despite the probable differences in their metabolism, the number of common proteins in both proteomes is considerably high, revealing the role of these modifications in the finely tuned balance between H2O2-based signal transduction and protection against overoxidation. Gene Ontology (GO) enrichment analysis of these proteins showed that several of these targets are associated with abiotic stress response-related GO terms, such as response to cadmium (170), metal ion (180), and zinc (12), response to oxidative stress (61), cellular response to oxidative stress (17), response to cold (57), response to heat (51), response to ROS (24), and response to H2O2 (11), among others (Fig. 2B). Included in these targets, three l-ascorbate peroxidases, four dehydroascorbate reductases, three glutaredoxins, 10 thioredoxins, two nitrate reductases, and numerous FAD/NAD(P)-oxidoreductases were found to be regulated by persulfidation and sulfenylation (see Table S1 at Zenodo https://zenodo.org/record/4727058), underlying the signaling role of these modifications in the activation of the antioxidant system against oxidative stress (Aroca et al., 2021b). In addition, GO enrichment showed that among those proteins regulated by persulfidation and sulfenylation, targets involved in response to biotic stress, hormones, signaling, other post-translational modifications, and transport were found (Fig. 2B).
Fig. 2.
Comparison of persulfidated and sulfenylated proteins. (A) Venn diagram showing the number of proteins. (B) Fold change enrichment of GO terms of common proteins modified by sulfenylation and persulfidation. Analysis was performed with PANTHER software. The numbers beside the bars indicate the number of proteins associated with each GO term for the input set.
Role of hydrogen sulfide in ABA signaling
In plants, precise mechanisms have been developed to perceive environmental stress. ABA, an important plant hormone, is involved in the regulation of growth and developmental processes, and defense against various environmental stresses. ABA is a central regulator that triggers complex signaling networks and is also involved in stomatal movement. Under certain conditions, ABA concentrations increase to activate these signaling pathways and, consequently, ABA binds to the ABA receptor protein family members Pyrabactin Resistance 1 (PYR1)/PYR1-Like (PYL)/Regulatory Component of ABA receptor (RCAR), and inhibits the activity of clade A protein phosphatases (PP2Cs) (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). This process then results in the release of sucrose non-fermenting 1 (SNF1)-related protein kinase 2s (SnRK2s) from suppression by the PP2Cs, enabling the activation of SnRK2 protein kinases. These kinases subsequently phosphorylate and activate dozens of downstream targets (Hauser et al., 2017).
Extensive and convincing evidence published in the last decade has shown a close inter-relationship between H2S and physiological processes regulated by the hormone ABA, suggesting that crosstalk occurs between both molecules in regulation and signaling in plants (Gotor et al., 2019; Aroca et al., 2020). It has been widely reported that H2S plays a role in stomatal closure, and the latest data are discussed in detail below. In addition to its role in plant growth and development, ABA plays a crucial role in plant responses to environmental stresses such as drought, salinity, osmotic stress, and heat stress, processes in which H2S has shown a protective effect, alleviating the oxidative stress associated with these adverse conditions (Gotor et al., 2019). There is a large amount of additional evidence that interconnects H2S signaling with other plant processes regulated by ABA beyond mere antioxidant defenses. For example, it has been observed that the response to drought or heat mediated by ABA induces the accumulation of intracellular H2S, and exogenous H2S addition increases plant tolerance to these stresses (Jin et al., 2011; Li and Jin, 2016). It has also been observed that ABA shows an opposite effect on the transcriptional regulation of the cytosolic l-cysteine desulfhydrase (DES1) that catalyzes the desulfuration of cysteine to generate H2S, depending on the tissue, inhibiting its transcription in mesophyll cells and increasing its transcription in guard cell-enriched tissues (Scuffi et al., 2014). In general, sulfate availability affects the ABA content and germination response to ABA and salt stress, highlighting the importance of sulfur for stress tolerance (Cao et al., 2014). From a molecular point of view, the most extensive proteomic analyses published to date on protein persulfidation has shown that several proteins involved in ABA signaling, such as PYR1 and PYL, SnRK2.2 protein kinase, and the protein phosphatase HAB2, are capable of being persulfidated (Aroca et al., 2017a; Jurado-Flores et al., 2021).
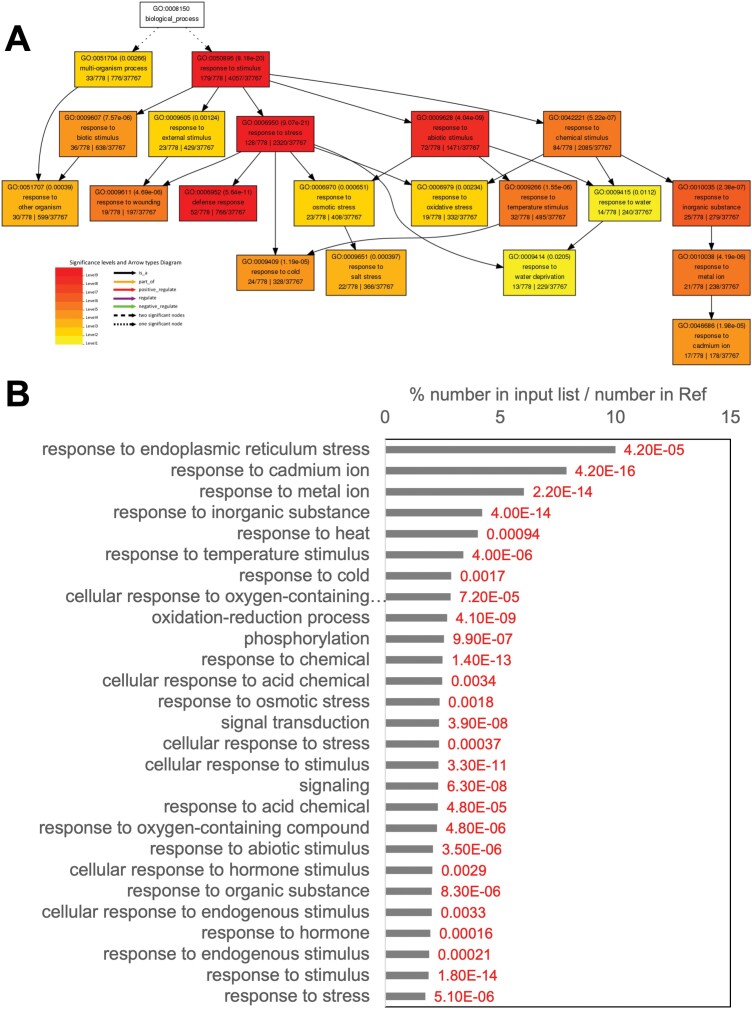
Recently, a publication about ABA-triggered persulfidation of proteins (Laureano-Marín et al., 2020) revealed nearly 800 proteins that undergo persulfidation in response to ABA treatment in comparison with an untreated control (see Table S2 at Zenodo). Data can be obtained from ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the identifier PXD019802. The GO enrichment data of the persulfidated proteins induced by ABA treatment were processed using AgriGO (Table S3 at Zenodo). The GO term associated with response to stimulus, which contained 778 proteins, was analyzed to identify the most enriched GO terms (Fig. 3A), and it included 23 proteins in response to osmotic stress, 32 in response to temperature stimulus, 19 in response to oxidative stress, 24 in response to cold, 22 in response to salt stress, and 13 in response to water deprivation. In addition, another 52, 19, and 36 proteins were involved in defense response, wounding, and biotic stimulus, respectively. All the ABA-induced persulfidated proteins involved in abiotic stress are listed in Table S4 at Zenodo.
Fig. 3.
Gene Ontology (GO) enrichment. (A) GO enrichment of ABA-induced persulfidated proteins involved in response to stimulus. (B) GO enrichment of persulfidated targets in response to ABA susceptible to sulfenylation. The P-value for each GO term is annotated in red numbers.
Overall, these results show that ABA treatment triggers persulfidation of a high number of proteins, and some of them aim to activate a cellular response to combat abiotic and biotic stresses. In addition, a total of 276 of these ABA-induced persulfidated proteins have been described as being sulfenylated. Further analysis of these targets shows that there are proteins involved in the response to abiotic stresses that are also susceptible to persulfidation and sulfenylation The GO enrichment data of the persulfidated proteins induced by ABA treatment, which can also be targets for sulfenylation, were processed using AgriGo tool (see Table S5 at Zenodo), and a selection of the GO-enriched terms associated with the stress response was constructed to identify the most enriched GO terms (Fig. 3B). Those GO terms most represented were response to endoplasmic reticulum stress (GO: 0034976) with a P-value of 0.000042 and a false discovery rate (FDR) of 0.00076, including six proteins in this GO term; response to cadmium (GO: 0046686) with scores of 4.2×10–16 and 8.8×10–14 for the P-value and FDR, respectively; and response to heat (GO: 0009408) and cold (GO: 0009409) with P-values of 0.00094 and 0.0017, and FDRs of 0.012 and 0.02, respectively. Nevertheless, as shown in Fig. 3B, other important GO terms, such as response to osmotic stress, signal transduction, and response to hormone are over-represented. These results highlight the existence of crosstalk between sulfenylation and persulfidation in response to certain abiotic stresses and that protein post-translational modifications play an important role in regulating these responses.
Role of hydrogen sulfide in guard cell ABA signaling
As pointed out previously, the activation of ABA signaling pathways induces downstream targets that, in conjunction with ROS, Ca2+, and Ca2+-dependent protein kinases (CDPKs), activate ion channels to mediate stomatal closure and reduce water loss from transpiration (Mustilli et al., 2002; Papanatsiou et al., 2015). The participation of H2S in stomatal closure has also been described previously (García-Mata and Lamattina, 2010; Jin et al., 2013). An initial study showed that ABA cannot induce the stomatal closure of des knockout mutants deficient in cytosolic DES1, which produces H2S in the cytosol (Alvarez et al., 2010), while the addition of an exogenous H2S donor restored the closure. Moreover, ABA-dependent stomatal closure was partially blocked by an inhibitor of l-cysteine desulfhydrase and a scavenger of H2S, dl-propargylglycine (PAG) and hypotaurine (HT), respectively, suggesting that H2S participates in ABA-triggered stomatal movement (Scuffi et al., 2014). Although DES1 is expressed at all growth stages, at the tissue level, green fluorescent protein (GFP) expression driven by the DES1 promoter is very high in guard cells (Laureano-Marín et al., 2014). It is also noteworthy that the DES1 gene expression level in the RNA extracts of epidermal cells was several-fold higher than that in the mesophyll cell-enriched samples upon ABA treatment (Scuffi et al., 2014), which provides a clue that the high expression level of DES1 in epidermal cells may largely be due to the proportion of guard cells. Recently, genetic evidence also indicated that guard cell-specific expressed DES1 is required for in situ H2S production and is sufficient for regulating ABA-induced stomatal closure (Zhang et al., 2020).
The synthesis of ABA is a central response to stress. Interestingly, guard cells contain the complete suite of ABA biosynthesis pathway components. The molybdenum cofactor sulfurase ABA3 that mediated ABA synthesis in guard cells is sufficient to induce stomatal closure and relieve leaf wilting (Bauer et al., 2013). Several studies have revealed that H2S is involved in ABA synthesis. Exogenous application of NaHS was shown to increase the transcript levels of ABA biosynthesis-related genes during polyethylene glycol (PEG) treatment in both wheat leaves and wheat roots (Ma et al., 2016). Consistently, it was found that the transcription of genes related to ABA biosynthesis, such as the 9-cis-epoxycarotenoid dioxygenases NCED2, NCED3, and NCED5, sharply increased in rice seedlings under drought stress conditions, and pre-treatment with NaHS further strengthened this inductive effect (Zhou et al., 2020). Recently, Zhang et al. (2021a) demonstrated that the accumulation of all of the transcripts involved in ABA synthesis in leaves increased in the wild type but not in des1 mutants, indicating that DES1 is essential for dehydration-induced ABA synthesis. Moreover, the H2S content was lower in the aba3 mutant than in the wild type, suggesting that DES1-produced H2S is regulated by ABA synthesis. In addition, H2S participates in NO- and ethylene-induced stomatal closure (Hou et al., 2013; Scuffi et al., 2014).
Proteomic analyses have revealed that various proteins involved in ABA signaling are susceptible to persulfidation and that ABA treatments trigger the persulfidation of a considerable number of protein targets (Aroca et al., 2017a; Laureano-Marín et al., 2020), thus suggesting that H2S regulates ABA signaling pathways through persulfidation of specific targets, including those within guard cells. In this way, the DES1-mediated guard cell ABA cascade is attributable to H2S signaling through persulfidation of open stomata 1 (OST1)/SNF1-RELATED PROTEIN KINASE 2.6 (SnRK2.6) at Cys131 and Cys137, which enhances ABA signaling (Chen et al., 2020). Remarkably, SnRK2.6/OST1 is also nitrosylated by NO at Cys137, leading to the inhibition of its activity and further negatively regulating guard cell ABA signaling (Wang et al., 2015). Crosstalk between H2S and NO has been previously described in the ABA signaling network in guard cells (Lisjak et al., 2010; Scuffi et al., 2014), and SnRK2.6/OST1 could be one of the targets driving this interplay.
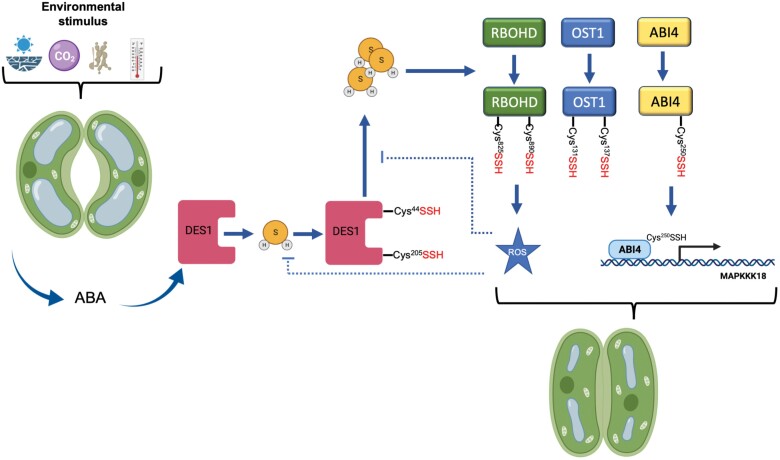
DES1 itself is also activated by H2S through autopersulfidation at Cys44 and Cys205, which leads to transient H2S overproduction and the amplification of H2S signals in guard cells (Shen et al., 2020). Activated DES1 persulfidates the NADPH oxidase RBOHD at Cys825 and Cys890 to rapidly induce a ROS burst that results in stomatal closure. Interestingly, persulfidation of both DES1 and RBOHD is redox dependent, and ROS accumulation at high levels induces persulfide oxidation, which inhibits the activity of these proteins, leading to ABA desensitization. The oxidized persulfides can be reduced back to thiol groups by thioredoxin and prevent the continuous activation of ABA signaling. Thus, these processes form a negative feedback loop through H2S- and ROS-mediated modification that finely tunes guard cell redox homeostasis and ABA signaling. In addition, the accumulation of ROS induced by H2S was also found to stimulate Ca2+ influx in guard cells (Wang et al., 2016). Another element involved in guard cell DES-mediated ABA signaling has been recently defined: the transcription factor ABA insensitive 4 (ABI4). The DES1-dependent H2S accumulation induced by ABA generates the persulfidation of ABI4 at Cys250, promoting the MAPKKK18 transactivation, and thus propagating the mitogen-activated protein kinase (MAPK) signaling cascade in response to ABA (Zhou et al., 2021a). Together, these findings hint at the complexity of the H2S signaling in stomatal movement.
The control of stomatal closure or opening relies on the activity of ion channels and ion transport proteins in the plasma and vacuolar membranes. The regulation of inward-rectifying K+ channels by H2S was shown in the sense that inactivation of the current associated with these channels induces stomatal closure by submicromolar concentrations of H2S (Papanatsiou et al., 2015). Proof was also provided of activation by a low concentration of H2S of S-type anion currents in guard cells, the process of which requires elevated free cytosolic Ca2+ levels and OST1 function (Wang et al., 2016). All these data highlight the complexity of the relationship between H2S and ion channels in the regulation of guard cell movement. Other secondary messengers that interact with H2S in the guard cell signaling network have been elucidated. In addition to the above-described ROS burst produced by NADPH oxidases, phospholipase D-derived phosphatidic acid is needed (Scuffi et al., 2018; Liu et al., 2021).
In summary, the H2S signaling network of stomatal movement is highly complex, and interactions among many different components of ABA-dependent signaling have been demonstrated (Pantaleno et al., 2021). Moreover, accumulative evidence indicates that the H2S molecular mechanism involves persulfidation (Fig. 4).
Fig. 4.
Graphical model of interconnections between H2S and ABA signaling networks in guard cells through the persulfidation of specific proteins. Under various environmental stress conditions, in guard cells, ABA concentrations increase and trigger the DES1 activity to induce the production of H2S to persulfidate specific protein targets. DES1 itself is persulfidated at Cys44 and Cys205, and causes the persulfidation of open stomata 1 (OST1) at Cys131 and Cys137, the NADPH oxidase RBOHD at Cys825 and Cys890, and ABI4 at Cys250. Persulfidated RBOHD produces a ROS burst that results in stomatal closure. Overaccumulation of ROS induces persulfide oxidation leading to ABA desensitivity. Persulfidated ABI4 promotes MAPKKK18 transactivation and MAPK signaling.
Conclusions and future perspectives
During past years, an immense number of plant studies describing the role of H2S in the regulation of essential processes have enabled H2S to be considered a signaling molecule of the same significance as NO and H2O2. Moreover, recent reports have permitted considerable insight into the molecular mechanism involved in H2S action in some specific processes such as ABA-dependent stomatal closure, and, importantly, to know the specific protein targets of persulfidation. Nevertheless, there is no doubt that the current challenges in the H2S field are, on one hand, to deepen knowledge of the molecular mechanism involved in H2S action and, on the other, to ascertain what is the bona fide sulfurating molecule that modifies the thiol group on proteins. Regarding the action of H2S, while an important effort has been made to establish chemical methods to label and enrich persulfidated proteins, and technical improvement of mass spectrometers have allowed the identification of an increasing number of plant proteins, mainly in Arabidopsis, knowledge of the H2S mechanism of action in a particular process is still scarce. As described above, insight into this mechanism has only been revealed to a certain extent in the ABA-dependent stomatal closure process, and in the regulation of autophagy by H2S, although, in the case of autophagy, the information is still limited.
With respect to the nature of the sulfurating species, this aspect is the subject of a great debate, even in animal systems. Due to the chemical nature of H2S, it cannot react directly with the thiols in proteins, and several scenarios leading to the formation of persulfidated proteins have been proposed. Thus, H2S can react with oxidized cysteine residues such as sulfenylated or nitrosylated cysteines or disulfides, and, therefore, under specific oxidative conditions, the sulfurating species can be H2S, or its ionic forms HS− and S2−. Other sulfurating molecules proposed are polysulfides, which contain the form of sulfur named sulfane with the oxidation state of 0 and which have the ability to attach reversibly to other sulfur atoms (Ida et al., 2014). We can hypothesize that depending on the specific condition/microenvironment of the target protein or the biological process in which the protein is involved, a particular sulfurating species or a mixture of them can be responsible for the protein persulfidation and it would be very difficult to differentiate between them. In addition, a prokaryotic and mammalian cysteinyl-tRNA synthetase has also been described that synthesizes persulfidated cysteine for direct incorporation to proteins (Akaike et al., 2017). Another interesting point is to correlate the protein persulfidation pattern/level in one specific tissue/condition with the level of the sulfurating molecules. Perhaps in the context of a high level of persulfidation, it would be possible to discriminate which sulfurating species is responsible for performing persulfidation.
Acknowledgements
This work was supported in part by the European Regional Development Fund through the Agencia Estatal de Investigación (grant nos PID2019-109785GB-I00 and RED2018-102397-T); Junta de Andalucía (grant nos P18-RT-3154 and US-1255781); the Marie Skłodowska-Curie Grant Agreement (grant no. 834120 to AA); the National Natural Science Foundation of China (grant no. 31607255 (L458) to YX); and the China Scholarship Council through the State Scholarship Fund (grant no. 201906850022 to JZ).
Glossary
Abbreviations
- ABA
abscisic acid
- CO
carbon monoxide
- H2O2
hydrogen peroxide
- NO
nitric oxide
- ROS
reactive oxygen species
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contributions
AA, JZ, and LCR: writing—original draft; YX and CG: review; CG: conceptualization and writing—editing.
Data availability
A list of common proteins susceptible to persulfidation and sulfenylation, ABA-induced persulfidated proteins involved in abiotic stress, persulfidated proteins in response to ABA susceptible to S-sulfenylation, persulfidated proteins identified in response to ABA treatments, and GO enrichment of ABA-induced persulfidated proteins are available at Zenodo https://zenodo.org/record/4727058; Aroca et al. (2021b).
References
- Abe K, Kimura H. 1996. The possible role of hydrogen sulfide as an endogenous neuromodulator. Journal of Neuroscience 16, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike T, Ida T, Wei FY, et al. 2017. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nature Communications 8, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez C, Bermudez MA, Romero LC, Gotor C, Garcia I. 2012. Cysteine homeostasis plays an essential role in plant immunity. New Phytologist 193, 165–177. [DOI] [PubMed] [Google Scholar]
- Alvarez C, Calo L, Romero LC, García I, Gotor C. 2010. An O-acetylserine(thiol)lyase homolog with l-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiology 152, 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Benito JM, Gotor C, Romero LC. 2017a. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. Journal of Experimental Botany 68, 4915–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Gotor C, Bassham DC, Romero LC. 2020. Hydrogen sulfide: from a toxic molecule to a key molecule of cell life. Antioxidants 9, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Gotor C, Romero LC. 2018. Hydrogen sulfide signaling in plants: emerging roles of protein persulfidation. Frontiers in Plant Science 9, 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Schneider M, Scheibe R, Gotor C, Romero LC. 2017b. Hydrogen sulfide regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant & Cell Physiology 58, 983–992. [DOI] [PubMed] [Google Scholar]
- Aroca Á, Serna A, Gotor C, Romero LC. 2015. S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiology 168, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Yruela I, Gotor C, Bassham DC. 2021a. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proceedings of National Academy of Sciences U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A, Zhang J, Xie Y, Romero LC, Gotor C. 2021b. Data from: Hydrogen sulfide signaling in plant adaptations to adverse conditions: molecular mechanisms. Zenodo https://zenodo.org/record/4727058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Ache P, Lautner S, et al. 2013. The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Current Biology 23, 53–57. [DOI] [PubMed] [Google Scholar]
- Boubeta FM, Bieza SA, Bringas M, Palermo JC, Boechi L, Estrin DA, Bari SE. 2020. Hemeproteins as targets for sulfide species. Antioxidants & Redox Signaling 32, 247–257. [DOI] [PubMed] [Google Scholar]
- Calderwood A, Kopriva S. 2014. Hydrogen sulfide in plants: from dissipation of excess sulfur to signaling molecule. Nitric Oxide 41, 72–78. [DOI] [PubMed] [Google Scholar]
- Cao MJ, Wang Z, Zhao Q, Mao JL, Speiser A, Wirtz M, Hell R, Zhu JK, Xiang CB. 2014. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. The Plant Journal 77, 604–615. [DOI] [PubMed] [Google Scholar]
- Chen S, Jia H, Wang X, Shi C, Wang X, Ma P, Wang J, Ren M, Li J. 2020. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Molecular Plant 13, 732–744. [DOI] [PubMed] [Google Scholar]
- Cooper CE, Brown GC. 2008. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. Journal of Bioenergetics and Biomembranes 40, 533–539. [DOI] [PubMed] [Google Scholar]
- Cuevasanta E, Denicola A, Alvarez B, Möller MN. 2012. Solubility and permeation of hydrogen sulfide in lipid membranes. PLoS One 7, e34562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. 2015. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. Journal of Biological Chemistry 290, 26866–26880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G, Zhou L, Wang Y, Zhang G, Chen X. 2020. Hydrogen sulfide acts downstream of jasmonic acid to inhibit stomatal development in Arabidopsis. Planta 251, 42. [DOI] [PubMed] [Google Scholar]
- De Smet B, Willems P, Fernandez-Fernandez AD, Alseekh S, Fernie AR, Messens J, Van Breusegem F. 2019. In vivo detection of protein cysteine sulfenylation in plastids. The Plant Journal 97, 765–778. [DOI] [PubMed] [Google Scholar]
- Dóka É, Ida T, Dagnell M, et al. 2020. Control of protein function through oxidation and reduction of persulfidated states. Science Advances 6, eaax8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic MR. 2015. Persulfidation (S-sulfhydration) and H2S. In: Moore PK, Whiteman M, eds. Chemistry, biochemistry and pharmacology of hydrogen sulfide. Cham: Springer International Publishing,29–59. [Google Scholar]
- Filipovic MR, Miljkovic JLj, Nauser T, Royzen M, Klos K, Shubina T, Koppenol WH, Lippard SJ, Ivanović-Burmazović I. 2012. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. Journal of the American Chemical Society 134, 12016–12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic MR, Zivanovic J, Alvarez B, Banerjee R. 2018. Chemical biology of H2S signaling through persulfidation. Chemical Reviews 118, 1253–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L. 2010. Hydrogen sulphide, a novel gasotransmitter involved in guard cell signalling. New phytologist 188, 977–984. [DOI] [PubMed] [Google Scholar]
- Gotor C, García I, Aroca Á, Laureano-Marín AM, Arenas-Alfonseca L, Jurado-Flores A, Moreno I, Romero LC. 2019. Signaling by hydrogen sulfide and cyanide through post-translational modification. Journal of Experimental Botany 70, 4251–4265. [DOI] [PubMed] [Google Scholar]
- Gotor C, García I, Crespo JL, Romero LC. 2013. Sulfide as a signaling molecule in autophagy. Autophagy 9, 609–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JT. 2019. Hydrogen sulfide and environmental stresses. Environmental and Experimental Botany 161, 50–56. [Google Scholar]
- Hauser F, Li Z, Waadt R, Schroeder JI. 2017. SnapShot: Abscisic acid signaling. Cell 171, 1708–1708.e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Wang L, Liu J, Hou L, Liu X. 2013. Hydrogen sulfide regulates ethylene-induced stomatal closure in Arabidopsis thaliana. Journal of Integrative Plant Biology 55, 277–289. [DOI] [PubMed] [Google Scholar]
- Huang J, Willems P, Wei B, et al. 2019. Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proceedings of the National Academy of Sciences, USA 116, 21256–21261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H, et al. 2014. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proceedings of the National Academy of Sciences, USA 111, 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen B, Fago A. 2018. Reactions of ferric hemoglobin and myoglobin with hydrogen sulfide under physiological conditions. Journal of Inorganic Biochemistry 182, 133–140. [DOI] [PubMed] [Google Scholar]
- Jia H, Chen S, Liu D, et al. 2018. Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Frontiers in Plant Science 9, 1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Hu Y, Fan T, Li J. 2015. Hydrogen sulfide modulates actin-dependent auxin transport via regulating ABPs results in changing of root development in Arabidopsis. Scientific Reports 5, 8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Shen J, Qiao Z, Yang G, Wang R, Pei Y. 2011. Hydrogen sulfide improves drought resistance in Arabidopsis thaliana. Biochemical and Biophysical Research Communications 414, 481–486. [DOI] [PubMed] [Google Scholar]
- Jin Z, Xue S, Luo Y, Tian B, Fang H, Li H, Pei Y. 2013. Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiology and Biochemistry 62, 41–46. [DOI] [PubMed] [Google Scholar]
- Johnston DT, Wolfe-Simon F, Pearson A, Knoll AH. 2009. Anoxygenic photosynthesis modulated Proterozoic oxygen and sustained Earth’s middle age. Proceedings of the National Academy of Sciences, USA 106, 16925–16929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado-Flores A, Romero LC, Gotor C. 2021. Label-free quantitative proteomic analysis of nitrogen starvation in Arabidopsis root reveals new aspects of H2S signaling by protein persulfidation. Antioxidants 10, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabil O, Banerjee R. 2010. Redox biochemistry of hydrogen sulfide. Journal of Biological Chemistry 285, 21903–21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Ok K, Shimberg GD, Bursac B, Marko L, Ivanovic-Burmazovic I, Michel SLJ, Filipovic MR. 2019. Direct zinc finger protein persulfidation by H2S is facilitated by Zn2. Angewandte Chemie 58, 7997–8001. [DOI] [PubMed] [Google Scholar]
- Laureano-Marín AM, Aroca Á, Pérez-Pérez ME, Yruela I, Jurado-Flores A, Moreno I, Crespo JL, Romero LC, Gotor C. 2020. Abscisic acid-triggered persulfidation of the Cys protease ATG4 mediates regulation of autophagy by sulfide. The Plant cell 32, 3902–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureano-Marín AM, García I, Romero LC, Gotor C. 2014. Assessing the transcriptional regulation of l-cysteine desulfhydrase 1 in Arabidopsis thaliana. Frontiers in Plant Science 5, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen S, Wang X, Shi C, Liu H, Yang J, Shi W, Guo J, Jia H. 2018. Hydrogen sulfide disturbs actin polymerization via S-sulfhydration resulting in stunted root hair growth. Plant Physiology 178, 936–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shi C, Wang X, Liu C, Ding X, Ma P, Wang X, Jia H. 2020. Hydrogen sulfide regulates the activity of antioxidant enzymes through persulfidation and improves the resistance of tomato seedling to copper oxide nanoparticles (CuO NPs)-induced oxidative stress. Plant Physiology and Biochemistry 156, 257–266. [DOI] [PubMed] [Google Scholar]
- Li Q, Lancaster JR Jr. 2013. Chemical foundations of hydrogen sulfide biology. Nitric Oxide 35, 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-G, Jin J-Z. 2016. Hydrogen sulfide partly mediates abscisic acid-induced heat tolerance in tobacco (Nicotiana tabacum L.) suspension cultured cells. Plant Cell, Tissue and Organ Culture 125, 207–214. [Google Scholar]
- Lisjak M, Srivastava N, Teklic T, Civale L, Lewandowski K, Wilson I, Wood ME, Whiteman M, Hancock JT. 2010. A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiology and Biochemistry 48, 931–935. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhou Y, Li H, Liu R, Wang W, Wu W, Yang N, Wang S. 2021. Osmotic stress-triggered stomatal closure requires phospholipase Dδ and hydrogen sulfide in Arabidopsis thaliana. Biochemical and Biophysical Research Communications 534, 914–920. [DOI] [PubMed] [Google Scholar]
- Ma D, Ding H, Wang C, Qin H, Han Q, Hou J, Lu H, Xie Y, Guo T. 2016. Alleviation of drought stress by hydrogen sulfide is partially related to the abscisic acid signaling pathway in wheat. PLoS One 11, e0163082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. 2009. H2S signals through protein S-sulfhydration. Science Signaling 2, ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. 2002. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. The Plant Cell 14, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offre P, Spang A, Schleper C. 2013. Archaea in biogeochemical cycles. Annual Review of Microbiology 67, 437–457. [DOI] [PubMed] [Google Scholar]
- Pantaleno R, Scuffi D, García-Mata C. 2021. Hydrogen sulphide as a guard cell network regulator. New Phytologist 230, 451–456. [DOI] [PubMed] [Google Scholar]
- Papanatsiou M, Scuffi D, Blatt MR, García-Mata C. 2015. Hydrogen sulfide regulates inward-rectifying K+ channels in conjunction with stomatal closure. Plant Physiology 168, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. 2018. Gasotransmitter hydrogen sulfide signaling in neuronal health and disease. Biochemical Pharmacology 149, 101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez ME, Zaffagnini M, Marchand CH, Crespo JL, Lemaire SD. 2014. The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 10, 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuffi D, Álvarez C, Laspina N, Gotor C, Lamattina L, García-Mata C. 2014. Hydrogen sulfide generated by l-cysteine desulfhydrase acts upstream of nitric oxide to modulate abscisic acid-dependent stomatal closure. Plant Physiology 166, 2065–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuffi D, Nietzel T, Di Fino LM, Meyer AJ, Lamattina L, Schwarzländer M, Laxalt AM, García-Mata C. 2018. Hydrogen sulfide increases production of NADPH oxidase-dependent hydrogen peroxide and phospholipase D-derived phosphatidic acid in guard cell signaling. Plant Physiology 176, 2532–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla F, Camejo D, Ortiz-Espín A, Calderón A, Lázaro JJ, Jiménez A. 2015. The thioredoxin/peroxiredoxin/sulfiredoxin system: current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. Journal of Experimental Botany 66, 2945–2955. [DOI] [PubMed] [Google Scholar]
- Shen J, Zhang J, Zhou M, et al. 2020. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. The Plant Cell 32, 1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitvitsky V, Miljkovic JL, Bostelaar T, et al. 2018. Cytochrome c reduction by H2S potentiates sulfide signaling. ACS Chemical Biology 13, 2300–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, Del-Toro N, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Research 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wan R, Shi Y, Xue S. 2016. Hydrogen sulfide activates S-type anion channel via OST1 and Ca2+ modules. Molecular Plant 9, 489–491. [DOI] [PubMed] [Google Scholar]
- Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu X, Tao WA, Song CP, Zhu JK. 2015. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proceedings of the National Academy of Sciences, USA 112, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. 2014. Gasotransmitters: growing pains and joys. Trends in Biochemical Sciences 39, 227–232. [DOI] [PubMed] [Google Scholar]
- Wei B, Willems P, Huang J, Tian C, Yang J, Messens J, Van Breusegem F. 2020. Identification of sulfenylated cysteines in Arabidopsis thaliana proteins using a disulfide-linked peptide reporter. Frontiers in Plant Science 11, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P, Van Breusegem F, Huang J. 2020. Contemporary proteomic strategies for cysteine redoxome profiling. Plant Physiology 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhang C, Lai D, Sun Y, Samma MK, Zhang J, Shen W. 2014. Hydrogen sulfide delays GA-triggered programmed cell death in wheat aleurone layers by the modulation of glutathione homeostasis and heme oxygenase-1 expression. Journal of Plant Physiology 171, 53–62. [DOI] [PubMed] [Google Scholar]
- Yuan S, Shen X, Kevil CG. 2017. Beyond a gasotransmitter: hydrogen sulfide and polysulfide in cardiovascular health and immune response. Antioxidants & Redox Signaling 27, 634–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Fermani S, Marchand CH, Costa A, Sparla F, Rouhier N, Geigenberger P, Lemaire SD, Trost P. 2019. Redox homeostasis in photosynthetic organisms: novel and established thiol-based molecular mechanisms. Antioxidants & Redox Signaling 31, 155–210. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhou H, Zhou M, Ge Z, Zhang F, Foyer CH, Yuan X, Xie Y. 2021a. The coordination of guard-cell autonomous ABA synthesis and DES1 function in situ regulates plant water deficit responses. Journal of Advanced Research 27, 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou M, Ge Z, et al. 2020. Abscisic acid-triggered guard cell l-cysteine desulfhydrase function and in situ hydrogen sulfide production contributes to heme oxygenase-modulated stomatal closure. Plant, Cell & Environment 43, 624–636. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhou M, Zhou H, et al. 2021b. Hydrogen sulfide, a signaling molecule in plant stress responses. Journal of Integrative Plant Biology 63, 146–160. [DOI] [PubMed] [Google Scholar]
- Zhou H, Chen Y, Zhai F, Zhang J, Zhang F, Yuan X, Xie Y. 2020. Hydrogen sulfide promotes rice drought tolerance via reestablishing redox homeostasis and activation of ABA biosynthesis and signaling. Plant Physiology and Biochemistry 155, 213–220. [DOI] [PubMed] [Google Scholar]
- Zhou M, Zhang J, Shen J, et al. 2021a. Hydrogen sulfide-linked persulfidation of ABI 4 controls ABA responses through the transactivation of MAPKKK18 in Arabidopsis. Molecular Plant 14, 921– 936. [DOI] [PubMed] [Google Scholar]
- Zhou M, Zhou H, Shen J, Zhang Z, Gotor C, Romero LC, Yuan X, Xie Y. 2021b. H2S action in plant life cycle. Plant Growth Regulation 94, 1–9. [Google Scholar]
- Zivanovic J, Kouroussis E, Kohl JB, et al. 2019. Selective persulfide detection reveals evolutionarily conserved antiaging effects of S-sulfhydration. Cell Metabolism 30, 1152–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A list of common proteins susceptible to persulfidation and sulfenylation, ABA-induced persulfidated proteins involved in abiotic stress, persulfidated proteins in response to ABA susceptible to S-sulfenylation, persulfidated proteins identified in response to ABA treatments, and GO enrichment of ABA-induced persulfidated proteins are available at Zenodo https://zenodo.org/record/4727058; Aroca et al. (2021b).