Abstract
Background:
Mendelian randomization (MR) analysis has become popular in inferring and estimating the causality of an exposure on an outcome due to the success of genome wide association studies. Many statistical approaches have been developed and each of these methods require specific assumptions.
Results:
In this article, we review the pros and cons of these methods. We use an example of high-density lipoprotein cholesterol on coronary artery disease to illuminate the challenges in Mendelian randomization investigation.
Conclusion:
The current available MR approaches allow us to study causality among risk factors and outcomes. However, novel approaches are desirable for overcoming multiple source confounding of risk factors and an outcome in MR analysis.
Keywords: Mendelian randomization, causality, summary statistics, confounding, instrumental variable
Author summary:
Mendelian randomization analysis is a popular approach to studying the causality of exposures on an outcome, and it shares similarities with randomized controlled trials. Since MR is based on observational data, it requires assumptions that are difficult to validate. We review the current developed MR approaches and the challenges in performing MR analysis and interpreting the results.
INTRODUCTION
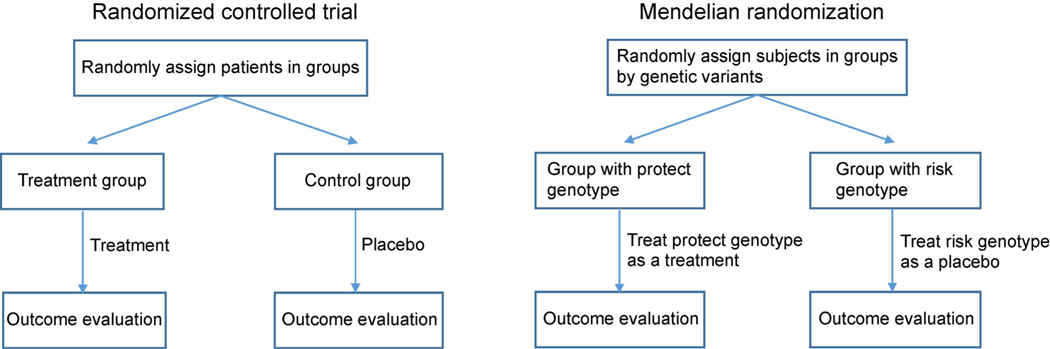
Randomized controlled trials (RCTs) are considered as the gold standard to establish a causal relationship between an exposure and an outcome in epidemiology studies. Many associations observed in epidemiological studies have failed to be replicated in RCTs, such as fiber and colon cancer [1], vitamin E, cardiovascular disease and lung cancer [2,3], and vitamin C and cardiovascular disease [4]. The failed replications in RCTs can be potentially attributed to confounding, reverse causation, and various biases [5,6]. Thus, RCTs are the primary tool to establish a causation between a risk factor and an outcome, but they come with a high cost. To circumvent the high cost in RCTs, Mendelian randomization (MR) has become a widely used epidemiological approach to infer causality of an exposure to a disease outcome [7–9]. This is benefitted from the rapid identifications of genetic variants associated with complex traits in large genome wide associations (GWAS) [10]. Intuitively, MR shares a similarity with RCTs (Fig. 1). In RCTs, the enrolled patients are randomly assigned to a treatment or a control group to eliminate potential confounding associated with both the treatment and the outcome. Therefore, causal effect can be estimated in an unbiased fashion. In contrast, MR assigns subjects based on their carried alleles, which are inherited from their parents. Since the alleles are transmitted from parents to offspring randomly, individuals are therefore divided into different groups randomly based on their genotypes. If the allele is associated with an exposure and the exposure causally affects the disease, we will observe different disease frequencies among different genotype groups. Otherwise, such an association will not be present if the exposure does not contribute to the disease. Furthermore, this association is independent of environment confounding because the allele predisposes to environment [8]. This idea was originally proposed by Katan [9] who focused on testing for the association of a genotype and an outcome rather than estimating the effect of an exposure to an outcome. Hence, the data on exposure in Katan’s approach is not required given that the association between the genotype and the exposure has been established. Recent development of MR focus on the causal effect estimation by using the genetic markers as “instrumental variables (IVs)” [8,11]. With that many large GWASs having already collected genomic data, the MR approach is an appealing method to estimate the causal effect between an exposure and an outcome with much less cost than RCTs.
Figure 1. Comparison between RCT and MR designs.
Early MR approach is based on having one genetic marker as an IV. To be a valid IV, the genetic marker must satisfy the following three IV conditions [8]: (IV1) the genetic marker is reliably associated with the exposure, (IV2) the genetic marker is associated with the outcome only through the exposure, and (IV3) the genetic marker is independent of unobserved confounders that affect the exposure and outcome after conditioning on observed confounders. The IV2 condition is also referred to as the exclusion restriction, and both IV2 and IV3 are difficult to be fully examined in reality [12]. VanderWeele et al. [12] listed multiple exclusion restriction violations in MR analysis when a single genetic marker is used for IV. The potential violations include: pleiotropic genetic instrument variable; measurement error; time-dependent exposure; reverse causation; sample selection bias; linkage disequilibrium between the genetic marker and a causal variant; and population structure. All the above violations will create a pathway from the genetic marker to the outcome that does not go through the exposure, therefore, bringing bias into the causal estimation. An IV with pleiotropic effect is a major concern in a MR analysis. In the past decade, GWASs have been successful in identifying genetic variants associated with complex traits (https://www.genome.gov/gwastudies/). A recent study showed that 90% of the identified variants are associated with multiple traits [13], a term that is characterized as cross phenotype association [14]. This also suggests pleiotropic variants are widely spread across traits. Such pleiotropic variants can be detected by analyzing multiple traits together [15], but the method itself does not indicate which variant is a pleiotropic variant. Thus, methods of using multiple genetic variants for IVs in MR analysis have been rapidly developed [16], and because the impact of some of the violations is alleviated when multiple IVs are applied.
In this review, I will discuss MR approaches using multiple genetic variants as IVs with summary statistics. I will also discuss the pros and cons for these methods (Table 1).
Table 1.
Summary of MR methods
| Methods | Conditions | Strengths | Weakness | Software Web address |
|---|---|---|---|---|
| IVW | Valid IVs or Balanced pleiotropy | Most efficient, Computationally fast | Large bias with invalid IVs | 1 |
| MR-Egger | InSIDE | Less requirement for IVs, Computationally fast | Less efficient, Sensitive to outliers | 1 |
| Weighted median | >50% valid IVs | Robust to outliers, Computationally fast | Sensitive to selecting IVs | 1 |
| MBE | ZEMPA* | Robust to outliers, Computationally fast | Less efficient, Depending on bandwidth parameter | 1 |
| MR-Robust | Robust to outliers | Efficient with valid IVs | Inflated FPR with invalid IVs | 2 |
| MR-Lasso | Robust to outliers | Efficient with valid IVs | Inflated FPR with invalid IVs | 2 |
| MRMix | ZEMPA* | Robust to outliers | Inflated FPR for some cases, Computationally slow | 3 |
| MR-PRESSO | >50% valid IVs | Detect pleiotropy variants | Inflated FPR with invalid IVs, Requires simulations/Computational intense | 4 |
| iMRP | >50% valid IVs | Identify pleiotropy variants, computationally fast | Inflated FPR with invalid IVs | 5 |
ZEMPA: the zero modal pleiotropy assumption
Available from the author
STATISTICAL METHODS FOR MR APPROACHES USING SUMMARY STATISTICS
IVW and MR-Egger
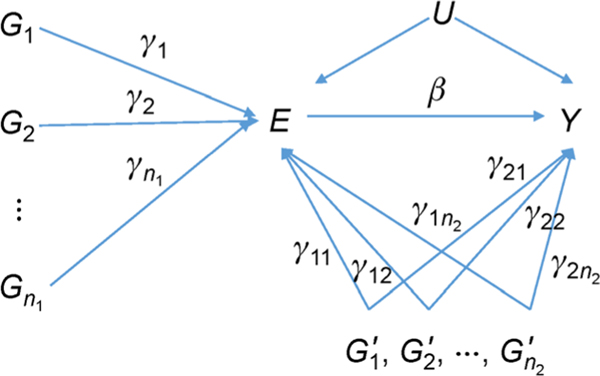
When individual level data is available, an MR analysis can be performed by using the standard two-stage least squares (TSLS) [17]. The same idea can be extended to the data where only summary statistics are available. We denote E and Y as the exposure and the outcome described in Fig. 2, respectively. The goal of MR is to establish the causal relationship from the exposure E to the outcome Y using independent genetic variants as the IVs. We assume that the association of an IV and the exposure has already been reliably established in GWAS. Because the genetic variants associated with the exposure are possibly associated with the outcome through different paths (Fig. 2), we use the following general models:
| (1) |
| (2) |
where γi is the direct contribution of the variant Gi to the exposure E, γ1i and γ2i are the direct contributions of variant to traits E and Y, β is the causal effect of E to trait Y, U represents confounding factors, and ε1 and ε2 are error terms, respectively. We use Gi and to separately represent two kinds of genetic IVs, that the contribution of Gi (i = 1,...,n1) to the outcome Y is mediated through the exposure E, and has a pleiotropic effect for the exposure and the outcome in independent paths (γ1i ≠ 0 and γ2i ≠ 0). In literature Gi and are often referred to as a vertical and a horizontal pleiotropy (Fig. 2). In the case of horizontal pleiotropy, there is a direct path from its genetic variant G to the outcome Y besides through the exposure E, which causes a biased causal effect estimate in an MR analysis.
Figure 2. A causal path diagram for multiple instrumental variables.
Let and be the estimated effect sizes of variants Gi and on the exposure E from the GWAS, respectively. Correspondingly, let and be the estimated effect sizes of variants Gi and on the outcome Y, respectively. From the Eqs. (1) and (2), we observe that for Gi, and for the variant .
In general, we can estimate the causal effect and its variance from the exposure to the outcome without differentiating Gi and by the following equation [18]
| (3) |
It is apparent that the causal estimate is unbiased based on Gi but biased based on . The bias for is induced by the nonzero effect γ2i, or horizontal pleiotropy. Because is inversely proportional to , smaller effect size of IV leads to a larger bias of the causal effect estimation. The MR analysis based on a single IV has a poor statistical power and potentially a large bias because
| (4) |
This is referred to as an inverse-variance weighted (IVW) estimate, which is calculated by assuming all the genetic variants are in linkage equilibrium. The IVW estimate can be calculated from summary statistics, an advantage over TSLS that requires individual-level data.
which comes from the average effect of the pleiotropic variants. If the pleiotropic effects happen to cancel out, the bias term tends to zero. When the pleiotropic effect γ2i on the outcome is independent of the effect γ1i on the exposure, or cov (γ2i,γ1i) = 0, the bias can approximate to 0. This condition is referred to as the InSIDE (Instrument Strength Independent of Direct Effect) assumption and can be viewed as a weaker version of the exclusion restriction assumption [22]. Asymptotically, the InSIDE assumption will guarantee the bias tends to zero. In reality, the InSIDE assumption can be difficult to satisfy. To solve this problem, Bowden et al. [22] adopted the idea of Egger’s test, which assesses small study bias in meta-analysis in epidemiology studies, into the MR analysis. They named this method as MR-Egger regression [23].
Without specifically distinguishing Gi and , we consider a linear regression of the coefficients on coefficients for a set of IVs,
| (5) |
where and represent the regression coefficients of the ith IV for the outcome and the exposure, respectively. The error term εi follows a normal distribution: . When regression model (5) constrains the intercept β0 = 0, the regression coefficient estimate of β corresponds to the IVW estimate. Alternatively, when regression model (5) includes the intercept term β0, the regression coefficient estimate of β corresponds to the MR-Egger estimate. Testing β0 = 0 also assesses the presence of pleiotropic variants. The itself is interpreted of the small variance explained by the IV. Therefore, using multiple IVs has been popular. Multiple IVs may partially mitigate the bias because it is possible to cancel the bias effect due to the nonzero effects of γ2i [8]. For multiple genetic variants, the causal effect of the exposure on the outcome is estimated by a weighted meta-analysis approach [19–21] given by as an estimate of the average pleiotropic effects [22]. When the InSIDE assumption is satisfied with balanced pleiotropy, referring to γ2i in Eq. (2) taking positive and negative values randomly, both IVW and MR-Egger approaches have unbiased causal estimate. In the presence of directional pleiotropy, the MR-Egger estimate is still consistent as long as the InSIDE assumption is satisfied, but IVW is not [22]. However, the MR-Egger estimator has a larger standard error than the IVW estimator. Intuitively, this is not surprising because an additional parameter is required for MR-egger, and the pleiotropic IVs will increase the uncertainty in the regression analysis. When sample sizes of GWAS are relatively small, MR-Egger estimator could be even worse [24].
Weighted median and mode-based estimate
An extension of the IVW is the weighted median estimator [25], which is less biased than IVW but more powerful than MR-Egger. The weighted median estimator takes the media of the of individual genetic variants in Eq. (3), either using equal weights or the inverse of the variance of the ratio estimates by . The weighted median estimator has a consistent causal estimator when less than 50% instrumental variables are invalid. Because the weighted median estimate is calculated by a single IV (median) when the number of IVs is odd and an average of two IVs when the number of IVs is even, more than 50% valid IVs is a necessary condition. Unlike IVW, the weighted median estimator is robust to outliers which depart from the true causal line.
Another extension is the weighted mode-based estimate (MBE) [26]. Let be the causal effect estimate in Eq. (3). The standard weights for the MBE is:
A simple MBE has . We define the normal kernel density function as
where b is the smoothing bandwidth parameter. The MBE causal effect estimate is . The magnitude of parameter b reflects a bias-variance trade-off. A larger b leads to a higher precision but also a larger bias. The bandwidth parameter is chosen according to the modified Silverman’s bandwidth rule [27,28]. The MBE relies on the assumption named the zero modal pleiotropy assumption (ZEMPA), that is, across all IVs, the most frequent value of γ2i in Eq. (2) is 0. The MBE is less biased and has lower type I error than the above mentioned methods under the null. The MBE is also less powerful in detecting causal effect than the IVW and weighted median methods, but it is more powerful than MR-Egger regression.
MR-Robust and MR-Lasso
In ordinary linear regression, one outlier can have a large impact to the regression coefficient estimate. Robust regression methods [29] have been recently applied to perform MR analysis. The current MR-Robust regression estimate by Rees et al. [30] is based on the MM-estimation approach by Koller and Stahel [31], which keeps asymptotic efficiency of the M-estimator and provides robustness against outliers. Lasso regression has been widely applied in high dimensional data by shrinking regression coefficients toward zero through a penalty term [32]. Recently, Lasso regression has been applied to MR analysis when individual level data is available [33,34]. Rees et al. [30] extended the Lasso regression to summary level data by modeling the pleiotropic effects γ2i in Eq. (2). MR-Lasso considers minimizing the following objective function by including a separate intercept coefficient for each genetic variant in the MR-Egger regression but with a Lasso-penalty term:
| (6) |
If β0i shrinks to 0 in Eq. (6), the genetic variant is considered as a valid IV. These genetic variants with a zero β0i are carried forward to perform the IVW analysis to estimate the causal effect β. The MR-Lasso shares some similarity with MR-PRESSO [18], which we will introduce later. When number of invalid IVs increases, both MR-Robust and MR-Lasso have inflated false positive rate and increased the bias of a causal effect estimate.
Mixture model MRmix
With substantial differences, Qi and Chatterjee developed a parametric mixture model (MRmix) by assuming bivariate effect-size distribution of the IVs across pairs of traits [35]. MRmix is an estimating equation approach that requires the residuals, , to follow a normal mixture model. The normal mixture model seems plausible when the genetic instruments include mediation variants, horizontal pleiotropic variants, as well as the genetic variants contributing to reverse causality. In order to achieve an unbiased causal estimate, MRmix requires the ZEMPA assumption, which is also required by MBE approach. When the sample size is large, MRmix usually shows a better trade-off between bias and variance than the approaches mentioned before, even more than when 50% IVs are invalid [35]. Similar to MR-Egger, MRmix did not performed well when the number of IVs is small.
MR-PRESSO
The MR-Lasso searches for potential outliers that may present pleiotropic effects [30]. Similarly, MR-PRESSO first identifies horizontal pleiotropic variants and then performs IVW to estimate the causal effect by removing the pleiotropic variants [18]. MR-PRESSO comprises of three steps: 1) testing whether horizontal pleiotropic variants are present through a global test; 2) performing an outlier test to detect pleiotropic variants; 3) comparing the causal estimates before and after removal of pleiotropic variants through a distortion test. The global test is based on a leave-one out approach which consists of 4 steps: 1) for each variant i, IVW regression is performed to obtain the causal effect after excluding the variant i; 2) the residual square is calculated by) and the global observed RSS is calculated by ; 3) The expected distribution of RSS under null hypothesis (no pleiotropic variants) is simulated by randomly drawing from a Gaussian distribution and from a Gaussian distribution , respectively. The expected RSS is ; 4) An empirical p-value is computed by taking the proportion of expected RSS greater than the observed RSS among K simulations in the third step. For the variant i, the p-value of the outlier test is calculated by taking the proportion of the expected RSSexp(i) greater than the observed RSSobs(i) among K simulations. Since MR-PRESSO estimates the causal effect after removing potential pleiotropic variants, it is less biased than IVW. However, MR-PRESSO is also biased when the InSIDE assumption fails. In addition, MR-PRESSO is computationally intensive because a large number of simulations are necessary, especially when the number of IVs is increasing.
Iterative Mendelian randomization and pleiotropy (IMRP)
It is easy to observe that for the mediation variant Gi, and for the pleiotropic variant . Note that the effect size of a pleiotropic variant to the outcome has an additional term γ2i besides the effect through the exposure. If we know the true β, we can test mediation against horizontal pleiotropy by testing the null hypothesis Γ = βγ using a test statistic
where and are the estimated effect sizes of a variant on E and Y, respectively. The statistic Tpleio asymptotically follows a standard normal distribution N(0,1) when mediation is true. Under the alternative hypothesis that a variant has a pleiotropic effect, TPleio departs from the mean of 0. The problem of this test is that the causal effect β is unknown. However, this problem can be solved through an iterative approach by combining the pleiotropy test and the MR analysis, which is named as iterative Mendelian randomization and pleiotropy (IMRP) [24]:
Performing MR-Egger analysis using all IVs to obtain the initial causal estimate of β, named as .
For all the IVs, performing pleiotropy test Tpleio by substituting β with at the kth iteration to determine which variant gi has a horizontal pleiotropic effect at a predefined significance level α.
Performing IVW analysis to obtain after removing the variants found to be significant in pleiotropy test at step 2;
Repeating the above steps 2 and 3 until there is no change in detected pleiotropic variants.
At step 2 above, the pleiotropy test statistic for a genetic variant is modified as
where the denominator variance is approximated by
where ρ is the correlation coefficient of the exposure E and the outcome Y, which can be estimated using GWAS summary statistics [15,36] or LD score regression [37]. Similarly to MR-PRESSO, the global test: , which approximately follows a chisquare distribution with n degrees of freedom for n independent IVs, is used to test for the presence of horizontal pleiotropic variants. A genetic variant with a Tpleio test p-value less than 0.05/n, is considered as a variant having a horizontal pleiotropic effect. Thus IMRP can perform both MR analysis and test horizontal pleiotropy simultaneously.
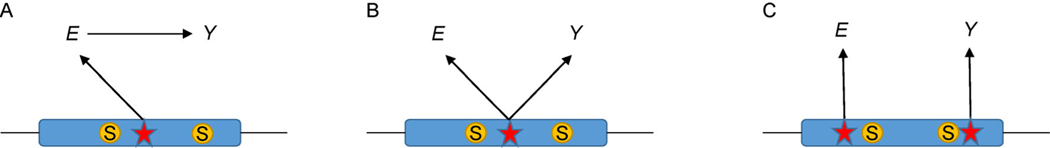
MR analysis often assumes that the IVs are in linkage equilibrium. At a single locus, multiple variants may contribute to exposure and the outcome in different ways, as illustrated in Fig. 3. Let and vectors representing the estimated effect sizes of outcome and exposure from GWAS of M variants, respectively. Let and . The test statistics Tpleio can be extended to M variants by , where is an M × M variance–covariance matrix [24]. If working on the standardized E, Y, and genotype values, Spleio can be simplified to
where R is the linkage disequilibrium (LD) matrix among the M variants. Under the null hypothesis of mediation, Spleio follows a chi-square distribution with M degrees of freedom.
Figure 3. The relationships of genetic variants, exposure and outcome.
(A) Mediation: the causal variant lies on the causal path to Y. (B) Horizontal pleiotropy: the causal variant affects both E and Y. (C) Colocalization: two different causal variants at one locus affect E and Y. The red star represents the causal variant and S represents genetic markers.
A significant difference between IMRP and MR-PRESSO is that IMRP removes horizontal pleiotropic variants step by step and re-estimates the causal effect accordingly, while MR-PRESSO removes horizontal pleiotropic variants in one step. Since the causal effect estimate is sensitive to outliers, an iterative method is less biased and can advantageously detect pleiotropic variants [24]. Because usually less than 10 iterations are sufficient, iMRP is more computationally efficient than other methods, such as MR-PRESSO, MRmix, MBE, MR-Lasso or MR-Robust. iMRP shares the same computational efficiency as IVW but is almost unbiased even if half of IVs are invalid [24]. iMRP can be easily extended to include multiple variants on a locus, whether they are in LD or not, thereby testing for pleiotropy and colocalization.
APPLICATIONS
In this review, it is not our intention to list all methods and software for performing MR analysis. Rather, we introduced the basic concept and strategy for performing MR analysis. In practice, inferring causality is extremely challenging because of many unmeasured confounders, reverse causality, and weak instrument bias. Burgess et al. recently published guidelines for performing MR in practice [38]. Multiple analytical steps have been suggested and these steps are categorized into: motivation and scope, data sources, choice of genetic variants, variant harmonization, primary analysis, supplementary and sensitivity analyses, data presentation, and interpretation [38]. These guidelines are useful for correctly inferring causality and unbiasedly estimating the causal effect of an exposure to an outcome, given more and more practices of MR analyses have been performed in this post genome wide association era. For example, it is clearly different to infer potential causality versus to estimate the causal effect, with the former focusing on hypothesis test and the latter focusing on the size of the causal effect. In the original MR analysis by Katan [9], testing for association of a genetic variant and an outcome will be sufficient to infer the causality, as long as the genetic variant is a valid IV. Such analysis does not require genotype data for the exposure, but statistical power for testing the association between the variant and the outcome is critical. In comparison, the intervention effect of an exposure to an outcome can be estimated from MR analysis, which is usually achieved by a randomized clinical trial. In this case, it will be important to know how well the genetic variant can proxy the true causal variant, whether the genetic variant has heterogeneous effects on exposure to outcome, for example due to gene-gene interactions, and whether the associations with exposure and outcome are obtained from the same population [38,39]. Mendelian randomization can be performed in either one sample with individual level data for both exposure and outcome or two samples with summary statistics for the exposure and the outcome in separate datasets. Although one sample with individual level data allows for flexible modeling and the exposure and outcome to be measured from the same individuals, the sample size is often small, leading to low statistical power. In comparison, two samples with summary statistics can often reach large sample sizes, but its dangers include potential different populations for the exposure and outcome, and different demographic information, which will make interpretation difficulty and invalidate causal inferences [40,41]. In GWAS meta-analysis, allele flips can cause loss of statistical power when meta-analyzing multiple datasets [42,43]. The same problem can occur in a two sample MR analysis when variant harmonization is a necessary step. In general, multiple analysis methods should be performed because these methods are valid under different assumptions, which have been reviewed before. As suggested by the guidelines [38], both IVW based methods and robust methods should be applied with additional sensitivity analysis. The iMRP method [24] can assess colocalization, which will help to identify more plausible genetic IVs for MR analysis. In addition, the interpretation of findings from MR analysis should always be cautious because of multiple untestable assumptions, as is illuminated in the guidelines [38,44].
Perhaps, which genetic variants should be included in a MR analysis might be the most important decision [38]. In MR analysis, only mediation genetic variants can satisfy the three IV conditions as valid instrumental variables [45]. In fact, there is a debate about whether MR can reliably identify causality between two traits given the widespread of pleiotropy or colocalization [46,47]. The variants with pleiotropic effects violate the IV2 condition for valid IVs. Detecting horizontal pleiotropy is challenging, as demonstrated that the recent HOPS approach has an inflated type I error [48], which may reduce the power of MR analysis compared with IMRP and MR-PRESSO.
AN MR ANALYSIS OF THE CAUSALITY OF HIGH-DENSITY LIPOPROTEIN CHOLESTEROL (HDL-C) ON CORONARY ARTERY DISEASE (CAD)
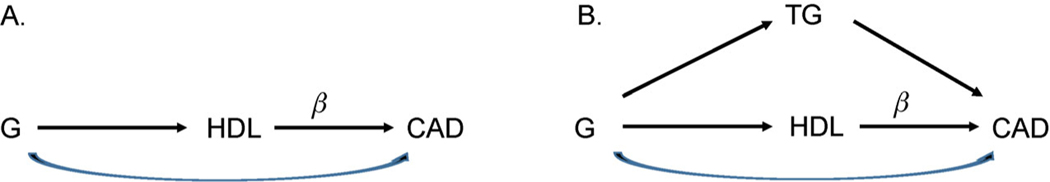
Many large-scale population studies have reported an inverse relationship between HDL-C and CAD [49,50]. A well cited MR analysis using 15 genetic variants as IVs suggested that there is no causal relationship between HDL-C and myocardial infarction [51], which was consistent with the evidence from a clinical trial study [52]. However, the MR study using more genetic variants by Holmes et al. [53] suggested uncertainty in a causal role for HDL-C on CVD risk. We downloaded the GWAS summary statistics of Global Lipids Genetics Consortium (cholesterol traits) (http://csg.sph.umich.edu/abecasis/public/lipids2013/) and CARDIoGRAMplusC4D Consortium (coronary artery disease) (http://www.cardiogramplusc4d.org/data-downloads/). We obtained 143 genome wide significant independent variants (p-value < 5 × 10−8) associated with HDL-C after pruning (r2 < 0.1) using the software Plink [54] on a 500 kb window size. We performed the MR analysis by assuming the path diagram in Fig. 4A, representing no confounders. Table 2 presents the causal estimates and p-values for the different MR methods, including IVW, MR-Egger, median, MBE, MRmix, MR_PRESSO and IMRP. In general, IVW, simple median, and MR_PRESSO had the largest protected causal effect estimates followed by IMRP and weighted median. MRmix and MBE had the smallest protected causal estimates. Interestingly, MR-Egger had a positive causal estimate, but it was not significant. We also observed that MR-Egger and MRmix had the largest standard errors, which suggested these approaches may lose statistical power. IVW, simple median, MR_PRESSO and IMRP also suggested significant causal effect of HDL-C on CAD, but the rest of the methods did not. The IMRP analysis also identified 10 pleiotropic variants for HDL-C and CAD.
Figure 4. The two path diagrams for HDL-C and CAD.
(A) There is no separate path through TG. (B) There is a separate path through TG.
Table 2.
Causal effect estimates of HDL on CAD using 143 IVs by different MR methods
| Method | Casual estimate | Standard error | 95% CI | p-value |
|---|---|---|---|---|
| IVW | −0.163 | 0.046 | (−0.252, −0.073) | 3.95 × 10−4 |
| MR-Egger | 0.06 | 0.084 | (−0.103, 0.224) | 0.47 |
| Simple median | −0.241 | 0.054 | (−0.348, −0.135) | 8.08 × 10−4 |
| Weighted median | −0.062 | 0.046 | (−0.152, 0.028) | 0.174 |
| MBE | −0.023 | 0.055 | (−0.130, 0.084) | 0.668 |
| MRmix | −0.04 | 0.086 | (−0.209, 0.129) | 0.642 |
| MR_PRESSO | −0.187 | 0.037 | (−0.260, −0.115) | 1.4 × 10−6 |
| IMRP | −0.075 | 0.027 | (−0.128, −0.022) | 5.19 × 10−3 |
| IMRP* | −0.062 | 0.031 | (−0.123, −0.001) | 0.043 |
IMRP analysis after dropping 60 pleiotropy variants.
It is known that HDL-C and triglycerides (TG) are correlated. The correlation between the summary statistics of HDL-C and TC was −0.235. It is then possible that a genetic instrumental variable (G) has a pleiotropic effect on HDL-C and TG, therefore, conditional on HDL-C, G can still affect CAD by the mediation of TG (Fig. 4B). The mediation of TG also led to a loss of statistical power to detect pleiotropic variants for HDL-C and CAD. Thus, we performed IMRP for HDL-C and TG to identify pleiotropic variants for these two correlated traits. We were able to identify 60 pleiotropic variants among the 143 IVs. After dropping these 60 variants, the OR of HDL-C on CAD estimated by IMRP was reduced to 0.94 (p = 0.043), suggesting that the initial significant causal effect of HDL-C on CAD was biased because of the correlated trait TG. This example also suggests the importance to examine the traits correlated with both the exposure and outcome.
FUTURE RESEARCH
We reviewed statistical approaches for performing a Mendelian Randomization analysis using summary statistics from genome wide associations. The application of HDL-C on CAD clearly suggested that there is no uniformly best approach. There is always a trade-off between bias and efficiency. Among the methods, MR-PRESSO and IMRP are able to both perform the MR analysis and detect pleiotropic variants, and IMRP is computationally 3 order faster than MR-PRESSO and is less biased [24]. MBE and MRmix are less biased, but they also lose statistical power. Although MR is easy to conduct due to increased availability of genome data, the challenges remain. First, one challenge is the identification of valid IVs, i.e., the genetic variants have no horizontal pleiotropic between exposure and outcome. Because of the modest contribution of a genetic variant to a trait, statistical power in detecting pleiotropy can be low and requires a large sample size. New statistical approaches will be welcomed for detecting horizontal pleiotropy. Second, most of the current MR approaches assess the causal effect of one exposure on one outcome. Traits are often correlated with shared genetic contributions [55]. The correlated traits can easily lead to confounding in MR analysis, which can create multiple independent paths from a genetic IV to an outcome without pathing through the exposure, as we observed in the MR analysis of HDL-C and CAD. Multivariate MR with multiple exposures and one outcome was less developed and the current multivariate MR is unable to deal with unknown pleiotropy [56], but has advantages to solve the problems of multiple correlated exposures. Third, a single genetic variant has little prediction power. To improve the power, polygenic risk score (PRS) has been used for predicting disease and inferring putative causal relationships among traits [57]. However its false positive rate is also inflated when inferring causality [58]. A PRS has less concern caused by weak instruments. Further methodological development is necessary to unbiasedly estimate and correctly interpret the causal effect estimation through a PRS. Fourth, large GWAS also demonstrated gene-environment/life style interactions contributing to phenotype variation, and these summary statistics are available [59,60]. It will be beneficial to develop MR approaches that can be applied to summary statistics from GWAS of gene-environment interaction studies, which can potentially provide better causal effect estimations. Lastly, current large GWASs have mainly been conducted in European ancestry populations. The sample sizes in other ancestry populations are much less, resulting less identifications of genetic variants. Utilizing the GWAS information from European ancestry populations to perform MR analysis in other ancestry populations needs innovative statistical approaches. In conclusion, current genomic advances provide an unprecedented opportunity to study casual relationships among risk factors and diseases via MR analysis, and the results will lay a foundation for future, well designed randomized control trials.
ACKNOWLEDGEMENTS
This work was supported by grants HG003054 and HG011052 (to X.Z.) from the National Human Genome Research Institute (NHGRI), USA.
Footnotes
COMPLIANCE WITH ETHICS GUIDELINES
The author Xiaofeng Zhu declare that he has no conflict of interests.
This article is a review article and does not contain any studies with human or animal subjects performed by the author.
REFERENCES
- 1.Schatzkin A, Lanza E, Corle D, Lance P, Iber F, Caan B,Shike M, Weissfeld J, Burt R, Cooper MR, et al. (2000) Lack of effect of a low-fat, high-fiber diet on the recurrence of colorectal adenomas. N. Engl. J. Med, 342, 1149–1155 [DOI] [PubMed] [Google Scholar]
- 2.The Heart Outcomes Prevention Evaluation Study Investigators.(2000) Vitamin E supplementation and cardiovascular events in high-risk patients. N. Engl. J. Med, 342, 154–160 [DOI] [PubMed] [Google Scholar]
- 3.Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group.(1994) The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N. Engl. J. Med, 330, 1029–1035 [DOI] [PubMed] [Google Scholar]
- 4.Sesso HD, Buring JE, Christen WG, Kurth T, Belanger C, MacFadyen J, Bubes V, Manson JE, Glynn RJ and Gaziano JM (2008) Vitamins E and C in the prevention of cardiovascular disease in men: the Physicians’ Health Study II randomized controlled trial. JAMA, 300, 2123–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey Smith G. and Ebrahim S. (2001) Epidemiology–is it time to call it a day? Int. J. Epidemiol, 30, 1–11 [DOI] [PubMed] [Google Scholar]
- 6.Fewell Z, Davey Smith G. and Sterne JA (2007) The impact of residual and unmeasured confounding in epidemiologic studies: a simulation study. Am. J. Epidemiol, 166, 646–655 [DOI] [PubMed] [Google Scholar]
- 7.Evans DM and Davey Smith G. (2015) Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu. Rev. Genomics Hum. Genet, 16, 327–350 [DOI] [PubMed] [Google Scholar]
- 8.Davey Smith G. and Hemani G. (2014) Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet, 23, R89–R98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katan MB (1986) Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet, 327, 507–508 [DOI] [PubMed] [Google Scholar]
- 10.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA and Yang J. (2017) 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet, 101, 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey Smith G. and Ebrahim S. (2003) ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol, 32, 1–22 [DOI] [PubMed] [Google Scholar]
- 12.VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M. andKraft P. (2014) Methodological challenges in Mendelian randomization. Epidemiology, 25, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe K, Stringer S, Frei O, Umićević Mirkov M, Polderman TJC, van der Sluis S, Andreassen OA, Neale BM and Posthuma D. (2018) A global view of pleiotropy and genetic architecture in complex traits. Nat. Genet, 51, 1339–1134 [DOI] [PubMed] [Google Scholar]
- 14.Solovieff N, Cotsapas C, Lee PH, Purcell SM and Smoller JW (2013) Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet, 14, 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Feng T, Tayo BO, Liang J, Young JH, Franceschini N, Smith JA, Yanek LR, Sun YV, Edwards TL, et al. (2015) Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am. J. Hum. Genet, 96, 21–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sleiman PM and Grant SF (2010) Mendelian randomization in the era of genomewide association studies. Clin. Chem, 56, 723–728 [DOI] [PubMed] [Google Scholar]
- 17.Angrist JD, Imbens GW and Rubin DB (1996) Identification of causal effects using instrumental variables. J. Am. Stat. Assoc, 91, 444–455 [Google Scholar]
- 18.Verbanck M, Chen CY, Neale B. and Do R. (2018) Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet, 50, 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S, Butterworth A. and Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol, 37, 658–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan N. and Thompson J. (2017) A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med, 36, 1783–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borenstein M, Hedges L, Higgins J. and Rothstein H. (2009) Generality of the basic inverse-variance method. In: Introduction to Meta-analysis. Wiley [Google Scholar]
- 22.Bowden J, Davey Smith G. and Burgess S. (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol, 44, 512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egger M, Davey Smith G, Schneider M. and Minder C. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ, 315, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Li X, Xu R, Wang T. (2020) An iterative approach to detect pleiotropy and perform Mendelian randomization analysis using GWAS summary statistics. Under review [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden J, Davey Smith G, Haycock PC and Burgess S(2016) Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol, 40, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartwig FP, Davey Smith G. and Bowden J. (2017) Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol, 46, 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bickel DR (2003) Robust and efficient estimation of the mode of continuous data: the mode as a viable measure of central tendency. J. Stat. Comput. Simul, 73, 899–912 [Google Scholar]
- 28.Bickel DR and Fruhwirth R. (2006) On a fast, robust estimator of the mode: Comparisons to other robust estimators with applications. Comput. Stat. Data Anal, 50, 3500–3530 [Google Scholar]
- 29.Huber PJ and Ronchetti E. (2009) Robust Statistics, 2nd edition, Hoboken, N.J. (ed.).Wiley [Google Scholar]
- 30.Rees JMB, Wood AM, Dudbridge F. and Burgess S. (2019) Robust methods in Mendelian randomization via penalization of heterogeneous causal estimates. PLoS One, 14, e0222362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koller M. and Stahel WA (2011) Sharpening Wald-type inference in robust regression for small samples. Comput. Stat. Data Anal, 55, 2504–2515 [Google Scholar]
- 32.Tibshirani R. (1996) Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B, 58, 267–288 [Google Scholar]
- 33.Kang H, Zhang AR, Cai TT and Small DS (2016) Instrumental variables estimation with some invalid instruments and its application to Mendelian randomization. J. Am. Stat. Assoc, 111, 132–144 [Google Scholar]
- 34.Windmeijer F, Farbmacher H, Davies N. and Davey Smith G(2019) On the use of the Lasso for instrumental variables estimation with some invalid instruments. J. Am. Stat. Assoc, 114, 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi G. and Chatterjee N. (2019) Mendelian randomization analysis using mixture models for robust and efficient estimation of causal effects. Nat. Commun, 10, 1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park H, Li X, Song YE, He KY and Zhu X. (2016) Multivariate analysis of anthropometric traits using summary statistics of genome-wide association studies from GIANT consortium. PLoS One, 11, e0163912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, the Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL and Neale BM (2015) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet, 47, 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, Hartwig FP, Holmes MV, Minelli C, Relton CL, et al. (2019) Guidelines for performing Mendelian randomization investigations. Wellcome Open Res, 4, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernán MA and Robins JM (2006) Instruments for causal inference: an epidemiologist’s dream? Epidemiology, 17, 360–372 [DOI] [PubMed] [Google Scholar]
- 40.Labrecque JA and Swanson SA (2019) Interpretation and potential biases of Mendelian randomization estimates with time-varying exposures. Am. J. Epidemiol, 188, 231–238 [DOI] [PubMed] [Google Scholar]
- 41.Swanson SA, Labrecque J. and Hernán MA (2018) Causal null hypotheses of sustained treatment strategies: What can be tested with an instrumental variable? Eur. J. Epidemiol, 33, 723–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, Tayo BO, Sun YV, Gottesman O, Adeyemo A, et al. (2013) Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am. J. Hum. Genet, 93, 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang J, Le TH, Edwards DRV, Tayo BO, Gaulton KJ, Smith JA, Lu Y, Jensen RA, Chen G, Yanek LR, et al. (2017) Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations. PLoS Genet, 13, e1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith GD, Davies NM, Dimou N, Egger M, Gallo V,Golub R, Higgins JPT, Langenberg C, Loder EW, Richards JB, et al. (2020) STROBE-MR: Guidelines for strengthening the reporting of Mendelian randomization studies. PeerJ Preprints 7, e27857v1 [Google Scholar]
- 45.Burgess S, Bowden J, Fall T, Ingelsson E. and Thompson SG(2017) Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology, 28, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickrell J. (2015) Fulfilling the promise of Mendelian randomization. bioRxiv, 018150 [Google Scholar]
- 47.Davey Smith G. (2015) Mendelian randomization: a premature burial? bioRxiv, 021386 [Google Scholar]
- 48.Jordan DM, Verbanck M. and Do R. (2019) HOPS: a quantitative score reveals pervasive horizontal pleiotropy in human genetic variation is driven by extreme polygenicity of human traits and diseases. Genome Biol, 20, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The Emerging Risk Factors Collaboration (2009) Major lipids, apolipoproteins, and risk of vascular disease. JAMA, 302, 1993–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewington S, Whitlock G, Clarke R, Sherliker P, Emberson J, Halsey J, Qizilbash N, Peto R, Collins R, Collins R, et al. (2007) Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet, 370, 1829–1839 [DOI] [PubMed] [Google Scholar]
- 51.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, et al. (2012) Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet, 380, 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geller AS, Polisecki EY, Diffenderfer MR, Asztalos BF, Karathanasis SK, Hegele RA and Schaefer EJ (2018) Genetic and secondary causes of severe HDL deficiency and cardiovascular disease. J. Lipid Res, 59, 2421–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmes MV, Asselbergs FW, Palmer TM, Drenos F,Lanktree MB, Nelson CP, Dale CE, Padmanabhan S, Finan C, Swerdlow DI, et al. (2015) Mendelian randomization of blood lipids for coronary heart disease. Eur. Heart J, 36, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet, 81, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortes A, Albers PK, Dendrou CA, Fugger L. and McVean G. (2020) Identifying cross-disease components of genetic risk across hospital data in the UK Biobank. Nat. Genet, 52, 126–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgess S. and Thompson SG (2015) Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol, 181, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies RB (1980) The distribution of a linear combination of χ2 random variables. J. R. Stat. Soc. C Appl. Stat, 29, 323–333 [Google Scholar]
- 58.Richardson TG, Harrison S, Hemani G. and Davey Smith G(2019) An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. eLife, 8, e43657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bentley AR, Sung YJ, Brown MR, Winkler TW, Kraja AT, Ntalla I, Schwander K, Chasman DI, Lim E, Deng X, et al. (2019) Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat. Genet, 51, 636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sung YJ, Winkler TW, de Las Fuentes L, Bentley AR,Brown MR, Kraja AT, Schwander K, Ntalla I, Guo X, Franceschini N, et al. (2018) A Large-scale multi-ancestry genome-wide study accounting for smoking behavior identifies multiple significant loci for blood pressure. Am. J. Hum. Genet, 102, 375–400 [DOI] [PMC free article] [PubMed] [Google Scholar]