ABSTRACT
Polybrominated diphenyl ethers (PBDEs) are persistent, highly toxic, and widely distributed environmental pollutants. The microbial populations and functional reductive dehalogenases (RDases) responsible for PBDE debromination in anoxic systems remain poorly understood, which confounds bioremediation of PBDE-contaminated sites. Here, we report a PBDE-debrominating enrichment culture dominated by a previously undescribed Dehalococcoides mccartyi population. A D. mccartyi strain, designated TZ50, whose genome contains 25 putative RDase-encoding genes, was isolated from the debrominating enrichment culture. Strain TZ50 dehalogenated a mixture of pentabrominated diphenyl ether (penta-BDE) and tetra-BDE congeners (total BDEs, 1.48 μM) to diphenyl ether within 2 weeks (0.58 μM Br−/day) via ortho- and meta-bromine elimination; strain TZ50 also dechlorinated tetrachloroethene (PCE) to vinyl chloride and ethene (260.2 μM Cl−/day). Results of native PAGE, proteomic profiling, and in vitro enzymatic activity assays implicated the involvement of three RDases in PBDE and PCE dehalogenation. TZ50_0172 (PteATZ50) and TZ50_1083 (TceATZ50) were responsible for the debromination of penta- and tetra-BDEs to di-BDE. TZ50_0172 and TZ50_1083 were also implicated in the dechlorination of PCE to trichloroethene (TCE) and of TCE to vinyl chloride/ethene, respectively. The other expressed RDase, TZ50_0090 (designated BdeA), was associated with the debromination of di-BDE to diphenyl ether, but its role in PCE dechlorination was unclear. Comparatively few RDases are known to be involved in PBDE debromination, and the identification of PteATZ50, TceATZ50, and BdeA provides additional information for evaluating debromination potential at contaminated sites. Moreover, the ability of PteATZ50 and TceATZ50 to dehalogenate both PBDEs and PCE makes strain TZ50 a suitable candidate for the remediation of cocontaminated sites.
IMPORTANCE The ubiquity, toxicity, and persistence of polybrominated diphenyl ethers (PBDEs) in the environment have drawn significant public and scientific interest to the need for the remediation of PBDE-contaminated ecosystems. However, the low bioavailability of PBDEs in environmental compartments typically limits bioremediation of PBDEs and has long impeded the study of anaerobic microbial PBDE removal. In the current study, a novel Dehalococcoides mccartyi strain, dubbed strain TZ50, that expresses RDases that mediate organohalide respiration of both PBDEs and chloroethenes was isolated and characterized. Strain TZ50 could potentially be used to remediate multiple cooccurring organohalides in contaminated systems.
KEYWORDS: brominated flame retardants, polybrominated diphenyl ethers, reductive debromination, Dehalococcoides, reductive dehalogenase (RDase)
INTRODUCTION
Pentabrominated diphenyl ethers (penta-BDEs) and tetra-BDEs are among the most widely distributed polybrominated diphenyl ether (PBDE) congeners in the environment. They have been used as flame retardants in a variety of manufactured products since the 1960s and can be produced as intermediates of physical and biological attenuation of octa- and deca-BDE mixtures (1). Penta- and tetra-BDEs are persistent organic pollutants with low solubility and bioavailability, high hydrophobicity and bioaccumulative potential, and high toxicity. The risks that penta- and tetra-BDEs pose to human populations and ecosystems have raised considerable public attention, and their use was banned by the United Nations Stockholm Convention in 2009 (2).
In situ bioremediation via reductive dehalogenation is an efficient and cost-effective approach for the removal of halogenated organic pollutants under anoxic conditions (3, 4). However, bioremediation of halogenated aromatic pollutants such as PBDEs is still limited by a lack of functional microorganisms for bioaugmentation and limited knowledge of the mechanisms involved in the dehalogenation of PBDEs, polychlorinated biphenyls, brominated bisphenols, and similar aromatic organohalides. Nevertheless, reductive dehalogenation is considered a promising technology for the remediation of halogenated aromatic pollutants, which tend to partition to anoxic soils and sediments and are resistant to aerobic degradation. Members of several bacterial genera, including Dehalococcoides, Dehalobacter, Desulfitobacterium, Acetobacterium, and Dehalogenimonas, are known to anaerobically debrominate PBDEs (5–8). However, anaerobic biological PBDE debromination is most often partial, resulting in the accumulation of tetra-, tri-, and di-BDEs, and relies heavily on supplementation with other halogenated compounds to induce debromination (9). Moreover, the presence of cometabolic processes and the extremely low abundance of debrominating populations in complex microbial consortia make unraveling the mechanisms of PBDE debromination a challenge. The only microorganism that has been reported to metabolically debrominate penta- and tetra-BDE congeners to diphenyl ether under anoxic conditions is Dehalococcoides mccartyi strain GY50. The genome of strain GY50 harbors 26 reductive dehalogenase homologous (rdh) genes, 3 (pbrA1, -2, and -3) of which encode different reductive dehalogenases (RDases) involved in the stepwise debromination of BDEs with one to five bromine substituents (7). It remains unclear whether these three enzymes are representative of all penta- and tetra-BDE RDases, but given the structural and chemical diversity of PBDE congeners, it seems most likely that other RDases capable of penta- and tetra-PBDE dehalogenation exist.
In the current study, we sought to investigate the distribution and identity of PBDE-degrading microbial populations in pristine environments (e.g., lake sediments) and environments exposed to PBDE contamination (e.g., e-waste recycling and dump sites and wetlands treating landfill leachate). Screening of microcosms and enrichment cultures derived from these various environments for anaerobic biological debromination of penta- and tetra-BDEs could shed light on the evolution of microbial populations in response to the presence of different PBDEs. Furthermore, the identification and characterization of functional PBDE-debrominating bacteria could broaden our understanding of anaerobic debromination, providing valuable data to support the development of improved strategies to implement and monitor PBDE debromination in anoxic soils and sediments.
RESULTS
PBDE debromination activity in established microcosms.
The biological debromination of a defined BDE mixture comprising two penta-BDEs (BDE-100 and BDE-99) and one tetra-BDE (BDE-47) was investigated in microcosms established with soil samples collected from multiple locations representing different ecological systems. Among 24 microcosms amended with ∼1.5 μM the penta- and tetra-BDE mixture, 7 exhibited partial or complete debromination within 90 days of incubation (Table 1). Neither the loss of BDE-100, BDE-99, or BDE-47 nor the production of daughter compounds was observed in the other 17 microcosms or the autoclaved control. Among the seven debrominating microcosms, only microcosm TZ-7 completely debrominated BDE-100, BDE-99, and BDE-47 to diphenyl ether. Microcosm TZ-5 debrominated BDE-47 to diphenyl ether, while the five other active microcosms showed partial debromination of at least one of the three BDE congeners in the defined mixture.
TABLE 1.
Debromination products generated in dehalogenating microcosms amended with a penta- and tetra-BDE mixture after 90 days of incubation
| Microcosm | Source | Concn of parent compound or debromination product (μM)a |
Presence of: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Penta-BDE | Tetra-BDE | Tri-BDE | Di-BDE | Mono-BDE | Diphenyl ether | D. mccartyi | pbrA1 | pbrA2 | pbrA3 | ||
| TZ-5 | Soil from electronic waste sites | 0.87 | ND | ND | ND | ND | 0.45 | + | − | − | + |
| TZ-7 | Soil from electronic waste sites | ND | ND | ND | ND | ND | 1.48 | + | − | − | − |
| ZJ-2 | Sediment from industrial waste discharge estuarine | 0.43 | 0.84 | ND | ND | ND | ND | + | − | − | + |
| LH-2 | Sediment from a constructed wetland to treat landfill leachate | 0.85 | 0.13 | ND | ND | ND | 0.33 | − | − | − | − |
| LH-3 | Sediment from a constructed wetland to treat landfill leachate | 0.74 | 0.12 | ND | 0.26 | 0.08 | 0.11 | − | − | − | − |
| WWTP-W | Concentrated mixed liquor from anoxic zone in a domestic wastewater treatment plant | 0.45 | 0.35 | 0.38 | 0.12 | ND | ND | + | + | − | + |
| WWTP-E | Concentrated mixed liquor from anoxic zone in a domestic wastewater treatment plant | 0.21 | 0.34 | 0.65 | 0.02 | 0.01 | ND | + | − | + | + |
| Control | Autoclaved soil-containing inoculum | 0.86 | 0.45 | ND | ND | ND | ND | ||||
ND, not determined.
There was no apparent pattern to the extent or range of PBDE debromination and the presence or absence of previously identified PBDE rdh genes (i.e., pbrA1, pbrA2, and pbrA3). Endpoint PCR detected pbrA3 in four of seven PBDE-degrading microcosms and either pbrA1 or pbrA2 in two of those four (WWTP-W and WWTP-E) (Table 1). None of the microcosms contained all three pbrA genes, and none of the pbrA genes were detected in three of the active microcosms (TZ-7, LH-2, and LH-3). None of the pbrA genes were detected in any nondebrominating microcosms. At least one Dehalococcoides population was detected in each active microcosm, except for LH-2 and LH-3, both of which originated from a constructed wetland.
Complete penta- and tetra-BDE debromination by the TZ50 enrichment culture.
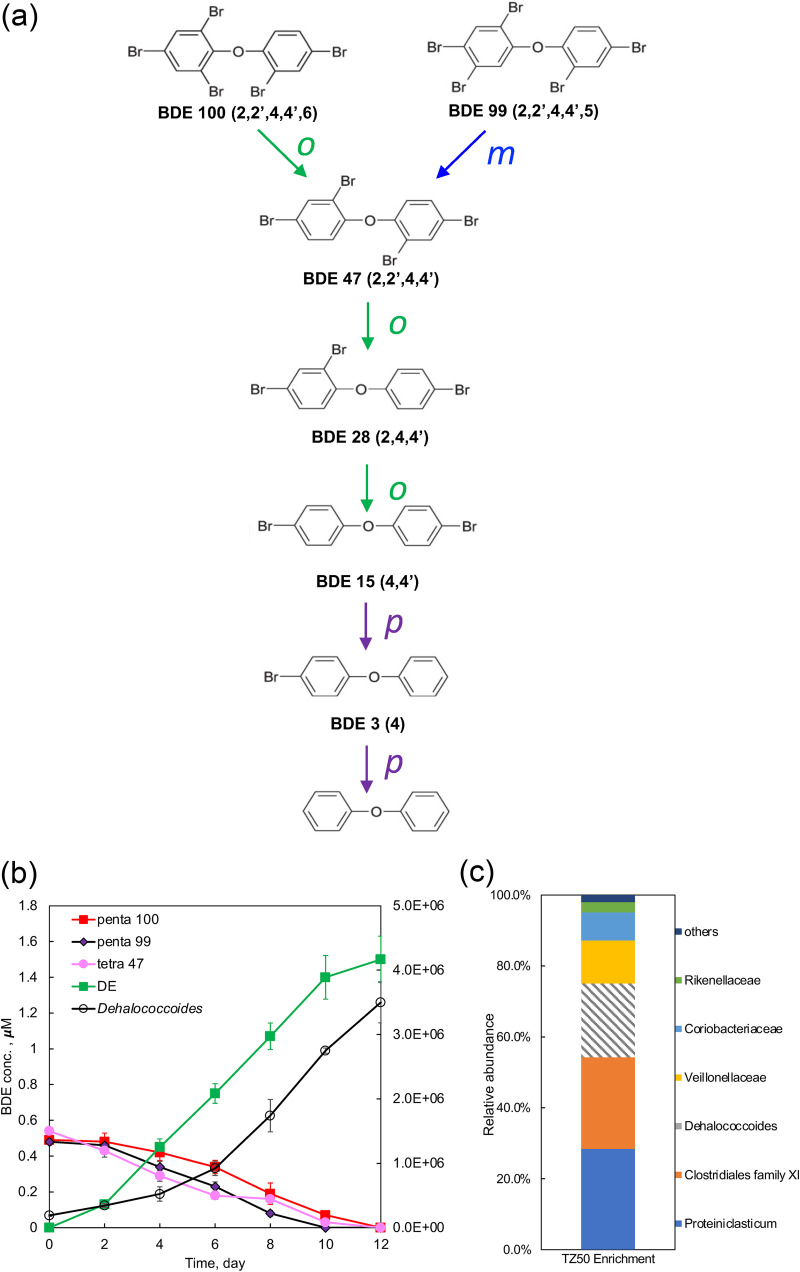
Among the debrominating microcosms, TZ-7 exhibited the most complete debromination of the penta- and tetra-BDE mixture and was selected for further study. A sediment-free culture generated by three consecutive transfers of microcosm TZ-7 debrominated the defined penta- and tetra-BDE mixture to diphenyl ether within 20 days, producing tri-BDE-28, di-BDE-15, and mono-BDE-3 as intermediates (see Fig. S1 in the supplemental material). This enrichment was designated TZ50 to indicate the debromination of penta-BDE to diphenyl ether (5 to 0 bromine moieties). Bromines in the ortho and meta positions were preferentially removed from penta- and tetra-BDEs, while the para substituent of BDE-15 and BDE-3 was attacked after all ortho- and meta-bromines had been removed (Fig. 1a). Further enrichment by continuous subculturing over 1 year led to a marked acceleration in the rate of debromination, which ultimately reached an average bromine removal rate of 0.58 μM Br−/day with no accumulation of intermediates (Fig. 1b).
FIG 1.
(a) Pathway analysis exhibits preferential removal of ortho- and meta-bromines from penta- and tetra-BDEs by the TZ50 enrichment culture. (b) Time course of penta- and tetra-BDE debromination by enrichment culture TZ50. DE, diphenyl ether. (c) Microbial community structure of the TZ50 enrichment culture as determined by 16S rRNA gene amplicon sequencing.
The microbial community of the TZ50 enrichment culture with marked acceleration rates and without accumulation of intermediates was elucidated via Illumina sequencing of the V4 region of the 16S rRNA gene. Six taxonomic groups, comprising two bacterial genera (Proteiniclasticum and Dehalococcoides) and four bacterial families (Clostridiales family IX, Veillonellaceae, Coriobacteriaceae, and Rikenellaceae), dominated the TZ50 enrichment culture, accounting for more than 98% of the entire community (Fig. 1c), of which Dehalococcoides (20.9% of the community) is the only taxonomic group known to dehalogenate organohalide compounds. The abundance of Dehalococcoides cells in the TZ50 enrichment culture increased 18.42-fold during debromination, from 1.90 × 105 ± 0.15 × 105 to 3.50 × 106 ± 0.32 × 106 16S rRNA gene copies/ml (Fig. 1a). Increases in Dehalococcoides abundance were positively correlated with bromine removal (R2 = 0.92), indicating the occurrence of Dehalococcoides-mediated organohalide respiration of the amended BDEs. The Dehalococcoides growth yield was 4.99 × 108 ± 0.26 × 108 cells/μmol Br− removed. There was no obvious change in the abundance of Dehalococcoides in the TZ50 enrichment culture in the absence of the penta- and tetra-BDE mixture after 90 days of incubation (data not shown).
Alternative electron acceptors accelerate enrichment of debrominating populations.
Chloroethenes, including tetrachloroethene (PCE), trichloroethene (TCE), and dichloroethene (DCE) isomers, were also dechlorinated by the TZ50 enrichment culture (Fig. 2; Fig. S2). Of these, PCE and TCE were rapidly dechlorinated to vinyl chloride (VC) at rates of 260.2 and 142.5 μM Cl−/day, respectively, with a gradual accumulation of ethene. The growth of Dehalococcoides in the TZ50 enrichment culture was coupled to chlorine removal from PCE and TCE to vinyl chloride, with cell yields of 2.25 × 108 ± 0.07 × 108 and 1.98 × 108 ± 0.26 × 108 cells/μmol Cl− released, respectively. The PCE and TCE dechlorination profiles suggested metabolic dechlorination of PCE and TCE and cometabolic dechlorination of VC to ethene. Debromination of PBDEs by cultures that had been amended with PCE for five consecutive transfers before amendment with the penta- and tetra-BDE mixture provided compelling evidence for the persistence of the debrominating population in the TZ50 enrichment culture when PCE was provided as an alternative electron acceptor.
FIG 2.
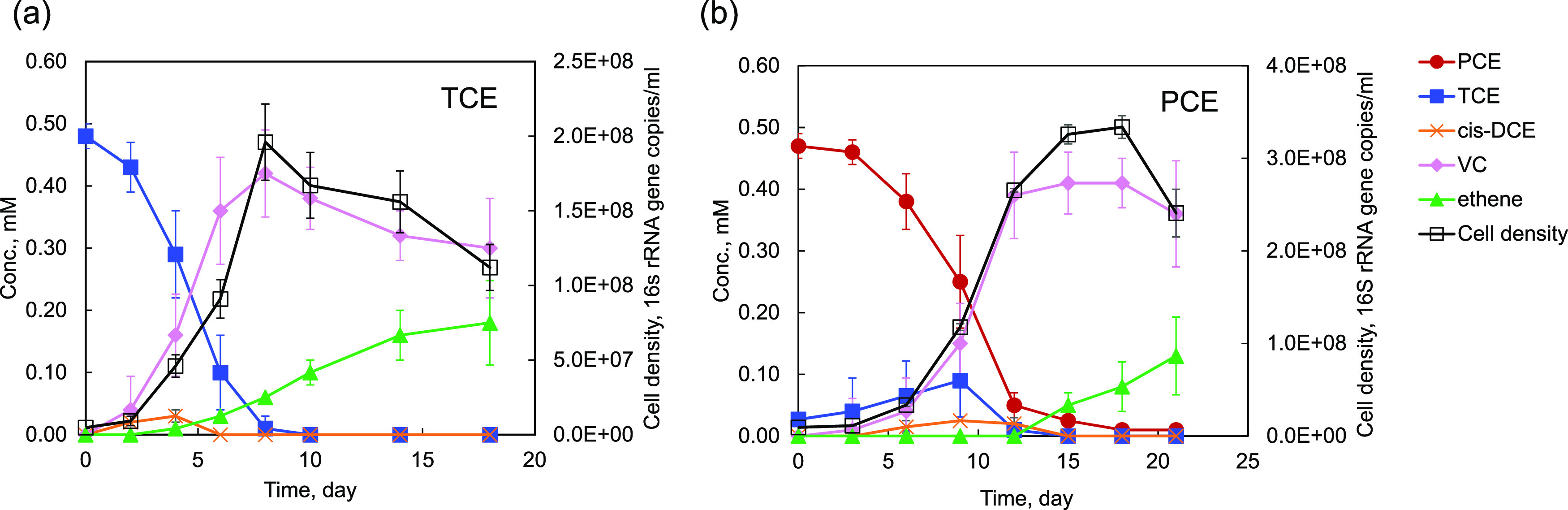
Dechlorination of trichloroethene (TCE) (a) and tetrachloroethene (PCE) (b) by D. mccartyi strain TZ50. VC, vinyl chloride.
In vitro enzymatic activity assays using crude cell extracts from TZ50 enrichment cultures amended with PCE showed the same debromination profiles, with the production of diphenyl ether and trace amounts of debrominating intermediates, as the TZ50 enrichment cultures amended with the penta- and tetra-PBDE mixture. Similarly, dechlorination of PCE to VC and ethene was observed in crude cell extracts from TZ50 enrichment cultures amended with the penta- and tetra-BDE mixture (data not shown). Data from in vitro assays supported the conclusion that the dehalogenating population(s) in the TZ50 enrichment culture could dehalogenate both PBDEs and chloroethenes.
Assembly of a D. mccartyi genome from the TZ50 enrichment culture metagenome.
The TZ50 enrichment culture was continuously subcultured with the penta- and tetra-BDE mixture as the sole electron acceptor but was amended with PCE for a single transfer to attain the biomass necessary to harvest sufficient DNA for metagenome sequencing. Paired-end reads from metagenomic sequencing were taxonomically classified against the minikraken2_v1_8GB database using Kraken2. Bracken estimated that a Dehalococcoides population accounted for 99.06% of the microbial community. The metagenomic reads were segregated into five bins, one of which contained reads related to Dehalococcoides. CheckM determined that the bin containing Dehalococcoides reads was 99.01% complete and had no strain heterogeneity (Fig. S3). Based on these results, the Dehalococcoides population in the TZ50 enrichment culture was designated D. mccartyi strain TZ50, and this bin was taken to represent the genome of the D. mccartyi population.
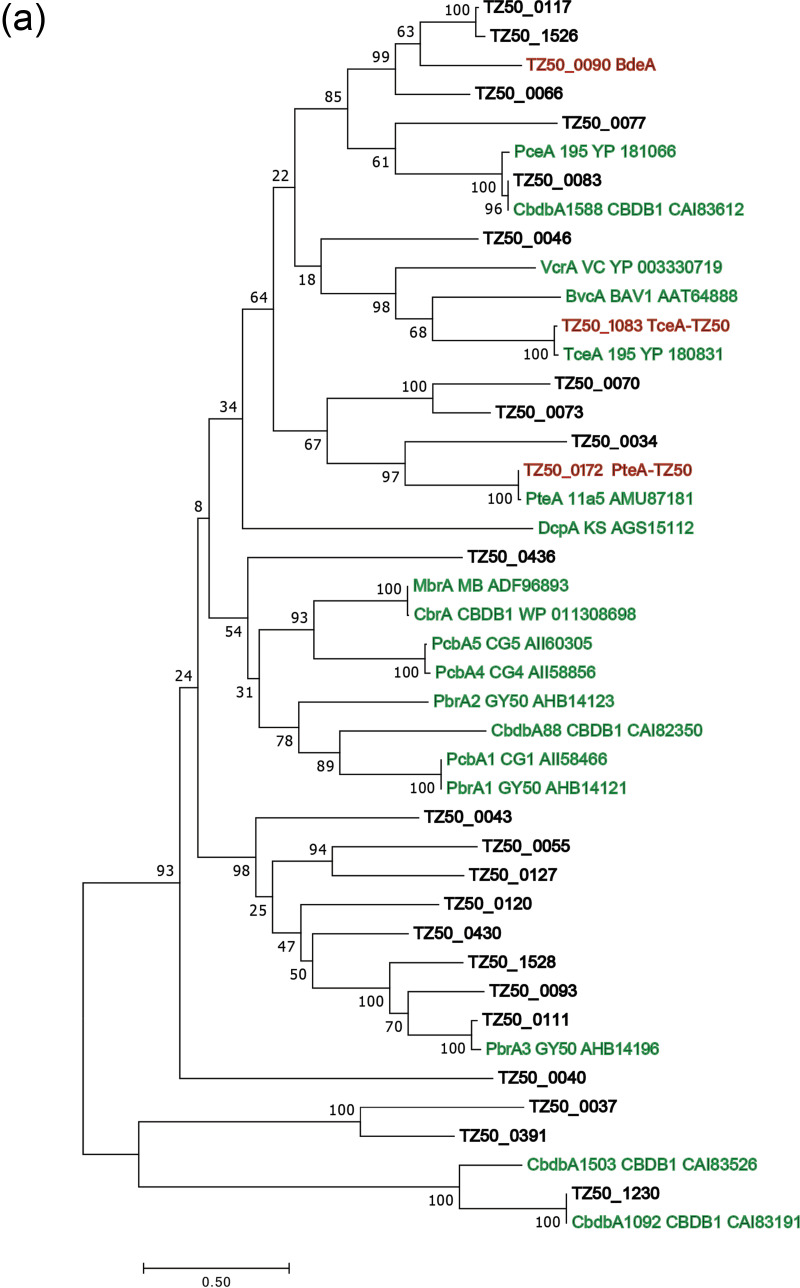
A draft genome of strain TZ50, comprising eight contigs (>1 kbp), was assembled from binned metagenomic reads (Table S1). The total length of the draft genome is 1.41 Mbp, and it contains 1,526 predicted open reading frames. Phylogenetic analysis based on the full-length 16S rRNA gene and average nucleotide identity comparison of the strain TZ50 draft genome with the genomes of other known Dehalococcoides strains placed strain TZ50 deep within the Pinellas subgroup (Fig. S4). Totals of 25 rdhA genes and 25 associated rdhB genes were annotated in the draft genome of strain TZ50 (Fig. 3a). The online RDase database (10) was used to place the 25 rdhA genes into orthologous groups: 23 were placed into existing RDase ortholog groups (RD_OGs), and 2 (TZ50_0040 and TZ50_0046) were grouped with existing ungrouped RdhAs, forming two new RD_OGs (Table S2). Five of the 25 RdhAs were assigned to groups containing functionally identified RDases (Table S2).
FIG 3.
(a) Maximum likelihood dendrogram of reductive dehalogenases (RDases) in D. mccartyi strain TZ50 and functionally characterized RDases in other D. mccartyi strains. The final tree is supported by 1,000 bootstrap replicates. Accession numbers and locus tags of amino acid sequences refer to the NCBI GenBank database. Green font indicates functionally characterized RDases; red font indicates RDases expressed in strain TZ50 during debromination of a defined penta- and tetra-BDE mixture (PBDE) and detected by proteomics analyses. Black font indicates RDases encoded in the draft genome of strain TZ50. (b) Relative abundances of reductive dehalogenase homologous (RdhA) proteins (relative to all expressed RdhAs) in D. mccartyi strain TZ50 grown on tetrachloroethene (PCE), trichloroethene (TCE), or the penta- and tetra-BDE mixture as measured by proteomics analysis.
Isolation of D. mccartyi strain TZ50.
The actively dehalogenating bacterium in the TZ50 enrichment culture was isolated by serial dilution to extinction in agar shakes amended with the penta- and tetra-BDE mixture; 3 of 5 colonies picked from the 10−5 dilution exhibited debromination identical to that of the TZ50 enrichment culture after 60 days of cultivation (data not shown) and likely represented isolates. One active culture was selected for further investigation. 16S rRNA gene-based denaturing gradient gel electrophoresis (DGGE) of DNA extracted from this culture and amplified using either a universal bacterial primer pair or a Dehalococcoides-specific primer pair resolved a single band (Fig. S5). In aggregate, metagenomic analysis of the TZ50 enrichment culture and 16S rRNA characterization of the isolate suggested the presence of a single Dehalococcoides strain. Cultures from all three colonies exhibited a chloroethene dechlorination profile identical to that of the TZ50 enrichment culture, indicating that the bacterial population in these cultures was the functional population in the TZ50 enrichment culture.
PBDE RDases and debromination pathways.
Prior to obtaining the isolate of strain TZ50, protein profiles of the TZ50 enrichment culture amended with different electron acceptors (PCE, TCE, or the penta- and tetra-BDE mixture) were elicited to investigate the abundances of different RDases during dehalogenation. Cells for proteomics analyses were cultivated with only pertinent halogenated substrates for at least six consecutive transfers (2%, vol/vol). Of the proteins detected in the proteome of the TZ50 enrichment culture grown with different substrates, 634 (PCE), 536 (TCE), and 40 (penta- and tetra-BDE mixture) corresponded to protein-coding sequences in the annotated draft genome of D. mccartyi strain TZ50. Notably, cultures amended with the penta- and tetra-BDE mixture yielded an abundance of detected proteins an order of magnitude lower than those of cultures amended with PCE or TCE, which is most likely due to the low biomass of cells cultivated with the penta- and tetra-BDE mixture. TZ50_0172 (PteA-like), TZ50_1083 (TceA-like), and TZ50_0090 represented the large majority of all expressed RDases and were present during the dehalogenation of all three halogenated substrates; other putative RDases (TZ50_0111 [PbrA3-like], TZ50_0117, TZ50_1526, TZ50_0070, and TZ50_1528) were present at lower abundance (<5%) (Fig. 3b).
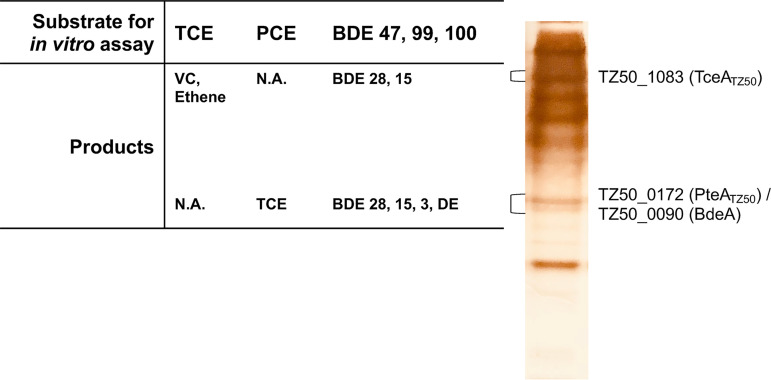
Further identification of PBDE RDases and their pathways was performed on isolate TZ50 to avoid any concealing effects in enzymatic assays and native PAGE by nondehalogenating proteins expressed in other bacterial populations. In vitro enzymatic assays of crude cell extracts of D. mccartyi strain TZ50 cultivated in PCE, TCE, or the penta- and tetra-BDE mixture produced the same dehalogenation end products (Fig. S6). Native PAGE assays were then performed using crude cell extracts of strain TZ50 cultivated with the penta- and tetra-BDE mixture to investigate the functions of TZ50_0172, TZ50_1083, and TZ50_0090 in the observed dehalogenation activity (Fig. 4). Three bands, an upper band corresponding to TZ50_1083 and two lower bands corresponding to TZ50_0172 and TZ50_0090, were apparent in the silver-stained counterparts of the native PAGE gel. The two lower bands could not be physically separated by excision in the native PAGE gel and so were assayed together. PBDE debromination was detected in in vitro activity assays of the upper and lower bands, but the observed debromination products differed from those generated by the culture itself (Fig. 4). The lower band produced TCE from PCE as well as BDE-28, BDE-15, and BDE-3 from the penta- and tetra-BDE mixture. The upper band produced VC and ethene from TCE as well as BDE-28 and BDE-15 from the penta- and tetra-BDE mixture. The proteins encoded by loci TZ50_1083 and TZ50_0172 were presumptively identified as TceA and PteA orthologs (TceATZ50 and PteATZ50), respectively, based on sequence similarity and chloroethene dechlorination profiles (Fig. S7), although penta- or tetra-BDE debromination by other characterized TceA and PteA orthologs has not been reported.
FIG 4.
In vitro activity assay of native PAGE bands corresponding to reductive dehalogenases (TZ50_1083, TZ50_0172, and TZ50_0090) in crude cell extracts of D. mccartyi strain TZ50 grown on a defined penta- and tetra-BDE mixture. Bands on the native PAGE gel were identified based on a silver-stained counterpart and excised for in vitro assays. Bands corresponding to TZ50_0172 and TZ50_0090 could not be separated by excision, so the two bands were excised and assayed together. N.A., not applicable.
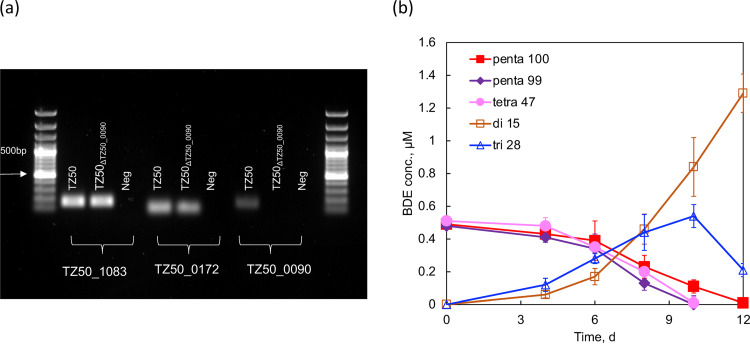
The proximity of TZ50_0172 and TZ50_0090 following electrophoresis precluded the differentiation of the activity of either individual band via in vitro analysis. However, the fortuitous emergence of a D. mccartyi strain TZ50 mutant without TZ50_0090 (TZ50ΔTZ50_0090) enabled functional characterization of TZ50_0172 and TZ50_0090 (Fig. 5a). It is possible that the continuous transfer of cultures before complete debromination may have resulted in the emergence of this mutant strain that could no longer debrominate BDE-15, but the reason for the emergence of this mutant was not investigated further. TZ50ΔTZ50_0090 maintained dechlorination of PCE and TCE, suggesting that it retained the functionality of TZ50_1083 and TZ50_0172. However, the debromination of the penta- and tetra-BDE mixture by TZ50ΔTZ50_0090 ceased at di-BDE-15 (Fig. 5b). These results suggested that TZ50_0090 was likely responsible for the removal of the para-bromine from di-BDE-15 but left the role of this gene product in PCE dechlorination unclear. Based on this functional characterization, TZ50_0090 was dubbed BdeA (debromination of brominated diphenyl ether).
FIG 5.
Loss of TZ50_0090 in a mutant of D. mccartyi strain TZ50 detected by endpoint PCR. Genomic DNAs derived from D. mccartyi strain TZ50 (TZ50) and the TZ50 mutant (TZ50ΔTZ50_0090) were used as the templates for PCR. (b) Debromination of the penta- and tetra-BDE mixture to BDE-28 and BDE-15 by strain TZ50ΔTZ50_0090.
DISCUSSION
Penta- and tetra-BDEs are highly toxic, and the complete removal of bromine substituents is crucial for mitigating toxicity. The slow and incomplete debromination of penta- and tetra-BDEs detected in the majority of microcosms established in the current study, yielding primarily tetra- and tri-BDEs as end products, is consistent with previous reports (6, 7, 11). Three functional PBDE-debrominating RDases have been described in Dehalococcoides (7), but none of these previously identified PBDE RDases were detected in three (TZ-7, LH-2, and LH-3) of seven penta- and tetra-BDE-debrominating microcosms in the current study. Notably, microbial community analyses revealed an absence of Dehalococcoides populations in microcosms LH-2 and LH-3, suggesting the involvement of a different bacterial lineage in the observed debromination. This inference is supported by other studies that have reported the possibility of anaerobic PBDE debromination by bacterial genera other than Dehalococcoides (5–8) and warrants further study. Microcosms TZ-5 and ZJ-2, which exhibited penta- and tetra-BDE debromination, harbored pbrA3 but exhibited dehalogenation patterns distinct from those that were previously described for PbrA3 (Table 1). The reason for this discrepancy is unclear, but it may be due to some combination of PbrA3 activity with that of other unknown functional RDases in these communities. Regardless, the debromination activity observed in the suite of microcosms established in the current study demonstrates that the array of currently described PBDE RDases cannot reliably indicate the debromination potential of the microbial communities in contaminated sediments.
The limited understanding of penta- and tetra-BDE-debrominating populations and the mechanisms involved has long impeded the study of the environmental fates of PBDEs (9, 12). D. mccartyi strain TZ50 harbors three rdhA genes that encode PBDE-debrominating RDases, including orthologs of pteA (TZ50_0172) and tceA (TZ50_1083), which dehalogenate penta- and tetra-BDEs to di-BDE, and bdeA (TZ50_0090), which has no functionally characterized ortholog and dehalogenates di-BDE-15 to diphenyl ether (5, 6, 13). These three genes expand the number of known PBDE-debrominating rdhA genes and contribute to the current understanding of microbial PBDE debromination. Additionally, strain TZ50 dechlorinated PCE, TCE, and DCE isomers to VC and ethene. Interestingly, dehalogenation of both aromatic and aliphatic organohalides by at least two of the PBDE-debrominating RDases in strain TZ50, PteATZ50 and TceATZ50, was observed. These RDases, which mediate the dehalogenation of both aromatic and aliphatic halogenated compounds, add to the growing number of RDases capable of dehalogenating structurally and chemically dissimilar compounds that have been described in Dehalococcoides, suggesting that this phenomenon could be more widespread than was initially thought (14–17). The mechanisms by which a single RDase can catalyze the dehalogenation of structurally and chemically dissimilar substrates and still maintain some measure of substrate specificity remain unclear (14–16, 18, 19). Identification and characterization of additional examples of this phenomenon can be used to obtain more comprehensive insights into the nature of this process.
Strain TZ50 harbors two RdhAs with a high degree of amino acid similarity to previously described RD_OGs that include functionally characterized RDases (TZ50_0111 [OG 10] [PbrA3] and TZ50_0083 [OG 30] [PceA]). Notably, while PbrA3 debrominates penta- and tetra-BDEs to di-BDE via the same pathways as those seen in strain TZ50, TZ50_0111 was not or at least was not dominantly expressed during dehalogenation of the penta- and tetra-BDE mixture. The low resolution of proteomics analysis, as indicated by the comparatively small number (40) of proteins detected in cultures amended with the penta- and tetra-BDE mixture, may have contributed to the low abundance of some proteins in this study. Similarly, the ortholog of PceA, which dechlorinates PCE to TCE, in the TZ50 genome, TZ50_0083, was also not detected in strain TZ50 during PCE dechlorination. Similar phenomena are commonly reported in Dehalococcoides, e.g., the lack of function of the PceA orthologs in the genomes of strains CG1, 11a5, BTF08, and GY50 during PCE dechlorination (see Fig. S7 in the supplemental material). Minor variations in primary amino acid sequence can significantly alter RDase functionality and expression. Differential expression of genes encoding RDases with high degrees of amino acid sequence identity in the presence of specific substrates was conclusively demonstrated in D. mccartyi strain BTF08, the genome of which encodes three separate chloroethene-dechlorinating RDases (PteAbtf08, VcrAbtf08, and TceAbtf08) (20). Furthermore, Dehalococcoides genomes often harbor multiple rdhA genes that have putative or empirically determined dehalogenation activity, but the presence of a potential halogenated compound does not necessarily imply that a given rdh gene will be expressed (21). Patterns of expression of different rdh genes in strain TZ50 are consistent with previous reports that the expression and activity of RDases in Dehalococcoides cannot be reliably inferred from sequence identity alone. The mechanisms underlying this selective expression remain unclear, and the identification of additional instances of this phenomenon can inform future research.
While previous work has demonstrated a role for the MarR-like regulation of some rdh genes in Dehalococcoides (22), the mechanisms underlying the selective expression of RDases under various conditions of substrate availability remain unclear. A greater degree of clarity regarding the expression of functional RDases would be beneficial to the implementation of effective Dehalococcoides-mediated bioremediation strategies. The ability of a single RDase to dehalogenate dissimilar environmental pollutants could be leveraged to increase the rate of PBDE debromination at sites contaminated with both PBDEs and PCE/TCE or to increase cell density via halopriming prior to augmentation.
MATERIALS AND METHODS
Chemicals.
A PBDE mixture containing 39 individual congeners (hepta- to mono-BDEs) at concentrations from 100 to 250 mg/liter (>98% purity) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA). The components of this mixture were used as standards to identify and quantify all BDEs for which individual calibration was not performed. Individual penta- and tetra-BDE congeners (BDE-100, BDE-99, and BDE-47) were purchased from Agilent Technologies, Inc. (Santa Clara, CA), at 5,000 ppm dissolved in 2,2,4-trimethylpentane. The penta- and tetra-BDE mixture was constructed by combining BDE-47, BDE-99, and BDE-100 in a 1:1:1 (vol/vol/vol) ratio. Other individual BDE congeners (BDE-28, BDE-15, and BDE-3) were purchased from AccuStandard (New Haven, CT, USA). All chloroethenes (>98% purity) were purchased from MilliporeSigma (Burlington, MA, USA).
Penta- and tetra-BDE debromination in environmental samples. (i) Sampling locations and microcosms.
A total of 24 samples were collected from various environments and used to establish microcosms (see Table S3 in the supplemental material). Microcosms were established by adding ∼5 g soil (collected 10 to 15 cm below the surface) or sediment to a 60-ml serum bottle containing 30 ml bicarbonate-buffered DCB-1 minimal salts medium, prepared as previously described (23). Lactate (10 mM) was added as the sole carbon source and electron donor. A total concentration of ∼1.4 μM penta- and tetra-BDEs was added as a 1:1:1 mixture of BDE-100, BDE-99, and BDE-47 (∼0.44, 0.44, and 0.51 μM, respectively) as electron acceptors. BDEs were amended to microcosms and enrichment cultures at a total concentration that is environmentally relevant but not necessarily representative of the total range of possible environmental concentrations (24). Abiotic controls without sediments or with autoclaved soils/sediments were established for each sampling location. All microcosms were incubated in the dark at 30°C.
(ii) Chemical analyses and quantification of PBDEs.
Samples (1 ml) were collected from cultures, and PBDEs were extracted with 2,2,4-trimethylpentane using decabromobiphenyl as an internal standard. The organic phase of extracted samples was analyzed on an Agilent gas chromatograph-mass spectrometer (GC6890-MSD5975) equipped with an Rxi-5ms column (15 m by 0.25 mm by 0.25 μm; Restek, Bellefonte, PA, USA) as previously described (25). The oven temperature was initially set at 110°C, increased to 310°C at a rate of 15°C/min, and held at 310°C for 3 min. PBDEs were quantified against five-point calibration curves prepared by amending defined concentrations of individual BDEs or a commercially available BDE mixture comprising 39 BDE congeners (Cambridge Isotope Laboratories, Inc.) in 60-ml serum bottles containing 30 ml sterilized DCB-1 medium. Individual calibration curves were constructed for diphenyl ether as well as for BDE-3, BDE-15, BDE-28, BDE-47, BDE-99, and BDE-100. Calibration for other BDE congeners was estimated by calculating the average response of all congeners in the 39-BDE mixture with the same number of bromine substituents (i.e., the average response of all tri-BDEs in the 39-BDE mixture was taken as the calibration value for tri-BDEs other than BDE-28, and so on). The detection limit for PBDE congeners was 20 ng/liter. Calibration curves were constructed in triplicate, and the mean response value was used for quantification. The standard deviation of each curve was within 20%, and the R2 of all curves was >0.98.
(iii) Prevalence of Dehalococcoides and PBDE rdh.
Samples (1 ml) were collected from all microcosms every 4 weeks, and DNA was extracted using the Qiagen DNeasy PowerSoil kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. Dehalococcoides was detected using endpoint PCR with genus-specific primer pair Dhc730F and Dhc1350R (26). The presence of pbrA1, pbrA2, and pbrA3 in each microcosm was assayed by PCR as previously described (7).
Enrichment of PBDE-degrading cultures. (i) Enrichment.
Microcosm TZ-7 exhibited complete debromination of the penta- and tetra-BDE mixture. An enrichment culture, dubbed TZ50 (described above), was obtained via consecutive subculturing (5%, vol/vol) of microcosm TZ-7 until a sediment-free culture was generated (five transfers), followed by five iterations of serial dilution to extinction (10−1 to 10−5). Subcultures and serial dilutions were cultivated in 30 or 10 ml (in 60- or 20-ml serum bottles, respectively) of DCB-1 medium amended with acetate (10 mM) as a carbon source, hydrogen (0.3 atm) as an electron donor, and the penta- and tetra-BDE mixture (1.5 μM). For dehalogenation and growth kinetics analyses, the TZ50 enrichment culture was grown in 160-ml serum bottles containing 100 ml DCB-1 medium amended with acetate, hydrogen, and halogenated substrates, as indicated.
(ii) Dechlorination of chlorinated ethenes.
Dechlorination of chloroethenes (PCE, TCE, DCE isomers, and VC at 0.5 mM) by the TZ50 enrichment culture was analyzed on Agilent 7890 gas chromatograph equipped with a flame ionization detector, as previously described (27).
(iii) Molecular and microbial community analyses.
DNA extracted from the TZ50 enrichment culture was sequenced on an Illumina MiSeq platform using the 515F and 806R primers (28) (Axil Scientific, Singapore). The resulting amplicons were analyzed using QIIME2 v2019.1.0 (29). Paired-end reads were joined, quality filtered, dereplicated, and de novo clustered at 97% identity into 1,214 operational taxonomic units (OTUs) (484,497 sequences) using the vsearch plug-in (30). Chimeric OTUs were identified (QIIME2 vsearch uchime-denovo plug-in) and removed, leaving 421,373 sequences in 515 OTUs. A phylogeny was constructed using FastTree, and taxonomy was assigned to OTUs against the Silva 128 database at a 97% similarity threshold. Default parameters were used unless stated otherwise. Growth of the total bacterial community and Dehalococcoides populations during PBDE debromination was monitored by quantitative real-time PCR (qPCR) with primers 338F/518R and Dhc-qF2/Dhc-qR (31), respectively.
Metagenome sequencing, binning, assembly, and annotation.
Following three additional enrichment transfers, DNA was extracted from the TZ50 enrichment culture (800-ml culture volume) amended with PCE to achieve sufficient DNA for metagenomic sequencing. DNA was extracted using a genomic DNA buffer kit with a 100-gauge Genomic-tip (Qiagen GmbH) according to the manufacturer’s instructions. Other than the culture used for outgrowth, enrichment culture TZ50 was cultivated in DCB-1 medium amended with the penta- and tetra-BDE mixture. The metagenome was sequenced on an Illumina HiSeq4000 platform at the Beijing Genomics Institute (Shenzhen, China), yielding 1,2030,146 paired-end reads. Reads were trimmed for quality and assembled with metaSPAdes v3.13.0 (32) into 5,640 contigs with a length of >500 bp (N50 = 15,698 bp); the largest assembled contig was 812,388 bp. The assembled contigs were fragmented to a maximum length of 10 kb, and reads were mapped back to the fragmented assembly using Bowtie2 v2.3.2 (33) to determine coverage depth. The fragmented contigs were automatically binned into individual genomes using MaxBin v2.2.6 (34), and the resulting bins were evaluated for completeness and integrity with CheckM v1.0.13 (35). A single bin containing reads belonging to Dehalococcoides was manually refined based on tetrad frequency, GC content, coding density, taxonomic identity, and sequencing depth. Contigs in the resulting bins were reassembled with SPAdes and submitted to the Rapid Annotation Using Subsystem Technology (RAST) server for gene calling and annotation using RASTk (36).
Taxonomic identification of metagenomic reads was performed with Kraken2 v.2.0.8-beta (37) using the prebuilt minikraken2_v1_8GB database, and taxonomic abundance was estimated using Bracken v2.1.0 (38) with a read length parameter of 150. Taxonomic classification of bins was determined based on the most abundant taxonomic assignment of the contigs in the bin and the 16S rRNA gene(s) present on binned contigs. A maximum likelihood phylogeny was constructed with Molecular Evolutionary Genetics Analysis 7 (MEGA7) (39) using available Dehalococcoides 16S rRNA gene sequences and the 16S rRNA gene sequence in the Dehalococcoides-containing metagenome bin. Sequences were aligned using ClustalW, and the final tree is supported by 1,000 bootstraps. Average nucleotide identity among selected Dehalococcoides genomes was calculated using the JSpeciesWS online service (40).
Enzyme assays and proteomics analysis.
Crude cell extracts of the TZ50 enrichment culture amended with PCE or the penta- and tetra-BDE mixture were used to assay alternative electron acceptor utilization by functional RDases in vitro as previously described (41). Briefly, cells were harvested by centrifugation (13,000 × g for 20 min at 4°C) and resuspended in degassed Tris-HCl buffer (100 mM; pH 7.0). Crude cell extracts were obtained by disrupting cells with a VCX 130 sonicator (130 W; 20% duty cycle; 3 min). Each in vitro activity assay was carried out in 4-ml vials containing 2 ml assay solution [2 mM methyl viologen, 1.5 mM titanium(III) citrate, 100 mM Tris-HCl buffer (pH 7.0)] inside an anaerobic chamber. Activity was measured after 72 h of incubation.
Native PAGE (10% resolving gel and 5% stacking gel) assays were performed with crude cell protein extracts of D. mccartyi strain TZ50 amended with the penta- and tetra-BDE mixture, as previously described (41). Briefly, each protein extract was run in four lanes on the gel: one lane for each sample was used for silver staining (ProteoSilver silver stain kit; Sigma-Aldrich, St. Louis, MO), and the other three were left unstained for in vitro activity analyses. Regions of unstained lanes corresponding to visible bands in the stained lanes were excised, and dechlorination (PCE and TCE, 3 μM each) and debromination (penta- and tetra-BDE mixture, 1.5 μM) in the excised bands were assayed. The activity in the gel slices was evaluated after 72 h of incubation; bands in the stained lane corresponding to gel slices exhibiting dehalogenation were then excised, digested with trypsin, and analyzed by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) on Eksigent nanoLC Ultra and ChiPLC-nanoflex instruments (Eksigent, Dublin, CA, USA) equipped with the Triple TOF 5600 system (AB Sciex, Foster City, CA, USA), as previously described (42). The MS/MS-based peptides and proteins were identified and visualized using ProteinPilot software 4.5 (Sciex). The MS/MS spectra were then searched using a customized database containing the draft genome of TZ50 as the database. The identification of peptides and proteins was validated if 99% probabilities were achieved by the Paragon algorithm.
For proteomics, cells from the TZ50 enrichment culture amended with either the penta- and tetra-BDE mixture (1.5 μM; 500-ml culture volume), PCE (0.6 mM; 300-ml culture volume), or TCE (0.6 mM; 300-ml culture volume) were harvested and concentrated by centrifugation at 13,000 × g for 10 min. Proteins were extracted on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with a nano-LC system (Dionex Ultimate 3000RSLC; Thermo Fisher Scientific), as previously described (43). Briefly, proteins were digested with trypsin and desalted using ZipTip-μC18 material (MilliporeSigma). Protein identification was conducted with Proteome Discoverer (v2.2; Thermo Fisher Scientific) using the Sequest HT search algorithm against a translated protein database derived from the metagenome bin containing reads corresponding to the D. mccartyi population. Protein and peptide abundances were calculated by label-free quantification based on area counts using the Minora node implemented in Proteome Discoverer.
Isolation of D. mccartyi strain TZ50.
A D. mccartyi population was isolated from the TZ50 enrichment culture via serial dilution to extinction and agar shake, as previously described (27). After 60 days of incubation, colonies with 0.2- to 0.3-mm radii were picked from the 10−5 dilution and immediately dispersed into liquid medium for serial dilution (10−1 to 10−5) to reduce or eliminate contamination from colocated colonies. The purity of cultures arising at the highest dilution was evaluated by DGGE (60°C, 30% to 60%, 120 V, and 12 h) using Dehalococcoides-specific and universal bacterial primers 1FGC/259R and 341FGC/518R (44, 45), respectively.
The presence of three PBDE RDase genes, TZ50_1083, TZ50_0172, and TZ50_0090, was identified using endpoint PCR by primer pair tceA1270F/1336R (46) and two other primer sets designed in this study (pteA848F [5′-CCA TAG GCA CCT TGG TAG CA]/pteA998R [5′-TCA CCA ATG CCT GCT AAC GT] and bdeA628F [5′-CTG CGT CAG GTA GTC CGT TT]/bdeA758R [5′-GCC GCT TCG TCT ACG TTT TC]).
Data availability.
The draft genome of D. mccartyi strain TZ50 and raw metagenomic sequencing data for this study were deposited in the NCBI database under BioProject accession number PRJNA564712. Raw proteomics data were deposited in the Proteomics Identification Database (PRIDE) under project accession number PXD024983.
ACKNOWLEDGMENTS
This study was supported by the Ministry of Education, Singapore, under Academic Research Fund Tier 2 of project MOE-000033-01 and Tier 1 project R302000239114.
We also acknowledge the help kindly provided by Lorenz Adrian from the Helmholtz Centre for Environmental Research for proteomics analysis.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Jianzhong He, Email: jianzhong.he@nus.edu.sg.
Alfons J. M. Stams, Wageningen University
REFERENCES
- 1.Darnerud PO, Eriksen GS, Jóhannesson T, Larsen PB, Viluksela M. 2001. Polybrominated diphenyl ethers: occurrence, dietary exposure, and toxicology. Environ Health Perspect 109(Suppl 1):49–68. 10.1289/ehp.01109s149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.United Nations Environment Programme. 2009. Stockholm Convention on persistent organic pollutants (POPs). https://www.env.go.jp/chemi/pops/treaty/treaty_en2009.pdf.
- 3.Major DW, McMaster ML, Cox EE, Edwards EA, Dworatzek SM, Hendrickson ER, Starr MG, Payne JA, Buonamici LW. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ Sci Technol 36:5106–5116. 10.1021/es0255711. [DOI] [PubMed] [Google Scholar]
- 4.Justicia-Leon SD, Higgins S, Mack EE, Griffiths DR, Tang S, Edwards EA, Löffler FE. 2014. Bioaugmentation with distinct Dehalobacter strains achieves chloroform detoxification in microcosms. Environ Sci Technol 48:1851–1858. 10.1021/es403582f. [DOI] [PubMed] [Google Scholar]
- 5.He J, Robrock KR, Alvarez-Cohen L. 2006. Microbial reductive debromination of polybrominated diphenyl ethers (PBDEs). Environ Sci Technol 40:4429–4434. 10.1021/es052508d. [DOI] [PubMed] [Google Scholar]
- 6.Robrock KR, Korytar P, Alvarez-Cohen L. 2008. Pathways for the anaerobic microbial debromination of polybrominated diphenyl ethers. Environ Sci Technol 42:2845–2852. 10.1021/es0720917. [DOI] [PubMed] [Google Scholar]
- 7.Ding C, Rogers MJ, Yang KL, He J. 2017. Loss of the ssrA genome island led to partial debromination in the PBDE respiring Dehalococcoides mccartyi strain GY50. Environ Microbiol 19:2906–2915. 10.1111/1462-2920.13817. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Wang C, Pan Y, Farzana SS, Tam NF. 2018. Biochar accelerates microbial reductive debromination of 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) in anaerobic mangrove sediments. J Hazard Mater 341:177–186. 10.1016/j.jhazmat.2017.07.063. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Rogers MJ, Ding C, He J. 2018. Reductive debromination of polybrominated diphenyl ethers—microbes, processes and dehalogenases. Front Microbiol 9:1292. 10.3389/fmicb.2018.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molenda O, Puentes Jácome LA, Cao X, Nesbø CL, Tang S, Morson N, Patron J, Lomheim L, Wishart DS, Edwards EA. 2020. Insights into origins and function of the unexplored majority of the reductive dehalogenase gene family as a result of genome assembly and ortholog group classification. Environ Sci Process Impacts 22:663–678. 10.1039/c9em00605b. [DOI] [PubMed] [Google Scholar]
- 11.Zhu HW, Wang Y, Wang XW, Luan TG, Tam NFY. 2014. Intrinsic debromination potential of polybrominated diphenyl ethers in different sediment slurries. Environ Sci Technol 48:4724–4731. 10.1021/es4053818. [DOI] [PubMed] [Google Scholar]
- 12.Xu G, Zhao X, Zhao S, Chen C, Rogers MJ, Ramaswamy R, He J. 2021. Insights into the occurrence, fate, and impacts of halogenated flame retardants in municipal wastewater treatment plants. Environ Sci Technol 55:4205–4226. 10.1021/acs.est.0c05681. [DOI] [PubMed] [Google Scholar]
- 13.Ding C, Chow WL, He J. 2013. Isolation of Acetobacterium sp. strain AG, which reductively debrominates octa- and pentabrominated diphenyl ether technical mixtures. Appl Environ Microbiol 79:1110–1117. 10.1128/AEM.02919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Chng KR, Chen C, Bedard DL, He J. 2015. Genomic characterization of Dehalococcoides mccartyi strain JNA that reductively dechlorinates tetrachloroethene and polychlorinated biphenyls. Environ Sci Technol 49:14319–14325. 10.1021/acs.est.5b01979. [DOI] [PubMed] [Google Scholar]
- 15.Yang C, Kublik A, Weidauer C, Seiwert B, Adrian L. 2015. Reductive dehalogenation of oligocyclic phenolic bromoaromatics by Dehalococcoides mccartyi strain CBDB1. Environ Sci Technol 49:8497–8505. 10.1021/acs.est.5b01401. [DOI] [PubMed] [Google Scholar]
- 16.Adrian L, Rahnenfuhrer J, Gobom J, Holscher T. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol 73:7717–7724. 10.1128/AEM.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, He J. 2018. Strategy for the rapid dechlorination of polychlorinated biphenyls (PCBs) by Dehalococcoides mccartyi strains. Environ Sci Technol 52:13854–13862. 10.1021/acs.est.8b03198. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Chng KR, Wilm A, Zhao S, Yang K-L, Nagarajan N, He J. 2014. Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc Natl Acad Sci U S A 111:12103–12108. 10.1073/pnas.1404845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao S, Rogers MJ, He J. 2020. Abundance of organohalide respiring bacteria and their role in dehalogenating antimicrobials in wastewater treatment plants. Water Res 181:115893. 10.1016/j.watres.2020.115893. [DOI] [PubMed] [Google Scholar]
- 20.Franke S, Seidel K, Adrian L, Nijenhuis I. 2020. Dual element (C/Cl) isotope analysis indicates distinct mechanisms of reductive dehalogenation of chlorinated ethenes and dichloroethane in Dehalococcoides mccartyi strain BTF08 with defined reductive dehalogenase inventories. Front Microbiol 11:1507. 10.3389/fmicb.2020.01507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hug LA. 2016. Diversity, evolution, and environmental distribution of reductive dehalogenase genes, p 377–393. In Adrian L, Löffler FE (ed), Organohalide-respiring bacteria. Springer, Berlin, Germany. 10.1007/978-3-662-49875-0_16. [DOI] [Google Scholar]
- 22.Krasper L, Lilie H, Kublik A, Adrian L, Golbik R, Lechner U. 2016. The MarR-type regulator RdhR1456 regulates rdh gene transcription in Dehalococcoides mccartyi strain CBDB1. J Bacteriol 198:3130–3141. 10.1128/JB.00419-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J, Holmes VF, Lee PKH, Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol 73:2847–2853. 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H, Tam NFY, Cheung SG, Wei P, Li S, Wu Q. 2019. Contamination of polybrominated diphenyl ethers (PBDEs) in watershed sediments and plants adjacent to e-waste sites. J Hazard Mater 379:120788. 10.1016/j.jhazmat.2019.120788. [DOI] [PubMed] [Google Scholar]
- 25.Lee LK, He J. 2010. Reductive debromination of polybrominated diphenyl ethers by anaerobic bacteria from soils and sediments. Appl Environ Microbiol 76:794–802. 10.1128/AEM.01872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunge M, Adrian L, Kraus A, Opel M, Lorenz WG, Andreesen JR, Görisch H, Lechner U. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357–360. 10.1038/nature01237. [DOI] [PubMed] [Google Scholar]
- 27.Zhao S, He J. 2019. Reductive dechlorination of high concentrations of chloroethenes by a Dehalococcoides mccartyi strain 11G. FEMS Microbiol Ecol 95:fiy209. 10.1093/femsec/fiy209. [DOI] [PubMed] [Google Scholar]
- 28.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeborn RA, West KA, Bhupathiraju VK, Chauhan S, Rahm BG, Richardson RE, Alvarez-Cohen L. 2005. Phylogenetic analysis of TCE-dechlorinating consortia enriched on a variety of electron donors. Environ Sci Technol 39:8358–8368. 10.1021/es048003p. [DOI] [PubMed] [Google Scholar]
- 32.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu YW, Tang YH, Tringe SG, Simmons BA, Singer SW. 2014. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, ThomasonJA, III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3:e104. 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a Web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Jiang L, Zhao X, Linag S, Yu M, Liu W, Peng S. 2016. The research on the solubility of decabromodiphenyl ether based on recycling of waste electronic plastic by solvent process. Procedia Environ Sci 31:827–831. 10.1016/j.proenv.2016.02.083. [DOI] [Google Scholar]
- 42.Zhao S, Ding C, He J. 2016. Genomic characterization of Dehalococcoides mccartyi strain 11a5 reveals a circular extrachromosomal genetic element and a new tetrachloroethene reductive dehalogenase gene. FEMS Microbiol Ecol 93:fiw235. 10.1093/femsec/fiw235. [DOI] [PubMed] [Google Scholar]
- 43.Seidel K, Kühnert J, Adrian L. 2018. The complexome of Dehalococcoides mccartyi reveals its organohalide respiration-complex is modular. Front Microbiol 9:1130. 10.3389/fmicb.2018.01130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duhamel M, Mo K, Edwards EA. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl Environ Microbiol 70:5538–5545. 10.1128/AEM.70.9.5538-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl Environ Microbiol 71:3866–3871. 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S7, Tables S1 to S3. Download AEM.00602-21-s0001.pdf, PDF file, 1.9 MB (1.9MB, pdf)
Data set. Download AEM.00602-21-s0002.xlsx, XLSX file, 0.4 MB (367.7KB, xlsx)
Data Availability Statement
The draft genome of D. mccartyi strain TZ50 and raw metagenomic sequencing data for this study were deposited in the NCBI database under BioProject accession number PRJNA564712. Raw proteomics data were deposited in the Proteomics Identification Database (PRIDE) under project accession number PXD024983.