Abstract
Samples of species close to Tremellafibulifera from China and Brazil are studied, and T.fibulifera is confirmed as a species complex including nine species. Five known species (T.cheejenii, T.fibulifera s.s., T. “neofibulifera”, T.lloydiae-candidae and T.olens) and four new species (T.australe, T.guangxiensis, T.latispora and T.subfibulifera) in the complex are recognized based on morphological characteristics, molecular evidence, and geographic distribution. Sequences of eight species of the complex were included in the phylogenetic analyses because T.olens lacks molecular data. The phylogenetic analyses were performed by a combined sequence dataset of the internal transcribed spacer (ITS) and the partial nuclear large subunit rDNA (nLSU), and a combined sequence dataset of the ITS, partial nLSU, the small subunit mitochondrial rRNA gene (mtSSU), the translation elongation factor 1-α (TEF1), the largest and second largest subunits of RNA polymerase II (RPB1 and RPB2). The eight species formed eight independent lineages with robust support in phylogenies based on both datasets. Illustrated description of the six species including Tremellafibulifera s.s., T. “neofibulifera” and four new species, and discussions with their related species, are provided. A table of the comparison of the important characteristics of nine species in the T.fibulifera complex and a key to the whitish species in Tremella s.s. are provided.
Keywords: Multi-gene, phylogeny, taxonomy, Tremellaceae
Introduction
Tremella Pers. is characterized by being parasitic on or associated with other fungi or lichens (Bandoni and Oberwinkler 1983; Chen 1998; Pippola and Kotiranta 2008; Malysheva et al. 2015; Zhao et al. 2019) and by having a haploid unicellular yeast stage and diploid stage in its life cycle (Boekhout et al. 2011; Weiss et al. 2014; Liu et al. 2015a; Zhao et al. 2019). Tremella s.l. includes approximately 90 species and about 50 are recognized as lichenicolous species (Kobayasi 1939; Bandoni 1958, 1987; Lowy 1971; Bandoni and Oberwinkler 1983; Diederich 1996; Chen 1998; Kirk et al. 2008; Pippola and Kotiranta 2008; Millanes et al. 2011, 2012, 2014, 2015, 2016; Malysheva et al. 2015; Zamora et al. 2016; Zhao et al. 2019). Tremella s.l. is polyphyletic and was divided into five groups by Chen (1998) including Mesenterica group, Fuciformis group, Indecorata group, Foliacea group and Aurantia group based on morphological characteristics and molecular data of ITS rDNA and nLSU rDNA sequencing. Then species in Mesenterica group and Fuciformis group were allocated to Tremella s.s., and species in Indecorata group, Foliacea group and Aurantia group were emended as Pseudotremella Xin Zhan Liu et al., Phaeotremella Rea and Naematelia Fr., respectively (Liu et al. 2015b; Wedin et al. 2016; Spirin et al. 2018). Besides, Tremella s.l. contained lichenicolous species that defined as Tremella clade I, clade II, clade III, and several single Tremella species lineages based on rDNA sequences (Millanes et al. 2011; Liu et al. 2015a, b).
Although Tremella s.l. was separated into several genera due to its polyphyletism, it is still somewhat confusing because taxonomic positions of some Tremella species are uncertain in Tremellales, especially some species recently described from lichens (Ariyawansa et al. 2015; Malysheva et al. 2015; Millanes et al. 2015; Zamora et al. 2016, 2018). These lichenicolous species were described as Tremella, but they were not clustered into Tremella s.s. in the phylogeny (Ariyawansa et al. 2015; Malysheva et al. 2015; Millanes et al. 2015; Zamora et al. 2016, 2018). Recently, Zhao et al. (2019) described four new Tremella species based on the phylogenetic relationship of 19 species in Tremella s.s., and Li et al. (2020) published a new yeast species of Tremella s.s. based on multi-gene analysis.
In this study, samples of species morphologically similar to Tremellafibulifera characterized by cerebriform whitish basidioma and abundant clamp complexes from China and Brazil are studied. Based on morphology, geographic distribution and phylogenetic analyses T.fibulifera is confirmed as a species complex, which was previously mentioned by Bandoni and Oberwinkler (1983) and Malysheva et al. (2015), and nine species are involved in the complex including five known species (T.cheejenii, T.fibulifera s.s., T. “neofibulifera”, T.lloydiae-candidae and T.olens) and four new species (T.australe, T.guangxiensis, T.latispora and T.subfibulifera) in the present study. The aim of this paper is to outline the T.fibulifera complex and describe two known species (T.fibulifera s.s., T. “neofibulifera”) and the four new species based on our collections.
Materials and methods
Sampling and morphological analysis
The studied specimens were collected from Rondônia and Pernambuco states in Brazil, Yunnan, Taiwan, Guangxi, Jilin Provinces in China. They are deposited at the herbaria of Beijing Forestry University (BJFC), Institute of Botânica in São Paulo (SP) and Universidade Federal de Pernambuco, Departamento de Micologia (URM). Macro-morphological illustrations refer to Chen (1998) and Zamora et al. (2017) and microscopic structures refer to Pippola and Kotiranta (2008) and Malysheva et al. (2015). Special color terms followed Petersen (1996). Handmade sections of dried basidioma were examined by a Nikon Eclipse 80i (Japan) microscope (magnification × 1000) after being mounted in 5% KOH for five minutes and treated with 1% Phloxine B (C20H4Br4Cl2K2O5). Microscopic structures were photographed using a Nikon Digital Sight DS-L3 (Japan) or Leica ICC50 HD (Japan) camera. Microscopic structures were examined and measured in the mix solution of 5% KOH and 1% Phloxine B. To represent variation in the size of spores, 5% of measurements were excluded from each end of the range, and are given in parentheses. Stalks were excluded for basidia measurements and apices were excluded for basidiospores measurements. Length and width of at least 30 basidia and basidiospores from each specimen were measured to micrometers.
Abbreviations as follows: L = mean length (arithmetic average of all basidia or spores length), W = mean width (arithmetic average of all basidia or spores width), Q = L/W ratio for each specimen studied, n (a/b) = number of spores (a) measured from given number of specimens (b).
Molecular phylogeny
Dry specimens were used to extract DNA after pretreatment using TissuePrep (Jie Ling, China) by CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd, Beijing) or directly using the DNA easy Plant Mini Kit (Qiagen, China), according to the manufacturer’s instructions with some modifications. The internal transcribed spacer regions (ITS), partial nuclear large subunit rDNA (nLSU), the translation elongation factor 1-α (TEF1), the largest and second largest subunits of RNA polymerase II (RPB1 and RPB2), the small subunit mitochondrial rRNA gene (mtSSU) sequences were amplified with primer pairs listed in the Table 1. All newly generated sequences were submitted to GenBank (Table 2).
Table 1.
Sequencing primers used in this study.
| Pairs of primer | Nucleotide sequence (5′–3′) | Reference |
|---|---|---|
| ITS | ||
| ITS5 | 5′–GGAAGTAAAAGTCGTAACAAGG–3′ | White et al. 1990 |
| ITS4 | 3′–TCCTCCGCTTATTGATATGC–5′ | |
| ITS1 | 5′–TCCGTAGGTGAACCTGCGG–3′ | |
| Partial nLSU | ||
| LR0R | 5′–ACCCGCTGAACTTAAGC–3′ | Hopple and Vilgalys 1994 |
| LR7 | 5′–TACTACCACCAAGATCT–3′ | |
| TEF1 | ||
| 983F | 5′–GCYCCYGGHCAYCGTGAYTTYAT–3′ | Rehner 2001 |
| 1567R | 3′–ACHGTRCCRATACCACCRATCTT–5′ | Rehner 2001 |
| 2218R | 3′–ATGACACCRACRGCRACRGTYTG–5′ | Rehner and Buckley 2005 |
| RPB1 | ||
| Af | 5′–GARTGYCCDGGDCAYTTYGG–3′ | Stiller and Hall 1997 |
| Cr | 3′–CCNGCDATNTCRTTRTCCATRTA–5′ | Matheny et al. 2002 |
| RPB2 | ||
| 5F | 5′–GAYGAYMGWGATCAYTTYGG–3′ | Matheny 2005 |
| 6F | 5′–TGGGGKWTGGTYTGYCCTGC–3′ | Matheny 2005 |
| 7R | 3′–CCCATWGCYTGCTTMCCCAT–5′ | Matheny 2005 |
| 7CR | 3′–CCCATRGCTTGYTTRCCCAT–5′ | Matheny 2005 |
| Fcrypto | 5′–TGGGGYATGGTTTGTCCKGC–3′ | Ye et al. 2012 |
| Rcrypto | 3′–CCCATGGCTTGTTTRCCCATYGC–5′ | Ye et al. 2012 |
| mtSSU | ||
| MS1 | 5′–CAGCAGTCAAGAATATTAGTCAATG–3′ | White et al. 1990 |
| MS2 | 3′–GCGGATTATCGAATTAAATAAC–5′ | White et al. 1990 |
Table 2.
Information on sequences used in this study.
| Species | Sample no. | GenBank accessions | References | |||||
|---|---|---|---|---|---|---|---|---|
| ITS | Partial nLSU | mtSSU | TEF1 | RPB1 | RPB2 | |||
| Tremellaaustral sp. nov. | Dai 11539 | MT445847 | – | – | MT445759 | – | – | Present study |
| T.austral sp. nov. | Wu 154 | MT445848 | MT425188 | MT483749 | MT445760 | – | MT445753 | Present study |
| T. basidiomaticola | CBS 8225 | MH712822 | MH712786 | – | – | – | – | Zhao et al. 2019 |
| T. basidiomaticola | CGMCC 2.5724T | MH712820 | MH712784 | – | – | – | – | Zhao et al. 2019 |
| T. basidiomaticola | CGMCC 2.5725 | MH712821 | MH712785 | – | – | – | – | Zhao et al. 2019 |
| T. brasiliensis | CBS 6966R | AF444429 | AF189864 | KF036694 | KF037200 | – | KF036938 | Liu et al. 2015a |
| T. brasiliensis | CBS 8212 | KY105674 | KY109886 | – | – | – | – | Vu et al. 2016 |
| T. cerebriformis | ZRL 20170101 | MH712823 | MH712787 | – | – | – | – | Zhao et al. 2019 |
| T. cerebriformis | ZRL 20170269 | MH712824 | MH712788 | – | – | – | – | Zhao et al. 2019 |
| T. cheejenii | GX 20172598 | MH712825 | MH712789 | – | – | – | – | Zhao et al. 2019 |
| T. cheejenii | GX 20172640 | MH712826 | MH712790 | – | – | – | – | Zhao et al. 2019 |
| T. dysenterica | LE 303447 | KP986509 | KP986542 | – | – | – | – | Malysheva et al. 2015 |
| T. dysenterica | VLA M 18599 | KP986531 | – | – | – | – | – | Malysheva et al. 2015 |
| T. erythrina | HMAS 255317 | MH712827 | MH712791 | – | – | – | – | Zhao et al. 2019 |
| T. erythrina | HMAS 279591 | MH712828 | MH712792 | – | – | – | – | Zhao et al. 2019 |
| T.fibulifera s.s. | SP 211759 | MT445850 | MT425190 | MT483750 | – | – | – | Present study |
| T.fibulifera s.s. | Alvarenga 471 | MT445851 | MT425191 | – | – | – | – | Present study |
| T. flava | CBS 8471R | KY105681 | KY105681 | KF036699 | KF037205 | KF036527 | KF036943 | Liu et al. 2015a |
| T. flava | CCJ 907 | AF042403 | AF042221 | – | – | – | – | Zhao et al. 2019 |
| T. fuciformis | CBS 6970R | KY105683 | AF075476 | KF036701 | KF037207 | KF036529 | – | Liu et al. 2015a |
| T. fuciformis | CCJ1080 | AF042410 | AF042228 | – | – | – | – | Malysheva et al. 2015 |
| T. globispora | CBS 6972R | AF444432 | AF189869 | KF036703 | KF037208 | KF036531 | KF036947 | Liu et al. 2015a |
| T. globispora | UBC 586 | AF042425 | AF042243 | – | – | – | – | Zhao et al. 2019 |
| T.guangxiensis sp. nov. | Wu 3 | MT445843 | MT425186 | MT483748 | MT445756 | MT445746 | MT445752 | Present study |
| T.guangxiensis sp. nov. | GX 20172028 | MH712829 | MH712793 | – | – | – | – | Zhao et al. 2019 |
| T.latispora sp. nov. | Dai 17574 | MT445852 | MT425192 | MT483751 | MT445761 | MT445750 | MT445754 | Present study |
| T.latispora sp. nov. | Dai 17568 | MT445853 | MT425193 | MT483752 | MT445762 | MT445751 | MT445755 | Present study |
| T.laurisilvae | S-F 102408(AM4) | JN053467 | JN043572 | – | – | – | – | Zhao et al. 2019 |
| T. lloydiae-candidae | VLA M 11702 | KP986536 | KP986559 | – | – | – | – | Malysheva et al. 2015 |
| T. lloydiae-candidae | VLA M 11703 | KP986559 | KP986560 | – | – | – | – | Malysheva et al. 2015 |
| T. mesenterica | CBS 6973R | AF444433 | AF444433 | KF036705 | KF037210 | KF036533 | KF036949 | Liu et al. 2015a |
| T. mesenterica | FO 24610 | AF042447 | AF042265 | – | – | – | – | Zhao et al. 2019 |
| T. “neofibulifera” | Wu 248 | MT445844 | MT425187 | – | MT445757 | MT445747 | – | Present study |
| T. “neofibulifera” | Wu 243 | MT445845 | – | – | – | MT445748 | – | Present study |
| T. “neofibulifera” | Wu 244 | MT445846 | – | – | MT445758 | MT445749 | – | Present study |
| T. “neofibulifera” | LE 303445 | KP986518 | KP986547 | – | – | – | – | Malysheva et al. 2015 |
| T. resupinata | CBS 8488T | AF042421 | AF042239 | KF036708 | KF037212 | KF036535 | KF036951 | Liu et al. 2015a |
| T. saccharicola | DMKU-SP23T | AB915385 | AB909021 | – | – | – | – | Khunnamwong et al. 2019 |
| T. saccharicola | DMKU-SP40 | AB915386 | AB909022 | – | – | – | – | Khunnamwong et al. 2019 |
| T. salmonea | GX 20172637 | MH712851 | MH712815 | – | – | – | – | Zhao et al. 2019 |
| T. samoensis | LE 303465 | KP986508 | KP986541 | – | – | – | – | Malysheva et al. 2015 |
| T. samoensis | VLA M 18603 | KP986532 | KP986555 | – | – | – | – | Malysheva et al. 2015 |
| T. shuangheensis | CBS 15561 | MK050285 | MK050285 | MK050285 | MK849087 | MK849223 | MK849362 | Li et al. 2020 |
| T.subfibulifera sp. nov. | Alvarenga 334 | MT445849 | MT425189 | – | – | – | – | Present study |
| T. taiwanensis | CBS 8479R | AF042412 | AF042230 | KF036709 | KF037213 | KF036536 | KF036952 | Liu et al. 2015a |
| T. taiwanensis | GX 20170625 | MH712854 | MH712818 | – | – | – | – | Zhao et al. 2019 |
| T. tropica | CBS 8483R | KY105697 | KY109908 | KF036710 | KF037214 | KF036537 | KF036953 | Liu et al. 2015a |
| T. tropica | CBS 8486 | KY105696 | KY109909 | – | – | – | – | Liu et al. 2015a |
| T. yokohamensis | JCM 16989 | HM222926 | HM222927 | – | – | – | – | Zhao et al. 2019 |
| T. yokohamensis | CBS 11776 | KY105698 | KY109910 | – | – | – | – | Malysheva et al. 2015 |
| Cryptococcus depauperatus | CBS 7841T | FJ534881 | FJ534911 | AJ568017 | KF037150 | KF036471 | – | Zhao et al. 2019 |
The samples used in this study are in bold.
Polymerase chain reaction (PCR) cycling schedule for ITS, mtSSU and TEF1 included an initial denaturation at 95 °C for 3 min, followed by 35 cycles at 95 °C for 40 s, 54–56 °C (ITS) and 56–58 °C (mtSSU, TEF1) for 45 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min, for RPB1 and RPB2 included an initial denaturation at 95 °C for 3 min, followed by 9 cycles at 94 °C for 45 s or 1 min , 58 °C for 45 s or 60 °C for 1 min and 72 °C for 1.5 min, then followed by 35 cycles at 95 °C for 1 min, 53 °C or 55 °C for 45 s and 72 °C for 1 min, and a final extension of 72 °C for 10 min, for partial nLSU included an initial denaturation at 94 °C for 1 min, followed by 34 cycles at 94 °C for 30 s, 50–52 °C for 1 min, 72 °C for 1.5 min, and a final extension at 72 °C for 10 min. PCR products were purified at the Beijing Genomics Institute (BGI), China or at the Plataforma Tecnológica de Genômica e Expressão Gênica do Centro de Biociências (UFPE) with the same primers.
Newly generated sequences in this study were aligned with additional related sequences downloaded from GenBank (Table 2) using MAFFT 7.0 online service with the Q-INS-i strategy (Katoh et al. 2019, http://mafft.cbrc.jp/alignment/server/). Prior to phylogenetic analysis, ambiguous sequences at the start and the end were deleted and gaps were manually adjusted to optimize the alignment using the default parameters in BioEdit (Hall 1999). Those positions deemed ambiguous to align were excluded manually. Multi-genes were concatenated as a combined file by Mesquite version 3.2. (Maddison and Maddison 2017). Phylogenetic analyses were applied to the ITS + partial nLSU dataset and the combined ITS+partial nLSU+mtSSU+TEF1+RPB1+RPB2 dataset. Sequences of Cryptococcusdepauperatus (Petch) Boekhoutet et al. were used as outgroup, which referred to Malysheva et al. (2015). The final concatenated sequence alignments were deposited in TreeBase (https://treebase.org/treebase-web/home.html; submission ID 28280 for ITS + partial nLSU dataset; submission ID 27553 for ITS + partial nLSU + mtSSU + TEF1 + RPB1 + RPB2 dataset) and the taxonomic novelties in MycoBank (http://www.MycoBank.org).
Phylogenetic constructions of Maximum likelihood (ML), Maximum parsimony (MP), and Bayesian analyses were performed in the CIPRES Science Gateway portal Version 3.3 (Miller et al. 2012) using tool of RAxML-HPC BlackBox 8.2.6, PAUP on XSEDE (4.a165) and Mrbayes on XSEDE 3.2.6 respectively. All characters were equally weighted, and gaps were treated as missing data. Trees were inferred using heuristic search option with TBR branch swapping and 1000 random sequence additions. MrModeltest 2.3 (Posada and Crandall 1998; Nylander 2004) was used to determine the best-fit evolution model for both datasets for Bayesian analyses using MrBayes3.1.2 (Ronquist and Huelsenbeck 2003). Four Markov chains were run for two runs from random starting trees for 3 million generations (ITS + nLSU) and for 5 million generations (ITS + partial nLSU + mtSSU + TEF1 + RPB1 + RPB2) until the split deviation frequency value < 0.01, and trees were sampled every 100 generation. The first quarter generations were discarded as burn-in. A majority rule consensus tree of all remaining trees was calculated.
Phylogenetic trees were viewed by FigTree v. 1.4.2 (Rambaut 2012) and edited by Adobe Illustrator CS6 (Guide 2012). Branches that received bootstrap support for Maximum parsimony (BP), Maximum likelihood (BS) and Bayesian posterior probabilities (BPP) greater than or equal to 50% (BP/BS) and 0.95 (BPP) were considered as significantly supported, respectively.
Results
Phylogeny
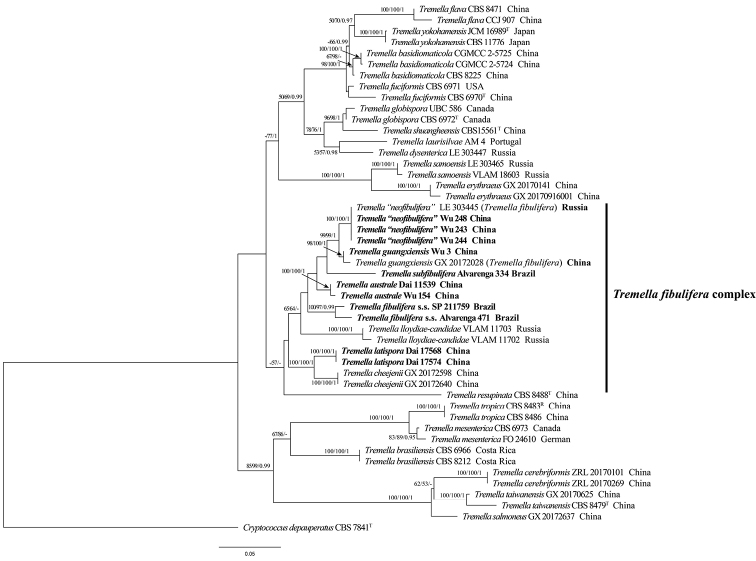
The ITS + partial nLSU dataset included 50 fungal specimens representing 27 species. The dataset has an aligned length of 2282 total characters including gaps, of which 1777 characters are constant, 128 variable characters are parsimony-uninformative, and 377 are parsimony-informative. MP analysis yielded four equally parsimonious trees (TL = 1394, CI = 0.529, RI = 0.792, RC = 0.419, HI = 0.471). The best model for the ITS + partial nLSU dataset estimated and applied in the BI analysis was GTR. BI and ML analyses generated similar topologies as MP analysis, with an average standard deviation of split frequencies = 0.002648 (BI). The best tree obtained from the ML analysis with bootstrap values for BP, BS and BPP is shown in Fig. 1. The phylogeny shows that eight species are clustered into the T.fibulifera complex and four new species form four independent lineages with robust support.
Figure 1.
The Maximum likelihood tree showing phylogenetic relationship of species in Tremella s.s. based on the ITS + partial nLSU dataset. Bootstrap support values for MP and ML greater than 50% and BI greater than 0.95 are given at each node respectively. The samples used in this study are in bold.
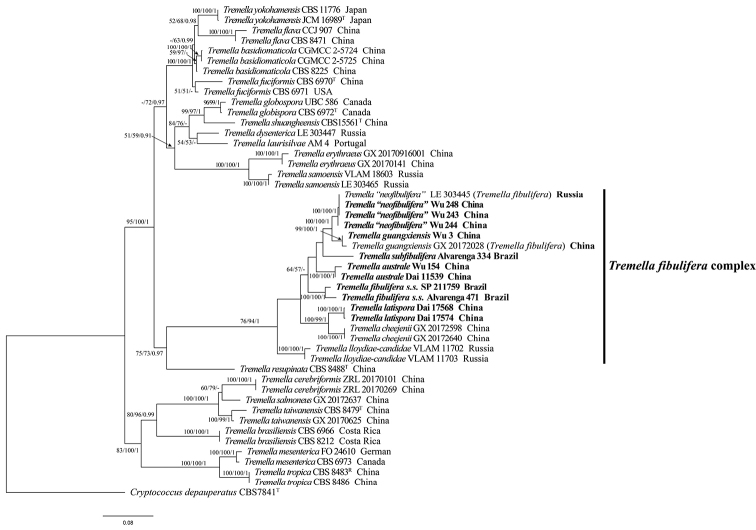
The combined dataset of ITS + partial nLSU + mtSSU + TEF1 + RPB1 + RPB2 has an aligned length of 5113 total characters including gaps, of which 3332 characters are constant, 383 variable characters are parsimony-uninformative, and 1398 are parsimony-informative. MP analysis yielded two equally parsimonious trees (TL = 4519, CI = 0.607, RI = 0.730, RC = 0.443, HI = 0.393). The best model for the combined dataset estimated and applied in the BI analysis was GTR+I+G. BI analysis generated similar topology to MP and ML analysis, with an average standard deviation of split frequencies = 0.008566 (BI). The best tree obtained from the ML analysis with bootstrap values for BP, BS and BPP is shown in Fig. 2. The phylogeny results in similar topology to the phylogeny based on the ITS + partial nLSU sequences, which supports four new species separated from T.fibulifera s.s. and T. “neofibulifera”.
Figure 2.
The Maximum likelihood tree showing phylogenetic relationship of species in Tremella s.s. based on the combined ITS + partial nLSU + mtSSU + TEF1 + RPB1 + RPB2 dataset. Bootstrap support values for MP and ML greater than 50% and BI greater than 0.95 are given at each node respectively. The samples used in this study are in bold.
Taxonomy
Tremella fibulifera
Möller, Botanische Mittheilungen aus den Tropen 8: 170 (1895)
084FF974-8F91-5300-BE30-11470F2DC526
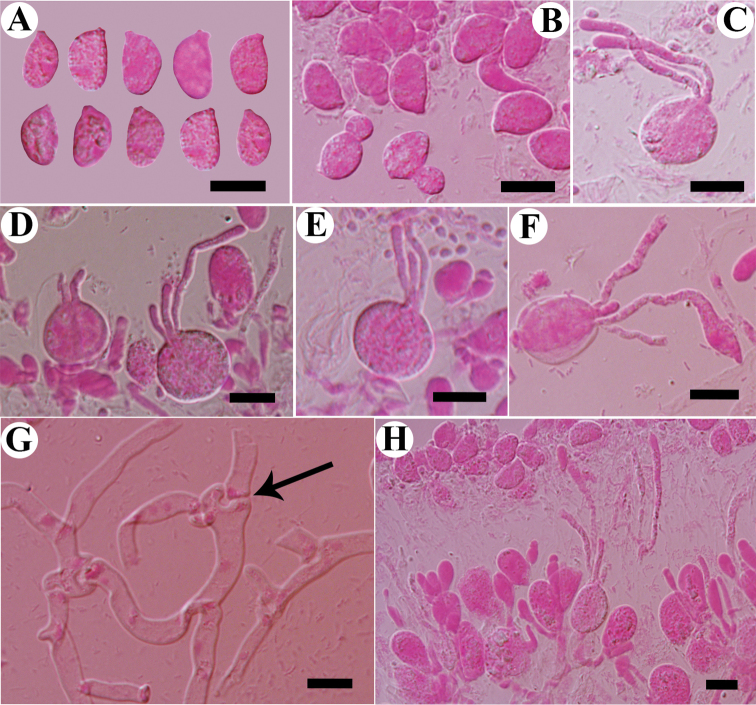
Figure 3.
Basidioma ATremellafibulifera (Alvarenga 471) BT.australe (Wu 154) CT.guangxiensis (Wu 3) DT.latispora (Dai 17568) ET. “neofibulifera” (Wu 244) FT.subfibulifera (Alvarenga 334). Scale bars: 1 cm (A–F).
Figure 4.
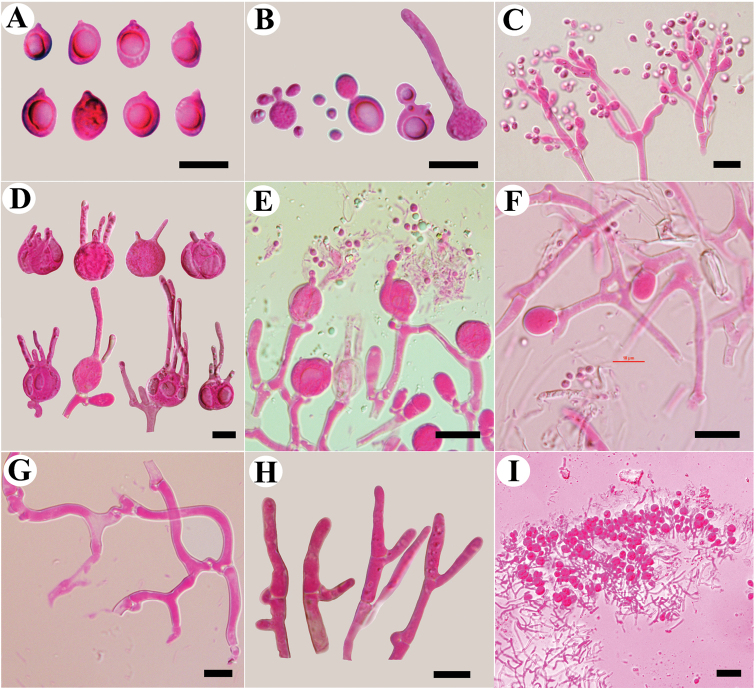
Microscopic structures of Tremellafibulifera s.s. (SP 211759) A basidiospores B germination tubes of basidiospores and secondary spores C–F basidia at different stages G hyphae with clamp connections and clamp complexes; H a section of hymenium. Scale bars: 10 μm (A–H).
Basidioma.
Sessile, when fresh gelatinous, pale whitish, lobed to irregularly cerebriform, becoming pale yellowish when dry, 0.5–2.5 cm in diameter, broadly attached to substratum.
Internal features.
Hyphae hyaline, smooth, thin- to thick-walled, 2.0–5.0 µm in diameter, branched, interwoven, with abundant clamp connections and medallion clamp connections (clamp complexes), thick-walled hyphae usually present near to base of basidioma; hyphidia hyaline, smooth, thin-walled, branched; swollen cells, vesicles and haustoria absent; mature basidia thin-walled, globose to subglobose, with a basal clamp connection, 13.0–18.0(–22.0) × 9.0–16.0 μm, L = 15.7 µm, W = 14.8 µm, Q = 1.06 (n = 30/1), sometimes their width greater than length, usually longitudinally septate, rarely obliquely septate, 2–4-celled, with obvious oil drops; sterigmata up to 100 μm long, 1.5–2.0 μm in diameter, slightly protuberant at apex; probasidia thin-walled, subglobose to ellipsoid, mostly proliferating directly from basidial clamps; basidiospores hyaline, thin-walled, mostly ellipsoid to slightly ovoid, apiculate, with oil drops, 7.0–10.0 × 6.0–7.0 μm, L = 8.4 µm, W = 6.5 µm, Q = 1.29–1.40 (n = 60/2), germinating by germ tubes or secondary spores; conidia occasionally present in cluster, originating from conidiophores, hyaline, thin-walled, ellipsoid to subglobose, 2.0–3.0 × 1.0–2.5 μm.
Specimens examined.
Brazil Rondônia, Municipality of Jaru, in mixed forest near the airport, 9°40'S, 61°50'W, on wood, associated with old pyrenomycete stromata and litter, 10 October 1986, M. Capelari & R. Maziero 944 (SP211759, duplicate BJFC028110); Pernambuco, Recife, Jardim Botânico do Recife, on angiosperm wood, 16 May 2017, R. L. M. Alvarenga 471 (URM).
Notes.
Tremellafibulifera was probably a species complex including T.olens originally from Australia and T.neofibulifera originally from Japan because they shared cerebriform whitish basidioma and abundant clamp complexes (Möller 1895; Bandoni and Oberwinkler 1983; Malysheva et al. 2015). Two specimens (SP211759, Alvarenga 471) from Brazil bearing the common feature of the complex formed a distinct lineage in our phylogenies (Figs 1, 2). Morphologically, the two specimens agree well with T.fibulifera except for the presence of conidia (Table 3). However, conidia are unstable in T.fibulifera. Möller (1895) described the anamorph of T.fibulifera, but the conidia were not observed when Bandoni and Oberwinkler (1983) re-described T.fibulifera based on the type designated by Möller (1895). Furthermore, T.fibulifera was originally described from Blumenau, Brazil, which is very close to the location of SP211759, Rondônia, Brazil. Therefore, we treat Alvarenga 471 and SP211759 as the representatives of T.fibulifera s.s. In addition, T.fibulifera s.s. are different from T.subfibulifera and T.australe by 8.51%, 9.87% sequence differences in the ITS sequences and 2.10%, 1.57% in the partial nLSU sequences respectively.
Table 3.
A morphological comparison of taxa in the Tremellafibulifera complex.
| Taxa | Basidia (µm) | Basidiospores (µm) | Conidia (µm) | Hyphidia | Distribution | Reference |
|---|---|---|---|---|---|---|
| T. fibulifera | 12.0–16.0 | 7.0–10.0 | 3.5 | Unknown | Brazil | Möller 1895 |
| T. fibulifera | 15.0–18.0 × 9.0–13.0 | 8.0–9.0 × 5.0–8.0 | Not observed | Unknown | Brazil | Bandoni and Oberwinkler 1983 |
| T.fibulifera s.s. | 13.0–18.0 × 9.0–16.0 | 7.0–10.0 × 6.0–7.0 | 2.0–3.0 × 1.0–2.5 | Branched | Brazil | Present study |
| T. australe | 14.0–19.0 × 13.0–17.0 | 8.0–10.0 × 6–8.0 | Absent | Present | China | Present study |
| T. cheejenii | 12.0–17.0 × 13.0–18.0 | 5.0–10.0 × 4.5–8.0 | 2.2–4.0 × 1.8–3.0 | Branched | China | Zhao et al. 2019 |
| T. guangxiensis | 14.0–17.0 × 14.0–16.0 | 8.0–9.5 × 6.0–7.5 | 2.0–3.2 × 1.8–3.0 | Branched | China | Present study |
| T. latispora | 17.2–24.0 × 17.0–23.0 | 10.1–11.8 × 9.9–11.4 | 2.8–3.6 × 1.8–3.0 | Present | China | Present study |
| T. lloydiae-candidae | 14.0–20.0 × 13.0–16.0 | 7.5–10 | Absent | Unknown | Japan, Russia | Malysheva et al. 2015 |
| T. olens | Unknown | 12.7–14.5 | Absent | Unknown | Australia | Hooker 1860 |
| T. neofibulifera | 13.2–15.5 × 9–10 | 5.5–8.5 × 4.5–5.5 | Absent | Unknown | Japan | Kobayasi 1939 |
| T. “neofibulifera” | 14.0–16.0 × 13.0–17.0 | 8.0–10.0 × 6.0–8.0 | Absent | Parallel | China, Russia | Present study |
| T. subfibulifera | 14.4–20.3 × 12.8–16.3 | 5.4–9.8 × 4.2–6.0 | 2.0–3.0 × 0.5–1.0 | Absent | Brazil | Present study |
Tremella australe
F. Wu, L.F. Fan & Y.C. Dai sp. nov.
E330B6AE-51B3-590D-87BE-B946D880128D
839825
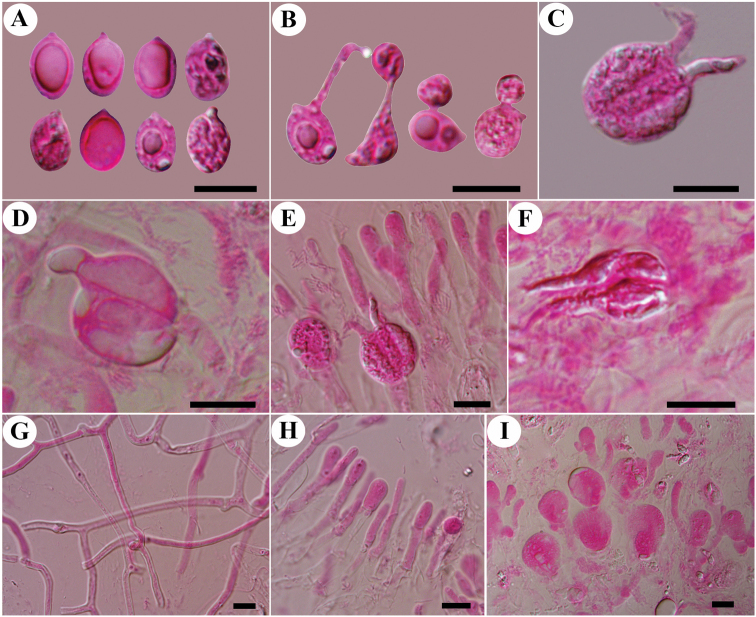
Figure 5.
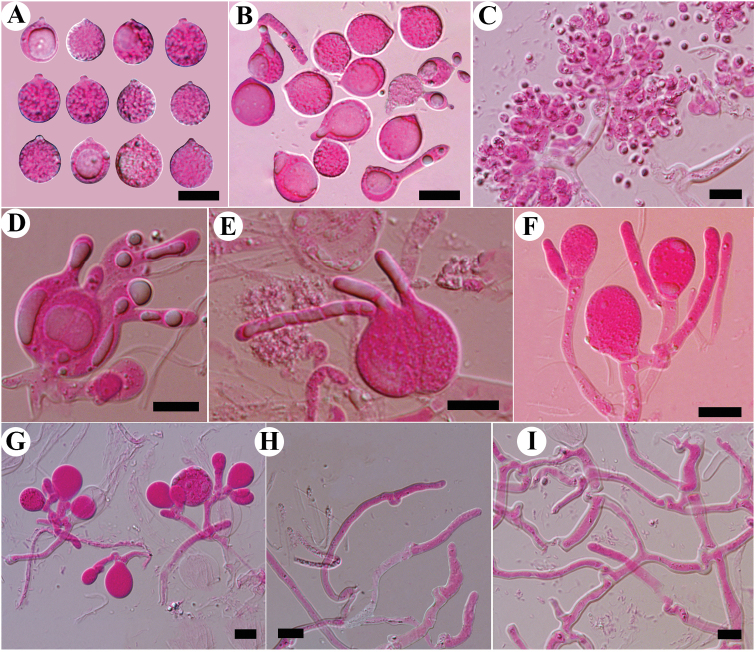
Microscopic structures of Tremellaaustrale (Wu 154) A basidiospores B germination tubes of basidiospores and secondary spores C–F basidia at different stages G hyphae with clamp connections and clamp complexes H hyphidia I a section of hymenium. Scale bars: 10 μm (A–I).
Holotype.
China Yunnan, Ruili, on fallen angiosperm branch, 23 April 2018, F. Wu 154 (BJFC028064).
Etymology.
Refers to the distribution of this species in South Asia.
Basidioma.
Sessile, when fresh soft gelatinous, creamy-white to beige, translucent, cerebriform, with thick and undulate lobes, up to 4.0 cm long, 2.0 cm broad and 2.0 cm high from base, distinctly shrinking into a film and becoming pale yellow when dry, broadly attached to substratum.
Internal features.
Hyphae hyaline, smooth, thin- to slightly thick-walled, 1.5–6.0 µm in diameter, branched, interwoven, with abundant clamp connections, clamp complexes and anastomoses, slightly thick-walled hyphae usually present near to base of basidioma and sometimes swollen up to 8.5 μm; hyphidia hyaline, smooth, thin-walled, usually derived from the same hyphae with basidia; swollen cells, vesicles and haustoria absent; mature basidia thin-walled, globose to subglobose, with a basal clamp connection, 14.0–19.0 × 13.0–17.0(–18.0) μm, L = 16.3 µm, W = 15.8 µm, Q = 1.03 (n = 30/1), sometimes their width greater than length, usually longitudinally septate, 2–4-celled, with obvious oil drops; sterigmata up to 20 μm long, 1.0–2.5 in diameter, slightly protuberant at apex; probasidia thin-walled, globose to subglobose, mostly proliferating directly from basidial clamps; basidiospores hyaline, thin-walled, broadly ellipsoid to ellipsoid, apiculate, with oil drops, 8.0–10.0 × 6.0–8.0 μm, L = 8.6 µm, W = 7.3 µm, Q = 1.18–1.28 (n = 60/2), germinating by germ tubes or secondary spores; conidia absent.
Additional specimen examined.
(paratype) China Taiwan, Yilan, Linmei Road, on fallen angiosperm branch, 20 June 2009, Y.C. Dai 11539 (BJFC007408).
Notes.
Tremellaaustrale formed an independent lineage with high support in our phylogenies (Figs 1, 2). The species is easily confused with T.guangxiensis by sharing whitish, translucent cerebriform basidioma and similar basidia and basidiospores, but T.guangxiensis has branched hyphidia and umbelliform conidiophores. Besides, T.australe are different from T.subfibulifera, T.guangxiensis and T. “neofibulifera” by 7.82%, 5.94% and 6.82% sequence differences in the ITS sequences and 2.13%, 3.43% and 1.25% in the partial nLSU sequences respectively.
Tremella guangxiensis
F. Wu, L.F. Fan & Y. C. Dai sp. nov.
BD36A161-CA43-56F5-A585-02532CFB2954
839827
Figure 6.
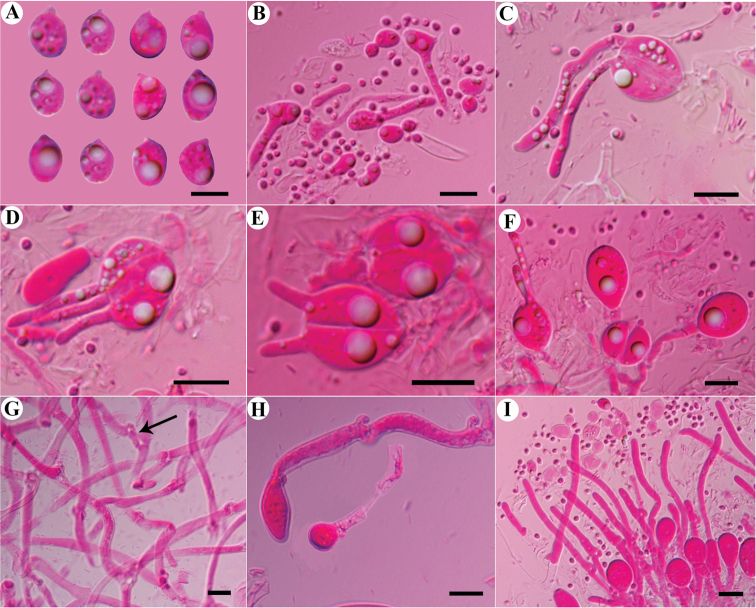
Microscopic structures of Tremellaguangxiensis (Wu 3) A basidiospores B germination tubes of basidiospores and secondary spores C conidia and conidiophores D basidia at different stages E, F probasidia G hyphae with clamp connections and clamp complexes H hyphidia I a section of hymenium. Scale bars: 10 μm (A–H); 20 μm (I).
Holotype.
China. Guangxi, Jinxiu, Dayao Mountain, on angiosperm tree, 15 July 2017, F. Wu 3 (BJFC026009).
Etymology.
Refers to the distribution of the species in Guangxi, China.
Basidioma.
Sessile, when fresh soft gelatinous, milky to creamy-white, translucent, pustulate to irregularly cerebriform, with thick and undulate lobes, up to 4.0 cm long, 4.0 cm broad and 1.5 cm high from base, distinctly shrinking into a film and becoming lightly yellowish when dry, broadly attached to substratum.
Internal features.
Hyphae hyaline, smooth, thin- to slightly thick-walled, 2.0–6.0 µm in diameter, branched, interwoven, with abundant clamp connections, clamp complexes and anastomoses, slightly thick-walled hyphae usually present near to base of basidioma and sometimes swollen up to 9.0 μm; hyphidia hyaline, smooth, thin-walled, branched; swollen cells present, hyaline, smooth and various in the shape, sometimes slightly concave; vesicles and haustoria absent; mature basidia thin-walled, globose to subglobose, with a basal clamp connection, 14.0–17.0 × (13.6–)14.0–16.0(–17.0) μm, L = 15.9 µm, W = 14.8 µm, Q = 1.07 (n = 30/1), sometimes their width greater than length, usually longitudinally septate, rarely obliquely septate, 2–4-celled, with obvious oil drops; sterigmata up to 60 μm long, 1.5–2.0 in diameter, slightly protuberant at apex; probasidia thin-walled, clavate to ellipsoid, proliferating from terminal hyphae; basidiospores hyaline, thin-walled, broadly ellipsoid to slightly ovoid, apiculate, with oil drops, (7.5–)8.0–9.5 × 6.0–7.5(–8.0) μm, L = 8.7 µm, W = 6.8 µm, Q = 1.28 (n = 30/1), germinating by germ tubes or secondary spores; conidia massively present, originating from umbelliform conidiophores, hyaline, thin-walled, ovoid to broadly ellipsoid or fusiform to cylindrical, 2.0–3.2 × 1.8–3.0 μm.
Notes.
Tremellaguangxiensis is closely related T. “neofibulifera” in our phylogenies (Figs 1, 2). The most distinctive characteristic of the species is branched hyphidia and umbelliform conidiophores, but T. “neofibulifera” has parallel hyphidia and lacks of conidia. In addition, T.guangxiensis are different from T.australe and T. “neofibulifera” by 6.35% and 5.09% sequence differences in the ITS sequences and 3.39% and 1.97% in the partial nLSU sequences respectively.
Tremella latispora
F. Wu, L.F. Fan & Y. C. Dai sp. nov.
AD8B8676-A659-5FB1-946D-E77E706D13AC
839828
Figure 7.
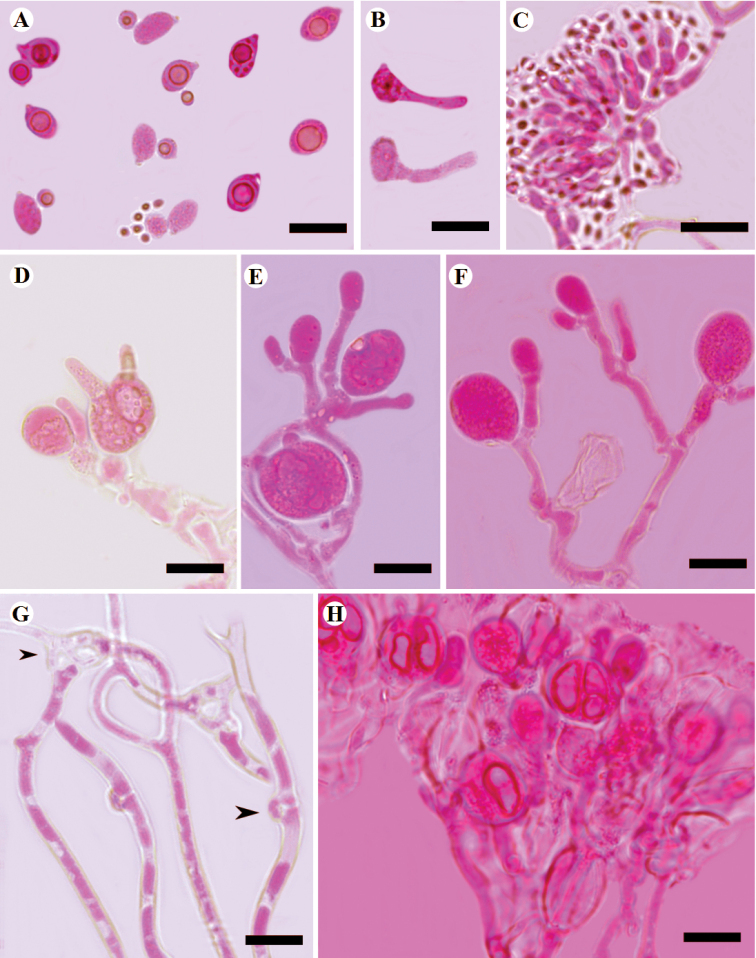
Microscopic structures of Tremellalatispora (Dai 17568) A basidiospores B germination tubes of basidiospores and secondary spores C conidia and conidiophores D, E basidia F, G probasidia H, I hyphae with clamp connections and clamp complexes. Scale bars: 10 μm (A–I).
Holotype.
China. Yunnan, Xinping, Shimenxia Park, on stump of Lithocarpus, 16 June 2017, Y.C. Dai 17574 (BJFC025106).
Etymology.
Refers to the species having wide basidiospores.
Basidioma.
Sessile, when fresh soft gelatinous, creamy-white to lvory, translucent, pustulate to irregularly cerebriform, with thick and undulate lobes, up to 4.0 cm long, 2.0 cm broad and 1.0 cm high from base, distinctly shrinking into a film and becoming whitish to pale yellow when dry, broadly attached to substratum.
Internal features.
Hyphae hyaline, smooth, thin- to thick-walled, 1.5–6.0 µm in diameter, branched, interwoven, with abundant clamp connections, clamp complexes and anastomoses, thick-walled hyphae usually present near to base of basidioma and sometimes swollen up to 7.5 μm; hyphidia hyaline, smooth, thin-walled, usually derived from the same hyphae with basidia; swollen cells, vesicles and haustoria absent; mature basidia thin-walled, globose to subglobose, with a basal clamp connection, 17.2–24.0(–27.0) × 17.0–23.0(–24.3) μm, L = 19.5 µm, W = 20.8 µm, Q = 0.94 (n = 30/1), commonly their width greater than length, usually longitudinally septate, occasionally obliquely septate, 2–4-celled, with obvious oil drops; sterigmata up to 60 μm long, 1.5–2.0 in diameter, slightly protuberant at apex; probasidia thin-walled, ellipsoid to subglobose, proliferating from terminal hyphae; basidiospores hyaline, thin-walled, globose to subglobose, apiculate, with oil drops, (9.0–)10.1–11.8(–12.0) × (9.6–)9.9–11.4(–11.7) μm, L = 11.0 µm, W = 10.7 µm, Q = 1.03 (n = 30/1), germination by germ tubes or secondary spores; conidia massively present, originating from umbelliform conidiophores, hyaline, thin-walled, ovoid to oblong or globose to subglobose, 2.8–3.6 × 1.8–3.0 μm.
Additional specimen examined.
(paratype) China Yunnan, Xinping, Shimenxia Park, on stump of Lithocarpus, 16 June 2017, Y.C. Dai 17568 (BJFC025100).
Notes.
Phylogenetically, Tremellalatispora formed a distinct lineage closely related to T.cheejenii (Figs 1, 2). Morphologically, the species has significantly larger basidia and basidiospores than T.cheejenii or other similar species (Table 3), and it has globose to subglobose basidiospores rather than more or less ellipsoid basidiospores in other species. And T.latispora are different from T.cheejenii and T.fibulifera s.s. by 4.63% and 5.09% sequence differences in the ITS sequences and 3.39% and 2.95% in the partial nLSU sequences respectively.
T. neofibulifera
Kobayasi, Scientific Report, Tokyo Bunrika Daigaku, Section 4: 15 (1939)
058D9F07-1D89-5218-994C-048A93A16083
Figure 8.
Microscopic structures of Tremella “neofibulifera” (Wu 248) A basidiospores B germination tubes of basidiospores and secondary spores C–E basidia at different stages F probasidia G hyphae with clamp connections and clamp complexes H vesicles I parallel hyphidia in hymenium. Scale bars: 10 μm (A–I).
Basidioma.
Sessile, when fresh soft gelatinous, creamy-white to pale yellowish, irregularly cerebriform or slightly foliose, with undulate lobes, up to 4.5 cm long, 2.0 cm broad and 2.5 cm high from base, becoming firmly gelatinous and invisible yellowish when dry, broadly attached to substratum.
Internal features.
Hyphae hyaline, smooth thin- to slightly thick-walled, 2.0–6.0 µm in diameter, branched, interwoven, with abundant clamp connections, clamp complexes and anastomoses, slightly thick-walled hyphae usually present near to base of basidioma, sometimes swollen up to 8.5 μm; hyphidia hyaline, smooth, thin-walled, arranged in cluster, usually parallel; vesicles infrequent, thick-walled; swollen cells and haustoria absent; mature basidia thin-walled, ovoid to subglobose, with a basal clamp connection, 14.0–16.0 × 13.0–17.0 μm, L = 14.9 µm, W = 14.8 µm, Q = 1.01 (n = 30/1), sometimes their width greater than length, usually longitudinally septate, rarely obliquely septate, 2–4-celled, with obvious oil drops; sterigmata up to 70 μm long, 1.5–2.0 in diameter, slightly protuberant at apex; probasidia thin-walled, ellipsoid to subglobose, usually proliferating from terminal hyphae; basidiospores hyaline, thin-walled, ellipsoid to broadly ellipsoid, apiculate, with oil drops, 8.0–10.0 × 6.0–8.0 μm, L = 8.9 µm, W = 6.5 µm, Q = 1.37 (n = 30/1), germination by germ tubes or secondary spores; conidia absent.
Specimens examined.
China Jilin, Helong, Quanshuidong Forest Farm, on stump of Quercus, 15 July 2017, F. Wu 243 (BJFC031046); F. Wu 244 (BJFC031047); F. Wu 248 (BJFC031051).
Notes.
Three specimens listed above from Northeast China together with LE303445 from Far East of Russia formed a distinct lineage closely related to T.guangxiensis in our phylogenies (Figs 1, 2). T.neofibulifera was originally described from Japan (Kobayasi 1939), and our studied East Asian samples have similar morphology to T.neofibulifera except bigger basidiospores (Table 3). We fail to loan the type of T.neofibulifera, and for the time being we treat our studied East Asia samples as T. “neofibulifera”. The current T. “neofibulifera” differs from other similar species of the Tremellafibulifera complex by the parallel hyphidia and the presence of vesicles. In addition, T. “neofibulifera” are different from T.guangxiensis, T.australe, T.subfibulifera and T.fibulifera s.s. by 3.15%, 5.25%, 7.14%, and 8.19% sequence differences in the ITS sequences and 2.04%, 1.32%, 3.18%, and 2.41% in the partial nLSU sequences respectively.
Tremella subfibulifera
Alvarenga, F. Wu, L.F. Fan & Y.C. Dai sp. nov.
6385C3CD-A50F-5D7A-9406-A5A84F19EF9B
839829
Figure 9.
Microscopic structures of Tremellasubfibulifera (Alvarenga 334) A basidiospores and secondary spores B germination tubes of basidiospores C conidia and conidiophores D–F basidia and probasidia G hyphae with clamp connections and clamp complexes H a section of hymenium. Scale bars: 10 μm (A–H).
Holotype.
Brazil. Pernambuco, Recife, Jardim Botânico do Recife, on angiosperm wood, 17 June 2016, R. L. M. Alvarenga 334 (URM).
Etymology.
Refers to the species being similar to Tremellafibulifera.
Basidioma.
Sessile, when fresh gelatinous, pale white, foliose to irregularly cerebriform, with undulate lobes, up to 3.0 cm long, 2.0 cm broad and 1.0 cm high from base, becoming firmly gelatinous and pale yellowish when dry, broadly attached to substratum.
Internal features.
Hyphae hyaline, smooth, slightly thick-walled, 2.0–4.0 μm in diameter, branched, interwoven, with abundant clamp connections, clamp complexes and anastomoses; hyphidia, swollen cells, vesicles and haustoria absent; mature basidia thin-walled, subglobose to broadly ellipsoid, with a basal clamp connection, (14.0–)14.4–20.3(–21.0) × (9.0–)12.8–16.3(–17.8) μm, L = 17.63 µm, W = 15.05 µm, Q = 1.17 (n = 30/1), sometimes their width greater than length, usually longitudinally or obliquely septate, 2–4-celled, with obvious oil drops; mature sterigmata often collapsed, juvenile sterigmata up to 15.0 µm long, 2.0–4.0 µm in diameter, slightly protuberant at apex; probasidia thin-walled, clavate to ellipsoid, guttulate, proliferating from terminal hyphae; basidiospores hyaline, thin-walled, ellipsoid apiculate, with oil drops, (5.0–)5.4–9.8(–10.0) × (4.0–)4.2–6.0(–6.4) μm, L = 8.0 µm, W = 5.3 µm, Q = 1.50 (n = 30/1); conidia massively present, originating from umbelliform conidiophores, hyaline, thin-walled, variously shaped, ellipsoid, fusiform to cylindrical, 2.0–3.0 × 0.5–1.0 μm.
Notes.
Tremellasubfibulifera nested in the clade of the T.fibulifera complex, and formed an independent lineage. It resembles T.fibulifera s.s., but T.fibulifera s.s. has larger basidiospores (7.0–10.0 × 6.0–7.0 μm vs. 5.4–9.8 × 4.2–6.0 μm) and the presence of branched hyphidia (Table 3). In addition, T.subfibulifera are different from T.australe and T.fibulifera s.s. by 6.19% and 7.85% sequence differences in the ITS sequences and 2.23% and 2.10% in the partial nLSU sequences respectively.
Discussion
Tremellafibulifera was originally described from Blumenau of Brazil (Möller 1895); later two similar species, T.olens and T.neofibulifera, were respectively described from Tasmania of Australia and Simotuke of Japan (Hooker 1860; Kobayasi 1939). The Russian Far East specimen LE303445 was identified as T.fibulifera by Malysheva et al. (2015). Our results demonstrated the Northeastern Chinese specimens and Russian Far East specimen formed an independent lineage, and this lineage is distantly related to the lineage formed by two Brazilian specimens, SP 211759 and Alvarenga 471 (Figs 1, 2). The location of SP 211759 is near to the type locality of T.fibulifera. So, we treat SP 211759 and Alvarenga 471 as representatives of T.fibulifera s.s. Molecular data are not available from type or type locality specimens of T.neofibulifera. Neither is its type re-examined, but the Northeastern Chinese specimens have more or less similar morphology as the description of T.neofibulifera, so we temporarily treat Northeast Chinese specimens and Russian Far East specimen as T. “neofibulifera”.
The Southern Chinese specimen GX20172028 was also identified as Tremellafibulifera by Zhao et al. (2019), but it clustered with another Southern Chinese specimen Wu 3 into a distinct lineage which is closely related to T. “neofibulifera” (Figs 1, 2). T.guangxiensis is different from T. “neofibulifera” by 5.09% sequence differences in the ITS sequences and 1.97% in the partial nLSU sequences respectively. In addition, the Southern Chinese specimens have translucent basidioma, branched hyphidia and umbelliform conidiophores, and they are readily distinguished from T. “neofibulifera”. So, these two specimens are identified as a new species T.guangxiensis.
Seven species, Tremellafibulifera, T.olens, T. “neofibulifera”, T.guangxiensis, T.australe, T.latispora and T.subfibulifera have cerebriform whitish basidioma and abundant clamp complexes, and they nested in the same clade. So, we treat these seven species as members of the T.fibulifera complex.
Tremellalloydiae-candidae Wojewoda and T.cheejenii Xin Zhan Liu & F.Y. Bai also have whitish basidioma and similar micro-morphology with T.fibulifera, but clamp complexes were not observed (Malysheva et al. 2015; Zhao et a. 2019), and we did not examine their types. Because these two species are nested in the same clade as other species of the T.fibulifera complex with robust support in our phylogenies (Figs 1, 2), we treat them as members of the T.fibulifera complex, too.
Currently, more than 30 morphological characteristics are applied for identification species of Tremella s.s. (Chen 1998; Zhao et al. 2019), and some features including basidioma color and basidia shape are variable at different stages. The shape and size of basidiospores are relatively stable characteristics for each species, but they are very similar among some species in the T.fibulifera complex; that is why several taxa were previously treated as T.fibulifera s.l. (Malysheva et al. 2015; Zhao et al. 2019). Consequently, combined morphology and molecular evidence are essential to distinguish species within the complex, and ITS + partial nLSU dataset are selected for species delimitation.
Key to the whitish species in Tremella s. s.
| 1 | Basidiospores > 10 μm long | 2 |
| – | Basidiospores < 10 μm long | 5 |
| 2 | Basidioma resupinate | T. resupinata |
| – | Basidioma pustulate to irregularly cerebriform or foliose | 3 |
| 3 | Basidiospores > 17 μm long | T. cerebriformis |
| – | Basidiospores < 17 μm long | 4 |
| 4 | Basidiospores > 12 μm long | T. olens |
| – | Basidiospores < 12 μm long | T. latispora |
| 5 | Basidia with stalks | 6 |
| – | Basidia without stalks | 8 |
| 6 | Basidia < 13 μm wide | T. yakohamensis |
| – | Basidia > 13 μm wide | 7 |
| 7 | Basidiospores mostly broader than long | T. globispora |
| – | Basidiospores mostly longer than broad | T. cheejenii |
| 8 | Basidia with sterigmata shorter than 35 μm | 9 |
| – | Basidia with sterigmata longer than 35 μm | 11 |
| 9 | Basidiospores < 6 μm wide | T. subfibulifera |
| – | Basidiospores > 6 μm wide | 10 |
| 10 | Hyphae with clamp complexes and anastomoses | T. australe |
| – | Hyphae without clamp complexes and anastomoses | T. lloydiae-candidae |
| 11 | Basidioma filamentous lobes, conjunctive as a ball | T. hainanensis |
| – | Basidioma pustulate to irregularly cerebriform or foliose | 12 |
| 12 | Basidiospores < 6 μm wide | T. fuciformis |
| – | Basidiospores > 6 μm wide | 13 |
| 13 | Hyphidia parallel; conidia absent | T. “neofibulifera ” |
| – | Hyphidia branched; conidia present | 14 |
| 14 | Basidioma pustulate to irregularly cerebriform; basidia with sterigmata up to 60 μm; conidia originating from umbelliform conidiophores | T. guangxiensis |
| – | Basidioma lobed to irregularly cerebriform; basidia with sterigmata up to 100 μm; conidia not originating from umbelliform conidiophores | T.fibulifera s.s. |
Supplementary Material
Acknowledgements
We sincerely thank Drs. Li-Wei Zhou (Shenyang, China), Jun-Zhi Qiu (Fujian, China), Bao-Kai Cui (Beijing, China), Shuang-Hui He (Beijing, China), Jun-Liang Zhou (Yunnan, China), Hai-Xia Ma (Hainan, China) and Adriana de Mello Gugliotta (Brazil) for collecting and providing specimens. The research was financed by the National Natural Science Foundation of China (Project Nos. 31701978, U1802231), CNPq (PQ 1D, 307601/2015-3, 302941/2019-3; ICMBio 421241/2017-9) and FACEPE (APQ 0003-2.03/18).
Citation
Fan L-F, Alvarenga RLM, Gibertoni TB, Wu F , Dai Y-C (2021) Four new species in the Tremella fibulifera complex (Tremellales, Basidiomycota). MycoKeys 82: 33–56. https://doi.org/10.3897/mycokeys.82.63241
Contributor Information
Fang Wu, Email: fangwubjfu2014@yahoo.com.
Yu-Cheng Dai, Email: yuchengd@yahoo.com.
References
- Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KWT, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R, Ghobad-Nejhad M, Niskanen T, Thambugala KM, Voigt K, Zhao RL, Li GJ, Doilom M, Boonmee S, Yang ZL, Cai Q, Cui YY, Bahkali AH, Chen J, Cui BK, Chen JJ, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, Hashimoto A, Hongsanan S, Jones EBG, Larsson E, Li WJ, Li QR, Liu JK, Luo ZL, Maharachchikumbura SSN, Mapook A, McKenzie EHC, Norphanphoun C, Konta S, Pang KL, Perera RH, Phookamsak R, Phukhamsakda C, Pinruan U, Randrianjohany E, Singtripop C, Tanaka K, Tian CM, Tibpromma S, Abdel-Wahab MA, Wanasinghe DN, Wijayawardene NN, Zhang JF, Zhang H, Abdel-Aziz FA, Wedin M, Westberg M, Ammirati JF, Bulgakov TS, Lima DX, Callaghan TM, Callac P, Chang CH, Coca LF, Dal-Forno M, Dollhofer V, Fliegerová K, Greiner K, Griffith GW, Ho HM, Hofstetter V, Jeewon R, Kang JC, Wen TC, Kirk PM, Kytövuori I, Lawrey JD, Xing J, Li H, Liu ZY, Liu XZ, Liimatainen K, Lumbsch HT, Matsumura M, Moncada B, Nuankaew S, Parnmen S, de Azevedo Santiago ALCM, Sommai S, Song Y, de Souza CAF, de Souza-Motta CM, Su HY, Suetrong S, Wang Y, Wei SF, Wen TC, Yuan HS, Zhou LW, Réblová M, Fournier J, Camporesi E, Luangsa-ard JJ, Tasanathai K, Khonsanit A, Thanakitpipattana D, Somrithipol S, Diederich P, Millanes AM, Common RS, Stadler M, Yan JY, Li XH, Lee HW, Nguyen TTT, Lee HB, Battistin E, Marsico O, Vizzini A, Vila J, Ercole E, Eberhardt U, Simonini G, Wen HA, Chen XH, Miettinen O, Spirin V. (2015) Fungal diversity notes 111–252 – Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75: 27–274. 10.1007/s13225-015-0346-5 [DOI] [Google Scholar]
- Bandoni RJ. (1958) Some tremellaceous fungi in the C.G. Lloyd collection. Lloydia 21: 137–151. [Google Scholar]
- Bandoni RJ. (1987) Taxonomic review of the Tremellales. Studies in Mycology 30: 87–110. [Google Scholar]
- Bandoni RJ, Oberwinkler F. (1983) On some species of Tremella described by Alfred Möller. Mycologia 75: 854–863. 10.1080/00275514.1983.12023760 [DOI] [Google Scholar]
- Boekhout T, Fonseca A, Sampaio JP, Bandoni RJ, Kwon-Chung KJ. (2011) Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In: Kurtzman CP, Fell JW, Boekhout T (Eds) The yeasts: a taxonomic study, 1339–1372. 10.1016/B978-0-444-52149-1.00100-2 [DOI]
- Chen CJ. (1998) Morphological and molecular studies in the genus Tremella. Bibliotheca Mycologica 174: 1–114. [Google Scholar]
- Diederich P. (1996) The lichenicolous heterobasidiomycetes. Bibliotheca Lichenologica 61: 1–198. [Google Scholar]
- Guide S. (2012) Adobe illustrator cs6. https://www.adobe.com/products/illustrator.html
- Hall TA. (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hopple JS, Vilgalys R. (1994) Phylogenetic relationships among coprinoid taxa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 86: 96–107. 10.1080/00275514.1994.12026378 [DOI] [Google Scholar]
- Hooker JD. (1860) Botany of the Antarctic Voyage. III Flora Tasmaniae 2: 1–422. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khunnamwong P, Surussawadee J, Ribeiro JRA, Hagler AN, Limtong S. (2019) Tremellasaccharicola f.a., sp. nov., a novel tremellaceous yeast species isolated from tropical regions. International Journal of Systematic and Evolutionary Microbiology 69: 2010–2016. 10.1099/ijsem.0.003420 [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PE, Minter DW, Stalpers JA. (2008) Dictionary of the fungi, 10th edition. CAB International, Wallingford, 696 pp. 10.1079/9780851998268.0000 [DOI] [Google Scholar]
- Kobayasi Y. (1939) On the genus Tremella and its allies from Japan. Science Report of the Tokyo Bunrika Diagaku 4: 1–26. [Google Scholar]
- Li AH, Yuan FX, Groenewald M, Bensch K, Yurkov AM, Li K, Guo LD, Aime MC, Sampaio JP, Jindamorakot S, Turchetti B, Inacio J, Fungsin B, Wang QM, Bai FY. (2020) Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Studies in Mycology 96: 17–140. 10.1016/j.simyco.2020.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Wang QM, Theelen B, Groenewald M, Bai FY, Boekhout T. (2015a) Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in Mycology 81: 1–26. 10.1016/j.simyco.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Wang QM, Göker M, Groenewald M, Kachalkin AV, Lumbsch HT, Millanes AM, Wedin M, Yurkov AM, Boekhout T, Bai FY. (2015b) Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in Mycology 81: 85–147. 10.1016/j.simyco.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy B. (1971) Flora neotropica monograph 6. Tremellales. Hafner Publishing Company, Inc. New York, 153 pp. [Google Scholar]
- Maddison WP, Maddison DR. (2017) Mesquite: a modular system for evolutionary analysis, Version 3.2. http://mesquiteproject.org
- Malysheva VF, Malysheva EF, Bulakh EM. (2015) The genus Tremella (Tremellales, Basidiomycota) in Russia with description of two new species and proposal of one nomenclatural combination. Phytotaxa 238: 40–70. 10.11646/phytotaxa.238.1.2 [DOI] [Google Scholar]
- Matheny PB. (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Molecular Phylogenetics and Evolution 35: 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Matheny PB, Liu YJ, Ammirati JF, Hall BD. (2002) Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany 89: 688–698. 10.3732/ajb.89.4.688 [DOI] [PubMed] [Google Scholar]
- Millanes AM, Diederich P, Ekman S, Wedin M. (2011) Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi). Molecular Phylogenetics and Evolution 61: 12–28. 10.1016/j.ympev.2011.05.014 [DOI] [PubMed] [Google Scholar]
- Millanes AM, Diederich P, Westberg M, Knutsson T, Wedin M. (2014) Tremellarhizocarpicola sp. nov. and other interesting lichenicolous Tremellales and Filobasidiales in the Nordic countries. Mycokeys 8: 31–41. 10.3897/mycokeys.8.8176 [DOI] [Google Scholar]
- Millanes AM, Diederich P, Westberg M, Pippola E, Wedin M. (2015) Tremellacetrariellae (Tremellales, Basidiomycota, Fungi), a new lichenicolous fungus on Cetrarielladelisei. Lichenologist 47: 359–368. 10.1017/S0024282915000377 [DOI] [Google Scholar]
- Millanes AM, Diederich P, Westberg M, Wedin M. (2016) Three new species in the Biatoropsisusnearum complex. Herzogia 29: 337–354. 10.13158/heia.29.2.2016.337 [DOI] [Google Scholar]
- Millanes AM, Westberg M, Wedin M, Diederich P. (2012) Tremelladiploschistina (Tremellales, Basidiomycota, Fungi), a new lichenicolous species growing on Diploschistes. Lichenologist 44: 321–332. 10.1017/S0024282911000788 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st conference of the extreme science and engineering discovery environment: bridging from the extreme to the campus and beyond, 1–8. 10.1145/2335755.2335836 [DOI]
- Möller A. (1895) Protobasidiomyceten. Botanische Mittheilungen aus den Tropen 8: 1–180. [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala.
- Petersen JH. (1996) Farvekort. The Danish Mycological Society’s colour-chart. Foreningen til Svampekundskabens Fremme, Greve, 1–6.
- Pippola E, Kotiranta H. (2008) The genus Tremella (Basidiomycota, Tremellales) in Finland. Annales Botanici Fennici 45: 401–489. 10.5735/085.045.0601 [DOI] [Google Scholar]
- Posada D, Crandall KA. (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree v1. 4.0. A graphical viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/
- Rehner S. (2001) Primers for elongation factor 1–α (EF1–α). http://ocid.nacse.org/research/deephyphae/EF1primer
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1–α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) Mrbayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sampaio JP, Agerer R, Piepenbring M, Blanz P. (2004) Diversity, phylogeny and classification of basidiomycetous yeasts. In: Agerer R, Piepenbring M, Blanz P. (Eds) Frontiers in basidiomycote mycology.Eching, 49–80.
- Stiller JW, Hall BD. (1997) The origin of red algae: implications for plastid evolution. Proceedings of the National Academy of Sciences 94: 4520–4525. 10.1073/pnas.94.9.4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D, Groenewald M, Szke S, Cardinali G, Eberhardt U, Stielow B, De Vries M, Verkleij GJM, Crous PW, Boekhout T, Robert V. (2016) DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Studies in Mycology 85: 91–105. 10.1016/j.simyco.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M, Bauer R, Sampaio JP, Oberwinkler F. (2014) Tremellomycetes and related groups. In: Mclaughlin DJ, Spatafora JW. (Eds) Systematics and evolution, The mycota 7.Berlin, 331–355. 10.1007/978-3-642-55318-9 [DOI]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: e134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed]
- Zamora JC, Diederich P, Millanes AM, Wedin M. (2017) An old familiar face: Tremellaanaptychiae sp. nov. (Tremellales, Basidiomycota). Phytotaxa 307: 254–262. 10.11646/phytotaxa.307.4.3 [DOI] [Google Scholar]
- Zamora JC, Millanes AM, Wedin M, Rico VJ, Pérez-Ortega S. (2016) Understanding lichenicolous heterobasidiomycetes: new taxa and reproductive innovations in Tremella s.l. Mycologia 108: 381–396. 10.3852/15-090 [DOI] [PubMed] [Google Scholar]
- Zamora JC, Millanes AM, Etayo J, Wedin M. (2018) Tremellamayrhoferi, a new lichenicolous species on Lecanoraallophana. Herzogia 31: 666–676. 10.13158/heia.31.1.2018.666 [DOI] [Google Scholar]
- Zhao Y, Liu XZ, Bai FY. (2019) Four new species of Tremella (Tremellales, Basidiomycota) based on morphology and DNA sequence data. Mycokeys 47: 75–95. 10.3897/mycokeys.47.29180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.