Abstract
The first night effect (FNE) is a type of sleep disturbance caused by an unfamiliar environment, which leads to difficulty falling asleep and reduced sleep duration. Previously, we reported that Lactobacillus fermentum PS150 (PS150) improves sleep conditions in a pentobarbital-induced sleep mouse model. In this study, we aimed to evaluate the effect of PS150 on the FNE in mice. Briefly, mice were implanted with electrodes and orally administered PS150 for four weeks, and then the FNE was induced by cage changing. Analysis of polysomnographic signals revealed that intervention with PS150 restored non-rapid eye movement (NREM) sleep length under the FNE. Compared to diphenhydramine, a commonly used sleep aid, PS150 had no unwanted side effects, such as rapid eye movement (REM) sleep deprivation and fragmented sleep. Moreover, temporal analysis revealed that PS150 efficiently reduced both sleep latency and time spent restoring normal levels of REM sleep. Taken together, these results suggest that PS150 efficiently ameliorates sleep disturbance caused by the FNE. Additionally, V3–V4 16S rRNA sequencing revealed significant increases in Erysipelotrichia, Actinobacteria, and Coriobacteriia in fecal specimens of the PS150-treated group, indicating that PS150 induces gut microbiota remodeling.
Subject terms: Circadian rhythms and sleep, Microbial communities, Microbiology, Neuroscience
Introduction
Insomnia, which comprises difficulties in maintaining or initiating sleep, is the most common sleep disorder in the general population. Greater than 20% of adults suffer from chronic insomnia1. Many factors that cause chronic insomnia, such as shift work, irregular work hours, jet lag, and stress, can be linked to modern lifestyles2. In addition to chronic factors, sudden changes in the environment, such as traveling or moving to a new place, also cause insomnia. This is called the first night effect (FNE), which can lead to various symptoms, including reduced total and rapid eye movement (REM) sleep length, reduced sleep efficiency, fragmented sleep, and increased sleep latency3,4. Therefore, FNE is considered a general sleep disturbance that potentially affects quality of life.
Insufficient sleep can lead to memory loss, irritability, depression, distraction, and fatigue. In addition to cognitive functions, sleep disorders are also correlated with metabolic syndromes, such as obesity, inflammation, diabetes, and cardiovascular diseases5,6. Although many drugs are readily available to treat insomnia, including benzodiazepine receptor agonists, antihistamines, melatonin receptor agonists, anxiolytics, antidepressants, and antipsychotics, the potential issue of drug dependence and abuse is concerning7. Moreover, these drugs are often accompanied by side effects, such as dizziness, headache, drowsiness, amnesia, and cognitive impairment, even leading to an increased risk of mortality1,8. Therefore, it is important to identify a potential hypnotic auxiliary to replace—or reduce the use of—these hypnotic drugs with safer options that improve sleep quality and efficiency without significant adverse effects.
Proposed by the FAO/WHO in 2002, probiotics are defined as living microorganisms that promote the health of the host9. Increasing evidence shows that long-term intake of probiotics improve health with no apparent side effects10–13. Therefore, numerous potential functions of probiotics have been proposed, even in the field of mental health. Possibly mediated by the immunoregulatory, neuroendocrine, or vagus nerve pathways, the microbiome-gut-brain axis (MGBA) explicitly refers to bidirectional signaling communication between the gastrointestinal and central nervous systems through the intestinal microbiome14–16. Probiotics that confer mental or behavioral benefits through the MGBA are called psychobiotics and have promising potential for future applications17.
Studies have shown that sleep disturbance leads to metabolic consequences accompanied by mild gut microbiota alterations18–20. Recently, some studies have reported preliminary evidence that the gut microbiota affects sleep conditions. Chronic antibiotic-treated mice exhibit disrupted sleep architecture accompanied by intestinal neurotransmitter alterations21. In addition, dietary prebiotics were found to effectively improve sleep conditions affected by stress22. Some novel microbial-dependent fecal metabolites have been proposed to improve sleep23. These results suggest the possible application of remodeling the microbiota to improve sleep quality of the host through the MGBA.
Numerous Lactobacillus spp. and Bifidobacterium spp. have been identified as effective psychobiotics against depression, anxiety, and even neurodegenerative disorders 24. Our previous studies demonstrated that oral administration of Lactobacillus fermentum PS150 (PS150) inhibits inflammation, reduces hypothalamic–pituitary–adrenal (HPA) axis activation, increases sleep duration, decreases sleep latency, and prevents caffeine-induced sleeplessness in experimental rodents25,26. These results suggest the potential application of PS150 as a daily supplement to alleviate sleep disturbance. Apart from the unstressed pentobarbital-induced sleep model in our previous study, we aimed to evaluate the effect of PS150 in a spontaneous sleep model with mild stress, which is more biologically representative. In this study, we applied electroencephalogram (EEG) and electromyogram (EMG) recordings to evaluate the effect of PS150 intervention on spontaneous sleep. Furthermore, since studies have shown a possible connection between the microbiota and sleep, 16S rRNA sequencing was performed to explore the possible effect of PS150 on the gut microbiota.
Results
PS150 prevents insomnia caused by FNE
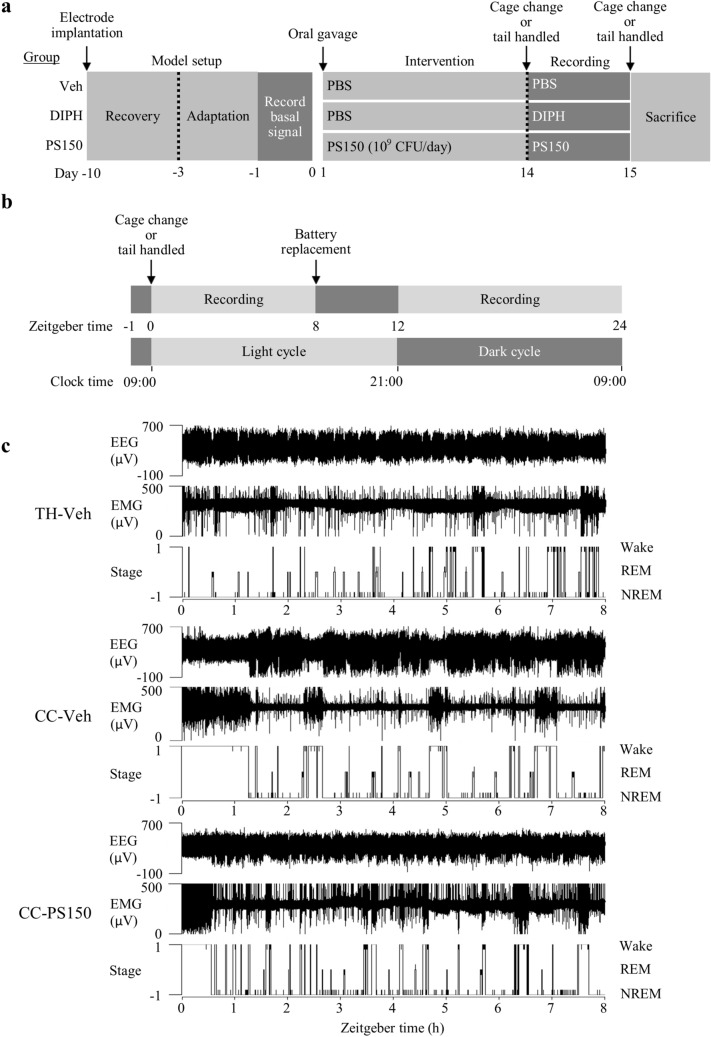
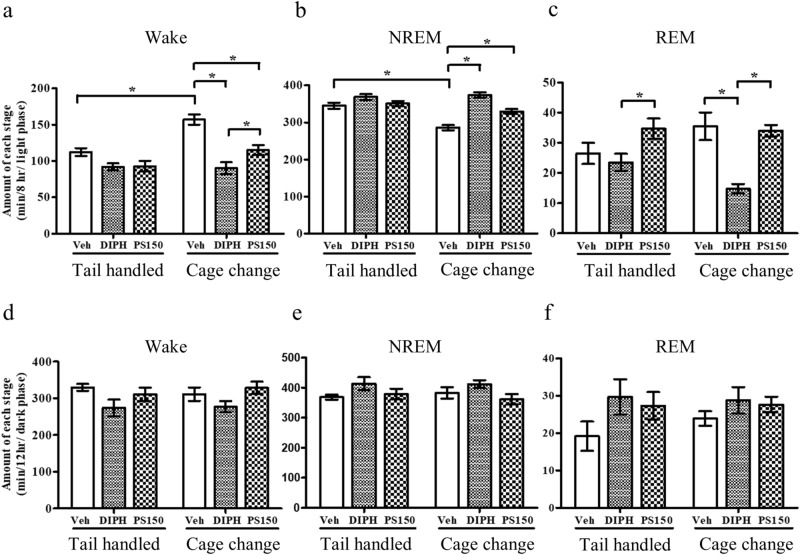
To minimize exterior alteration, a wireless recording system and a noninvasive cage change model were applied to evaluate the effect of PS150 on FNE (Fig. 1a). Briefly, mice were orally administered vehicle (phosphate-buffered saline), diphenhydramine (DIPH), or PS150 immediately before the cage change procedure. Subsequently, mice in each group were divided into cage change (CC-Veh, CC-DIPH, and CC-PS150) and tail-handled (TH-Veh, TH-DIPH, and TH-PS150) groups. The cage change procedure and polysomnographic recordings were performed at Zeitgeber time (ZT) 0 to record the vigilance stages of mice (Fig. 1b). Two periods (ZT 0–8 and ZT 12–24) were recorded and fast Fourier transformed for further analysis (Fig. 1c). To provide an overview of the sleep condition, we calculated the total length of all three vigilance states of sleep during the two recording periods. The first recording period (ZT 0–8) lies in the light phase, which is the inactive time for mice in their normal circadian rhythm. The cage change group exhibited significantly increased wakefulness and reduced non-rapid eye movement (NREM) sleep, suggesting a successful mimic of the FNE in this animal model (Fig. 2a,b). Intervention using DIPH, an antihistamine used to treat insomnia, efficiently ameliorated the effect of FNE. More importantly, the PS150 group exhibited prolonged NREM sleep in response to cage change stress (Fig. 2b). Interestingly, DIPH treatment significantly reduced REM sleep, which was not affected in the PS150 group (Fig. 2c). The distribution of wake and sleep bouts was also affected by FNE. The number of wake bouts longer than 300 s was significantly increased in the CC-Veh group. However, this effect was efficiently ameliorated by DIPH and PS150 (Fig. S1a). Additionally, we discovered that treatment with either DIPH or PS150 led to an increase in the number of shorter sleep bouts (Fig. S1b). These results suggest that administration of PS150 alleviates the effects of FNE. We further analyzed the second recording period (ZT 12–24), which lies mostly in the dark phase. None of the states were significantly different between any of the groups in the dark phase (Fig. 2d–f).
Figure 1.
Schematic view of the experimental design for polysomnographic recording. (a) Experimental flow chart of the animal model. The probiotic intervention was performed 10 days after electrode implantation and proceeded for 14 days. The cage change procedure was performed before polysomnographic recording on day 14 and repeated before sacrifice on day 15. (b) Experimental flow chart of polysomnographic recording on day 14. Zeitgeber time (ZT) 0 was defined as 0900 h when the light was turned on. (c) Representative polysomnographs. Continuous analysis of representative data recorded at ZT 0–8 from tail-handled control (TH-Veh), cage change control (CC-Veh), and cage change PS150-treated (CC-PS150) groups are shown.
Figure 2.
Total time spent in vigilance and sleep states during the light or dark phase. (a–c) The total amount of wake, non-rapid eye movement (NREM), and REM states in the 8-h recording period of the light phase (ZT 0–ZT 8). (d–f) The total amount of wake, NREM, and REM states in the 12 h recording period of the dark phase (ZT 12–ZT 24). Data are expressed as the mean ± SEM (n = 8–9) and were analyzed by two-way ANOVA with Bonferroni correction (*p < 0.05).
PS150 and DIPH modify sleep architecture in different manners
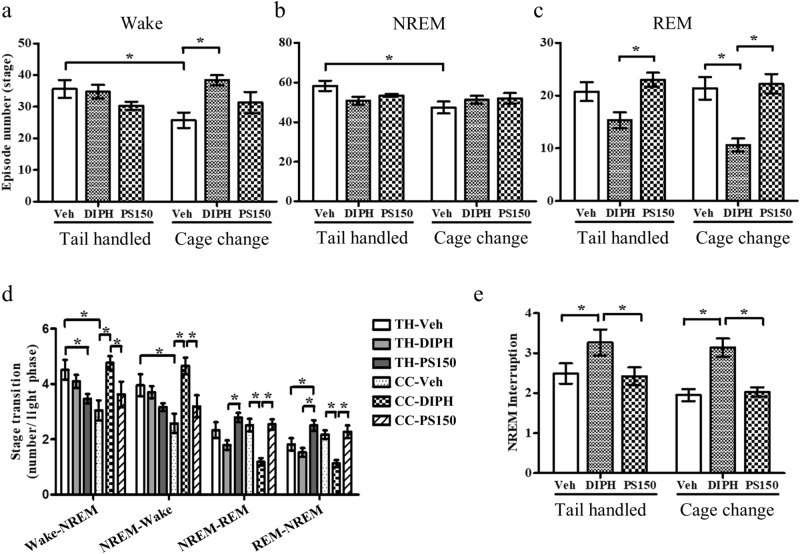
To explore the differences in sleep architecture between oral administration of DIPH and PS150 in the cage change model, we evaluated the number of episodes, interruption times, and state transitions during the light phase. The cage change procedure significantly reduced the number of wakefulness and NREM sleep events (Fig. 3a,b). This effect was partially reversed by the DIPH intervention, which significantly increased the amount of wakefulness (Fig. 3a). Furthermore, the number of REM sleep episodes was affected by DIPH in which the cage change (CC-DIPH) group exhibited a significant decrease compared to the control group (CC-Veh) under FNE (Fig. 3c). In contrast, no significant alteration in episode number was caused by PS150 compared to the vehicle group. The reduced wakefulness (Fig. 2a) and increased number of wake stages (Fig. 3a) indicate shorter average wakefulness caused by DIPH in the cage change group, indicating that DIPH and PS150 may modulate sleep architecture by distinct mechanisms, not only with respect to the length of NREM and REM sleep but also the transition between vigilance and sleep stages.
Figure 3.
Total number of vigilance and sleep states during the light phase. (a–c) The total number of wakefulness, non-rapid eye movement (NREM), and REM states. (d) The average number of transitions per hour between each state. (e) The total number of NREM sleep interruptions observed. Data are expressed as the mean ± SEM (n = 7–9) and were analyzed by two-way ANOVA with Bonferroni correction (*p < 0.05).
PS150 retains a normal stage transition under FNE
To clarify the differential effects of PS150 on vigilance and sleep state transition, we analyzed the state transition number between each state. The cage change procedure led to a significant decrease in the number of transitions between wakefulness and NREM sleep. However, it did not affect the NREM–REM transition. In contrast, the NREM–REM transition in the DIPH group was significantly reduced, which was not observed in the PS150 group (Fig. 3d). The reduced total length, number of transitions, and number of REM sleep episodes (Figs. 2c, and 3c) suggest that DIPH leads to REM sleep deprivation; however, PS150 did not affect REM sleep status. For the DIPH group under FNE, we observed a significantly increased frequency of wake–NREM transitions, consistent with our previous observation (Fig. 3d). Moreover, the frequency of NREM interruption in response to FNE was significantly increased by DIPH intervention, suggesting worse sleep quality (Fig. 3e). Collectively, these results demonstrate that PS150 does not affect sleep architecture. In contrast, DIPH treatment induces REM sleep deprivation and NREM sleep fragmentation, ultimately leading to unstable sleep architecture.
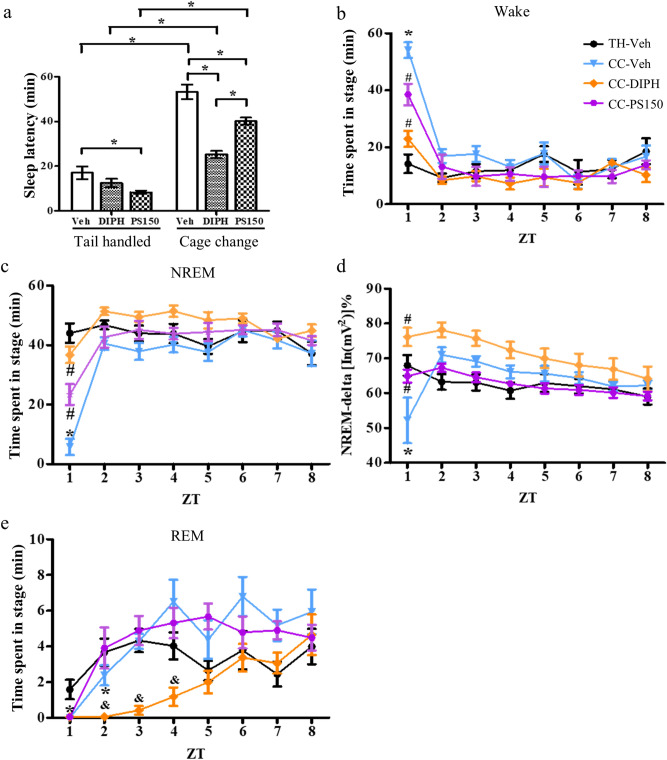
Oral administration of PS150 reduces sleep latency
Previously, we reported significantly reduced sleep latency in response to PS150 treatment in a pentobarbital-induced sleep mouse model. Similar to our previous discovery, we observed that PS150 intervention significantly reduced sleep latency in both the cage change and tail-handled groups. (Fig. 4a). Therefore, we inferred that PS150 may enhance sleep initiation. Subsequently, we plotted the accumulated time of vigilance and sleep stages at each hour to verify the temporal alteration caused by PS150. The results showed that mice under FNE stayed almost fully awake during the first hour (Fig. 4b), whereas both PS150 and DIPH intervention ameliorated this effect. A similar phenomenon was also observed in NREM sleep, where both PS150 and DIPH intervention significantly increased the length of NREM sleep during the first hour (Fig. 4c). Notably, as FNE leads to a major decrease in NREM-delta power, DIPH and PS150 treatment restored the amount of delta power to a normal level (Fig. 4d). Complete loss of REM sleep during the first hour was observed in all cage change groups. Intervention with PS150 revealed a more rapid recovery of REM sleep than that in the vehicle control. REM sleep in the control group returned to normal levels in the third hour, whereas the PS150-treated group returned to normal levels in the second hour. In contrast, the reduction in REM sleep persisted to the fourth hour in the DIPH-treated group (Fig. 4e).
Figure 4.
Time-dependent alterations in vigilance and sleep stages during the light phase. (a) Sleep latency of each group. Latency was defined as the time between the cage change procedure and the first non-rapid eye movement (NREM) sleep that lasted longer than 5 min. (b,c) The amount of wake and NREM states in each hour. Data from the unchanged control (TH-Veh), cage change control (CC-Veh), cage change positive control (CC-DIPH), and cage change PS150-treated (CC-PS150) groups are colored black, blue, orange, and violet, respectively. (d) The relative intensity of NREM-delta power in each hour. (e) Accumulated amount of REM states in each hour. Data are expressed as the mean ± SEM (n = 7–9). For sleep latency, data were analyzed using two-way ANOVA with Bonferroni correction. For the effect of treatment in each hour, data were analyzed using one-way ANOVA with Tukey’s post hoc test. (*p < 0.05, compared to TH-Veh; #p < 0.05, compared to CC-Veh. &p < 0.05, compared to CC-Veh and CC-PS150).
PS150 may improve sleep conditions through a noncanonical pathway
Our results clearly demonstrated the beneficial effect of PS150 on FNE in a spontaneous sleep model. Next, we aimed to improve our understanding of the underlying mechanism of PS150. Psychobiotics have been reported to decrease proinflammatory cytokines and suppress HPA activity17. Therefore, we determined levels of corticosterone in the serum to determine whether PS150 promotes sleep efficiency by affecting HPA axis activity. The results revealed that PS150 reduced corticosterone levels in the tail-handled group. However, no significant difference was found in the cage change group, suggesting that PS150 is unlikely to improve sleep conditions by suppressing the HPA axis (Fig. S2). Since PS150 treatment prolongs NREM sleep, increases NREM-delta power, and shortens sleep latency, we focused on the adenosine and histamine pathways, as they have been reported to be related to NREM sleep and wakefulness. Transcription levels of adenosine A1 receptor (A1R), adenosine A2A receptor (A2AR), ecto-5′-nucleotidase (NT5e), and histamine1 receptor (Hist1R) were analyzed by quantitative PCR (Table S1). Oral administration of PS150 significantly increased mRNA expression of A1R in the hypothalamus rather than the basal forebrain in the tail-handled group. However, this trend was not observed in the cage change group. No significant alteration was found for Hist1R.
PS150 alters fecal microbiota composition in the cage change group
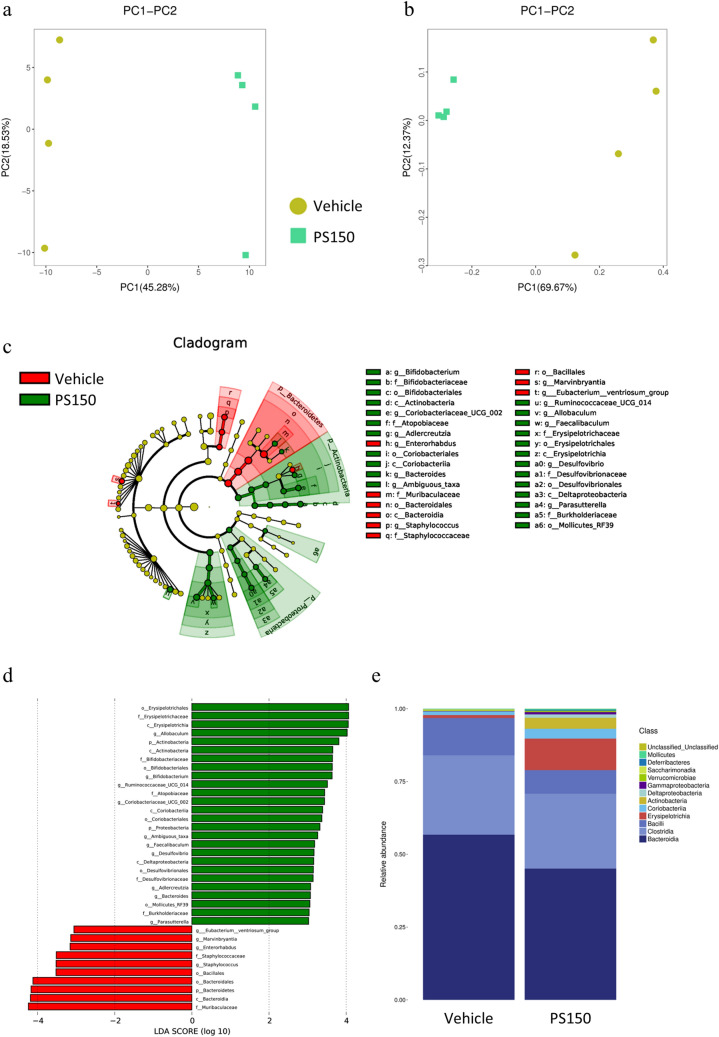
To explore possible roles of the gut microbiota in the hypnotic effect of PS150, we applied 16S rRNA sequencing to verify the effect of PS150 in the cage change group. Sequencing was performed with good quality (Table S2) and sufficient read depth (Figure S3a). The results showed that PS150 treatment significantly elevated the Chao1 index, suggesting a major increase in observed species diversity (Table 1). Furthermore, both principal component analysis (PCA) (Fig. 5a) and principal coordinate analysis (PCoA) (Fig. 5b) showed distinct clustering of fecal microbiota composition from either the PS150-administered or control group. The change in beta diversity was further confirmed using both Adonis (p = 0.035) and ANOSIM (p = 0.037) analysis. In addition to beta diversity, we performed linear discriminant analysis effect size (LEfSE) to identify differentially presented taxa. At the class level, we observed that the abundance of Bacteroidia was decreased in the PS150-treated group. In contrast, Erysipelotrichia, Coriobacteriia, and Actinobacteria were significantly increased in the PS150-treated group (Fig. 5c,d). Notably, the three enriched taxa in the PS150-treated group accounted for approximately 15% of the total microbiota (Fig. 5e), suggesting that PS150 intervention created a competitive advantage for their growth in the gut environment.
Table 1.
Alpha diversity indexes.
| Model | Groups | Chao1 | Shannon | Source | ||
|---|---|---|---|---|---|---|
| Mean | P-value | Mean | P-value | |||
| The first-night effect | PS150 | 222 ± 6.1 | < 0.001 | 5.4 ± 0.3 | 0.624 | This study |
| Veh | 190 ± 5.5 | 5.2 ± 0.6 | ||||
|
Pentobarbital induced sleep |
PS150 | 288 ± 8.6 | < 0.001 | 5.5 ± 0.4 | 0.03 | 27 |
| Veh | 237 ± 8.2 | 5.8 ± 0.2 | ||||
Statistical significance was calculated using the t-test.
Figure 5.
Differential microbiota composition of the fecal specimen from the cage change group. (a) PCA and (b) PCoA plot of fecal microbiota. (c) Cladogram from linear discriminant analysis effect size (LEfSE) and enriched taxonomy in either the control or PS150 group are colored red and green, respectively. (d) LDA score from LEfSE analysis. (e) The stacked plot of relative microbial distribution at the order level.
PS150 alters fecal microbiota composition in naïve mice
Although we observed a significant alteration in the fecal microbiota, this result was hampered by the small sample size (n = 4 in each group). To eliminate this concern, we reanalyzed the fecal samples obtained in our previous study 25. Briefly, 19 naïve mice were treated with either vehicle (n = 8) or PS150 (n = 11) for 4 weeks, and their feces were collected for subsequent 16S rRNA sequencing. Overall, all samples were sequenced with sufficient quality (Table S2) and read depth (Fig. S3b). In contrast to our observation under a cage change background, the naïve mice exhibited a significant increase in alpha diversity in both the major and observed species diversity (Table 1). In line with our expectation, both PCA (Fig. S4a) and PCoA (Fig. S4b) analysis revealed distinct differences in microbiota composition. Moreover, both Adonis (p = 0.005) and ANOSIM (p = 0.001) analyses were statistically significant. We further confirmed the alteration in microbiota using LEfSE analysis (Fig. S4c and S4d). Several taxa in the fecal microbiota were enriched in response to PS150 administration. Similar to previous observations, we identified a significant increase in Erysipelotrichia, Actinobacteria, and Coriobacteriia. The three increased taxa accounted for 19% of the total microbiota (Fig. S4e). In contrast to the aforementioned results, the relative abundance of Bacterioidia was increased in PS150-treated naïve mice. A significant decrease in Clostridia was also observed, which was not affected by PS150 treatment under a cage change background. The occupancy of Erysipelotrichia was highest among the three enriched taxa, and the relative abundance in PS150-treated mice increased by approximately tenfold compared to that in the control group (1.4% to 13.8%). In contrast, enrichment of Actinobacteria was the most dramatic, increasing by more than 40-fold (0.05% to 2.3%). We also observed a threefold increase in Coriobacteriia in the PS150-treated group (1% to 3.2%). Taken together, these results confirm that PS150 intervention induced similar microbiota alterations in two independent animal models.
PS150 selectively induces microbiota enrichment
Because we observed similar microbiota alterations in response to PS150 administration in both studies, we aimed to inspect altered microorganisms in depth through a comparative study of data from two models. We applied Metastats analysis to explore the differentially presented taxa down to the genus level (Table S3). For data from the naïve mouse model, we applied an FDR cutoff of 0.05 for positive discovery, whereas a p-value cutoff of 0.05 was applied for data from this study. A total of 9 and 10 genera were significantly enriched in our previous study and this study, respectively. The genera Allobaculum, Coriobacteriaceae UCG-002, and Bifidobacterium, which belong to Erysipelotrichia, Coriobacteriia, and Actinobacteria, respectively, were consistently elevated in both studies. Notably, the three genera were the dominant taxa in the altered microbiota, which occupied 69% ± 25% of Erysipelotrichia, 70% ± 8% of Coriobacteriia, and 99% ± 1% of Actinobacteria in the PS150-treated groups in both studies. Collectively, our work demonstrated that PS150 administration selectively enriches microbial growth in the host fecal microbiota.
The remodeled microbiota exhibits distinctive metabolic capability
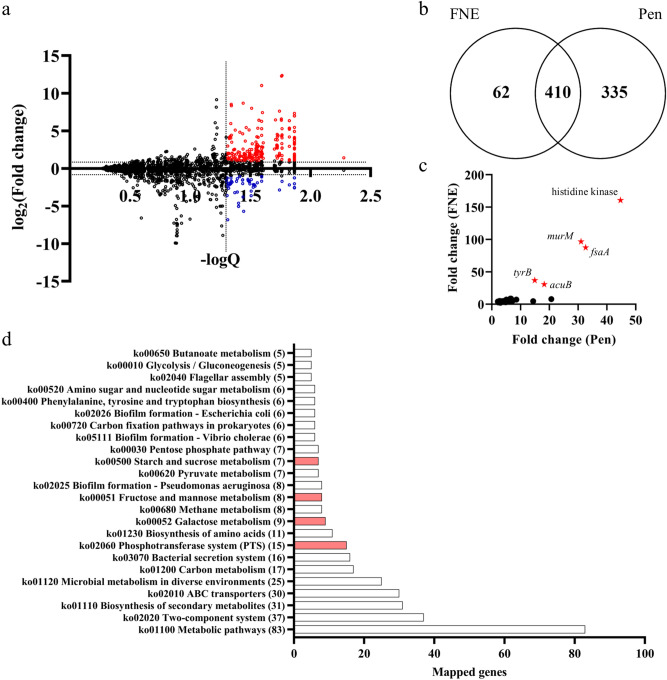
To predict the possible functional correlation of the remodeled microbiota, we conducted Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis and interpreted the results using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. A total of nearly 4000 functional orthologs were present in the predicted metagenome, and a large portion of them were significantly enriched/reduced in the PS150-treated groups compared to the control groups. We filtered the predicted metagenome profile using the estimated fold difference in copy numbers (> twofold) and FDR-adjusted p-values (Q < 0.05) to identify significantly altered genes in the PS150-remodeled microbiota compared to the control group (Fig. 6a). We focused only on enriched genes for the predictively enhanced function of the remodeled microbiota. The results revealed a total of 472 and 745 enriched genes in the remodeled microbiota from the FNE and pentobarbital-induced sleep mouse models, respectively (Fig. 6b). Moreover, the estimated gene composition to the total metagenome (> 100 ppm) was used as a criterion to identify enriched genes with higher abundance. A total of 45 abundant genes were commonly enriched in the microbiota from both mouse models (Fig. 6c). The top 5 most enriched genes were histidine kinase, serine/alanine adding enzyme (murM), fructose-6-phosphate aldolase 1 (fsaA), aromatic-amino-acid transaminase (tyrB), and acetoin utilization protein (acuB). Furthermore, the 410 commonly enriched genes in both models (Fig. 6b) were related to multiple pathways, including amino acid metabolism, carbohydrate metabolism, gene regulation, chemical transportation, and biofilm formation (Fig. 6d).
Figure 6.
Differential predictive metagenome composition of the fecal microbiota. (a) Overall distribution of the predictive metagenome composition. The fold change (PS150/control) of predicted gene copy numbers from the cage change group was plotted against statistical significance (Q value). Significantly enriched and reduced genes in the PS150 group are colored red and blue, respectively. (b) Venn diagram of significantly enriched genes from this study (FNE) and our previous pentobarbital-induced sleep model (Pen). (c) Abundant genes enriched in the microbiota of the PS150 group from both mouse models. Only genes with an estimated composition > 100 ppm of the metagenome were plotted. The top five coenriched genes are labeled with a red star, including histidine kinase, serine/alanine adding enzyme (murM), fructose-6-phosphate aldolase 1 (fsaA), aromatic-amino-acid transaminase (tyrB), and acetoin utilization protein (acuB). (d) Pathway mapping of the 410 coenriched genes from both models using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Phosphotransferase (PTS) systems and the related carbohydrate metabolic pathways are colored red.
Discussion
Lactobacillus fermentum is considered a safe food supplement, and many strains have been shown to have health-improving properties27. L. fermentum PCC reduces severe gastrointestinal disorders and respiratory symptoms caused by weight-bearing training in athletes28. L. fermentum NS9 reduces symptoms of intestinal inflammation caused by antibiotics and reduces anxiety-like behavior and memory impairment29. Recently, the use of probiotics has been extended to improving sleep conditions. Heat-killed Lactobacillus brevis SBC8803 restored wakefulness and increased NREM sleep during the night in a stress-induced chronic sleep disorder mouse model30. Lactobacillus casei Shirota improved sleep quality under psychological stress31. Previously, we demonstrated a strain-specific effect caused by PS150 intervention on sleep latency using a chemically induced sleep mouse model25. In this study, we not only confirmed this effect in a spontaneous sleep model but also extended our understanding of the hypnotic effect of PS150 on sleep architecture. Furthermore, the effect of PS150 was compared to that of DIPH, a drug commonly used in over-the-counter sleep aids. Despite their low cost and availability, first-generation antihistamines, such as DIPH, have been reported to inhibit REM sleep, promote daytime sleep, and impair cognitive function32. In agreement with previous studies, we discovered that DIPH leads to REM sleep deprivation and fragmented NREM sleep in response to FNE (Figs. 2c and 3e). In particular, DIPH treatment led to a prolonged REM recovery compared to that in the control group, in accordance with the reported side effects (Fig. 4e). In contrast, none of the unwanted effects were induced by PS150, which indicates that it may be a better sleep aid than DIPH.
Excessive stress is considered to affect sleep efficiency and cause insomnia33. Many animal models, such as social defeat stress and novel environment exposure, are used to study the relationship between stress and sleep34,35. In addition, stressful events activate the HPA axis, which leads to the secretion of corticosteroids. Generally, hyperactivation of the HPA axis is considered to result in lighter and more fragmented sleep36. The bidirectional relationship between the HPA axis and the gut microbiota has been established 37. In this study, we found that administration of PS150 reduced serum corticosterone in the tail-handled group, suggesting a less active HPA axis. However, no significant HPA axis suppression was found under acute stress caused by cage changes, indicating that the hypnotic effect of PS150 may not result from reduced sensitivity to stress. Thus, we concluded that the hypnotic effect is achieved through pathways other than the HPA axis.
Elevated extracellular adenosine levels in the basal forebrain and hypothalamus promote slow-wave sleep and sleep duration38,39. Previous studies have indicated that adenosine inhibits the waking action of orexins and histaminergic neurons through A1R in the lateral hypothalamus and caudolateral hypothalamus40–42. Histaminergic neurons project from the tuberomammillary nucleus to the central nervous system and have been shown to promote wakefulness through Hist1R43. In addition, inosine produced by the gut microbiota can trigger A2AR signaling44,45. Although current studies are limited to the T-cell response, it still affects brain function because it efficiently passes through the blood–brain barrier46,47. In line with our previous study, we found that PS150 administration increased mRNA expression of A1R in the hypothalamus. However, the increased expression levels of A1R vanished under acute stress caused by cage changes. As there was no significant alteration in adenosine production (NR5e), adenosine signaling (A1R and A2AR), or histamine signaling (Hist1R), we concluded that PS150 may not function through either histamine- or adenosine-related pathways.
Studies have suggested a bidirectional relationship between the microbiota and host metabolism48,49. Furthermore, the diurnal rhythmicity of host gene expression was found to be regulated by the gut microbiota50. Evidence has shown that expression levels of host clock genes could be affected by several common microbial metabolites, including secondary bile acids, hydrogen sulfide, and short-chain fatty acids51. However, most studies have focused on the metabolic consequences and impact on microbiota caused by circadian dysregulation or sleep disruption52. Few studies have focused on the effect of altered microbiota on sleep quality. In this study, we demonstrated that the administration of PS150 leads to enrichment of the genera Allobaculum, Bifidobacterium, and Coriobacteriaceae UCG-002. Notably, Bifidobacterium is known to carry bile salt hydrolase genes (bsh) that convert host bile acids into secondary bile acids. In addition, Allobaculum belongs to Erysipelotrichaceae, which also generally carries bsh51,53. In contrast, only a portion of members of Coriobacteriaceae possess BSH activity54. Unfortunately, given the resolution limit of V3–V4 sequencing, we were unable to identify increased taxa at the species level. Therefore, we are unsure whether the increased Coriobacteriaceae produces secondary bile acids. The gut microbiota has been proven to modulate the host bile acid pool55. Furthermore, gut microbial bsh was found to regulate the transcription of key genes involved in lipid metabolism, cholesterol metabolism, gastrointestinal homeostasis, and circadian rhythm56,57. Taken together, we speculate that the remodeled microbiota may change the bile acid pool through BSH activity and that secondary bile acids could then affect the physiological activities of the host.
Prediction of the metagenome based on PICRUSt analysis revealed many enriched functional genes in the PS150-remodeled microbiota (Fig. 6b). In agreement with the significantly remodeled microbiota, the metagenomic profile of the PS150-treated groups was distinct from that of the control groups. One of the most enriched genes in the remodeled microbiota is tyrB, which encodes aromatic-amino-acid transaminase that catalyzes the reversible reaction to synthesize glutamate and aromatic oxo acids using 2-oxoglutarate and aromatic amino acids58,59. Enrichment of tyrB in the gut microbiota may potentially affect systemic aromatic amino acids, since a previous study demonstrated that oral administration of Escherichia coli expressing amino acid-degrading enzymes affects systemic phenylalanine availability in the host60. In addition, glutamate levels may also be affected by the enrichment of tyrB. This is supported by the fact that glutamate ABC transporter genes were also significantly increased in the remodeled microbiota (Fig. 6d). Glutamate receptors and transporters have been found to be universally expressed along the gastrointestinal tract and play an important role in the regulation of intestinal functions61,62. Moreover, supplementation with monosodium glutamate has been shown to regulate cognitive function, suggesting a possible effect of dietary glutamate on the brain63. However, since colonic glutamate makes nearly no contribution to the systemic glutamate concentration, the microbiota should only affect local glutamate levels in the colonic lumen64. Colonic luminal glutamate is a precursor of functional bacterial secondary metabolites, such as short-chain fatty acids65 and gamma-aminobutyric acid (GABA)66. As a result, the remodeled microbiota may affect the host through glutamate-dependent secondary metabolites. However, further studies are still required to determine the actual effect.
In addition to genes related to amino acid metabolism, genes related to carbohydrate metabolism, including galactose, fructose, mannose, amino sugars, and polysaccharide utilization pathways, were significantly enriched, as were their corresponding phosphotransferase (PTS) systems (Fig. 6d). Additionally, fsaA, which encodes fructose-6-phosphate aldolase and catalyzes the reversible reaction of synthesizing fructose 6-phosphate from dihydroxyacetone and D-glyceraldehyde 3-phosphate67, was one of the most enriched genes (Fig. 6c). These results suggest that the remodeled microbiota exhibit differential fermentation capability to carbon sources. Collectively, the predicted metagenome of the PS150-remodeled microbiota exhibited potential effects on the host metabolite pool and differential utilization of carbohydrates. To our knowledge, there is no direct evidence linking our functional prediction to the hypnotic effect. Additionally, further experimental evidence is required to elucidate the actual metabolic activity of the remodeled microbiota and its effect on host metabolism. It is still unclear how the remodeled microbiota may affect host behavior.
In conclusion, L. fermentum PS150 increases NREM sleep and reduces sleep latency. As a safe probiotic strain, PS150 effectively induces the hypnotic effect without the unwanted side effects of hypnotics, such as DIPH. Additionally, it can remodel the host microbiota, which may lead to changes in the host metabolite pool. Collectively, these data suggest that PS150 represents a promising hypnotic probiotic that may serve as a replacement for over-the-counter hypnotic drugs.
Materials and methods
Animals
All experimental mice used in this study were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and housed in the animal center at National Yang-Ming University. All experiments were performed following relevant guidelines and regulations and were approved by the Institutional Animal Care and Use Committee of National Yang-Ming University (IACUC No. 1070311). Adult male C57BL/6J mice (6 weeks old) were used for all experiments. After the mice arrived at the Laboratory Animal Center of National Yang-Ming University, they were quarantined in a pathogen-free room for 2 weeks and checked for diseases. Mice were provided with water and chow ad libitum (Lab Diet Autoclavable Rodent Diet 5010; PMI Nutrition International, Brentwood, MO, USA). The room was soundproofed and kept at 22 ± 1 °C, 55%–65% humidity, and under a 12:12 h light/dark cycle. The light was turned on at 0900 h, which was defined as Zeitgeber time (ZT) 0, whereas 2100 h was defined as ZT 12.
Preparation of L. fermentum PS150
PS150 used in the animal study was cultured in Lactobacilli Man Rogosa Sharpe (MRS) broth (BD Difco, MD, USA), anaerobically at 37 °C for 18 h, and then harvested by centrifugation at 4 °C and 10,000×g for 10 min. The supernatant was discarded, and the pellet was resuspended in MRS broth plus 12.5% glycerol to a final concentration of 5 × 109 colony-forming units (CFU)/mL. The mixture was then aliquoted in tubes and stored at –20 °C until use. Each tube of the PS150 stock was then dissolved and washed twice using sterile PBS before use.
Electrode implantation
Eight-week-old mice were intraperitoneally anesthetized using 50 mg/kg sodium pentobarbital and mounted on a stereotaxic apparatus before the surgery. Briefly, the dorsal surface of the skull was exposed, cleaned, and implanted with EEG and EMG electrodes for polysomnography. For the EEG electrode, three stainless-steel screws were driven bilaterally into the skull overlying the frontal lobe (+ 1 mm anterior to and –1 mm lateral to the bregma) and parietal cortex (1 mm posterior to and ± 1 mm lateral to the bregma) to fix the electrode device. A major electrode was inserted into the skull above the cortex (frontal lobe + 1 mm anterior to and + 1 mm lateral to bregma) as well as a reference electrode 2 mm caudal to the lambda. For the EMG electrode, two thin-strand stainless-steel microwires were inserted into the dorsal neck muscles to record the EMG. All electrodes were fixed onto the skull using dental cement, and the wounds were closed. After surgery, the mice were given antibiotics (chlortetracycline) and housed individually for 1 week to allow recovery. Wireless physiological sensor units were then placed on the head electrodes two days before the basal signal recording to allow adaptation. Polysomnographic recordings were performed, and subjects that failed to return normal signals were euthanized.
Polysomnographic recording and analysis
Polysomnographic recordings were performed using a wireless physiological record sensor system (KY1C; K&Y Lab, Taiwan), as previously described68. Briefly, EEG and EMG signals were continuously recorded for 8 h (ZT 0 to ZT 8), and then the batteries were replaced. The next recording was performed for 12 h (ZT 12 to ZT 24) until the batteries ran out. Both EEG and EMG signals were amplified 1000-fold. For EEG, the signals were bandpass filtered over a frequency range of 0.16–53.05 Hz and digitalized using a 12-bit analog–digital converter with a 125 Hz sampling rate. For EMG, the signals were filtered at 0.72–112.88 Hz, and digitalized with a 250 Hz sampling rate. The digital signals were then wirelessly transmitted to a data recorder. EEG signals were then converted by fast Fourier transformation using a time window of 16 s with 50% overlap, and the intensities were integrated into delta (0.16–4 Hz), theta (6–9 Hz), sigma (10–14 Hz), and beta (14–32 Hz) powers69. EMG signals were converted by FFT using a time window of 2 s and were integrated according to designated frequencies (32–64 Hz). For determination of sleep–wake stages in each time epoch (8 s), three vigilance states were defined based on the product of sigma and theta powers (sigma*theta), delta power, and EMG power: wake (high EMG activity and low sigma*theta), NREM sleep (low EMG activity, high sigma*theta and high delta activity), and REM sleep (low EMG, high sigma*theta and low delta activity). As a final step, defined sleep–wake stages were visually examined and regulated if necessary.
Cage change procedure
Mice were divided into six groups and orally administered 0.2 mL of PBS (TH-Veh, CC-Veh, TH-DIPH, or CC-DIPH) or 109 CFU PS150 (TH-PS150 and CC-PS150) for 13 consecutive days. On day 14, mice were orally administered PBS (TH-Veh and CC-Veh), 4 mg/kg diphenhydramine hydrochloride (TH-DIPH and CC-DIPH), or PS150 (TH-PS150 and CC-PS150) immediately before the cage change procedure. The cage change procedure was performed at ZT 0 similarly as described70,71. For the cage change groups (CC-Veh, CC-DIPH, and CC-PS150), mice were transferred to a new cage with fresh bedding and food. For the tail-handled groups (TH-Veh, TH-DIPH, and TH-PS150), mice were removed by the tail and returned to their original cages to mimic transfer. Polysomnographic recordings were performed immediately after the cage change procedure. After completion of the recording on day 15, the cage change procedure was repeated, and mice were sacrificed. Tissue samples were stored at –80 °C until further use. This animal study was checked using SYRCLE’s risk of bias tool72 (Table S4) and was reported in compliance with the ARRIVE guidelines73.
Corticosterone measurement
Blood samples were collected for estimation of corticosterone levels after treatment administration and repeated cage changes or tail handling on day 15. After centrifuging the blood at 3000 × g for 10 min, the serum was collected. We used a commercial corticosterone EIA kit (Cayman Chemical, Michigan, MI, USA) with diluted serum to analyze corticosterone concentrations following the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from the brain tissue of mice using an RNeasy mini kit (Qiagen, Germantown, MD, USA) and was reverse transcribed to cDNA using a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher, Waltham, MA, USA) for real-time PCR. cDNA samples were amplified using specific primers (Table S5) and KAPA SYBR FAST ABI Prism (KAPA Biosystems, Woburn, MA, USA) using the StepOnePlus Real-Time PCR System (Applied Biosystems, CA, USA). The target threshold cycle (Ct) was subtracted from the Ct for GAPDH to calculate ΔCt, and relative quantification analysis was performed using the 2−ΔΔCT method25.
Fecal sample collection, DNA extraction, and 16S rRNA library construction
For fecal sample collection, each mouse was placed in a clean cage until defecation. The feces were immediately collected into a 1.5 ml microtube containing 0.3 mL of RNAlater solution (Invitrogen) using sterilized forceps and placed on ice. The feces were then homogenized and washed twice with PBS. Fecal samples were then lysed with 0.2 mm glass beads using a bead beating (FastPrep) system, and metagenomic DNA was extracted using phenol–chloroform extraction. The quality of the DNA extracts was validated using nanodrops. For library construction, the prokaryotic rRNA V3–V4 hypervariable region was amplified using an adaptor containing a region-specific primer set. After amplification, index PCR was performed using amplified DNA to generate an indexed library for next-generation sequencing.
V3–V4 16S rRNA sequencing and data analysis
Next generation sequencing and data processing were carried out by our service provider (Genewiz, South Plainfield, NJ, USA). For raw data collection, 2 × 300 bp paired-end (PE) sequencing was performed using a MiSeq v3 600 cycles kit on an Illumina MiSeq instrument following the manufacturer’s instructions. The adaptor sequences were removed using Cutadapt74 (v1.9.1), and the QIIME75 analysis package (v1.9.1) was used for 16S rRNA data analysis. PE reads were joined using overlapping sequences. The joined sequences were then trimmed to remove low-quality bases (Qphred score < 20) from both the 5’ and 3’ ends. Raw reads with overlapping regions less than 20 bp or a total length shorter than 200 bp were discarded. SILVA 132 was used as the reference database, and chimera sequences were removed using UCHIME76,77. The resulting clean reads were subjected to operational taxonomic unit (OTU) clustering using VSEARCH78 (1.9.6) at a 97% threshold. For beta diversity analysis, beta distances between each sample were calculated based on Bray–Curtis and the UniFrac distance matrix. Intergroup differences were explored using either PCA or PCoA, and statistical significance was calculated by Adonis and Anosim analysis using the R package vegan79,80. Differential analysis of community composition was performed using the linear discriminant analysis effect size (LEfSE) method81. Metagenome prediction was performed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis82. Differential analysis of the predictive metagenome was performed using Metastats83. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used for interpretation and pathway mapping of the predictive metagenome84.
Statistical analysis
Data were analyzed using GraphPad Prism software and are expressed as the mean ± standard error of the mean (SEM). We used one-way analysis of variance (ANOVA) with Tukey’s post hoc test for different groups. For comparison of effects from both the FNE and treatments, we conducted two-way ANOVA with Bonferroni correction. p < 0.05 was considered statistically significant.
Supplementary Information
Acknowledgements
We thank all staff members and Ph.D. students who contributed to the preparation, design, and performance of this project.
Author contributions
A.L. and C.T.S. initiated the project, designed and performed the experiments, analyzed the data, and performed the experiments; A.L., C.T.S., C.W.C., Y.T.C., and H.F.C. contributed to technical support and data interpretation; A.L. and C.T.S. wrote the original draft; C.C.W. and H.F.C. performed review and editing; C.H.Y. and Y.C.T. supervised the project, obtained funding, and finalized the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the Research and Development Department of Bened Biomedical Co., Ltd., Taipei, Taiwan. The bacterial strain Lactobacillus fermentum PS150 was provided by Bened. C.C.W. is employed by Bened. The funder had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Data availability
The V3–V4 16S rRNA sequencing data have been uploaded to NCBI BioProject (PRJNA689879).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alexander Lin and Ching-Ting Shih.
Contributor Information
Cheryl C. H. Yang, Email: cchyang@ym.edu.tw
Ying-Chieh Tsai, Email: tsaiyc@ym.edu.tw.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95659-3.
References
- 1.Morin CM, Benca R. Chronic insomnia. Lancet. 2012;379:1129–1141. doi: 10.1016/s0140-6736(11)60750-2. [DOI] [PubMed] [Google Scholar]
- 2.Porkka-Heiskanen T, Zitting KM, Wigren HK. Sleep, its regulation and possible mechanisms of sleep disturbances. Acta Physiol. (Oxf.) 2013;208:311–328. doi: 10.1111/apha.12134. [DOI] [PubMed] [Google Scholar]
- 3.McCall C, McCall WV. Objective vs. subjective measurements of sleep in depressed insomniacs: First night effect or reverse first night effect? J. Clin. Sleep Med. 2012;8:59–65. doi: 10.5664/jcsm.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamaki M, Nittono H, Hayashi M, Hori T. Examination of the first-night effect during the sleep-onset period. Sleep. 2005;28:195–202. doi: 10.1093/sleep/28.2.195. [DOI] [PubMed] [Google Scholar]
- 5.Depner CM, Stothard ER, Wright KP. Metabolic consequences of sleep and circadian disorders. Curr. Diab. Rep. 2014 doi: 10.1007/s11892-014-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sofi F, et al. Insomnia and risk of cardiovascular disease: A meta-analysis. Eur. J. Prev. Cardiol. 2014;21:57–64. doi: 10.1177/2047487312460020. [DOI] [PubMed] [Google Scholar]
- 7.Buysse DJ. Insomnia. JAMA. 2013;309:706–716. doi: 10.1001/jama.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsaik AK, et al. Mortality associated with anxiolytic and hypnotic drugs-A systematic review and meta-analysis. Aust. N. Z. J. Psychiatry. 2016;50:520–533. doi: 10.1177/0004867415616695. [DOI] [PubMed] [Google Scholar]
- 9.FAO WHO. Health and nutritional properties of probiotics in food including powder milk with live lactic: Report of a joint FAO/WHO expert consultation on acid bacteria expert consultation on evaluation of health and nutritional properties of probitics in food including powder milk with live lactic acid bacteria. in Technical Report 1–56 (2001).
- 10.Kazemi A, Noorbala AA, Azam K, Eskandari MH, Djafarian K. Effect of probiotic and prebiotic vs placebo on psychological outcomes in patients with major depressive disorder: A randomized clinical trial. Clin. Nutr. 2019;38:522–528. doi: 10.1016/j.clnu.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Kechagia M, et al. Health benefits of probiotics: A review. ISRN Nutr. 2013;481651–481651:2013. doi: 10.5402/2013/481651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George Kerry R, et al. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018;26:927–939. doi: 10.1016/j.jfda.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalesi S, et al. A review of probiotic supplementation in healthy adults: Helpful or hype? Eur. J. Clin. Nutr. 2019;73:24–37. doi: 10.1038/s41430-018-0135-9. [DOI] [PubMed] [Google Scholar]
- 14.Burokas A, Moloney RD, Dinan TG, Cryan JF. Microbiota regulation of the mammalian gut-brain axis. Adv. Appl. Microbiol. 2015;91:1–62. doi: 10.1016/bs.aambs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar A, et al. Psychobiotics and the manipulation of bacteria-gut-brain signals. Trends Neurosci. 2016;39:763–781. doi: 10.1016/j.tins.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinan TG, Cryan JF. The microbiome-gut-brain axis in health and disease. Gastroenterol. Clin. N. Am. 2017;46:77–89. doi: 10.1016/j.gtc.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Dinan TG, Stanton C, Cryan JF. Psychobiotics: A novel class of psychotropic. Biol. Psychiatry. 2013;74:720–726. doi: 10.1016/j.biopsych.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, et al. Acute sleep-wake cycle shift results in community alteration of human gut microbiome. mSphere. 2020 doi: 10.1128/mSphere.00914-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poroyko VA, et al. Chronic sleep disruption alters gut microbiota, induces systemic and adipose tissue inflammation and insulin resistance in mice. Sci. Rep. 2016;6:35405. doi: 10.1038/srep35405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson JR, et al. A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med. 2017;38:104–107. doi: 10.1016/j.sleep.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa Y, et al. Gut microbiota depletion by chronic antibiotic treatment alters the sleep/wake architecture and sleep EEG power spectra in mice. Sci. Rep. 2020;10:19554. doi: 10.1038/s41598-020-76562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson RS, et al. Dietary prebiotics and bioactive milk fractions improve NREM sleep, enhance REM sleep rebound and attenuate the stress-induced decrease in diurnal temperature and gut microbial alpha diversity. Front. Behav. Neurosci. 2017 doi: 10.3389/fnbeh.2016.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson RS, et al. Dietary prebiotics alter novel microbial dependent fecal metabolites that improve sleep. Sci. Rep. 2020;10:3848. doi: 10.1038/s41598-020-60679-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng LH, Liu YW, Wu CC, Wang S, Tsai YC. Psychobiotics in mental health, neurodegenerative and neurodevelopmental disorders. J. Food Drug Anal. 2019;27:632–648. doi: 10.1016/j.jfda.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin A, et al. Hypnotic effects of Lactobacillus fermentum PS150TM on pentobarbital-induced sleep in mice. Nutrients. 2019 doi: 10.3390/nu11102409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y-W, et al. Lactobacillus fermentum PS150 showed psychotropic properties by altering serotonergic pathway during stress. J. Funct. Foods. 2019;59:352–361. doi: 10.1016/j.jff.2019.05.043. [DOI] [Google Scholar]
- 27.Sue Barlow AC, Collins JD, Dybing E, Flynn A, Fruijtier-Pölloth C, Hardy A, Knaap A, Kuiper H, Neindre PL, Schans J, Schlatter J, Silano V, Skerfving S, Vannier P. Introduction of a qualified presumption of safety (QPS) approach for assessment of selected microorganisms referred to EFSA. EFSA J. 2007;587:1–85. [Google Scholar]
- 28.West NP, et al. Lactobacillus fermentum (PCC(R)) supplementation and gastrointestinal and respiratory-tract illness symptoms: A randomised control trial in athletes. Nutr. J. 2011;10:30. doi: 10.1186/1475-2891-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang T, et al. Lactobacillus fermentum NS9 restores the antibiotic induced physiological and psychological abnormalities in rats. Benef. Microbes. 2015;6:707–717. doi: 10.3920/BM2014.0177. [DOI] [PubMed] [Google Scholar]
- 30.Higo-Yamamoto SYS, Miyazaki K, Nakakita Y, Kaneda H, Takata Y, Nakamura T, Oishi K. Dietary heat-killed Lactobacillus brevis SBC8803 attenuates chronic sleep disorders induced by psychophysiological stress in mice. J. Nutr. Sci. Vitaminol. (Tokyo) 2019;65:164–170. doi: 10.3177/jnsv.65.164. [DOI] [PubMed] [Google Scholar]
- 31.Takada M, et al. Beneficial effects of Lactobacillus casei strain Shirota on academic stress-induced sleep disturbance in healthy adults: A double-blind, randomised, placebo-controlled trial. Benef. Microbes. 2017;8:153–162. doi: 10.3920/BM2016.0150. [DOI] [PubMed] [Google Scholar]
- 32.Church MK, et al. Risk of first-generation H(1)-antihistamines: A GA(2)LEN position paper. Allergy. 2010;65:459–466. doi: 10.1111/j.1398-9995.2009.02325.x. [DOI] [PubMed] [Google Scholar]
- 33.Sanford LD, Suchecki D, Meerlo P. Stress, arousal, and sleep. Curr. Top. Behav. Neurosci. 2015;25:379–410. doi: 10.1007/7854_2014_314. [DOI] [PubMed] [Google Scholar]
- 34.Meerlo P, Turek FW. Effects of social stimuli on sleep in mice: Non-rapid-eye-movement (NREM) sleep is promoted by aggressive interaction but not by sexual interaction. Brain Res. 2001;907:84–92. doi: 10.1016/s0006-8993(01)02603-8. [DOI] [PubMed] [Google Scholar]
- 35.Tang X, Xiao J, Parris BS, Fang J, Sanford LD. Differential effects of two types of environmental novelty on activity and sleep in BALB/cJ and C57BL/6J mice. Physiol. Behav. 2005;85:419–429. doi: 10.1016/j.physbeh.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Buckley TM, Schatzberg AF. On the interactions of the hypothalamic-pituitary-adrenal (HPA) axis and sleep: Normal HPA axis activity and circadian rhythm, exemplary sleep disorders. J. Clin. Endocrinol. Metab. 2005;90:3106–3114. doi: 10.1210/jc.2004-1056. [DOI] [PubMed] [Google Scholar]
- 37.Farzi A, Fröhlich EE, Holzer P. Gut microbiota and the neuroendocrine system. Neurotherapeutics. 2018;15:5–22. doi: 10.1007/s13311-017-0600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basheer R, Strecker RE, Thakkar MM, McCarley RW. Adenosine and sleep-wake regulation. Prog. Neurobiol. 2004;73:379–396. doi: 10.1016/j.pneurobio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Liu ZW, Gao XB. Adenosine inhibits activity of hypocretin/orexin neurons by the A1 receptor in the lateral hypothalamus: A possible sleep-promoting effect. J. Neurophysiol. 2007;97:837–848. doi: 10.1152/jn.00873.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma R, Sahota P, Thakkar MM. Role of adenosine and the orexinergic perifornical hypothalamus in sleep-promoting effects of ethanol. Sleep. 2014;37:525–533. doi: 10.5665/sleep.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cun Y, et al. Orexin A attenuates the sleep-promoting effect of adenosine in the lateral hypothalamus of rats. Neurosci. Bull. 2014;30:877–886. doi: 10.1007/s12264-013-1442-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CR, et al. Paeoniflorin promotes non-rapid eye movement sleep via adenosine A1 receptors. J. Pharmacol. Exp. Ther. 2016;356:64–73. doi: 10.1124/jpet.115.227819. [DOI] [PubMed] [Google Scholar]
- 43.Oishi Y, Huang ZL, Fredholm BB, Urade Y, Hayaishi O. Adenosine in the tuberomammillary nucleus inhibits the histaminergic system via A1 receptors and promotes non-rapid eye movement sleep. Proc. Natl. Acad. Sci. U S A. 2008;105:19992–19997. doi: 10.1073/pnas.0810926105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He B, Hoang TK, Tran DQ, Rhoads JM, Liu Y. Adenosine A(2A) receptor deletion blocks the beneficial effects of Lactobacillus reuteri in regulatory T-deficient scurfy mice. Front. Immunol. 2017;8:1680–1680. doi: 10.3389/fimmu.2017.01680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mager LF, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 46.Isakovic AJ, Abbott NJ, Redzic ZB. Brain to blood efflux transport of adenosine: Blood-brain barrier studies in the rat. J. Neurochem. 2004;90:272–286. doi: 10.1111/j.1471-4159.2004.02439.x. [DOI] [PubMed] [Google Scholar]
- 47.Parkinson FE, et al. Molecular biology of nucleoside transporters and their distributions and functions in the brain. Curr. Top. Med. Chem. 2011;11:948–972. doi: 10.2174/156802611795347582. [DOI] [PubMed] [Google Scholar]
- 48.Parekh PJ, Oldfield EC, Challapallisri V, Ware CJ, Johnson DA. Sleep disorders and inflammatory disease activity: chicken or the egg? Am. J. Gastroenterol. 2015;110:484–488. doi: 10.1038/ajg.2014.247. [DOI] [PubMed] [Google Scholar]
- 49.Voigt RMFC, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. Circadian disorganization alters intestinal microbiota. PLoS ONE. 2014;21:e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thaiss CA, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510.e1412. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Parkar SG, Kalsbeek A, Cheeseman JF. Potential role for the gut microbiota in modulating host circadian rhythms and metabolic health. Microorganisms. 2019 doi: 10.3390/microorganisms7020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Hao Y, Fan F, Zhang B. The role of microbiome in insomnia, circadian disturbance and depression. Front. Psych. 2018;9:669. doi: 10.3389/fpsyt.2018.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Labbe A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn's, ulcerative colitis and type 2 diabetes metagenomes. PLoS ONE. 2014;9:e115175. doi: 10.1371/journal.pone.0115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wegner K, et al. Rapid analysis of bile acids in different biological matrices using LC-ESI-MS/MS for the investigation of bile acid transformation by mammalian gut bacteria. Anal. Bioanal. Chem. 2017;409:1231–1245. doi: 10.1007/s00216-016-0048-1. [DOI] [PubMed] [Google Scholar]
- 55.Fiorucci S, Distrutti E. Bile acid-activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol. Med. 2015;21:702–714. doi: 10.1016/j.molmed.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Joyce SA, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eggink HM, et al. Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiol. Int. 2017;34:1339–1353. doi: 10.1080/07420528.2017.1363226. [DOI] [PubMed] [Google Scholar]
- 58.Mavrides C, Orr W. Multispecific aspartate and aromatic amino acid aminotransferases in Escherichia coli. J. Biol. Chem. 1975;250:4128–4133. doi: 10.1016/S0021-9258(19)41395-1. [DOI] [PubMed] [Google Scholar]
- 59.Kuramitsu S, Inoue K, Ogawa T, Ogawa H, Kagamiyama H. Aromatic amino acid aminotransferase of Escherichia coli: Nucleotide sequence of the tyrB gene. Biochem. Biophys. Res. Commun. 1985;133:134–139. doi: 10.1016/0006-291x(85)91851-0. [DOI] [PubMed] [Google Scholar]
- 60.Isabella VM, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018;36:857–864. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 61.Filpa V, et al. Role of glutamatergic neurotransmission in the enteric nervous system and brain-gut axis in health and disease. Neuropharmacology. 2016;111:14–33. doi: 10.1016/j.neuropharm.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 62.Baj A, et al. Glutamatergic signaling along the microbiota-gut-brain axis. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20061482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holton KF, Hargrave SL, Davidson TL. Differential effects of dietary MSG on hippocampal dependent memory are mediated by diet. Front. Neurosci. 2019;13:968. doi: 10.3389/fnins.2019.00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tome D. The roles of dietary glutamate in the intestine. Ann. Nutr. Metab. 2018;73(Suppl 5):15–20. doi: 10.1159/000494777. [DOI] [PubMed] [Google Scholar]
- 65.Blachier F, Boutry C, Bos C, Tomé D. Metabolism and functions of L-glutamate in the epithelial cells of the small and large intestines. Am. J. Clin. Nutr. 2009;90:814s–821s. doi: 10.3945/ajcn.2009.27462S. [DOI] [PubMed] [Google Scholar]
- 66.Mazzoli R, Pessione E. The neuro-endocrinological role of microbial glutamate and GABA signaling. Front. Microbiol. 2016 doi: 10.3389/fmicb.2016.01934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schurmann M, Sprenger GA. Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J. Biol. Chem. 2001;276:11055–11061. doi: 10.1074/jbc.M008061200. [DOI] [PubMed] [Google Scholar]
- 68.Kuo TB, Lai CT, Chen CY, Yang YC, Yang CC. The high-frequency component of heart rate variability during extended wakefulness is closely associated with the depth of the ensuing sleep in C57BL6 mice. Neuroscience. 2016;330:257–266. doi: 10.1016/j.neuroscience.2016.05.060. [DOI] [PubMed] [Google Scholar]
- 69.Veasey SC, et al. An automated system for recording and analysis of sleep in mice. Sleep. 2000;23:1–16. doi: 10.1093/sleep/23.8.1c. [DOI] [PubMed] [Google Scholar]
- 70.Xu Q, et al. A mouse model mimicking human first night effect for the evaluation of hypnotics. Pharmacol. Biochem. Behav. 2014;116:129–136. doi: 10.1016/j.pbb.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 71.Kaushik MK, Aritake K, Takeuchi A, Yanagisawa M, Urade Y. Octacosanol restores stress-affected sleep in mice by alleviating stress. Sci. Rep. 2017;7:8892. doi: 10.1038/s41598-017-08874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hooijmans CR, et al. SYRCLE's risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014;14:43. doi: 10.1186/1471-2288-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Percie du Sert N, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLOS Biol. 2020;18:3000410. doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Nucleic Acids Res. 2011;17(3):2011. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 75.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics (Oxford, England) 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team. R: A language and environment for statistical computing. https://www.R-project.org/ (R Foundation for Statistical Computing, Vienna, Austria, 2020).
- 80.Jari Oksanen, F. G. B. et al. vegan: Community ecology package. https://cran.r-project.org, https://github.com/vegandevs/vegan (2020).
- 81.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paulson JN, Pop M, Bravo HC. Metastats: An improved statistical method for analysis of metagenomic data. Genome Biol. 2011;12:P17. doi: 10.1186/gb-2011-12-s1-p17. [DOI] [Google Scholar]
- 84.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The V3–V4 16S rRNA sequencing data have been uploaded to NCBI BioProject (PRJNA689879).