Abstract
Despite new combination therapies improving survival of breast cancer patients with estrogen receptor α (ER+) tumors, the molecular mechanisms for endocrine-resistant disease remain unresolved. Previously we demonstrated that expression of the RNA binding protein and N6-methyladenosine (m6A) reader HNRNPA2B1 (A2B1) is higher in LCC9 and LY2 tamoxifen (TAM)-resistant ERα breast cancer cells relative to parental TAM-sensitive MCF-7 cells. Here we report that A2B1 protein expression is higher in breast tumors than paired normal breast tissue. Modest stable overexpression of A2B1 in MCF-7 cells (MCF-7-A2B1 cells) resulted in TAM- and fulvestrant- resistance whereas knockdown of A2B1 in LCC9 and LY2 cells restored TAM and fulvestrant, endocrine-sensitivity. MCF-7-A2B1 cells gained hallmarks of TAM-resistant metastatic behavior: increased migration and invasion, clonogenicity, and soft agar colony size, which were attenuated by A2B1 knockdown in MCF-7-A2B1 and the TAM-resistant LCC9 and LY2 cells. MCF-7-A2B1, LCC9, and LY2 cells have a higher proportion of CD44+/CD24−/low cancer stem cells (CSC) compared to MCF-7 cells. MCF-7-A2B1 cells have increased ERα and reduced miR-222–3p that targets ERα. Like LCC9 cells, MCF-7-A2B1 have activated AKT and MAPK that depend on A2B1 expression and are growth inhibited by inhibitors of these pathways. These data support that targeting A2B1 could provide a complimentary therapeutic approach to reduce acquired endocrine resistance.
Keywords: breast cancer, endocrine resistance, ER, tamoxifen, fulvestrant
1. Introduction
Most (~80%) breast tumors express estrogen receptor α (ERα, ESR1) [1] which has been successfully targeted by selective ER modulators (SERMs), e.g., the partial ER agonist/antagonist tamoxifen (TAM), and by aromatase inhibitors (AI) e.g., letrozole. Despite the increase in disease-free and overall survival (DFS, OS) of early stage breast cancer patients in response to these endocrine therapies, approximately 20% of patients taking these oral antiestrogens develop endocrine-resistant metastatic disease [2]. Multiple mechanisms contributing to endocrine resistance have been evaluated including somatic activating mutations within the ligand binding domain (LBD) of ESR1 driven by long term AI treatment in ~ 20–29% of patients [3–5]. Aberrant gene expression profiles in primary breast tumors have been widely studied [6–9] and mutations in genes encoding epigenetic remodelers, e.g., histone methyltransferases and demethylases, are associated with endocrine resistance (reviewed in [2]). Mutations in metastatic breast cancer include alterations in ERBB2, NF1, EGFR, KRAS, MYC, CTCF, FOXA1, and TBX3 in ~ 22% of ER+ post-endocrine therapy tumors [10]. However, a full understanding of the molecular mechanisms regulating the loss of response to endocrine therapies leading to an increased metastatic phenotype remain to be fully elucidated.
The contribution of epitranscriptomics to drug resistance in breast and other cancers is being actively investigated [11]. The most common post-transcriptional mRNA modification is N(6)-methyladenosine (m6A) [12, 13]. A recent report observed higher METTL14 and m6A levels in breast tumors versus adjacent normal tissue that was associated with tumor infiltration and size [14]. m6A is recognized by protein “Readers” including YTHDF1, YTHDF2, YTHDF3, ELF3, HNRNPC, and HNRNPA2B1 (A2B1) [15–17]. Immunohistochemical staining (IHC) of a breast tumor tissue array (TMA) revealed higher YTHDF1, YTHDF2, YTHDF3, HNRNPC, and A2B1 in breast tumors compared to normal breast tissue [18]. The processing of selected pri-miRNA to pre-miRNA is stimulated by m6A recognition by A2B1 which interacts with the DROSHA complex protein DGCR8 [19]. In addition, A2B1 regulates mRNA stability and alternative splicing, DNA repair, and telomere maintenance [20]. A2B1 protein expression is higher in breast tumors compared to normal breast tissue [18, 21, 22]. Knockdown of A2B1 inhibited the proliferation of MCF-7 and MDA-MB-231 breast cancer cells [21]. We reported that A2B1 expression was higher in two TAM-resistant, ERα+ breast cancer cell lines LCC9 and LY2 compared to parental MCF-7 luminal A breast cancer cells [23]. Further, higher A2B1 expression was significantly associated with reduced relapse-free survival (RFS) up to 150 months after primary breast tumor resection according to data in KM Plotter [23]. Transient (48 h) modest overexpression of A2B1 (~ 5.4-fold) in MCF-7 cells reduced sensitivity to growth inhibition by 4-hydroxytamoxifen (4-OHT, an active TAM metabolite in vivo [24]) and fulvestrant, a selective ER degrader (SERD), suggesting that increased A2B1 plays a role in TAM and fulvestrant resistant cell proliferation. Using miRNA-seq, we identified global changes in mature miRNA transcripts in MCF-7 cells when A2B1 was transiently overexpressed. The enrichment analysis of the A2B1-regulated miRome in MCF-7 cells identified the GO/ KEGG pathway “steroid hormone-mediated signaling pathway” and “response to estradiol” as significantly altered pathways [23].
The aim of this study was to determine if stable A2B1 overexpression in endocrine-sensitive MCF-7 cells at a level similar to that detected in TAM-resistant LCC9 and LY2 cells would result in a phenotype of endocrine-resistance. Concordantly, we hypothesized that knockdown of A2B1 in LCC9 and LY2 cells, with higher endogenous A2B1 (~4-fold higher compared to MCF-7 cells [23]) would have the opposite effect, thereby restoring antiestrogen sensitivity. Our goal was to characterize the cellular phenotype of the MCF-7-A2B1 stable cell line, its response to TAM and fulvestrant, and identify proteins and pathways altered by A2B1 expression. This study provides evidence that higher expression of A2B1 plays a role in endocrine-resistance, increases cell motility, and acquisition of cancer stem cell properties. These findings suggest that identifying ways to inhibit A2B1 may be a therapeutic target in ERα+ breast cancer patients who relapse on endocrine therapies.
2. Materials and Methods
2.1. Chemicals
4-hydroxytamoxifen (4-OHT) and Wortmannin (PI3K inhibitor) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Dihydrotestosterone (DHT) was purchased from Steraloids, Inc. (Newport, RI, USA). PD98059 (MEK1/2 inhibitor) and fulvestrant (a selective estrogen receptor degrader (SERD)) were purchased from Tocris Bioscience (Ellisville, MO, USA). All were dissolved in DMSO or ethanol (EtOH) which were used as the respective vehicle control in experiments.
2.2. Immunohistochemistry (IHC) for HNRNPA2B1
IHC was performed using a commercial breast cancer tissue microarray (TMA) paired with metastatic tumors (BC27 from Reveal Biosciences, San Diego, CA, USA). The TMA contained 48 primary breast tumors: 12 paired primary breast tumors and normal breast tissue, and 36 primary breast tumors paired with lymph node (LN) metastases. The TMA was dewaxed and rehydrated, followed by antigen retrieval in citric acid buffer (pH 6.0). Sections were then blocked in 5% goat serum and incubated with 1:200 dilution of HNRNPA2B1 (B1 epitope-specific: IBL # 18941, IBL America, Minneapolis, MN USA) antibody overnight at 4°C. Slides were subsequently incubated with 1:500 dilution of horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (Invitrogen, Cat No. 32260) for 1 h at room temperature, washed, and incubated in a 1:15 3,3’-diaminobenzidine (DAB) stain (Vector Laboratories, Berlingame, CA, USA). The stained slides were counterstained with hematoxylin. Images were captured using a Pannoramic Flash Desk DX slide scanner (3D Histotech, Budapest, Hungary) or a Nikon Eclipse Ci microscope with dedicated Nikon DS-Fi3 microscope camera (Tokyo, Japan) for higher magnification. Semi-quantitative measures were performed by two independent observers. Nuclear expression of HNRNPA2B1 was evaluated using the Allred score which combines intensity and the percentage of positive cells [25]. Scoring of percent of positive cells was as described [26, 27]: 0 (<1% negative), 1 (1%–25% positive), 2 (>25%–75% positive), and 3 (>75% positive).
2.3. Cell culture and treatments
MCF-7, T47D, MDA-MB-231, MDA-MB-468, and HCC1806 breast cancer cells were purchased from American Type Tissue Collection (ATCC, Manassas, VA, USA) and were used within nine passages from ATCC. LCC9 and LY2 breast cancer cells were generously provided by Dr. Robert Clarke, Georgetown University Medical Center [28–30] and were maintained as previously reported [31]. T47D cells were maintained in RPMI 1640 (Gen Clone, Genesee Scientific, El Cajon, CA, USA) supplemented with 5% FBS, 6 μg/ml insulin (Sigma-Aldrich), and 1% Pen/Strep [32]. MDA-MB-231 and MDA-MB-468 triple negative breast cancer (TNBC) cells were maintained in DMEM (Corning, Corning, NY, USA), supplemented with 10% FBS and 1% Pen/Strep. HCC1806 TNBC cells were maintained in RPMI 1640 supplemented witih 10% FBS and 1% Pen/Strep. All cell lines were verified by short tandem repeat (STR) genotyping (Genetica, LabCorp, Burlington, NC, USA). Genomic DNA was extracted from each cell line using QIAmp DNA Mini Kit (Qiagen, Germantown, MD, USA). STR profiles were compared with publicly available profiles using Cellosaurus STR (ExPASy).
MCF-7 cells were maintained as described previously [33] prior to transfection with pcDNA3.1+C-DYK into which HNRNPA2B1 was cloned (GenScript, Piscataway, NJ, USA) as described [23]. After 24 h, the medium was changed to IMEM + 10% FBS + 1% penicillin/streptomycin (Pen/Strep-Invitrogen). After 48 h, the medium was changed to IMEM + 5% FBS + 1% Pen/Strep and clones were selected for stable HNRNPA2B1 expression in 400 μg/ml G418 (Geneticin, ThermoFisher Scientific, Waltham MA, USA) for > 6 months. The MCF-7-A2B1 stable cells are maintained in medium containing 200 μg/ml G418. The MCF-7-A2B1 cell line STR profile matched MCF-7 cells.
2.4. Transient transfection
Silencer™Select siHNRNPA2B1 (catalog #4390824) and Silencer™ Select Negative Control No. 1 (catalog # 4390843) were purchased from ThermoFisher. Prior to transfection, cells were grown in phenol red-free Opti-MEM (cat. # 11058021, ThermoFisher) for ~ 18 h. Transfection used Lipofectamine RNAiMAX reagent (cat # 13778075 ThermoFisher) following the manufacturer’s protocol.
2.5. RNA extraction and quantitative real-time PCR (qPCR)
RNA was extracted using the RNeasy Mini Kit (Qiagen, Gaithersburg, MD, USA). miRNA was isolated using the miRNeasy Mini Kit RNA (Qiagen). RNA concentration and quality were assessed using a NanoDrop spectrophotometer (Thermo Scientific, Rockford, IL, USA). The TaqMan® MicroRNA Reverse Transcription Kit and the High Capacity cDNA Reverse Transcription Kit for RNA (both from ThermoFisher) were used to make cDNA from miRNA and mRNA, respectively. Quantitative real-time PCR (qPCR) for HNRNPA2B1 and ESR1 was performed using TaqMan assays (ThermoFisher). 18S rRNA and GAPDH were used as normalizers. qPCR for miR-222–3p used TaqMan assays and were normalized to RNU6B (ThermoFisher). qPCR was performed using an ABI Viia 7 Real-Time PCR system (LifeTechnologies) with each reaction run in triplicate. The comparative threshold cycle (CT) method was used to determine ΔCT, ΔΔCT, and fold-change, log 2, relative to control [34].
2. 6. Western blots
Whole cell extracts (WCE) were prepared as described [35]. WCE were separated on 10% SDS-PAGE, transferred to PVDF membranes (BioRad) and immunoblotted with antibodies: HNRNPA2B1 (B1 epitope-specific: IBL # 18941, IBL America (Minneapolis, MN USA); ERα (D8H8, cat #8644), HNRNPA2B1 (recognizes B1 and A2 variants, Proteintech monoclonal # 67445–1-Ig (Rosemont, IL, USA), phospho-ser-473-AKT (cat #4051), AKT (cat #9272), phospho (p44/42)-MAPK (ERK1/2, cat # 9101L), E-cadherin (cat #3195) from Cell Signaling Technology, Danvers, MA, USA; GAPDH (cat.# sc-365062), MAPK/ERK2 (cat.# sc-154), vimentin (cat # sc-32322) from Santa Cruz Biotechnology (Dallas, TX); α-tubulin (ThermoFisher Scientific # MS-81-P1). Blots were stained with Ponceau S for additional quantification [36, 37]. Blots were imaged in a Bio-Rad ChemiDoc™ XRS+ System with Image Lab™ Software (Bio-Rad Laboratories Inc., Hercules, CA).
2.7. MTT assays
MTT assays were performed in 96-well plates with each treatment in quadruplicate. Cells were ‘serum starved’ in phenol red-free IMEM supplemented with 5% dextran-coated charcoal-stripped (DCC) FBS (here after referred to as SS (serum starved) medium (SS medium)) prior to 48 h treatment with DMSO or EtOH (vehicle controls), 100 nM or 1 μM 4-OHT, or 100 nM fulvestrant for the number of h or days indicated in the Figure legend. CellTiter (Promega) was used to quantitate cell viability.
2.8. Cell transwell migration assay
MCF-7, MCF-7-A2B1, LCC9, and LY2 cells were grown in SS medium for 72 h prior to being seeded at 25,000 cells/polyethylene terephthalate (PET) membrane (VWR, 8 μm pore size) insert in 24-well plates in SS medium and 10% FBS-IMEM was used as the chemoattractant. Cells were incubated at 37°C and 5% CO2 for 24 h. Cells remaining within the inner chamber were removed with cotton swabs. The filter with the migrated cells on the underside were fixed and stained with a 5% crystal violet solution. Cell images were captured using an EVOS microscope, and quantified using ImageJ2 software [38].
2.9. Cell invasion assay
MCF-7, MCF-7-A2B1, LCC9, and LY2 cells were grown in ‘serum starved’ medium for 72 h followed by treatment for 24h with either Vehicle (EtOH) or 100 nM 4-OHT. Likewise, following 72 h in ‘serum starved’ medium, MCF −7 and MCF-7-A2B1 stable cells were treated with vehicle control (EtOH), 100 nM fulvestrant, 50 μM PD98059 (MEK1/2 inhibitor), or 100 nM Wortmannin (PI3K inhibitor). Concentrations of PD98059 and Wortmannin were selected to minimize potential off target effects [39]. Cells (25,000/insert) were plated in Corning Matrigel Invasion Chamber (VWR Cat #62405–744) following treatment, and incubated 18 h using 10% FBS-IMEM as the chemoattractant in the lower chamber. The inserts were removed, scrubbed, fixed, stained with 0.5% Crystal Violet, and allowed to dry overnight. Images were captured on an EVOS microscope (4x) and quantified using ImageJ2 software [38].
2.10. Soft agar colony formation assay
For the base layer, 1.5 ml of 0.5% Noble agar (Thermo Scientific) in DMEM medium was added per well in 6 well plates and allowed to polymerize. The top layer was prepared by preparing 0.35% agar (as above) in the same medium, and was cooled to ~ 40°C and then 50,000 cells were mixed in 1.5 mL of top layer agar and plated over the base layer. Plates were allowed to solidify prior to incubation at 37°C in 5% CO2. Colony formation and growth on soft agar was monitored daily by microscopic observation. Images were captured by EVOS microscope at days 1, 7, 14, and 21 after plating. Data were analyzed by one way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test in GraphPad Prism. Two-way ANOVA as used to assess the main effect of HNRNPA2B1 (control or stable overexpression; siControl or siHNRNPA2B1 transfection) and antiestrogen treatment (DMSO vehicle control vs. 4-OHT or fulvestrant treated sample groups) and the HNRNPA2B1 × antiestrogen treatment interaction. Two-way ANOVA was also used to examine the main effect of HNRNPA2B1 stable overexpression and interaction with time (days) in the soft agar assay. For two-way ANOVA, the Bonferroni post-hoc test was used for analysis in GraphPad Prism.
2.11. Clonogenic survival assay [40, 41]
Cells were seeded at 5,000 (MCF-7-A2B1) or 2,500 (MCF-7, LCC9, and LY2) cells/ well in 6-well plate in IMEM + 10% FBS + 1% P/S. After 12 h, the medium was replaced with SS medium and treatments (DMSO or 100 nM 4-OHT) every 48 h. After 14 d, the cells were stained with 0.5 % crystal violet, 25% methanol and images were captured with a Canon PowerShot SX700 camera and converted to TIFF files in Adobe Photoshop for analysis using ImageJ2 software [38]. The number of colonies and % colony forming efficiency (% CFE = (Number of cells counted/number of cells seeded) X100) [42] was analyzed by Image J.
2.12. Fluorescent-activated cell sorting (FACs) for cancer stem cell (CSC) makers:
Cells were washed once with phosphate-buffered saline (PBS) and harvested using Cellstripper (25–056-Cl; Corning, Corning, NY). Detached cells were washed with PBS and resuspended in stain buffer (106 cells/100 μl) (cat# 554656; BD Biosciences, San Diego, CA). A combination of fluorochrome-conjugated monoclonal antibodies against human CD44 (mouse allophycocyanin (APC) anti-CD44 antibody cat# 559942; BD Biosciences) and human CD24 (mouse PE anti-CD24 antibody cat# 555428; BD Biosciences) were used to stain single-cell suspensions at 4°C in the dark for 30 minutes. Control samples were unstained. The labeled and control cells were washed in stain buffer, fixed in PBS-containing 4% paraformaldehyde, and analyzed using BD LSRFortessa™ Flow Cytometer (BD Biosciences, San Diego, CA). For each cell line, gating was performed on unstained cells. The percentage of CD44+/CD24−/low cells were calculated for each cell line based on gated unstained control population.
2.13. Statistical Evaluation
Prism™ software (version 8, GraphPad Inc.) was used to perform data transformations and plotting, linear and nonlinear regression, and statistical analyses. For plotting and statistical analyses, the pooled sample from (n) total experiments were used as the data set. Where two data sets are compared, Student’s two-tailed t-test was used. Where more than two data sets were compared, one way ANOVA followed by Tukey’s multiple comparison post-hoc test or two way ANOVA followed by Bonferroni post-hoc test were performed. P values are provided in Figures with p < 0.05 considered significant.
3. RESULTS
3.1. IHC examination of HNRNPA2B1 (A2B1) in a breast TMA
Higher expression of A2B1 in primary breast tumors compared to normal breast has been reported by several [18, 21, 22, 43], but not all [44] investigators. A2B1 transcript expression is higher in breast tumors compared to normal breast in the GEPIA database [45] (Supplementary Fig. 1A). Higher A2B1 protein expression is significantly associated with reduced OS in all breast cancer patients and in those with ER+ breast tumors in KM plotter analysis (Supplementary Fig. 1B and C) [46]. IHC was performed to examine A2B1 staining in a TMA containing primary breast tumors paired with normal breast tissue or lymph node metastases (LNM) from the same patient (Figure 1 and Supplementary Fig. 2). A2B1 staining was nuclear. Strong nuclear A2B1 staining was identified in > 50 % of the epithelial cells in the invasive ductal carcinomas (IDC) on the TMA. A2B1 expression was higher in the ER+ IDC tumors than the paired normal ER+ breast tissue (Table 1). All 13 primary ER+ IDC tumors paired with ER+ LNM showed equivalent staining (Figure 1E, Table 1). We detected higher A2B1 staining in HER2+/ER− tumors compared to their paired LNM (Table 1). We did not detect any difference in A2B1 staining between ER+/HER2+ or triple negative breast cancer (TNBC) and their matched LMN (Table 1).
Figure 1: IHC staining for HNRNPA2B1 (A2B1) in ER+ invasive breast carcinoma (IDC) tumors paired with normal breast or lymph node (LN) metastasis from the same patient.
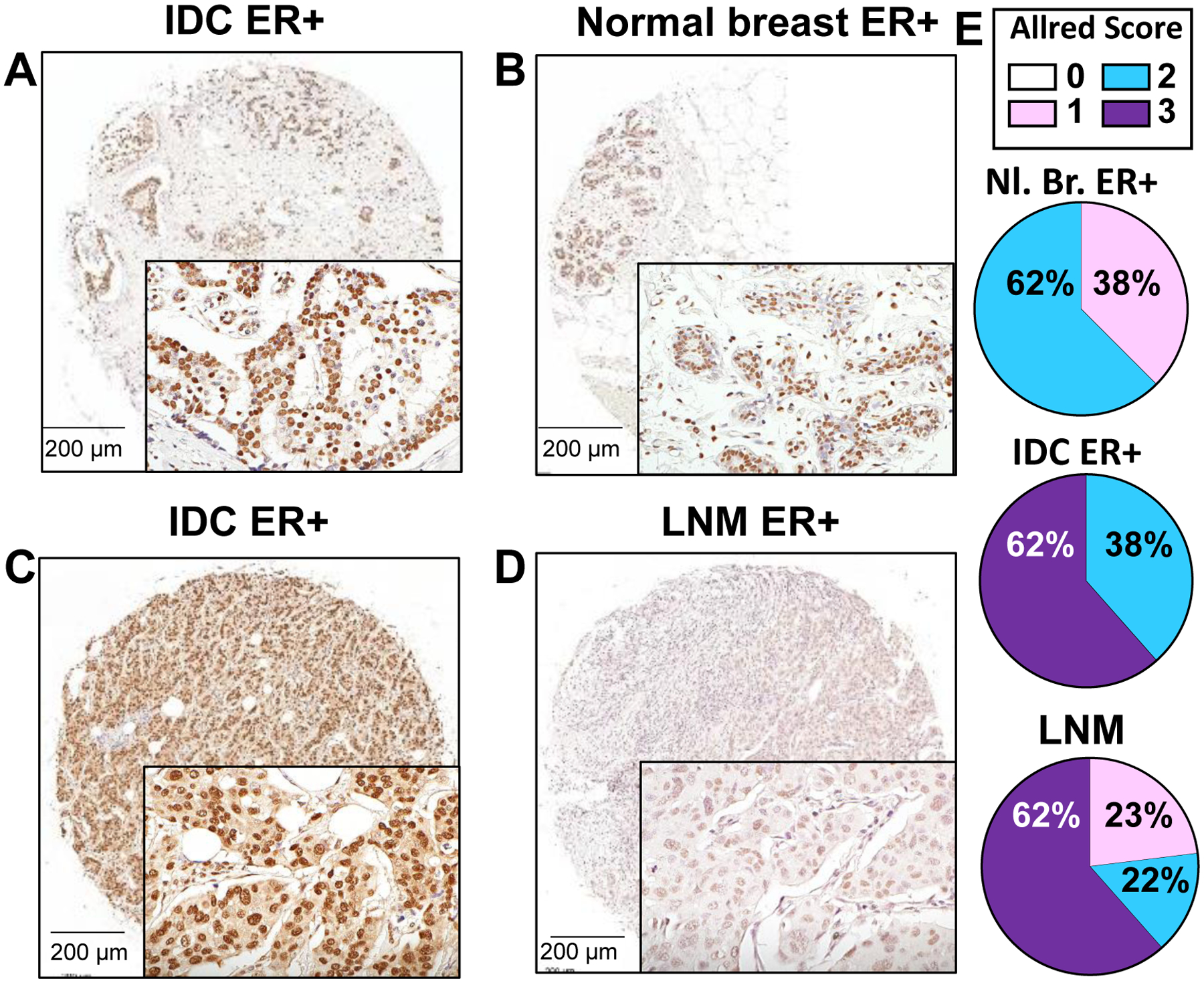
Shown are representative images from a stained TMA. The tumor in A (Allred score 3) is paired with normal breast tissue (B) from the same patient (Allred score 2). The tumor in C (Allred score 3) is paired with its LN metastasis in D (Allred score 1). Bar is 200 μm. Images were taken under 20x magnification. E) Quantification of Allred scores based on intensity and percent of cells stained are indicated as the percent total number of that sample in the TMA (Nl. Breast ER+ = 8; IDC ER+ with matched LNM ER+ = 13).
Table 1: Allred scores from IHC HNRNPA2B1 of the breast TMA with paired breast tumor-normal breast tissue and breast tumor-lymph node metastasis (LNM).
All primary breast tumors were classified as invasive ductal carcinomas (IDC). ER and HER2 staining data were provided by the supplier (Reveal Biosciences, see Materials and Methods). Overall statistical differences was calculated using the Kruskal-Wallace test followed by Dunn’s multiple comparison or Mann Whitney non-parametric tests.
| Sample | IDC vs. Normal Breast | IDC vs. LNM | Normal Breast vs. LNM |
|---|---|---|---|
| ER+/PR+ | 0.0360* | 0.5645 | 0.5312 |
| HER2+, ER− | NA | 0.0024* | NA |
| HER2+, ER+, PR+/− | NA | 0.7669 | NA |
| TNBC | NA | 0.3636 | NA |
statistically different.
NA = Not applicable because the TMA did not include normal matched breast tissue from these primary breast tumors.
3.2. Characterization of stable HNRNPA2B1-overexpressing MCF-7 cells (MCF-7-A2B1)
A2B1 protein expression is ~ 4-fold higher in TAM-resistant LY2 and LCC9 breast cancer cells compared to parental MCF-7 cells [23] (Supplementary Fig. 3). To investigate if higher A2B1 expression contributes to a loss of growth inhibition by TAM, we established a stable MCF-7 cell line with modest A2B1 overexpression (MCF-7-A2B1) similar to levels in LCC9 and LY2 cells. The expression of A2B1 was confirmed by qRT-PCR (Fig. 2A), and western blot, showing an ~ 4.7-fold increase in 37 kDa A2B1, relative to MCF-7 cells (Fig. 2B,C). This stable overexpression reflects the chronic upregulation of A2B1 observed in breast tumors [18, 21, 22, 43]. In addition to the 37kDa A2B1, a band of ~ 128 kDa was also detected in MCF-7-A2B1 cells (Fig. 2B). A2B1 dimerizes upon recognition of viral DNA to activate and amplify type I interferon responses [47], forms tetramers in 40S nuclear ribonuleoprotein particles [20], and self-aggregates via its intrinsic prion-like domain [48]. Thus, the higher MW band may be an A2B1 aggregate.
Figure 2: Stable overexpression of HNRNPA2B1 (A2B1) in MCF-7 cells.
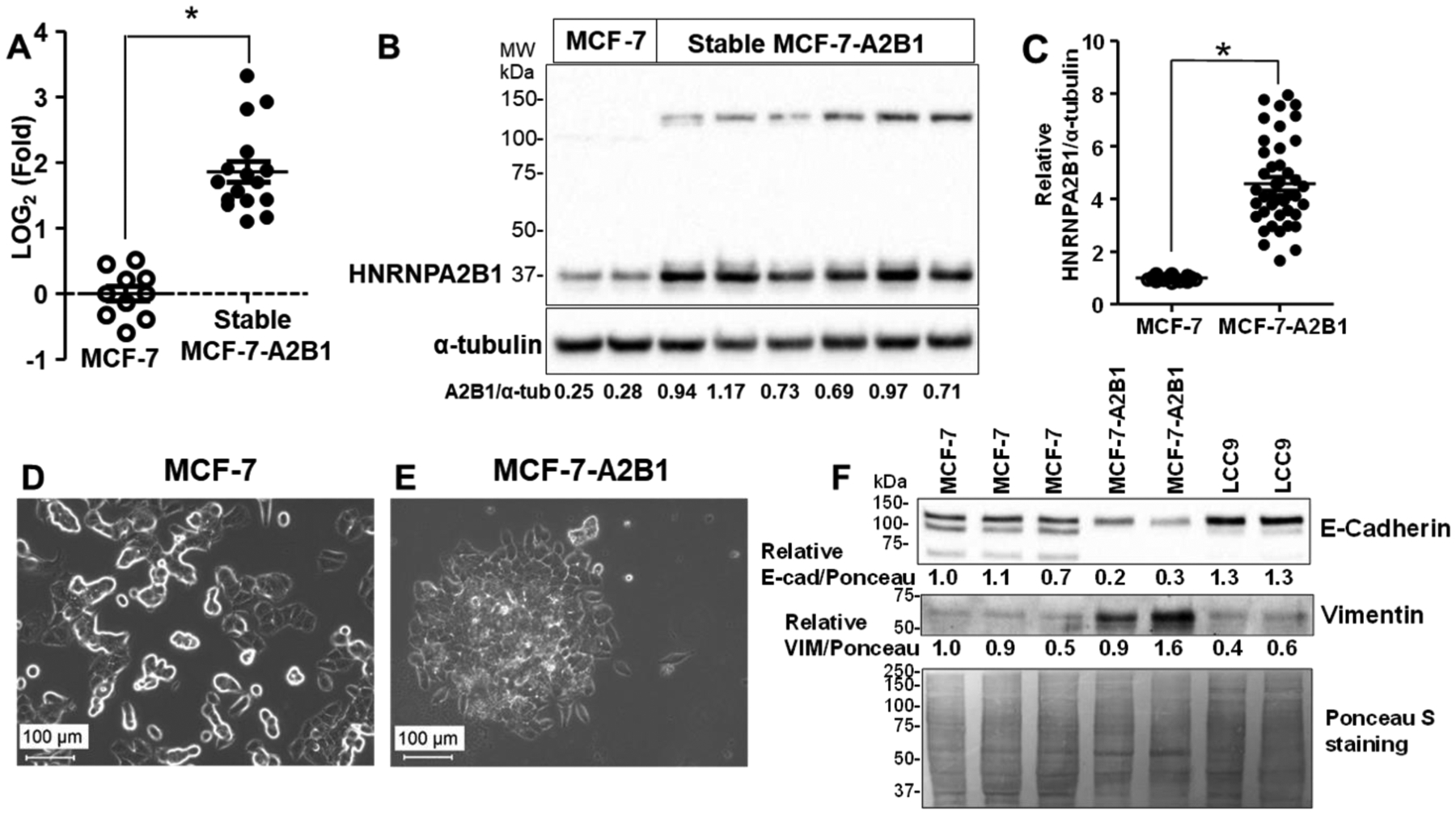
A2B1 transcript (A) and protein (B and C) levels were measured in the G418-selected MCF-7-A2B1 cells and MCF-7 cells. GAPDH and α-tubulin were was used as a normalization controls for qPCR and western blots respectively. C) Summary of 16 separate western blots, each with 3–8 separate preparations of WCE. *p < 0.0001, Student’s two tailed t-test. Phase contrast images of MCF-7 (D) and MCF-7-A2B1 cells (E) at 20X phase contrast imaging, scale bar = 100 μm (additional images in Supplementary Figure 4). F) Representative western blot showing E-cadherin and vimentin in WCE from the indicated cell lines. Quantification is relative to Ponceau S staining with the MCF-7 sample in the first lane set to 1.0 for relative quantification.
MCF-7-A2B1 cells aggregate more than MCF-7 cells (Fig. 2D,E; Supplementary Fig. 4), characteristic of some invasive breast cancer cells [49]. Similar changes in some TAM-resistant MCF-7 cells created by prolonged exposure to 4-OHT have been reported and termed “mesenchymal phenotype” [50]. We detected reduced E-cadherin and increased vimentin in MCF-7-A2B1 cells (Fig. 2F), supporting the possibility, that overexpression of A2B1 promotes an apparent epithelial-to-mesenchymal transition (EMT) in MCF-7 cells. LCC9 TAM-resistant cells have not undergone EMT, serving with MCF-7 as a negative control.
3.2. Stable A2B1 expression reduces MCF-7 cell viability and attenuates endocrine-sensitivity to ER antagonists
We compared the effect of 4-OHT and fulvestrant on cell viability in MCF-7, MCF-7-A2B1, and LCC9 cells (Fig. 3). As expected based on previous reports [51, 52], 4-OHT and fulvestrant inhibited MCF-7 viability whereas neither 4-OHT nor fulvestrant inhibited LCC9 cell viability. MCF-7-A2B1 showed reduced viability compared to MCF-7 cells with similar viability as LCC9 cells (Fig. 3A). As seen in LCC9 cells, MCF-7-A2B1 cells showed no growth inhibition by either 4-OHT or fulvestrant. While the concentrations of 4-OHT and fulvestrant used in cell treatments are higher than the serum levels of ~ 5–6 nM [53–55] and 4–16 nM [56, 57], respectively, 4-OHT in tissues are 10–60-fold higher, from 29–165 nM to 174 −990 nM (8,9). These concentrations are identical or lower than those used in studies of MCF-7 and other breast cancer cell lines [58–63].
Figure 3: Stable overexpression of A2B1 in MCF-7 cells reduces cell viability and reduces growth inhibition by 4-OHT and fulvestrant.
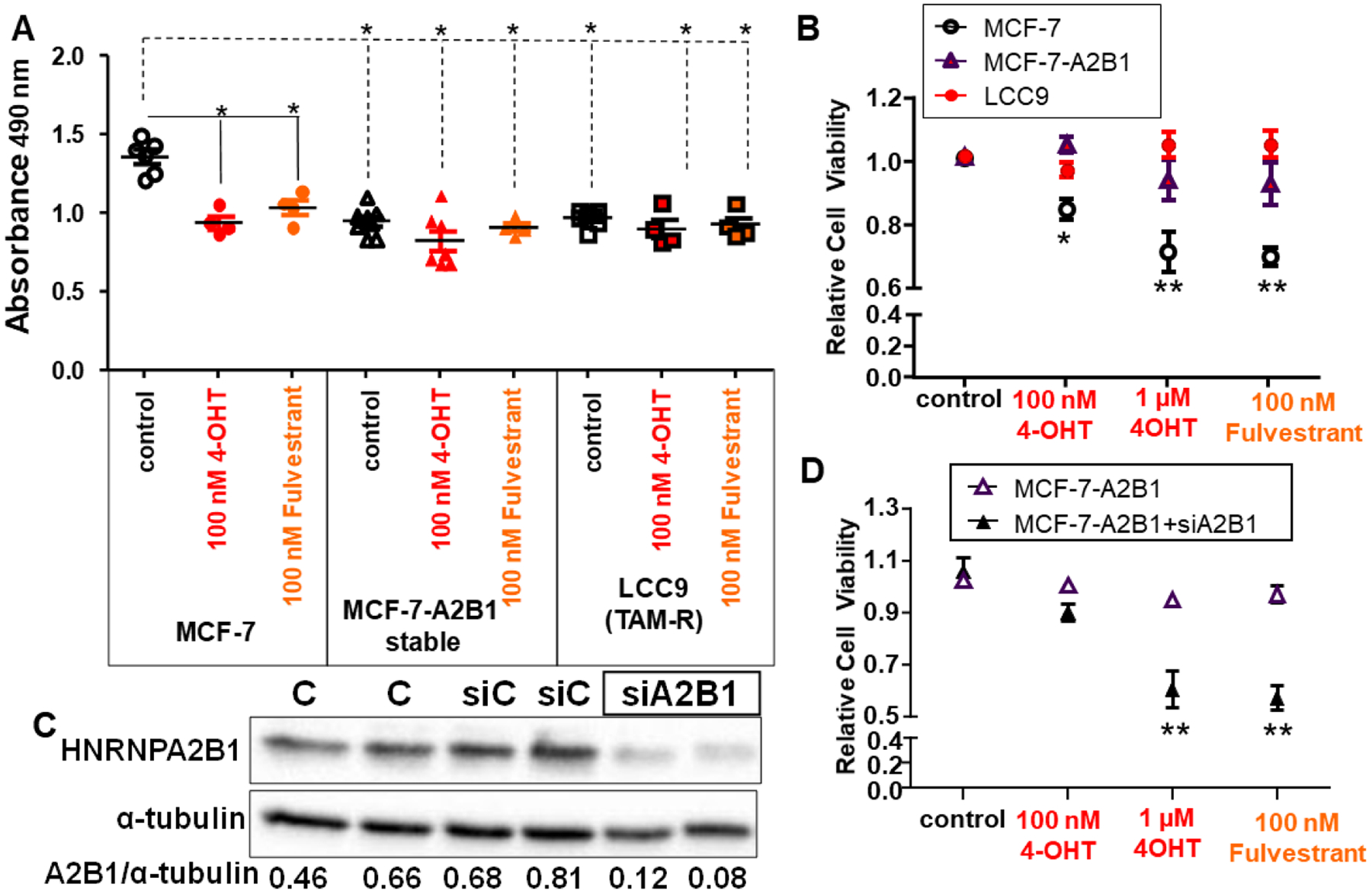
MCF-7, MCF-7-A2B1, and LCC9 (TAM-resistant (R)) cells were grown in SS medium for 48 h prior to treatment with vehicle control (DMSO), 100 nM or 1 μM 4-OHT, or 100 nM Fulvestrant for 4 days. A) Absorbance values are from one experiment in quadruplicate wells. *p < 0.05 one-way ANOVA, Tukey’s post hoc multiple comparisons test. B) Values were normalized to control within each cell line and are the mean ± SEM from 8 independent MTT assays. *p ≤ 0.01 and **p < 0.0001 two-way ANOVA, Bonferroni post hoc test. C) MCF-7-A2B1 cells were not transfected (C) or transfected with siControl (siCont) or siHNRNPA2B1 (siA2B1) in OPTI-MEM 48 h. Cells were grown in SS medium for an additional 24 h prior to isolation of WCE. The blot was stripped and re-probed for α-tubulin. The A2B1/α-tubulin ratios are shown. D) MCF-7-A2B1 cells were transfected with siControl (−) or siA2B1 for 48 h prior to counting and replating for 5 d MTT assays. Treatments: C = EtOH control, Fulv. = Fulvestrant. Values are the mean ± SEM from 6 experiments. **p < 0.0001 two-way ANOVA, Bonferroni post hoc test.
Further independent cell viability experiments were performed using 100 nM or 1 μM 4-OHT to model the concentrations of 4-OHT detected in breast cancer tissues when patients received 5 or 20 mg tamoxifen citrate/day [64] and 100 nM fulvestrant [29]. Two-way ANOVA followed by Bonferroni post-hoc tests indicate that stable A2B1 expression in MCF-7 cells reduced 4-OHT and fulvestrant inhibition of cell viability (Fig. 3B; Supplementary Fig. 5). These data agree with the reduced endocrine sensitivity detected in A2B1-transiently transfected MCF-7 cells [23] and support a role for increased A2B1 expression in loss of sensitivity to 4-OHT and fulvestrant in MCF-7 cells. Transfection of MCF-7-A2B1 cells with siA2B1 reduced A2B1 protein by ~85% (Fig. 3C). Two-way ANOVA followed by Bonferroni post-hoc tests indicate that siA2B1 transfection of MCF-7-A2B1 cells increased 1 μM 4-OHT and 100 nM fulvestrant inhibition of cell viability (Fig. 3D). These data suggest that A2B1 promotes resistance to antiestrogens and reduction of A2B1 expression restores antiestrogen-sensitivity in these ER+ breast cancer cells. Knockdown of A2B1 in MCF-7 cells did not affect the growth inhibition by 4-OHT or fulvestrant (Supplementary Fig. 6), as expected because MCF-7 have lower A2B1 than the antiestrogen-resistant cell lines.
3.3. A2B1 knockdown increases endocrine-sensitivity in LCC9 and LY2 cells
To examine A2B1’s role on the endocrine-sensitivity of LCC9 and LY2 TAM-resistant cells, each cell line was transfected with siControl or siA2B1 resulting in an average 73 and 80 % decrease in A2B1 protein expression, respectively (Fig. 4A,B,C). Depletion of A2B1 did not affect basal LCC9 or LY2 cell viability (Supplementary Fig. 7). A mixed-design, two-way ANOVA identified a significant interaction between siA2B1 and 4-OHT or fulvestrant treatment for LCC9 (F= 58.41, P < 0.001; Fig. 5D) and LY2 cell (F = 83, P < 0.001; Fig. 5E). Post hoc test analysis indicated that both doses of 4-OHT and fulvestrant were significantly different from DMSO control in both cell lines. In addition, the higher concentration of 4-OHT and 100 nM fulvestrant were significantly different from 100 nM 4-OHT in both cells. Importantly, these data indicate that inhibition of cell viability by 4-OHT and fulvestrant in both LCC9 and LY2 cells was dependent on HNRNPA2B1 knockdown (Fig. 4D, E). These data suggest that A2B1 suppression re-sensitized LCC9 and LY2 to antiestrogen therapies.
Figure 4: Transient knockdown of A2B1 enhanced antiestrogen-sensitivity of LCC9, LY2, and T47D cells.
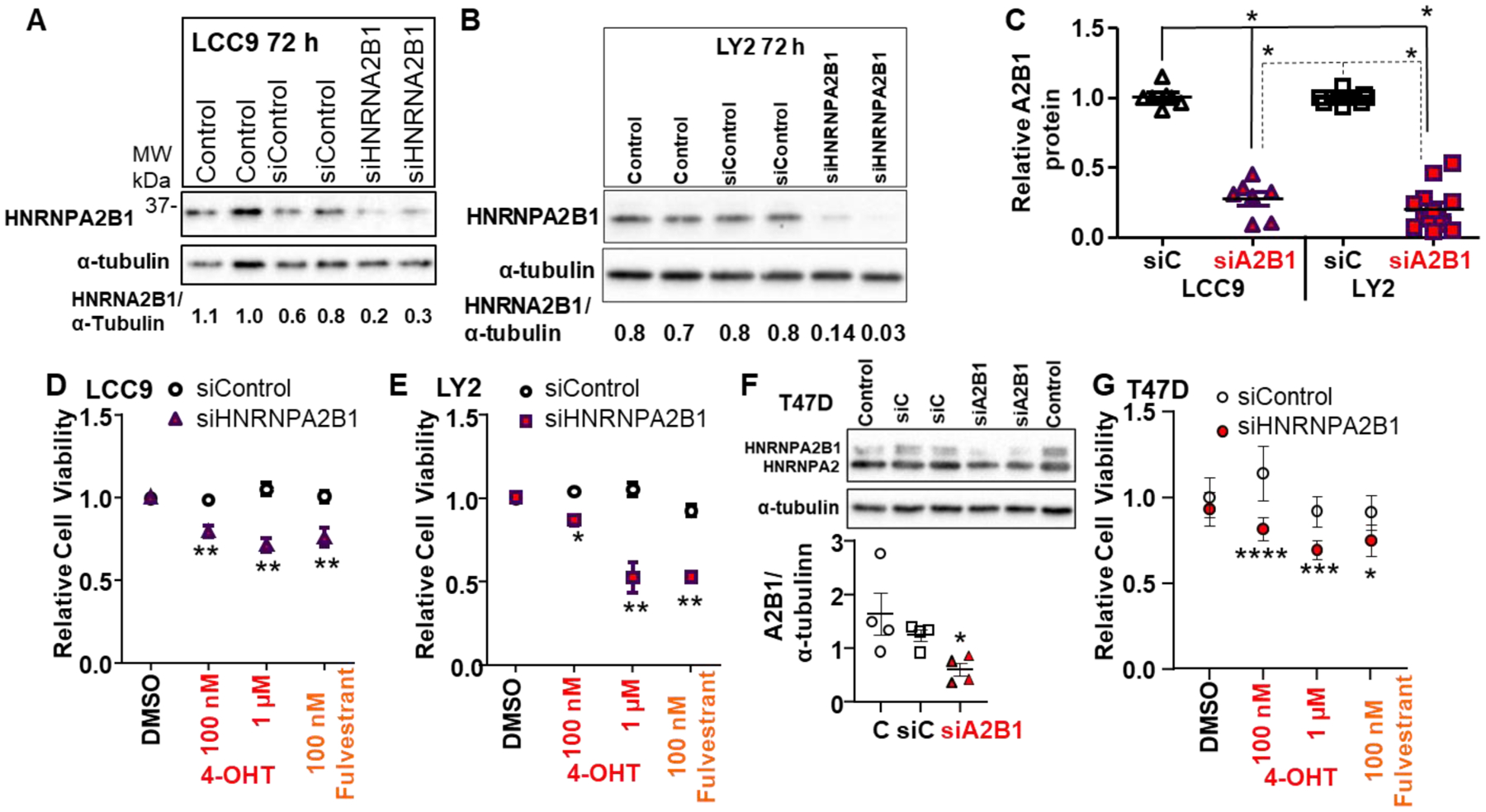
For A,B, and F the indicated cell lines were either not transfected (Control) or were transfected with siControl or siA2B1 for 48 h in Opti-MEM prior to 36 h in SS medium. For the blots shown, 30 μg WCE was loaded/lane. C: Summary of western blots of A2B1 relative to α-tubulin in LCC9 (n = 7) and LY2 (n = 13) separate WCE. *p < 0.0001 for one-way ANOVA followed by Tukey’s post hoc multiple comparisons test. D, E, and G summarize 8 (LCC9), 10 (LY2), and 4 (T47D) separate, independent MTT assays. After transfection and ~ 16 h in OPTI-MEM, cells were incubated for an additional ~ 32 h in SS medium prior to treatment with 100 nM or 1 μM 4-OHT or 100 nM fulvestrant, with medium changed every 48h, for 4 d. Values are the relative mean viability compared DMSO (vehicle control) ± SEM. *p < 0.05, ** < 0.01,***p < 0.001,***p < 0.0001 versus siControl DMSO-treated cells in two-way ANOVA, Tukey’s multiple comparison post hoc test. F shows a representative western blot for A2B1 knockdown in T47D cells using a monoclonal antibody that recognizes both the B1 and A2 splice variants of HNRNPA2B1. The graph shows data from 4 experiments, *p = 0.039 in one-way ANOVA followed by Tukey’s post hoc test.
Figure 5: A2B1 enhances MCF-7 cell migration and invasion.
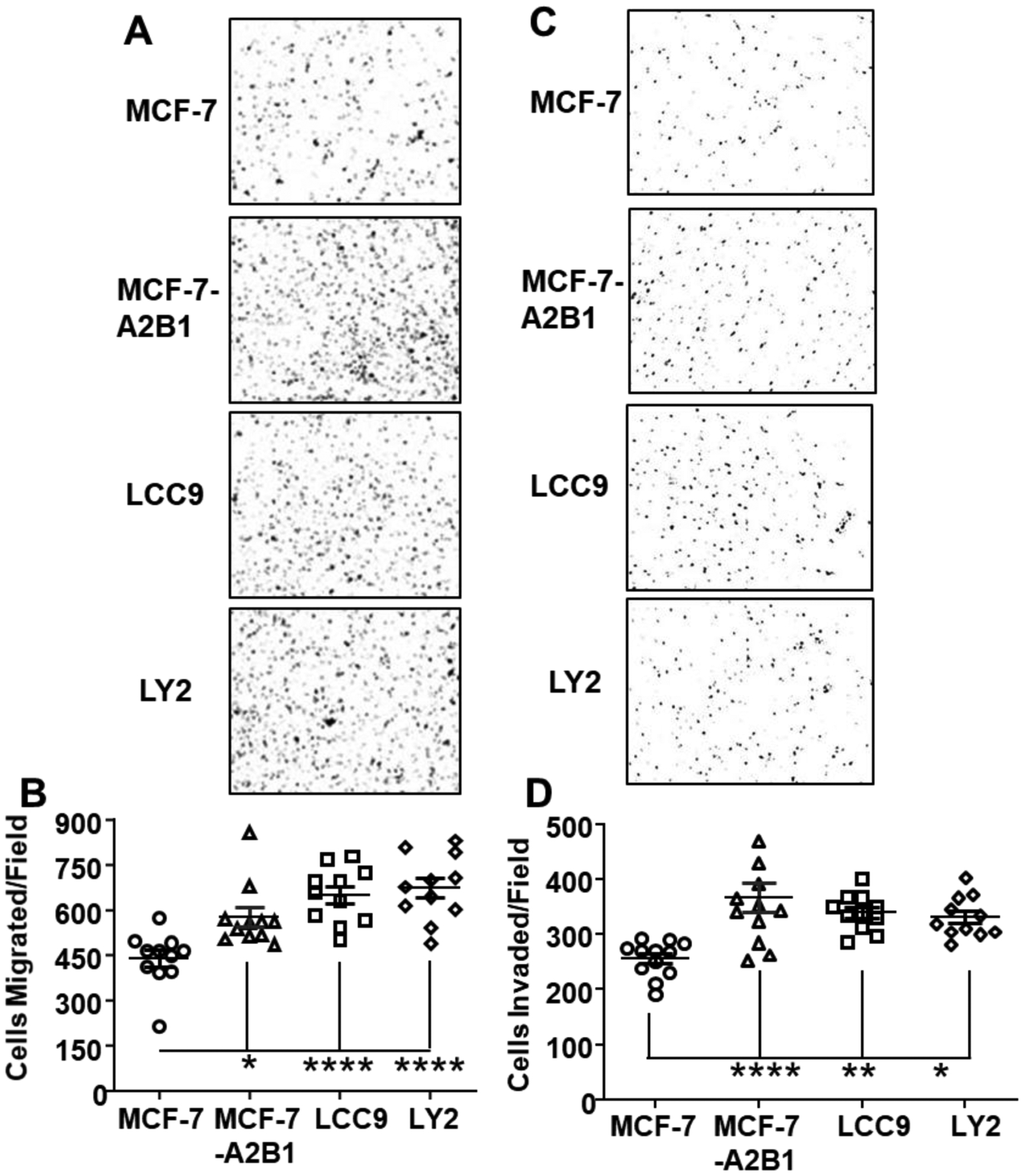
Boyden chamber assays comparing the migration and invasion of MCF-7, MCF-7-A2B1, LCC9, and LY2 cells were performed using FBS-containing medium as the attractant. Cells were grown in SS medium for 72 h prior to plating. Migrated (A) and invaded (C) cells were stained with crystal violet and quantified using ImageJ software. A) Representative images of migrated cells. B) Quantification of cell migration by Image J. C) Representative images of invaded cells. D) Quantification of cell invasion. Values in C and D are from individual wells with 3 wells/treatment. *p < 0.05, **P < 0.001,****p < 0.0001 in one-way ANOVA followed by Tukey’s post hoc multiple com.
3.4. A2B1 knockdown in T47D cells increases inhibition by antiestrogens
To address whether A2B1 regulates antiestrogen responses in another luminal A breast cancer cell line, we first examined the expression of A2B1 in T47D cells. We observed that the B1 splice variant of HNRNPA2B1 is lower in T47D cells compared to MCF-7 cells, but similar levels of the A2 splice variant (Supplementary Fig. 8). We then tested if knockdown of A2B1 would alter antiestrogen-sensitivity in T47D cells (Fig. 4F and G). A mixed-design, two-way ANOVA identified a significant interaction between siA2B1 transfection and 4-OHT or fulvestrant treatment for T47D cells (F= 5.813, P = 0.0047; Fig. 4G). Post hoc test analysis indicated that both doses of 4-OHT and fulvestrant were significantly different from DMSO-treated siControl-transfected T47D cells. These data suggest that siA2B1 enhanced antiestrogen-sensitivity in T47D cells.
3.5. HNRNPA2 is higher than HNRNPA2B1 in TNBC
Because TNBC tumors lack ERα, these tumors are ‘intrinsically resistant’ to endocrine therapies [65, 66]. We examined A2B1 expression by western blot analysis in three TNBC cell lines: MDA-MB-231 (Basal B, KRASG13D mutation), MDA-MB-468 (Basal A, PTEN homo deletion), and HCC1806 (Basal-like, BL2 subtype) (Supplementary Fig. 9). We observed that the levels of the B1 splice variant of HNRNPA2B1 is lower in TNBC cells compared to MCF-7 cells (Supplementary Fig. 9B, C). TNBC cells also have lower A2 splice variant protein compared to MCF-7 cells (Supplementary Fig. 9C).
3.6. A2B1 stimulates cell migration and invasion
To investigate the effect of A2B1 on cell migration, we performed Boyden Chamber assays for MCF-7, MCF-7-A2B1, LCC9, and LY2 cells (Fig. 5). The number of migrating cells was significantly higher in MCF-7-A2B1, LCC9, and LY2 cells compared to MCF-7 cells (Fig. 5B). Invasion assays revealed that more MCF-7-A2B1, LCC9, and LY2 cells invaded through the Matrigel layer than MCF-7 cells (Fig. 5D). 4-OHT did not affect the cell migration or invasion of any of these four cell lines (Supplementary Fig. 10). These observations are in agreement with previous reports that 4-OHT does not impair MCF-7 cell migration and invasion [67, 68]. Together these data suggest that increased A2B1 promotes MCF-7 cell migration and invasion as seen in the endocrine-resistant LCC9 and LY2 cells.
3.7. A2B1 knockdown inhibits LCC9 and MCF-7-A2B1 migration and invasion
To further test the role of A2B1 in cell migration and invasion, we transfected LCC9 or MCF-7-A2B1 cells with siControl or siA2B1 and examined cell migration and invasion (Fig. 6). Knockdown of A2B1 inhibited the migration and invasion of LCC9 and MCF-7-A2B1 cells (Fig. 6). Knockdown of A2B1 sensitized LCC9 cells to inhibition of cell migration by 4-OHT (Supplementary Fig. 11). These data suggest that A2B1 plays a role in the migration and invasion activities of LCC9 and MCF-7-A2B1 cells.
Figure 6: A2B1 knockdown reduces LCC9 and MCF-7-A2B1 cell migration and invasion.
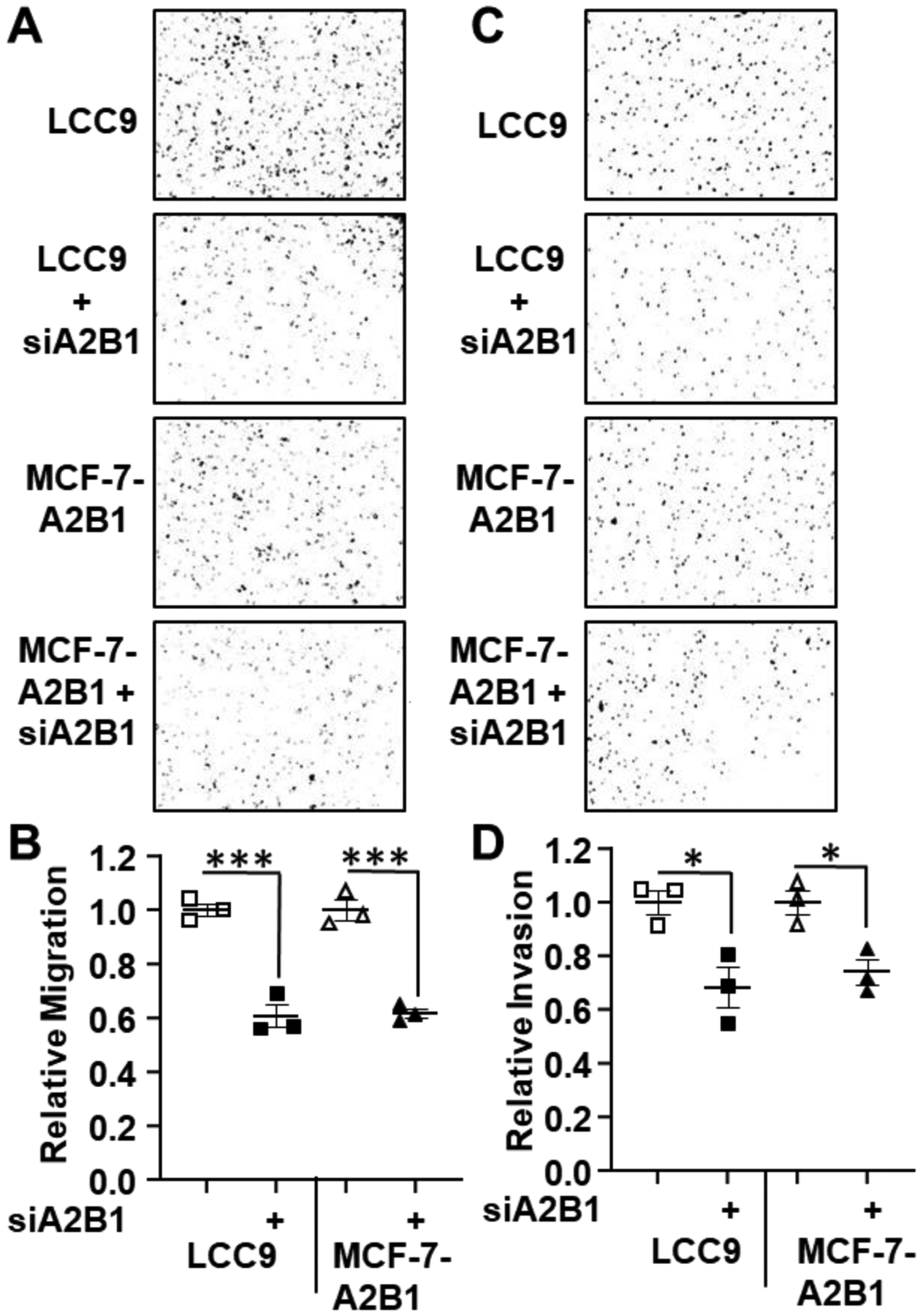
LCC9 and MCF-7-A2B1 cells were transfected with siControl or siHNRNPA2B1 (siA2B1) for 48 h prior to plating for cell migration and invasion assays as in Figure 5. Migrated (A) and invaded (C) cells were stained with crystal violet and quantified using ImageJ software. B and D) Quantification of cell migration and invasion by Image J. Values are relative migration or invasion/well (n = 3 wells/transfection). ***p < 0.001, *p < 0.05 in one-way ANOVA followed by Tukey’s multiple comparison post hoc test.
3.8. A2B1 knockdown inhibits clonogenic MCF-7-A2B1 and MCF-7 cell survival
Clonogenic assays are used to examine the ability of a single cell to grow into a colony, thus testing the proliferative capacity of the cells [69]. We examined the clonogenicity of MCF-7-A2B1 and LCC9 cells (Fig. 7). Transfection of siA2B1 inhibited clonogenicity in MCF-7-A2B1 cells (Fig. 7C) and the size of colonies formed for both MCF-7-A2B1 and LCC9 cells (Fig. 7D).
Figure 7: A2B1 stimulates clonogenicity in MCF-7-A2B1 cells.
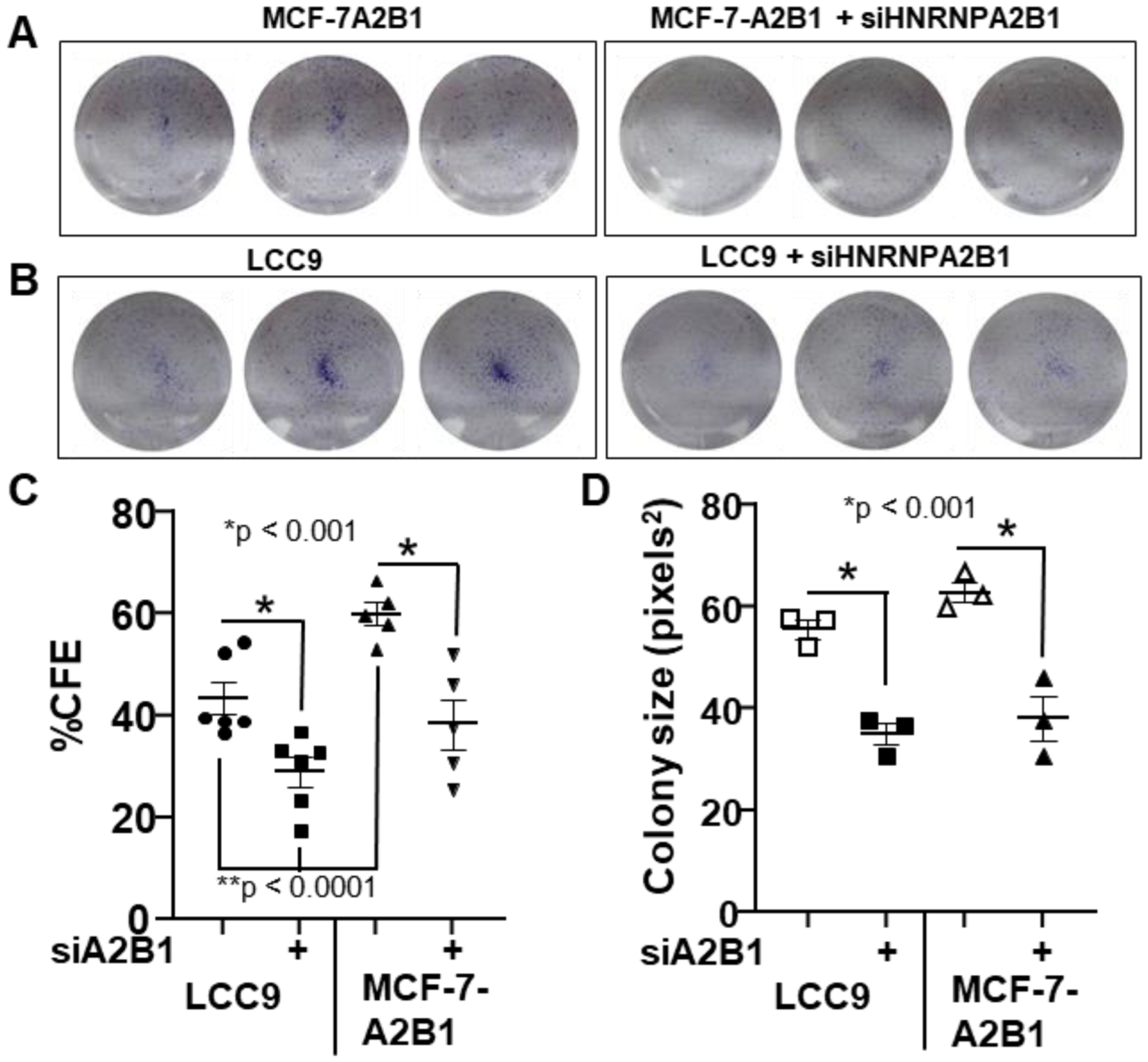
(A-B) Cells were transfected with siControl or siA2B1 for 24 h, then counted and re-plated at a density of 5,000/well (MCF-7-A2B1) or 2,500/well (LCC9) with 6 wells/treatment in six-well plates. After 5 d, cell images were captured and % colony forming efficiency (% CFE in C) and colony size (D) was analyzed by Image J. Data were analyzed by one-way ANOVA followed by Tukey’s post hoc multiple comparisons test;* p values of significance are indicated.
3.9. MCF-7-A2B1 cells form larger colonies in soft agar and siA2B1 inhibits soft agar colony growth in LCC9 and MCF-7-A2B1 cells
Soft agar colony formation assays are used to evaluate the anchorage-independent growth ability of cells as a hallmark of tumorigenesis and metastatic potential [70]. To address the impact of A2B1 on tumorigenic potential, we evaluated the total colony number and size of cell colonies of MCF-7 versus MCF-7-A2B1 after 1, 7, 14, and 21 days of incubation (Fig. 8A). There was no significant increase in colony number after plating (data not shown). Notably, MCF-7-A2B1 cells formed significantly larger colonies compared to MCF-7-parental cells (Fig. 8B). This suggests that modest A2B1 overexpression supports anchorage-independent growth.
Figure 8: A2B1 increases MCF-7 soft agar colony size and A2B1 knockdown inhibits soft agar colony growth.
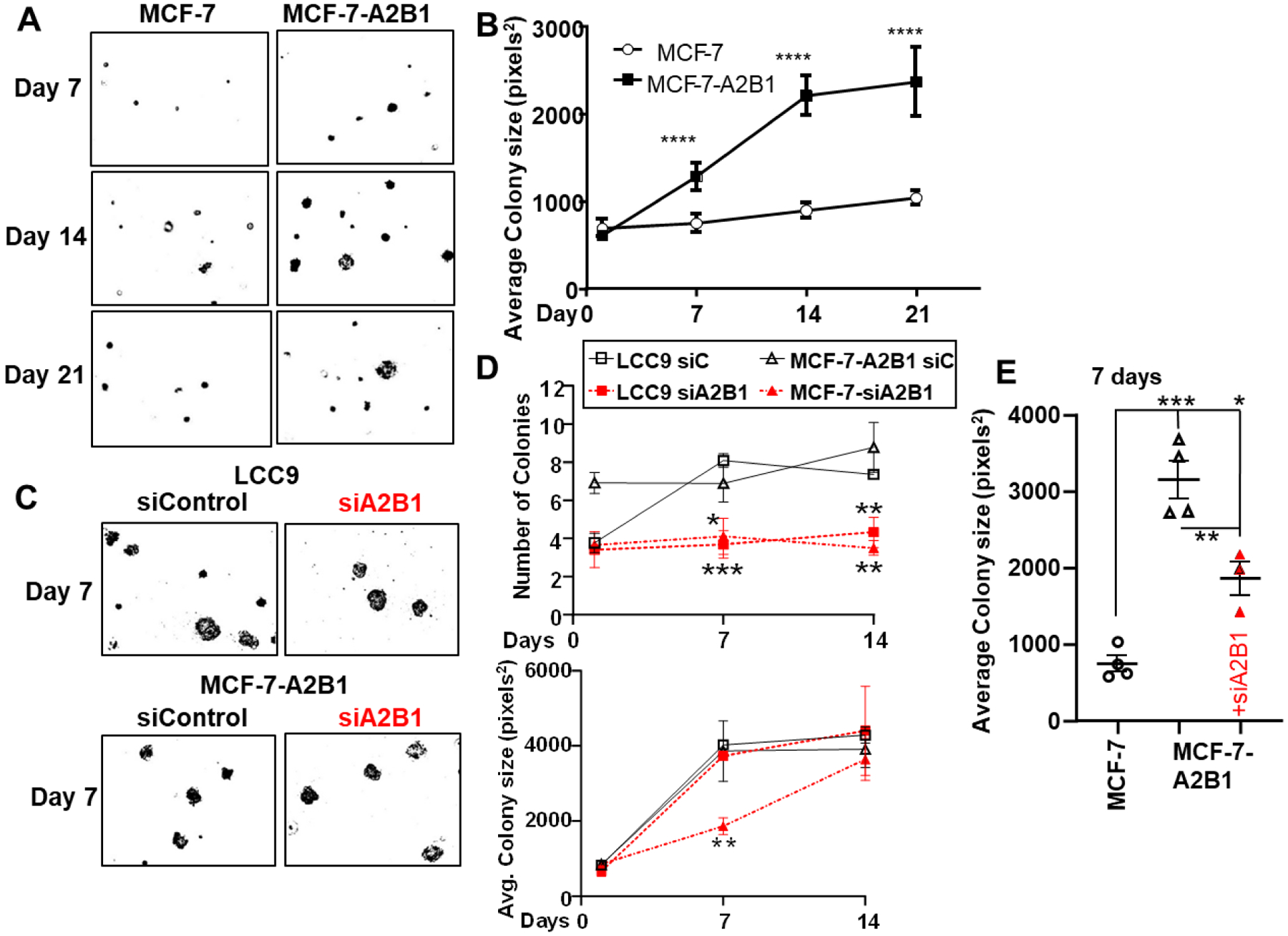
A) MCF-7 and MCF-7-A2B1 stable cells were plated 50,000 cells/60mm plate. For data analysis: cell images were captured after 7, 14, and 21 d at 10X on an EVOS microscope. The number of colonies and colony size (area of selection in pixels2) was analyzed by Image J. B) colony size values were averaged from 3 plates/sample and displayed as mean ±SEM. Data were analyzed by two way ANOVA followed by Bonferroni’s post test. *p < 0.0001 C) LCC9 and MCF-7-A2B1 cells were transfected with siControl or siA2B1 for 48 h prior to plating in three separate 60mm plates for soft agar assay/treatment. Data were analyzed by two way ANOVA followed by Bonferroni’s post test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. E) Colony sizes at 7 d for the MCF-7 and MCF-7-A2B1 +/− siA2B1 transfection were analyzed by one-way ANOVA followed by Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.0001
To address the specific role of A2B1 on the anchorage-independent growth of LCC9 and MCF-7A2B1 cells, we transfected LCC9 and MCF-7-A2B1 cells with siControl or siA2B1 for 48 h prior to plating for soft agar colony growth evaluated colony number and size after 1, 7, and 14, and 21 days (Fig. 8C and Supplementary Fig. 12). Like MCF-7-A2B1 cells, LCC9 formed significantly larger colonies compared to MCF-7-parental cells. Knockdown of A2B1 inhibited colony formation in LCC9 and MCF-7-A2B1 cells (Fig. 8D). These data support a role for A2B1 in anchorage-independent growth. We observed a significant decrease in colony size in siA2B1-transfected LCC9 cells treated with 4-OHT (Supplementary Fig. 12B). These data suggest that siA2B1 sensitizes LCC9 to inhibition of colony growth by 4-OHT, similar to the sensitization to antiestrogens seen in the cell viability assay (Fig. 4D). Knockdown of A2B1 in MCF-7-A2B1 cells reduced colony size at 7d, but not at 14 d (Fig. 8E, Supplemental Fig. 12B). We suggest that the transfected siA2B1 is reduced over time with cell replication.
3.10. Increased population of CD44+/CD24−/low cancer stem cells (CSC) in MCF-7-A2B1 cells
Cancer stem cells (CSC) are a small proportion of the cellular heterogeneity with a breast tumor that have the capacity for unlimited self-renewal and generate morphologically diverse progeny cells that form metastases [71]. Breast CSC are detected as a CD44+/CD24−/low cell subpopulation using FACS [72]. We examined the expression of the cell surface antigen markers CD44 and CD24 by FACS (Fig. 9A and Supplementary Fig.13). We observed ~ MCF-7-A2B1 showed a higher CSC population as assessed by CD44+/CD24−/low expression compared to MCF-7, LCC9, and LY2 cells (Fig. 9B). LCC9 and LY2 cells showed a higher population of CD44+/CD24−/low cells compared to MCF-7 cells (Fig. 9B). These data demonstrate that the maintenance of a CSC population in endocrine-resistant breast cancer cells associates with higher A2B1 expression.
Figure 9: FACS analysis of CD24 and CD44.
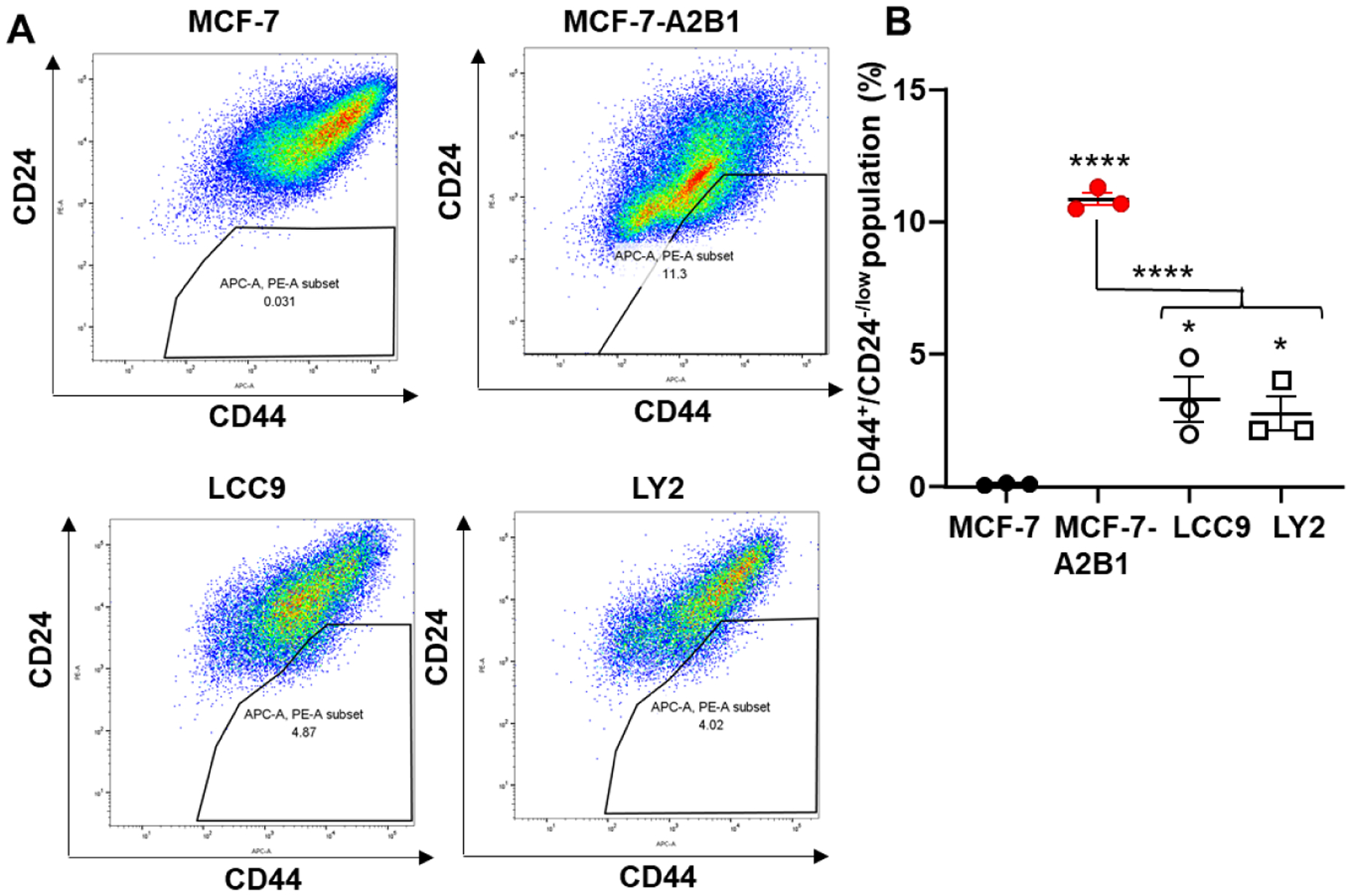
A) MCF-7, MCF-7-A2B1, LCC9 and LY2 cells were stained with anti-CD44 APC and anti-CD24 PE. The percent of CD44+/CD24−low cells were analyzed by flow cytometry. Data were quantified from three separate biological replicates of each cell line and plotted % CD44+/CD24−low. Values are mean ± SEM. ****p < 0.001, * p < 0.05 in one-way ANOVA followed by Tukey’s post hoc multiple comparisons test.
3.11. A2B1-mediated endocrine-resistance involves multiple pathways
Accumulating evidence supports a key role for PI3K/AKT/mammalian target of rapamycin (mTOR), and cyclin-dependent kinase (CDK) 4/6 activation in endocrine resistance and targeted therapies are in clinical use and being developed to block these pathways [73, 74]. Activation of AKT contributes to antiestrogen-resistance in LCC9 cells [75]. AKT1, AKT2, and AKT3 are key genes in antiestrogen/AI-resistant breast tumors [76]. AKT1 transcript levels are higher in LCC9 compared to MCF-7 cells and are higher than AKT2 or AKT3 transcript expression (Supplementary Fig. 14). Activation of AKT signaling is required for MCF-7 cell aggregation [77] and inhibition of PI3K/mTOR signaling inhibits TNBC cell soft agar colony formation in vitro [78]. The PI3K/AKT/MTOR pathway also plays a role in CSC maintenance [79]. To address whether the larger colony size of MCF-7-A2B1 cells (Fig. 7 and 8) associates with increased AKT signaling, we examined P-ser473-AKT/AKT by western blotting (Fig. 10). We observed higher P-ser473-AKT/AKT in LCC9 and MCF-7-A2B1 cells compared to MCF-7 cells (Fig. 10A). Knockdown of A2B1 resulted in a significant reduction of P-AKT/AKT in MCF-7-A2B1 and LCC9 (Fig. 10B, 10C). These data suggest a role for increased A2B1 in activation of AKT in MCF-7-A2B1 and LCC9 cells.
Figure 10: AKT and MAPK activation in MCF-7-A2B1 and LCC9 cells.
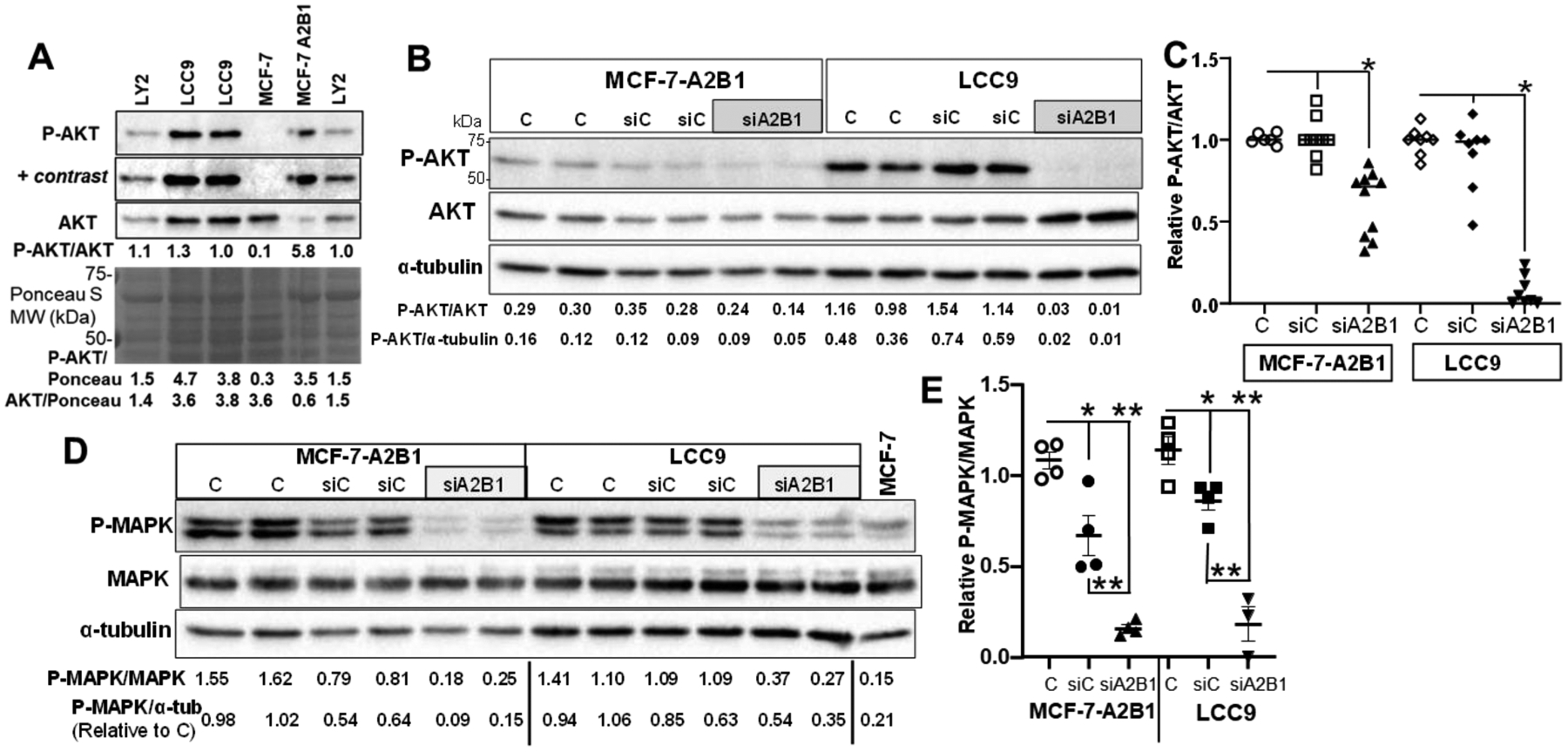
A) Western blot of phospho-ser-473 AKT/AKT/Ponceau S in the indicated cells grown in regular medium. + contrast indicates that the image was adjusted using ‘auto-contrast’ in Adobe Photoshop. B-D) MCF-7-A2B1 and LCC9 cells were transfected with siControl (siC) or siHNRNPA2B1 (siA2B1) in OPTI-MEM 48 h and then grown in 5% DCC-FBS medium for an additional 24 h prior to isolation of WCE for western blotting. Non-transfected cells grown in parallel, including medium changes are Control (C). For B and D) Values of the ratio of P-AKT/AKT, P-AKT/α-tubulin, P-MAPK/MAPK, or P-MAPK α-tubulin are shown for each blot. C and E) The P-AKT/AKT or P-MAPK/MAPK ratios for control cells in each cell line was set to 1 within each experiment. Data are from 3 separate experiments. E) * P < 0.001 from C and siC. E) * P < 0.01 and ** P < 0.001 versus C or siC, as indicated. Statistical analysis: one-way ANOVA followed by Tukey’s multiple comparisons test.
Activation of MAPK signaling is involved in TAM-resistance in MCF-7 cells [80] and alterations in genes in the MAPK pathway are enriched in ER+ breast tumors without ESR1 mutations in patients who developed metastatic disease [10]. We observed higher P-MAPK/MAPK in MCF-7-A2B1 and LCC9 cells compared to MCF-7 cells (Fig. 10D). Knockdown of A2B1 reduced of P-MAPK/MAPK in MCF-7-A2B1 and LCC9 cells (Fig. 10E). Because MAPK phosphorylates ERα on ser118, activating ERα in the absence of ligand [81], we examined P-ser118-ERα as a marker of MAPK activation. Knockdown of A2B1 reduced the P-ser118-ERα/ERα ratio more in MCF-7-A2B1 than LCC9 cells (Supplementary Fig. 15).
Since MCF-7-A2B1 show activation of MAPK and AKT, we tested the ability of MEK1/2 inhibitor PD98059 and PI3K inhibitor wortmannin to inhibit soft agar colony growth using each inhibitor at a concentration selective for respective pathway inhibition [82, 83]. Neither PD98059 nor wortmannin affected MCF-7 soft agar colony formation (Fig. 11A, C); however, both reduced MCF-7-A2B1 colony number at 14 d (Fig. 11B). In addition, wortmannin reduced MCF-7-A2B1 colony size (Fig. 11C). These data suggest that the gain of function of anchorage-independent colony growth in MCF-7-A2B1 cells depends on PI3K pathway activation. In agreement with previous reports for MCF-7 cells [84, 85], both wortmannin and PD98059 inhibited MCF-7, MCF-7-A2B1, and LCC9 cell viability after 5 d (Supplementary Fig. 16).
Figure 11: Inhibition of MEK and PI3K activity attenuates MCF-7-A2B1 soft agar colony growth.
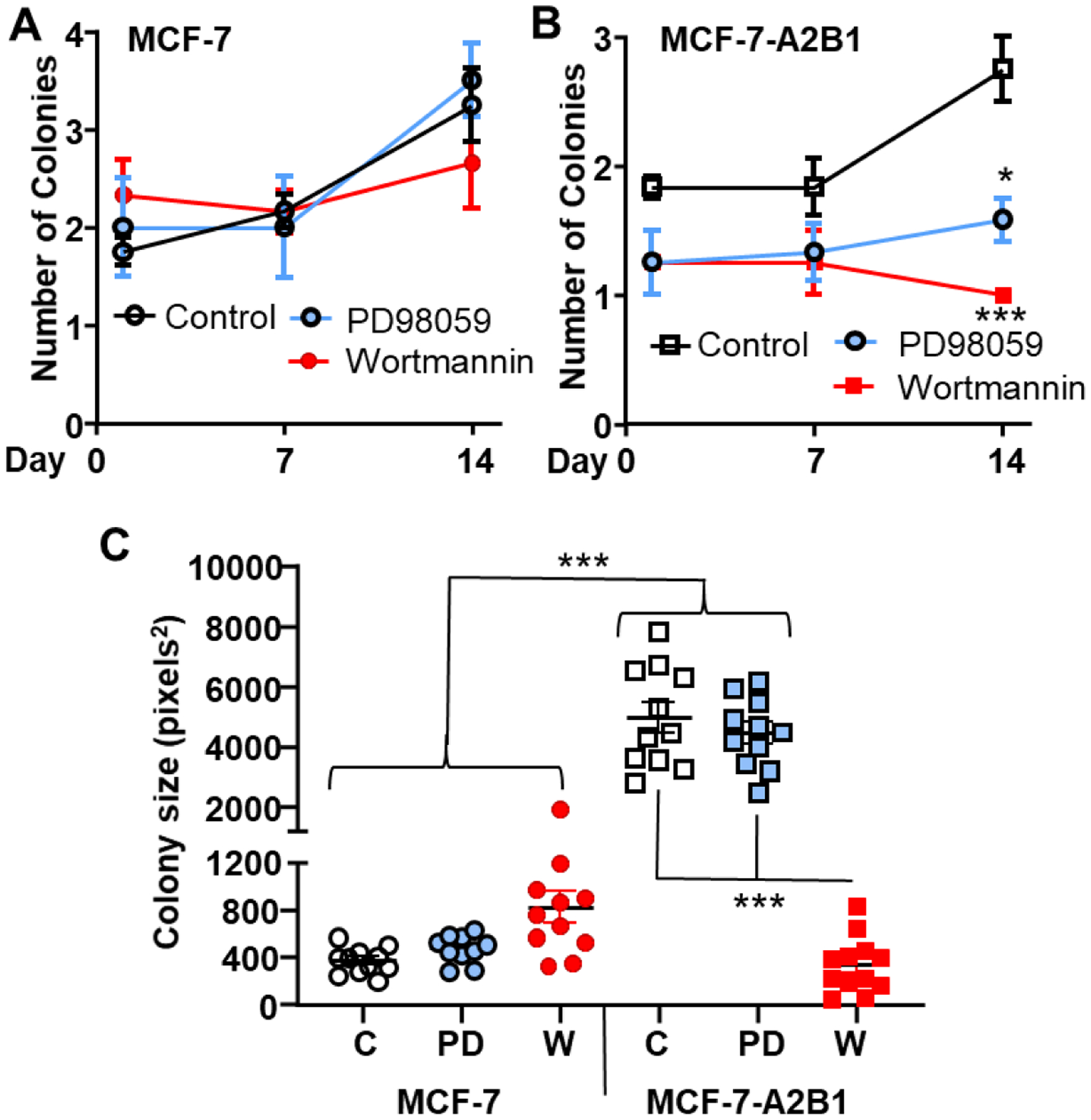
MCF-7 and MCF-7-A2B1 stable cells were treated with vehicle control (EtOH, C), 50 μM PD98059, or 100 nM wortmannin and plated 50,000 cells/60mm plate. The medium, with respective treatment, was replenished every 4 d. Cell images were captured and analyzed after 1, 7, and 14 d. Average ± SEM values from 3 replicate plates are plotted (A and B) for the number of colonies counted. Data were analyzed by one-way ANOVA followed by Tukey’s multiple comparison test * p < 0.05, ***P < 0.001 compared to the control-treated cells on the same day. C) Mean individual colony size values at 14 d ± SEM. Data were analyzed by one-way ANOVA followed by Tukey’s post hoc multiple comparison test, ***p < 0.0001.
Paradoxically, increased ERα plays a role in endocrine-resistance in breast cancer [86–88]. Since MCF-7-A2B1 cells show reduced sensitivity to antiestrogens (Fig. 3B), we examined ERα expression. MCF-7-A2B1 cells show increased ESR1 transcript levels (Fig. 12A) and ERα protein (Fig. 12B). We reported that transient A2B1 overexpression reduced miR-222–3p in MCF-7 cells [23]. ESR1//ERα is bona fide target of miR-222–3p [89]. Here we observed that MIR222–3p levels were significantly lower in MCF-7-A2B1 compared to MCF-7 cells (Fig. 12C). There was a significant inverse Pearson correlation between ESR1 and miR-222–3p expression in MCF-7-A2B1 cells (r = −0.97, p = 0.001, Supplementary Fig. 17).
Figure 12: ERα is increased in MCF-7-A2B1 cells and siA2B1 reduces ERα in LCC9, LY2, and MCF-7-A2B1 cells.
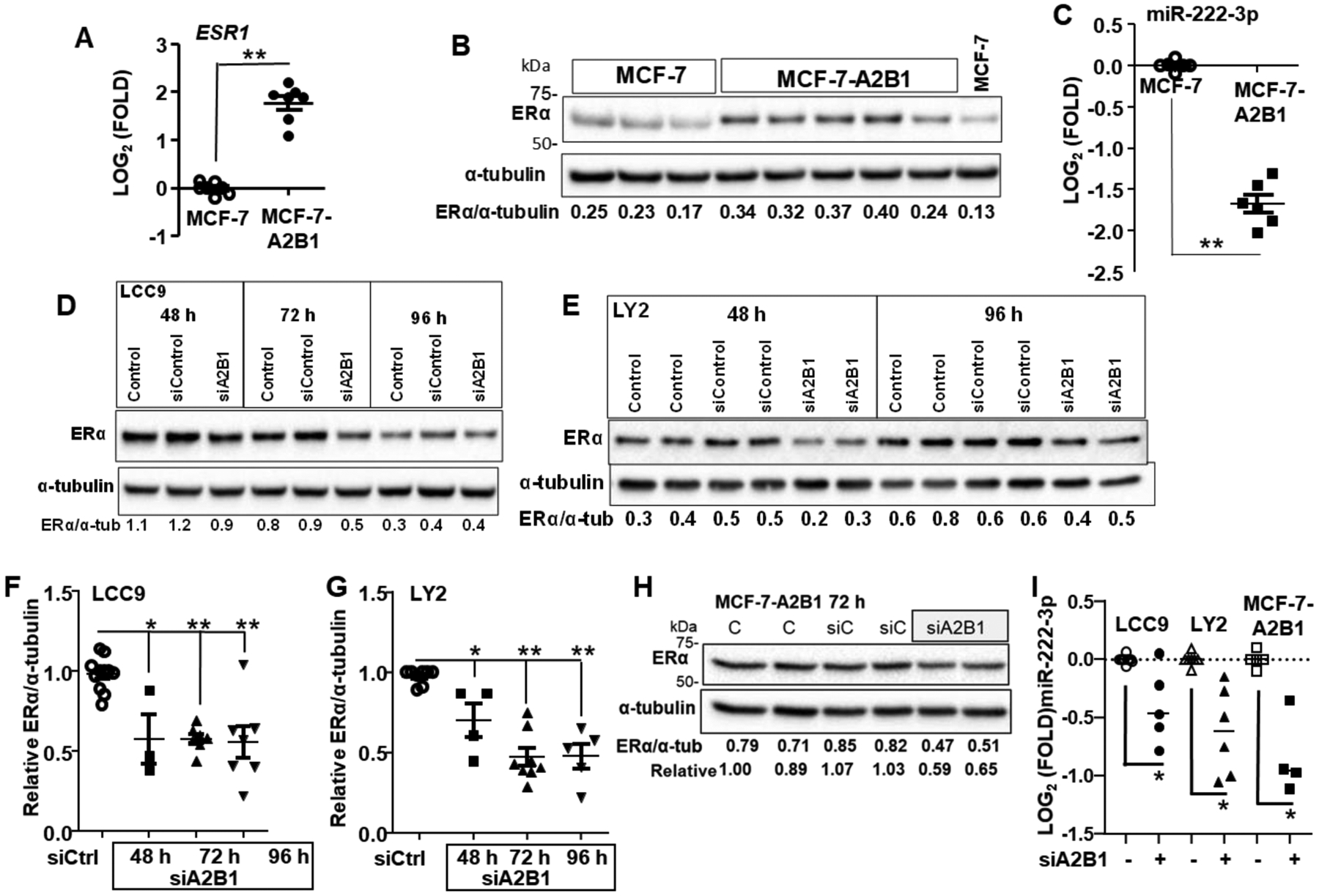
A) Data are qPCR of ESR1 relative to GAPDH and normalized to MCF-7 control cells. Values are from 8 (A) separate miRNA extractions. ** p < 0.001 in a two-tailed Student’s t-test. For B, D, E, and H WCE were isolated from the indicated cell lines. Cells were not transfected (B or Control (C) in panels D,E, and H) or transfected with siControl, siA2B1 for the indicated amount of time. The blots were probed for ERα, then stripped and re-probed for α–tubulin for normalization. Representative blots are shown. Data F and G) are summarized from biological replicate ERα westerns with different cell lysates from 3–4 separate transfection experiments/cell line. ERα/α-tubulin were normalized to siControl (set to 1) in each experiment for comparisons between separate western blots (Relative ERα). *P < 0.05, **P < 0.001 vs. siControl in a one-way ANOVA followed by Tukey’s post hoc test. C and I) qPCR of MIR221–3p relative to SNORD48 and normalized to control cells. For I, the cells were transfected with siControl or siHNRNPA2B1 for 72 h prior to miRNA extraction and qPCR. *P < 0.0025 vs. siControl in a one-way ANOVA followed by Tukey’s post hoc test.
Endocrine resistant LCC9 and LY2 cells express ERα [51]. In LCC9 cells, ERα plays a role in antiestrogen-resistance and ERα knockdown sensitized LCC9 cells to fulvestrant [90]. We examined if the observed restoration of antiestrogen-sensitivity with A2B1 knockdown (Fig. 4D, E) is accompanied by a decrease in ERα (Fig. 12D,E). Knockdown of A2B1 led to a significant decrease in ERα in LCC9, LY2, and MCF-7-A2B1 cells (Fig. 12F–H). Knockdown of A2B1 had no significant effect on steady state levels of ESR1 transcript expression in any cell line (Supplementary Fig. 18A). Knockdown of A2B1 had no significant effect on ERα in MCF-7 cells (Supplementary Fig. 18B, C). These data suggest that the higher endogenous levels of A2B1 in endocrine-resistant LCC9 and LY2 cells and in MCF-7-A2B1 cells support ERα protein expression.
4. Discussion:
It is well documented that endocrine resistance inevitably occurs in ERα+ metastatic breast cancer and involves somatic, epigenetic, and tumor microenvironment changes [2]. In this study, we demonstrated that a modest stable ~ 4.2-fold overexpression of A2B1 in MCF-7 cells reduced sensitivity to 4-OHT and fulvestrant. Conversely, transient A2B1 knockdown in TAM-resistant LCC9 and LY2 cells sensitized each cell line to growth inhibition by 4-OHT and fulvestrant that was dependent on the reduction in A2B1. These data suggest a role for A2B1 in endocrine resistance in vitro.
We have summarized the direct findings attributable to A2B1 reported here and connected them to TAM-resistance in Figure 13. The MCF-7-A2B1 cells share features in established TAM- and fulvestrant-resistant LCC9 and LY2 cells that were independently derived for endocrine-resistance in vivo and in vitro, respectively [29, 91, 92]. Like LCC9 and LY2 cells, MCF-7-A2B1 have higher ERα, activation of MAPK and AKT. Knockdown of A2B1 restores TAM and fulvestrant sensitivity to LCC9, LY2, and MCF-7-A2B1 cells confirming a role for A2B1 in resistance to these endocrine therapies. Further, A2B1 knockdown in MCF-7-A2B1, LCC9, and LY2 cells reverses most of the observed growth and movement phenotypes and markers. However, there are differences, e.g., knockdown of A2B1 in MCF-7-A2B1 reduces their clonogenicity, but had no effect on the clonogenicity of either LCC9 or LY2 cells. These observations indicate that A2B1 is one of several regulatory proteins whose expression is increased in endocrine-resistant breast cancer cells and whose expression correlates with reduced DFS and OS in breast cancer patients [23].
Figure 13: A model of how upregulation of HNRNPA2B1 may contribute to TAM-resistant (TAM-R) breast cancer.
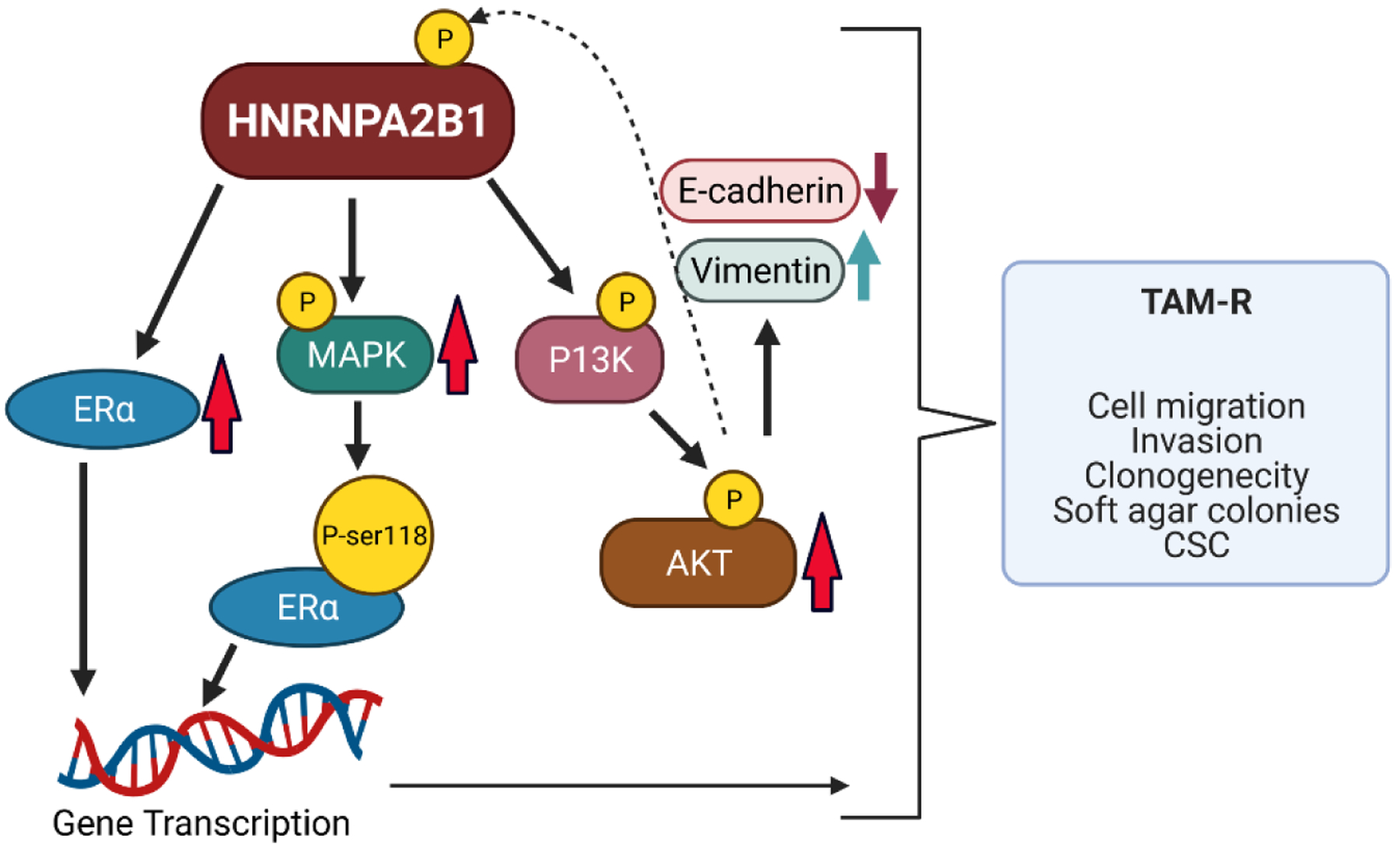
Shown are the proteins altered by increased A2B1 expression in this study that contribute to hallmarks of TAM-resistance. Up arrows indicate increased expression and down arrows indicate reduced expression. The dotted line indicates that AKT may phosphorylate A2B1 and modulate its activity as reported for other cancers [93]. The model was created with BioRender.com.
Both 4-OHT and fulvestrant compete with E2 or other estrogens for binding to ERα and cause a conformational shift that precludes coactivator recruitment, instead allowing corepressors to interact, thus blocking ERα transcriptional activity [94]. In contrast to the stabilization of nuclear ERα when occupied by 4-OHT, fulvestrant stimulates proteasome-mediated ERα degradation [95]. However, fulvestrant-induced ERα degradation is saturable and is not coupled to its antagonist activity [96]. It was also reported that the efficacy of fulvestrant and experimental SERDs is not directly related to the extent of ERα turnover [97]. Fulvestrant increased ERα chromatin binding sites in MCF-7 cells and immobilized ERα at those sites which did not show marks of increased chromatin accessibility and were transcriptionally inert, thus indicating that ERα degradation does not account for the antagonism by fulvestrant [98]. Others reported that fulvestrant-occupied ERα was bound by the coactivator p300/CBP, was recruited to gene promoters, and induced transcription in MCF-7 cells [99]. Explanations for the cross-resistance to TAM and fulvestrant include activation of growth factor signaling pathways [52, 100–102], increased ERα, activation of NFkB [103], elevation of the unfolded protein response (UPR) [63, 90, 104, 105], and decreased EGR1 with activation of the glutamine and arachidonic pathways [106]. More recent studies reported that the coactivator MED1 is activated by ERK and AKT phosphorylation and is recruited to fulvestrant-occupied ERα, blocking recruitment of corepressors, thus activating ERα target genes and stimulating breast cancer cell proliferation and tumor growth in vivo [107–109]. Coactivator recruitment to fulvestrant-occupied ERα is a plausible mechanism involved in fulvestrant-resistance commensurate with the observed activation of AKT and MAPK (ERK) in LCC9 and MCF-7-A2B1 cells. In a recent phase Ib clinical trial, fulvestrant-resistant breast cancer patients were treated with a combination of fulvestrant and the proteasome inhibitor ixazomib (MLN9708) because increased proteasome activity has been suggested to contribute to fulvestrant-resistance by stimulating the UPR [110].
Mechanisms involved in AI-resistance include AI-driven mutations in ESR1 and mutations in PIK3CA, PTEN, AKT1, HER2, FOXA1, MAP2KA, KRAS, ARID1A, CTCR, MYC, NF1, BRAF, MLH1/3, PMS1/2, and ESR1 fusion genes; alterations (amplification) of CYP19A1, EGFR, and PRR11 reviewed in [2]. Models of AI-resistance in BC cells employ stable human CYP19A1 aromatase-overexpression in MCF-7 cells [111, 112]. Fulvestrant inhibits while TAM, letrozole, and anastrozole stimulate the migration of the AI-resistant cell lines [113]. Similarly, MCF-7 cells selected for AI-resistance by growth in testosterone + letrozole or exemestane are sensitive to growth inhibition by fulvestrant, but not TAM [114]. These data suggest differences in mechanisms underlie resistance to tamoxifen and fulvestrant versus AI resistance.
MCF-7-A2B1 cells form larger sized colonies in soft agar assays, suggesting greater tumorigenic and metastatic potential. This was reflected in enhanced migration, invasion, upregulation of vimentin, and downregulation of E-cadherin (hallmarks of EMT) in MCF-7-A2B1 cells. We also observed a higher percent of CD44+/CD24−/low CSC in MCF-7-A2B1, LCC9, and LY2 cells relative to the parental MCF-7 cells. Similar results were reported for other TAM-resistant MCF-7 cell lines [115, 116]. Endocrine therapies and chemotherapies promote the selection of CSC populations in human breast tumors in vivo [117, 118]. Previous studies correlate increased soft agar colony size and the CSC population in HIF-2α-overexpressing MCF-7 cells that are resistant to Paclitaxel [119]. This finding is not merely in vitro: TAM-resistant patient derived xenografts (PDX) had higher CSC activity [120] and a mouse model of breast cancer showed that fulvestrant increased CSC activity in mammary tumors [121].
Mechanistically, we observed higher P-ser473-AKT/AKT and P-MAPK/MAPK in MCF-7-A2B1 and LCC9 cells that was ablated by A2B1 knockdown. The reduction in AKT and MAPK activation with siA2B1 corresponds to the restoration of endocrine-sensitivity seen with knockdown of A2B1 in MCF-7-A2B1 and LCC9 cells. Increased expression of growth factors along with activation of their tyrosine kinase receptors activate MAPK, PI3K-AKT, JNK signaling and are widely implicated in the development of acquired endocrine-resistance [122, 123]. Genetic, epigenetic, and epitranscriptomic alterations in genes in the PI3K and MEK-MAPK signaling pathways drive the metastatic cascade [124–126].
TAM-resistant breast cancer cells have lower mitochondrial bioenergetic capacity and evidence of mitochondrial stress [35, 127]. In response to mitochondrial stress, AKT phosphorylates HNRNPA2, enhancing its recruitment to gene promotors in murine C2C12 skeletal myoblasts where it acts as a transcriptional coactivator [128]. This suggests that activation of AKT in TAM-resistant cells may increase A2B1 phosphorylation and alter its interaction with transcription factors, thus changing gene transcription. We have included this suggestion in the model of the role of A2B1 in TAM-resistance (Fig. 13), although verification is beyond the current report.
Similar to previous reports [18, 21, 22, 43], we identified higher A2B1 expression in primary breast tumors compared to normal breast. We did not observe a significant lower A2B1 expression in LN metastasis compared to primary paired breast IDC tumors except for HER2+/ER− breast tumors. Our findings differ from a previous TMA study that reported ~ 6% lower A2B1 IHC staining in LMN from paired IDC tumors [44]. However, that study did not consider the tumor status of ER, HER2, or TNBC, nor the expression of these markers in the LNM. In addition, a different A2B1 antibody was used [44]. A search in HCMDB: the human cancer metastasis database [129] revealed that A2B1 expression is higher in breast tumors with metastasis compared to breast tumors without metastasis (Supplementary Fig. 19A and B). A2B1 was higher in brain metastasis compared with LN metastasis (Supplementary Fig. 19C). While A2B1 was higher in liver, lung, and spleen metastasis compared to the primary tumor, values were not different from LN metastasis in another data set (Supplementary Fig. 19D). It will be important to examine A2B1 levels in distant metastasis from primary ER+ breast tumors in patients treated with AI, TAM, or fulvestrant and examine the impact of cotreatment with CDK4/6 or PI3K inhibitors.
Increased expression of A2B1 has been detected in other cancers, e.g., melanoma [130], pancreatic ductal carcinoma (PDAC) [131], prostate cancer [132], and colorectal cancer (CRC) [44] where it contributes to metastasis. Similar to findings reported here, A2B1 promoted EMT by down-regulating E-cadherin and up-regulating vimentin, and also stimulated the invasion capacity of PDAC cell lines [131]. A complex of A2B1 with the m6A-modified lncRNA RP11–138 J23.1 (RP11, LINC02598) was reported to accelerate the degradation of mRNA transcripts of two E3 ligases, SIAH1 and FBXO45 in CRC, thus preventing the proteolytic degradation of ZEB1 and leading to EMT, e.g., decreased E-cadherin and increased cell invasion [133]. Similarly, we observed decreased E-cadherin and increased vimentin in MCF-7-A2B1 cells. A2B1 was increased in human small cell and non-small cell lung tumors and knockdown of A2B1 inhibited migration and invasion of DLKP-M squamous cell carcinoma cells in vitro [134]. Together these data indicate a role for A2B1 in promoting EMT and invasion in multiple cancer types.
MCF-7-A2B1 cells showed an ~ 50% increase in the number of migrating cells. Our findings are in contrast with a report that CRISPR/CAS9 knockout of A2B1 increased MCF-7 and MDA-MB-231 breast cancer cell migration and invasion [44]. We do not know why our results differ from this report of HNRNPA2B1 ablation in MCF-7 cells. Global knockout likely has a more profound effect on the cells compared to overexpression of A2B1 equivalent to levels detected in endocrine-resistant LCC9 and LY2 cells. It is also possible that there may have been off-target effects of the CRISPR/CAS9 knockout [135]. It is noteworthy that A2B1 expression was increased in lung metastasis after tail vein injection of human breast cancer patient-derived circulating tumor cells in mice [136]. Together, these reports indicate that the role for A2B1 in breast cancer metastases requires further investigation.
A2B1 plays a role in transcript splicing, trafficking, and stability [137, 138]. Aaron Johnson’s group used eCLIP to identify transcripts bound by A2B1 in MCF-7 cells (GEO # GSE103165) [139]. We downloaded these data from GEO (in bigwig format) and analyzed them using the UCSC genome browser to determine if A2B1 interacts with ESR1 transcripts. A number of A2B1 binding peaks upstream and within the ESR1 gene, including intronic regions (Supplementary Fig. 20), were observed, supporting the suggestion that A2B1 may regulate ERα by interacting with the ESR1 transcript for stabilization, increased splicing efficiency, or trafficking to the cytoplasm for translation. Whether this is causative in the increase in ERα protein in MCF-7-A2B1 cells is unknown, but we demonstrated that siA2B1 reduced ERα protein in both MCF-7-A2B1 and LCC9 cells, suggesting a regulatory interplay between A2B1 and ERα protein.
5. Conclusions
We demonstrated that modest stable overexpression of A2B1 in MCF-7 endocrine-sensitive luminal A cells endowed the cells with phenotypic features of acquired endocrine resistance, including loss of growth inhibition by 4-OHT and fulvestrant, increased migration, invasion, clonogenicity, larger colony size in soft agar, increased CD44+/CD24−/low CSC, altered expression of EMT markers, and activation of AKT and MAPK, making them similar to LCC9 and LY2 cells. Conversely, knockdown of A2B1 in two independent models of endocrine-resistance: LCC9 and LY2 cells, resulted in a ‘restoration’ of TAM and fulvestrant sensitivity. These data suggest a role for HNRNPA2B1 in promoting the initiation of acquired endocrine-resistance by activating ser/thr kinase growth factor signaling pathways regulating downstream targets. Selective inhibition of HNRNPA2B1 may be a target to prevent acquisition of endocrine therapy resistance, but not to treat established metastatic disease.
Supplementary Material
Highlights.
HNRNPA2B1 (A2B1) is more highly expressed in breast tumors than normal breast and is upregulated in tamoxifen (TAM)-resistant LCC9 and LY2 breast cancer cells
Stable A2B1 expression in TAM-sensitive MCF-7 cells (MCF-7-A2B1) blocks growth inhibition by TAM and fulvestrant and results in mesenchymal-like properties
Knockdown of A2B1 restores TAM and fulvestrant sensitivity in TAM-resistant LCC9 and LY2 cells
A2B1 confers properties of endocrine resistance including activated AKT and MAPK and increased cancer stem cells (CSC)
Acknowledgements:
We thank Stephanie Metcalf for assistance with the Boyden chamber transwell migration assays, Emily Duderstadt for help with Kruskal-Wallace analysis, and Dr. Michal Hetman for his comments on the cell images. We thank Dr. David Tieri for assistance with bigWig analysis and interpretation of the eCLIP data in GSE103165. We thank Dr. Barbara J. Clark for her review and comments on this manuscript.
Funding Sources:
This work was supported by NIH R21CA212952 to C.M.K. B.J.P. was supported by a fellowship from NIH T32 ES011564. G.C.S.W. and A.E.W. were supported by a fellowship from NIH T35 DK072923 to C.M.K. C.C.P. was supported by a fellowship from NIH R25 CA134283.
Abbreviations:
- 4-OHT
4-hydroxytamoxifen
- AI
aromatase inhibitor
- A2B1
HNRNPA2B1
- CFE
colony forming efficiency
- CSC
cancer stem cells
- DFS
disease free survival
- DHT
dihydrotestosterone
- ERα
estrogen receptor alpha
- FACS
Fluorescent-activated cell sorting
- FBS
fetal bovine serum
- IHC
immunohistochemistry
- LBD
ligand binding domain
- KM
Kaplan-Meier
- m6A
N(6)-methyladenosine
- OE
overexpression
- OS
overall survival
- PDAC
pancreatic ductal carcinoma
- RFS
relapse-free survival
- SERMs
selective estrogen receptor modulators
- SS medium
‘serum-starved’, 5% dextran-coated charcoal stripped FBS-containing, phenol red-free IMEM medium (+ 1% pen/step)
- TAM
tamoxifen
- TNBC
triple negative breast cancer
- UPR
unfolded protein response
- WCE
whole cell extract
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors have no conflicts of interest to declare.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A, Siegel RL, Breast cancer statistics, 2019, CA. Cancer J. Clin, 69 (2019) 438–451. [DOI] [PubMed] [Google Scholar]
- [2].Hanker AB, Sudhan DR, Arteaga CL, Overcoming Endocrine Resistance in Breast Cancer, Cancer Cell, 37 (2020) 496–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R, ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer, Nat Rev Clin Oncol, 12 (2015) 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chandarlapaty S, Chen D, He W, et al. , Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: A secondary analysis of the bolero-2 clinical trial, JAMA Oncology, 2 (2016) 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rani A, Stebbing J, Giamas G, Murphy J, Endocrine Resistance in Hormone Receptor Positive Breast Cancer–From Mechanism to Therapy, Frontiers in Endocrinology, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale A-L, Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications, PNAS, 98 (2001) 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH, Gene expression profiling predicts clinical outcome of breast cancer, Nature, 415 (2002) 530–536. [DOI] [PubMed] [Google Scholar]
- [8].Creighton CJ, Cordero KE, Larios JM, Miller RS, Johnson MD, Chinnaiyan AM, Lippman ME, Rae JM, Genes regulated by estrogen in breast tumor cells in vitro are similarly regulated in vivo in tumor xenografts and human breast tumors, Genome Biol, 7 (2006) R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Varešlija D, Priedigkeit N, Fagan A, Purcell S, Cosgrove N, O’Halloran PJ, Ward E, Cocchiglia S, Hartmaier R, Castro CA, Zhu L, Tseng GC, Lucas PC, Puhalla SL, Brufsky AM, Hamilton RL, Mathew A, Leone JP, Basudan A, Hudson L, Dwyer R, Das S, O’Connor DP, Buckley PG, Farrell M, Hill ADK, Oesterreich S, Lee AV, Young LS, Transcriptome Characterization of Matched Primary Breast and Brain Metastatic Tumors to Detect Novel Actionable Targets, JNCI: Journal of the National Cancer Institute, 111 (2019) 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, Penson A, Shen R, Pareja F, Kundra R, Middha S, Cheng ML, Zehir A, Kandoth C, Patel R, Huberman K, Smyth LM, Jhaveri K, Modi S, Traina TA, Dang C, Zhang W, Weigelt B, Li BT, Ladanyi M, Hyman DM, Schultz N, Robson ME, Hudis C, Brogi E, Viale A, Norton L, Dickler MN, Berger MF, Iacobuzio-Donahue CA, Chandarlapaty S, Scaltriti M, Reis-Filho JS, Solit DB, Taylor BS, Baselga J, The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers, Cancer Cell, 34 (2018) 427–438.e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xu Z, Peng B, Cai Y, Wu G, Huang J, Gao M, Guo G, Zeng S, Gong Z, Yan Y, N6-methyladenosine RNA modification in cancer therapeutic resistance: Current status and perspectives, Biochem. Pharmacol, 182 (2020) 114258. [DOI] [PubMed] [Google Scholar]
- [12].Pan T, N6-methyl-adenosine modification in messenger and long non-coding RNA, Trends Biochem. Sci, 38 (2013) 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fu L, Amato NJ, Wang P, McGowan SJ, Niedernhofer LJ, Wang Y, Simultaneous Quantification of Methylated Cytidine and Adenosine in Cellular and Tissue RNA by Nano-Flow Liquid Chromatography-Tandem Mass Spectrometry Coupled with the Stable Isotope-Dilution Method, Anal. Chem, 87 (2015) 7653–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, Lu S, Xu D, Wu Y, Chen Q, Ding X, Guo H, Li Y, Wang Y, Fu B, Yao W, Wei M, Wu H, LNC942 promoting METTL14-mediated m6A methylation in breast cancer cell proliferation and progression, Oncogene, 39 (2020) 5358–5372. [DOI] [PubMed] [Google Scholar]
- [15].Licht K, Jantsch MF, Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications, J. Cell Biol, 213 (2016) 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C, N6-methyladenosine-dependent regulation of messenger RNA stability, Nature, 505 (2014) 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Deng X, Su R, Weng H, Huang H, Li Z, Chen J, RNA N6-methyladenosine modification in cancers: current status and perspectives, Cell Res, 28 (2018) 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, Ren F, Cui G, Sun R, N6-methyladenosine-related Genomic Targets are Altered in Breast Cancer Tissue and Associated with Poor Survival, J Cancer, 10 (2019) 5447–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Alarcón Claudio R., Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie Sohail F., HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events, Cell, 162 (2015) 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu Y, Shi S-L, The roles of hnRNP A2/B1 in RNA biology and disease, WIREs RNA, n/a (2020) e1612. [DOI] [PubMed] [Google Scholar]
- [21].Hu Y, Sun Z, Deng J, Hu B, Yan W, Wei H, Jiang J, Splicing factor hnRNPA2B1 contributes to tumorigenic potential of breast cancer cells through STAT3 and ERK1/2 signaling pathway, Tumour Biol, 39 (2017) 1010428317694318. [DOI] [PubMed] [Google Scholar]
- [22].Ma Y, Yang L, Li R, HnRNPA2/B1 Is a Novel Prognostic Biomarker for Breast Cancer Patients, Genetic testing and molecular biomarkers, 24 (2020) 701–707. [DOI] [PubMed] [Google Scholar]
- [23].Klinge CM, Piell KM, Tooley CS, Rouchka EC, HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells, Scientific reports, 9 (2019) 9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Katzenellenbogen BS, Miller MA, Eckert RL, Sudo K, Antiestrogen pharmacology and mechanism of action., J. Steroid Biochem, 19 (1983) 59–68. [PubMed] [Google Scholar]
- [25].Allred DC, Harvey JM, Berardo M, Clark GM, Prognostic and predictive factors in breast cancer by immunohistochemical analysis, Mod. Pathol, 11 (1998) 155–168. [PubMed] [Google Scholar]
- [26].Fitzgibbons PL, Goldsmith JD, Souers RJ, Fatheree LA, Volmar KE, Stuart LN, Nowak JA, Astles JR, Nakhleh RE, Analytic Validation of Immunohistochemical Assays: A Comparison of Laboratory Practices Before and After Introduction of an Evidence-Based Guideline, Arch. Pathol. Lab. Med, 141 (2017) 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fitzgibbons PL, Dillon DA, Alsabeh R, Berman MA, Hayes DF, Hicks DG, Hughes KS, Nofech-Mozes S, Template for Reporting Results of Biomarker Testing of Specimens From Patients With Carcinoma of the Breast, Arch. Pathol. Lab. Med, 138 (2013) 595–601. [DOI] [PubMed] [Google Scholar]
- [28].Crawford AC, Riggins RB, Shajahan AN, Zwart A, Clarke R, Co-Inhibition of BCL-W and BCL2 Restores Antiestrogen Sensitivity through BECN1 and Promotes an Autophagy-Associated Necrosis, PLoS ONE, 5 (2010) e8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brunner N, Boysen B, Jirus S, Skaar TC, Holst-Hansen C, Lippman J, Frandsen T, Spang-Thomsen M, Fuqua SA, Clarke R, MCF7/LCC9: an antiestrogen-resistant MCF-7 variant in which acquired resistance to the steroidal antiestrogen ICI 182,780 confers an early cross-resistance to the nonsteroidal antiestrogen tamoxifen., Cancer Res, 57 (1997) 3486–3493. [PubMed] [Google Scholar]
- [30].Davidson NE, Bronzert DA, Chambon P, Gelmann EP, Lippman ME, Use of two MCF-7 cell variants to evaluate the growth regulatory potential of estrogen-induced products, Cancer Res, 46 (1986) 1904–1908. [PubMed] [Google Scholar]
- [31].Muluhngwi P, Krishna A, Vittitow SL, Napier JT, Richardson KM, Ellis M, Mott JL, Klinge CM, Tamoxifen differentially regulates miR-29b-1 and miR-29a expression depending on endocrine-sensitivity in breast cancer cells, Cancer Lett, 388 (2017) 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Litchfield LM, Riggs KA, Hockenberry AM, Oliver LD, Barnhart KG, Cai J, Pierce WM Jr., Ivanova MM, Bates PJ, Appana SN, Datta S, Kulesza P, McBryan J, Young LS, Klinge CM, Identification and Characterization of Nucleolin as a COUP-TFII Coactivator of Retinoic Acid Receptor β Transcription in Breast Cancer Cells, PLoS ONE, 7 (2012) e38278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schultz DJ, Muluhngwi P, Alizadeh-Rad N, Green MA, Rouchka EC, Waigel SJ, Klinge CM, Genome-wide miRNA response to anacardic acid in breast cancer cells, PLOS ONE, 12 (2017) e0184471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative C(T) method, Nat Protoc, 3 (2008) 1101–1108. [DOI] [PubMed] [Google Scholar]
- [35].Radde BN, Ivanova MM, Mai HX, Alizadeh-Rad N, Piell K, Van Hoose P, Cole MP, Muluhngwi P, Kalbfleisch TS, Rouchka EC, Hill BG, Klinge CM, Nuclear respiratory factor-1 and bioenergetics in tamoxifen-resistant breast cancer cells, Exp. Cell Res, 347 (2016) 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Romero-Calvo I, Ocón B, Martínez-Moya P, Suárez MD, Zarzuelo A, Martínez-Augustin O, de Medina FS, Reversible Ponceau staining as a loading control alternative to actin in Western blots, Anal. Biochem, 401 (2010) 318–320. [DOI] [PubMed] [Google Scholar]
- [37].Moritz CP, Tubulin or Not Tubulin: Heading Toward Total Protein Staining as Loading Control in Western Blots, Proteomics, 17 (2017) 1600189. [DOI] [PubMed] [Google Scholar]
- [38].Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW, ImageJ2: ImageJ for the next generation of scientific image data, BMC Bioinformatics, 18 (2017) 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Teng Y, Radde BN, Litchfield LM, Ivanova MM, Prough RA, Clark BJ, Doll MA, Hein DW, Klinge CM, Dehydroepiandrosterone Activation of G-protein-coupled Estrogen Receptor Rapidly Stimulates MicroRNA-21 Transcription in Human Hepatocellular Carcinoma Cells, J. Biol. Chem, 290 (2015) 15799–15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Munshi A, Hobbs M, Meyn RE, Clonogenic cell survival assay, Methods Mol Med, 110 (2005) 21–28. [DOI] [PubMed] [Google Scholar]
- [41].Liu J, Sareddy GR, Zhou M, Viswanadhapalli S, Li X, Lai Z, Tekmal RR, Brenner A, Vadlamudi RK, Differential Effects of Estrogen Receptor beta Isoforms on Glioblastoma Progression, Cancer Res, 78 (2018) 3176–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Singh-Kaw P, Zarnegar R, Siegfried JM, Stimulatory effects of hepatocyte growth factor on normal and neoplastic human bronchial epithelial cells, Am J Physiol Lung Cell Mol Physiol, 268 (1995) L1012–1020. [DOI] [PubMed] [Google Scholar]
- [43].Zhou J, Allred DC, Avis I, Martinez A, Vos MD, Smith L, Treston AM, Mulshine JL, Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer, Breast Cancer Res. Treat, 66 (2001) 217–224. [DOI] [PubMed] [Google Scholar]
- [44].Liu Y, Li H, Liu F, Gao L-B, Han R, Chen C, Ding X, Li S, Lu K, Yang L, Tian H-M, Chen B-B, Li X, Xu D-H, Deng X-L, Shi S-L, Heterogeneous nuclear ribonucleoprotein A2/B1 is a negative regulator of human breast cancer metastasis by maintaining the balance of multiple genes and pathways, EBioMedicine, 51 (2020) 102583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z, GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses, Nucleic Acids Res, 45 (2017) W98–w102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ősz Á, Lánczky A, Győrffy B, Survival analysis in breast cancer using proteomic data from four independent datasets, medRxiv, (2020) 2020.2012.2003.20242065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang L, Wen M, Cao X, Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses, Science, 365 (2019) eaav0758. [DOI] [PubMed] [Google Scholar]
- [48].Paul KR, Molliex A, Cascarina S, Boncella AE, Taylor JP, Ross ED, Effects of Mutations on the Aggregation Propensity of the Human Prion-Like Protein hnRNPA2B1, Mol. Cell. Biol, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hughes L, Malone C, Chumsri S, Burger AM, McDonnell S, Characterisation of breast cancer cell lines and establishment of a novel isogenic subclone to study migration, invasion and tumourigenicity, Clin. Exp. Metastasis, 25 (2008) 549–557. [DOI] [PubMed] [Google Scholar]
- [50].Kumari K, Kumar S, Parida DK, Mishra SK, EZH2 knockdown in tamoxifen-resistant MCF-7 cells unravels novel targets for regaining sensitivity towards tamoxifen, Breast Cancer, (2020). [DOI] [PubMed] [Google Scholar]
- [51].Riggs KA, Wickramasinghe NS, Cochrum RK, Watts MB, Klinge CM, Decreased Chicken Ovalbumin Upstream Promoter Transcription Factor II Expression in Tamoxifen-Resistant Breast Cancer Cells, Cancer Res, 66 (2006) 10188–10198. [DOI] [PubMed] [Google Scholar]
- [52].Riggins RB, Zwart A, Nehra R, Clarke R, The nuclear factor kappa B inhibitor parthenolide restores ICI 182,780 (Faslodex; fulvestrant)-induced apoptosis in antiestrogen-resistant breast cancer cells, Mol Cancer Ther, 4 (2005) 33–41. [PubMed] [Google Scholar]
- [53].Helland T, Henne N, Bifulco E, Naume B, Borgen E, Kristensen VN, Kvaloy JT, Lash TL, Alnaes GIG, van Schaik RH, Janssen EAM, Hustad S, Lien EA, Mellgren G, Soiland H, Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients, Breast Cancer Res, 19 (2017) 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Helland T, Hagen KB, Haugstøyl ME, Kvaløy JT, Lunde S, Lode K, Lind RA, Gripsrud BH, Jonsdottir K, Gjerde J, Bifulco E, Hustad S, Jonassen J, Aas T, Lende TH, Lien EA, Janssen EAM, Søiland H, Mellgren G, Drug monitoring of tamoxifen metabolites predicts vaginal dryness and verifies a low discontinuation rate from the Norwegian Prescription Database, Breast Cancer Res. Treat, 177 (2019) 185–195. [DOI] [PubMed] [Google Scholar]
- [55].Helland T, Naume B, Hustad S, Bifulco E, Kvaløy JT, Sætersdal AB, Synnestvedt M, Lende TH, Gilje B, Mjaaland I, Weyde K, Blix ES, Wiedswang G, Borgen E, Hertz DL, Janssen EAM, Mellgren G, Søiland H, Low Z-4OHtam concentrations are associated with adverse clinical outcome among early stage premenopausal breast cancer patients treated with adjuvant tamoxifen, Molecular Oncology, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Robertson JFR, Harrison M, Fulvestrant: pharmacokinetics and pharmacology, Br. J. Cancer, 90Suppl 1 (2004) S7–S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rocca A, Maltoni R, Bravaccini S, Donati C, Andreis D, Clinical utility of fulvestrant in the treatment of breast cancer: a report on the emerging clinical evidence, Cancer management and research, 10 (2018) 3083–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rechoum Y, Rovito D, Iacopetta D, Barone I, Ando S, Weigel NL, O’Malley BW, Brown PH, Fuqua SA, AR collaborates with ERalpha in aromatase inhibitor-resistant breast cancer, Breast Cancer Res. Treat, 147 (2014) 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bhat-Nakshatri P, Song E-K, Collins NR, Uversky VN, Dunker AK, O’Malley BW, Geistlinger TR, Carroll JS, Brown M, Nakshatri H, Interplay between estrogen receptor and AKT in estradiol-induced alternative splicing, BMC medical genomics, 6 (2013) 21–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bhatt S, Stender JD, Joshi S, Wu G, Katzenellenbogen BS, OCT-4: a novel estrogen receptor-[alpha] collaborator that promotes tamoxifen resistance in breast cancer cells, Oncogene, 35 (2016) 5722–5734. [DOI] [PubMed] [Google Scholar]
- [61].Harrod A, Fulton J, Nguyen VTM, Periyasamy M, Ramos-Garcia L, Lai CF, Metodieva G, de Giorgio A, Williams RL, Santos DB, Gomez PJ, Lin ML, Metodiev MV, Stebbing J, Castellano L, Magnani L, Coombes RC, Buluwela L, Ali S, Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer, Oncogene, 36 (2017) 2286–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Buck MB, Pfizenmaier K, Knabbe C, Antiestrogens Induce Growth Inhibition by Sequential Activation of p38 Mitogen-Activated Protein Kinase and Transforming Growth Factor-{beta} Pathways in Human Breast Cancer Cells, Mol. Endocrinol, 18 (2004) 1643–1657. [DOI] [PubMed] [Google Scholar]
- [63].Hu R, Warri A, Jin L, Zwart A, Riggins RB, Fang H-B, Clarke R, NF-κB Signaling Is Required for XBP1 (Unspliced and Spliced)-Mediated Effects on Antiestrogen Responsiveness and Cell Fate Decisions in Breast Cancer, Mol. Cell. Biol, 35 (2015) 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kisanga ER, Gjerde J, Guerrieri-Gonzaga A, Pigatto F, Pesci-Feltri A, Robertson C, Serrano D, Pelosi G, Decensi A, Lien EA, Tamoxifen and Metabolite Concentrations in Serum and Breast Cancer Tissue during Three Dose Regimens in a Randomized Preoperative Trial, Clin. Cancer Res, 10 (2004) 2336–2343. [DOI] [PubMed] [Google Scholar]
- [65].Ademuyiwa FO, Ellis MJ, Ma CX, Neoadjuvant therapy in operable breast cancer: application to triple negative breast cancer, Journal of oncology, 2013 (2013) 219869–219869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cardoso F, Paluch-Shimon S, Senkus E, Curigliano G, Aapro MS, André F, Barrios CH, Bergh J, Bhattacharyya GS, Biganzoli L, Boyle F, Cardoso MJ, Carey LA, Cortés J, El Saghir NS, Elzayat M, Eniu A, Fallowfield L, Francis PA, Gelmon K, Gligorov J, Haidinger R, Harbeck N, Hu X, Kaufman B, Kaur R, Kiely BE, Kim SB, Lin NU, Mertz SA, Neciosup S, Offersen BV, Ohno S, Pagani O, Prat A, Penault-Llorca F, Rugo HS, Sledge GW, Thomssen C, Vorobiof DA, Wiseman T, Xu B, Norton L, Costa A, Winer EP, 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5), Ann. Oncol, 31 (2020) 1623–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Thompson EW, Reich R, Shima TB, Albini A, Graf J, Martin GR, Dickson RB, Lippman ME, Differential Regulation of Growth and Invasiveness of MCF-7 Breast Cancer Cells by Antiestrogens, Cancer Res, 48 (1988) 6764–6768. [PubMed] [Google Scholar]
- [68].Chen Y, Hong DY, Wang J, Ling‐Hu J, Zhang YY, Pan D, Xu YN, Tao L, Luo H, Shen XC, Baicalein, unlike 4-hydroxytamoxifen but similar to G15, suppresses 17β-estradiol-induced cell invasion, and matrix metalloproteinase-9 expression and activation in MCF-7 human breast cancer cells, Oncology letters, 14 (2017) 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C, Clonogenic assay of cells in vitro, Nature protocols, 1 (2006) 2315. [DOI] [PubMed] [Google Scholar]
- [70].Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, Winn RA, The soft agar colony formation assay, Journal of visualized experiments : JoVE, (2014) e51998–e51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Peiris-Pagès M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP, Cancer stem cell metabolism, Breast Cancer Research, 18 (2016) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Alferez DG, Simoes BM, Howell SJ, Clarke RB, The Role of Steroid Hormones in Breast and Effects on Cancer Stem Cells, Current stem cell reports, 4 (2018) 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Araki K, Miyoshi Y, Mechanism of resistance to endocrine therapy in breast cancer: the important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer, Breast Cancer, 25 (2018) 392–401. [DOI] [PubMed] [Google Scholar]
- [74].Verret B, Cortes J, Bachelot T, Andre F, Arnedos M, Efficacy of PI3K inhibitors in advanced breast cancer, Ann. Oncol, 30 (2019) x12–x20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O’Brien K, Wang Y, Hilakivi-Clarke LA, Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling, Oncogene, 22 (2003) 7316–7339. [DOI] [PubMed] [Google Scholar]
- [76].Zhang YW, Nasto RE, Varghese R, Jablonski SA, Serebriiskii IG, Surana R, Calvert VS, Bebu I, Murray J, Jin L, Johnson M, Riggins R, Ressom H, Petricoin E, Clarke R, Golemis EA, Weiner LM, Acquisition of estrogen independence induces TOB1-related mechanisms supporting breast cancer cell proliferation, Oncogene, 35 (2016) 1643–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Morimoto-Kamata R, Yui S, Insulin-like growth factor-1 signaling is responsible for cathepsin G-induced aggregation of breast cancer MCF-7 cells, Cancer Sci, 108 (2017) 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Cai J, Xia J, Zou J, Wang Q, Ma Q, Sun R, Liao H, Xu L, Wang D, Guo X, The PI3K/mTOR dual inhibitor NVP-BEZ235 stimulates mutant p53 degradation to exert anti-tumor effects on triple-negative breast cancer cells, FEBS open bio, 10 (2020) 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Xia P, Xu X-Y, PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application, American journal of cancer research, 5 (2015) 1602–1609. [PMC free article] [PubMed] [Google Scholar]
- [80].Hutcheson IR, Knowlden JM, Madden TA, Barrow D, Gee JM, Wakeling AE, Nicholson RI, Oestrogen receptor-mediated modulation of the EGFR/MAPK pathway in tamoxifen-resistant MCF-7 cells, Breast Cancer Res. Treat, 81 (2003) 81–93. [DOI] [PubMed] [Google Scholar]
- [81].Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P, Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase., Science, 270 (1995) 1491–1494. [DOI] [PubMed] [Google Scholar]
- [82].Chen J, An B-S, Cheng L, Hammond GL, Leung PCK, Gonadotropin-Releasing Hormone-Mediated Phosphorylation of Estrogen Receptor-{alpha} Contributes to fosB Expression in Mouse Gonadotrophs, Endocrinology, 150 (2009) 4583–4593. [DOI] [PubMed] [Google Scholar]
- [83].Lobenhofer EK, Huper G, Iglehart JD, Marks JR, Inhibition of Mitogen-activated Protein Kinase and Phosphatidylinositol 3-Kinase Activity in MCF-7 Cells Prevents Estrogen-induced Mitogenesis, Cell Growth Differ, 11 (2000) 99–110. [PubMed] [Google Scholar]
- [84].Akter R, Hossain M, Kleve M, Gealt M, Wortmannin induces MCF-7 breast cancer cell death via the apoptotic pathway, involving chromatin condensation, generation of reactive oxygen species, and membrane blebbing Breast Cancer: Targets and Therapy, 4 (2012) 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhao Y, Ge C-C, Wang J, Wu X-X, Li X-M, Li W, Wang S-S, Liu T, Hou J-Z, Sun H, Fang D, Xie S-Q, MEK inhibitor, PD98059, promotes breast cancer cell migration by inducing β-catenin nuclear accumulation, Oncol. Rep, 38 (2017) 3055–3063. [DOI] [PubMed] [Google Scholar]
- [86].Riggins RB, Bouton AH, Liu MC, Clarke R, Antiestrogens, Aromatase Inhibitors, and Apoptosis in Breast Cancer, Vitam. Horm, 71 (2005) 201–237. [DOI] [PubMed] [Google Scholar]
- [87].Santen RJ, Song RX, Zhang Z, Yue W, Kumar R, Adaptive hypersensitivity to estrogen: mechanism for sequential responses to hormonal therapy in breast cancer, Clin. Cancer Res, 10 (2004) 337S–345S. [DOI] [PubMed] [Google Scholar]
- [88].Kim S-S, Lee M-H, Lee M-O, Histone methyltransferases regulate the transcriptional expression of ERα and the proliferation of tamoxifen-resistant breast cancer cells, Breast Cancer Res. Treat, 180 (2020) 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhao J-J, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ, MicroRNA-221/222 negatively regulates ERalpha and associates with tamoxifen resistance in breast cancer, J. Biol. Chem, 283 (2008) 31079–31086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [90].Cook KL, Clarke PAG, Parmar J, Hu R, Schwartz-Roberts JL, Abu-Asab M, Wärri A, Baumann WT, Clarke R, Knockdown of estrogen receptor-α induces autophagy and inhibits antiestrogen-mediated unfolded protein response activation, promoting ROS-induced breast cancer cell death, The FASEB Journal, 28 (2014) 3891–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Brunner N, Boulay V, Fojo A, Freter CE, Lippman ME, Clarke R, Acquisition of hormone-independent growth in MCF-7 cells is accompanied by increased expression of estrogen-regulated genes but without detectable DNA amplifications, Cancer Res, 53 (1993) 283–290. [PubMed] [Google Scholar]
- [92].Bronzert DA, Greene GL, Lippman ME, Selection and characterization of a breast cancer cell line resistant to the antiestrogen LY 117018, Endocrinology, 117 (1985) 1409–1417. [DOI] [PubMed] [Google Scholar]
- [93].Gupta A, Yadav S, Pt A, Mishra J, Samaiya A, Panday RK, Shukla S, The HNRNPA2B1–MST1R–Akt axis contributes to epithelial-to-mesenchymal transition in head and neck cancer, Lab. Invest, 100 (2020) 1589–1601. [DOI] [PubMed] [Google Scholar]
- [94].Huang HJ, Norris JD, McDonnell DP, Identification of a negative regulatory surface within estrogen receptor alpha provides evidence in support of a role for corepressors in regulating cellular responses to agonists and antagonists., Mol. Endocrinol, 16 (2002) 1778–1792. [DOI] [PubMed] [Google Scholar]
- [95].Wijayaratne AL, McDonnell DP, The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators, J. Biol. Chem, 276 (2001) 35684–35692. [DOI] [PubMed] [Google Scholar]
- [96].Wardell SE, Marks JR, McDonnell DP, The turnover of estrogen receptor α by the selective estrogen receptor degrader (SERD) fulvestrant is a saturable process that is not required for antagonist efficacy, Biochem. Pharmacol, 82 (2011) 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wardell SE, Yllanes AP, Chao CA, Bae Y, Andreano KJ, Desautels TK, Heetderks KA, Blitzer JT, Norris JD, McDonnell DP, Pharmacokinetic and pharmacodynamic analysis of fulvestrant in preclinical models of breast cancer to assess the importance of its estrogen receptor-α degrader activity in antitumor efficacy, Breast Cancer Res. Treat, 179 (2020) 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Guan J, Zhou W, Hafner M, Blake RA, Chalouni C, Chen IP, De Bruyn T, Giltnane JM, Hartman SJ, Heidersbach A, Houtman R, Ingalla E, Kategaya L, Kleinheinz T, Li J, Martin SE, Modrusan Z, Nannini M, Oeh J, Ubhayakar S, Wang X, Wertz IE, Young A, Yu M, Sampath D, Hager JH, Friedman LS, Daemen A, Metcalfe C, Therapeutic Ligands Antagonize Estrogen Receptor Function by Impairing Its Mobility, Cell, 178 (2019) 949–963.e918. [DOI] [PubMed] [Google Scholar]
- [99].Jaber BM, Gao T, Huang L, Karmakar S, Smith CL, The Pure Estrogen Receptor Antagonist ICI 182,780 Promotes a Novel Interaction of Estrogen Receptor-{alpha} with the 3’,5’-Cyclic Adenosine Monophosphate Response Element-Binding Protein-Binding Protein/p300 Coactivators, Mol. Endocrinol, 20 (2006) 2695–2710. [DOI] [PubMed] [Google Scholar]
- [100].Chen J, Gomes AR, Monteiro LJ, Wong SY, Wu LH, Ng T-T, Karadedou CT, Millour J, Ip Y-C, Cheung YN, Sunters A, Chan KYK, Lam EWF, Khoo U-S, Constitutively Nuclear FOXO3a Localization Predicts Poor Survival and Promotes Akt Phosphorylation in Breast Cancer, PLoS ONE, 5 (2010) e12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Woo YM, Shin Y, Lee EJ, Lee S, Jeong SH, Kong HK, Park EY, Kim HK, Han J, Chang M, Park JH, Inhibition of Aerobic Glycolysis Represses Akt/mTOR/HIF-1alpha Axis and Restores Tamoxifen Sensitivity in Antiestrogen-Resistant Breast Cancer Cells, PLoS One, 10 (2015) e0132285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Nicholson R, Hutcheson I, Hiscox S, Knowlden J, Giles M, Barrow D, Gee J, Growth factor signalling and resistance to selective oestrogen receptor modulators and pure anti-oestrogens: the use of anti-growth factor therapies to treat or delay endocrine resistance in breast cancer, Endocr Relat Cancer, 12 (2005) S29–36. [DOI] [PubMed] [Google Scholar]
- [103].Yde CW, Emdal KB, Guerra B, Lykkesfeldt AE, NFkappaB signaling is important for growth of antiestrogen resistant breast cancer cells, Breast Cancer Res. Treat, 135 (2012) 67–78. [DOI] [PubMed] [Google Scholar]
- [104].Shajahan-Haq A, Cook K, Schwartz-Roberts J, Eltayeb A, Demas D, Warri A, Facey CO, Hilakivi-Clarke L, Clarke R, MYC regulates the unfolded protein response and glucose and glutamine uptake in endocrine resistant breast cancer, Molecular Cancer, 13 (2014) 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Cook KL, Soto-Pantoja DR, Clarke PA, Cruz MI, Zwart A, Warri A, Hilakivi-Clarke L, Roberts DD, Clarke R, Endoplasmic Reticulum Stress Protein GRP78 Modulates Lipid Metabolism to Control Drug Sensitivity and Antitumor Immunity in Breast Cancer, Cancer Res, 76 (2016) 5657–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Shajahan-Haq AN, Boca SM, Jin L, Bhuvaneshwar K, Gusev Y, Cheema AK, Demas DD, Raghavan KS, Michalek R, Madhavan S, Clarke R, EGR1 regulates cellular metabolism and survival in endocrine resistant breast cancer, Oncotarget, 8 (2017) 96865–96884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zhang L, Cui J, Leonard M, Nephew K, Li Y, Zhang X, Silencing MED1 sensitizes breast cancer cells to pure anti-estrogen fulvestrant in vitro and in vivo, PloS one, 8 (2013) e70641–e70641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Chen Q, Weng Z, Lu Y, Jia Y, Ding L, Bai F, Ge M, Lin Q, Wu K, An Experimental Analysis of the Molecular Effects of Trastuzumab (Herceptin) and Fulvestrant (Falsodex), as Single Agents or in Combination, on Human HR+/HER2+ Breast Cancer Cell Lines and Mouse Tumor Xenografts, PloS one, 12 (2017) e0168960–e0168960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Weber H, Garabedian MJ, The mediator complex in genomic and non-genomic signaling in cancer, Steroids, 133 (2018) 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Schwartz G, Shee K, Romo B, Marotti J, Kisselev A, Lewis L, Miller T, Phase Ib Study of the Oral Proteasome Inhibitor Ixazomib (MLN9708) and Fulvestrant in Advanced ER+ Breast Cancer Progressing on Fulvestrant, Oncologist, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Gilani RA, Kazi AA, Shah P, Schech AJ, Chumsri S, Sabnis G, Jaiswal AK, Brodie AH, The importance of HER2 signaling in the tumor-initiating cell population in aromatase inhibitor-resistant breast cancer, Breast Cancer Res. Treat, 135 (2012) 681–692. [DOI] [PubMed] [Google Scholar]
- [112].Zhou DJ, Pompon D, Chen SA, Stable expression of human aromatase complementary DNA in mammalian cells: a useful system for aromatase inhibitor screening, Cancer Res, 50 (1990) 6949–6954. [PubMed] [Google Scholar]
- [113].Smollich M, Götte M, Fischgräbe J, Macedo LF, Brodie A, Chen S, Radke I, Kiesel L, Wülfing P, ETAR antagonist ZD4054 exhibits additive effects with aromatase inhibitors and fulvestrant in breast cancer therapy, and improves in vivo efficacy of anastrozole, Breast Cancer Res. Treat, 123 (2010) 345–357. [DOI] [PubMed] [Google Scholar]
- [114].Hole S, Pedersen AM, Hansen SK, Lundqvist J, Yde CW, Lykkesfeldt AE, New cell culture model for aromatase inhibitor-resistant breast cancer shows sensitivity to fulvestrant treatment and cross-resistance between letrozole and exemestane, Int. J. Oncol, 46 (2015) 1481–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Dubrovska A, Hartung A, Bouchez LC, Walker JR, Reddy VA, Cho CY, Schultz PG, CXCR4 activation maintains a stem cell population in tamoxifen-resistant breast cancer cells through AhR signalling, Br. J. Cancer, 107 (2012) 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Uchiumi K, Tsuboi K, Sato N, Ito T, Hirakawa H, Niwa T, Yamaguchi Y, S.-i. Hayashi, Cancer stem-like properties of hormonal therapy-resistant breast cancer cells, Breast Cancer, 26 (2019) 459–470. [DOI] [PubMed] [Google Scholar]
- [117].Sansone P, Ceccarelli C, Berishaj M, Chang Q, Rajasekhar VK, Perna F, Bowman RL, Vidone M, Daly L, Nnoli J, Santini D, Taffurelli M, Shih NNC, Feldman M, Mao JJ, Colameco C, Chen J, DeMichele A, Fabbri N, Healey JH, Cricca M, Gasparre G, Lyden D, Bonafe M, Bromberg J, Self-renewal of CD133hi cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer, Nat Commun, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhang M, Lee AV, Rosen JM, The Cellular Origin and Evolution of Breast Cancer, Cold Spring Harbor perspectives in medicine, 7 (2017) a027128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yan Y, Liu F, Han L, Zhao L, Chen J, Olopade OI, He M, Wei M, HIF-2α promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating Wnt and Notch pathways, Journal of experimental & clinical cancer research : CR, 37 (2018) 256–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Simoes Bruno M., O’Brien Ciara S., Eyre R, Silva A, Yu L, Sarmiento-Castro A, Alférez Denis G., Spence K, Santiago-Gómez A, Chemi F, Acar A, Gandhi A, Howell A, Brennan K, Rydén L, Catalano S, Andó S, Gee J, Ucar A, Sims Andrew H., Marangoni E, Farnie G, Landberg G, Howell Sacha J., Clarke Robert B., Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity, Cell Reports, 12 (2015) 1968–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Shea MP, O’Leary KA, Fakhraldeen SA, Goffin V, Friedl A, Wisinski KB, Alexander CM, Schuler LA, Antiestrogen Therapy Increases Plasticity and Cancer Stemness of Prolactin-Induced ERα+ Mammary Carcinomas, Cancer Res, 78 (2018) 1672–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Clarke R, Tyson JJ, Dixon JM, Endocrine resistance in breast cancer – An overview and update, Mol. Cell. Endocrinol, 418, Part 3 (2015) 220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nunnery SE, Mayer IA, Targeting the PI3K/AKT/mTOR Pathway in Hormone-Positive Breast Cancer, Drugs, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Clarke R, Kraikivski P, Jones BC, Sevigny CM, Sengupta S, Wang Y, A systems biology approach to discovering pathway signaling dysregulation in metastasis, Cancer Metastasis Rev, 39 (2020) 903–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].AlFakeeh A, Brezden-Masley C, Overcoming endocrine resistance in hormone receptor-positive breast cancer, Current oncology (Toronto, Ont.), 25 (2018) S18–s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Petri BJ, Klinge CM, Regulation of breast cancer metastasis signaling by miRNAs, Cancer Metastasis Rev, 39 (2020) 837–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Tomková V, Sandoval-Acuña C, Torrealba N, Truksa J, Mitochondrial fragmentation, elevated mitochondrial superoxide and respiratory supercomplexes disassembly is connected with the tamoxifen-resistant phenotype of breast cancer cells, Free Radic. Biol. Med, (2019). [DOI] [PubMed] [Google Scholar]
- [128].Guha M, Tang W, Sondheimer N, Avadhani NG, Role of calcineurin, hnRNPA2 and Akt in mitochondrial respiratory stress-mediated transcription activation of nuclear gene targets, Biochimica et Biophysica Acta (BBA) - Bioenergetics, 1797 (2010) 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Zheng G, Ma Y, Zou Y, Yin A, Li W, Dong D, HCMDB: the human cancer metastasis database, Nucleic Acids Res, 46 (2018) D950–d955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Li T, Gu M, Deng A, Qian C, Increased expression of YTHDF1 and HNRNPA2B1 as potent biomarkers for melanoma: a systematic analysis, Cancer Cell Int, 20 (2020) 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yan-Sanders Y, Hammons GJ, Lyn-Cook BD, Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells, Cancer Lett, 183 (2002) 215–220. [DOI] [PubMed] [Google Scholar]
- [132].Singh AN, Sharma N, Quantitative SWATH-Based Proteomic Profiling for Identification of Mechanism-Driven Diagnostic Biomarkers Conferring in the Progression of Metastatic Prostate Cancer, Frontiers in oncology, 10 (2020) 493–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, Li J, An P, Lu L, Luo N, Du J, Shan H, Liu H, Wang H, m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1, Molecular Cancer, 18 (2019) 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Dowling P, Pollard D, Larkin A, Henry M, Meleady P, Gately K, O’Byrne K, Barr MP, Lynch V, Ballot J, Crown J, Moriarty M, O’Brien E, Morgan R, Clynes M, Abnormal levels of heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) in tumour tissue and blood samples from patients diagnosed with lung cancer, Molecular bioSystems, 11 (2015) 743–752. [DOI] [PubMed] [Google Scholar]
- [135].Marx V, Guide RNAs: it’s good to be choosy, Nat. Methods, 17 (2020) 1179–1182. [DOI] [PubMed] [Google Scholar]
- [136].Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia A, Truesdell S, Wiley DF, Wesley B, Li S, Mai A, Aceto N, Vincent-Jordan N, Szabolcs A, Chirn B, Kreuzer J, Comaills V, Kalinich M, Haas W, Ting DT, Toner M, Vasudevan S, Haber DA, Maheswaran S, Micalizzi DS, Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis, Science, 367 (2020) 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Han SP, Friend LR, Carson JH, Korza G, Barbarese E, Maggipinto M, Hatfield JT, Rothnagel JA, Smith R, Differential subcellular distributions and trafficking functions of hnRNP A2/B1 spliceoforms, Traffic, 11 (2010) 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Golan-Gerstl R, Cohen M, Shilo A, Suh S-S, Bakàcs A, Coppola L, Karni R, Splicing Factor hnRNP A2/B1 Regulates Tumor Suppressor Gene Splicing and Is an Oncogenic Driver in Glioblastoma, Cancer Res, 71 (2011) 4464. [DOI] [PubMed] [Google Scholar]
- [139].Nguyen ED, Balas MM, Griffin AM, Roberts JT, Johnson AM, Global Profiling of hnRNP A2/B1-RNA Binding on Chromatin Highlights LncRNA Interactions, RNA Biology, 15 (2018) 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


