SUMMARY
The enteric nervous system (ENS) coordinates diverse functions in the intestine, but has eluded comprehensive molecular characterization due to the rarity and diversity of cells. Here, we develop two methods to profile the ENS of adult mice and humans at single cell resolution: RAISIN RNA-Seq, for profiling intact nuclei with ribosome-bound mRNA, and MIRACL-Seq, for label-free enrichment of rare cell types by droplet-based profiling. The 1,187,535 nuclei in our mouse atlas include 5,068 neurons from the ileum and colon, revealing extraordinary neuron diversity. We highlight circadian expression changes in enteric neurons, show that disease-related genes are dysregulated with aging, and identify differences between the ileum and proximal/distal colon. In human, we profile 436,202 nuclei recovering 1,445 neurons and identify conserved and species-specific transcriptional programs, and putative neuro-epithelial, neuro-stromal, and neuro-immune interactions. The human ENS expresses risk genes for neuropathic, inflammatory, and extraintestinal diseases, suggesting neuronal contributions to disease.
Graphical Abstract
INTRODUCTION
The enteric nervous system (ENS) coordinates intestinal motility, digestion, and barrier defense (Yoo and Mazmanian, 2017). In humans, the ENS rivals the spinal cord in complexity (Furness, 2012), and is divided into the myenteric and submucosal plexuses (Sasselli et al., 2012), with differences within ganglia, across intestinal regions, and among species (Furness, 2012). Other factors proposed to contribute to ENS heterogeneity include age (Bernard et al., 2009), sex (Li et al., 2018), circadian phase (Scheving, 2000), and disease (e.g., functional motility disorders) (De Giorgio et al., 2016).
A broad range of intestinal and extraintestinal diseases are associated with ENS dysfunction. These include monogenic neuropathies that directly affect enteric neurons (e.g., Hirschsprung’s disease) (Furness, 2012), and complex neurologic disorders with early-onset gut dysmotility (e.g., autism spectrum disorders (Chaidez et al., 2014) and Parkinson’s disease (Pfeiffer, 2003)). The latter may involve neuroinflammatory circuits, such as ENS activation of type 2 innate lymphoid cells (ILCs) (Klose et al., 2017; Talbot et al., 2020; Wallrapp et al., 2017; Xu et al., 2019).
Our characterization of the ENS is largely incomplete due to longstanding technical challenges, because enteric neurons are rare, fragile, and challenging to isolate from surrounding tissue. This has restricted our ability to enrich populations for genomic profiling, especially in human, and restricted most work to rodent models (Grider, 2003). Recent efforts to profile the ENS using single cell RNA-Seq (scRNA-seq) have been limited to embryonic or early postnatal mice (Lasrado et al., 2017; Zeisel et al., 2018).
Here, we overcome these challenges to generate a map of the adult ENS at single cell resolution, spanning species, age, sex, region, and circadian phase (Figure 1A). We develop two methods to enable single nucleus RNA-seq (“snRNA-seq”) of the ENS and other tissues: RAISIN RNA-seq, for the isolation of intact nuclei and ribosome-bound RNA, and MIRACL-Seq, for label-free profiling of rare cell types, such as enteric neurons. With these methods, we profiled 1.6 million nuclei from the mouse ileum, mouse colon, and human colon, yielding 5,068 mouse and 1,445 human neurons. Our analysis reveals dozens of transcriptionally distinct neuron subsets, anatomical, age, and circadian sources of ENS variation, conserved programs across species, putative ENS cell-cell circuits, and risk genes for intestinal and extraintestinal diseases with early gut dysmotility that are enriched in the ENS.
Figure 1: RAISIN RNA-Seq provides an ENS reference map of adult mice and humans.
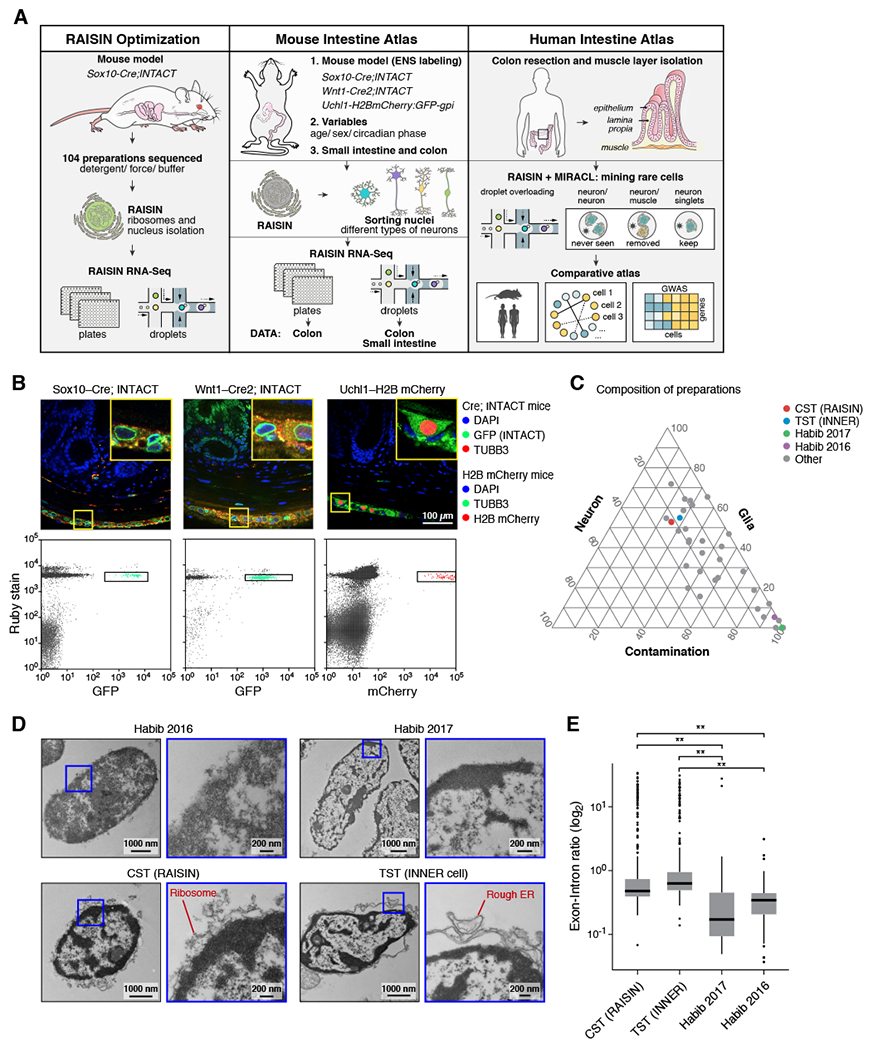
(A) Study design. (B) Validation of ENS reporter mice. Top: Representative cross-sections of muscularis propria (bottom) and mucosa (top), with TUBB3+ neurons. Bottom: Representative FACS plots. (C) Proportion of neurons, glia, and other cell subsets (triangle edges) from each extraction condition (n = 36 conditions, 104 experiments; dots), with select conditions labeled (legend). (D) RAISIN and INNER Cell yield nuclei with attached ribosomes and rough ER. Ultra-thin section TEM of published extractions (top; n = 2 experiments) vs. RAISIN and INNER Cell (bottom; n = 3 experiments). (E) Higher exon-intron ratios for RAISIN and INNER Cell. Distribution of exon-intron ratios (y axis) following snRNA-seq from each condition (x axis). All comparisons of RAISIN or INNER Cell vs. published methods significant (Wilcoxon test, p-value < 10−10); boxplots: 25%, 50%, and 75% quantiles; error bars: SD. See also Figure S1 and Table S1.
RESULTS
Validation of murine models to label the ENS
Because neurons comprise less than 1% of colon cells, we devised a strategy to enrich for the mouse ENS using three transgenic models, which label the pre-migratory neural crest (Wnt1-Cre), migratory neural crest (Sox10-Cre), or mature enteric neurons (Uchl1-H2B mCherry). The Wnt1 and Sox10 Cre lines (Debbache et al., 2018) were each mated to INTACT reporter mice (Mo et al., 2015), which label nuclei with GFP following Cre activity; specifically, we used the corrected Wnt1-Cre2 driver (Lewis et al., 2013) and a well-characterized Sox10-Cre mouse (Matsuoka et al., 2005, (Laranjeira et al., 2011). Additionally, to enrich for mature neurons, we used Uchl1-Histone2BmCherry:GFP-gpi (“Uchl1-H2B mCherry”) mice, which label neurons and potentially neuroendocrine cells (Wiese et al., 2013) (we did not observe the latter; below).
For all transgenic lines, we validated nuclear labeling of TUBB3+ neurons and confirmed that we could enrich for labeled nuclei with FACS (Figure 1B). For the Wnt1-Cre2 driver, we found signal in the colon mucosa (Figure 1B), which we mapped to epithelial cells using sn-RNA-seq (below). For the Uchl1-H2B mCherry mice, we observed labeling of neurons but not enteroendocrine cells by both histology (Figure 1B) and snRNA-seq (below).
We next applied two published snRNA-seq protocols (Habib et al., 2016, 2017) to FACS-enriched labeled nuclei from the colon of Sox10-Cre mice. Unfortunately, neither protocol performed well on ENS nuclei from the colon, in contrast to their performance in the brain (Figure S1A). These limitations raised the need to develop new snRNA-seq approaches.
Systematic optimization of nucleus extraction enables single nucleus profiling of colon ENS
To develop snRNA-seq methods that are compatible with a broader range of tissues, including the intestine, we optimized nucleus extraction conditions for the colon of adult Sox10-Cre mice, systematically varying the detergent (NP40, CHAPS, Tween, Digitonin), detergent concentration, buffer (HEPES, Tris, Tricine), mechanical extraction method (dounced, chopped, ground tissue), and added modifiers (e.g., salts) (STAR Methods), and compared these to published protocols (Habib et al., 2016, 2017). In total, we profiled 5,236 nuclei isolated from 104 preparations, spanning 36 extraction conditions (~145 nuclei per condition) using plate-based SMART-Seq2 (Table S1, STAR Methods). We scored conditions by (1) neuron and glia recovery rates relative to other cells (i.e. damaged or contaminating cells); (2) the number of genes detected per nucleus; and (3) an ENS signature score of the expression of literature-curated ENS marker genes (Figure 1C, Figure S1B-F, Table S1; STAR Methods).
Detergent type and concentration, buffer, and extraction method impacted all quality metrics (Figure S1B-F) and we identified two extractions with high ENS recovery, low contamination (~20% neurons and 55% glia, Figure 1C), and nuclei enriched in the ENS signature score (Figure S1E,F). We named these extractions “CST” (0.49% CHAPS detergent, Salts, Tris buffer) and “TST” (0.03% Tween-20 detergent, Salts, Tris buffer). Both extractions yielded significantly more detected genes per nucleus than published methods (CST: 2,486, TST: 2,542, published: 1,502; P < 10−10 for both comparisons; Wilcoxon test).
Preservation of ribosomes or rough endoplasmic reticulum on the nuclear envelope enables mature mRNA capture
To understand the basis for these differences, we examined nucleus structure using ultra-thin section transmission electron microscopy (TEM) (Figure 1D; STAR Methods). As expected, published methods yielded intact nuclei (Figure 1D). In contrast, CST retained the nuclear envelope and ribosomes on the outer nuclear membrane (Figure 1D) (Prunuske and Ullman, 2006); we named this profiling method, RAISIN (Ribosomes And Intact Single Nucleus) RNA-seq. TST also retained the nuclear envelope and attached ribosomes, as well as the rough ER (Figure 1D); we named this profiling method “INNER Cell” (Intact Nucleus and ER from a Cell) RNA-seq. Consistent with the TEM results, RAISIN and INNER Cell RNA-seq yielded higher exon-intron ratios than the published methods (Figure 1E), suggesting greater mRNA recovery. We used RAISIN RNA-seq to profile the mouse and human ENS, because it captured more neurons and fewer contaminating cells than INNER Cell RNA-seq (Figure 1C; Figure S1B-D).
RAISIN RNA-Seq of the ENS from adult mice identifies 21 neuron and 3 glia subsets
We used RAISIN RNA-Seq with SMART-Seq2 to profile 5,696 high-quality transcriptomes from the ENS of 29 adult mice, spanning a range of ages (11–104 weeks), both sexes, three transgenic models, and morning and evening phases of the day (Table S2). To capture differences along the proximal-distal axis, we divided each colon specimen into four equal segments (STAR Methods).
In total, we profiled 2,657 neurons and 3,039 glia (7,369 and 4,758 genes detected per nucleus, respectively; STAR Methods), which clustered into 21 neuron and 3 glia subsets (Figure 2A,B, Figure S2) and were enriched for known ENS markers (below) (Haber et al., 2017; Lasrado et al., 2017). Neurons and glia largely grouped by cell subset rather than by mouse, region, or other technical covariates (Figure S2A,B). After adjusting the numbers of FACS-sorted nuclei by neuron proportions in each sample (Figure S2C), we estimate that neurons comprise < 1% of all nuclei in the murine colon.
Figure 2: Reference map of the mouse colon ENS reveals 21 neuron and 3 glia subsets.
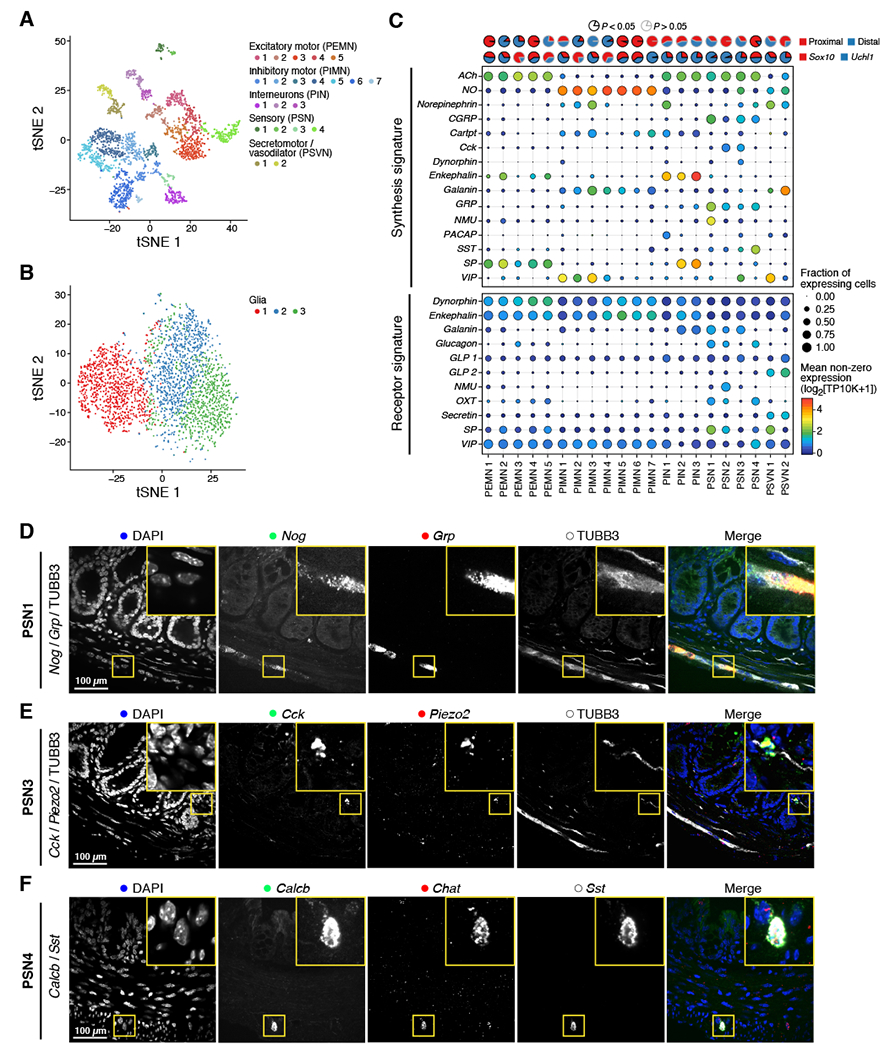
(A-C) Reference map of ENS in adult mouse colon (n = 102 samples collected from 29 total mice). (A,B) t-stochastic neighborhood embedding (t-SNE) of 2,657 neurons (A) or 3,039 glia (B) (dots) colored by subset and annotated post-hoc (legend). (C) Neuron subsets vary by location, transgenic model, and signaling. Top: Proportions of neuron subsets (columns) isolated from each colon region (upper pie chart) or transgenic model (lower pie chart). Bottom: Fraction of nuclei (dot size) in each subset expressing synthesis or receptor genes for signaling pathways, and their mean expression level in expressing cells (dot color). SP: Substance P, GLP: glucagon-like peptide. (D-F) Validation of gene expression in situ. Representative smFISH (n = 3 biological replicates per stain) for Nog and Grp showing co-expression of PSN1 markers with TUBB3 immunostaining (D), Cck and Piezo2 showing co-expression of PSN3 markers with TUBB3 immunostaining (E), and Calcb and Sst showing co-expression of PSN4 markers with Chat smFISH staining (F). Scale bar: 100 μm. See also Figures S2–S4, and Tables S2 and S3.
Neuron and glia subsets are distinguished by expression of canonical neurotransmitters
We annotated neuron subsets post-hoc using canonical ENS markers and differentially expressed (DE) genes between neuron subsets (Figure 2C and S3; Table S3; STAR Methods), and validated their co-expression in situ (Figure S4A-K). Broadly, neurons partitioned into either cholinergic (Chat+; acetylcholine (ACh) producing) or nitrergic (Nos1+; nitric oxide (NO) producing) subsets, except for two subsets that co-expressed both Chat and Nos1 (validated in situ, Figure 2C and S4A).
Based on the expression of known marker genes, we annotated 21 subsets (Figure 2A,C and S3; Table S3; STAR Methods) across five major groups: (1) Chat+Tac1+ putative excitatory motor neurons (PEMNs; 5 subsets) and (2) Nos1+ putative inhibitory motor neurons (PIMNs; 7 subsets: 4 subsets are Nos1+Vip+), which together coordinate muscle contraction and relaxation; (3) CGRP+ putative sensory neurons (PSNs; 4 subsets), which sense and respond to chemical and mechanical stimuli in the intestine; (4) Penk+ putative interneurons (PINs; 3 subsets), which relay signals between neurons; and (5) Glp2r+ putative secretomotor/vasodilator neurons (PSVNs; 2 subsets), which trigger secretions and fluid movement in other cell types.
In addition to these shared chemical codes, each neuron subset also expressed unique marker genes (Figure S3C; Table S3), including the D2 dopamine receptor (Drd2; PEMN3), adrenomedullin (Adm; PIMN3), prolactin receptor (Prlr; PIN2), melatonin receptor (Mtnr1a; PIN2), follistatin (Fst; PSVN1), Lgr5 (validated in situ; Figure S4I), and Csf2 receptor (Csf2rb and Csf2rb2; PSVN2). Interestingly, putative motor neurons express the mechanosensitive ion channel, Piezo1 (Figure S3B; confirmed in situ, Figure S4J), suggesting they may directly sense distention.
In contrast to the transcriptional diversity of neurons, glia clustered into three subsets (Figure 2B), each characterized by differentially expressed receptors and transporters (Figure S2E and Table S2). For example, Glia1 were enriched for the GDNF receptor, Gfra2; Glia2 for the monoamine transporter, Slc18a2; and Glia3 for the neurotensin receptor, Ntsr1 (Figure S2E).
Putative sensory neurons express key regulators of tissue homeostasis and immunity
We annotated four PSN subsets based on their expression of calcitonin gene-related peptide (CGRP) (Figure 2A,C and S3; Table S3). CGRP is encoded by two genes, Calca and Calcb. While all subsets express Calcb, only PSN1 and PSN3 were enriched for Calca (Figure S3B and S4B,H).
PSN subsets were largely distinguished by the expression of distinct sensory and effector genes. For example, PSN3 expresses cholecystokinin (Cck) and vasoactive intestinal polypeptide (Vip) (Figure 2C), markers of intestinofugal neurons (Furness et al., 2004), as well as brain-derived neurotrophic factor (Bdnf; Figure S3C), which is elevated in patients with irritable bowel syndrome (Yu et al., 2012), and Piezo2, a mechanosensitive ion channel that may regulate smooth muscle tone (validated in situ, Figure 2E and S3B) (Szurszewski et al., 2002). PSN4s were strongly enriched for somatostatin (Sst, validated in situ, Figure 2C,F). Notably, PSN1s were distinguished by expression of Noggin (Nog) and Neuromedin U (Nmu) (validated in situ, Figure 2C,D, S3B, and S4H). Noggin is a BMP antagonist needed to maintain the intestinal stem cell niche (Haramis et al., 2004), but its cell of origin has not been identified. Noggin expression by PSN1 suggests it may regulate the patterning or differentiation of stem cells. In addition, NMU production by enteric neurons activates ILCs, supporting a PSN1-ILC circuit. Indeed, PSN1s express other genes that may mediate signaling with ILCs, including CGRP, the IL-13 receptor (Il4ra/Il13ra1), and IL-7 (Il7), a major regulator of ILCs and T cells (Satoh-Takayama et al., 2010) (Figure 2C and S3B,D).
ENS composition varies by transgenic model and intestinal location
To systematically assess ENS heterogeneity, we examined the effects of transgenic background and intestinal location on ENS composition (i.e. the relative proportions of neuron subsets). Transgenic background had profound effects on neuron composition (Figure 2C). In particular, two sensory neuron subsets (PSN1, PSN2) were nearly absent from Sox10-Cre mice (Figure 2C), suggesting they may arise from distinct lineages (Betancur et al., 2010; Matsuoka et al., 2005). ENS composition also varied significantly along the proximal-distal axis of the colon (Figure 2C). For example, PSN1 and PSN2 were enriched in the proximal colon (p < 10−22 and 10−6, respectively; Fisher’s exact test), while subsets of putative motor neurons were enriched either proximally (e.g., PEMN1, PIMN2) or distally (e.g., PEMN2, PIMN4).
ENS expression programs influenced by intestinal location, circadian phase, and age
We next used a regression framework to identify genes differentially expressed with intestinal location, circadian phase, sex, and age (STAR Methods; Table S3).
First, after controlling for ENS composition (which itself varies by location; Figure 2C), neurons from the distal colon had higher expression of many neurotransmitter receptors, including those for serotonin (Htr3a, Htr3b), glutamate (Gria3, Grid1), and ACh (Chrna7, Chrm1) (Figure 3A). Next, we combined the synthesis/receptor genes for each neurotransmitter and neuropeptide, and tested for differences in expression between regions (Figure 3B). Sst expression was higher in the proximal colon, whereas chromogranin B (Chgb), enkephalin (Penk), norepinephrine (NE), secretogranin II (Scg2) and Vip were higher in the distal colon.
Figure 3: Mouse colon ENS varies with intestinal location, circadian phase, and age.
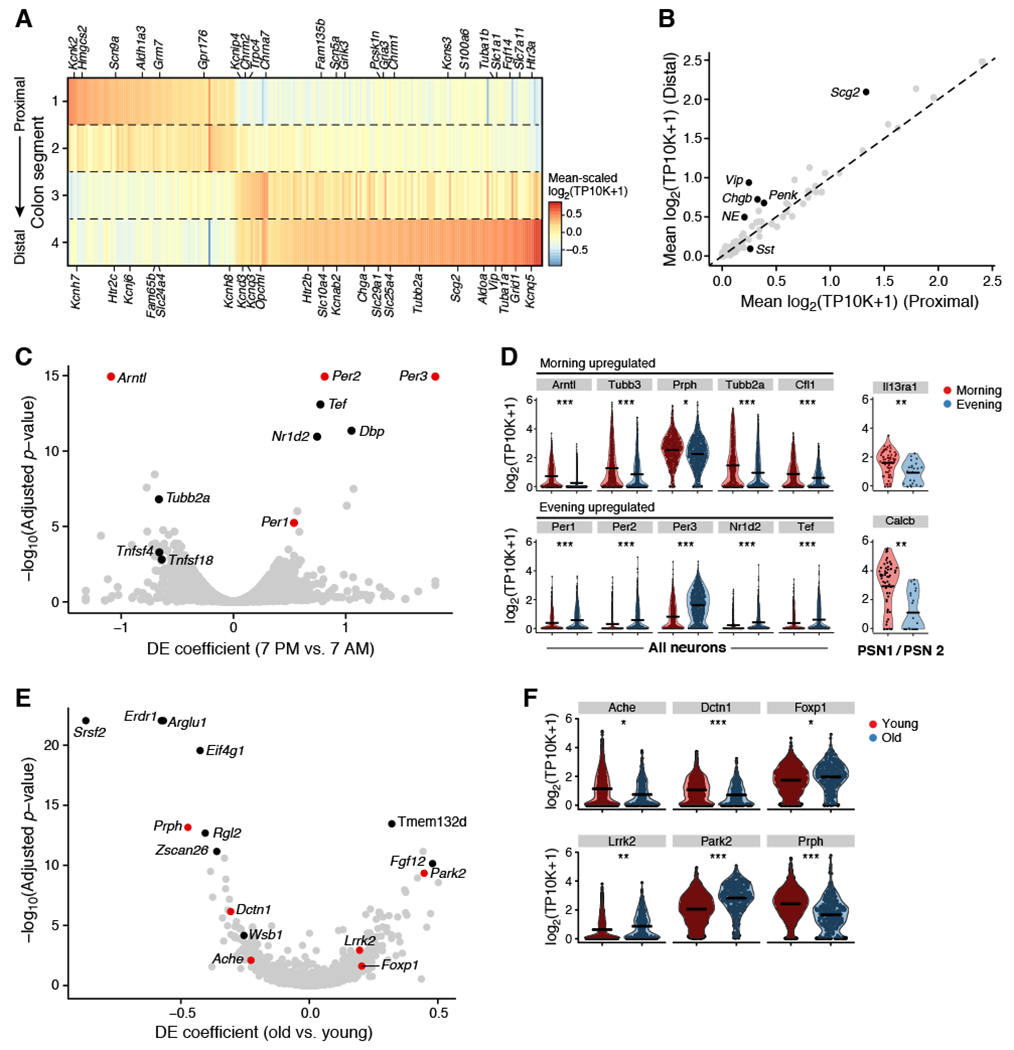
(A,B) Regional changes in ENS gene expression. (A) Mean expression levels (color bar) of DE genes for neurons from regions 1, 2, 3, and 4 (n = 682, 742, 506, and 657 neurons per region), sorted by peak expression. (B) Mean expression levels of synthesis and receptor genes for neurotransmitters and neuropeptides in distal (y axis, n = 1,163) vs. proximal (x axis, n = 1,424) colon. Dashed line: identity. Select genes highlighted in black. (C,D) Circadian changes in ENS gene expression. (C) DE genes between neurons from evening (n = 1,432 neurons; 11 mice) vs. morning (n = 1,170 neurons; 13 mice) showing effect size (x axis) and significance (y axis). Red dots: clock genes. (D) Distribution of gene expression levels (y axis) of select genes (x axis) upregulated in morning (red) or evening (blue) across all neurons (left) or in PSN1 and PSN2 (right). MAST regression (discrete term), adjusted p, * = 0.05, ** = 0.01, *** = 0.001. (E,F) Aging changes in ENS gene expression. (E) DE genes between neurons from aged (n = 434 neurons; 7 mice) vs. young (n = 2,223 neurons; 22 mice) mice, showing effect size (x axis) and significance (y axis). (F) Distribution of gene expression levels (y axis) of select risk genes (x axis) for neurodegenerative diseases. MAST regression (discrete term), adjusted p, * = 0.05, ** = 0.01, *** = 0.001. See also Table S3.
We found evidence for circadian regulation of the ENS, with core clock regulators among the most differentially expressed genes during morning (Arntl) and evening (Per1, Per2, Per3) (Figure 3C,D; Table S3). Genes for ENS signaling (e.g., Scg2, Pcsk1n), neuro-immune signaling (e.g., Calcb, Il13ra1 in PSN1 and PSN2 (Assas et al., 2014; Junttila, 2018)), and the cytoskeleton (e.g., Tubb2a, Prph) were induced in the morning, the latter suggesting circadian control of structural remodeling (Herrero et al., 2017) (Figure 3D; Table S3). In the evening, transcription factors were enriched, including Nr1d2, Tef, and Dbp (Figure 3C,D; Table S3), many of which are known circadian regulators (Yan et al., 2008).
Neurons from 2 year old mice showed dysregulation of many genes whose human orthologs are associated with CNS diseases (Figure 3E,F; Table S3). For example, Park2 and Lrrk2 are among the most upregulated genes in old vs. young mice, and mutations in these genes are some of the leading causes of early-onset Parkinson’s disease (Lesage and Brice, 2009). Similarly, genes linked to autism spectrum disorder (Ache, Foxp1) and amyotrophic lateral sclerosis (ALS) (e.g., Prph, Dctn1) were dysregulated with aging.
MIRACL-Seq: Massively parallel RAISIN-Seq for rare cell types in mouse and human
While our approach enabled profiling the ENS of transgenic mice, it is not feasible for unlabeled cells in mouse or human. While droplet-based profiling of unsorted nuclei could provide an alternative, it is inefficient for rare cell types.
We therefore developed MIRACL-Seq (“Mining Rare Cells”) to efficiently profile rare cell types from a complex tissue by overloading droplets with single nuclei/cells. Overloading captures more cells in one experiment, but also increases the probability of droplets containing multiple cells (multiplets). However, we estimated that overloaded neurons (or other rare cells) will only form heterotypic multiplets containing distinct cell types, which we can computationally identify and remove. In contrast, homotypic multiplets containing cells of the same type (e.g., multiple neurons) are very unlikely for rare cell types (< 1 neuron-neuron doublet per channel, Table S6; STAR Methods). MIRACL-Seq should thus enrich for enteric neurons and other rare cell types without yielding homotypic multiplets.
We used MIRACL-Seq to profile the ENS of the mouse colon, mouse ileum, and human colon at a range of overloading coefficients (Table S2). Because multiplet detection methods did not perform well, we developed a new method for detecting contaminated droplets (Figure S5A-D; STAR Methods). As expected, the cellular recovery and contamination rate increased with the extent of overloading (Figure 4A). For high levels of overloading, contamination rates were within established ranges; however, for low levels of overloading, contamination rates exceeded the expected co-encapsulation rate, consistent with increased ambient RNA (Figure 4A). We confirmed that neurons and glia from MIRACL-Seq expressed known ENS markers (Figure 4B). Notably, because droplet based RAISIN RNA-Seq generalizes to other tissues, including mouse lung, esophagus, and heart, (Figure S5E,F; Table S2), it should be possible to enrich for rare cells in other tissues with MIRACL-Seq.
Figure 4: MIRACL-Seq enables efficient droplet-based profiling of the mouse ENS in the ileum and colon.
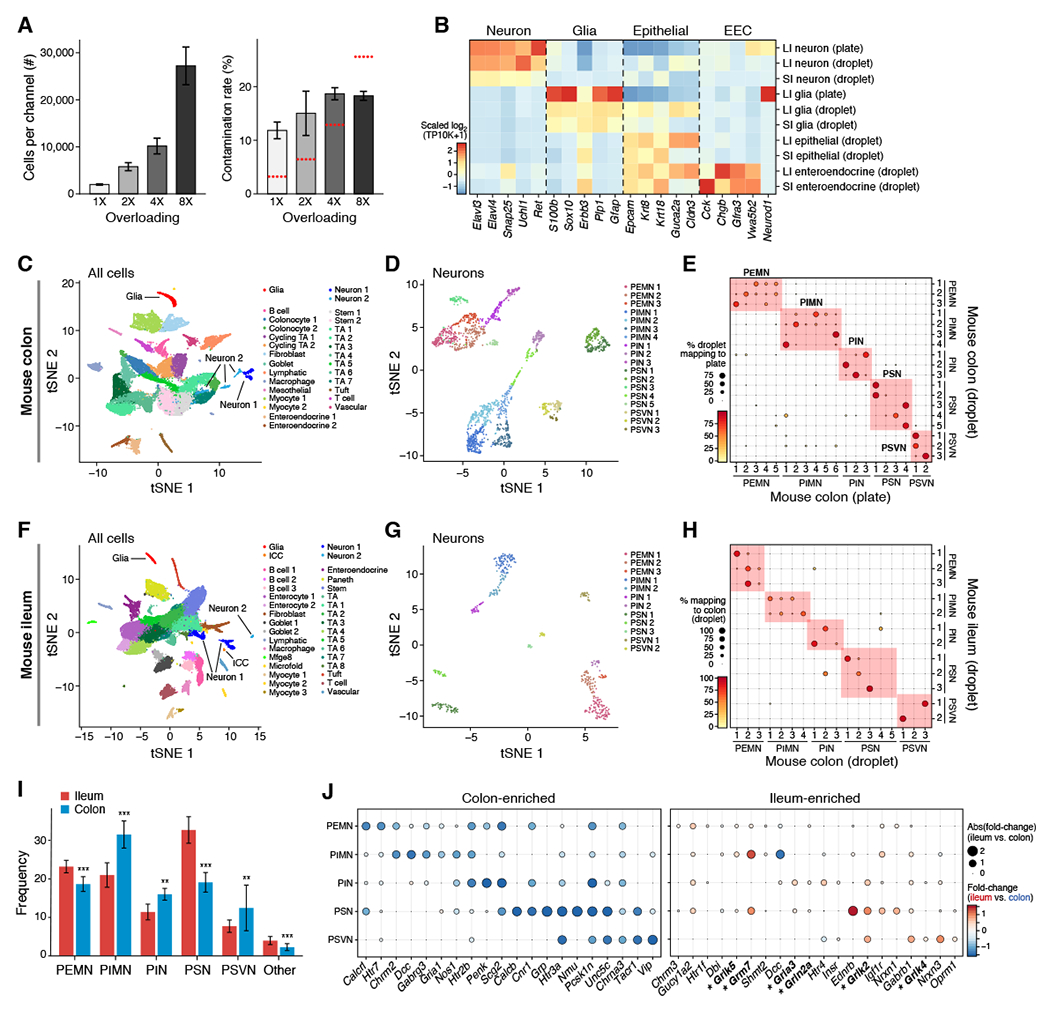
(A) Cell and doublet recovery using MIRACL-Seq. Number of nuclei per channel (left, y axis) and contamination rate (right, y axis) for a range of overloading coefficients (x axis). Error bars: SEM. Red line: expected multiplet rate. (B) MIRACL-Seq recovers high quality ENS. Mean expression levels (color bar) of select hallmark genes (columns) across cell subsets (rows). (C-E) MIRACL-Seq recapitulates plate-based colon atlas. (C) t-SNE of 343,000 highest-quality mouse colon nuclei (dots; n = 16 channels, 6 mice) colored by subset and annotated post-hoc. (D) t-SNE of 1,938 mouse colon neurons (dots) colored by subset and annotated with classifier. (E) Percent of neurons (dot size and color) from droplet-based subsets (rows) that map onto plate-based subsets (columns) using classifier. (F-H) Congruent subsets in ileum and colon. (F) t-SNE of 79,293 highest-quality mouse ileum nuclei (dots; n = 7 channels, 4 mice) colored by subset and annotated post-hoc. (G) t-SNE of 473 mouse ileum neurons (dots) colored by subset and annotated with classifier. (H) Percent of neurons (dot size and color) from droplet-based ileum subsets (rows) that map onto droplet-based colon subsets (columns) using classifier. (I-J) ENS differences between ileum and colon. (I) Frequencies (y axis) of neuron types (x axis) in ileum (n = 7; red) and colon (n = 16; blue). Dirichlet-multinomial regression, adjusted p, * = 0.05, ** = 0.01, *** = 0.001; error bars: SEM. (J) Fraction of nuclei (dot size) in neuron types (rows) expressing select genes (columns) that were enriched in the colon (left) or ileum (right), and their fold-change in colon vs. ileum (dot color; blue: colon-enriched, red: ileum-enriched) See also Figures S5-S7, and Tables S2, S4, and S6.
To rigorously test MIRACL-Seq, we applied it to the mouse colon and compared the results to the plate-based SMART-Seq2 atlas. We profiled 704,314 mouse colon nuclei by MIRACL-Seq, but filtered them to the 343,000 highest-quality nuclei for visualization and downstream analyses (STAR Methods). MIRACL-Seq yielded diverse epithelial, stromal (endothelial, fibroblast, glia), muscle, and immune (myeloid, lymphoid) cell lineages (Figure 4C), including 1,938 neurons (Figures 4D and S6A; 18 subsets; 3,508 genes detected per nucleus) and 1,620 glia (Figure S6B; 3 subsets; 1,616 genes detected per nucleus). We identified marker genes for each subset (Table S4) and used supervised classification (Peng et al., 2019) to map the 21 plate-based subsets onto the 18 subsets from MIRACL-Seq (Figure 4E; STAR Methods). Neuron subsets were congruent between the two methods, with each plate-based subset found in the MIRACL-Seq data and strong correlation of mean expression levels between neurons from both methods (Spearman’s ρ = 0.87; p < 10−15). MIRACL-Seq thus generates a comprehensive, detailed, and accurate colon ENS atlas in adult mice without labeling or sorting.
ENS atlas of the mouse ileum reveals 12 neuron subsets and key differences with colon
We next applied MIRACL-Seq to profile 483,221 unsorted nuclei from the mouse ileum (filtered to the 79,293 highest-quality nuclei for visualization and downstream analyses; STAR Methods), recovering diverse cell subsets (Figure 4F). These include 473 neurons (Figure 4G and S6C; 12 subsets; 2,310 genes detected per nucleus), 429 glia (Figure S6D; 2 subsets; 1,801 genes detected per nucleus), and other rare cell types, including 43 interstitial cells of Cajal (ICCs; Figure 4F).
Although neuron subsets from the ileum and colon were congruent (Figure 4E,H), they differed significantly in both their proportions and aspects of their gene expression programs (Figure 4I,J). The ileum was enriched in PSNs and depleted in PSVNs relative to the colon (Figure 4I; both q < 10−3, Dirichlet regression). Ileum neurons had higher expression of multiple glutamate receptors (Figure 4J; q < 10−4, Fisher test of DE genes; e.g., Grm7 in PIMNs, Gria3 in PINs, Grik2 in PSNs), while colon PSNs were enriched for Grp (Figure 4J; q < 10−25, MAST DE test). These differences may reflect gut physiology: glutamate is absorbed in the ileum, whereas colon regulation of fluid balance may require PSVNs.
A transcriptional atlas of human enteric neurons with MIRACL-Seq
To construct an ENS atlas of the human colon, we used MIRACL-Seq to profile unsorted nuclei from cancer-adjacent regions of colon resections from 16 colorectal cancer patients, spanning both sexes and a range of ages (35 – 90 years; Table S2). We dissected and profiled the muscularis propria, which contains more neurons than the mucosa/submucosa (submucosa yielded few neurons; Table S2).
We profiled 436,202 nuclei (filtered to the 146,442 highest-quality nuclei; STAR Methods), revealing diverse cell types: neurons, glia, adipocytes, endothelial cells, fibroblasts, immune cells, ICCs, and myocytes (Figure 5A; Table S4). We also found patient-enriched subsets (Figure 5A), which may reflect rare cell types or differences in location or cellular state.
Figure 5: Human colon ENS atlas reveals conserved expression programs across species.
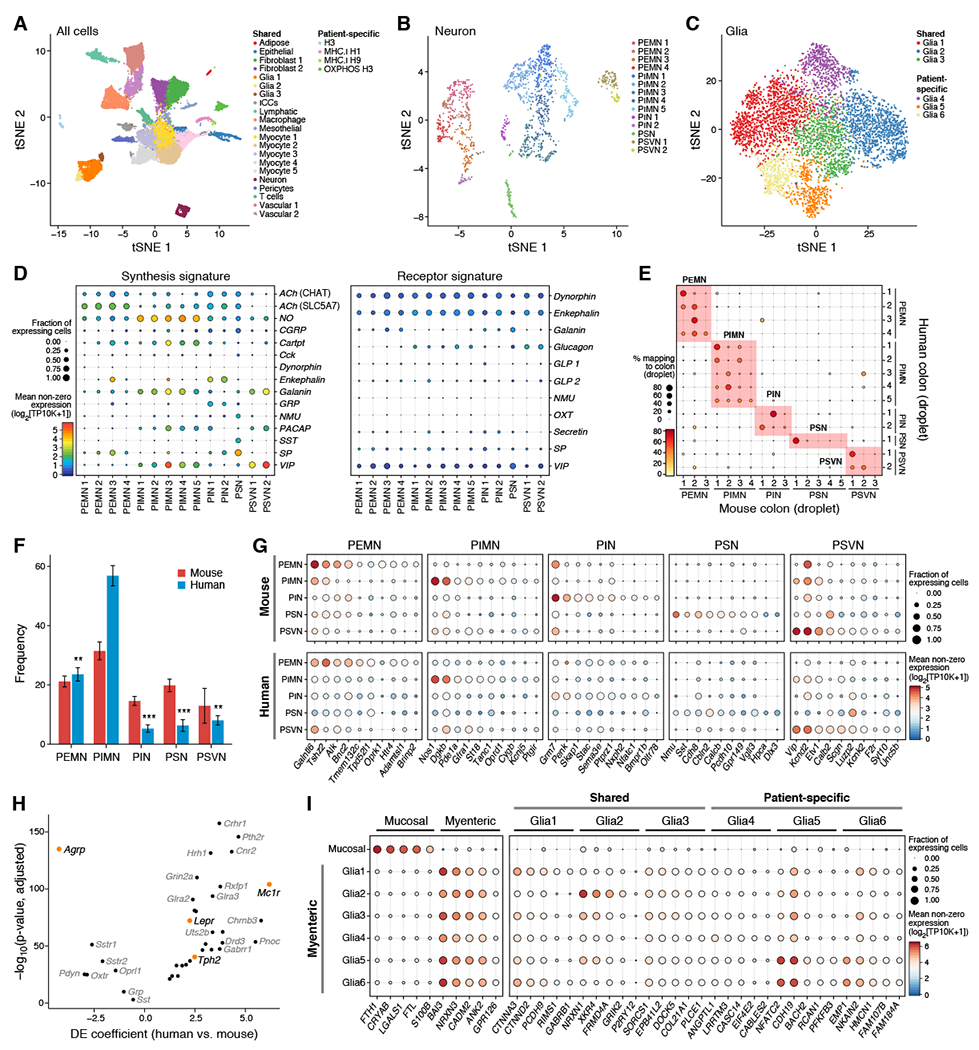
(A-E) Reference map of human colon muscularis ENS. (A) t-SNE of 146,442 highest-quality human colon nuclei (dots; n = 52 channels, 16 patients) colored by subset and annotated post-hoc. (B) t-SNE of 1,445 human colon neurons (dots) colored by subset and annotated with classifier. (C) t-SNE of 6,054 human colon glia (dots) colored by shared or unique subsets (legend). (D) Human colon neurons express diverse signaling genes. Fraction of cells (dot size) in subsets (columns) expressing synthesis (left) or receptor (right) genes for signaling molecules (rows), and their mean expression level in expressing cells (dot color). (E) Percent of neurons (dot size and color) from droplet-based human colon subsets (rows) that map onto droplet-based mouse colon subsets (columns) using classifier. (F) Differences in ENS composition between species. Frequencies (y axis) of neuron types in mouse (n = 16; red) or human (n = 14; blue) colon. Dirichlet-multinomial regression, adjusted p, * = 0.05, ** = 0.01, *** = 0.001); error bars: SEM. (G) Conserved expression programs between species. Fraction of nuclei (dot size) in neuron types (rows) expressing select genes (columns) from conserved transcriptional programs, and their mean expression level in expressing cells (dot color), for mouse (top) and human (bottom). (H) DE genes between colon neurons from human (n = 1,445) vs. mouse (n = 1,938) showing effect size (x axis) and significance (y axis). (I) Fraction of cells (dot size) in each glia subset (rows) expressing select marker genes (columns) and their mean expression level in expressing cells (dot color), for mucosal vs. myenteric glia (left) or myenteric subsets (right). See also Figures S4–S7, and Tables S2 and S4.
The 1,445 human neurons clustered into 14 subsets: 4 PEMN, 5 PIMN, 2 PIN, 1 PSN, and 2 PSVN subsets (4,302 genes detected per nucleus; Figure 5B,D; Table S4; STAR Methods). While we detect hallmark neurotransmitters, CHAT was lowly expressed (Figure 5D), possibly due to low nuclear expression in human (we did not find an alternative 3’ end (Wallrapp et al., 2017)). However, SLC5A7, a choline transporter that is necessary for ACh synthesis in the mouse CNS (Ferguson et al., 2004), was detected (Figure 5D). Moreover, the mean expression levels of CHAT and SLC5A7 were strongly correlated across neuron subsets in both species (Spearman’s ρ = 0.74), and in mouse, Slc5a7 levels perfectly predicted cholinergic vs. noncholinergic subsets (AUC = 1). We thus use SLC5A7 expression as additional evidence of ACh synthesis in human neurons.
Human ENS subsets share core transcriptional programs with mouse
Comparing the 14 subsets of human colon neurons with the 18 subsets from mouse (Figure 5E) revealed strong congruence between species, while also highlighting compositional differences. PEMNs and PIMNs were enriched in human, and PSNs, PINs, and PSVNs were depleted with less observed diversity (Figure 5F). Notably, we did not find ACh+ PSVNs in human (Figure 5D), potentially due to submucosa removal, lower PSVN proportions, or species-specific features.
We leveraged the human-mouse mapping to identify conserved transcriptional programs for each neuron type (Figure 5G, Table S4; STAR Methods). For example, the shared program for PEMNs (n = 51 genes) includes ACh, neurotransmitter receptors (e.g., Gfra2, Oprk1, Htr4), transporters (e.g., Slc5a7), TFs (e.g., Casz1), and Colq, which tethers acetylcholinesterase in the neuromuscular junction (Legay, 2000) (Figure 5G, Table S4). We also defined shared programs for PIMNs, (n = 42 genes; e.g., Nos1, Gfra1, Etv1, Oprd1, Dgkb), PINs (n = 59 genes; e.g., Penk, Adra2a, Bmpr1b), PSNs (n = 149 genes; e.g., Calcb, Nmu, Nog, Sst, Vipr2), and PSVNs (n = 48 genes; e.g., Vip, Gal, Scgn, Calb2) (Figure 5G, Table S4).
Expression differences between species (Figure 5H; Table S4) included the melanocortin system, a central regulator of energy homeostasis (Garfield et al., 2009). Both human and mouse neurons were strongly enriched for the melanocortin receptor (MC1R) and its antagonist (Agrp), respectively (Figure 5H,I). The leptin receptor (LEPR) and the enzyme for serotonin synthesis (TPH2) we strongly expressed in human neurons, but not in mouse (Figure 5H,I). We validated the absence of Tph2 from the mouse colon in situ, using targeted brain regions as negative and positive controls, and supported this finding with bulk RNA-seq (Söllner et al., 2017) (Figure S7). Expression differences in the melanocortin, leptin, and serotonin pathways, which suppress appetite, may reflect adaptations to feeding behavior in the human and mouse ENS.
Transcriptionally diverse subsets of myenteric and mucosal human glia
Four glia types have been identified based on morphology and localization (Boesmans et al., 2015). We profiled 6,054 human colon glia, which clustered into 3 shared and 3 patient-enriched subsets (1,497 genes detected per nucleus; Figure 5C and S6F). Combining our muscularis glia with 1,326 glia from the colon mucosa (Smillie et al., 2018), we derived markers for each glial subset (Figure 5I; Table S4). Mucosal glia were enriched for ferritin genes (e.g., FTH1, FLT), myenteric glia expressed cell adhesion molecules (e.g., NRXN1, CADM2) (Figure 5I), and each myenteric subset expressed unique genes (Figure 5I), such as the purinergic receptor, P2Y12R in Glia2 cells, which may explain varying responses to adenosine triphosphate (Boesmans et al., 2015).
Enteric neurons express genes for interaction with diverse cell types in the colon
To examine interactions between the ENS and other colon cell types, we mapped thousands of receptor-ligand pairs onto our data, along with 115,517 single cells from the human colon mucosa (Smillie et al., 2018). These data span diverse ENS, epithelial, stromal, and immune cell lineages.
Supporting the validity of this approach, putative cell-cell interactions recapitulate the best-studied ENS circuit, the peristaltic reflex (Grider, 2003; Kunze and Furness, 1999) (Figure 6A), beginning with enteroendocrine cell production of 5HT (Tph1), which signals to 5HT receptors on sensory neurons. Sensory neurons relay this stimulus via CGRP (Calcb) to interneurons, which modulate motor neuron activity via ACh and Penk signaling. Motor neurons then activate or inhibit muscle contraction via release of ACh and tachykinin (Tac1) or NO, resulting in peristaltic movement. Our analysis also suggests new interactions in this circuit. For example, PSN3 and PIN1 express Cck and its receptor (Cckar), respectively, while PIMN expresses the glutamate synthesis enzyme, Gls, and myocytes express its receptor.
Figure 6: Cell-cell interactions connect the ENS and diverse cell subsets in mouse and human.
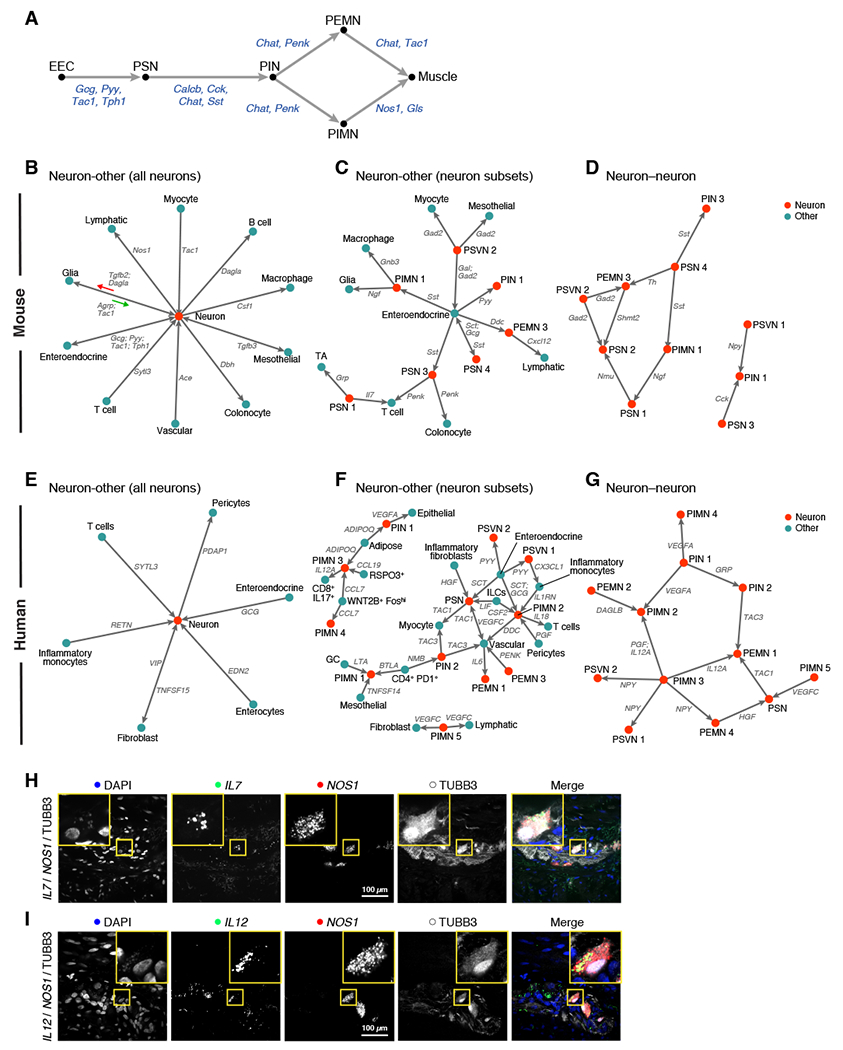
(A) Recovery of peristaltic circuit in mouse colon. Interactions (arrows) from ligand- to receptor-expressing cell subsets involved in the peristaltic circuit. Black: cell subset; Blue: ligand; EEC: enteroendocrine cell. (B-G) ENS interactions in mouse and human colon. For mouse (B-D) and human (E-G) colon, interactions (arrows) from ligand- to receptor-expressing cell subsets, between all neurons and other subsets (B, E), neuron subsets and other subsets (C,F), or among neuron subsets (D,G). Grey: ligand; Red: neuron subset; Blue: other subset. (H,I) Expression validation for putative neuro-immune interactions. Representative smFISH (n = 3 biological replicates per stain) for IL7 and NOS1 showing expression of IL-7 in nitrergic neurons (H) and IL12A and NOS1 showing expression of IL-12 in nitrergic neurons (I), with TUBB3 immunostaining. Scale bar: 100 μm. See also Table S5.
We next generated ENS interaction networks for cases where both a ligand and its receptor were strongly enriched in their respective cell subsets (Figure 6B-G). (These interactions only comprise a subset of all putative predicted ENS interactions; see Table S5 for complete list). Human and mouse enteroendocrine cells signaled to distinct neuron subsets via the neuropeptides glucagon (GCG-GCGR/GLP2R), peptide YY (PYY-NPY1/2/5R), and secretin (SCT-SCTR), while in mouse they also signaled via Sst (Sstr1/2/3/5) (Figure 6B-G). We also mapped putative interactions between neurons and other cell types, such as mouse PEMN3 and lymphatic cells (Cxcl12-Ackr3), mouse PSN1 and transit amplifying cells (Grp-Grpr), human PIN1 and adipocytes (ADIPOQ-ADIPOR1), and human PIMNs and RSPO3+ fibroblasts (CCL7/19-CCR10) (Figure 6C,F).
In particular, we found known and novel neuro-immune interactions. Among known ones are murine Csf1+ neurons that promote the development of Csf1r+ muscularis macrophages (Muller et al., 2014) (Figure 6B). Among new predictions are interactions between mouse neurons and B cells via 2-Arachidonoylglycerol (Figure 6B; Dagla-Cnr2); mouse PIN3 and T cells via enkephalin (Figure 6C; Penk-Oprm1); human PIMN3 and CD8+IL-17+ T cells via IL-12 (IL12A-IL12RB1; validated in situ, Figure 6I), human PIMN2 and T cells via IL-18 (IL18-IL18R1), and human PSVN1 and inflammatory monocytes via fractalkine (CX3CL1-CX3CR1) (Figure 6F).
Human enteric neurons express risk genes for intestinal and extraintestinal diseases
Finally, we interrogated ENS contributions to disease by testing whether enteric neurons expressed risk genes for diseases with suspected ENS involvement. These included: Hirschsprung’s disease (HSCR), autism spectrum disorders (ASD) and Parkinson’s disease (PD), both associated with gut dysmotility at early stages of disease progression (Bondurand and Southard-Smith, 2016; Chang et al., 2017; Sanders et al., 2015), and inflammatory bowel disease (IBD) because the ENS may regulate inflammation (Klose et al., 2017) and ENS disruption is a feature of Crohn’s disease (Dvorak and Silen, 1985; Steinhoff et al., 1988).
Mapping 185 disease risk genes (STAR Methods) onto cells from the human mucosa (Figure 7A) and muscularis (Figure 7B) revealed genes that were enriched in the ENS, even relative to the cortex (Figure 7C) (Hodge et al., 2019). For example, most HSCR risk genes were specifically expressed by enteric neurons, including RET, PHOX2B, GFRA1, and ECE1 (Figure 7B; RET and PHOX2B validated in situ; Figure S4L,M); two exceptions, EDN3 and EDNRB, are expressed in the neural crest (Pla and Larue, 2003). While most IBD risk genes were enriched in mucosal cell subsets, a few were enriched in enteric neurons: BTBD8, GRP, JAZF1, NDFIP1, PLA2R1, and CNTNAP2 (GRP validated in situ; Figure 7B and S4N).
Figure 7: Human ENS expresses risk genes for intestinal and extraintestinal diseases.
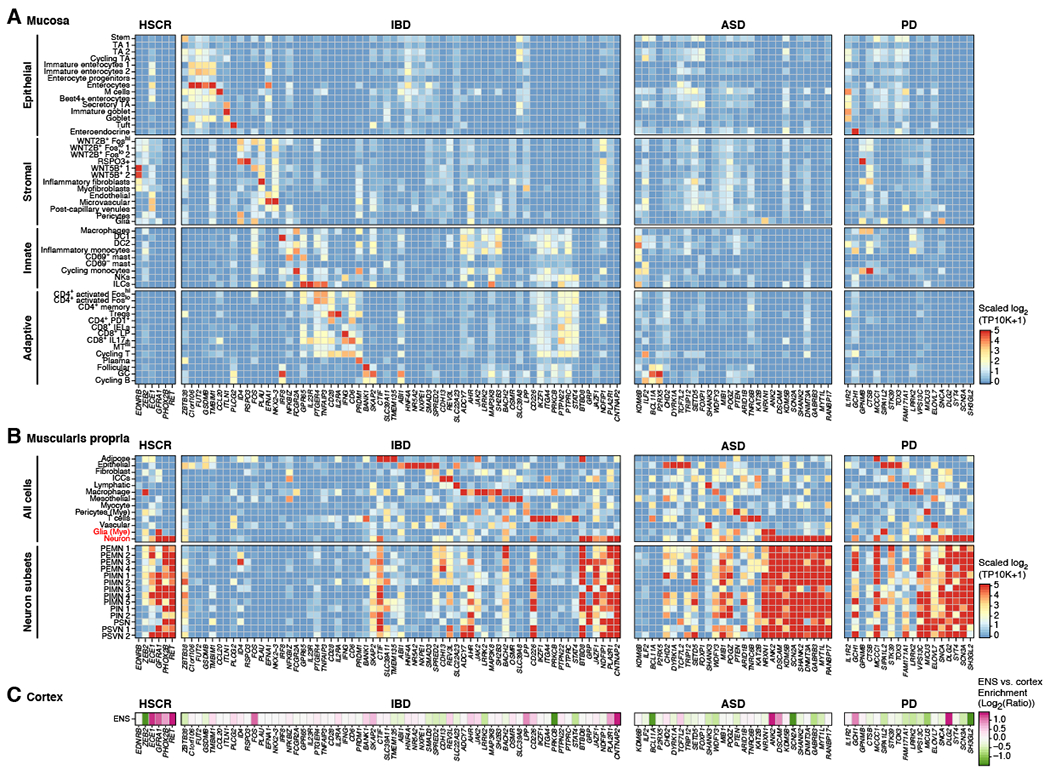
(A,B) Mean expression levels across cell subsets (rows) of risk genes (columns) implicated by genome-wide association studies for Hirschsprung’s disease (HRSC), inflammatory bowel disease (IBD), autism spectrum disorders (ASD), and Parkinson’s disease (PD), which are enriched in cell subsets from either the human colon mucosa (A) or muscularis propria (B). (C) Fold enrichment (log2) of the expression of disease risk genes in colon vs. cortex neurons. See also Figure S4.
Risk genes for ASD and PD were mainly expressed in the ENS relative to other cells in the colon (Figure 7B), including NRXN1, ANK2, DSCAM, and GABRB3 for ASD, and DLG2, SNCA, and SCN3A for PD (DSCAM and SNC3A validated in situ, Figure 7B and S4O,P). A few, including DSCAM and DLG2, were enriched in enteric vs. cortical neurons (Figure 7C), while others were enriched in glia (e.g., NRXN1, ANK2).
DISCUSSION
To study the ENS at single cell resolution despite major technical limitations, we developed RAISIN RNA-seq and MIRACL-Seq to profile 5,068 mouse and 1,445 human enteric neurons and diverse gut cell types from both species. We provide in situ validations of our findings, and demonstrate the broad utility of these methods in other tissues (heart, lung, and esophagus). These methods can be applied to fresh and frozen tissue specimens, enabling study of the ENS and archived frozen tissue samples, and can be used to profile other rare cell types in a range of tissues. Indeed, MIRACL-Seq enriched not only for enteric neurons, but also other rare cell types, including 43 murine ICCs (0.05% frequency) and 1,103 human ICCs (0.75% frequency). Even the multiplet profiles in MIRACL-Seq can be utilized, for example using decompression techniques (Cleary et al., 2017).
We found dozens of mouse and human neuron subsets and characterized sources of heterogeneity. We show circadian regulation of the ENS, motivating work into circadian control of ENS function, nutrient absorption, and metabolism. We also highlight regional differences in the ENS along the mouse intestine, including in the ileum and proximal/distal colon. Future work may profile other ENS regions, including the mouse duodenum and jejunum and the human small intestine. Examination of old mice revealed genes that are perturbed with aging, including Lrrk2 and Park2; future work can dissect the role of the ENS in age-related diseases.
We derived conserved transcriptional programs across species, highlighting processes that can be modeled in mouse; for example, PSN1 in both species expresses NMU, which activates ILCs, and Noggin, a BMP inhibitor that supports the intestinal stem cell niche. We also mapped differences in gene expression between human and mouse in the melanocortin, leptin, and serotonin pathways. Taken together, these data generate hypotheses allowing experimental dissection of ENS function.
We inferred putative interactions between the ENS and diverse parenchymal, stromal, and immune cell types. The ENS was enriched for the expression of several cytokines and cytokine receptors, which may signal to myeloid, B, or T cells to shape mucosal immunity. For example, PIMN2 production of IL-18 may signal to its receptor on T cells (Figure 6F), in addition to goblet cells (Jarret et al., 2020). Future work on neuro-immune interactions, including IL-7, IL-12, IL-13, and IL-18 circuits, may reveal a broad capacity for the ENS to interact with the immune environment.
Finally, our results are consistent with a model where ENS dysfunction can lead to immune dysregulation, which can exacerbate inflammation and related pathologies. For example, many IBD risk genes are enriched in the ENS, raising the need to characterize the role of the ENS in intestinal inflammation. In addition, dozens of ASD and PD risk genes are enriched in the ENS, suggesting a mechanism for gut dysmotility. Recent associations between the gut microbiota and diseases, such as autoimmune disorders (Vatanen et al., 2016) and cancers and cancer therapies (Gopalakrishnan et al., 2018), suggest that immune modulation in the gut can have systemic effects. Proper immune function is necessary for CNS maintenance and repair, with immune dysregulation contributing to neurodegeneration (Deczkowska and Schwartz, 2018). Thus, the ENS may be a nexus linking the gut, the immune system, and the brain, and dysfunction in the ENS may exacerbate diseases of the CNS.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Aviv Regev (aregev@broadinstitute.org or regev.aviv@gene.com).
Materials availability
This study did not generate unique reagents.
Data and code availability
FASTQ files for mouse and processed data for mouse and human were deposited in the Single Cell Portal (SCP1038). Human FASTQ files were deposited in the Broad Data Use Oversight System (DUOS) (“ColonENS-RegevXavier-CRCAdjacentNormal-Broad-snRNASeq-UnsortedCells”).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human donors and tissue samples
All colon resection samples were obtained from colon cancer patients after informed consent at either the Dana Farber Cancer Institute, Boston (IRB 03-189; ORSP 3490) or Massachusetts General Hospital, Boston (IRB 02-240; ORSP 1702). Metadata for the samples are provided in Table S4. Normal colon located proximal to tumor was placed into conical tubes containing Roswell Park Memorial Institute (RPMI) media supplemented with 2% human serum and placed on ice for transport to the Broad Institute, Boston. Upon arrival, the muscularis propria was dissected from the remainder of the tissue (e.g., submucosa), divided into pieces (~ 20-120 mg), which were placed into cryo-vials, frozen on dry-ice and stored at −80°C. When possible, a portion of the tissue was fixed overnight in 4% paraformaldehyde at 4°C for histology.
Mouse models
All animal work was performed under the guidelines of the Division of Comparative Medicine, in accordance with the Institutional Animal Care and Use Committees (IACUC) relevant guidelines at the Broad Institute and MIT, and consistent with the Guide for Care and Use of Laboratory Animals, National Research Council, 1996 (institutional animal welfare assurance no. A4711-01), with protocol 0122-10-16. Mice were housed under specific-pathogen-free (SPF) conditions at the Broad Institute vivarium.
Briefly, we took two approaches to label the murine ENS. In the first approach, we used Cre/LoxP technology to label ENS nuclei with a nuclear GFP, which allowed for subsequent FACS enrichment. The nuclear GFP label (known as the INTACT allele) is a conditional GFP allele that is only expressed following Cre recombinase-mediated excision of a transcriptional stop cassette from the genome (Mo et al., 2015) and is a permanent change in all daughter cells. The INTACT strain was crossed with mouse lines that express Cre recombinase during the development and/or migration of the neural crest; these are the Wnt1-Cre2 and Sox10-Cre mouse lines, which are well-established and previously published lines (Lewis et al., 2013; Matsuoka et al., 2005). The progeny of these mice contained labeled enteric neurons and glia, which we profiled. For the second approach, we used a fluorescent reporter line to label enteric neuron nuclei with mCherry, which allowed for subsequent FACS enrichment. This line expresses mCherry fused to a histone (nuclear localization) under the Uchl1 regulatory region (Wiese et al., 2013); Uchl1 marks enteric neurons (Schofield et al., 1995) and this line thus has mCherry expression in enteric neurons. We validated these lines using histology and with RAISIN RNA-seq prior to using them in the atlas. We note that the Sox10-Cre mouse labeled both neurons and oligodendrocytes in the brain (Figure S1A), whereas we anticipated recovering only brain oligodendrocytes (Simon et al., 2012).
The following strains were used:
| Strain | Jackson Laboratory (Bar Harbor, ME) catalog number | Reference | Acknowledgement |
|---|---|---|---|
| C57BL/6J | 000664 | ||
| B6;CBA-Tg(Sox10-cre)1Wdr/J | 025807 | (Matsuoka et al., 2005) | We thank William Richardson for donating animals to a publicly available repository. |
| 129S4.Cg-E2f1Tg(Wnt1-cre)2Sor/J | 022137 | (Lewis et al., 2013) | We thank Philippe Soriano for donating animals to a publicly available repository. |
| B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J | 021039 | (Mo et al., 2015) | We thank Jeremy Nathans for donating animals to a publicly available repository. |
| Tg(Uchl1-HIST2H2BE/mCherry/EGFP*)FSout/J | 016981 | (Wiese et al., 2013) | We thank Michelle Southard-Smith for donating animals to a publicly available repository. |
The following mouse crosses were performed:
| Strain 1 | Strain 2 |
|---|---|
| B6;CBA-Tg(Sox10-cre)1Wdr/J | B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J |
| 129S4.Cg-E2f1Tg(Wnt1-cre)2Sor/J (Note: did not label neurons and was not eventually used in the atlas) | B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J |
| did not label neurons and was not eventually used in the atlas) | Sun1/sfGFP)Nat/J |
| Tg(Uchl1-HIST2H2BE/mCherry/EGFP*)FSout/J | C57BL/6J |
METHOD DETAILS
Tissue collection for snRNA-seq
For snRNA-seq optimization, tissue was collected from 11-14 week animals. For the ENS atlas, tissue was collected from 11-14 week old and 50-52 week old mice at either 7-8am or 7-8pm. Each colon was isolated and rinsed in ice cold PBS. Next, the colon was opened longitudinally and separated into four equally-sized sections, which were frozen in a 1.5 mL tube on dry ice. For brain collection, the brain was removed, quartered and frozen in a 1.5 mL tube on dry ice. Frozen tissue was stored at −80°C until subsequent tissue processing.
Tissue collection and preparation for RNA fluorescence in situ hybridization and immunohistochemistry
For RNA fluorescence in situ hybridization (RNA FISH) and Immunohistochemistry (IHC), isolated colon was cut into four sections of equal size and processed as described (Drokhlyansky et al., 2017). Briefly, tissue was fixed in 4% paraformaldehyde overnight at 4°C. Then, tissue was sequentially passed through PBS containing 7.5%, 15% and 30% (w/v) sucrose at 4°C. Tissue was then embedded in O.C.T. (23-730-571, Fisher Scientific, Hampton, NH) and stored at −80°C. Tissue was cut at 25 micron thick sections onto Superfrost Plus microscope slides (22-037-246, Fisher Scientific) using a Leica CM1950 Cryostat (Leica Biosystems Inc., Buffalo Grove, IL).
Immunofluorescence (IF)
Slides with tissue sections were washed three times in PBS for 10 minutes, blocked 1 hour in CAS-Block Histochemical Reagent (00-8120, Thermo Fisher Scientific), incubated with primary antibodies overnight at 4°C, washed three times in PBS for 10 minutes, and then incubated with secondary antibodies at for 1 hour at room temperature. Slides were then washed twice in PBS for 10 minutes and then for 10 minutes with a PBS containing DAPI (D9542, Sigma-Aldrich). Lastly, slides were mounted using Southern Biotech Fluoromount-G (010001, VWR) and sealed. Antibodies used for IF: Rabbit anti-Tubb3 (1:1000, AB18207, Abcam), Chicken anti-mCherry (1:1000, AB356481, EMD Millipore), Goat anti-Ass1 (1:1000, ab77590, Abcam), Rabbit anti-Celsr3 (1:500, SAB4500707-100UG, Sigma), Rabbit anti-Uchl1 (1:800, 13179T, Cell Signaling), Rabbit anti-ASL (1:200, ab97370, Abcam), Rabbit anti-GRP (1:1000, ab22623, Abcam), Rabbit anti-Prph (1:2000, ab4666, Abcam), Goat anti-Phox2b (5 μg/mL, AF4940, Novus Biologicals) and Alexa Fluor 488-, 594-, and 647-conjugated secondary antibodies (Life Technologies) were used.
Single-molecule fluorescence in situ hybridization (smFISH)
RNAScope Multiplex Fluorescent Kit (Advanced Cell Diagnostics) was used per manufacturer’s recommendations for fresh-frozen samples with the following alterations. All Wash Buffer times were increased to 5 minutes and, following final HRP-Block step, slides were washed for 10 minutes with PBS containing DAPI (Sigma-Aldrich) followed by mounting with Southern Biotech Fluoromount-G (VWR) and sealed. Probes used for smFISH (Advanced Cell Diagnostics): Calca (417961), Calcb (425511), Cck (402271), Chat (408731-C2), Grp (317861-C2), Htr4 (408241), Lgr5 (312171), Nmu (446831), Nog (467391), Nos1 (437651-C3), Piezo1 (500511), Piezo2 (400191-C3), Sst (404631-C3), Tph2 (318691-C2), Vip (415961 and 415961-C2), ANO1 (349021-C2), CHAT (450671 and 450671-C2), DSCAM (810481), GUCY1A3 (425831), IL7 (424251), IL12A (402061), KIT (606401-C3), and NOS1 (506551-C2), RET (424871), GRP (465261), NR5A2 (490261) and SCN3A (460121) were used.
Combined smFISH and IF
smFISH was performed as described above, with the following changes. After the final HRP-Block step, tissue sections were incubated with primary antibodies overnight at 4°C, washed in TBST for 5 minutes, twice, and then incubated with secondary antibodies for 30 min at room temperature. Slides were then washed in TBST for 5 minutes, twice, followed by a 10 minute wash with containing DAPI (Sigma-Aldrich) before mounting with Southern Biotech Fluoromount-G (VWR) and sealed.
Confocal microscopy and image analysis
Images were taken using a Nikon TI-E microscope with a Yokohama W1 spinning disk, 405/488/561/640 lasers, and a Plan Apo 60X/1.4 objective. Images were visualized and overlaid using FIJI (Pietzsch et al., 2012; Schindelin et al., 2012, 2015; Schneider et al., 2012). The Bio-Formats plugin (Linkert et al., 2010) was used to import all images. All histology validations were performed in at least three independent experiments. As controls, we used probe-minus reactions, which gave no signal in the tissue, and epithelial targets, which gave no signal in the ENS. All applied probes were highly specific with no background labeling of cells within tissue sections.
Nucleus extractions
The following nucleus extractions were performed from either mouse colon or brain and subsequently processed for profiling:
Dounce homogenization:
Nuclei were extracted using either dounce homogenization followed by sucrose gradient centrifugation as described (Habib et al., 2016), or using the Nuclei EZ Prep (NUC101-1KT, Sigma-Aldrich) as described (Habib et al., 2017), with the following modifications. The tissues were dounce homogenized with a 7 mL Dounce Tissue Grinder (VWR 22877-280) (20 times pestle A, 20 times pestle B) and buffer volumes were increased to 5 mL for homogenization.
Tissue grinding:
Fresh-Frozen tissues were crushed into a fine powder with a mortar and pestle (89038-144 and 89038-160, VWR) over a bath of liquid nitrogen. The powder was briefly resuspended in 2 mL of liquid nitrogen for transfer to a 50 mL conical tube, where liquid nitrogen was allowed to evaporate. The tissue powder was resuspended in 5 mL of Nuclei EZ Prep reagent (NUC101-1KT, Sigma-Aldrich) or NST (NP-40, Salts and Tris; see Table S1) and transferred to a 7 mL Dounce Tissue Grinder. For the Nuclei EZ Prep kit, all subsequent steps were as described (Habib et al., 2017). For NST, the tissue was dounce homogenized with a 7 mL Dounce Tissue Grinder (VWR 22877-280) (20 times pestle A, 20 times pestle B), filtered through a 40 μm strainer (Falcon), and flow-through was spun at 500g for 5 minutes at 4°C. The pellet was resuspended in 0.5-3 mL of ST (Salts: 146 mM NaCl, 1 mM CaCl2, 21 mM MgCl2; Tris; Table S1).
Chopping extraction:
Fresh-frozen tissues were disaggregated in 1 mL of custom nuclear extraction buffer (see Table S1 for all combinations used) with mild chopping by Tungsten Carbide Straight 11.5 cm Fine Scissors (14558-11, Fine Science Tools, Foster City, CA) for 10 minutes on ice. Large debris were removed with a 40 μm strainer (Falcon). An additional 1 mL of buffer was used to wash the filter before proceeding to fluorescence-activated cell sorting (FACS). For droplet-based RNA-Seq, nuclei were isolated as described above, but with the addition of 3 ml of ST (Salts and Tris; Table S1) to extracted nuclei. Nuclei were then pelleted at 500g for 5 mins at 4°C. Supernatant was discarded and the nuclei pellet was resuspended in 100-500 μL of ST buffer (Salts and Tris; Table S1) before filtering through a 40 μm strainer-capped round bottom tube (Falcon).
For the ENS atlas, the CST (“RAISIN”) composition was: 0.49% (w/v) CHAPS (Cat#220201-1GM, EMD Millipore), 146 mM NaCl (Cat#S6546-1L, Sigma-Aldrich), 1mM CaCl2 (Cat#97062-820, VWR), 21 mM MgCl2 (Cat#M1028-10X1ML, Sigma-Aldrich), 10 mM Tris pH 8.0 (CAT#AM9855G, Thermo Fisher Scientific). The TST (“INNER Cell”) composition was: 146 mM NaCl (Cat#S6546-1L, Sigma Aldrich), 1 mM CaCl2 (Cat#97062-820, VWR), 21 mM MgCl2 (Cat#M1028-10X1ML, Sigma Aldrich), 10 mM Tris pH 8.0 (CAT#AM9855G, Thermo Fisher Scientific).
Fluorescence-activated cell sorting (FACS)
Prior to sorting, isolated nuclei and RAISINs were stained with Vybrant DyeCycle Ruby Stain (V-10309, Thermo Fisher Scientific). Sorting was performed on a MoFlo Astrios EQ Cell Sorter (Beckman Coulter) using 488nm (GFP, 513/26 filter) or 561nm (mCherry 614/20 filter), and 640nm (Vybrant DyeCycle Ruby, 671/30 filter) lasers. Single nuclei were sorted into the wells of a 96-well PCR plate containing 5 μl of TCL buffer (1031576, Qiagen) with 1% β-mercaptoethanol. The 96 well plate was sealed tightly with a Microseal F and centrifuged at 800g for 3 minutes before being frozen on dry ice. Frozen plates were stored at −80°C until whole-transcriptome amplification, library construction, sequencing, and processing.
Whole-transcriptome amplification, library construction, sequencing, and processing
Libraries from isolated single nuclei and RAISINs were generated using SMART-seq2 as described (Shekhar et al., 2016), with the following modifications. RNA from individual wells was first purified with Agencourt RNAClean XP beads (A63987, Beckman Coulter) prior to oligo-dT primed reverse transcription with Maxima reverse transcriptase (EP0753, Thermo Fisher Scientific) and locked TSO oligonucleotide, which was followed by 21 cycles of PCR amplification using KAPA HiFi HotStart ReadyMix (NC0295239, KAPA Biosystems). cDNA was purified twice using Agencourt AMPure XP beads (A63881, Beckman Coulter) as described (Shekhar et al., 2016). The Nextera XT Library Prep kit (FC-131-1096, Illumina, San Diego, CA) with custom barcode adapters (sequences available upon request) was used for library preparation. Libraries from 384 wells (nuclei/RAISINs) with unique barcodes were combined and sequenced using a NextSeq 500 sequencer (FC-404-2005, Illumina).
Droplet-based RAISIN RNA-Seq
Single RAISINs were processed through the GemCode Single Cell Platform using the GemCode Gel Bead kit (V2 chemistry), Chip and Library Kits (10X Genomics, Pleasanton, CA), following the manufacturer’s protocol. RAISINs were resuspended in ST buffer (Salt and Tris; table S1). An input of 7,000 RAISINs was added to each channel of a chip. The RAISINs were then partitioned into Gel Beads in Emulsion (GEMs) in the GemCode instrument, where lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5′ adaptor and sample index attachment. Libraries were sequenced on an Illumina NextSeq 500.
Using MIRACL-Seq to efficiently profile rare cell types from complex tissues
Droplet-based profiling of unsorted nuclei is a costly and inefficient, albeit effective, method for profiling the ENS. However, we reasoned that because enteric neurons are rare, comprising fewer than 1% of all cells in the intestine, we could potentially “overload” nuclei into droplets without generating homotypic neuron-neuron doublets. Using the Multiplexing Cost Calculator (https://satijalab.org/costpercell), we estimated the number of neurons, doublets, and other cells that would be generated for a range of overloading coefficients (Table S6), finding that we could potentially achieve 3-fold cost reductions with < 1 predicted neuron-neuron doublet per channel. We tested MIRACL-Seq for a range of overloading coefficients between 2x and 8x (see “Droplet-based RAISIN RNA-Seq”), finding that it produces high-quality data with no detectable homotypic doublets (based on comparison of MIRACL-Seq vs. plate-based data; Figure 4E). We developed methods to remove heterotypic doublets from the resulting data (see “Identifying and removing heterotypic doublets using MIRACL-Seq”).
Transmission electron microscopy (TEM)
Extracted nuclei and RAISINs were pelleted and fixed at 4°C overnight in 2.5% Glutaraldehyde and 2% Paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). The pellet was then washed in 0.1M cacodylate buffer, and post-fixed with 1% Osmiumtetroxide (OsO4) and 1.5% Potassiumferrocyanide (KFeCN6) for 1 hour. Next, the pellet was washed in water 3 times and incubated in 1% aqueous uranyl acetate for 1 hour followed by 2 washes in water and subsequent dehydration in grades of alcohol (10 minutes each; 50%, 70%, 90%, 100%, and 100%). The pellet was then put in propyleneoxide for 1 hour and infiltrated overnight in a 1:1 mixture of propyleneoxide and TAAB Epon (Marivac Canada Inc. St. Laurent, Canada). The following day the samples were embedded in TAAB Epon and polymerized at 60oC for 48 hours.
Ultrathin sections (about 60nm) were cut on a Reichert Ultracut-S microtome, picked up on to copper grids stained with lead citrate and examined in a JEOL 1200EX Transmission electron microscope and images were recorded with an AMT 2k CCD camera.
Processing FASTQ reads into gene expression matrices
For SMART-seq2, FASTQ files were demultiplexed and aligned to a reference transcriptome (see “Mouse and human reference transcriptomes”), and transcripts were quantified using RSEM, as previously described (Li and Dewey, 2011). For droplet-based scRNA-Seq, Cell Ranger v2.0 was used to demultiplex the FASTQ reads, align them to a reference transcriptome, and extract their “cell” and “UMI” barcodes. The output of each pipeline is a digital gene expression (DGE) matrix for each sample, which records the number of transcripts or UMIs for each gene that are associated with each cell barcode. DGE matrices were filtered to remove low quality nuclei, defined as nuclei with fewer than 250 detected genes. To facilitate downstream analyses for nuclei profiled with RAISIN RNA-Seq and MIRACL-Seq, a separate dataset of “highest-quality” cells was constructed, using a more stringent cutoff of 1,000 detected genes per nucleus. This cutoff was designed to enrich for enteric neurons, which typically have a large number of genes (mean = 3,508 and 4,302 detected genes per nucleus for mouse and human, respectively), but for other cell types is likely overly conservative. The total numbers of nuclei in each dataset is summarized below. To account for differences in sequencing depth across cells, DGE counts were normalized by the total number of transcripts or UMIs per nucleus and converted to transcripts-per-10,000 (henceforth “TP10K”).
| Dataset | Total nuclei | High-quality nuclei |
|---|---|---|
| Mouse colon (droplet) | 704,314 | 343,000 |
| Mouse ileum (droplet) | 483,221 | 79,293 |
| Human colon (droplet) | 436,202 | 146,442 |
| Total | 1,623,737 | 568,735 |
Mouse and human reference transcriptomes
For the optimization of nucleus extraction conditions, reads were aligned to the mm10 reference transcriptome. However, for the mouse and human ENS atlases, we augmented the reference transcriptomes with introns, thus allowing pre-mRNAs to be mapped along with mature mRNAs. Both the mm10 and hg19 reference transcriptomes were modified according to the instructions provided by the 10X Genomics website (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references). Briefly, we converted the standard GTF files into pre-mRNA GTF files by changing all “transcript” feature tags to “exon” feature tags. Using these modified GTF files, we then constructed Cell Ranger compatible references using the Cell Ranger “mkref” command. These modified GTF files were used for both the Cell Ranger pipeline and for our SMART-seq2 data (i.e. mouse ENS atlas).
QUANTIFICATION AND STATISTICAL ANALYSIS
Cell clustering overview
To cluster single cells into distinct cell subsets, we followed the general procedure we have previously outlined in (Haber et al., 2017) with additional modifications. This workflow includes the following steps: the selection of variable genes, batch correction, dimensionality reduction by PCA, and clustering. In all cases, clustering was performed twice: first, to separate neurons and glia from other cells, and then, to sub-cluster the neurons and glia to obtain high-resolution clusters within each group.
Partitioning cells into neuron, glia, and “other” compartments
Cells were partitioned into neuron, glia, and non-ENS compartments based on their expression of known marker genes (see “Gene signatures”). Signature scores were calculated as the mean log2(TP10K+1) across all genes in the signature. Each cluster was assigned to the compartment of its maximal score and all cluster assignments were inspected to ensure the accurate segregation of cells. Neurons and glia were then assembled into two separate DGE matrices for further analysis.
Variable gene selection
To identify variable genes within a sample, we first calculated the mean (μ) and the coefficient of variation (CV) of expression of each gene. Genes were then grouped into 20 equal-frequency bins (ventiles) according to their mean expression levels. LOESS regression was used to fit the relationship, log(CV) ~ log(μ), and the 1,500 genes with the highest residuals were evenly sampled across these expression bins. To extend this approach to multiple samples, we performed variable gene selection separately for each sample to prevent “batch” differences between samples from unduly impacting the variable gene set. A consensus list of 1,500 variable genes was then formed by selecting the genes with the greatest recovery rates across samples, with ties broken by random sampling. This consensus gene set was then pruned through the removal of all ribosomal, mitochondrial, immunoglobulin, and HLA genes, which were found to induce unwanted batch effects in some samples in downstream clustering steps.
Batch correction
We observed substantial variability between cells that had been obtained from different mice or different individuals, which likely reflects a combination of technical and biological differences. In some cases, these “batch effects” led to cells clustering first by mouse or individual, rather than by cell type or cell state. To control for these batch differences, we ran ComBat (Johnson et al., 2007) with default parameters on the log2(TP10K+1) expression matrix, allowing cells to be clustered by cell type or cell state. Importantly, these batch-corrected data were only used for the PCA and other steps relying on PCA (e.g. clustering, t-SNE visualization); all other analyses (e.g. differential expression analysis) were based on the original expression data. Note that we tested two additional methods for batch correction – one based on Canonical Correlation Analysis (Butler et al., 2018) and another on a k-nearest neighbors (k-NN) approach (Shekhar et al., 2016) – but did not obtain any enhancement in performance (data not shown).
Dimensionality reduction, graph clustering, and t-SNE visualization
Cells were clustered at two stages of the analysis: first, to initially partition the cells into neuron, glia, and “other” compartments, and second, to sub-cluster neurons and glia into different subsets. In all cases, we ran low-rank PCA on the variable genes of the batch-corrected log2(TP10K+1) expression matrix. We then applied Leiden clustering (Traag et al., 2019) or Phenograph (Levine et al., 2015) to the k-NN graph defined using the first n PCs and k nearest neighbors, which were separately estimated for each dataset. Leiden was used to cluster nuclei from droplet-based data (due to its speed), while Phenograph was used for clustering of all ENS cells. To estimate n, we calculated the number of “significant” PCs using a permutation test. Because this test may underestimate the number of PCs, we conservatively increased this number (i.e. to 15 or 30; see table below) to ensure that most of the variability in the dataset was captured. Next, to estimate k, we considered a range of clustering solutions with varying values of k, and calculated the marker genes for each set of clusters. We selected k based on inspection of the data. When clustering data from multiple cell types, we tried to select k such that the major cell types (e.g. neurons, glia, and muscle) were split, without fragmenting them into several sub-clusters. When clustering neurons and glia, we tried to select a k yielding the highest granularity clusters that were still biologically distinct, determined by close examination of the marker gene lists. Finally, the Barnes-Hut t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm was run on the selected PCs with perplexity = 20 and for 1,000 iterations to produce two-dimensional embeddings of the data for visualization.
| Dataset | Cell type | # Sig PCs | Used PCs | k-NN |
|---|---|---|---|---|
| Optimization | All cells | 13 | 15 | 250 (Phenograph) |
| Mouse colon (plate) | All cells | 16 | 30 | 250 (Phenograph) |
| Mouse colon (plate) | Neurons | 15 | 30 | 15 (Phenograph) |
| Mouse colon (plate) | Glia | 9 | 15 | 500 (Phenograph) |
| Mouse colon (droplet) | All cells | 19 | 30 | 10 (Leiden) |
| Mouse colon (droplet) | Neurons | 13 | 30 | 25 (Phenograph) |
| Mouse colon (droplet) | Glia | 6 | 15 | 750 (Phenograph) |
| Mouse ileum (droplet) | All cells | 20 | 30 | 10 (Leiden) |
| Mouse ileum (droplet) | Neurons | 10 | 30 | 10 (Phenograph) |
| Mouse ileum (droplet) | Glia | 9 | 15 | 250 (Phenograph) |
| Human colon (droplet) | All cells | 20 | 30 | 10 (Leiden) |
| Human colon (droplet) | Neurons | 7 | 30 | 25 (Phenograph) |
| Human colon (droplet) | Glia | 8 | 15 | 750 (Phenograph) |
Scoring nucleus extraction conditions
To identify optimal conditions for snRNA-seq of the ENS, we performed nucleus extractions while systematically varying the detergent (CHAPS, Digitonin, EZ, NP40, Tween), buffer (HEPES, Tricine, Tris), mechanical extraction conditions (Dounce, Grind, Chop), and additional modifiers (e.g. polyamines, RNAse inhibitors) (Table S1). In total, 104 different extraction conditions were examined. For each extraction, we profiled single nuclei transcriptomes by SMART-Seq2 and clustered the resulting RNA into neurons, glia, and “other” (i.e. non-ENS or low quality) clusters (see “Cell clustering overview”). To compare extractions, we calculated several quality metrics for each condition: (1) the proportion of recovered neurons, glia, and “other” cells, (2) the mean number of detected genes per cell, and (3) the mean ENS signature score (derived from markers of neurons and glia; see “Cell type signatures”). Conditions that yielded high-quality nuclei enriched in the ENS signature score were then identified.
Cell lineage dendrogram
As an auxiliary tool, the plate-based enteric neuron subsets from the mouse colon were organized on a dendrogram according to their transcriptional similarities (Figure S2D). To construct this dendrogram, we performed complete linkage clustering on the distance matrix corresponding to the mean transcriptional distances among all cell subsets, calculated using the variable genes from the log2(TP10K+1) expression matrix. These calculations were performed using the “hclust” and “dist” functions in R with default parameters.
Enteric neuron annotation and classification
We employed the following markers and considerations in annotating enteric neurons post hoc.
Broad segmentation of the mouse ENS
Neurons segmented into two major divisions comprising either cholinergic or nitrergic subsets. This broad division was correlated with several other genes. For example, expression of glial cell line-derived neurotrophic factor (GDNF) family receptors α1 (Gfra1) and α2 (Gfra2) segregate Nos1 and Chat expressing neurons, respectively. Gfra1 and Gfra2 are co-receptors for the GDNF receptor, Ret, which is necessary for ENS formation (Rossi et al., 1999; Schuchardt et al., 1996). Similarly, Chat and Nos1 expressing subsets also differentially expressed the transcription factors, Casz1 and Etv1.
Annotating mouse excitatory motor neurons
We annotated 5 subsets of putative excitatory motor neurons (PEMNs) based on co-expression of Chat and Tac1 (Brookes et al., 1991a) and their clustering within one subtree of the dendrogram (Figure S2D). PEMN subsets express the endogenous opioid, enkephalin (Penk), which is found in motor neurons (Brookes et al., 1991a), and/or the myenteric motor neuron marker, calretinin (Calb2) (Brookes et al., 1991b).
Annotating mouse inhibitory motor neurons
We annotated 7 subsets of putative inhibitory motor neurons (PIMNs) based on co-expression of Nos1 and Vip (Grider, 2003; Sang and Young, 1996), and their clustering on the dendrogram (Figure S2D). In total, 73% of Vip-positive neurons co-express Nos1, which is consistent with the previously reported estimate of 75% (Grider, 2003; Sang and Young, 1996).
Annotating mouse interneurons
Enteric interneurons (INs) relay sensory information and coordinate motor neuron activity. Six subtypes are known: (1) descending INs that signal via ACh, 5HT, and ATP, (2) descending Nos1+Vip+Grp+Chat- INs, (3) descending Vip+Chat+Nos1+ INs with ATP signaling, (4) descending Chat+Sst+ INs, (5) descending Penk+ INs (responsive to Sst), and (6) ascending Chat+Penk+ INs with ATP signaling (Furness, 2012; Grider, 1994, 2003; Young et al., 1995).
Some of these subsets (3, 5, 6) are at least partly matched as discrete clusters in our data, whereas others (1, 2, 4) are not clearly observed in our atlas. PIMN7 is a potential candidate for the descending Vip+Chat+Nos1+ INs with ATP signaling (3 above), based on co-expression of Vip, Chat, Nos1, and ATP transporters (e.g. SLC28a1, Slc28a2, Slc28a3, Slc29a1, Slc29a2, Slc29a3, Slc29a4) (Brookes et al., 1991a). PSN3 also expresses these genes, but their expression of Cck, Calca, and Calcb makes it unlikely they are interneurons. Three subsets of Chat+Penk+ putative INs (PIN1-3) may reflect either descending Penk+ INs (5 above; responsive to Sst), or ascending Chat+Penk+ INs with ATP signaling (6 above). Because all express combinations of Sst receptors, they may be descending INs. However, given the substantial number of additional receptors expressed by all PINs (5HT, VIP, GAL, GLP, prolactin, prostaglandin E2, EGF, BMP) or subsets of PINs (e.g., catecholamine synthetic enzymes), they may not be INs. Finally, there was little to no evidence for other IN subtypes: we did not detect any serotonergic (5HT) neurons (1 above) in our sampling, consistent with previous observations (Sang and Young, 1996); found no discernible cluster of Nos1+Vip+Grp+Chat- cells; and the only Chat+Sst+ neurons we observed were the Calcb+ PSN4 subset, which we interpret as a sensory neuron, not INs.
Annotating mouse secretomotor and vasodilator neurons
We annotated two subsets of Glp2r+ putative secretomotor/vasodilator (PSVNs) in one subtree of the dendrogram (Figure S2D), including Vip+ non-cholinergic and Chat+ cholinergic subsets (PSVN1 and PSVN2, respectively). PSVN2 also expresses Galanin (Gal), expressed in neurons that innervate the epithelium and arterioles (Furness et al., 1987), and neuropeptide Y (Npy), expressed in secretomotor neurons (Furness, 2012). Also, some neurons in PSVN2 express glutamate decarboxylase 2 (Gad2), possibly forming cholinergic/GABAergic neurons.
Annotating human interneuron subtype 2
Human PIN2s express two markers of mouse sensory neurons, Calcb and Grp, suggesting they may be mislabeled sensory neurons. Another possibility is that PIN2s represent multiple neuron subtypes that cannot be resolved with the number of neurons we profiled. Consistent with this possibility, Penk and Calcb are mutually exclusive within this subset (3 of 34 co-positive cells; expected = 7.24; Fisher test, p < 0.001).
Power analysis to determine the number of cells to profile
We performed a power analysis to determine our statistical power for detecting neuron subsets, based on the number of cells we profiled from each tissue, using the “How Many Cells” tool (https://satijalab.org/howmanycells) (Haber et al., 2017). For all such analyses, we assume that we want to detect a minimum of 10 neurons per cluster (which is sufficient to recover distinguishable clusters in this context). The total number of cells we would need to have a 95% probability of detecting the rarest cell type is provided below, as function of the total number of cell types (ranging from 20 to 30) and the minimum fraction of the rarest cell type within the enteric neurons (between 1% and 5% of ENs).
| Number of enteric neuron subsets | Minimum fraction (rarest cell type within enteric neurons) | Total number of enteric neurons required | Relevant datasets |
|---|---|---|---|
| 30 | 1% | > 2,123 | Mouse colon (plate-based) |
|
30 |
1.5% |
> 1,447 |
Mouse colon
(droplet-based) Human colon (droplet-based) |
| 30 | 3% | > 721 | |
| 25 | 4% | > 532 | |
| 20 | 5% | > 417 | Mouse ileum (droplet-based) |
Based on this analysis, we estimate that the plate-based mouse colon dataset (n = 2,657 neurons) can detect 30 neuron clusters with a minimum frequency of < 1% for the rarest subset; the droplet-based mouse colon dataset (n = 1,938 neurons) can detect 30 neuron clusters with a minimum frequency of 1.5%; the droplet-based ileum dataset (n = 472 neurons) can detect 20 neuron clusters with a minimum frequency of < 5%; and the droplet-based human dataset (n = 1,445 neurons) can detect 30 neuron clusters with a minimum frequency of 1.5%.
Differential expression analysis
Differential expression (DE) tests were performed using MAST (Finak et al., 2015), which fits a hurdle model to the expression of each gene, consisting of logistic regression for the zero process (i.e. whether the gene is expressed) and linear regression for the continuous process (i.e. the expression level). For the mouse atlas, this regression model included terms to capture the effects of the cell subset, age, sex, colon location, circadian phase, mouse model, and cell complexity. For the human atlas, this regression model only included terms for cell subset and cell complexity.
For the mouse colon atlas, we fit the regression formula, Yi ~ X + A + C + L + S + T + N, where Yi is the standardized log2(TP10K+1) expression vector for gene i across cells, X is a variable reflecting cell subset membership (e.g. PSNs vs. non-PSNs), A is the age associated with each cell (adult vs. aged), C is the circadian phase for each cell (morning vs. evening), L is the location for each cell (proximal vs. distal), S is the sex for each cell (male vs. female), T is the mouse model for each cell (Sox10 vs. Uchl1), and N is the scaled number of genes for each cell (i.e. cell complexity). For the mouse ileum, mouse colon, and human colon droplet-based data, we used the regression formula, Yi ~ X + N, with X and N defined as above. To identify genes that are either “conserved” or “species-specific” in human and mouse, we used the regression formula, Yi ~ X + D, with X defined above and where D is the species associated with each cell (see “Identifying core and species-specific transcriptional programs for major neuron classes”).
Additionally, two heuristics were used to increase the speed of the tests: we required all tested genes to have a minimum fold change of 1.2 and to be expressed by at least 1% of the cells within the group of interest. In all cases, the discrete and continuous coefficients of the model were retrieved and p-values were calculated using the likelihood ratio test in MAST. Q-values were separately estimated for each cell subset comparison using the Benjamini-Hochberg correction. Unless otherwise indicated, all reported DE coefficients and q-values correspond to the discrete component of the model (i.e. the logistic regression).
Identifying statistically significant differences in cell proportions
In identifying changes in neuron proportions between intestinal locations (Figures 4I and 5F), a major concern is that they are not independent of each other. Because all proportions sum to 1, an increase in the proportion of one subset will necessarily lead to a decrease in the proportions of other subsets. To account for these dependencies, we used a Dirichlet-multinomial regression, which tests for differences in cell composition between conditions (e.g., proximal vs. distal), while accounting for the proportions of all of the other cell subsets. This regression model and its associated p-values were calculated with the “DirichReg” function in the DirichletReg package.
Acquisition and scoring of gene signatures
The ENS signature score reflects the mean expression levels of marker genes for neurons and glia. We used this score to assess neuron and glia nucleus quality across nucleus preparations, in cases where the proportions of neurons and glia may not be sufficient. For example, if a nucleus preparation yields a high proportion of low-quality neurons, it will score high for total neuron yield, but will have a low ENS signature score, because the neurons themselves are low quality. The ENS signature score was therefore used as a secondary quality control to ensure that we selected the best preparations. To construct the ENS signature score, we compiled the following lists of marker genes for enteric neurons and glia from the literature (Lasrado et al., 2017). These gene signatures were then combined to construct an overall “ENS” signature score (Figure S1F).
Neurons:
Tubb3, Elavl4, Ret, Phox2b, Chrnb4, Eml5, Smpd3, Tagln3, Snap25, Gpr22, Gdap1l1, Stmn3, Chrna3, Scg3, Syt4, Ncan, Crmp1, Adcyap1r1, Elavl3, Dlg2, Cacna2d.
Glia:
Erbb3, Sox10, Fabp7, Plp1, Gas7, Nid1, Qk, Sparc, Mest, Nfia, Wwtr1, Gpm6b, Rasa3, Flrt1, Itpripl1, Itga4, Lama4, Postn, Ptprz1, Pdpn, Col18a1, Nrcam.
To prevent highly expressed genes from dominating a gene signature score, we scaled each gene vector of the log2(TP10K+1) expression matrix by its root mean squared expression across cells (using the ‘scale’ function in R with center = FALSE). The signature score for each cell was then computed as the mean scaled expression across all genes in the signature.
Estimation of false discovery rate
Unless otherwise specified, false discovery rates were estimated with the Benjamini-Hochberg correction (Benjamini and Hochberg, 1995), using the “p.adjust” R function with the “fdr” method.
Identifying and removing heterotypic (contaminated) droplets from MIRACL-seq data
To identify “contaminated droplets” (due to co-encapsulation or ambient RNA) in droplet-based MIRACL-seq data, we first scored each cell for several distinct cell type signatures (e.g., neuron, glia, muscle, fibroblast, and epithelial signatures; Figure S5), using the mean log2(TP10K+1) expression across all genes in the signature. We hypothesized that cells expressing the signatures for multiple cell types represent contaminated droplets. For example, we identified clusters of human neurons and glia that were strongly enriched also for muscle, epithelial, fibroblast, or glial signatures (Figure S5A,C); these are putative doublets. We applied published methods, including Scrublet (Wolock et al., 2019), Doublet Finder (McGinnis et al., 2019; Wolock et al., 2019), and Solo (Bernstein et al., 2019) to these data (Figure S5B,D). However, while these methods were accurate for some cell types, none of them worked for enteric neurons and glia. In particular, Scrublet failed to identify many neuron-muscle and neuron-epithelial doublets, while Doublet Finder and Solo classified nearly all neurons as putative doublets (Figure S5A,B). We therefore sought to develop a method for contaminated droplet that would work for enteric neurons and glia, using cell type signature scores.
This method first uses a DE test to construct a gene signature for each annotated cell type (see “Differential expression analysis”), and scores each single cell transcriptome for these signatures (see “Acquisition and scoring of gene signatures”). We note that these gene signature scores are approximately normally distributed. The probability that cell type i expresses a gene signature j at level xij is thus: , where μij and σij are the mean and standard deviation of signature j in cell type i. We assume that for a contaminated droplet d of cell types i and j, the probability of expressing a signature j at a level xdj is: . Therefore, for a given cell type i, we can construct a likelihood ratio test to evaluate the competing hypotheses that a gene signature score was generated by a contaminated droplet vs. a pure singlet, incorporating a prior that reflects the probability that the cell is a contaminated droplet, . Here, we used an odds ratio cutoff of 8, which conservatively separates contaminated droplet vs. a pure across all examined cell types (e.g., neurons and glia, Figure S5B,D). However, because these contaminated droplets were included in our initial estimation of cell type signatures, we further improve our results by iteratively re-running this inference procedure, each time removing predicted contaminated droplets, re-estimating cell type signatures, and re-running the contaminated droplet test. For the mouse and human datasets, we ran this procedure with an odds ratio cutoff of 8 for 16 iterations, until our predictions converged onto a final set of contaminated droplets and the total number of contaminated droplets increased by less than 5%.
Matching human and mouse subsets
To map neurons onto their plate-based or droplet-based counterparts (Figures 4E,H, and 5E), we trained a Random Forest classifier to distinguish each subset of reference neurons using the log2(TP10K+1) expression matrix of mouse variable genes that had human orthologs (see “Variable gene selection”). The Random Forest model was built with the R “randomForest” package using default parameters with the following exception: to account for class imbalances, we down-sampled each neuron class to the median class size, while constructing each tree (implemented using the “sampsize” argument). In total, the “out of bag” estimate of the error rate (which estimates test rather than training error) was 8.8%, indicating that we can accurately distinguish among major neuron classes. Next, to extend this model to droplet-based neurons, we used this model to predict the class for each droplet-based neuron using expression data for the variable genes (note: for human neurons, we used the expression of their 1:1 mouse orthologs). All class assignments were then manually examined to ensure accurate predictions for all cells. Note that we also tested an approach using a variational autoencoder (VAE) (Lopez et al., 2018), but did not observe a noticeable improvement in performance (data not shown).
Identifying core and species-specific transcriptional programs for major neuron classes
To identify conserved and species-specific transcriptional signatures for each neuron class (i.e., PEMN, PIMN, PIN, PSN, PSVN), we mapped all mouse genes to their human orthologs (using only 1:1 orthologs), and combined both expression matrices according to these genes. We next calculated DE orthologs that were either shared by both species (e.g., PEMNs vs. non-PEMNs) or unique to each species (e.g., human PEMNs vs. mouse PEMNs), within each neuron class (see “Differential expression analysis”) (Table S4). We required that “conserved” marker genes be present in > 20% of expressing cells and have a log2 fold-change > 1 for both human and mouse. Similarly, we required that “specific” marker genes be present in > 20% of expressing cells.
Using receptor-ligand pairs to infer cell-cell interactions
To identify cell-cell interactions, we mapped the FANTOM5 database of literature-supported receptor-ligand interactions (Ramilowski et al., 2015) onto our lists of cell subset markers. Following a recent approach (CellPhoneDB (Vento-Tormo et al., 2018)), we filtered this database to remove all integrins (defined using the HUGO “Integrin” gene group), which were involved in many non-specific cell-cell interactions. We further required cell subset markers to be expressed in at least 10% of all cells within the subset, with a minimum expression level of 0.5 log2(TP10K+1). To identify highly specific interactions, we required that the expression level of both the ligand and the receptor in their respective cell subsets to be at least 2-fold higher than their expression levels in all other cells. For network visualization (Figure 6B-G), we removed edges in which a single ligand participates in interactions with over 4 distinct receptors; however, all receptor-ligand interactions were included in Table S5.
Defining disease risk genes
We compiled lists of genes that have been implicated by human genetics studies as contributing to risk for the following diseases: Hirschsprung’s disease (HRSC), inflammatory bowel disease (IBD), autism spectrum disorders (ASDs), and Parkinson’s disease (PD). Because human genetics studies do not always pinpoint a causative risk gene, we used the literature to identify sets of genes that are particularly likely to contribute to disease risk, including: 9 HRSC-associated genes (Bondurand and Southard-Smith, 2016), 106 IBD-associated genes (Smillie et al., 2018), 28 ASD-associated genes (Sanders et al., 2015), and 29 PD-associated genes (Chang et al., 2017). To compare the expression levels of risk genes in the colon vs. brain, we measured the log2-fold-change of the mean expression level of each gene in neurons from the human colon vs. human cortex (Hodge et al., 2019) (Figure 7C).
Supplementary Material
(A) Single nucleus RNA-seq (snRNA-seq) of GFP+ nuclei from Sox10-Cre; INTACT mice with previous protocols. Fraction (y axis) of identified cell types (x axis) in samples of the brain (grey) and colon (black) using two previously published snRNA-seq methods (Habib et al., 2016, 2017). (B-D) Quality metrics across all tested nucleus extractions. Cell type composition, number of detected genes per nucleus, and ENS signature score (columns, x axes) across nucleus extraction parameters (y axes), including detergent concentration (B), sample handling method (C), and buffer (D). (E) Decision tree for selection of optimal nucleus extraction conditions. (F) Quality metrics for each nucleus extraction. Mean number of genes per nucleus (x axis) and mean ENS signature score (y axis) for the nuclei from each extraction method (dots). (n = 5,236 GFP+ sorted nuclei across all protocols). Error bars: SEM. Select nucleus extractions are marked in color (legend).
Fraction of expressing cells (dot size) and mean levels in expressing (non-zero) cells (dot color) of genes (columns) in neurons vs. other cells (top panel) or individual neuron subsets (bottom panel) (rows) for (A) selected neurotransmitters and neuropeptides, (B) genes functionally important in the ENS, (C) data-driven marker genes that are uniquely enriched in each neuron subset, and (D) cytokines, chemokines, and their receptors. Outline: significant DE coefficient (discrete coefficient of MAST hurdle model; STAR Methods).
(A,B) Reproducible cell subset distributions across transgenic mouse models and individual mice. t-SNE of RAISIN RNA-Seq transcriptomes of 2,567 neurons (A) and 3,039 glia (B) colored by either subset, age, mouse model, sex, mouse, the number of detected genes per nucleus, or circadian time point. (C) Estimated proportion of enteric neurons in the mouse colon. Percent of all sorted cells in the colon that are neurons (y axis) as estimated by FACS (transgene-expressing nuclei vs. unlabeled nuclei) and post-hoc adjustment based on RAISIN RNA-Seq data. (D) Hierarchical relationships among neuron subset profiles. Dendrogram of hierarchical clustering by transcriptional similarities (STAR Methods) of neuron subsets (leaves). (E) Glia subset markers. Effect size within each glia subset (i.e. discrete DE coefficient, x axis) and associated significance (y axis) for each gene (dots) in each subset. Color: gene specificity (fold-change relative to the next highest expressing glial subset; color bar). Dashed line: p < 0.05.
(A-D) Comparison of doublet detection methods for neurons and glia from the human colon. (A,B) Representative t-SNEs of 2,258 human colon neuron profiles colored by (A) signature scores for muscle, epithelial cells, fibroblasts, and glia (putative doublets have high scores for non-neuron cell types), or (B) doublet predictions by our new method (left) vs. Scrublet, Doublet Finder, and Solo (right). (C,D) Representative t-SNEs of 18,721 human colon glia colored by (C) signature scores for muscle, epithelial cells, fibroblasts, and glia or (D) doublet predictions by our new method (left) vs. Scrublet, Doublet Finder, and Solo (right). (E,F) MIRACL-Seq of mouse esophagus, heart, and lung. t-SNE of MIRACL-Seq profiles (dots) from mouse esophagus (11,550 nuclei; 19 subsets), heart (8,343 nuclei; 15 subsets), and lung (8,441 nuclei; 18 subsets), colored by tissue and sample of origin (E) or cell subset (F), annotated post-hoc using data-derived marker genes (STAR Methods).
t-SNEs of MIRACL-Seq RAISIN RNA-seq profiles (dots) of neurons and glia from the mouse ileum (A,B), mouse colon (C,D), and human colon (E,F), colored by cell subset (left), mouse or human identifier (middle), and the number of detected genes per nucleus (right).
Representative in situ validations of DAPI staining (blue) with combined smFISH and immunostaining (green, red, and white), showing the co-expression of selected genes in mouse (A-K) and human (L-P) colon neurons. Individual channels for each probe in greyscale (left) with merged channels in color (right). Representative images selected from a total of 3 experiments per validation (STAR Methods). Scale bar: 50 μm.
(A) Schematic of coronal brain section. Raphe nuclei containing serotonergic (Tph2+) neurons served as a positive control. The pontine reticular nucleus lacking Tph2-expressing neurons served as a negative control. (B, C) Representative images of smFISH for Tph2 in the mouse brain (B) and colon (C) of Sox10-Cre; INTACT (GFP) mice (n = 2 animals; 12 colon sections). (D, E) Representative images of smFISH for Tph2 in the mouse brain (D) and colon (E) of wild-type C57BL/6J mice. (n = 2 animals; 12 colon sections). (F) Analysis of bulk RNA-seq data from several tissues of C57BL/6 mice (Söllner et al., 2017). RNA expression of Tph1 and Tph2 from the brain, colon and small intestine. RNA expression independently analyzed in three mice per tissue is indicated 1–3. Scale bar: 100 μm.
Description and statistics for nuclei extractions, aggregated either by sample or condition. Includes descriptions of the buffers, detergents, detergent concentrations, salts, and modifiers profiled, and provides various statistics, including exon:intron ratios, the number of genes per nucleus, and ENS compositions.
Description of each mouse and human sample profiled in this study, including model, age, sex, circadian time point, intestinal location, quality metrics (number of genes and reads/UMIs per nucleus), and cell type frequencies.
Differential expression statistics summarizing the expression of ligands and receptors, and lists of receptor-ligand pairs, in droplet-based cell subsets from the mouse and human colon.
Differential expression statistics summarizing the marker genes for each cell subset and the effects of location, age, mouse model, sex, and circadian phase on the gene expression programs of mouse colon neurons and glia.
Statistics showing the expected numbers of nuclei, neurons, and multiplets for varying levels of overloading, and estimated cost reductions.
Differential expression statistics summarizing the marker genes for each cell subset, the conserved transcriptional programs shared by human and mouse neurons, and differences between the mouse ileum vs. colon ENS and the human vs. mouse ENS.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| Rabbit anti-Tubb3 | Abcam | Cat# AB18207 RRID:AB_444319 |
|
| Chicken anti-mCherry | EMD Millipore | Cat#AB356481 | |
| Goat anti-Ass1 | Abcam | Cat# ab77590 RRID:AB_2060467 | |
| Rabbit anti-Celsr3 | Sigma-Aldrich | Cat# SAB4500707–100UG RRID:AB_10744051 | |
| Rabbit anti-Uchl1 | Cell Signaling | Cat#13179T RRID:AB_2798141 | |
| Rabbit anti-ASL | Abcam | Cat# ab97370 RRID:AB_10680261 | |
| Rabbit anti-GRP | Abcam | Cat# ab22623 RRID:AB_447204 | |
| Rabbit anti-Prph | Abcam | Cat# ab4666 RRID:AB_449340 | |
| Goat anti-Phox2b | Novus Biologicals | Cat#AF4940 | |
| Mm-Calca | ACDBio | Cat#417961 | |
| Mm-Calcb | ACDBio | Cat#425511 | |
| Mm-Cck | ACDBio | Cat#402271 | |
| Mm-Chat-C2 | ACDBio | Cat#408731-C2 | |
| Mm-GRP | ACDBio | Cat#317861-C2 | |
| Mm-Htr4 | ACDBio | Cat#408241 | |
| Mm-Lgr5 | ACDBio | Cat#312171 | |
| Mm-Nmu | ACDBio | Cat#446831 | |
| Mm-Nog | ACDBio | Cat#467391 | |
| Mm-Nos1-C3 | ACDBio | Cat#437651-C3 | |
| Mm-Piezo1 | ACDBio | Cat#500511 | |
| Mm-Piezo2-C3 | ACDBio | Cat#400191-C3 | |
| Mm-Sst-C3 | ACDBio | Cat#404631-C3 | |
| Mm-Tph2-C2 | ACDBio | Cat#318691-C2 | |
| Mm-Vip/Mm-Vip-C2 | ACDBio | Cat#415961/415961-C2 | |
| Hs-ANO1-C2 | ACDBio | Cat#349021-C2 | |
| Hs-CHAT/Hs-CHAT-C2 | ACDBio | Cat#450671/450671-C2 | |
| Hs-DSCAM | ACDBio | Cat# | |
| Hs- GUCY1A3 | ACDBio | Cat#425831 | |
| Hs-IL7 | ACDBio | Cat#424251 | |
| Hs-IL12A | ACDBio | Cat#402061 | |
| Hs-KIT-C3 | ACDBio | Cat#606401-C3 | |
| Hs-NOS1-C2 | ACDBio | Cat#506551-C2 | |
| Hs-RET | ACDBio | Cat#424871 | |
| Hs-GRP | ACDBio | Cat#465261 | |
| Hs-SCN3A | ACDBio | Cat#460121 | |
| Biological Samples | |||
| Human cancer-proximal colon tissue from resections | Dana Farber Cancer Institute, Boston (IRB
03–189; ORSP 3490); Massachusetts General Hospital, Boston (IRB 02–240; ORSP 1702) |
Table S1 | |
| Chemicals, Peptides, and Recombinant Proteins | |||
| CHAPS, Molecular Biology Grade | EMD Millipore | Cat#220201–1GM | |
| Nonidet™ P40 Substitute, Ultrapure, Affymetrix/USB | Fisher Scientific | Cat#AAJ19628AP | |
| Tween®-20 Solution, 10% | Teknova | VWR-100216–360 | |
| Triton X-100 | Sigma-Aldrich | Cat#T8787–100ML | |
| Digitonin | Promega | Cat#G9441 | |
| Hexylene Glycol | Sigma-Aldrich | Cat#112100–500GT | |
| Tricine Buffer, Ultra Pure Grade | VWR | Cat#E170–100G | |
| HEPES, 1M Solution, pH 7.3, Molecular Biology Grade, Ultrapure | Fisher Scientific | Cat# AAJ16924AE | |
| Tris (1 M), pH 8.0, Rnase-Free | Thermo Fisher Scientific | Cat#AM9855G | |
| Sodium Chloride Solution | Sigma-Aldrich | Cat#S6546–1L | |
| CACL2 1M ST BIOTECH GRD | VWR | Cat#97062–820 | |
| Magnesium Chloride Solution | Sigma-Aldrich | Cat#M1028–10X1ML | |
| Potassium Chloride Solution | Sigma-Aldrich | Cat#60142–100ML-F | |
| Spermidine (1g) | Sigma-Aldrich | Cat#S2626–1G | |
| Spermine (1g) | Sigma-Aldrich | Cat#S3256–1G | |
| BSA, Molecular Biology Grade | New England Biolabs | Cat#B9000S | |
| Sucrose, RNase & DNase Free, Ultra Pure Grade | VWR | Cat#97061–432 | |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#11836170001 | |
| Cyclohexamide | Cell Signaling Technology | Cat#2112S | |
| Cytochalasin D | Sigma-Aldrich | Cat#C2618–200UL | |
| Recombinant RNase Inhibitor | Takara | Cat#2313B | |
| Magnesium Acetate Solution | Sigma-Aldrich | 63052–100ML | |
| B-Mercaptoethanol | Sigma-Aldrich | Cat#M3148–100ML | |
| Buffer TCL (125 mL) | Qiagen | Cat#1031576 | |
| Deoxynucleotide (dNTP) Solution Mix | New England Biolabs | Cat#N0447L | |
| Maxima H Minus Reverse Transcriptase (200 U/uL) | Thermo Fisher Scientific | Cat#EP0753 | |
| KAPA HiFi HotStart ReadyMix PCR Kit | Roche | Cat#KK2602 | |
| tousimis® FORMALDEHYDE 20% | tousimis | Cat#1008A | |
| TSA Plus Cyanine 5 and Flourescein System | PerkinElmer | Cat#NEL754001KT | |
| TSA Plus Cyanine 3 Evaluation Kit | PerkinElmer | Cat#NEL744E001KT | |
| Pierce™ 20X TBS Tween™ 20 Buffer | Thermo Fisher Scientific | Cat#28360 | |
| DAPI | Sigma-Aldrich | Cat#D9542–10MG | |
| Tissue-Tek O.C.T. compound 4583 | VWR | Cat#25608–930 | |
| Vybrant DyeCycle Ruby Stain | Thermo Fisher Scientific | Cat#V-10309 | |
| CAS-Block™ Histochemical Reagent | Thermo Fisher Scientific | Cat#008120 | |
| RNAscope® Probe Diluent | ACD | Cat#300041 | |
| Critical Commercial Assays | |||
| Agencourt RNAClean (RNA SPRI beads) | Beckman Coulter | Cat#A63987 | |
| Nuclei EZ Prep | Sigma-Aldrich | Cat#NUC101–1KT | |
| AMPureXP SPRI (DNA SPRI beads) | Beckman Coulter | Cat#A63881 | |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131–1096 | |
| NextSeq 500 high output kit V2, 75 cycles | Illumina | Cat#FC-404–2005 | |
| RNAscope® Fluorescent Multiplex Reagent Kit | ACD | Cat#320850 | |
| 10X Chromium Single Cell 3’ Kit | 10X Genomics | Cat#120237 | |
| Deposited Data | |||
| Genome Reference Consortium Mouse Build GRCm38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/ | |
| Genome References Consortium Human Build GRCh37 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ | |
| TcoF-DB v2 human transcription factors | Schmeier et al., 2016 | http://compbio.massey.ac.nz/apps/tcof/home/ | |
| PRRDB human pattern recognition receptors | Lata et al., 2008 | http://crdd.osdd.net/raghava/prrdb/ | |
| FANTOM5 receptor-ligand database | Ramilowski et al., 2015 | http://fantom.gsc.riken.jp/5 | |
| Mouse SS2 – FASTQ files and processed data | This paper | Single cell portal accession SCP1038 (https://portals.broadinstitute.org/single_cell) | |
| Mouse 10X – FASTQ files and processed data | This paper | Single cell portal accession SCP1038 (https://portals.broadinstitute.org/single_cell) | |
| Human 10X - FASTQ files | This paper | Broad DUOS project “ColonENS-RegevXavier-CRCAdjacentNormal-Broad-snRNASeq-UnsortedCells” (https://duos.broadinstitute.org) | |
| Human 10X – processed data | This paper | Single cell portal accession SCP1038 (https://portals.broadinstitute.org/single_cell) | |
| Experimental Models: Organisms/Strains | |||
| C57BL/6J | The Jackson Laboratory | Cat#000664 RRID:IMSR_JAX:000664 | |
| B6;CBA-Tg(Sox10-cre)1Wdr/J | The Jackson Laboratory | Cat#025807 RRID:IMSR_JAX:025807 | |
| 129S4.Cg-E2f1Tg(Wnt1-cre)2Sor/J | The Jackson Laboratory | Cat#022137 RRID:IMSR_JAX:022137 | |
| B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J | The Jackson Laboratory | Cat#021039 RRID:IMSR_JAX:021039 | |
| Tg(Uchl1-HIST2H2BE/mCherry/EGFP*)FSout/J | The Jackson Laboratory | Cat#016981 RRID:IMSR_JAX:016981 | |
| Oligonucleotides | |||
| Reverse Transcription DNA oligonucleotide primer (RNase-free, 100 mM) 5’ -AAGCAGTGGTATCAACGCAGAGTACT(30)VN-3’ | IDT | N/A | |
| SMARTER TSO (with LNA) 5’ -AAGCAGTGGTATCAACGCAGAGTACrGrG+G-3’ | Exiqon | N/A | |
| PCR oligonucleotide primer 5’ -AAGCAGTGGTATCAACGCAGAGT-3’ | IDT | N/A | |
| Recombinant DNA | |||
| Software and Algorithms | |||
| CellRanger v2.0 | 10X Genomics | https://github.com/10XGenomics/cellranger | |
| sva (R package) | Leek et al., 2014 | https://www.bioconductor.org/packages/devel/bioc/html/sva.html | |
| Infomap clustering algorithm | Rosvall et al., 2008 | http://igraph.org/ | |
| Barnes-Hut t-SNE algorithm | van der Maaten et al., 2014 | https://cran.r-project.org/web/packages/Rtsne/ | |
| Scanpy | Wolf et al., 2018 | https://github.com/theislab/scanpy | |
| MAST | Finak et al., 2015 | https://github.com/RGLab/MAST | |
| Rtsne | CRAN | https://cran.r-project.org/web/packages/Rtsne/ | |
| PhenoGraph | Levine et al., 2015 | https://github.com/jacoblevine/PhenoGraph | |
| MAGIC | van Dijk et al., 2018 | https://github.com/KrishnaswamyLab/MAGIC | |
| nlme (R package) | CRAN | https://cran.r-project.org/web/packages/nlme/index.html | |
| rsvd (R package) | CRAN | https://cran.r-project.org/web/packages/rsvd/index.html | |
| Seurat (R toolkit) | Butler et al., 2018 | https://satijalab.org/seurat | |
| Analysis code | This paper | https://github.com/klarman-cell-observatory/ens_atlas | |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
| Antibodies | |||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 | |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 | |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A | |
| Bacterial and Virus Strains | |||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
|
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A | |
| Cowpox virus Brighton Red | BEI Resources | NR-88 | |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A | |
| Staphylococcus aureus | ATCC | ATCC 29213 | |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 | |
| Biological Samples | |||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 | |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ | |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ | |
| Chemicals, Peptides, and Recombinant Proteins | |||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350–13-2 | |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433–59-5 (free base) | |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124–87-8 | |
| Human TGF-β | R&D | 240-B; GenPept: P01137 | |
| Activated S6K1 | Millipore | Cat#14–486 | |
| GST-BMAL1 | Novus | Cat#H00000406-P01 | |
| Critical Commercial Assays | |||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC | |
| CaspaseGlo 3/7 | Promega | G8090 | |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202–1012 | |
| Deposited Data | |||
| Raw and analyzed data | This paper | GEO: GSE63473 | |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 | |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ | |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 | |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 | |
| Experimental Models: Cell Lines | |||
| Hamster: CHO cells | ATCC | CRL-11268 | |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 | |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A | |
| Human: HUES 8 hESC line (NIH approval
number NIHhESC-09–0021) |
HSCI iPS Core | hES Cell Line: HUES-8 | |
| Experimental Models: Organisms/Strains | |||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 | |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 | |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 | |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 | |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 | |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207–1 | |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A | |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 | |
| Oligonucleotides | |||
| siRNA targeting sequence: PIP5K I alpha #1:
ACACAGUACUCAGUUGAUA |
This paper | N/A | |
| Primers for XX, see Table SX | This paper | N/A | |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A | |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 | |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 | |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A | |
| Recombinant DNA | |||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 | |
| Plasmid: GFP-Nito | This paper | N/A | |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 | |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A | |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 | |
| Software and Algorithms | |||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ | |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ | |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA | |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 | |
| Other | |||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu | |
| Resource website for the AML31
publication |
This paper | https://github.com/chrisamiller/aml31SuppSite | |
ACKNOWLEDGMENTS
We thank Karin Pelka, Jonathan Chen, and Nir Hacohen for discussions and donor tissue transport and banking; Asaf Rotem and Isaac Wakiro for help with donor tissue transport; Louise Trakimas and Margaret Coughlin (HMS Electron Microscopy Facility) for help with TEM; Patricia Rogers, Chelsea Ortis, and Stephanie Saldi (Broad Flow Cytometry Core) for help with FACS; Leslie Gaffney and Anna Hupalowska for help with figures and illustrations; Fei Chen for microscope access; Leah Caplan for help with microscopy; and Vijay Kuchroo and Ana Anderson for discussions. Work was supported by the Klarman Cell Observatory, HHMI, Manton Foundation, Broadnext10, DK114784, DK043351, DK114784, DK117263, Helmsley Charitable Trust, Crohn’s and Colitis Foundation, and Food Allergy Science Initiative.
Footnotes
DECLARATION OF INTERESTS
A.R. is a co-founder and equity holder of Celsius Therapeutics, equity holder of Immunitas, and until August, 2020, SAB member of ThermoFisher Scientific, Syros Pharmaceuticals, Neogene Therapeutics, and Asimov. A.R. is an employee of Genentech Inc. R.J.X. is a co-founder and equity holder of Celsius Therapeutics and Jnana Therapeutics and consultant for Novartis. E.D. is an employee of Bristol-Myers Squibb. E.D., C.S., G.E., O.R.R., R.X., and A.R. are co-inventors on PCT/US2019/055894, which includes methods of this manuscript.
REFERENCES
- Assas BM, Pennock JI, and Miyan JA (2014). Calcitonin gene-related peptide is a key neurotransmitter in the neuro-immune axis. Front. Neurosci. 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300. [Google Scholar]
- Bernard CE, Gibbons SJ, Gomez-Pinilla PJ, Lurken MS, Schmalz PF, Roeder JL, Linden D, Cima RR, Dozois EJ, Larson DW, et al. (2009). Effect of age on the enteric nervous system of the human colon. Neurogastroenterol. Motil. 21, 746–e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein N, Fong N, Lam I, Roy M, Hendrickson DG, and Kelley DR (2019). Solo: doublet identification via semi-supervised deep learning. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, and Sauka-Spengler T (2010). Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. U. S. A. 107, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesmans W, Lasrado R, Vanden Berghe P, and Pachnis V (2015). Heterogeneity and phenotypic plasticity of glial cells in the mammalian enteric nervous system. Glia 63, 229–241. [DOI] [PubMed] [Google Scholar]
- Bondurand N, and Southard-Smith EM (2016). Mouse models of Hirschsprung disease and other developmental disorders of the enteric nervous system: Old and new players. Dev. Biol. 417, 139–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes SJ, Steele PA, and Costa M (1991a). Identification and immunohistochemistry of cholinergic and non-cholinergic circular muscle motor neurons in the guinea-pig small intestine. Neuroscience 42, 863–878. [DOI] [PubMed] [Google Scholar]
- Brookes SJ, Steele PA, and Costa M (1991b). Calretinin immunoreactivity in cholinergic motor neurones, interneurones and vasomotor neurones in the guinea-pig small intestine. Cell Tissue Res. 263, 471–481. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaidez V, Hansen RL, and Hertz-Picciotto I (2014). Gastrointestinal problems in children with autism, developmental delays or typical development. J. Autism Dev. Disord. 44, 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, International Parkinson’s Disease Genomics Consortium, 23andMe Research Team, Kerchner GA, Ayalon G, et al. (2017). A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 49, 1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary B, Cong L, Cheung A, Lander ES, and Regev A (2017). Efficient Generation of Transcriptomic Profiles by Random Composite Measurements. Cell 171, 1424–1436.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbache J, Parfejevs V, and Sommer L (2018). Cre-driver lines used for genetic fate mapping of neural crest cells in the mouse: An overview. Genesis 56, e23105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deczkowska A, and Schwartz M (2018). Targeting neuro-immune communication in neurodegeneration: Challenges and opportunities. J. Exp. Med. 215, 2702–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giorgio R, Bianco F, Latorre R, Caio G, Clavenzani P, and Bonora E (2016). Enteric neuropathies: Yesterday, Today and Tomorrow. Adv. Exp. Med. Biol. 891, 123–133. [DOI] [PubMed] [Google Scholar]
- Drokhlyansky E, Göz Aytürk D, Soh TK, Chrenek R, O’Loughlin E, Madore C, Butovsky O, and Cepko CL (2017). The brain parenchyma has a type I interferon response that can limit virus spread. Proc. Natl. Acad. Sci. U. S. A. 114, E95–E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak AM, and Silen W (1985). Differentiation between Crohn’s disease and other inflammatory conditions by electron microscopy. Ann. Surg. 201, 53–63. [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Bazalakova M, Savchenko V, Tapia JC, Wright J, and Blakely RD (2004). Lethal impairment of cholinergic neurotransmission in hemicholinium-3-sensitive choline transporter knockout mice. Proc. Natl. Acad. Sci. U. S. A. 101, 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB (2012). The enteric nervous system and neurogastroenterology. Nat. Rev. Gastroenterol. Hepatol. 9, 286–294. [DOI] [PubMed] [Google Scholar]
- Furness JB, Costa M, Rökaeus A, McDonald TJ, and Brooks B (1987). Galanin-immunoreactive neurons in the guinea-pig small intestine: their projections and relationships to other enteric neurons. Cell Tissue Res. 250, 607–615. [DOI] [PubMed] [Google Scholar]
- Furness JB, Jones C, Nurgali K, and Clerc N (2004). Intrinsic primary afferent neurons and nerve circuits within the intestine. Prog. Neurobiol. 72, 143–164. [DOI] [PubMed] [Google Scholar]
- Garfield AS, Lam DD, Marston OJ, Przydzial MJ, and Heisler LK (2009). Role of central melanocortin pathways in energy homeostasis. Trends Endocrinol. Metab. 20, 203–215. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, and Wargo JA (2018). The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 33, 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grider JR (1994). Interplay of somatostatin, opioid, and GABA neurons in the regulation of the peristaltic reflex. Am. J. Physiol. 267, G696–G701. [DOI] [PubMed] [Google Scholar]
- Grider JR (2003). Neurotransmitters mediating the intestinal peristaltic reflex in the mouse. J. Pharmacol. Exp. Ther. 307, 460–467. [DOI] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, and Regev A (2016). Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramis A-PG, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJA, and Clevers H (2004). De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686. [DOI] [PubMed] [Google Scholar]
- Herrero A, Duhart JM, and Ceriani MF (2017). Neuronal and Glial Clocks Underlying Structural Remodeling of Pacemaker Neurons in. Front. Physiol. 8, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarret A, Jackson R, Duizer C, Healy ME, Zhao J, Rone JM, Bielecki P, Sefik E, Roulis M, Rice T, et al. (2020). Enteric Nervous System-Derived IL-18 Orchestrates Mucosal Barrier Immunity. Cell 180, 50–63.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila IS (2018). Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front. Immunol. 9, 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CSN, Mahlakõiv T, Moeller JB, Rankin LC, Flamar A-L, Kabata H, Monticelli LA, Moriyama S, Putzel GG, Rakhilin N, et al. (2017). The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze WA, and Furness JB (1999). The enteric nervous system and regulation of intestinal motility. Annu. Rev. Physiol. 61, 117–142. [DOI] [PubMed] [Google Scholar]
- Laranjeira C, Sandgren K, Kessaris N, Richardson W, Potocnik A, Vanden Berghe P, and Pachnis V (2011). Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J. Clin. Invest. 121, 3412–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasrado R, Boesmans W, Kleinjung J, Pin C, Bell D, Bhaw L, McCallum S, Zong H, Luo L, Clevers H, et al. (2017). Lineage-dependent spatial and functional organization of the mammalian enteric nervous system. Science 356, 722–726. [DOI] [PubMed] [Google Scholar]
- Legay C (2000). Why so many forms of acetylcholinesterase? Microsc. Res. Tech. 49, 56–72. [DOI] [PubMed] [Google Scholar]
- Lesage S, and Brice A (2009). Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 18, R48–R59. [DOI] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, and Bush JO (2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev. Biol. 379, 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkert M, Rueden CT, Allan C, Burel J-M, Moore W, Patterson A, Loranger B, Moore J, Neves C, Macdonald D, et al. (2010). Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R, Regier J, Cole MB, Jordan MI, and Yosef N (2018). Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, McMahon AP, and Koentges G (2005). Neural crest origins of the neck and shoulder. Nature 436, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 8, 329–337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres M-L, Hashimoto D, Mortha A, Leboeuf M, Li X-M, Mucida D, et al. (2014). Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 158, 300–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y-R, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer RF (2003). Gastrointestinal dysfunction in Parkinson’s disease. Lancet Neurol. 2, 107–116. [DOI] [PubMed] [Google Scholar]
- Pietzsch T, Preibisch S, Tomancák P, and Saalfeld S (2012). ImgLib2--generic image processing in Java. Bioinformatics 28, 3009–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla P, and Larue L (2003). Involvement of endothelin receptors in normal and pathological development of neural crest cells. Int. J. Dev. Biol. 47, 315–325. [PubMed] [Google Scholar]
- Prunuske AJ, and Ullman KS (2006). The nuclear envelope: form and reformation. Curr. Opin. Cell Biol. 18, 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, et al. (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J, Luukko K, Poteryaev D, Laurikainen A, Sun YF, Laakso T, Eerikäinen S, Tuominen R, Lakso M, Rauvala H, et al. (1999). Retarded growth and deficits in the enteric and parasympathetic nervous system in mice lacking GFR alpha2, a functional neurturin receptor. Neuron 22, 243–252. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, et al. (2015). Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Q, and Young HM (1996). Chemical coding of neurons in the myenteric plexus and external muscle of the small and large intestine of the mouse. Cell Tissue Res. 284, 39–53. [DOI] [PubMed] [Google Scholar]
- Sasselli V, Pachnis V, and Burns AJ (2012). The enteric nervous system. Dev. Biol. 366, 64–73. [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CAJ, and Di Santo JP (2010). IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207, 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheving LA (2000). Biological clocks and the digestive system. Gastroenterology 119, 536–549. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, and Eliceiri KW (2015). The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield JN, Day IN, Thompson RJ, and Edwards YH (1995). PGP9.5, a ubiquitin C-terminal hydrolase; pattern of mRNA and protein expression during neural development in the mouse. Brain Res. Dev. Brain Res. 85, 229–238. [DOI] [PubMed] [Google Scholar]
- Schuchardt A, D’Agati V, Pachnis V, and Costantini F (1996). Renal agenesis and hypodysplasia in ret-k- mutant mice result from defects in ureteric bud development. Development 122, 1919–1929. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al. (2018). Rewiring of the cellular and inter-cellular landscape of the human colon during ulcerative colitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner JF, Leparc G, Hildebrandt T, Klein H, Thomas L, Stupka E, and Simon E (2017). An RNA-Seq atlas of gene expression in mouse and rat normal tissues. Sci Data 4, 170185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff MM, Kodner IJ, and DeSchryver-Kecskemeti K (1988). Axonal degeneration/necrosis: a possible ultrastructural marker for Crohn’s disease. Mod. Pathol. 1, 182–187. [PubMed] [Google Scholar]
- Szurszewski JH, Ermilov LG, and Miller SM (2002). Prevertebral ganglia and intestinofugal afferent neurones. Gut 51 Suppl 1, i6–i10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J, Hahn P, Kroehling L, Nguyen H, Li D, and Littman DR (2020). Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature 579, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen A-M, et al. (2016). Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 1551. [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J-E, Stephenson E, Polański K, Goncalves A, et al. (2018). Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour R-EE, Nyman J, Dionne D, Hofree M, Cuoco MS, Rodman C, Farouq D, et al. (2017). The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese CB, Fleming N, Buehler DP, and Southard-Smith EM (2013). A Uchl1-Histone2BmCherry:GFP-gpi BAC transgene for imaging neuronal progenitors. Genesis 51, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock SL, Lopez R, and Klein AM (2019). Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Ding J, Porter CBM, Wallrapp A, Tabaka M, Ma S, Fu S, Guo X, Riesenfeld SJ, Su C, et al. (2019). Transcriptional Atlas of Intestinal Immune Cells Reveals that Neuropeptide α-CGRP Modulates Group 2 Innate Lymphoid Cell Responses. Immunity 51, 696–708.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, and Shao C (2008). Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput. Biol. 4, e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BB, and Mazmanian SK (2017). The Enteric Network: Interactions between the Immune and Nervous Systems of the Gut. Immunity 46, 910–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HM, Furness JB, and Povey JM (1995). Analysis of connections between nitric oxide synthase neurons in the myenteric plexus of the guinea-pig small intestine. J. Neurocytol. 24, 257–263. [DOI] [PubMed] [Google Scholar]
- Yu Y-B, Zuo X-L, Zhao Q-J, Chen F-X, Yang J, Dong Y-Y, Wang P, and Li Y-Q (2012). Brain-derived neurotrophic factor contributes to abdominal pain in irritable bowel syndrome. Gut 61, 685–694. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Single nucleus RNA-seq (snRNA-seq) of GFP+ nuclei from Sox10-Cre; INTACT mice with previous protocols. Fraction (y axis) of identified cell types (x axis) in samples of the brain (grey) and colon (black) using two previously published snRNA-seq methods (Habib et al., 2016, 2017). (B-D) Quality metrics across all tested nucleus extractions. Cell type composition, number of detected genes per nucleus, and ENS signature score (columns, x axes) across nucleus extraction parameters (y axes), including detergent concentration (B), sample handling method (C), and buffer (D). (E) Decision tree for selection of optimal nucleus extraction conditions. (F) Quality metrics for each nucleus extraction. Mean number of genes per nucleus (x axis) and mean ENS signature score (y axis) for the nuclei from each extraction method (dots). (n = 5,236 GFP+ sorted nuclei across all protocols). Error bars: SEM. Select nucleus extractions are marked in color (legend).
Fraction of expressing cells (dot size) and mean levels in expressing (non-zero) cells (dot color) of genes (columns) in neurons vs. other cells (top panel) or individual neuron subsets (bottom panel) (rows) for (A) selected neurotransmitters and neuropeptides, (B) genes functionally important in the ENS, (C) data-driven marker genes that are uniquely enriched in each neuron subset, and (D) cytokines, chemokines, and their receptors. Outline: significant DE coefficient (discrete coefficient of MAST hurdle model; STAR Methods).
(A,B) Reproducible cell subset distributions across transgenic mouse models and individual mice. t-SNE of RAISIN RNA-Seq transcriptomes of 2,567 neurons (A) and 3,039 glia (B) colored by either subset, age, mouse model, sex, mouse, the number of detected genes per nucleus, or circadian time point. (C) Estimated proportion of enteric neurons in the mouse colon. Percent of all sorted cells in the colon that are neurons (y axis) as estimated by FACS (transgene-expressing nuclei vs. unlabeled nuclei) and post-hoc adjustment based on RAISIN RNA-Seq data. (D) Hierarchical relationships among neuron subset profiles. Dendrogram of hierarchical clustering by transcriptional similarities (STAR Methods) of neuron subsets (leaves). (E) Glia subset markers. Effect size within each glia subset (i.e. discrete DE coefficient, x axis) and associated significance (y axis) for each gene (dots) in each subset. Color: gene specificity (fold-change relative to the next highest expressing glial subset; color bar). Dashed line: p < 0.05.
(A-D) Comparison of doublet detection methods for neurons and glia from the human colon. (A,B) Representative t-SNEs of 2,258 human colon neuron profiles colored by (A) signature scores for muscle, epithelial cells, fibroblasts, and glia (putative doublets have high scores for non-neuron cell types), or (B) doublet predictions by our new method (left) vs. Scrublet, Doublet Finder, and Solo (right). (C,D) Representative t-SNEs of 18,721 human colon glia colored by (C) signature scores for muscle, epithelial cells, fibroblasts, and glia or (D) doublet predictions by our new method (left) vs. Scrublet, Doublet Finder, and Solo (right). (E,F) MIRACL-Seq of mouse esophagus, heart, and lung. t-SNE of MIRACL-Seq profiles (dots) from mouse esophagus (11,550 nuclei; 19 subsets), heart (8,343 nuclei; 15 subsets), and lung (8,441 nuclei; 18 subsets), colored by tissue and sample of origin (E) or cell subset (F), annotated post-hoc using data-derived marker genes (STAR Methods).
t-SNEs of MIRACL-Seq RAISIN RNA-seq profiles (dots) of neurons and glia from the mouse ileum (A,B), mouse colon (C,D), and human colon (E,F), colored by cell subset (left), mouse or human identifier (middle), and the number of detected genes per nucleus (right).
Representative in situ validations of DAPI staining (blue) with combined smFISH and immunostaining (green, red, and white), showing the co-expression of selected genes in mouse (A-K) and human (L-P) colon neurons. Individual channels for each probe in greyscale (left) with merged channels in color (right). Representative images selected from a total of 3 experiments per validation (STAR Methods). Scale bar: 50 μm.
(A) Schematic of coronal brain section. Raphe nuclei containing serotonergic (Tph2+) neurons served as a positive control. The pontine reticular nucleus lacking Tph2-expressing neurons served as a negative control. (B, C) Representative images of smFISH for Tph2 in the mouse brain (B) and colon (C) of Sox10-Cre; INTACT (GFP) mice (n = 2 animals; 12 colon sections). (D, E) Representative images of smFISH for Tph2 in the mouse brain (D) and colon (E) of wild-type C57BL/6J mice. (n = 2 animals; 12 colon sections). (F) Analysis of bulk RNA-seq data from several tissues of C57BL/6 mice (Söllner et al., 2017). RNA expression of Tph1 and Tph2 from the brain, colon and small intestine. RNA expression independently analyzed in three mice per tissue is indicated 1–3. Scale bar: 100 μm.
Description and statistics for nuclei extractions, aggregated either by sample or condition. Includes descriptions of the buffers, detergents, detergent concentrations, salts, and modifiers profiled, and provides various statistics, including exon:intron ratios, the number of genes per nucleus, and ENS compositions.
Description of each mouse and human sample profiled in this study, including model, age, sex, circadian time point, intestinal location, quality metrics (number of genes and reads/UMIs per nucleus), and cell type frequencies.
Differential expression statistics summarizing the expression of ligands and receptors, and lists of receptor-ligand pairs, in droplet-based cell subsets from the mouse and human colon.
Differential expression statistics summarizing the marker genes for each cell subset and the effects of location, age, mouse model, sex, and circadian phase on the gene expression programs of mouse colon neurons and glia.
Statistics showing the expected numbers of nuclei, neurons, and multiplets for varying levels of overloading, and estimated cost reductions.
Differential expression statistics summarizing the marker genes for each cell subset, the conserved transcriptional programs shared by human and mouse neurons, and differences between the mouse ileum vs. colon ENS and the human vs. mouse ENS.
Data Availability Statement
FASTQ files for mouse and processed data for mouse and human were deposited in the Single Cell Portal (SCP1038). Human FASTQ files were deposited in the Broad Data Use Oversight System (DUOS) (“ColonENS-RegevXavier-CRCAdjacentNormal-Broad-snRNASeq-UnsortedCells”).


