Abstract
Fungal pathogens secrete effector proteins that regulate host immunity and can suppress basal defence mechanisms against colonization in plants. Verticillium dahliae is a widespread and destructive soilborne fungus that can cause vascular wilt disease and reduces plant yields. However, little is currently known about how the effectors secreted by V. dahliae function. In this study, we analysed and identified 34 candidate effectors in the V. dahliae secretome and found that Vd424Y, a glycoside hydrolase family 11 protein, was highly upregulated during the early stages of V. dahliae infection in cotton plants. This protein was located in the nucleus and its deletion compromised the virulence of the fungus. The transient expression of Vd424Y in Nicotiana benthamiana induced BAK1‐ and SOBIR1‐dependent cell death and activated both salicylic acid and jasmonic acid signalling. This enhanced its resistance to the oomycetes Phytophthora capsici in a way that depended on its nuclear localization signal and signal peptides. Our results demonstrate that Vd424Y is an important effector protein targeting the host nucleus to regulate and activate effector‐triggered immunity in plants.
Keywords: cell death, effector, nuclear localization signal, Verticillium dahliae, virulence
Vd424Y is an important effector protein targeting the host nucleus to regulate and activate effector‐triggered immunity in plants.

1. INTRODUCTION
Pathogenic microorganisms are responsible for a variety of diseases affecting plants. Despite the wide range of defence strategies that plants have developed to trigger immune responses, pathogenic microorganisms still represent a serious threat to agriculture. These defence strategies include pathogen‐associated molecular patterns (PAMPs)‐triggered immunity (PTI), a process through which PAMPs are recognized by plasma membrane‐bound receptors located at the cell surface, inducing primary defence response (Akira et al., 2006). This process can be suppressed by pathogen‐delivered effector proteins, which can also cause infections (Houterman et al., 2008; de Jonge et al., 2010; Stergiopoulos & de Wit, 2009; Stergiopoulos et al., 2012). Plants have acquired a second layer of immune response known as effector‐triggered immunity (ETI), a process through which host cells detect the presence of a pathogen's effectors via R proteins. These two kinds of immunity are involved in the accumulation of reactive oxygen species (ROS), callose deposition, and the regulation of hormone signalling genes (Dodds & Rathjen, 2010; Tsuda & Katagiri, 2010). Immune signalling contains PTI and ETI and is initiated from either the extracellular space or the cytoplasm of the cell through the interaction of the pathogen molecule and plant receptor, though this signalling typically must be transduced into the plant nucleus to activate the expression of defence genes (Eulgen & Somssich, 2007). The barley MLA10 protein can recognize the effector AVRA10 and induce nuclear association between the receptor and WRKY transcription factors, which is needed to activate ETI (Shen et al., 2007). In Fusarium oxysporum, the effector AVR2 triggers ETI via recognition by the resistance protein I‐2 and subsequent translocation into the plant nucleus (Ma et al., 2013). In Arabidopsis, the PTI signalling pathway also relies on the function of transcription factors (Eulgen & Somssich, 2007).
Previous studies have demonstrated that pathogen effectors can attenuate ETI (Chisholm et al., 2006; Petre & Kamoun, 2014), making it important to study effector functions to better understand pathogenic mechanisms. Biotrophic and necrotrophic pathogens are responsible for delivering apoplastic and intracellular effector proteins that regulate innate host immunity. This includes RxLR effectors secreted from oomycetes (Qiao et al., 2013; Wu et al., 2019; Yang et al., 2017) that can directly hijack plant resistance pathways (Du, Mpina, et al., 2015; King et al., 2014) and interfere with endoplasmic reticulum stress (Jing et al., 2016) and epigenetic regulation (Hou et al., 2019; Kong et al., 2017; Qiao et al., 2013).
The fungus Verticillium dahliae is a hemibiotrophic pathogen that progresses through biotrophic and necrotrophic stages during infection. V. dahliae is a soilborne vascular pathogen that infects plants through the roots and colonizes the xylem vessels of different plants species, such as cotton, potato, pepper, Arabidopsis, tomato, and Nicotiana benthamiana (Lo Presti et al., 2015; Pantelides et al., 2010; Wang, Cai, et al., 2004). V. dahliae is similar to other pathogens in that it uses apoplastic and intracellular effector proteins to combat plant immunity and successfully colonize the host (Lo Presti et al., 2015). Examples include the following: Ave 1 is the first cloned V. dahliae effector and is recognized by a membrane‐bound receptor‐like protein in tomatoes (de Jonge et al., 2012); Vdlcs1 is an isochorismate synthase able to suppress salicylic acid (SA) signalling during host colonization (Liu et al., 2014); VdSCP41 is located in the nucleus and can suppress plant immunity by disrupting the transcriptional activity of host genes (Qin et al., 2018); VdCUT11 is an apoplastic effector that induces cell death and triggers defence responses in N. benthamiana, cotton, and tomato (Gui et al., 2018). These studies demonstrated that V. dahliae effectors have evolved to target host regulatory proteins and disrupt the host defence signal network during the infection process.
The secretome of V. dahliae is composed of nearly 800 proteins (Klosterman et al., 2011). However, relatively few of these secreted proteins have been functionally characterized, and a better understanding of how V. dahliae facilitates plant immunity could provide insights into ways to improve host plant resistance. In this study, we assessed the variation of the Vd991 strain and compared the published Vd991 genome with our de novo assembly of the V. dahliae 991 genome (unpublished data), finding that they were highly conserved. We also analysed 34 candidate effectors from secreted proteins and identified a potential secreted protein, Vd424Y, that induces cell death in tobacco, Arabidopsis, and pepper leaves. Vd424Y is a member of the glycoside hydrolase family 11 (GH11) and targeted the nucleus of the host cells. Its presence was necessary for V. dahliae to be fully virulent in cotton. Moreover, we observed that whether or not Vd424Y triggered cell death depended on its nuclear localization signal (NLS) and signal peptide (SP), and that its ability to induce cell death was dependent on BAK1 and SOBIR1. Finally, we demonstrated that a Vd424Y‐induced immune response in N. benthamiana leaves could depend on its translocation from the plant cell apoplastic space to the nucleus.
2. RESULTS
2.1. Identification of a V. dahliae secreted protein that elicits plant cell death
Few proteins secreted by V. dahliae capable of inducing cell death have been reported. We analysed the genomes of V. dahliae strain 991 (Vd991) and 34 effectors with a signal peptide (SP), which were individually cloned into potato virus X (PVX) vector pGR107 (Table S1). We then performed transient expression in N. benthamiana to screen potential candidate V. dahliae effectors capable of inducing cell death. Green fluoresecnt protein (GFP) and Bcl‐2‐associated X protein (BAX) were used as negative and positive controls, respectively. Agrobacterium carrying each of the effectors was injected into the leaves of 4‐week‐old N. benthamiana plants. Seven days after infiltration, we identified a candidate effector EVM0004916 (hereafter designated Vd424Y) that induced intense cell death (Figure 1a). Western blot analysis demonstrated that both GFP and Vd424Y were expressed (Figure 1c), with a significant increase in the electrolyte leakage around the region of Vd424Y expression compared to regions of GFP expression. This is consistent with the cell death phenotype (Figure 1b). Additionally, the Agrobacterium‐mediated transient expression of Vd424Y in pepper and Arabidopsis thaliana induced cell death, which did not occur in the GFP control (Figure S1). These findings highlighted the ability of Vd424Y to induce cell death in various plant species.
FIGURE 1.
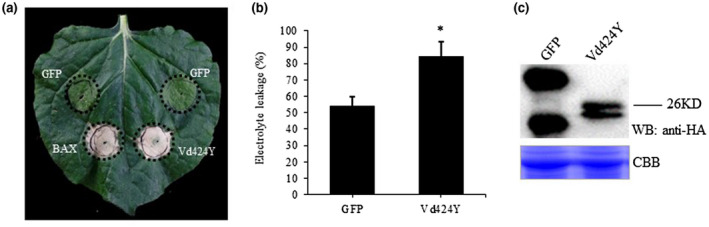
Vd424Y is an elicitor of cell death. (a) Vd424Y induces cell death in Nicotiana benthamiana. Four‐week‐old plants were used to express PVX‐GFP‐HA, PVX‐Vd424Y‐HA, and PVX‐BAX. Photographs were taken 7 days after Agrobacterium infiltration. (b) Quantification analysis of cell death via electrolyte leakage measurement. The data shown represent the mean ± SE estimated from three biological replicates. Significant p values (p < .05) for a Student's t test are represented by *. (c) Western blotting (WB) analysis of protein levels in N. benthamiana transiently expressing green fluorescent protein (GFP) control (left) and Vd424Y fused with HA tag (right). Proteins were stained with Coomassie brilliant blue R‐250 (CBB) to confirm equal loading
Vd424Y encodes a 223 amino acid protein with a conserved glycoside hydrolase domain (Figure S2), and has a signal peptide (SP) encoded by the first 60 nucleotides (Figure S2) along with a predicted nuclear localization signal (NLS) (Dingwall & Laskey, 1991) encoded by 33 nucleotides (Figure S3). This indicates that Vd424Y could have secretory functioning and target the host protein in the nucleus. As shown in Figure S3, only Avr1bSP and Vd424YSP constructs grew well on the YPRAA medium. However, the empty vector pSUC2 and the YTK12 strain did not grow well. These results indicate that the SP of Vd424Y is essential for secretory functioning.
2.2. The NLS and SP are required for Vd424Y‐induced cell death
Subcellular localization demonstrated that Vd424Y is predominantly located in the plant nuclei (Figures 2c and S4a). This suggests it could be closely related to the presence of a putative NLS (Figure S2). To investigate whether the NLS had a significant effect on the Vd424Y‐induced cell death due to its location in the nucleus, we performed NLS mutation analysis and fused a nuclear export signal (NES) to the effector (Vd424Y‐nls). Three vectors were individually expressed in N. benthamiana via Agrobacterium tumefaciens‐mediated infiltration, and yellow fluorescent protein (YFP) fluorescence was detected in the infiltrated leaves using a confocal microscope at 48 hr postinfiltration (hpi). These vectors included Vd424Y‐nls‐YFP, in which the NLS residues were mutated to alanines (Figures 2a and S4), Vd424Y‐nls‐NES‐YFP, in which Vd424Y‐nls was fused with a NES (Du, Berg, et al., 2015, 2015) at the C‐terminus (Figure 2a), and Vd424Y‐NES‐YFP, in which Vd424Y was fused with an NES at the C‐terminus (Figure 2a). As shown in Figure 2c, the YFP signal intensity in the nuclei expressing Vd424Y‐nls was weaker than the signalling intensity in the nuclei expressing Vd424Y‐YFP. Vd424Y‐YFP also displayed a significantly stronger YFP signal intensity in the nucleus than both Vd424Y‐nls‐NES and Vd424Y‐NES. These results indicate that the predicted NLS in Vd424Y affects nuclear localization, and that the NES is capable of guiding Vd424Y out of the nucleus. Additionally, we performed a transient expression assay demonstrating that Vd424Y‐nls, Vd424Y‐nls‐NES, and Vd424Y‐NES were unable to induce intense cell death (Figure 2b). This suggests that NLS‐mediated nuclear localization is required for Vd424Y to induce cell death.
FIGURE 2.
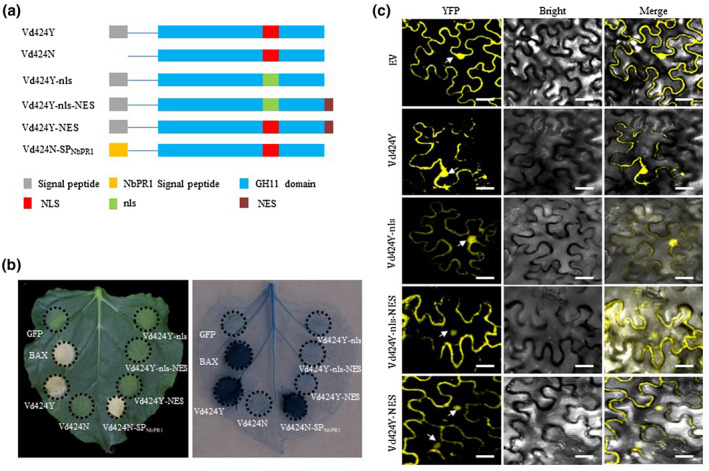
Signal peptide and nuclear localization signal are required for Vd424Y‐induced cell death. (a) Schematic illustration of the Vd424Y deletion mutants: Vd424Y, the full‐length sequence of the candidate effector; Vd424N, effector variant lacking the signal peptide (SP) sequence; Vd424Y‐nls, effector variant where the predicted nuclear localization signal (NLS) was mutated to alanine residues; Vd424Y‐nls‐NES, the variant Vd424Y‐nls was fused with a NES at C‐terminus; Vd424Y‐NES, effector variant where the NES at C‐terminus was tagged to the wildtype Vd424Y allele; Vd424N‐SPNbPR1, effector variant with a fused SP of Nicotiana benthamiana pathogenesis‐related protein (NbPR1). (b) N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens carrying Vd424Y, Vd424N, Vd424Y‐nls, Vd424Y‐nls‐NES, Vd424Y‐NES, Vd424N‐SPNbPR1 , positive control BAX, and control green fluorescent protein (GFP). Representative photographs of N. benthamiana leaves were taken after 7 days. (c) Subcellular localizations of Vd424Y‐YFP, Vd424Y‐nls‐YFP, Vd424Y‐nls‐NES‐YFP, and Vd424Y‐NES‐YFP in N. benthamiana on A. tumefaciens‐mediated transient expression. Fluorescence was detected by confocal microscopy 48 hr postinfiltration. Bars, 40 µm
To determine whether or not Vd424Y‐induced cell death depends on Vd424Y secretion, we constructed an SP‐deleted (amino acids 1–20; Figure S2) mutant (Vd424Y21‐223, hereafter designated Vd424N; Figure 2a) and fused the SP of N. benthamiana pathogenesis‐related protein (NbPR1) to produce Vd424N‐SPNbPR1 (Figure 2a). Both of these Vd424Y variants were expressed in N. benthamiana. Immunoblot analysis confirmed the presence of Vd424N and Vd424N‐SPNbPR1 (Figure S5). We found that Vd424N did not induce cell death in N. benthamiana, while the expression of Vd424N‐SPNbPR1 did (Figure 2b). This suggests that SP is required for Vd424Y‐induced cell death and that Vd424Y must target the extracellular space for cell death to occur. These results demonstrate that NLS and SP are necessary for Vd424Y to induce intense cell death, and that a Vd424Y‐induced immune response in N. benthamiana leaves depends on its translocation from the plant's apoplastic space to the nucleus.
2.3. Vd424Y triggers plant immunity responses
To determine whether Vd424Y‐activated cell death was associated with plant immune response, we examined a hypersensitive response (HR)‐specific marker gene (NbHIN1) (Takahashi et al., 2004) in N. benthamiana leaves following Vd424Y infiltration via Agrobacterium. Analysis of expression levels demonstrated that 3 days after infiltration NbHIN1 was significantly activated by Vd424Y but not by Vd424N or Vd424Y‐nls (Figure 3). To confirm whether Vd424Y‐trigged immunity involved hormone signalling pathways, we used quantitative reverse transcription PCR (RT‐qPCR) analysis to detect the SA‐dependent immunity marker genes NbPR1a and NbPR2 (Dean et al., 2005), and the jasmonic acid (JA)‐dependent immunity gene NbPR4 (Asai & Yoshioka, 2009; Rodriguez et al., 2014). These marker genes were significantly induced in N. benthamiana leaves expressing Vd424Y, and their expression levels were higher than in leaves expressing the variants Vd424N and Vd424Y‐nls (Figure 3). Furthermore, we demonstrate that Vd424Y (but not its variants Vd424N and Vd424Y‐nls) was able to induce the expression of PTI‐related marker genes, including NbCYP71D20 (Heese et al., 2007), NbPti5, NbWRKY7, and NbWRKY8 (Ishihama et al., 2011; Nguyen et al., 2010) (Figure 4). This confirms that Vd424Y is a microbial PAMP. These results suggest that Vd424Y can be recognized by N. benthamiana and activate plant immunity in a manner dependent on its NLS and SP.
FIGURE 3.
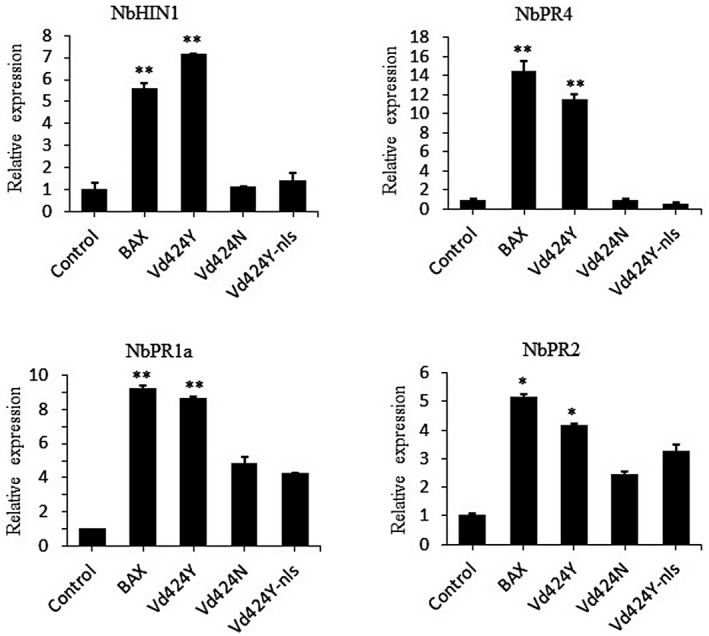
Vd424Y induces plant defence responses in Nicotiana benthamiana. Relative expression of hypersensitive‐response‐specific and defence‐related marker genes in N. benthamiana infiltrated with Agrobacterium tumefaciens carrying Vd424Y, Vd424N, and Vd424Y‐nls. At 3 days postinfiltration (dpi), total RNA was extracted and transcript levels were detected by quantitative reverse transcription PCR. NbActin was used as the internal reference gene. The data shown represents the mean across three independent experiments. Bars indicate ± SE. Significance levels p < 0.05 and p < 0.01 are represented by * and **, respectively
FIGURE 4.
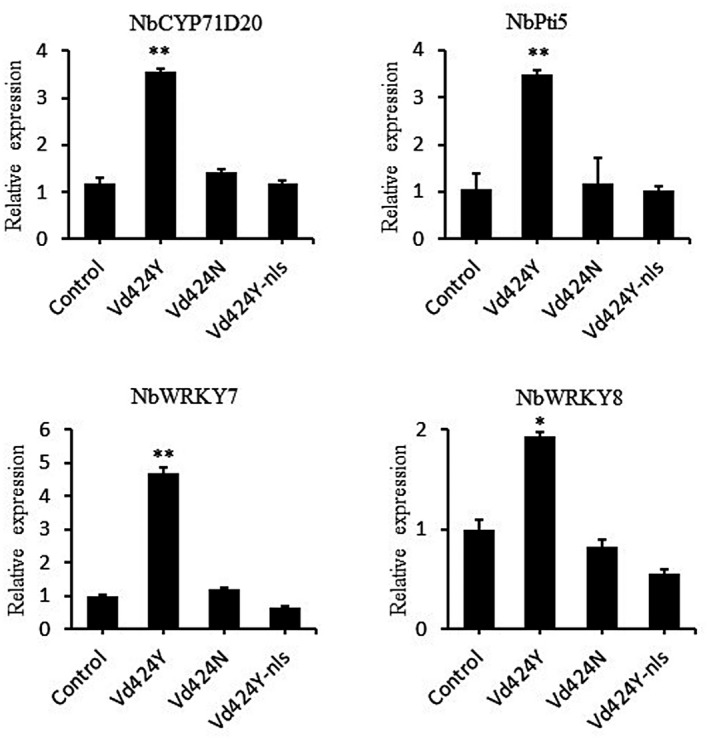
Vd424Y promotes transcription of PAMP‐PTI marker genes in Nicotiana benthamiana. Relative transcript levels of NbCYP71D20, NbPti5, NbWRKY7, and NbWRKY8 were analysed in N. benthamiana infiltrated with Agrobacterium tumefaciens carrying Vd424Y, Vd424N, and Vd424Y‐nls. At 3 days postinfiltration, total RNA was extracted and transcript levels were detected by quantitative reverse transcription PCR. NbActin was used as the internal reference gene. The data shown represent the mean across three independent experiments. Significance levels p < 0.05 and p < 0.01 are represented by * and **, respectively
2.4. NbBAK1 and NbSOBIR1 are required for Vd424Y‐induced cell death in N. benthamiana
BAK1 and SOBIR1 are receptor‐associated kinases that participate in various PAMP‐triggered immune responses and are activated by cell death‐inducing proteins secreted by fungi or oomycetes (Heese et al., 2007; Ma et al., 2015; Nie et al., 2019). Vd424Y properties that induce cell death in the extracellular space are dependent on the SP, so we tested whether BAK1 and SOBIR1 mediate Vd424Y‐induced cell death in N. benthamiana. We performed virus‐induced gene silencing (VIGS) to silence NbBAK1 and NbSOBIR1 in N. benthamiana leaves, and used RT‐qPCR to confirm the silencing of NbBAK1 and NbSOBIR1 21 days after VIGS‐mediated gene silencing (Figure 5b). These leaves were then agroinfiltrated with Vd424Y, Vd424Y variants (Vd424N, Vd424Y‐nls, and Vd424N‐SPNbPR1), and BAX. Immunoblotting analysis confirmed that Vd424Y and its variants were expressed in N. benthamiana leaves inoculated with pTRV2:BAK1, pTRV2:SOBIR1, and pTRV2:GFP (Figure 5c) but did not induce cell death in either NbBAK1‐ or NbSOBIR1‐silenced plants. However, the positive control BAX retained its ability to induce cell death (Figure 5a). In contrast, Vd424Y and Vd424N‐SPNbPR1 were able to activate cell death in control plants (Figure 5a). These results indicate that Vd424Y‐mediated plant cell death depends on NbBAK1 and NbSOBIR1. We further investigated whether Vd424Y‐triggered cell death involved other signalling components by silencing NbNDR1 and NbEDS1 in N. benthamiana leaves (Figure S6). Although immunoblotting analysis confirmed that Vd424Y and its variants were expressed in leaves inoculated with pTRV2:NDR1, pTRV2:EDS1, and pTRV2:GFP, the silencing of either NbNDR1 or NbEDS1 did not affect Vd424Y‐induced cell death (Figure S6).
FIGURE 5.
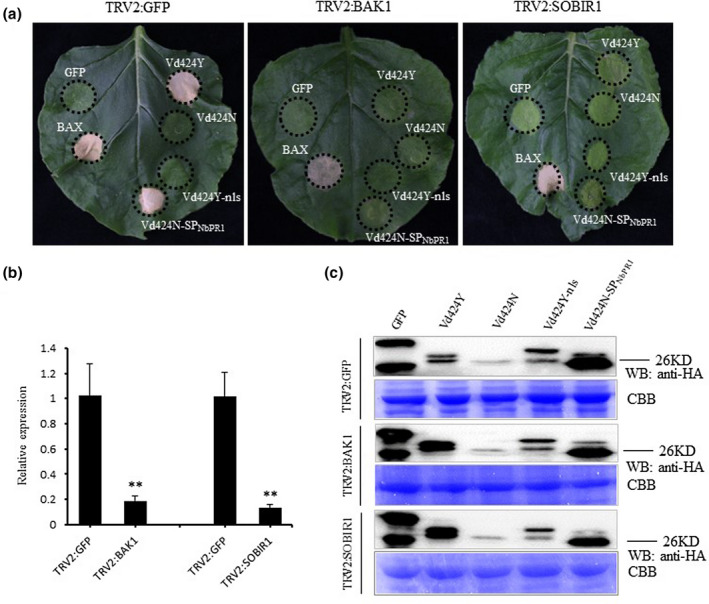
NbBAK1 and NbSOBIR1 are required for Vd424Y‐induced cell death. (a) Virus‐induced gene silnecing (VIGS) technology was used to silence NbBAK1 and NbSOBIR1 by inoculation with TRV constructs (pTRV2:GFP, pTRV2:NbBAK1, and NbSOBIR1) in Nicotiana benthamiana plants. Three weeks after inoculation, GFP, BAX, Vd424Y, and Vd424Y mutations were transiently expressed in NbBAK1‐ and NbSOBIR1‐silenced N. benthamiana plant leaves. Photographs were taken 7 days after agroinfiltration. The experiment was carried out three times with five plants for each TRV construct. (b) The expression levels of NbBAK1 and NbSOBIR1 after VIGS treatment as evaluated by quantitative reverse transcription PCR. NbActin was used as the internal reference gene. Mean and SE were calculated from three independent experiments. Bars indicate ± SE. Significance level p < 0.01 is represented by *. (c) Western blot (WB) analysis of green fluorescent protein (GFP), Vd424Y, and Vd424Y mutations protein fused with HA tags after transient expression in N. benthamiana leaves. Proteins were stained with Coomassie brilliant blue (CBB) to confirm equal loading
2.5. Vd424Y enhances N. benthamiana resistance to oomycete pathogens
To investigate whether Vd424Y modulates plant resistance to the oomycete pathogen Phytophthora capsici, we transiently expressed Vd424Y, Vd424N, and Vd424Y‐nls in N. benthamiana leaves. P. capsici zoospores were inoculated onto the infiltrated area 24 hr after Agrobacterium infiltration (Figure 6a), and the size of the resulting lesions was recorded at 48 hr postinoculation (hpi). As shown in Figure 6b,c, the biomass and lesion size of Phytophthora in leaves expressing Vd424Y were both significantly lower than in leaves expressing Vd424N, Vd424Y‐nls, and an empty vector control. These results strongly demonstrate that Vd424Y triggers plant defence responses and alters plant immunity, enhancing N. benthamiana resistance to oomycete pathogens. We also found that both SP and NLS are essential for enhancing Vd424Y’s resistance in N. benthamiana.
FIGURE 6.
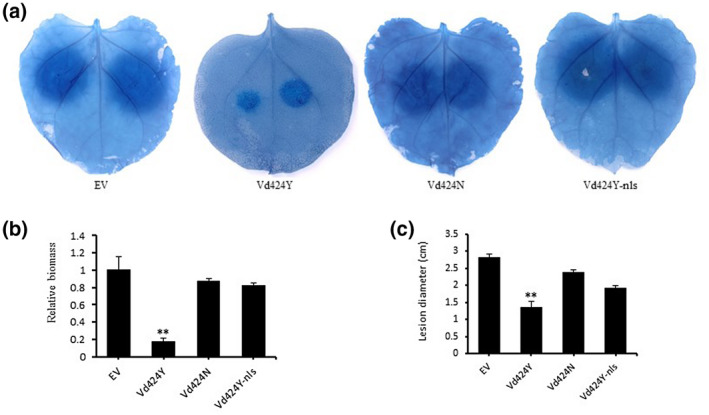
Expression of Vd424Y in Nicotiana benthamiana enhanced resistance to an oomycete pathogen. Leaf regions transiently expressing Vd424Y, Vd424N, and Vd424Y‐nls were inoculated with zoospores of Phytophthora capsici strain 35. (a) Resulting lesions visualized using trypan blue staining. (b) Quantitative PCR analysis of relative Phytophthora biomass following P. capsici infection. Infected leaves (n = 10) were collected 48 hr after infection, after which DNA was isolated and qPCR analysis was performed. (c) Size of the lesions caused by P. capsici infection on plant leaves expressing Vd424Y and Vd424Y mutations
2.6. Vd424Y contributes to V. dahliae virulence
To evaluate the expression profile of Vd424Y during the early stages of V. dahliae infections in cotton, we inoculated cotton roots with fresh V. dahliae spores and collected the infected cotton root samples at different time points. RT‐qPCR analysis indicated that Vd424Y’s expression was strongly upregulated during the early stages of cotton infection by V. dahliae (Figure S7), reaching its peak at 3 hpi. This strong induction indicated that Vd424Y could promote V. dahliae infection.
To determine whether Vd424Y is involved in fungal virulence, we generated V. dahliae mutants by replacing the Vd424Y gene with a hygromycin resistance gene (Figure S8) according to methods previously described (Gao et al., 2010; Gui et al., 2017). There were similarities in the growth rate and colony morphology of Vd424Y knockout mutants (ΔVd424Y) and the wild type Vd991 (Figure 7a,b). However, compared to the wild type, ΔVd424Y mutants caused fewer disease symptoms and had a lower disease index 26 days after pathogen inoculation in cotton plants (Figure 7c,d). We found that complementing ΔVd424Y mutants resulted in disease symptoms and colony morphology similar to the wild type Vd991 (Figure 7a,d), highlighting the important role that Vd424Y plays in V. dahliae virulence during cotton infection.
FIGURE 7.
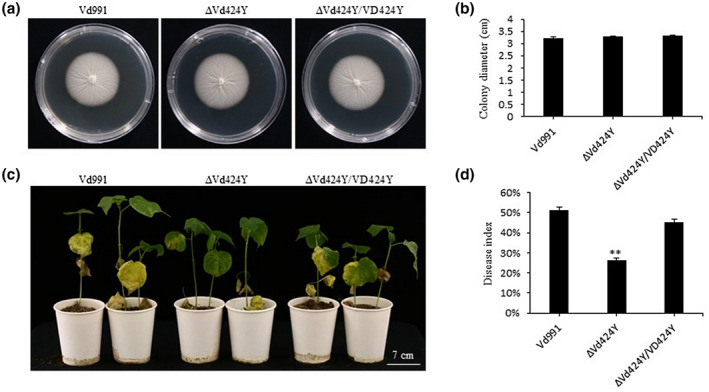
Vd424Y contributes to the virulence of Verticillium dahliae. (a) Images of Vd991 (wild type), ΔVd424Y mutant, and ΔVd424Y/VD424Y‐complementation transformants cultured on potato dextrose agar plates at 25 °C for 7 days in the dark. (b) Colony diameter of Vd991, ΔVd424Y mutant, and ΔVd424Y/VD424Y transformants. The data shows the mean across three independent replicates. (c) Images of disease symptoms on cotton 26 days postinoculation (dpi). The cotton cultivar Jimian 11 was inoculated with Vd991, ΔVd424Y mutant, and ΔVd424Y/VD424Y transformants. Images are representative of three independent experiments. (d) Disease index of the cotton plants at 26 dpi. The disease index (DI) was calculated as the following formula: DI = [(Ʃdisease grades × number of infected plants)/(total checked plants × 4)] × 100%. Seedlings were classified into five grades (0, 1, 2, 3, and 4). The data represent the mean across three independent experiments
3. DISCUSSION
V. dahliae is a soilborne fungal pathogen that destroys the xylem of host cells and causes injury to plants (Fradin & Thomma, 2006; Klosterman et al., 2009). Relatively few secreted effector proteins have been found to play a role in virulence. Therefore, it is necessary to identify virulence‐related effectors and determine their potential roles and pathogenic mechanisms. In this study, we analysed transient expression in N. benthamiana leaves to identify the toxic effectors that induced intense cell death. One of the effectors secreted by V. dahliae, Vd424Y, can trigger strong cell death in N. benthamiana, pepper, and Arabidopsis (Figures 1 and S1). This indicates that Vd424Y could act as a PAMP during host plant infection.
After assessing the yeast signal trap assay system, we found that the fusion of the signal peptide of Vd424Y into the invertase gene resulted in the secretion of invertase in yeast (Figure S3). This indicates that Vd424Y was most probably secreted into the extracellular space during infection of the host plant. The transient expression of Vd424Y lacking the signal peptide, Vd424N, did not induce cell death (Figure 2b), suggesting that the extracellular secretion of Vd424Y was needed to induce cell death in N. benthamiana. These effector‐mediated cell death effects have been previously reported in numerous pathogens. PsXEG1, a glycoside hydrolase family 12 (GH12) protein, acts as a PAMP in soybean and N. benthamiana, in which SP was required for PsXEG1‐induced cell death by targeting the apoplast (Ma et al., 2015). VmE02 exhibited cell‐death‐inducing activity in N. benthamiana, while an SP‐mediated apoplastic location is required to fully induce death of the cell (Nie et al., 2019).
Pathogenic effectors located in the cell nuclei are recognized by their host and have been identified in pathogenic bacteria, fungi, oomycetes, and mutualistic fungi (Kloppholz et al., 2011; Szurek et al., 2001). PSR1 was identified in oomycetes and targeted the RNAi component PINP1 to disturb the defence mechanism in plants (Qiao et al., 2013, 2015). A VdSCP7‐induced immune response was dependent on its nuclear localization (Zhang et al., 2017). Similar research found that Vd424Y‐induced immunity depends on its nuclear localization, while defence signalling is probably initiated in the host nucleus. Our research demonstrated that certain V. dahliae effectors with SP and NLS are secreted into extracellular space and subsequently enter the plant cell nucleus, altering plant immunity. This study assessed Vd424Y‐activated innate immune responses in N. benthamiana, pepper, and Arabidopsis, including the upregulation of HR‐specific marker genes and the activation of SA‐ and JA‐mediated resistance pathways (Figure 3). Therefore, we speculated that Vd424Y is a potential PAMP. Consistent with our hypothesis, Vd424Y dramatically activated the expression of PTI marker genes (Figure 4). Two PRR‐RLKs, BAK1 and SOBIR1, were shown to be necessary for Vd424Y‐triggered cell death in N. benthamiana (Figure 5), further confirming Vd424Y to be a PAMP. This is similar to the case of VdCUT11 and VmE02: VdCUT11‐induced plant defence responses in N. benthamiana were dependent on both BAK1 and SOBIR1 (Gui et al., 2018) and VmE02 triggered cell death in N. benthamiana depending on the presence of BAK1, SOBIR1, HSP90, and SGT1 (Nie et al., 2019). We observed that SP and NLS are essential for Vd424Y‐trigged immune responses, because they activate the expression of marker genes involved in SA and JA signalling and PTI, while mutations in the SP and the NLS weaken expression of the above marker genes (Figures 3 and 4). These results demonstrated that immune responses in N. benthamiana leaves that are induced by the Vd424Y effector protein depend on its translocation from the plant's apoplatic space to the nucleus. Phylogenetic analysis demonstrated that Vd424Y is conserved in different Verticillium species and that its homologs are present in several fungi species (Figure S9), suggesting that Vd424Y may have a conserved function in triggering plant immunity.
We also performed V. dahliae infection assays in cotton plants and found that Vd424Y was highly expressed at the early stages of cotton infection by V. dahliae (Figure S7). This indicates that Vd424Y is closely related to the virulence of V. dahliae when it has infected cotton. Vd424Y knockout mutants did not affect the growth rate and colony morphology of Vd991 but weakened its virulence. Similar findings were reported in Fusarium graminearum (Yang et al., 2021), where the secreted ribonuclease Fg12 was significantly induced at the early stages of F. graminearum infection in soybean. Deleting the secreted ribonuclease Fg12 reduced the virulence of F. graminearum. In conclusion, our results demonstrate that Vd424Y can efficiently activate of plant immunity.
4. EXPERIMENTAL PROCEDURES
4.1. Strains and plant growth conditions
The wildtype V. dahliae strain 991 (Vd991) was grown in the dark on potato dextrose agar (PDA) at 25 ℃. The P. capsici strain 35 (Table S2) was also grown in the dark in 10% V8 juice agar at 25 ℃. Escherichia coli DH5α, which was cultured in a Luria‐Bertani (LB) medium at 37 ℃, was used for plasmid construction, whereas A. tumefaciens GV3101 (pSoup), which was grown in LB medium at 28 ℃, was used for the agroinfiltration of plants. N. benthamiana, Capsicum annuum, and A. thaliana were grown in a greenhouse under a 16 hr light/8 hr dark cycle at 22 ± 1 ℃. Cotton plants were also grown in a greenhouse under a 16 hr light/8 hr dark cycle at 26 ± 1 ℃.
4.2. Plasmids construction
The V. dahliae candidate effector EVM0004916 (Vd424) with SP (Vd424Y), was amplified from a Vd991 cDNA library with gene‐specific primers (Table S3) using TKS Gflex DNA polymerase (Takara). The fragment was then cloned into the PVX vector pGR107‐HA (Yang et al., 2021) using the ClonExpress Ⅱ One Step Cloning Kit (Vazyme Biotech Co. Ltd). The variants Vd424N (without SP), Vd424Y‐nls (with the mutated NLS), and Vd424N‐NbPR1‐SP (with the SP of NbPR1 fused into Vd424N) were cloned into pGR107. For subcellular localization, the Vd424Y, Vd424Y‐nls, Vd424Y‐nls‐NES (Vd424Y‐nls fused with the NES), and Vd424Y‐NES (Vd424Y fused with the NES) variants were cloned into the Gateway entry vector QBV3 and then into the destination expression vector pEG101 (Earley et al., 2006). For Phytophthora infection assays, the QBV3‐Vd424Y, QBV3‐Vd424N, and QBV3‐Vd424Y‐nls were ligated into the pEG100 vector. For VIGS assays, NbBAK1, NbSOBIR1, NbEDS1, NbNDR1, GFP, and phytoene desaturase (PDS) were cloned into pTRV2 (Dong et al., 2007).
4.3. Yeast signal sequence trap
A yeast secretion system was performed to validate the function of the predicted SP (Yin et al., 2018). Specifically, Vd424Y’s predicted SP was cloned into pSUC2T7M13ORI (pSUC2) using specific primers (Table S2), and the resulting plasmid was then transformed into the YTK12, an invertase mutant yeast strain (Oh et al., 2009). The positive colonies were screened on a CMD−W medium (0.67% yeast N base without amino acids, 0.075% W dropout supplement, 2% sucrose, 0.1% glucose, 2% agar) and incubated on YPRAA medium (1% yeast extract, 2% peptone, 2% raffinose, 2 mg/ml antimycin A) to assay invertase secretion. The empty pSUC2 and pSUC2‐Avr1bSP vectors were used as negative and positive controls, respectively.
4.4. VIGS in N. benthamiana
In TRV‐mediated gene silencing assays, the plasmid constructs pTRV1, pTRV2‐NbBAK1, pTRV2‐NbSOBIR1, pTRV2‐NbEDS1, pTRV2‐NbNDR1, pTRV2‐GFP, and pTRV2‐PDS were introduced into A. tumefaciens GV3101. The two primary leaves of four‐leaf‐stage N. benthamiana seedlings were injected with a mixture (1:1 ratio) of A. tumefaciens culture (OD600 = 1.8) containing pTRV1 and pTRV2‐genes. The pTRV2‐PDS was used to evaluate VIGS efficiency, while the pTRV2‐GFP served as control. The RNA extracted from leaves was used to validate the efficiency of gene silencing by RT‐qPCR.
4.5. Infection assays
To infect cotton plants with V. dahliae, 4‐week‐old Jimian 11 plants were inoculated with 10 ml conidial suspensions following previously published methods (Gong et al., 2017). The disease index (DI) was calculated according to previously described protocols (Gong et al., 2017). The infected plants were classified into five grades (grade 0, 1, 2, 3, and 4) according to the symptoms on the cotyledons and new leaves (Wang, Chen, et al., 2004; Zhang et al., 2012).
To infect N. benthamiana with P. capsici, 4‐week‐old N. benthamiana plants expressing the empty vector pEG100 (used as control), Vd424Y, Vd424N, and Vd424Y‐nls were inoculated with P. capsici strain 35 using a previously published protocol (Zhang et al., 2019). Disease development was evaluated by staining lesions with trypan blue (Xiong et al., 2014) and assessing the biomass of P. capsici (Zhang et al., 2019). This procedure was repeated twice for validation and provided similar results.
4.6. RT‐qPCR analysis
Total RNA was isolated using Tiangen RNAprep Pure Plant Plus Kit (DP441), quantified and used as a template for reverse transcription with the PrimeScript RT reagent Kit (Takara). The RT‐qPCR assays were performed using the 2× Wiz Universal SYBR Green qPCR Master Mix (BCS, No. SPE00005). The NbActin gene was used as internal control. The sequences of each of the primers used in the different RT‐qPCR assays are listed in Table S2.
4.7. Subcellular localization assays
Recombinant plasmids were transformed into A. tumefaciens GV3101. The resulting strains were then expressed in 4‐week‐old N. benthamiana leaves using a previously described protocol (Qiao et al., 2015). Forty‐eight hours after Agrobacterium‐infiltration, yellow fluorescence from YFP was detected using a LSM780 confocal microscope.
4.8. Protein extraction and western blotting
For transient expression analysis, the protein was extracted from the leaves of 4‐week‐old N. benthamiana plants after Agrobacterium‐infiltration. The infiltrated leaves were subsequently collected after 48 hr, ground in liquid nitrogen, and mixed with an equal volume of cold protein isolation buffer (1 mM EDTA pH 8.0, 20 mM Tris‐HCl pH 7.5, 5 mM dithiothreitol, 150 mM NaCl, 0.1% sodium dodecyl sulphate [SDS], 10% glycerol, and 1 × protease inhibitor cocktail [Sigma‐Aldrich]). The mixture was centrifuged at 4 ℃ for 10 min at 13,000 × g, transferred to a new tube, and boiled in protein sample buffer for 5 min. Proteins were analysed by SDS‐polyacrylamide gel electrophoresis (PAGE) and electroblotted onto polyvinylidene fluoride (PVDF) membranes (GE Healthcare).
4.9. Bioinformatics analysis
SP prediction was implemented using the SignalP‐5.0 Server (http://www.cbs.dtu.dk/services/SignalP/). The putative bipartite NLS (Ding et al., 1991) was predicted using the cNLS Mapper (nls‐mapper.iab.keio.ac.jp/cgi‐bin/NLS_Mapper_form.cgi).
AUTHOR CONTRIBUTIONS
X.Y.G., L.S.L., F.G.L., and Y.J.L. planned and designed the research. L.S.L., Z.H.W., and J.N.L. performed the research. L.S.L., Z.H.W., J.N.L., Y.W., J.C.Y., J.J.Z., and P.W. analysed the data. L.S.L., X.Y.G. F.G.L., and Y.J.L. wrote the paper. All authors commented on the article before submission.
Supporting information
FIGURE S1 Hypersensitive cell death induced by Vd424Y in (a) pepper and (b) Arabidopsis leaves. Leaves were infiltrated with Agrobacterium tumefaciens carrying Vd424Y. Photographs were taken 12 days postinfiltration. BAX and GFP were used as positive control and negative control, respectively
FIGURE S2 Protein sequences of Vd424Y with the different regions marked
FIGURE S3 The signal peptide (SP) of Vd424Y is functional. The validation of the function of Vd424YSP with yeast signal trap assay. The YTK12 yeast strain containing pSUC2 is able to grow on a CMD−W medium without tryptophan, but not on YPRAA medium. Vd424YSP can grow on both CMD−W and YPRAA media. The SP of Avr1b was used as positive control
FIGURE S4 Subcellular localization of Vd424Y. (a) Yellow fluorescent protein (YFP)‐tagged Vd424Y when transiently coexpressed with H2B in Nicotiana benthamiana. The fluorescence was detected by confocal microscopy. (b) YFP‐tagged Vd424Y was transiently expressed in N. benthamiana. The fluorochrome FM4‐64 was used for staining the cell membrane
FIGURE S5 Western blot of Vd424Y and its mutations. Immunoblot of proteins from Nicotiana benthamiana leaves transiently expressing Vd424Y and its mutations. Proteins were stained with Coomassie brilliant blue (CBB) to confirm equal loading
FIGURE S6NbNDR1 and NbEDS1 are not required for Vd424Y‐induced cell death. (a) Nicotiana benthamiana plants were subjected to virus‐induced gene silencing (VIGS) by inoculation with TRV constructs (pTRV2:GFP, pTRV2:NbNDR1 and pTRV2:NbEDS1). Three weeks after inoculation, GFP, BAX, Vd424Y, and Vd424Y mutations were transiently expressed in NbNDR1‐ and NbEDS1‐silenced N. benthamiana plant leaves. Photographs were taken 7 days postagroinfiltration. The experiment was performed three times with five plants for each TRV construct. (b) The expression levels of NbNDR1 and NbEDS1 after VIGS treatment as evaluated by quantitative reverse transcription PCR. NbActin was used as the internal reference gene. Mean and SE were calculated from three independent experiments. Bars indicate ±SE. Significance level p < .01 is represented by *. (c) Immunoblot analysis of green fluorescent protein (GFP), Vd424Y, and Vd424Y mutations protein fused with HA tags after transient expression in N. benthamiana leaves. Proteins were stained with Coomassie brilliant blue (CBB) to confirm equal loading
FIGURE S7 Expression profile of Verticillium dahliae secreted Vd424Y during infection. The Vd424Y relative transcript levels during different infection stages of V. dahliae 991 were confirmed by quantitative reverse transcription PCR. The cotton roots were inoculated and harvested at 3, 9, 12, 16, 24, 36, and 48 hr postinoculation (hpi). Mycelia (0 hr) was used as control. VdGAPDH was used as internal reference. The data shows the mean across three independent experiments
FIGURE S8 Vd424Y knockout by targeted gene replacement and gene complementation. (a) Organization of the Vd424Y locus before and after homologous recombination in the wild type Vd991. (b, c) PCR analysis of wild type Vd991 and respective mutants. The genomic DNA of each strain was used to verify the targeted and HPH genes
FIGURE S9 The phylogeny of Vd424Y and its homologous sequences from selected species. Bootstrap percentage support for each branch is indicated. The scale bar represents 50% weighted sequence divergence
FILE S1 The homologs of 34 effectors in Vd Ls. 17
TABLE S1Verticillium dahliae Vd genes screened for targeting cell death in this study
TABLE S2 Microbial strains and plasmids used in this study
TABLE S3 Primers used in this study
ACKNOWLEDGEMENTS
The authors thank Professor Suomeng Dong (Nanjing Agriculture University) for providing the pGR107 vector and Professor Yongli Qiao (Shanghai Normal University) for providing the P. capsici strain 35, as well as the pEG100 and pEG101 vectors. The authors are also grateful to Professor Yule Liu (Tsinghua University) for kindly providing the pTRV plasmids, Professor Heng Jian (China Agricultural University) for providing the YTK12 strain and the pSUC2 vector, and Associate Professor Xiaofeng Su (Biotechnology Research Institute of Chinese Academy of Agricultural Sciences) for providing the pCHG vector. This research was financially supported by “Seven Crop Breeding” National Major Project (grant no. 2016YFD0101006), Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, China Agriculture Research System of MOF and MARA (CARS‐15‐02).
Liu, L., Wang, Z., Li, J., Wang, Y., Yuan, J., Zhan, J., et al (2021) Verticillium dahliae secreted protein Vd424Y is required for full virulence, targets the nucleus of plant cells, and induces cell death. Molecular Plant Pathology, 22, 1109–1120. 10.1111/mpp.13100
Lisen Liu and Zhaohan Wang contributed equally to this work.
Contributor Information
Yongjun Lin, Email: yongjunlin@mail.hzau.edu.cn.
Fuguang Li, Email: aylifug@caas.cn.
Xiaoyang Ge, Email: gexiaoyang@caas.cn.
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Akira, S., Uematsu, S. & Takeuchi, O. (2006) Pathogen recognition and innate immunity. Cell, 124 783–801. [DOI] [PubMed] [Google Scholar]
- Asai, S. & Yoshioka, H. (2009) Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana . Molecular Plant‐Microbe Interactions, 22 619–629. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B. & Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124 803–814. [DOI] [PubMed] [Google Scholar]
- Dean, J., Goodwin, P. & Hsiang, T. (2005) Induction of glutathione S‐transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. Journal of Experimental Botany, 56, 1525–1533. [DOI] [PubMed] [Google Scholar]
- Dingwall, C. & Laskey, R.A. (1991) Nuclear targeting sequences—a consensus? Trends in Biochemical Science, 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Dodds, P. & Rathjen, J. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews Genetics, 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dong, Y., Burch‐Smith, T., Liu, Y., Mamillapalli, P. & Dinesh‐Kumar, S. (2007) A ligation‐independent cloning tobacco rattle virus vector for high‐throughput virus‐induced gene silencing identifies roles for NbMADS4‐1 and ‐2 in floral development. Plant Physiology, 145, 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y., Berg, J., Govers, F. & Bouwmeester, K. (2015) Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytologist, 207, 735–747. [DOI] [PubMed] [Google Scholar]
- Du, Y., Mpina, M.H., Birch, P.R.J., Bouwmeester, K. & Govers, F. (2015) Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiology, 169, 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K. et al. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. The Plant Journal, 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. & Somssich, I.E. (2007) Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology, 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Fradin, E.F. & Thomma, B.P. (2006) Physiology and molecular aspects of Verticillium wilt disease caused by V. dahliae and V. albo‐atrum . Molecular Plant Pathology, 7, 71–86. [DOI] [PubMed] [Google Scholar]
- Gao, F., Zhou, B.‐J., Li, G.‐Y., Jia, P.‐S., Li, H., Zhao, Y.‐L. et al. (2010) A glutamic acid‐rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS One, 5, e15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Q., Yang, Z., Wang, X., Butt, H., Chen, E., He, S. et al. (2017) Salicylic acid‐related cotton (Gossypium arboretum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae . BMC Plant Biology, 17, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui, Y., Chen, J., Zhang, D., Li, N., Li, T., Zhang, W. et al. (2017) Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate‐binding module 1. Environmental Microbiology, 19, 1914–1932. [DOI] [PubMed] [Google Scholar]
- Gui, Y., Zhang, W., Zhang, D., Zhou, L., Short, D., Wang, J. et al. (2018) A Verticillium dahliae extracellular cutinase modulated plant immune responses. Molecular Plant‐Microbe Interactions, 2, 260–273. [DOI] [PubMed] [Google Scholar]
- Heese, A., Hann, D.R., Gimenez‐Ibanez, S., Jones, A.M.E., He, K., Li, J. et al. (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plant. Proceedings of the National Academy of Sciences of the United States of America, 104, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., Zhai, Y.I., Feng, L.I., Karimi, H.Z., Rutter, B.D., Zeng, L. et al. (2019) A Phytophthora effector suppresses trans‐kingdom RNAi to promote disease susceptibility. Cell Host & Microbe, 25, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman, P., Cornelissen, B. & Rep, M. (2008) Suppression of plant resistance gene‐based immunity by a fungal effector. PLoS Pathogens, 4, e1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama, N., Yamada, R., Yoshioka, M., Katou, S. & Yoshioka, H. (2011) Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. The Plant Cell, 23, 1153–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, M., Guo, B., Li, H., Yang, B.O., Wang, H., Kong, G. et al. (2016) A Phytophthora sojae effector suppresses endoplasmic reticulum stress‐mediated immunity by stabilizing plant binding immunoglobulin proteins. Nature Communications, 7, 11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge, R., Peter van Esse, H., Kombrink, A., Shinya, T., Desaki, Y., Bours, R. et al. (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. [DOI] [PubMed] [Google Scholar]
- de Jonge, R., Peter van Esse, H., Maruthachalam, K., Bolton, M.D., Santhanam, P., Saber, M.K. et al. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proceedings of the National Academy of Sciences of the United States of America, 109 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.R.F., McLellan, H., Boevink, P.C., Armstrong, M.R., Bukharova, T., Sukarta, O. et al. (2014) Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKK epsilon to suppress plant immune signaling. The Plant Cell, 26, 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppholz, S., Kuhn, H. & Requena, N. (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Current Biology, 21, 1204–1209. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J., Atallah, Z.K., Vallad, G.E. & Subbarao, K.V. (2009) Diversity, pathogenicity, and management of Verticillium species. Annual Review of Phytopathology, 47, 39–62. [DOI] [PubMed] [Google Scholar]
- Klosterman, S.J., Subbarao, K.V., Kang, S., Veronese, P., Gold, S.E., Thomma, B.P.H.J. et al. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathogens, 7, e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L., Qiu, X., Kang, J., Wang, Y., Chen, H., Huang, J. et al. (2017) A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Current Biology, 27, 981–991. [DOI] [PubMed] [Google Scholar]
- Liu, T., Song, T., Zhang, X., Yuan, H., Su, L., Li, W. et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nature Communications, 5, 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M. et al. (2015) Fungal effectors and plant susceptibility. Annual Review of Plant Biology, 66, 513–545. [DOI] [PubMed] [Google Scholar]
- Ma, L., Cornelissen, B. & Takken, F. (2013) A nuclear localization for Avr2 from Fusarium oxysporum is required to activate the tomato resistance protein I‐2. Frontiers in Plant Science, 4, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y. et al. (2015) A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. The Plant Cell, 27, 2057–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H.P., Chakravarthy, S., Velásquez, A.C., McLane, H.L., Zeng, L., Nakayashiki, H. et al. (2010) Methods to study PAMP‐triggered immunity using tomato and Nicotiana benthamiana . Molecular Plant‐Microbe Interactions, 23, 991–999. [DOI] [PubMed] [Google Scholar]
- Nie, J., Yin, Z., Li, Z., Wu, Y. & Huang, L. (2019) A small cysteine‐rich protein from two kingdoms of microbes is recognized as a novel pathogen‐associated molecular pattern. New Phytologist, 222, 995–1011. [DOI] [PubMed] [Google Scholar]
- Oh, S., Young, C., Lee, M., Oliva, R., Bozkurt, T., Cano, L. et al. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi‐blb2 . The Plant Cell, 21, 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelides, I., Tjamos, S. & Paplomatas, E. (2010) Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae . Molecular Plant Pathology, 11, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre, B. & Kamoun, S. (2014) How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biology, 12, e1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y., Liu, L., Xiong, Q., Flores, C., Wong, J., Shi, J. et al. (2013) Oomycete pathogens encode RNA silencing suppressors. Nature Genetics, 45, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y., Shi, J., Zhai, Y., Hou, Y. & Ma, W. (2015) Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proceedings of the National Academy of Sciences of the United States of America, 112, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, J., Wang, K., Sun, L., Xing, H., Wang, S., Li, L. et al. (2018) The plant‐specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. eLife, 7, e34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, P., Stam, R., Warbroek, T. & Bos, J.I.B. (2014) Mp10 and Mp42 from the aphid species Myzus persocae trigger plant defense in Nicotiana benthamiana through different activities. Molecular Plant‐Microbe Interactions, 27, 30–39. [DOI] [PubMed] [Google Scholar]
- Shen, Q.‐H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B. et al. (2007) Nuclear activity of MLA immune receptors links isolate‐specific and basal disease‐resistance responses. Science, 315, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. & de Wit, P.J. (2009) Fungal effector proteins. Annual Review of Phytopathology, 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I., Kourmpetis, Y.A., Slot, J.C., Bakker, F.T., de Wit, P.J. & Rokas, A. (2012) In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins. Molecular Biology and Evolution, 29, 3371–3384. [DOI] [PubMed] [Google Scholar]
- Szurek, B., Marois, E., Bonas, U. & Van den Ackerveken, G. (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. The Plant Journal, 26, 523–534. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Berberich, T., Yamashita, K., Uehara, Y., Miyazaki, A. & Kusano, T. (2004) Identification of tobacco HIN1 and two closely related genes as spermine‐responsive genes and their differential expression during the Tobacco mosaic virus‐induced hypersensitive response and during leaf‐ and flower‐senescence. Plant Molecular Biology, 54, 613–622. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. & Katagiri, F. (2010) Comparing signaling mechanisms engaged in pattern‐triggered and effector‐triggered immunity. Current Opinion in Plant Biology, 13, 459–465. [DOI] [PubMed] [Google Scholar]
- Wang, J., Cai, Y., Gou, J., Mao, Y., Xu, Y., Jiang, W. et al. (2004) VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Applied and Environmental Microbiology, 70, 4989–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.Q., Chen, D.J., Wang, D.M., Huang, Q.S., Yao, Z.P., Liu, F.J. et al. (2004) Over‐expression of Gastrodia anti‐fungal protein enhances Verticillium wilt resistance in coloured cotton. Plant Breeding, 123, 454–459. [Google Scholar]
- Wu, D., von Roepenack‐Lahaye, E., Buntru, M., de Lange, O., Schandry, N., Pérez‐Quintero, A.L. et al. (2019) A plant pathogen type III effector protein subverts translational regulation to boost host polyamine levels. Cell Host & Microbe, 26, 638–649. [DOI] [PubMed] [Google Scholar]
- Xiong, Q., Ye, W., Choi, D., Wong, J., Qiao, Y., Tao, K. et al. (2014) Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana . Molecular Plant‐Microbe Interactions, 27, 1379–1389. [DOI] [PubMed] [Google Scholar]
- Yang, B., Wang, Q., Jing, M., Guo, B., Wu, J., Wang, H. et al. (2017) Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytologist, 214, 361–375. [DOI] [PubMed] [Google Scholar]
- Yang, B., Wang, Y., Tian, M., Dai, K., Zheng, W., Liu, Z. et al. (2021) Fg12 ribonuclease secretion contributes to Fusarium graminearum virulence and induces plant cell death. Journal of Integrative Plant Biology, 63, 365–377. [DOI] [PubMed] [Google Scholar]
- Yin, W., Wang, Y., Chen, T., Lin, Y. & Luo, C. (2018) Functional evaluation of the signal peptides of secreted proteins. Bio‐Protocols, 8, e2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Yang, Y., Chen, T., Yu, W., Liu, T., Li, H. et al. (2012) Island cotton Gbve1 gene encoding a receptor‐like protein confers resistance to both defoliating and non‐defoliating isolated of Verticillium dahliae . PLoS One, 7, e51091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Ni, H., Du, X., Wang, S., Ma, X., Nürnberger, T. et al. (2017) The Verticillium‐specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytologist, 215, 368–381. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Jia, Y., Shi, J., Chen, C., Ye, W., Wang, Y. et al. (2019) The WY domain in the Phytophthora effector PSR1 is required for infection and RNA silencing suppression activity. New Phytologist, 223, 839–852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Hypersensitive cell death induced by Vd424Y in (a) pepper and (b) Arabidopsis leaves. Leaves were infiltrated with Agrobacterium tumefaciens carrying Vd424Y. Photographs were taken 12 days postinfiltration. BAX and GFP were used as positive control and negative control, respectively
FIGURE S2 Protein sequences of Vd424Y with the different regions marked
FIGURE S3 The signal peptide (SP) of Vd424Y is functional. The validation of the function of Vd424YSP with yeast signal trap assay. The YTK12 yeast strain containing pSUC2 is able to grow on a CMD−W medium without tryptophan, but not on YPRAA medium. Vd424YSP can grow on both CMD−W and YPRAA media. The SP of Avr1b was used as positive control
FIGURE S4 Subcellular localization of Vd424Y. (a) Yellow fluorescent protein (YFP)‐tagged Vd424Y when transiently coexpressed with H2B in Nicotiana benthamiana. The fluorescence was detected by confocal microscopy. (b) YFP‐tagged Vd424Y was transiently expressed in N. benthamiana. The fluorochrome FM4‐64 was used for staining the cell membrane
FIGURE S5 Western blot of Vd424Y and its mutations. Immunoblot of proteins from Nicotiana benthamiana leaves transiently expressing Vd424Y and its mutations. Proteins were stained with Coomassie brilliant blue (CBB) to confirm equal loading
FIGURE S6NbNDR1 and NbEDS1 are not required for Vd424Y‐induced cell death. (a) Nicotiana benthamiana plants were subjected to virus‐induced gene silencing (VIGS) by inoculation with TRV constructs (pTRV2:GFP, pTRV2:NbNDR1 and pTRV2:NbEDS1). Three weeks after inoculation, GFP, BAX, Vd424Y, and Vd424Y mutations were transiently expressed in NbNDR1‐ and NbEDS1‐silenced N. benthamiana plant leaves. Photographs were taken 7 days postagroinfiltration. The experiment was performed three times with five plants for each TRV construct. (b) The expression levels of NbNDR1 and NbEDS1 after VIGS treatment as evaluated by quantitative reverse transcription PCR. NbActin was used as the internal reference gene. Mean and SE were calculated from three independent experiments. Bars indicate ±SE. Significance level p < .01 is represented by *. (c) Immunoblot analysis of green fluorescent protein (GFP), Vd424Y, and Vd424Y mutations protein fused with HA tags after transient expression in N. benthamiana leaves. Proteins were stained with Coomassie brilliant blue (CBB) to confirm equal loading
FIGURE S7 Expression profile of Verticillium dahliae secreted Vd424Y during infection. The Vd424Y relative transcript levels during different infection stages of V. dahliae 991 were confirmed by quantitative reverse transcription PCR. The cotton roots were inoculated and harvested at 3, 9, 12, 16, 24, 36, and 48 hr postinoculation (hpi). Mycelia (0 hr) was used as control. VdGAPDH was used as internal reference. The data shows the mean across three independent experiments
FIGURE S8 Vd424Y knockout by targeted gene replacement and gene complementation. (a) Organization of the Vd424Y locus before and after homologous recombination in the wild type Vd991. (b, c) PCR analysis of wild type Vd991 and respective mutants. The genomic DNA of each strain was used to verify the targeted and HPH genes
FIGURE S9 The phylogeny of Vd424Y and its homologous sequences from selected species. Bootstrap percentage support for each branch is indicated. The scale bar represents 50% weighted sequence divergence
FILE S1 The homologs of 34 effectors in Vd Ls. 17
TABLE S1Verticillium dahliae Vd genes screened for targeting cell death in this study
TABLE S2 Microbial strains and plasmids used in this study
TABLE S3 Primers used in this study
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.


