Abstract
Continuous exposure of the skin to environmental, mechanical and chemical stress necessitates constant self‐renewal of the epidermis to maintain its barrier function. This self‐renewal ability is attributed to epidermal stem cells (EPSCs), which are long‐lived, multipotent cells located in the basal layer of the epidermis. Epidermal homeostasis – coordinated proliferation and differentiation of EPSCs – relies on fine‐tuned adaptations in gene expression which in turn are tightly associated with specific epigenetic signatures and metabolic requirements. In this review, we will briefly summarize basic concepts of EPSC biology and epigenetic regulation with relevance to epidermal homeostasis. We will highlight the intricate interplay between mitochondrial energy metabolism and epigenetic events – including miRNA‐mediated mechanisms – and discuss how the loss of epigenetic regulation and epidermal homeostasis manifests in skin disease. Discussion of inherited epidermolysis bullosa (EB) and disorders of cornification will focus on evidence for epigenetic deregulation and failure in epidermal homeostasis, including stem cell exhaustion and signs of premature ageing. We reason that the epigenetic and metabolic component of epidermal homeostasis is significant and warrants close attention. Charting epigenetic and metabolic complexities also represents an important step in the development of future systemic interventions aimed at restoring epidermal homeostasis and ameliorating disease burden in severe skin conditions.
Keywords: epidermal stem cells, epidermolysis bullosa, epigenetics, keratinocytes, miRNAs, mitochondria
1. INTRODUCTION
The skin has a very high cellular turnover rate that, through epigenetic and transcriptional re‐programming, swiftly adapts to environmental stressors such as wounding and barrier disruption. Epidermal homeostasis – the balance between proliferation, differentiation and loss of cells in the stratified epithelium of the skin – sustains tissue integrity and function.1, 2 Within the skin, epidermal stem cells (EPSCs) reside in the basal layer of the epidermis where they are attached to the basal membrane, which separates the epidermis from the underlying dermis. As stem cells differentiate, they move upward through the different layers of the epidermis towards the surface of the skin.1
The rate by which adult epidermal stem cells renew themselves and yield daughter cells depends on developmental stage, external injury, steady‐state tissue turnover and remodelling. Several models of epidermal differentiation and regeneration have been posited to explain the nature and behaviour of EPSCs located within the basal layer of the epidermis.3, 4 The hierarchical model of epidermal homeostasis proposes the existence of a limited number of slow‐cycling long‐term stem cells within the basal layer that self‐renew and give rise to fast‐cycling transit‐amplifying cells.5 According to the stochastic model, on the other hand, all basal cells have equal potential to either divide or directly differentiate.3, 6 The existence of slow‐ and fast‐cycling stem cells that occupy spatially distinct skin regions and are capable of producing unique differentiated lineages suggests yet another possibility.7 Recent data in human 3D cultures suggest that there is a striking variety of signalling processes in the basal layers of the epidermis despite the relatively stable architecture of the terminally differentiated layers.8 Which of the different models of stem cell differentiation and regeneration most accurately describes EPSC behaviour in vivo is still a subject of ongoing research.9, 10 Combining cell labelling and linage tracing experiments Piedrafita et al. found compelling evidence for the stochastic model of epidermal homeostasis. Their data suggest a state of neutral clonal competition where a population of cells with balanced stochastic cell fate generates, on average, one proliferating and one differentiating daughter cell.10 Consistent with earlier reports on grafting experiments in immune compromised mice,11 clones develop into widely varying sizes and arise from any point in the basal layer. Importantly, it seems likely that EPSC behaviour in animal models only partially recapitulates the situation observed in the human epidermis. Moreover, deliberate in vivo lineage tracing in humans is not feasible. Nonetheless, recent epidermal grafting studies12 provided important mechanistic understanding of epidermal regeneration in humans (discussed below).
Through control of gene expression and homeostasis, aspects of the epigenome regulate almost every biological process, from cellular differentiation and maintenance of phenotypes to onset of disease and ageing.13, 14 Epigenetic mechanisms such as DNA methylation, histone tail modifications, chromatin accessibility and changes in DNA architecture are tightly correlated with normal cellular function, while their dysregulation manifests in aberrant gene expression and disease.15 According to a contemporary definition, epigenomics is defined as “the study of molecules and mechanisms that can perpetuate alternative gene activity states in the context of the same DNA sequence”.14 Because of their essential role in establishing specific transcriptional configurations, epigenetic mechanisms govern many aspects of EPSC proliferation, as well as differentiation of their descendants.16, 17, 18, 19 Uncharacteristic epigenetic modifications often associate with a loss of transcriptional fidelity, unchecked proliferation, de‐differentiation, and malignant epidermal to mesenchymal transition.20 At the same time, pronounced changes in the epigenetic landscape often accompany, and are critical for, resolving challenges to epidermal homeostasis induced by changes in the local microenvironment or external stimuli, such as injury.21
Ageing and a variety of diseases, such as chronic inflammation or cancer, manifest themselves in characteristic changes of the epigenetic profile.22, 23, 24 A multitude of other factors, ranging from DNA damage to dietary‐ or drug‐induced metabolic changes, are known to affect the epigenetic status as well. For example, exposure to high altitude,25, 26 cancer‐associated elevated concentration of lactate,27 and increased uptake of dietary methionine28, 29 have been linked to epigenetic changes and thus highlight the intricate connection between metabolic events and alterations in the epigenome. The pivotal role of mitochondrial energy metabolism in regulating epigenetic events and epidermal homeostasis will be discussed below.
The epigenome is structured into distinct, but interconnected layers ranging from overall chromatin structure and organization to specific histone and DNA modifications. Histones are predominantly modified by methylation, acetylation and phosphorylation, but they can be adapted by other modifications such as ubiquitination, sumoylation, ribosylation and citrullination.30 While DNA methylation is the most prominent and studied epigenetic modification (see Box 1|Mechanisms of DNA methylation), other aspects of the epigenome include RNA methylation,31 and the expression of coding and non‐coding RNAs, most notable the expression of microRNAs (miRNAs, discussed below).32 In recent years, research in epigenomics has been enormously propelled by a multitude of large consortia including the NIH Roadmap Epigenomics Mapping Consortium,33, 34 International Human Epigenome Consortium,35 ENCODE project,36, 37, 38 the Genotype‐Tissue Expression (GTEx) project,39, 40 the Human Biomolecular Atlas Program (HuBMAP)41 or the 4D nucleome project.42 The EWAS data hub – comprising normalized DNA methylation array data from 75 K samples – is now available for epigenome‐wide association studies (EWAS).43
BOX 1. Mechanisms of DNA methylation.
DNA methylation is a biochemical process denoted by the addition of a methyl group to cytosines in DNA. Cytosine is methylated at the 5’ position of the pyrimidine ring to form 5‐methylcytosine (5mC). In mammals, DNA methylation almost exclusively occurs in CpG dinucleotides, with the cytosines on both strands being methylated. The human genome contains 56 million CpG sites of which about 70–80% are methylated.44 Methylated CpGs are predominantly associated with repetitive elements. Clusters of unmethylated CpG sites – so‐called CpG islands (CGIs) – are associated with promoter and enhancer regions. Importantly, most cell types display relatively stable DNA methylation patterns and dynamic regulation occurs for only about 20% of autosomal CpGs.45 These CpGs participate in the genomic regulation of key lineage‐specific factors. Cell‐, tissue‐ and condition‐specific differences in methylation define so‐called differentially methylated regions (DMR).44 Hypermethylation generally refers to an increase in methylation and can be found in regions where most cytosines are methylated, like in heterochromatin. Hypomethylation denotes a loss of methylation. Regions where most cytosines are non‐methylated are found in euchromatin and active gene promoters. Cancers generally exhibit global hypomethylation, whereas regional hypermethylation in the promoter regions of tumour suppressor genes is frequently observed.
Methyl groups are added to cytosines by DNA methyltransferase (DNMTs). DNMT1 predominantly methylates hemimethylated CpGs and therefore is crucial for maintaining methylation during DNA replication.46 Although DNMT1 displays a very high fidelity, there is an inevitable global loss of methylation with each cell division. DNMT3a and DNMT3b are de novo methyltransferases that can methylate both unmethylated and hemimethylated DNA, and orchestrate the establishment of DNA methylation patterns early in development.47 Conversely, TET (Ten eleven translocation) enzymes actively remove methyl groups from DNA by oxidation with the production of 5‐hydroxymethylation as an intermediate.48
2. EPIDERMAL HOMEOSTASIS IN HEALTH AND DISEASE
2.1. Epigenetic regulation of epidermal homeostasis
There is ample evidence that epigenetic mechanisms, such as DNA methylation, histone modifications or changes in DNA topology, contribute to epidermal homeostasis and differentiation49, 50, 51, 52 (summarized in Figure 1 and Table 1). Epigenetic regulation has also been analysed in wound healing and functional links between chromatin architecture and gene expression in keratinocytes have been found.53, 54, 55, 56
FIGURE 1.
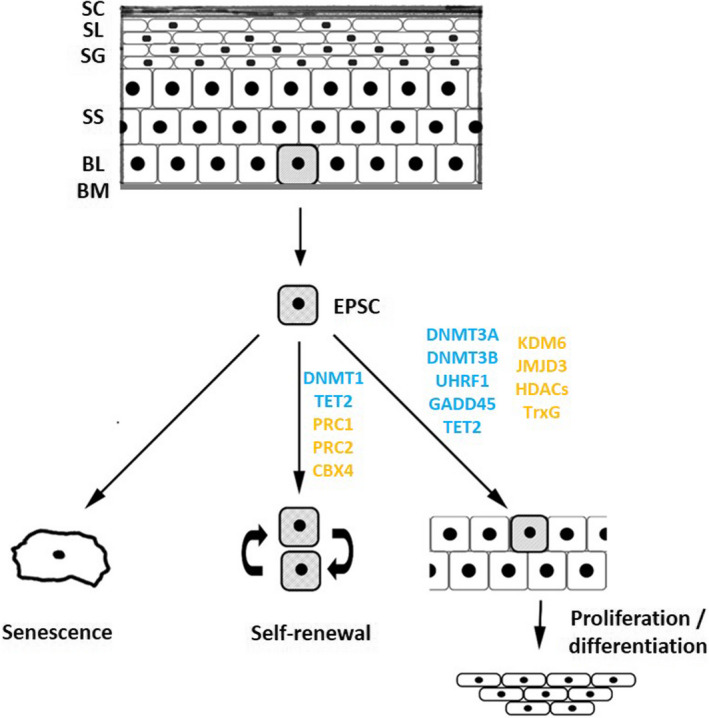
Epigenetic effectors and epidermal homeostasis. Maintenance and differentiation of EPSCs critically governs epidermal homeostasis. Individual, proliferating EPSCs (indicated as shaded cells) are located in the basal layer. As cells differentiate, they progressively move upward through the various layers of the epidermis. Eventually, these cells lose their nuclei before forming the layers of the outermost stratum corneum. Multiple epigenetic effectors regulate EPSC self‐renewal, proliferation and differentiation. These factors control DNA methylation (indicated in blue), histone modification and chromatin remodelling (indicated in orange). Abbreviations: BL, basal layer; BM, basement membrane; DNMT, DNA methyltransferase; EPSC, epidermal stem cell; HDAC, histone deacetylases; PRC, polycomb repressive complex; SC, stratum corneum; SG, stratum granulosum; SL, stratum lucidum; SS, stratum spinosum; TET, ten‐eleven translocation; TrxG, trithorax group proteins
TABLE 1.
Epigenetic factors in skin homeostasis
| Epigenetic effector | Main activity | Major phenotypes of loss of function | Reference |
|---|---|---|---|
| DNA modifiers | |||
| DNMT1 | Maintains methylation of CpGs (‘maintenance DNMT’) | Defects in EPSC maintenance and proliferation; disrupted epidermal stratification and hair follicle development; development of alopecia | 46, 153, 154, 155 |
| DNMT3A | De novo methylation of CpGs (‘de novo DNMT’) | Defects in EPSC differentiation; Cutaneous tumourigenesis; squamous transformation; skin ageing | 59, 60, 156, 157 |
| DNMT3B | De novo methylation of CpGs (‘de novo DNMT’) | Defects in EPSC differentiation; squamous transformation; skin ageing | 59, 60, 157 |
| TET1 | Demethylation of CpGs | Dysregulated EPSC kinetics | 158 |
| TET2 | Demethylation of CpGs | Dysregulated EPSC kinetics; defects in EPSC proliferation and migration; Skin ageing | 59, 157, 158, 159 |
| UHRF1 | Co‐factor, binds hemi‐methylated DNA and recruits DNMT1 | Defects in epidermal differentiation | 46, 160 |
| Gadd45A/B | Co‐factor, involved in DNA demethylation | Defects in epidermal differentiation | 46 |
| Histone modifiers | |||
| KDM6B | H3K27 demethylase | Epidermal differentiation | 161 |
| JMJD3 | H3K27 demethylase | Delayed wound healing | 162 |
| Histone demethylases | hypomethylation of histone H3K4/9/27me3 | impaired epithelial cell differentiation | 50 |
| HDAC1/2 | Suppression of gene expression | Decrease SC proliferation, impaired stratification, alopecia | 163, 164 |
| Trichostatin‐A | HDAC inhibitor | HFSC, IFE proliferation, block of terminal differentiation | 165, 166 |
| Chromatin remodelers | |||
| PRC1 | Suppression of gene expression | Defects in EPSC differentiation | 167, 168, 169, 170 |
| PRC2 | Suppression of gene expression | Defects in EPSC differentiation | 167, 171 |
| BMI1 | Component of PRC1, mediates monoubiquitination of H2AK119 | EPSC maintenance and proliferation | 172 |
| CBX4 | Component of PRC1, mediates monoubiquitination of H2AK119 | EPSC maintenance and proliferation | 168 |
| EZH1 | Component of PRC2, catalyzes methylation of H3 K27 | EPSC maintenance and proliferation | 173 |
| EZH2 | Component of PRC2, catalyzes methylation of H3 K9 and H3 K27 | EPSC maintenance and proliferation | 167, 173 |
| SUZ12 | Component of PRC2, catalyzes methylation of H3 K9 and H3 K27 | EPSC maintenance | 171 |
| TrxG | Activation of gene expression | Defects in EPSC differentiation | 174 |
Abreviations: DNMT, DNA methyltransferase; HDAC, Histone deacetylases; PRC, polycomb repressive complex; TET, Ten‐eleven translocation; TrxG, trithorax group proteins
Disrupted chromatin regulation, prompted by the loss of PRC1, results in impaired epidermal tissue integrity and blistering skin resembling human skin fragility syndromes.49 In regards to histone modifications, chemical inhibition of histone demethylases impairs differentiation of inter‐follicular stem cells and delays injury repair.50 Chronic sun exposure is associated with distinct histone acetylation changes and altered gene expression in human photodamaged skin.57 Histone acetyltransferase (HAT) activity is dependent on zinc and depletion of zinc results in decreased HAT activity. The epithelial zinc transporter ZIP10 epigenetically regulates human epidermal homeostasis by modulating zinc availability and histone acetyltransferase activity.58 Reduced ZIP10 activity or depletion of zinc leads to reduced HAT activity and decreased expression of genes, such as filaggrin or metallothionein, involved in epidermal homeostasis.58
Likewise, dynamic epigenetic regulation of DNA methylation (see Box 1|Mechanisms of DNA methylation) is critical for the maintenance of EPSC status and proliferative capacity. A progressive loss of DNA methylation patterns caused by forced depletion of DNMT1 in the epidermis leads to failure of EPSC self‐renewal and tissue regeneration.46 Consistently, DNMT1 expression is normally restricted to the basal layers of the epidermis containing the EPSC population, and mostly absent in the outer differentiated layers. The de novo DNMTs, DNMT3A and DNMT3B, also critically contribute to EPSC homeostasis by controlling enhancer methylation and active chromatin conformation of stem cell relevant genes.59 Specifically, co‐localization of DNMT3A and TET‐2 at target enhancers results in 5‐hmC formation and gene activation.59 Interestingly, DNMT3A and DNMT3B also seem to protect the epidermis from tumourigenesis since the loss of these genes in the mouse epidermis promotes squamous transformation.60 In atopic dermatitis, DNA methylation patterns from patients differ significantly from those of healthy controls.61, 62, 63 Moreover, epigenetic dysregulation caused by diminished TET‐1 and TET‐2 expression and concomitant reduction of 5‐hmC marks leads to unbalanced EPSCs proliferation and maturation in psoriasis.64 Although the cause of diminished TET expression in psoriasis remains unresolved, reconstitution of TET expression increases 5‐hmC levels and results in normalized EPSCs kinetics.
The P16ink4/Rb signalling pathway further highlights the critical role of epigenetic regulation in maintaining epidermal homeostasis. P16ink4 is a potent inhibitor of the G1/S phase transition and therefore a tumour suppressor gene and entry point to cellular senescence. P16ink4 is also crucial for controlling EPSC behaviour and is in turn an important target of multiple epigenetic regulatory processes involving DNMTs, TET enzymes, Polycomb group proteins and Jumanji protein families.65 Interestingly, epigenetic drift or disrupted epigenetic regulation, respectively, have also been linked to loss of epidermal homeostasis in skin ageing and rare skin conditions (see Box 2|Epigenetic drift and skin ageing and Box 3|Epigenetic regulation in disorders of cornification).
BOX 2. Epigenetic drift and skin ageing.
The observation that global DNA methylation marks stochastically change with age, led to the idea that the methylation status of a distinctive and – compared to the entire methylome – narrow set of CpG sites could be used to predict the chronological and biological age of an organism. Thus, global assessment of age‐related DNA methylation changes can be used to configure so‐called epigenetic clocks for highly accurate age prediction. Since the first development of a DNA methylation age estimator, the predictive power of epigenetic clocks has been constantly improving and contemporary epigenetic clocks are considered the most accurate biomarkers of ageing available. In skin ageing, methylation data have been used to predict the chronological age of sample donors with high accuracy. One of the most recent iterations of an epigenetic clock, the Skin&Bood clock, is based on assessing the methylation status of 391 CpG sites and predicts the chronological age of subjects from human fibroblasts, keratinocytes, buccal cells, whole skin, blood and saliva samples with high precision.
Interestingly, accelerated epigenetic ageing is observed in disease and cancer. A hallmark of ageing is the increased cell‐to‐cell variability in epigenetic marks and gene expression. This epigenetic drift invariably leads to a decline in stem cell number and function and entails the onset of age‐associated illnesses. Changes in methylation variability were accompanied by reduced connectivity of transcriptional networks. These findings thus define the loss of epigenetic regulatory fidelity as a key feature of the ageing epigenome.66
BOX 3. Epigenetic regulation in disorders of cornification.
Dominant‐negative mutations in KRT9 cause diffuse palmoplantar keratoderma (PPK), a debilitating genodermatosis for which there is no effective treatment.67 The disease phenotype of PPK is limited to palmoplantar surfaces where KRT9 protein is expressed, while there is little KRT9 expression in other body locations. Notably, previous work has shown that the site‐specific Homeobox protein Hox‐A13 (HOXA13) in fibroblasts can be modulated by Aza‐C, a DNA methylation inhibitor, and implicated the presence of HOXA13‐expressing fibroblasts in palmoplantar skin to be important for site‐specific KRT9 expression via Wnt family member 5A (WNT5A) in these body locations.68
Ichthyosis vulgaris (IV), characterized by generalized dry skin and scaling, is the most common monogenic genodermatosis. It is caused by mutations in the profilaggrin/filaggrin gene (FLG). It is well‐known that IV families also have a high incidence of atopic dermatitis (AD), a common inflammatory skin disease with often severe itching and association with hay fever and asthma. However, it remains unknown why some family members in IV families develop both, IV and AD, and others display IV only. One study reported a lack of correlation between methylation in the FLG gene promoter and allergic phenotypes.69 Conversely, DNA methylation within the FLG gene, specifically within the CpG site ‘cg07548383’ was reported to significantly interact with FLG sequence variants on the risk for eczema,70 although this study did not provide direct evidence of DNA methylation modulating FLG expression. Furthermore, another genome‐wide study revealed differences in DNA‐methylation in lesional AD as compared to healthy control skin.63 In this publication, differences in DNA‐methylation are described for genes that are involved in regulating epidermal homeostasis and innate immunity, i.e. KRT6A OAS2, S100A and LRRC8C, the latter with expression probes in trans with CD36. In turn, CD36 was shown to be increased in states of skin barrier disruptions71 and mutations in CD36 cause ichthyosis prematurity syndrome72 with skin barrier abnormalities and disturbances in epidermal lipid metabolism.73 In contrast, the level of demethylation of FOX3i1 in circulating regulatory T cells (Tregs) is similar between AD and control subjects.74 However, the demethylation of the FCER1G promoter in monocytes75 and the TSLP promoter in keratinocytes showed differences.76 Thus, current knowledge implies epigenetic regulation of epidermal homeostasis in AD and may account for the association between IV and AD.
2.2. The role of miRNAs in epidermal homeostasis
The overall contribution of miRNAs to skin homeostasis was demonstrated in functional studies in which conditional epidermal knockout of key elements of the miRNA processing machinery in murine embryos, namely Dicer77, 78 and Dgcr8,79 resulted in a severe skin phenotype, characterized by follicular dysplasia, epidermal hyperproliferation, and defects in barrier function, accompanied by a failure to thrive and early postnatal lethality. Numerous miRNAs have now been assigned specific roles in skin morphogenesis, homeostasis and tissue regeneration (as reviewed in80, 81, 82). Under normal physiological circumstances, miRNAs are predicted to mediate the post‐transcriptional control of up to 60% of all expressed genes.32 Additionally, they are intricately interconnected in epigenetic networks. Their expression can be affected by the classic epigenetic modifications of promoter DNA methylation and histone acetylation, and they themselves can control the epigenetic machinery by directly targeting individual enzymatic components.83, 84 Moreover, reports of miRNAs co‐localizing to specific promoter regions as components of different DNA‐binding complexes indicate potentially active roles in chromatin remodelling.84 This places miRNAs at the core of epigenetic/miRNA regulatory circuits that can significantly impact a plethora of cell functions.85
MiRNome profiling, coupled with functional validation of candidates, continues to drive our understanding of miRNA regulation of prominent skin processes, both in health and disease contexts (see Table 2). In the context of epidermolysis bullosa (EB) – a rare genetic disorder of the skin discussed in more detail below – the role of miRNAs in disease pathogenesis is beginning to surface. To date, three miRNAs have been described to modulate EB‐associated complications such as fibrosis (miR‐29b,86, 87 miR‐14588, 89) and cancer (miR‐10b90). The repetitive destabilization of the extracellular matrix that accompanies recessive dystrophic EB (RDEB) upon injury results in progressive soft tissue fibrosis with debilitating consequences, such as tumour development.91 miR‐145‐5p was shown to be upregulated in RDEB‐fibroblasts, which typically exhibit more contractile features than their wild type counterparts, indicating a potential correlation between RDEB severity and miR‐145‐5p levels, by contributing to skin fibrosis.88 Indeed, inhibition of miR‐145‐5p resulted in a downregulation of α‐SMA, TAGLN and JAG1, all of which are contractile markers, leading to a reduction of fibrotic traits.88 Another miRNA, miR‐29, which directly targets the disease‐causing gene COL7A1, as well as the essential COL7A1 expression regulator SP1, was found to be downregulated in RDEB fibroblasts.87 Furthermore, in a complex network, TGF‐ß was shown to be a further activator of COL7A1 expression and at the same time reduces miR‐29 levels via SMAD phosphorylation.87, 92, 93 Apart from COL7A1 regulation, miR‐29 family members were also shown to influence DNA methylation by targeting distinct DNA methyl transferases94 and proteins involved in DNA demethylation.95
TABLE 2.
Selected microRNAs with function in skin biology
| MicroRNA | Function | Reference |
|---|---|---|
| Homeostasis and morphogenesis | ||
| miR‐34a | Induces differentiation. Anti‐proliferative function | 175 |
| miR‐34c | Suppresses differentiation, involved in senescence | 176 |
| miR‐125b | Represses stem cell differentiation and promotes stem cell renewal | 177 |
| miR‐184 | KC differentiation | 178 |
| miR‐203 | Repressor of p63. Regulator of keratinocyte differentiation | 179 |
| miR‐205 | Enhances migration | 180 |
| miR‐210 | Pro‐angiogenic. Cell‐cycle regulation. Hypoxa‐miR | 181 |
| Epigenetic targets | ||
| miR‐29 family | DNMT3A‐B, indirectly DNMT1 | 182 |
| miR‐145 | HDAC2 | 183 |
| miR‐200b | PCGF4 | 184 |
| miR‐200c | PCGF4 | 185 |
| miR‐221 | HDAC6 | 186 |
| Cancer | ||
| miR‐10b | Confers of stemness features to tumour cells in cSCCs | 90 |
| miR‐21 | OncomiR, anti‐apoptotic, pro‐survival | 187, 188 |
| miR‐27a | Targets EGFR | 189 |
| miR‐34a | Tumour‐suppressor miRNA, targets HMGB1 | 190 |
| Ageing | ||
| miR‐146a | Involved in fibroblast senescence via regulation of Smad4 | 191 |
| miR‐181a,b | Involved in senescence in keratinocytes and fibroblasts | 192, 193 |
| Fibrosis | ||
| miR‐21 | Fibroblast proliferation and transdifferentiation | 194 |
| miR‐29b | Regulator of collagen expression | 86, 87 |
| miR‐145 | Regulation of myofibroblast differentiation | 88, 89 |
Abbreviations: HDAC, Histone deacetylase; PCGF4, Polycomb group RING finger protein 4.
Patients suffering from RDEB are particularly prone to developing exceptionally aggressive squamous cell carcinomas (SCCs). In this context, overexpression of miR‐10b has been attributed a role in conferring stemness to tumour cells, specifically by increasing cell adhesion in 2D and 3D functional models. While miR‐10b is the first miRNA described to be associated with RDEB‐SCCs,90 the role of miRNAs in tumourigenesis is generally well‐accepted, and has been described for several tumour entities, among them cutaneous SCCs, affecting diverse mechanisms like migration and proliferation.96, 97, 98
2.3. Mitochondrial control of epidermal homeostasis
Emerging evidence suggests that mitochondria are vital regulators of skin physiology.99 Epidermal progenitor/stem cells do not rely on the mitochondrial respiratory chain, but still require a functional dynamic mitochondrial compartment.100 One main task of keratinocytes is corneocyte renewal and production of stratum corneum‐specific proteins and lipids needed for a functional skin barrier. These processes require high amounts of energy, which is normally generated by oxidative phosphorylation (OXPHOS). During differentiation of keratinocytes in the skin, mitochondrial membrane potential declines and mitochondria undergo phenotypic changes in an apoptosis‐like process.101, 102, 103 A decline in mitochondrial energy production in favour of glycolysis might contribute to the production of lactate in the stratum corneum. Lactate production of keratinocytes is important to skin barrier function as well as the maintenance of skin flexibility.104
Mitochondria are the major intracellular source of reactive oxygen species (ROS), predominantly generated via complex I and III of the OXPHOS system.105 ROS can inflict oxidative damage on biomolecules, resulting in loss of catalytic and/or structural integrity. With ageing, ROS‐damaged proteins accumulate and OXPHOS activity declines. Accordingly, mitochondrial oxidative stress limits epidermal cell proliferation and stem cell numbers, leading to reduced wound healing in older mice. Interestingly, in young mice, mitochondrial oxidative stress actually accelerates wound healing.106 Both naive and differentiating progenitor stem cells (PSCs) activate OXPHOS, whereas primed PSCs rely on glycolysis.
Several mitochondriopathies are associated with skin manifestations, including hair abnormalities, rashes, pigmentation disorders, hypertrichosis and acrocyanosis.99 Cytochrome oxidase (complex IV of OXPHOS) activity is greatly reduced in allergic contact dermatitis and ichthyosis, indicating diminished aerobic respiration.107 Mutations in the plectin 1 (PLEC1) gene cause epidermolysis bullosa simplex (EBS) with muscular dystrophy (EBS‐MD).108 PLEC1B, which localizes in the outer mitochondrial membrane, helps to maintain organelle shape and network formation by tethering mitochondria to intermediate filaments.109 PLEC1‐deficient cells show a disorganized intermediate filament network and severe mitochondrial dysfunction.110, 111 Furthermore, in keratinocytes of patients with EBS caused by a mutation of keratin (KRT) 5 or KRT14, abnormal mitochondrial distribution has been reported.112
Interpreting the crosstalk between the nuclear epigenome and mitochondria in both normal physiological function and different diseases is an advancing research topic.113, 114, 115, 116 The field distinguishes between anterograde (nucleus to mitochondria) and retrograde (mitochondria to nucleus) signalling. Both communicate intracellular requirements or a need to compensate for a dysfunction to maintain epidermal homeostasis. Pertaining to anterograde signalling, growing evidence suggests that nuclear regulators, including transcription factors, DNMTs and TET demethylases, as well as non‐coding RNAs may be exported from the nucleus and directly impact transcription of the mitochondrial genome.116 Moreover, a recently discovered link between toxin‐induced promoter hypomethylation and mitochondrial biogenesis in skin cancer development further highlights the importance of coordinated epigenetic regulation and mitochondrial function.117 When it comes to retrograde signalling, an increasing number of studies have identified potential alterations in the epigenetic landscape of the nuclear genome as a consequence of mitochondrial dysfunction.114 For example, depletion of mtDNA results in significant changes in methylation pattern of a number of genes. The methylation changes are reversed by the restoration of mtDNA.118 In addition, numerous mitochondrially‐derived metabolites serve as regulators or substrates for epigenetic marks, e.g. S‐adenosylmethionine (SAM) is required as a substrate for methylation of many histone proteins but also DNA‐methylation.119
2.4. Epidermal homeostasis in epidermolysis bullosa
Recently, our understanding of EPSC biology and epidermal homeostasis has been fuelled by advances in treating rare skin conditions such as inherited epidermolysis bullosa (EB), caused by loss of adhesion and cohesion of the skin. EB manifests itself as a wide spectrum of clinically heterogeneous phenotypes. The type of mutated gene, position and nature of the mutation within the respective gene, as well as mode of inheritance, predict the particular subtype of EB.120 Phenotypic variability amongst patients with the same or similar mutations, however, remains often unexplained. Frequently, the same mutation results in intra‐ and interfamilial disease variability. Siblings with the same mutation in COL7A1, for example, can present with different clinical phenotypes. Even monozygotic twins can show pronounced phenotypic variation for diverse traits, including disease susceptibility and progression. In other rare diseases, discordant phenotypes between monozygotic twins can often be attributed to different epigenetic states and aberrant epigenetic regulation.121 Epigenetic modifications in EB remain underexplored at large and inference of the importance of epigenetic mechanisms in influencing disease progression is largely circumstantial. For now, the only published disease modifiers in EB include genes associated with TGF‐β pathway inhibition122 and members of the matrix metalloproteinase family (MMP‐1),123, 124 although the importance of MMP‐1 as a modifier gene remains unclear.125, 126 In a mouse model of junctional EB (JEB), featuring a hypomorphic mutation in Lamc2, Col17a1 acts as a strong disease modifier.127
Through the application of combined ex vivo cell and gene therapy, almost the entire epidermis of an EB patient can be reconstituted by genetically corrected long‐lived EPSCs.12, 128, 129 In a series of therapeutic skin transplantations, we discovered that, apart from technical issues, the outcome of the procedure depends on the anatomical site of the initial biopsy, the age of the patient, the genes involved, and, perhaps more importantly, on the microenvironment characterizing the receiving wound bed. The contribution of age is common knowledge in the field and has also been observed by us128 in the case of a 49‐year‐old patient vs. a 7‐year‐old patient.12 The intrinsic ageing processes of the skin have been revealed to depend on cytoskeletal proteins (e.g. keratins; cytoskeletal proteins including desmosomes, microtubules and microfilaments)130 and other cellular processes, like cell cycle control, inflammatory response, signalling and metabolism.131, 132, 133 Moreover, EB per se is a disease not only of skin attachment, but it also displays an ageing phenotype exemplified by a specific gene expression signature.134
In the case of JEB, this has been related to dysregulation of the YAP/TAZ pathway, which causes progressive, age‐related depletion of stem cells.135 We provided evidence that the reduction of clonogenic potential and the loss of stem cells in primary JEB keratinocytes is associated with perturbation of the YAP/TAZ signalling which renders ex vivo gene therapy cumbersome.135 The Hippo signalling pathway, better known for its function in organ size control through its effectors Yes‐associated protein (YAP) and WW domain‐containing transcription regulator 1 (commonly listed as TAZ), has been demonstrated to play a pivotal role in regulating tissue homeostasis and regeneration in skin.135, 136, 137, 138, 139 The transcriptional regulators YAP and TAZ localize to the nucleus in the basal layer of human and mouse epidermis135, 139, 140 and are elevated during wound healing.136, 137, 138, 139 Skin specific deletion of both YAP and TAZ in adult mice leads to hair loss and impairs regeneration after wounding.136 YAP expression correlates with stem cell content and it has been reported that nuclear YAP progressively declines with age and correlates with the proliferative potential of epidermal progenitors.135, 139
Compared to those derived from healthy donors, EPSCs from EB patients are often difficult to culture ex vivo. Repeated wounding and sustained proliferative stress may contribute to decreased plasticity and increased exhaustion of EPSCs in EB patients. There are distinct differences in clonogenic ability and proliferation potential in LAMB3‐ and COL7‐deficient keratinocytes. In LAMB3‐deficient keratinocytes, both properties are severely altered, but they can be rescued by transduction with a LAMB3‐expressing vector (Figure 2). This does not hold true for COL7‐ or COL17‐deficient keratinocytes, which have a proliferative potential similar to that of normal keratinocytes. Therefore, competition between untransduced vs. transduced patient keratinocytes might occur in transplanted areas of dystrophic EB patients, hampering full therapeutic success (M De Luca, JW Bauer, unpublished observation). The cell‐ and molecular‐biological reasons for this constellation have been only partially elucidated.135
FIGURE 2.
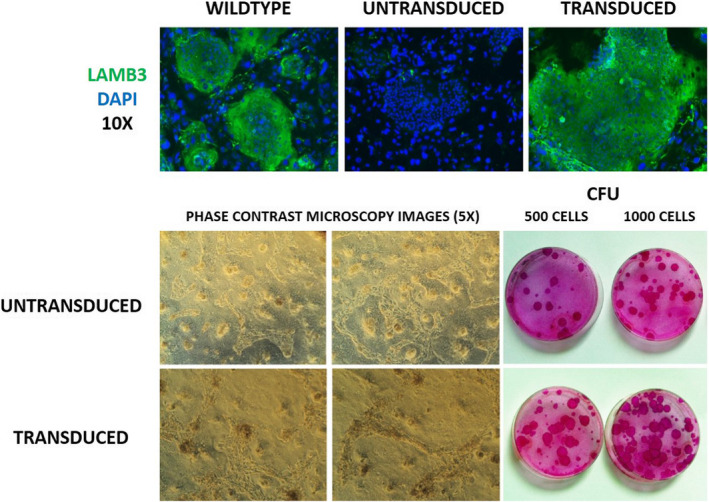
Proliferation potential of primary keratinocytes from EB patients. Keratinocytes from a 49‐year‐old JEB patient were transfected to re‐express LAMB3 protein. Upper panel: LAMB3‐deficient patient keratinocytes in cell culture. After transfection, there is improved clonal potency as can be seen by the increased number and size of red‐stained clones in the lower panel (M. De Luca et al, unpublished results). CFE: Colony‐forming units
Most likely, characteristic transcriptional and epigenetic anomalies beyond the causal EB mutation promote the observed differences in proliferative capacities of EPSCs. In general, proper interaction of EPSCs with the basement membrane assures their maintenance and propagation. LAMA3, LAMB3 and LAMC2 encode subunits of laminin‐332, which is crucial for anchoring epithelial cells to the basement membrane. Reduced or absent expression of functional laminin‐332, caused by mutations in the corresponding genes, accounts for the majority of cases with JEB. Beyond its structural role, Laminin‐332 influences EPSC differentiation and its absence in JEB leads to stem cell depletion.12, 128, 135, 141 Aberrant laminin‐332 expression has also been connected to tumour progression. LAMB3 features a CpG poor promoter region. How the methylation status of non‐CpG island promoters affects gene expression is generally not well defined. Correspondingly, the influence of LAMB3 promoter methylation on gene expression is somewhat ambiguous. Epigenetic silencing of the LAMB3 gene has been linked to certain cancers142, 143, 144 and resistance to cisplatin treatment.145 A different study, however, found promoter hypomethylation and up‐regulated expression of LAMB3 in gastric cancer.146 To our knowledge, however, there are currently no published reports on prospective epigenetic differences of EPSCs from EB patients and age‐matched healthy donors.
In light of high somatic mutation rates, stem cell competition further appears to be an important factor in maintaining tissue homeostasis by keeping propagation of stem cell clones with cancer‐causing mutations and abnormal cellular behaviour in check.147 At the same time, stem cell competition may also account for the phenomenon of revertant phenotypes in JEB caused by inherited mutations in COL17A1,148, 149, 150 and ichthyosis with confetti caused by mutations in KRT1 or 10.151, 152
3. CONCLUSION
The contribution of epigenetic, miRNA‐mediated and mitochondrial events during epidermal homeostasis in health and disease is profound. Substantial progress has been made in understanding the underlying molecular details. Even so and despite the success of progressive treatment options, like ex vivo stem cell/‐gene therapy, our understanding of several aspects of basic skin biology is still incomplete. It is clear that the regulation of self‐renewal and proliferative potential of EPSCs is strongly determined by their genetic background, epigenetic signatures, metabolic state and the tissue microenvironment. The combination of these factors may impinge on the clinical outcome of advanced stem cell therapies. Since genetic material is often integrated into the genome of the patient during the procedure, it should be feasible to include supplemental components for manipulating these factors and thus improve the chances for therapeutic success. At the same time, continuing research and a deeper mechanistic understanding of skin homeostasis will likely reveal novel avenues for therapeutics and regenerative medicine in the field of genodermatoses.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
RNW, JWB conceptualized the manuscript. RNW, JPH, VW, BK, MS, LDR, MDL, JWB performed literature review and wrote different paragraphs. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
We thank Dr. Rudolf Hametner for his help in preparing Figure 1.
Funding information
Helga Kellerhals Foundation (to RNW, JWB), DEBRA Austria (to JPH, VW), Austrian Science Fund I4229, Hans Gröber Foundation, Walter Schaar Foundation (to MS).
REFERENCES
- 1.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10(3):207‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge Y, Fuchs E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet. 2018;19(5):311‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rompolas P, Mesa KR, Kawaguchi K, et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science. 2016;352(6292):1471‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez‐Celeiro M, Zhang B, Hsu YC. Fate by chance, not by choice: epidermal stem cells go live. Cell Stem Cell. 2016;19(1):8‐10. [DOI] [PubMed] [Google Scholar]
- 5.Mascre G, Dekoninck S, Drogat B, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489(7415):257‐262. [DOI] [PubMed] [Google Scholar]
- 6.Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446(7132):185‐189. [DOI] [PubMed] [Google Scholar]
- 7.Sada A, Jacob F, Leung E, et al. Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol. 2016;18(6):619‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roshan A, Murai K, Fowler J, Simons BD, Nikolaidou‐Neokosmidou V, Jones PH. Human keratinocytes have two interconvertible modes of proliferation. Nat Cell Biol. 2016;18(2):145‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dekoninck S, Hannezo E, Sifrim A, et al. Defining the design principles of skin epidermis postnatal growth. Cell. 2020;181(3):604‐620 e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piedrafita G, Kostiou V, Wabik A, et al. A single‐progenitor model as the unifying paradigm of epidermal and esophageal epithelial maintenance in mice. Nat Commun. 2020;11(1):1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghazizadeh S, Taichman LB. Organization of stem cells and their progeny in human epidermis. J Invest Dermatol. 2005;124(2):367‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch T, Rothoeft T, Teig N, et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551(7680):327‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487‐500. [DOI] [PubMed] [Google Scholar]
- 14.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571(7766):489‐499. [DOI] [PubMed] [Google Scholar]
- 15.Holtzman L, Gersbach CA. Editing the epigenome: reshaping the genomic landscape. Ann Rev Genom Hum Genet. 2018;19:43‐71. [DOI] [PubMed] [Google Scholar]
- 16.Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY. Epigenetic regulation of gene expression in keratinocytes. J Invest Dermatol. 2012;132(11):2505‐2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohler F, Rodriguez‐Paredes M. DNA methylation in epidermal differentiation, aging, and cancer. J Invest Dermatol. 2020;140(1):38‐47. [DOI] [PubMed] [Google Scholar]
- 18.Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E. Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med. 2014;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang S, Chovatiya G, Tumbar T. Epigenetic control in skin development, homeostasis and injury repair. Exp Dermatol. 2019;28(4):453‐463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latil M, Nassar D, Beck B, et al. Cell‐type‐specific chromatin states differentially prime squamous cell carcinoma tumor‐initiating cells for epithelial to mesenchymal transition. Cell Stem Cell. 2017;20(2):191‐204.e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miroshnikova YA, Cohen I, Ezhkova E, Wickstrom SA. Epigenetic gene regulation, chromatin structure, and force‐induced chromatin remodelling in epidermal development and homeostasis. Curr Opin Genet Dev. 2019;55:46‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field AE, Robertson NA, Wang T, Havas A, Ideker T, Adams PD. DNA methylation clocks in aging: categories, causes, and consequences. Mol Cell. 2018;71(6):882‐895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gronniger E, Weber B, Heil O, et al. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010;6(5):e1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childebayeva A, Harman T, Weinstein J, et al. DNA methylation changes are associated with an incremental ascent to high altitude. Front Genet. 2019;10:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Childebayeva A, Jones TR, Goodrich JM, et al. LINE‐1 and EPAS1 DNA methylation associations with high‐altitude exposure. Epigenetics. 2019;14(1):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, Tang Z, Huang H, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575‐580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Yip LY, Lee JHJ, et al. Methionine is a metabolic dependency of tumor‐initiating cells. Nat Med. 2019;25(5):825‐837. [DOI] [PubMed] [Google Scholar]
- 29.Gao X, Sanderson SM, Dai Z, et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature. 2019;572(7769):397‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harbor Perspect Biol. 2015;7(9):a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature. 2012;485(7397):201‐206. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 2010;28(10):1045‐1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roadmap Epigenomics C, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stunnenberg HG, International Human Epigenome C , Hirst M. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167(5):1145‐1149. [DOI] [PubMed] [Google Scholar]
- 36.Consortium EP . The ENCODE (ENCyclopedia of DNA elements) Project. Science. 2004;306(5696):636‐640. [DOI] [PubMed] [Google Scholar]
- 37.Consortium EP . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Consortium EP, Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799‐816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium GT . Human genomics. The Genotype‐Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648‐660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium GT , Laboratory DA , Coordinating Center ‐Analysis Working G . Coordinating Center ‐Analysis Working G, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu BC. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature. 2019;574(7777):187‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dekker J, Belmont AS, Guttman M, et al. The 4D nucleome project. Nature. 2017;549(7671):219‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong Z, Li M, Yang F, et al. EWAS Data Hub: a resource of DNA methylation array data and metadata. Nucleic Acids Res. 2020;48(D1):D890‐D895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziller MJ, Gu H, Muller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477‐481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self‐renewing somatic tissue. Nature. 2010;463(7280):563‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang ZM, Lu R, Wang P, et al. Structural basis for DNMT3A‐mediated de novo DNA methylation. Nature. 2018;554(7692):387‐391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5‐methylcytosine to 5‐hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930‐935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen I, Zhao D, Menon G, et al. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev. 2019;33(1–2):55‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang S, Long K, Wang S, Sada A, Tumbar T. Histone H3 K4/9/27 trimethylation levels affect wound healing and stem cell dynamics in adult skin. Stem cell reports. 2020;14(1):34‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez‐Redondo P, Izpisua Belmonte JC. Tailored chromatin modulation to promote tissue regeneration. Semin Cell Dev Biol. 2020;97:3‐15. [DOI] [PubMed] [Google Scholar]
- 52.Shue YT, Lee KT, Walters BW, et al. Dynamic shifts in chromatin states differentially mark the proliferative basal cells and terminally differentiated cells of the developing epidermis. Epigenetics. 2020;1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao X, Rubin AJ, Qu K, et al. A novel ATAC‐seq approach reveals lineage‐specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015;16:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rapisarda V, Malashchuk I, Asamaowei IE, et al. p63 Transcription factor regulates nuclear shape and expression of nuclear envelope‐associated genes in epidermal keratinocytes. J Invest Dermatol. 2017;137(10):2157‐2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb‐mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10(8):881‐886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plikus MV, Guerrero‐Juarez CF, Treffeisen E, Gay DL. Epigenetic control of skin and hair regeneration after wounding. Exp Dermatol. 2015;24(3):167‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding S, Chen J, Zeng Q, et al. Chronic sun exposure is associated with distinct histone acetylation changes in human skin. Br J Dermatol. 2018;179(1):110‐117. [DOI] [PubMed] [Google Scholar]
- 58.Bin BH, Lee SH, Bhin J, et al. The epithelial zinc transporter ZIP10 epigenetically regulates human epidermal homeostasis by modulating histone acetyltransferase activity. Br J Dermatol. 2019;180(4):869‐880. [DOI] [PubMed] [Google Scholar]
- 59.Rinaldi L, Datta D, Serrat J, et al. Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016;19(4):491‐501. [DOI] [PubMed] [Google Scholar]
- 60.Rinaldi L, Avgustinova A, Martin M, et al. Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR‐gamma. Elife. 2017;6:e21697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boorgula MP, Taub MA, Rafaels N, et al. Replicated methylation changes associated with eczema herpeticum and allergic response. Clin Epigenetics. 2019;11(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olisova OY, Nikolay KG, Kayumova LN, Zavarykina TM, Dmitriev AA, Asanov AY. Skin DNA methylation profile in atopic dermatitis patients: a case‐control study. Exp Dermatol. 2020;29(2): 184‐189. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez E, Baurecht H, Wahn AF, et al. An integrated epigenetic and transcriptomic analysis reveals distinct tissue‐specific patterns of DNA methylation associated with atopic dermatitis. J Invest Dermatol. 2014;134(7):1873‐1883. [DOI] [PubMed] [Google Scholar]
- 64.Li F, Yuan CW, Xu S, et al. Loss of the epigenetic mark 5‐hmC in psoriasis: implications for epidermal stem cell dysregulation. J Invest Dermatol. 2020;140(6):1266‐1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D'Arcangelo D, Tinaburri L, Dellambra E. The role of p16INK4a pathway in human epidermal stem cell self‐renewal, aging and cancer. Int J Mol Sci. 2017;18(7):1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bormann F, Rodriguez‐Paredes M, Hagemann S, et al. Reduced DNA methylation patterning and transcriptional connectivity define human skin aging. Aging Cell. 2016;15(3):563‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonifas JM, Matsumura K, Chen MA, et al. Mutations of keratin 9 in two families with palmoplantar epidermolytic hyperkeratosis. J Invest Dermatol. 1994;103(4):474‐477. [DOI] [PubMed] [Google Scholar]
- 68.Rinn JL, Wang JK, Allen N, et al. A dermal HOX transcriptional program regulates site‐specific epidermal fate. Genes Dev. 2008;22(3):303‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan HT, Ellis JA, Koplin JJ, et al. Methylation of the filaggrin gene promoter does not affect gene expression and allergy. Pediatr Allergy Immunol. 2014;25(6):608‐610. [DOI] [PubMed] [Google Scholar]
- 70.Ziyab AH, Karmaus W, Holloway JW, Zhang H, Ewart S, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss‐of‐function variants. J Eur Acad Dermatol Venereology. 2013;27(3):e420‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmuth M, Ortegon AM, Mao‐Qiang M, Elias PM, Feingold KR, Stahl A. Differential expression of fatty acid transport proteins in epidermis and skin appendages. J Invest Dermatol. 2005;125(6):1174‐1181. [DOI] [PubMed] [Google Scholar]
- 72.Klar J, Schweiger M, Zimmerman R, et al. Mutations in the fatty acid transport protein 4 gene cause the ichthyosis prematurity syndrome. Am J Hum Genet. 2009;85(2):248‐253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamamoto H, Hattori M, Chamulitrat W, Ohno Y, Kihara A. Skin permeability barrier formation by the ichthyosis‐causative gene FATP4 through formation of the barrier lipid ω‐O‐acylceramide. Proc Natl Acad Sci USA. 2020;117(6):2914‐2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moosbrugger‐Martinz V, Gruber R, Ladstätter K, et al. Filaggrin null mutations are associated with altered circulating Tregs in atopic dermatitis. J Cell Mol Med. 2019;23(2):1288‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang Y, Wang P, Zhao M, et al. Demethylation of the FCER1G promoter leads to FcεRI overexpression on monocytes of patients with atopic dermatitis. Allergy. 2012;67(3):424‐430. [DOI] [PubMed] [Google Scholar]
- 76.Luo Y, Zhou B, Zhao M, Tang J, Lu Q. Promoter demethylation contributes to TSLP overexpression in skin lesions of patients with atopic dermatitis. Clin Exp Dermatol. 2014;39(1):48‐53. [DOI] [PubMed] [Google Scholar]
- 77.Andl T, Murchison EP, Liu F, et al. The miRNA‐processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16(10):1041‐1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yi R, O'Carroll D, Pasolli HA, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38(3):356‐362. [DOI] [PubMed] [Google Scholar]
- 79.Yi R, Pasolli HA, Landthaler M, et al. DGCR8‐dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA. 2009;106(2):498‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Botchkareva NV. The molecular revolution in cutaneous biology: noncoding RNAs: new molecular players in dermatology and cutaneous biology. J Invest Dermatol. 2017;137(5):e105‐e111. [DOI] [PubMed] [Google Scholar]
- 81.Horsburgh S, Fullard N, Roger M, et al. MicroRNAs in the skin: role in development, homoeostasis and regeneration. Clin Sci. 2017;131(15):1923‐1940. [DOI] [PubMed] [Google Scholar]
- 82.Yi R, Fuchs E. MicroRNA‐mediated control in the skin. Cell Death Differ. 2010;17(2):229‐235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moutinho C, Esteller M. MicroRNAs and epigenetics. Adv Cancer Res. 2017;135:189‐220. [DOI] [PubMed] [Google Scholar]
- 84.Zardo G, Ciolfi A, Vian L, et al. Transcriptional targeting by microRNA‐polycomb complexes: a novel route in cell fate determination. Cell Cycle (Georgetown, Tex). 2012;11(19):3543‐3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. 2019;51:11‐17. [DOI] [PubMed] [Google Scholar]
- 86.Harmanci D, Erkan EP, Kocak A, Akdogan GG. Role of the microRNA‐29 family in fibrotic skin diseases. Biomed Reports. 2017;6(6):599‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vanden Oever M, Muldoon D, Mathews W, McElmurry R, Tolar J. miR‐29 regulates type VII collagen in recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2016;136(10):2013‐2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Condorelli AG, Logli E, Cianfarani F, et al. MicroRNA‐145‐5p regulates fibrotic features of recessive dystrophic epidermolysis bullosa skin fibroblasts. Br J Dermatol. 2019;181(5):1017‐1027. [DOI] [PubMed] [Google Scholar]
- 89.Yang S, Cui H, Xie N, et al. miR‐145 regulates myofibroblast differentiation and lung fibrosis. FASEB J. 2013;27(6):2382‐2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wimmer M, Zauner R, Ablinger M, et al. A cancer stem cell‐like phenotype is associated with miR‐10b expression in aggressive squamous cell carcinomas. Cell Commun Signal. 2020;18(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nyström A, Bruckner‐Tuderman L. Injury‐ and inflammation‐driven skin fibrosis: The paradigm of epidermolysis bullosa. Matrix Biol. 2018;68–69:547‐560. [DOI] [PubMed] [Google Scholar]
- 92.Fritsch A, Loeckermann S, Kern JS, et al. A hypomorphic mouse model of dystrophic epidermolysis bullosa reveals mechanisms of disease and response to fibroblast therapy. J Clin Investig. 2008;118(5):1669‐1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mittapalli VR, Madl J, Loffek S, et al. Injury‐driven stiffening of the dermis expedites skin carcinoma progression. Cancer Res. 2016;76(4):940‐951. [DOI] [PubMed] [Google Scholar]
- 94.Cui Y, Li T, Yang D, Li S, Le W. miR‐29 regulates Tet1 expression and contributes to early differentiation of mouse ESCs. Oncotarget. 2016;7(40):64932‐64941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I. miR‐29 represses the activities of DNA methyltransferases and DNA demethylases. Int J Mol Sci. 2013;14(7):14647‐14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agostini M, Ganini C, Candi E, Melino G. The role of noncoding RNAs in epithelial cancer. Cell Death Discov. 2020;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of microRNA expression in cancer. Int J Mol Sci. 2020;21(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.García‐Sancha N, Corchado‐Cobos R, Pérez‐Losada J, Cañueto J. MicroRNA dysregulation in cutaneous squamous cell carcinoma. Int J Mol Sci. 2019;20(9):2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feichtinger RG, Sperl W, Bauer JW, Kofler B. Mitochondrial dysfunction: a neglected component of skin diseases. Exp Dermatol. 2014;23(9):607‐614. [DOI] [PubMed] [Google Scholar]
- 100.Baris OR, Klose A, Kloepper JE, et al. The mitochondrial electron transport chain is dispensable for proliferation and differentiation of epidermal progenitor cells. Stem Cells (Dayton, Ohio). 2011;29(9):1459‐1468. [DOI] [PubMed] [Google Scholar]
- 101.Allombert‐Blaise C, Tamiji S, Mortier L, et al. Terminal differentiation of human epidermal keratinocytes involves mitochondria‐ and caspase‐dependent cell death pathway. Cell Death Differ. 2003;10(7):850‐852. [DOI] [PubMed] [Google Scholar]
- 102.Feichtinger RG, Kofler B. Peculiarities and pitfalls of quantifying mitochondrial energy metabolism in the skin. Exp Dermatol. 2016;25(2):101‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tamiji S, Beauvillain JC, Mortier L, et al. Induction of apoptosis‐like mitochondrial impairment triggers antioxidant and Bcl‐2‐dependent keratinocyte differentiation. J Invest Dermatol. 2005;125(4):647‐658. [DOI] [PubMed] [Google Scholar]
- 104.Hornig‐Do HT, von Kleist‐Retzow JC, Lanz K, et al. Human epidermal keratinocytes accumulate superoxide due to low activity of Mn‐SOD, leading to mitochondrial functional impairment. J Invest Dermatol. 2007;127(5):1084‐1093. [DOI] [PubMed] [Google Scholar]
- 105.Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol. 2013;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Velarde MC, Demaria M, Melov S, Campisi J. Pleiotropic age‐dependent effects of mitochondrial dysfunction on epidermal stem cells. Proc Natl Acad Sci USA. 2015;112(33):10407‐10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Walzer C, Frenk E. Cytochemical expression of epidermal peroxidase and cytochrome oxidase activities in pathological skin conditions of man. Arch Dermatol Res. 1986;278(6):460‐464. [DOI] [PubMed] [Google Scholar]
- 108.Bauer JW, Rouan F, Kofler B, et al. A compound heterozygous one amino‐acid insertion/nonsense mutation in the plectin gene causes epidermolysis bullosa simplex with plectin deficiency. Am J Pathol. 2001;158(2):617‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Winter L, Abrahamsberg C, Wiche G. Plectin isoform 1b mediates mitochondrion‐intermediate filament network linkage and controls organelle shape. J Cell Biol. 2008;181(6):903‐911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maselli RA, Arredondo J, Cagney O, et al. Congenital myasthenic syndrome associated with epidermolysis bullosa caused by homozygous mutations in PLEC1 and CHRNE. Clin Genet. 2011;80(5):444‐451. [DOI] [PubMed] [Google Scholar]
- 111.Schroder R, Kunz WS, Rouan F, et al. Disorganization of the desmin cytoskeleton and mitochondrial dysfunction in plectin‐related epidermolysis bullosa simplex with muscular dystrophy. J Neuropathol Exp Neurol. 2002;61(6):520‐530. [DOI] [PubMed] [Google Scholar]
- 112.Uttam J, Hutton E, Coulombe PA, et al. The genetic basis of epidermolysis bullosa simplex with mottled pigmentation. Proc Natl Acad Sci USA. 1996;93(17):9079‐9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fcl A. Mitochondrial metabolism and DNA methylation: a review of the interaction between two genomes. Clin Epigenetics. 2020;12(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Minocherhomji S, Tollefsbol TO, Singh KK. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics. 2012;7(4):326‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Santos JH. Mitochondria signaling to the epigenome: A novel role for an old organelle. Free Radic Biol Med. 2020;S0891‐5849(20):31628‐31632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weinhouse C. Mitochondrial‐epigenetic crosstalk in environmental toxicology. Toxicology. 2017;391:5‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanyal T, Paul M, Bhattacharjee S, Bhattacharjee P. Epigenetic alteration of mitochondrial biogenesis regulatory genes in arsenic exposed individuals (with and without skin lesions) and in skin cancer tissues: A case control study. Chemosphere. 2020;258:127305. [DOI] [PubMed] [Google Scholar]
- 118.Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG, Singh KK. A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer Biol Ther. 2008;7(8):1182‐1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mentch SJ, Locasale JW. One‐carbon metabolism and epigenetics: understanding the specificity. Ann N Y Acad Sci. 2016;1363:91‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fine JD, Bruckner‐Tuderman L, Eady RA, et al. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J Am Acad Dermatol. 2014;70(6):1103‐1126. [DOI] [PubMed] [Google Scholar]
- 121.Castillo‐Fernandez JE, Spector TD, Bell JT. Epigenetics of discordant monozygotic twins: implications for disease. Genome Med. 2014;6(7):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Odorisio T, Di Salvio M, Orecchia A, et al. Monozygotic twins discordant for recessive dystrophic epidermolysis bullosa phenotype highlight the role of TGF‐beta signalling in modifying disease severity. Hum Mol Genet. 2014;23(15):3907‐3922. [DOI] [PubMed] [Google Scholar]
- 123.Bodemer C, Tchen SI, Ghomrasseni S, et al. Skin expression of metalloproteinases and tissue inhibitor of metalloproteinases in sibling patients with recessive dystrophic epidermolysis and intrafamilial phenotypic variation. J Invest Dermatol. 2003;121(2):273‐279. [DOI] [PubMed] [Google Scholar]
- 124.Titeux M, Pendaries V, Tonasso L, Decha A, Bodemer C, Hovnanian A. A frequent functional SNP in the MMP1 promoter is associated with higher disease severity in recessive dystrophic epidermolysis bullosa. Hum Mutat. 2008;29(2):267‐276. [DOI] [PubMed] [Google Scholar]
- 125.Garza‐Gomez J, Cerda‐Flores RM, Gomez‐Flores M, et al. An investigation into the MMP1 gene promoter region polymorphism–1607 2G with recessive dystrophic epidermolysis bullosa disease severity in northeastern Mexican patients. Int J Dermatol. 2014;53(8):985‐990. [DOI] [PubMed] [Google Scholar]
- 126.Kern JS, Gruninger G, Imsak R, et al. Forty‐two novel COL7A1 mutations and the role of a frequent single nucleotide polymorphism in the MMP1 promoter in modulation of disease severity in a large European dystrophic epidermolysis bullosa cohort. Br J Dermatol. 2009;161(5):1089‐1097. [DOI] [PubMed] [Google Scholar]
- 127.Sproule TJ, Bubier JA, Grandi FC, et al. Molecular identification of collagen 17a1 as a major genetic modifier of laminin gamma 2 mutation‐induced junctional epidermolysis bullosa in mice. PLoS Genet. 2014;10(2):e1004068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bauer JW, Koller J, Murauer EM, et al. Closure of a large chronic wound through transplantation of gene‐corrected epidermal stem cells. J Invest Dermatol. 2017;137(3):778‐781. [DOI] [PubMed] [Google Scholar]
- 129.Prodinger CM, Reichelt J, Bauer JW, Laimer M. Current and future perspectives of stem cell therapy in dermatology. Ann. Dermatol. 2017;29(6):667‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oender K, Trost A, Lanschuetzer C, et al. Cytokeratin‐related loss of cellular integrity is not a major driving force of human intrinsic skin aging. Mech Ageing Dev. 2008;129(10):563‐571. [DOI] [PubMed] [Google Scholar]
- 131.Laimer M, Kocher T, Chiocchetti A, et al. Proteomic profiling reveals a catalogue of new candidate proteins for human skin aging. Exp Dermatol. 2010;19(10):912‐918. [DOI] [PubMed] [Google Scholar]
- 132.Lener T, Moll PR, Rinnerthaler M, Bauer J, Aberger F, Richter K. Expression profiling of aging in the human skin. Exp Gerontol. 2006;41(4):387‐397. [DOI] [PubMed] [Google Scholar]
- 133.Rinnerthaler M, Duschl J, Steinbacher P, et al. Age‐related changes in the composition of the cornified envelope in human skin. Exp Dermatol. 2013;22(5):329‐335. [DOI] [PubMed] [Google Scholar]
- 134.Breitenbach JS, Rinnerthaler M, Trost A, et al. Transcriptome and ultrastructural changes in dystrophic Epidermolysis bullosa resemble skin aging. Aging. 2015;7(6):389‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Rosa L, Secone Seconetti A, De Santis G, et al. Laminin 332‐dependent YAP dysregulation depletes epidermal stem cells in junctional epidermolysis bullosa. Cell Rep. 2019;27(7):2036‐2049 e2036. [DOI] [PubMed] [Google Scholar]
- 136.Beverdam A, Claxton C, Zhang X, James G, Harvey KF, Key B. Yap controls stem/progenitor cell proliferation in the mouse postnatal epidermis. J Invest Dermatol. 2013;133(6):1497‐1505. [DOI] [PubMed] [Google Scholar]
- 137.Elbediwy A, Vincent‐Mistiaen ZI, Thompson BJ. YAP and TAZ in epithelial stem cells: A sensor for cell polarity, mechanical forces and tissue damage. BioEssays. 2016;38(7):644‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schlegelmilch K, Mohseni M, Kirak O, et al. Yap1 acts downstream of alpha‐catenin to control epidermal proliferation. Cell. 2011;144(5):782‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang H, Pasolli HA, Fuchs E. Yes‐associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci USA. 2011;108(6):2270‐2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Walko G, Woodhouse S, Pisco AO, et al. A genome‐wide screen identifies YAP/WBP2 interplay conferring growth advantage on human epidermal stem cells. Nat Commun. 2017;8:14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yamada T, Hasegawa S, Miyachi K, et al. Laminin‐332 regulates differentiation of human interfollicular epidermal stem cells. Mech Ageing Dev. 2018;171:37‐46. [DOI] [PubMed] [Google Scholar]
- 142.Sathyanarayana UG, Maruyama R, Padar A, et al. Molecular detection of noninvasive and invasive bladder tumor tissues and exfoliated cells by aberrant promoter methylation of laminin‐5 encoding genes. Cancer Res. 2004;64(4):1425‐1430. [DOI] [PubMed] [Google Scholar]
- 143.Sathyanarayana UG, Padar A, Huang CX, et al. Aberrant promoter methylation and silencing of laminin‐5‐encoding genes in breast carcinoma. Clin Cancer Res. 2003;9(17):6389‐6394. [PubMed] [Google Scholar]
- 144.Sathyanarayana UG, Padar A, Suzuki M, et al. Aberrant promoter methylation of laminin‐5‐encoding genes in prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2003;9(17):6395‐6400. [PubMed] [Google Scholar]
- 145.Chang X, Monitto CL, Demokan S, et al. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 2010;70(7):2870‐2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kwon OH, Park JL, Kim M, et al. Aberrant up‐regulation of LAMB3 and LAMC2 by promoter demethylation in gastric cancer. Biochem Biophys Res Commun. 2011;406(4):539‐545. [DOI] [PubMed] [Google Scholar]
- 147.Brown S, Pineda CM, Xin T, et al. Correction of aberrant growth preserves tissue homeostasis. Nature. 2017;548(7667):334‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pasmooij AM, Nijenhuis M, Brander R, Jonkman MF. Natural gene therapy may occur in all patients with generalized non‐Herlitz junctional epidermolysis bullosa with COL17A1 mutations. J Invest Dermatol. 2012;132(5):1374‐1383. [DOI] [PubMed] [Google Scholar]
- 149.van den Akker PC, Pasmooij AMG, Joenje H, Hofstra RMW, Te Meerman GJ, Jonkman MF. A "late‐but‐fitter revertant cell" explains the high frequency of revertant mosaicism in epidermolysis bullosa. PLoS One. 2018;13(2):e0192994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kowalewski C, Bremer J, Gostynski A, et al. Amelioration of junctional epidermolysis bullosa due to exon skipping. Br J Dermatol. 2016;174(6):1375‐1379. [DOI] [PubMed] [Google Scholar]
- 151.Choate KA, Lu Y, Zhou J, et al. Frequent somatic reversion of KRT1 mutations in ichthyosis with confetti. J Clin Invest. 2015;125(4):1703‐1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Suzuki S, Nomura T, Miyauchi T, et al. Revertant mosaicism in ichthyosis with confetti caused by a frameshift mutation in KRT1. J Invest Dermatol. 2016;136(10):2093‐2095. [DOI] [PubMed] [Google Scholar]
- 153.Khavari DA, Sen GL, Rinn JL. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle (Georgetown, Tex). 2010;9(19):3880‐3883. [DOI] [PubMed] [Google Scholar]
- 154.Li J, Jiang TX, Hughes MW, et al. Progressive alopecia reveals decreasing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. J Invest Dermatol. 2012;132(12):2681‐2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bock C, Beerman I, Lien WH, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47(4):633‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Davies HR, Hodgson K, Schwalbe E, et al. Epigenetic modifiers DNMT3A and BCOR are recurrently mutated in CYLD cutaneous syndrome. Nat Commun. 2019;10(1):4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Qian H, Xu X. Reduction in DNA methyltransferases and alteration of DNA methylation pattern associate with mouse skin ageing. Exp Dermatol. 2014;23(5):357‐359. [DOI] [PubMed] [Google Scholar]
- 158.Li F, Yuan CW, Xu S, et al. Loss of the epigenetic Mark 5‐hmC in psoriasis: implications for epidermal stem cell dysregulation. J Invest Dermatol. 2020;140(6):1266‐1275.e1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang J, Yang C, Wang C, et al. AGE‐induced keratinocyte MMP‐9 expression is linked to TET2‐mediated CpG demethylation. Wound Repair Regen. 2016;24(3):489‐500. [DOI] [PubMed] [Google Scholar]
- 160.Mulder KW, Wang X, Escriu C, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14(7):753‐763. [DOI] [PubMed] [Google Scholar]
- 161.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self‐renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22(14):1865‐1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Na J, Lee K, Na W, et al. Histone H3K27 Demethylase JMJD3 in Cooperation with NF‐κB Regulates Keratinocyte Wound Healing. J Invest Dermatol. 2016;136(4):847‐858. [DOI] [PubMed] [Google Scholar]
- 163.LeBoeuf M, Terrell A, Trivedi S, et al. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19(6):807‐818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hughes MW, Jiang TX, Lin SJ, et al. Disrupted ectodermal organ morphogenesis in mice with a conditional histone deacetylase 1, 2 deletion in the epidermis. J Invest Dermatol. 2014;134(1):24‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Frye M, Fisher AG, Watt FM. Epidermal stem cells are defined by global histone modifications that are altered by Myc‐induced differentiation. PLoS One. 2007;2(8):e763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Connelly JT, Mishra A, Gautrot JE, Watt FM. Shape‐induced terminal differentiation of human epidermal stem cells requires p38 and is regulated by histone acetylation. PLoS One. 2011;6(11):e27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue‐specific stem cells. Cell. 2009;136(6):1122‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Luis NM, Morey L, Mejetta S, et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb‐ dependent and ‐independent functions of Cbx4. Cell Stem Cell. 2011;9(3):233‐246. [DOI] [PubMed] [Google Scholar]
- 169.Mardaryev AN, Liu B, Rapisarda V, et al. Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol. 2016;212(1):77‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cohen I, Zhao D, Bar C, et al. PRC1 Fine‐tunes gene repression and activation to safeguard skin development and stem cell specification. Cell Stem Cell. 2018;22(5):726‐739.e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. Dissecting the roles of polycomb repressive complex 2 subunits in the control of skin development. J Invest Dermatol. 2016;136(8):1647‐1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lee K, Adhikary G, Balasubramanian S, et al. Expression of Bmi‐1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol. 2008;128(1):9‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25(5):485‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hopkin AS, Gordon W, Klein RH, et al. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012;8(7):e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C. Transcriptional repression of miR‐34 family contributes to p63‐mediated cell cycle progression in epidermal cells. J Invest Dermatol. 2010;130(5):1249‐1257. [DOI] [PubMed] [Google Scholar]
- 176.Zhou BR, Guo XF, Zhang JA, et al. Elevated miR‐34c‐5p mediates dermal fibroblast senescence by ultraviolet irradiation. Int J Biol Sci. 2013;9(7):743‐752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang L, Stokes N, Polak L, Fuchs E. Specific microRNAs are preferentially expressed by skin stem cells to balance self‐renewal and early lineage commitment. Cell Stem Cell. 2011;8(3):294‐308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Richardson A, Powell AK, Sexton DW, Parsons JL, Reynolds NJ, Ross K. microRNA‐184 is induced by store‐operated calcium entry and regulates early keratinocyte differentiation. J Cell Physiol. 2020;235:6854‐6861. [DOI] [PubMed] [Google Scholar]
- 179.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness'. Nature. 2008;452(7184):225‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yu J, Peng H, Ruan Q, Fatima A, Getsios S, Lavker RM. MicroRNA‐205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010;24(10):3950‐3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Biswas S, Roy S, Banerjee J, et al. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci USA. 2010;107(15):6976‐6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Garzon R, Liu S, Fabbri M, et al. MicroRNA‐29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411‐6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Matsui M, Chu Y, Zhang H, et al. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41(22):10086‐10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wu WR, Sun H, Zhang R, et al. Methylation‐associated silencing of miR‐200b facilitates human hepatocellular carcinoma progression by directly targeting BMI1. Oncotarget. 2016;7(14):18684‐18693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA‐200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bae HJ, Jung KH, Eun JW, et al. MicroRNA‐221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J Hepatol. 2015;63(2):408‐419. [DOI] [PubMed] [Google Scholar]
- 187.Li X, Huang K, Yu J. Inhibition of microRNA‐21 upregulates the expression of programmed cell death 4 and phosphatase tensin homologue in the A431 squamous cell carcinoma cell line. Oncol Lett. 2014;8(1):203‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yin S, Lin X. MicroRNA‐21 contributes to cutaneous squamous cell carcinoma progression via mediating TIMP3/PI3K/AKT signaling axis. Int J Gen Med. 2021;14:27‐39. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 189.Wang Y, Deng X, Dai Y, Niu X, Zhou M. miR‐27a downregulation promotes cutaneous squamous cell carcinoma progression via targeting EGFR. Front in Oncol. 2019;9:1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li S, Luo C, Zhou J, Zhang Y. MicroRNA‐34a directly targets high‐mobility group box 1 and inhibits the cancer cell proliferation, migration and invasion in cutaneous squamous cell carcinoma. Exp Ther Med. 2017;14(6):5611‐5618. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 191.Li W, Zhou BR, Hua LJ, Guo Z, Luo D. Differential miRNA profile on photoaged primary human fibroblasts irradiated with ultraviolet A. Tumour Biol. 2013;34(6):3491‐3500. [DOI] [PubMed] [Google Scholar]
- 192.Mancini M, Saintigny G, Mahé C, Annicchiarico‐Petruzzelli M, Melino G, Candi E. MicroRNA‐152 and ‐181a participate in human dermal fibroblasts senescence acting on cell adhesion and remodeling of the extra‐cellular matrix. Aging. 2012;4(11):843‐853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.di Val R, Cervo P, Lena AM, et al. p63‐microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci USA. 2012;109(4):1133‐1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu Y, Li Y, Li N, et al. TGF‐beta1 promotes scar fibroblasts proliferation and transdifferentiation via up‐regulating MicroRNA‐21. Sci Rep. 2016;6:32231. [DOI] [PMC free article] [PubMed] [Google Scholar]


