Abstract
At dawn of a scorching summer day, land plants must anticipate upcoming extreme midday temperatures by timely establishing molecular defences that can keep heat‐labile membranes and proteins functional. A gradual morning pre‐exposure to increasing sub‐damaging temperatures induces heat‐shock proteins (HSPs) that are central to the onset of plant acquired thermotolerance (AT). To gain knowledge on the mechanisms of AT in the model land plant Physcomitrium patens, we used label‐free LC–MS/MS proteomics to quantify the accumulated and depleted proteins before and following a mild heat‐priming treatment. High protein crowding is thought to promote protein aggregation, whereas molecular chaperones prevent and actively revert aggregation. Yet, we found that heat priming (HP) did not accumulate HSP chaperones in chloroplasts, although protein crowding was six times higher than in the cytosol. In contrast, several HSP20s strongly accumulated in the cytosol, yet contributing merely 4% of the net mass increase of heat‐accumulated proteins. This is in poor concordance with their presumed role at preventing the aggregation of heat‐labile proteins. The data suggests that under mild HP unlikely to affect protein stability. Accumulating HSP20s leading to AT, regulate the activity of rare and specific signalling proteins, thereby preventing cell death under noxious heat stress.
Keywords: heat‐priming, heat‐shock proteins, heat‐shock response, HSP20s‐heat‐shock proteins, molecular crowding, Physcomitrium patens, proteomics, RNA‐Seq, thermotolerance
Short abstract
Quantitative proteomics showed that a mild heat pre‐treatment leading to acquired thermotolerance in moss strongly induces the accumulation of cytosolic HSP20s, albeit in low amounts compared to other HSPs. The cytosol was found to be the least crowded cellular compartment, suggesting that in addition to their general anti‐aggregation function, cytosolic HSP20s carry specific signalling functions to regulate noxious heat‐induced apoptosis.
1. INTRODUCTION
During a typical summer day, land plants living in arid continental environments often need to cope with an increase of 20°C in the ambient temperature, between sunrise and early afternoon. Early in the morning, they need to anticipate the extent of excessive temperatures that will culminate at midday and timely establish appropriate molecular defences to secure their survival until evening relief. At the molecular level, excessively high temperatures can cause the hyper‐fluidization of membranes, leading to ion leakage and the production of reactive oxygen species, which in turn induce programmed cell death (Niu & Xiang, 2018; Schöffl, Irina, Phillip, & Friedrich, 2006). Excessive heat may also denature heat‐labile proteins that spontaneously convert into inactive aggregates with abnormally exposed hydrophobic surfaces that can compromise membrane integrity and may become toxic (Lashuel, Hartley, Petre, Walz, & Lansbury, 2002; Sharma, De Los Rios, & Goloubinoff, 2011).
Moss and higher plant cells harbour several heat sensors that possibly act in parallel. Alongside the family of heat‐responsive cyclic nucleotide gated calcium channels (CNGCs) in the plasma membrane, which were shown to act as membrane fluidity thermosensors (Finka, Cuendet, Maathuis, Saidi, & Goloubinoff, 2012; Gao et al., 2012; Tunc‐Ozdemir et al., 2013), an unfolded protein response (UPR) sensor was found in the endoplasmic reticulum and another in the cytosol and nucleus (Che et al., 2010; Deng et al., 2011; Sugio, Dreos, Aparicio, & Maule, 2009). A putative heat‐responsive retrograde pathway was also reported in chloroplasts (Yu et al., 2012). In addition, a heat‐responsive histone variant sensor was suggested to operate in the nucleus (Cortijo et al., 2017; Kumar & Wigge, 2010). Ultimately, these various heat‐sensing and signalling pathways coalesce into the effective hyper‐phosphorylation, oligomerization and migration to the nucleus of heat shock transcription factors (HSFs) (Chan‐Schaminet, Baniwal, Bublak, Nover, & Scharf, 2009) that activate in the nucleus the transcription of genes encoding for protective heat‐induced proteins, generally called heat‐shock proteins (HSPs). This mechanism is called the heat shock response (HSR). HSFs binding to HSE cis‐elements in the promoters of HSP genes must first command chromatin remodelling complexes to evict bound histones that otherwise repress HSP transcription at low temperature (Kumar & Wigge, 2010). In parallel, the bound HSFs can recruit RNA polymerase to transcribe the histone‐free HSP genes (Guo et al., 2016; Kurshakova et al., 2007; Rohner et al., 2013), ultimately leading to the rapid accumulation of HSPs and the onset of acquired thermotolerance.
Many, but not all of the heat‐induced genes, encode for members of the conserved chaperones families HSP20s, HSP70s, HSP100 and HSP90s (Finka et al., 2012; Gao et al., 2012; Peng, Zhang, Li, & Zhao, 2019; Saidi et al., 2009). Importantly, a mild heat priming (HP) causing no, or only minimal damages in proteins and membranes, should not be confused with a noxious heat shock that can cause reversible and irreversible damages in labile macromolecules. By translating an initial moderate temperature increment into an effective heat‐priming response that leads to the establishment of HSP‐ and osmolyte‐based molecular defences (Hasanuzzaman, Nahar, Alam, Roychowdhury, & Fujita, 2013; Larkindale & Vierling, 2008), plant cells can prevent molecular damages caused by an upcoming noxious stress and repair them thereafter.
Here, we used a high‐throughput quantitative proteomic approach, complemented by RNAseq, in an attempt to gain new unbiased information on the identity, the cellular concentrations and the subcellular localization of most of the significantly accumulated and depleted proteins, following a mild heat‐priming treatment, typically leading to AT. The moss Physcomitrium patens was chosen as a model land plant because bryophytes, which with the liverworts were the first to colonize land, needed to acquire new mechanisms to sense and anticipate extreme diurnal variations of the ambient temperature and timely build‐up effective defences against upcoming noxious heat stresses.
Because AT results from the integrated heat‐induced accumulation of HSPs in the various compartments of the plant cell, we first established a bioinformatic pipeline in an attempt to annotate with a predicted function and a cellular localization all the Physcomitrium patens proteins that we could significantly quantify. High protein crowding is thought to increase the aggregation‐propensity of nascent polypeptides, especially under heat‐stress. We therefore aimed at estimating the protein crowding in various cellular compartments, expecting that the effectiveness of AT will depend on the prior accumulation of anti‐aggregation HSP chaperones in the most crowded, and therefore the expectedly most aggregation‐prone compartments of the cell. We found that a mild heat priming unlikely to cause protein aggregations, did not accumulate HSPs in the chloroplasts that were six times more crowded than the cytosol. In contrast, three HSP20s were strongly accumulated, albeit in low absolute amounts, in the cytosol, suggesting that alongside their known anti‐aggregation function, HSP20s possibly repress the onset of programmed cell death during and following a noxious heat shock, thereby contributing to AT.
2. MATERIALS AND METHODS
2.1. Plant materials, growth conditions and treatments
The moss Physcomitrium patens transgenic lines HSP‐GUS (Saidi et al., 2005) was grown on moss solid minimal medium overlaid with a cellophane disk as described by (Saidi et al., 2005). For heat treatment, 7 days‐old HSP‐GUS protonemal tissues were transferred to liquid minimal medium in Costar multiwell plates (Corning) and heated in a temperature‐controlled chamber at 38°C for 4 hrs or 30 min, respectively for LFQ approach and RNAseq.
2.2. Total protein isolation
Total protonemata proteins were isolated with a protocol adapted from (Finka, Schaefer, Saidi, Goloubinoff, & Zryd, 2007; Saidi et al., 2005). Total protein concentrations in soluble extracts were quantified using the Bradford method. Six biological replicates were done, both in control and heat‐stress conditions. Protein content was estimated using Bradford method.
2.3. Western blotting
Twenty microgram of moss soluble protein extracts were separated on 12% SDS‐PAGE. Proteins were transferred to a nitrocellulose membrane (BioRad) by electroblotting and incubated with a rabbit‐derived polyclonal antibody (Molecular probes) against HSP17 and diluted 1/1000 (vol/vol). The blot was then incubated with HRP‐conjugated anti‐rabbit IgG (BioRad) diluted 1/5000 (vol/vol). Immune complexes were visualized using the chemiluminescent ImmunstarTM Kit (BioRad) according to manufacturer's instructions.
2.4. Measurement of PSII activity
The Fv/fm ratio is the maximal photochemical efficiency of Photosystem II (PSII). PSII activity measurements were made with the Plant Efficiency Analyser PEA from Hansatech Instruments Ltd, UK as detailed by (Strasser & Strasser, 1995). Measurements were taken following a brief (2 s) strong illumination of protonemata that have been previously dark‐adapted for 2 min.
2.5. Label‐free quantitative proteome analysis
Eluted peptides were analysed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) operated in data‐dependent mode, controlled by Xcalibur software (version 3.0.63). Full survey scans were performed at a 120,000 resolution, and a top speed precursor selection strategy was applied to maximize acquisition of peptide tandem MS spectra with a maximum cycle time of 3 s. HCD fragmentation mode was used at a normalized collision energy of 32%, with a precursor isolation window of 1.6 m/z and MS/MS spectra were acquired in the ion trap. Peptides selected for MS/MS were excluded from further fragmentation during 60 s. Data collected by the mass spectrometer were processed for protein identification and quantification using MaxQuant version 1.5.3.30, using the Andromeda search engine set to search the UniProt database restricted to the Physcomitrium patens proteome (UniProt proteome ID: UP000006727, November 2016). Trypsin (cleavage after K,R) was used as the enzyme definition, allowing two missed cleavages. Carbamidomethylation of cysteine was specified as a fixed modification, while N‐terminal acetylation of protein and oxidation of methionine were specified as variable modifications. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021959.
All data post‐processing and statistical analyses were performed using custom Matlab scripts. iBAQ and LFQ data were used as the basis for quantification. Instead of only using raw iBAQ intensities, we took advantage of the additional normalization introduced by the LFQ method (Cox et al., 2014) to re‐calculate ‘normalized iBAQs’ by dividing LFQ intensities by the number of theoretically observable tryptic peptides, as specified in the original iBAQ publication (Schwanhäusser et al., 2011). Since normalized iBAQs are proportional to protein molar quantities, protein mass fractions were obtained as where I i is the normalized IBAQ intensity of protein i, M i its molecular weight and the index k runs over all identified proteins. Then, the corresponding micromolar quantities c i were derived using an estimated total intracellular protein concentration of C T = 162 mg/ml: . The estimated total intracellular protein concentration (162 mg/ml) was calculated as , where C extract is the total protein concentration in the soluble extract as determined by a Bradford assay, V extract is the volume of quantified extract, N cells is the number of cells in the extract (counted by microscopy) and V cell = 3.14 × 10−8 ml is the estimated volume of an individual cell as estimated from morphometric analysis (Furt, Lemoi, Tuzel, & Vidali, 2012).
Statistical analyses using six biological replicates were then performed, first to determine which proteins were significantly quantified (i.e., had a mass fraction significantly larger than zero). This was done using t tests with a post‐hoc Benjamini‐Hochberg FDR‐controlling procedure to account for multiple testing, at a FDR threshold of 0.01. Finally, significant differences were determined using two‐sample t tests followed by Benjamini‐Hochberg procedures using an FDR cut‐off of 0.05.
2.6. Annotation pipeline
To identify the sequences of all putative unknowns or uncharacterized proteins, BLAST searches (http://www.expasy.org/tools/blast/) with these protein sequences were performed on the UniProt Knowledgebase (UniProtKB, http://www.uniprot.org/uniprot) to find their homologs. Functional categorization of the identified proteins was based on annotations in UniProtKB and previous studies on their homologs. Subcellular locations of the identified proteins were determined according to the annotation in UniProtKB and predicted using Loctree3 program (Goldberg et al., 2014).
2.7. Total RNA extraction
Total RNA was extracted from 7‐day protonemata with the Nucleospin RNA plant kit (Macherey‐Nagel‐Germany) and DNAse treated with the kit following the manufacturer's instructions for RNA‐Seq analyses. Total extracted RNA was quantified using a spectrophotometer (nanodrop) and the integrity was verified on 2% agarose gel and using a BioAnalyzer.
2.8. RNAseq experiment
Library preparation and sequencing was performed at the Lausanne Genomic Technologies Facility, University of Lausanne, Switzerland (https://www.unil.ch/gtf/en/home.html). After sequencing, purity‐filtered reads were adapted and quality trimmed with Cutadapt v. 1.8. Reads matching ribosomal RNA sequences were removed with fastq_screen (v. 0.9.3). Reads were aligned against Physcomitrium patens 318 v3.3 genome and transcriptome using STAR v. 2.5.2b (Dobin et al., 2013), and the estimation of transcript abundance in TPM was computed using RSEM v. 1.2.31 (Li & Dewey, 2011). The RNAseq data have been deposited to the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) repository with the dataset identifier PRJNA666761.
3. RESULTS
3.1. Protein quantification
Using a gene‐centric label‐free LC–MS/MS proteomic analysis, we identified 7,097 proteins in the six biological replicates of the control and six biological replicates of heat‐treated moss cells (Figure 1b). Following the merging of isoforms, 1,514 proteins were found to be significantly quantified (p < 0.01) in the six controls and the six heat‐treated samples. Whereas the moss genome is encoding for ~34′000 ORFs (Lang et al., 2018), the 1,514 significantly quantified proteins were expectedly the most abundant one: Although being less than 5% of the moss ORFs, they summed up into being 96.6% of total protein mass of the moss protonema cells, suggesting that the remaining ~32′000 ORF are only expressed in very low copy number and at particular developmental stages, or only in response to various stresses (Figure 1b,c). Following a mild heat‐treatment of 4 hrs at 38°C, 91 proteins were found to be significantly accumulated and 140 significantly depleted, compared to the 22°C control (Figure 1b).
FIGURE 1.
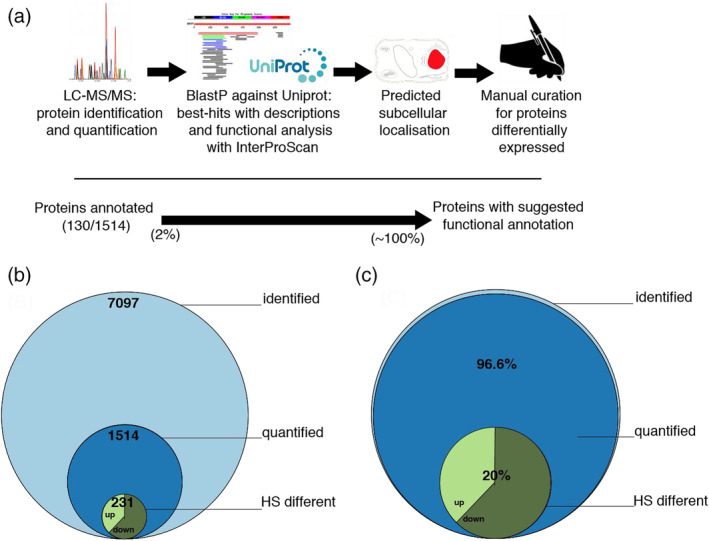
Workflow for the identification and annotation of Physcomitrium patens proteins. Illustration of the pipeline to produce the annotations used in this study (a). Stacked Venn diagram showing the number of different proteins identified, significantly quantified and significantly differentially accumulated proteins in physcomitrium patens under heat‐priming (b) and in mass fraction of the significantly quantified (c) [Colour figure can be viewed at wileyonlinelibrary.com]
Whereas gene expression under two different conditions is classically expressed as a fold change (FC) of a challenged sample, from an unchallenged control, here, we chose to also express the quantitative differences between heat‐treated and control conditions with a net mass difference parameter, in which the measured amount (expressed in mass fraction of the total) of a given protein before heat‐treatment was subtracted from that after heat‐treatment, referred to as ‘delta’. Delta values showed that 60 proteins became significantly heat‐accumulated and 89 proteins significantly heat‐depleted. Noticeably, 47 of the heat‐accumulated and 81 of the heat‐depleted proteins were common to both FC and delta ranking methods, highlighting the importance of using both criteria to optimally capture the molecular variations between the two conditions (Figure S1 and Table S1). The remaining ~1,200 significantly quantified proteins were expressed at non‐significantly different levels, under both conditions.
3.2. Workflow for moss gene and protein identification and annotation
Thus far, only ~2% of the Physcomitrium patens ORFs were annotated with predicted names, cellular localizations and putative functions. Here, we designed a pipeline in an attempt to increase the functional annotation, in particular of the 1,514 most abundant moss proteins that we significantly quantified (Figure 1a). Briefly, the moss protein sequences were blasted against the plant Uniprot database. Hits with a putative description, mostly deduced from the extensive research on Arabidopsis thaliana, were merged and using InterProScan, and assigned with a putative function. The LocTree program was then used to predict their subcellular localization. The differentially expressed ORFs under mild heat treatment were manually curated, to confirm sequence homologies with known gene products in higher plants, with annotated functions and to reinforce their predicted cellular localization. Thus, a putative function and a predicted cellular localization were assigned to all the 1,514 significantly quantified Physcomitrium patens proteins (Figure 1a).
3.3. Measured stoichiometric ratios in protein complexes
To validate our quantitative proteomic approach, the cellular concentration of chosen proteins were converted into micromolar concentrations, based on their specific molecular weights and a total protein concentration 162 mg/ml (± ~10%), which was experimentally determined using the Bradford method. The stoichiometry of subunits in known abundant protein complexes was addressed (Figure 2). In the case of the cytosolic CCT chaperonin complex, which in yeast and animals is forming two octameric rings, made of eight slightly different orthologous 60 kDa subunits (Finka & Goloubinoff, 2013; Lin & Sherman, 1997), we found a near‐equimolar stoichiometry for the moss CCTs, each of the eight orthologous subunits averaging reasonably well around the theoretical value of 12.5% from the total CCT mass (Figure 2a). Likewise, the RubisCO complex, which, in cyanobacteria, Algae and plants, is composed of eight chloroplast‐encoded large subunits and eight small subunits, the latter in Physcomitrium patens being encoded by 18 different nuclear‐encoded RbcS genes. The quantitative proteomic analysis found a near equimolar 1:1 stoichiometry between large and small subunits, as expected from the L8S8 structure of the RubisCO oligomer (Figure 2b). Similarly, a near equimolar stoichiometry was found for the photosystem II core, between D1 (PsbA), D2 (PsbD), CP43 (PsbC) and CP47 (PsbB). A near equimolar stoichiometry was also found within the photosystem I core between P700 chlorophyll apoprotein A1 and A2 (PsaA and PsaB) (Figure 2c,d). Noticeably, PSII reaction centres were 3–4 times more abundant than PSI reaction centres, as reported earlier to be the case for higher plant chloroplasts (Jia, Ito, & Tanaka, 2016; Koskela et al., 2020). This highlights the potential for this data to be used as a basis for further functional exploration of complex enzymatic processes that require knowledge of enzyme abundances and stoichiometries, possibly at the systems biology scale.
FIGURE 2.
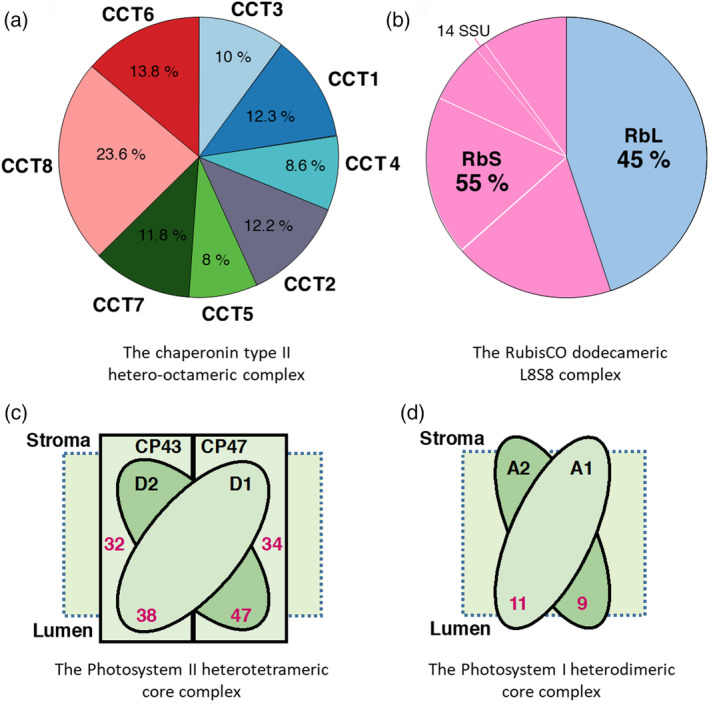
Quality control: Estimated stoichiometry of known biological complexes. Composition and subunits stoichiometry of (a) the chaperonin folding complex (CCTs). (b) the ribulose‐1,5‐bisphosphate carboxylase/oxygenase (RubisCO) large subunit (RbL, pale blue), which is chloroplast‐encoded and small subunits (RbS, pink), which are encoded by eighteen nuclear encoded genes. (c) Concentrations in micromolars (red) of photosystem II core subunits D1 (PsbA), D2 (PsbD), CP43 (PsbC) and CP47 (PsbB). (d) Concentrations in micromolars (red) of photosystem I core subunits, P700 chlorophyll a apoprotein A1 and A2 (PsaA and PsaB) [Colour figure can be viewed at wileyonlinelibrary.com]
3.4. Cellular compartmentation of significantly quantified proteins
The new annotation assigned a probable cellular compartment to most of the significantly quantified proteins. Chloroplast and mitochondrial proteins summed up to be ~64% and ~4% of the total protein mass, respectively. The cytosolic, nuclear, ER, vacuolar and peroxisomal proteins were together the remaining ~32% of the total protein mass (Figure 3a). Noticeably, chloroplasts occupy 20% and mitochondria 3% of the cell volume in typical P. patens protonemata cells (Furt et al., 2012), whereas the remaining cytosol, nucleus, ER, peroxisomes and the vacuoles together occupy 77% of the cell volume (Figure 3b). This implies dramatic differences of protein crowding values between cellular compartments: As much as 500 mg/ml in the case of chloroplasts, which is a very compact, near solid, protein concentration, likely owing to the great compactness of the grana lamella of the thylakoids. 235 mg/ml were estimated for the mitochondria, which is in the range of crowding values measured for bacteria (Cayley, Lewis, Guttman, & Record, 1991; Zimmerman & Trach, 1991). As low as ~75 mg/ml was estimated to be the average protein crowding value for the remaining volume occupied by the cytosol, the nucleus, the ER, peroxisomes and the vacuoles. Noticeably vacuoles are small and fragmented at this early developmental stage of the moss protonema cells and do not contribute to most of the cellular volume, as it is the case for differentiated mesophyll cells of higher plants (Figure 3c).
FIGURE 3.
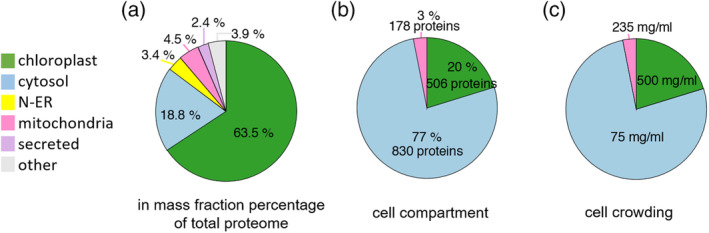
Distribution of significantly quantified proteins according to their predicted protein subcellular location (a) Protein distribution expressed in mass fraction percentage of total proteome within different predicted cellular compartments. (b) Distribution of identified proteins by estimated cellular compartment volume. (c) Molecular crowding of proteome based on the estimated volumes of cellular compartments. Here, cytoplasm included also organelles (nucleus‐endoplasmic reticulum, vacuoles and plasma membrane). Chloroplast is in green, cytoplasm in blue and mitochondrion in pink [Colour figure can be viewed at wileyonlinelibrary.com]
Increased protein crowding is thought to increase the aggregation‐propensity of nascent proteins (Martin & Hartl, 1997; van den Berg, Wain, Dobson, & Ellis, 2000), especially under heat shock. We therefore expected that a mild heat‐priming treatment would call for the accumulation of HSP‐chaperones, as part of a mechanism to prevent and revert heat‐induced protein aggregations, in the most crowded and therefore the most aggregation‐prone subcellular compartments: the chloroplasts and the mitochondria.
3.5. Mass fraction differences between heat‐priming treatment and control temperature
Without yet applying a statistical significance threshold, mass fraction bulk differences of average values between heat‐treated and control cells showed 716 proteins that became heat‐accumulated and contributed to a net mass gain of 7% and 798 proteins that became heat‐depleted that contributed to a reciprocal net mass loss of 7%. This indicates that, as expected, the mild 4 hrs heat‐priming treatment did not change the overall protein crowding of the various cellular compartments (Figure 4a). When applying a stringent significance threshold of p < 0.05 (applying multiple‐testing correction), 91 proteins remained significantly accumulated and 140 significantly depleted, summing into an equilibrated mass gain and mass loss of 4% (Figure 4b). Confirming earlier RNA expressomic observations (Finka & Goloubinoff, 2013), and in opposition to the general interchangeable use in the literature of the terms ‘heat‐shock proteins’ with ‘molecular chaperones’, only 11% of the heat‐accumulated proteins in the chloroplast belonged to members of the ‘chaperome’ (chaperone and co‐chaperones) category (Figure 4b). The other, non‐chaperome heat‐inducible proteins of the chloroplast belonged to various categories, mostly enzymes acting on small‐molecule metabolites.
FIGURE 4.
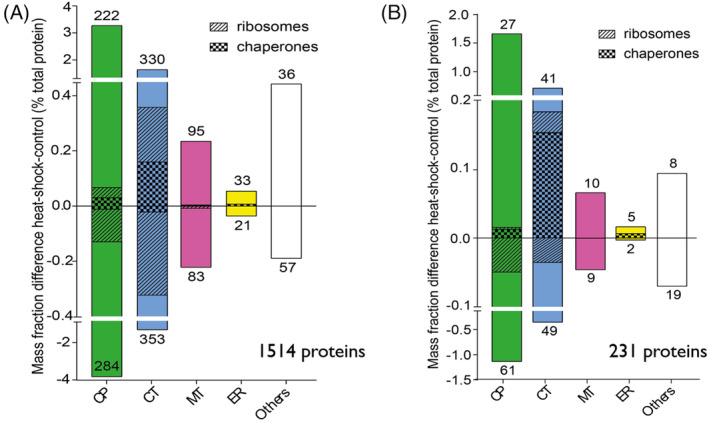
Mass fraction differences between HS/control by cellular compartments. (a) Mass fraction of the whole set of 1,514 quantified proteins. Proteins annotated as chaperones are represented as bars with diagonal hatches; and ribosomal proteins with vertical hatches. Other proteins are shown in white. The respective number of up‐ and downregulated proteins in each compartment are shown above and below each bar, respectively. (b) Mass fraction differences (expressed in % of the total proteome) between heat‐primed and control cells within the different cellular compartments calculated from the 231 proteins set which significantly change under HS [Colour figure can be viewed at wileyonlinelibrary.com]
Similarly, out of five significantly heat‐accumulated proteins in the ER, HSP70 (HSPA5) was the sole member of the chaperome. In the mitochondria, none of the ten significantly heat‐accumulated proteins were molecular chaperones or co‐chaperones. Together, the net mass increase by chaperome members in chloroplast, ER and mitochondria, was 1.1% of the total net mass increase in all heat‐accumulated proteins in these respective compartments. In contrast, three members of the HSP20s became strongly heat‐priming accumulated in the cytosol (including also the nucleus). While strongly repressed at basal temperature, three HSP20s (HSP17.4A, HSP17.7 and HSP17.8) made up 11.2% of the accumulated mass of all heat‐induced proteins in the cytoplasm+nucleus alone. This was independently observed by western‐blot (Figure S2). Hence, unexpectedly, the heat‐induced molecular chaperones were mainly found in the least crowded cellular compartment, where proteins were expected to be the least aggregation‐prone under heat‐shock. It should be noticed that the nuclear and cytosolic localizations were merged in our analysis. It is thus possible that HSP20s, which may diffuse into the more crowded nucleoplasm, might protect labile proteins there. Yet, the mild, harmless heat‐priming treatment addressed here, which unlikely causes protein aggregation, must be considered as dissimilar to the noxious heat‐shock treatments typically described previously in plants, which were likely more harmful (Larkindale & Vierling, 2008; Sun et al., 2020).
Here, the mild heat‐treatment impacted on the protein composition of the cytosolic and the chloroplast ribosomes. In chloroplasts, more ribosomal proteins were heat‐depleted than heat‐accumulated, leading to a net mass decrease of 0.06% of the total proteome, which does not necessarily indicates a decrease of protein synthesis capacity. We also observed the heat‐depletion, or the non‐replenishment, of members of the photosynthesis machinery, PSII in particular. In the cytosol, many ribosomal proteins were heat‐depleted and others became heat‐accumulated with no significant net abundance change (Figure 4b).
3.6. Distribution of differentially expressed proteins by subcellular localization
In terms of the classic FC parameter, the most dramatically heat‐induced proteins were localized in the cytosol (Figure 5). Whereas in terms of FC, the HSP17.4A and HSP17.7 were dramatically heat‐accumulated, their net increased abundance was remarkably low, according to their average molarity (Figure S3). In contrast, abundant chloroplast proteins, such as PSBE, a member of the photosystem machinery and CB2, which in FC were only slightly increased by heat, were strongly increased in abundance (delta mass). Thus, whereas HSP17.8 was induced by HP 5.7‐fold, its mass was increased only by 0.011% of the total protein mass, while the mass increase of PSBE in HP mosses was 0.35% of the total protein mass (i.e., ~32 times more than HSP17.8), despite a mere FC of 1.35 due to its high basal abundance. Thus, delta analysis allowed capturing significant changes that would have been overlooked using FC only, in the case of proteins that are already abundant in the control condition.
FIGURE 5.
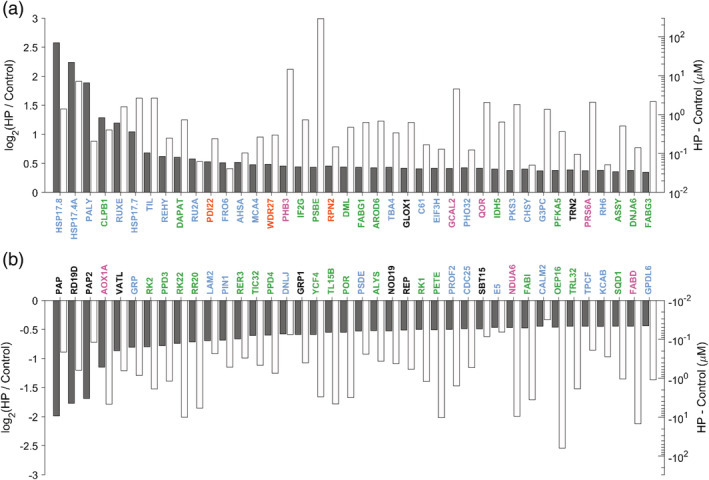
Ranking of the 40 most differentially accumulated or degraded protein (expressed in log2FC) with their quantity gained or lost after heat priming (expressed in molarity). Ranking of the 40 most differentially accumulated (a) or degraded protein (b) (expressed in log2FC) with their quantity gained or lost after heat priming (expressed in molarity). The gray bars represents the fold change state. The gain or loss in protein quantity following heat priming is represented into each bar, in white. Subcellular localization is represented in green for chloroplast, pink for mitochondrion, yellow for endoplasmic reticulum, blue for cytoplasm and nucleus and in dark for others [Colour figure can be viewed at wileyonlinelibrary.com]
When sorting the 40 most differentially accumulated or degraded proteins (expressed in log2‐FC), alongside their quantity gained or lost after the heat‐treatment (expressed in log molarity), according their intracellular concentration (Figure 5), the HSP20s were readily recognized as the most heat‐responsive, while their absolute mass gain was low, compared to other heat‐accumulated non‐chaperone proteins, such as the temperature‐induced lipocalin (TIL) or peroxiredoxin (REHY) (Figure 5a). PSBE is a special case as its absolute amounts were increased by about 200 μM (in the chloroplast) under heat, compared to the control (which was 906 μM), but because its basal abundance was extremely high, the fold‐change increase wrongly appeared to be nearly‐negligible. According to both delta abundance and fold‐changes, most of the heat‐depleted proteins were localized in the chloroplast (Figure 5b). Among them, were subunits of the photosystem machinery, such as YCF4, the outer plastid envelope protein OEP16, which is an amino‐acid selective channel, and the NADPH‐dependent protochlorophyllide oxidoreductase POR, which is also important for protein import. Proteins involved in lipid metabolism, such as FABI in the chloroplast, or FADB, in the mitochondrion were drastically heat‐depleted, suggesting a possible slowing down of fatty acid synthesis under heat‐priming conditions.
In many plant species, a heat‐stress can affect the photosynthetic efficiency of PSII (Havaux, 1992). To address this independently, the photosynthetic activity of protonema moss cells was measured before and after heat treatment with a PAM fluorometer. The maximum quantum efficiency of PSII (Fv/fm) was found to be indeed slightly, but significantly reduced following 4 hrs at 38°C. As a control, the Fv/fm was found to be dramatically reduced following a noxious 4 hrs heat shock at 45°C (Figure S4).
3.7. Heat‐induced changes in mRNA versus proteins
The net accumulation of new HSP proteins requires a prior accumulation of HSP mRNAs and their subsequent translation at a rate exceeding that of protein degradation. In contrast, the depletion of proteins may not obligatorily require a prior depletion/degradation of their mRNA. The preferred translation of new HSP mRNA following heat‐priming by the same amount (or possibly slightly less) of ribosomes, may suffice to slow down the rate of replenishment of the remaining non‐HSP proteins, while their rate of degradation can remain unchanged (Finka, Sood, Quadroni, Rios, & Goloubinoff, 2015). In an attempt to address the causes for protein depletion during heat‐priming, we performed an RNAseq analysis, in search for correlations between heat‐induced transcript abundances and MS‐derived protein abundances. We used four biological replicates of control and four replicates of 30 min 38°C‐treated cells, and obtained RNA derived from moss protonema (Dataset PRJNA666761). The relatively early time point of 30 min heat‐treatment was chosen based on the reasoning that heat‐induced mRNA production, which obligatorily precedes heat‐induced protein accumulation, becomes arrested beyond 30–60 min and starts subsiding thereafter despite the ongoing heat treatment (Finka & Goloubinoff, 2014). To correlate RNA and protein abundances, genes detected by both analyses were identified. Not all detected proteins and the corresponding transcripts could be correlated because the unlabelled MS method used was limited to the ~1,500 most abundant proteins of the cell. Hence, a correlation analysis was possible with only 88 heat‐induced genes and 136 heat‐depleted genes. Fold‐change in RNA and protein between control and heat‐treated mosses (Figure 6) showed an expected moderate positive correlation in the case of heat‐accumulated proteins, with a Pearson correlation coefficient of 0.52. When considering only the heat‐induced molecular chaperones (Figures 6a and S5a), the RNA‐protein fold‐change correlation was much more significant, with a Pearson correlation coefficient of 0.84. It confirmed that expectedly, the heat‐accumulated chaperones resulted from a prior heat‐induced accumulation of their corresponding mRNA. This was not the case of heat‐depleted proteins, for which most of the corresponding mRNAs were not depleted. Some mRNAs were even slightly accumulated, whereas their proteins became depleted during the heat‐treatment (Figures 6b and S5B). This indicates that the heat‐depletion of the proteins did not necessarily result from the heat‐induced degradation of their mRNA. Protein depletion would have as well resulted from their less effective replenishment during the heat‐treatment, by the ribosomes that become over‐flooded by an excess new HSP mRNAs.
FIGURE 6.
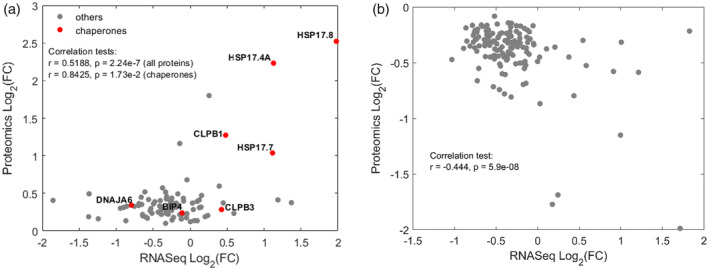
Scatter Plot showing a correlation in fold‐change expression, between 88 heat‐upregulated (a) and 136 downregulated (b) mRNAs and their corresponding significantly quantified proteins. Chaperones are in red. Pearson test are indicated. Whereas heat‐accumulated chaperone proteins expectedly result from the heat‐induced accumulation of their corresponding mRNA (a), the heat‐depleted proteins are not a consequence of the depletion of their mRNA [Colour figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
The growing impact of global warming and its adverse effects on crop yields is urging for more research on the mechanisms by which plants may cope with increasingly frequent extreme heat waves (Bokszczanin et al., 2013; Cramer, Urano, Delrot, Pezzotti, & Shinozaki, 2011; Lobell, Schlenker, & Costa‐Roberts, 2011). Physcomitrium patens (previously called Physcomitrella patens—see [Rensing, Goffinet, Meyberg, Wu, & Bezanilla, 2020]) is a bryophyte model for the study of land plant biology, as it may best highlight the mechanisms by which green algae that were initially protected by the buffering effect of large surrounding water bodies, acquired new abilities to cope with the much harsher terrestrial habitats, including high free oxygen, excess sun light and UV radiation, severe water shortage, dehydration and extreme diurnal variations in the ambient temperature (Rensing et al., 2008).
4.1. The advantages of LFQ‐based protein quantitation
High throughput microarray analyses have initially shown wide spectra of responses in Arabidopsis and tobacco to noxious heat shock treatments that may typically cause the denaturation of labile proteins and compromise membrane integrity and induce programmed cell death (Finka, Mattoo, & Goloubinoff, 2011; Larkindale & Vierling, 2008; Marsoni et al., 2010; Swindell, Huebner, & Weber, 2007; Vacca et al., 2006). The transcripts that were reported to be significantly up‐regulated under strong heat stress primarily encompass those of heat‐shock induced chaperones, HSFs and enzymes responsible for the synthesis of osmolytes, such as trehalose synthase (Glatz et al., 2016; Slama, Abdelly, Bouchereau, Flowers, & Savoure, 2015), of saturated lipids and of enzymes that detoxify reactive oxygen species (ROS) (Mittler, Finka, & Goloubinoff, 2012). Such approaches have been used in the systematic study of responses to a wide range of abiotic stresses (Ahmad et al., 2016; Kosova, Vitamvas, Urban, Prasil, & Renaut, 2018). Here, in contrast, we aimed at exploring the mechanisms by which a model land plant can sense increasingly warmer, albeit still non‐damaging temperatures and timely build effective molecular defences in anticipation of upcoming more noxious temperatures. The four hours of mild heat‐priming treatment at 38°C that were applied, should not be equated to a noxious HS: the protonema cells remained alive and resumed growth after this mild heat treatment (data not shown), suggesting that the integrity of proteins and membranes was minimally compromised, while effectively conferring acquired thermotolerance.
In view of the limitations of traditional two‐dimensional electrophoresis techniques, a number of high throughput alternatives with improved sensitivity and reproducibility have been applied (Fauvet et al., 2018; Finka et al., 2011; Goloubinoff, Sassi, Fauvet, Barducci, & De Los Rios, 2018). The quantitative proteomic approach used here offered a powerful means to unravel new proteins and pathways involved in the plant response to a mild HP condition typically leading to the onset of AT. Moreover, information at the protein level has thus far not provided absolute quantities of heat‐induced HSPs and the specific quantitative differences between the heat‐primed and control condition, nor about the cellular localization of HSPs, limiting our understanding of the molecular mechanisms leading to plant AT.
4.2. Protein annotation and quantification
To analyse the data from label‐free quantitative proteomic analysis (LFQ) of Physcomitrium patens cells without and following HP, we used both principal component analysis and hierarchical clustering to obtain an initial overview of the datasets (Figures S6 and S7). In both cases, it provided evidence for a significant reorganization of the moss proteome in response to heat priming. The LFQ method using six biological replicates per condition was necessary because it could easily generate estimates of absolute amounts, at least for the most abundant proteins and using less replicates as generally done in proteomic studies for economic reasons, would have strongly reduced the number of the significantly differentially expressed proteins (Cox & Mann, 2008; Goloubinoff et al., 2018). 7,097 proteins were thus identified in both control and heat‐priming conditions (Figure 1b) but the LFQ proteomics approach could not detect very low‐abundance proteins, such as transcription factors (Cox & Mann, 2008). When a stringent statistical filtering was applied on the six biological replicates per condition, only 1,514 proteins were retained with ascertained abundances above zero. While appearing at first conservative, this LFQ analysis showed that they nevertheless represented >96% of the total proteome by mass (Figure 1c).
In order to provide a mechanistic insight into plant heat responses, the OMICs analysis required the quantified protein IDs to be functionally annotated and presumably assigned to a given cellular compartment. Using BLASTp‐based sequence similarity and loctree3 annotation (Goldberg et al., 2014), the pipeline that combined automated and manual curation could assign probable functions and a subcellular localization to all 1,514 identified and quantified proteins of moss cells (Figure 1). The resulting absolute protein quantifications were validated by translating them in relative cellular concentrations that confirmed expected stoichiometries of subunits in well‐known complexes.
4.3. Quantifying treatment effects using abundance ratios and differences
With data from DNA microarrays (Larsson, Wahlestedt, & Timmons, 2005; Tusher, Tibshirani, & Chu, 2001), or label‐based proteomic MS (Ong et al., 2002; Thompson et al., 2003), ‘fold‐change’ (FC) analysis is the only way to express differences between treatment and control conditions. However, the systematic division of the treatment value by the control value, and its expression in the form of a single fold‐change value results in an effective loss of precious information. FC values may be misleading when abundances in the control condition are zero or very near to zero. Therefore, in addition to using the FC in protein abundances (HP/Control), we also quantified protein abundance differences (HP minus Control), which was rendered possible by the LFQ method and referred to as ‘deltas’ (Fauvet et al., 2018; Finka et al., 2015; Finka & Goloubinoff, 2013) (Figure S1 and Table S1). More than half (128) of the 231 significantly different proteins between HP and control were identified using both FC and delta values. 82 more proteins were significantly varying only in fold‐change, and 21 only had significant deltas more than FCs (Figure S1). Using the combination of fold‐change and delta values, it appeared that the most strongly heat‐induced or heat‐depleted proteins according to fold changes were not necessarily those with the largest change in absolute abundance (deltas) (Figure 5 and Table S1). In particular, the HSP20s, which according to FC appeared strongly induced, but contributed only mildly to the increased absolute abundance, compared to other HSPs that were much less induced in FCs but contributed much more to the delta masses.
4.4. Effects of mild heat on chloroplast functions
The majority of heat‐depleted proteins were in the most crowded cellular compartment, the chloroplast (Figure 3). Their identity suggested that the photosynthesis machineries, as well as protein and lipid synthesis machineries became slightly depleted during the mild heat treatment (Figures 4, 5, 6 and S3). Photosynthesis is usually the first metabolic mechanism to become affected by thermal variation and photosystem II (PSII) is apparently the most thermosensitive component (Mathur, Agrawal, & Jajoo, 2014). Indeed, the measured Fv/fm ratio after the mild heat‐treatment showed an inhibition of Photosystem II (PSII) that could derive from the observed mild depletion of protein subunits of Photosystem II and in the thylakoid antennae (Figure S4). We found that PSII reaction centers were more abundant than PSI reaction centers. The stoichiometry between PSII and PSI was initially thought to be ~1:1 (Whitmarsh & Ort, 1984). However, later studies determined a ratio closer to two (Fan et al., 2008). It is also known that the extent of PSII excess is dependent upon the plant species as well as on the light regime (Chow, Melis, & Anderson, 1990; Dietzel, Brautigam, & Pfannschmidt, 2008; Fan et al., 2008). Moreover, the ratio in cyanobacteria is 5:1 and quite variable ‐ leading to state changes and regulation of movement of complexes (Fujita, Murakami, Aizawa, & Ohki, 1994). Therefore, we interpret the four‐fold excess of PSII core over that of PSI that was found here, as possibly related to archaic nature of the chloroplasts in P. patens protonemata cells, as well as on the growth conditions. Photosynthesis rates generally decrease when the temperature exceeds the optimum specific for each plant species (Mathur et al., 2014). Moderate heat stress stimulates photoinhibition, resulting in increased ROS in the chloroplast, leading to increased ROS in the cytosol (Berry & Bjorkman, 1980). The main ROS scavenging enzymatic systems include superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX) and glutathione peroxidase (GPX) (Apel & Hirt, 2004). Thus, it is reasonable to assume that HS‐accumulated levels of ROS are transiently well tolerated in plants, playing a role in the mediation of HSR signalling and being rapidly eliminated by ROS scavenging enzymes.
HS has also been shown to slow down cellular protein synthesis by sequestering mRNAs encoding house‐keeping proteins in stress granules. These mRNAs are also in part outcompeted by the newly accumulated HSP mRNAs for slightly lesser amount of ribosomes, and possibly also because of a general slowing down of translation (Shalgi et al., 2013). The overall depletion of chloroplast ribosomal proteins could imply a general slowing down of the translation machinery. Yet, it should be noted that ribosomes are ancient RNA‐based enzymes that became increasingly regulated by peripheral proteins in the course of evolution (Fox, 2010). Plant ribosomes in particular, are highly heterogeneous in their protein composition (Giavalisco et al., 2005) and subunits may vary as a function of leaf development (Horiguchi et al., 2011) or in response to nutrient availability (Hummel et al., 2012), and here also in response to heat‐priming.
4.5. The lesson from comparing mRNA amounts and heat‐accumulated and depleted protein amounts
Relatively few studies compared global heat‐induced proteins and their corresponding transcript expression (Fu et al., 2009; Irmler et al., 2008; Pavelka et al., 2008). RNAseq for measurements of mRNA levels show only a very poor correlation with protein expression levels (Schwanhäusser et al., 2011). Regarding crowding, it should be noted that that the number of mRNA molecules can be three to four orders of magnitude lower that their protein they produce (de Sousa Abreu, Penalva, Marcotte, & Vogel, 2009). Hence, during heat priming, the total (RNA + protein) cellular crowding is not expected to significantly increase; and the flooding of the pre‐existing ribosomes with newly synthesized HSP mRNAs results in the production of new specialized proteins, such as the HSP20s, without globally increasing protein crowding. A delicate equilibrium between rate of protein degradation and protein‐replenishment must exist for each of the constitutively‐expressed house‐keeping protein of the cell. However, during heat‐priming, the massive production of a few heat‐induced proteins may require much higher mRNA molecules per newly produced HSP. These abundant new mRNAs of ~ a hundred HSPs are expected to compete with the many thousands of house‐keeping mRNAs, which are in very low copy numbers, thereby leading to the net accumulation of new HSPs, at the cost of a general, mild, across‐the‐board depletion owing to an expectedly slower replenishment, of the non‐HSPs house‐keeping proteins (Finka et al., 2015). In our study, the relative transcript and protein expression were reasonably well correlated only in the case of the most heat‐enriched proteins and RNAs (Finka et al., 2011). We found that more RNA was needed to make more HSP proteins, but in contrast, RNA degradation (or lower transcription) was not needed for depletion of proteins. This strongly indicates that only several heat‐accumulated proteins (HSP20s in particular) apparently act as specific key players in the onset of AT, whereas the heat‐depleted proteins are more likely to be non‐specific bystanders that do not play a direct role in the onset of AT, their depletion being the consequence of being out‐competed by HSP‐specific mRNAs and some of their polypeptides of having high intrinsic degradation rates.
4.6. The HSR takes place in less‐crowded cellular compartments
Microarray and RNAseq analysis have shown that in all organisms, less than a third of the heat‐inducible proteins (HSPs) are bona fide members of the ‘chaperome’ (HSP100, HSP90, HSP70, HSP60, co‐chaperones and foldases). This notwithstanding, chaperome encoding genes are in general ~20 times more likely to be also heat‐inducible, than non‐chaperome encoding genes (Finka et al., 2011, 2015). Our proteomic data confirmed that chaperome proteins are 14‐times more likely to be also heat‐inducible, than the other heat‐inducible non‐chaperome proteins (Figures 4, 5, 6, S3, and S5). An unexpected finding was that the mild heat treatment that typically leads to the onset of AT, did not involve an increase in the amount of molecular chaperones in the chloroplast, although the chloroplast is the most crowded compartment of the cell (Figures 4, 5, S3, S5). The chloroplast‐localized small heat‐shock protein HSP21 has been reported to act as a chaperone to maintain the stability of PSII in Arabidopsis under heat stress (Bernfur, Rutsdottir, & Emanuelsson, 2017; Chen, He, Chen, & Guo, 2017). Whereas RNAseq of mosses treated 30 min at 38°C indeed showed that the moss HSP21 ortholog (Pp3c7_25570) was significantly upregulated 1.26 fold (p value < .005), the low resolution of our quantitative proteomic analysis could not confirm a corresponding mass difference for this chloroplast sHSP. Instead, as initially shown by several gene‐expression data in different plant species (Cohen & Leach, 2019; Kang et al., 2020; Liu et al., 2015; Rahmati Ishka et al., 2018; Wang et al., 2019), the quantitative proteomic data confirmed that the heat‐treatment response is primarily taking place in the cytoplasm (Figures 4, 5, 6, and S3), which we found to be the least crowded compartment (~75 mg/ml vs. over 500 mg/ml total protein in the chloroplast, Figure 3). High protein crowding is thought to be detrimental to protein integrity under stress, such as heat shock, because this generates misfolded proteins with a high tendency to aggregate (Hartl & Hayer‐Hartl, 2002). Aggregation is a concentration‐dependent process where higher protein crowding would be associated to faster aggregation (van den Berg, Ellis, & Dobson, 1999). To counteract this, one would expect a large fold increase of chaperones in the chloroplast; however this was not the case (Figures 4, 5, and S3). There is evidence that the seemingly hostile environment of a highly crowded organelle might protect native proteins against stress‐induced unfolding and misfolding. Previous studies have documented the stabilizing effects of elevated molecular crowding on the equilibrium thermodynamics of biomolecular folding (Benton, Smith, Young, & Pielak, 2012; Roque, Ponte, & Suau, 2007; Zhou, Rivas, & Minton, 2008). For the crowding calculation, the presence of vacuoles in the cytosol may have contributed to the resulting very low crowding value of the cytosol. Yet, young Physcomitrium protonema cells contain only few small vacuoles that do not occupy most of the cellular volume as in the case of differentiated higher plant cells (Furt et al., 2012). Surprisingly, it was in the cytosol that we found most of the heat‐accumulated molecular chaperones, and especially HSP20s (Figures 4, 5, and S3), suggesting that under priming heat treatment, cytosolic proteins are more susceptible to misfolding. This argument is valid only if we assume that misfolding and aggregation is the problem of some cytosolic proteins during mild heat‐priming that the HSP20s could resolve. Yet, if no significant protein misfolding and aggregation was to take place in the cytosol, this would raise the question: what essential function are the HSP20s carrying out that during HP that leads to AT?
4.7. The central role of HSP20 in plant AT
Cytosolic members of the HSP20 family, also called the small HSPs (Haslbeck & Vierling, 2015) are molecular chaperones, whose expression in plants is repressed at non‐stressful basal temperature and become dramatically accumulated under heat‐shock (Larkindale & Vierling, 2008; Lindquist & Craig, 1988). There is an experimentally established correlation between the over‐expression of HSPs, among which are the HSP101, HSP70 and HSP20s chaperones, and the successful onset of plant acquired thermotolerance. Reversely, loss of function mutations in heat‐inducible molecular chaperones has been shown to carry defective acquired thermotolerance phenotypes (Hong, Lee, & Vierling, 2003; Kim, Lee, Small, des Francs‐Small, & Vierling, 2012; McLoughlin et al., 2016; Queitsch, Hong, Vierling, Lindquist, & Goloubinoff, 2000) (Waters & Vierling, 2020). Without stress, HSP20s often form oligomers, which can transiently dissociate in response to a HS, exposing hydrophobic surfaces to bind misfolded protein intermediates, thereby inhibiting aggregation (Nakamoto & Vígh, 2007). The rapid accumulation of HSP20s following a mild non‐damaging rise of temperature is one of the most sensitive molecular responses to an environmental cue (Saidi et al., 2010). HSP20s are able to prevent the aggregation of artificially unfolded polypeptides or of stress‐denaturing polypeptides through the ATP‐dependent HSP70/90 chaperone system (Kotak et al., 2007; Liberek, Lewandowska, & Zietkiewicz, 2008), whereas their apparent membrane localization indicates where they can act as ‘membrane‐stabilizing factors’ by influencing the integrity of the cell and thylakoid membranes during stress (Horvath et al., 2012). In this study, we detected by proteomic three different HSP20s and observed a massive (according to fold changes) up‐regulation of HSP20s in cytoplasm, the least crowded compartment (Figure S2), expected to be the least prone to protein aggregation. In the RNAseq data, we detected 20 HSP20s but only the same three from the proteomic analysis were strongly induced. This counter‐intuitive massive upregulation of HSP20s in relatively low net mass amounts in the cytosol may be indicative of a specialized, rather than a generalist function to this subset of HSP20s, which could target for example rare specific heat‐labile cytosolic proteins and counter their aggregation. Moreover, these HSP20s could act as specific repressors of signal proteins mediating programmed cell death, otherwise taking place inexorably during and following a noxious damaging heat‐shock. Indeed, indicative of an apoptotic signal, plants that were not heat‐primed, that once exposed to a noxious heat‐shock do not readily show large‐scale cellular damages may nevertheless die within a few days after the noxious heat stress (Reape & McCabe, 2010; Reape, Molony, & McCabe, 2008). It was shown that an overexpression of David's Lily HSP20 in Arabidopsis transgenic plants resulted in increased transcript and enzymatic activities of superoxide dismutases and catalases, the latter being effective scavengers for ROS, which in turn protect cells from oxidative damage (Mu et al., 2013) and from ROS‐mediated apoptosis. Hence, mammalian HSP20 homologs have been shown to have anti‐apoptotic activities (Kamradt, Chen, & Cryns, 2001; Mao, Liu, Xiang, & Li, 2004; Mehlen, Schulze‐Osthoff, & Arrigo, 1996; Nahomi, DiMauro, Wang, & Nagaraj, 2015). In animals, HSP27 was shown to prevent TNF‐α‐induced apoptosis and to interact with Bax, a pro‐apoptotic protein, by inhibiting its translocation into mitochondria (Mao et al., 2004; Mehlen, Kretz‐Remy, Preville, & Arrigo, 1996). A possible similar anti‐apoptotic effect and involvement in signalling pathway, remains to be explored in plants. Nevertheless, the transcriptomic data showed that an apoptotic signal (Reape & McCabe, 2010) composed of upregulated metacaspases occurs. By over‐expressing actors that can inhibit apoptotic signals, such as HSP20, peroxidases or anti‐apoptotic Bax inhibitor‐1 family protein, priming may act as a transient safe‐keeping mechanism when land plants are more severely heat‐stressed for several hours, around noon, in order to prevent the onset of a default stress‐induced programed cell death.
Proteomics and transcriptomics show that also in moss, not all heat‐induced proteins are molecular chaperones and conversely, not all molecular chaperones become accumulated in response of a heat‐treatment. Hence, in higher plants, the enzyme ascorbate peroxidase is one of the most heat‐accumulated proteins, although its cellular function and mechanism is to detoxify H2O2 (Caverzan et al., 2012) and not to prevent and avert protein misfolding, as most chaperones do (Mattoo & Goloubinoff, 2014). In our study, we found other heat‐accumulated proteins, such as the temperature‐induced lipocalin (TIL), which is required for AT (Chi, Fung, Liu, Hsu, & Charng, 2009) by acting against lipid peroxidation induced by severe HS or peroxiredoxin (REHY) and by scavenging hydrogen peroxide and lipid peroxides (Cheng et al., 2016) (Figure 5, S3).
Using omic approaches, we have obtained unbiased new information on the identity, cellular concentration and subcellular localization of the main heat‐accumulated proteins, following a typical mild heat‐priming treatment leading to the onset of AT. It suggested putative new functions for cytosolic HSP20s and indicated that the heat‐depletion of some abundant proteins, many of which from the photosynthetic apparatus, unlikely contributed to the onset mechanism of AT, but was rather a counterbalancing consequence of the heat‐priming accumulation of various HSPs, in particular the HSP20s with apoptotic actions which are central to the onset of AT.
5. OUTLOOK
At dawn of a summer day, land plants must sense, respond and adapt within a few hours to upcoming excessive afternoon temperatures. Plants respond to a rising heat through the specific up regulation of genes encoding, per definition, for heat‐shock proteins, which must be balanced by an across‐the board slowing down in the synthesis of non‐heat shock protein genes, many of which house‐keeping genes. Proteins are the carrier of most cellular functions. The law of mass action proposes that biochemical reactions depend both on the stoichiometry and on the absolute concentrations of substrate and the product molecules, as well as on the absolute concentration of the enzyme, which is accelerating that reaction in the cell. In an attempt to decipher this network (thermotolerance, HSR), quantitative values, alongside classic fold change ratios must be introduced into the equations. This research generated new insights on the mechanism leading to plant acquired thermotolerance. It confirmed the central role of HSP20s in AT albeit not so much, as generally thought, in the preventing heat‐induced protein aggregations that unlikely take place under heat‐priming conditions and are dramatically accumulated, in FC, during an HS, but mass‐wise it is not substantial. When a protein is very abundant, it is likely to be very important for basal house‐keeping cellular functions. When however, the expression of a protein is strongly repressed by the cell without heat, and it is specifically de‐repressed under heat‐, it accumulates but only in minute amounts, it is likely to carry a very important role in cellular defences against stress damages, albeit only transiently. The fact that evolution has chosen to strongly repress the expression of HSP20s without stress, when they are not needed, suggest that HSP20s carry an important stress‐protection function whose fitness cost to maintain constitutively without heat‐stress would be excessively high. Our data suggest that rather than, or in addition to the undertaking of a non‐specific aggregation‐preventing function, the accumulated cytosolic HSP20s may also act on a minority of rare signalling and/or regulatory proteins that control heat‐induced apoptosis and scavenging for ROS, which therefore lead to the onset of moss AT. Further research is needed to address a comprehensive analyse of responses to priming and acquired thermotolerance. Metabolomics is to be integrated to the transcriptomic and proteomic approaches. Indeed plant cells may accumulate metabolites and osmoprotectants such as proline, betaine or trehalose, acting as chemical chaperones that can mitigate heat stress damages in protein (Diamant, Eliahu, Rosenthal, & Goloubinoff, 2001) and membrane integrity (Bownik, Szabelak, Kulinska, & Waleka, 2019; H. W. Li, Zang, Deng, & Wang, 2011).
Supporting information
Table S1. List of the proteins identified and quantified. Abundance values in CONTROL and HP are expressed as mass fractions (percentage of the total protein mass) or molarity (as micromolars).
Figure S1. Venn diagram showing the number of overlapping and not overlapping proteins identified, significantly quantified and significantly differentially accumulated proteins in Physcomitrium patens under heat priming in fold‐change and in delta (differences of mass).
Figure S2. Immunoblot showing the relative expression levels of HSP17.7 in Physcomitrium patens HSP‐GUS line following 4 hrs priming at 38°C and at 22°C. The coomassie gel showing the large Rubisco subunit (~53 kDa) was used as a loading control.
Figure S3. Distribution of differentially expressed proteins by subcellular localization. MA‐plot showing log ratio versus abundance plots in molarity for heat treated moss protonemata compared to control. Each protein is represented as a dot and is mapped according to its mass fraction in fold change on the abscissa axis (x) and its fold change on the ordinate axis (y). Proteins considered as significantly regulated (p value < .05) are plotted in full dot; or grey dot if they are not significant. Subcellular localization is represented in green for chloroplast, pink for mitochondrion, yellow for endoplasmic reticulum, blue for cytoplasm and nucleus and in dark for others.
Figure S4. Bar diagram showing the effect of 4 hrs at various temperature on photosynthesis activity by calculating Fv/fm ratio. Each black dot represents biological replicate and the asterisks designate the significance of changes from their resultant control (****p < .001).
Figure S5. ScatterPlot showing a correlation between upregulated proteins in Parts Per Million (ppm) mapped against RNAseq data in Transcripts Per Million (tpm) at 22°C (a) and under heat priming (b), and between downregulated proteins in ppm mapped against RNAseq data in tpm at 22°C (c) and under heat priming (d). Chaperones are shown as red dots.
Figure S6. Principal component analysis. (a) PCA biplot obtained from the abundances of the 1,514 significantly quantified proteins, with the corresponding scree plot in (b). (c) PCA biplot obtained from the abundances of the 231 significantly varying proteins between CT and HP, with the corresponding scree plot in (d). Note that the GUS transgene was excluded from the data before performing PCAs.
Figure S7. Clustered heatmaps from the standardized, log‐transformed protein mass fractions obtained either from (a) the 1,514 significantly quantified proteins, or (b) from the 231 significantly varying proteins between CT and HP. Hierarchical clustering was computed using Euclidean distances and average linkages. Red and blue colours indicate standardized abundances above and below the per‐protein average, respectively.
ACKNOWLEDGMENTS
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This work was supported and funded by the University of Lausanne and the Swiss National Fund grant 31003A_175453.
Guihur A, Fauvet B, Finka A, Quadroni M, Goloubinoff P. Quantitative proteomic analysis to capture the role of heat‐accumulated proteins in moss plant acquired thermotolerance. Plant Cell Environ. 2021;44:2117–2133. 10.1111/pce.13975
Funding information Swiss National Fund, Grant/Award Number: 31003A_175453; University of Lausanne
REFERENCES
- Ahmad, P., Abdel Latef, A. A., Rasool, S., Akram, N. A., Ashraf, M., & Gucel, S. (2016). Role of proteomics in crop stress tolerance. Frontiers in Plant Science, 7, 1336. 10.3389/fpls.2016.01336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, K., & Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology, 55, 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Benton, L. A., Smith, A. E., Young, G. B., & Pielak, G. J. (2012). Unexpected effects of macromolecular crowding on protein stability. Biochemistry, 51, 9773–9775. 10.1021/bi300909q [DOI] [PubMed] [Google Scholar]
- Bernfur, K., Rutsdottir, G., & Emanuelsson, C. (2017). The chloroplast‐localized small heat shock protein Hsp21 associates with the thylakoid membranes in heat‐stressed plants. Protein Science, 26, 1773–1784. 10.1002/pro.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, J., & Bjorkman, O. (1980). Photosynthetic response and adaptation to temperature in higher plants. Annual Review of Plant Physiology, 31, 491–543. 10.1146/annurev.pp.31.060180.002423 [DOI] [Google Scholar]
- Bokszczanin, K. L., Fragkostefanakis, S., Bostan, H., Bovy, A., Chaturvedi, P., Chiusano, M. L., … Winter, P. (2013). Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Frontiers in Plant Science, 4, 315. 10.3389/fpls.2013.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownik, A., Szabelak, A., Kulinska, M., & Waleka, M. (2019). Effects of L‐proline on swimming parameters of Daphnia magna subjected to heat stress. Journal of Thermal Biology, 84, 154–163. 10.1016/j.jtherbio.2019.06.012 [DOI] [PubMed] [Google Scholar]
- Caverzan, A., Passaia, G., Rosa, S. B., Ribeiro, C. W., Lazzarotto, F., & Margis‐Pinheiro, M. (2012). Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genetics and Molecular Biology, 35, 1011–1019. 10.1590/s1415-47572012000600016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayley, S., Lewis, B. A., Guttman, H. J., & Record, M. T., Jr. (1991). Characterization of the cytoplasm of Escherichia coli K‐12 as a function of external osmolarity. Implications for protein‐DNA interactions in vivo. Journal of Molecular Biology, 222, 281–300. 10.1016/0022-2836(91)90212-o [DOI] [PubMed] [Google Scholar]
- Chan‐Schaminet, K. Y., Baniwal, S. K., Bublak, D., Nover, L., & Scharf, K. D. (2009). Specific interaction between tomato HsfA1 and HsfA2 creates hetero‐oligomeric superactivator complexes for synergistic activation of heat stress gene expression. The Journal of Biological Chemistry, 284, 20848–20857. 10.1074/jbc.M109.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che, P., Bussell, J. D., Zhou, W., Estavillo, G. M., Pogson, B. J., & Smith, S. M. (2010). Signaling from the endoplasmic reticulum activates Brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Science Signaling, 3, ra69. 10.1126/SCISIGNAL.2001140 [DOI] [PubMed] [Google Scholar]
- Chen, S. T., He, N. Y., Chen, J. H., & Guo, F. Q. (2017). Identification of core subunits of photosystem II as action sites of HSP21, which is activated by the GUN5‐mediated retrograde pathway in Arabidopsis. The Plant Journal, 89, 1106–1118. 10.1111/tpj.13447 [DOI] [PubMed] [Google Scholar]
- Cheng, F., Yin, L. L., Zhou, J., Xia, X. J., Shi, K., Yu, J. Q., … Foyer, C. H. (2016). Interactions between 2‐Cys peroxiredoxins and ascorbate in autophagosome formation during the heat stress response in Solanum lycopersicum . Journal of Experimental Botany, 67, 1919–1933. 10.1093/jxb/erw013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, W. T., Fung, R. W., Liu, H. C., Hsu, C. C., & Charng, Y. Y. (2009). Temperature‐induced lipocalin is required for basal and acquired thermotolerance in Arabidopsis. Plant, Cell & Environment, 32, 917–927. 10.1111/j.1365-3040.2009.01972.x [DOI] [PubMed] [Google Scholar]
- Chow, W. S., Melis, A., & Anderson, J. M. (1990). Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 87, 7502–7506. 10.1073/pnas.87.19.7502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. P., & Leach, J. E. (2019). Abiotic and biotic stresses induce a core transcriptome response in rice. Scientific Reports, 9, 6273. 10.1038/s41598-019-42731-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo, S., Charoensawan, V., Brestovitsky, A., Buning, R., Ravarani, C., Rhodes, D., … Wigge, P. A. (2017). Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in Arabidopsis. Molecular Plant, 10, 1258–1273. 10.1016/j.molp.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., & Mann, M. (2014). Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & Cellular Proteomics, 13, 2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J., & Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nature Biotechnology, 26, 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cramer, G. R., Urano, K., Delrot, S., Pezzotti, M., & Shinozaki, K. (2011). Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biology, 11, 163. 10.1186/1471-2229-11-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Abreu, R., Penalva, L. O., Marcotte, E. M., & Vogel, C. (2009). Global signatures of protein and mRNA expression levels. Molecular BioSystems, 5, 1512–1526. 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y., Humbert, S., Liu, J.‐X., Srivastava, R., Rothstein, S. J., & Howell, S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 108, 7247–7252. 10.1073/pnas.1102117108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant, S., Eliahu, N., Rosenthal, D., & Goloubinoff, P. (2001). Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. The Journal of Biological Chemistry, 276, 39586–39591. 10.1074/jbc.M103081200 [DOI] [PubMed] [Google Scholar]
- Dietzel, L., Brautigam, K., & Pfannschmidt, T. (2008). Photosynthetic acclimation: State transitions and adjustment of photosystem stoichiometry–functional relationships between short‐term and long‐term light quality acclimation in plants. The FEBS Journal, 275, 1080–1088. 10.1111/j.1742-4658.2008.06264.x [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., … Gingeras, T. R. (2013). STAR: Ultrafast universal RNA‐seq aligner. Bioinformatics, 29, 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, D.‐Y., Hope, A. B., Smith, P. J., Jia, H., Pace, R. J., Anderson, J. M., & Chow, W. S. (2008). The stoichiometry of photosystem II to photosystem I in higher plants. Dordrecht: Springer. [Google Scholar]
- Fauvet, B., Finka, A., Castanié‐Cornet, M.‐P., Cirinesi, A.‐M., Genevaux, P., Quadroni, M., & Goloubinoff, P. (2018). Bacterial Hsp90 mediates the degradation of aggregation‐prone Hsp70‐Hsp40 substrates preferentially by HslUV proteolysis. bioRxiv, 451989. 10.1101/451989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka, A., Cuendet, A. F. H., Maathuis, F. J. M., Saidi, Y., & Goloubinoff, P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired Thermotolerance. Plant Cell, 24, 3333–3348. 10.1105/tpc.112.095844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka, A., & Goloubinoff, P. (2013). Proteomic data from human cell cultures refine mechanisms of chaperone‐mediated protein homeostasis. Cell Stress & Chaperones, 18, 591–605. 10.1007/s12192-013-0413-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka, A., & Goloubinoff, P. (2014). The CNGCb and CNGCd genes from Physcomitrella patens moss encode for thermosensory calcium channels responding to fluidity changes in the plasma membrane. Cell Stress and Chaperones, 19, 83–90. 10.1007/s12192-013-0436-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka, A., Mattoo, R. U. H., & Goloubinoff, P. (2011). Meta‐analysis of heat‐ and chemically upregulated chaperone genes in plant and human cells. Cell Stress & Chaperones, 16, 15–31. 10.1007/s12192-010-0216-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka, A., Schaefer, D. G., Saidi, Y., Goloubinoff, P., & Zryd, J. P. (2007). In vivo visualization of F‐Actin structures during the development of the moss Physcomitrella patens . The New Phytologist, 174, 63–76. 10.1111/j.1469-8137.2007.01989.x [DOI] [PubMed] [Google Scholar]
- Finka, A., Sood, V., Quadroni, M., Rios, P. D. L., & Goloubinoff, P. (2015). Quantitative proteomics of heat‐treated human cells show an across‐the‐board mild depletion of housekeeping proteins to massively accumulate few HSPs. Cell Stress and Chaperones, 20, 605–620. 10.1007/s12192-015-0583-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, G. E. (2010). Origin and evolution of the ribosome. Cold Spring Harbor Perspectives in Biology, 2, a003483. 10.1101/cshperspect.a003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Fu, N., Guo, S., Yan, Z., Xu, Y., Hu, H., … Khaitovich, P. (2009). Estimating accuracy of RNA‐Seq and microarrays with proteomics. BMC Genomics, 10, 161. 10.1186/1471-2164-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y., Murakami, A., Aizawa, K., & Ohki, K. (1994). Short‐term and long‐term adaptation of the photosynthetic apparatus: Homeostatic properties of thylakoids. In Bryant D. A. (Ed.), The molecular biology of cyanobacteria (pp. 677–692). Dordrecht: Springer Netherlands. [Google Scholar]
- Furt, F., Lemoi, K., Tuzel, E., & Vidali, L. (2012). Quantitative analysis of organelle distribution and dynamics in Physcomitrella patens protonemal cells. BMC Plant Biology, 12, 70. 10.1186/1471-2229-12-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F., Han, X., Wu, J., Zheng, S., Shang, Z., Sun, D., … Li, B. (2012). A heat‐activated calcium‐permeable channel–Arabidopsis cyclic nucleotide‐gated ion channel 6–Is involved in heat shock responses. The Plant Journal, 70, 1056–1069. 10.1111/j.1365-313X.2012.04969.x [DOI] [PubMed] [Google Scholar]
- Giavalisco, P., Wilson, D., Kreitler, T., Lehrach, H., Klose, J., Gobom, J., & Fucini, P. (2005). High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Molecular Biology, 57, 577–591. 10.1007/s11103-005-0699-3 [DOI] [PubMed] [Google Scholar]
- Glatz, A., Pilbat, A. M., Nemeth, G. L., Vince‐Kontar, K., Josvay, K., Hunya, A., … Torok, Z. (2016). Involvement of small heat shock proteins, trehalose, and lipids in the thermal stress management in Schizosaccharomyces pombe. Cell Stress & Chaperones, 21, 327–338. 10.1007/s12192-015-0662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, T., Hecht, M., Hamp, T., Karl, T., Yachdav, G., Ahmed, N., … Rost, B. (2014). LocTree3 prediction of localization. Nucleic Acids Research, 42, W350–W355. 10.1093/nar/gku396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloubinoff, P., Sassi, A. S., Fauvet, B., Barducci, A., & De Los Rios, P. (2018). Chaperones convert the energy from ATP into the nonequilibrium stabilization of native proteins. Nature Chemical Biology, 14, 388–395. 10.1038/s41589-018-0013-8 [DOI] [PubMed] [Google Scholar]
- Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., & Lu, M. H. (2016). The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Frontiers in Plant Science, 7, 114. 10.3389/fpls.2016.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, F. U., & Hayer‐Hartl, M. (2002). Molecular chaperones in the cytosol: From nascent chain to folded protein. Science, 295, 1852–1858. 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R., & Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. International Journal of Molecular Sciences, 14, 9643–9684. 10.3390/ijms14059643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck, M., & Vierling, E. (2015). A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. Journal of Molecular Biology, 427, 1537–1548. 10.1016/j.jmb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux, M. (1992). Stress tolerance of photosystem II in vivo: Antagonistic effects of water, heat, and photoinhibition stresses. Plant Physiology, 100, 424–432. 10.1104/pp.100.1.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.‐W., Lee, U., & Vierling, E. (2003). Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiology, 132, 757–767. 10.1104/pp.102.017145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi, G., Molla‐Morales, A., Perez‐Perez, J. M., Kojima, K., Robles, P., Ponce, M. R., … Tsukaya, H. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. The Plant Journal, 65, 724–736. 10.1111/j.1365-313X.2010.04457.x [DOI] [PubMed] [Google Scholar]
- Horvath, I., Glatz, A., Nakamoto, H., Mishkind, M. L., Munnik, T., Saidi, Y., … Vigh, L. (2012). Heat shock response in photosynthetic organisms: Membrane and lipid connections. Progress in Lipid Research, 51, 208–220. 10.1016/j.plipres.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Hummel, M., Cordewener, J. H., de Groot, J. C., Smeekens, S., America, A. H., & Hanson, J. (2012). Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MSE proteomics. Proteomics, 12, 1024–1038. 10.1002/pmic.201100413 [DOI] [PubMed] [Google Scholar]
- Irmler, M., Hartl, D., Schmidt, T., Schuchhardt, J., Lach, C., Meyer, H. E., … Beckers, J. (2008). An approach to handling and interpretation of ambiguous data in transcriptome and proteome comparisons. Proteomics, 8, 1165–1169. 10.1002/pmic.200700741 [DOI] [PubMed] [Google Scholar]
- Jia, T., Ito, H., & Tanaka, A. (2016). Simultaneous regulation of antenna size and photosystem I/II stoichiometry in Arabidopsis thaliana . Planta, 244, 1041–1053. 10.1007/s00425-016-2568-5 [DOI] [PubMed] [Google Scholar]
- Kamradt, M. C., Chen, F., & Cryns, V. L. (2001). The small heat shock protein alpha B‐crystallin negatively regulates cytochrome c‐ and caspase‐8‐dependent activation of caspase‐3 by inhibiting its autoproteolytic maturation. The Journal of Biological Chemistry, 276, 16059–16063. 10.1074/jbc.C100107200 [DOI] [PubMed] [Google Scholar]
- Kang, W. H., Sim, Y. M., Koo, N., Nam, J. Y., Lee, J., Kim, N., … Yeom, S. I. (2020). Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum L. Scientific Data, 7, 17. 10.1038/s41597-020-0352-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Lee, U., Small, I., des Francs‐Small, C. C., & Vierling, E. (2012). Mutations in an Arabidopsis mitochondrial transcription termination factor‐related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. The Plant Cell, 24, 3349–3365. 10.1105/tpc.112.101006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela, M. M., Brunje, A., Ivanauskaite, A., Lopez, L. S., Schneider, D., DeTar, R. A., … Mulo, P. (2020). Comparative analysis of thylakoid protein complexes in state transition mutants nsi and stn7: Focus on PSI and LHCII. Photosynthesis Research, 145, 15–30. 10.1007/s11120-020-00711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova, K., Vitamvas, P., Urban, M. O., Prasil, I. T., & Renaut, J. (2018). Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Frontiers in Plant Science, 9, 122. 10.3389/fpls.2018.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak, S., Larkindale, J., Lee, U., von Koskull‐Döring, P., Vierling, E., & Scharf, K.‐D. (2007). Complexity of the heat stress response in plants. Current Opinion in Plant Biology, 10, 310–316. 10.1016/j.pbi.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Kumar, S. V., & Wigge, P. A. (2010). H2A.Z‐containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell, 140, 136–147. 10.1016/j.cell.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Kurshakova, M. M., Krasnov, A. N., Kopytova, D. V., Shidlovskii, Y. V., Nikolenko, J. V., Nabirochkina, E. N., … Georgieva, S. G. (2007). SAGA and a novel drosophila export complex anchor efficient transcription and mRNA export to NPC. The EMBO Journal, 26, 4956–4965. 10.1038/sj.emboj.7601901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, D., Ullrich, K. K., Murat, F., Fuchs, J., Jenkins, J., Haas, F. B., … Rensing, S. A. (2018). The Physcomitrella patens chromosome‐scale assembly reveals moss genome structure and evolution. The Plant Journal, 93, 515–533. 10.1111/tpj.13801 [DOI] [PubMed] [Google Scholar]
- Larkindale, J., & Vierling, E. (2008). Core genome responses involved in acclimation to high temperature. Plant Physiology, 146, 748–761. 10.1104/pp.107.112060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, O., Wahlestedt, C., & Timmons, J. A. (2005). Considerations when using the significance analysis of microarrays (SAM) algorithm. BMC Bioinformatics, 6, 129. 10.1186/1471-2105-6-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T., & Lansbury, P. T., Jr. (2002). Neurodegenerative disease: Amyloid pores from pathogenic mutations. Nature, 418, 291. 10.1038/418291a [DOI] [PubMed] [Google Scholar]
- Li, B., & Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. W., Zang, B. S., Deng, X. W., & Wang, X. P. (2011). Overexpression of the trehalose‐6‐phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta, 234, 1007–1018. 10.1007/s00425-011-1458-0 [DOI] [PubMed] [Google Scholar]
- Liberek, K., Lewandowska, A., & Zietkiewicz, S. (2008). Chaperones in control of protein disaggregation. The EMBO Journal, 27, 328–335. 10.1038/sj.emboj.7601970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, P., & Sherman, F. (1997). The unique hetero‐oligomeric nature of the subunits in the catalytic cooperativity of the yeast Cct chaperonin complex. Proceedings of the National Academy of Sciences, 94, 10780–10785. 10.1073/pnas.94.20.10780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist, S., & Craig, E. A. (1988). The heat‐shock proteins. Annual Review of Genetics, 22, 631–677. 10.1146/annurev.ge.22.120188.003215 [DOI] [PubMed] [Google Scholar]
- Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., & Sun, Q. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biology, 15, 152. 10.1186/s12870-015-0511-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell, D. B., Schlenker, W., & Costa‐Roberts, J. (2011). Climate trends and global crop production since 1980. Science, 333, 616–620. 10.1126/science.1204531 [DOI] [PubMed] [Google Scholar]
- Mao, Y. W., Liu, J. P., Xiang, H., & Li, D. W. (2004). Human alphaA‐ and alphaB‐crystallins bind to Bax and Bcl‐X(S) to sequester their translocation during staurosporine‐induced apoptosis. Cell Death and Differentiation, 11, 512–526. 10.1038/sj.cdd.4401384 [DOI] [PubMed] [Google Scholar]
- Marsoni, M., Cantara, C., De Pinto, M. C., Gadaleta, C., De Gara, L., Bracale, M., & Vannini, C. (2010). Exploring the soluble proteome of tobacco bright Yellow‐2 cells at the switch towards different cell fates in response to heat shocks. Plant, Cell & Environment, 33, 1161–1175. 10.1111/j.1365-3040.2010.02137.x [DOI] [PubMed] [Google Scholar]
- Martin, J., & Hartl, F.‐U. (1997). The effect of macromolecular crowding on chaperonin‐mediated protein folding. Proceedings of the National Academy of Sciences, 94, 1107–1112. 10.1073/pnas.94.4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, S., Agrawal, D., & Jajoo, A. (2014). Photosynthesis: Response to high temperature stress. Journal of Photochemistry and Photobiology. B, 137, 116–126. 10.1016/j.jphotobiol.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Mattoo, R. U., & Goloubinoff, P. (2014). Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins. Cellular and Molecular Life Sciences, 71, 3311–3325. 10.1007/s00018-014-1627-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin, F., Basha, E., Fowler, M. E., Kim, M., Bordowitz, J., Katiyar‐Agarwal, S., & Vierling, E. (2016). Class I and II Small heat shock proteins together with HSP101 protect protein translation factors during heat stress. Plant Physiology, 172, 1221–1236. 10.1104/pp.16.00536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen, P., Kretz‐Remy, C., Preville, X., & Arrigo, A. P. (1996). Human hsp27, drosophila hsp27 and human alphaB‐crystallin expression‐mediated increase in glutathione is essential for the protective activity of these proteins against TNFalpha‐induced cell death. The EMBO Journal, 15, 2695–2706. [PMC free article] [PubMed] [Google Scholar]
- Mehlen, P., Schulze‐Osthoff, K., & Arrigo, A. P. (1996). Small stress proteins as novel regulators of apoptosis. Heat shock protein 27 blocks Fas/APO‐1‐ and staurosporine‐induced cell death. The Journal of Biological Chemistry, 271, 16510–16514. 10.1074/jbc.271.28.16510 [DOI] [PubMed] [Google Scholar]
- Mittler, R., Finka, A., & Goloubinoff, P. (2012). How do plants feel the heat? Trends in Biochemical Sciences, 37, 118–125. 10.1016/j.tibs.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Mu, C., Zhang, S., Yu, G., Chen, N., Li, X., & Liu, H. (2013). Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS One, 8, e82264. 10.1371/journal.pone.0082264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahomi, R. B., DiMauro, M. A., Wang, B., & Nagaraj, R. H. (2015). Identification of peptides in human Hsp20 and Hsp27 that possess molecular chaperone and anti‐apoptotic activities. The Biochemical Journal, 465, 115–125. 10.1042/BJ20140837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto, H., & Vígh, L. (2007). The small heat shock proteins and their clients. Cellular and Molecular Life Sciences, 64, 294–306. 10.1007/s00018-006-6321-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Y., & Xiang, Y. (2018). An overview of biomembrane functions in plant responses to high‐temperature stress. Frontiers in Plant Science, 9. 10.3389/fpls.2018.00915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., & Mann, M. (2002). Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics, 1, 376–386. 10.1074/mcp.m200025-mcp200 [DOI] [PubMed] [Google Scholar]
- Pavelka, N., Fournier, M. L., Swanson, S. K., Pelizzola, M., Ricciardi‐Castagnoli, P., Florens, L., & Washburn, M. P. (2008). Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Molecular & Cellular Proteomics: MCP, 7, 631–644. 10.1074/mcp.M700240-MCP200 [DOI] [PubMed] [Google Scholar]
- Peng, X., Zhang, X., Li, B., & Zhao, L. (2019). Cyclic nucleotide‐gated ion channel 6 mediates thermotolerance in Arabidopsis seedlings by regulating nitric oxide production via cytosolic calcium ions. BMC Plant Biology, 19, 368. 10.1186/s12870-019-1974-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch, C., Hong, S.‐W., Vierling, E., Lindquist, S., & Goloubinoff, P. (2000). Heat shock protein 101 plays a crucial role in Thermotolerance in Arabidopsis. The Plant Cell, 12, 479–492. 10.1105/tpc.12.4.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmati Ishka, M., Brown, E., Weigand, C., Tillett, R. L., Schlauch, K. A., Miller, G., & Harper, J. F. (2018). A comparison of heat‐stress transcriptome changes between wild‐type Arabidopsis pollen and a heat‐sensitive mutant harboring a knockout of cyclic nucleotide‐gated cation channel 16 (cngc16). BMC Genomics, 19, 549. 10.1186/s12864-018-4930-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reape, T. J., & McCabe, P. F. (2010). Apoptotic‐like regulation of programmed cell death in plants. Apoptosis, 15, 249–256. 10.1007/s10495-009-0447-2 [DOI] [PubMed] [Google Scholar]
- Reape, T. J., Molony, E. M., & McCabe, P. F. (2008). Programmed cell death in plants: Distinguishing between different modes. Journal of Experimental Botany, 59, 435–444. 10.1093/jxb/erm258 [DOI] [PubMed] [Google Scholar]
- Rensing, S. A., Goffinet, B., Meyberg, R., Wu, S. Z., & Bezanilla, M. (2020). The moss physcomitrium (Physcomitrella) patens: A model organism for non‐seed plants. Plant Cell, 32, 1361–1376. 10.1105/tpc.19.00828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., … Boore, J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science, 319, 64–69. 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- Rohner, S., Kalck, V., Wang, X., Ikegami, K., Lieb, J. D., Gasser, S. M., & Meister, P. (2013). Promoter‐ and RNA polymerase II‐dependent hsp‐16 gene association with nuclear pores in Caenorhabditis elegans . The Journal of Cell Biology, 200, 589–604. 10.1083/jcb.201207024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque, A., Ponte, I., & Suau, P. (2007). Macromolecular crowding induces a molten globule state in the C‐terminal domain of histone H1. Biophysical Journal, 93, 2170–2177. 10.1529/biophysj.107.104513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi, Y., Finka, A., Chakhporanian, M., Zryd, J. P., Schaefer, D. G., & Goloubinoff, P. (2005). Controlled expression of recombinant proteins in Physcomitrella patens by a conditional heat‐shock promoter: A tool for plant research and biotechnology. Plant Molecular Biology, 59, 697–711. 10.1007/s11103-005-0889-z [DOI] [PubMed] [Google Scholar]
- Saidi, Y., Finka, A., Muriset, M., Bromberg, Z., Weiss, Y. G., Maathuis, F. J., & Goloubinoff, P. (2009). The heat shock response in moss plants is regulated by specific calcium‐permeable channels in the plasma membrane. Plant Cell, 21, 2829–2843. 10.1105/tpc.108.065318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi, Y., Peter, M., Finka, A., Cicekli, C., Vigh, L., & Goloubinoff, P. (2010). Membrane lipid composition affects plant heat sensing and modulates Ca(2+)‐dependent heat shock response. Plant Signaling & Behavior, 5, 1530–1533. 10.4161/psb.5.12.13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl, R. A. V., Irina, I. P., Phillip, M. M., & Friedrich, S. (2006). Heat stress‐induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Molecular Biology, 61, 733–746. 10.1007/s11103-006-0045-4 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., … Selbach, M. (2011). Global quantification of mammalian gene expression control. Nature, 473, 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Shalgi, R., Hurt, J. A., Krykbaeva, I., Taipale, M., Lindquist, S., & Burge, C. B. (2013). Widespread regulation of translation by elongation pausing in heat shock. Molecular Cell, 49, 439–452. 10.1016/j.molcel.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S. K., De Los Rios, P., & Goloubinoff, P. (2011). Probing the different chaperone activities of the bacterial HSP70‐HSP40 system using a thermolabile luciferase substrate. Proteins, 79, 1991–1998. 10.1002/prot.23024 [DOI] [PubMed] [Google Scholar]
- Slama, I., Abdelly, C., Bouchereau, A., Flowers, T., & Savoure, A. (2015). Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Annals of Botany, 115, 433–447. 10.1093/aob/mcu239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, B. J., & Strasser, R. J. (1995). Measuring fast fluorescence transients to address environmental questions: the jip‐test. In Mathis P. (Ed.), Photosynthesis: from Light to Biosphere vol. 5 (pp. 977–980). Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Sugio, A., Dreos, R., Aparicio, F., & Maule, A. J. (2009). The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. The Plant Cell, 21, 642–654. 10.1105/TPC.108.062596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M., Huang, D., Zhang, A., Khan, I., Yan, H., Wang, X., … Huang, L. (2020). Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full‐length transcriptome sequencing. BMC Plant Biology, 20, 323–323. 10.1186/s12870-020-02530-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell, W. R., Huebner, M., & Weber, A. P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non‐heat stress response pathways. BMC Genomics, 8, 125. 10.1186/1471-2164-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A., Schafer, J., Kuhn, K., Kienle, S., Schwarz, J., Schmidt, G., … Hamon, C. (2003). Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical Chemistry, 75, 1895–1904. 10.1021/ac0262560 [DOI] [PubMed] [Google Scholar]
- Tunc‐Ozdemir, M., Tang, C., Ishka, M. R., Brown, E., Groves, N. R., Myers, C. T., … Harper, J. F. (2013). A cyclic nucleotide‐gated channel (CNGC16) in pollen is critical for stress tolerance in pollen reproductive development. Plant Physiology, 161, 1010–1020. 10.1104/pp.112.206888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G., Tibshirani, R., & Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America, 98, 5116–5121. 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca, R. A., Valenti, D., Bobba, A., Merafina, R. S., Passarella, S., & Marra, E. (2006). Cytochrome c is released in a reactive oxygen species‐dependent manner and is degraded via caspase‐like proteases in tobacco bright‐yellow 2 cells en route to heat shock‐induced cell death. Plant Physiology, 141, 208–219. 10.1104/pp.106.078683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, B., Ellis, R. J., & Dobson, C. M. (1999). Effects of macromolecular crowding on protein folding and aggregation. The EMBO Journal, 18, 6927–6933. 10.1093/emboj/18.24.6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, B., Wain, R., Dobson, C. M., & Ellis, R. J. (2000). Macromolecular crowding perturbs protein refolding kinetics: Implications for folding inside the cell. The EMBO Journal, 19, 3870–3875. 10.1093/emboj/19.15.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Jiang, B., Liu, W., Lin, Y., Liang, Z., He, X., & Peng, Q. (2019). Transcriptome analyses provide novel insights into heat stress responses in Chieh‐qua (Benincasa hispida Cogn. Var. Chieh‐qua how). International Journal of Molecular Sciences, 20. 10.3390/ijms20040883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, E. R., & Vierling, E. (2020). Plant small heat shock proteins – Evolutionary and functional diversity. New Phytologist, 227, 24–37. 10.1111/nph.16536 [DOI] [PubMed] [Google Scholar]
- Whitmarsh, J., & Ort, D. R. (1984). Stoichiometries of electron transport complexes in spinach chloroplasts. Archives of Biochemistry and Biophysics, 231, 378–389. 10.1016/0003-9861(84)90401-6 [DOI] [PubMed] [Google Scholar]
- Yu, H.‐D., Yang, X.‐F., Chen, S.‐T., Wang, Y.‐T., Li, J.‐K., Shen, Q., … Guo, F.‐Q. (2012). Downregulation of chloroplast RPS1 negatively modulates nuclear heat‐responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genetics, 8, e1002669. 10.1371/journal.pgen.1002669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H.‐X., Rivas, G., & Minton, A. P. (2008). Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annual Review of Biophysics, 37, 375–397. 10.1146/annurev.biophys.37.032807.125817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, S. B., & Trach, S. O. (1991). Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli . Journal of Molecular Biology, 222, 599–620. 10.1016/0022-2836(91)90499-v [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of the proteins identified and quantified. Abundance values in CONTROL and HP are expressed as mass fractions (percentage of the total protein mass) or molarity (as micromolars).
Figure S1. Venn diagram showing the number of overlapping and not overlapping proteins identified, significantly quantified and significantly differentially accumulated proteins in Physcomitrium patens under heat priming in fold‐change and in delta (differences of mass).
Figure S2. Immunoblot showing the relative expression levels of HSP17.7 in Physcomitrium patens HSP‐GUS line following 4 hrs priming at 38°C and at 22°C. The coomassie gel showing the large Rubisco subunit (~53 kDa) was used as a loading control.
Figure S3. Distribution of differentially expressed proteins by subcellular localization. MA‐plot showing log ratio versus abundance plots in molarity for heat treated moss protonemata compared to control. Each protein is represented as a dot and is mapped according to its mass fraction in fold change on the abscissa axis (x) and its fold change on the ordinate axis (y). Proteins considered as significantly regulated (p value < .05) are plotted in full dot; or grey dot if they are not significant. Subcellular localization is represented in green for chloroplast, pink for mitochondrion, yellow for endoplasmic reticulum, blue for cytoplasm and nucleus and in dark for others.
Figure S4. Bar diagram showing the effect of 4 hrs at various temperature on photosynthesis activity by calculating Fv/fm ratio. Each black dot represents biological replicate and the asterisks designate the significance of changes from their resultant control (****p < .001).
Figure S5. ScatterPlot showing a correlation between upregulated proteins in Parts Per Million (ppm) mapped against RNAseq data in Transcripts Per Million (tpm) at 22°C (a) and under heat priming (b), and between downregulated proteins in ppm mapped against RNAseq data in tpm at 22°C (c) and under heat priming (d). Chaperones are shown as red dots.
Figure S6. Principal component analysis. (a) PCA biplot obtained from the abundances of the 1,514 significantly quantified proteins, with the corresponding scree plot in (b). (c) PCA biplot obtained from the abundances of the 231 significantly varying proteins between CT and HP, with the corresponding scree plot in (d). Note that the GUS transgene was excluded from the data before performing PCAs.
Figure S7. Clustered heatmaps from the standardized, log‐transformed protein mass fractions obtained either from (a) the 1,514 significantly quantified proteins, or (b) from the 231 significantly varying proteins between CT and HP. Hierarchical clustering was computed using Euclidean distances and average linkages. Red and blue colours indicate standardized abundances above and below the per‐protein average, respectively.


