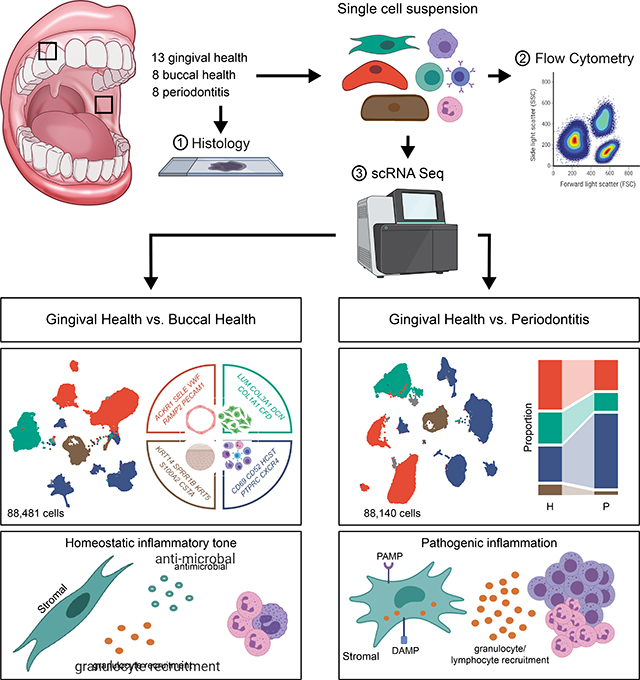
Summary
The oral mucosa remains an understudied barrier tissue. This is a site of rich exposure to antigens and commensals, and a tissue susceptible to one of the most prevalent human inflammatory diseases, periodontitis. Towards understanding tissue-specific pathophysiology, we compile a single cell transcriptome atlas of human oral mucosa in health individuals and patients with periodontitis. We uncover the complex cellular landscape of oral mucosal tissues and identify epithelial and stromal cell populations with inflammatory signatures that promote antimicrobial defenses and neutrophil recruitment. Our findings link exaggerated stromal cell responsiveness with enhanced neutrophil and leukocyte infiltration in periodontitis. Our work provides a resource characterizing the role of tissue stroma in regulating mucosal tissue homeostasis and disease pathogenesis.
Keywords: Oral mucosal immunity, human oral mucosal single-cell atlas, periodontitis single-cell atlas
Graphical Abstract
eTOC
A single-cell atlas of human oral mucosa connects inflammatory signatures in epithelial and stromal cells with neutrophil infiltration in periodontitis and identifies cell-specific expression patterns of periodontitis susceptibility genes.
Introduction
The oral mucosa is a site of first encounters (Gaffen and Moutsopoulos, 2020). Life-sustaining substances such as food, water, and air pass through the oral cavity, carrying along allergens, microbes, and food particles as they enter the lower gastrointestinal tract or respiratory system. In addition to constant exposure to microbes, allergens, and other antigens, tissues in the oral cavity must withstand frequent microdamage from masticatory forces. Despite – or perhaps because of – this tumultuous environment, the oral barrier displays minimal infection at mucosal surfaces and no overt inflammation at steady state. Moreover, the oral mucosa is remarkably efficient at wound healing (Iglesias-Bartolome et al., 2018), as most injuries heal without infection or scar formation. Yet, the mechanisms by which oral mucosal tissues interpret and respond to environmental cues are still largely unappreciated, which renders the study of this remarkably exposed and resilient tissue of broad interest.
The oral mucosal barrier is a multilayer squamous cell epithelium. However, different areas within the oral cavity have acquired unique anatomical characteristics adapted to the functions and specialization of their specific microenvironment (Moutsopoulos and Konkel, 2018). The majority of the oral mucosal barrier consists of a multilayer squamous epithelium with minimal keratinization (lining epithelium) which lines the inside of the cheeks (buccal mucosa), lips, floor of the mouth and the back of the throat, and protects vital structures. The rest of the oral mucosa is specialized. The hard palate, tongue dorsum and external area of gingiva are fully keratinized to withstand the constant forces of mastication (masticatory epithelia), while the tongue contains taste buds that mediate taste perception. A particularly unique area of the oral barrier is the tooth-associated mucosa, termed the gingiva. The external area of the gingiva is formed by masticatory epithelium, while the internal side (gingival crevice) is a non-keratinized squamous epithelium, which becomes progressively thin (down to 3–4 layers at its base). This thin mucosa is exposed to the complex tooth-adherent microbiome (Mark Welch et al., 2016).
In fact, a major distinction in oral epithelia can be made between buccal and gingival mucosae. Indeed, the commensal microbiome communities are particularly rich and diverse at the tooth interface compared to microbial communities found in mucosal oral sites (Human Microbiome Project, 2012; Proctor and Relman, 2017).. Furthermore, disease susceptibilities within the oral mucosa are most distinct between the lining and gingival mucosa. As such, ulcerative disorders and infections are most commonly present at the lining mucosa, while bullous dermatological diseases are encountered at the external gingiva (Moutsopoulos and Moutsopoulos, 2018). The gingiva is also the location involved in one of the most common inflammatory human diseases, periodontitis.
Periodontitis is a highly prevalent human disease, affecting in its moderate-to-severe forms approximately 8% of the general population (Eke et al., 2015). In periodontitis, a dysbiotic microbiome on the tooth surface is thought to trigger exaggerated inflammation within the tooth associated mucosa (gingiva) in susceptible individuals (Lamont et al., 2018; Loos and Van Dyke, 2020). This dysregulated inflammatory response leads to destruction of tooth-supporting structures, including mucosal tissue and tooth-supporting bone. To date the pathogenic mechanisms involved in periodontitis are incompletely understood.
Herein, we have compiled a human cell atlas of oral mucosal tissues in health and periodontal disease. Our work entails a comprehensive characterization of cell populations and their transcriptional signatures in clinical health and inflammatory disease and aims to uncover cell types and functions associated with promoting homeostasis and disease susceptibility at this important barrier. Indeed, in this atlas we identify populations of stromal cells promoting neutrophil migration in health that expand in disease. Furthermore, evaluation of cellular expression of genes linked to periodontal disease susceptibility reveals stromal and immune cell (particularly neutrophil) involvement in disease susceptibility. Collectively, our work provides a cellular census of distinct oral mucosal tissues in health and disease and suggests unique stromal-immune responsiveness mediating mucosal homeostasis which becomes dysregulated in mucosal inflammatory disease.
Results
Single-cell sequencing of ~88,000 cells identifies 4 major tissue compartments in healthy oral mucosa
To create a comprehensive single-cell transcriptome atlas of the human oral mucosa, we obtained biopsies of the buccal and gingival mucosa (Figure 1A) from healthy volunteers (n=20, Demographic data indicated in Table S1), meticulously screened for systemic and oral health. Biopsies were processed immediately into a single-cell suspension by mild enzymatic digestion and mechanical separation and cells were profiled using the 10X Genomics Chromium Droplet platform.. After quality control to remove low-quality cells expressing high mitochondrial gene signatures and exclude doublets, our dataset included ~88,000 cells (gingival mucosa [n=13, 53,482 cells] and buccal mucosa [n=8, 34,999 cells]) (Figure S1A–C; see Methods).
Figure 1: Major cell types in healthy adult oral mucosal tissues.
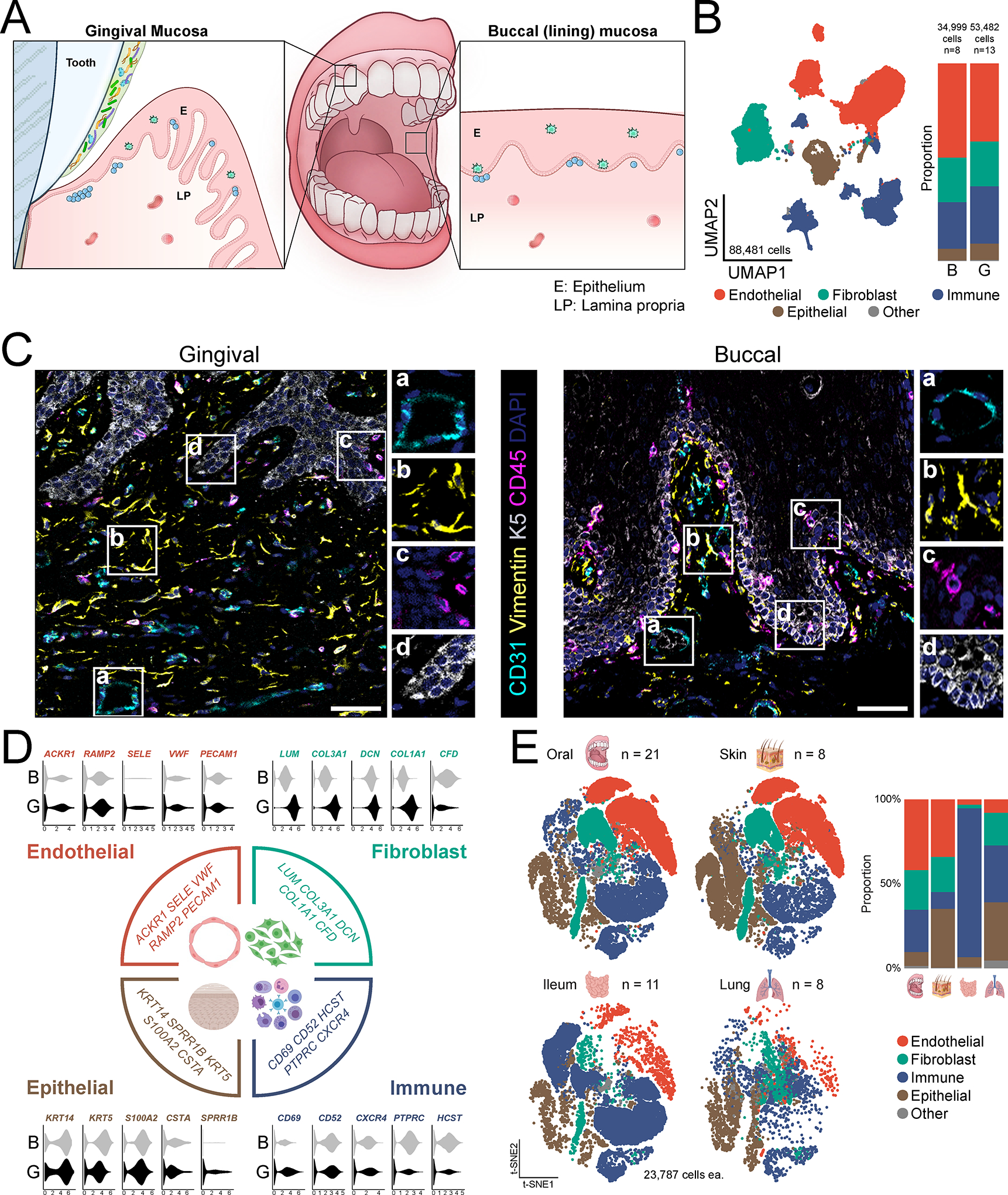
A. Schematic of the oral mucosal tissues biopsied with locations indicated (E-epithelium, LP- lamina propria). B. UMAP representation of major cell types identified by scRNA seq (n=21, 88,481 cells, left) and bar graph of relative cell proportions by tissue type (B-buccal, G-gingival) (right). C. Immunofluorescence depicting major cell types in healthy gingival and buccal mucosa. Scale bar: 41 μm. Stains indicated and color coded. D. Violin plots of selected markers for major cell populations and relative expression per tissue type. X-axis represents normalized average expression. E. t-SNE (left) and proportion plots (right) showing representation of major cell types in oral (n=21), skin (n=8), ileum (n=11), and lung (n=8) barrier tissues. All datasets were truncated to ensure equal number of cells (23,787) were compared. See also Table S1–S3, Figure S1.
Our analysis revealed 4 major cell compartments in oral mucosa comprised of epithelial, endothelial, fibroblast and immune cells, which were present in both gingiva and buccal mucosa of healthy adults (Figure 1B). Seurat batch correction (Stuart et al., 2019) showed good mixing of cells across donors (Table S2). Histological imaging confirmed the presence of the aforementioned 4 compartments within oral mucosal tissues and provided insight into the local tissue architecture (Figure 1C).
Analysis of the transcriptomic signatures for the major cell types documented differential expression of cell-defining genes for the 4 major cell compartments, which were generally conserved across mucosal sites. Identified endothelial (ACKR1, RAMP2, SELE, VWF, PECAM1), fibroblast (LUM, COL3A1, DCN, COL1A1, CFD), immune (CD69, CD52, CXCR4, PTPRC, HCST), and epithelial (KRT14, KRT5, S100A2, CTSA, SPRR1B) marker genes were cell-type specific and consistent with classical, well-established markers for each respective cell population (Figure 1D, Figure S1D).
Finally, interrogation of published data sets from healthy barrier tissues across the human body (oral, skin (He et al., 2020), ileum (Martin et al., 2019), lung (Reyfman et al., 2019)) demonstrated that these 4 major populations are also the main cellular compartments in all human barrier tissues evaluated (Figure 1E, Table S3). However, the relative representation of each major cell population differed between tissue types. Representation of these 4 main building blocks of barrier tissues was most comparable between the oral mucosa and skin barriers, although the oral mucosa contained an expanded immune compartment while the skin harbored a considerably larger epithelial compartment by proportion (Figure 1E).
Identification of stromal and epithelial subpopulations with distinct immune functionality
Louvain clustering demonstrates great diversity in stromal and epithelial cell populations in human oral mucosal tissues. Within the endothelial cell compartment, we defined 4 health- associated (H) venular endothelial cell subsets (H.VEC), vascular smooth muscle cells (H.SMC) and lymphatic endothelial cells (H.LEC), all of which were represented in both buccal and gingival mucosa (Figure 2A, Table S4). H.SMC and H.LEC clusters expressed genes related to muscle contraction (such as MYL9, ACTA2) and LEC fate commitment (PROX1) (Figure 2B, Figure S2A, Table S4). Venular endothelial cell subpopulations expressed genes consistent with distinct functions. H.VEC 1.1, 1.3, and 1.4 were all linked to immune functions. H.VEC 1.1 expressed a gene signature consistent with antigen presentation (HLADMA, CD74), while 1.3 and 1.4 appeared to promote immune and inflammatory responses (such as response to LPS and IFN signaling) and expressed genes relevant to neutrophil recruitment and IFN signaling (CSF3, CXCL2, MX1) (Figure 2B, Figure S2A). H.VEC 1.2 cells displayed a gene signature consistent with active pathways for endothelium development (such as FLT1) (Figure 2B, Figure S2A, Table S4).
Figure 2: Stromal and epithelial subpopulations in healthy oral mucosa.
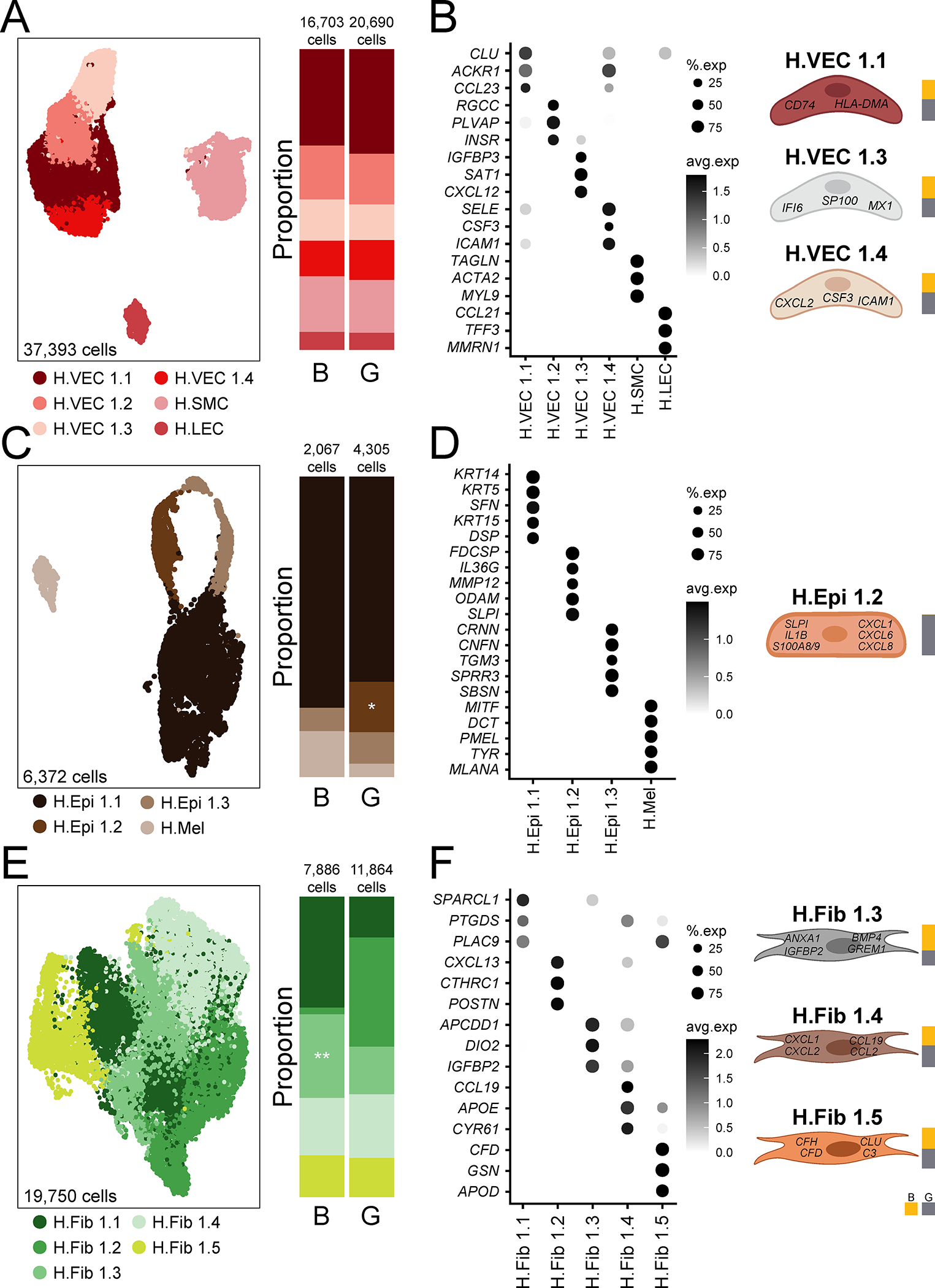
A,C,E. UMAP (left) and proportion plots (right) of endothelial (37,393 cells), epithelial (6,372 cells) and fibroblast (19,750 cells) populations, respectively, in gingival (G) and buccal (B) mucosa (n=13 and n=8, respectively). Refer to methods for statistical tests used. *p < 0.05, **p <0.01. B,D,F. Dot plots depicting the expression of cluster-defining genes and percentage of cells expressing each gene for endothelial, epithelial, and fibroblast populations, respectively. Expression values are normalized and scaled averages. Cartoon on the right illustrate cell subtypes associated with immune activation/recruitment. Proportion plots to the right of each illustration depict the contribution of each tissue type to each cell subtype. Clusters were generated using a resolution of 1 prior to subsetting into major cell types and only epithelial clusters were reclustered at a resolution of 0.1. See also Table S4, Figure S2.
Within the epithelial compartment, we identified 4 subtypes in healthy oral mucosa; three subtypes of keratinocytes (H.Epi 1.1,1.2,1.3) and a melanocyte population (H.Mel) (Figure 2C,D, Table S4). H.Mel was the most distal epithelial cluster by UMAP (Figure 2C) and clearly represented melanocytes based on significant expression of pigmentation genes (PMEL, MLANA) (Figure 2D, Figure S2B). Based on gene expression, H.Epi 1.1 cells were further categorized as basal layer/differentiating keratinocytes with expression of KRT5, KRT14 and KRT15, while H.Epi 1.3 expressed genes involved in keratinization and cornification (SPRR3, CRNN, CNFN) typically encountered in the outmost layers of the epithelium (Figure 2D, Figure S2B). Whereas H.Epi 1.1 and 1.3 were present in both buccal and gingival mucosa, H.Epi 1.2 was only found in the gingiva and featured a gene expression profile consistent with inflammatory responses. In fact, top expressed genes included antimicrobial and inflammatory factors such as IL1B, SLPI, and S100A8/9, and a top pathway was leukocyte chemotaxis, which reflected primarily recruitment of neutrophils as evident by the expression of CXCL1, CXCL6, CXCL8 (Figure 2D, Figure S2B, Table S4).
Fibroblasts separated into 5 cell clusters which displayed gene signatures consistent with distinct inferred functions (Figure 2E, Table S4). Cluster 1.1 displayed active translation and cluster 1.2 displayed a gene signature corresponding to the prototypic functions of the cell type, including collagen synthesis and tissue remodeling (COL1A1, COL1A2, MMP2) (Fig 2F, Figure S2C, Table S4). Clusters H.Fib 1.3, 1.4, and 1.5 expressed gene signatures consistent with inflammatory responses, such as regulation of leukocyte proliferation (ANXA1, IGFBP2), granulocyte migration (CXCL1, 2, 8) and complement activation (C3, CFD) (Figure 2F, Figure S2C, Table S4).
Overall, our data reveal a complex landscape of epithelial and stromal cells present in oral mucosal tissues with diverse inferred functions, particularly related to inflammation, antimicrobial defense and recruitment of neutrophils. Additionally, an epithelial population with a transcriptional program related to neutrophil recruitment was unique to the gingiva compared to the buccal mucosa.
T lymphocyte and myeloid cells are dominant in healthy oral mucosa
Within the immune compartment of the healthy oral mucosa we found a diverse population of cells. Generally, immune cells divided into 5 major clusters of T/NK, B/plasma, and granulocyte/myeloid cells, which were defined accordingly by classic cell-specific marker gene expression (Figure 3A, Table S5). Immune cells were annotated using SingleR (Aran et al., 2019) and manual annotation, which resulted in 14 cell types that were confirmed based on gene expression for marker genes (Figure 3A–D). In both buccal and gingival mucosa, the majority of immune cells were T cells, which were subdivided into αβ CD4+, TH17, mucosal- associated invariant T (MAIT), αβ CD8+, yδ T, Treg, and NK/T cells. The second largest overall population was myeloid cells, which included myeloid dendritic cells (mDC), macrophages and neutrophils. We also observed a population of mast cells, and small populations of plasmacytoid DC (pDC), B and plasma cells (Figure 3A–D). The buccal mucosa harbored an increased proportion of CD8+ T cells whereas proportions of Treg, mast, pDC and B cells were significantly elevated in the gingiva. Consistent with the sequencing data, flow cytometry on biopsies from an independent cohort of volunteers (n=10–14, Table S1) revealed an overall enriched immune compartment in the gingiva with significantly increased proportion and number of CD45+ cells in gingival biopsies (Figure 3E). Flow cytometry also confirmed the presence of major immune cell populations in healthy oral mucosa including CD45+CD3+ T cells, CD45+CD19+ B cells, CD45+HLADR+ antigen presenting cells (APC), HLADR−CD15+SSChigh and HLADR− CD117+SSChigh granulocytes (Figure 3F–H, Figure S3A). Increased inflammatory cell numbers in the gingival mucosa were primarily attributed to the increased presence of granulocytes (mast cells and neutrophils).
Figure 3: Immune populations in healthy oral mucosa.
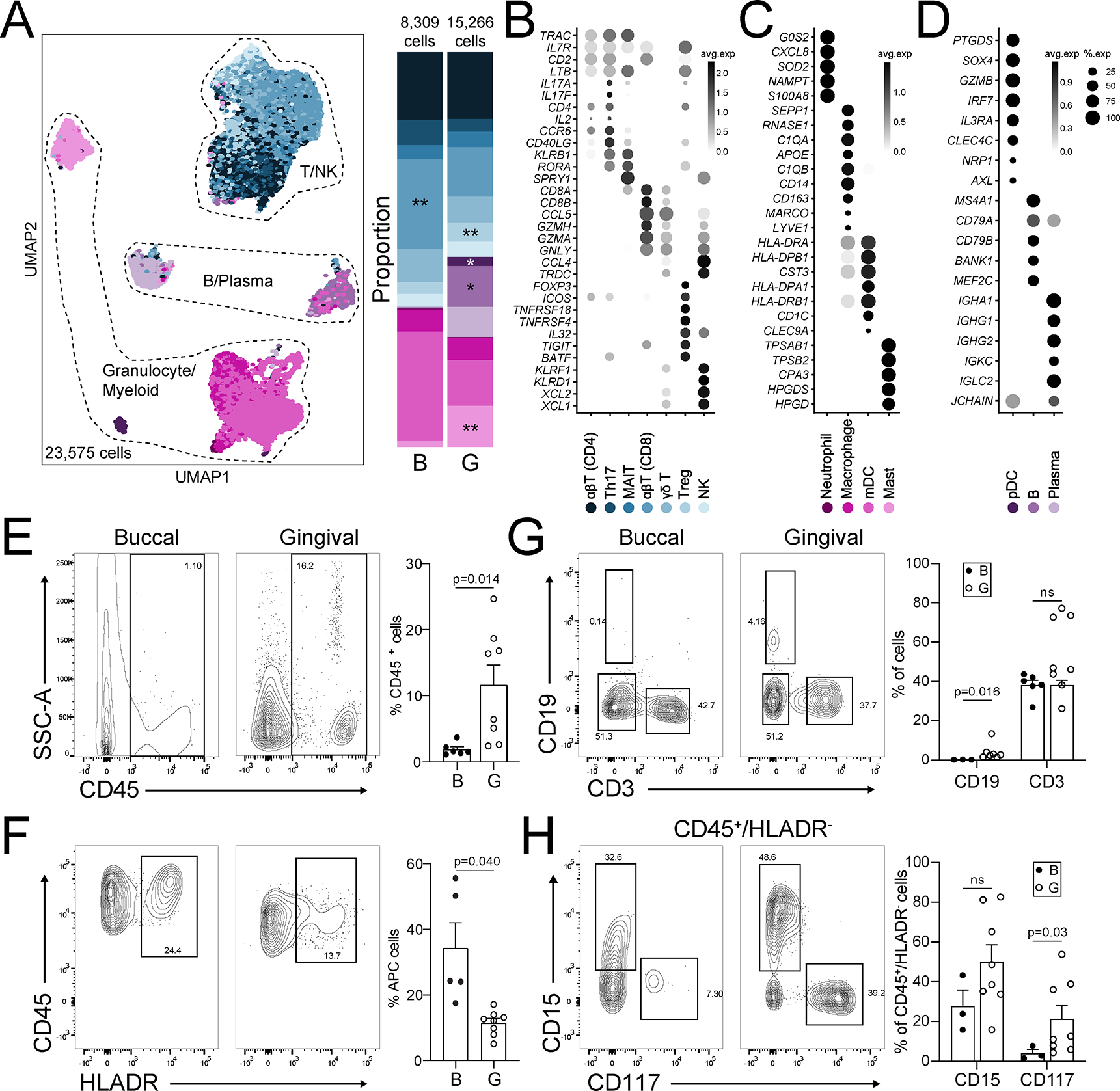
A. UMAP (left) and proportion plots (right) of immune cell populations in oral mucosa (23,575 cells; gingival (G, n=13), buccal (B, n=8)). Immune cells were annotated using a combination of reference based (SingleR) and manual annotation. Refer to methods for statistical tests used. *p < 0.05, **p<0.01. B-D. Dot plots depicting expression of cell type marker genes. Expression values are normalized and scaled averages. E-H. Representative flow cytometry scatter plots from buccal (n=3–8) and gingival (n=8) biopsies. Cells were gated from Single/Live,CD45 (E) and CD45/HLADR (F), CD45+ cells were further gated for CD19/CD3 (G) and HLADR−SSChighCD15/CD117 (H). Bar graphs (E-H) demonstrate % expression for each marker indicated, each dot represents an individual biopsy. Refer to methods for statistical tests used. p values indicated on each graph. See also Table S1, S3, S5, and Figure S3.
Finally, we interrogated immune cell populations in published databases of healthy barrier tissues across the human body (oral, ileum, skin, lung; Figure 1E) and minor salivary glands (Huang et al., 2021) (Table S3). We found representation of the immune cell subsets present in the oral mucosa scRNA seq dataset across other barriers, yet with varying proportions. Interestingly, we documented a significantly higher proportion of neutrophils at the oral mucosa compared to other barriers, suggesting active innate immune activity during health at this barrier, consistent with transcriptome of neutrophil chemoattractant in stromal/epithelial cells of oral mucosal tissues (Figure S3B,C).
Altogether, this data illustrates the complex cellular constituency of the oral mucosa at steady state and reveals an environment specialized towards neutrophil recruitment. Moreover, cell- type specific transcriptomes in gingiva reflect the anatomical characteristics of this mucosal site, which is subject to microbial stimulation and constant mechanical triggering.
Immune cell infiltration of mucosal tissues is the hallmark of periodontitis
Host responses at the gingiva become exaggerated and destructive in the setting of the common human inflammatory disease, periodontitis. Therefore, we next aimed to characterize cell populations and their transcriptomes in periodontitis at a single-cell level. For these studies, we profiled gingival mucosal biopsies from healthy adults and patients with untreated, severe periodontitis using scRNA seq (n=21, ~88,000 cells) alongside complementary histology and flow cytometry studies (Figure 4A). With these independent approaches, we document profound infiltration of inflammatory cells in the mucosal tissues of periodontitis compared to healthy adult mucosa, reflecting mucosal tissue destruction in periodontitis (Figure 4B–E). H&E and immunofluorescence staining in healthy and diseased tissues revealed infiltration of mucosal tissues with CD45+ leukocytes in the setting of disease (Figure 4B,C). Significant inflammatory cell infiltration was also confirmed by flow cytometry in an independent group of healthy and diseased individuals (n=6/group) (Figure 4D). Proportional changes in cell populations as well as changes in absolute cell numbers in a subsample are reported (Figure 5E, Figure S5).
Figure 4: Periodontal disease is an inflammatory disease at the gingival mucosa.
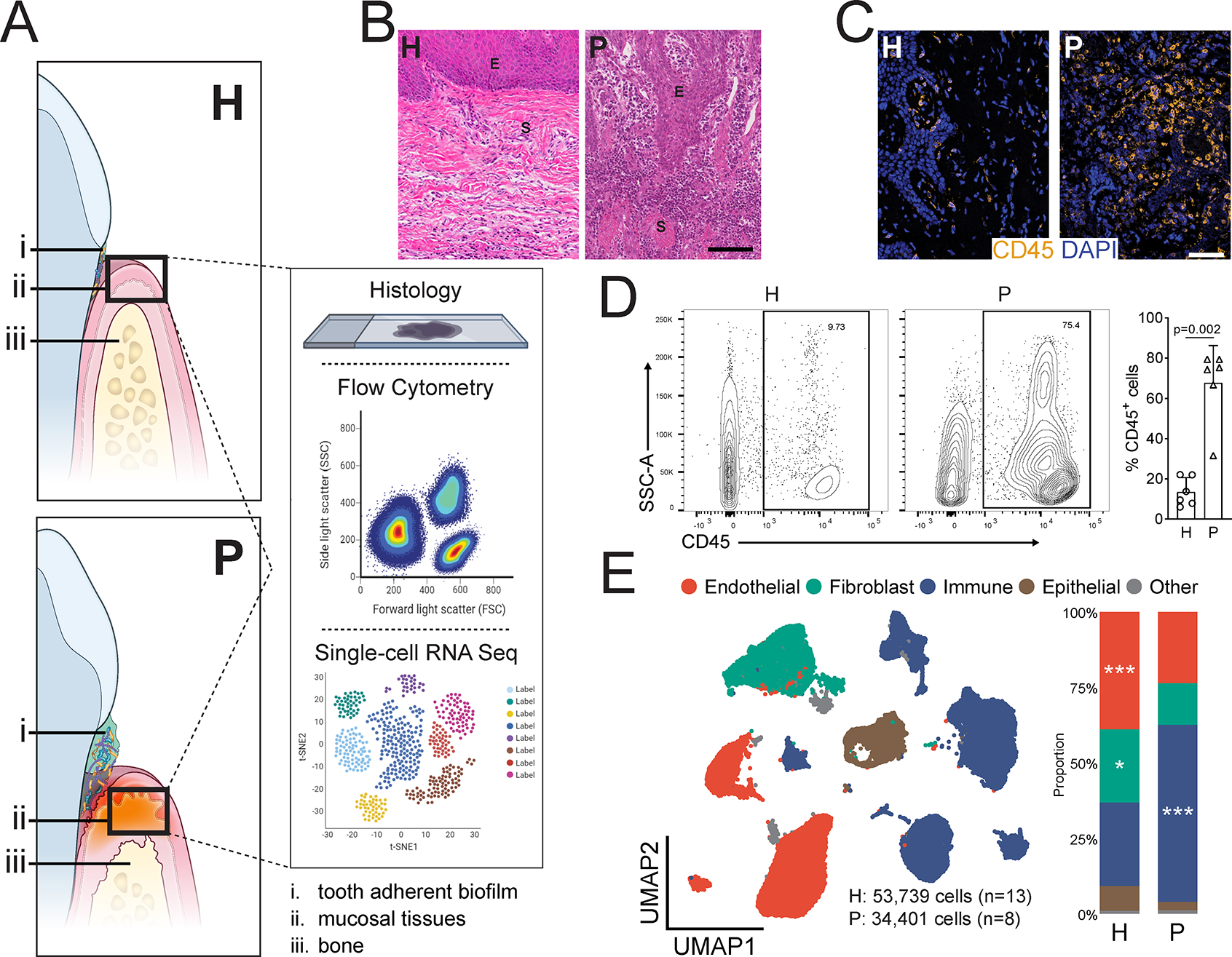
A. Illustration of the gingival mucosa in health (H, top) and periodontal disease (P, bottom). A schematic of methods for tissue analysis is presented (right). B. H&E staining of representative gingival health (left) and periodontitis (right) sections. E-epithelium, S-stroma. Scale bar: 100μm. C. Immunofluorescence staining for CD45+ lymphocytes. Scale bar: 50μm. D. Representative flow cytometry scatter plots (left). Cells were gated from Single/Live and stained with CD45, graph indicates CD45 % expression (one dot per patient, n=6/group. Refer to methods for statistical tests used, p value indicated). E. UMAP representation of major cell populations in health and periodontal disease (left; H: 53,739 cells, n=13; P: 34,401 cells, n=8). Graph demonstrates the proportion of each cell type in health and disease (right). Refer to methods for statistical tests used. *p < 0.05, ***p<0.001. See also Table S1.
Figure 5: Immune cell subpopulations in periodontal disease.
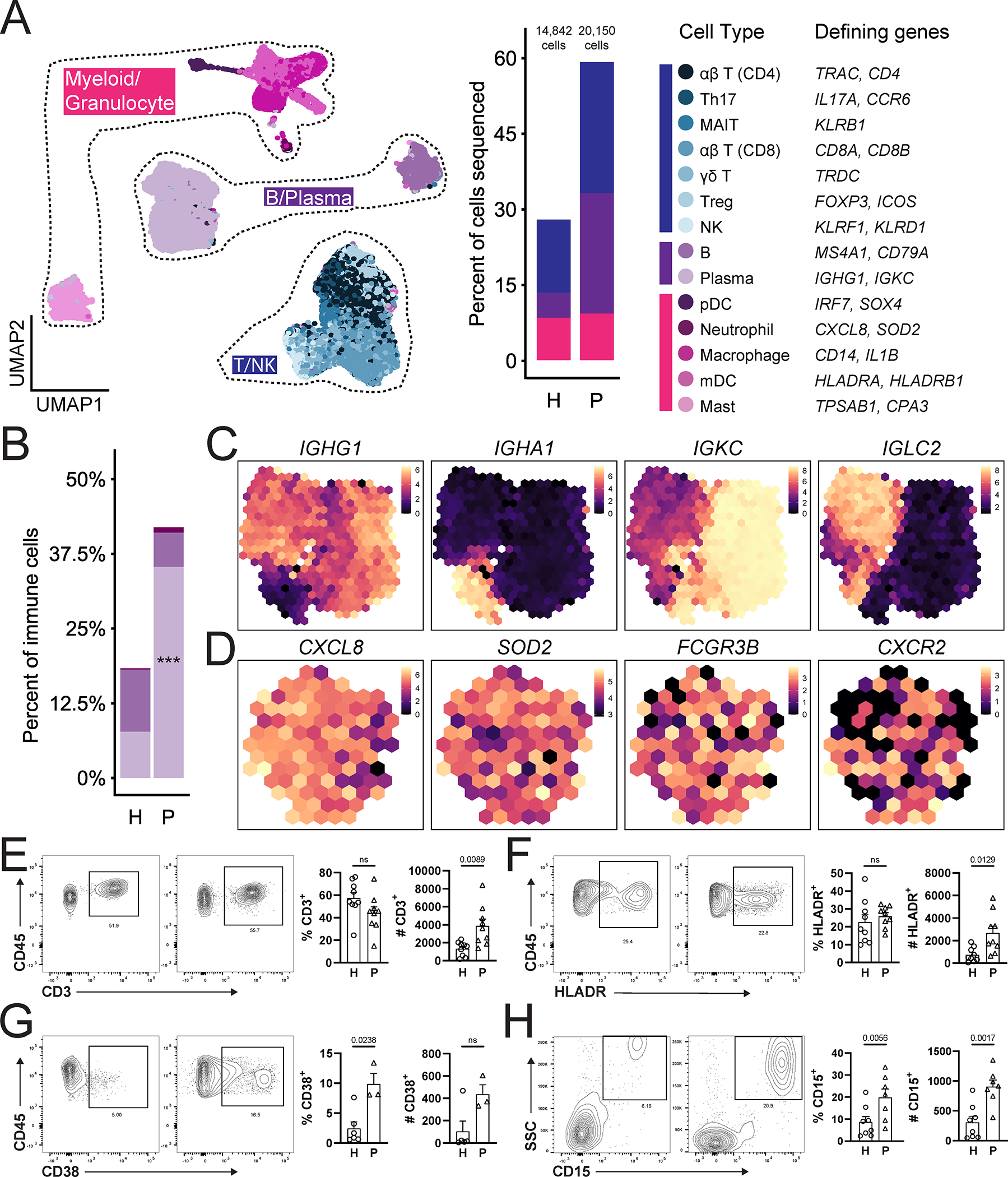
A. UMAP (left) showing immune cell subpopulations from both health (n=13) and disease (n=8) divided spatially into three main categories (T/NK, B/Plasma and Myeloid/Granulocyte). Proportion plot (middle). Colors indicate the cell type annotated using SingleR and manual annotation and validated by cell-type specific gene expression (right). B. Bar graph depicts proportion of B, plasma cells and neutrophils in health (H) and periodontitis (P). Refer to methods for statistical tests used. ***p<0.001. C,D. Normalized average expression of plasma cell- (C) and neutrophil- (D) specific markers visualized in low-dimensional space with schex. Each area containing cells on the UMAP was divided into hexagonal areas and gene expression values within each area were averaged across cells present. E-H. Representative flow cytometry scatter plots from an independent health and periodontitis cohort. Cells were gated from Single/Live cells and CD45/CD3 (E), CD45/HLADR (F), CD45/CD38 (G), CD45/CD15 (H). Bar graphs demonstrate % expression (one dot per individual, n=3–9/group). Refer to methods for statistical tests used. P values are indicated on each graph. See also Table S1, Figure S4, S5.
Investigation of the immune cell types in the integrated scRNA seq datasets from health and disease demonstrated the presence of the same immune cell subsets previously seen in health, albeit with varying proportions in disease. Similar to health, T cells remained the major immune cell population with representation of ap CD4+, TH17, MAIT, αβ CD8+, yδ T, Treg, and NK/T cells in disease (Figure 5A). The next largest populations by proportion were B and plasma cells, of which plasma cells were significantly expanded in periodontitis compared to health (Figure 5A,B). Finally, myeloid/granulocyte cell populations were also present in disease, with expansion of neutrophils (p=0.1586) (Figure 5A,B). Focusing on the cell types that expand in disease, we further define gene signatures for plasma cells and neutrophils in periodontitis. A majority of plasma cells expressed IgG and either k or ⋋ light chains while a small but distinct population expressed IgA (Figure 5C). Differential expression analysis did not identify further heterogeneity in the neutrophil cluster, although cells in this cluster uniformly expressed classical neutrophil markers such as CXCL8, CXCR2 and FCGR3B (Figure 5D).
Flow cytometry in an independent patient cohort (n=3–9 individuals/group) demonstrated significantly increased numbers of all major cell types, including CD3+ T cells, CD38+ plasma cells, HLADR+ antigen presenting cells, and HLADR−SSChighCD15+ neutrophils in periodontitis (Figure 5E–H). However, only plasma cells and neutrophils were increased in proportion with disease, consistent with data from single-cell sequencing. Of interest, while scRNA seq revealed an increase in neutrophil numbers in periodontitis, it did not capture the magnitude of neutrophil infiltration seen by flow and histology (Figure 5H, Figure S4), reflecting a relative limitation of this method in capturing neutrophil transcriptomes (Hoogendijk et al., 2019).
Stromal and epithelial cells display transcriptional signatures of inflammation in periodontitis
We next investigated changes in the stromal and epithelial compartments in disease. Overall, both histological and scRNA seq analysis showed that epithelial and stromal cell proportions decreased in disease (Figure 4B,E). Joint embedding of the stromal/epithelial cell states in both health and periodontitis are in close proximity in UMAP space, suggesting transcriptional similarity (Figure 6A–C) with shifts in select cell subsets in disease (Figure S5, S6A–F). Overall, the gene expression profile of epithelial and stromal cells shifted to an inflammatory state in disease. Specifically, endothelial cells upregulated pathways related to cell adhesion, lymphocyte adhesion and chemokine signaling. Epithelial cells and fibroblasts upregulated pathways related to antimicrobial responses (response to LPS and cell response to bacterial molecule) and cytokine biosynthesis (Figure 6D). Of interest, genes associated with antimicrobial processes upregulated in both fibroblasts and epithelial cells in disease reflected active neutrophil recruitment (CXCL1, CXCL8) (Table S6). This data suggests unique wiring of the epithelial and stromal compartment in periodontitis particularly geared towards neutrophil recruitment.
Figure 6: Stromal cell populations and stromal- immune interactome in periodontitis.
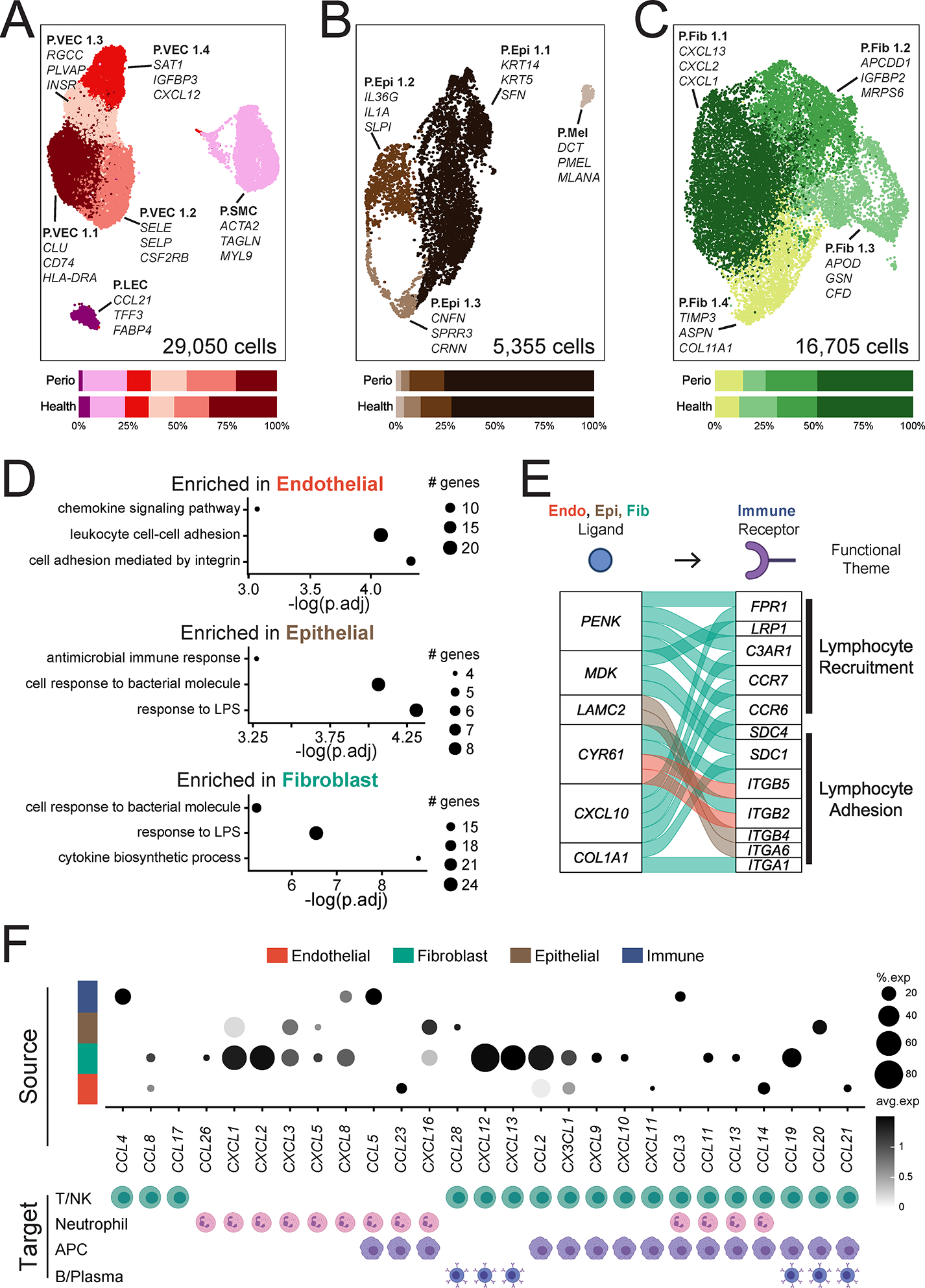
A-C. UMAP representation (top) and proportion plots (bottom) for endothelial (A; 29,050 cells), epithelial (B; 5,355 cells) and fibroblast (C; 16,705 cells) populations, respectively, in the combined health (n=13) and periodontitis (n=8) dataset. Gene labels indicate cluster-defining genes. Refer to methods for statistical tests used. D. Dot plots of pathways enriched in periodontal disease per major cell population. Some GO terms truncated for brevity, original terms and gene list in Table S6. The size of each dot represents the number of genes from each pathway. E. Alluvial plot showing selected ligand-receptor pairs significantly over-represented in periodontal disease determined by NicheNetR. For this analysis, only interactions of immune cells (receptors) with all other cell types, and only ligands and receptors that are differentially expressed (p<0.05), are considered. F. Chemokine/chemokine receptor interactions in health and periodontal disease. Dot plot indicating expression of chemokines in major cell types (top) and the immune cell(s) in the scRNA seq dataset that express cognate receptors (bottom). Expression values are normalized and scaled averages. Refer to Figure S6 for expression data of chemokine receptors. Clusters were generated using a resolution of 1 prior to sub-setting into major cell types and were not reclustered. See also Table S6, Figure S5, S6.
Stromal-immune cell interactions are potential drivers of periodontal inflammation
To further investigate potential stromal- and epithelial-immune interactions in the context of periodontitis we used NicheNet to infer cell-cell interactions with a prior model of ligand-target regulatory potential. We identified the top predicted immune receptor-ligand interactions based on gene expression in our scRNA seq dataset that were differentially regulated in disease. In periodontitis, there was extensive inferred communication between stromal and immune cells (Figure S6G–I). Of interest and consistent with pathways upregulated in disease, stromal and epithelial cells appeared to promote adhesion of immune cells, while fibroblasts displayed a potential towards recruitment of inflammatory cells (Figure 6E). Overall, based on gene expression signatures and predicted interactions, our data indicates an active role for stromal cells in the recruitment of immune cells to the site of disease.
To further explore the overall potential of stromal cells to recruit immune cells, we specifically interrogated expression of chemokine and chemokine receptor pairs in our dataset (Figure 6F, Figure S6J). Our data showed broad expression of chemokine ligands by all cell types in disease. In line with pathway analysis (Figure 6D), fibroblasts were particularly transcriptionally active in the production of chemokines. Fibroblasts expressed a broad array of chemokine ligands exclusive in their potential to recruit neutrophils (CXCL1, 2, 5, 8) as well as chemokines with the potential of recruiting several types of leukocytes (e.g. CXCL12, CXCL13, CCL19) (Figure 6F). Collectively, this data suggests that stromal cells utilize intercellular signaling to drive immune cell recruitment and tissue transmigration in periodontitis.
Expression of periodontitis susceptibility genes in immune and stromal cells
To begin to understand cell contribution to disease pathogenesis, we next investigated cellular expression of genes linked to periodontitis susceptibility. Select Mendelian genetic diseases are clearly linked to periodontal disease susceptibility (Betts et al., 2015; Hajishengallis and Moutsopoulos, 2016; Hart and Atkinson, 2007) and recent GWAS studies have uncovered single-nucleotide polymorphisms associated with periodontal disease (Morelli et al., 2020; Nibali et al., 2019). However, the cellular expression patterns and role of the relevant genetic determinants in periodontitis remains unclear. Our scRNA seq atlas uniquely enabled us to globally assess the expression of 15 extant periodontal disease genes within the gingival mucosa. Mendelian disease genes associated with periodontitis were almost exclusively expressed in the immune cell compartment with the exception of C1S and C1R (expressed exclusively in fibroblasts) indicating that immune, and possibly fibroblast, cell dysfunction is a critical driver of monogenic periodontitis. Neutrophil-related defects (HAX1, LYST, ITGB2, FPR1) are particularly linked to susceptibility for periodontitis in genetic syndromes. In contrast, disease-associated genes identified by GWAS (Divaris et al., 2012; Divaris et al., 2013; Offenbacher et al., 2016; Sanders et al., 2017; Shaffer et al., 2014; Shang et al., 2015; Shungin et al., 2019) were more broadly expressed, including by stromal, epithelial, and immune cells, demonstrating a complex interplay between immune and non-immune cells in periodontitis (Figure 7A). Our exploration of disease-associated genes at the tissue level suggests diverse cell populations are involved in common forms of periodontitis, while neutrophils and fibroblasts likely drive the pathogenesis of Mendelian forms of periodontitis.
Figure 7: Genes associated with periodontal disease susceptibility, microbe and damage sensing in the oral mucosa.
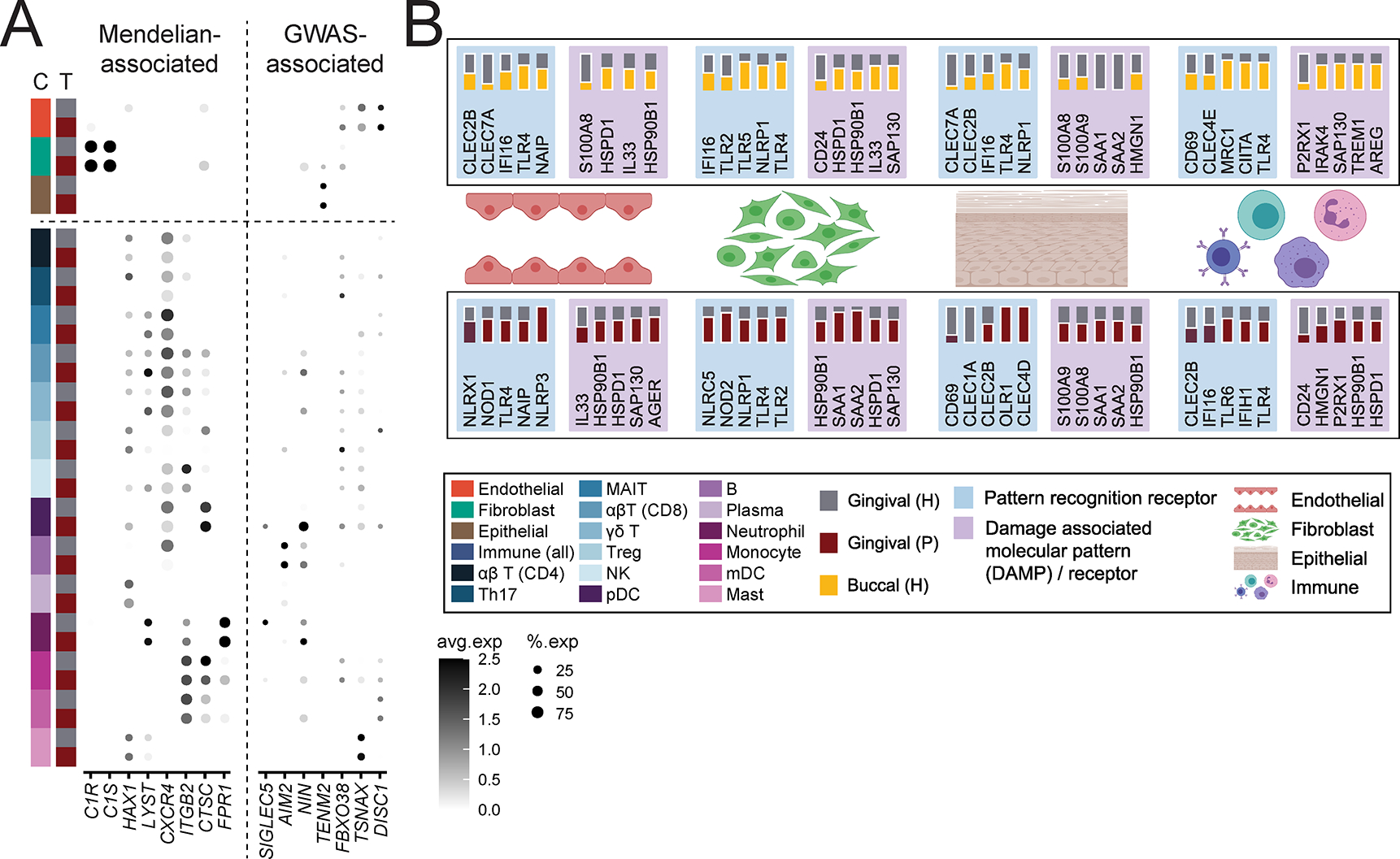
A. Dot plot depicting the normalized and scaled average expression of genes and percentage of expressing cells for genes associated with Mendelian forms of periodontitis (left) and genes identified by GWAS to be associated with periodontitis (right). B. Summary of select pattern recognition receptors and damage-associated molecular pattern and associated receptors expression in buccal and gingival healthy mucosa (top) and expression in gingival health and periodontitis (bottom). Expression is shown by major tissue type (Endothelial, Fibroblast, Epithelial, Immune). A solid white border indicates statistical significance and the tissue that had higher expression. Significance determined by non-parametric Wilcoxon rank sum test. See also Table S7, Figure S7.
Microbiome and damage sensing may underlie stromal hyperresponsiveness in periodontitis
Finally, to investigate triggering of stromal-immune cell interactions linked to periodontitis we utilized the atlas to investigate expression of microbial and cell-damage receptors and sensors within our oral cell database in health and disease. As such we explored the cellular distribution of pattern recognition receptors, including Toll like receptors (TLR), C-type lectin receptors (CLR), nucleotide-binding oligomerization domain-like receptors (NLR), RIG-I-like receptors (RLR), viral entry factors, damage associated molecular patterns (DAMP) and associated receptors (DAMP-R) (Figure 7B, Figure S7, Table S7). We document diverse expression of microbial and damage receptors in epithelial, endothelial, fibroblast, and immune cells within oral tissues in health consistent with microbial and/or damage triggering of immune responsiveness in the stromal and immune compartments. Furthermore, we find significantly increased expression of NLR, CLR, TLR, DAMP and DAMP-R molecules in disease, particularly within stromal and epithelial cells, suggesting increased triggering of these cellular populations in disease.
Collectively, our data promote the concept that stromal cell populations in the oral mucosa are constantly triggered via microbial and damage sensing and acquire an immune specification focused toward the neutrophil recruitment. Moreover, exaggerated activation of stromal cells in periodontal disease is linked to excessive recruitment of immune cell populations and destructive inflammatory immune responses.
Discussion
Herein we present a robust atlas of human oral mucosal tissues in health and in the setting of the common oral inflammatory disease periodontitis. Our work details the complex landscape of oral mucosal cell types and provides comprehensive insights into cell functionality and disease susceptibility at this unique barrier.
A strength of our study is the meticulous characterization of patient cohorts with a strict definition of oral health and periodontal disease. Indeed, our healthy volunteer cohort was screened for medical history, medication, tobacco/alcohol/drug use and also evaluated through blood work and detailed oral examination. This study design has enabled the creation of an atlas of systemic and oral health that can serve as a community resource. Our health cohort also serves as a normative baseline against which we can begin to explore changes in cell populations and their transcriptomes in the setting of oral diseases. Furthermore, our evaluation of both tooth-associated (gingival) and lining (buccal) oral mucosa provides a baseline towards evaluation of diseases associated with these two distinct sites which typically present with unique disease susceptibilities (Porter et al., 2017; Schifter et al., 2010).
In the current study, we explored shifts in cell populations and their transcriptomes in the setting of untreated, severe periodontitis, a disease of the gingival mucosa. Periodontitis is a prevalent inflammatory disease and is considered an independent risk factor for multiple systemic inflammatory diseases (Hajishengallis, 2015). However, mechanisms underlying disease susceptibility and pathogenesis are not fully understood and no targeted treatment is currently available. Our work here details an atlas of cell populations, transcriptomes and inferred interactions providing insights into cell functionality in disease susceptibility and pathogenesis, opening avenues for further mechanistic exploration of this complex disease.
Our findings demonstrate that the gingival mucosa is a particularly active tissue site with unique immunological wiring and enhanced inflammatory responsiveness of stromal cell populations. We document increased representation of immune cells at this site (particularly granulocytes/neutrophils) but also enhanced proinflammatory and antimicrobial gene expression from specific clusters of epithelial cells, consistent with increased stimulation from the commensal microbiota (Abusleme et al., 2013) and continuous mechanical stimulation through mastication (Dutzan et al., 2017).
The atlas reveals a previously unappreciated diversity of stromal and epithelial cells at the oral barrier. We find that several stromal/epithelial populations support prototypic cell functions, including keratinization in the epithelium, matrix accumulation and turnover in fibroblasts and angiogenesis/cell transmigration in endothelial cells. In addition, we document stromal cell populations with transcriptional programs that suggest immune functionality. Overall, in health we observe that oral stromal cells, particularly in the gingiva, are wired towards the induction of inflammatory responses and neutrophil recruitment. Indeed, the gingival mucosa is a site of constant neutrophil transmigration (Moutsopoulos and Konkel, 2018). Importantly, we find that stromal cells become increasingly activated in the setting of periodontitis and stimulate immune cell recruitment and migration into disease lesions. In disease, fibroblasts and other stromal cells promote the specific recruitment of neutrophils but also express chemokines known to attract other leukocytes, including lymphocytes. These findings suggest a previously unacknowledged role for the mesenchymal compartment in immune responsiveness in oral homeostasis and disease pathogenesis, similar to recent findings that identify a mesenchymal- inflammatory axis in diseases of other tissue compartments (Croft et al., 2019; Koliaraki et al., 2020). Importantly, an unreported aspect of stromal hyper-responsiveness in gingival mucosal health and disease is the specification of gingival stromal cells towards the recruitment of neutrophils.
Our findings also further reinforce the critical role of neutrophils in oral mucosal immunity. We document that many of the immune-related pathways upregulated in stromal and epithelial cells in gingival health and disease are related to neutrophil homing. In fact, some of the top upregulated genes in gingival stromal and epithelial cells include CSF3, CXCL1, CXCL2, and CXCL8 which are specific for neutrophil recruitment. Indeed, at steady state, neutrophils are the immune cell type with significantly higher representation by scRNA seq in oral mucosa compared to salivary glands as well as all other healthy human barrier tissues investigated. We hypothesize that these active antimicrobial defenses, mediated by neutrophils, are critical not only for immune surveillance at this barrier, but also proactively contribute to immune regulation. Indeed, patients with genetic neutrophil defects present with severe inflammatory diseases of the oral mucosa (Moutsopoulos et al., 2017; Silva et al., 2019).
In periodontal disease, we further document significant recruitment of neutrophils in tissue lesions by flow cytometry, as previously reported (Dutzan et al., 2016; Fine et al., 2016). scRNA seq captures the increase in tissue neutrophils in periodontitis, albeit without reaching statistical significance. This indicates a technical limitation in capturing tissue neutrophil responses, likely due to the abrupt decline in transcriptional activity of mature neutrophils after leaving the bone marrow, as well as the sensitivity of neutrophils to tissue preparation and isolation steps (Hoogendijk et al., 2019). Finally, expression of known disease susceptibility genes within oral tissues reflects a predominant expression of Mendelian-disease associated genes within tissue neutrophils. Indeed, it is well recognized that patients with genetic neutrophil defects commonly develop dominant severe oral phenotypes including aggressive periodontitis in children and recurrent oral ulcerations in lining mucosa (Halai et al., 2020; Hart and Atkinson, 2007). Furthermore, our atlas allows us to evaluate the expression of genes related to periodontal susceptibility in common forms of periodontitis, identified through GWAS studies. Expression of susceptibility genes within the tissue of interest reveals GWAS-associated gene expression in a variety of cell populations and opens avenues for investigation of diverse pathways of pathogenesis in common forms of disease
Finally, we utilized our atlas to explore expression of microbial receptors and damage sensing molecules in the oral mucosa. At this barrier, a rich commensal microbiome is present in health which may participate in tissue homeostasis and is decidedly involved in the pathogenesis of common oral mucosal diseases including periodontitis, candidiasis and oral ulcers, suggesting a primary role for host-microbial interaction in oral tissue pathophysiology. Furthermore, as multiple commensals and pathogens are first encountered at the oral mucosa prior to entry into the gastrointestinal and respiratory tracts, understanding initial recognition of microbial elements at this barrier is particularly important. Indeed, oral fibroblasts have been shown to upregulate inflammatory programs in response to microbial byproducts in vitro (Buskermolen et al., 2017; Jonsson et al., 2009; Muller et al., 2016; Wang and Ohura, 2002). In addition, the masticatory mucosa, which includes the gingiva, is continuously exposed to injury through mastication (Groeger and Meyle, 2019). We therefore created a reference for expression of major damage, bacterial, fungal, and viral receptors/sensing molecules within the cell populations of the oral mucosa. We determined distinct receptor expression at buccal and gingival mucosal sites, potentially reflecting unique disease and infection susceptibilities at each site. Furthermore, modulation of microbial expression in periodontitis suggests altered host-microbial interactions in the presence of inflammation. Importantly, the expression of microbial sensing and damage receptors within stromal cells and further upregulation of such receptors in the setting of disease supports the concept that the stromal compartment receives diverse microbial and damage signals to promote homeostatic inflammation, but upon exacerbation causes inflammatory tissue destruction and disease.
Interestingly, many of the features identified by this human atlas are consistent with findings in the ligature-induced experimental model of periodontitis (LIP). As such, in the LIP model, disease is microbiome-induced and characterized by upregulation of PRRs and other inflammatory molecules with significant migration of neutrophils to the tissue (Bao et al., 2020; Maekawa et al., 2014) which have a pathogenic role in experimental periodontitis (Dutzan et al., 2018). These parallels between the human data and experimental model, suggest that aspects of human disease are recapitulated in the LIP model, allowing for further mechanistic experimentation in the animal setting.
Taken together, our oral mucosal tissue atlas provides a comprehensive characterization of cell populations and states in health and periodontitis and provides insights into cell functionality and intercellular interactions in disease pathogenesis. Furthermore, our work reveals unique functionality of stromal cell populations participating in homeostatic immunity and inflammation at this barrier.
Limitations of the study
Our single cell census of the oral mucosa will provide a valuable resource for the research and clinical communities. However, we recognize the limitations of our study primarily focused on single cell transcriptome profiling of dissociated tissue and have therefore complemented our analysis with flow cytometry for protein level validation and histological analysis for in situ validation and tissue architecture. Further studies combining transcriptomics, proteomics and imaging will be essential to fully define cell populations within the tissue microenvironment.
Our substantive dataset (~120,000 cells) increases the likelihood of identifying rare cells for comprehensive cell capture. Nevertheless, neutrophils are challenging to profile by scRNA-seq and are thus under-represented in our dataset. Previous studies have characterized significant infiltration of neutrophils in periodontitis tissues in humans and mice (Dutzan et al., 2018; Nicu and Loos, 2016) and we demonstrate neutrophil prominence in periodontitis tissue by flow cytometry analysis.
Our study is focused on human tissue analysis and presents a snapshot in health and disease. We sample a single time point per individual and therefore cannot evaluate temporal changes due to age or progression of disease. Furthermore, the sample size of our cohort does not allow us to thoroughly evaluate the contribution of age, sex and race in our dataset. Additionally, because we control for known environmental factors such as smoking and disease comorbidities, we cannot currently assess their contribution to the disease process. Future analysis incorporating contribution of age/sex/race, environmental exposures and temporal changes relating to disease evolution will further enrich understanding of this mucosal tissue microenvironment in health and disease. Further mechanistic studies in animal models are also necessary to establish cell-specific functionality in disease susceptibility and pathogenesis.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Niki Moutsopoulos (nmoutsop@mail.nih.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Raw and processed single-cell RNA sequencing datasets have been deposited in the NCBI GEO database under accession GSE164241 and can be accessed via the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164241. Code to reproduce findings in this manuscript is available at: https://github.com/williamsdrake/oralAtlas. An interactive analytics portal to further explore gene expression is available at https://oral.cellatlas.io.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patient recruitment and ethical approval
All healthy volunteers and patients participating in this study were enrolled in an IRB approved clinical protocol at the NIH Clinical Center and provided written informed consent for participation in this study. All participants were deemed systemically healthy based on detailed medical history and select laboratory work up.
Inclusion criteria for this study were: ≥ 18 years of age, a minimum of 20 natural teeth, and in good general health. Exclusion criteria were: history of Hepatitis B or C, HIV, prior radiation therapy to the head or neck, active malignancy, treatment with systemic chemotherapeutics or radiation therapy within 5 years, pregnant or lactating, diagnosis of diabetes and/or HbA1C level >6%, > 3 hospitalizations in the last 3 years, autoimmune disorders. Additional exclusion criteria included the use of any of the following in the 3 months before study enrollment: systemic antibiotics, systemic or inhaled corticosteroids or other immunosuppressants, chemotherapeutic agents, large doses of commercial probiotics, or use of tobacco products (including e-cigarettes) within 1 year of screening. In addition to systemic screening, oral health was assessed and only participants with no soft tissue lesions, no signs/symptoms of oral/dental infection and no/minimal gingival inflammation were considered for the health group. Inclusion criteria for periodontal disease group were moderate to severe periodontitis (>5mm probing depth in more than 4 interproximal sites) and visible signs of tissue inflammation including erythema/edema, and bleeding upon probing. A total of 13 biopsies were included in the gingival health group, 8 biopsies were included in the buccal health group, and 8 biopsies were included in the gingival disease (periodontitis) group. Demographic information for each patient in the study including age, sex, and ethnicity can be found in Table S1.
Biopsy collection
During study visits participants received detailed intraoral soft tissue and periodontal examination, which included full mouth probing depth and clinical attachment loss measurements (PD, measures of bone destruction) and bleeding on probing (BOP, measure of mucosal inflammation). Standardized 4mm long x 2mm wide gingival collar biopsies and/or 4mm buccal punch biopsies were obtained under local anesthesia. Health group buccal and gingival biopsies were obtained from individuals that met criteria for oral health and in areas without BOP and with PD <3mm. Biopsies of periodontitis patients were obtained from areas of severe inflammation and bone loss (BOP positive and PD >5mm). Each biopsy was analyzed separately and not pooled. Biopsies were either placed into RPMI medium to be processed into single cell suspensions or fixed with 10% zinc formalin (American MasterTech Scientific) for histology. Donor information can be found in Table S1.
METHOD DETAILS
Histology
For histology, tissues were placed in zinc formalin for 18–24 hours and then formalin was replaced with 70% ethanol (Sigma). Formalin-fixed tissues were embedded in paraffin and sectioned into 5^m sections, deparaffinized, and rehydrated, followed by Hematoxylin & Eosin staining (H&E) and immunofluorescence staining. Immunofluorescence was performed via heat- mediated antigen retrieval with a Tris-EDTA buffer (10mM Tris, 1mM EDTA, pH 9.0) in a Retriever 2100 pressure cooker (Electron Microscopy Sciences). Slides were incubated overnight at 4°C with the following primary antibodies: anti-vimentin (abcam, ab24525, 1:100 dilution), anti-CD45 (Cell Signaling, 13917, 1:100), anti-CD31 (Cell Signaling, 3528, 1:250), and anti-keratin 5 (LSBio, LS-C22715, 1:500). Alexa Fluor-conjugated secondary antibodies were used to detect the primary antibodies (ThermoFisher, Alexa 488, 546, 594, & 647, all secondary antibodies at 1:300 dilution), and nuclei stained with DAPI (ThermoFisher, 1μg/mL) prior to covering with ProLong Gold Antifade Mountant (ThermoFisher). Slides were imaged on the Leica TCS SP8 X (Leica) located in the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) Light Imaging Core and processed on LAS X software (Leica).
Flow Cytometry
Biopsy preparation into single cell suspension
Human gingival tissues were minced and digested for 50 minutes at 37°C with Collagenase IV (Gibco) and DNase (Sigma). A single-cell suspension was then generated by mashing digested samples through a 7μm filter (Falcon, Corning), as previously described (Dutzan et al., 2016).
Flow cytometry of human oral mucosal tissue samples
Single-cell suspensions from gingival tissues were incubated with mouse serum (Jackson ImmunoResearch Lab) and fluorochrome-conjugated antibodies against surface markers in PBS with 2.5% FBS, for 20 minutes at 4°C in the dark, and then washed. Dead cells were excluded with Live/Dead fixable dye (Amcyan, 1:100, Invitrogen). Anti-human antibody information used for staining is described in the Key Resources Table. Cell acquisition was performed on a BD LSRFortessa machine using FACSDiVa software (BD Biosciences). Data were analyzed with FlowJo software (TreeStar).
KEY RESOURCES TABLE.
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| CD45 (AF-700), Clone HI30, 1:50 | Biolegend | Cat#: 304024; RRID: AB_493761 |
| CD3 (FITC), Clone SK7, 1:20 | Invitrogen | Cat#: 11-0036-42; RRID: AB_1272072 |
| HLADR (BV711), Clone G46-6, 1:20 | BD Bioscience | Cat#: 563696; RRID: AB_2738378 |
| CD38 (Pacific Blue), Clone HB7, 1:20 | eBioscience | Cat#: 48-0388-42; RRID: AB_11040389 |
| CD15 (PerCP-Cy 5.5), Clone HI98, 1:20 | Invitrogen | Cat#: 46-159-42; RRID: AB_1834387 |
| CD117 (PE-Cy 7), Clone 104D2, 1:20 | Invitrogen | Cat#: 25-1178-42; RRID: AB_10718535 |
| CD19 (BV650), Clone SJ25C1, 1:20 | BD Horizon | Cat#: 563226; RRID: AB_2744313 |
| Live/Dead Stain (AmCyan), 1:100 | Invitrogen | Cat#: L34966 |
| Chicken anti-Vimentin, polyclonal, 1:100 | Abcam | Cat#: ab24525; RRID: AB_778824 |
| Guinea pig anti-KRT5, polyclonal, 1:500 | LSBio | Cat#: LS-C22715; RRID: AB_903255 |
| Rabbit anti-CD45, Clone D9M8I, 1:100 | Cell Signaling | Cat#: 13917; RRID: AB_2750898 |
| Mouse anti-CD31, Clone 89C2, 1:250 | Cell Signaling | Cat#: 3528; RRID: AB_2160882 |
| Rabbit anti-MPO, polyclonal, 1:500 | Abcam | Cat#: ab9535; RRID: AB_307322 |
| Goat anti-Chicken IgY Alexa Fluor 488 | ThermoFisher | Cat#: A32931; RRID: AB_2762843 |
| Goat anti-Rabbit IgG Alexa Fluor 594 | ThermoFisher | Cat#: A11037; RRID: AB_2534095 |
| Goat anti-Mouse IgG Alexa Fluor 546 | ThermoFisher | Cat#: A11030; RRID: AB_2534089 |
| Goat anti-Guinea Pig IgG Alexa Fluor 647 | ThermoFisher | Cat#: A21450; RRID: AB_2735091 |
| ImmPRESS HRP Horse Anti-Rabbit IgG Polymer Detection Kit, Peroxidase | Vector Laboratories | Cat#: MP-7401; RRID: AB_2336529 |
|
| ||
| Biological samples | ||
|
| ||
| Human oral biopsies | NIH Clinical Center | See Table S1 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| DNase I | Sigma | Cat#: DN25-1G |
| Collagenase II | Worthington Biochemical Corp. | Cat#: LS004176 |
| Collagenase IV | GIBCO | Cat#: 17104-019 |
| RPMI | Lonza/BioWhittaker | Cat#: 12-167F |
| 10% Zinc Formalin | American MasterTech Scientific | Cat#: FXZFO10GAL |
| Mouse Serum | Millipore Sigma | Cat#: NS03L |
|
| ||
| Critical commercial assays | ||
|
| ||
| Preparation of single-cell suspensions from human nasal mucosa using the gentleMACS Dissociator | Miltenyi Resources | https://www.miltenyibiotec.com/GB-en/resources/resource-finder.html?countryRedirected=1 |
| Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1 | 10X Genomics | Cat# 1000121 |
| Single Index Kit T Set A | 10X Genomics | Cat# PN-1000213 |
|
| ||
| Deposited data | ||
|
| ||
| Single cell RNA-Seq data generated in this study (oral) | This study | GEO: GSE164241 |
| Single cell RNA-Seq skin data | He et al., 2020 | GEO: GSE147424 |
| Single cell RNA-Seq lung data | Reyfman et al., 2019 | GEO: GSE122960 |
| Single cell RNA-Seq ileum data | Martin et al., 2019 | GEO: GSE134809 |
| Single cell RNA-Seq salivary gland data | Huang et al., 2021 | https://www.covid19cellatlas.org/ |
|
| ||
| Software and algorithms | ||
|
| ||
| Cell Ranger 3.0.1 | 10X Genomics | https://www.10xgenomics.com/ |
| Seurat 3.2.2 | Stuart et al., 2019 | https://github.com/satijalab/seurat/ |
| SingleR 1.0.6 | Aran et al., 2019 | https://github.com/dviraran/SingleR |
| Schex | N/A | https://github.com/SaskiaFreytag/schex |
| Gsfisher 0.2 | N/A | https://github.com/sansomlab/gsfisher/ |
| NicheNet 1.0.0 | Browaeys et al., 2020 | https://github.com/saeyslab/nichenetr |
| FlowJo 10.6.1 | FlowJo | https://flowjo.com |
| LAS X 3.7.0.20979 | Leica Microsystems | https://www.leica-microsystems.com |
| Prism 8 | GraphPad | https://www.graphpad.com |
| FACSDiva 9.0 | BD Biosciences | https://www.bdbiosciences.com/en-us |
|
| ||
| Other | ||
|
| ||
| LSRFortessa | BD Biosciences | https://www.bdbiosciences.com/en-us |
| NextSeq 500 | Illumina | https://www.illumina.com/ |
| Leica TCS SP8 X | Leica Microsystems | https://www.leica-microsystems.com |
| gentleMACS Dissociator | Miltenyi Biotec | https://www.miltenyibiotec.com/GB-en/ |
| Cellometer Auto 2000 | Nexcelom | https://www.nexcelom.com/ |
| 70 μm filter | Falcon/Corning | Cat#: 352350 |
Single Cell RNA Sequencing
Biopsy preparation into single cell suspension
To prepare single cell suspensions biopsies were minced and digested using Collagenase II (Worthington Biochemical Corporation) and DNase (Sigma) and processed through the gentleMACS Dissociator (Miltenyl) utilizing a nasal mucosa protocol for processing (Derycke et al., 2012). Following tissue dissociation, cells were passed through a 70pm filter (Falcon, Corning), washed, and counted with a Cellometer Auto 2000 (Nexcelom).
10X Genomics Sequencing
Single-cell suspensions were loaded onto a 10X Chromium Controller (10X Genomics) and library preparation was performed according to the manufacturer’s instructions for the 10X Chromium Next GEM Single Cell Library kit v3 (10X Genomics). Libraries were then pooled in groups of 4 and sequenced on 4 lanes on a NextSeq500 sequencer (Illumina) using 10X Genomics recommended reads configuration.
Data alignment
Read processing was performed using the 10X Genomics workflow. Briefly, the Cell Ranger Single-Cell Software Suite v3.0.1 was used for demultiplexing, barcode assignment, and unique molecular identifier (UMI) quantification (http://software.10xgenomics.com/single-cell/overview/welcome). The reads were aligned to the hg38 human reference genome (Genome Reference Consortium Human Build 38) using a pre-built annotation package obtained from the 10X Genomics website (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references). All 4 lanes per sample were merged using the ‘cellranger mkfastq’ function and processed using the ‘cellranger count’ function.
Data import
For oral datasets, filtered feature barcode matrices generated by the cellranger pipeline were used as input into a Seurat-compatible format using the function ‘Read10X’ and ‘CreateSeuratObject’.
For previously published skin (He et al., 2020), lung (Reyfman et al., 2019) and ileum (Martin et al., 2019) datasets, files downloaded from GEO Accession Viewer were extracted and imported into a Seurat-compatible object. Salivary gland data (Huang et al., 2021; Huang et al., 2020) was downloaded from covid19cellatlas.org and imported into a Seurat-compatible object. All quality control, normalization, and downstream analysis was performed using the R package Seurat (Stuart et al., 2019) (ver. 3.2.2, https://github.com/satijalab/seurat) unless otherwise noted.
Data quality control, transformation and integration
In order to account for the experimental artefact of background/ambient RNA, we ran the Cell Ranger pipeline with ‘EmptyDrops’, to select for good quality cells/barcodes, which estimates the profile of the ambient RNA pool and tests each barcoded droplet for deviations from the ambient RNA profile using a Dirichlet-multinomial model of UMI count sampling (Lun et al., 2019). Barcodes with significant deviations were considered to be real cells (and were retained), while barcodes that were similar to the ambient RNA profile were removed. Within the Seurat workflow, low quality and artefact cells were excluded by removing any cells that expressed fewer than 200 genes, more than 5,000 genes and those with more than 15% mitochondrial gene expression. Gene expression matrices were transformed for better interpretability using the Seurat function ‘NormalizeData’.
For cross-tissue and cross-condition data integration and batch correction, ‘FindIntegrationAnchors’ and ‘IntegrateData’ were applied to data in Seurat, following the data integration vignette (https://satijalab.org/seurat/archive/v3.2/immunealignment.html). A total of four unique dataset integrations were performed for data analysis by supplying a list of Seurat objects containing the following datasets: 1. buccal and gingival health, 2. gingival health and disease, 3. oral (buccal and gingival health), skin, lung, and ileum, and 4. buccal, gingival and salivary gland health. For the third and fourth group of samples described above, all cohorts were randomly down sampled using the base R function ‘sample’ such that each group contained the same number of cells for global comparison.
Dimensionality reduction, cell clustering and identification
To mitigate effects of cell cycle heterogeneity on cell clustering, each sequenced cell was given a cell cycle phase ‘score’ based on expression of canonical markers using the Seurat function ‘CellCycleScoring’, with the Seurat object as input in addition to G1/S- and G2/M-phase specific genes (Tirosh et al., 2016). During data scaling (‘ScaleData’), scores were used to ‘regress out’ heterogeneity due to cell cycle. Louvain clustering was performed on all cells with the ‘FindClusters’ function using the first 50 PCs and a resolution of 1. Differential gene expression was used to guide manual annotation of clusters (‘FindAllMarkers’ function). Clusters defined by expression of hemaglobin genes were considered to be contaminated with red blood cells and were excluded from further analysis. All annotations of previously published datasets were performed in this manner following integration and clustering with oral mucosal datasets, unless otherwise noted in figure legends. The transcriptomic signature of immune cells was further investigated using the Monaco reference transcriptomic database (Monaco et al., 2019) of FACS purified cell types via SingleR (Aran et al., 2019) (ver. 1.0.6, https://github.com/dviraran/SingleR). To eliminate overlap of individual cells which may obfuscate gene expression patterns, UMAPs for select cell subtypes were tessellated by a regular grid of hexagons, the number of cells within each hexagon were counted, and the average gene expression across cells within each hexagon was represented by a color ramp via schex (ver. 1.0.55, https://github.com/SaskiaFreytag/schex).
Pathway analysis and receptor-ligand signaling inference
Gene set over-representation analysis of significantly upregulated cluster-defining genes was performed using the gsfisher R package (ver. 0.2, https://github.com/sansomlab/gsfisher/), and filtered to include only ‘biological process’ gene sets obtained from the GO database.
To predict cell-cell interactions using expression data, we utilized NicheNet (ver. 1.0.0, https://aithub.com/saevslab/nichenetr) which combined gene expression data of cells from our sequencing cohort with a database of prior knowledge on signaling and gene regulatory networks. The integrated Seurat object containing healthy and diseased gingival tissue was used as input into the NicheNet Seurat wrapper (‘nichenet_seuratobj_aggregate’). In addition, receiver cells were defined as all immune cells, sender cells were defined as endothelial, epithelial and fibroblast cells, and the experimental condition was disease state. Ligands expressed by one or more sender cells were ranked based on the Pearson correlation coefficient between the ligand’s target predictions and observed transcriptional response. Receiver cell receptors were inferred based on the NicheNet prebuilt prior model, which utilizes several curated ligand-receptor and signaling databases to predict links between sender ligands, receiver receptors and downstream target genes (Browaeys et al., 2020).
QUANTIFICATION AND STATISTICAL ANALYSIS
To assess significance of flow cytometry results, if both sample groups passed the Shapiro-Wilk normality test, an unpaired t-test with Welch’s correction was used. If one or more groups did not pass the normality test, a Mann-Whitney test was used. When assessing differentially expressed genes with Seurat, a non-parametric Wilcoxon rank sum test was used. To determine significance of proportion changes across tissue types and disease states, an unpaired t-test was used. The minimum threshold for significance was defined as p < 0.05.
ADDITIONAL RESOURCES
Patient samples were collected as part of clinical trial #NCT01568697: https://clinicaltrials.gov/ct2/show/NCT01568697
Supplementary Material
Figure S1 (related toFigure 1). A. Violin plots showing the number of features, RNA counts and percent mitochondrial transcripts found in gingival health (GM), buccal health (BM), and periodontal disease (PD) patient samples prior to quality control. B. Violin plots showing the number of features, RNA counts and percent mitochondrial transcripts following quality control. C. Bar graphs showing the number of cells that were originally sequenced (preQC) and the number of cells included for analysis (postQC) for each patient sample. D. Heatmap showing the average expression of the top differentially regulated genes for the major cell types identified in healthy gingival and buccal tissues.
Figure S2 (related toFigure 2). A-C. Heatmaps depicting average gene expression for each gene associated with the GO term on the right that was enriched in each individual endothelial (A), epithelial (B) and fibroblast (C) cluster.
Figure S3 (related toFigure 3). A. Example gating strategy for flow cytometry analysis in Figure 3H. B. Proportion plots showing the percentage of each immune cell type in each barrier tissue. **p<0.01 based on a one-way ANOVA and Dunnett’s multiple comparisons test. C. Summary of analysis comparing salivary gland (n=5) to subsampled buccal (n=8) and gingival (n=13) mucosa single cell RNA sequencing. UMAP representation (left) and proportion plots (right) for all cells (21,423 cells) in the combined datasets. Gene labels indicate cluster-defining genes. The shading on the UMAP and cluster proportions matches the major cell types seen in the main figures, thus endothelial clusters are shaded in red, fibroblast in green, epithelial in brown, and immune in blue. Salivary gland specific clusters are shaded in yellow.
Figure S4 (related toFigure 5). A,B. Immunohistochemistry of healthy (A) and periodontitis (B) tissues stained with myeloperoxidase, an enzyme abundantly expressed by neutrophils. Scale bar: 100μm.
Figure S5 (related toFigure 5,6). Summary of comparison between gingiva health and periodontitis cohorts. The gingival health group was randomly subsampled such that both groups contained the same number of cells (33,994 each). Each bar graph represents the percentage of each cluster within the four major cell types (endothelial, epithelial, fibroblast and immune). The actual number of cells per cluster in each tissue is present above their respective bar graphs.
Figure S6 (related toFigure 6). A,C,E. Dot plots depicting the expression of cluster-defining genes and percentage of cells expressing each gene for endothelial (A), epithelial (C), and fibroblast (E) populations, respectively, in health (n=13) and disease (n=8). Expression values are normalized and scaled averages. B, D, F. UMAP of sequenced cells from health (top) and periodontitis (bottom) of endothelial (B), epithelial (D) and fibroblast (F) populations. G. Dot plot showing periodontitis immune cell expression of receptors identified by NicheNet. Expression values are normalized and scaled averages. H. Dot plot showing tissue- and cell-type specific expression of top ligands identified by NicheNet. Expression values are normalized and scaled averages. I. Heatmap of prior interaction potential between receptors and ligands identified by NicheNet and used to generate alluvial plots in Figure 6G. J. Dot plots of chemokine receptor expression in immune cells in health and disease (left) and expression of chemokine ligand expression in general cell types in health and disease (right). Colored lines extending from receptors to ligands represent known interactions. Expression of chemokine receptors was simplified in Figure 6H based on this dot plot. Expression values are normalized and scaled averages.
Figure S7 (related toFigure 7). A,B. Dot plots showing expression of TLRs (green), NLRs (brown), CLR/RLRs (orange), DAMPs (blue), DAMP receptors (cyan), SARS-CoV-2 viral entry factors (red), and viral entry factors for viruses that lead to oral pathology (purple) in major cell types of gingival and buccal health (A) and in gingival health and disease (B). Expression values are normalized and scaled.
Table S1 (Related toFigure 1–7): Demographic information of all patients analyzed with single cell RNA sequencing and flow cytometry
Table S2 (Related toFigure 1): Distribution of major cell types by patient following batch correction
Table S3 (Related toFigure 1,3): Cell and source information for comparative single cell sequencing analysis of barrier tissues and salivary gland
Table S4 (Related toFigure 2): List of cluster defining genes for endothelial, epithelial and fibroblasts
Table S5 (Related toFigure 3): List of cluster defining genes for major immune cell types identified
Table S6 (Related toFigure 6): List of gene ontology pathway information associated with pathways enriched in periodontitis
Table S7 (Related toFigure 7): Wilcoxon rank sum test results for expression of pattern recognition receptors and viral entry factors. Comparison between healthy gingival/ buccal tissues and healthy gingival/periodontitis tissues.
Highlights.
A single-cell atlas of human oral mucosa in individuals with or without periodontitis
Diverse stromal and immune cell populations promote barrier immunity
Stromal cell inflammatory profile is linked to neutrophil recruitment
Identification of cell-specific expression patterns of periodontitis susceptibility genes
Acknowledgements
This study was funded in part by the intramural programs of NIH/NIDCR (NMM) and NIH/NIAMS (MM), by extramural grants NIH/NIDCR DE025046 (KD), DE029436 and DE028561 (GH), FONDECYT #11180389 through the National Agency of Research and Development (ANID), Chile (ND), and a PhD studentship from the Barbour Foundation (SW). MH is funded by Wellcome (WT107931/Z/15/Z), The Lister Institute for Preventive Medicine and Newcastle NIHR Biomedical Research Centre (BRC). This work was also made possible through funds from the NIDCD Division of Intramural Research/NIH (ZIC DC000086 to GCBC) and Combined Technical Research Core (NIDCR, ZIC DE000729-09). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). Figure illustrations were created in part with Biorender.com and by Alan Hoofring and Erina He (NIH Medical Arts). The authors also thank Dr. Tassos Sfondouris for clinical support and Dave Horsfall for web portal setup. This publication is part of the Human Cell Atlas - www.humancellatlas.org/publications.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, and Diaz PI (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7, 1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature immunology 20, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K, Li X, Kajikawa T, Toshiharu A, Selevsek N, Grossmann J, Hajishengallis G, and Bostanci N (2020). Pressure Cycling Technology Assisted Mass Spectrometric Quantification of Gingival Tissue Reveals Proteome Dynamics during the Initiation and Progression of Inflammatory Periodontal Disease. Proteomics 20, e1900253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts K, Abusleme L, Freeman AF, Sarmadi M, Fahle G, Pittaluga S, Cuellar- Rodriguez J, Hickstein D, Holland SM, Su H, et al. (2015). A 17-year old patient with DOCK8 deficiency, severe oral HSV-1 and aggressive periodontitis - a case of virally induced periodontitis? J Clin Virol 63, 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys R, Saelens W, and Saeys Y (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 17, 159–162. [DOI] [PubMed] [Google Scholar]
- Buskermolen JK, Roffel S, and Gibbs S (2017). Stimulation of oral fibroblast chemokine receptors identifies CCR3 and CCR4 as potential wound healing targets. Journal of cellular physiology 232, 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft AP, Campos J, Jansen K, Turner JD, Marshall J, Attar M, Savary L, Wehmeyer C, Naylor AJ, Kemble S, et al. (2019). Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derycke L, Zhang N, Holtappels G, Dutre T, and Bachert C (2012). IL-17A as a regulator of neutrophil survival in nasal polyp disease of patients with and without cystic fibrosis. J Cyst Fibros 11, 193–200. [DOI] [PubMed] [Google Scholar]
- Divaris K, Monda KL, North KE, Olshan AF, Lange EM, Moss K, Barros SP, Beck JD, and Offenbacher S (2012). Genome-wide association study of periodontal pathogen colonization. Journal of dental research 91, 21S–28S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divaris K, Monda KL, North KE, Olshan AF, Reynolds LM, Hsueh WC, Lange EM, Moss K, Barros SP, Weyant RJ, et al. (2013). Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum Mol Genet 22, 2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzan N, Abusleme L, Bridgeman H, Greenwell-Wild T, Zangerle-Murray T, Fife ME, Bouladoux N, Linley H, Brenchley L, Wemyss K, et al. (2017). On-going Mechanical Damage from Mastication Drives Homeostatic Th17 Cell Responses at the Oral Barrier. Immunity 46, 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzan N, Kajikawa T, Abusleme L, Greenwell-Wild T, Zuazo CE, Ikeuchi T, Brenchley L, Abe T, Hurabielle C, Martin D, et al. (2018). A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzan N, Konkel JE, Greenwell-Wild T, and Moutsopoulos NM (2016). Characterization of the human immune cell network at the gingival barrier. Mucosal immunology 9, 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD, and Genco RJ (2015). Update on Prevalence of Periodontitis in Adults in the United States: NHANES 2009 to 2012. Journal of periodontology 86, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine N, Hassanpour S, Borenstein A, Sima C, Oveisi M, Scholey J, Cherney D, and Glogauer M (2016). Distinct Oral Neutrophil Subsets Define Health and Periodontal Disease States. Journal of dental research 95, 931–938. [DOI] [PubMed] [Google Scholar]
- Gaffen SL, and Moutsopoulos NM (2020). Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci Immunol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger S, and Meyle J (2019). Oral Mucosal Epithelial Cells. Front Immunol 10, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G (2015). Periodontitis: from microbial immune subversion to systemic inflammation. Nature reviews Immunology 15, 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, and Moutsopoulos NM (2016). Role of bacteria in leukocyte adhesion deficiency-associated periodontitis. Microb Pathog 94, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halai H, Somani C, Donos N, and Nibali L (2020). Periodontal status of children with primary immunodeficiencies: a systematic review. Clin Oral Investig 24, 1939–1951. [DOI] [PubMed] [Google Scholar]
- Hart TC, and Atkinson JC (2007). Mendelian forms of periodontitis. Periodontol 2000 45, 95–112. [DOI] [PubMed] [Google Scholar]
- He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, Kameyama N, Estrada Y, Der E, Krueger JG, et al. (2020). Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. The Journal of allergy and clinical immunology 145, 1615–1628. [DOI] [PubMed] [Google Scholar]
- Hoogendijk AJ, Pourfarzad F, Aarts CEM, Tool ATJ, Hiemstra IH, Grassi L, Frontini M, Meijer AB, van den Biggelaar M, and Kuijpers TW (2019). Dynamic Transcriptome- Proteome Correlation Networks Reveal Human Myeloid Differentiation and Neutrophil-Specific Programming. Cell Rep 29, 2505–2519 e2504. [DOI] [PubMed] [Google Scholar]
- Huang N, Perez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Conde CD, Gasmi B, Stein S, Beach M, et al. (2021). SaRs-CoV-2 infection of the oral cavity and saliva. Nature medicine. 25 March. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Perez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Dominguez Conde C, Gasmi B, Stein S, Beach M, et al. (2020). Integrated Single-Cell Atlases Reveal an Oral SARS-CoV-2 Infection and Transmission Axis. medRxiv. [Google Scholar]
- Human Microbiome Project C (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Bartolome R, Uchiyama A, Molinolo AA, Abusleme L, Brooks SR, Callejas- Valera JL, Edwards D, Doci C, Asselin-Labat ML, Onaitis MW, et al. (2018). Transcriptional signature primes human oral mucosa for rapid wound healing. Sci Transl Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson D, Amisten S, Bratthall G, Holm A, and Nilsson BO (2009). LPS induces GROalpha chemokine production via NF-kappaB in oral fibroblasts. Inflamm Res 58, 791–796. [DOI] [PubMed] [Google Scholar]
- Koliaraki V, Prados A, Armaka M, and Kollias G (2020). The mesenchymal context in inflammation, immunity and cancer. Nature immunology 21, 974–982. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Koo H, and Hajishengallis G (2018). The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 16, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos BG, and Van Dyke TE (2020). The role of inflammation and genetics in periodontal disease. Periodontol 2000 83, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, participants in the 1st Human Cell Atlas, J., and Marioni JC (2019). EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol 20, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Abe T, Hajishengallis E, Hosur KB, DeAngelis RA, Ricklin D, Lambris JD, and Hajishengallis G (2014). Genetic and intervention studies implicating complement C3 as a major target for the treatment of periodontitis. Journal of immunology 192, 6020–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, and Borisy GG (2016). Biogeography of a human oral microbiome at the micron scale. Proceedings of the National Academy of Sciences of the United States of America 113, E791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang LS, Nayar S, Greenstein AJ, et al. (2019). Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508 e1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carre C, Burdin N, Visan L, Ceccarelli M, Poidinger M, et al. (2019). RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep 26, 16271640 e1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli T, Agler CS, and Divaris K (2020). Genomics of periodontal disease and tooth morbidity. Periodontol 2000 82, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, and Konkel JE (2018). Tissue-Specific Immunity at the Oral Mucosal Barrier. Trends in immunology 39, 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, and Moutsopoulos HM (2018). The oral mucosa: A barrier site participating in tissue-specific and systemic immunity. Oral diseases 24, 22–25. [DOI] [PubMed] [Google Scholar]
- Moutsopoulos NM, Zerbe CS, Wild T, Dutzan N, Brenchley L, DiPasquale G, Uzel G, Axelrod KC, Lisco A, Notarangelo LD, et al. (2017). Interleukin-12 and Interleukin-23 Blockade in Leukocyte Adhesion Deficiency Type 1. The New England journal of medicine 376, 1141–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HD, Cvikl B, Lussi A, and Gruber R (2016). Chemokine expression of oral fibroblasts and epithelial cells in response to artificial saliva. Clin Oral Investig 20, 1035–1042. [DOI] [PubMed] [Google Scholar]
- Nibali L, Bayliss-Chapman J, Almofareh SA, Zhou Y, Divaris K, and Vieira AR (2019). What Is the Heritability of Periodontitis? A Systematic Review. Journal of dental research 98, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicu EA, and Loos BG (2016). Polymorphonuclear neutrophils in periodontitis and their possible modulation as a therapeutic approach. Periodontol 2000 71, 140–163. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Divaris K, Barros SP, Moss KL, Marchesan JT, Morelli T, Zhang S, Kim S, Sun L, Beck JD, et al. (2016). Genome-wide association study of biologically informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum Mol Genet 25, 2113–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SR, Mercadante V, and Fedele S (2017). Oral manifestations of systemic disease. British dental journal 223, 683–691. [DOI] [PubMed] [Google Scholar]
- Proctor DM, and Relman DA (2017). The Landscape Ecology and Microbiota of the Human Nose, Mouth, and Throat. Cell Host Microbe 21, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen CI, Ren Z, et al. (2019). Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med 199, 1517–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AE, Sofer T, Wong Q, Kerr KF, Agler C, Shaffer JR, Beck JD, Offenbacher S, Salazar CR, North KE, et al. (2017). Chronic Periodontitis Genome-wide Association Study in the Hispanic Community Health Study / Study of Latinos. Journal of dental research 96, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifter M, Yeoh SC, Coleman H, and Georgiou A (2010). Oral mucosal diseases: the inflammatory dermatoses. Aust Dent J 55 Suppl 1, 23–38. [DOI] [PubMed] [Google Scholar]
- Shaffer JR, Polk DE, Wang X, Feingold E, Weeks DE, Lee MK, Cuenco KT, Weyant RJ, Crout RJ, McNeil DW, et al. (2014). Genome-wide association study of periodontal health measured by probing depth in adults ages 18–49 years. G3 (Bethesda) 4, 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Dong L, Zeng L, Yang R, Xu J, Wu Y, Xu R, Tao H, and Zhang N (2015). Two-stage comprehensive evaluation of genetic susceptibility of common variants in FBXO38, AP3B2 and WHAMM to severe chronic periodontitis. Scientific reports 5, 17882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shungin D, Haworth S, Divaris K, Agler CS, Kamatani Y, Keun Lee M, Grinde K, Hindy G, Alaraudanjoki V, Pesonen P, et al. (2019). Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nature communications 10, 2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LM, Brenchley L, and Moutsopoulos NM (2019). Primary immunodeficiencies reveal the essential role of tissue neutrophils in periodontitis. Immunological reviews 287, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, and Ohura K (2002). Porphyromonas gingivalis lipopolysaccharide signaling in gingival fibroblasts-CD14 and Toll-like receptors. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists 13, 132–142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (related toFigure 1). A. Violin plots showing the number of features, RNA counts and percent mitochondrial transcripts found in gingival health (GM), buccal health (BM), and periodontal disease (PD) patient samples prior to quality control. B. Violin plots showing the number of features, RNA counts and percent mitochondrial transcripts following quality control. C. Bar graphs showing the number of cells that were originally sequenced (preQC) and the number of cells included for analysis (postQC) for each patient sample. D. Heatmap showing the average expression of the top differentially regulated genes for the major cell types identified in healthy gingival and buccal tissues.
Figure S2 (related toFigure 2). A-C. Heatmaps depicting average gene expression for each gene associated with the GO term on the right that was enriched in each individual endothelial (A), epithelial (B) and fibroblast (C) cluster.
Figure S3 (related toFigure 3). A. Example gating strategy for flow cytometry analysis in Figure 3H. B. Proportion plots showing the percentage of each immune cell type in each barrier tissue. **p<0.01 based on a one-way ANOVA and Dunnett’s multiple comparisons test. C. Summary of analysis comparing salivary gland (n=5) to subsampled buccal (n=8) and gingival (n=13) mucosa single cell RNA sequencing. UMAP representation (left) and proportion plots (right) for all cells (21,423 cells) in the combined datasets. Gene labels indicate cluster-defining genes. The shading on the UMAP and cluster proportions matches the major cell types seen in the main figures, thus endothelial clusters are shaded in red, fibroblast in green, epithelial in brown, and immune in blue. Salivary gland specific clusters are shaded in yellow.
Figure S4 (related toFigure 5). A,B. Immunohistochemistry of healthy (A) and periodontitis (B) tissues stained with myeloperoxidase, an enzyme abundantly expressed by neutrophils. Scale bar: 100μm.
Figure S5 (related toFigure 5,6). Summary of comparison between gingiva health and periodontitis cohorts. The gingival health group was randomly subsampled such that both groups contained the same number of cells (33,994 each). Each bar graph represents the percentage of each cluster within the four major cell types (endothelial, epithelial, fibroblast and immune). The actual number of cells per cluster in each tissue is present above their respective bar graphs.
Figure S6 (related toFigure 6). A,C,E. Dot plots depicting the expression of cluster-defining genes and percentage of cells expressing each gene for endothelial (A), epithelial (C), and fibroblast (E) populations, respectively, in health (n=13) and disease (n=8). Expression values are normalized and scaled averages. B, D, F. UMAP of sequenced cells from health (top) and periodontitis (bottom) of endothelial (B), epithelial (D) and fibroblast (F) populations. G. Dot plot showing periodontitis immune cell expression of receptors identified by NicheNet. Expression values are normalized and scaled averages. H. Dot plot showing tissue- and cell-type specific expression of top ligands identified by NicheNet. Expression values are normalized and scaled averages. I. Heatmap of prior interaction potential between receptors and ligands identified by NicheNet and used to generate alluvial plots in Figure 6G. J. Dot plots of chemokine receptor expression in immune cells in health and disease (left) and expression of chemokine ligand expression in general cell types in health and disease (right). Colored lines extending from receptors to ligands represent known interactions. Expression of chemokine receptors was simplified in Figure 6H based on this dot plot. Expression values are normalized and scaled averages.
Figure S7 (related toFigure 7). A,B. Dot plots showing expression of TLRs (green), NLRs (brown), CLR/RLRs (orange), DAMPs (blue), DAMP receptors (cyan), SARS-CoV-2 viral entry factors (red), and viral entry factors for viruses that lead to oral pathology (purple) in major cell types of gingival and buccal health (A) and in gingival health and disease (B). Expression values are normalized and scaled.
Table S1 (Related toFigure 1–7): Demographic information of all patients analyzed with single cell RNA sequencing and flow cytometry
Table S2 (Related toFigure 1): Distribution of major cell types by patient following batch correction
Table S3 (Related toFigure 1,3): Cell and source information for comparative single cell sequencing analysis of barrier tissues and salivary gland
Table S4 (Related toFigure 2): List of cluster defining genes for endothelial, epithelial and fibroblasts
Table S5 (Related toFigure 3): List of cluster defining genes for major immune cell types identified
Table S6 (Related toFigure 6): List of gene ontology pathway information associated with pathways enriched in periodontitis
Table S7 (Related toFigure 7): Wilcoxon rank sum test results for expression of pattern recognition receptors and viral entry factors. Comparison between healthy gingival/ buccal tissues and healthy gingival/periodontitis tissues.
Data Availability Statement
Raw and processed single-cell RNA sequencing datasets have been deposited in the NCBI GEO database under accession GSE164241 and can be accessed via the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164241. Code to reproduce findings in this manuscript is available at: https://github.com/williamsdrake/oralAtlas. An interactive analytics portal to further explore gene expression is available at https://oral.cellatlas.io.


