Summary
The continuous development and application of technology for genetic improvement is a key element for advancing sheep production in the United States. The US sheep industry has contracted over time but appears to be at a juncture where a greater utilization of technology can facilitate industry expansion to new markets and address inefficiencies in traditional production practices. Significant transformations include the increased value of lamb in relation to wool, and a downtrend in large‐scale operations but a simultaneous rise in small flocks. Additionally, popularity of hair breeds not requiring shearing has surged, particularly in semi‐arid and subtropical US environments. A variety of domestically developed composite breeds and newly established technological approaches are now widely available for the sheep industry to use as it navigates these ongoing transformations. These genetic resources can also address long‐targeted areas of improvement such as growth, reproduction and parasite resistance. Moderate progress in production efficiency has been achieved by producers who have employed estimated breeding values, but widespread adoption of this technology has been limited. Genomic marker panels have recently shown promise for reducing disease susceptibility, identifying parentage and providing a foundation for marker‐assisted selection. As the ovine genome is further explored and genomic assemblies are improved, the sheep research community in the USA can capitalize on new‐found information to develop and apply genetic technologies to improve the production efficiency and profitability of the sheep industry.
Keywords: genetic diversity, genetic selection, genomics, Ovis aries, production systems
Introduction
Genetic technology is changing livestock production across the globe (Georges et al. 2019; Rexroad et al. 2019) and will be an important element for the US sheep industry (Lupton 2008). The US sheep industry is evolving as it continuously responds to challenges while simultaneously exploring growth opportunities. Genetic technologies that improve production and welfare under an environmentally sustainable format will propel the industry forward in the twenty‐first century. We summarize current and planned genomic research activities, including marker exploration for production and health traits, the development of genomic enhanced estimated breeding values (EBV), and the ongoing formation of national resource flocks. This review will present both the historical perspective on what has come to define the current US sheep industry and how genetic research can provide a foundation for future opportunities.
Industry overview
In 1942, the US sheep inventory was 56 million animals but it has since contracted to approximately 5 million sheep today (National Research Council 2008; USDA National Agriculture Statistics Service 2020a). The reasons for the decline are multifaceted and include, for example, legislative actions and changing markets. Historically, wool production was the primary source of revenue for sheep producers, but in the 1950s the advent of more cheaply made synthetic fibers reduced wool consumption (National Research Council 2008). Predation of domestic sheep by wildlife (principally coyotes) has long been a significant challenge and remains a critical issue as predation accounts for 36–43% of all lamb deaths per year (USDA Animal & Plant Health Inspection Service 2014), despite costly management practices by producers to prevent these losses. Sheep grazing on publicly owned lands has a rich history in the western states, but this use is not without controversy that originated over a century ago (USDA 1903; Adams 1916; Kelso 1947). During the 1970s additional legislation placed constraints upon the grazing of public lands (National Environmental Policy Act of 1969, Endangered Species Act of 1973, Clean Water Act of 1972 and Federal Land Policy and Management Act of 1976). The phasing‐out of the National Wool Act of 1954 removed wool and mohair price support in the 1990s, causing market volatility; without the subsidy’s price stabilization additional producers exited the industry.
The net result of these and other challenges is that today’s US sheep inventory (Fig. 1), and number of producers (Table 1), has reduced from that 70 years ago. The inventory of breeding ewes has also shifted among major geographic regions; there has been an increase in the Midwest and eastern areas and a proportional decrease in Texas/New Mexico, whereas the western proportion has remained stable (Fig. 1). The advancement of genetic technology, specifically the advent of accurate genomic selection, is a critical element in increasing production efficiency, improving environmental adaptability and ultimately re‐invigorating the US sheep industry. To better contextualize how genomic research might be utilized in developing biological solutions, it is necessary to have a basis of understanding of the changing landscape of the US sheep industry.
Figure 1.
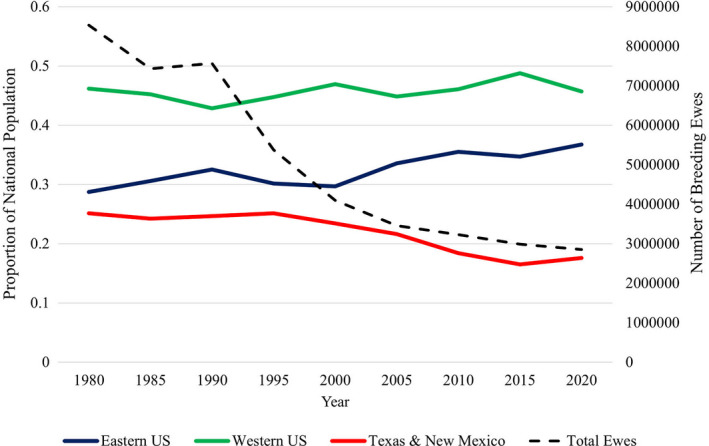
Total inventory of breeding ewes (dashed line) and proportional regional distribution for the eastern USA, western USA and Texas and New Mexico from 1980 to 2020 (USDA NASS 2020a).
Table 1.
Number of sheep operations by inventory size and year according to the US Census of Agriculture (https://usda.library.cornell.edu/concern/publications/000000018).
| Inventory class1 | Census year, n (%) | ||
|---|---|---|---|
| 1959 | 1987 | 2017 | |
| <25 | 169 421 (49.5) | 45 827 (49.5) | 70 455 (69.5) |
| 25–99 | 117 546 (34.4) | 31 254 (33.8) | 24 089 (23.8) |
| 100–299 | 36 938 (10.8) | 9740 (10.5) | 4750 (4.7) |
| 300–999 | 12 522 (3.7) | 3713 (4.0) | 1438 (1.4) |
| 1000–4999 | 4986 (1.4) | 1717 (1.9) | 548 (0.5) |
| ≥5000 | 539 (0.2) | 238 (0.3) | 107 (0.1) |
| Total | 341 952 | 92 489 | 101 387 |
Inventory size includes number of sheep and lambs of all ages.
Climate, landscape and production systems
The geographical scope of USA sheep production is broad, including arid, semi‐arid, temperate and subtropical climatic regions. Many of the arid and semi‐arid regions, found in the western USA, are public lands (ranging from 30 to 80% of total land area in western states). Within these climatic regions, sheep are primarily raised in mixed crop–livestock and extensive grazing systems and a smaller proportion in industrial production systems (de Haan et al. 1997). Extensive grazing systems are partitioned into those occurring on public lands (where transhumance is still practiced) or on privately owned lands. The use of industrial production systems is limited in the USA to temperate climates. Nationally, a high proportion of lambs destined for slaughter are fed high‐concentrate diets in an industrial production system setting until they reach a targeted weight.
The USA has consistently been the largest importer of sheep milk cheese in the world, which sparked the development of a domestic dairy industry and importation of improved European dairy germplasm in the 1990s (Thomas et al. 2014). Nevertheless, dairy sheep make up less than 1% of the national inventory (USDA APHIS 2014) and the primary commodities of US sheep systems are lamb and, to a lesser extent, wool.
Sheep production systems in the USA operate on different enterprise scales. Flocks with more than 1000 animals are generally managed more extensively, whereas those with fewer than 300 are typically managed in a mixed crop–livestock system or as an intensively managed farm flock. The number of producers in extensive production systems is small but they generally own a disproportionately larger (>1000 animals per producer) part of the national inventory. Conversely, more than 93% of producers have fewer than 100 animals (Table 1). Generally, extensive grazing systems have utilized Rambouillet, or its derivative breeds, which produce fine‐diameter wool and heavier fleeces and thereby capture higher sale prices than wool traditionally produced in mixed crop–livestock systems, $5.29/kg vs. $1.76/kg respectively (USDA NASS 2020a). In mixed crop–livestock systems, there is greater lamb production per ewe on average, 1.28 vs. 1.09, than in the extensive grazing system (USDA NASS 2020a). Current revenue ratios range from 76 to 83% and from 6 to 13% for lamb and wool respectively (Livestock Marketing Information Center 2016). These figures are in line with biologically based proportions for the costs of producing lamb and wool (Dickerson 1970; Blackburn & Pittroff 1999).
Despite an overall reduction in wool demand, extensive production systems are still dependent on wool revenues, with fiber diameter and fleece weight important breeding priorities (USDA Economic Research Service 2004). Improving growth rates have also driven selection within the industry. During the 1970s market lambs were typically slaughtered at 45–50 kg, but current slaughter weight averages have increased to 61 kg (USDA ERS 2004; USDA NASS 2020b). Lambs are predominantly marketed by weight, and thus faster growing and larger individuals capture greater individual returns, but as a result of selecting for growth in the lamb, there is a correlated increase in mature ewe body size (Borg et al. 2009). Recent work (Posbergh and Huson 2021) used GWAS to explore genomic associations for body size that might serve as a starting point for optimizing this trait in mature sheep. Reproductive traits have remained at the forefront of selection, particularly in production settings where nutritional resources are adequate.
Persistent and emerging issues
Longstanding biological and management issues exist and are generally based around increasing production efficiency, saving labor and developing a product that meets consumer demand. For example, the industry did not meet the goal of a 150% lamb crop by 2020 (ASI Roadmap; https://www.lambresourcecenter.com/roadmap), but it is still a worthy objective to reduce production costs (Dickerson 1970).
Regarding flock health, common diseases diagnosed or suspected in at least 20% of USA sheep operations include footrot, small ruminant lentivirus (ovine progressive pneumonia), caseous lymphadenitis, enterotoxaemia, coccidiosis and contagious ecthyma (USDA APHIS 2014). Identifying genetic markers for these and other health‐related traits has been, and continues to be, a priority of USA sheep researchers. Notably, Heaton et al. (2012) first described variants within TMEM154 associated with reduced susceptibility to lentivirus and Mousel et al. (2015) identified significant SNP in SLC2Ap and near NLN affiliated with entropion. More recently two research groups have been part of the effort to identify genomic regions affiliated with Mycoplasma ovipneumoniae (Mousel et al. 2021) and the degenerative condition ovine Johne’s disease (Y. Yaman, in review). Genetic markers for scrapie have resulted in reduced susceptibility using associated variants of PRNP at codons 136, 141, 154 and 171 (Hunter et al. 1994; Westaway et al. 1994; Belt et al. 1995; Moum et al. 2005). According to the National Scrapie Eradication Program of the US Department of Agriculture (USDA), the prevalence of scrapie‐positive animals at harvest was less than 0.0001%. This low incidence may be attributed to the implication and adoption of genomic testing over two decades (Westaway et al. 1994). Gastrointestinal nematodes (GIN), specifically Haemonchus contortus, are a drain on production efficiency and flock health in the USA, and resistance to anthelmintics is becoming more prevalent (Howell et al. 2008). Improving sheep resistance or tolerance to GIN is a high priority for genomics‐based research.
The availability of summer grazing of public lands is foundational to the feasibility of ranching in the western United States. Despite much research supporting sheep as an effective tool for rangeland improvement (Havstad 1994; Frost & Launchbaugh 2003), there is debate over whether sheep are beneficial or harmful to the natural ecosystems on public land. A perceived concern about disease transmission between domestic and wild (Ovis canadensis) sheep has been raised (Onderka & Wishart 1988; Lawrence et al. 2010; Besser et al. 2013), further reducing public land available for grazing (Hendrickson 2015). There is strong evidence that the immunopathologic response to M. ovipneumoniae differs between domestic and wild sheep (Grossman et al. 2019), often resulting in more detrimental effects of the pathogen to wild sheep. Recently, Mousel et al. (2021) has presented research that could aid in solving the problem of disease transmission between wild and domestic sheep.
With production scenarios in the USA evolving, producer priorities for animal selection are also becoming more diverse. The decline in sheep numbers in the extensive southwest systems and subsequent increase in temperate and subtropical mixed crop systems suggest that new strategies for selection will be needed for smaller farm flocks with fewer than 100 animals. A shift toward smaller flock sizes of diverse breeds presents a challenge for genomic research and implementation, which is predicted to realize the greatest benefit in large flocks with well‐defined phenotypes (Casellas & Piedrafita 2015).
Climate change presents a range of challenges and opportunities for genetic enhancement. Across USA ecosystems, climate change will have varying effects, such as increased variability in precipitation, increased temperatures and shifts in composition of native plant species upon which sheep forage (Holechek et al. 2020). Snowder et al. 2001 first described directional selection of sheep for the consumption of sagebrush, a woody plant common to the western United States. With climate change shifting plant species composition to less desirable forages (Morgan et al. 2007), the opportunity for genomic research into grazing habits of sheep could result in the control of brush and invasive plant species.
Genetic diversity
A country’s genetic resource base and variability governs the ability to alter animal productivity. Genomic research has facilitated the quantification of differences among our populations and these populations in turn are critical in planning and executing future genomic research. To address many of the concerns previously mentioned, USA producers have long imported sheep from other areas of the world and owing to the array of climatic regions and production systems in the USA, there have always been niches for a wide variety of breeds. Currently, the database of the Food and Agriculture Organization, Domestic Animal Diversity – Information System (http://www.fao.org/dad‐is/en/) indicates there are 50 US sheep breeds, originating from Asia, Africa, Europe and Oceania, all with diverse phenotypes and characteristics.
Generally, and unlike other livestock species, sheep tend to have a weak population substructure (Kijas et al. 2009; Groeneveld et al. 2010). Levels of heterozygosity for USA breeds tend to be similar to those in Europe and other parts of the world (Lawson‐Handley et al. 2007; Peter et al. 2007; Dalvit et al. 2008; Blackburn et al. 2011a). Previous analyses have shown how breeds from close to centers of domestication tend to have more within‐breed genetic diversity (Bruford et al. 2003; Tapio et al. 2010; Sulaiman et al. 2011). However, when US breeds were combined into one group and compared with sheep near the center of domestication, similar levels of genetic diversity were present (Blackburn et al. 2011b), suggesting that as a country, substantial genetic variation exists. Similar findings among populations near the center of domestication and the USA were reported for goats (Paim et al. 2019) and Davenport et al. (2018) have suggested that relatively high recombination rates in sheep, and subsequently low LD, may be a contributor to genetic differentiation.
There is evidence of subpopulations within breeds formed by genetic drift, natural and/or artificial selection and geographical constraints. For example, Texel, Suffolk, Dorper, Dorset, St Croix and Blackbelly Barbados have shown within‐breed differences, via calculations for F ST (range = 0.025–0.082) and/or PCA (Kijas et al. 2009; Paiva et al. 2011). Kijas et al. (2012) used F ST, PCA, and allele‐sharing metrics to characterize two different subpopulations of the Gulf Coast Native and suggested that the two subpopulations should be classified as separate breeds. Within the USA, Davenport et al. (2020) quantified subpopulation differences in Suffolk raised in either semi‐arid or temperate environments (F ST = 0.07). It has also been demonstrated that breeders frequently admixed two prominent terminal sire breeds, Hampshire and Suffolk, to the point where genetic structure is lacking between the two (Blackburn et al. 2011a; Davenport et al. 2020).
When evaluating genetic relatedness across breeds, previous population admixture, principal component and differentially selected region analyses found that US breeds tend to group within generic phenotypic descriptors (Blackburn et al. 2011a; Zhang et al. 2013; Davenport et al. 2020). Some example breeds and their classifications are Lincoln, Leicester Longwool, Cotswold and Romney as longwool; Hampshire and Suffolk as meat; St Croix and Barbados Blackbelly as hair; and Rambouillet, Targhee and Columbia as fine wool. More recent importations of Dorper and Romanov tended not to be associated with the groupings mentioned. The composite breeds Columbia, Targhee and Katahdin were placed intermediate to the progenitor breeds in PCA, as has been demonstrated with cattle (Paim et al. 2020). Zhang et al. (2013) also suggested that genetic differences between Rambouillet, Columbia and Targhee were small after identifying differentially selected regions common to the three breeds, and that these breeds could be considered one population.
No official government statistics are maintained on breed inventory, therefore breed association registrations per year are a proxy, which is also the case for cattle and swine. These records suggest that Suffolk, Hampshire and Dorset registrations have decreased in recent decades, whereas those of Katahdin and Dorper have grown and surpassed these breeds (Fig. S1). There tends to be a geographic preference for the hair breeds, with Katahdin predominating in the Midwest and east and Dorper in the southwestern region of the country, where they are displacing Rambouillet (Table 2). Further suggesting that there is a transition from Rambouillet to Dorper in Texas, the largest sheep‐producing state (USDA NASS 2020a), is the rising number of hair‐breed lambs being marketed, as shown in Fig. 2 (Waldron et al. 2016).
Table 2.
Breeds commonly utilized in various production systems and climates in the USA
| Production system/climate1 | Arid | Semi‐arid | Temperate | Subtropical |
|---|---|---|---|---|
| Mixed crop–livestock | – |
Dorper Suffolk Columbia Hampshire Dorset |
Suffolk Hampshire Dorset Southdown Columbia Polypay Finnsheep Katahdin Texel Romanov Blackbelly Barbados Minor breeds2 |
Katahdin St Croix Dorper Blackbelly Barbados Gulf Coast Native |
| Extensive grazing |
Rambouillet Columbia Targhee Navajo Churro Dorper |
Rambouillet Columbia Targhee Dorper Polypay Navajo Churro Romney |
– |
Rambouillet Dorper |
Via the Köppen climate classification system.
Indicative minor breeds: Shropshire, Oxford, Lincoln, Leceister Longwool and Cotswold.
Figure 2.
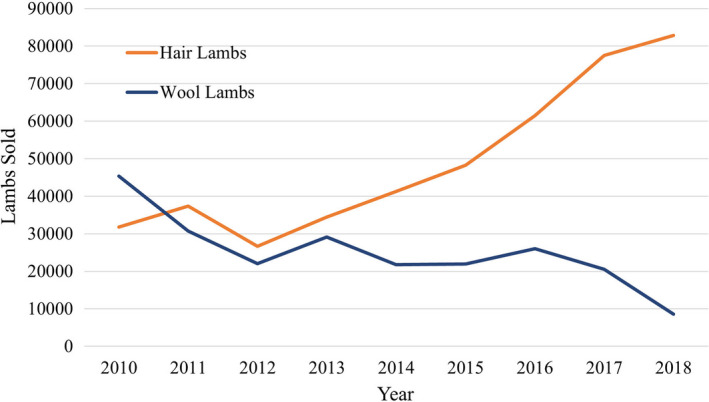
Lambs sold at Producers Livestock Auction in San Angelo, Texas classified by hair or wool phenotype, based upon Waldron et al. (2016) and updated. Hair lambs are predominantly Dorper and Dorper crossbreeds.
American producers have always been willing to develop new composite breeds to take advantage of heterosis and breed complementarity. For example, Columbia (in 1917) and Targhee (in 1938) were developed for use in extensive grazing in arid and semi‐arid environments (Terrill 1947). In 1969, the Polypay was developed at the same location with equal contributions of Rambouillet, Dorset, Finnsheep and Targhee (Rasali et al. 2006). With its higher prolificacy, the Polypay is prominent in mixed crop–livestock systems in temperate environments (Hulet et al. 1984). The Katahdin was created to be a fast‐growing and prolific hair breed with reduced susceptibility to GIN (Wildeus 1997) and is rapidly gaining in popularity (Fig. S1).
Genetic experimentation
Following the development of breeding techniques and genetic advancements in the beef and dairy industries, a genetics research agenda took place at various public institutions and federal facilities (Table 3). This laid the foundation for numerous recommendations to the sheep industry concerning selection for multiple economically relevant traits. Public institutions also started centralized ram performance tests, predominantly with Rambouillet, where wool and growth traits were evaluated. The Texas A&M Agriculture Experiment Station in Sonora, TX started the first such test in 1949; followed by the University of Wyoming, North and South Dakota State Universities, Montana State University and Virginia Tech University. This ‘early’ form of genetic measurement improved fleece and growth performance in extensively raised Rambouillet (Fig. 3; Shelton 1979; Burton et al. 2015).
Table 3.
Exemplary breeding programs, and their predominant research goals and outcomes, conducted at universities and various US Department of Agriculture (USDA) research facilities.
| Institution/status | Goal | Primary result | Breed(s) | Citation |
|---|---|---|---|---|
| Cornell University, terminated | Accelerated lambing | Intensively managed STAR lambing system | Dorset | Lewis et al. (1996) |
| Montana State University, terminated | Selection for prolificacy | Divergent selection for reproductive performance | Rambouillet | Burfening et al. (1993) |
| Texas A&M University, terminated | Centralized testing | Improved fiber and increased growth rate | Rambouillet | Shelton (1979) |
| University of California Davis, terminated | Quantify prolificacy/litter size | Development of high‐prolificacy line of sheep | Targhee | Bradford et al. (1986) |
| University of Minnesota, terminated | Finnsheep and Prolificacy evaluation | Quantified Finnsheep and Finn‐cross reproduction and lamb survival | Finnsheep Suffolk Targhee Minnesota‐100 | Oltenacu & Boylan (1981) |
| University of Nevada—Reno, ongoing | Wool improvement | Development of a superior line of fine‐wool sheep | Merino | Wuliji et al. (2019) |
| University of Wisconsin Madison, terminated | Dairy | Improved milk quantity and quality | East Friesian Lacaune | Thomas et al. (2014), Murphy et al. (2017) |
| University of Wyoming, terminated | Centralized testing | Improved fiber and increased growth rate | Rambouillet | Burton et al. (2015) |
| Virginia Tech University, terminated | Seasonality | Altered breeding season | Dorset Rambouillet Finnsheep | al‐Shorepy & Notter (1997) |
| USDA‐ARS‐Booneville, ongoing | GIN Resistance | Improved resistance in hair breeds |
Katahdin St Croix Dorper |
Burke & Miller (2004) |
| USDA‐ARS‐Dubois, ongoing | Breed development | Development of Polypay breed | Rambouillet Dorset Finnsheep | Hulet et al. (1984) |
| USDA‐ARS‐Dubois, terminated | Long‐term effects of inbreeding | Quantified economic impact of inbreeding | Rambouillet Targhee Columbia | Ercanbrack & Knight (1991, 1993) |
| USDA‐ARS‐Meat Animal Research Center, ongoing | Breed development | Developed terminal and maternal composites | Composites I–IV | Leymaster (1991), Murphy et al. (2020) |
| USDA‐ARS‐Meat Animal Research Center, ongoing | Breed characterization | Quantify breed differences for terminal and maternal traits | Multiple1 | Dickerson et al. (1972), Casas et al. (2004), Casas et al. (2005), Freking & Leymaster, (2005) |
Including Suffolk, Hampshire, Texel, Rambouillet, Targhee, Columbia, Polypay, Romanov, Finnsheep, Corriedale, Dorset, Montadale, Dorper, Katahdin, and Navajo Churro. ARS =Agricultural Research Service
Figure 3.
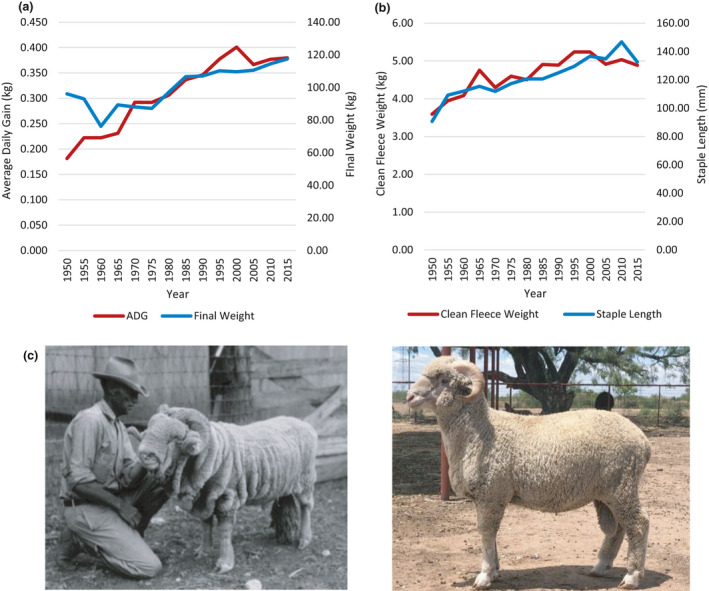
The Texas A&M central ram test has evaluated more than 12 000 rams from 1942 to 2018. (a) Average daily gain and final weight of rams from 1950 to 2015. (b) Average clean fleece weight and staple length of rams from 1950 to 2015. (c) Phenotypic differences among Rambouillet rams, 1950 (left) and 2020 (right). Historic records for the Texas A&M ram test can be viewed at https://sanangelo.tamu.edu/performance‐tests/ram/.
A transition to programs using EBV occurred with the inception of the National Sheep Improvement Program (NSIP) in the 1980s. This program initially calculated within‐flock EBVs for ewe reproduction, lamb body weight and wool traits using single‐trait animal models (Wilson & Morrical 1991). Over time, sufficient genetic connectedness was established among participating flocks, since the mid‐1990s across‐flock EBVs have been made available for participating breeders (Notter 1998).
Today, NSIP offers producers EBVs on body weight, growth, GIN resistance, wool traits and reproduction. In addition, several multiple‐trait indices including the western range index, maternal indices for hair and wool breeds and a carcass index for use with sire breeds have been developed (Borg et al. 2007; Vanimisetti et al. 2007). Figure 4 is illustrative of the reproductive and GIN resistance improvements made by producers using both the USA Hair Index and fecal egg count EBV.
Figure 4.
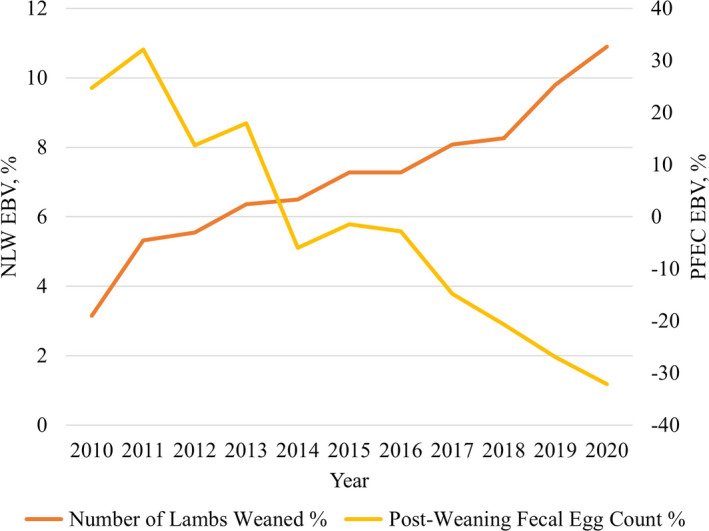
Progress made within the Katahdin breed for lamb production and parasite resistance using the National Sheep Improvement Program (NSIP) from 2010 to 2020. Displayed are the average EBVs for number of lambs weaned percentage (NLW%) and post‐weaning fecal egg count percentage (PFEC%) by birth year of over 46 000 Katahdins enrolled in NSIP during the last decade.
As of August 2020, there were 294 flocks enrolled in NSIP across 39 US states. From 2015 to 2019, approximately 77 000 individual lamb records were processed through the program. Currently, there are 24 different breeds enrolled, and as a percentage of the enrolled sheep, Katahdin (26.9%), Polypay (17.7%), Suffolk (11.2%), Targhee (9.9%) and Rambouillet (6.5%) are the most common.
Through the use of NSIP, genetic gains have been observed in many economically important traits. Despite this, relative to the number of US sheep producers, the program is still limited in scope. Improvement in the accuracy of EBVs earlier in the animal’s life, specifically for hard to measure traits such as carcass quality and reproduction, could increase the importance of genetic technology to US sheep producers. Efforts to incorporate genomic technology into NSIP EBVs are underway, including establishing suitable reference populations and exploring additional complex traits. These reference populations, managed by USDA Agricultural Research Service (ARS) or public institutions, along with a database of genetic and phenotypic data, should improve the accuracy of breeding strategies for a wide variety of traits. Genetic connectedness to these resource flocks will be especially beneficial to producers with smaller flock sizes where accuracy of the EBV is often limited. The design and management of large reference populations have also been critical to the progress of genomic‐based research in other countries (van der Werf et al. 2010).
Progression of genomics
New breeds and utilization of EBV have resulted in production progress, but genomic technology has the capability to further this, as has been witnessed in other species (Georges et al. 2019). In addition, using US dairy, swine and poultry genomic utilizations as a model will provide ample lessons for the sheep industry. For example, until recently, the cost of utilizing SNP arrays (in relation to revenue) has led to slower adoption of this technology by sheep producers than in other species. Even though applications to the sheep industry have been slower to come given financial and human resource limitations, our goal for the past 30 years has been to use the technology to better understand the genetic mechanisms of the sheep.
Early efforts with genomic technologies focused on creating linkage maps with several short tandem di‐ or tri‐nucleotide repeats (microsatellites). The United States Meat Animal Research Center (USMARC) and other US researchers were involved and contributed to the international mapping flock to create linkage maps (Crawford et al. 1995; de Gortari et al. 1998; Maddox et al. 2001) with bacterial artificial chromosome clones also being used to define the relative chromosomal location of microsatellite markers. This in turn facilitated the identification of location and mutation causing hypertrophy in skeletal muscles, termed callipyge, (Cockett et al., 1994; Freking et al., 2002). Microsatellite markers were used with the CHORI‐243 BAC library to develop one of the first sheep genome maps (Dalrymple et al. 2007; Ratnakumar et al. 2010). This map of ordered microsatellites for each chromosome in the sheep genome was converted into a virtual sheep genome in 2006 using a comparative genomics approach based on methods used for the human genome (Dalrymple et al. 2007). However, the combined information generated from BAC and radiation hybrid panels still needed improvement, requiring refined resolution for each chromosome. In general, research commenced with chromosomes that were previously identified as containing regions associated with traits of biological interest to sheep producers (Tetens et al. 2007; Wu et al. 2007; Drögemüller et al. 2008; Wu et al. 2008; Goldammer et al. 2009, 2009,2009, 2009).
A high‐quality reference genome has been a research goal for the last decade (Archibald et al. 2010), with the first ovine genome reference sequence being released in 2012 (http://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.1/). This assembly employed Illumina short read sequence technology generated from a Texel ram and ewe (Jiang et al. 2014). The ram that was used for this reference genome was from the USMARC population and the same animal used in the generation of the CHORI BAC library. These sequence data were anchored using sheep‐specific physical maps to assemble the genome, which was ultimately improved using data generated from long‐read sequencing technology (from PacBio) for gap filling (Liu et al. 2016). Specifically, the sequence length of the shortest of 50% of the genome (contig N50) more than doubled from 70 kb in the reference genome assembly Oar_v3.1 to 150 kb in the next assembly, Oar_v4.0. The contig L50 (the minimum number of contigs to contain half the assembly) was also reduced from over half a million to ~5000 as shown in Table 4 (http://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2/).
Table 4.
Progression of ovine reference genome assemblies
| Reference genome | Breed of sheep | Contig N50 (Mb) | LG50 (contigs) | Number of contigs | Release year(s) |
|---|---|---|---|---|---|
| Oar_v3.1 | Texel | 0.07 | 545 914 | 2 352 347 | 2012–2014 |
| Oar_v4.0 | Texel | 0.15 | 5008 | 48 482 | 2012–2015 |
| Oar_rambouillet_v1.0 | Rambouillet | 2.57 | 313 | 7486 | 2017 |
| ARS_UI_ramb_v2.0 | Rambouillet | 43.18 | 24 | 1226 | 2021 |
| Romanov | Romanov | 78.2 | 499 | (in progress) | |
| White Dorper | White Dorper | 82.5 | 1157 | (in progress) |
The development of long‐read sequencing technology that can form the basis of the assembly was game changing, as exemplified by genomic research in goats (Bickhart et al. 2017). Using 66‐fold PacBio and long‐read sequence and Hi‐C technique for scaffolding, a de novo sheep reference assembly, Oar_rambouillet_v1.0, of a mature Rambouillet ewe (Benz‐2616) was developed in 2017 (Worley 2018; Salavati et al. 2020). This assembly has a contig N50 of approximately 2.6 Mb. Keeping pace with the genome quality of other livestock species, a more recent de novo assembly (ARS‐UI_ramb_v2.0) has been produced incorporating OxNan PromethIONsequence also generated from the lung DNA of Benz‐2616. This new reference assembly was just released and is a dramatic improvement in comparison with Oar_rambouillet_v1.0, with contig N50 increasing 20‐fold and L50 reduced more than 12‐fold.
A new, alternative strategy for genome assembly that was developed with cattle and other species and holds promise for future sheep research is the Trio Binning method (Koren et al. 2018). This approach uses the F1 offspring of two genetically diverse breeds to generate two separate genome assemblies. Trio Binning is an efficient and cost‐effective approach to generate reference‐quality genomes using a single F1 interbreed cross to provide two individual haplotype‐resolved assemblies for both input breeds. Given the array of genetically distinct breeds in the USA, this approach should be highly useful for the Ovine pangenome and in developing new composite populations to fit a broad environmental and production system landscape.
Resources for future advances
The Functional Annotation of Animal Genomes (FAANG) consortium is an international collaborative effort to improve the annotation of reference genomes for multiple livestock species (Andersson et al. 2015). Members of the sheep genetics research community from the USA are actively involved in the Ovine FAANG portion of this effort. The latest reference genome, ARS‐UI_Rambouillet v2.0, will be used to annotate the functional elements such as gene promoters and enhancers. This consortium first identified tissue‐specific gene expression using tissue collected from Benz‐2616, and then identified active promoters and transcription start sites using cap analysis of gene expression workflow (Salavati et al. 2020). Recent reports by Salavati et al. (2020) indicate that, on average, for each of 56 tissues examined, including all major organs, more than 11 000 novel transcription start sites were identified. Findings from this work will be beneficial for a better understanding of gene transcription regulation in sheep. The newly funded research effort to construct an Ovine pangenome will look to further ‘fill in the gaps’ by adding genome assembly information from other sheep breeds.
The US sheep research community has also been involved in the formation of a number of genomic tools, including the 1.5K pilot sheep SNP array through the virtual sheep genome project (https://www.sheephapmap.org/Kijas_et_al_2009.pdf), the development and design of Illumina 50K SNP chip, the sheep parentage panel (Heaton et al. 2014), the high‐density SNP chip (Anderson 2014) and Flock54, a low‐density genotype by sequencing panel (Job et al. 2019). US producers have begun more widely utilizing markers from these tools to capture resistance/susceptibility for a number of diseases/conditions including scrapie, lentivirus, spider‐lamb syndrome, callipyge and more recently dwarfism. Amongst the growing number of small producers in the USA, there are also flocks of ‘heritage’ and other less common breeds such as the Navajo Churro, Gulf Coast Native and Jacob Sheep. In these systems selection for coat color or unique phenotypes, such as polycerate, often occurs. Kijas et al. (2016) identified markers for a four‐horned phenotype, which may be of interest to this niche area of the industry. Many of the causative mutations for diseases/conditions are more thoroughly described in the online mendelian inheritance of animals library (https://www.omia.org/home/). Whereas cost was a limiting factor to adoption of genomic technology previously, many of these panels are now priced lower, making them more affordable to the industry.
Following the model of the 1000 Bull Genomes project, members of the International Sheep Genomics Consortium, including US researchers, have contributed to SheepGenomesDB (https://sheepgenomesdb.org/), a publicly available database that contains WGSs from over 50 different breeds (Daetwyler et al. 2017). Furthermore, the National Animal Genome Research Program has developed and supported the collections of QTL and GWASs for sheep in the Animal QTL database (https://www.animalgenome.org/cgi‐bin/QTLdb/OA/index) and the CorrDB animal trait correlation database (https://www.animalgenome.org/cgi‐bin/CorrDB/index).
Geneticists at the USMARC and the US Sheep Experiment Station are in the process of establishing reference populations, with connectedness to industry flocks, for Katahdin, Suffolk, Rambouillet and Polypay sheep with EBVs calculated from phenotypic records in addition to genotypic data from medium‐ and high‐density SNP arrays. Researchers at USDA‐ARS Boonville, in collaboration with NSIP technical leadership, are pioneering efforts in the USA to develop genomic EBVs in Katahdin. With USDA and public institutions leading the effort in establishing genomic EBVs and resource flocks, this lays the groundwork for expanded use by the industry. In addition, validation of previously discovered markers in other populations and continuing the advancement of marker‐assisted selection strategies for both production and health‐related traits are prioritized goals of establishing reference populations.
To further increase genetic resources for future use, the USMARC has also developed the Composite IV line of sheep, based on combining coat shedding, prolificacy and maternal ability of Katahdin (25%), White Dorper (25%) and Romanov (50%) breeds. Favorable variants at TMEM154 and PRNP for improved disease resistance are also a noted breed component. To support this research, an F1 cross between a White Dorper ram and a Romanov ewe was created, the trio binning approach was applied and haploid assemblies are near ready, with contig N50s approximately 63 Mb each (B. Rosen and T. Smith, unpublished data).
To satisfy regional producer needs, the US Sheep Experiment Station recently developed a new terminal sire composite line enhanced for performance in the extensive rangeland production systems common in the western United States. The new composite was formed from Suffolk (3/8), Columbia (3/8) and Texel (1/4) breeds. The Suffolk breed was used for superior growth rates, Columbia for range hardiness and white pelt, and Texel for superior carcass traits and lack of expression of the myostatin gene.
With breed development expanding and directional change within breeds is progressing, there is also a need for an archive of genetics that span the variety of animals crucial to the industry. The National Animal Germplasm Program serves as a genetic security resource for stakeholders and the research community. Collection information (germplasm samples, phenotypes, genotypes and management systems) and requests for samples can be accessed via the database Animal‐GRIN at: https://agrin.ars.usda.gov/database_collaboration_page_dev?alert=Y. The NAGP employs a multidisciplinary approach using quantitative and molecular genetics, reproductive biology, cryopreservation, evaluation of live animal populations and information systems to strengthen the collection of genetic diversity in the USA. To date, the sheep collection contains 69 818 semen, embryo and blood samples from 3328 animals that represent 47 breeds. Major and minor breeds are represented in the collection, as well as research populations that have utility for future research efforts, correcting breeder mistakes and reconstituting whole breeds in the event of catastrophic events.
Future applications of genomics
Genomic technology can combat many of the inter‐related challenges facing the US sheep industry. Regarding genetic diversity, Rambouillet and its derivative breeds, plus various hair breeds, should have sufficient genetic variability to facilitate selection for any number of environmental challenges associated with climate change. In addition, the sheep’s smaller metabolic body size and faster passage rate of nutrients through the rumen, compared with cattle, may be a positive attribute in lowering methane emissions from the livestock sector and therefore offers a unique opportunity for genetic modification. Hair breeds also provide the potential to adapt to climate change in terms of increased resistance to GIN, broader grazing patterns and a smaller metabolic size, enabling them to shed heat load more efficiently (McManus et al. 2020). Selection for resistance to GIN can be achieved with most breeds; however a negative correlated response with prolificacy may be observed (Woolaston 1992). Recently, Estrada‐Reyes et al. (2019) documented directional signatures of selection for genes associated with immune protection from H. contortus among US hair breeds. By identifying genes of interest, it may be more feasible to employ selection for GIN resistance without soliciting a negative correlated response for prolificacy. In a collaborative effort between multiple US sheep researchers, genomic exploration is underway to identify strategies for selection of GIN‐resistant animals within the Katahdin breed. Publications from this effort have already reported potential SNP for identification of allelic variants associated with resistance to GIN (Becker et al. 2020).
With the broad array of production settings and breeds, precision breeding programs that incorporate ‘genetics × environment × production system interactions will be paramount for the sheep industry to capitalize on new genomic technologies (Rexroad et al. 2019). Making progress in this area will require the increased accumulation of both phenotypic and genotypic data across the sheep industry. Furthermore, information extrapolated from research efforts such as FAANG and the Ovine pangenome projects will be valuable in understanding how sheep genomes result in different phenotypic traits.
Conclusion
The US sheep industry has been able to make important adjustments to production given myriad market and policy challenges. Despite such issues, there may be cause for optimism. The rate of decline in ewe numbers has slowed in recent years, perhaps signaling an end to reductions in the sheep inventory. The adoption of genomic tools and strategies being employed by other livestock species will also serve as a guide for implementation of this biotechnology into the US sheep industry. An increasing array of genetic tools and resources are becoming more available for all sectors of the industry for a variety of breed or trait improvements. In addition to a continuously improving reference genome owing to the efforts of Ovine FAANG and USDA‐ARS, key infrastructures such as NSIP and NAGP are in place and can be utilized to advance the industry’s genetic goals. Importantly, a cadre of scientists, with a broad array of expertise, engaged in sheep research have developed a comprehensive multi‐institutional network for collaboration capable of leveraging resources to bring resolution to the challenges facing the sheep industry. We believe these factors and resources are poised to converge in the twenty‐first century for a vibrant US sheep industry.
Supporting information
Figure S1. Number of sheep registered for five popular breeds from 2000 to 2017.
Acknowledgements
The authors would like to acknowledge the individuals who provided consultation on the preparation of this manuscript, including Rusty Burgett with the National Sheep Improvement Program and Bill Thompson with Texas A&M AgriLife Economics. We additionally want to thank the multiple members of international sheep genetics community for their innumerable contributions to the advancements reported. Hatch grant #IDA01566 from USDA National Institute of Food supported Dr Murdoch’s involvement.
Mention of a trade name or proprietary product does not constitute a guaranty or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable. USDA Agricultural Research Service, Plains and Pacific West Areas are equal opportunity affirmative action employers. All agency services are available without discrimination.
Contributor Information
B. M. Murdoch, Email: bmurdoch@uidaho.edu.
H. D. Blackburn, Email: harvey.blackburn@usda.gov.
References
- Adams R. (1916) Public range lands—a new policy needed. American Journal of Sociology 22, 324–51. [Google Scholar]
- al‐Shorepy S.A. & Notter D.R. (1997) Response to selection for fertility in a fall‐lambing sheep flock. Journal of Animal Science 75, 2033–40. [DOI] [PubMed] [Google Scholar]
- Anderson R. (2014) Development of a high density (600 K) Illumina Ovine SNP Chip and its use to fine map the yellow fat locus. Plant and Animal Genome XXII Conference. San Diego, CA.
- Andersson L., Archibald A.L., Bottema C.D.et al. (2015) Coordinated international action to accelerate genome‐to‐phenome with FAANG, the Functional Annotation of Animal Genomes project. Genome Biology 16, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald A.L., Cockett N.E., Dalrymple B.P.et al. (2010) The sheep genome reference sequence: a work in progress. Animal Genetics 41, 449–53. [DOI] [PubMed] [Google Scholar]
- Becker G.M., Davenport K.M., Burke J.M., Lewis R.M., Miller J.M., Morgan J.L.M., Notter D.M. & Murdoch B.M. (2020) Genome‐wide association study to identify genetic loci associated with gastrointestinal nematode resistance in Katahdin sheep. Animal Genetics 51, 330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belt P.B.G.M., Muileman I.H., Schreuder B.E.C., Ruijter J.B., Gielkens A.L.J. & Smits M.A. (1995) Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. Journal of General Virology 76, 509–17. [DOI] [PubMed] [Google Scholar]
- Besser T.E., Cassirer E.F., Highland M.A., Wolff P., Justice‐Allen A., Mansfield K., Davis M. & Foreyt W. (2013) Bighorn sheep pneumonia: sorting out the cause of a polymicrobial disease. Preventative Veterinary Medicine 108, 85–93. [DOI] [PubMed] [Google Scholar]
- Bickhart D., Rosen B., Koren S.et al. (2017) Single‐molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nature Genetics 49, 643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn H.D., Pavia S.R., Wildeus S.et al. (2011a) Genetic structure and diversity among sheep breeds in the United States: identification of the major gene pools. Journal of Animal Science 89, 2336–48. [DOI] [PubMed] [Google Scholar]
- Blackburn H.D. & Pittroff W. (1999) Biologically based coefficients for portioning lamb and wool production costs. Journal of Animal Science 77, 1353–63. [DOI] [PubMed] [Google Scholar]
- Blackburn H.D., Toishibekov Y., Toishibekov M., Welsh C.S., Spiller S.F., Brown M. & Paiva S.R. (2011b) Genetic diversity of Ovis aries populations near domestication centers and in the New World. Genetica 139, 1169–78. [DOI] [PubMed] [Google Scholar]
- Borg R.C., Notter D.R. & Kott R.W. (2009) Phenotypic and genetic associations between lamb growth traits and adult ewe body weights in western range sheep. Journal of Animal Science 87, 3506–14. [DOI] [PubMed] [Google Scholar]
- Borg R.C., Notter D.R., Kuehn L.A. & Kott R.W. (2007) Breeding objectives for Targhee sheep. Journal of Animal Science 85, 2815–29. [DOI] [PubMed] [Google Scholar]
- Bradford G.E., Quirke J.F. & Famula T.R. (1986) Fertility, embryo survival and litter size in lines of Targhee sheep selected for weaning weight or litter size. Journal of Animal Science 62, 895–904. [DOI] [PubMed] [Google Scholar]
- Bruford M., Bradley D. & Luikart G. (2003) DNA markers reveal the complexity of livestock domestication. Nature Review Genetics 4, 900–10. [DOI] [PubMed] [Google Scholar]
- Burfening P.J., Kachman S.D., Hanford K.J. & Rossi D. (1993) Selection for reproductive rate in Rambouillet sheep: estimated genetic change in reproductive rate. Small Ruminant Research 10, 317–30. [Google Scholar]
- Burke J.M. & Miller J.E. (2004) Relative resistance to gastrointestinal nematode parasites in Dorper, Katahdin, and St. Croix lambs under conditions encountered in the southeastern region of the United States. Small Ruminant Research 54, 43–51. [Google Scholar]
- Burton D.J., Ludden P.A., Stobart R.H. & Alexander B.M. (2015) 50 years of the Wyoming ram test: how sheep have changed. Journal of Animal Science 93, 1327–31. [DOI] [PubMed] [Google Scholar]
- Casas E., Freking B.A. & Leymaster K.A. (2004) Evaluation of Dorset, Finnsheep, Romanov, Texel, and Montadale breeds of sheep: II. Reproduction of F1 ewes in fall mating seasons. Journal of Animal Science 82, 1280–9. [DOI] [PubMed] [Google Scholar]
- Casas E., Freking B.A. & Leymaster K.A. (2005) Evaluation of Dorset, Finnsheep, Romanov, Texel, and Montadale breeds of sheep: V. Reproduction of F1 ewes in spring mating seasons. Journal of Animal Science 83, 2743–51. [DOI] [PubMed] [Google Scholar]
- Casellas J. & Piedrafita J. (2015) Accuracy and expected genetic gain under genetic or genomic evaluation in sheep flocks with different amounts of pedigree, genomic and phenotypic data. Livestock Science 182, 58–63. [Google Scholar]
- Cockett N.E., Jackson S.P., Shay T.L., Nielsen D.N., Moore S.S., Steele M.R., Barendse W., Green R.D. & Georges M. (1994) Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers. Proceedings of the National Academy of Sciences of the United States of America 91, 3019–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford A.M., Dodds K.G., Ede A.J., Pierson C.A., Montgomery G.W., Garmonsway H.G., Beattie A.E., Davies K., Maddox J.F. & Kappes S.W. (1995) An autosomal genetic linkage map of the sheep genome. Genetics 140, 703–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H.D., Brauning R., Chamberlain A.J., McWilliams S., McCulloch A., Vander Jagt C.J., Sunduimijid B., Hayes B.J. & Kijas J.W. (2017) 1000 Bull Genomes and SheepGenomeDB projects: enabling cost‐effective sequence level analyses globally. Proceedings Association for Advancement of Animal Breeding and Genetics 22, 201–4. [Google Scholar]
- Dalrymple B.P., Kirkness E.F., Nefedov M.et al. (2007) Using comparative genomics to reorder the human genome sequence into a virtual sheep genome. Genome Biology 8, R152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvit C., Saccà E., Cassandro M., Gervaso M., Pastore E. & Piasentier E. (2008) Genetic diversity and variability in Alpine sheep breeds. Small Ruminant Research 80, 45–51. [Google Scholar]
- Davenport K.M., Hiemke C., McKay S.D., Thorne J.W., Lewis R.M., Taylor T. & Murdoch B.M. (2020) Genetic structure and admixture in sheep from terminal breeds in the United States. Animal Genetics 51, 284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport K.M., McKay S., Fahey A.G., Gill C. & Murdoch B.M. (2018) Meiotic recombination differences in rams from three breeds of sheep in the U.S. Cytogenetic Genome Research 156, 106–16. [DOI] [PubMed] [Google Scholar]
- de Gortari M.J., Freking B.A., Cuthbertson R.P., Kappes S.M., Keele J.W., Stone R.T., Leymaster K.A., Dodds K.G., Crawford A.M. & Beattie C.W. (1998) A second‐generation linkage map of the sheep genome. Mammalian Genome 9, 204–9. [DOI] [PubMed] [Google Scholar]
- de Haan C., Steinfeld H., Blackburn H. (1997) Livestock & the Environment: Finding a Balance. Food and Agriculture Organization, United States Agency for International Development and the World Bank. 2002. https://www.researchgate.net/profile/Cornelis_De_Haan5/publication/44550726_Livestock_the_environment_finding_a_balance_Cees_de_Haan_Henning_Steinfeld_Harvey_Blackburn/links/5786514408aec5c2e4e2e64f/Livestock‐the‐environment‐finding‐a‐balance‐Cees‐de‐Haan‐Henning‐Steinfeld‐Harvey‐Blackburn.pdf [Google Scholar]
- Dickerson G. (1970) Efficiency of animal production – molding the biological components. Journal of Animal Science 30, 849–59. [Google Scholar]
- Dickerson G.E., Glimp H.A., Tuma H.J. & Gregory K.E. (1972) Genetic resources for efficient meat production in sheep. growth and carcass characteristics of ram lambs of seven breeds. Journal of Animal Science 34, 940–51. [Google Scholar]
- Drögemüller M., Tetens J., Dalrymple B., Goldammer T., Wu C.H., Cockett N.E., Leeb T. & Drögemüller C. (2008) A comparative radiation hybrid map of sheep chromosome 10. Cytogenetic Genome Research 121, 35–40. [DOI] [PubMed] [Google Scholar]
- Ercanbrack S.K. & Knight A.D. (1991) Effects of inbreeding on reproduction and wool production of Rambouillet, Targhee, and Columbia ewes. Journal of Animal Science 69, 4734–44. [DOI] [PubMed] [Google Scholar]
- Ercanbrack S.K. & Knight A.D. (1993) Ten‐year linear trends in reproduction and wool production among inbred and non‐inbred lines of Rambouillet, Targhee, and Columbia sheep. J. Anim. Sci. 71, 341–354. 10.2527/1993.712341x [DOI] [PubMed] [Google Scholar]
- Estrada‐Reyes Z.M., Tsukahara Y., Amadeu R.R., Goetsch A.L., Gipson T.A., Sahlu T., Puchala R., Wang Z., Hart S.P. & Mateescu R.G. (2019) Signatures of selection for resistance to Haemonchus contortus in sheep and goats. BMC Genomics 20, 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freking B.A. & Leymaster K.A. (2005) Evaluation of Dort, Finnsheep, Romanov, Texel, and Montadale breeds of sheep: IV. Survival, growth, and carcass traits of F1 lambs. Journal of Animal Science 82, 3144–53. [DOI] [PubMed] [Google Scholar]
- Freking B.A., Murphy S., Wylie A., Jirtle R.L., Rhodes S., Keele J.W., Leymaster K.A. & Smith T.P.L (2002) Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 12, 1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost R.A. & Launchbaugh K.L. (2003) Prescription grazing for rangeland weed management: a new look at an old tool. Rangelands 25, 43–7. [Google Scholar]
- Georges M., Charlier C. & Hayes B. (2019) Harnessing genomic information for livestock improvement. Nature Reviews Genetics 20, 135–56. [DOI] [PubMed] [Google Scholar]
- Goldammer T., Brunner R.M., Rebl A., Wu C.H., Nomura K., Hadfield T., Gill C., Dalrymple B.P., Womack J.E. & Cockett N.E. (2009) A high‐resolution radiation hybrid map of sheep chromosome X and comparison with human and cattle. Cytogenetic Genome Research 125, 40–5. [DOI] [PubMed] [Google Scholar]
- Goldammer T., Brunner R.M., Rebl A., Wu C.H., Nomura K., Hadfield T., Maddox J.F. & Cockett N.E. (2009) Cytogenic anchoring of radiation hybrid and virtual maps of sheep chromosome X and comparison of X chromosomes in sheep, cattle and human. Chromosome Research 17, 497–506. [DOI] [PubMed] [Google Scholar]
- Groeneveld L.F., Lenstra J.A., Eding H.et al. (2010) Genetic diversity in farm animals – a review. Animal Genetics 41, 6–31. [DOI] [PubMed] [Google Scholar]
- Grossman PC, Schneider DA, Knowles DP & Highland MA (2019) Differential pulmonary immunopathologic response of domestic sheep (Ovis aries) and bighorn sheep (Ovis canadensis) to Mycoplasma ovipneumoniae infection: a retrospective study. BioRxiv. 10.1101/703249 [DOI] [PubMed] [Google Scholar]
- Havstad K.M. (1994) Sheep grazing as a range improvement tool. Sheep and Goat Research Journal 10, 72–8. [Google Scholar]
- Heaton M.P., Clawson M.L., Chitko‐Mckown C.G.et al. (2012) Reduced Lentivirus susceptibility in sheep with TMEM154 mutations. PLOS Genetics 8, e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton M.P., Leymster K.A., Kalbfleisch T.S.et al. (2014) SNPs for parentage testing and traceability in globally diverse breeds of sheep. PLOS ONE 9, e94851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson A.W. (2015) A paradigm shift in use and management of United States public lands for livestock grazing. Animal Frontiers 5, 36–42. [Google Scholar]
- Holechek J.L., Geli M.E., Cibils A.F. & Sawalhah M.N. (2020) Climate change, rangelands, and sustainability of ranching in the western United States. Sustainability 12, 4942. [Google Scholar]
- Howell S.B., Burke J.M., Miller J.E., Terrill T.T., Valencia E., Williams M.J., Williamson L.H., Zajac A.M. & Kaplan R.M. (2008) Prevalence of anthelmintic resistance on sheep and goat farms in the southeastern United States. Journal Animal Veterinary Medical Association 233, 1913–9. [DOI] [PubMed] [Google Scholar]
- Hulet C.V., Ercanbrack S.K. & Knight A.D. (1984) Development of the Polypay breed of sheep. Journal of Animal Science 58, 15–24. [DOI] [PubMed] [Google Scholar]
- Hunter N., Goldmann W., Smith G. & Hope J. (1994) The association of a codon 136 PrP gene variant with the occurrence of natural scrapie. Archives of Virology 137, 171–7. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Xie M., Chen W.et al. (2014) The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344, 6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job R.J., Duan M., Hunter S.S., Davenport K.M., Rodriguez A.M., Eidman L. & Murdoch B.M. (2019) Development of Flock54: a targeted genotyping panel for the sheep industry. Plant and Animal Genome XXII Conference. San Diego, CA.
- Kelso M.M. (1947) Current issues in federal land management in the western United States. Journal of Farm Ecology 29, 1295–313. [Google Scholar]
- Kijas J.W., Hadfield T., Sanchez M.N. & Cockett N. (2016) Genome‐wide association reveals the locus responsible for four‐horned ruminant. Animal Genetics 47, 258–62. [DOI] [PubMed] [Google Scholar]
- Kijas J.W., Miller J.E., Hadfield T., McCulloch R., Garcia‐Gamez E., Porto Neto L.R. & Cockett N. (2012) Tracking the emergence of a new breed using 49,034 SNP in sheep. PLOS ONE 7, e41508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijas J.W., Townley D., Dalrymple B.P.et al. (2009) A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLOS ONE 4, e4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Rhie A., Walenz B., Dilthey A.T., Bickhart D.M., Kingan S.B., Hiendleder S., Williams J.L., Smith T.P.L. & Phillippy A.M. (2018) De novo assembly of haplotype‐resolved genomes with trio binning. Nature Biotechnology 36, 1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P.K., Shanthalingam S., Dassanayake R.P.et al. (2010) Transmission of Mannheimia haemoloytica from domestic sheep (Ovis aries) to Bighorn sheep (Ovis canadensis): unequivocal demonstration with green fluorescent protein‐tagged organisms. Journal of Wildlife Diseases 46, 706–17. [DOI] [PubMed] [Google Scholar]
- Lawson‐Handley L., Byrne K., Santucci F., Townsend S., Taylor M., Bruford M.W. & Hewitt G.M. (2007) Genetic structure of European sheep breeds. Heredity 99, 620–31. [DOI] [PubMed] [Google Scholar]
- Lewis R.M., Notter D.R., Hogue D.E. & Magee B.H. (1996) Ewe fertility in the STAR accelerated lambing system. Journal of Animal Science 74, 1511–22. [DOI] [PubMed] [Google Scholar]
- Leymaster K.A. (1991) Straightbred comparison of a composite population and the Suffolk breed for performance traits of sheep. Journal of Animal Science 69, 993–9. [DOI] [PubMed] [Google Scholar]
- Livestock Marketing Information Center (2016) Sheep cost of production study. http://lmic.info/page/cost‐sheep‐production‐budget‐sponsored‐american‐sheep‐industry
- Liu Y., Murali S.C., Harris R.A.et al. (2016) Sheep reference genome sequence updates: texel improvements and Rambouillet progress. Journal of Animal Science 94, 18–9. [Google Scholar]
- Lupton C.J. (2008) ASAS Centennial Paper: impacts of animal science research on United States sheep production and predictions for the future. Journal of Animal Science 86, 3252–74. [DOI] [PubMed] [Google Scholar]
- Maddox J.F., Davies K., Crawford A.M.et al. (2001) An enhanced linkage map of the sheep genome comprising more than 1000 loci. Genome Research 11, 1275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C.M., Faria D.A., Lucci C.M., Louvandini H., Pereira A. & Paiva S.R. (2020) Heat stress effects on sheep: are hair sheep more heat resistant? Theriogenology 155, 157–67. [DOI] [PubMed] [Google Scholar]
- Morgan J.A., Milchunas D.G., LeCain D.R., West M. & Mosier A.R. (2007) Carbon dioxide enrichment alters plant community structure and accelerates shrub growth in the shortgrass steppe. Proceedings of the National Academy of Sciences of the United States of America 104, 14724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moum T., Olsaker I., Hopp P., Moldal T., Valheim M., Moum T. & Benestad S.L. (2005) Polymorphisms at codons 141 and 154 in the ovine prion protein gene are associated with scrapie Nor98 cases. Journal of General Virology 86, 231–5. [DOI] [PubMed] [Google Scholar]
- Mousel M.R., Reynolds J.O. & White S.N. (2015) Genome‐wide association identifies SLC2A9 and NLN gene regions as associated with entropion in domestic sheep. PLOS ONE 10, e0128909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousel M.R., White S.N., Herndon M.K., Herndon D.R., Taylor J.B., Becker G.M. & Murdoch B.M. (2021) Genomic regions associated with Mycoplasma ovipneumoniae presence in nasal secretions of domestic sheep. bioRxiv 2021.02.04.429710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.W., Berger Y.M., Holman P.W., Baldin M., Burgett R.L. & Thomas D.L. (2017) Estimates of genetic parameters, genetic trends, and inbreeding in a crossbred dairy sheep research flock in the United States. Journal of Animal Science 95, 4300–9. [DOI] [PubMed] [Google Scholar]
- Murphy T.W., Freking B.A., Bennett G.L. & Keele J.W. (2020) Development, selection criteria, and performance of composite IV sheep at the U.S. Meat Animal Research Center. Journal of Animal Science 98, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (2008) Changes in the Sheep Industry in the United States: Making the Transition from Tradition. Washington, DC: National Academy Press. [Google Scholar]
- Notter D.R. (1998) The U.S. National Sheep Improvement Program: across‐flock genetic evaluations and new trait development. Journal of Animal Science 76, 2324–30. [DOI] [PubMed] [Google Scholar]
- Oltenacu E.A.B., & Boylan W.J. (1981) Productivity of purebred and crossbred Finnsheep. I. reproductive traits of ewes and lamb survival. Journal of Animal Science 52, 989–97. [DOI] [PubMed] [Google Scholar]
- Onderka D.K. & Wishart W.D. (1988) Experimental contact transmission of Pasteurella haemolytica from clinically normal domestic sheep causing pneumonia in Rocky Mountain Bighorn sheep. Journal of Wildlife Diseases 24, 663–7. [DOI] [PubMed] [Google Scholar]
- Paim T.P., Faria D.A., Hay E.H.et al. (2019) New world goat populations are a genetically diverse reservoir for future use. Scientific Reports 9, 1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paim T.P., Hay E.H.A., Wilson C., Thomas M.G., Kuehn L.A., Paiva S.R., McManus C. & Blackburn H.D. (2020) Dynamics of genomic architecture during composite breed development in cattle. Animal Genetics 51, 224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S.R., Mariante A.S. & Blackburn H.D. (2011) Combining U.S. and Brazilian microsatellite data for a meta‐analysis of sheep (Ovis aries) breed diversity: facilitating the FAO Global Plan of Action for conserving animal genetic resources. Journal of Heredity 102, 697–704. [DOI] [PubMed] [Google Scholar]
- Peter C., Bruford M., Perez T., Dalamitra S., Hewitt G. & Erhardt G. (2007) Genetic diversity and subdivision of 57 European and Middle‐Eastern sheep breeds. Animal Genetics 38, 37–44. [DOI] [PubMed] [Google Scholar]
- Posbergh C.J. & Huson H.J. (2021) All sheeps and sizes: a genetic investigation of mature body size across sheep breeds reveals a polygenic nature. Animal Genetics 52, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasali D.P., Shrestha J.N.B. & Crow G.H. (2006) Development of composite sheep breeds in the world: a review. Canadian Journal of Animal Science 86, 1–24. [Google Scholar]
- Ratnakumar A., Kirkness E.F. & Dalrymple B.P. (2010) Quality control of the sheep bacterial artificial chromosome library, CHORI‐243. BMC Research Notes 3, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexroad C., Vallet J., Matukumalli L.K.et al. (2019) Genome to phenome: improving animal health, production, and well‐being – a new USDA blueprint for animal genome research 2018–2027. Frontiers in Genetics 10, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati M., Caulton A., Clark R.et al. (2020) Global analysis of transcription start sites in the new ovine reference genome (oar rambouillet v1.0). Frontiers Genetics 11, 580580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton M. (1979) Estimation of genetic change in a performance testing program for sheep. Journal of Animal Science 48, 26–31. [Google Scholar]
- Snowder G.D., Walker J.W., Launchbaugh K.L. & Dale Van Vleck L. (2001) Genetic and phenotypic parameters for dietary selection of mountain big sagebrush in Rambouillet sheep. Journal of Animal Science 79, 486–92. [DOI] [PubMed] [Google Scholar]
- Sulaiman Y., Wu C.X. & Zao C.J. (2011) Phylogeny of 19 indigenous sheep populations in Northwestern China inferred from mitochondrial DNA control region. Asian Journal of Animal and Veterinary Advances 6, 71–9. [Google Scholar]
- Tapio M., Ozerov M., Tapio I., Toro M.A., Marzanov N., Cinkulov M., Goncharenko G., Kiselyova T., Murawski M. & Kantenen J. (2010) Microsatellite‐based genetic diversity and population structure of domestic sheep in northern Eurasia. BMC Genetics 11, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrill C.E. (1947) Breed Crosses used in the development of Targhee sheep. Journal of Animal Science 6, 83–92. [DOI] [PubMed] [Google Scholar]
- Tetens J., Goldammer T., Maddox J.F., Cockett N.E., Leeb T. & Drögemüller C. (2007) A radiation hybrid map of sheep chromosome 23 based on ovine BAC‐end sequences. Animal Genetics 38, 132–40. [DOI] [PubMed] [Google Scholar]
- Thomas D.L., Berger Y.M., McKusick B.C. & Mikolayunas C.M. (2014) Dairy sheep production research at the University of Wisconsin‐Madison, USA – a review. Journal of Animal Science and Biotechnology 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA (1903) Nineteenth Annual Report of the Bureau of Animal Husbandry: Sheep Ranching in the Western States. Washington D.C.: Government Printing Office. [Google Scholar]
- USDA, Animal Plant Health Inspection Service (2014) Sheep 2011. https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/monitoring‐and‐surveillance/nahms/nahms_sheep_studies [DOI] [PubMed]
- USDA, Economic Research Service (2004) Trends in the U.S. sheep industry. https://downloads.usda.library.cornell.edu/usda‐esmis/files/kw52j804p/08612r694/gm80hz92n/aib787.pdf
- USDA, National Agriculture Statistics Service (2020a) Sheep and goats. https://downloads.usda.library.cornell.edu/usda‐esmis/files/000000018/n296xf83n/m900pb410/shep0120.pdf
- USDA, National Agriculture Statistics Service (2020b) Livestock slaughter. https://downloads.usda.library.cornell.edu/usda‐esmis/files/rx913p88g/0z709q707/j6731z16z/lstk0221.pdf
- van der Werf JHJ, Kinghorn B.P. & Banks R.G. (2010) Design and role of an information nucleus in sheep breeding programs. Animal Production Science 50, 998–1003. [Google Scholar]
- Vanimisetti H.B., Notter D.R. & Kuehn L.A. (2007) Genetic (co)variance components for ewe productivity traits in Katahdin sheep. Journal of Animal Science 85, 60–8. [DOI] [PubMed] [Google Scholar]
- Waldron D.F., Thompson W.J. & Hogan R.J. (2016) Factors affecting price differences between wool and hair lambs n San Angelo, TX. Sheep & Goat Research Journal 31, 9–16. [Google Scholar]
- Westaway D., Zuliai V., Cooper C.M., Da Costa M., Neuman S., Jenny A.L., Detwiler L. & Prusiner S.B. (1994) Homozygosity for prion protein alleles encoding glutamine‐171 renders sheep susceptible to natural scrapie. Genes & Development 8, 959–69. [DOI] [PubMed] [Google Scholar]
- Wildeus S. (1997) Hair sheep genetic resources and their contribution to diversified small ruminant production in the United States. Journal of Animal Science 75, 630–40. [DOI] [PubMed] [Google Scholar]
- Wilson D.E. & Morrical D.G. (1991) The National Sheep Improvement Program: a review. Journal of Animal Science 69, 3872–81. [DOI] [PubMed] [Google Scholar]
- Woolaston R.R. (1992) Selection of Merino sheep for increased and decreased resistance to Haemonchus contortus: peri‐parturient effects on faecal egg counts. International Journal of Parasitology 22, 947–53. [DOI] [PubMed] [Google Scholar]
- Worley K.C. (2018) Rambouillet Sheep Genome and Annotation Resources. Abstract W152 Plant and Animal Genome Conference XXVI. San Diego, CA.
- Wu C.H., Nomura K., Goldammer T., Hadfield T., Dalrymple B.P., McWilliam S., Maddox J.F., Womack J.E. & Cockett N.E. (2008) A high‐resolution comparative radiation hybrid map of ovine chromosomal regions that are homologous to human chromosome 6 (HSA6). Animal Genetics 39, 459–67. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Nomura K., Goldammer T., Hadfield T., Womack J.E. & Cockett N.E. (2007) An ovine whole‐genome radiation hybrid panel used to construct an RH map of ovine chromosome 9. Animal Genetics 38, 534–6. [DOI] [PubMed] [Google Scholar]
- Wuliji T., Wuri L., Glimp H. & Filbin T. (2019) Merino breeding program improves wool quality in US wool sheep flocks. In: Advances in Fibre Production Science in South American Camelids and other Fibre Animals (Ed. Gerken M., Renieri C., Allain D., Galbraith H., Gutierrez J. P., Mckenna L., Niznikowski R., Wurzinger M.), pp. 135–47. University of Gottingen, Germany. 10.17875/gup2019-1158 [DOI] [Google Scholar]
- Zhang L., Mousel M.R., Wu X., Michal J.J., Zhou X., Ding B., Dodson M.V., El‐Halawany N.K., Lewis G.S. & Jiang Z. (2013) Genome‐wide genetic diversity and differentially selected regions among Suffolk, Rambouillet, Columbia, Polypay, and Targhee sheep. PLOS ONE 8, e65942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Number of sheep registered for five popular breeds from 2000 to 2017.


