Abstract
QM/MM simulations have become an indispensable tool in many chemical and biochemical investigations. Considering the tremendous degree of success, including the recognition by a 2013 Nobel Prize in Chemistry, are there still “burning challenges” in QM/MM methods, especially for biomolecular systems? In this short Perspective, we discuss several issues that we believe greatly impact the robustness and quantitative applicability of QM/MM simulations to many, if not all, biomolecules. We highlight these issues with observations and relevant advances from recent studies in our group and others in the field. Despite such limited scope, we hope the discussions are of general interest and will stimulate additional developments that help push the field forward in meaningful directions.
Graphical Abstract
1. Introduction
Following many years of development and calibration, hybrid quantum mechanical/molecular mechanical (QM/MM) methods1–4 have become an essential tool5–8 in chemical and biochemical research. Applications of QM/MM simulations to enzymes include not only dissection of reaction mechanisms, elucidation of various factors that contribute to catalysis, but also design of enzyme inhibitors, analysis and improvement of designed enzymes as well as understanding evolutionary relations between enzymes.9–16 Indeed, a search using the keyword “QM/MM” or “ONIOM”17 in the Web of Science points to >800 articles published in 2019–2020 alone. Numerous molecular simulation packages such as CHARMM, AMBER, NAMD, OpenMM and Gromacs all have QM/MM functionalities, some with build-in QM modules, while others providing interfaces with popular QM packages such as Gaussian, Q-Chem, Turbomole, Psi4 and ORCA; similarly, many quantum chemistry packages such as Gaussian, ORCA, C2PK and DFTB+ offer coupling with external MM models; there are also platforms that focus entirely on QM/MM calculations, such as the celebrated Chemshell environment18 pioneered by Sherwood and the late Walter Thiel,19 who has made numerous important contributions to the field in terms of both method developments and systematic analyses of factors that impact the accuracy of QM/MM simulations.
In light of such tremendous success, including the recognition by the 2013 Nobel Prize in Chemistry, one is tempted to ask: are there still “burning issues” for QM/MM methods, especially for biomolecular systems? If so, are the corresponding challenges fundamental in nature, or should they be better thought as establishing the best practice and enabling most efficient execution for realistic applications? The answers to such questions are clearly highly subjective, and in this short Perspective article, we discuss our views based on the recent research interest and developments in the group. Despite such limited scope, we hope the discussions are of general interest and will stimulate additional developments that help push the field forward in meaningful directions.
Before turning to these discussions, it is worth noting that application of QM/MM methods to solid/liquid interfaces,20,21 for example in the important context of electrochemistry22 and heterogeneous catalysis,23 has been much more limited compared to biomolecular studies. This is perhaps due mainly to the less obvious scheme to divide the solid into QM and MM regions without considerable perturbation to the electronic structure of the solid, especially for metallic systems (e.g., the electrode). Therefore, efforts have focused primarily on pure QM based embedding approaches, such as those pioneered by Carter and co-workers based on orbital-free DFT,24 which have been successfully applied to many materials problems. Nevertheless, these pure QM based embedding approaches remain computationally expensive for solid/liquid interfaces, for which an adequate sampling of the interfacial solvent and ions (i.e., the electric double layer25) is expected to be essential. Therefore, further development, implementation and calibration of robust QM/MM methods for solid/liquid interfaces, especially under the condition of constant potential,26,27 ought to be considered an important area of research. Similar to the studies of reactions in solution, QM/MM simulations with explicit solvent28 and advanced implicit solvent models29,30 are expected to be complementary to each other for the mechanistic analysis of electrochemical reactions.
2. Several “Burning Issues”
In the following, we discuss several “burning issues” that we believe greatly impact the robustness and quantitative applicability of QM/MM simulations to many, if not all, biomolecules. We group these issues into several general topics related to QM/MM simulations, and we highlight them with observations and relevant advances from recent studies in our group and others in the field.
2.1. Potential Function: QM and MM Selections
The choices for QM/MM partitioning and the relevant QM and MM methods are largely system-dependent. Nevertheless, given a considerable number of recent studies and sometimes passionate debates31–40 regarding these choices, it is worth discussing several issues explicitly.
2.1.1. The value of semi-empirical and machine learning potentials
Since adequate sampling is important to biomolecular applications, the need of an efficient QM potential is compelling, especially when the proper QM region contains at least hundreds of atoms. Along this line, approximate QM approaches such as semi-empirical QM methods41,42 and the empirical valence bond (EVB) method43 remain particularly attractive in many applications. They need to be carefully calibrated to generate meaningful results; once calibrated (e.g., using spline fits to high-level reference reactions44), they are uniquely powerful for the analysis of systems and/or experimental observables that demand extensive sampling. For example, as discussed further below, DFTB3/MM simulations enable the sampling of multi-dimensional free energy surfaces, which are required to analyze complex reaction pathways and to characterize the coupling between different processes (e.g., mechanochemical coupling). On the other hand, EVB based simulations have been instrumental to the computation of experimental observables that require extensive sampling to achieve the necessary numerical precision, such as activation entropy45 and linear free energy relations,46 which are difficult to compute otherwise. These examples include >150 ns of DFTB3/MM simulations47 and thousands of independent EVB free energy profile simulations,48 which are indeed very difficult, if not currently impossible, to reach with ab initio QM/MM simulations, despite impressive progress in computational algorithm as well as hardware.49,50
Motivated by these considerations, a “burning issue” is to improve the accuracy and applicability of semi-empirical QM methods; work along this line has been reviewed fairly recently51 and will not be repeated here. A notable advance since then is the development of the extended tight binding model of Grimme et al. (xTB),52 which has been parameterized for the entire periodic table. While the model was developed mainly for capturing structures and vibrational frequencies, it gives encouraging results also for energetics, especially for non-covalent interactions.53 In Fig.1a, we compare the performance of several popular semi-empirical methods for computing the second virial coefficients of small molecules of different levels of polarity; in Fig.1b, we compare the two-body and three-body interactions of water molecules by DFTB3 and xTB using the dataset of Paesani and co-workers.54 Evidently, despite the minimal basis nature of these methods, the performance is quite respectable even for three-body interactions of water, and these tight binding methods outperforms several NDDO methods, even those including empirical corrections for hydrogen-bonding interactions (see Supporting Information). On the other hand, there is clearly room for improvements, which would also require analysis in the condensed phase environment,55 such as the computation of solvation free energy and binding affinity.
Figure 1:
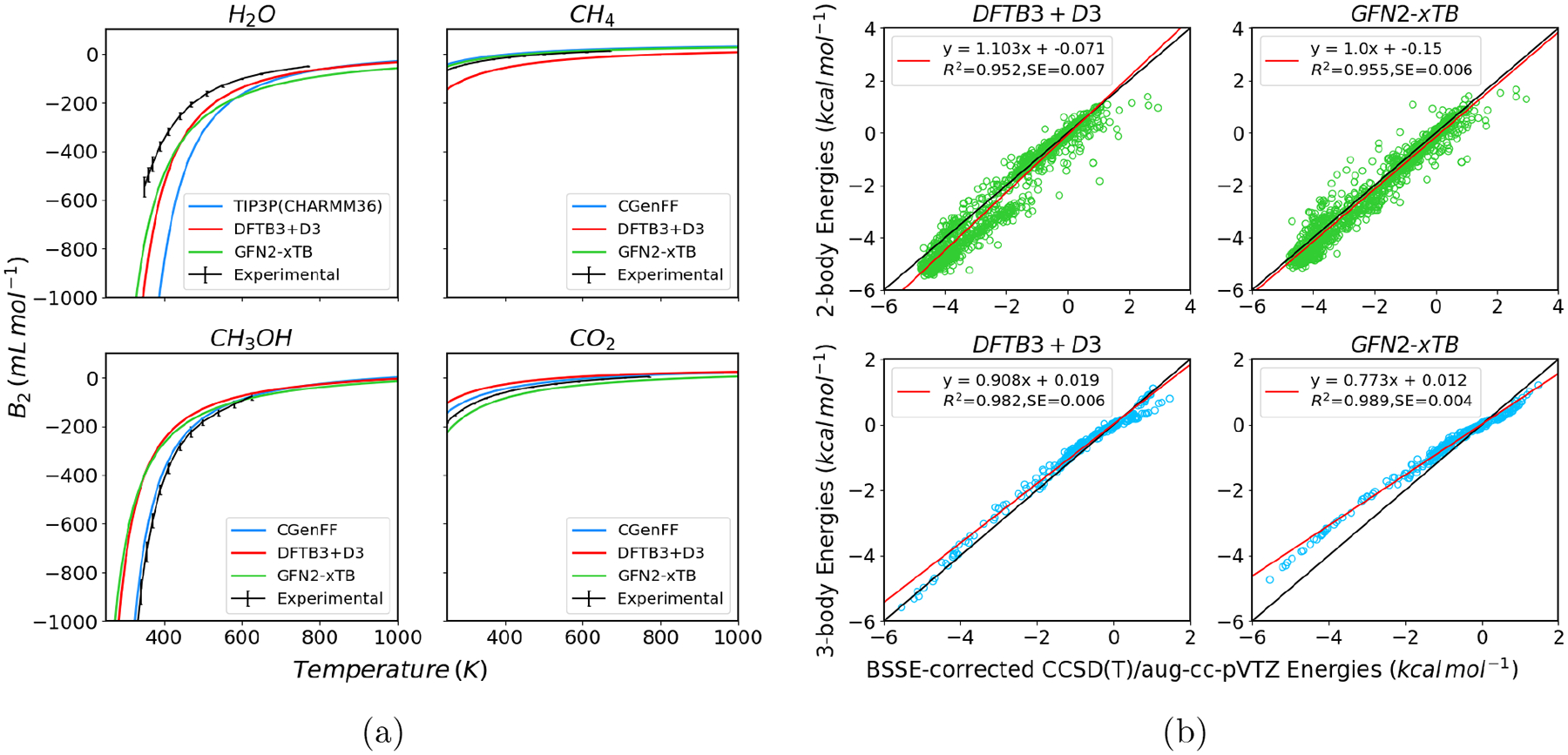
Non-covalent interaction with two density functional tight binding methods. (a) Second virial coefficients (B2) for several small molecules, in comparison with CHARMM-CGenFF56 and experimental results; (b) 2,3-body interactions of water in comparison with the BSSE corrected CCSD(T)/aug-cc-pVTZ database of Paesani and co-workers.54 For additional results, see Supporting Information.
A major remaining challenge for the semi-empirical QM methods (and ab initio QM methods) is the treatment of transition metal ions, which are common in enzymes. The highly localized nature of the d and f electrons requires a reliable treatment of electron correlation, both dynamic and static. For structural properties, both DFTB3 and xTB appear to be able to provide adequate equilibrium parameters,51,53 although the energy landscape has not been thoroughly analyzed. Analysis of the electronic structure using Natural Bonding Orbital analysis57,58 found that DFTB3 provides physically sound descriptions for different bonding scenarios, including those exhibiting pseudo Jahn-Teller effects (see Fig.2a as an example). The degree of ligand to metal charge transfer and ionic character of certain bonds are overestimated, likely reflecting the minimal basis nature of DFTB3, and certain orbital interactions, such as geminal interactions, were observed to be grossly overestimated by DFTB3 for several transition metal compounds. Energetics are generally less satisfying, especially when considering different spin states, since interactions among d/f electrons are treated in an averaged fashion in the current tight-binding models. One possible improvement is to treat the d/f electrons separately, along the line of the DFT+U model in materials science59 or the ligand-field model in the inorganic chemistry literature.60,61 In a recent explorative study,62 together with collaborators we have implemented a preliminary version of the DFTB3+U model,
| (1) |
in which PU(PI) are the spin unpolarized (polarized) density matrix elements involving the d/f electrons and the usual two-electron integrals are parameterized in terms of the Slater integrals or the Racah parameters.60 It was found62 that the additional U contribution indeed improved the splitting between the low-spin and high-spin states in a series of Ni(II) and Ni(III) compounds as well as the populations of the 3d orbitals.63 Further self-consistent tuning of the electronic parameters in the model will be informative regarding the degree of transferability and expected accuracy for the +U model, and whether a multi-determinant formulation of the DFTB3 model (e.g., in the framework of ensemble64 or multi-state65 DFT) is warranted.
Figure 2:
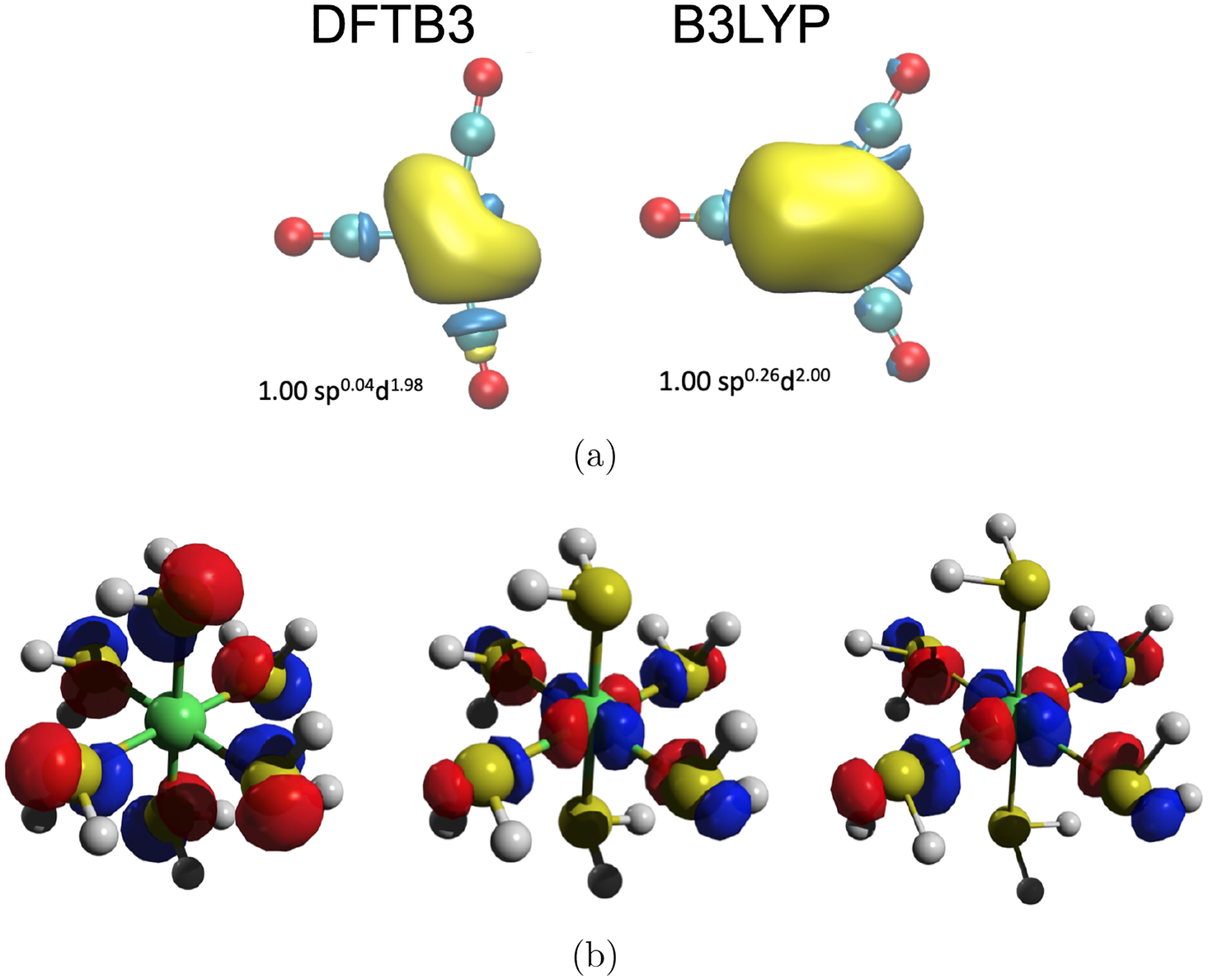
Description of Ni compounds with DFTB3. (a) Comparison of the nickel lone valence hybrid in [Ni(CO)3]2+ with DFTB3/3OB and B3LYP based Natural Bonding Orbital analyses;57,58 DFTB3 and B3LYP favor D3h and C2v symmetry, respectively. (b) Examples of frontier orbital comparisons between DFTB3, DFTB3+U and PBE calculations for high-spin [Ni(H2S)6]2+. Including the +U correction in DFTB3/3OB62 improves various properties such as d orbital populations, nature of frontier orbitals, ligand field splitting and energy difference between low/high-spin states.
Another promising direction for improving approximate (tight-binding and DFT) methods is to employ machine learning (ML) techniques,66 which have seen explosive progress in recent years for developing potential energy functions,67 predicting molecular/materials properties68 as well as enhancing conformational sampling.69 Specific for metal ions, for example, ML approaches have been used to analyze spin state properties70 as well as diagnostics for static electron correlations.71 ML models have also been used to improve the accuracy of mean-field models such as tight-binding and Hartree-Fock, with features ranging from simply atomic coordinates to Fock matrix elements.72,73 For realistic condensed phase QM/MM applications, the judicious combination of a physical reference QM level and ML is required to ensure the optimal balance of accuracy, robustness and transferability. Further developments along this line, so as to enable stable, energy-conserving MD simulations,74,75 will be exciting and transformative in terms of the type of problems that QM/MM simulations are able to tackle.
2.1.2. Going beyond popular additive MM models
Most QM/MM applications to biomolecules use popular fixed-charge MM force fields. The general success of these applications suggests that this combination is adequate for many problems, an observation also made in several comparisons of QM/MM results using both fixed-charge and polarizable force fields.76 Nevertheless, the need to include explicit electronic polarization for certain problems has also been well documented, such as for the prediction of absorption spectra,77,78 reduction potential79 and pKa values;80 i.e., for situations where there is a very large change in the electrostatic properties (e.g., either the net charge or dipole moment) of the QM region. While more ad hoc models such as charge scaling80 or atomic polarizability models81 were used in earlier studies, more systematic efforts that integrate QM with well calibrated polarizable force fields such as AMOEBA, Effective Fragment Potential and MB-pol have been reported.79,82–84
A systematic comparison between QM/MM simulations with different MM models will be informative for better identifying problems for which an explicit treatment of electronic polarization makes a qualitative difference; for example, it was suggested that to properly capture the electric field in enzyme active site for the purpose of guiding design, including electronic polarization is essential.85 Along this line, we highlight that a meaningful comparison will require extensive sampling. In part, this is because electrostatic interactions between groups in a biomolecule are screened quite effectively;86,87 this is especially the case for many enzyme active sites, which are buried but not far from the protein/solvent interface. To properly capture the screening effect, it is important to consider reorientation of the protein and solvent dipoles during a chemical reaction (this contrasts to the situation of ultrafast spectroscopy,78,83 for which electronic dielectric response of the environment tends to be particularly important due to the limited degree of dipolar reorientation during the short time scale); without adequate sampling of such dipolar reorientations, the effect of distant groups can be overestimated. A somewhat extreme illustration is that contributions from distant charged residues to the pKa of a titratable group are significantly overestimated (>10 kcal/mol) when analyzed in a perturbative fashion,88,89 i.e., by computing the change of QM/MM interaction energies without re-sampling the MD trajectory after the MM partial charges on a charged residue are turned off. For a similar reason, without adequate sampling, the effect of altering the description (MM vs. QM) of distal residues can be exaggerated, thus the issue of sampling is also highly pertinent to the discussion of QM region size in QM/MM simulations.31–39
The sampling challenge is particularly significant for the prediction of properties that involve a net change of total charge, such as reduction potential and pKa values, since the corresponding “charging” process is likely coupled with non-trivial changes in the protein structure and local hydration levels.90,91 Along this line, recent studies suggested that the lack of explicit electronic polarization leads to errors more than 1 eV for the reduction potential of flavins in proteins;79 this important observation is somewhat surprising as fixed-charge force field models have been quite successful at capturing relative reduction potential and pKa values based on alchemical free energy simulations,92–94 provided that adequate sampling is done; one possible explanation is that the average polarization effect is empirically included in the fixed-charge force fields. Therefore, thorough comparison of different QM/MM models for both absolute and relative free energies with sufficient sampling is of great value. To enable such comparisons, advanced sampling techniques, including grand canonical Monte Carlo approaches for efficiently sampling local hydration level, are expected to be essential; subtle technical issues such as the effect of net charge change in Ewald summation, which are well understood,95 should be considered when comparing results from different set ups, as many ab initio QM/MM simulations, especially those employ advanced MM models, still do not use periodic boundary condition while many semi-empirical QM/MM simulations do.96,97
Another deviation from standard QM/MM setups is to employ coarse-grained (CG) models for at least part of the MM environment,98–100 which was motivated by applications that involve slow processes such as large-scale structural transitions of the biomolecule101 or protein design. Considering that QM/MM interaction terms are crucial to the accuracy of QM/MM simulations in general, it is challenging to construct reliable QM/CG models that are transferable among distinct conformational states, although solid progress have been reported.98
2.2. Local Sampling: Multi-dimensional and Multi-level free energy simulations
For effective sampling, depending on the nature of the bottleneck, different strategies are required. Thus we separately discuss several “burning issues” related to local sampling and remote (allosteric) effects in this and subsequent subsections, respectively. We focus on issues most relevant to biomolecular QM/MM applications, since excellent reviews are available on the general topic of enhanced sampling for biomolecules.102,103
2.2.1. Multi-dimensional free energy simulations: competitive pathways and causal relations among different processes
One could argue that to capture the key features of the chemical step(s) in most enzymes, standard QM/MM free energy simulations (e.g., umbrella sampling and metadynamics) using one or two collective variables (CVs) are likely adequate. For more complex transformations that explicitly involve multiple catalytic groups, such as ATP hydrolysis in molecular motors, finite temperature string methods104 that parameterize the minimum free energy path with a large number of CVs can be very effective.105,106 A direction of major interest is to develop the finite temperature analog of automated and exhaustive reaction path searches107,108 on potential energy surfaces such that catalytic mechanism analysis for multi-step reactions in enzymes can be done in an automated fashion without prior bias by human intuition.
On the other hand, some problems require alternative approaches. For example, one hall-mark that distinguishes biomolecules from artificial catalysts is that the chemical activities in the former are often tightly coupled to other processes. As a result, chemical reactions can be used in biology to drive other events such as conformational transitions or pumping ions across the cell membrane, forming the basis of energy and signal transductions in cells.109 As the other side of the coin, the level of chemical activity in biomolecules can be significantly perturbed by processes that occur either proximal or distal to the active site, giving rise to a multitude of regulatory mechanisms of enzyme catalysis. Therefore, understanding the physical principles that govern the coupling between the chemical step and other events, such as penetration of water molecules into the active site,110–112 recruitment of transient metal ions,113,114 or conformational rearrangements near and afar,115–119 is of great fundamental and practical importance.
For such problems, the most revealing approach that can clearly elucidate the causality among different processes is to conduct multi-dimensional free energy simulations. This is computationally demanding and therefore particularly requires a balance between computational accuracy and efficiency; semi-empirical QM/MM methods, perhaps augmented with ML corrections, are uniquely appropriate in this context. Even with inexpensive QM/MM methods, it is essential to ensure optimal efficiency in building up the free energy surface and allocating computational resources for the sampling of the relevant CV space; the selection of the appropriate set of CVs is not unique to QM/MM simulations and has been discussed extensively in the literature.69,103
Along this line, ML approaches can again offer simple and effective solutions. For example, Zhang et al.121 developed a reinforcement learning approach in which a neural network ensemble is used to learn the multi-dimensional free energy surface. The approach takes advantage of the numerical flexibility of neural networks to parameterize high-dimensional functions; in addition, the use of an ensemble of neural networks makes it straightforward to identify regions of the CV space that have been undersampled, so that computational resources are automatically re-allocated accordingly. The approach can be straightforwardly applied to QM/MM simulations (Fig. 3) to analyze competition of multiple reaction pathways as well as coupling between the chemical step and other local processes, such as metal ion trafficking. In the current form, the approach does not actively enhance the sampling of degrees of freedom not included in the CV space, which again is not a unique challenge to QM/MM simulations.
Figure 3:
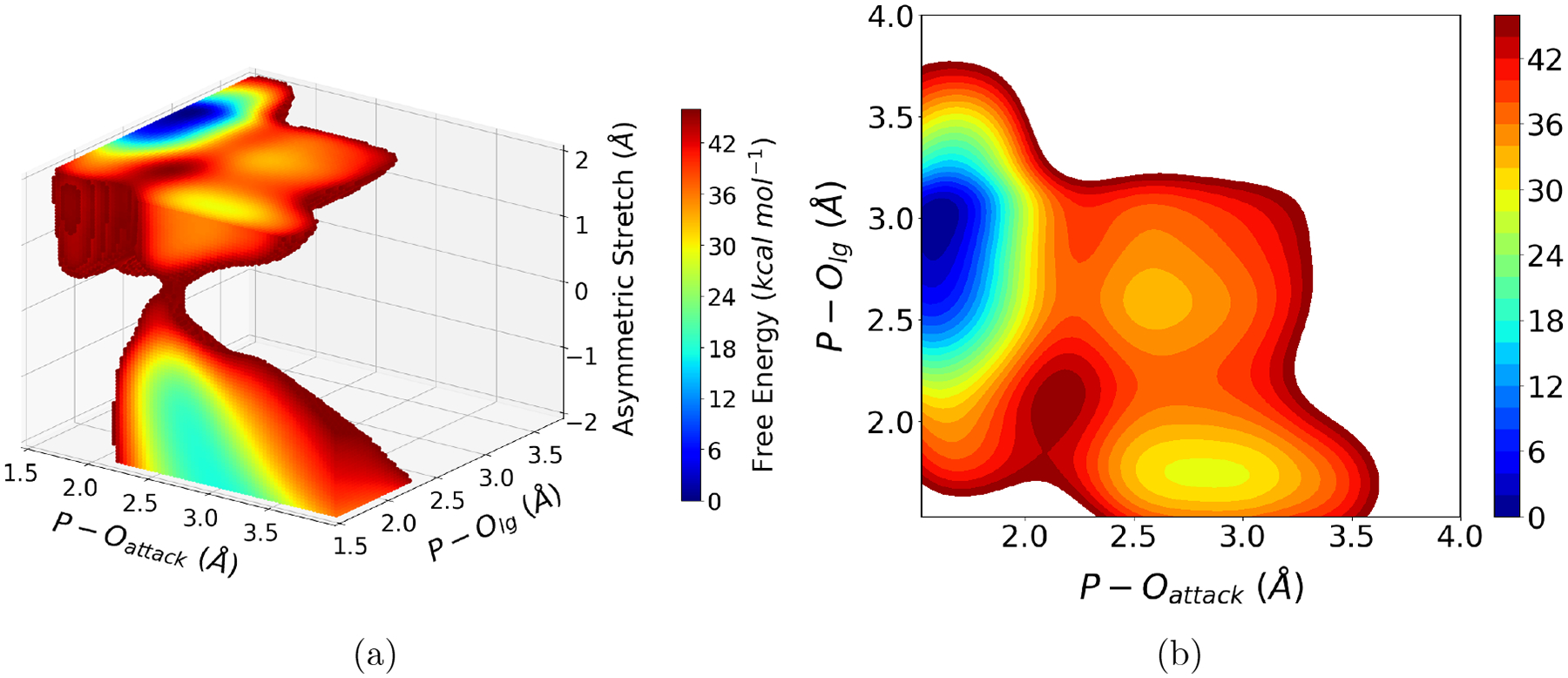
Results from an automated reinforcement learning driven free energy simulation of methyl phosphate hydrolysis in solution. The three coordinates are the nucleophilic attack P-O distance, the leaving group P-O distance and the antisymmetric O-H-O stretch that describes the proton transfer from the nucleophile (water) to the phosphate oxygen. (a) The three-dimensional PMF converges after 42 iterations of automated restrained MD-reinforcement learning cycles; the results indicate that with the current DFTB3/MM model, the solvent-assisted pathway120 is not the dominant mechanism. (b) The two-dimensional PMF cut after the proton transfer is complete indicates a dissociative pathway that involves a loosely bound metaphosphate species. Further refinement of the QM/MM energetics will provide insights into this prototypical phosphoryl transfer reaction at an unprecedented level of detail.
2.2.2. Multi-level free energy simulations: bridging the distributions at different levels of theory
In quantum chemistry, it is routine to combine different levels of theory to focus on structure and energetics, respectively.122 It is therefore natural to pursue “multi-level” free energy simulations123,124 in which an inexpensive QM/MM potential is used to explore the conformational space and a more accurate QM/MM potential is used to improve the energetics. The fundamental challenge for condensed phase simulations at a finite temperature is that the configurations with high Boltzmann weights may differ significantly at different levels of theory, thus merely re-weighting of samples collected from a low-level simulation with high-level energies125 is unlikely to lead to proper convergence,126 especially when the QM region is large (for example, 100–250 atoms, as in many semi-empirical QM/MM simulations127).
As discussed extensively in the literature, the success of “multi-level” free energy simulations depends critically on the overlap of the configurational distributions; specifically in the framework of free energy perturbation upon changing potential energy, one useful metric is the overlap in the energy gap (i.e., ΔULH(X) = UH(X) − UL(X)) distribution at the two levels (L, H) of theory (ρL(ΔULH) vs. ρH(ΔULH)), as reflected by the well-established identity,128
| (2) |
in which ΔALH is the free energy difference between the two levels of theory; more quantitative criteria for the degree of overlap have been proposed in the literature,129–131 many of which are based on the work of Kofke and co-workers for free energy perturbation in general.132 Therefore, the key challenge is to establish protocols that ensure adequate overlap during the L to H transformation while maintaining computational efficiency, i.e., to minimize the amount of computations at the H level.
One natural approach is to improve the L level towards the H level, by either system-specific reparameterization of L,133 learning the difference between L and H on the fly via ML,72 or switching from L to H via many short, non-equilibrium simulations134 in which L and H potentials are mixed explicitly. Alternatively, one can systematically identify the degrees of freedom that lead to poor energy gap overlaps at L/H levels, and treat these “problematic” degrees of freedom separately. Several studies of systems that included relatively small QM regions suggested that the bonded, stiff degrees of freedom tend to be problematic,131,135–137 as even small structural differences between L and H can have significant energetic consequences and thus leading to a poor overlap in energy gap distributions. Different approaches have been developed accordingly, which involve either refitting these degrees of freedom via, for example, force-matching,131,138 or ignoring them so as to focus on interaction energies between the QM and MM degrees of freedom.139,140
In the “staged transformation” approach explored by us recently,141 these problematic degrees of freedom are subject to constraints or restraints, such that the conversion from L to H models is done through a thermodynamic route that ensures favorable distribution overlaps along the way. The free energy components associated with different steps are mostly evaluated explicitly, thus the final result can be compared to the rigorous free energy difference between the two levels of theory with limited and well-defined approximations. Importantly, the additional free energy component calculations involve simulations at the low level of theory and therefore do not incur high computational costs. The approach has been illustrated with solution systems with encouraging results, although further tests and improvements are required for treating more complex systems.
For systems that involve larger QM regions, however, it is not clear at all that only bonded degrees of freedom are problematic, as small errors associated with non-bonded degree of freedom can accumulate quickly, leading to poor energy gap overlaps. Therefore, additional developments are sorely needed to automatically identify the “problematic” degrees of freedom and mitigate their impact on the computed free energies. Along this line, we note that broadening (as opposed to narrowing, as commonly done in previous work) the distribution at the L level and/or introducing intermediate distributions that bridge the L/H distributions with generalized ensemble based sampling102,142 or generative ML methods143 are interesting alternative directions to pursue.
2.3. Remote Effects: Allostery, Enzyme Evolution and Design
As alluded to above, one fascinating aspect about biomolecules is the existence of coupling, or co-operativity, among distant sites. Although the mechanism of allostery in biomolecules has been studied for decades,144–149 it remains difficult to precisely predict residues that dictate the long-range co-operativity; this incomplete level of understanding limits our ability to engineer allostery into biomolecules, although sporadic successful examples have been reported in the literature.149–151 Specifically in the context of enzyme catalysis, it has been well documented that remote mutations may have a significant impact on catalysis,118 leading to hypotheses and debates about the roles of enzyme motions in catalysis. In directed evolution studies of designed enzymes, it is often observed that improvement of catalytic proficiency or expansion of substrate scope involves residues not in the active site.118,152–154 In enzyme evolution studies,155 fitness landscape of several enzymes has been shown to be determined by residues distributed throughout the enzyme structure,154 again highlighting the holistic nature of protein function.
In some cases, the roles of remote residues can be intuitively understood at a structural level;119 i.e., through a domino or Goldberg machine fashion, remote mutation(s) perturb the dominant conformation of active site residues156,157 and therefore the chemical activity therein, including both catalytic proficiency and substrate scope. The challenge for any computational analysis is thus to capture such “population shifts” due to distal mutations. Several compelling examples have been reported recently using enhanced sampling techniques, such as path-variable based metadynamics simulations103 for variants of Tryptophan synthase subunit B (TrpB);158 the simulations showed that the populations of several conformational states of TrpB were modulated by distal mutations, providing a molecular level rational for the emergence of these mutation in directed evolution studies159,160 that engineered TrpB into a standalone enzyme in the absence of the neighboring α subunit.
However, in other cases, the effects of distal changes appear more indirect. For example, in our recent analysis of ATP hydrolysis of myosin,106 we studied two models for the motor domain that have almost identical conformations for residues in the nucleotide binding site but differ in distal regions and hydration of the critical R238-E459 salt bridge (Fig. 5a). String based DFTB3/MM free energy simulations found that activation free energy and exergonicity in the two models differ by as much as 9 kcal/mol, highlighting that the average ground state configurations of catalytic residues are not the only determinant. Indeed, the distal residues can instead modify the fluctuations of active site residues and water molecules, and thus the reorganization energy or entropic contributions to the activation free energy.45 In a different context, elegant EVB analysis of Aqvist and co-workers have clearly shown that flexibility of surface protein residues plays a major role in modulating the enthalpic and entropic components of activation free energy,161 leading to different temperature adaptations for mesophilic and psychrophilic enzymes.48
Figure 5:
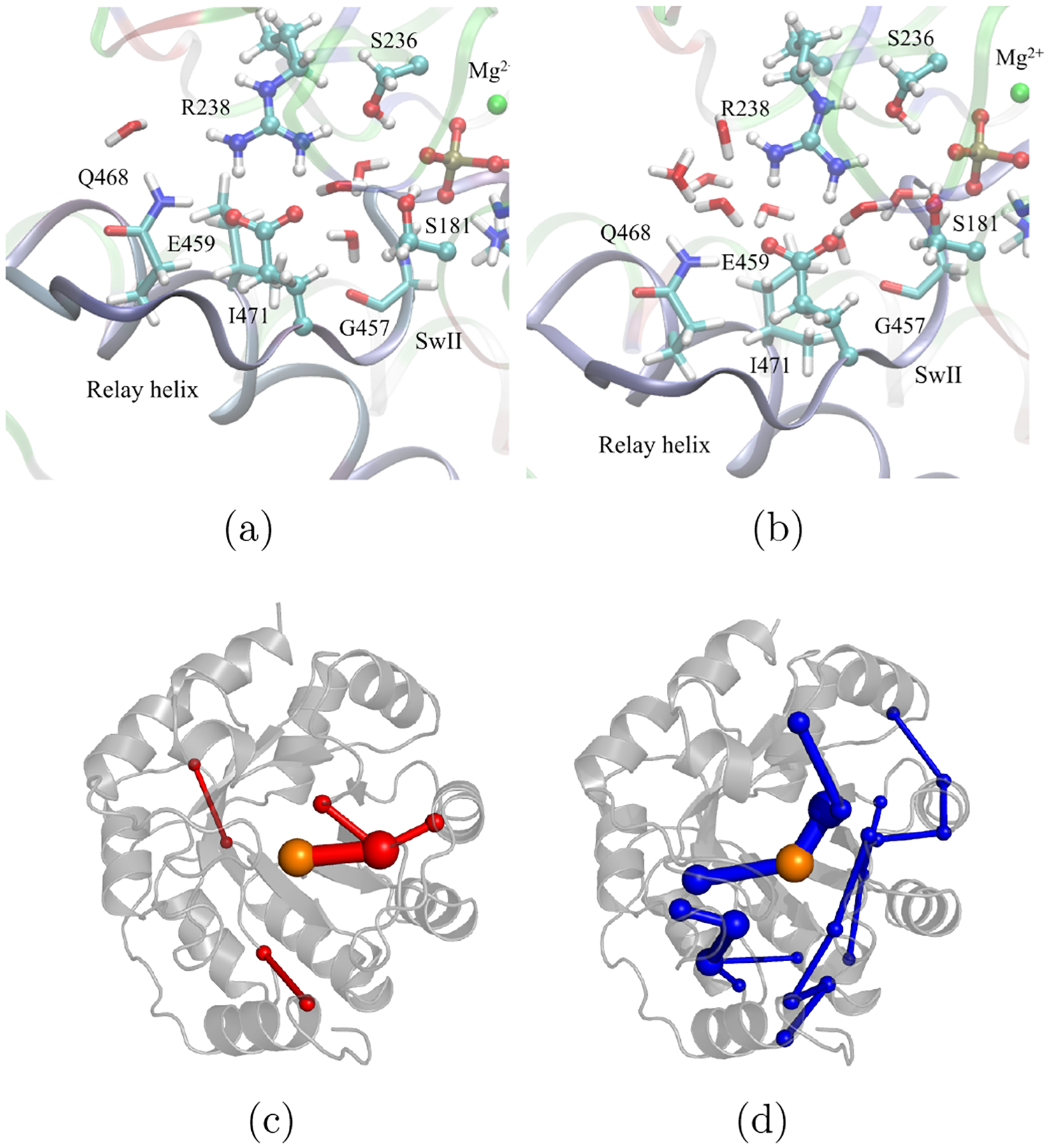
Examples of distal contributions to enzyme catalysis from computational analyses. (a-b) ATP hydrolysis in two models of the myosin motor domain may differ by ~9 kcal/mol in activation free energy although the nucleotide binding site residues have almost comparable average configurations;106 (c-d) A de novo designed enzyme for Kemp elimination and its more efficient variant following directed evolution feature rather different collective motions;173 the latter also features a negative activation heat capacity.174 Panels c-d have been provided by Drs. A. Bunzel and A. Mulholland.
The temperature dependence of enzymes has led to many fascinating discussions regarding the mechanisms that govern enzyme stability and catalysis.48,162–165 A particularly intriguing phenomenon noted in recent literature concerns non-linearity in the Arrhenius plot at high temperatures,165–167 for which quantum mechanical tunneling of nuclei11,168,169 is unlikely the cause. Two different models have been proposed to explain the non-linear Arrhenius plot: conformational redistribution170,171 vs. activation heat capacity.166,172 In the former model, the equilibrium between catalytically active and inactive conformations is perturbed by temperature, leading to different populations of the active conformation and therefore different apparent rate constants. In the latter case, a non-vanishing heat capacity difference between the ground and transition states () makes the activation free energy temperature-dependent, leading also to deviation of the Arrhenius plot from linearity. Both thermodynamic models can be used to fit available experimental kinetic data, and for different systems, both models have been supported with microscopic analysis based on extensive molecular dynamics simulations,167,171–173 respectively. As discussed by Aqvist and co-workers,171 the two models do lead to significantly different predictions about activation enthalpy and entropy for broader temperatures, for which no experimental data are yet available. On the other hand, it has been suggested that the different proposals consistently point toward a general picture of more than one reactant state before passing the transition state.165
A particularly interesting case concerns the de novo designed enzyme for catalyzing the Kemp elimination reaction.174 While the designed enzyme, which has low catalytic proficiency, features a linear Arrhenius plot and therefore zero activation heat capacity, its more efficient variant following several generations of directed evolution exhibits a non-linear Arrhenius plot, which can be fitted with a negative . Mulholland and co-workers173 showed that extensive (~5 μs) classical MD simulations with a MM model for the transition state indeed recapitulated a negative , which was estimated based on the fluctuation of the protein energy in the MD simulations. Further analysis of the trajectories found that the negative in the more active enzyme variant was likely the result of a more compact active site in the transition state due to a higher population of an active site loop in its closed conformation. The more compact structure is also congruent with extensive correlated motions throughout the protein (Fig.5b). Therefore, an interesting notion from the analysis is that to further enhance catalytic proficiency of designed enzymes, one strategy might be to enhance correlated motions that implicate both active site and distal regions; this point was also made in the aforementioned study of TrpB,158 in which the distal mutation sites observed in the directed evolution study were also identified with a motional network analysis for the wild type enzyme.
Evidently, the causal relation between correlated motion, negative and catalytic proficiency remains to be further analyzed. Nevertheless, these recent studies further highlight the significance of better understanding collective motions of enzymes, their thermodynamic signature and factors that control the nature and functional implication of such motions. Along this line, close integration of extensive classical and QM/MM studies with recent breakthrough in continuous evolution techniques,175–177 which makes it straightforward to evolve enzymes for many (>100) generations in independent trajectories,178 is poised to lead to significant insights into new strategies that can be used to enhance the catalytic proficiency of designed enzymes.
3. Concluding Remarks
In this short perspective, we discussed a number of “burning issues” related to biomolecular QM/MM simulations. Although most of these discussions are motivated primarily by our own research interest, we believe many of the discussions are relevant to biomolecular QM/MM applications in general. Many aspects of the discussed issues are related to establishing “best practice” for realistic applications, such as choices of the QM region size, QM and MM potential functions, and methodologies that best integrate physical methods and established machine learning techniques for efficient free energy simulations. There are, nevertheless, in our opinion, fundamental conceptual issues, such as developing efficient methods for better treating transition metal ions,179,180 pursuing generally applicable machine learning models for both short-range and long-range interactions,67,181 identifying problems for which the explicit inclusion of electronic polarization makes a qualitative difference, and clearly dissecting and defining the roles of distal residues and specific motions in enzyme catalysis.
In terms of connections with experiments, while it remains important to compute multiple observables that characterize physical properties of biomolecules, such as kinetic isotope effects, free energy relations and various spectroscopies,182 an emerging opportunity is to integrate computations with high-throughput experiments. For the analysis of protein allostery, for example, deep mutation studies183 have revealed a rather different picture for the distribution of “hotspot” residues that are essential to co-operativity.184 Similarly, continuous evolution studies are starting to generate large datasets on mutation effects on catalytic properties.178 Providing a molecular level understanding for these rich datasets and making testable predictions with multi-faceted computational analysis is a formidable yet exciting challenge. To paraphrase one of the ten big ideas from the National Science Foundation, developing novel methodologies that can truly embrace “the data revolution” is of great urgency. With further development and integration with data science/machine learning techniques, QM/MM methods will play increasingly essential roles in the mechanistic analysis and rational design of novel biomolecular functions.119,185
Before closing, we note that exciting developments in ab initio quantum chemistry methodologies continue to take place at a pace as rapid as ever.186 We also can’t resist mentioning that major strides are being made in the areas of quantum Monte Carlo187 and related ML methods188 as well as quantum computing,189,190 in terms of both fundamental algorithms and hardware. While these more advanced and expensive methodologies are yet to directly impact biophysical and biochemical applications, it is clear that the list of powerful tools available to multi-scale computations will continue to expand rapidly and therefore the future is undoubtedly bright.
Supplementary Material
Figure 4:
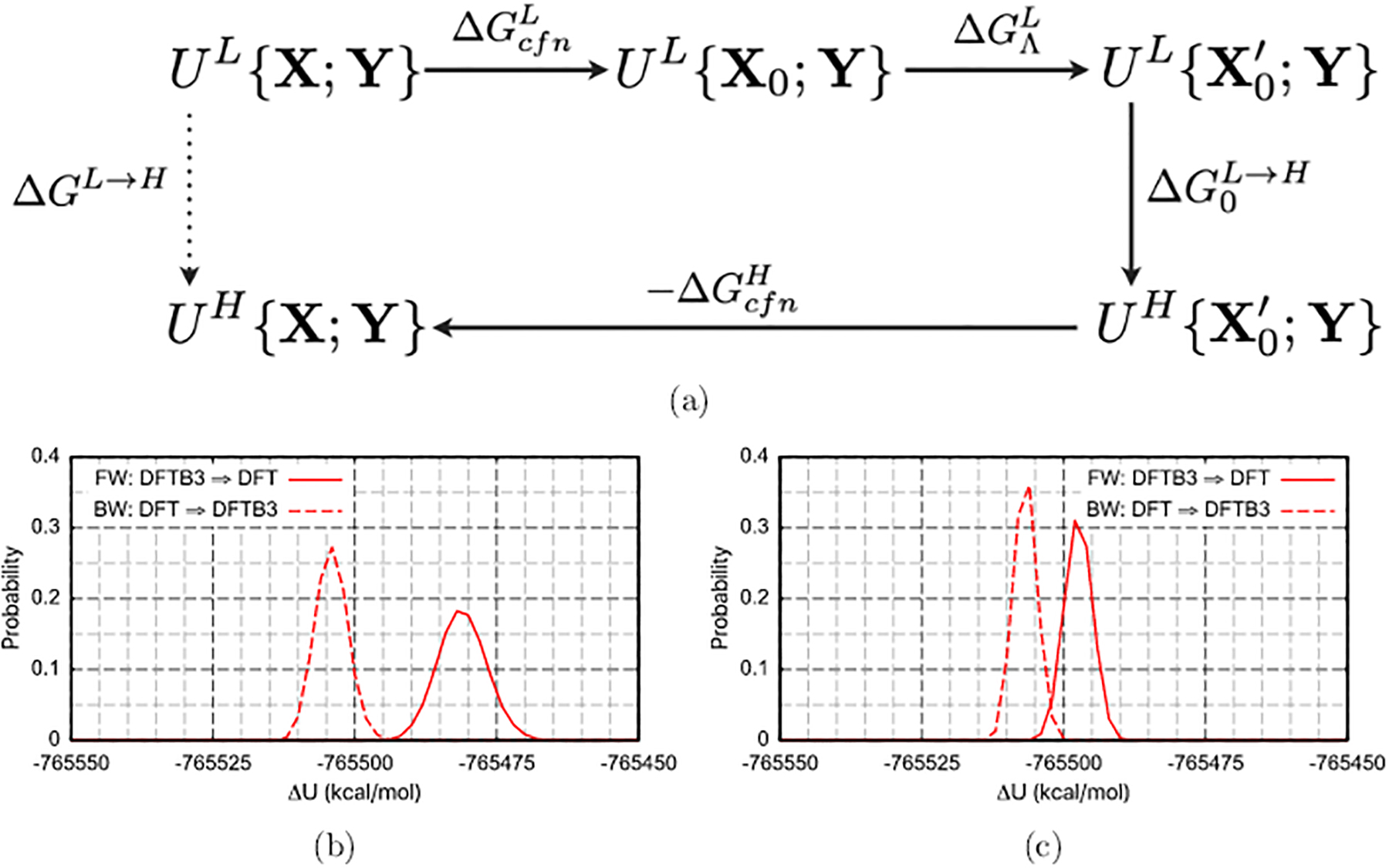
A staged transformation approach141 for computing the free energy difference at two levels (L/H) of theory, ΔGL→H. (A) The staged thermodynamic path treats selected degrees of freedom (X) separately from the rest (Y); X represents the degrees of freedom that lead to a large gap in the ΔULH distribution. Assuming that the free energy costs for confining X to values at (or near) the free energy minima are similar at the L and H levels, ΔGL→H is given by the sum of , which converges readily since the sampling involves only Y, and the “reorganization free energy”, , which is the free energy cost of changing X0 to at the low level of theory. (b-c) Illustration of the impact of bond and angle restraints on the ΔULH distribution for a methyl diphosphate, which is treated with either DFTB3 (L) or B3LYP (H), solvated by TIP3P water.
Acknowledgement
This work was supported by a grant from the NIH (R01 GM106443) to Q.C. L.X. was supported through the BU RISE program. T. P. acknowledges discussions with Prof. F. Paesani’s group regarding second virial coefficient calculations and for sharing the CCSD(T)/CBS data for 2,3-body energies for water, and Prof. W. E and Dr. H. Wang for discussion on reinforcement learning based free energy simulations and sharing of their code. Q.C. acknowledges Dr. A. Bunzel and Prof. A. Mulholland for providing figures 5c–d. Computational resources from the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF grant number OCI-1053575, are greatly appreciated; part of the computational work was performed on the Shared Computing Cluster which is administered by Boston University’s Research Computing Services (URL: www.bu.edu/tech/support/research/).
Biography
Qiang Cui
Qiang Cui is a professor of Chemistry at Boston University and also affiliated with Departments of Physics and Biomedical Engineering. He received B.S. in Chemical Physics from the University of Science and Technology of China in 1993, and Ph. D. in Theoretical Chemistry in 1997 from Emory University under the tutelage of Professor Keiji Morokuma. He conducted postodctoral research with Professor Martin Karplus at Harvard University and, in 2001, moved to the University of Wisconsin, Madison as an assistant professor of Chemistry. After spending almost seventeen wonderful years in Madison, he moved back to Boston in 2018. His current research interest includes quantum chemistry and statistical mechanics and their application to various chemical, biological and materials problems.
Tanmoy Pal
Tanmoy Pal recieved his B.S. in Chemistry from the Indian Institute of Science, Bengaluru in 2016. Currently he is a doctoral candidate in the Cui group at Boston University. Tanmoy’s research involves classical and QM/MM simulations of biomolecular systems. His recent interests include studies of ionic liquid-interface systems and development of machine learning based techniques to improve the accuracy and efficiency of multidimensional free energy calculations.
Luke Xie
Luke Xie is a high school senior from Newport High School, Washington. He spent the summer of 2019 in the Cui group at Boston University as a part of the RISE Internship program. He is interested in studying computer science.
Footnotes
Supporting Information Available
Additional data for the second virial coefficients for small molecules and 2-/3-body energies for water at different levels of NDDO and tight-binding methods, and calculation details for the 3-dimensional free energy surface of methyl phosphate hydrolysis are included. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- (1).Warshel A; Levitt M Theoretical Studies of Enzymic Reactions - Dielectric, Electrostatic and Steric Stabilization of Carbonium-Ion in Reaction of Lysozyme. J. Mol. Biol 1976, 103, 227–249. [DOI] [PubMed] [Google Scholar]
- (2).Field MJ; Bash PA; Karplus M A Combined Quantum-Mechanical and Molecular Mechanical Potential for Molecular-Dynamics Simulations. J. Comput. Chem 1990, 11, 700–733. [Google Scholar]
- (3).Lipkowitz KB, Boyd DB, Eds. Gao J, In Reviews in Computational Chemistry VII; VCH: New York, 1995; p 119. [Google Scholar]
- (4).Senn HM; Thiel W QM/MM methods for biomolecular systems. Angew. Chem. Int. Ed 2009, 48, 1198–1229. [DOI] [PubMed] [Google Scholar]
- (5).Friesner RA; Guallar V Ab intio QM and QM/MM methods for studying enzyme catalysis. Annu. Rev. Phys. Chem 2005, 56, 389–427. [DOI] [PubMed] [Google Scholar]
- (6).Hu H; Yang WT Free Energies of Chemical Reactions in Solution and in Enzymes with Ab Initio Quantum Mechanics/Molecular Mechanics Methods. Annu. Rev. Phys. Chem 2008, 59, 573–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kamerlin SCL; Haranczyk M; Warshel A Progress in Ab Initio QM/MM Free-Energy Simulations of Electrostatic Energies in Proteins: Accelerated QM/MM Studies of pK(a), Redox Reactions and Solvation Free Energies. J. Phys. Chem. B 2009, 113, 1253–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lu X; Fang D; Ito S; Okamoto Y; Ovchinnikov V; Cui Q QM/MM Free Energy Simulations: Recent Progress and Challenges. Mol. Simul. (Special Issue on Free Energy Simulations) 2016, 42, 1056–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Gao JL; Ma SH; Major DT; Nam K; Pu JZ; Truhlar DG Mechanisms and free energies of enzymatic reactions. Chem. Rev 2006, 106, 3188–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Alexandrova AN; Rothlisberger D; Baker D; Jorgensen WL Catalytic Mechanism and Performance of Computationally Designed Enzymes for Kemp Elimination. J. Am. Chem. Soc 2008, 130, 15907–15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gao JL; Truhlar DG Quantum mechanical methods for enzyme kinetics. Annu. Rev. Phys. Chem 2002, 53, 467–505. [DOI] [PubMed] [Google Scholar]
- (12).Merz KM Jr., Using Quantum Mechanical Approaches to Study Biological Systems. Acc. Chem. Res 2014, 47, 2804–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Shaik S; Cohen S; Wang Y; Chen H; Kumar D; Thiel W P450 Enzymes: Their Structure, Reactivity, and Selectivity-Modeled by QM/MM Calculations. Chem. Rev 2010, 110, 949–1017. [DOI] [PubMed] [Google Scholar]
- (14).Lonsdale R; Mulholland A QM/MM Modelling of Drug-Metabolizing Enzymes. Curr. Top. Med. Chem 2014, 14, 1339–1347. [DOI] [PubMed] [Google Scholar]
- (15).Brunk E; Rothlisberger U Mixed Quantum Mechanical/Molecular Mechanical Molecular Dynamics Simulations of Biological Systems in Ground and Electronically Excited States. Chem. Rev 2015, 115, 6217–6263. [DOI] [PubMed] [Google Scholar]
- (16).Amaro RE; Mulholland AJ Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat. Rev. Chem 2018, 2, 0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chung LW; Sameera WMC; Ramozzi R; Page AJ; Hatanaka M; Petrova GP; Harris TV; Li X; Ke ZF; Liu FY et al. , The ONIOM Method and Its Applications. Chem. Rev 2015, 115, 5678–5769. [DOI] [PubMed] [Google Scholar]
- (18).Metz S; Kästner J; Sokol AA; Keal TW; Sherwood P ChemShell—a modular software package for QM/MM simulations. WIREs Comput. Mol. Sci 2013, 4, 101–110. [Google Scholar]
- (19).Fürstner A; List B; Ritter T; Schüth F; Neese F Walter Thiel (1949–2019). Angew. Chem. Int. Ed 2020, 59, 1382–1383. [DOI] [PubMed] [Google Scholar]
- (20).Sokol AA; Bromley ST; French SA; Richard C; Catlow A; Sherwood P Hybrid QM/MM Embedding Approach for the Treatment of Localized Surface States in Ionic Materials. Int. J. Quant. Chem 2004, 99, 695–712. [Google Scholar]
- (21).Golze D; Iannuzzi M; Nguyen MT; Passerone D; Hutter J Simulation of Adsorption Processes at Metallic Interfaces: An Image Charge Augmented QM/MM Approach. J. Chem. Theory Comput 2013, 9, 5086–5097. [DOI] [PubMed] [Google Scholar]
- (22).Magnussen OM; Gross A Toward an atomic-scale understanding of electrochemical interface structure and dynamics. J. Am. Chem. Soc 2019, 141, 4777–4790. [DOI] [PubMed] [Google Scholar]
- (23).Bruix A; Margraf JT; Andersen M; Reuter K First-principles-based multiscale modelling of heterogeneous catalysis. Nat. Cata 2019, 2, 659–670. [Google Scholar]
- (24).Huang P; Carter EA Advances in correlated electronic structure methods for solids, surfaces, and nanostructures. Annu. Rev. Phys. Chem 2008, 59, 261–290. [DOI] [PubMed] [Google Scholar]
- (25).Sakong S; Gross A The electric double layer at metal-water interfaces revisited based on a charge polarization scheme. J. Chem. Phys 2018, 149, 084705. [DOI] [PubMed] [Google Scholar]
- (26).Sakong S; Gross A Water structures on a Pt(111) electrode from ab initio molecular dynamic simulations for a variety of electrochemical conditions. Phys. Chem. Chem. Phys 2020, 22, 10431. [DOI] [PubMed] [Google Scholar]
- (27).Le JB; Cheng J Modeling electrochemical interfaces from ab initio molecular dynamics: water adsorption on metal surfaces at potential of zero charge. Curr. Opin. Electrochem 2020, 19, 129–136. [Google Scholar]
- (28).Liang DY; Hong JW; Dong F; Bennett JW; Mason SE; Hamers RJ; Cui Q Analysis of Conformational Properties of Amine Ligands at the Gold/Water Interface with QM, MM and QM/MM simulations. Phys. Chem. Chem. Phys 2018, 20, 3349–3362. [DOI] [PubMed] [Google Scholar]
- (29).Heenen HH; Gauthier JA; Kristoffersen HH; Ludwig T; Chan K Solvation at metal/water interfaces: An ab initio molecular dynamics benchmark of common computational approaches. J. Chem. Phys 2020, 152, 144703. [DOI] [PubMed] [Google Scholar]
- (30).Ringe S; Oberhofer H; Hille C; Matera S; Reuter K Function-Space-Based Solution Scheme for the Size-Modified Poisson-Boltzmann Equation in Full-Potential DFT. J. Chem. Theory Comput 2016, 12, 4052–4066. [DOI] [PubMed] [Google Scholar]
- (31).Sumowski CV; Ochsenfeld C A Convergence Study of QM/MM Isomerization Energies with the Selected Size of the QM Region for Peptidic Systems. J. Phys. Chem. A 2009, 113, 11734–11741. [DOI] [PubMed] [Google Scholar]
- (32).Flaig D; Beer M; Ochsenfeld C Convergence of Electronic Structure with the Size of the QM Region: Example of QM/MM NMR Shieldings. J. Chem. Theory Comput 2012, 8, 2260–2271. [DOI] [PubMed] [Google Scholar]
- (33).Hu L; Söderhjelm P; Ryde U On the Convergence of QM/MM Energies. J. Chem. Theory Comput 2011, 7, 761–777. [DOI] [PubMed] [Google Scholar]
- (34).Roßbach S; Ochsenfeld C Influence of Coupling and Embedding Schemes on QM Size Convergence in QM/MM Approaches for the Example of a Proton Transfer in DNA. J. Chem. Theory Comput 2017, 13, 1102–1107. [DOI] [PubMed] [Google Scholar]
- (35).Kulik HJ; Zhang J; Klinman JP; Martínez TJ How Large Should the QM Region Be in QM/MM Calculations? The Case of Catechol O-Methyltransferase. J. Phys. Chem. B 2016, 120, 11381–11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jindal G; Warshel A Exploring the Dependence of QM/MM Calculations of Enzyme Catalysis on the Size of the QM Region. J. Phys. Chem. B 2016, 120, 9913–9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kulik HJ Large-scale QM/MM free energy simulations of enzyme catalysis reveal the influence of charge transfer. Phys. Chem. Chem. Phys 2018, 20, 20650–20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Das S; Nam K; Major DT Rapid Convergence of Energy and Free Energy Profiles with Quantum Mechanical Size in Quantum Mechanical–Molecular Mechanical Simulations of Proton Transfer in DNA. J. Chem. Theory Comput 2018, 14, 1695–1705. [DOI] [PubMed] [Google Scholar]
- (39).Mehmood R; Kulik HJ Both Configuration and QM Region Size Matter: Zinc Stability in QM/MM Models of DNA Methyltransferase. J. Chem. Theory Comput 2020, 16, 3121–3134. [DOI] [PubMed] [Google Scholar]
- (40).Watanabe H; Cui Q Quantitative analysis and correction of QM/MM boundary artifacts in adaptive QM/MM methods. J. Chem. Theory Comput 2019, 15, 3917–3928. [DOI] [PubMed] [Google Scholar]
- (41).Thiel W Perspectives on semiempirical molecular orbital theory. Adv. Chem. Phys 1996, 93, 703–757. [Google Scholar]
- (42).Gaus M; Cui Q; Elstner M Density Functional Tight Binding (DFTB): Application to organic and biological molecules. WIREs Comput. Mol. Sci 2014, 4, 49–61. [Google Scholar]
- (43).Warshel A Computer Modeling of Chemical Reactions in Enzymes and Solution; Wiley, New York, 1991. [Google Scholar]
- (44).Ruiz-Pernìa JJ; Silla E; Tuñón I; Martì S Hybrid Quantum Mechanics/Molecular Mechanics Simulations with Two-Dimensional Interpolated Corrections: Application to Enzymatic Processes. J. Phys. Chem. B 2006, 110, 17663–17670. [DOI] [PubMed] [Google Scholar]
- (45).Aqvist J; Kazemi M; Isaksen GV; Brandsdal BO Entropy and Enzyme Catalysis. Acc. Chem. Res 2017, 50, 199–207. [DOI] [PubMed] [Google Scholar]
- (46).Kulkarni YS; Amyes TL; Richard J; Kamerlin SCL Uncovering the Role of Key Active-Site Side Chains in Catalysis: An Extended Bronsted Relationship for Substrate Deprotonation Catalyzed by Wild-Type and Variants of Triosephosphate Isomerase. J. Am. Chem. Soc 2019, 141, 16139–16150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Nomura Y; Roston D; Montemayor EJ; Cui Q; Butcher SE Structural and Mechanistic Basis for Preferential Deadenylation of U6 snRNA by Usb1. Nuc. Acids Res 2018, 46, 11488–11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Aqvist J; Isaksen GV; Brandsdal BO Computation of enzyme cold adaptation. Nat. Rev. Chem 2017, 1, 0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kussmann J; Beer M; Ochsenfeld C Linear-scaling self-consistent field methods for large molecules. WIREs Comput. Mol. Sci 2013, 3, 614–636. [Google Scholar]
- (50).Seritan S; Bannwarth C; Fales BS; Hohenstein EG; Kokkila-Schumacher SIL; Leuhr N; Snyder JW; Song CC; Titov AV; Ufimtsev IS et al. , TeraChem: Accelerating electronic structure and ab initio molecular dynamics with graphical processing units. J. Chem. Phys 2020, 152, 224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Christensen AS; Kubar T; Cui Q; Elstner M, Semi-empirical Quantum Mechanical Methods for Non-covalent Interactions for Chemical and Biochemical Applications. Chem. Rev 2016, 116, 5301–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Bannwarth C; Ehlert S; Grimme S GFN2-xTB-An Accurate and Broadly Parametrized Self-Consistent Tight-Binding Quantum Chemical Method with Multipole Electrostatics and Density-Dependent Dispersion Contributions. J. Chem. Theory Comput 2019, 15, 1652–1671. [DOI] [PubMed] [Google Scholar]
- (53).Bannwarth C; Caldeweyher E; Ehlert S; Hansen A; Pracht P; Seibert J; Spicher S; Grimme S Extended tight-binding quantum chemistry methods. WIREs Comput. Mol. Sci 2020, e01493. [Google Scholar]
- (54).Redders GR; Babin V; Paesani F A Critical Assessment of Two-Body and Three-Body Interactions in Water. J. Chem. Theory Comput 2013, 9, 1103–1114. [DOI] [PubMed] [Google Scholar]
- (55).Christensen AS; Kromann JC; Jensen JH; Cui Q Intermolecular Interactions in the Condensed Phase: Evaluation of Semi-empirical Quantum Mechanical Methods. J. Chem. Phys 2017, 147, 161704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov I et al. , CHARMM General Force Field (CGenFF): A force field for drug-like molecules compatible with the CHARMM allatom additive biological force fields. J. Comput. Chem 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Weinhold F; Landis CR Valency and Bonding; Cambridge University Press, 2005. [Google Scholar]
- (58).Lu X; Duchimaza-Heredia J; Cui Q Analysis of Density Functional Tight Binding (DFTB) with Natural Bonding Orbitals (NBOs). J. Phys. Chem. A 2019, 123, 7439–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kulik HJ Perspective: Treating electron over-delocalization with the DFT+U method. J. Chem. Phys 2016, 142, 240901. [DOI] [PubMed] [Google Scholar]
- (60).Griffith JS The Theory of Transition-Metal Ions; Cambridge University Press, 1963. [Google Scholar]
- (61).Singh SK; Eng J; Atanasov M; Neese F Covalency and chemical bonding in transition metal complexes: An ab initio based ligand field perspective. Coord. Chem. Rev 2017, 344, 2–25. [Google Scholar]
- (62).Stepanovic S; Lai R; Elstner M; Gruden M; Garcia-Fernandez P; Cui Q Improvement of d-d interactions in Density Functional Tight Binding for Transition Metal Ions with a Ligand Field Model: Assessment of a DFTB3+U model on Nickel coordination compounds. Phys. Chem. Chem. Phys 2020, In press ( 10.1039/D0CP04694A). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Vujovic M; Huynh M; Steiner S; Garcia-Fernandez P; Elstner M,; Cui Q; Gruden M Exploring the Applicability of Density Functional Tight Binding to Transition Metal Ions: Parametrization for Nickel with the Spin-polarized DFTB3 model. J. Comput. Chem 2019, 40, 400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Filatov M Spin-restricted ensemble-referenced Kohn-Sham method: basic principles and application to strongly correlated ground and excited states of molecules. WIREs Comput. Mol. Sci 2015, 5, 146–167. [Google Scholar]
- (65).Grofe A; Chen X; Liu WJ; Gao JL Spin-Multiplet Components and Energy Splittings by Multistate Density Functional Theory. J. Phys. Chem. Lett 2017, 8, 4838–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Noe F; Tkatchenko A; Müller KR; Clementi C Machine Learning for Molecular Simulation. Annu. Rev. Phys. Chem 2020, 71, 361–390. [DOI] [PubMed] [Google Scholar]
- (67).Unke OT; Chmiela S; Sauceda HE; Gastegger M; Poltavsky I; Schẗt KT; Tkatchenko A; Müller KR Machine Learning Force Fields. arXiv:2010.07067v1 2020, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).von Lilienfeld OA; Müller K-R; Tkatchenko A Exploring chemical compound space with quantum-based machine learning. Nat. Rev. Chem 2020, 4, 347–358. [DOI] [PubMed] [Google Scholar]
- (69).Wang YH; Ribeiro JML; Tiwary P Machine learning approaches for analyzing and enhancing molecular dynamics simulations. Curr. Opin. Struct. Biol 2020, 61, 139–145. [DOI] [PubMed] [Google Scholar]
- (70).Janet JP; Kulik HJ Predicting electronic structure properties of transition metal complexes with neural networks. Chem. Sci 2017, 8, 5137–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Duan CR; Liu F; Nandy A; Kulik HJ Semi-supervised Machine Learning Enables the Robust Detection of Multireference Character at Low Cost. J. Phys. Chem. Lett 2020, 11, 6640–6648. [DOI] [PubMed] [Google Scholar]
- (72).Shen L; Yang WT Molecular Dynamics Simulations with Quantum Mechanics/Molecular Mechanics and Adaptive Neural Networks. J. Chem. Theory. Comput 2018, 14, 1442–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Qiao Z; Welborn M; Anandkumar A; Manby FR; Miller III TF, OrbNet: Deep Learning for Quantum Chemistry Using Symmetry-Adapted Atomic-Orbital Features. J. Chem. Phys 2020, 153, 124111. [DOI] [PubMed] [Google Scholar]
- (74).Zhang L; Han J; Wang H; Car R; E W Deep Potential Molecular Dynamics: A Scalable Model with the Accuracy of Quantum Mechanics. Phys. Rev. Lett 2018, 120, 143001. [DOI] [PubMed] [Google Scholar]
- (75).Wang H; Zhang L; Han J; E W DeePMD-kit: A deep learning package for many-body potential energy representation and molecular dynamics. Comput. Phys. Comm 2018, 228, 178–184. [Google Scholar]
- (76).Ganguly A; Boulanger E; Thiel W Importance of MM Polarization in QM/MM Studies of Enzymatic Reactions: Assessment of the QM/MM Drude Oscillator Model. J. Chem. Theory Comput 2017, 13, 2954–2961. [DOI] [PubMed] [Google Scholar]
- (77).Wanko M; Hoffmann M; Strodel P; Koslowski A; Thiel W; Neese F; Frauenheim T; Elstner M Calculating absorption shifts for retinal proteins: Computational challenges. J. Phys. Chem. B 2005, 109, 3606–3615. [DOI] [PubMed] [Google Scholar]
- (78).Mennucci B Modeling environment effects on spectroscopies through QM/classical models. Phys. Chem. Chem. Phys 2013, 15, 6583–6594. [DOI] [PubMed] [Google Scholar]
- (79).Tazhigulov RN; Gurunathan PK; Kim Y; Slipchenko LV; Bravaya KB Polarizable embedding for simulating redox potentials of biomolecules. Phys. Chem. Chem. Phys 2019, 21, 11642–11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Goyal P; Lu J; Yang S; Gunner MR; Cui Q Changing hydration level in an internal cavity modulates the proton affinity of a key glutamate in Cytochrome c Oxidase. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 18886–18891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Wanko M; Hoffmann M; Frahmcke J; Frauenheim T; Elstner M Effect of polarization on the opsin shift in rhodopsins. 2. empirical polarization models for proteins. J. Phys. Chem. B 2008, 112, 11468–11478. [DOI] [PubMed] [Google Scholar]
- (82).Loco D; Lagardere L; Cisneros GA; Scalmani G; Frisch M; Lipparini F; Mennucci B; Piquemal JP Towards large scale hybrid QM/MM dynamics of complex systems with advanced point dipole polarizable embeddings. Chem. Sci 2019, 10, 7200–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Bondanza M; Nottoli M; Cupellini L; Lipparini F; Mennucci B Polarizable embedding QM/MM: the future gold standard for complex (bio)systems? Phys. Chem. Chem. Phys 2020, 22, 14433–14448. [DOI] [PubMed] [Google Scholar]
- (84).Lambros E; Lipparini F; Cisneros GA; Paesani F A Many-Body, Fully Polarizable Approach to QM/MM Simulations. J. Chem. Theory Comput 2020, In press, ( 10.1021/acs.jctc.0c00932). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Welborn VV; Pestana LR; Head-Gordon T Computational optimization of electric fields for better catalysis design. Nat. Cata 2018, 1, 649–655. [Google Scholar]
- (86).Simonson T; Perahia D Internal and interfacial dielectric properties of cytochrome c from molecular dynamics in aqueous solution. Proc. Natl. Acad. Sci. U.S.A 1995, 92, 1082–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Warshel A Computer simulations of enzyme catalysis: Methods, Progress and Insights. Annu. Rev. Biophys. Biomol. Struct 2003, 32, 425–443. [DOI] [PubMed] [Google Scholar]
- (88).Riccardi D; Cui Q pKa analysis for the zinc-bound water in Human Carbonic Anhydrase II: benchmark for ”multi-scale” QM/MM simulations and mechanistic implications. J. Phys. Chem. A 2007, 111, 5703–5711. [DOI] [PubMed] [Google Scholar]
- (89).Ghosh N; Cui Q pKa of residue 66 in Staphylococal nuclease: insights from QM/MM simulations with conventional sampling. J. Phys. Chem. B 2008, 112, 8387–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Chimenti MS; Castaneda CA; Majumdar A; Garcia-Moreno EB Structural Origins of High Apparent Dielectric Constants Experienced by Ionizable Groups in the Hydrophobic Core of a Protein. J. Mol. Biol 2011, 405, 361–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Zheng YQ; Cui Q Microscopic Mechanisms that Govern the Titration Response and pKa Values of Buried Residues in Staphylococcal Nuclease Mutants. Proteins: Struct., Funct., & Bioinf 2017, 85, 268–281. [DOI] [PubMed] [Google Scholar]
- (92).Simonson T; Carlsson J; Case DA Proton binding to proteins: pK(a) calculations with explicit and implicit solvent models. J. Am. Chem. Soc 2004, 126, 4167–4180. [DOI] [PubMed] [Google Scholar]
- (93).Goh GB; Knight JL; Brooks III CL, Constant pH Molecular Dynamics Simulations of Nucleic Acids in Explicit Solvent. J. Chem. Theo. Comp 2012, 8, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Huang YD; Chen W; Wallace JA; Shen J All-Atom Continuous Constant pH Molecular Dynamics With Particle Mesh Ewald and Titratable Water. J. Chem. Theory Comput 2016, 12, 5411–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Lin YL; Aleksandrov A; Simonson T; Roux B An Overview of Electrostatic Free Energy Computations for Solutions and Proteins. J. Chem. Theory Comput 2014, 10, 2690–2709. [DOI] [PubMed] [Google Scholar]
- (96).Riccardi D; Schaefer P; Cui Q pKa calculations in solution and proteins with QM/MM free energy perturbation simulations. J. Phys. Chem. B 2005, 109, 17715–17733. [DOI] [PubMed] [Google Scholar]
- (97).Nam K; Gao JL; York DM An efficient linear-scaling Ewald method for long-range electrostatic interactions in combined QM/MM calculations. J. Chem. Theo. Comp 2005, 1, 2–13. [DOI] [PubMed] [Google Scholar]
- (98).Sinitskiy AV; Voth GA Quantum mechanics/coarse-grained molecular mechanics (QM/CG-MM). J. Chem. Phys 2018, 148, 014102. [DOI] [PubMed] [Google Scholar]
- (99).Valdez CE; Morgenstern A; Eberhart ME; Alexandrova AN Predictive methods for computational metalloenzyme redesign - a test case with carboxypeptidase A. Phys. Chem. Chem. Phys 2016, 18, 31744–31756. [DOI] [PubMed] [Google Scholar]
- (100).Sokkar P; Boulanger E; Thiel W; Sanchez-Garcia E Hybrid Quantum Mechanics/Molecular Mechanics/Coarse Grained Modeling: A Triple-Resolution Approach for Biomolecular Systems. J. Chem. Theory Comput 2015, 11, 1809–1818. [DOI] [PubMed] [Google Scholar]
- (101).Hocky GM; Dannenhoffer-Lafage T; Voth GA Coarse-Grained Directed Simulation. J. Chem. Theory Comput 2017, 13, 4593–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Zuckerman DM Equilibrium Sampling in Biomolecular Simulations. Annu. Rev. Biophys 2011, 40, 41–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Valsson O; Tiwary P; Parrinello M Enhancing Important Fluctuations: Rare Events and Metadynamics from a Conceptual Viewpoint. Annu. Rev. Phys. Chem 2016, 67, 159–184. [DOI] [PubMed] [Google Scholar]
- (104).E W; Vanden-Eijnden E Transition-Path Theory and Path-Finding Algorithms for the Study of Rare Events. Annu. Rev. Phys. Chem 2010, 61, 391–420. [DOI] [PubMed] [Google Scholar]
- (105).Rosta E; Nowotny M; Yang W; Hummer G Catalytic Mechanism of RNA Back-bone Cleavage by Ribonuclease H from Quantum Mechanics/Molecular Mechanics Simulations. J. Am. Chem. Soc 2011, 133, 8934–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Lu X; Ovchinnikov V; Roston DR; Demapan D; Cui Q Regulation and Plasticity of Catalysis in Enzymes: Insights from Analysis of Mechanochemical Coupling in Myosin. Biochem. 2017, 56, 1482–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Maeda S; Ohno K; Morokuma K Systematic exploration of the mechanism of chemical reactions: the global reaction route mapping (GRRM) strategy using the ADDF and AFIR methods. Phys. Chem. Chem. Phys 2013, 15, 3683–3701. [DOI] [PubMed] [Google Scholar]
- (108).Dewyer AL; Arguelles AJ; Zimmerman PM Methods for exploring reaction space in molecular systems. WIREs Comput. Mol. Sci 2018, 8, e1354. [Google Scholar]
- (109).Nicholls DG; Ferguson SJ Bioenergetics, 3rd ed.; Academic Press: New York, 2002. [Google Scholar]
- (110).Son CY; Yethiraj A; Cui Q Cavity Hydration Dynamics in Cytochrome c Oxidase and Functional Implications. Proc. Natl. Acad. Sci. USA 2017, 114, E8830–E8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Liang RB; Swanson JMJ; Peng YX; Wikström M; Voth GA Multiscale simulations reveal key features of the proton-pumping mechanism in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2016, 113, 7420–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Chakrabarty S; Warshel A Capturing the energetics of water insertion in biological systems: The water flooding approach. Proteins: Struct., Funct., & Bioinf 2013, 81, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Yang W; Weng PJ; Gao Y A new paradigm of DNA synthesis: three-metal-ion catalysis. Cell Biosci. 2016, 6, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Samara NL; Yang W Cation trafficking propels RNA hydrolysis. Nat. Struct. Mol. Biol 2018, 25, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Benkovic SJ; Hammes GG; Hammes-Schiffer S Free energy landscape of enzyme catalysis. Biochem. 2008, 47, 3317–3321. [DOI] [PubMed] [Google Scholar]
- (116).Nashine VC; Hammes-Schiffer S; Benkovic SJ Coupled motions in enzyme catalysis. Curr. Opin. Chem. Biol 2010, 14, 644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Tokuriki N; Tawfik DS Protein Dynamics and Evolvability. Science 2009, 324, 203–207. [DOI] [PubMed] [Google Scholar]
- (118).Lee J; Goodey NM Catalytic Contributions from Remote Regions of Enzyme Structure. Chem. Rev 2011, 111, 7595–7624. [DOI] [PubMed] [Google Scholar]
- (119).Crean RM; Gardner JM; Kamerlin SCL Harnessing Conformational Plasticity to Generate Designer Enzymes. J. Am. Chem. Soc 2020, 142, 11324–11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Duarte F; Åqvist J; Williams NH; Kamerlin SCL Resolving Apparent Conflicts between Theoretical and Experimental Models of Phosphate Monoester Hydrolysis. J. Am. Chem. Soc 2015, 137, 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Zhang LF; Wang H; E WN Reinforced dynamics for enhanced sampling in large atomic and molecular systems. J. Chem. Phys 2018, 148, 124113. [DOI] [PubMed] [Google Scholar]
- (122).Helgaker T; Jorgensen P; Olsen J Molecular Electronic Structure Theory; J. Wiley & Sons: West Sussex, England, 2000. [Google Scholar]
- (123).Gao J Absolute Free Energy of Solvation from Monte Carlo Simulations Using Combined Quantum and Molecular Mechanical Potentials. J. Phys. Chem 1992, 96, 537–540. [Google Scholar]
- (124).Luzhkov V; Warshel A Microscopic Models for Quantum Mechanical Calculations of Chemical Processes in Solutions: LD/AMPAC and SCAAS/AMPAC Calculations of Solvation Energies. J. Comp. Chem 1992, 13, 199–213. [Google Scholar]
- (125).König G; Hudson PS; Boresch S; Woodcock HL Multiscale free energy simulations: An efficient method for connecting classical MD simulations to QM or QM/MM free energies using Non-Boltzmann Bennett reweighting schemes. J. Chem. Theo. Comp 2014, 10, 1406–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Ryde U How Many Conformations Need To Be Sampled To Obtain Converged QM/MM Energies? The Curse of Exponential Averaging. J. Chem. Theory Comput 2017, 13, 5745–5752. [DOI] [PubMed] [Google Scholar]
- (127).Roston D; Demapan D; Cui Q Extensive Free Energy Simulations Identify Water as the Base in Nucleotide Addition by DNA Polymerase. Proc. Natl. Acad. Sci. USA 2019, 116, 25048–25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Pohorille A; Jarzynski C; Chipot C Good Practices in Free Energy Calculations. J. Phys. Chem. B 2010, 114, 10235–10253. [DOI] [PubMed] [Google Scholar]
- (129).Boresch S; Woodcock HL Convergence of single-step free energy perturbation. Mol. Phys 2017, 115, 1200–1213. [Google Scholar]
- (130).König G; Brooks BR; Thiel W; York DM On the convergence of multi-scale free energy simulations. Mol. Simulat 2018, 44, 1062–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Giese TJ; York DM Development of a Robust Indirect Approach for MM → QM Free Energy Calculations That Combines Force-Matched Reference Potential and Bennett’s Acceptance Ratio Methods. J. Chem. Theory Comput 2019, 15, 5543–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Lu N; Kofke DA Accuracy of free-energy perturbation calculations in molecular simulation. II. Heuristics. J. Chem. Phys 2001, 115, 6866–6875. [Google Scholar]
- (133).Zhou Y; Ojeda-May P; Nagaraju M; Pu J Toward Determining ATPase Mechanism in ABC Transporters: Development of the Reaction Path-Force Matching QM/MM Method. Methods in Enzymol. 2016, 577, 185–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Kearns FL; Hudson PS; Woodcock HL; Boresch S Computing Converged Free Energy Differences between Levels of Theory via Nonequilibrium Work Methods: Challenges and Opportunities. J. Comput. Chem 2017, 38, 1376–1388. [DOI] [PubMed] [Google Scholar]
- (135).Kearns FL; Warrensford L; Boresch S; Woodcock HL The Good, the Bad, and the Ugly: “HiPen”, a New Dataset for Validating (S)QM/MM Free Energy Simulations. Molecules 2019, 24, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).König G; Brooks BR Correcting for the free energy costs of bond or angle constraints in molecular dynamics simulations. Biochim. Biophys. Acta 2015, 1850, 932–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Heimdal J; Ryde U Convergence of QM/MM free-energy perturbations based on molecular-mechanics or semiempirical simulations. Phys. Chem. Chem. Phys 2012, 14, 12592. [DOI] [PubMed] [Google Scholar]
- (138).Hudson PS; Boresch S; Rogers DM; Woodcock HL Accelerating QM/MM Free Energy Computations via Intramolecular Force Matching. J. Chem. Theory Comput 2018, 14, 6327–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Hudson PS; Woodcock HL; Boresch S Use of Interaction Energies in QM/MM Free Energy Simulations. J. Chem. Theory Comput 2019, 15, 4632–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Olsson MA; Soderhjelm P; Ryde U Converging Ligand-Binding Free Energies Obtained with Free-Energy Perturbations at the Quantum Mechanical Level. J. Comp. Chem 2016, 37, 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Ito S; Cui Q Multi-level Free Energy Simulation with a Staged Transformation Approach. J. Chem. Phys 2020, 153, 044115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Andricioaei I; Straub JE Generalized simulated annealing algorithms using Tsallis statistics: Application to conformational optimization of a tetrapeptide. Phys. Rev. E 1996, 53, R3055–R3058. [DOI] [PubMed] [Google Scholar]
- (143).Noé F; Olsson S; Köhler J; Wu H Boltzmann generators: Sampling equilibrium states of many-body systems with deep learning. Science 2019, 365, eaaw1147. [DOI] [PubMed] [Google Scholar]
- (144).Cui Q; Karplus M Allostery and cooperativity revisited. Protein Science 2008, 17, 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Nussinov R; Tsai CJ Allostery in Disease and in Drug Discovery. Cell 2013, 153, 293–305. [DOI] [PubMed] [Google Scholar]
- (146).Changeux JP Allostery and the Monod-Wyman-Changeux Model After 50 Years. Annu. Rev. Biophys 2012, 41, 103–133. [DOI] [PubMed] [Google Scholar]
- (147).Motlagh HN; Wrabl JO; Li J; Hilser VJ The ensemble nature of allostery. Nature 2014, 508, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Marzen S; Garcia HG; Phillips R Statistical Mechanics of Monod-Wyman-Changeux (MWC) Models. J. Mol. Biol 2013, 425, 1433–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Raman AS; White KI; Ranganathan R Origins of allostery and evolvability in proteins: a case study. Cell 2016, 166, 468–480. [DOI] [PubMed] [Google Scholar]
- (150).Taylor ND; Garruss AS; Moretti R; Chan S; Arbing MA; Cascio D; Rogers JK; Isaacs FJ; Kosuri S; Baker D et al. , Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 2016, 13, 177+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Kuhlman B; Bradley P Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol 2019, 20, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Yang G; Hong N; Baier F; Jackson CJ; Tokuriki N Conformational tinkering drives evolution of a promiscuous activity through indirect mutational effects. Biochem. 2016, 55, 4583–4593. [DOI] [PubMed] [Google Scholar]
- (153).Oue S; Okamoto A; Yano T; Kagamiyama H Redesigning the substrate specifity of an enzyme by cumulative effects of the mutations of non-active site residues. J. Biol. Chem 1999, 274, 2344–2349. [DOI] [PubMed] [Google Scholar]
- (154).Wrenbeck EE; Azouz LR; Whitehead TA Single-mutation fitness landscapes for an enzyme on multiple substrates reveal specificity is globally encoded. Nat. Comm 2016, 8, 15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Romero A; Arnold FH Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol 2009, 10, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Hong NS; Petrovic D; Lee R; Gryn’ova G; Purg M; Saunders J; Bauer P; Carr PD; Lin CY; Mabbitt PD et al. , The evolution of multiple active site configurations in a designed enzyme. Nat. Comm 2018, 9, 3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Campbell E; Kaltenbach M; Correy GJ; Carr PD; Porebski BT; Living-stone EK; Afriat-Jurnou L; Buckle AM; Weik M; Hollfelder F et al. , The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol 2016, 12, 944. [DOI] [PubMed] [Google Scholar]
- (158).Maria-Solano MA; Iglesias-Fernandez J; Osuna S Deciphering the Allosterically Driven Conformational Ensemble in Tryptophan Synthase Evolution. J. Am. Chem. Soc 2019, 141, 13049–13056. [DOI] [PubMed] [Google Scholar]
- (159).Buller AR; Brinkmann-Chen S; Romney DK; Herger M; Murciano-Calles J; Arnold FH Directed evolution of the tryptophan synthase β-subunit for standalone function recapitulates allosteric activation. Proc. Natl. Acad. Sci. USA 2015, 112, 14599–14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Buller AR; van Roye P; Cahn JKB; Scheele RA; Herger M; Arnold FH Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble. J. Am. Chem. Soc 2018, 140, 7256–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Isaksen GV; Aqvist J; Brandsdal BO Enzyme surface rigidity tunes the temperature dependence of catalytic rates. Proc. Natl. Acad. Sci. USA 2016, 113, 7822–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (162).Bae E; Phillips GN Jr., Roles of static and dynamic domains in stability and catalysis of adenylate kinase. Proc. Natl. Acad. Sci. USA 2006, 103, 2132–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Daily MD; Phillips GN; Cui Q Interconversion of Functional Motions between Mesophilic and Thermophilic Adenylate Kinases. PLoS Comput. Biol 2011, 7, e1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Nguyen V; Wilson C; Hemberger M; Stiller JB; Agafonov RV; Kutter S; English J; Theobald DL; Kern D Evolutionary drivers of thermoadaptation in enzyme catalysis. Science 2017, 355, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Arcus VL; Mulholland AJ Temperature, Dynamics, and Enzyme-Catalyzed Reaction Rates. Annu. Rev. Biophys 2020, 49, 163–180. [DOI] [PubMed] [Google Scholar]
- (166).Arcus VL; Prentice EJ; Hobbs JK; Mulholland AJ; van der Kamp MW,; Pudney CR; Parker EJ; Schipper LA On the Temperature Dependence of Enzyme-Catalyzed Rates. Biochem. 2016, 55, 1681–1688. [DOI] [PubMed] [Google Scholar]
- (167).Socan J; Purg M; Aqvist J Computer simulations explain the anomalous temperature optimum in a cold-adapted enzyme. Nat. Comm 2020, 11, 2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Klinman JP; Kohen A Hydrogen Tunneling Links Protein Dynamics to Enzyme Catalysis. Annu. Rev. Biochem 2013, 82, 471–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Benkovic SJ; Hammes-Schiffer S A perspective on enzyme catalysis. Science 2003, 301, 1196–1202. [DOI] [PubMed] [Google Scholar]
- (170).Daniel RM; Danson MJ A new understanding of how temperature affects the catalytic activity of enzymes. Trends Biochem. Sci 2010, 35, 584–591. [DOI] [PubMed] [Google Scholar]
- (171).Aqvist J; Socan J; Purg M Hidden Conformational States and Strange Temperature Optima in Enzyme Catalysis. Biochem. 2020, 59, 3844–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).van der Kamp MW,; Prentice EJ; Kraakman KL; Connolly M; Mulholland AJ; Arcus VL Dynamical origins of heat capacity changes in enzymecatalysed reactions. Nat. Comm 2018, 9, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Bunzel HA; Anderson JLR; Hilvert D; Arcus VL; van der Kamp MW,; Mulholland AJ Evolution of dynamical networks enhances catalysis in a designer enzyme. bioRxiv 2020, 10.1101/2020.08.21.260885. [DOI] [PubMed] [Google Scholar]
- (174).Bunzel HA; Kries H; Marchetti L; Zeymer C; Mittl PRE; Mulholland AJ; Hilvert D Emergence of a Negative Activation Heat Capacity during Evolution of a Designed Enzyme. J. Am. Chem. Soc 2019, 141, 11745–11748. [DOI] [PubMed] [Google Scholar]
- (175).Packer MS; Liu DR Methods for the directed evolution of proteins. Nat. Rev. Genet 2015, 16, 379–394. [DOI] [PubMed] [Google Scholar]
- (176).Chen K; Arnold FH Engineering new catalytic activities in enzymes. Nat. Cata 2020, 3, 203–213. [Google Scholar]
- (177).Ravikumar A; Arzumanyan GA; Obadi MKA; Javanpour AA; Liu CC Scalable, Continuous Evolution of Genes at Mutation Rates Above Genomic Error Thresholds. Cell 2018, 175, 1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Rix G; Watkins-Dulaney EJ; Almhjell PJ; Boville CE; Arnold FH; Liu CC Scalable, continuous evolution for the generation of diverse enzyme variants encompassing promiscuous activities. Nat. Comm 2020, 11, 5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Gagliardi L; Truhlar DG; Li Manni G,; Carlson RK; Hoyer CE; Bao JWL Multiconfiguration Pair-Density Functional Theory: A New Way To Treat Strongly Correlated Systems. Acc. Chem. Res 2017, 50, 66–73. [DOI] [PubMed] [Google Scholar]
- (180).Chan GKL; Sharma S The Density Matrix Renormalization Group in Quantum Chemistry. Annu. Rev. Phys. Chem 2011, 62, 465–481. [DOI] [PubMed] [Google Scholar]
- (181).Grisafi A; Ceriotti M Incorporating long-range physics in atomic-scale machine learning featured. J. Chem. Phys 2019, 151, 204105. [DOI] [PubMed] [Google Scholar]
- (182).Cui Q Quantum Mechanical Methods in Biochemistry and Biophysics. J. Chem. Phys 2016, 145, 140901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Raman S Systems Approaches to Understanding and Designing Allosteric Proteins. Biochem. 2018, 57, 376–382. [DOI] [PubMed] [Google Scholar]
- (184).Leander M; Yuan YC; Mager A; Cui Q; Raman S Functional Plasticity and Evolutionary Adaptation of Allosteric Regulation. Proc. Natl. Acad. Sci. USA 2020, 117, 25445–25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Swiderek K; Tuñón I; Moliner V; Bertran J Computational strategies for the design of new enzymatic functions. Arch. Biochem. Biophys 2015, 582, 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Sherrill CD; Manolopoulos DE; Martinez TJ; Michaelides A Electronic structure software. J. Chem. Phys 2020, 153, 070401. [DOI] [PubMed] [Google Scholar]
- (187).Zen A; Brandenburg JG; Klimes J; Tkatchenko A; Alfe D; Michaelides A Fast and accurate quantum Monte Carlo for molecular crystals. Proc. Natl. Acad. Sci. USA 2018, 115, 1724–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Hermann J; Schatzle Z; Noe F Deep-neural-network solution of the electronic Schrodinger equation. Nat. Chem 2020, 12, 891. [DOI] [PubMed] [Google Scholar]
- (189).Google AI Quantum and Collaborators, Hartree-Fock on a superconducting qubit quantum computer. Science 2020, 369, 1084–1089. [DOI] [PubMed] [Google Scholar]
- (190).McArdle S; Endo S; Aspuru-Guzik A; Benjamin SC; Yuan X Quantum computational chemistry. Rev. Mod. Phys 2020, 92, 015003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


