Abstract
During last few decades, role of microbiota and its importance in several diseases has been a hot topic for research. The microbiota is considered as an accessory organ for maintaining normal physiology of an individual. These microbiota organisms which normally colonize several epithelial surfaces are known to secrete several small molecules leading to local and systemic effects on normal biological processes. The role of microbiota is also established in carcinogenesis as per several recent findings. The effects of microbiota on cancer is not only limited to their contribution in oncogenesis, but the overall susceptibility for oncogenesis and its subsequent progression, development of coinfections, and response to anticancer therapy is also found to be affected by microbiota. The information about microbiota and subsequent contributions of microbes in anticancer response motivated researchers in development of microbes-based anticancer therapeutics. We provided current status of microbiota contribution in oncogenesis with special reference to their mechanistic implications in different aspects of oncogenesis. In addition, the mechanistic implications of bacteria in anticancer therapy are also discussed. We conclude that several mechanisms of microbiota-mediated regulation of oncogenesis is known, but approaches must be focused on understanding contribution of microbiota as a community rather than single organisms-mediated effects.
Keywords: Carcinogenesis, Microbiome, Pharmacomicrobiomics, Anticancer, Infection
Introduction
Recently a large number of studies are accumulating linking role of microbiota with normal host physiology and subsequent disease progression. The disruption of normal microbiota, known as microbiota dysbiosis contributes to development of several clinical conditions related to physical to behavioral aspects [1, 2]. Plethora of literature is supporting the role of microbiota in several physical abnormalities including, gastrointestinal, neurological to cardiovascular, etc. [3, 4]. Moreover, studies are linking role of microbiota with cancer and supporting its contribution in almost all aspect of carcinogenesis ranging from cancer susceptibility and progression to response for anticancer therapy [5, 6]. Literature is suggesting that correction of microbiota dysbiosis through beneficial probiotic bacteria can be beneficial in a large number of cancer types [7, 8]. In addition, several microbes have been found with anticancer potential and are being developed for effective anticancer therapy [9].
Though, several complexities are creating bottlenecks in integrating such therapeutic modalities in routine anticancer management practices, multiple strategies are devised to utilize information obtained from microbiota-related experiments for management of cancer. These complexities including, geographical and inter individual variability of microbiota, identification of key microbiota component affecting carcinogenic process, and identification of proper mechanistic information leading to microbiota-mediated oncogenic transformation. Herein, we draw attention to the mechanistic implications of microbiota in influencing different aspects of oncogenesis and its subsequent influence on development of anticancer management strategies.
Microbiota and oncogenesis
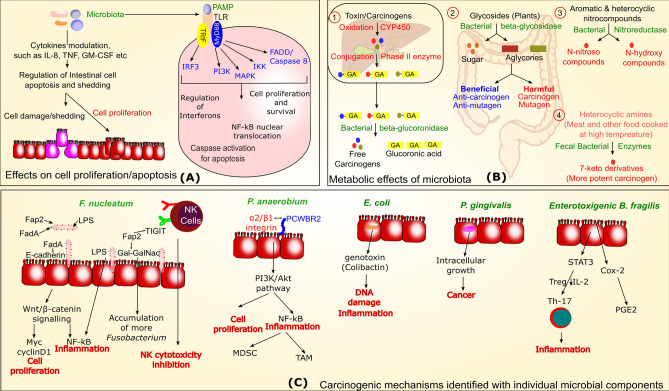
The role of microbiota in oncogenesis is widely recognized in a number of studies. It is estimated that more than 20% of cancer cases are associated with infectious agents [10]. Several microorganisms are specifically linked with development of cancer, like H, pylori which is declared as a class I carcinogen by the World Health Organization and involved in gastric adenocarcinoma and mucosa associated lymphoid tissue (MALT) lymphoma [11]. Chlamydia trachomatis, Escherichia coli, Salmonella enterica are also suspiciously involved in cervical, colorectal, and gallbladder cancer, respectively [12–17]. In addition, microbiota dysbiosis involving modulation of several microbiota organisms is also linked with carcinogenesis [18]. The antibiotics-mediated microbial dysbiosis is also known to play important role in tumorigenesis in some recent studies [19]. Several mechanisms of microbiota dysbiosis-mediated tumorigenesis are also proposed recently (Fig. 1), though the collective information about contribution of each microbiota components and their subsequent involvement in carcinogenesis is still lacking. Several studies are available indicating causal role of microbiota dysbiosis and cancer. For example, the microbiota of cancer-associated area is found to be different from microbiota occupying nearby healthy mucosa [20, 21]. Several articles are available on types of microbiota components modulation in different cancer, but their specific links with carcinogenesis needs more attention to understand their mechanistic implications. Following section indicates some mechanisms possibly involved in microbiota-mediated cancer etiology. Table 1 also lists some specific microbial components and their mechanistic involvement in carcinogenesis.
Fig. 1.
Role of microbiota and individual microbiota components in carcinogenesis. The figure is divided in three sections and cancer-associated mechanisms are generally shown with red color while microbial components are shown with green color. Section A indicates influence of microbiota on cell growth or apoptosis related process, while section B and C indicate towards metabolic influence of microbiota and mechanisms associated with individual microbes on carcinogenesis, respectively
Table 1.
The mechanistic implication of microbes in carcinogenesis
| Sr. no | Bacteria | Cancer | Suggested key mechanisms | References |
|---|---|---|---|---|
| 1 | Chlamydophila psittaci | Ocular adenxal MALT lymphoma |
Clonal selection of MALT for lymphoma development, chromosomal aberration caused by either genetic instability or oxidative DNA damage, affecting NF-kB pathway leading to anti-apoptotic effects Additional risk factor including autoimmune diseases |
[84, 85] |
| 2 | Porphyromonas gingivalis | Oral cancer | Receptor upregulation on OSCC cells, EMT transition of normal oral epithelial cells, activation of IL-8 and MMP-9, inhibition of apoptosis and acceleration of cell cycle, conversion of ethanol to carcinogenic acetaldehyde | [86] |
| 3 | Helicobacter pylori | Gastric cancer | Inflammation induction through epithelial cell death and consequent repair of remaining cells leading to increase cell survival and proliferation and resultant precancerous lesions. Direct effects through bacterial effectors, such as cagA, vacA, and omp activating cell signaling pathways including PI3K/Akt, Ras, Raf, ERK, JAK/STAT, etc., leading to uncontrolled cell proliferation | [87, 88] |
| 4 | Mycobacterium tuberculosis | Lung cancer | DNA damage, production of epidermal growth factor (epiregulin). PD-1/PD-L1 pathway modulating T cell immune response mediating tumor metastasis | [78, 89] |
| 5 | Chlamydia (Chlamydophila) pneumoniae | Lung cancer | Inflammation mediated cell and DNA damage and consequent repair of cell injury contributes to increased cell proliferation and cancer. Superoxide radicals, TNF, IL-8, and IL1β secreted by monocytes in response to infection also contributes to cell and DNA damage and resultant carcinogenesis | [90] |
| 6 | Salmonella typhi | Gallbladder cancer | Chronic inflammation, typhoid toxin causes DNA damage and cell cycle alterations | [79] |
| 7 | Streptococcus bovis | Colon cancer | Inflammatory cytokines (such as IL1β, IL-8, TNF-alpha, and IL-6) may lead to formation of free radicals causing DNA alterations and cancer. Bacteria also degrade anticancer substances, such as tannic acid present in diet and contribute to cancer | [91, 92] |
| 8 | Campylobacter jejuni | Intestinal lymphoma | The CDT of bacteria cause dsDNA breaks in germinal centre B cells. Mutational events involving Pax5 and other oncogenes can lead to neoplastic changes | [93] |
| 9 | Fusobacterium nucleatum | Colorectal cancer | Induction of cell proliferation through Wnt/β-catenin signaling. Inflammation regulation and inhibition of natural killer cell cytotoxicity | [94] |
| 10 | Bacteroides fragilis | Colon cancer | BFT (B. fragilis toxin) cleaves cell surface protein E-cadherin and its cytoplasmic domain associate with β-catenin. The loss of E cadherin stimulate β-catenin signaling, induce c-myc and IL-8. Cause oxidative DNA damage, epithelial barrier damage, STAT3/TH17 immune response | [95] |
| 11 | Citrobacter rodentium | Colorectal cancer | Colonic crypt hyperplasia is regulated through Wnt/β-catenin, PI3K, and Notch pathway. MEK/ERK/NF-kB regulates inflammation. Other related mutagenic effects contribute to neoplasia | [96] |
| 12 | Escherichia coli | Colorectal cancer | Production of genotoxin leading to DNA damage and cell cycle modulation. Chronic inflammation and possibility to affect DNA repair | [13] |
| 13 | Chlamydia trachomatis | Cervical cancer | Acts as a cofactor with HPV for cancer development. Induces cell proliferation and inhibition of apoptosis | [12, 97] |
*EMT-epithelial to mesenchymal transition, IL-8 Interleukin8, MMP9 Mettaloproteinase 9, CDT Cytolethal Distending Toxin, OSCC oral squamous cell carcinoma,
Metabolic influence
The gut microbiota is primarily involved in providing energy from non-digested food substances and therefore the dietary components are a major factor deciding microbiota composition. Under certain situations, these microbiota-mediated metabolic products have ability to influence carcinogenicity and these effects range from activation and synthesis of carcinogen, to removal of carcinogen [22]. Several bacterial enzymes are involved in cancer and indicate towards involvement of microbiota in carcinogenesis. Figure 1B represents microbiota enzymes and their metabolic influence on carcinogenesis. It is found that prebiotics are known to increase beneficial Bifidobacteria and suppress these carcinogen metabolizing enzymes activities [23]. The probiotics are beneficial bacteria known for their ability to provide several anticancer effects and a number of articles are available for anticancer effects of probiotics [7, 8]. Therefore increasing the number of beneficial bacteria through fecal microbiota transplant is a well-known strategy for management of several gastrointestinal ailments [24]. The role of metabolic influence of microbiota is also evident through strong influence of dietary pattern on carcinogenesis. The excess energy intake in comparison to normal requirement is suggested to be linked with human cancer [25] and microbiota plays important role in this process through regulation of metabolic process [26].
Influence on cell growth
Development of cancer is directly linked with abnormal cell proliferation and it can be ranged from increased cellular proliferation to inhibition of cell death. The microbiota is known to regulate cell proliferation and regeneration under various conditions [27]. It has been found a long time ago that addition of dietary fiber in rat increases intestinal cell proliferation [28]. In addition, restriction of intestinal nutrients is known to induce intestinal atrophy even after providing nutrients through parenteral route [29]. The role of microbiota in digestion of dietary fiber and production of necessary metabolites indicate towards their involvement in regulating cell proliferation through this mechanism. It is known that certain bacterial fermentative metabolites, such as short chain fatty acid (SCFA) produced from nutrients are having ability to regulate cell proliferation [30, 31]. Several bacteria are known to produce certain proteins known as nucleomodulins with the ability to alter normal nuclear function and thereby influence cell growth. These nucleomodulins are also identified in bacteria suspiciously associated with cancer, thereby indicate towards potential involvement of these proteins in carcinogenesis [32]. In contrast, microbiota also has ability to both promote and inhibit cell death through regulating apoptosis and therefore contributing in intestinal mucosal epithelial cell inflammation and integrity [33]. It has been identified that germ free newborn mice has reduced interleukin 1β, tumor necrosis factor (TNF) and altered cell death [34]. Though this study was designed to evaluate effect of microbiota on brain development, but their results demonstrating microbiota-mediated regulation of major apoptotic regulator TNF demonstrate its ability to regulate cell death. Supporting this notion, probiotics bacteria Lactobacillus rhamnosus GG is able to inhibit cytokine-mediated apoptosis through activation of anti-apoptotic Akt/protein kinase B and inhibition of pro-apoptotic TNF, p38/MAPK, IL-1α or IFN-γ [35]. L. rhamnosus GG is also known to produce two proteins (p75 and p40) which activate Akt and promote colon epithelial cell growth and reduce TNF-mediated epithelial cell damage [36].
The pathogen associated molecular patterns (PAMP) of microbiota components are recognized through Toll like receptor (TLR) present on enterocytes and other surfaces to mount immunologic response. The toll-like receptor are known to show modulated expression during carcinogenesis and this is also considered to contribute in infectious complications among cancer patients [37]. The vice versa modulated TLR expression in response to different microbiota composition can also regulate cellular proliferation and death in multiple ways. The central TLR signaling pathways involving MAPK and PI3K play important role in regulating cell proliferation through TLR [38]. Figure 1A indicates different mechanisms through which microbiota can regulate cell proliferation and apoptosis.
Cancer therapy-associated microbiota modulation and complications
Anticancer therapy used to manage cancer also creates several effects on host microbiota and therefore generate several associated complications. These effects are mediated through multiple mechanisms, including dietary alterations, necessary surgical interventions and use of antibiotics for preventing post-surgical and other infections in addition to effects caused by anticancer drugs [39]. Therefore the reverse effect of microbiota modulation on anticancer therapy is also gaining importance and studies are conducted to understand and utilize this attribute for improving anticancer therapy treatment outcomes. The recent concept of pharmacomicrobiomics is considering effects of microbiota in drug response including anticancer therapy [40]. A common antimetabolite methotraxate is used in a number of clinical diseases including cancers, it has been found to inhibit several representative gut bacteria and alter microbiota composition [41]. An in vivo study on gut microbiota of colorectal cancer mouse model showed that anticancer drug 5-Fluorouracil (5-FU) can change microbiota diversity and composition. In addition, antibiotics-mediated gut microbiota disruption also contributed to reduced antitumor efficacy of 5-FU [42]. An important anticancer drug cyclophosphamide is also known to alter composition of microbiota in intestine and promote certain gram positive bacterial translocation to secondary lymphoid organs, where these bacteria stimulate production of pathogenic Th17 cells Th1 response [43]. Several other anticancer drugs are known to cause diarrhea through multiple mechanisms including killing of beneficial bacteria helping in digestion [44]. In contrast, microbiota components are also known to alter the response toward chemotherapeutic drugs and have been reviewed extensively. Microbiota components are able to modulate anticancer therapy outcomes through multiple mechanisms including resistance to anticancer drugs and immune check points inhibitors, modulation of metabolism, etc. Therefore it is suggested that these microbiota organisms can also serve as a prognostic and diagnostic marker for cancer [45]. Several microbial components itself possess anticancer activity and it is mediated by multiple mechanisms. Some microbes are known for production of anticancer substances [46], activation of antitumor immune response, and modulation of signaling pathways, etc. [6, 47]. Few bacteria known to possess anticancer activity are mentioned in Table 2 with their mechanistic implications.
Table 2.
Mechanistic implications of bacteria in prevention of cancer
| Sr. no | Bacteria | Suggested key anticancer mechanism | References |
|---|---|---|---|
| 1 | Listeria monocytogenes | The intracellular growth without extracellular cell to cell spread makes it an ideal vector for anticancer therapy. Acts as an immunomodulator to enhance anticancer T cell immune response | [98, 99] |
| 2 | Bifidobacterium spp. | Biotrasformation and production of antitumor metabolites from nutrition and drugs, competitive advantage in cancer microenvironment by exclusion of harmful microbiota and pathogens, modulation of cancer-associated cytokines and genes | [100] |
| 3 | Salmonella typhimurium | Attenuated bacterium is suggested as a delivery vehicle for anticancer therapy due to selective tumor targeting and anticancer response | [101] |
| 4 | Streptococcus pyogenes | Direct tumor cell lysis and activation of immune response by bacterial components leads to cancer cell death | [102] |
| 5 | Clostridium novyi | Activation of host antitumor immune response and direct destruction of tumor cell. Works on hypoxic area (which is generally difficult to treat with conventional radio and chemotherapy) due to anaerobic nature of bacteria | [103, 104] |
| 6 | Bacillus calmette Guerin | Used as immunotherapy for prevention of cancer relapse and progression. Elevate the level of IL-2 and mediate anticancer activity by type 1 immune response | [105] |
| 7 | Serratia marcescens | Produce several metabolites such as, extracellular metalloproteinase serralysin, and prodigiosin with anticancer potential | [106, 107] |
| 8 | Corynebacterium diptheriae | Diptheria exotoxin possess antitumor activity and used as a immunotoxins consisting toxin with additional targeting element | [108] |
Microbiota, inflammation and cancer-associated signaling pathways
In addition to above mentioned mechanisms, microbiota is also known to alter several important signaling pathways leading to progression or inhibition of cancer. Some of these signaling pathways are already discussed in earlier section, but some other bacteria-specific signaling events leading to carcinogenesis are mentioned in Fig. 1C. The signaling pathways are regulating multiple mechanisms contributing to carcinogenesis including immune regulation. Microbiota is known to mediate development and regulation of immune system and therefore it can influence outcome of anticancer immunotherapy [48]. The role of microbiota and probiotics are also proposed in management of inflammatory and cellular immune response in COVID-19 [49, 50]. Moreover, the role of immune regulation in cancer and its management is a widely studied aspect. Antibodies against CTLA-4 are successfully used in anticancer immunotherapy but its efficacy depends on gut microbiota organisms Bacteroides spp. [51]. The role of commensal bacteria in influencing anticancer activity of immunotherapy is also reviewed [52] and it was suggested that normal intestinal microbiota supports anticancer therapy while this attribute is missing with dysbiotic microbiota and inflammation regulation is considered as an important mechanisms in this process [53]. For more than one century, inflammation is considered as a key mechanism for carcinogenesis after detection of leukocytes in cancer tissues [54]. Recent studies are also finding strong links between chronic inflammation and cancer risk [55]. The microbiota components are known to up regulate several inflammatory cytokines and it has been reviewed in other articles [56]. Human functional genomics project indicated that fungal and bacterial agents are significantly associated with inflammatory cytokine response [57]. It is found that microbiota can promote cytokine production through engaging microbial metabolite sensor receptors that are highly expressed on inflammatory cells [58]. The study of antibiotic-mediated microbiota reduction and subsequent effects on pancreatic, colon, and melanoma tumor models revealed that microbiota depletion increases IFNγ producing T cell and reduces IL17A and IL10 producing T cell and reduces tumor burden in all models except Rag1-knockout mice [59]. IL-17 and IL-10 are considered as major immune regulators in a variety of carcinogenesis with a range of observations [60, 61], the role of microbiota in regulating these cytokines indicate towards their involvement in this process. Targeting of harmful microbiota components through antibiotics further boosts immune system and aids in suppression of cancer development. Several other bacteria are studied for their potential involvement in cancer with varied mechanisms. Among these, Escherichia coli is also found to be linked with colorectal cancer with a range of observations. It is found that increase of genotoxin producing microorganisms including E. coli can contribute to chronic inflammation-mediated colorectal cancer. It is suggested that colibactin induced host DNA damage contribute to colorectal cancer etiology [62]. Moreover, mutY is a DNA repair gene in E. coli and its human homolog MUTYH is linked with colorectal cancer. This homology of both DNA repair genes and intracellular localization of E. coli in colonic epithelium during chronic inflammation raises suspicion that both DNA repair homologous proteins can compete and affect host cell DNA repair and therefore hypothesized for E. coli-mediated colorectal cancer [16]. Another well-studied bacteria for its involvement in carcinogenesis is Fusobacterium nucleatum, which is found to be implicated in colorectal cancer [63, 64], gastric cancer [65], esophageal cancer [66], and head and neck squamous cell carcinoma [67]. F. nucleatum is having certain regulators known to mediate cancerous changes, for example Fap2 protein of F. nucleatum interacts with TIGIT receptor present on natural killer (NK) cells and inhibit their cytotoxicity and subsequently contribute to tumor evasion [68]. Moreover, Fap2 also leads to Fusobacterium abundance in CRC which over-express Gal-GalNAc and binds to Fap2 protein, even though hematogenous route [69] and further contribute to its carcinogenic ability. Another F. nucleatum protein FadA bind to E-cadherin on CRC cells and induces β -catenin signaling leading to regulation of inflammatory and oncogenic response. This study also identified that synthetic peptide inhibit FadA—E cadherin-mediated CRC progression, inflammatory and oncogenic response and the level of FadA is also found to be higher in adenomas and adenocarcinoma than normal colon tissues [70]. The role of F. nucleatum in carcinogenesis is also reviewed and it has been suggested that its LPS can stimulate inflammatory cytokines leading to proinflammatory environment promoting tumor progression. The activation of ß-catenin signaling also activates Wnt signaling, Myc and cyclin D1 oncogenes contributing to cancer cell proliferation [71].
Another anaerobic bacterium Peptostreptococcus anaerobius selectively enriched in CRC microbiota. It is known to bind α2/β1 integrin (overexpressed on CRC cells) through putative cell wall binding repeat 2 (PCWBR2) surface protein present on bacteria. This interaction activate PI3K-Akt pathway through phospho-focal-adhesion kinase and stimulate increased cell proliferation, NF-kB activation and resultant proinflammatory events, such as interleukin-10, interferon-γ production and involvement of tumour-associated macrophages (TAM), myeloid-derived suppressor cells (MDSC), and granulocytic tumour-associated neutrophils (GTAM) [72]. The role of PI3K-Akt pathway is already suggested in several other microbes-associated cancer, such as Salmonella typhi is suggested to be involved in gallbladder carcinoma through this important pathway in addition to several other events [73].
Enteotoxigenic Bacteroides fragilis (ETBF) is another bacterium which produces BF toxin (BFT) contributing to diarrhea, inflammatory bowel disease and colon cancer [74]. BFT produced by ETBF promotes inflammation through expression of cyclooxygenase (COX-2) releasing PGE2, activation of STAT3 signaling, and degradation of E-cadherin leading to upregulation of spermine oxidase-mediated DNA damage and carcinogenesis [75].
Porphyromonas gingivalis is found to be associated with increased pancreatic cancer risk. Though it is an oral bacteria but detected in human pancreatic cancer as an intracellular pathogen. Its intracellular residence is enhanced by hypoxia, which is an important characteristic of pancreatic cancer and intracellular persistence is directly related to increased tumor cell proliferation [76]. Some studies have indicated role of chronic Mycobacterium tuberculosis (Mtb) infection in lung cancer through induction of several mechanisms including inflammation [77]. It is demonstrated that Mtb induces lung specific cell dysplasia, squamous cell carcinoma. The Mtb-infected macrophages induces DNA damage in surrounding tissue and produces epiregulin (an epidermal growth factor) contributing to tumorigenesis [78]. Salmonella enterica subsp. enterica serovar Typhi (S. typhi) is also found to be linked with gall bladder carcinoma risk with its ability to form biofilm and production of toxin contributing to DNA damage and cell cycle alterations [79]. Similarly, Helicobacter pylori is a well-known bacteria recognized as carcinogen and contribute to carcinogenesis through multiple mechanisms. H. pylori induces inflammation, DNA damage, DNA methylation, epithelial mesenchymal transition, and direct cell proliferation through activation of various signaling events, like PI3K/Akt pathway [80]. These organisms are linked to certain diseases and do not form components of general microbiota, but their potentials in carcinogenesis provide indication of bacterial involvement in cancer progression.
Conclusion
The role of microbiota in cancer is a relatively new aspect and need more investigations to understand collective effect of microbiota components in carcinogenesis process. Discrete studies are arising understanding role of individual microbiota component in cancer progression, prevention and management. In contrast, microbiota works as an ecosystem where each component is contributing to the process of affecting normal host physiology. Therefore, we need a model to understand contribution of microbiota as a community in addition to understanding role of individual microbiota component in carcinogenic process. The recent development of next generation sequencing technologies with omics analysis can help us to develop such models. Several recent approaches such as microarray, metagenomics analysis coupled with meta-trascriptomics analyses can indicate about microbial community composition in addition to their influence on normal host physiology and subsequent effects on carcinogenesis. System biological approaches of host–pathogen interactions analyses can also aid in these objectives through reducing time and labor required for such large-scale analyses of microbial community and their influence on disease progression [81–83]. Nonetheless, current findings and observations are indicating that microbiota can contribute to carcinogenesis through a variety of mechanisms and understanding of these mechanisms can help us to plan suitable preventive and therapeutic strategy for cancer. In addition, mechanistic identification of anticancer activity of microbes is paving the way to develop suitable therapeutics for management of cancer. Summarily, identification of mechanistic implication of microbiota in carcinogenesis and its prevention can provide us interventions for prevention and management of a variety of cancer.
Author contributions
AAK conceptualization; data curation; formal analysis; project administration; resources; visualization; roles/writing—original draft; writing—review and editing, ATS data curation; formal analysis; investigation; roles/writing—review and editing, HS conceptualization; data curation; formal analysis; roles/writing—writing—review and editing, PC formal analysis; project administration; resources; supervision; validation; roles/writing—review and editing.
Funding
None.
Declarations
Conflict of interest
The authors have declared no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Data availability
All relevant data are available with manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Afzal M, Mazhar SF, Sana S, Naeem M, Rasool MH, Saqalein M, et al. Neurological and cognitive significance of probiotics: a holy grail deciding individual personality. Future Microbiol. 2020;15:1059–1074. doi: 10.2217/fmb-2019-0143. [DOI] [PubMed] [Google Scholar]
- 2.Markowiak P, Slizewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017 doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazemian N, Mahmoudi M, Halperin F, Wu JC, Pakpour S. Gut microbiota and cardiovascular disease: opportunities and challenges. Microbiome. 2020;8(1):36. doi: 10.1186/s40168-020-00821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hills RD, Jr, Pontefract BA, Mishcon HR, Black CA, Sutton SC, Theberge CR. Gut microbiome: profound implications for diet and disease. Nutrients. 2019 doi: 10.3390/nu11071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome. 2018;6(1):92. doi: 10.1186/s40168-018-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan AA, Shrivastava A, Khurshid M. Normal to cancer microbiome transformation and its implication in cancer diagnosis. Biochem Biophys Acta. 2012;1826(2):331–337. doi: 10.1016/j.bbcan.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Khan AA, Nema V, Khan Z. Current status of probiotics for prevention and management of gastrointestinal cancers. Expert Opin Biol Therapy. 2020 doi: 10.1080/14712598.2021.1828858. [DOI] [PubMed] [Google Scholar]
- 8.Khan AA, Khurshid M, Khan S, Alshamsan A. Gut microbiota and probiotics: current status and their role in cancer therapeutics. Drug Dev Res. 2013;74(6):365–375. doi: 10.1002/ddr.21087. [DOI] [Google Scholar]
- 9.Sedighi M, Zahedi Bialvaei A, Hamblin MR, Ohadi E, Asadi A, Halajzadeh M, et al. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 2019;8(6):3167–3181. doi: 10.1002/cam4.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zur HH. The search for infectious causes of human cancers: where and why. Virology. 2009;392(1):1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench. 2015;8(Suppl 1):S6–S14. [PMC free article] [PubMed] [Google Scholar]
- 12.Khan AA, Abuderman AA, Ashraf MT, Khan Z. Protein-protein interactions of HPV-Chlamydia trachomatis-human and their potential in cervical cancer. Future Microbiol. 2020;15:509–520. doi: 10.2217/fmb-2019-0242. [DOI] [PubMed] [Google Scholar]
- 13.Khan AA, Khan Z, Malik A, Kalam MA, Cash P, Ashraf MT, et al. Colorectal cancer-inflammatory bowel disease nexus and felony of Escherichia coli. Life Sci. 2017;180:60–67. doi: 10.1016/j.lfs.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Khan AA, Khan Z, Malik A, Shrivastava A, Jain SK, Alshamsan A. Computational prediction of Escherichia coli proteins host subcellular targeting and their implications in colorectal cancer etiology. Cancer Lett. 2015;364(1):25–32. doi: 10.1016/j.canlet.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Khan AA. In silico prediction of Escherichia coli proteins targeting the host cell nucleus, with special reference to their role in colon cancer etiology. J Comput Biol J Comput Mol Cell Biol. 2014;21(6):466–475. doi: 10.1089/cmb.2014.0001. [DOI] [PubMed] [Google Scholar]
- 16.Khan AA, Cash P. E. coli and colon cancer: is mutY a culprit? Cancer Lett. 2013;341(2):127–131. doi: 10.1016/j.canlet.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Tewari M, Mishra RR, Shukla HS. Salmonella typhi and gallbladder cancer: report from an endemic region. Hepat Pancreat Dis Int HBPD INT. 2010;9(5):524–530. [PubMed] [Google Scholar]
- 18.Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16(10):406. doi: 10.1007/s11912-014-0406-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins SV, Robeson MS, 2nd, Griffin RJ, Quick CM, Siegel ER, Cannon MJ, et al. Gastrointestinal tract dysbiosis enhances distal tumor progression through suppression of leukocyte trafficking. Can Res. 2019;79(23):5999–6009. doi: 10.1158/0008-5472.CAN-18-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, et al. Microbial dysbiosis is associated with human breast cancer. PLoS ONE. 2014;9(1):e83744. doi: 10.1371/journal.pone.0083744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowland RHIR. Metabolic activities of the gut microflora in relation to cancer. Microb Ecol Health Dis. 2000;12(2):179–185. doi: 10.1080/089106000750060431. [DOI] [Google Scholar]
- 23.Saito Y, Takano T, Rowland I. Effects of soybean oligosaccharides on the human gut microflora in in vitro culture. Microb Ecol Health Dis. 1992;5(2):105–110. doi: 10.3109/08910609209141296. [DOI] [Google Scholar]
- 24.Gupta S, Allen-Vercoe E, Petrof EO. Fecal microbiota transplantation: in perspective. Ther Adv Gastroenterol. 2016;9(2):229–239. doi: 10.1177/1756283X15607414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willett WC. Nutrition and cancer. Salud Publ Mex. 1997;39(4):298–309. doi: 10.1590/s0036-36341997000400008. [DOI] [PubMed] [Google Scholar]
- 26.Heiss CN, Olofsson LE. Gut microbiota-dependent modulation of energy metabolism. J Innate Immun. 2018;10(3):163–171. doi: 10.1159/000481519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abo H, Chassaing B, Harusato A, Quiros M, Brazil JC, Ngo VL, et al. Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration. Nat Commun. 2020;11(1):513. doi: 10.1038/s41467-019-14258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs LR. Effects of dietary fiber on mucosal growth and cell proliferation in the small intestine of the rat: a comparison of oat bran, pectin, and guar with total fiber deprivation. Am J Clin Nutr. 1983;37(6):954–960. doi: 10.1093/ajcn/37.6.954. [DOI] [PubMed] [Google Scholar]
- 29.Shaw D, Gohil K, Basson MD. Intestinal mucosal atrophy and adaptation. World J Gastroenterol. 2012;18(44):6357–6375. doi: 10.3748/wjg.v18.i44.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakata T, von Engelhardt W. Stimulatory effect of short chain fatty acids on the epithelial cell proliferation in rat large intestine. Comp Biochem Physiol A Comp Physiol. 1983;74(2):459–462. doi: 10.1016/0300-9629(83)90631-x. [DOI] [PubMed] [Google Scholar]
- 31.Kaunitz JD, Akiba Y. Control of intestinal epithelial proliferation and differentiation: the microbiome, enteroendocrine L cells, telocytes, enteric nerves, and GLP. Too Dig Dis Sci. 2019;64(10):2709–2716. doi: 10.1007/s10620-019-05778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan AA, Khan Z. Bacterial nucleomodulins and cancer: an unresolved enigma. Transl Oncol. 2021;14(1):100922. doi: 10.1016/j.tranon.2020.100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausmann M. How bacteria-induced apoptosis of intestinal epithelial cells contributes to mucosal inflammation. Int J Inflamm. 2010;2010:574568. doi: 10.4061/2010/574568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castillo-Ruiz A, Mosley M, George AJ, Mussaji LF, Fullerton EF, Ruszkowski EM, et al. The microbiota influences cell death and microglial colonization in the perinatal mouse brain. Brain Behav Immun. 2018;67:218–229. doi: 10.1016/j.bbi.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002;277(52):50959–50965. doi: 10.1074/jbc.M207050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132(2):562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan AA, Khan Z, Warnakulasuriya S. Cancer-associated toll-like receptor modulation and insinuation in infection susceptibility: association or coincidence? Ann Oncol. 2016;27(6):984–997. doi: 10.1093/annonc/mdw053. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Jiang S, Tapping RI. Toll-like receptor signaling in cell proliferation and survival. Cytokine. 2010;49(1):1–9. doi: 10.1016/j.cyto.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pouncey AL, Scott AJ, Alexander JL, Marchesi J, Kinross J. Gut microbiota, chemotherapy and the host: the influence of the gut microbiota on cancer treatment. Ecancermedicalscience. 2018;12:868. doi: 10.3332/ecancer.2018.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doestzada M, Vila AV, Zhernakova A, Koonen DPY, Weersma RK, Touw DJ, et al. Pharmacomicrobiomics: a novel route towards personalized medicine? Protein Cell. 2018;9(5):432–445. doi: 10.1007/s13238-018-0547-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letertre MPM, Munjoma N, Wolfer K, Pechlivanis A, McDonald JAK, Hardwick RN, et al. A two-way interaction between methotrexate and the gut microbiota of male sprague-dawley rats. J Proteome Res. 2020;19(8):3326–3339. doi: 10.1021/acs.jproteome.0c00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan L, Zhang S, Li H, Yang F, Mushtaq N, Ullah S, et al. The influence of gut microbiota dysbiosis to the efficacy of 5-fluorouracil treatment on colorectal cancer. Biomed Pharmacother. 2018;108:184–193. doi: 10.1016/j.biopha.2018.08.165. [DOI] [PubMed] [Google Scholar]
- 43.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boussios S, Pentheroudakis G, Katsanos K, Pavlidis N. Systemic treatment-induced gastrointestinal toxicity: incidence, clinical presentation and management. Ann Gastroenterol. 2012;25(2):106–118. [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng WY, Wu CY, Yu J. The role of gut microbiota in cancer treatment: friend or foe? Gut. 2020;69(10):1867–1876. doi: 10.1136/gutjnl-2020-321153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shrivastava A, Khan AA, Jain SK, Singhal PK, Jain S, Marotta F, et al. Biotechnological advancement in isolation of anti-neoplastic compounds from natural origin: a novel source of L-asparaginase. Acta bio-medica Atenei Parm. 2010;81(2):104–108. [PubMed] [Google Scholar]
- 47.Karpinski TM, Adamczak A. Anticancer activity of bacterial proteins and peptides. Pharmaceutics. 2018 doi: 10.3390/pharmaceutics10020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer. 2019;7(1):108. doi: 10.1186/s40425-019-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lugito N, Kurniawan A, Damay V, Chyntya H, Sugianto N. The role of gut microbiota in SARS-CoV-2 infection: focus on angiotensin-converting enzyme 2. Curr Med Issues. 2020;18(3):261–263. doi: 10.4103/cmi.cmi_80_20. [DOI] [Google Scholar]
- 50.Kurian SJ, Unnikrishnan MK, Miraj SS, Bagchi D, Banerjee M, Reddy BS, et al. Probiotics in prevention and treatment of COVID-19: current perspective and future prospects. Arch Med Res. 2021 doi: 10.1016/j.arcmed.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conejo-Garcia JR, Rutkowski MR. Small but mighty: selected commensal bacterial species determine the effectiveness of anti-cancer immunotherapies. Immunity. 2015;43(6):1037–1039. doi: 10.1016/j.immuni.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 53.Daillere R, Derosa L, Bonvalet M, Segata N, Routy B, Gariboldi M, et al. Trial watch : the gut microbiota as a tool to boost the clinical efficacy of anticancer immunotherapy. Oncoimmunology. 2020;9(1):1774298. doi: 10.1080/2162402X.2020.1774298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Virchow R. An address on the value of pathological experiments. BMJ. 1881;2(1075):198–203. doi: 10.1136/bmj.2.1075.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Francescone R, Hou V, Grivennikov SI. Microbiome, inflammation, and cancer. Cancer J. 2014;20(3):181–189. doi: 10.1097/PPO.0000000000000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167(4):1125–36 e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendes V, Galvao I, Vieira AT. Mechanisms by which the gut microbiota influences cytokine production and modulates host inflammatory responses. J Interferon Cytokine Res Off J Int Soc Interferon Cytokine Res. 2019;39(7):393–409. doi: 10.1089/jir.2019.0011. [DOI] [PubMed] [Google Scholar]
- 59.Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, et al. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology. 2018;155(1):33–76. doi: 10.1053/j.gastro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao J, Chen X, Herjan T, Li X. The role of interleukin-17 in tumor development and progression. J Exper Med. 2020 doi: 10.1084/jem.20190297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 2015;367(2):103–107. doi: 10.1016/j.canlet.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fukugaiti MH, Ignacio A, Fernandes MR, Ribeiro Junior U, Nakano V, Avila-Campos MJ. High occurrence of fusobacterium nucleatum and clostridium difficile in the intestinal microbiota of colorectal carcinoma patients. Braz J microbial Publ Braz Soc Microbiol. 2015;46(4):1135–1140. doi: 10.1590/S1517-838246420140665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, et al. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. 2018;8(1):158. doi: 10.1038/s41598-017-18596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22(22):5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 67.Shin JM, Luo T, Kamarajan P, Fenno JC, Rickard AH, Kapila YL. Microbial communities associated with primary and metastatic head and neck squamous cell carcinoma—a high fusobacterial and low streptococcal signature. Sci Rep. 2017;7(1):9934. doi: 10.1038/s41598-017-09786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother Biomed pharmacother. 2017;89:918–925. doi: 10.1016/j.biopha.2017.02.102. [DOI] [PubMed] [Google Scholar]
- 72.Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4(12):2319–2330. doi: 10.1038/s41564-019-0541-3. [DOI] [PubMed] [Google Scholar]
- 73.Khan AA, Bano Y. Salmonella enterica subsp. enterica host-pathogen interactions and their implications in gallbladder cancer. Microb Pathog. 2021;157:105011. doi: 10.1016/j.micpath.2021.105011. [DOI] [PubMed] [Google Scholar]
- 74.Zamani S, Taslimi R, Sarabi A, Jasemi S, Sechi LA, Feizabadi MM. Enterotoxigenic bacteroides fragilis: a possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front Cell Infect Microbiol. 2020 doi: 10.3389/fcimb.2019.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng WT, Kantilal HK, Davamani F. The mechanism of bacteroides fragilis toxin contributes to colon cancer formation. Malays J Med Sci MJMS. 2020;27(4):9–21. doi: 10.21315/mjms2020.27.4.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gnanasekaran J, Binder Gallimidi A, Saba E, Pandi K, Eli Berchoer L, Hermano E, et al. Intracellular porphyromonas gingivalis promotes the tumorigenic behavior of pancreatic carcinoma cells. Cancers. 2020;12(8):2331. doi: 10.3390/cancers12082331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shiels MS, Albanes D, Virtamo J, Engels EA. Increased risk of lung cancer in men with tuberculosis in the alpha-tocopherol, beta-carotene cancer prevention study. Cancer Epidemiol Biomark Prev. 2011;20(4):672. doi: 10.1158/1055-9965.EPI-10-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nalbandian A, Yan BS, Pichugin A, Bronson RT, Kramnik I. Lung carcinogenesis induced by chronic tuberculosis infection: the experimental model and genetic control. Oncogene. 2009;28(17):1928–1938. doi: 10.1038/onc.2009.32. [DOI] [PubMed] [Google Scholar]
- 79.Di Domenico EG, Cavallo I, Pontone M, Toma L, Ensoli F. Biofilm producing salmonella typhi: chronic colonization and development of gallbladder cancer. Int J Mol Sci. 2017 doi: 10.3390/ijms18091887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Servetas SL, Bridge DR, Merrell DS. Molecular mechanisms of gastric cancer initiation and progression by Helicobacter pylori. Curr Opin Infect Dis. 2016;29(3):304–310. doi: 10.1097/QCO.0000000000000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khan AA, Khan Z, Kalam MA, Khan AA. Inter-kingdom prediction certainty evaluation of protein subcellular localization tools: microbial pathogenesis approach for deciphering host microbe interaction. Brief Bioinform. 2018;19(1):12–22. doi: 10.1093/bib/bbw093. [DOI] [PubMed] [Google Scholar]
- 82.Khan AA, Khan Z. COVID-2019-associated overexpressed Prevotella proteins mediated host-pathogen interactions and their role in coronavirus outbreak. Bioinformatics. 2020;36(13):4065–4069. doi: 10.1093/bioinformatics/btaa285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khan AA, Khan Z. Comparative host-pathogen protein-protein interaction analysis of recent coronavirus outbreaks and important host targets identification. Brief Bioinform. 2021;22(2):1206–1214. doi: 10.1093/bib/bbaa207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collina F, De Chiara A, De Renzo A, De Rosa G, Botti G, Franco R. Chlamydia psittaci in ocular adnexa MALT lymphoma: a possible role in lymphomagenesis and a different geographical distribution. Infect Agents Cancer. 2012;7:8. doi: 10.1186/1750-9378-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferreri AJM, Dolcetti R, Du MQ, Doglioni C, Giordano Resti A, Politi LS, et al. Ocular adnexal MALT lymphoma: an intriguing model for antigen-driven lymphomagenesis and microbial-targeted therapy. Ann Oncol. 2008;19(5):835–846. doi: 10.1093/annonc/mdm513. [DOI] [PubMed] [Google Scholar]
- 86.Olsen I, Yilmaz O. Possible role of porphyromonas gingivalis in orodigestive cancers. J Oral Microbiol. 2019;11(1):1563410. doi: 10.1080/20002297.2018.1563410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Díaz P, Valenzuela Valderrama M, Bravo J, Quest AFG. Helicobacter pylori and gastric cancer: adaptive cellular mechanisms involved in disease progression. Front Microbiol. 2018 doi: 10.3389/fmicb.2018.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alipour M. Molecular mechanism of helicobacter pylori-induced gastric cancer. J Gastrointest Cancer. 2020 doi: 10.1007/s12029-020-00518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cao S, Li J, Lu J, Zhong R, Zhong H. Mycobacterium tuberculosis antigens repress Th1 immune response suppression and promotes lung cancer metastasis through PD-1/PDl-1 signaling pathway. Cell Death Dis. 2019;10(2):44. doi: 10.1038/s41419-018-1237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Littman AJ, Jackson LA, Vaughan TL. Chlamydia pneumoniae and lung cancer: epidemiologic evidence. Cancer Epidemiol Biomark Prev. 2005;14(4):773. doi: 10.1158/1055-9965.EPI-04-0599. [DOI] [PubMed] [Google Scholar]
- 91.Abdulamir AS, Hafidh RR, Abu BF. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exper Clin Cancer Res CR. 2011;30:11. doi: 10.1186/1756-9966-30-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oehmcke-Hecht S, Mandl V, Naatz LT, Dühring L, Köhler J, Kreikemeyer B, et al. Streptococcus gallolyticus abrogates anti-carcinogenic properties of tannic acid on low-passage colorectal carcinomas. Sci Rep. 2020;10(1):4714. doi: 10.1038/s41598-020-61458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al-Saleem T, Al-Mondhiry H. Immunoproliferative small intestinal disease (IPSID): a model for mature B-cell neoplasms. Blood. 2005;105(6):2274–2280. doi: 10.1182/blood-2004-07-2755. [DOI] [PubMed] [Google Scholar]
- 94.Sun CH, Li BB, Wang B, Zhao J, Zhang XY, Li TT, et al. The role of Fusobacterium nucleatum in colorectal cancer: from carcinogenesis to clinical management. Chronic Dis Transl Med. 2019;5(3):178–187. doi: 10.1016/j.cdtm.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haghi F, Goli E, Mirzaei B, Zeighami H. The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC Cancer. 2019;19(1):879. doi: 10.1186/s12885-019-6115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Umar S. Citrobacter infection and wnt signaling. Curr Colorectal Cancer Rep. 2012 doi: 10.1007/s11888-012-0143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karim S, Souho T, Benlemlih M, Bennani B. Cervical cancer induction enhancement potential of chlamydia trachomatis: a systematic review. Curr Microbiol. 2018;75(12):1667–1674. doi: 10.1007/s00284-018-1439-7. [DOI] [PubMed] [Google Scholar]
- 98.Tangney M, Gahan CG. Listeria monocytogenes as a vector for anti-cancer therapies. Curr Gene Ther. 2010;10(1):46–55. doi: 10.2174/156652310790945539. [DOI] [PubMed] [Google Scholar]
- 99.Morrow ZT, Powers ZM, Sauer J-D. Listeria monocytogenes cancer vaccines: bridging innate and adaptive immunity. Curr Clin Microbiol Rep. 2019;6(4):213–224. doi: 10.1007/s40588-019-00133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei H, Chen L, Lian G, Yang J, Li F, Zou Y, et al. Antitumor mechanisms of bifidobacteria. Oncol Lett. 2018;16(1):3–8. doi: 10.3892/ol.2018.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Broadway KM, Scharf BE. Salmonella typhimurium as an anticancer therapy: recent advances and perspectives. Curr Clin Microbiol Rep. 2019;6(4):225–239. doi: 10.1007/s40588-019-00132-5. [DOI] [Google Scholar]
- 102.Maletzki C, Linnebacher M, Kreikemeyer B, Emmrich J. Pancreatic cancer regression by intratumoural injection of live Streptococcus pyogenes in a syngeneic mouse model. Gut. 2008;57(4):483–491. doi: 10.1136/gut.2007.125419. [DOI] [PubMed] [Google Scholar]
- 103.Staedtke V, Roberts NJ, Bai RY, Zhou S. Clostridium novyi-NT in cancer therapy. Genes Dis. 2016;3(2):144–152. doi: 10.1016/j.gendis.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feng X, He P, Zeng C, Li YH, Das SK, Li B, et al. Novel insights into the role of clostridium novyi-NT related combination bacteriolytic therapy in solid tumors. Oncol Lett. 2021;21(2):110. doi: 10.3892/ol.2020.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Magno C, Melloni D, Gali A, Mucciardi G, Nicocia G, Morandi B, et al. The anti-tumor activity of bacillus Calmette-Guerin in bladder cancer is associated with an increase in the circulating level of interleukin-2. Immunol Lett. 2002;81(3):235–238. doi: 10.1016/s0165-2478(02)00040-8. [DOI] [PubMed] [Google Scholar]
- 106.Araghi A, Hashemi S, Sepahi AA, Faramarzi MA, Amin M. Purification and study of anti-cancer effects of Serratia marcescens serralysin. Iran J Microbiol. 2019;11(4):320–327. [PMC free article] [PubMed] [Google Scholar]
- 107.Li D, Liu J, Wang X, Kong D, Du W, Li H, et al. Biological potential and mechanism of prodigiosin from Serratia marcescens Subsp. lawsoniana in human choriocarcinoma and prostate cancer cell lines. Int J Mol Sci. 2018 doi: 10.3390/ijms19113465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shafiee F, Aucoin MG, Jahanian-Najafabadi A. Targeted diphtheria toxin-based therapy: a review article. Front Microbiol. 2019;10:2340. doi: 10.3389/fmicb.2019.02340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are available with manuscript.