Abstract
The gut microbiota directly impacts the pathophysiology of different human body districts. Consequently, microbiota investigation is an hot topic of research and its in vitro culture has gained extreme interest in different fields. However, the high sensitivity of microbiota to external stimuli, such as sampling procedure, and the physicochemical complexity of the gut environment make its in vitro culture a challenging task. New engineered microfluidic gut‐on‐a‐chip devices have the potential to model some important features of the intestinal structure, but they are usually unable to sustain culture of microbiota over an extended period of time. The integration of gut‐on‐a‐chip devices with bioreactors for continuous bacterial culture would lead to fast advances in the study of microbiota‐host crosstalk. In this review, we summarize the main technologies for the continuous culture of microbiota as upstream systems to be coupled with microfluidic devices to study bacteria‐host cells communication. The engineering of integrated microfluidic platforms, capable of sustaining both anaerobic and aerobic cultures, would be the starting point to unveil complex biological phenomena proper of the microbiota‐host crosstalks, paving to way to multiple research and technological applications.
Keywords: anaerobiosis, bioreactors, gut‐brain axis, microfluidic systems, organ‐on‐a‐chip
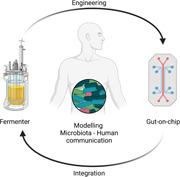
In the last decades, the interest about the impact of the microbiota on body districts and diseases rose exponentially, leading huge attention and stimulating interdisciplinary studies. In this review, technologies that enable the coculture of bacteria with cell are described. The engineering of advanced microfluidic platform, allowing for long coculture time and modeling anaerobic environment, is a promising strategy for the research in the complex field of the microbiota‐human cells communication.
1. INTRODUCTION
1.1. Gut environment and microbiota‐host interaction
The gut microbiota refers to the whole of symbiotic microorganisms colonizing the human intestine and growing in physiological synergy with cells (a condition defined as eubiosis) (Bäckhed, 2011; Dieterich et al., 2018; Quigley, 2013).
The gastrointestinal (GI) tract (Figure 1), where gut microbiota resides, has a complex structure which is divided into three regions: the stomach, the small intestine (duodenum, jejunum, and ileum) and the colon (ascending, transverse and descending). Each region has physicochemical peculiarities influencing bacterial composition and strain abundance (Arrieta & Finlay, 2012; Boeri et al., 2019; Chambers et al., 2015; Conlon & Bird, 2015; Keshavarzian et al., 2012; Kim et al., 2014, 2016; Manach et al., 2004; Morrison & Preston, 2016; Stokholm et al., 2016). Indeed, the concentration of bacteria per gram content increases along the GI tract: 101 bacteria/g in the stomach; 103 bacteria/g in the duodenum; 104 bacteria/g in the jejunum; 107 bacteria/g in the ileum; 1012 bacteria/g in the colon (Dieterich et al., 2018; Drissi et al., 2017; Shah et al., 2020). However, the exact composition of the bacterial species of the GI is still discussed and its detection depends on the experimental analytic techniques used (e.g., culturomics, metagenomics). Recently, targeted studies have investigated these aspects, even if the results are not yet generalizable (Rinninella et al., 2019). One example is a work by Maihle and collegues where they found 110 species in the stomach and 235 in the left colon by using culturomics as detection technique (Mailhe et al., 2018). Microbiota community also varies along the transversal section of the intestine, from the gut wall to the internal lumen. This depends on chemical and nutrient gradients, antimicrobial immune response, epithelial activity, and morphological features of the gut walls (Donaldson et al., 2015).
Figure 1.
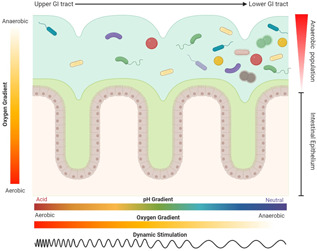
The gastrointestinal tract is a complex system both from a physicochemical and biological point of view. Moving from the upper to the lower GI tract, there are different parameters that vary accordingly to the site, such as oxygen partial pressure, pH and intensity of the dynamic stimuli, as well as an increased number and diversity of bacteria shaping the microbiota. Regardless from the tract considered, the distribution of bacterial strains is influenced by the oxygen gradient. Figure made with biorender (https://biorender.com) [Color figure can be viewed at wileyonlinelibrary.com]
Moreover, the composition and abundance of the gut microbiota is related to sex, age, health, mental conditions and diet of the host, among others (Sekirov et al., 2010). Other parameters like pH, flow rate, transit time of digesta, thickness of the mucus layer, activity of immune cells, enzyme secretion, bile acid concentration, redox potential and oxygen concentration are also crucial to determine microbiota features along the different GI tracts (Espey, 2013; Tuohy & Scott, 2015). The stomach has the lowest bacterial concentration due to its unfavorable physicochemical properties, (pH 2), high concentrations of pancreatic enzymes and bile acids. The gastric extreme environment actively filters the ingested bacteria, allowing for the survival and/or successful passage into the small intestine of only few microbial strains (e.g., Helicobacter pylori, E. coli, Salmonella, and Shigella) (Tuohy & Scott, 2015). Along the three small intestine regions the pH raises from 6 to 7.7 (Hillman et al., 2017; Koziolek et al., 2015). In these tracts, the high flow rate of digesta, high micronutrient digestion and adsorbtion, and the intense reactivity of immune cells demonstrated to mainly influence the residing micriobiota (Judkins et al., 2020). The continuous excretion of digestive enzymes and bile acids, and a short transit time cause continuous bacterial renewal and, hence, a dynamic microbiota pattern (Zoetendal et al., 2012). Reactive immune cells residing in the Peyer's patches along the walls of the small intestine, contribute to gut homeostasis and microbiota colonization (Tuohy & Scott, 2015). Differently, the colon is characterized by the most stable microbiota community, accounting for more than 400 species according to culturomics analysis (Mailhe et al., 2018). This is mainly due to less critical pH values, low transit time of digesta and the availability of food‐derived energy. Since the ascending colon is a site of intense bacterial fermentation, the lumen becomes slightly acidic in the proximal portion, while turning neutral in the distal one (Tuddenham & Sears, 2015; Tuohy & Scott, 2015).
Oxygen concentration is one of the key factors influencing bacterial composition in the whole GI tract, since it determines the survival and growth of facultative or obligate anaerobes. An oxygen gradient exists along both the radial and longitudinal axes of the intestine (Zheng et al., 2015). Within the radial section, the oxygen partial pressure (PO2) decreases precipitously from the submucosa (80–100 mmHg) to the lumen (<10 mmHg) (Espey, 2013). Cell heterogeneity and distribution along the gut villi mirror oxygen requirements: the cells consuming oxygen at higher rates (i.e., Paneth cells and intestinal stem cells) are located at the basis of the intestinal crypt, while the cells consuming oxygen at lower rates (i.e., bacteria and enterocytes) are located at the lumen interface (Clevers, 2013). Along the longitudinal axis, PO2 decreases gradually while descending the GI tract. It reduces from ~77 mmHg in the stomach to <1 mmHg in the rectum (Espey, 2013). These oxygen gradients are mainly due to the structural anatomy and blood vessel distribution. For instance, at the basis of the gut villus the blood flow and nutrient exchange is maximum and it decreases towards the lumen (Zheng et al., 2015).
Such a complex environment influences the gut microbiota, and the communication between bacteria and host‐cells. This interplay has recently attracted the attention of researchers, due to the increasing awareness of its direct or indirect effect on the state of health of different body districts, even if anatomically distant from the GI tract. Increasing resources were devoted to model the gut enviroinment and to investigate microbiota‐human cell communication to elucidate different biological questions. In this view, one of the most exciting examples of microbiota‐host interaction is the microbiota‐gut‐brain axis as a new paradigm for neuroscience research. Several hypotheses have been proposed to describe the microbiota‐gut‐brain axis (such as neuroanatomical and neuroendocrine pathways) and were reviewed in detail in other works (Cryan et al., 2019; Serra et al., 2019; Wang & Wang, 2016).
The gut microbiota influences brain development and functionality through neural networks (i.e., enteric nervous system [ENS] or vagus nerve), and the immunomodulatory effects of microbial factors, which can be released both at peripheral and central level (Braniste et al., 2014; Dinan & Cryan, 2017a, 2017b; Fung et al., 2017; Heijtz et al., 2011; Michel & Prat, 2016; Neufeld et al., 2011; Raimondi et al., 2020; Rea et al., 2016; Rook et al., 2014; Silva et al., 2020; Williams et al., 2014; Yunes et al., 2016). The shift of microbiota population towards a pathologic profile is defined as dysbiosis and its role in neurological and neurodegenerative diseases has been deeply investigated (Brial et al., 2018; Keightley et al., 2015; Leclercq et al., 2014; Moser et al., 2018; Patterson et al., 2014; Tilg et al., 2020). Emerging data suggest that dysbiosis could trigger inflammation by acting on the permeability of both gut and blood–brain barrier (BBB), thus affecting disease onset and progression (Pellegrini et al., 2018; Quigley, 2017; Roy Sarkar & Banerjee, 2019; Sampson et al., 2016; Spielman et al., 2018; Vogt et al., 2017).
In the last decades, based on transgenic animal models research, the hypothesis of a connection between microbiota alterations and Alzheimer's disease (AD), Parkinson's disease (PD) or other neurodegenerative disorders was introduced (Cattaneo et al., 2017; Forsyth et al., 2011; Harach et al., 2017; Nair et al., 2018; Tufekci et al., 2011).
Despite in vivo studies were fundamental in highlighting the possible presence of the microbiota‐gut‐brain axis, they were unable to fully describe the series of biological events underlying this relationship. For this reason, growing resources were dedicated to study in vitro the axis, and in particular the first step of the microbial‐host cells molecular dialogue. For instance, it is worthy to notice that the European Research Council funded more than 60 microbiota‐related projects in <10 years. Among them, the ERC‐CoG‐MINERVA project aims at developing an innovative technological platform to investigate the relationship between intestinal microflora and brain functionality, in healthy and pathological conditions (Raimondi et al., 2019, 2020).
Modeling bacteria‐host communication in vitro is not a trivial task. Researchers have used two different approaches: to produce and purify microbiota‐derived molecules, they have applied bioreactors able to sustain the culture of human microbiota over an extended period (i.e. fermenters), while to study the effect of bacteria‐derived molecules on human cells they have exploited advanced in vitro gut‐on‐a‐chip devices (Holmes et al., 2020; Kim et al., 2012; Mailhe et al., 2018; Sontheimer‐Phelps et al., 2020; Villa et al., 2020). Apart from the specific focus of the study, these systems face similar issues, such as the optimization of the sample preparation and modeling the complexity of intestinal environment. Indeed, human microbiota is a very variable bio‐ecosystem requiring stringent technical specifications to be efficiently cultured. In addition, the faithful reproduction of its chemical‐physical and structural features is a difficult task, particularly because dynamic stimulation should be included to model physiological phenomena like intestinal peristalsis and biological fluids flows. In addition, microbiota cultivation is higly dependent on the sampling procedure (Table 1). At the same time, from an engineering point of view, the tools allowing for a prolonged microbiota culture with in vivo‐like conditions are based on complex technologies (Marzorati et al., 2014; Venema, 2015). In the following paragraphs, we will focus on the main technologies available for long‐lasting human microbiota culturing and the most advanced systems to model in vitro the human gut. In particular, the advantages and disadvantages of recent in vitro approaches to achieve a reliable representation of bacteria‐host communication will be discussed.
Table 1.
Possible methods for gut microbiota sampling for in vitro culture
| Methods | Pros | Cons | Main application | |
|---|---|---|---|---|
| FECES | Patient collect their stool, usually in bulk and at home. Commercial containers (e.g., Fisherbrand™ Commode Specimen Collection System, EasySampler®, Fe‐Col® or BioCollector™) limit their discomfort, prevent sample contamination from urine, toilet water and sanitizers, and provide closed containers for transportation |
‐ Simplicity, repeatability on daily basis, affordability and noninvasiveness ‐ Guidelines and protocols for donor selection, stool collection, transportation and DNA extraction (Wu et al., 2019) |
‐ Difficult collection for diarrhea or baby stool (Videnska et al., 2019) ‐ Use of protectant media (Bellali et al., 2019, 2020; Lagier et al., 2015; Martínez et al., 2019; Million et al., 2020) to maintain anaerobic bacteria ‐ Daily interindividual variations lead to greater differences in microbiota analysis than stool handling (e.g., homogenization or freezing; Cheng & Ning, 2019; Hsieh et al., 2016; Lozupone et al., 2012; Rinninella et al., 2019; Scepanovic et al., 2019; Vandeputte et al., 2016; Zarrinpar et al., 2014) ‐ Possible incomplete removal of fecal bacteria from intestinal flora (Tang et al., 2020) ‐ Samples treated with stabilization buffers (e.g., RNALater®, OMNIgene®·GUT, and FTA cards) to prevent DNA/RNA degradation are unsuitable for culturing and continuous colonic fermentation models (Chan et al., 2016; Morjaria et al., 2019) |
‐ Study of the more transient luminal bacteria of the large intestine. Unsuitable to recapitulate the spatial organization of gut microbiota communities (Jones et al., 2018) |
| ENDOSCOPIC PROCEDURES |
A) Mucosal biopsy: A flexible endoscopy reaches the sampling site from the mouth or the anus. With respect to standard forceps, the Brisbane Aseptic Biopsy Device reduces sample contamination and diversity (Shanahan et al., 2016). In this device, a membrane covers the tip of the endoscope. The biopsy forceps penetrate the membrane, that retracts when the forceps is advanced |
‐ Suitable for the diagnosis of disease type in IBDs (Salvatori et al., 2012) |
‐ Unfriendly, possible bleeding ‐ Bacteria in not‐sampling sites are dragged to sampling sites ‐ Small sampling area, leading to sample deviation ‐ Not enough DNA, RNA or proteins for multi‐omics technologies ‐ Large amounts of contaminated host DNA |
‐ Assess the composition of mucosal microbiota in different GI (Tang et al., 2020; Zhang et al., 2017) |
|
B) Luminal brushing: Initially developed to sample lung microbiota (Wimberley et al., 1979) it couples mucosal biopsy to protected specimen brushing. A plug and sheath protect the sample in the colonoscope working channel (Lavelle et al., 2013) |
‐ Reduced bleeding and infection, more representative mucosal samples with respect to biopsy ‐ Large ratio of bacterial to host DNA |
‐ The general ones of endoscopic procedures |
‐ Study of luminal‐associated microbiota (e.g., spatial variations between luminal and mucosal microbiota) |
|
|
C) Laser capture microdissection: An infrared laser beam allows for the adhesion of the tissue section on the surface of a biopsy to a thin, transparent film. Then, the film is removed and the sample (e.g., DNA, RNA, proteins) is treated Emmert‐Buck et al.,1996) |
‐ Isolation of pure sections ‐ Punctual analysis (a small focal region is transferred to the film) ‐ Easy, specific and accurate sampling (resolution: 5 µm, (Nava et al., 2011) |
‐ Biopsy samples are required ‐ Biopsy preparation before laser treatment (Baarlen et al., 2008) ‐ Nuclei acid degradation and low sample amount ‐ Not for large‐scale analyses, but suitable for precision medicine |
‐ Study of mucosal‐ and crypt‐associated microbial communities (Nava et al., 2011; Pédron et al., 2012; Richard et al., 2018; Wang et al.,) ‐ Study of mucus layers and mucus‐associated bacteria (Chassaing & Gewirtz, 2019; Lavelle et al., 2015) |
|
| ASPIRATION OF INTESTINAL FLUID |
A) Capsules: For example, stainless steel capsules connected to a negative‐pressure source by a tube: (Shiner, 1963) |
‐ Prevention of contamination (after collection, the sample is isolated from the external environment) |
‐ Technically challenging |
‐ Culture of small bowel microflora for the diagnosis of SIBO (Choung et al., 2011) ‐ In vitro evaluation of orally administered drug products (Litou et al., 2020) |
|
B) Tubes: For example, tubes for enteral feeding and ingestible tubes (Lavelle et al., 2010; Minekus et al., 1999) |
‐ Prevention of contamination |
‐ Difficult and time‐consuming ‐ Possible occlusions due to the viscosity of intestinal fluid |
||
|
C) Endoscopic aspiration: (Rao & Bhagatwala, 2019) |
‐ Alternative to endoscopic biopsy |
‐ Patient discomfort ‐ Contamination of the endoscopic channel by oropharyngeal and GI content ‐ Time‐consuming ‐ Possible unsuccessful suction due to the sparseness of fluid aspirates |
‐ Most popular method for intestinal fluid collection |
|
| CAPSULE ENDOSCOPY | Swallowable pills move actively or passively through the GI tract by peristalsis. They offer recognition, anchoring and bio‐sensing capabilities (Amoako‐Tuffour et al., 2014; Hale et al., 2014). Some of them can also collect tissue samples (at least 400 µm in size or aspirate intestinal fluid (> 200 µl) (Cui et al., 2008; Park et al., 2008) |
‐ Low invasiveness ‐ Accurate location of sampling points ‐ Reduction of patient comfort ‐ High technological content (e.g., locomotion mechanisms; wireless connection; temperature, pH, pressure, oxygenation, oxidation/reduction, conductivity sensors; multi‐axial accelerometers and gyroscopes for inertial navigation and positioning) |
‐ Risk of capsule aspiration and retention ‐ Possible sample contamination by intestinal fluid from noncollected sites (Cui et al., 2008b) ‐ High costs of the medical procedures ‐ High costs for fabrication, but a reduction is possible with 3D printing (Rezaei Nejad et al., 2019) ‐ Commercial products, but they do not perform biopsy (Pan et al., 2019) |
‐ Study of small bowel diseases (e.g., celiac disease, CD, cancers), assess mucosal activity ‐ Intestinal microbiome sampling and preservation (Koziolek et al., 2015) ‐ Imaging the distal duodenum, jejunum and ileum (Moglia et al., 2009) ‐ Drug release (Cui et al., 2008b) |
Abbreviations: CD, Crohn's disease; DBE, double‐balloon enteroscopy; IBD, inflammatory bowel diseases; GI, gastrointestinal; MEMS, microelectromechanical; SBE, single‐balloon enteroscopy; SIBO, small intestinal bacterial overgrowth.
1.2. Long‐term culture of human microbiota
To study microbiota biodiversity, culture‐independent and ‐dependent methods can be exploited. Culture‐independent approaches, whose advantages and disadvantage were already described (Su et al., 2012; Weinstock, 2012) directly characterize native microbiota by molecuar techniques, without the need of bacteria culturing (Gupta et al., 2019; Su et al., 2012; Weinstock, 2012; Woo et al., 2008). Culture‐dependent approaches rely on growing microbiota samples in different media formulations Before characterization. In this case, the first challenge is the selection of the inoculation method. An intrinsic issue arising when culturing microbiota is the difference among in vitro and in vivo conditions (i.e., pH, transit time, carbohydrate content in the medium, variations in oxygen levels, absence of mucosal binding sites or selective pressure), which may result in compositional discrepancies between native and cultured microbiota (McDonald et al., 2013; O'Donnell et al., 2016; Payne, et al., 2012). Hence, the isolated microbes may not be representative of the most abundant taxa in situ, leading to ineffective inoculation, regardless of the experimental set‐up used for the culture (Stefani et al., 2015). Moreover, from a technical point of view, bacteria can be inoculated in liquid form or immobilized in polymer beads (de Vos, 2015). Even though Silverman et al. (2018) showed that gut microbiota composition was stable over long periods of time, the immobilization reproduces both motile and immobilized microbes, and it helps to ensure bacterial diversity at high cell densities for long periods in continuous intestinal reactors (Cinquin et al., 2004, 2006b).
Regardless of the application, numerous complications should be taken into account when investigating microbiota effect on human health in vitro (Payne, et al., 2012; Sommer, 2015). First, the presence of cells, modeling the human body, inhibits the use of specific bacterial culture broths imposing the use of cell culture medium, which may affect bacterial growth due to a suboptimal culture condition (Ito et al., 2019; Jalili‐Firoozinezhad et al., 2019; Yousi et al., 2019). Second, when chronic phenomena such as gut inflammation and its possible role in neurodegeneration are considered, bacterial culture should be maintained for long time frames. This is important not only to accurately model the in vivo situation, but also to reach the microbiota metabolic equilibrium in vitro and, hence, the cellular reaction in time to the steady state condition (Peebo & Neubauer, 2018; Webb, 2017). Short‐term in vitro cocultures of bacteria and cells are excellent tools to study acute phenomena, such as bacterial adhesion and infection of the intestinal walls (Jalili‐Firoozinezhad et al., 2019). However, they are not able to model chronic pathological conditions yet, which are typical features of neurodegenerative diseases (Kelly et al., 2017). For this reason, bioreactors for long‐term bacterial culture have become particularly attractive even outside their classic fields of use, in particular as suitable upstream systems for the coculture of bacteria with cells in microfluidic devices (Marzorati et al., 2014).
Long‐term culture of the microbiota was firstly assessed in fed‐batch conditions, where the medium is poured into the culture vessel hosting the biomass, thus avoiding nutrients depletion (Rumney & Rowland, 1992; Venema & Van Den Abbeele, 2013). The biomass can be composed of a single cell type as well as complex microbial communities, usually cultured with a specific medium in a closed glass vessel (Coimbra et al., 2020; Khalil et al., 2014; Mou & Cooney, 1983). The two main factors influencing the biomass growth‐profile in a fed‐batch bioreactor are the inoculum density and the substrate depletion rate (Modak et al., 1986; Rumney & Rowland, 1992). When properly combined, these parameters induce a shift of the growth curve from the typical quadriphasic trend of the batch culture (i.e., lag, logarithmic, stationary and death phases) to enhanced logarithmic and stationary phases, as demonstrated both experimentally and theoretically (Figure 2a) (Goudar, 2012; Landa et al., 2001; Modak et al., 1986). The possibility to culture the same bacteria population for prolonged times in a single procedure brings some advantages, such as the minimal incidence of mutations and multi‐compound fermentation (Kubitschek & Bendigkeit, 1964; Mora‐Villalobos et al., 2020). Moreover, the batch and fed‐batch conditions can be coupled to operate in nearly stationary regimen, maximizing the control of fermentation conditions before nutritive supplies. Even though these advantages are unquestionable, the fed‐batch is a closed system, which drags at least two limitations. First, the logarithmic growth is sustained until fresh medium is supplied (Pompei et al., 2008). This may result in unfavorable temporal constraint, especially for weeks‐long experiments like monitoring of the microbiota metabolism over time (Kaarel Adamberg et al., 2020; Cleusix et al., 2008). Second, it is not possible to couple the bioreactor with other downstream systems, which is a sine qua non priority when microbiota‐host or bacterial‐cellular crosstalk is investigated.
Figure 2.
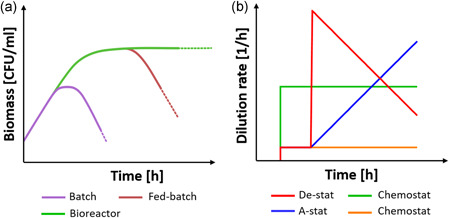
(a) The typical growth curves of batch and fed‐batch cultures are compared to the continuous culture performed by a bioreactor, such as a chemostat, where the presence of a medium inflow and outflow allow for a potentially infinite stationary phase. (b) The different approaches for the continuous culture in a bioreactor with reference to the chosen method to control D. In particular, the increasing or decreasing of the dilution rate corresponds to the accelerostat or decelerostat condition (red and blu line respectively). Differently, a constant D defines the chemostat approach (in the figure, two different D are represented by the green and orange line). Figure made with biorender [Color figure can be viewed at wileyonlinelibrary.com]
To overcome these limitations, closed fed‐batch bioreactors evolved to include medium outflow (Rumney & Rowland, 1992). The balance between nutrient availability and waste removal allows for the prolonged maintenance of a univocal correspondence between biomass and environmental conditions (i.e., steady state condition) (Shachrai et al., 2010; Wang et al.,). Only in steady state the changes of molecules’ concentration, and hence the physiological state of cells, are negligible and an unaltered biological process can be assumed in time (Korem Kohanim et al., 2018). The control over experimental parameters (e.g., temperature, pH, oxygen concentration and flow rates) and the possibility of their modification with reasonable resources are mandatory prerequisites for continuous culture.
The control of medium flow is a standard strategy to reach the steady state condition. Culture systems are classified accordingly to the method used to dilute the culture by considering the dilution rate,D, which is a parameter defined as the value of the medium flow rate divided by the total culture volume (Figure 2b) (Adamberg et al., 2015). The chemostat approach consists in maintaining a constant D over time. In this case, D should be set not to exceed the maximum specific growth rate of the biomass, thus not diluting the bacterial population faster than its growth capability and leading to a constant depopulation (Bhunia, 2014). Importantly, when the steady state is reached, D can be assumed as numerically equivalent to the specific growth rate of the biomass (Doble & Kumar, 2005).
Hence, the control over the process parameters (in/out flows) can be used to engineer important fermentation conditions, such as the biomass in the culture. However, this is not a trivial process. For instance, increasing D to maximize the growth rate is extremely time‐consuming, as it requires culture stabilization after each D variation step (Adamberg et al., 2015; Gresham & Hong, 2015). Control over the culture can be obtained by the changestat approach, where the environmental parameters are constantly tuned during the culture while preserving the steady state condition (Lahtvee et al., 2011). If D is increased or decreased over time, then the changestat is defined as accelerostat or decelerostat (A‐stat or B‐stat, respectively). In both cases, D variations should be not too high to disrupt the bioprocess equilibrium, but not too low to lead specific phenotypical mutations. Differently, if D is maintained constant and other environmental parameters are changed (e.g., pH, oxygen concentration, temperature, medium additives), then the system is defined as dilution rate‐stat (D‐stat) (Adamberg et al., 2015). The A‐/De‐stat bioreactors are particularly suitable for culture condition and bioprocess optimization. For example, changestats were used in microbiota continuous cultures, revealing that D has a substantial effect on microbial prevalence (Feria‐Gervasio et al., 2011), molecule production (Child et al., 2006; Macfarlane et al., 1998) and biodiversity maintenance (Tottey et al., 2017). Moreover, the De‐stat approach has enormous benefits in terms of time required for reching the targed culture condition (reduction up to 94% compared to chemostat) with only 5% of accuracy loss (Hoekema et al., 2006). Even though these strategies are different, the available bioreactors can switch from one approach to another; for example, passing from a fed‐batch pre‐stabilization phase to a chemostat/chagestat culture (Fehlbaum et al., 2015; Tanner et al., 2014; Zihler Berner et al., 2013). Hence, the aforementioned classification is useful to define the experimental ratio, but it is limited in the description of the experimental set‐up. Indeed, the bioreactors for long‐term culture of microbiota were designed in different ways, though maintaining the common aspect of medium flowing in the system (Figure 3). The number of serial vessels (reservoirs and waste excluded) composing the bioreactor is the main classification parameter. To date, single‐, double‐, triple‐ and multi‐stage bioreactors have been described (Payne et al., 2012) (Table 2). Single‐stage bioreactors consist in a single glass vessel directly connected to the medium reservoir and the waste container (Cinquin et al., 2004). They can be used to optimize experimental procedures, such as the reproducibility of a new inoculum type or the definition of the best dilution conditions (Adamberg & Adamberg, 2018; McDonald et al., 2013). Also, the effect of certain target compounds, such as ions, fibers, and other nondietary molecules, can be assessed with single‐stage chemostats or changestats after a suitable stabilization time. The complexity of the vessel containing the microbiota increases accordingly to the aim of the study and can be designed to include hydraulic accessories to recapitulate relevant biological processes as peristalsis, water absorption and salt dialysis (Minekus et al., 1999). In two‐stage bioreactors, a second vessel receives the outflow from a first vessel. The two vessels sustain the culture in different conditions of pH, retention time and working volume to model two different GI tracts, usually the proximal colon and the transverse or descending colon (Fehlbaum et al., 2015; Zihler Berner et al., 2013). This set‐up allows investigating the effect of an in vivo‐like environment on microbiota metabolism, composition and other age‐dependent bioprocesses (Dostal et al., 2013; Tanner et al., 2014; Zihler Berner et al., 2013). Two‐stage bioreactors are more flexible than single‐stage ones and open to the possibility of multi‐treatment analysis, as well as the presence of a control‐culture by substituting the second vessel with multiple vessels in parallel (Fehlbaum et al., 2015). The addition of a third vessel, modeling the remaining colonic tract, represents the three‐stage approach (Poeker et al., 2018). Similarly to two‐stage bioreactors, the medium is pumped by peristaltic pumps through three vessels bearing different environmental conditions each (Feria‐Gervasio et al., 2014). Three‐stage systems are the main type of bioreactors for long‐time culture of microbiota, as they more accurately model the in vivo modularity (Gibson & Fuller, 2000; Payne et al., 2012). In this way, it is possible not only to study the effect of a certain treatment on microbiota composition, biodiversity and metabolism, but also (at the same time) to distinguish the combined effects of each colonic sub‐environment on biomass features (Venema & Van Den Abbeele, 2013b). Three‐stage bioreactors for long‐term microbiota culture were applied in different fields, including –without being exhaustive—the effects of dietary fibers, age‐dependent metabolism, probiotics, prebiotics and mechanisms of bacterial infection (Cinquin et al., 2006a; Macfarlane & Macfarlane, 2007; Payne et al., 2012; Zihler et al., 2010). However, they are more complex than single‐ or two‐stage bioreactors, as they require advanced control systems and empowered resource accessibility (Gibson & Fuller, 2000).
Figure 3.
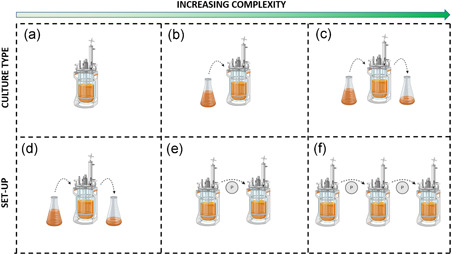
Schematic representation of the microbiota culture methods with increasing level of complexity, including batch (a), fed‐batch (b), and continuous culture (d). The bioreactors for continuous culture of microbiota were classified accordingly to the number of vessels: single‐, double‐ and triple‐stage bioreactors (d, e and f, respectively). Figure made with biorender (https://biorender.com) [Color figure can be viewed at wileyonlinelibrary.com]
Table 2.
Main studies exploiting long terms culture of bacteria and their definition in terms of the experimental ratio and set up
| Type of control | Experimental set‐up | inoculum | Preculture stability (d) | Experiment duration (d) | Main application | Ref. |
|---|---|---|---|---|---|---|
| Chemostat | Single stage | Immobilized/slurry | 16 | 54 | Effect of the inoculum type on the microbiota biodiversity | Cinquin et al. (2004) |
| Immobilized | 13 | 71 | Changes in the production of SCFAs with four different prebiotic fructans | Le Blay et al. (2010) | ||
| 12 | 62 | Combined effect of glycerol and Limosilactobacillus reuteri on microbiota composition | Cleusix et al. (2008) | |||
| Slurry | 4 | 48 | Stability of microbiota culture with different inoculum techniques and replicates | McDonald et al. (2013) | ||
| Variable | Variable | Differentiation of the microbiota accordingly to different dilution rate | Adamberg and Adamberg (2018) | |||
| Variable | Variable | Microbiota stability with different dilution rates and dietary fibers | Adamberg et al. (2020) | |||
| 2 | > 62.5 | Monitoring of the mictrobiota steady‐state metabolism at different dilution rates | Feria‐Gervasio et al. (2011) | |||
| Two stage | Immobilized | 13 | 38 | Changes induced by pH variation to the microbiota ecology | Zihler Berner et al. (2013) | |
| 11 | 54 | Swine microbiota composition and diversity in new poly‐fermenter system | Tanner et al. (2014) | |||
| 14 | 80 | Differences in elderly microbiota composition cultured in multiple environmental condition | Fehlbaum et al. (2015) | |||
| 3 | 70 | Impact of Fe availability on the child microbiota composition and metabolism | Dostal et al. (2013) | |||
| Three stage | Slurry | 48 | 120 | Effect of mucin on the dissimilatory sulfate reduction and CH4 production | Gibson et al. (1988) | |
| Immobilized | 12 | 29 | FISH counting efficacy compared to CFUs. Stability of the microbiota culture | Cinquin et al. (2006b) | ||
| > 3 | 33 | EPS and FOS influence on the human infant microbiota | Cinquin et al. (2006a) | |||
| 2 | 42 | Metabolic adaptation of child microbiota to obese, normal‐weighed and anorectic dietary | Payne, Chassard, et al. (2012) | |||
| 10 | 65 | Probiotics effect on salmonella infection and E. coli colonization | Zihler et al. (2010) | |||
| Changestat | Three stage | Slurry | o.n. | > 16 | Monitoring of the mictrobiota metabolism and composition at different retention time | Macfarlane et al. (1998) |
| > 10 | Variable | Effect of the transient time on the microbiota metabolism and composition | Tottey et al. (2017) |
Microbiota culture conditions were further optimized by adding other districts of the GI tract (Venema, 2015). In this way, bioreactors were not only able to model the environment where microbiota is prevalently found in vivo (colon), but also to include other host‐related biological processes, such as digestion and bile secretion (Venema, 2015). For example, the TIM1 and TIM2 bioreactors modeled the upper and lower intestine, respectively, and were coupled to investigate the effect of inulin, galacto‐oligosaccharides and other dietary fermentation products (Maathuis et al., 2012; Macfarlane et al., 1992; Van Nuenen et al., 2003). Similarly, the SHIME aims at modeling all the compartments of the GI tract (Venema, 2015). These bioreactors were designed with several improvements, such as the possibility to run multiple experiments in parallel or include simple mucus models (García‐Villalba et al., 2017; Van Den Abbeele et al., 2013). Despite the relevance of gut multi‐stage bioreactors, they will not be described in this review, since already presented in other works (Ceppa et al., 2020).
Regardless of the design, scope and objectives, the culture of microbiota is performed in anaerobic conditions. A modified three‐stage chemostat exploited microbiota metabolism to produce different ratios of CO2 and H2, thus self‐generating anaerobiosis (Feria‐Gervasio et al., 2014). However, fluxes of N2, CO2, H2 or their combination (e.g., 90% N2, 5% CO2, 5% H2) remain the preferred strategy to reduce oxygen concentration. Before the experiment, the gases are flushed to remove oxygen from the tubes and vessels; then, the same flux is maintained during the whole culture to guarantee anaerobic conditions (Gaci et al., 2017; McDonald et al., 2013).
The presence of strict anaerobic bacteria in the microbiota is one of the main limitations impairing cocultures with cells. However, the combination of new technologies with increasing interest on microbiota effect on human health has boosted the design of bioengineered platforms able to finely control oxygen concentration.
1.3. Advanced culture systems to study microbiota‐host interaction
In vitro systems that sustain the coculture of obligate anaerobic bacteria with cells are as important as challenging. In the last decade, considerable efforts were employed to engineer complex in vitro models with an anoxic‐oxic interface (AOI) inside microphysiological environments (Figure 4 and Table 3). As the intestinal oxygen gradient contributes to the definition and the maintenance of microbiota heterogeneity and distribution, the AOI should be considered a necessary component to be implemented in advanced gut‐on‐a‐chip devices aiming at supporting microbial biodiversity (Zheng et al., 2015). To engineer the AOI, the simplest approach relies on a membrane separating a basal chamber, hosting a model of the intestinal epithelium (usually based on Caco‐2 cells), from an apical anaerobic chamber (Figure 4). In this way, the two chambers are ideally decoupled, allowing bacteria to survive in anaerobic conditions, while the oxygen flux coming from the aerobic chamber ensures cell survival. These models are static and indicated as apical anaerobic coculture systems and can be obtained with different levels of complexity (Maier et al., 2018; Ulluwishewa et al., 2015). From a technical point of view, the simplest configuration adds a oxygen‐impermeable shell to the standard Transwell® chambers, which is conventionally used to model biological barriers, thus creating anaerobic environment in the apical chamber (Figure 4a). This approach was able to sustain epithelial cell function, but also demonstrated the beneficial effects of bacteria (e.g., Faecalibacterium prausnitzii) on certain epithelial features and activity, as increased transepithelial electrical resistance (TEER), and modulation of anti‐inflammatory activity. However, the porous membrane fails to finely control oxygen gradient, because the gas may diffuse across the cell monolayer from the basal chamber (permeable to oxygen), eventually saturating the microbial apical side. For this reason, those system demonstrated to sustain anaerobic bacterial survival no longer than 8 h (Ulluwishewa et al., 2015). Even though the coculture of anaerobic bacteria and cells was significantly improved if compared to previous pure oxygen‐containing systems, these engineered Transwell® systems are not suitable for prolonged microbiota‐host commumunication investigation (von Martels et al., 2017).
Figure 4.
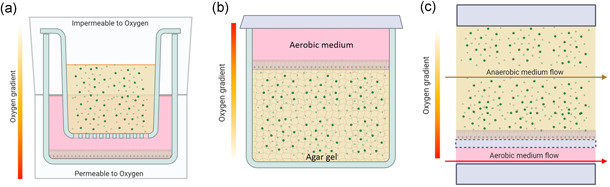
The different approaches used to develop in vitro AOI in a crescendo of complexity. The gradient of oxygen was statically engineered by the presence of O2 permeable/impermeable environments (a) or agar‐based gels (b). Differently, the aerobic/anaerobic medium flows allowed modeling AOI in dynamic gut‐on‐a‐chip systems (c). Figure made with biorender (https://biorender.com) [Color figure can be viewed at wileyonlinelibrary.com]
Table 3.
Different fluidic tools used for the modeling of the gut environment and the AOI interface
| System type | Pros | Cons | Main application | Ref. |
|---|---|---|---|---|
| Apical anaerobic coculture systems |
‐ Consolidate intestinal epithelium model ‐ Automated measurements of epithelial barrier integrity |
‐ Impossibility to control the oxygen tension gradient in time ‐ Coculture duration limited by Caco‐2 viability for prolonged periods |
‐ Faecalibacterium prausnitzii altered the immunomodulatory action of Caco‐2 cells |
Ulluwishewa et al. (2015, 2015); Maier et al. ()2017 |
| HoxBan |
‐ Use of compact and user‐friendly culture system made of classic centrifuge tubes ‐ Use of solid agar medium for the coculture of anaerobic gut bacteria ‐ Generation of a physiological steep oxygen tension gradient |
‐ Need of anaerobic culture environment ‐ Absence of dynamic perfusion for controlled metabolites exchange and mechanical stimulation ‐ Bacteria overgrowth and gases saturation |
‐ Demonstration of the metabolic mutualism between Caco2 cells and F. prausnitzii |
Sadabad et al. (2015) |
| HMI |
‐ Dynamic fluidic system ‐ Control on cell seeding and bacteria inoculation through separate culture chambers ‐ Complex microbiota culture up to 48 h ‐ Possibility of interfacing with human gastrointestinal tract simulators (i.e. SHIME) |
‐ Limited control on fluid velocities, shear forces and molecules concentration profiles due to the macro scale dimensions. |
‐ Enterocytes exposed to complex microbial community for prolonged time showed to stay vital and sustain the coculture for more than 24 h |
Marzorati et al. (2014) |
| Humix |
‐ Dynamic fluidic system ‐ Intermediate perfusion chamber separates human cells from the microbiota with in vivo‐like distance and smooth oxygen tension profile |
‐ Difficult measurements of epithelial barrier integrity ‐ Impossibility of interfacing with a SHIME‐like bioreactor |
‐ Individual transcriptional responses from human epithelial cells cocultured with LGG are comparable with in vivo findings |
Shah et al. (2016) |
| Gut‐on‐a‐chip |
‐ Microfluidics allows for fine tuning of flow velocity profiles and shear stress distribution as well as transport of molecules ‐ Mechanical actuation mimicking the intestinal peristalsis |
‐ Caco‐2 cells are put in direct contact with anaerobic bacteria through a permeable membrane ‐ Bacteria overgrowth in a micrometric space ‐ Need geometrical adaptation to host anaerobic bacteria. |
‐ Intestinal barrier dysbiosis triggers the onset of intestinal inflammation ‐ The barrier integrity is needed and sufficient to suppress the pro‐inflammatory cascades mediated by the host‐microbiome cross‐talk |
Shin and Kim (2018) |
| Anoxic‐oxic interface‐on‐a‐chip |
‐ no need of complex apparatus to maintain the anoxic condition (e.g. incubators, glove box) ‐ Geometry adapted for efficient physiological transepithelial anoxic gradient |
‐ Bacteria overgrowth in a micrometric space ‐ Impossibility of interfacing with a SHIME‐like bioreactor |
The transepithelial anoxic gradient is necessary and sufficient to coculture obligate anaerobic B. adolescentis and E. hallii with aerobic host epithelial cells | Shin et al. (2019) |
The Human oxygen‐Bacteria anaerobic (HoxBan) coculture system is an example of a compact and user‐friendly system for modeling the AOI in vitro (Sadabad et al., 2015). The aerobic and anaerobic compartments are recreated inside a 50 ml centrifuge tube. Caco‐2 cells are seeded on the top of a glass coverslip, which is exploited as a separatory element between the two chambers. On the basal level, anaerobic conditions were obtained by a 1% agar gel, with or without mucin, where the bacteria were ( 80% of the total) guarantees the generation of a steep oxygen gradient inside the gel, resembling the in vivo situation. Indeed, the oxygen was demonstrated to gradually diffuse into the agar bulk, without reaching the lowest part of the tube, where the obligate anaerobic bacteria grow.
The HoxBan successfully sustained the coculture of Caco‐2 cells and F. prausnitzii for one day, showing a mutual communication between intestinal epithelial cells and obligate anaerobic bacteria. However, the simplicity of the system is unable to recapitulate the complex gut environment, also because of the absence of important physiological stimuli like mucus flow and mechanical deformations. Moreover, culturing bacteria in the confined volume of a centrifuge tube leads to either bacteria overgrowth or saturation of gases derived from bacterial and cellular metabolism.
All these limitations are mitigated by ventilated and dynamic systems, where the presence of a fluid flow enables material renewal and dynamic stimulation with physiological levels of shear forces (few units of mPa) acting on bacteria (Kim et al., 2016; Marzorati et al., 2014).
Fluidic systems like the HMI module (Marzorati et al., 2014) and the HuMix device (Shah et al., 2016) showed to successfully sustain the AOI interface for suitable strict anaerobic coculture with cells. These systems represent suitable models of dynamic host‐microbiota interaction. The concept at the basis of fluidic systems consists of parallel chambers forming the luminal and basolateral compartments for bacteria and cell culture, respectively, intermediated by semipermeable membranes. In this configuration, the presence of an inlet and outlet in each chamber allows for a controlled seeding of cells and bacteria inoculation, while not impairing sample collection. In this way, the cocultures can be monitored in real‐time by biological analysis, including cytokine assays or metabolomics profiling (Shah et al., 2016). Another advantage of the decoupled perfusion is the possibility to inflow different media and gasses in the chambers (e.g., anoxic and/or oxic gasses), establishing a controlled gradient of oxygen from the aerobic environment of the cellular compartment (21% pO2) to the anaerobic condition for anaerobic bacteria culture (0.8% pO2) (P. Shah et al., 2016; Shin et al., 2019). In this way, the AOI interface is generated between the two chambers, recreating microaerophilic conditions within the mucosal layer. Additionally, dynamic fluid perfusion guarantees continuous metabolite intake and catabolite removal, while avoiding bacteria overgrowth. The achievement of a correct balance between the flow rate, molecule and metabolite diffusion, and the dynamic steady state of the culture requires a careful optimization of the process parameters. Therefore, the maintenance of the AOI interfaces increases the complexity of the experimental set‐up. For instance, the absence of oxygen in the anoxic medium needs to be finely controlled by constantly and continuously bubbling nitrogen, which must not interfere in the maintainace of the equilibrium in the diffusion of oxygen coming from the aerobic compartments across the permeable membrane (Shah et al., 2016). To this purpose, the platform is usually provided of sensors for the continuous monitoring of oxygen concentration and, ideally, of an automated control line to restore this equilibrium (Shah et al., 2016).
The dimensions of the dynamic systems can be at the macro‐ or microscale. The fabrication technique and the final application are determinant for the selection of the dimensions of the fluidic channels. Macro‐bioreactors show flow chambers with dimensions in the order of centimetres (e.g., the HMI module has a chamber of 10 × 6 cm). Such dimensions allow for interfacing with bioreactors for the long‐term culture of bacteria, which require the displacement of consistent volumes of media at relatively high flow rates (in the order of ml/min) (Marzorati et al., 2014). Reducing the dimensions of the fluidic channels brings several advantages, such as a more accurate control over the velocity profiles in the medium, the shear forces exerted on cells and bacteria, and the concentration profiles of the target molecules. Indeed, at the microscale, the characteristic length (L) of the system is micrometric and its contribution to inertial forces is reduced of three orders of magnitude if compared to macroscale bioreactors. This implies the fall in Reynolds number while increasing the laminarity of the flow (Brennan et al., 2014).
In microfluidic systems, the flow experiences minimal mixing due to the lack of turbulence and the prevailing of viscous forces. The modulation of the advection/diffusion ratio is necessary to control and enhance oxygen diffusion for stable and tailored oxygen tension profiles, which can be controlled by acting on the flow velocity along the principal channel direction (Rivera et al., 2019):
Microfabrication techniques (e.g., soft lithography) allow to fabricate fluidic devices with more than two microchambers. The HuMix device shows a central perfusion chamber between the upper microbial chamber and the lower human cells chamber. The presence of the central space separating human enterocytes and the microbial population recreates the typical distance (about 0.5–1 mm) between epithelial cells and microbiota found in healthy, intact epithelial barriers (Shah et al., 2016). This optimized structure is further coupled with the simultaneous perfusion of ‐oxic and ‐anoxic media through the micro‐chambers, which leads to the establishment and maintenance of an oxygen gradient close to the in vivo situation.
An advanced gut‐on‐a‐chip device supporting the growth of microbes and cells was presented by Kim and collaborators (Kim et al., 2012). They cultured the intestinal Lactobacillus rhamnosus GG (LGG) on the apical surface of a Caco‐2 cell monolayer in a microfluidic device. They demonstrated that the intestinal epithelium could coexist in direct contact with bacteria in a dynamic device with peristaltic‐like motion, showing enhanced and constant functional TEER with values around 4 kΩcm2, and the presence of morphological features (villi structure). Peristaltic‐like deformation is applied by with a controlled vacuum source, which induces the uniaxial stretching of a membrane, reaching cyclic 10% cell strain at the frequency of 0.15 Hz like in the human physiology (Deloose et al., 2012; Kim et al., 2012). In this example, the maintenance of anaerobic conditions inside the microchannel was not an issue, as LGG is a facultative anaerobic bacterium.
Most recently, the so called AOI‐on‐a‐chip was the first example of a microfluidic device where obligate anaerobic bacteria were successfully integrated in a chip for the study of host‐microbiota interaction (Shin et al., 2019). In this case, the AOI interface was optimized by tuning the height ratio between the upper and the lower channel. The increased height of the luminal microchamber was exploited to achieve the equilibrium between shear stress values, cell/bacterial viability and flow rates to recreate the physiological transepithelial AOI. Indeed, the authors demonstrated that the presence of a flow of preconditioned anaerobic medium at specific flow rates was sufficient to establish a proper oxygen gradient, without any other equipment or procedure. This not only established a steady‐state AOI in vitro, but also supported the growth of anaerobic human gut microbiomes, such as B. adolescentis and E. hallii, improving the total culture period up to 72 h (Shin et al., 2019). Gut‐on‐a‐chip devices with bacteria in direct contact with cells were suitable to investigate microbiota‐host crosstalk, exposing either synergy or antagonism. Still, further improvements are needed to achieve long‐term cultures and avoid overgrowth in the range of weeks for more robust recpaitulation of microbiota‐gut interactions. The main limitation of these systems when used alone is their impossibility to maintain bacterial cultures for more than few hours, because bacteria overgrowth occurs independently from the dynamic condition of the set‐up. This causes bacteria suffering and damage and results in metabolic alterations.
The use of bioreactors for microbiota fermentation—upstream to the gut‐on‐a‐chip device—is a valid tool to overcome this limitation, as demonstrated by the HMI module. Indeed, this gut‐on‐a‐chip device was successfully connected to a SHIME‐like reactor, where the fecal samples of patients were processed and then inoculated into the module (Marzorati et al., 2014). The combination of bacteria bioreactors and gut‐on‐a‐chip devices allowed to study the effects of microbiota‐related molecules on the modulation of the host response.
Even though the gut‐on‐a‐chip devices described above have the advantage of introducing a dynamic stimulation, they separate anaerobic bacteria from the epithelial cell layer with a synthetic membrane. This strategy protects the host cells from direct exposure to microbiota infections, but fails in recapitulating the real microbiota‐gut interface, for example including mucus. Indeed, the mucus is not only a biological structure with complex architecture, but it is also strongly dependent on the microbiota‐host crosstalk itself (Sardelli et al., 2019). For this reason, gut‐on‐a‐chip systems evolved to coculture bacteria into the luminal channel, in direct contact with the mucus of intestine epithelial layer, while keeping the microenvironment perfused (Kim et al., 2012; Shin et al., 2019; Sontheimer‐Phelps et al., 2020).
Up to date, the AOI has been developed in single organ in vitro models, i.e., gut‐on‐a‐chip devices. However, even though these systems demonstrated to sustain dynamic models of the intestinal barrier, they show some limitations when applied to the study of the intestinal microenvironment (Ceppa et al., 2020). To investigate microbiota relationship with distant organs (e.g., gut‐brain axis), AOI in vitro models need to be integrated in multiorgan platforms, defined as body‐on‐chips (BOC). Generally, they are made up of multi‐chamber devices connected in series and kept in communication by fluidic circuits (Kimura et al., 2018). They were efficiently used to determine the absorption process of drugs, by combining in vitro models of intestine and liver, or to investigate the distribution and excretion of molecules by modeling the blood circulation and glomerular filtration (Imura et al., 2010, 2013; Kimura et al., 2015). However, engineering AOI compartments in BOC represents a challenging technological issue. Indeed, it requires not only the high control over flows to guarantee the correct mass transport among the different body districts, but also to provide the oxygen gradients for the coculture of bacteria and cells. This is particularly difficult when long biological processes are considered, because the presence of continuous bacterial cultures must be integrated into the system without leading to undesired infections. The development of BOC that can cope with AIO, long term bacterial growth and multi‐organs models may represens the opportunity for a significative step forward towards more reliable models of complex biological processes, such as molecular axis involving microbiota metabolism, immune response and neurodegeneration (Raimondi et al., 2019).
2. CONCLUSIONS
Continuous bioreactors allow for the culture of bacteria for months whent the proper control of the experimental parameters is guaranteed. However, their application has mainly regarded the analysis of the fermentation products, and not the investigation of microbiota‐host cell crosstalk. This is due to several limitations. First, culturing in vitro the human microbiota is intrinsically difficult: microbiota isolated from patients’ samples differs in terms biodiversity and concentration from the in vivo situation, being heavily manipulated by the inoculation process. Moreover, the experimental set‐up is extremely elaborate and expensive, reaching the highest complexity in the case of three‐stage bioreactors, where a deep technical knowledge is needed for the correct control of the culture parameters (e.g., chemostats or changestat configuration). Second, the available gut‐on‐a‐chip devices are not designed to sustain long‐term cocultures of bacteria and cells. Although relevant progresses has been achieved in integrating bacteria into cellular microdevices, the small sizes, low flow rates and the complexity required to maintain an AOI made these systems suitable only for limited applications. For instance, they are valid and efficient tools for the analysis of infection mechanisms and the modulation of gut permeability. However, they do not allow investigating the mechanisms behind complex biological phenomena, such as molecular axis between microbiota products and distant organs homeostatis. A considerable step forward in this field could be achieved by coupling bioreactors for bacterial culture with multicomponent microfluidic systems. Indeed, this new engineered approach may strengthen the advantages of bacterial bioreactors (i.e. long‐term cultures, indispensable to recapitulate chronic diseases), with those of microfluidic devices, like the deep control over experimental parameters. In this way, complex phenomena such as the gut‐brain axis would be potentially unveiled, opening new medical paradigms and therapeutic scenarios.
AUTHOR CONTRIBUTIONS
Lorenzo Sardelli: original draft preparation (lead), visualization, conceptualization (supporting), review and editing (equal); Simone Perottoni: original draft preparation (equal), review and editing (equal); Marta Tunesi: original draft preparation (equal), review and editing (equal); Lucia Boeri: original draft preparation (equal), review and editing (equal); Federica Fusco: original draft preparation (equal), review and editing (equal); Paola Petrini: conceptualization (lead), review and editing (lead); Diego Albani: conceptualization (lead), review and editing (lead), supervision; Carmen Giordano: conceptualization (lead), review and editing (lead), supervision, funding acquisition.
ACKNOWLEDGMENTS
This study was funded by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (Grant agreement No. 724734‐MINERVA). The results reflect only the authors' views and the Agency is not responsible for any use that may be made of the information contained.
Sardelli, L., Perottoni, S., Tunesi, M., Boeri, L., Fusco, F., Petrini, P., Albani, D., & Giordano, C. (2021). Technological tools and strategies for culturing human gut microbiota in engineered in vitro models. Biotechnology Bioengineering. 118, 2886–2905. 10.1002/bit.27816
REFERENCES
- Adamberg, K., & Adamberg, S. (2018). Selection of fast and slow growing bacteria from fecal microbiota using continuous culture with changing dilution rate. Microbial Ecology in Health and Disease, 29(1), 1549922. 10.1080/16512235.2018.1549922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamberg, K., Raba, G., & Adamberg, S. (2020). Use of changestat for growth rate studies of gut microbiota. Frontiers in Bioengineering and Biotechnology, 8, 1–12. 10.3389/fbioe.2020.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamberg, K., Valgepea, K., & Vilu, R. (2015). Advanced continuous cultivation methods for systems microbiology. Microbiology, 161(9), 1707–1719. 10.1099/mic.0.000146 [DOI] [PubMed] [Google Scholar]
- Amoako‐Tuffour, Y., Jones, M. L., Shalabi, N., Labbé, A., Vengallatore, S., & Prakash, S. (2014). Ingestible gastrointestinal sampling devices: State‐of‐the‐art and future directions. Critical Reviews in Biomedical Engineering, 42(1), 1–15. 10.1615/CritRevBiomedEng.2014010846 [DOI] [PubMed] [Google Scholar]
- Arrieta, M. C., & Finlay, B. B. (2012). The commensal microbiota drives immune homeostasis. Frontiers in Immunology, 3(1), 33. 10.3389/fimmu.2012.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarlen, P. V., Troost, F. J., Hemert, S. V., Meer, C. V. D., & Vos, W. M. D. (2008). Differential NF‐ B pathways induction by Lactobacillus plantarum in the duodenum of healthy. Proceedings of the National Academy of Sciences of the United States of America, 106(7), 2371–2376. 10.1073/pnas.0809919106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellali, S., Bou Khalil, J., Fontanini, A., Raoult, D., & Lagier, J.‐C. (2020). A new protectant medium preserving bacterial viability after freeze drying. Microbiological Research, 236, 126454. 10.1016/j.micres.2020.126454 [DOI] [PubMed] [Google Scholar]
- Bellali, S., Lagier, J. C., Raoult, D., & Khalil, J. B. (2019). Among live and dead bacteria, the optimization of sample collection and processing remains essential in recovering gut microbiota components. Frontiers in Microbiology, 10, 1–9. 10.3389/fmicb.2019.01606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunia, P. (2014). Fundamentals of biological treatment. Comprehensive Water Quality and Purification, 3, 47–73. 10.1016/B978-0-12-382182-9.00048-7 [DOI] [Google Scholar]
- Boeri, L., Izzo, L., Sardelli, L., Tunesi, M., Albani, D., & Giordano, C. (2019). Advanced organ‐on‐a‐chip devices to investigate liver multi‐organ communication: Focus on gut, microbiota and brain. Bioengineering, 6(4), 91. 10.3390/bioengineering6040091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braniste, V., Al‐Asmakh, M., Kowal, C., Anuar, F., Abbaspour, A., Tóth, M., Korecka, A., Bakocevic, N., Guan, N. L., Kundu, P., Gulyás, B., Halldin, C., Hultenby, K., Nilsson, H., Hebert, H., Volpe, B. T., Diamond, B., & Pettersson, S. (2014). The gut microbiota influences blood‐brain barrier permeability in mice. Science Translational Medicine, 6(263), 263. 10.1126/scitranslmed.3009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan, M. D., Rexius‐Hall, M. L., Elgass, L. J., & Eddington, D. T. (2014). Oxygen control with microfluidics. Lab on a Chip, 14(22), 4305–4318. 10.1039/c4lc00853g [DOI] [PubMed] [Google Scholar]
- Brial, F., Le Lay, A., Dumas, M. E., & Gauguier, D. (2018). Implication of gut microbiota metabolites in cardiovascular and metabolic diseases. Cellular and Molecular Life Sciences, 75(21), 3977–3990. 10.1007/s00018-018-2901-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed, F. (2011). Programming of host metabolism by the gut microbiota. Annals of Nutrition and Metabolism, 58(Suppl. 2), 44–52. 10.1159/000328042 [DOI] [PubMed] [Google Scholar]
- Cattaneo, A., Cattane, N., Galluzzi, S., Provasi, S., Lopizzo, N., Festari, C., Ferrari, C., Guerra, U. P., Paghera, B., Muscio, C., Bianchetti, A., Volta, G. D., Turla, M., Cotelli, M. S., Gennuso, M., Prelle, A., Zanetti, O., Lussignoli, G., Mirabile, D., … Frisoni, G. B. (2017). Association of brain amyloidosis with pro‐inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiology of Aging, 49, 60–68. 10.1016/j.neurobiolaging.2016.08.019 [DOI] [PubMed] [Google Scholar]
- Ceppa, F. A., Izzo, L., Sardelli, L., Raimondi, I., Tunesi, M., Albani, D., & Giordano, C. (2020). Human gut‐microbiota interaction in neurodegenerative disorders and current engineered tools for its modeling. Frontiers in Cellular and Infection Microbiology, 10, 297. 10.3389/fcimb.2020.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, E. S., Viardot, A., Psichas, A., Morrison, D. J., Murphy, K. G., Zac‐Varghese, S. E., MacDougall, K., Preston, T., Tedford, C., Finlayson, G. S., Blundell, J. E., Bell, J. D., Thomas, E. L., Mt‐Isa, S., Ashby, D., Gibson, G. R., Kolida, S., Dhillo, W. S., Bloom, S. R., … Frost, G. (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut, 64(11), 1744–1754. 10.1136/gutjnl-2014-307913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K. S., Lee, W. Y., & Yu, W. L. (2016). Coexisting cytomegalovirus infection in immunocompetent patients with Clostridium difficile colitis. Journal of Microbiology, Immunology and Infection, 49(6), 829–836. 10.1016/j.jmii.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Chassaing, B., & Gewirtz, A. T. (2019). Identification of inner mucus‐associated bacteria by laser capture microdissection. Cmgh, 7(1), 157–160. 10.1016/j.jcmgh.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, M., & Ning, K. (2019). Stereotypes about enterotype: the old and new ideas. Genomics, Proteomics and Bioinformatics, 17(1), 4–12. 10.1016/j.gpb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child, M. W., Kennedy, A., Walker, A. W., Bahrami, B., Macfarlane, S., & Macfarlane, G. T. (2006). Studies on the effect of system retention time on bacterial populations colonizing a three‐stage continuous culture model of the human large gut using FISH techniques. FEMS Microbiology Ecology, 55(2), 299–310. 10.1111/j.1574-6941.2005.00016.x [DOI] [PubMed] [Google Scholar]
- Choung, R. S., Ruff, K. C., Malhotra, A., Herrick, L., Locke, G. R., Harmsen, W. S., Zinsmeister, A. R., Talley, N. J., & Saito, Y. A. (2011). Clinical predictors of small intestinal bacterial overgrowth by duodenal aspirate culture. Alimentary Pharmacology and Therapeutics, 33(9), 1059–1067. 10.1111/j.1365-2036.2011.04625.x [DOI] [PubMed] [Google Scholar]
- Cinquin, C., Le Blay, G., Fliss, I., & Lacroix, C. (2004). Immobilization of infant fecal microbiota and utilization in an in vitro colonic fermentation model. Microbial Ecology, 48(1), 128–138. 10.1007/s00248-003-2022-7 [DOI] [PubMed] [Google Scholar]
- Cinquin, C., Le Blay, G., Fliss, I., & Lacroix, C. (2006a). Comparative effects of exopolysaccharides from lactic acid bacteria and fructo‐oligosaccharides on infant gut microbiota tested in an in vitro colonic model with immobilized cells. FEMS Microbiology Ecology, 57(2), 226–238. 10.1111/j.1574-6941.2006.00118.x [DOI] [PubMed] [Google Scholar]
- Cinquin, C., Le Blay, G., Fliss, I., & Lacroix, C. (2006b). New three‐stage in vitro model for infant colonic fermentation with immobilized fecal microbiota. FEMS Microbiology Ecology, 57(2), 324–336. 10.1111/j.1574-6941.2006.00117.x [DOI] [PubMed] [Google Scholar]
- Cleusix, V., Lacroix, C., Vollenweider, S., & Le Blay, G. (2008). Glycerol induces reuterin production and decreases Escherichia coli population in an in vitro model of colonic fermentation with immobilized human feces. FEMS Microbiology Ecology, 63(1), 56–64. 10.1111/j.1574-6941.2007.00412.x [DOI] [PubMed] [Google Scholar]
- Clevers, H. (2013). The intestinal crypt, a prototype stem cell compartment. Cell, 154(2), 274. 10.1016/j.cell.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Coimbra, J. M., Cristina dos Reis, K., Schwan, R. F., & Silva, C. F. (2020). Effect of the strategy of molasses supplementation in vinasse to high scp production and rose flavor compound. Waste and Biomass Valorization, 12, 359–369. 10.1007/s12649-020-00961-2 [DOI] [Google Scholar]
- Conlon, M. A., & Bird, A. R. (2015). The impact of diet and lifestyle on gut microbiota and human health. Nutrients, 7, 17–44. 10.3390/nu7010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan, J. F., O'riordan, K. J., Cowan, C., Sandhu, K. V., Bastiaanssen, T., Boehme, M., Codagnone, M. G., Cussotto, S., Fulling, C., Golubeva, A. V., Guzzetta, K. E., Jaggar, M., Long‐Smith, C. M., Lyte, J. M., Martin, J. A., Molinero‐Perez, A., Moloney, G., Morelli, E., Morillas, E., … Dinan, T. G. (2019). The microbiota‐gut‐brain axis. Physiological Reviews, 99(4), 1877–2013. 10.1152/physrev.00018.2018 [DOI] [PubMed] [Google Scholar]
- Cui, J., Zheng, X., Hou, W., Zhuang, Y., Pi, X., & Yang, J. (2008). The study of a remote‐controlled gastrointestinal drug delivery and sampling system. Telemedicine and E‐Health, 14(7), 715–719. 10.1089/tmj.2007.0118 [DOI] [PubMed] [Google Scholar]
- Deloose, E., Janssen, P., Depoortere, I., & Tack, J. (2012). The migrating motor complex: Control mechanisms and its role in health and disease. Nature Reviews Gastroenterology & Hepatology, 9(5), 271–285. 10.1038/nrgastro.2012.57 [DOI] [PubMed] [Google Scholar]
- Dieterich, W., Schink, M., & Zopf, Y. (2018). Microbiota in the gastrointestinal tract. Medical Sciences, 6(4), 116. 10.3390/medsci6040116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan, T. G., & Cryan, J. F. (2017a). Gut instincts: Microbiota as a key regulator of brain development, ageing and neurodegeneration. Journal of Physiology, 595(2), 489–503. 10.1113/JP273106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan, T. G., & Cryan, J. F. (2017b). Brain‐gut‐microbiota axis and mental health. Psychosomatic Medicine, 79(8), 920–926. 10.1097/PSY.0000000000000519 [DOI] [PubMed] [Google Scholar]
- Doble, M., & Kumar, A. (2005). Mathematical models. In Doble M., & Kumar A. (Eds.), Biotreatment of industrial effluents (pp. 39–53). Butterworth‐Heinemann: Elsevier. 10.1016/B978-075067838-4/50005-1 [DOI] [Google Scholar]
- Donaldson, G. P., Lee, S. M., & Mazmanian, S. K. (2015). Gut biogeography of the bacterial microbiota. Nature Reviews Microbiology, 14(1), 20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal, A., Fehlbaum, S., Chassard, C., Zimmermann, M. B., & Lacroix, C. (2013). Low iron availability in continuous in vitro colonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS Microbiology Ecology, 83(1), 161–175. 10.1111/j.1574-6941.2012.01461.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi, F., Raoult, D., & Merhej, V. (2017). Metabolic role of lactobacilli in weight modification in humans and animals. Microbial Pathogenesis, 106, 182–194. 10.1016/j.micpath.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Emmert‐Buck, M. R., Bonner, R. F., Smith, P. D., Chuaqui, R. F., Zhuang, Z., Goldstein, S. R., Weiss, R. A., & Liotta, L. A. (1996). Laser capture microdissection. Science, 274(5289), 998–1001. 10.1126/science.274.5289.998 [DOI] [PubMed] [Google Scholar]
- ERC Funded Projects | ERC: European Research Council . (n.d.). Retrieved July 16, 2020, from https://erc.europa.eu/projects-figures/erc-funded-projects
- Espey, M. G. (2013). Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radical Biology and Medicine, 55, 130–140. 10.1016/j.freeradbiomed.2012.10.554 [DOI] [PubMed] [Google Scholar]
- Fehlbaum, S., Chassard, C., Haug, M. C., Fourmestraux, C., Derrien, M., & Lacroix, C. (2015). Design and investigation of PolyFermS in vitro continuous fermentation models inoculated with immobilized fecal microbiota mimicking the elderly colon. PLOS One, 10(11), 0142793. 10.1371/journal.pone.0142793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feria‐Gervasio, D., Denis, S., Alric, M., & Brugère, J. F. (2011). In vitro maintenance of a human proximal colon microbiota using the continuous fermentation system P‐ECSIM. Applied Microbiology and Biotechnology, 91(5), 1425–1433. 10.1007/s00253-011-3462-5 [DOI] [PubMed] [Google Scholar]
- Feria‐Gervasio, D., Tottey, W., Gaci, N., Alric, M., Cardot, J. M., Peyret, P., Martin, J. F., Pujos, E., Sébédio, J. L., & Brugère, J. F. (2014). Three‐stage continuous culture system with a self‐generated anaerobia to study the regionalized metabolism of the human gut microbiota. Journal of Microbiological Methods, 96(1), 111–118. 10.1016/j.mimet.2013.11.015 [DOI] [PubMed] [Google Scholar]
- Forsyth, C. B., Shannon, K. M., Kordower, J. H., Voigt, R. M., Shaikh, M., Jaglin, J. A., Estes, J. D., Dodiya, H. B., & Keshavarzian, A. (2011). Increased intestinal permeability correlates with sigmoid mucosa alpha‐synuclein staining and endotoxin exposure markers in early Parkinson's disease. PLOS One, 6(12), 28032. 10.1371/journal.pone.0028032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, T. C., Olson, C. A., & Hsiao, E. Y. (2017). Interactions between the microbiota, immune and nervous systems in health and disease. Nature Neuroscience, 20, 145–155. 10.1038/nn.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaci, N., Chaudhary, P. P., Tottey, W., Alric, M., & Brugère, J.‐F. (2017). Functional amplification and preservation of human gut microbiota. Microbial Ecology in Health and Disease, 28(1), 1308070. 10.1080/16512235.2017.1308070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Villalba, R., Vissenaekens, H., Pitart, J., Romo‐Vaquero, M., Espín, J. C., Grootaert, C., Selma, M. V., Raes, K., Smagghe, G., Possemiers, S., Van Camp, J., & Tomas‐Barberan, F. A. (2017). Gastrointestinal simulation model TWIN‐SHIME shows differences between human urolithin‐metabotypes in gut microbiota composition, pomegranate polyphenol metabolism, and transport along the intestinal tract. Journal of Agricultural and Food Chemistry, 65(27), 5480–5493. 10.1021/acs.jafc.7b02049 [DOI] [PubMed] [Google Scholar]
- Gibson, G. R., & Fuller, R. (2000). Aspects of in vitro and in vivo research approaches directed toward identifying probiotics and prebiotics for human use. The Journal of Nutrition, 130(2), 391S–395S. 10.1093/jn/130.2.391S [DOI] [PubMed] [Google Scholar]
- Gibson, G. R., Cummings, J. H., & Macfarlane, G. T. (1988). Use of a three‐stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Applied and Environmental Microbiology, 54(11), 2750–2755. 10.1128/aem.54.11.2750-2755.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudar, C. T. (2012). Computer programs for modeling mammalian cell batch and fed‐batch cultures using logistic equations. Cytotechnology, 64(4), 465–475. 10.1007/s10616-011-9425-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham, D., & Hong, J. (2015). The functional basis of adaptive evolution in chemostats. FEMS Microbiology Reviews, 39(1), 2–16. 10.1111/1574-6976.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Mortensen, M. S., Schjørring, S., Trivedi, U., Vestergaard, G., Stokholm, J., Bisgaard, H., Krogfelt, K. A., & Sørensen, S. J. (2019). Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Communications Biology, 2(1), 1–7. 10.1038/s42003-019-0540-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, M. F., Sidhu, R., & McAlindon, M. E. (2014). Capsule endoscopy: Current practice and future directions. World Journal of Gastroenterology, 20(24), 7752–7759. 10.3748/wjg.v20.i24.7752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harach, T., Marungruang, N., Duthilleul, N., Cheatham, V., Mc Coy, K. D., Frisoni, G., Neher, J. J., Fåk, F., Jucker, M., Lasser, T., & Bolmont, T. (2017). Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Scientific Reports, 7, 41802. 10.1038/srep41802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijtz, R. D., Wang, S., Anuar, F., Qian, Y., Björkholm, B., Samuelsson, A., Hibberd, M. L., Forssberg, H., & Pettersson, S. (2011). Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America, 108(7), 3047–3052. 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman, E. T., Lu, H., Yao, T., & Nakatsu, C. H. (2017). Microbial ecology along the gastrointestinal tract. Microbes and Environments, 32(4), 300–313. 10.1264/jsme2.ME17017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema, S., Douma, R. D., Janssen, M., Tramper, J., & Wijffels, R. H. (2006). Controlling light‐use byRhodobacter capsulatus continuous cultures in a flat‐panel photobioreactor. Biotechnology and Bioengineering, 95(4), 613–626. 10.1002/bit.20907 [DOI] [PubMed] [Google Scholar]
- Holmes, Z. C., Silverman, J. D., Dressman, H. K., Wei, Z., Dallow, E. P., Armstrong, S. C., Seed, P. C., Rawls, J. F., & David, L. A. (2020). Short‐chain fatty acid production by gut microbiota from children with obesity differs according to prebiotic choice and bacterial community composition. mBio, 11(4), 1–15. 10.1128/mbio.00914-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, Y. H., Peterson, C. M., Raggio, A., Keenan, M. J., Martin, R. J., Ravussin, E., & Marco, M. L. (2016). Impact of different fecal processing methods on assessments of bacterial diversity in the human intestine. Frontiers in Microbiology, 7, 1–11. 10.3389/fmicb.2016.01643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imura, Y., Sato, K., & Yoshimura, E. (2010). Micro total bioassay system for ingested substances: assessment of intestinal absorption, hepatic metabolism, and bioactivity. Analytical Chemistry, 82(24), 9983–9988. 10.1021/ac100806x [DOI] [PubMed] [Google Scholar]
- Imura, Y., Yoshimura, E., & Sato, K. (2013). Microcirculation system with a dialysis part for bioassays evaluating anticancer activity and retention. Analytical Chemistry, 85(3), 1683–1688. 10.1021/ac302938q [DOI] [PubMed] [Google Scholar]
- Ito, T., Sekizuka, T., Kishi, N., Yamashita, A., & Group, F. (2019). Conventional culture methods with commercially available media unveil the presence of novel culturable bacteria. Gut Microbes, 10(1), 77–91. 10.1080/19490976.2018.1491265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili‐Firoozinezhad, S., Gazzaniga, F. S., Calamari, E. L., Camacho, D. M., Fadel, C. W., Bein, A., Swenor, B., Nestor, B., Cronce, M. J., Tovaglieri, A., Levy, O., Gregory, K. E., Breault, D. T., Cabral, J. M. S., Kasper, D. L., Novak, R., & Ingber, D. E. (2019). A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nature Biomedical Engineering, 3, 520–531. 10.1038/s41551-019-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R. B., Zhu, X., Moan, E., Murff, H. J., Ness, R. M., Seidner, D. L., Sun, S., Yu, C., Dai, Q., Fodor, A. A., Azcarate‐peril, M. A., & Shrubsole, M. J. (2018). Inter‐niche and inter‐individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Scientific Reports, 8, 4139. 10.1038/s41598-018-22408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judkins, T. C., Archer, D. L., Kramer, D. C., & Solch, R. J. (2020). Probiotics, nutrition, and the small intestine. Current Gastroenterology Reports, 22(1), 1–8. 10.1007/s11894-019-0740-3 [DOI] [PubMed] [Google Scholar]
- Keshavarzian, A., Green, S. J., Engen, P. A., Voigt, R. M., Naqib, A., Forsyth, C. B., Mutlu, E., & Shannon, K. M. (2015). Colonic bacterial composition in Parkinson's disease. Movement Disorders, 30(10), 1351–1360. 10.1002/mds.26307 [DOI] [PubMed] [Google Scholar]
- Keightley, P. C., Koloski, N. A., & Talley, N. J. (2015). Pathways in gut‐brain communication: Evidence for distinct gut‐to‐brain and brain‐to‐gut syndromes. Australian and New Zealand Journal of Psychiatry, 49(3), 207–214. 10.1177/0004867415569801 [DOI] [PubMed] [Google Scholar]
- Kelly, J. R., Minuto, C., Cryan, J. F., Clarke, G., & Dinan, T. G. (2017). Cross talk: The microbiota and neurodevelopmental disorders. Frontiers in Neuroscience, 11, 1–31. 10.3389/fnins.2017.00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, N. A., Walton, G. E., Gibson, G. R., Tuohy, K. M., & Andrews, S. C. (2014). In vitro batch cultures of gut microbiota from healthy and ulcerative colitis (UC) subjects suggest that sulphate‐reducing bacteria levels are raised in UC and by a protein‐rich diet. International Journal of Food Sciences and Nutrition, 65(1), 79–88. 10.3109/09637486.2013.825700 [DOI] [PubMed] [Google Scholar]
- Kim, C. H., Park, J., & Kim, M. (2014). Gut microbiota‐derived short‐chain fatty acids, T cells, and inflammation. Immune Network, 14(6), 277–288. 10.4110/in.2014.14.6.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. J., Huh, D., Hamilton, G., & Ingber, D. E. (2012). Human gut‐on‐a‐chip inhabited by microbial flora that experiences intestinal peristalsis‐like motions and flow. Lab on a Chip, 12(12), 2165–2174. 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- Kim, H. J., Li, H., Collins, J. J., & Ingber, D. E. (2016). Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut‐on‐a‐chip. Proceedings of the National Academy of Sciences of the United States of America, 113(1), E7–E15. 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Qie, Y., Park, J., & Kim, C. H. (2016). Gut microbial metabolites fuel host antibody responses. Cell Host and Microbe, 20(2), 202–214. 10.1016/j.chom.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H., Ikeda, T., Nakayama, H., Sakai, Y., & Fujii, T. (2015). An on‐chip small intestine‐liver model for pharmacokinetic studies. Journal of Laboratory Automation, 20, 4–6. 10.1177/2211068214557812 [DOI] [PubMed] [Google Scholar]
- Kimura, H., Sakai, Y., & Fujii, T. (2018). Drug metabolism and pharmacokinetics organ/body‐on‐a‐chip based on micro fl uidic technology for drug discovery. Drug metabolism and pharmacokinetics, 33(1), 43–48. 10.1016/j.dmpk.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Korem Kohanim, Y., Levi, D., Jona, G., Towbin, B. D., Bren, A., & Alon, U. (2018). A bacterial growth law out of steady state. Cell Reports, 23(10), 2891–2900. 10.1016/j.celrep.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Koziolek, M., Grimm, M., Becker, D., Iordanov, V., Zou, H., Shimizu, J., Wanke, C., Garbacz, G., & Weitschies, W. (2015). Investigation of pH and temperature profiles in the GI tract of fasted human subjects using the Intellicap® system. Journal of Pharmaceutical Sciences, 104(9), 2855–2863. 10.1002/jps.24274 [DOI] [PubMed] [Google Scholar]
- Kubitschek, H. E., & Bendigkeit, H. E. (1964). Mutation in continuous cultures. I. Dependence of mutational response upon growth‐limiting factors. Mutation Research ‐ Fundamental and Molecular Mechanisms of Mutagenesis, 1(2), 113–120. 10.1016/0027-5107(64)90013-2 [DOI] [PubMed] [Google Scholar]
- Lagier, J. C., Edouard, S., Pagnier, I., Mediannikov, O., Drancourt, M., & Raoult, D. (2015). Current and past strategies for bacterial culture in clinical microbiology. Clinical Microbiology Reviews, 28(1), 208–236. 10.1128/CMR.00110-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahtvee, P. J., Adamberg, K., Arike, L., Nahku, R., Aller, K., & Vilu, R. (2011). Multi‐omics approach to study the growth efficiency and amino acid metabolism in Lactococcus lactis at various specific growth rates. Microbial Cell Factories, 10(1), 12. 10.1186/1475-2859-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa, B. B., Navas‐Cortés, J. A., Hervás, A., & Jiménez‐Díaz, R. M. (2001). Influence of temperature and inoculum density of Fusarium oxysporum f. sp. ciceris on suppression of fusarium Wilt of chickpea by rhizosphere bacteria. Phytopathology, 91(8), 807–816. 10.1094/PHYTO.2001.91.8.807 [DOI] [PubMed] [Google Scholar]
- Lavelle, A., Lennon, G., O'Sullivan, O., Docherty, N., Balfe, A., Maguire, A., Mulcahy, H. E., Doherty, G., O'Donoghue, D., Hyland, J., Ross, R. P., Coffey, J. C., Sheahan, K., Cotter, P. D., Shanahan, F., Winter, D. C., & O'Connell, P. R. (2015). Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut, 64(10), 1553–1561. 10.1136/gutjnl-2014-307873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle, A., Lennon, G., Docherty, N., Balfe, A., Mulcahy, H. E., Doherty, G., O'Donoghue, D., Hyland, J. M., Shanahan, F., Sheahan, K., Coffey, J. C., Winter, D. C., & O'Connell, P. R. (2013). Depth‐dependent differences in community structure of the human colonic microbiota in health. PLOS One, 8(11), 78835. 10.1371/journal.pone.0078835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle, A., Lennon, G., O'Sullivan, O., Docherty, N., Balfe, A., Maguire, A., Mulcahy, H. E., Doherty, G., O'Donoghue, D., Hyland, J. M., Ross, R. P., Coffey, J. C., Sheahan, K., Cotter, P. D., Shanahan, F., Winter, D. C., O'Connell, P. R., Sheahan, K., Coffey, J. C., & Kleerebezem, M. (2010). Investigation of pH and temperature profiles in the GI tract of fasted human subjects using the Intellicap® system. The New England Journal of Medicine, 20(1), 1–8. 10.1038/ismej.2011.212 [DOI] [Google Scholar]
- Le Blay, G., Chassard, C., Baltzer, S., & Lacroix, C. (2010). Set up of a new in vitro model to study dietary fructans fermentation in formula‐fed babies. British Journal of Nutrition, 103(3), 403–411. 10.1017/S0007114509991796 [DOI] [PubMed] [Google Scholar]
- Leclercq, S., Matamoros, S., Cani, P. D., Neyrinck, A. M., Jamar, F., Stärkel, P., Windey, K., de Tremaroli, V., Bäckhed, F., Verbeke, K., Timary, P., & Delzenne, N. M. (2014). Intestinal permeability, gut‐bacterial dysbiosis, and behavioral markers of alcohol‐dependence severity. Proceedings of the National Academy of Sciences of the United States of America, 111(42), E4485–E4493. 10.1073/pnas.1415174111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litou, C., Psachoulias, D., Vertzoni, M., Dressman, J., & Reppas, C. (2020). Measuring pH and buffer capacity in fluids aspirated from the fasted upper gastrointestinal tract of healthy adults. Pharmaceutical Research, 37(3), 42. 10.1007/s11095-019-2731-3 [DOI] [PubMed] [Google Scholar]
- Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., & Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature, 489(7415), 220–230. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis, A. J. H., van den Heuvel, E. G., Schoterman, M. H. C., & Venema, K. (2012). Galacto‐oligosaccharides have prebiotic activity in a dynamic in vitro colon model using a 13C‐labeling technique. The Journal of Nutrition, 142(7), 1205–1212. 10.3945/jn.111.157420 [DOI] [PubMed] [Google Scholar]
- Macfarlane, G. T., & Macfarlane, S. (2007). Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Current Opinion in Biotechnology, 18(2), 156–162. 10.1016/j.copbio.2007.01.011 [DOI] [PubMed] [Google Scholar]
- Macfarlane, G. T., Gibson, G. R., & Cummings, J. H. (1992). Comparison of fermentation reactions in different regions of the human colon. Journal of Applied Bacteriology, 72(1), 57–64. 10.1111/j.1365-2672.1992.tb04882.x [DOI] [PubMed] [Google Scholar]
- Macfarlane, G. T., Macfarlane, S., & Gibson, G. R. (1998). Validation of a three‐stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microbial Ecology, 35(2), 180–187. 10.1007/s002489900072 [DOI] [PubMed] [Google Scholar]
- Maier, E., Anderson, R., & Roy, N. (2017). Live Faecalibacterium prausnitzii Does Not Enhance Epithelial Barrier Integrity in an Apical Anaerobic Co‐Culture Model of the Large Intestine. Nutrients, 9(12), 1349 10.3390/nu9121349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, E., Anderson, R. C., Altermann, E., & Roy, N. C.. (2018). Live Faecalibacterium prausnitzii induces greater TLR2 and TLR2/6 activation than the dead bacterium in an apical anaerobic co‐culture system. Cellular Microbiology, 20(2), e12805. 10.1111/cmi.12805 [DOI] [PubMed] [Google Scholar]
- Mailhe, M., Ricaboni, D., Vitton, V., Gonzalez, J. M., Bachar, D., Dubourg, G., Cadoret, F., Robert, C., Delerce, J., Levasseur, A., Fournier, P. E., Angelakis, E., Lagier, J. C., & Raoult, D. (2018). Repertoire of the gut microbiota from stomach to colon using culturomics and next‐generation sequencing. BMC Microbiology, 18(1), 157. 10.1186/s12866-018-1304-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Manach, C., Scalbert, A., Morand, C., Rémésy, C., & Jiménez, L. (2004). Polyphenols: Food sources and bioavailabilit. The American Journal of Clinical Nutrition, 79(5), 727–747. https://academic.oup.com/ajcn/article-abstract/79/5/727/4690182 [DOI] [PubMed] [Google Scholar]
- Martínez, N., Hidalgo‐Cantabrana, C., Delgado, S., Margolles, A., & Sánchez, B. (2019). Filling the gap between collection, transport and storage of the human gut microbiota. Scientific Reports, 9(1), 1–8. 10.1038/s41598-019-44888-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzorati, M., Vanhoecke, B., De Ryck, T., Sadaghian Sadabad, M., Pinheiro, I., Possemiers, S., Van Den Abbeele, P., Derycke, L., Bracke, M., Pieters, J., Hennebel, T., Harmsen, H. J., Verstraete, W., & Van De Wiele, T. (2014). The HMITM module: A new tool to study the Host‐Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiology, 14(1), 133. 10.1186/1471-2180-14-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J. A. K., Schroeter, K., Fuentes, S., Heikamp‐deJong, I., Khursigara, C. M., de Vos, W. M., & Allen‐Vercoe, E. (2013). Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. Journal of Microbiological Methods, 95(2), 167–174. 10.1016/j.mimet.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Michel, L., & Prat, A. (2016). One more role for the gut: Microbiota and blood brain barrier. Annals of Translational Medicine, 4(1), 15. 10.3978/j.issn.2305-5839.2015.10.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million, M., Armstrong, N., Khelaifia, S., Guilhot, E., Richez, M., Lagier, J. C., Dubourg, G., Chabriere, E., & Raoult, D. (2020). The antioxidants glutathione, ascorbic acid and uric acid maintain butyrate production by human gut clostridia in the presence of oxygen in vitro. Scientific Reports, 10(1), 1–11. 10.1038/s41598-020-64834-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minekus, M., Smeets‐Peeters, M., Havenaar, R., Bernalier, A., Fonty, G., Marol‐Bonnin, S., Alric, M., Marteau, P., & Huis In't Veld, J. H. J. (1999). A computer‐controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Applied Microbiology and Biotechnology, 53(1), 108–114. 10.1007/s002530051622 [DOI] [PubMed] [Google Scholar]
- Modak, J. M., Lim, H. C., & Tayeb, Y. J. (1986). General characteristics of optimal feed rate profiles for various fed‐batch fermentation processes. Biotechnology and Bioengineering, 28(9), 1396–1407. 10.1002/bit.260280914 [DOI] [PubMed] [Google Scholar]
- Moglia, A., Menciassi, A., Dario, P., & Cuschieri, A. (2009). Capsule endoscopy: Progress update and challenges ahead. Nature Reviews Gastroenterology and Hepatology, 6(6), 353–361. 10.1038/nrgastro.2009.69 [DOI] [PubMed] [Google Scholar]
- Mora‐Villalobos, J. A., Montero‐Zamora, J., Barboza, N., Rojas‐Garbanzo, C., Usaga, J., Redondo‐Solano, M., Schroedter, L., Olszewska‐Widdrat, A., & Lopez‐Gomez, J. P. (2020). Multi‐product lactic acid bacteria fermentations. Fermentation, 6, 1–21. [Google Scholar]
- Morjaria, S., Schluter, J., Taylor, B. P., Littmann, E. R., Carter, R. A., Fontana, E., Peled, J. U., van den Brink, M. R. M., Xavier, J. B., & Taur, Y. (2019). Antibiotic‐induced shifts in fecal microbiota density and composition during hematopoietic stem cell transplantation. Infection and Immunity, 87(9):e00206‐19. 10.1128/IAI.00206-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, D. J., & Preston, T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes, 7(3), 189–200. 10.1080/19490976.2015.1134082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, G., Fournier, C., & Peter, J. (2018). Intestinale Mikrobiom‐Darm‐Hirn‐Achse und Reizdarmsyndrom. Wiener Medizinische Wochenschrift, 168(3–4), 62–66. 10.1007/s10354-017-0592-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, D‐G., & Cooney, C. L. (1983). Growth monitoring and control through computer‐aided on‐line mass balancing in a fed‐batch penicillin fermentation. Biotechnology and Bioengineering, 25(1), 225–255. 10.1002/bit.260250118 [DOI] [PubMed] [Google Scholar]
- Nair, A. T., Ramachandran, V., Joghee, N. M., Antony, S., & Ramalingam, G. (2018). Gut microbiota dysfunction as reliable non‐invasive early diagnostic biomarkers in the pathophysiology of Parkinson's disease: A critical review. Journal of Neurogastroenterology and Motility, 24(1), 30–42. 10.5056/jnm17105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava, G. M., Friedrichsen, H. J., & Stappenbeck, T. S. (2011). Spatial organization of intestinal microbiota in the mouse ascending colon. ISME Journal, 5(4), 627–638. 10.1038/ismej.2010.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, K. M., Kang, N., Bienenstock, J., & Foster, J. A. (2011). Reduced anxiety‐like behavior and central neurochemical change in germ‐free mice. Neurogastroenterology and Motility, 23(3), 255–264. 10.1111/j.1365-2982.2010.01620.x [DOI] [PubMed] [Google Scholar]
- O'Donnell, M. M., Rea, M. C., O'Sullivan, Ó., Flynn, C., Jones, B., McQuaid, A., Shanahan, F., & Ross, R. P. (2016). Preparation of a standardised faecal slurry for ex‐vivo microbiota studies which reduces inter‐individual donor bias. Journal of Microbiological Methods, 129, 109–116. 10.1016/j.mimet.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Pan, X., Ma, T., Li, P., Jiang, X., Song, S., & Max, M. Q. H. (2019). A novel intestinal microcapsule endoscope robot with biopsy function. 2018 IEEE International Conference on Cyborg and Bionic Systems, CBS 2018, 308–312. 10.1109/CBS.2018.8612210 [DOI]
- Park, S., Koo, K. I., Bang, S. M., Park, J. Y., Song, S. Y., & Cho, D. (2008). A novel microactuator for microbiopsy in capsular endoscopes. Journal of Micromechanics and Microengineering, 18(2), 025032. 10.1088/0960-1317/18/2/025032 [DOI] [Google Scholar]
- Patterson, E., Cryan, J. F., Fitzgerald, G. F., Ross, R. P., Dinan, T. G., & Stanton, C. (2014). Gut microbiota, the pharmabiotics they produce and host health. Proceedings of the Nutrition Society, 760(4), 477–489. 10.1017/S0029665114001426 [DOI] [PubMed] [Google Scholar]
- Payne, A. N., Chassard, C., Banz, Y., & Lacroix, C. (2012). The composition and metabolic activity of child gut microbiota demonstrate differential adaptation to varied nutrient loads in an in vitro model of colonic fermentation. FEMS Microbiology Ecology, 80(3), 608–623. 10.1111/j.1574-6941.2012.01330.x [DOI] [PubMed] [Google Scholar]
- Payne, A. N., Zihler, A., Chassard, C., & Lacroix, C. (2012). Advances and perspectives in in vitro human gut fermentation modeling. Trends in Biotechnology, 30(1), 17–25. 10.1016/j.tibtech.2011.06.011 [DOI] [PubMed] [Google Scholar]
- Peebo, K., & Neubauer, P. (2018). Application of continuous culture methods to recombinant protein production in microorganisms. Microorganisms, 6(3), 56. 10.3390/microorganisms6030056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, C., Antonioli, L., Colucci, R., Blandizzi, C., & Fornai, M. (2018). Interplay among gut microbiota, intestinal mucosal barrier and enteric neuro‐immune system: a common path to neurodegenerative diseases? Acta Neuropathologica, 136(3), 345–361. 10.1007/s00401-018-1856-5 [DOI] [PubMed] [Google Scholar]
- Poeker, S. A., Geirnaert, A., Berchtold, L., Greppi, A., Krych, L., Steinert, R. E., Wouters, T. D., & Lacroix, C. (2018). Understanding the prebiotic potential of different dietary fibers using an in vitro continuous adult fermentation model (PolyFermS). Scientific Reports, 8, 4318. 10.1038/s41598-018-22438-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompei, A., Cordisco, L., Raimondi, S., Amaretti, A., Pagnoni, U. M., Matteuzzi, D., & Rossi, M. (2008). In vitro comparison of the prebiotic effects of two inulin‐type fructans. Anaerobe, 14(5), 280–286. 10.1016/j.anaerobe.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Pédron, T., Mulet, C., Dauga, C., Frangeul, L., Chervaux, C., Grompone, G., & Sansonettia, P. J. (2012). A crypt‐specific core microbiota resides in the mouse colon. mBio, 3(3), 1–7. 10.1128/mBio.00116-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, E. M. M. (2013). Gut bacteria in health and disease. Gastroenterology and Hepatology, 9(9), 560–569. [PMC free article] [PubMed] [Google Scholar]
- Quigley, E. M. M. (2017). Microbiota‐brain‐gut axis and neurodegenerative diseases. Current Neurology and Neuroscience Reports, 17(12), 94. 10.1007/s11910-017-0802-6 [DOI] [PubMed] [Google Scholar]
- Raimondi, I., Izzo, L., Tunesi, M., Comar, M., Albani, D., & Giordano, C. (2020). Organ‐on‐a‐chip in vitro models of the brain and the blood‐brain barrier and their value to study the microbiota‐gut‐brain axis in neurodegeneration. Frontiers in Bioengineering and Biotechnology, 7, 435. 10.3389/fbioe.2019.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi, M. T., Albani, D., & Giordano, C. (2019). An organ‐on‐a‐chip engineered platform to study the microbiota‐gut‐brain axis in neurodegeneration. Trends in Molecular Medicine, 25(9), 737–740. 10.1016/j.molmed.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Rao, S. S. C., & Bhagatwala, J. (2019). Small intestinal bacterial overgrowth: Clinical features and therapeutic management. Clinical and Translational Gastroenterology, 10(10), e00078. 10.14309/ctg.0000000000000078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, K., Dinan, T. G., & Cryan, J. F. (2016). The microbiome: A key regulator of stress and neuroinflammation. Neurobiology of Stress, 4, 23–33. 10.1016/j.ynstr.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei Nejad, H., Oliveira, B. C. M., Sadeqi, A., Dehkharghani, A., Kondova, I., Langermans, J. A. M., Guasto, J. S., Tzipori, S., Widmer, G., & Sonkusale, S. R. (2019). Ingestible osmotic pill for in vivo sampling of gut microbiomes. Advanced Intelligent Systems, 1(5), 1900053. 10.1002/aisy.201900053 [DOI] [Google Scholar]
- Richard, M. L., Liguori, G., Lamas, B., Brandi, G., da Costa, G., Hoffmann, T. W., Pierluigi Di Simone, M., Calabrese, C., Poggioli, G., Langella, P., Campieri, M., & Sokol, H. (2018). Mucosa‐associated microbiota dysbiosis in colitis associated cancer. Gut Microbes, 9(2), 131–142. 10.1080/19490976.2017.1379637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinninella, E., Raoul, P., Cintoni, M., Franceschi, F., Miggiano, G. A. D., Gasbarrini, A., & Mele, M. C. (2019). What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms, 7(1), 14. 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera, K. R., Yokus, M. A., Erb, P. D., Pozdin, V. A., & Daniele, M. (2019). Measuring and regulating oxygen levels in microphysiological systems: Design, material, and sensor considerations. Analyst, 144(10), 3190–3215. 10.1039/c8an02201a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, G. A. W., Raison, C. L., & Lowry, C. A. (2014). Microbiota, immunoregulatory old friends and psychiatric disorders. Advances in Experimental Medicine and Biology, 817, 319–356. 10.1007/978-1-4939-0897-4_15 [DOI] [PubMed] [Google Scholar]
- Roy Sarkar, S., & Banerjee, S. (2019). Gut microbiota in neurodegenerative disorders. Journal of Neuroimmunology, 328, 98–104. 10.1016/j.jneuroim.2019.01.004 [DOI] [PubMed] [Google Scholar]
- Rumney, C. J., & Rowland, I. R. (1992). In Vivo and In Vitro Models of the Human Colonic Flora. Critical Reviews in Food Science and Nutrition, 31(4), 299–331. 10.1080/10408399209527575 [DOI] [PubMed] [Google Scholar]
- Silverman, J. D., Durand, H. K., Bloom, R. J., Mukherjee, S., & David, L. A. (2018). Dynamic linear models guide design and analysis of microbiota studies within artificial human guts. Microbiome, 6(1), 10.1186/s40168-018-0584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadabad, M. S., Von Martels, J. Z. H., Khan, M. T., Blokzijl, T., Paglia, G., Dijkstra, G., Harmsen, H. J. M., & Faber, K. N. (2015). A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco‐2 cells. Scientific Reports, 5, 1–9. 10.1038/srep17906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori, F., Siciliano, S., Maione, F., Esposito, D., Masone, S., Persico, M., & De Palma, G. D. (2012). Confocal laser endomicroscopy in the study of colonic mucosa in IBD patients: A review. Gastroenterology Research and Practice, 2012, 525098. 10.1155/2012/525098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, T. R., Debelius, J. W., Thron, T., Janssen, S., Shastri, G. G., Ilhan, Z. E., Challis, C., Schretter, C. E., Rocha, S., Gradinaru, V., Chesselet, M.‐F., Keshavarzian, A., Shannon, K. M., Krajmalnik‐Brown, R., Wittung‐Stafshede, P., Knight, R., & Mazmanian, S. K. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell, 167(6), 1469–1480. 10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardelli, L., Pacheco, D. P., Ziccarelli, A., Tunesi, M., Caspani, O., Fusari, A., Briatico Vangosa, F., Giordano, C., & Petrini, P. (2019). Towards bioinspired in vitro models of intestinal mucus. RSC Advances, 9(28), 15887–15899. 10.1039/c9ra02368b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scepanovic, P., Hodel, F., Mondot, S., Partula, V., Byrd, A., Hammer, C., Alanio, C., Bergstedt, J., Patin, E., Touvier, M., Lantz, O., Albert, M. L., Duffy, D., Quintana‐Murci, L., & Fellay, J. (2019). A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals. Microbiome, 7(1), 130. 10.1186/s40168-019-0747-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov, I., Russell, S. L., Caetano, M., Antunes, L., & Finlay, B. B. (2010). Gut microbiota in health and disease. Physiological Reviews, 90(3), 859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- Serra, D., Almeida, L. M., & Dinis, T. C. P. (2019). The impact of chronic intestinal inflammation on brain disorders: the microbiota‐gut‐brain axis. Molecular Neurobiology, 56(10), 6941–6951. 10.1007/s12035-019-1572-8 [DOI] [PubMed] [Google Scholar]
- Shachrai, I., Zaslaver, A., Alon, U., & Dekel, E. (2010). Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Molecular Cell, 38(5), 758–767. 10.1016/j.molcel.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Shah, P., Fritz, J. V., Glaab, E., Desai, M. S., Greenhalgh, K., Frachet, A., Niegowska, M., Estes, M., Jäger, C., Seguin‐Devaux, C., Zenhausern, F., & Wilmes, P. (2016). A microfluidics‐based in vitro model of the gastrointestinal human‐microbe interface. Nature Communications, 7, 11535. 10.1038/ncomms11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, R. M., McKenzie, E. J., Rosin, M. T., Jadhav, S. R., Gondalia, S. V., Rosendale, D., & Beale, D. J. (2020). An integrated multi‐disciplinary perspective for addressing challenges of the human gut microbiome. Metabolites, 10(3), 94. 10.3390/metabo10030094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan, E. R., Zhong, L., Talley, N. J., Morrison, M., & Holtmann, G. (2016). Characterisation of the gastrointestinal mucosa‐associated microbiota: A novel technique to prevent cross‐contamination during endoscopic procedures. Alimentary Pharmacology and Therapeutics, 43(11), 1186–1196. 10.1111/apt.13622 [DOI] [PubMed] [Google Scholar]
- Shin, W. & Kim H. J. (2018). Intestinal barrier dysfunction orchestrates the onset of inflammatory host–microbiome cross‐talk in a human gut inflammation‐on‐a‐chip. Proceedings of the National Academy of Sciences, 115(45). 10.1073/pnas.1810819115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, W., Wu, A., Massidda, M. W., Foster, C., Thomas, N., Lee, D. W., Koh, H., Ju, Y., Kim, J., & Kim, H. J. (2019). A robust longitudinal co‐culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic‐oxic interface‐on‐a‐chip. Frontiers in Bioengineering and Biotechnology, 7, 13. 10.3389/fbioe.2019.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiner, M. (1963). A capsule for obtaining sterile samples of gastrointestinal fluids. The Lancet, 281(7280), 532–533. 10.1016/S0140-6736(63)91328-X [DOI] [PubMed] [Google Scholar]
- Silva, Y. P., Bernardi, A., & Frozza, R. L. (2020). The role of short‐chain fatty acids from gut microbiota in gut‐brain communication. Frontiers in Endocrinology, 11, 25. 10.3389/fendo.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, M. O. A. (2015). Advancing gut microbiome research using cultivation. Current Opinion in Microbiology, 27, 127–132. 10.1016/j.mib.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Sontheimer‐Phelps, A., Chou, D. B., Tovaglieri, A., Ferrante, T. C., Duckworth, T., Fadel, C., Frismantas, V., Sutherland, A. D., Jalili‐Firoozinezhad, S., Kasendra, M., Stas, E., Weaver, J. C., Richmond, C. A., Levy, O., Prantil‐Baun, R., Breault, D. T., & Ingber, D. E. (2020). Human colon‐on‐a‐chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cmgh, 9(3), 507–526. 10.1016/j.jcmgh.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman, L. J., Gibson, D. L., & Klegeris, A. (2018). Unhealthy gut, unhealthy brain: The role of the intestinal microbiota in neurodegenerative diseases. Neurochemistry International, 120, 149–163. 10.1016/j.neuint.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Stefani, F. O. P., Bell, T. H., Marchand, C., de la Providencia, I. E., El Yassimi, A., St‐Arnaud, M. & Hijri, M. (2015). Culture‐dependent and ‐independent methods capture different microbial community fractions in hydrocarbon‐contaminated soils. PLOS ONE, 10(6), 1–16. 10.1371/journal.pone.0128272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokholm, J., Thorsen, J., Chawes, B. L., Schjørring, S., Krogfelt, K. A., Bønnelykke, K., & Bisgaard, H. (2016). Cesarean section changes neonatal gut colonization. Journal of Allergy and Clinical Immunology, 138(3), 881–889. 10.1016/j.jaci.2016.01.028 [DOI] [PubMed] [Google Scholar]
- Su, C., Lei, L., Duan, Y., Zhang, K. Q., & Yang, J. (2012). Culture‐independent methods for studying environmental microorganisms: Methods, application, and perspective. Applied Microbiology and Biotechnology, 93(3), 993–1003. 10.1007/s00253-011-3800-7 [DOI] [PubMed] [Google Scholar]
- Tang, Q., Jin, G., Wang, G., Liu, T., Liu, X., & Wang, B. (2020). Current sampling methods for gut microbiota: A call for more precise devices. Frontiers in Cellular and Infection Microbiology, 10, 151. 10.3389/fcimb.2020.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, S. A., Berner, A. Z., Rigozzi, E., Grattepanche, F., Chassard, C., & Lacroix, C. (2014). In vitro continuous fermentation model (PolyFermS) of the swine proximal colon for simultaneous testing on the same gut microbiota. PLOS One, 9(4), 94123. 10.1371/journal.pone.0094123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg, H., Zmora, N., Adolph, T. E., & Elinav, E. (2020). The intestinal microbiota fuelling metabolic inflammation. Nature Reviews Immunology, 20(1), 40–54. 10.1038/s41577-019-0198-4 [DOI] [PubMed] [Google Scholar]
- Tottey, W., Feria‐Gervasio, D., Gaci, N., Laillet, B., Pujos, E., Martin, J. F., Sebedio, J. L., Sion, B., Jarrige, J. F., Alric, M., & Brugère, J. F. (2017). Colonic transit time is a driven force of the gut microbiota composition and metabolism: In vitro evidence. Journal of Neurogastroenterology and Motility, 23(1), 124–134. 10.5056/jnm16042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuddenham, S., & Sears, C. L. (2015). The intestinal microbiome and health. Current Opinion in Infectious Diseases, 28(5), 464–470. 10.1097/QCO.0000000000000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufekci, K. U., Genc, S., & Genc, K. (2011). The endotoxin‐induced neuroinflammation model of Parkinson's disease. Parkinson's Disease, 2011, 1–25. 10.4061/2011/487450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuohy, K. M., & Scott, K. P. (2015). The microbiota of the human gastrointestinal tract: A molecular view. In Tuohy K, & Rio D. D. (Eds.), Diet‐microbe interactions in the gut: Effects on human health and disease (pp. 1–15). Elsevier Inc. 10.1016/B978-0-12-407825-3.00001-0 [DOI] [Google Scholar]
- Ulluwishewa, D., Anderson, R. C., Young, W., McNabb, W. C., van Baarlen, P., Moughan, P. J., Wells, J. M., & Roy, N. C. (2015). Live Faecalibacterium prausnitzii in an apical anaerobic model of the intestinal epithelial barrier. Cellular Microbiology, 17(2), 226–240. 10.1111/cmi.12360 [DOI] [PubMed] [Google Scholar]
- Van Den Abbeele, P., Belzer, C., Goossens, M., Kleerebezem, M., De Vos, W. M., Thas, O., De Weirdt, R., Kerckhof, F. M., & Van De Wiele, T. (2013). Butyrate‐producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME Journal, 7(5), 949–961. 10.1038/ismej.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nuenen, M. H. M. C., Meyer, P. D., & Venema, K. (2003). The effect of various inulins and clostridium difficile on the metabolic activity of the human colonic microbiota in vitro. Microbial Ecology in Health and Disease, 15(2–3), 137–144. 10.1080/08910600310018959 [DOI] [Google Scholar]
- Vandeputte, D., Falony, G., Vieira‐Silva, S., Tito, R. Y., Joossens, M., & Raes, J. (2016). Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut, 65(1), 57–62. 10.1136/gutjnl-2015-309618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, K. (2015). The impact of food bioactives on health. In (Eds.) Verhoeckx, K., Cotter, P., López‐Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D. & Wichers, H., The impact of food bioactives on health: In vitro and ex vivo models. Springer International Publishing. 10.1007/978-3-319-16104-4 [DOI] [PubMed] [Google Scholar]
- Venema, K., & Van Den Abbeele, P. (2013). Experimental models of the gut microbiome. Best Practice and Research: Clinical Gastroenterology, 27(1), 115–126. 10.1016/j.bpg.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Videnska, P., Smerkova, K., Zwinsova, B., Popovici, V., Micenkova, L., Sedlar, K., & Budinska, E. (2019). Stool sampling and DNA isolation kits affect DNA quality and bacterial composition following 16S rRNA gene sequencing using MiSeq Illumina platform. Scientific Reports, 9(1), 1–14. 10.1038/s41598-019-49520-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, M. M., Bloom, R. J., Silverman, J. D., Durand, H. K., Jiang, S., Wu, A., Dallow, E. P., Huang, S., You, L., & David, L. A. (2020). Interindividual variation in dietary carbohydrate metabolism by gut bacteria revealed with droplet microfluidic culture. mSystems, 5(3), 1–17. 10.1128/msystems.00864-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, N. M., Kerby, R. L., Dill‐McFarland, K. A., Harding, S. J., Merluzzi, A. P., Johnson, S. C., Carlsson, C. M., Asthana, S., Zetterberg, H., Blennow, K., Bendlin, B. B., & Rey, F. E. (2017). Gut microbiome alterations in Alzheimer's disease. Scientific Reports, 7(1), 13537. 10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Martels, J. Z. H., Sadaghian Sadabad, M., Bourgonje, A. R., Blokzijl, T., Dijkstra, G., Faber, K. N., & Harmsen, H. J. M. (2017). The role of gut microbiota in health and disease: In vitro modeling of host‐microbe interactions at the aerobe‐anaerobe interphase of the human gut. Anaerobe, 44, 3–12. 10.1016/j.anaerobe.2017.01.001 [DOI] [PubMed] [Google Scholar]
- de Vos, W. M. (2015). Microbial biofilms and the human intestinal microbiome. npj Biofilms and Microbiomes, 1(1). 10.1038/npjbiofilms.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. X., & Wang, Y. P. (2016). Gut microbiota‐brain axis. Chinese Medical Journal, 129(19), 2373–2380. 10.4103/0366-6999.190667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Antonopoulos, D. A., Zhu, X., Harrell, L., Hanan, I., Alverdy, J. C., Meyer, F., Musch, M. W., Young, V. B., & Chang, E. B. (2010). Laser capture microdissection and metagenomic analysis of intact mucosa‐associated microbial communities of human colon. Applied Microbiology and Biotechnology, 88(6), 1333–1342. 10.1007/s00253-010-2921-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, C. (2017). Design aspects of solid state fermentation as applied to microbial bioprocessing. Journal of Applied Biotechnology & Bioengineering, 4(1), 511–532. 10.15406/jabb.2017.04.00094 [DOI] [Google Scholar]
- Weinstock, G. M. (2012). Genomic approaches to studying the human microbiota. Nature, 489(7415), 250–256. 10.1038/nature11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. B., Van Benschoten, A. H., Cimermancic, P., Donia, M. S., Zimmermann, M., Taketani, M., Ishihara, A., Kashyap, P. C., Fraser, J. S., & Fischbach, M. A. (2014). Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host and Microbe, 16(4), 495–503. 10.1016/j.chom.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberley, N., Faling, L. J., & Bartlett, J. G. (1979). A fiberoptic bronchoscopy technique to obtain uncontaminated lower airway secretions for bacterial culture. American Review of Respiratory Disease, 119(3), 337–343. 10.1164/arrd.1979.119.3.337 [DOI] [PubMed] [Google Scholar]
- Woo, P. C. Y., Lau, S. K. P., Teng, J. L. L., Tse, H., & Yuen, K. Y. (2008). Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clinical Microbiology and Infection, 14(10), 908–934. 10.1111/j.1469-0691.2008.02070.x [DOI] [PubMed] [Google Scholar]
- Wu, W. K., Chen, C. C., Panyod, S., Chen, R. A., Wu, M. S., Sheen, L. Y., & Chang, S. C. (2019). Optimization of fecal sample processing for microbiome study—The journey from bathroom to bench. Journal of the Formosan Medical Association, 118(2), 545–555. 10.1016/j.jfma.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Yousi, F., Kainan, C., Junnan, Z., Chuanxing, X., Lina, F., Bangzhou, Z., Jianlin, R., & Baishan, F. (2019). Evaluation of the effects of four media on human intestinal microbiota culture in vitro. AMB Express, 9(1), 69. 10.1186/s13568-019-0790-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunes, R. A., Poluektova, E. U., Dyachkova, M. S., Klimina, K. M., Kovtun, A. S., Averina, O. V., Orlova, V. S., & Danilenko, V. N. (2016). GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe, 42, 197–204. 10.1016/j.anaerobe.2016.10.011 [DOI] [PubMed] [Google Scholar]
- Zarrinpar, A., Chaix, A., Yooseph, S., & Panda, S. (2014). Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metabolism, 20(6), 1006–1017. 10.1016/j.cmet.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Cao, X., & Huang, H. (2017). Sampling strategies for three‐dimensional spatial community structures in IBD microbiota research. Frontiers in Cellular and Infection Microbiology, 7, 51. 10.3389/fcimb.2017.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Kelly, C. J., & Colgan, S. P. (2015). Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: Cellular responses to hypoxia. American Journal of Physiology‐Cell Physiology, 309(6), C350–C360. 10.1152/ajpcell.00191.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihler, A., Gagnon, M., Chassard, C., Hegland, A., Stevens, M. J. A., Braegger, C. P., & Lacroix, C. (2010). Unexpected consequences of administering bacteriocinogenic probiotic strains for Salmonella populations, revealed by an in vitro colonic model of the child gut. Microbiology, 156(11), 3342–3353. 10.1099/mic.0.042036-0 [DOI] [PubMed] [Google Scholar]
- Zihler Berner, A., Fuentes, S., Dostal, A., Payne, A. N., Vazquez Gutierrez, P., Chassard, C., Grattepanche, F., de Vos, W. M., & Lacroix, C. (2013). Novel Polyfermentor intestinal model (PolyFermS) for controlled ecological studies: validation and effect of pH. PLOS One, 8(10), 2–11. 10.1371/journal.pone.0077772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal, E. G., Raes, J., Van Den Bogert, B., Arumugam, M., Booijink, C. C., Troost, F. J., Bork, P., Wels, M., De Vos, W. M., & Kleerebezem, M. (2012). The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME Journal, 6(7), 1415–1426. 10.1038/ismej.2011.212 [DOI] [PMC free article] [PubMed] [Google Scholar]


