Abstract
Purpose of review:
Intestinal stem cells, the most rapidly proliferating adult stem cells, are exquisitely sensitive to extrinsic dietary factors. Uncontrolled regulation of intestinal stem cells is closely linked to colon tumorigenesis. This review focuses on how dietary and microbial derived cues regulate intestinal stem cell functionality and colon tumorigenesis in mouse models by targeting the aryl hydrocarbon receptor (AhR).
Recent findings:
AhR, a ligand activated transcription factor, can integrate environmental, dietary and microbial cues to modulate intestinal stem cell proliferation, differentiation and their microenvironment, affecting colon cancer risk. Modulation of AhR activity is associated with many chronic diseases, including inflammatory bowel diseases where AhR expression is protective.
Summary:
AhR signaling controls the maintenance and differentiation of intestinal stem cells, influences local niche factors, and plays a protective role in colon tumorigenesis. Mounting evidence suggests that extrinsic nutritional/dietary cues which modulate AhR signaling may be a promising approach to colon cancer chemoprevention.
Keywords: aryl hydrocarbon receptor, colonic stem cells, colon cancer
Introduction
Over the past decade, exciting advances have been made in the identification of stem cells, which replenish the intestinal epithelium every 3–5 days [1]. Lgr5 (leucine-rich-repeat-containing G-protein-coupled receptor 5, also known as Gpr49), a receptor for R-spondins [2], marks a long-lived pool of rapidly cycling stem cells in the small intestine and colon (approximately 6 per crypt) [3]. The distribution of Lgr5+ cells within stem cell-derived adenomas indicates that a stem cell/progenitor cell hierarchy is maintained in early neoplastic lesions [4]. By crossing stem-cell-specific Lgr5-EGFP-IRES-creERT2 knockin mice to Apcflox/flox mice, Barker et al unequivocally demonstrated that crypt stem cells are the cells-of-origin of intestinal cancer [4]. Indeed, with respect to irrefutable stem cell characterization, a single Lgr5+ intestinal stem cell can generate a continuously expanding, self-organizing epithelial structure reminiscent of normal gut [5]. In addition, it has been demonstrated that intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5+ stem cells [6]. Consistent with findings that tissue stem cells act as tumor-initiating “cancer stem cells”, cancer therapy has been used to promote the elimination of Lgr5+ stem cells inappropriately activated by oncogenic events [7]. Collectively, these data provide evidence indicating that Lgr5+ is a marker of adult stem cells, and that perturbations in their behavior drives cancer initiation and/or progression by regulating tumor growth and metastasis [8]. Interestingly, some of these properties are shared by other slow cycling crypt cells expressing Musashi-1, DCAMKL1, BMI1 and Lrig1[9].
Over the last several years, emerging data have shown that signals from both the local stem cell niche/microenvironment as well as circulating, systemic factors contribute to the regulation of stem cells [10, 11]. In this context, intestinal stem cell/ cancer stem cell fate, i.e., the balance between self-renewal and differentiation, is influenced by both the intrinsic metabolic state and extrinsic nutritional/dietary cues. In depth reviews on the metabolic requirements of stem cells and cancer stem cells have previously been reported [12–15]. With respect to nutritional and metabolic control of stem cell fate, recent studies have examined the effects of calorie restriction/fasting/ketogenic diet, obesigenic high fat diet, vitamin D and Ca2+; vitamin C, dietary fiber/butyrate and curcumin/omega-3 fatty acids [16–20]. One of the novel findings of these studies is the fact that rapidly cycling Lgr5+ stem cells are exquisitely sensitive to extrinsic dietary factors that modulate colon cancer risk. For example, dietary curcumin combined with omega-3 fatty acids synergistically reduced carcinogen-induced nuclear β-catenin levels in aberrant crypt foci, in part, by promoting p53-dependent signaling and targeted apoptosis in damaged colonic Lgr5+ stem cells at the cancer initiation stage [19]. In addition, mice with DNA-damaged Lgr5+ stem cells were highly responsive to this combination diet compared with DNA-damaged differentiated cells [19].
We recently demonstrated that some of the effects of diet on stem cells in the gut, e.g., modulation of Lgr5+ energy metabolism and cell number, are mediated by ligand induced activation of the aryl hydrocarbon receptor (AhR) [21, 22]. Thus, this review will focus on recent studies describing the effects of AhR signaling and precision nutrition on colonic stem cells and colon tumorigenesis.
AhR signaling pathway: direct vs indirect effects of diet
AhR is a ligand activated basic helix-loop-helix transcription factor that senses environmental xenobiotics and dietary- or microbiota-derived small molecules. Generally, AhR ligands are generated by combustion and chemical synthesis and are produced by plants and microorganisms. The synthetic AhR ligands, including halogenated aromatic hydrocarbons, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), and polycyclic aromatic hydrocarbons, such as 3-methylcholanthrene (3MC), are generated from incomplete combustion or are formed as industrial by-products, and TCDD binds with high affinity to the AhR [23]. Naturally occurring ligands include phytochemicals such as polyphenolics, tryptophan metabolites, indole-3 carbinol related compounds, gut microbial metabolites, and photoproducts derived from tryptophan. Naturally occurring AhR ligands typically exhibit modest AhR binding affinities (KD ~ 10−6 M) with variable metabolic half-lives; however, high physiological concentrations of some ligands, such as indole, indole-3-aldehyde, and kynurenine, can compensate for the low binding affinities resulting in AhR activation [24–26].
In the absence of ligands, the AhR is bound to several chaperone proteins in the cytoplasm including heat shock proteins 90 (hsp90), p23 and immunophilin related protein XAP2. Upon ligand binding, XAP2 dissociates from the cytosolic AhR complex, and the AhR-ligand complex is then translocated into the nucleus. Once in the nucleus, hsp90 and p23 are displaced by the AhR nuclear translocator (ARNT) [27, 28] to form a heterodimeric AhR-ARNT complex, which then interacts with cis-acting dioxin response elements (DREs) with a core sequence of 5’-TNGCGTG-3’ or 5’-CACGCNA-3’ on promoters of AhR-responsive genes, including several forms of cytochrome P450 (CYP1A1, CYP1A2, CYP1B1), AhR repressor (AHRR), glucuronosyl transferases (UGT1A1) and other phase II drug metabolizing enzymes [29–31]. CYP1A1, CYP1A2, and CYP1B1 limit the availability of AhR ligands by catalyzing their metabolism. AHRR can competitively bind to ARNT, preventing AhR-ARNT formation. Following transcription, AhR is then exported from the nucleus and degraded by the cytoplasmic proteasome [32]. The AhR can also regulate expression of genes that lack canonical DREs in their promoter regions, such as plasminogen activator inhibitor 1 (PAI-1) [33], independent of ARNT. For example, the recognition of non-canonical DREs in the PAI-1 promoter requires the interaction between AhR and the Kruppel-like factor (KLF) family member KLF6 [34]. In addition, AhR may act as an E3 ligase mediating protein degradation, of which, β-catenin is one of the most characterized targets, although this remains controversial [35–37]. Since β-catenin plays a pivotal role in Wnt signaling and upregulated Wnt signaling is commonly observed in colon cancer, ongoing studies are probing the relationship between AhR and β-catenin.
AhR mediated regulation of colonic stem cells
Trillions of microorganisms (microbiota), including bacteria, archaea and fungus, inhabit the large intestine [38], and produce many bioactive metabolites, including short chain fatty acids and tryptophan metabolites, which serve as endogenous AhR ligands [39]. Reduced levels of AhR ligands, such as indole-3-acetic acid and indole-3-sulfate, are observed in the feces and blood of germ free or antibiotic treated mice compared to control mice [40, 41]. Due to close proximity, gut microbiota-derived tryptophan metabolites are readily absorbed by colonic stem cells, thereby regulating their self-renewal, differentiation and functionality (Figure 1A). Importantly, recent findings suggest that intestinal epithelial cells serve as a gatekeeper, regulating the availability of AhR ligands to the host [42]. However, at present it is not known whether differentiated epithelial cells further metabolize gut microbiota derived AhR ligands to create a physiological concentration gradient along the crypt axis, including the colonic stem cell zone.
Figure 1.
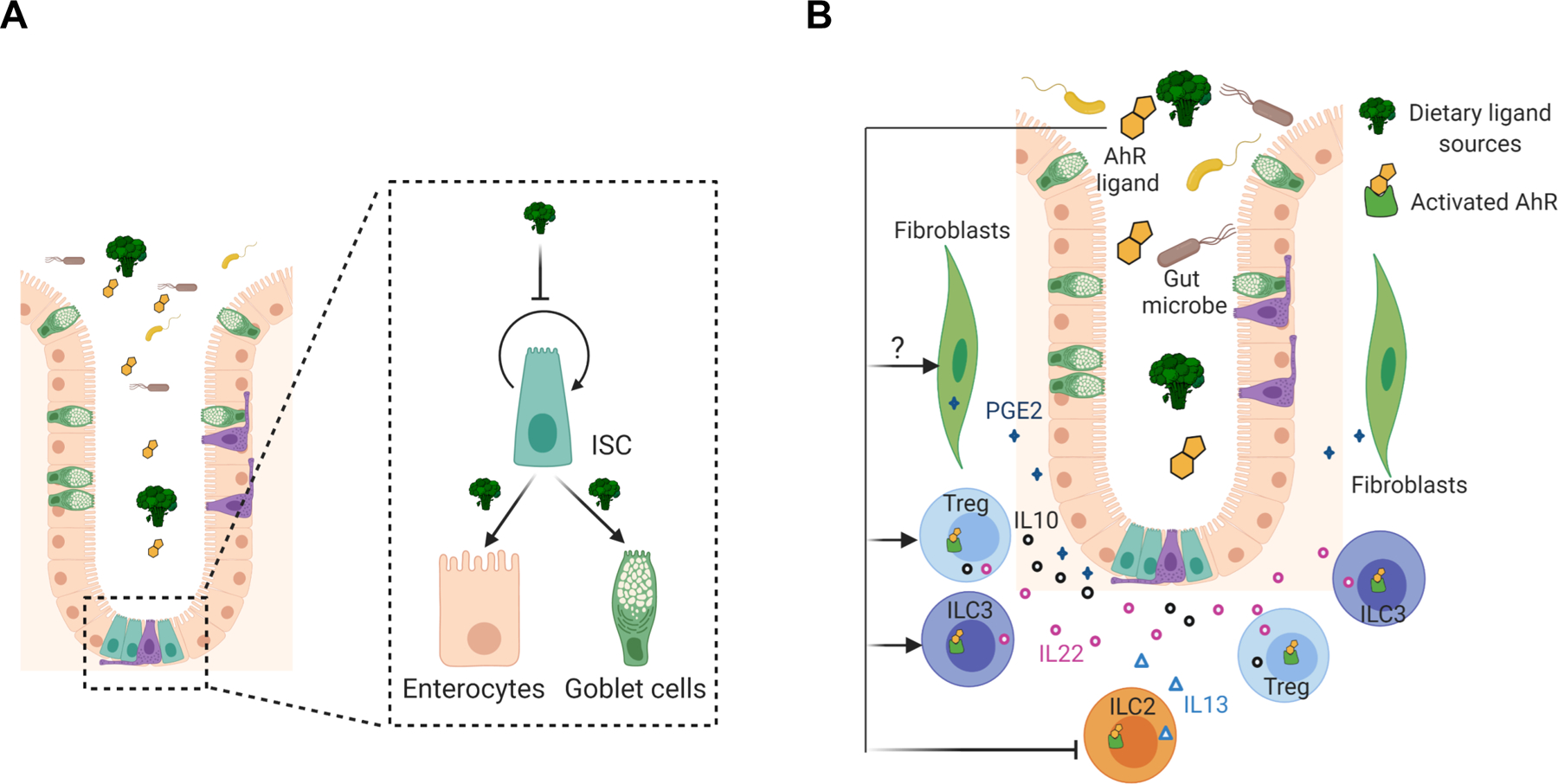
AhR mediated regulation of intestinal stem cells. (A) Direct regulation of intestinal stem cells by AhR signaling. AhR activation by dietary or gut microbiota derived metabolites/ligands controls intestinal stem cell (ISC) proliferation and self-renewal and promotes their differentiation into enterocytes or goblet cells. (B) AhR mediated indirect regulation of intestinal stem cells by controlling the production of their niche factors. For example, AhR activation promotes the production of IL10, IL22 and PGE2, and inhibits the secretion of IL13. These cytokines and PGE2 serve as growth mediators, capable of modulating the maintenance of intestinal stem cells.
Colonic stem cells, marked by Lgr5, reside at the bottom region of crypts, are intermingled with Reg4+ deep crypt secretory cells [43]. Lgr5-EGFP-IRES-CreERT2 knockin mice are a commonly used mouse model to study colonic stem cells, where green fluorescent protein (GFP) is a proxy for colonic stem cells. However, Lgr5-GFP reporter mice are mosaic and not all colonic stem cells are marked with GFP, sometimes causing inaccurate or even confusing assessment of treatment-related effects on colonic stem cells. To reduce the confounding effect of mosacism, our lab introduced a temporal controlled Rosa26LSL-Tdtomato reporter into these mice. Cell lineage tracing revealed that AhR activation by TCDD (a high affinity AhR ligand) reduced the percentage of colonic stem cells, while intestinal stem cell specific AhR deletion promoted the expansion of colonic stem cells, which was consistent with a previous study [44]. Moreover, similar conclusions were drawn using an organoid culture model, which is deprived of submucosal stromal and immune cells, demonstrating that colonic stem cells are intrinsically regulated by AhR signaling.
In complementary experiments, AhR signaling modulated the functionality of colonic stem cells. For example, AhR activation decreased the clonogenic capacity and organoid growth of colonic stem or progenitor cells, while AhR KO promoted their stemness and inhibited differentiation of colonic stem cells toward goblet cells and enterocytes (Figure 1A) [44, 22]. Further analysis revealed that intestinal specific AhR KO promoted colonic stem cell proliferation and cell cycle progression [22]. From a mechanistic perspective, AhR acted as a transcriptional suppressor of FoxM1, a master regulator of cell proliferation and cell cycle progression. Inhibition of FoxM1 phenocopied the effects of AhR activation and attenuated AhR KO mediated phenotypes. It is noteworthy that 3,3’-diindolylmethane (DIM), an AhR agonist [45], effectively downregulates FoxM1 in various breast cancer cell lines and inhibits breast cancer cell growth [46]. In terms of the effects of AhR on stem cell maintenance, Metidji et al found that AhR KO potentiated Wnt signaling by downregulating the expression of Znrf3 and Rnf43 [44], which act as E3 ubiquitin ligases to target Wnt receptors for degradation [47]. Furthermore, AhR may serve as an E3 ligase for β-catenin degradation, suppressing Wnt signaling [36]. These observations are noteworthy, because Wnt signaling is required for maintanance of colonic stem cell renewal [48]. Interestingly, we did not detect altered Wnt signaling and β-catenin levels in colonic Lgr5+ stem cells following AhR activation or AhR KO, which is consistent with other groups [49, 37]. The discrepancies observed with respect to AhR and Wnt signaling pathways may be explained by differences in cell context, e.g., normal vs malignant transformed state. Future work is needed to determine whether AhR signaling directly regulates Wnt signaling and/or how AhR signaling affects Wnt signaling in the context of normal regenerative homeostasis vs tumorigenesis.
Even though AhR is a highly conserved transcription factor between mouse and human (82% similarity in sequence), some species related differences have been observed. For example, human AhR contains an alanine to valine substitution at codon 375 in the ligand binding domain, compared with murine AhRb1 isoform, and this substitution results in a lower binding affinity to some ligands, including TCDD [50]. Importantly, it is noted that human AhR regulates different gene expression profiles [51]. Therefore, caution should be taken when translating mouse related data to humans, particularly with respect to AhR downstream targets. For example, inherent physiological differences between human and mouse immune cells can influence how endogenous ligands to AhR can modulate lymphocyte responses and anti-inflammatory sequelae in the intestine [52]. Consistent with mouse based studies, AhR can directly regulate the expression of FoxM1, and AhR activation reduces the percentage of human colonic stem cells and decreases the clonogenic capacity and organoid growth of human colonocytes, which are independent of β-catenin and pERK1/2 levels [22]. This is noteworthy, because lower levels of AhR ligands are observed in individuals with inflammatory bowel diseases, metabolic syndrome and obesity [53, 41]. It will be interesting to determine whether reduced AhR activity can affect the expansion of colonic stem cells and barrier permeability due to compromized cell differentiation and whether impaired AhR signaling accelerates premature colonic stem cell exhaustion in the aged. In addition, emerging studies have reported that AhR signaling plays an important role in modulating the dynamics and functionality of other stem cells, including hematopoietic stem cells (HSCs), neural stem cells and hepatic stem cells [54–56], implying that AhR broadly functions to regulate multiple stem cell populations in the body.
The regulation of colonic stem cell niche factors
Colonic stem cells are in close proximity to gut immune cells and mucosal stroma cells. This is noteworthy, because neighboring cells in the stem cell niche play an important role in shaping colonic AhR signaling. In particular, how AhR signaling regulates soluble mediators, such as IL22, IL10, and PGE2, has attracted significant attention (Figure 1B). For example, AhR acts as a key transcription factor in controlling the differentiation of Th17, Treg cells, group 2 innate lymphoid cells (ILC2s) and group 3 innate lymphoid cells (ILC3s), in which Treg and ILC3 are two major IL22-producing immune cells in the gut, and Foxp3+ Treg cells are the major source of IL10 secretion [57]. Specifically, AhR activation by TCDD favorably induces Treg cell differentiation by directly promoting Foxp3 expression [58], a key lineage-specific transcription factor of Treg cells. Similarly, AhR signaling is required for ILC3 generation, even though the molecular mechanism remains unknown [59, 60]. Recently, it has been demonstrated that AhR activation intrinsically inhibits ILC2 cell differentiation and function [61]. AhR regulates chromatin accessibility at select gene loci, such as AhR and Interleukin 1 Receptor Like 1 (Il1rl1), which encodes IL33 receptor ST2. Moreover, AhR can bind to Il1rl1 promoter and directly suppress its expression, as evidenced by upregulated Il1rl1 mRNA in AhR−/− ILC2s. The production of type 2 cytokines, such as IL5 and IL13, is partially dependent on the IL33-ST2 pathways [61]. Importantly, ILC2 derived IL13 has been shown to promote the differentiation of intestinal stem cells towards tuft and goblet cells [62].
AhR can directly control the production of IL10 and IL22. Upon binding to DREs in the IL22 promoter, AhR cooperatively interacts with RORγt and STAT3 to promote IL22 expression [63, 64] and the AhR synergistically interacts with c-Maf to directly control expression of IL10[65, 66]. Consequently, IL10 and IL22 derived from gut immune cells can modulate intestinal stem cell renewal and differentiation. For example, IL10 treatment or coculture of Treg cells with intestinal stem cells (ISCs) promotes the expansion of ISCs within organoids [57]. The regulation of ISC by IL22 remains controversial, where some studies indicate that IL22 treatment promotes ISC expansion and organoid growth [67, 57], while other groups report IL22 actually inhibits ISC expansion [68, 69]. Interestingly, in addition to IL10 and IL22, other cytokines have been shown to affect ISCs. For example, proinflammatory cytokines, such as IL17A, IFNγ and IL13, promote ISC differentiation [57]. However, little is known regarding how crypt neighboring immune cells or their cytokines, such as IL10 and IL22, influence other cell types, such as progenitor cells, and how they contribute to epithelial plasticity and crypt regeneration in the context of inflammation.
Several studies suggest that prostaglandin E2 (PGE2) plays an important role in tumorigenesis, crypt regeneration and inflammation [70–72]. AhR activation increases the production of PGE2 in the colon, and inhibition of PGE2 production abrogates the beneficial effects of TCDD on DSS-induced colitis [73]. The production of PGE2 is primarily synthesized by pericryptal fibroblasts and/or colonic mesenchymal stem cells (cMSCs) [71, 70]. It will be interesting to explore whether and how AhR signaling regulates fibroblasts or cMSCs to enhance PGE2 synthesis in the colon. Interestingly, AhR activation promotes the expression of prostaglandin-endoperoxide synthase 2 (COX-2) and microsomal prostaglandin E synthase-1 (mPGES) to enhance PGE2 production in human lung fibroblasts [74]. However, little in known regarding whether PGE2 signaling affects intestinal stem cells. Evidence from recent studies show that PGE2 drives the expansion of Sca-1+ reserve-like stem cells by promoting yes-associated protein 1 (YAP) signaling, which promotes colon cancer stem cell expansion by activating NF-κB [71, 75]. In contrast, PGE2-receptor 4 (EP4) signaling reduces Lgr5+ intestinal stem cell pools both in human and mouse organoids [76, 77] and modulates intestinal stem cells, progenitor cells, and immature enterocytes to form wound associated epithelial cells [77]. Similarly, our lab demonstrated that PGE2 signaling had no effect on the clonogenic capacity and organoid growth from sorted colonic stem cells [22]. Thus, PGE2 exerts diverse effects on different) intestinal reserve noncycling vs rapidly cycling and normal vs cancer) stem cells.
Colon cancer
Since AhR signaling closely regulates colonic stem cells and their niches, and dysregulated intestinal stem cells are the cells-of-origin of intestinal cancer [4], it is not surprising that AhR signaling modulates colon tumorigenesis. Currently, three types of colon tumor models are utilized to assess the role of AhR signaling in regulating colon tumorigenesis (Table 1).
Table 1.
Summary of effects of AhR signaling on intestinal tumorigenesis in mouse models
| Colon cancer models | AhR status | Effects | References |
|---|---|---|---|
| Global KO | Spontaneously develop cecum tumors | [36] | |
| ApcMin/+ | Global KO or heterozygotes | Accelerates cecum tumorigenesis and promotes intestinal tumor multiplicity | [36] |
| Activation by I3C and DIM | Indole-3-carbinol (I3C) and 3,3’-Diindoylmethane (DIM) delay cecum tumorigenesis and inhibits intestinal tumor multiplicity | [36] | |
| AOM | Intestinal specific KO | Increases aberrant crypt foci or colon tumor multiplicity | [44, 85] |
| AOM/DSS | Global KO | Azoxymethane (AOM) and dextran sodium sulphate (DSS) combination promotes colon tumor multiplicity | [49] |
| Activation by I3C | I3C reduces colon tumor incidence Inhibits colon tumor multiplicity | [49] | |
| Intestinal specific KO | Promotes colon tumor incidence, multiplicity, and size | [22, 44] |
i. Genetically induced gastrointestinal tumor models.
Several colon genetic tumor models are utilized to recapitulate human hereditary and/or sporadic colorectal tumorigenesis [78, 79]. Typically, the adenomatous polyposis coli (Apc) gene is targeted since ~80% of sporadic colorectal tumors contain Apc mutations [80]. Apcmin/+ mice encode a nonsense mutation at codon 850, and are predisposed to developing intestinal adenomas [79]. Interestingly, Kawajiri et al reported that spontaneous cecal tumors were observed in AhR null mice and haploinsufficiency of AhR accelerated tumorigenesis in cecum and small intestinal in AhR+/−; Apcmin/+ mice. The increased tumorigenesis in AhR+/−; Apcmin/+ was associated with increased stability of β-catenin [36]. Interestingly, AhR activation by feeding dietary AhR ligands, e.g., indole-3-carbinol (I3C) and 3,3’-diindoylmethane (DIM), robustly decreased β-catenin level and significantly reduced cecal tumorigenesis and tumor burdens in Apcmin/+ mice [36].
Contradictory claims have surfaced regarding the functions of the AhR as an E3 ligase to degrade β-catenin [36, 37]. For example, AhR does not interact with β-catenin even in the presence of AhR interacting proteins, e.g. Arnt, CUL4B and DDB1, and AhR activation has no effect on TCF/β-catenin dependent transcription in human colon cancer cell lines [37]. Additional findings indicate that AhR null mice do not spontaneously develop tumors in the cecum or colon, and β-catenin and its target c-myc expression are not altered [49]. Contributing factors for these distinct outcomes include model related differences in AhR exon deletion (exon 1 vs exon 2), and the potential contribution of distinct communities of gut microbiota, since gut microbiota plays an important role in colon tumorigenesis [81, 82]. Moreover, germ free AhR null mice or mice lacking both the AhR and apoptosis-associated speck-like protein containing a CARD (ASC), exhibit a reduction in cecal tumorigenesis compared with conventional AhR KO mice [83], implying that inflammation plays a role in cecal and colon tumorigenesis in AhR null mice. In the future, it will be interesting to determine the epithelial role of AhR in different genetic colon tumor models, and whether AhR ligand supplementation suppresses colon tumorigenesis.
ii. Carcinogen induced colon tumorigenesis.
Azoxymethane (AOM) is one of the most commonly used inducers of colorectal cancer. AOM induced colorectal tumors recapitulate the multistage progression and histopathological characteristics of human colorectal cancer [84]. Intestinal specific AhR KO promotes stem cell proliferation and AOM induced aberrant crypt foci, premalignant colon tumor lesions, and colon tumors [85, 44]. Further analysis revealed that accumulated nuclear β-catenin is observed in AhR KO tumor masses. However, it is unclear if this accumulated nuclear β-catenin is the cause or result of more tumors in AhR KO mice. Metidji et al reported that intestinal specific KO or depletion of AhR ligands by overexpressing Cyp1a1 also increased the expression of IL-6, implying that impaired ligand-dependent AhR signaling in epithelial cells enhances gut inflammation [44].
iii. Colitis associated colon tumorigenesis.
Compared with carcinogen induced colon tumorigenesis, the preclinical colitis associated tumor model (AOM with DSS) accelerates the progression and multiplicity of colon tumors. In this context, AhR signaling also plays a protective role. For example, global AhR knockout promotes colitis-associated colon tumorigenesis, and AhR activation by feeding I3C decreases tumor incidence [49]. From a mechanistic perspective, the protective role of AhR in this colitis associated tumor model is not associated with altered Wnt signaling, but with decreased DNA damage and DSS induced inflammation [49]. Interestingly, global AhR KO mice are deficient with respect to IL22 production [42, 64], and IL22 signaling is required to initiate efficient DNA damage response after exposure to AOM [86]. In addition, studies using intestinal specific AhR KO mice unequivocally show that the loss of AhR in the gut epithelium increases colon tumor incidence and multiplicity [22, 44]. Metidji et al found that AhR KO potentiated Wnt signaling, as evidenced by increased β-catenin levels and Wnt target genes. In addition, dietary I3C supplementation decreased tumor multiplicity in R26LSL-Cyp1a1; Villin-Cre mice [44], in which overexpression of Cyp1a1 in intestinal epithelial cells potentially depleted AhR ligands, effectively suppressing AhR activation [42]. Interestingly, I3C has no effect on colon tumorigenesis in intestinal specific AhR KO mice [44], in which I3C can potentially activate AhR in gut immune cells. In complementary studies, it has been demonstrated that AhR activation suppresses DSS induced inflammation and increases the production of IL22 and IL10, which can act on intestinal epithelial cells to suppress colon tumorigenesis [87–89, 86]. Hence, it is important to determine whether epithelial AhR KO indirectly affects AhR activation in non-epithelial immune cells.
Precision nutrition
AhR ligands, such as dietary- and intestinal microbiota-derived compounds may uniquely modulate gastrointestinal stem cells and the immune system. Importantly, the chronic reduction in cellular AhR activation has been linked to the suppression in AhR ligand production in patients with numerous chronic diseases, including inflammatory bowel disease, obesity, Type 2 diabetes and high blood pressure [87, 90, 53]. The defect in AhR agonist production appears to be in part, the result of an impaired capacity of gut microbiota to metabolize tryptophan into AhR agonists in mice and humans [53, 90]. These findings suggest that the substantial inter-individual variability in AhR-mediated response to dietary exposures is likely the result of microbiome influences. Moreover, polymorphisms of human AhR may also contribute to individual sensitivity to AhR ligand exposure [91–93], and several AhR single nucleotide polymorphisms (SNPs) have been significantly associated with many diseases [94–96]. Since “Precision Nutrition” aspires to offer individual tools to personalize dietary and lifestyle practices for optimal health [97], future research should explore relationships between host AhR biology and microbiome changes in response to specific probiotics and dietary prebiotic intervention.
Conclusion
Intestinal epithelial cells co-exist with gut microbiota and are frequently exposed to external environmental stimuli, where bioactive components from diet or gut microbiota derived metabolites can uniquely modulate crypt stem cells. AhR acts as an environmental sensor that can integrate environmental, dietary and gut microbial cues to modulate intestinal stem cells and their microenvironment, affecting colon cancer risk. AhR signaling regulates intestinal stem cells both intrinsically and extrinsically and constitutes an important axis to mediate interaction between gut microbiota, intestinal stem cells and immune cells in the stem cell niche. However, precisely how AhR signaling regulates this multi-cellular tripartite interaction in the context of homeostasis and inflammatory pathogenesis is still under investigation. In summary, current studies provide rationale for AhR as a potential therapeutic target to optimize and reduce the burden of chronic disease, and this is particularly true for the colon.
Acknowledgements
Funding was provided by Texas AgriLife Research, the Sid Kyle Chair Endowment, the Allen Endowed Chair in Nutrition & Chronic Disease Prevention, the Cancer Prevention Research Institute of Texas (RP160589), and the National Institutes of Health (R01-ES025713, R01-CA202697, R01-AT01282, R35-CA197707 and T32-CA090301). The illustrative figures were created using BioRender.com.
Footnotes
Conflict of Interest: Huajun Han, Arul Jayaraman, Stephen Safe and Robert Chapkin declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Barker N, Clevers H. Tracking down the stem cells of the intestine: strategies to identify adult stem cells. Gastroenterology. 2007;133(6):1755–60. doi: 10.1053/j.gastro.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 2.Carmon KS, Lin Q, Gong X, Thomas A, Liu Q. LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/beta-catenin signaling. Molecular and cellular biology. 2012;32(11):2054–64. doi: 10.1128/MCB.00272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, Clevers H. Lineage tracing in the intestinal epithelium. Current protocols in stem cell biology. 2010;Chapter 5:Unit5A.4. doi: 10.1002/9780470151808.sc05a04s13. [DOI] [PubMed] [Google Scholar]
- 4.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 6.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143(1):134–44. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Qiu W, Wang X, Leibowitz B, Liu H, Barker N, Okada H et al. Chemoprevention by nonsteroidal anti-inflammatory drugs eliminates oncogenic intestinal stem cells via SMAC-dependent apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(46):20027–32. doi: 10.1073/pnas.1010430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung C, Tan SH, Barker N. Recent Advances in Lgr5(+) Stem Cell Research. Trends Cell Biol. 2018;28(5):380–91. doi: 10.1016/j.tcb.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Kim C-K, Yang VW, Bialkowska AB. The Role of Intestinal Stem Cells in Epithelial Regeneration Following Radiation-Induced Gut Injury. Curr Stem Cell Rep. 2017;3(4):320–32. doi: 10.1007/s40778-017-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer. 2014;14(7):468–80. doi: 10.1038/nrc3744. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy N, Kraiczy J, Shivdasani RA. Cellular and molecular architecture of the intestinal stem cell niche. Nature Cell Biology. 2020;22(9):1033–41. doi: 10.1038/s41556-020-0567-z. [DOI] [PubMed] [Google Scholar]
- 12.García-Prat L, Sousa-Victor P, Muñoz-Cánoves P. Proteostatic and Metabolic Control of Stemness. Cell Stem Cell. 2017;20(5):593–608. doi: 10.1016/j.stem.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Ren R, Ocampo A, Liu GH, Izpisua Belmonte JC. Regulation of Stem Cell Aging by Metabolism and Epigenetics. Cell Metab. 2017;26(3):460–74. doi: 10.1016/j.cmet.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Schell JC, Wisidagama DR, Bensard C, Zhao H, Wei P, Tanner J et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nature cell biology. 2017;19(9):1027–36. doi: 10.1038/ncb3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batlle E, Clevers H. Cancer stem cells revisited. Nature Medicine. 2017;23(10):1124–34. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 16.Alonso S, Yilmaz ÖH. Nutritional Regulation of Intestinal Stem Cells. Annual Review of Nutrition. 2018;38(1):273–301. doi: 10.1146/annurev-nutr-082117-051644. [DOI] [PubMed] [Google Scholar]
- 17.Lee Chong T, Ahearn EL, Cimmino L. Reprogramming the Epigenome With Vitamin C. Frontiers in Cell and Developmental Biology. 2019;7(128). doi: 10.3389/fcell.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing PY, Pettersson S, Kundu P. Microbial Metabolites and Intestinal Stem Cells Tune Intestinal Homeostasis. PROTEOMICS. 2020;20(5–6):1800419. doi: 10.1002/pmic.201800419. [DOI] [PubMed] [Google Scholar]
- 19.Kim E, Davidson LA, Zoh RS, Hensel ME, Salinas ML, Patil BS et al. Rapidly cycling Lgr5(+) stem cells are exquisitely sensitive to extrinsic dietary factors that modulate colon cancer risk. Cell Death Dis. 2016;7(11):e2460. doi: 10.1038/cddis.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E, Wright GA, Zoh RS, Patil BS, Jayaprakasha GK, Callaway ES et al. Establishment of a multicomponent dietary bioactive human equivalent dose to delete damaged Lgr5+ stem cells using a mouse colon tumor initiation model. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP). 2019;28(5):383–9. doi: 10.1097/cej.0000000000000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan YY, Davidson LA, Callaway ES, Wright GA, Safe S, Chapkin RS. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am J Physiol Gastrointest Liver Physiol. 2015;309(1):G1–9. doi: 10.1152/ajpgi.00052.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Han H, Davidson LA, Fan Y-Y, Goldsby JS, Yoon G, Jin U-H et al. Loss of aryl hydrocarbon receptor potentiates FoxM1 signaling to enhance self-renewal of colonic stem and progenitor cells. The EMBO Journal. 2020:e104319. doi: 10.15252/embj.2019104319. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that AhR signaling directly controls the expression of FoxM1 to regulate the functionality of colonic stem/progenitor cells, thus affecting colon tumorigenesis.
- 23.Bradfield CA, Poland A. A competitive binding assay for 2,3,7,8-tetrachlorodibenzo-p-dioxin and related ligands of the Ah receptor. Mol Pharmacol. 1988;34(5):682–8. [PubMed] [Google Scholar]
- 24.Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG et al. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin UH, Karki K, Cheng Y, Michelhaugh SK, Mittal S, Safe S. The aryl hydrocarbon receptor is a tumor suppressor-like gene in glioblastoma. J Biol Chem. 2019;294(29):11342–53. doi: 10.1074/jbc.RA119.008882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soshilov A, Denison MS. Role of the Per/Arnt/Sim domains in ligand-dependent transformation of the aryl hydrocarbon receptor. The Journal of biological chemistry. 2008;283(47):32995–3005. doi: 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the Same but Different: Promiscuity and Diversity in the Molecular Mechanisms of Action of the Aryl Hydrocarbon (Dioxin) Receptor. Toxicological Sciences. 2011;124(1):1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poland A, Glover E. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem. 1976;251(16):4936–46. [PubMed] [Google Scholar]
- 30.Lucier GW, McDaniel OS, Hook GE, Fowler BA, Sonawane BR, Faeder E. TCDD-induced changes in rat liver microsomal enzymes. Environmental health perspectives. 1973;5:199–209. doi: 10.1289/ehp.7305199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Pei X, Zhang W, Xie HQ, Zhao B. Functional analysis of the dioxin response elements (DREs) of the murine CYP1A1 gene promoter: beyond the core DRE sequence. Int J Mol Sci. 2014;15(4):6475–87. doi: 10.3390/ijms15046475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davarinos NA, Pollenz RS. Aryl Hydrocarbon Receptor Imported into the Nucleus following Ligand Binding Is Rapidly Degraded via the Cytosplasmic Proteasome following Nuclear Export. Journal of Biological Chemistry. 1999;274(40):28708–15. doi: 10.1074/jbc.274.40.28708. [DOI] [PubMed] [Google Scholar]
- 33.Huang G, Elferink CJ. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol. 2012;81(3):338–47. doi: 10.1124/mol.111.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson SR, Joshi AD, Elferink CJ. The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J Pharmacol Exp Ther. 2013;345(3):419–29. doi: 10.1124/jpet.113.203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446(7135):562–6. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 36.Kawajiri K, Kobayashi Y, Ohtake F, Ikuta T, Matsushima Y, Mimura J et al. Aryl hydrocarbon receptor suppresses intestinal carcinogenesis in ApcMin/+ mice with natural ligands. Proc Natl Acad Sci U S A. 2009;106(32):13481–6. doi: 10.1073/pnas.0902132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Shiizaki K, Kido K, Mizuta Y. Insight into the relationship between aryl-hydrocarbon receptor and β-catenin in human colon cancer cells. PLOS ONE. 2019;14(11):e0224613. doi: 10.1371/journal.pone.0224613. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides follow-up evidence that AhR does not act as a E3 ligase to mediate β-catenin degradation in human colon cancer cell lines.
- 38.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLOS Biology. 2016;14(8):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A et al. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014;85(5):777–88. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nature Medicine. 2016;22:586. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ et al. Feedback control of AHR signalling regulates intestinal immunity. Nature. 2017;542:242. doi: 10.1038/nature21080. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides evidence that intestinal epithelial cells act as a gatekeeper to control the availabilty of dietary- or gut microbiota- derived AhR ligands.
- 43.Sasaki N, Sachs N, Wiebrands K, Ellenbroek SI, Fumagalli A, Lyubimova A et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc Natl Acad Sci U S A. 2016. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E et al. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity. 2018. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that AhR signaling controls the maintenance and differentiation of intestinal stem cells by modulating Wnt/β-catenin signaling, and ameliorates carcinogen-associated colon tumorigenesis.
- 45.Okino ST, Pookot D, Basak S, Dahiya R. Toxic and Chemopreventive Ligands Preferentially Activate Distinct Aryl Hydrocarbon Receptor Pathways: Implications for Cancer Prevention. Cancer Prevention Research. 2009;2(3):251–6. doi: 10.1158/1940-6207.Capr-08-0146. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad A, Ali S, Wang Z, Ali AS, Sethi S, Sakr WA et al. 3,3’-Diindolylmethane enhances taxotere-induced growth inhibition of breast cancer cells through downregulation of FoxM1. International journal of cancer. 2011;129(7):1781–91. doi: 10.1002/ijc.25839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hao H-X, Jiang X, Cong F. Control of Wnt Receptor Turnover by R-spondin-ZNRF3/RNF43 Signaling Module and Its Dysregulation in Cancer. Cancers. 2016;8(6):54. doi: 10.3390/cancers8060054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205):1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 49.Diaz-Diaz CJ, Ronnekleiv-Kelly SM, Nukaya M, Geiger PG, Balbo S, Dator R et al. The Aryl Hydrocarbon Receptor is a Repressor of Inflammation-associated Colorectal Tumorigenesis in Mouse. Ann Surg. 2016. doi: 10.1097/SLA.0000000000001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ema M, Ohe N, Suzuki M, Mimura J, Sogawa K, Ikawa S et al. Dioxin binding activities of polymorphic forms of mouse and human arylhydrocarbon receptors. J Biol Chem. 1994;269(44):27337–43. [PubMed] [Google Scholar]
- 51.Flaveny CA, Murray IA, Perdew GH. Differential gene regulation by the human and mouse aryl hydrocarbon receptor. Toxicol Sci. 2010;114(2):217–25. doi: 10.1093/toxsci/kfp308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goettel JA, Gandhi R, Kenison JE, Yeste A, Murugaiyan G, Sambanthamoorthy S et al. AHR Activation Is Protective against Colitis Driven by T Cells in Humanized Mice. Cell Rep. 2016;17(5):1318–29. doi: 10.1016/j.celrep.2016.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Natividad JM, Agus A, Planchais J, Lamas B, Jarry AC, Martin R et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018;28(5):737–49 e4. doi: 10.1016/j.cmet.2018.07.001. [DOI] [PubMed] [Google Scholar]; Provides evidence that reduced gut microbiota-derived AhR ligands are observed in individuals with metabolic syndrome.
- 54.Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE et al. Aryl Hydrocarbon Receptor Antagonists Promote the Expansion of Human Hematopoietic Stem Cells. Science. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrill JA, Parks BB, Wauthier E, Rowlands JC, Reid LM, Thomas RS. Lineage-dependent effects of aryl hydrocarbon receptor agonists contribute to liver tumorigenesis. Hepatology. 2015;61(2):548–60. doi: 10.1002/hep.27547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tofighi R, Wan Ibrahim WN, Rebellato P, Andersson PL, Uhlén P, Ceccatelli S. Non–Dioxin-like Polychlorinated Biphenyls Interfere with Neuronal Differentiation of Embryonic Neural Stem Cells. Toxicological Sciences. 2011;124(1):192–201. doi: 10.1093/toxsci/kfr221. [DOI] [PubMed] [Google Scholar]
- 57.Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A et al. T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell. 2018;175(5):1307–20.e22. doi: 10.1016/j.cell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453(7191):65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 59.Kiss EA, Diefenbach A. Role of the Aryl Hydrocarbon Receptor in Controlling Maintenance and Functional Programs of RORgammat(+) Innate Lymphoid Cells and Intraepithelial Lymphocytes. Front Immunol. 2012;3:124. doi: 10.3389/fimmu.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, Bostick JW, Zhou L. Regulation of Innate Lymphoid Cells by Aryl Hydrocarbon Receptor. Frontiers in immunology. 2018;8:1909–. doi: 10.3389/fimmu.2017.01909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Li S, Bostick JW, Ye J, Qiu J, Zhang B, Urban JF Jr. et al. Aryl Hydrocarbon Receptor Signaling Cell Intrinsically Inhibits Intestinal Group 2 Innate Lymphoid Cell Function. Immunity. 2018;49(5):915–28.e5. doi: 10.1016/j.immuni.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demontrates that AhR signaling regulates the gut balance of ILC2 and ILC3 and supresses the number and functionality of ILC2 in a cell-intrinsic fashion.
- 62.Hayakawa Y, Wang TC. The Tuft Cell-ILC2 Circuit Integrates Intestinal Defense and Homeostasis. Cell. 2018;174(2):251–3. doi: 10.1016/j.cell.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 63.Yeste A, Mascanfroni ID, Nadeau M, Burns EJ, Tukpah A-M, Santiago A et al. IL-21 induces IL-22 production in CD4+ T cells. Nature Communications. 2014;5:3753. doi: 10.1038/ncomms4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36(1):92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. 2010;11(9):854–61. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Gutierrez-Vazquez C, Quintana FJ. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity. 2018;48(1):19–33. doi: 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive review of current findings linking AhR signaling and host immune response.
- 67.Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–4. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwarycz B, Gracz AD, Rivera KR, Williamson IA, Samsa LA, Starmer J et al. IL22 Inhibits Epithelial Stem Cell Expansion in an Ileal Organoid Model. Cellular and Molecular Gastroenterology and Hepatology. 2019;7(1):1–17. doi: 10.1016/j.jcmgh.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zha J-M, Li H-S, Lin Q, Kuo W-T, Jiang Z-H, Tsai P-Y et al. Interleukin 22 Expands Transit-Amplifying Cells While Depleting Lgr5+ Stem Cells via Inhibition of Wnt and Notch Signaling. Cellular and Molecular Gastroenterology and Hepatology. 2019;7(2):255–74. doi: 10.1016/j.jcmgh.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jain U, Lai CW, Xiong S, Goodwin VM, Lu Q, Muegge BD et al. Temporal Regulation of the Bacterial Metabolite Deoxycholate during Colonic Repair Is Critical for Crypt Regeneration. Cell Host Microbe. 2018;24(3):353–63.e5. doi: 10.1016/j.chom.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roulis M, Kaklamanos A, Schernthanner M, Bielecki P, Zhao J, Kaffe E et al. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature. 2020;580(7804):524–9. doi: 10.1038/s41586-020-2166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Montrose DC, Nakanishi M, Murphy RC, Zarini S, McAleer JP, Vella AT et al. The role of PGE2 in intestinal inflammation and tumorigenesis. Prostaglandins Other Lipid Mediat. 2015;116–117:26–36. doi: 10.1016/j.prostaglandins.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takamura T, Harama D, Matsuoka S, Shimokawa N, Nakamura Y, Okumura K et al. Activation of the aryl hydrocarbon receptor pathway may ameliorate dextran sodium sulfate-induced colitis in mice. Immunol Cell Biol. 2010;88(6):685–9. doi: 10.1038/icb.2010.35. [DOI] [PubMed] [Google Scholar]
- 74.Martey CA, Baglole CJ, Gasiewicz TA, Sime PJ, Phipps RP. The aryl hydrocarbon receptor is a regulator of cigarette smoke induction of the cyclooxygenase and prostaglandin pathways in human lung fibroblasts. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2005;289(3):L391–L9. doi: 10.1152/ajplung.00062.2005. [DOI] [PubMed] [Google Scholar]
- 75.Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology. 2015;149(7):1884–95.e4. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y, Soendergaard C, Bergenheim FH, Aronoff DM, Milne G, Riis LB et al. COX-2-PGE2 Signaling Impairs Intestinal Epithelial Regeneration and Associates with TNF Inhibitor Responsiveness in Ulcerative Colitis. EBioMedicine. 2018;36:497–507. doi: 10.1016/j.ebiom.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyoshi H, VanDussen KL, Malvin NP, Ryu SH, Wang Y, Sonnek NM et al. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. The EMBO journal. 2017;36(1):5–24. doi: 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jackstadt R, Sansom OJ. Mouse models of intestinal cancer. The Journal of Pathology. 2016;238(2):141–51. doi: 10.1002/path.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Washington K, Zemper AED. Apc-related models of intestinal neoplasia: a brief review for pathologists. Surgical and Experimental Pathology. 2019;2(1):11. doi: 10.1186/s42047-019-0036-9. [DOI] [Google Scholar]
- 80.Joseph R, Little P, Hayes DN, Lee MS. Characterization of the number and site of APC mutations in sporadic colorectal cancer. Journal of Clinical Oncology. 2017;35(4_suppl):630–. doi: 10.1200/JCO.2017.35.4_suppl.630. [DOI] [Google Scholar]
- 81.Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S et al. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Reports. 2020;31(1):107471. doi: 10.1016/j.celrep.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoon K, Kim N. The Effect of Microbiota on Colon Carcinogenesis. J Cancer Prev. 2018;23(3):117–25. doi: 10.15430/JCP.2018.23.3.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ikuta T, Kobayashi Y, Kitazawa M, Shiizaki K, Itano N, Noda T et al. ASC-associated inflammation promotes cecal tumorigenesis in aryl hydrocarbon receptor-deficient mice. Carcinogenesis. 2013;34(7):1620–7. doi: 10.1093/carcin/bgt083. [DOI] [PubMed] [Google Scholar]
- 84.De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9–. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia-Villatoro EL, DeLuca JAA, Callaway ES, Allred KF, Davidson LA, Hensel ME et al. Effects of high-fat diet and intestinal aryl hydrocarbon receptor deletion on colon carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2020;318(3):G451–g63. doi: 10.1152/ajpgi.00268.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86••.Gronke K, Hernandez PP, Zimmermann J, Klose CSN, Kofoed-Branzk M, Guendel F et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature. 2019;566(7743):249–53. doi: 10.1038/s41586-019-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates that the production of IL22 by AhR signaling mainly in ILC3, protects intestinal stem cells from genotoxic insults, and reduces carcinogen-associated colon tumorigenesis.
- 87.Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141(1):237–48, 48 e1. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 88•.Busbee PB, Menzel L, Alrafas HR, Dopkins N, Becker W, Miranda K et al. Indole-3-carbinol prevents colitis and associated microbial dysbiosis in an IL-22–dependent manner. JCI Insight. 2020;5(1). doi: 10.1172/jci.insight.127551. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides evidence that supplementation of the AhR ligand precusor indole-3-carbinol prevents colitis-associated microbial dysbiosis by induction of IL22.
- 89.Kawai S, Iijima H, Shinzaki S, Hiyama S, Yamaguchi T, Araki M et al. Indigo Naturalis ameliorates murine dextran sodium sulfate-induced colitis via aryl hydrocarbon receptor activation. Journal of gastroenterology. 2017;52(8):904–19. doi: 10.1007/s00535-016-1292-z. [DOI] [PubMed] [Google Scholar]
- 90.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bin P, Leng S, Cheng J, Dai Y, Huang C, Pan Z et al. Association of Aryl Hydrocarbon Receptor Gene Polymorphisms and Urinary 1-Hydroxypyrene in Polycyclic Aromatic Hydrocarbon–Exposed Workers. Cancer Epidemiology Biomarkers & Prevention. 2008;17(7):1702–8. doi: 10.1158/1055-9965.Epi-07-2812. [DOI] [PubMed] [Google Scholar]
- 92.Kovalova N, Manzan M, Crawford R, Kaminski N. Role of aryl hydrocarbon receptor polymorphisms on TCDD-mediated CYP1B1 induction and IgM suppression by human B cells. Toxicol Appl Pharmacol. 2016;309:15–23. doi: 10.1016/j.taap.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wong JM, Okey AB, Harper PA. Human aryl hydrocarbon receptor polymorphisms that result in loss of CYP1A1 induction. Biochemical and biophysical research communications. 2001;288(4):990–6. doi: 10.1006/bbrc.2001.5861. [DOI] [PubMed] [Google Scholar]
- 94.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gu A, Ji G, Long Y, Zhou Y, Shi X, Song L et al. Assessment of an association between an aryl hydrocarbon receptor gene (AHR) polymorphism and risk of male infertility. Toxicol Sci. 2011;122(2):415–21. doi: 10.1093/toxsci/kfr137. [DOI] [PubMed] [Google Scholar]
- 96.Huang S, Shui X, He Y, Xue Y, Li J, Li G et al. AhR expression and polymorphisms are associated with risk of coronary arterial disease in Chinese population. Sci Rep. 2015;5:8022. doi: 10.1038/srep08022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodgers GP, Collins FS. Precision Nutrition-the Answer to “What to Eat to Stay Healthy”. Jama. 2020. doi: 10.1001/jama.2020.13601. [DOI] [PubMed] [Google Scholar]


