Abstract
Background
Immunotherapy yields survival benefit for some advanced stage non-small cell lung cancer (NSCLC) patients. Because highly predictive biomarkers of immunotherapy response are an unmet clinical need, we used pretreatment radiomics and clinical data to train and validate a parsimonious model associated with survival outcomes among NSCLC patients treated with immunotherapy.
Methods
Three cohorts of NSCLC patients treated with immunotherapy were analyzed: training (n = 180), validation 1 (n = 90), and validation 2 (n = 62). The most informative clinical and radiomic features were subjected to decision tree analysis, which stratified patients into risk groups of low, moderate, high, and very high risk of death after initiation of immunotherapy. All statistical tests were 2-sided.
Results
The very high-risk group was associated with extremely poor overall survival (OS) in validation cohorts 1 (hazard ratio [HR] = 5.35, 95% confidence interval [CI] = 2.14 to 13.36; 1-year OS = 11.1%, 95% CI = 1.9% to 29.8%; 3-year OS = 0%) and 2 (HR = 13.81, 95% CI = 2.58 to 73.93; 1-year OS = 47.6%, 95% CI = 18.2% to 72.4%; 3-year OS = 0%) when compared with the low-risk group (HR = 1.00) in validation cohorts 1 (1-year OS = 85.0%, 95% CI = 60.4% to 94.9%; 3-year OS = 38.9%, 95% CI = 17.1% to 60.3%) and 2 (1-year OS = 80.2%, 95% CI = 40.3% to 94.8%; 3-year OS = 40.1%, 95% CI = 1.3% to 83.5%). The most informative radiomic feature, gray-level co-occurrence matrix (GLCM) inverse difference, was positively associated with hypoxia-related carbonic anhydrase 9 using gene-expression profiling and immunohistochemistry.
Conclusion
Utilizing standard-of-care imaging and clinical data, we identified and validated a novel parsimonious model associated with survival outcomes among NSCLC patients treated with immunotherapy. Based on this model, clinicians can identify patients who are unlikely to respond to immunotherapy.
Checkpoint blockade immunotherapy demonstrates durable and long-term survival benefit in 20%-50% patients with advanced stage non-small cell lung cancer (NSCLC) (1-6). Because patterns of immunotherapy response (7) are complex, including rapid disease progression (8), hyperprogression (9), and acquired resistance (10), there is an urgency to identify highly predictive biomarkers that can stratify patients into distinct risk groups of survival and progression. Tumor programmed cell death ligand-1 (PD-L1) expression by immunohistochemistry (IHC) is a standard-of-care biomarker; however, recent clinical trials demonstrated statistically significant survival benefit for patients irrespective of tumor PD-L1 expression (6,11). Although tumor mutational burden (TMB) (12) has also shown to be a predictor of immunotherapy response (13–15), tumor specimens have to be sufficient in both quantity and quality to assess TMB (14), and laboratory methods to calculate TMB can be timely and expensive. As such, complimentary biomarkers that are predictive, noninvasive, and measured in a timely fashion using standard-of-care modalities would have direct translational implications.
Quantitative image-based features, or radiomics, reflect the underlying pathophysiology and tumor heterogeneity (16) and have advantages over tissue-based biomarkers as they can be rapidly extracted using standard-of-care medical images and can capture data from the entire region of interest (eg, tumor) rather than a small portion of the tumor that is biopsied and assayed. Therefore, in this study, we used pretreatment clinical data and radiomic features extracted from pretreatment computed tomography (CT) scans to develop a risk model that is associated with survival outcomes among NSCLC patients treated with immunotherapy. The biological underpinnings of the radiomics signature were assessed by gene expression and IHC analyses.
Methods
Immunotherapy-Treated Lung Cancer Patients
This study was approved by the University of South Florida institutional review board, and a waiver of informed consented was obtained. Three cohorts of NSCLC patients treated with immunotherapy were analyzed: training (Moffitt Cancer Center [MCC]1, n = 180), validation 1 (MCC2, n = 90), and validation 2 (Veterans Health Administration [VA], n = 62). Patients in MCC1 and MCC2 cohorts were treated at the H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida, and patients in the VA cohort were treated at James A. Haley Veterans’ Hospital, Tampa, Florida. All patients were treated with PD-1 or PD-L1 therapy or combinations of PD-L1 or PD-1 with cytotoxic T-lymphocyte–associated protein 4 doublet therapy. Inclusion criteria included pretreatment CT or positron emission tomography-CT imaging acquired less than 90 days prior to initiation of immunotherapy and having at least 1 measurable lung lesion defined by Response Evaluation Criteria in Solid Tumors (RECIST) criteria. Additional details about the lung cancer patients and patient data are provided in the Supplementary Methods (available online).
Radiogenomics Dataset
A previously described (17) patient dataset of 103 surgically resected adenocarcinomas (Lung1) who had CT radiomics and tumor gene-expression profiling was used to identify biological underpinnings of the most informative radiomic feature; additional details are provided in the Supplementary Methods (available online).
Immunohistochemistry
To validate the radiogenomics data, IHC of CAIX was performed on 16 NSCLC patient samples that had a presurgical CT. The IHC staining was quantified by automated evaluation of positive staining percentage defined by the Aperio ImageScope (http://www.leicabiosystems.com) and by H-scoring metric (18) quantified by a board-certified pathologist (JS) who was masked to radiomics and automated IHC scoring data.
Prognostic Datasets
The most informative radiomics was analyzed in 4 independent datasets of NSCLC patients not treated with immunotherapy. Overall survival (OS) was the dependent variable. The first dataset (Lung1) was the radiogenomics dataset described above. The second dataset (Lung2) included 234 patients diagnosed with incidental lung cancer from the National Lung Screening Trial (16,19). The third dataset (Lung3) included 62 patients who had presurgical CTs and underwent resection at MCC (20,21). The fourth dataset (Lung4) (20,21) included 47 patients who had presurgical CT scans and underwent surgical resection at the Maastricht Radiation Oncology Clinic (MAASTRO).
Radiomic Feature Extraction
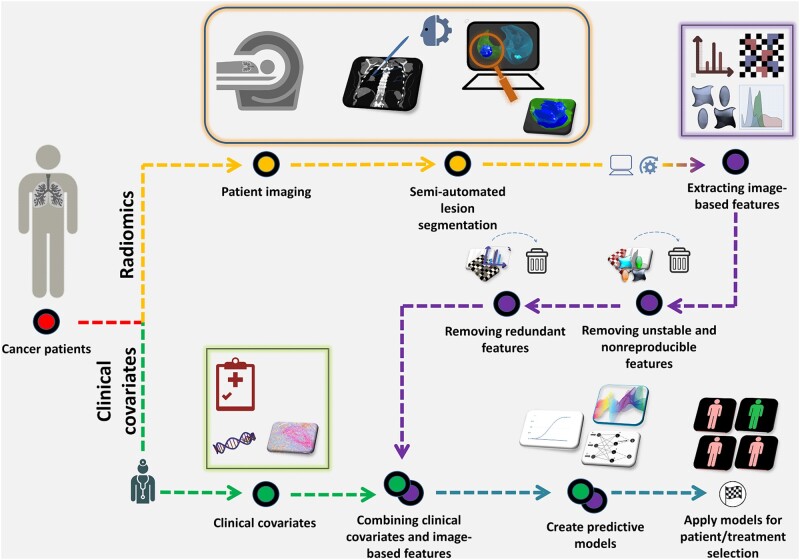
Figure 1 is an overview of our radiomics pipeline. Pretreatment thoracic CT scans were retrieved from the picture archiving and communication system and loaded into the HealthMyne software (http://www.healthmyne.com) for segmentation. Using the HealthMyne software, 2 radiologists (YT and JQ) with more than 10 years of clinical experience initialized the 3D segmentation algorithm on the largest lung lesion. The radiologists either confirmed or edited the segmentations as necessary. The tumor masks were imported into in-house radiomic feature extraction toolboxes created in MATLAB 2015b (The Mathworks Inc, Natick, MA) and C++ (https://isocpp.org). A total of 213 radiomic features were extracted from the intratumoral (122 features) and the peritumoral (3 mm outside of tumor boundary) regions (91 features) using standardized algorithms defined by the Image Biomarker Standardization Initiative (22). Stable and reproducible radiomic features were identified using previously described methods (23); details are included in the Supplementary Methods (available online).
Figure 1.
The radiomics pipeline. After pretreatment (ie, baseline), imaging, and patient data are obtained, radiomic features are extracted from standard-of-care imaging studies (yellow). Radiologists mark target the lesions, and lesions are automatically (or semi-automatically) segmented. Radiomic features are then extracted from the region of interest (purple). Unstable, nonreproducible and correlated radiomic features are removed. The remaining features are combined with the pretreatment clinical covariates (green), and model building approaches are applied, which can be used for patient stratification and/or treatment selection.
Statistical Analysis
Statistical analyses were performed using Stata/MP 14.2 (StataCorp LP, College Station, TX) and R Project for Statistical Computing version 3.4.3 (http://www.r-project.org/). All statistical tests were 2-sided, and a P value of less than .05 was considered statistically significant. Differences between cohorts for clinical covariates were tested using Fisher exact test for categorical variables and Mann-Whitney U test and Student’s t test for continuous variables. Survival analyses were performed using Cox regression, Kaplan-Meier survival estimates, and log-rank test. OS and progression-free survival (PFS) were the 2 dependent variables. Index date was the date of initiation of immunotherapy. For OS, an event was defined as death. For PFS, an event was defined as death or either clinical- or RECIST-based progression of cancer, and the data were right censored at 36 months.
Rigorous model building approaches were employed to reduce the number of variables and identify the most informative clinical covariates and radiomic features associated with OS in the training cohort (MCC1). The most informative clinical covariates and radiomic features that were identified in the training cohort were then tested in 2 validation cohorts (MCC2 and VA). For the clinical covariates, univariable Cox regression identified variables statistically significantly associated with OS. To generate a parsimonious clinical model, statistically significant clinical covariates from the univariable analyses were included in a stepwise backward elimination model using a threshold of P less than .01 for inclusion. For the radiomic features, we removed radiomic features correlated with tumor volume using a Pearson correlation coefficient of 0.80 or higher; among intracorrelated features using an absolute Pearson correlation coefficient of 0.80 or higher, the feature with the lowest P value in univariable analysis was retained, and the other(s) was removed. Among the remaining features, univariable Cox regression identified those statistically significantly associated with OS. Among the remaining features, a parsimonious model was identified using stepwise backward elimination using a threshold of P less than .01 for inclusion. Bonferroni-Holm method was used for multiple testing corrections.
The variables from the parsimonious clinical and radiomics models were used as inputs into a survival Classification and Regression Tree (CART) (24), which yielded risk groups associated with OS in MCC1 and then was validated in MCC2 and VA. Time-dependent area under the curve (AUC) values were calculated for 6, 12, 24, and 36 months.
Radiomics Quality Score (RQS)
RQS was calculated using an established metric that assesses the quality of a radiomics study (25). The RQS contains 16 items with a maximum score of 36.
Results
Patient Demographics
Type of checkpoint inhibitor, Eastern Cooperative Oncology Group (ECOG) performance status, number of previous lines of therapy, serum albumin, lymphocyte counts, and neutrophils to lymphocytes ratio were statistically significantly different between MCC1 and MCC2 (Table 1). Median age, sex, smoking status, stage, type of checkpoint inhibitor, ECOG performance status, lymphocyte counts, and neutrophils to lymphocytes ratio were statistically significantly different between MCC1 and VA.
Table 1.
Patient characteristics of the training and 2 validation cohorts
| Characteristicsb,c | Training | Validation | P b ,c | Validation | P b ,d |
|---|---|---|---|---|---|
| MCC1 cohort | MCC2 cohort | VA cohort | |||
| (n = 180) | (n = 90) | (n = 62) | |||
| Age at initiation of treatment, No. (%) | |||||
| Dichotomized | |||||
| <65 y | 68 (37.8) | 37 (41.1) | 15 (24.2) | ||
| ≥65 y | 112 (62.2) | 53 (58.9) | .60 | 47 (75.8) | .06 |
| Median (95% CI), y | 67 (65 to 68) | 67 (64 to 69) | .78 | 68 (67 to 71) | .03 |
| Sex, No. (%) | |||||
| Female | 95 (52.8) | 43 (47.8) | 3 (4.8) | ||
| Male | 85 (47.2) | 47 (52.2) | .44 | 59 (95.2) | <.001 |
| Smoking status, No. (%)d | |||||
| Never smoker | 30 (16.7) | 16 (17.8) | 2 (3.2) | ||
| Ever smoker | 146 (81.1) | 74 (82.2) | .87 | 60 (96.8) | .004 |
| Unknown/Missing | 4 (2.2) | 0 (0) | 0 (0) | ||
| Stage, No. (%) | |||||
| III | 6 (3.3) | 4 (4.4) | 13 (21.0) | ||
| IV | 174 (96.7) | 86 (95.6) | .74 | 49 (79.0) | <.001 |
| Histology, No. (%) | |||||
| Adenocarcinoma/others | 137 (76.1) | 71 (78.9) | 43 (69.3) | ||
| Squamous cell carcinoma | 43 (23.9) | 19 (21.1) | .65 | 19 (30.7) | .14 |
| Checkpoint inhibitors, No. (%) | |||||
| Anti–PD-L1 | 48 (26.6) | 18 (20.0) | 8 (12.9) | ||
| Anti–PD-1 | 57 (31.7) | 69 (76.7) | 54 (87.1) | ||
| Doublet | 75 (41.7) | 3 (3.3) | <.001 | 0 (0) | <.001 |
| ECOG performance status, No. (%) | |||||
| 0 | 39 (21.7) | 10 (11.1) | 12 (19.4) | ||
| 1 | 141 (78.3) | 67 (74.4) | 39 (62.9) | ||
| 2 | 0 (0) | 13 (14.4) | <.001 | 11 (17.7) | <.001 |
| Previous lines of therapy on current diagnosis, No. (%) | |||||
| None | 70 (43.9) | 21 (23.3) | N/A | ||
| 1 | 48 (26.7) | 47 (52.2) | N/A | ||
| ≥2 | 62 (34.4) | 22 (24.4) | <.001 | N/A | — |
| Number of metastatic sites, No. (%) | |||||
| 1 | 82 (46.6) | 51 (56.7) | 25 (40.3) | ||
| ≥2 | 98 (54.4) | 39 (43.3) | .09 | 37 (59.7) | .55 |
| EGFR mutational status, No. (%)e | |||||
| Not detected | 107 (59.4) | 37 (41.1) | N/A | ||
| Detected | 25 (13.9) | 5 (5.6) | .36 | N/A | — |
| Missing/Inconclusive | 48 (26.7) | 48 (53.3) | N/A | ||
| KRAS mutational status, No. (%)e | |||||
| Not detected | 61 (33.9) | 20 (22.2) | N/A | ||
| Detected | 29 (16.1) | 12 (13.3) | .66 | N/A | — |
| Missing/Inconclusive | 90 (50.0) | 58 (64.4) | N/A | ||
| Hematology, median (95% CI) | |||||
| Serum albumin, g/dL | 4.0 (2.8 to 4.9) | 3.8 (2.3 to 4.7) | < 001 | 3.9 (2.9 to 4.6) | .09 |
| Lymphocytes, 1x109/L | 1.3 (0.3 to 3.6) | 1.0 (0.3 to 3.4) | <.001 | 1.0 (0.2 to 3.2) | .01 |
| WBC, 1x109/L | 7.1 (3.2 to 61.5) | 7.7 (1.4 to 45.1) | .25 | 7.5 (1.8 to 19.4) | .38 |
| Neutrophils, 1x109/L | 4.8 (1.6 to 31.7) | 5.3 (0.4 to 40.2) | .13 | 5.6 (1.1 to 15.1) | .33 |
| Ratio of neutrophils: Lymphocytes | 3.7 (1.1 to 30.4) | 5.2 (0.5 to 53.1) | .002 | 5.3 (0.8 to 34.3) | .004 |
The majority of the MCC1 (95.3%) and the MCC2 cohorts (86.7%) were self-reported White race and majority of MCC1 cohort (97.0%) and the MCC2 cohort (88.9%) were self-reported non-Hispanic. For the VA cohort, racial and ethnicity data were not available. CI = confidence interval; ECOG = Eastern Cooperative Oncology Group; EGFR = estimated glomerular filtration rate; MCC = Moffitt Cancer Center; PD-L1 = programmed cell death ligand-1; VA = Veterans Health Administration; WBC = white blood cell; N/A = not available (i.e., these covariates were not available in the VA validation cohort).
P values for continuous variables were calculated using Mann-Whitney test and Fisher exact test for categorical variables. All P values are 2-sided. P values were not calculated for N/A cells and an em dash was denoted.
P values were calculated comparing MCC1 and MCC2 cohorts.
P values were calculated comparing MCC1 and VA cohorts.
P values for smoking status, EGFR mutational status, and KRAS mutational status were calculated excluding missing or inconclusive data.
The OS and PFS rates for all cohorts are presented in Supplementary Table 1 (available online), and the Kaplan-Meier survival curves are presented in Supplementary Figure 1 (available online). MCC1 and MCC2 had statistically significantly different OS (36-month OS = 32.6% vs 19.2%; P < .001) and PFS (36-month PFS = 20.8% vs 9.5%; P = .003). PFS was not available for the VA cohort.
Covariate Reduction
Among the 16 covariates (Table 1) considered for the clinical model, 5 covariates (serum albumin, number of metastatic sites, previous lines of therapy, white blood cell, neutrophil count) were statistically significant in univariable analysis in MCC1 (Supplementary Table 2, available online). The parsimonious clinical model included 2 clinical covariates that were statistically significantly associated with OS: serum albumin (hazard ratio [HR] = 0.33, 95% confidence interval [CI] = 0.20 to 0.52) and number of metastatic sites (HR = 2.14, 95% CI = 1.48 to 3.11).
Among the 213 intratumoral and peritumoral radiomic features, 67 were found to be stable and reproducible (Figure 2, A). Of 67 features, 8 were correlated with tumor volume and removed. Ten of the remaining features were statistically significantly associated with OS in univariable analysis (Supplementary Table 3, available online), and 2 features remained after removing correlated features (Figure 2, B). Among the 2 remaining features (gray level co-occurrence matrix [GLCM] inverse difference and peritumoral quartile coefficient of dispersion), stepwise backward elimination identified a parsimonious model with 1 feature: GLCM inverse difference (HR = 1.41, 95% CI = 1.19 to 1.67).
Figure 2.
The heat map of concordance correlation coefficients, the correlation matrix for the “avatar” feature, and the Classification and Regression Tree (CART). A) The heat map plots the concordance correlation coefficients (CCC) of the radiomic features acquired by different segmentations and image acquisitions. Each column in the heat map represents a radiomic feature from the indicated feature group and region of interest (eg, intratumoral or peritumoral). The features are compared between different segmentation algorithms (ALG), different initial parameters (IP), and test-retest scans (RIDER). The green boxes represent higher (CCC > 0.95), blue boxes represent moderate (CCC ≥ 0.75 and CCC ≤ 0.95), and red boxes represent lower (CCC < 0.75) CCCs. B) The correlation matrix plots the radiomic features that were statistically significantly associated with overall survival in the univariable analysis. The most informative radiomic feature (gray-level co-occurrence matrix [GLCM] inverse difference) was correlated with 7 other features. C) CART analysis was used to identify patient risk groups based on a decision tree containing 1 radiomic feature and 2 clinical features. Patients were grouped from low-risk to very high-risk based on overall survival outcomes.
CART Analysis
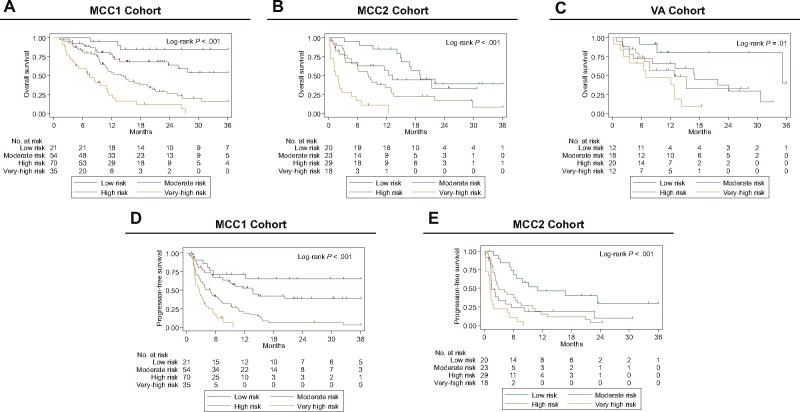
Using the 2 covariates from the parsimonious clinical model and 1 radiomic feature from the parsimonious radiomic model as inputs, CART analysis identified a novel tree structure (Figure 2, C) and grouped patients into 6 risk groups associated with OS (Supplementary Figure 2, available online). Of the original 6 groups, groups 2 and 3 were combined and groups 4 and 5 were combined as the hazard ratios for groups 2 and 3 and groups 4 and 5 were not statistically significantly different, resulting in 4 risk groups: low risk, moderate risk, high risk, and very high risk (Figure 3, A). The risk groups from the MCC1 cohort were also analyzed in both validation cohorts (MCC2 and VA; Figure 3, B and C). Similar findings were observed when the risk groups were analyzed for PFS for MCC1 and MCC2 (Figure 3, D and E). PFS data were not available for VA.
Figure 3.
Kaplan-Meier survival curves for the 4 risk groups identified by CART analysis. Overall survival is presented for (A) MCC1 cohort, (B) MCC2 cohort, and (C) VA cohort. Progressive-free survival (PFS) is presented for (D) MCC1 cohort and (E) MCC2 cohort. PFS was not available for the VA cohort. Risk groups 2 and 3 were combined, and risk groups 4 and 5 were combined (as shown in Supplementary Figure 2, A, available online) to create the moderate-risk and the high-risk groups, respectively. The log-rank test was used to calculate 2-sided P values. MCC = Moffitt Cancer Center; VA = Veterans Health Administration.
Time-dependent AUCs demonstrated that the final model (Supplementary Figure 3, A, available online) consistently had higher AUCs compared with the clinical only model (Supplementary Figure 3, B, available online).
Multivariable Analyses
Multivariable Cox regression analyses demonstrate that the main effects for the risk groups were consistent when adjusting for clinical covariates (Table 1) that were statistically significantly different between MCC1 and MCC2. The multivariable models revealed the very high-risk group was associated with extremely poor OS in validation cohort 1 (HR = 5.35, 95% CI = 2.14 to 13.36; 1-year OS = 11.1%, 95% CI = 1.9% to 29.8%; 3-year OS = 0%) and validation cohort 2 (HR = 13.81, 95% CI = 2.58 to 73.93; 1-year OS = 47.6%, 95% CI = 18.2% to 72.4%; 3-year OS = 0%) when compared with the low-risk group (HR = 1.00) in validation cohort 1 (1-year OS = 85.0%, 95% CI = 60.4% to 94.9%; 3-year OS = 38.9%, 95% CI = 17.1% to 60.3%) and validation cohort 2 (1-year OS = 80.2%, 95% CI = 40.3% to 94.8%; 3-year OS = 40.1%, 95% CI = 1.3% to 83.5%; Table 2; Supplementary Tables 1 and 4, available online). The results were similar for PFS (Table 2; Supplementary Table 1, available online). Additionally, results were consistent for OS in multivariable analyses adjusting for clinical covariates that were statistically significantly different between MCC1 and VA cohorts (Supplementary Tables 1 and 4, available online).
Table 2.
Univariable and multivariable Cox regression analysis for overall survival and progression-free survival
| Characteristics | MCC1 cohort (n = 180) |
MCC2 cohort (n = 90) |
|||
|---|---|---|---|---|---|
| Univariable modela | Multivariable modelb | Multivariable modelc | Univariable modela | Multivariable modelb | |
| HR (95% CI) | HR (95% CI) | HR (95% CI) | HR (95% CI) | HR (95% CI) | |
| Overall survival | |||||
| Risk groupd | |||||
| Low risk | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) |
| Moderate risk | 3.79 (1.13 to 12.68)e | 3.08 (0.89 to 10.66) | 3.56 (1.02 to 12.48)e | 1.70 (0.75 to 3.87) | 1.51 (0.66 to 3.51) |
| High risk | 8.02 (2.47 to 26.09)e | 7.87 (2.38 to 25.97)e | 6.98 (2.10 to 23.18)e | 2.73 (1.33 to 5.63)e | 3.33 (1.57 to 7.05)e |
| Very high risk | 19.32 (5.80 to 64.32)e | 17.33 (5.11 to 58.72)e | 17.24 (5.09 to 58.36)e | 10.52 (4.58 to 24.17)e | 5.35 (2.14 to 13.36)e |
| ECOG | — | 1.22 (0.70 to 2.11) | 1.20 (0.69 to 2.07) | — | 2.63 (1.47 to 4.68)e |
| Pr. treatment | — | — | 1.36 (1.01 to 1.81)e | — | — |
| Lymphocytes | — | 1.04 (0.74 to 1.46) | . | — | 0.73 (0.45 to 1.17) |
| WBC | — | — | 0.98 (0.88 to 1.09) | — | — |
| Neutrophils | — | — | 1.10 (0.89 to 1.34) | — | — |
| NLR | — | 1.01 (0.97 to 1.06) | 0.98 (0.92 to 1.05) | — | 1.05 (1.02 to 1.08)e |
| Progression-free survival | |||||
| Risk groupd | |||||
| Low risk | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) |
| Moderate risk | 2.02 (0.89 to 4.64) | 2.05 (0.88 to 4.76) | 2.36 (1.00 to 5.58)e | 2.96 (1.43 to 6.14)e | 2.80 (1.34 to 5.85)e |
| High risk | 5.15 (2.33 to 11.36)e | 5.55 (2.46 to 12.49)e | 4.89 (2.15 to 11.14)e | 2.58 (1.29 to 5.14)e | 3.05 (1.50 to 6.18)e |
| Very high risk | 9.62 (4.12 to 22.44)e | 9.03 (3.77 to 21.63)e | 8.79 (3.66 to 21.11)e | 7.13 (3.31 to 15.35)e | 3.95 (1.56 to 8.54)e |
| ECOG | — | 1.09 (0.68 to 1.74) | 1.05 (0.66 to 1.68) | — | 2.33 (1.35 to 4.03)e |
| Pr. treatment | — | — | 1.32 (1.04 to 1.67)e | — | — |
| Lymphocytes | — | 0.83 (0.63 to 1.09) | — | — | 0.88 (0.59 to 1.33) |
| WBC | — | — | 1.00 (0.90 to 1.11) | — | — |
| Neutrophils | — | — | 1.04 (0.86 to 1.26) | — | — |
| NLR | — | 1.04 (0.99 to 1.09) | 1.01 (0.95 to 1.07) | — | 1.05 (1.02 to 1.08)e |
The main effects for each risk group with the low-risk group as the referent category (ie, HR = 1.00). Hazard ratios were not calculated for the cells with the em dash. CI = confidence interval; ECOG = Eastern Cooperative Oncology Group; HR = hazard ratio; MCC = Moffitt Cancer Center; NLR = neutrophils to lymphocytes ratio; PFS = progression-free survival; Pr. treatment = previous lines of treatments at current diagnosis; WBC = white blood cell.
These models included the clinical covariates that were found to be statistically significant different between the MCC1 and MCC2 cohorts (Table 1). The low risk group was the referent category.
These models included the clinical covariates that were found to be statistically significantly different between the Classification and Regression Tree risk groups in Supplementary Table 5 (available online).
Low-risk group refers to patients who have low gray level co-occurrence matrix (GLCM) inverse difference (≤0.43) and lower number of metastatic sites (n = 1). The moderate risk group refers to patients who either have low GLCM inverse difference (≤0.43) and higher number of metastatic sites (≥2), or patients who have higher GLCM inverse difference (>0.43), higher serum albumin (≥3.9), and lower number of metastatic sites (1). The high-risk group refers to either patients who have higher GLCM inverse difference (>0.43), higher serum albumin (≥3.9), and higher number of metastatic sites (≥2), or patients who have higher GLCM inverse difference (>0.43), lower serum albumin (<3.9), and lower number of metastatic sites (n = 1). The very high-risk group refers to patients who have higher GLCM inverse difference (>0.43), lower serum albumin (<3.9), and lower number of metastatic sites (≥2).eHazard ratios are statistically significant.
Multivariable Cox regression was also performed adjusting for clinical covariates that were statistically significant between the CART risk groups (Supplementary Table 4, available online) but did not appreciably change the hazard ratios (Table 2). Serum albumin and number of metastatic sites were not included as these covariates were already part of the CART structure.
As noted above, 8 features were found to be correlated with tumor volume. Three (longest diameter, minor axis length, and least axis length) of the 8 features were statistically significantly associated with OS. However, the hazard ratios for these 3 features were not statistically significant when they were included in a multivariable model containing the most informative clinical covariates and radiomic features.
Other Model Building Approaches
Other model approaches were explored and yielded identical results (data not shown). First, all radiomic and clinical covariates that were statistically significant in univariable analyses (Supplementary Table 6, available online) were subjected to CART analysis. Second, we substituted least absolute shrinkage and selection operator for the stepwise backward approach, then applied CART analysis. Lastly, we included the features correlated with volume and applied the original model-building approach. These extensive experiments demonstrated that our model is robust and reproducible by different modeling approaches.
Radiogenomics Analyses
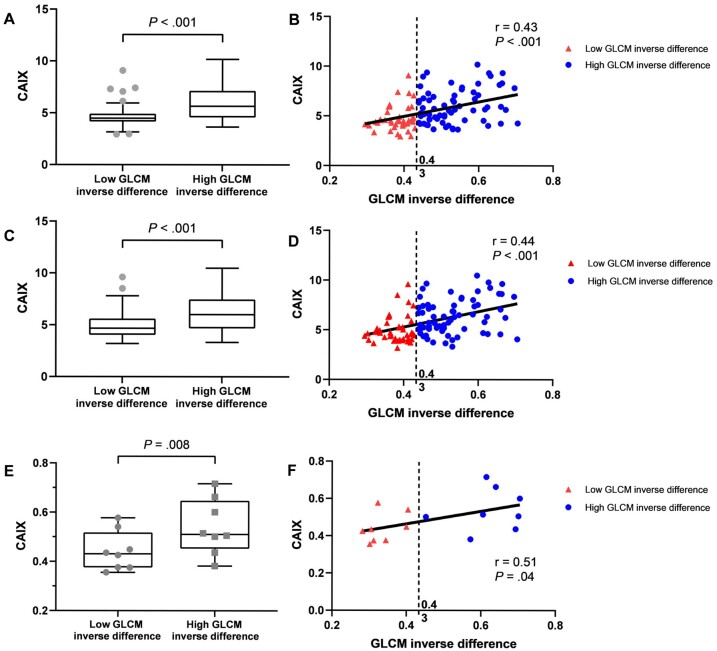
The most informative radiomic feature was compared with every gene probeset using 2 different approaches: 2-group analysis and correlation analysis. For the 2-group analyses, GLCM inverse difference was dichotomized at the cut-point (0.43) determined by CART. Correlation and 2-group analyses identified 123 statistically significant probesets representing 91 unique genes that were associated with the GLCM inverse difference radiomic feature (Supplementary Table 7, available online). Pathway analysis found no statistically significant enrichment after correcting by false discovery rate. Gene Ontology Biological Process enrichment of the gene set identified terms including regulation of cardiac conduction, sodium ion export across plasma membrane, and membrane depolarization during action potential. Only 3 probesets (representing 2 genes) were positively associated with GLCM inverse difference: CAIX and family with sequence similarity 83 member F (FAM83F). For both gene probesets, CAIX gene expression was statistically significantly different by GLCM inverse difference, and CAIX gene expression was positively correlated with GLCM inverse difference (Figure 4, A-D).
Figure 4.
Gray level co-occurrence matrix (GLCM) inverse difference radiomic feature and CAIX expression. A-D) The association between GLCM inverse difference and CAIX expression based off 2 different probesets: merck2-DQ892208_at is presented in panels A and B, and merck-NM_001216_at is presented in panels C and D. E) The association between the automated pathology IHC scoring for CAIX and GLCM inverse difference. F) The correlation between the automated pathology IHC scoring for CAIX and GLCM inverse difference. In panels A, C, and E, the Mann-Whitney U test was used to calculate 2-sided P values, and the error bars depict Tukey whiskers (fences). In panels B, D, and F, Pearson correlation coefficient was used to calculate 2-sided P values.
Immunohistochemistry Analyses
The automated pathology scoring was correlated with the pathologist-scored H-score (r = 0.629; P = .009). CAIX IHC by automated pathology scoring was statistically significantly different by GLCM inverse difference (Mann-Whitney U test P = .008; Figure 4, E), and CAIX IHC by automated pathology scoring was modestly correlated with GLCM inverse difference (r = 0.514; P = .04; Figure 4, F).
Prognostic Datasets
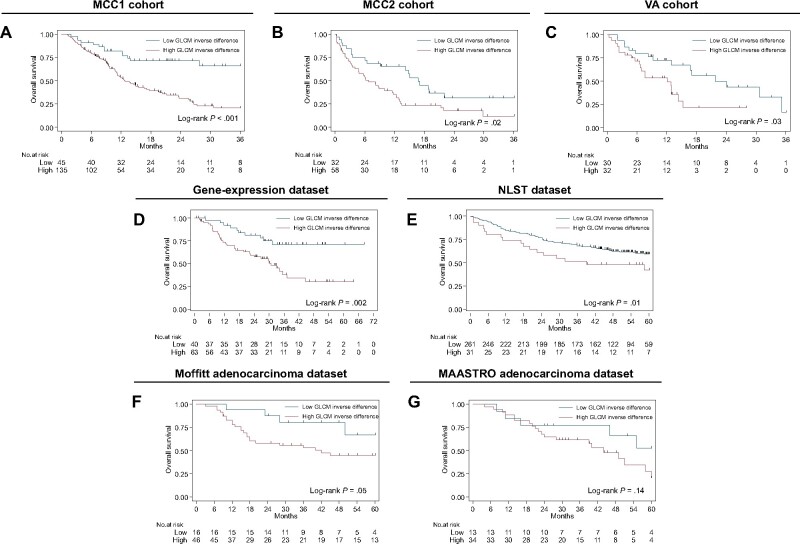
GLCM inverse difference was statistically significantly associated with OS in 3 independent NSCLC cohorts (Lung1, Lung2, and Lung3 in Figure 5) using the identified CART cut-point (0.43). Although this a priori cut-point was not statistically significant in Lung4, GLCM inverse difference as a continuous covariate was statistically significantly associated with OS in univariable analysis (HR = 2.74, 95% CI = 1.04 to 7.24).
Figure 5.
Kaplan-Meier survival curves (overall survival) for the dichotomized radiomics feature (gray level co-occurrence matrix [GLCM] inverse difference). The same dichotomized cut-point found in the MCC1 training cohort was used for the survival analyses (A-C) in the training and 2 validation cohorts, respectively, and (D-G) the 4 prognostic datasets (ie, gene-expression dataset, National Lung Screening Trial (NLST) dataset, Moffitt adenocarcinoma dataset, and Maastricht Radiation Oncology Clinic (MAASTRO) adenocarcinoma dataset, respectively). The log-rank test was used to calculate 2-sided P values.
Radiomics Quality Score
This study yields a radiomic quality score of 22 out of a possible maximum of 36 (Supplementary Table 8, available online).
Discussion
Predictive biomarkers that identify lung cancer patients who will experience rapid and lethal outcomes in the setting of immunotherapy are critical unmet needs as such patients could avoid ineffective therapy. In this study, we used a rigorous radiomics pipeline to identify a parsimonious clinical-radiomic risk model that stratified into 4 risk groups based on risk of patient death (ie, overall survival). The very high-risk group was associated with extremely poor OS and PFS in the training cohort and validated in internal and external cohorts (Figure 3, A-E). These results suggest very high-risk patients should either avoid immunotherapy altogether or utilize upfront combination treatments that may yield an improved response. The 4 risk groups identified were derived from a decision tree structure based on 1 radiomic feature (GLCM inverse difference) and 2 clinical covariates (baseline number of metastatic sites and serum albumin). The most informative radiomic feature, GLCM inverse difference, was positively associated with CAIX, which is related to tumor hypoxia.
We classify GLCM inverse difference as an “avatar” radiomic feature as it is correlated with 7 other radiomic features (Figure 2, B). Higher GLCM inverse difference, which is associated with poor survival, was found in dense and uniform lesions (Supplementary Figure 4, available online). Radiogenomics and IHC analyses revealed that the avatar feature was positively associated with CAIX expression, which is an important pH regulatory enzyme upregulated in hypoxic tumors leading to an acidic tumor microenvironment (26). Hypoxia is associated with angiogenesis, tumor growth, invasiveness, metastases, therapy resistance by inducing cell quiescence and an immunosuppressive phenotype (27), and poor survival in cancer patients including NSCLC (28–31). Because our results demonstrate that GLCM inverse difference is associated with tumor hypoxia, this biomarker should have wider reaching implications beyond immunotherapy. Thus, it is not surprising that GLCM inverse difference was associated with OS in the prognostic datasets suggesting this avatar feature could have broad clinical relevance beyond immunotherapy. Importantly, GLCM inverse difference was not associated with routinely obtained clinical covariates (other than ECOG; P = .04) implicating an independent effect (Supplementary Table 9, available online).
The most informative clinical covariates demonstrate the utility of standard-of-care information to predict patient outcomes. A higher number of metastatic sites increase patient disease burden and may lead to mixed immunotherapy response, where some lesions are regressing and others are progressing, which ultimately leads to patient-level progressive disease. The other clinical covariate, serum albumin, has been shown to be associated with survival in NSCLC patients (32,33) and is used in existing cancer prognostic scores [ie, Royal Marsden Hospital prognostic score (34) and MD Anderson risk score (35)]. Low serum albumin is an indicator of malnutrition, inflammation, and hepatic dysfunction. The cut-point of 3.9 identified by CART was within the normal limits of serum albumin (lower limit = 3.5), which may suggest a potentially novel threshold for survival outcomes in immunotherapy.
Emerging evidence demonstrates the utility of radiomics as a noninvasive approach to predict lung cancer treatment response of tyrosine kinase inhibitors (36,37), platinum-based chemotherapy (36), neo-adjuvant chemoradiation (38,39), stereotactic body radiation therapy (40,41), and immunotherapy (8,42,43). We previously (8) demonstrated that pretreatment radiomic models predict rapid disease progression phenotypes, including hyperprogression (AUCs ranging 0.804-0.865) among 228 NSCLC patients treated with immunotherapy. Sun et al. (42) developed a radiomic signature that predicts CD8 cell infiltration and used the signature to predict immunotherapy outcomes among different cancers (AUC = 0.67); however, NSCLCs were only a small subset (22%) of the dataset. Utilizing 123 NSCLC patients treated with immunotherapy, Trebeschi et al. (43) developed a machine learning model that discriminates progressive disease from stable disease and responsive disease (AUC = 0.83). This current study represents one of the largest radiomics study of NSCLC patients treated with immunotherapy.
We do acknowledge some limitations of this study. TMB information was not available for any of the patients, and PD-L1 IHC was only available for 8 patients in MCC1 and 29 patients in MCC2. Thus, we are unable to verify the performance of TMB or PD-L1 expression related to outcomes. However, recent studies have shown that patients respond to immunotherapy regardless of PD-L1 expression (6,11), so inclusion of PD-L1 status may add little or no improvement to our models. Although MCC1 and MCC2 were curated from the same cancer center, the OS of the 2 cohorts were statistically significantly different (Supplementary Figure 1, available online). These differences in OS may be attributed to the MCC1 cohort comprised of clinical trial patients who typically have higher performance status, and the majority of patients in the MCC2 cohort were treated with standard-of-care immunotherapy. However, the hazard ratios were not appreciably modified in multivariable analysis adjusting for covariates that differed between the 2 cohorts. Lastly, clinical covariates were not available to validate the clinical-radiomic model for the prognostic datasets that were not treated with immunotherapy. Despite these modest limitations, this study yields one of the highest radiomic quality scores of 22 (Supplementary Table 8, available online), which is a stringent metric that quantifies the quality of analysis and the potential clinical relevance of a radiomics study (25).
In conclusion, using standard-of-care imaging and clinical covariates, we identified and validated a novel parsimonious risk model that is associated with survival outcomes among NSCLC patients treated with immunotherapy. The most informative radiomic feature was found to be positively associated with CAIX, an important enzyme upregulated in hypoxic and acidotic tumors, which is related to treatment resistance. The potential clinical application of this work is that baseline radiomics and clinical covariates can identify patients who are unlikely to respond to immunotherapy and eliminate unnecessary treatments. Confirmation is required on the biological tumor-hypoxia underpinnings of GLCM inverse difference.
Funding
This work was supported by the National Cancer Institute (NCI) (U01-CA143062 to Drs Gillies and Schabath), the NCI Early Detection Research Network (U01-CA200464 to Drs Gillies and Schabath), and CA186145 and CA196405. Additional support was provided by the Cancer Center Support Grant (CCSG) at the H. Lee Moffitt Cancer Center and Research Institute, an NCI-designated Comprehensive Cancer Center [grant number P30-CA76292]. Also, this material is the result of work supported with resources and the use of facilities at the Moffitt Cancer Center and the James A. Haley Veterans’ Hospital.
Footnotes
Role of the funders: The funders had no role in the design of the study; collection, management, analysis, and interpretation of the data; writing, review, or approval of the manuscript; and the decision to submit for publication.
Disclosures: JE Gray reports receiving commercial research grants from AstraZeneca, Merck, Array, Epic Sciences, Genentech, Bristol-Myers Squibb, BI, Trovagene, and Novartis and is a consultant/advisory board member for AstraZeneca, Janssen, Genentech, Eli Lilly, Celgene, and Takeda, and other remuneration from Genentech, AstraZeneca, Merck, and Lilly/Genenech. RJ Gillies is an investor and member of the Advisory Board at HealthMyne, Inc, and has Research support form Helix Biopharma. No potential conflicts of interest were disclosed by the other authors.
Author contributions: Conceptualization: IT, JEG, RJG, MBS. Data curation: IT, YT, EK, SAE, JS, HJWLA, TB, JQ, JEG, RJG, MBS. Formal analysis: IT, SAE, TB, MBS. Funding acquisition: RJG, MBS. Investigation: IT, JEG, RJG, MBS. Methodology: IT, JEG, RJG, MBS. Project administration: JEG, RJG, MBS. Supervision: JEG, AG, RJG, MBS. Writing-original draft: IT, JEG, EK, AG, RJG, MBS. Writing-review & editing: IT, JEG, AG, RJG, MBS.
Acknowledgements: We would like to thank to the Quantitative Imaging Shared Resources Core for their assistance in implementing the image biomarker standardisation initiative (IBSI) radiomic feature set. The authors dedicate this manuscript to Dr Pierre Pascal Massion (March 28, 1963, to April 4, 2021).
Disclaimer: Contents of this research do not represent the views of the Department of Veterans Affairs or the US government.
Data Availability
The data and data analysis code will be available upon reasonable request from the corresponding author (MBS). The MATLAB scripts to create peritumoral masks derived from intratumoral masks are publicly available (https://github.com/TunaliIlke/peritumoral_regions/). The radiomic features were extracted using algorithms from the IBSI (22). The algorithms were written in C++ and MATLAB v2015a, and these codes are available upon request from the corresponding author (MBS).
Supplementary Material
Contributor Information
Ilke Tunali, Department of Cancer Physiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA; Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey.
Yan Tan, Department of Radiology, First Hospital of Shanxi Medical University, Taiyuan, Shanxi Province, China.
Jhanelle E Gray, Department of Thoracic Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA.
Evangelia Katsoulakis, Department of Radiation Oncology, James A. Haley Veterans’ Hospital, Tampa, FL, USA.
Steven A Eschrich, Department of Biostatistics and Bioinformatics, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA.
James Saller, Department of Molecular Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA.
Hugo J W L Aerts, Department of Radiation Oncology, Brigham and Women’s Hospital, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA.
Theresa Boyle, Department of Molecular Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA.
Jin Qi, Department of Cancer Physiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA.
Albert Guvenis, Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey.
Robert J Gillies, Department of Cancer Physiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA.
Matthew B Schabath, Department of Thoracic Oncology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA; Department of Cancer Epidemiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA.
References
- 1.Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. [DOI] [PubMed] [Google Scholar]
- 2.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387(10027):1540–1550. [DOI] [PubMed] [Google Scholar]
- 5.Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389(10066):255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi L, Rodriguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. [DOI] [PubMed] [Google Scholar]
- 7.Borcoman E, Nandikolla A, Long G, et al. Patterns of response and progression to immunotherapy. Am Soc Clin Oncol Educ Book. 2018;38:169–178. [DOI] [PubMed] [Google Scholar]
- 8.Tunali I, Tan Y, Gray JE, et al. Clinical-radiomic models predict overall survival among non-small cell lung cancer patients treated with immunotherapy. J Thoracic Oncol. 2019;14(11):S1129. [Google Scholar]
- 9.Champiat S, Dercle L, Ammari S, et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clin Cancer Res. 2017;23(8):1920–1928. [DOI] [PubMed] [Google Scholar]
- 10.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonia SJ, Villegas A, Daniel D, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med. 2017;377(20):1919–1929. [DOI] [PubMed] [Google Scholar]
- 12.Yarchoan M, Hopkins A, Jaffee EM.. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843–852.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411):eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawkins S, Wang H, Liu Y, et al. Predicting malignant nodules from screening CT scans. J Thorac Oncol. 2016;11(12):2120–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35(24):3209–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizzardi AE, Johnson AT, Vogel RI, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Wang H, Li Q, et al. Radiologic features of small pulmonary nodules and lung cancer risk in the national lung screening trial: a nested case-control study. Radiology. 2018;286(1):298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove O, Berglund AE, Schabath MB, et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS One. 2015;10(3):e0118261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tunali I, Stringfield O, Guvenis A, et al. Radial gradient and radial deviation radiomic features from pre-surgical CT scans are associated with survival among lung adenocarcinoma patients. Oncotarget. 2017;8(56):96013–96026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwanenburg A, Vallieres M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tunali I, Hall LO, Napel S, et al. Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med Phys. 2019;46(11):5075–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breiman L.Classification and Regression Trees. New York, NY: Kluwer Academic Publishers; 1984. [Google Scholar]
- 25.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 26.Innocenti A, Pastorekova S, Pastorek J, et al. The proteoglycan region of the tumor-associated carbonic anhydrase isoform IX acts as an intrinsic buffer optimizing CO2 hydration at acidic pH values characteristic of solid tumors. Bioorg Med Chem Lett. 2009;19(20):5825–5828. [DOI] [PubMed] [Google Scholar]
- 27.Muz B, de la Puente P, Azab F, et al. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl). 2015;3(9):83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris AL.Hypoxia–a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. [DOI] [PubMed] [Google Scholar]
- 29.Chan DA, Giaccia AJ.. Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 2007;26(2):333–339. [DOI] [PubMed] [Google Scholar]
- 30.Ilie M, Hofman V, Zangari J, et al. Response of CAIX and CAXII to in vitro re-oxygenation and clinical significance of the combined expression in NSCLC patients. Lung Cancer. 2013;82(1):16–23. [DOI] [PubMed] [Google Scholar]
- 31.Pastorek J, Pastorekova S.. Hypoxia-induced carbonic anhydrase IX as a target for cancer therapy: from biology to clinical use. Semin Cancer Biol. 2015;31:52–64. [DOI] [PubMed] [Google Scholar]
- 32.Espinosa E, Feliu J, Zamora P, et al. Serum albumin and other prognostic factors related to response and survival in patients with advanced non-small cell lung cancer. Lung Cancer. 1995;12(1-2):67–76. [DOI] [PubMed] [Google Scholar]
- 33.Miura K, Hamanaka K, Koizumi T, et al. Clinical significance of preoperative serum albumin level for prognosis in surgically resected patients with non-small cell lung cancer: comparative study of normal lung, emphysema, and pulmonary fibrosis. Lung Cancer. 2017;111:88–95. [DOI] [PubMed] [Google Scholar]
- 34.Garrido-Laguna I, Janku F, Vaklavas C, et al. Validation of the Royal Marsden Hospital prognostic score in patients treated in the phase I clinical trials program at the MD Anderson Cancer Center. Cancer. 2012;118(5):1422–1428. [DOI] [PubMed] [Google Scholar]
- 35.Wheler J, Tsimberidou AM, Hong D, et al. Survival of 1,181 patients in a phase I clinic: the MD Anderson Clinical Center for targeted therapy experience. Clin Cancer Res. 2012;18(10):2922–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khorrami M, Khunger M, Zagouras A, et al. Combination of peri- and intratumoral radiomic features on baseline CT scans predicts response to chemotherapy in lung adenocarcinoma. Radiol Artif Intell. 2019;1(2):180012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aerts HJWL, Grossmann P, Tan YQ, et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC (vol 6, 33860, 2016). Sci Rep. 2017;6(1):33860–33865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bibault JE, Giraud P, Housset M, et al. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer (vol 8, 12611, 2018). Sci Rep. 2018;8(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coroller TP, Agrawal V, Huynh E, et al. Radiomic-based pathological response prediction from primary tumors and lymph nodes in NSCLC. J Thorac Oncol. 2017;12(3):467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huynh E, Coroller TP, Narayan V, et al. Associations of radiomic data extracted from static and respiratory-gated CT scans with disease recurrence in lung cancer patients treated with SBRT. PLoS One. 2017;12(1):e0169172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattonen SA, Palma DA, Johnson C, et al. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol Biol Phys. 2016;94(5):1121–1128. [DOI] [PubMed] [Google Scholar]
- 42.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180–1191. [DOI] [PubMed] [Google Scholar]
- 43.Trebeschi S, Drago SG, Birkbak NJ, et al. Predicting response to cancer immunotherapy using non-invasive radiomic biomarkers. Ann Oncol. 2019;30(6):998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and data analysis code will be available upon reasonable request from the corresponding author (MBS). The MATLAB scripts to create peritumoral masks derived from intratumoral masks are publicly available (https://github.com/TunaliIlke/peritumoral_regions/). The radiomic features were extracted using algorithms from the IBSI (22). The algorithms were written in C++ and MATLAB v2015a, and these codes are available upon request from the corresponding author (MBS).