Significance
Biosynthesis of sterols requires oxygen. This study identifies a previously unknown evolutionary adaptation in a eukaryote, which enables anaerobic growth in absence of exogenous sterols. A squalene–hopene cyclase, proposed to have been acquired by horizontal gene transfer from an acetic acid bacterium, is implicated in a unique ability of the yeast Schizosaccharomyces japonicus to synthesize hopanoids and grow in anaerobic, sterol-free media. Expression of this cyclase in Saccharomyces cerevisiae confirmed that at least one of its hopanoid products acts as sterol surrogate. These observations provide leads for research into the structure and function of eukaryotic membranes and into the development of sterol-independent yeast cell factories for application in anaerobic processes.
Keywords: sterols, yeast, anaerobic, hopanoids, Schizosaccharomyces
Abstract
Biosynthesis of sterols, which are key constituents of canonical eukaryotic membranes, requires molecular oxygen. Anaerobic protists and deep-branching anaerobic fungi are the only eukaryotes in which a mechanism for sterol-independent growth has been elucidated. In these organisms, tetrahymanol, formed through oxygen-independent cyclization of squalene by a squalene–tetrahymanol cyclase, acts as a sterol surrogate. This study confirms an early report [C. J. E. A. Bulder, Antonie Van Leeuwenhoek, 37, 353–358 (1971)] that Schizosaccharomyces japonicus is exceptional among yeasts in growing anaerobically on synthetic media lacking sterols and unsaturated fatty acids. Mass spectrometry of lipid fractions of anaerobically grown Sch. japonicus showed the presence of hopanoids, a class of cyclic triterpenoids not previously detected in yeasts, including hop-22(29)-ene, hop-17(21)-ene, hop-21(22)-ene, and hopan-22-ol. A putative gene in Sch. japonicus showed high similarity to bacterial squalene–hopene cyclase (SHC) genes and in particular to those of Acetobacter species. No orthologs of the putative Sch. japonicus SHC were found in other yeast species. Expression of the Sch. japonicus SHC gene (Sjshc1) in Saccharomyces cerevisiae enabled hopanoid synthesis and stimulated anaerobic growth in sterol-free media, thus indicating that one or more of the hopanoids produced by SjShc1 could at least partially replace sterols. Use of hopanoids as sterol surrogates represents a previously unknown adaptation of eukaryotic cells to anaerobic growth. The fast anaerobic growth of Sch. japonicus in sterol-free media is an interesting trait for developing robust fungal cell factories for application in anaerobic industrial processes.
Sterols are key constituents of canonical eukaryotic membranes, in which they influence integrity, permeability, and fluidity (1, 2). The core pathway for sterol biosynthesis is highly conserved, but the predominant final products differ for animals (cholesterol), plants (phytosterols), and fungi (ergosterol) (3). Multiple reactions in sterol biosynthesis require molecular oxygen, and no evidence for anaerobic sterol pathways has been found in living organisms or in the geological record (3). The first oxygen-dependent conversion in sterol synthesis is the epoxidation of squalene to oxidosqualene by squalene monooxygenase. Oxidosqualene is subsequently cyclized to lanosterol, the first tetracyclic intermediate in sterol biosynthesis, in an oxygen-independent conversion catalyzed by oxidosqualene cyclase (OSC; SI Appendix, Fig. S1). Molecular oxygen is also required for multiple subsequent demethylation and desaturation steps (4). In fungi, synthesis of a single molecule of ergosterol from squalene requires 12 molecules of oxygen.
Deep-branching fungi belonging to the Neocallimastigomycota phylum are the only eukaryotes that have been unequivocally demonstrated to naturally exhibit sterol-independent growth under strictly anaerobic conditions. These anaerobic fungi contain a squalene–tetrahymanol cyclase (STC; SI Appendix, Fig. S1), which catalyzes oxygen-independent cyclization of squalene to tetrahymanol (5, 6). This pentacyclic triterpenoid acts as a sterol surrogate, and acquisition of a bacterial STC gene by horizontal gene transfer is considered a key evolutionary adaptation of Neocalllimastigomycetes to the strictly anaerobic conditions of the gut of large herbivores (7). The reaction catalyzed by STC resembles oxygen-independent conversion of squalene to hopanol and/or other hopanoids by squalene–hopene cyclases (8) (SHC; SI Appendix, Fig. S1), which are found in many bacteria (9, 10). Some bacteria synthesize tetrahymanol by ring expansion of hopanol, in a reaction catalyzed by tetrahymanol synthase (THS) for which the precise mechanism has not yet been resolved (11).
Already in the 1950s, anaerobic growth of the industrial yeast and model eukaryote Saccharomyces cerevisiae was shown to strictly depend on sterol supplementation (12) or use of intracellular stores of this anaerobic growth factor (13). Similarly, fast anaerobic growth of S. cerevisiae, which is a key factor in its large-scale application in bioethanol production, wine fermentation, and brewing (14, 15), requires availability of unsaturated fatty acids (UFAs). Biosynthesis of UFAs by yeasts requires an oxygen-dependent acyl-CoA desaturase (16), and in anaerobic laboratory studies, the sorbitan oleate ester Tween 80 is commonly used as UFA supplement (13, 17).
Per gram of yeast biomass, ergosterol and UFA synthesis require only small amounts of oxygen. Studies on these oxygen requirements therefore require extensive measures to prevent unintended oxygen entry into cultures. Even though most yeast species readily ferment sugars to ethanol under oxygen-limited conditions, only very few grow anaerobically on sterol- and UFA-supplemented media when such precautions are taken (18, 19). As opposed to Neocallimastigomycetes, no evolutionary adaptations to sterol-independent anaerobic growth have hitherto been reported for yeasts or for ascomycete and basidiomycete fungi in general.
We recently demonstrated that expression of an STC gene from the ciliate Tetrahymena thermophila–supported tetrahymanol synthesis and sterol-independent growth of S. cerevisiae (20). This result inspired us to re-examine a 1971 publication in which Bulder (21) reported sterol- and UFA-independent growth of the dimorphic fission yeast Schizosaccharomyces japonicus. Sch. japonicus was originally isolated from fermented fruit juices (22, 23), and its potential for wine fermentation is being explored (24, 25). It shows marked genetic and physiological differences with other fission yeasts (26, 27) and has gained interest as a model for studying cell division dynamics and hyphal growth (28–30). Sch. japonicus grows well at elevated temperatures and rapidly ferments glucose to ethanol (31). A low sterol content, control of membrane fluidity via chain length of saturated fatty acids (SFAs), and respiratory deficiency may all reflect adaptations of Sch. japonicus to low-oxygen environments (21, 31–33). However, the report by Bulder (21) stating that Sch. japonicus can grow anaerobically without sterol supplementation has not been confirmed or further investigated.
S. cerevisiae is able to synthesize sterols at nanomolar concentrations of oxygen (34). This high affinity for oxygen complicates experimental analysis of oxygen requirements for sterol synthesis in yeast cultures (20, 35, 36) and warranted a reassessment of the sterol requirements of Sch. japonicus. If confirmed, an ability of Sch. japonicus to grow in the absence of an exogenous supply of sterols would raise urgent questions on the molecular and evolutionary basis for this trait, which is extremely rare among eukaryotes. Based on theoretical grounds, it has been proposed that oxygen-independent sterol synthesis may be possible (37). Alternatively, sterol-independent growth of Sch. japonicus might depend on synthesis of an as yet unidentified sterol surrogate or on membrane adaptations that involve neither sterols nor sterol surrogates. In addition to these fundamental scientific questions, independence of anaerobic growth factors is a relevant trait for large-scale industrial applications of yeasts, as exemplified by “stuck” brewing fermentations caused by depletion of intracellular sterols and/or UFA reserves (38, 39).
The goals of the present study were to reinvestigate the reported ability of Sch. japonicus to grow anaerobically without sterol supplementation and to elucidate its molecular basis. In view of reported challenges in avoiding oxygen contamination in laboratory cultures of yeasts (19, 20, 35, 36), we first reassessed anaerobic growth and lipid composition of Sch. japonicus in the presence and absence of ergosterol. After identifying a candidate SHC gene in Sch. japonicus, we investigated its role in anaerobic growth by its expression in S. cerevisiae. In addition, we tested the hypothesis of Bulder (40) that Sch. japonicus is able to synthesize UFAs in an oxygen-independent pathway.
Results
Anaerobic Growth of Sch. japonicus without Ergosterol and UFA Supplementation.
To reassess conclusions from an early literature report on sterol- and UFA-independent anaerobic growth of Sch. japonicus (21), we performed serial transfer experiments in an anaerobic chamber (36, 41) using phosphate-buffered synthetic medium with glucose as carbon source (SMPD), with and without supplementation of ergosterol and/or Tween 80 as source of UFAs. Parallel experiments with the S. cerevisiae laboratory strain CEN.PK113-7D were included to assess low-level contamination with oxygen, as reflected by residual slow growth in sterol-free media (41).
To deplete reserves of sterols and/or UFAs in aerobically pregrown cells, anaerobic precultures were grown on SMPD with an increased glucose concentration (50 g ⋅ L−1) and lacking ergosterol and Tween 80. In these precultures, growth of S. cerevisiae CEN.PK113-7D ceased after 4.8 doublings (Fig. 1A), when less than one-half of the glucose had been consumed (SI Appendix, Table S1). Under the same conditions, Sch. japonicus CBS5679 completed 6.1 doublings (Fig. 1B), and while full glucose depletion was intentionally avoided to prevent excessive flocculation and sporulation, it had consumed almost 90% of the glucose (SI Appendix, Table S1). Samples from the anaerobic precultures were transferred to SMPD (20 g ⋅ L−1 glucose) supplemented with different combinations of ergosterol and/or Tween 80. Consistent with earlier reports (36), S. cerevisiae showed extremely slow, nonexponential growth on SMPD without ergosterol (Fig. 1A), which indicated a minor entry of oxygen into the anaerobic chamber. In contrast, Sch. japonicus showed maximum specific growth rates of 0.26 to 0.30 h−1 and reached optical densities of 4 to 5 in all media tested (Fig. 1B and SI Appendix, Table S2). This anaerobic growth was sustained upon two consecutive transfers in SMPD lacking either Tween 80, ergosterol, or both (Fig. 1B). These results confirmed Bulder’s (21) conclusion that Sch. japonicus can grow anaerobically without sterol and UFA supplementation. Remarkably, Sch. japonicus grew slower in aerobic cultures (0.19 h−1; SI Appendix, Fig. S2) than in anaerobic cultures grown on the same medium (0.24 to 0.26 h−1; Fig. 1B and SI Appendix, Table S2).
Fig. 1.
Anaerobic growth of S. cerevisiae CEN.PK113-7D and Sch. japonicus CBS5679 with different ergosterol and UFA (Tween 80) supplementation in a dark anaerobic chamber. Anaerobic precultures on SMPD on 50 g ⋅ L−1 glucose (closed circles, gray shading) were grown until the end of the exponential phase. (A) After the anaerobic preculture, S. cerevisiae was transferred to SMPD (20 g ⋅ L−1 glucose) supplemented with either Tween 80 and ergosterol (open circles), Tween 80 only (closed squares), or neither Tween 80 nor ergosterol (open squares). (B) Sch. japonicus was grown on the same media as S. cerevisiae and additionally on medium containing ergosterol but not Tween 80 (closed triangles). Sch. japonicus cultures supplemented with only Tween 80, only ergosterol, and those without supplements were serially transferred to fresh media with the same composition in the anaerobic chamber. Data are represented as average ± SEM of measurements on independent duplicate cultures for each combination of yeast strain and medium composition.
Absence of Ergosterol and UFAs in Anaerobically Grown Sch. japonicus.
To further investigate anaerobic growth of Sch. japonicus in sterol- and UFA-free media, lipid fractions were isolated from anaerobic cultures and analyzed by gas chromatography with flame ionization detection (GC-FID). UFAs were detected in aerobically grown biomass but not in anaerobic cultures grown on SMPD without Tween 80 supplementation (Fig. 2 and Dataset S1). These results showed that fast anaerobic growth of Sch. japonicus on UFA-free medium did not, as suggested by Bulder (40), reflect oxygen-independent UFA synthesis. Total fatty acid contents of aerobically and anaerobically grown biomass were similar, but anaerobically grown biomass showed higher contents of FA 10:0, FA 16:0, and FA 18:0 and lower contents of FA 26:0. In aerobically grown Sch. japonicus biomass, no FA 16:1 was detected, and levels of FA 18:1 were higher than in Tween 80–supplemented anaerobic cultures (Fig. 2).
Fig. 2.
Quantification of fatty acids in Sch. japonicus CBS5679 biomass. Sch. japonicus CBS5679 was grown in SMPD with 20 g ⋅ L−1 glucose. Under anaerobic conditions, cultures were supplemented with Tween 80 and ergosterol (TE), only ergosterol (E), only Tween 80 (T), or neither of those supplements (−/−). Data are shown for the first anaerobic culture following the anaerobic preculture. Aerobic cultures of Sch. japonicus were grown in SMPD without supplements (−/−). Data are represented as average ± SEM of measurements on independent duplicate cultures for each cultivation condition. Detailed information on data presented in this figure and additional anaerobic transfers are provided in Dataset S1.
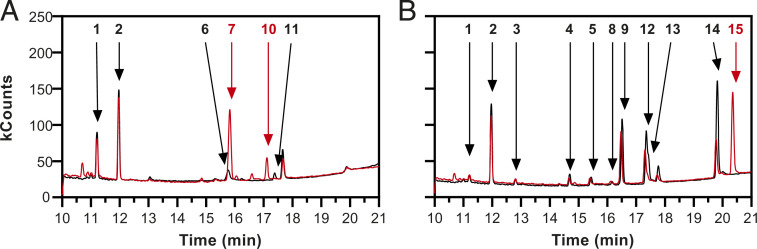
S. cerevisiae biomass, grown anaerobically on SMPD with Tween 80 and ergosterol, was used as a reference for analysis of triterpenoid compounds. In addition to squalene and ergosterol, lipid samples contained small amounts of lanosterol (Fig. 3A). Lipid samples from extremely slow-growing S. cerevisiae cultures on SMPD supplemented with only Tween 80 contained squalene and a small amount of lanosterol but not ergosterol (Dataset S1). Presence of lanosterol was attributed to de novo synthesis, enabled by a minor entry of oxygen into the anaerobic chamber.
Fig. 3.
GC-MS analysis of triterpenoid fractions of anaerobically grown yeast biomass. Anaerobic cultures were harvested in early stationary phase. Triterpenoids were extracted for GC-MS analysis and injected immediately (black lines) or after silylation (red lines). (A) S. cerevisiae CEN.PK113-7D grown anaerobically on medium supplemented with Tween 80 and ergosterol, and (B) Sch. japonicus CBS5679 was grown on medium supplemented with only Tween 80. Numbers indicate the following compounds: 1, squalene; 2, 5α-cholestane (internal standard); 3, squalene epoxide; 4, hop-17(22)-ene; 5, 22-Hydroxy-21αH-hopane; 6, ergosterol; 7, ergosterol-TMS-ether; 8, adian-5-ene; 9, fern-7-ene; 10, lanosterol-TMS-ether; 11, lanosterol; 12, hop-22(29)-ene (diploptene); 13, hop-21(22)-ene; 14, hopan-22-ol (diplopterol); and 15, hopan-22-ol-TMS-ether.
Similarly prepared triterpenoid fractions of anaerobic Sch. japonicus cultures that were supplemented with only Tween 80 did not contain detectable amounts of ergosterol or lanosterol. Instead, in addition to squalene, gas chromatography–mass spectrometry (GC-MS) revealed several compounds that were not observed in anaerobically grown S. cerevisiae (Fig. 3B). A prominent appearance of the ions m/z 367 and 395 in mass spectra of the detected compounds was consistent with a five-membered ring system with loss of either a methyl or a propyl group (42). We therefore hypothesized that these compounds had a pentacyclic structure. Tetrahymanol, which has not been found in wild-type yeasts but does occur in several other eukaryotes (6, 43, 44), did not match any of the detected peaks based on its relative retention time (RRT, with cholestane as ref. 20).
Sch. japonicus Synthesizes Hopanoids.
Hopanoids are triterpenoids with a pentacyclic backbone that occur in bacteria, plants, and fungi (9) but have not previously been found in yeasts. GC-MS analysis of biomass samples yielded eight distinct analytes that were detected in Sch. japonicus (Fig. 3B) but not in S. cerevisiae CEN.PK113-7D (Fig. 3A). Squalene epoxide (compound 3) was identified based on RRT and spectral matching with authentic standard material as previously described (45). Presence of squalene epoxide but not sterols was attributed to inadvertent entry of small amounts of oxygen into the anaerobic chamber. Hop-22(29)-ene (diploptene, compound 12) was identified based on synthetic reference material (Dataset S2). To investigate whether any of the remaining six unidentified components in Sch. japonicus (Fig. 3B) were also hopanoids, their mass spectra (Dataset S2) were compared with literature data (11, 42, 46, 47) (SI Appendix, Table S3 and Dataset S2). A fragment ion with a mass–charge ratio (m/z) of 191, which frequently occurs as base peak in mass spectra of hopanoids (42, 46), was detected for compounds 4, 5, 13, and 14 (Dataset S2). Based on comparison with published data, we tentatively identified compounds 4, 13, and 14 as hop-17(21)-ene, hop-21(22)-ene, and hopan-22-ol (diplopterol), respectively (11, 42, 46). Mass and retention-time shifts caused by silylation (48) were investigated (Fig. 3) and confirmed presence of the hydroxy group of diplopterol (Fig. 3B; 14 and 15) in the Sch. japonicus biomass, as well as those of the sterols (Fig. 3A; 6 and 7 and 10 and 11) in the S. cerevisiae samples. A small peak at the retention time of unsilylated diplopterol was tentatively attributed to steric hindrance by the tertiary-alcohol context of its hydroxy group. In the chromatograms representing silylated triterpenoids of S. cerevisiae, small additional peaks at retention times of 16.0 and 16.6 min were attributed to ergosta-5,7,22-trien-3β-ol (also detected in the commercial ergosterol preparation used for supplementation of anaerobic growth media; SI Appendix, Fig. S3) and fecosterol, an intermediate in ergosterol biosynthesis (49), respectively (Fig. 3A).
While the base peak of compound 5 was at m/z 367, the spectrum showed the same characteristic peak of m/z 191 and a molecular ion of m/z 428. Although this observation points toward a hydroxylated structure analogous to diplopterol, compound 5 was not readily silylated. A tentative identification of compound 5 as hydroxy-21αH-hopane is consistent with its almost identical mass spectrum relative to that of diplopterol and its different retention time (42). Moreover, its lower signal intensity and different stereochemistry at position 21 could explain its higher resistance to silylation. For compounds 8 and 9, the ion at m/z 410 suggests an unsubstituted triterpene which is unaffected by silylation, with retention times similar to those of other identified hopenes (Fig. 3B and Dataset S2). The corresponding base peaks of m/z 259 and 243 were previously reported for polycyclic triterpenoids with a different backbone configuration than those of substances 4, 12, and 13 (42). Although compounds 8 and 9 could not be identified with a high degree of confidence, they may be adian-5-ene and fern-7-ene, respectively. These two pentacyclic triterpenoids only differ from other hopanoids by their stereochemistry and by the position of methyl groups.
The identified compounds were quantified by GC-FID analysis in biomass samples of anaerobic Sch. japonicus cultures supplemented with different combinations of Tween 80 and ergosterol. Presence of ergosterol in biomass from ergosterol-supplemented anaerobic cultures indicated that Sch. japonicus is able to import sterols (Fig. 4A). In anaerobic cultures, the content of diplopterol, the major hopanoid detected in Sch. japonicus, was similar to the ergosterol content of sterol-supplemented anaerobic cultures (close to a 1:1 molar ratio; Fig. 4A and Dataset S1).
Fig. 4.
Quantification of triterpenoids in yeast biomass. (A) Triterpenoid content of cultures of Sch. japonicus CBS5679 grown in SMPD with 20 g ⋅ L−1 glucose. Under anaerobic conditions, cultures were supplemented with Tween 80 and ergosterol (TE), only ergosterol (E), only Tween 80 (T), or neither of these supplements (−/−). Data are shown for the first anaerobic culture following the anaerobic preculture. Aerobic cultures of Sch. japonicus were grown in SMPD without supplements (−/−). (B) Triterpenoid composition of anaerobic cultures of S. cerevisiae IMX2616 (sga1∆::Sjshc1; Left) and IMX2629 (sga1∆::Sjshc1 X-2::Maths; Right) grown in SMPD with 20 g ⋅ L−1 glucose and Tween 80 (T) supplementation. Data are represented as average ± SEM of data from two independent duplicate cultures for each cultivation condition. Detailed information on data presented in this figure and additional anaerobic transfers are provided in Dataset S1.
Except for the presence and absence of ergosterol, the triterpenoid composition of anaerobically grown biomass was not markedly affected by the supplementation of ergosterol and/or Tween 80. To investigate whether hopanoid synthesis in Sch. japonicus is affected by oxygen availability, additional analyses were performed on aerobically grown cultures. These analyses confirmed the ability of Sch. japonicus strain CBS5679 to synthesize ergosterol (Fig. 4A). Aerobically grown biomass showed a 3.5-fold higher squalene content than biomass grown in anaerobic cultures without Tween 80 and ergosterol, while its hopanoid content was fourfold lower (Fig. 4A and Dataset S1). These observations suggest that oxygen availability may regulate triterpenoid synthesis in Sch. japonicus.
Predicted Sch. japonicus Proteins Resemble Bacterial SHCs.
In bacteria, plants, and fungi, hopanoid synthesis occurs by cyclization of squalene by SHCs (47). To explore whether the Sch. japonicus genome encodes an SHC, amino acid sequences of characterized representatives of three related classes of triterpene cyclases were used as queries to search the predicted proteomes of Sch. japonicus strains yFS275 (26) and CBS5679. Specifically, sequences of an OSC from Schizosaccharomyces pombe (50) (SpErg7; Universal Protein Resource [UniProt] accession No. Q10231), an SHC from Acidocaldarius alicyclobacillus (51) (AaShc; P33247), and an STC from T. thermophila (5, 20) (TtThc1; Q24FB1), were used for a homology search based on Hidden Markov Models using HMMER (52). For each Sch. japonicus strain, this search yielded a sequence with significant homology to OSC and another with significant homology to SHC (Table 1). Consistent with the lipid analysis results, neither strain yielded a clear STC homolog.
Table 1.
Homology search results using amino acid sequences of characterized triterpenoid cyclases against Sch. japonicus proteomes
| Query | Accession of subject sequence | Query coverage (%) | E-value | Identity (%) |
| Subject proteome: Sch. japonicus yFS275 | ||||
| SpErg7* (Q10231) | B6JW54 | 99.7 | 0.0 | 65.9 |
| AaShc† (P33247) | B6K412 | 99.2 | 2.7 × 10−141 | 38.1 |
| Subject proteome: Sch. japonicus CBS5679 | ||||
| SpErg7* (Q10231) | SCHJC_A005630 (SjErg7) | 99.7 | 0.0 | 65.7 |
| AaShc† (P33247) | SCHJC_C003990 (SjShc1) | 98.9 | 2.5 × 10−139 | 37.8 |
To explore the phylogeny of the Sch. japonicus OSC and SHC homologs, SpErg7, AaSHC, and TtThc1 were used as queries for HMMER searches against all eukaryotic and all bacterial protein sequences in UniProt reference proteomes. A set of selected cyclase homologs (Dataset S3; see SI Appendix, Table S4 for detailed information) were subjected to multiple sequence alignment and used to generate a maximum-likelihood phylogenetic tree (Fig. 5 and Dataset S4). This phylogenetic analysis showed that the putative Sch. japonicus SHC sequences (SCHJC_C003990 from CBS5679, and B6K412 from yFS275) are related to bacterial SHCs, with sequences from Acetobacter spp. (A0A0D6N754 from Acetobacter indonesiensis 5H-1, and A0A0D6NG57 from Acetobacter orientalis 21F-2) as closest relatives (Fig. 5). To check if this conclusion was biased by the selection of sequences from species of interest, the putative SHC sequence SCHJC_C003990 from Sch. japonicus CBS5679 was used as query for a second HMMER search of either the eukaryotic or the bacterial databases described above. The resulting E-values distribution (Fig. 5A and Dataset S5) showed a strong overrepresentation of low E-values among bacterial sequences. Sequences of two Acetobacter species, A0A0D6NG57 from A. orientalis 21F-2 and A0A0D6N754 from A. indonesiensis 5H-1, showed 67.9 and 66.9% sequence identity, respectively, and yielded zero E-values in this search. In contrast, E-value distributions obtained with the putative OSC sequence SCHJC_A005630 from Sch. japonicus CBS5679 as query showed an overrepresentation of low E-values among eukaryotic sequences (Fig. 5B). Horizontal gene transfer from Acetobacter species which, like other members of the order Rhodospirillales, are well known to synthesize hopanoids (10, 53), is therefore a highly probable origin of SHC in Sch. japonicus. No SHC homologs were found in the predicted proteomes of Schizosaccharomycetes other than Sch. japonicus nor in those of 371 Saccharomycotina yeast species included in the eukaryotic UniProt database.
Fig. 5.
Maximum-likelihood phylogenetic tree of selected triterpenoid cyclases. The colored bar indicates different types of cyclases: green, SHCs; red, STCs; and purple, OSCs. Sequences were obtained from a systematic homology search using the characterized cyclases marked with an asterisk (A. alicyclobacillus AaShc, P33247, SHC; T. thermophila, TtThc1, Q24FB1, STC; Schizosaccharomyces pombe SpErg7, Q10231, OSC) as queries. Eukaryotic and bacterial sequences are indicated by black and gray bars, respectively. Clades were collapsed when having three or more members from the same taxonomic division (third level below Bacteria or Eukaryota according to NCBI taxonomy) with the exception of Neocallimastogomycetes and Schizosaccharomycetes. A total of 100 bootstrap replicates were performed, values above 70 are shown on the corresponding branches. All sequences and the final tree are provided in Datasets S3 and S4, respectively. The tree was midrooted, visualized, and made available in iTOL (https://itol.embl.de/tree/19319025314539181623851833). (A) Distribution of HMMER E-values obtained with Sch. japonicus SHC (SCHJC_C003990) as query against a bacterial sequence database (gray bars) and a eukaryotic database (black bars). (B) Distribution of HMMER E-values obtained with Sch. japonicus OSC (SCHJC_A005630) as query against a bacterial sequence database (gray bars) and a eukaryotic database (black bars).
In bacteria, hopanoids can be methylated or decorated with other side chains by enzymes encoded by hpn genes (54). To explore whether homologs of bacterial hopanoid-modifying enzymes occur in Sch. japonicus, a tblastn (55) homology search was performed on the genome sequences of strains CBS5679 and yFS275 with amino acid sequences of HpnG, HpnH, HpnI, HpnJ, HpnK, HpnO, HpnP, and HpnR from relevant bacterial species, including A. orientalis (SI Appendix, Table S5), as queries. Of these queries, only HpnO yielded two significant hits (alignment length of greater than 75% and E-value of lower than 1 × 10−5). However, a tblastn search of the genome of S. cerevisiae CEN.PK113-7D, which does not synthesize hopanoids, with HpnO as also yielded two hits. These corresponded to L-ornithine aminotransferase (Car2) and acetylornithine aminotransferase (Arg8) and showed coverages of >99% and amino acid identities of 60 and 41%, respectively, with the two Sch. japonicus sequences. Homology searches therefore did not provide a clear indication for occurrence of known hopanoid-modifying enzymes in Sch. japonicus.
Expression of Sch. japonicus SHC Stimulates Anaerobic Growth of S. cerevisiae in the Absence of Sterol Supplementation.
To investigate if the putative SHC gene of Sch. japonicus CBS5679 (Sjshc1) was responsible for hopanoid synthesis, its coding sequence was codon optimized and expressed in the Cas9-expressing S. cerevisiae strain IMX2600. Growth and triterpenoid production of the resulting strain IMX2616 (sga1Δ::Sjshc1; SI Appendix, Table S7) was studied in anaerobic shake flask cultures. After an anaerobic preculture for depletion of cellular reserves of sterols and/or hopanoids, neither the reference strain S. cerevisiae CEN.PK113-7D nor the strain carrying the Sjshc1 expression cassette grew on SMPD without ergosterol and Tween 80 (Fig. 6). On SMPD with only Tween 80, S. cerevisiae CEN.PK113-7D reached an optical density of 0.7 after 33 h (Fig. 6A), at which point ∼70% of the initially present glucose remained unused (SI Appendix, Table S6). In contrast, S. cerevisiae IMX2616 (sga1Δ::Sjshc1) reached an optical density of 2.1 after the same time period (Fig. 6B), at which point 98% of the initially added glucose had been consumed (SI Appendix, Table S6), and showed sustained anaerobic growth upon transfer to a second flask containing the same medium (Fig. 6B). Upon termination of the experiments after 58 h, the optical density at 600 nm (OD600) of the S. cerevisiae CEN.PK113-7D cultures had increased to 1.1.
Fig. 6.
Anaerobic growth of S. cerevisiae strains in sterol-free media. S. cerevisiae cultures were inoculated from an anaerobic preculture on SMPD (50 g ⋅ L−1 glucose) to fresh SMPD (20 g ⋅ L−1 glucose), either supplemented with Tween 80 (closed circles), or lacking UFAs and sterols (open circles). (A) Reference strain CEN.PK113-7D. (B) S. cerevisiae strain IMX2616 (sga1∆::Sjshc1). (C) S. cerevisiae strain IMX2629 (sga1∆::Sjshc1 X-2::Maths). Cultures supplemented with Tween 80 represented in panels (B) and (C) were transferred to fresh medium of the same composition (closed circles) during exponential phase. Data are represented as average ± SEM of measurements on independent duplicate cultures for each yeast strain.
To investigate whether S. cerevisiae IMX2616 (sga1Δ::Sjshc1) produced the same hopanoid compounds as Sch. japonicus CBS5679, biomass was harvested from anaerobic shake flask cultures grown on SMDP with Tween 80. Analysis of the triterpenoid fraction by GC-MS and GC-FID showed the same hopanoids that were detected and identified in Sch. japonicus strain CBS5679 (SI Appendix, Fig. S4), albeit in smaller amounts, while squalene contents were higher in the S. cerevisiae cultures (Fig. 5B and Dataset S1). The only sterol identified in these samples was lanosterol (Fig. 5B). Presence of this first tetracyclic intermediate of ergosterol biosynthesis and very slow, nonexponential growth of the reference strain S. cerevisiae CEN.PK113-7D in sterol-free medium (Fig. 6) were attributed to a minor entry of oxygen into the anaerobic chamber (20).
In some bacteria, SHC is involved in a pathway for tetrahymanol production, in which a THS converts hopene into tetrahymanol (11). To investigate whether such a two-step pathway for tetrahymanol synthesis can be engineered in S. cerevisiae, a codon-optimized expression cassette for the Methylomicrobium alcaliphilum (11) gene encoding THS (locus tag MEALZ_1626; referred to as Maths) was integrated at the X-2 locus (56) in strain IMX2616 (sga1∆::Sjshc1), yielding strain IMX2629 (sga1Δ::Sjshc1 X-2::Maths; SI Appendix, Table S7). Anaerobic growth and sugar consumption rates of these two S. cerevisiae strains were similar (Fig. 6 B and C and SI Appendix, Table S6). Tetrahymanol was detected in anaerobically grown biomass of strain IMX2629 but not of strain IMX2616 (Fig. 4B and SI Appendix, Fig. S5). Together, these results confirm that Sjshc1 encodes a bona fide SHC, at least one of whose hopanoid products can act as sterol surrogate in anaerobic yeast cultures.
Discussion
The notion that sterols are indispensable components of all eukaryotic membranes was first dispelled by research on ciliates of the genus Tetrahymena, in which tetrahymanol acts as a sterol surrogate (43, 57). Subsequent research showed that fungi belonging to the phylum Neocallimastigomycota linked their ability to maintain membrane integrity in the absence of oxygen or exogenous sterols to tetrahymanol synthesis (6, 44). Acquisition of a bacterial STC-encoding DNA sequence has been proposed as a key event in their adaptation to an anaerobic lifestyle (7, 58). Inspired by a half-century-old, intriguing publication by Bulder (21) on the yeast Sch. japonicus, the present study uncovered hopanoid production as a similar but different eukaryotic adaptation to minimize or eliminate oxygen requirements for sterol biosynthesis. Because efficient procedures for genetic modification of Sch. japonicus CBS5679 are not yet available (59), the role of one or more hopanoids as sterol surrogates was confirmed by expression in a heterologous host. Expression of Sjshc1 in S. cerevisiae stimulated its anaerobic growth in the absence of ergosterol supplementation (Fig. 6) and illustrated how the mere acquisition of an SHC gene by horizontal gene transfer may have benefited an ancestor of Sch. japonicus in severely oxygen-limited or anaerobic environments.
SHC enzymes and hopanoid synthesis have been found in ferns (60, 61), and putative SHC proteins have been identified in several filamentous fungi (62, 63) (Fig. 5). Although Sch. japonicus is therefore not unique among eukaryotes in containing an SHC, hopanoid synthesis has not previously been found in yeasts or associated with sterol-independent anaerobic growth of eukaryotes. Putative SHC genes were neither found in other Schizosaccharomyces species nor in more distantly related yeasts. Confinement to a single yeast species and a strong sequence similarity with putative SHC sequences from Acetobacter species identifies horizontal gene transfer as a highly plausible evolutionary origin of Sjshc1 (62). Independent acquisition of SHC and STC genes by phylogenetically distant eukaryotes represents a remarkable case of convergent evolution toward an anaerobic eukaryotic lifestyle.
The hopanoids identified in anaerobically grown Sch. japonicus CBS5679 (Fig. 3 and SI Appendix, Table S3) were also detected in an Sjshc1-expressing S. cerevisiae strain (Fig. 5 and SI Appendix, Fig. S4). These results indicate that the observed product diversity originated from the SjShc1 enzyme itself, rather than from additional enzyme-catalyzed modifications. Formation of multiple products is consistent with reports on triterpenoid extracts of bacterial hopanoid producers and product spectra of purified bacterial SHCs (64–66). For example, in addition to diploptene and diplopterol, analysis of triterpenoids in Zymomonas mobilis biomass revealed a number of minor hopene variants whose synthesis was attributed to deviation from the regular cyclization process (67). Alternatively, minor structural variations could represent artifacts of lipid isolation and derivatization.
In bacteria, hopanoids produced by SHC are often modified to generate side-chain–extended hopanepolyols (54). Molecular dynamics simulation of lipid bilayers showed that sterols and bacteriohopanetetrol are correctly oriented in membranes due to their polar moieties, whereas unsubstituted diploptene molecules cannot be properly inserted and instead accumulate horizontally in the hydrophobic core (68). In the present study, diplopterol (hopan-22-ol), the major hopanoid detected in Sch. japonicus, is therefore the best candidate for a role as membrane-active triterpenoid due to its hydroxyl group. It was previously demonstrated that its potential to influence order and phase behavior of lipid bilayers resembles that of cholesterol (69). While the scan range of the GC-MS analyses (m/z 100 to 600) would have allowed for identification of methylated or double-hydroxylated hopanoids, our analyses do not exclude presence of more elaborately modified hopanoids in Sch. japonicus. Absence of clear homologs of bacterial genes involved in hopanoid modification (70, 71) from the Sch. japonicus genome and the high diplopterol content of the lipid fraction of anaerobically grown biomass may, however, argue against this possibility.
The ability, unique among yeasts, of Sch. japonicus to grow fast in anaerobic cultures without supplementation of sterols or UFAs is likely to reflect a specialized composition of its membranes. A recent comparison of the membrane properties of Sch. japonicus showed remarkable differences with those of the closely related yeast species Sch. pombe, including a higher membrane stiffness, denser lipid packing, and generally shorter and more saturated fatty acid residues (33). In our study, Sch. japonicus showed two- to threefold lower squalene contents during anaerobic growth in sterol-free media than in S. cerevisiae strains expressing Sjshc1 (Fig. 6) or a tetrahymanol cyclase gene from T. thermophila (20). High squalene contents have been implicated in suboptimal membrane function of S. cerevisiae (72) and may therefore at least be partially responsible for the slow anaerobic growth of these engineered S. cerevisiae strains (Fig. 5). Cultivation of Sch. japonicus in UFA-free medium led to a reduction of the average chain length of SFAs (Fig. 2). This change is, however, unlikely to directly affect effectiveness of hopanoids as sterol surrogates since UFA supplementation did not affect anaerobic growth rates of Sch. japonicus on sterol-free media (Fig. 1 and SI Appendix, Table S2).
Further research should resolve how hopanoids interact with other membrane components, including squalene, acyl lipids, and proteins, and thereby influence membrane functionality of Sch. japonicus. In bacteria, hopanoids have been implicated in tolerance to external stresses such as nonoptimal temperature and pH and the presence of antimicrobials (73–75). Elucidating how sterol surrogates interact with other membrane components, including proteins, and thereby influence membrane functionality can contribute to a deeper insight in microbial adaptation to anaerobic environments and to physicochemical stress factors. In addition, such studies will contribute to the design of membrane engineering strategies aimed at the construction of robust industrial strains of S. cerevisiae and other yeasts for application in anaerobic fermentation processes.
Materials and Methods
Strains, Media, and Maintenance.
Sch. japonicus CBS5679 was obtained from the Westerdijk Institute. S. cerevisiae strains used and constructed in this study belonged to the CEN.PK lineage (76, 77) and are listed in SI Appendix, Table S7. Yeast strains were stored at −80 °C as described previously (20). To avoid sexual coflocculation (78) of Sch. japonicus CBS5679, carbon source depletion in precultures was prevented and buffered synthetic media were used in all growth studies. Yeasts were grown on synthetic medium with ammonium as nitrogen source (79) with an increased concentration of KH2PO4 [14.4 g ⋅ L−1, SMP (80)]. Unless otherwise indicated, glucose was added from a concentrated stock solution, separately autoclaved at 110 °C, to a concentration of 20 g ⋅ L−1 (SMPD). Concentrated stock solutions (800-fold) of Tween 80 (polyethylene glycol sorbitan monooleate; Merck), ergosterol (≥95%; Sigma-Aldrich), or a combination of both were prepared with pure ethanol as solvent (36). Where indicated, SMPD was supplemented with Tween 80, ergosterol, or both from these stock solutions to reach final concentrations of 420 and 10 mg ⋅ L−1, respectively. Bacto agar (BD Biosciences, 20 g ⋅ L−1) was added to prepare solid media. The counter selectable amdS-marker was used as described previously (81). Strains with geneticin, hygromycin, or nourseothricin resistance were selected by supplementing yeast extract peptone dextrose medium (YPD) with 200 mg ⋅ L−1 geneticin (G418), 100 mg ⋅ L−1 hygromycin B (hygB) or 100 mg ⋅ L−1 nourseothricin (ClonNAT), respectively.
Molecular Biology Techniques and Plasmid and Strain Construction.
Open-reading frames of a putative SHC (SCHJC_C003990) from Sch. japonicus CBS5679 and of a Methylomicrobium alkaliphilum THS gene (GenBank Accession No. CCE23313) were codon optimized for use in S. cerevisiae with the GeneOptimizer algorithm (GeneArt) (82). A detailed description of the construction of plasmids pUDE1059 (pTEF1-Sjshc1-tCYC1) and pUDE1060 (pTDH3-Maths-tADH1) is provided in SI Appendix. Plasmids and oligonucleotides are listed in SI Appendix, Tables S8 and S9, respectively. Construction of S. cerevisiae strains IMX2616 (sga1Δ::Sjshc1) and IMX2629 (sga1Δ::Sjshc1 X-2::Maths) is described in detail in SI Appendix. In brief, expression cassettes harboring Sjshc1 or Maths coding sequences were amplified from plasmids pUDE1059 and pUDE1060 and used for Cas9-mediated genomic integration into a Cas9-expressing S. cerevisiae platform strain (83).
Aerobic Shake Flask Cultivation.
Aerobic cultivation on SMPD was performed in 500-mL shake flasks with a working volume of 100 mL. A preculture was inoculated from a frozen stock culture and, after overnight incubation, transferred to a second preculture. Upon reaching midexponential phase, cells were transferred to fresh SMPD, and optical density was monitored at 660 nm. All experiments were performed in duplicate. Light-induced flocculation of Sch. japonicus (84) was prevented by wrapping flasks in aluminum foil.
Anaerobic Shake Flask Cultivation.
Anaerobic chamber experiments were performed as previously described (36), using 100-mL shake flasks containing 80 mL medium. The anaerobic chamber was placed in a mobile darkroom that was only illuminated with red light-emitting diodes (85) during sampling. An aerobic preculture on SMPD was used to inoculate an anaerobic preculture on SMPD with an increased glucose concentration of 50 g ⋅ L−1, which was grown until the end of the exponential phase. Samples from these precultures were then used to inoculate experiments on regular SMPD supplemented with either Tween 80 and ergosterol, only Tween 80 or ergosterol, or neither. Cultures of S. cerevisiae CEN.PK113-7D on SMPD without Tween 80 or ergosterol were included in all anaerobic chamber experiments to assess low-level oxygen contamination (41). All growth experiments were performed by monitoring optical density at 600 nm in independent duplicate cultures.
Analytical Methods.
Extracellular metabolite concentrations were analyzed by high-performance liquid chromatography (86). Extraction and quantification of fatty acids, sterols, and tetrahymanol by GC-FID was performed as previously described (20, 36). A 6-point calibration of the GC-FID system with hop-22(29)-ene (Sigma-Aldrich, 0.1 mg ⋅ mL−1 in isooctane) was used for quantification of hop-22(29)-ene and other detected hopanoid compounds. Detailed methods for detection and identification of hopanoid compounds by GC-MS are described in SI Appendix. Optical density was measured at 600 nm for anaerobic cultures with an Ultrospec 10 cell density meter (Biochrom, Harvard Biosience) placed in the anaerobic chamber. For aerobic cultures, optical density at 660 nm was measured on a Jenway 7200 spectrophotometer (Bibby Scientific).
Genome Sequencing and Assembly.
The genome of Sch. japonicus CBS5679 was sequenced using short-read and long-read sequencing technologies. Genomic DNA was isolated with a Qiagen genomic DNA 100/G kit (Qiagen), with minor modifications to the manufacturer’s instructions. Detailed information on DNA isolation and library preparation is provided in SI Appendix. Short-read whole-genome sequencing was performed on a MiSeq platform (Illumina). Long-read sequencing was performed using the MinION platform (Oxford Nanopore Technologies). Base-calling was performed using Guppy version 2.1.3 (Oxford Nanopore Technologies) using dna_r9.4.1_450bps_flipflop.cfg. Genome assembly was performed using Flye version 2.7.1-b167359 (87). Flye contigs were polished using Pilon version 1.18 (88). The polished assembly was annotated with Funannotate version 1.7.1 (89) using RNAseq data from bioprojects PRJNA53947 and PRJEB30918 as evidence of transcription, adding functional information with Interproscan version 5.25-64.0 (90).
Sequence Homology Search and Phylogenetic Analyses.
Eukaryotic and bacterial amino acid sequence databases were built from UniProtKB reference proteomes (Release 2019_02) using the taxonomic divisions (taxids) shared with databases from the National Center for Biotechnology Information (NCBI). Only amino acid sequences from reference or representative organisms having genome assemblies at chromosome or scaffold level according to the NCBI genomes database (Release 2019_03) were included. Amino acid sequences of an OSC from Sch. pombe (50) (SpErg7; UniProt accession Q10231), an SHC from A. alicyclobacillus (51) (AaShc; P33247), and an STC from T. thermophila (5, 20) (TtThc1; Q24FB1) were used as queries for a HMMER3 (52) homology search. HMMER hits with an E-value below 1 × 10−5, and a total alignment length (query coverage) exceeding 75% of the query sequence were considered significant. A total number of 128 selected sequences (SI Appendix, Table S4 and Dataset S6) were subjected to multiple sequence alignment using MAFFT version 7.402 (91) in “einsi” mode. Alignments were trimmed using trimAl version 1.2 (92) in “gappyout” mode and used to build a phylogenetic tree with RAxML-NG version 0.8.1 (93) using 10 random and 10 parsimony starting trees, 100 Felsestein Bootstrap replicates, and PROTGTR + FO model. The final, midrooted tree provided in Dataset S4 was visualized using iTOL (94).
Supplementary Material
Acknowledgments
This work was funded by an Advanced Grant of the European Research Council to J.T.P. (Grant 694633). We gratefully acknowledge Jasmijn Hassing for constructing strain IMX2600 and Nicolò Baldi for the construction of plasmid pUDR538. We thank our colleagues in the Industrial Microbiology group of Delft University of Technology for stimulating discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105225118/-/DCSupplemental.
Data Availability
Whole-genome sequencing data for Sch. japonicus CBS5679 have been deposited under the BioProject Accession PRJNA698797 in NCBI. All other study data are included in the article and/or supporting information.
References
- 1.Parks L. W., Smith S. J., Crowley J. H., Biochemical and physiological effects of sterol alterations in yeast—A review. Lipids 30, 227–230 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Lingwood D., Simons K., Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Summons R. E., Bradley A. S., Jahnke L. L., Waldbauer J. R., Steroids, triterpenoids and molecular oxygen. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 951–968 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J. F., Xia J. J., Nie K. L., Wang F., Deng L., Outline of the biosynthesis and regulation of ergosterol in yeast. World J. Microbiol. Biotechnol. 35, 98 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Saar J., Kader J.-C., Poralla K., Ourisson G., Purification and some properties of the squalene-tetrahymanol cyclase from Tetrahymena thermophila. Biochim. Biophys. Acta 1075, 93–101 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Kemp P., Lander D. J., Orpin C. G., The lipids of the rumen fungus Piromonas communis. J. Gen. Microbiol. 130, 27–37 (1984). [DOI] [PubMed] [Google Scholar]
- 7.Murphy C. L., et al., Horizontal gene transfer as an indispensable driver for evolution of Neocallimastigomycota into a distinct gut-dwelling fungal lineage. Appl. Environ. Microbiol. 85, e00988–e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe I., Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep. 24, 1311–1331 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Ourisson G., Rohmer M., Poralla K., Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Annu. Rev. Microbiol. 41, 301–333 (1987). [DOI] [PubMed] [Google Scholar]
- 10.Rohmer M., Bouvier-Nave P., Ourisson G., Distribution of hopanoid triterpenes in prokaryotes. J. Gen. Microbiol. 130, 1137–1150 (1984). [Google Scholar]
- 11.Banta A. B., Wei J. H., Welander P. V., A distinct pathway for tetrahymanol synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 112, 13478–13483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreasen A. A., Stier T. J. B., Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J. Cell. Comp. Physiol. 41, 23–36 (1953). [DOI] [PubMed] [Google Scholar]
- 13.Andreasen A. A., Stier T. J. B., Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for growth in a defined medium. J. Cell. Comp. Physiol. 43, 271–281 (1954). [DOI] [PubMed] [Google Scholar]
- 14.Holm Hansen E., Nissen P., Sommer P., Nielsen J. C., Arneborg N., The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 91, 541–547 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Jansen M. L. A., et al., Saccharomyces cerevisiae strains for second-generation ethanol production: From academic exploration to industrial implementation. FEMS Yeast Res. 17, fox044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin C. E., Oh C.-S., Jiang Y., Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 1771, 271–285 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Verduyn C., Postma E., Scheffers W. A., van Dijken J. P., Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136, 395–403 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Barnett J. A., Payne R. W., Yarrow D., A guide to identifying and classifying yeasts. Mycologia 72, 440 (1980). [Google Scholar]
- 19.Visser W., Scheffers W. A., Batenburg-van der Vegte W. H., van Dijken J. P., Oxygen requirements of yeasts. Appl. Environ. Microbiol. 56, 3785–3792 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiersma S. J., Mooiman C., Giera M., Pronk J. T., Squalene-tetrahymanol cyclase expression enables sterol- independent growth of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 86, e00672–e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulder C. J. E. A., Anaerobic growth, ergosterol content and sensitivity to a polyene antibiotic, of the yeast Schizosaccharomyces japonicus. Antonie van Leeuwenhoek 37, 353–358 (1971). [DOI] [PubMed] [Google Scholar]
- 22.Yukawa M., Maki T., Regarding the new fission yeast Schizosaccharomyces japonicus. Kyushu Daigaku Kiyou 4, 218–226 (1931). [Google Scholar]
- 23.Wickerham L. J., Duprat E., A remarkable fission yeast, Schizosaccharomyces versatilis nov. sp. J. Bacteriol. 50, 597–607 (1945). [DOI] [PubMed] [Google Scholar]
- 24.Domizio P., et al., Evaluation of the yeast Schizosaccharomyces japonicus for use in wine production. Am. J. Enol. Vitic. 69, 266–277 (2018). [Google Scholar]
- 25.Benito S., The impacts of Schizosaccharomyces on winemaking. Appl. Microbiol. Biotechnol. 103, 4291–4312 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Rhind N., et al., Comparative functional genomics of the fission yeasts. Science 332, 930–936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niki H., Schizosaccharomyces japonicus: The fission yeast is a fusion of yeast and hyphae. Yeast 26, 545–551 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Kinnaer C., Dudin O., Martin S. G., Yeast-to-hypha transition of Schizosaccharomyces japonicus in response to environmental stimuli. Mol. Biol. Cell 30, 975–991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klar A. J. S., Schizosaccharomyces japonicus yeast poised to become a favorite experimental organism for eukaryotic research. G3 (Bethesda) 3, 1869–1873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alfa C. E., Hyams J. S., Distribution of tubulin and actin through the cell division cycle of the fission yeast Schizosaccharomyces japonicus var. versatilis: A comparison with Schizosaccharomyces pombe. J. Cell Sci. 96, 71–77 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Kaino T., Tonoko K., Mochizuki S., Takashima Y., Kawamukai M., Schizosaccharomyces japonicus has low levels of CoQ10 synthesis, respiration deficiency, and efficient ethanol production. Biosci. Biotechnol. Biochem. 82, 1031–1042 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Bulder C. J. E. A., Weijers C., Absence of cyanide-insensitive respiration in Schizosaccharomyces japonicus. FEMS Microbiol. Lett. 15, 145–147 (1982). [Google Scholar]
- 33.Makarova M., et al., Delineating the rules for structural adaptation of membrane-associated proteins to evolutionary changes in membrane lipidome. Curr. Biol. 30, 367–380.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldbauer J. R., Newman D. K., Summons R. E., Microaerobic steroid biosynthesis and the molecular fossil record of Archean life. Proc. Natl. Acad. Sci. U.S.A. 108, 13409–13414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Costa B. L. V., et al., Forever panting and forever growing: Physiology of Saccharomyces cerevisiae at extremely low oxygen availability in the absence of ergosterol and unsaturated fatty acids. FEMS Yeast Res. 19, foz054 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Dekker W. J. C., Wiersma S. J., Bouwknegt J., Mooiman C., Pronk J. T., Anaerobic growth of Saccharomyces cerevisiae CEN.PK113-7D does not depend on synthesis or supplementation of unsaturated fatty acids. FEMS Yeast Res. 19, foz060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirschvink J. L., Kopp R. E., Palaeoproterozoic ice houses and the evolution of oxygen-mediating enzymes: The case for a late origin of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2755–2765 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David M. H., Kirsop B. H., Yeast growth in relation to the dissolved oxygen and sterol content of wort. J. Inst. Brew. 79, 20–25 (1973). [Google Scholar]
- 39.Varela C., Torrea D., Schmidt S. A., Ancin-Azpilicueta C., Henschke P. A., Effect of oxygen and lipid supplementation on the volatile composition of chemically defined medium and Chardonnay wine fermented with Saccharomyces cerevisiae. Food Chem. 135, 2863–2871 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Bulder C. J. E. A., Reinink M., Unsaturated fatty acid composition of wild type and respiratory deficient yeasts after aerobic and anaerobic growth. Antonie van Leeuwenhoek 40, 445–455 (1974). [DOI] [PubMed] [Google Scholar]
- 41.Mooiman C., et al., Critical parameters and procedures for anaerobic cultivation of yeasts in bioreactors and anaerobic chambers. FEMS Yeast Res. 21, foab035 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiojima K., et al., Mass spectra of pentacyclic triterpenoids. Chem. Pharm. Bull. (Tokyo) 40, 1683–1992 (1992). [Google Scholar]
- 43.Mallory F. B., Gordon J. T., Conner R. L., The isolation of a pentacyclic triterpenoid alcohol from a protozoan. J. Am. Chem. Soc. 85, 1362–1363 (1963). [Google Scholar]
- 44.Takishita K., et al., Microbial eukaryotes that lack sterols. J. Eukaryot. Microbiol. 64, 897–900 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Giera M., Plössl F., Bracher F., Fast and easy in vitro screening assay for cholesterol biosynthesis inhibitors in the post-squalene pathway. Steroids 72, 633–642 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Sessions A. L., et al., Identification and quantification of polyfunctionalized hopanoids by high temperature gas chromatography-mass spectrometry. Org. Geochem. 56, 120–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoshino T., Sato T., Squalene-hopene cyclase: Catalytic mechanism and substrate recognition. Chem. Commun. (Camb.) 21, 291–301 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Müller C., Binder U., Bracher F., Giera M., Antifungal drug testing by combining minimal inhibitory concentration testing with target identification by gas chromatography-mass spectrometry. Nat. Protoc. 12, 947–963 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Giera M., Müller C., Bracher F., Analysis and experimental inhibition of distal cholesterol biosynthesis. Chromatographia 78, 343–358 (2014). [Google Scholar]
- 50.Corey E. J., Matsuda S. P., Baker C. H., Ting A. Y., Cheng H., Molecular cloning of a Schizosaccharomyces pombe cDNA encoding lanosterol synthase and investigation of conserved tryptophan residues. Biochem. Biophys. Res. Commun. 219, 327–331 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Ochs D., Kaletta C., Entian K. D., Beck-Sickinger A., Poralla K., Cloning, expression, and sequencing of squalene-hopene cyclase, a key enzyme in triterpenoid metabolism. J. Bacteriol. 174, 298–302 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mistry J., Finn R. D., Eddy S. R., Bateman A., Punta M., Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kannenberg E. L., Poralla K., Hopanoid biosynthesis and function in bacteria. Naturwissenschaften 86, 168–176 (1999). [Google Scholar]
- 54.Belin B. J., et al. , Hopanoid lipids: From membranes to plant–bacteria interactions. Nat. Rev. Microbiol. 16, 304–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gertz E. M., Yu Y. K., Agarwala R., Schäffer A. A., Altschul S. F., Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 4, 41 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mikkelsen M. D., et al., Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 14, 104–111 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Conner R. L., et al., Ergosterol replacement of tetrahymanol in Tetrahymena membranes. Biochem. Biophys. Res. Commun. 44, 995–1000 (1971). [DOI] [PubMed] [Google Scholar]
- 58.Youssef N. H., et al., The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 79, 4620–4634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furuya K., Niki H., Isolation of heterothallic haploid and auxotrophic mutants of Schizosaccharomyces japonicus. Yeast 26, 221–233 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Shinozaki J., Shibuya M., Masuda K., Ebizuka Y., Squalene cyclase and oxidosqualene cyclase from a fern. FEBS Lett. 582, 310–318 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Shinozaki J., Nakene T., Takano A., Squalene cyclases and cycloartenol synthases from Polystichum polyblepharum and six allied ferns. Molecules 23, 1843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frickey T., Kannenberg E., Phylogenetic analysis of the triterpene cyclase protein family in prokaryotes and eukaryotes suggests bidirectional lateral gene transfer. Environ. Microbiol. 11, 1224–1241 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Racolta S., Juhl P. B., Sirim D., Pleiss J., The triterpene cyclase protein family: A systematic analysis. Proteins 80, 2009–2019 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Pale-Grosdemange C., Feil C., Rohmer M., Poralla K., Occurrence of cationic intermediates and deficient control during the enzymatic cyclization of squalene to hopanoids. Angew. Chem. Int. Ed. Engl. 37, 2237–2240 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Syrén P. O., Henche S., Eichler A., Nestl B. M., Hauer B., Squalene-hopene cyclases-evolution, dynamics and catalytic scope. Curr. Opin. Struct. Biol. 41, 73–82 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Howard D. L., Simoneit B. R. T., Chapman D. J., Triterpenoids from lipids of Rhodomicrobium vanniellii. Arch. Microbiol. 137, 200–204 (1984). [Google Scholar]
- 67.Douka E., Koukkou A., Drainas C., Grosdemange-Billiard C., Rohmer M., Structural diversity of the triterpenic hydrocarbons from the bacterium Zymomonas mobilis: The signature of defective squalene cyclization by the squalene/hopene cyclase. FEMS Microbiol. Lett. 199, 247–251 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Poger D., Mark A. E., The relative effect of sterols and hopanoids on lipid bilayers: When comparable is not identical. J. Phys. Chem. B 117, 16129–16140 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Sáenz J. P., Sezgin E., Schwille P., Simons K., Functional convergence of hopanoids and sterols in membrane ordering. Proc. Natl. Acad. Sci. U.S.A. 109, 14236–14240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bradley A. S., Pearson A., Sáenz J. P., Marx C. J., Adenosylhopane: The first intermediate in hopanoid side chain biosynthesis. Org. Geochem. 41, 1075–1081 (2010). [Google Scholar]
- 71.Koch B., Schmidt C., Daum G., Storage lipids of yeasts: A survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol. Rev. 38, 892–915 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Csáky Z., et al., Squalene lipotoxicity in a lipid droplet-less yeast mutant is linked to plasma membrane dysfunction. Yeast 37, 45–62 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Schmerk C. L., Bernards M. A., Valvano M. A., Hopanoid production is required for low-pH tolerance, antimicrobial resistance, and motility in Burkholderia cenocepacia. J. Bacteriol. 193, 6712–6723 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Welander P. V., et al., Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 191, 6145–6156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brenac L., Baidoo E. E. K., Keasling J. D., Budin I., Distinct functional roles for hopanoid composition in the chemical tolerance of Zymomonas mobilis. Mol. Microbiol. 112, 1564–1575 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Entian K. D., Kötter P., 25 yeast genetic strain and plasmid collections. Methods Microbiol. 36, 629–666 (2007). [Google Scholar]
- 77.Nijkamp J. F., et al., De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Fact. 11, 36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miyata M., Doi H., Miyata H., Johnson B. F., Sexual co-flocculation by heterothallic cells of the fission yeast Schizosaccharomyces pombe modulated by medium constituents. Antonie van Leeuwenhoek 71, 207–215 (1997). [DOI] [PubMed] [Google Scholar]
- 79.Verduyn C., Postma E., Scheffers W. A., Van Dijken J. P., Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517 (1992). [DOI] [PubMed] [Google Scholar]
- 80.Jensen N. B., et al., EasyClone: Method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 14, 238–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Solis-Escalante D., et al., amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res. 13, 126–139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raab D., Graf M., Notka F., Schödl T., Wagner R., The GeneOptimizer algorithm: Using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst. Synth. Biol. 4, 215–225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mans R., et al., CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 15, fov004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Itoh S., et al., Effect of light on sexual flocculation in Schizosaccharomyces japonicus. Plant Cell Physiol. 17, 1355–1358 (1976). [Google Scholar]
- 85.Okamoto S., Furuya K., Nozaki S., Aoki K., Niki H., Synchronous activation of cell division by light or temperature stimuli in the dimorphic yeast Schizosaccharomyces japonicus. Eukaryot. Cell 12, 1235–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verhoeven M. D., et al., Mutations in PMR1 stimulate xylose isomerase activity and anaerobic growth on xylose of engineered Saccharomyces cerevisiae by influencing manganese homeostasis. Sci. Rep. 7, 46155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kolmogorov M., Yuan J., Lin Y., Pevzner P. A., Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 88.Walker B. J., et al., Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Love J., et al., Funannotate (v1.7.1, Zenodo, 2019).
- 90.Jones P., et al., InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A., RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Letunic I., Bork P., Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44 (W1), W242-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sievers F., Higgins D. G., Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequencing data for Sch. japonicus CBS5679 have been deposited under the BioProject Accession PRJNA698797 in NCBI. All other study data are included in the article and/or supporting information.