Heat stress redefines priorities for translating circadian mRNAs in a time-of-day-dependent context in Arabidopsis thaliana.
Abstract
The circadian clock helps organisms to anticipate and coordinate gene regulatory responses to changes in environmental stimuli. Under growth limiting temperatures, the time of the day modulates the accumulation of polyadenylated mRNAs. In response to heat stress, plants will conserve energy and selectively translate mRNAs. How the clock and/or the time of the day regulates polyadenylated mRNAs bound by ribosomes in response to heat stress is unknown. In-depth analysis of Arabidopsis thaliana translating mRNAs found that the time of the day gates the response of approximately one-third of the circadian-regulated heat-responsive translatome. Specifically, the time of the day and heat stress interact to prioritize the pool of mRNAs in cue to be translated. For a subset of mRNAs, we observed a stronger gated response during the day, and preferentially before the peak of expression. We propose previously overlooked transcription factors (TFs) as regulatory nodes and show that the clock plays a role in the temperature response for select TFs. When the stress was removed, the redefined priorities for translation recovered within 1 h, though slower recovery was observed for abiotic stress regulators. Through hierarchical network connections between clock genes and prioritized TFs, our work provides a framework to target key nodes underlying heat stress tolerance throughout the day.
Introduction
Fine-tuning of an organism’s metabolism, growth, and behavior in response to daily and seasonal changes in environmental conditions depends heavily on the circadian clock (Dodd, 2005; McClung, 2006; Nagel and Kay, 2012). When external cues that synchronize the clock such as temperature, occur outside the ambient limits, the clock helps organisms to anticipate and respond to the perceived stress. For instance, in response to heat stress (>37°C), the Arabidopsis clock can control the magnitude of change in transcript abundance based on the time of the day, a phenomenon referred to as gating (Fowler et al., 2005; Hotta et al., 2007; Wilkins et al., 2010; Blair et al., 2019). The core clock components, CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION1 (TOC1), show differential transcriptional responses to heat stress depending on the time of the day (Blair et al., 2019). At the post-transcriptional level, clock genes are alternatively spliced in response to heat stress (Kwon et al., 2014; Filichkin et al., 2015). In addition, the clock gene ZEITLUPE (ZTL) plays a functional role in the post-translational regulation of select proteins in response to heat stress (Gil et al., 2017). Whether the time of the day plays a role in the accumulation of alternative spliced isoforms or protein degradation remains unclear.
In plants, a lesser understood and critically important level of regulatory control is the circadian translatome, as this will most likely inform the modulation of protein rhythms and abundance, in response to heat stress. Transcripts corresponding to genes acting in thermoresponsive mechanisms need to be translated, despite the reduction of polysomes at high temperatures (Yángüez et al., 2013; Merret et al., 2017). To manage the accumulation of misfolded proteins under heat stress, the unfolded protein and heat shock responses are activated (Richter et al., 2010; Bao and Howell, 2017). Heat shock factors (HSFs) are rapidly produced to enhance the synthesis of heat shock proteins (HSPs). An HSF-independent pathway involving REVEILLE (RVE) 4 and RVE8 circadian-regulated genes has recently been described, and their dual role in plant thermotolerance has been shown to be gated by the clock (Li et al., 2019). However, interconnections between the clock and translational regulation of thermotolerance mechanisms remain obscure. With the increase in the frequencies, intensities, and duration of heat waves due to climate change, identifying and defining the mechanisms of prioritization for translation in a time-of-day-dependent context is critically important (Sun et al., 2019).
In this study, we investigated the circadian translatome responsiveness to heat stress in a time-of-day-dependent context. Using translating ribosome affinity purification (TRAP), we isolated translating polyadenylated mRNAs and subjected them to deep sequencing in Arabidopsis (Zanetti et al., 2005). We found a statistically significant influence of the time of the day, including a gated response on the translatome in response to the stress. For core stress regulators such as HSPs and DREBs, the time of the day significantly alters the level of mRNA bound by ribosomes. Furthermore, following removal of the stress, the majority of the circadian translatome recovered within 1 h, while others are much slower. We compared multiple levels (transcriptome, translatome, and proteome) of regulation, which had not been previously possible and hypothesized a broader role for post-translational regulation of clock-controlled genes. Moreover, we uncovered and connected putative nodes in the heat stress response network, which included TFs involved in growth regulation (CYCLING DOF FACTORS, CDFs; myeloblastosis-related, MYB-related; and B-box proteins, BBXs), and showed that a functional clock and the time of the day are important in regulating their heat response.
Understanding how the priority to access ribosomes changes depending on the time of the day in response to heat stress will reveal how organisms coordinately regulate networks of general and time-specific heat-responsive genes to optimize fitness.
Results
The specific rhythmicity of the circadian translatome
We first analyzed the dynamics of the clock-regulated transcriptome and translatome under normal conditions (Figure 1A). Following 10 days of entrainment (light/dark cycles), and 2 days in free-running conditions (constant light and temperature at 22°C, days 11–12), seedlings were collected every 3 h during a complete 24 h time course (ZT48, beginning of the light, to ZT72, end of the subjective night) on day 13 (Figure 1A). Total mRNAs and ribosome-associated (TRAP) mRNAs were extracted from seedlings and mRNA-Seq and TRAP-Seq were performed, respectively (Supplemental Data Set S1). For TRAP, we used an Arabidopsis line containing N-termini His-FLAG epitope tagged RPL18 driven by the 35S promoter, Pro35S:HF-RPL18 (Zanetti et al., 2005). To analyze circadian oscillations, a robust analysis of rhythmic expression was performed using the integrated R package Metacycle (Supplemental Figure S1 and Supplemental Data Set S2). Clock components that are expressed from morning to evening, exhibited oscillations at the expected times of the day (Figure 1, B and C; Supplemental Figure S2; Mockler et al., 2007; Hsu and Harmer, 2012; Romanowski et al., 2020). At the genome scale, this analysis identified 8,028 cycling transcripts in total mRNAs, consistent with previous data sets that were obtained in similar conditions, and with conserved calculated phases (Figure 1D;Supplemental Figures S1 and S2 and Supplemental Data Set S2; Mockler et al., 2007; Hsu and Harmer, 2012; Romanowski et al., 2020). At the translatome level (TRAP mRNAs), 10,657 transcripts exhibited circadian oscillations including 64% shared with the cycling transcriptome, corresponding to genes involved in photosynthesis and response to stimuli, among other processes (Figure 1, D–F; Supplemental Data Set S3). Circadian transcripts peaked at multiple times during the day, including 12% and 11% peaking at dawn in total and TRAP mRNAs, respectively (i.e. phase 0, Figure 1, G and H). A phase comparison between the two mRNA populations revealed a high conservation of the timing of oscillations between the circadian transcriptome and translatome (88% with a phase difference ≤1.5 h; Figure 1I).
Figure 1.
The Arabidopsis circadian transcriptome and translatome. A, Experimental design. B, Simplified circadian clock model showing connections and timing of expression of the clock components. C, Transcript profiles of circadian clock genes in total mRNAs. Data are from RNA-seq, and are scaled (means, n = 3). A spline function was used to connect time points. D, Heatmaps of the 8,028 and 10,657 transcripts exhibiting significant circadian oscillations in total and TRAP mRNAs, respectively. Data are scaled by row and are ordered by phase (timing of peak of abundance). The color scale represents the normalized transcript level, with yellow and purple representing high and low transcript abundance, respectively. E, Venn diagram depicting the overlapping circadian transcripts between total and TRAP mRNAs. F, Selected over-represented GO biological processes in the list of 6,840 circadian transcripts described in (E). For all enriched terms, see Supplemental Data Set S3. G, Diagram defining the phase as the time of peak abundance, a phase of 0 and 12 corresponding to a peak abundance occurring at dawn and on the evening, respectively. H, Circular plots representing the phase distribution of the circadian total and TRAP mRNAs. I, Histogram showing the phase difference between total and TRAP mRNAs for the 6,840 shared circadian transcripts. The dashed brown rectangle highlights circadian transcripts having a phase difference of 1.5 h or less between the two mRNA populations, which correspond to 88% of the shared transcripts. In A–C, G–H, gray areas represent the subjective night.
From the 3,817 TRAP-specific cycling transcripts (Figure 1E), 667 (17%) were not detected in previous circadian transcriptomes and suggest specific control of rhythmicity at the translatome level (Figure 2, A and B). The clock control of mRNA ribosome loading over the diel cycle and the proteome rhythmicity have been previously reported in Arabidopsis (Missra et al., 2015; Krahmer et al., 2019). Genes exhibiting oscillations from the mRNA to protein level are mainly involved in clock-regulated processes such as photosynthesis, response to abiotic stimulus, and metabolism-related processes (Supplemental Data Set S3). In addition, several components of the translation initiation complex also show circadian regulation across these three levels of regulation. Though we found a significant overlap between our circadian transcriptome and translatome and these two data sets, rhythmicity of the 667 TRAP-specific circadian mRNAs is specifically observed in our study (Figure 2A;Supplemental Figure S3). In this context, assessing the circadian translatome provides a powerful and comprehensive method to distinguish between post-transcriptional and translational control. Interestingly, an over-representation of cell cycle genes within the 667 TRAP-specific cycling transcripts supports previous findings of the translational control of mitotic genes and the common rhythmicity of the circadian clock and the cell cycle machinery (Figure 2, C and D; Hunt and Sassone-Corsi, 2007; Tanenbaum et al., 2015). Technical limitations of proteomic methods to detect low abundant proteins and those involved in specific processes could contribute to an underestimation of what is rhythmic at the protein level. It can therefore be inferred that future circadian proteomic data sets will provide further resolution on clock control at these different levels of gene regulation (Patole and Bindschedler, 2019).
Figure 2.
Identification of translatome-specific circadian transcripts. A, Venn diagrams showing the overlapping circadian transcripts identified in DIURNAL data sets (Mockler et al., 2007, Hsu and Harmer 2012; Romanowski et al., 2020), and the circadian total and TRAP mRNAs identified in Figure 1D; and the overlaps between the 667 TRAP-specific circadian transcripts and a diel translatome (Missra et al., 2015) and rhythmic proteome data sets (Krahmer et al., 2019). Red font and red shading highlight the 667 TRAP-specific circadian transcripts. Additional comparisons are shown in Supplemental Figure S3. B and C, Heatmap (B) and over-represented GO biological processes (C) of the 667 TRAP-specific circadian transcripts. In (B), data are scaled by row and ordered by phase. The color scale represents the normalized transcript level, with yellow and purple representing high and low transcript abundance, respectively. D, Transcript profiles of selected TRAP-specific circadian genes involved in the cell cycle (means ± sd, n = 3). Grey areas represent the subjective night. All panels correspond to data obtained at 22°C
The circadian transcriptome and translatome uncover different magnitudes of response to heat stress
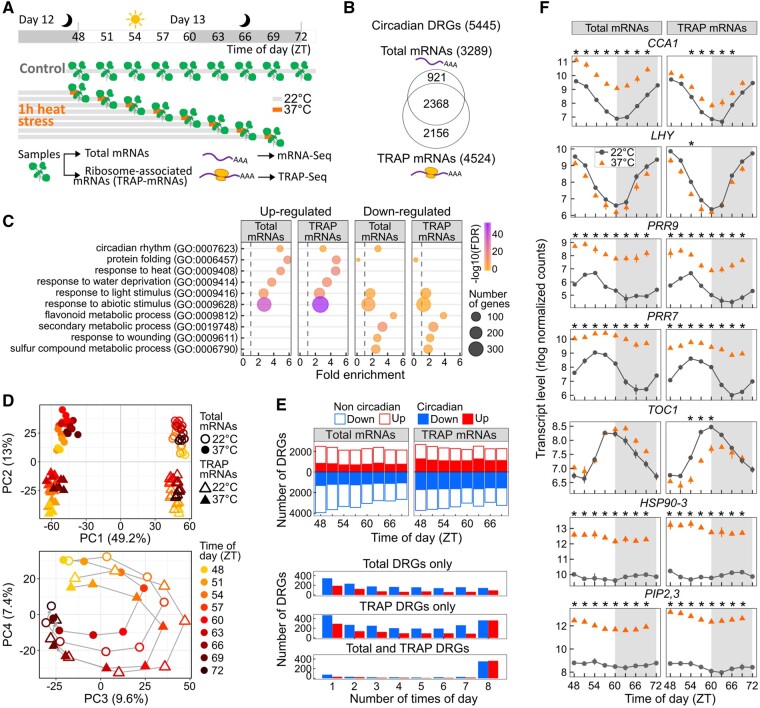
To next dissect how the clock- and circadian-controlled genes are coordinately regulated by heat stress during the day at the translatome level in plants, we examined the response of polyadenylated mRNAs bound by ribosomes at different times of the day in Arabidopsis. When performing the 24 h time course described in Figure 1, a 1-h heat stress at 37°C was applied on different sets of seedlings every 3 h during the 24 h time course (ZT48, beginning of the light, to ZT69, 3 h before the end of the subjective night, Figure 3A). Total (mRNA-Seq) and ribosome-associated (TRAP-Seq) mRNAs were analyzed from stressed (37°C) seedlings and data were compared to the control condition (22°C, Figure 3A;Supplemental Figure S4). In response to heat stress, 3,289 and 4,524 circadian transcripts were differentially accumulated at one or multiple times of the day in total and TRAP mRNAs, respectively, corresponding to 5,445 circadian differentially regulated genes (DRGs; Figure 3B;Supplemental Data Set S4). Processes such as protein folding, responses to light, abiotic stimulus, heat, and water deprivation were activated, while metabolic processes and response to biotic stresses were downregulated, highlighting the critical role of abiotic stress-related genes for plant thermotolerance under heat stress (Figure 3C;Supplemental Data Set S3).
Figure 3.
Heat stress regulates circadian genes at both transcriptional and translational levels. A, Experimental design. B, Venn diagram depicting the overlapping circadian DRGs between total and TRAP mRNAs. C, Selected enriched biological processes in the sets of upregulated and downregulated circadian DRGs. For all enriched terms, see Supplemental Data Set S3. D, Principal component analysis (PCA) of the 5,445 circadian DRGs. PC1 and PC2 separate temperatures and mRNA populations, respectively, and PC3 and PC4 separate times of the day. Gray lines link times of the day, from early morning to late night. E, Bar plots representing numbers of DRGs in Total and TRAP mRNAs at the different times of the day (upper plots), and numbers of DRGs responding to heat stress at a single time of day (1) to all times of day (8) and either specifically in total or TRAP mRNAs, or both (lower plots). F, Transcript abundance of major circadian clock genes, HSP90-3 and PIP2,3, in total and TRAP mRNAs (means ± sd, n = 3). Stars above the plots indicate a significant difference between measurements taken at 37°C and 22°C (FDR <0.05 and log2 fold change > |1|). In (A) and (F), gray areas represent the subjective night
Multidimensional analysis showed that temperature, the time of the day, and mRNA population contributed to the variations in transcript abundance of circadian DRGs (Figure 3D). Interestingly, the number of circadian DRGs remained constant throughout the day (Figure 3E). At first glance, clock gene profiles revealed consistent responses throughout the day at both transcriptome and translatome levels, with CCA1, PRR7, and PRR9 upregulated and LHY downregulated, which we also validated by quantitative reverse transcription polymerase chain reaction (RT-qPCR) (Figure 3F;Supplemental Figure S5). However, TOC1, a core component of the plant clock showed a specific downregulation in TRAP mRNAs, possibly reflecting an increase in unproductive transcripts that are then targeted by nonsense mediated decay (Figure 3F;Supplemental Figure S5; Kwon et al., 2014). In addition, comparing the lists of upregulated and downregulated DRGs in total and TRAP mRNAs revealed that genes specifically responding to heat stress at a single time point are specific to the transcriptome or the translatome (Figure 3E). However, comparing lists of DRGs obtained from multiple pairwise comparisons may lead to misinterpretation as the results are highly dependent on the thresholds used in this study to define DRGs (i.e. false discovery rate [FDR] and log2 fold change cutoffs). For example, LHY was significantly downregulated in TRAP mRNAs at ZT54 but showed an obvious downregulation under heat stress at most time points (Figure 3F). To statistically compare the responses observed at the transcriptome and translatome levels, a statistical analysis considering the whole data set and looking at the interaction between the effects of temperature and mRNA population was performed (Supplemental Figure S6A). This analysis revealed that 41% of the circadian DRGs including CCA1, PRR7, and PRR9 showed significant changes in magnitude of response to heat stress between transcriptome and translatome levels, despite strong correlations between the two mRNA populations (Supplemental Figure S6, A–C and Supplemental Data Set S4). We speculate that a reduced response at the translatome versus transcriptome level suggests potential mRNA decay or sequestration in stress granules and processing bodies (Supplemental Figure S6D; Merret et al., 2015; Chantarachot and Bailey-Serres, 2018). The presence of Upstream Open Reading Frames (uORFs) in this category of DRGs might correlate with the decrease in translation efficiency under heat stress (Supplemental Figure S6, E and F). Conversely, genes involved in water transport and several heat shock proteins such as PIP2,3 and HSP90-3, respectively, showed a greater upregulation at the translatome level (Figure 3F). Differences in the magnitude of response between the two mRNA populations mirror the changes in preference for translation under heat stress.
Timing of peak expression for circadian translating mRNAs in response to heat stress
As heat mostly occurs during the early afternoon in natural conditions, many heat-responsive genes were assumed to be expressed around that time. In fact, 79% of the upregulated TRAP DRGs including GI significantly responded when heat stress occurred before or after but not during the peak of transcript abundance (Figure 4A;Supplemental Figure S7). To confirm this observation, we compared the proportions of phases associated with each list of DRGs (obtained at the different times of the day) to the proportions of phases identified in all circadian genes (Figure 4B). These phase enrichment analyses were performed at the translatome and transcriptome levels (Figure 4C and Supplemental Figure S8, respectively; Supplemental Data Set S5). For example, when a heat stress was applied at ZT48 (early morning), an overrepresentation of genes oscillating from midday to the evening was observed within upregulated TRAP DRGs (i.e. phases 6, 10.5, and 12 were significantly enriched, Figure 4C). This analysis revealed that genes with expression peaking during the day were not preferentially upregulated when heat stress occurred at that time, but rather when it occurred at night (Figure 4C;Supplemental Figure S8). In downregulated DRGs, late night and morning expressed DRGs were always overrepresented (Figure 4C;Supplemental Figure S8). In addition, a high proportion (43%) of the downregulated DRGs showed a significant downregulation during the mRNA peak accumulation (i.e. phase), as observed for defense response-related genes, such as CCG-BINDING PROTEIN 1 (CBP1; Figure 4A;Supplemental Figure S7). Taken together, these results suggest that priorities for transcription and translation are redefined under heat stress. When the stress occurs during their lowest expression (i.e. trough), genes acting in the heat response are selectively transcribed and translated, while genes peaking (highest) at that time of the day but not directly involved in the stress response are not. It is also possible that a shift in the timing of peak transcript abundance as a result of the acute heat stress occurred for a significant proportion of circadian DRGs (Gil et al., 2017).
Figure 4.
Timing effect of heat stress on the circadian translatome. A, Transcript abundance of selected genes in total and TRAP mRNAs (means ± sd, n = 3). Stars above the plots indicate a significant difference between 37°C and 22°C (FDR <0.05 and log2 fold change > |1|). B, Example representing how the phase enrichment is calculated. C, Circular plots showing the phase enrichment at each time of the day (ZT48–ZT69) of the 4,524 circadian TRAP DRGs, as compared to all circadian TRAP mRNAs (10,657). Highlighting that significant upregulation occur primarily when heat stress occurred before or after but not during the peak of transcript abundance. At each time of the day, the number of circadian TRAP DRGs is indicated in parentheses. In (A) to (C), gray areas represent the subjective night. D, Histograms representing the proportions of DRGs with a similar response to heat at all times of the day
Time of the day influences translating circadian mRNAs under heat stress
To determine whether there is a preference for which mRNAs are cued to be translated, and if the heat stress response is also gated (i.e. time-of-day-specific changes in magnitude), we compared the response of DRGs at the different times of the day. First, general heat-responsive genes (i.e. upregulated or downregulated throughout the day) accounted for 24%–26% and 37%–41% of the downregulated and upregulated circadian DRGs, respectively, depending on the mRNA population (Figure 4D). These DRGs included 179 TFs of diverse families such as HSF, PLATZ, Pseudo, bZIP, C2C2-DOF (upregulated), and ARF, HMG, TCP (downregulated; Supplemental Figure S9 and Supplemental Data Set S6). When only considering the nature of the response (higher or lower transcript level at 37°C versus 22°C), up to 90% of the DRGs were similarly impacted by heat stress (Figure 4D), suggesting that the time of the day significantly influenced the magnitude of changes in transcript abundance, as observed for clock genes (Figure 3F). To test this hypothesis, a robust statistical analysis looking at the interaction between the effects of temperature and the time of the day was performed from data obtained in all conditions and in both mRNA populations. This revealed that 1614 (36%) circadian TRAP DRGs showed different magnitudes of response based on the time of the day (Figure 5A;Supplemental Data Set S7). In other words, the time of the day gated the heat stress response of these DRGs. Similar proportions were obtained with circadian total DRGs (40%, Supplemental Data Set S7). These genes included all described clock genes except LHY.
Figure 5.
Time of the day influences the magnitude of response to heat. A, Groups of circadian TRAP DRGs whose magnitude of response to heat stress is dependent (groups 3–4) or not (groups 1 and 2) on the time of the day. A likelihood ratio test looking at the statistical interaction between the effects of temperature and time of the day was used for this analysis (Supplemental Data Set S4). For each group, phase distributions are represented, bars representing the number of TRAP DRGs for each phase. Phase proportions were compared to proportions in all circadian TRAP mRNAs (10,657). Below phase plots, transcript abundance profiles of two selected genes within the group are shown (means ± sd, n = 3). Stars above the plots indicate a significant difference between 37°C and 22°C (FDR < 0.05 and log2 fold change > |1|). Data correspond to TRAP mRNAs. Gray areas from ZT60 to ZT72 represent the subjective night. Numbers in parentheses indicate the total number of DRGs within the groups. B, Model representing a hierarchy between groups of transcripts to access ribosomes under heat stress
To identify groups of transcripts with the highest versus lowest priorities to access ribosomes under heat stress, we focused on genes always upregulated or downregulated when a heat stress occurs, and whose response is or is not gated by the time of the day (Figure 5A). Interestingly, 27% (435 of 1614) of the gated TRAP DRGs are always upregulated under heat stress, such as DREB2B and TIP2 (Group 3, Figure 5A). In comparison, 288 (of 2,910, 10%) TRAP DRGs were always upregulated but their response to heat stress is not gated by the time of the day, such as HSFB4 and a PLATZ TF (group 1, Figure 5A). This result suggests that most general heat-responsive genes may need to be translated to a certain level when a heat stress occurs, whether during the day or at night. In addition, group 3 contained significant proportions of transcripts peaking in the midday and evening that potentially regulate heat responses, with functions related to protein refolding and water transport (Figure 5A;Supplemental Table S1). Conversely, group 4 contained highly downregulated DRGs, exhibiting oscillations at dawn and including a significant proportion involved in metabolic-related processes such as ECI3 (Figure 5A;Supplemental Table S1). It is worth noting that even though groups 1 and 2 did not exhibit changes in the magnitude of response to heat stress, the time of the day influenced the transcript accumulation of these DRGs at 37°C, as it followed the oscillations observed at 22°C (Figure 5A).
We defined an order of priority to access ribosomes under heat stress based on the generated groups (Figure 5B). Groups 3 and 1 contain transcripts highly prioritized throughout the day, whereas transcripts in groups 4 and 2 showed the opposite responses, respectively, and therefore have the lowest priority to access ribosomes under heat (Figure 5B). Between these two categories, other TRAP DRGs that were not classified in groups 1–4 displayed smaller changes in priorities to associate with ribosomes under heat stress as compared to control conditions. These other genes were significant at more specific times of the day (Supplemental Data Set S7). Even though these genes were not described in detail here, they may also play a significant role in the plant temperature responses.
We further investigated the influence of day (light) versus night (dark) on the gating response, by performing a similar statistical analysis as described above during the day (ZT51–ZT57) and night (ZT63–ZT69). We showed that the time of the day preferentially gates the response of circadian TRAP mRNAs during the light period as compared to the dark (Supplemental Figure S10). An overrepresentation of genes peaking in the evening has been observed in day-specific gated DRGs, while night-specific gated DRGs peaked in the early morning (Supplemental Figure S10). This result suggests that the gating response occurs before the transcript peak abundance, rather than after. Interestingly, the GO term “circadian rhythm” was specifically enriched in DRGs that are specifically gated during the day, while “photosynthesis/light harvesting in photosystem I” was specifically enriched in genes gated at night (Supplemental Figure S10). These observations support the anticipatory role of the clock, and that the dynamic regulation of translation by the time of the day is critical for temporal coordination of key biological processes to environmental stimuli. These results provide comprehensive insights of how the time of the day influences translating mRNAs in response to a stress.
The clock regulates the gating response of heat-responsive TFs
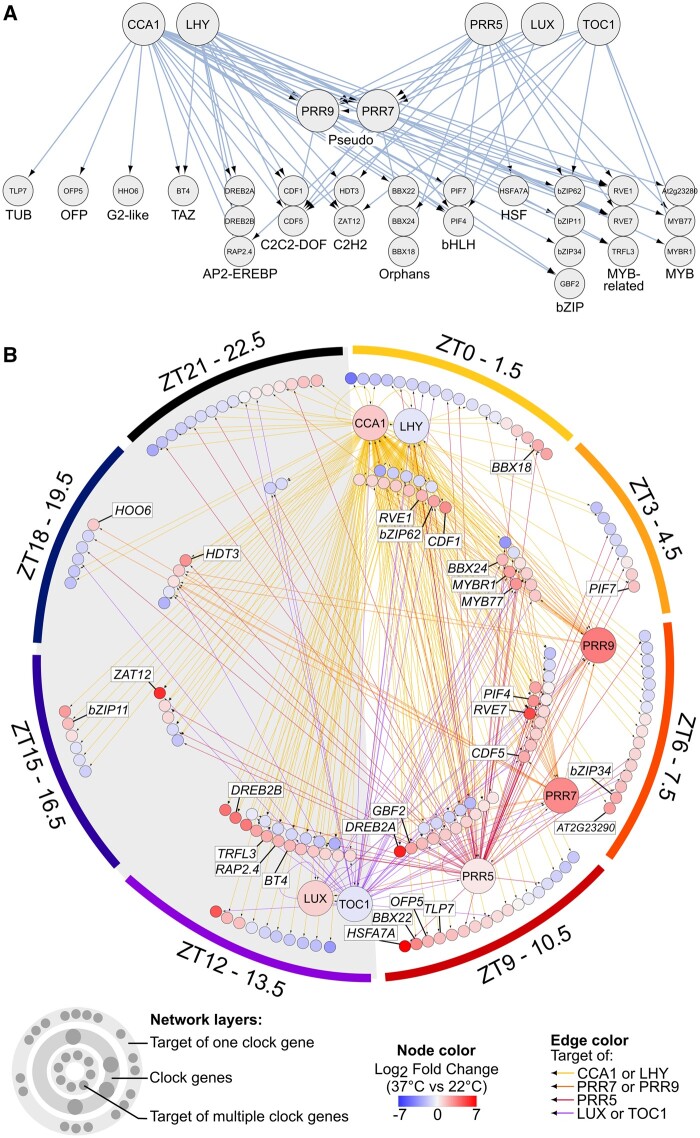
As TFs are key regulators of abiotic stress-responsive genes, they represent interesting targets for crop improvement. From the 52 TFs identified in group 3 with the highest priority to access ribosomes, 29 were previously identified as direct targets of clock genes in ChIP-Seq analyses (Figure 6A;Supplemental Figure S11 and Supplemental Table S1). These TFs include regulators of photomorphogenesis (BBX18, BBX22, BBX24, PIF4, PIF7), response to water deprivation (DREB2A, DREB2B, RAP2-4, MYB44, HDT3), and salt stress (ZAT12, BBX24, RAP2-4, MYB44, HDT3, BT4). The high induction of major and well-characterized TFs such as dehydration-responsive genes DREB2A and DREB2B strongly support the prime role of this subgroup of genes in the heat stress response, primarily during the day period. In fact, integrating these TFs in a larger network of clock-controlled TFs highlights the dominance of morning to evening oscillating genes, but also the connection with TFs expressed at night, suggesting a coordinated role of the clock and these regulators throughout the day to respond to heat stress (Figure 6B). In addition, our analysis implicates the CDF, Myb-related, and BBX TF families as important regulatory hubs or new nodes in the heat stress response pathways. As their function in growth-related processes has been best defined, we speculate that the members of these TF families might modulate the dynamics of growth in response to heat stress depending on the time of the day.
Figure 6.
Circadian networks of TFs translationally regulated by heat stress. Network of TFs exhibiting circadian oscillations and differentially regulated under heat stress in TRAP mRNAs; and identified as targets of proteins encoded by clock genes in published ChIP-Seq analyses were selected to build the networks. Edges correspond to interactions between clock proteins and their targets. In (A), all nodes except CCA1, LHY, PRR5, LUX, and TOC1 belong to group 3 presented in Figure 5A and clock gene targets are sorted by TF family. In (B), target nodes correspond to all clock targets that are TFs exhibiting circadian oscillations and differentially regulated under heat stress at the translatome level. The accession numbers for all genes represented in the network are provided in Supplemental Data Set S11. Edge colors differentiate targets of the different clock proteins. Nodes on the external layer (e.g. PIF7) correspond to TFs targeted by one clock protein and nodes on the internal layer (e.g. CDF1) are targeted by at least two clock proteins. Nodes are sorted by the timing of peak abundance of the corresponding transcript, from ZT0 (early morning) to ZT22.5 (end of night). Node colors reflect the heat stress response of the corresponding transcript and correspond to the average of log2 fold change values obtained in TRAP mRNAs. The grey area represents the subjective night. Names of TFs connected in the network presented in (A) are highlighted (e.g. DREB2A)
To determine the influence of the clock in the heat stress response, we analyzed using RT-qPCR in WT (Pro35:HF-RPL18) and cca1 lhy/Pro35:HF-RPL18 seedlings the transcript abundance of two highly induced TFs, CDF1, expressed in the morning and involved in the control of photoperiodic flowering, and the Myb-related TRFL3, expressed in the evening and with a putative function in genome stability (Figure 7, A and B;Supplemental Figure S12). Because the cca1 lhy double mutant exhibits a short period, we did not specifically compare the heat stress responses between genotypes. When considering only a single time point, changes in transcript abundance in response to heat stress were compared within each genotype. Overall, the upregulation of CDF6 and TRFL3 in both genotypes in response to heat stress is consistent with the data observed in the total and TRAP-seq analysis (Figure 7, A and B). However, to further analyze the influence of the clock mutation on the temperature response of these selected TFs at more than one time point, we performed a multiway ANOVA, to assess the influence of temperature, the time of the day, genotype, mRNA population, and the interaction between these factors (Figure 7, B and C;Supplemental Table S2). The observed significance of the temperature effect confirms the results observed in the RNA-Seq data sets (Figure 7C). For both genes, a significant interaction between the effects of genotype and the time of the day has been observed, possibly reflecting the shorter period in the cca1 lhy/Pro35:HF-RPL18, and consequently a change in the timing of peak expression. However, for CDF1, the significant interaction between temperature and genotype is not dependent on the time of the day (Figure 7C). Tukey’s honestly significant difference (HSD) tests were performed to compare levels of the interaction Temperature: Genotype for CDF1 and revealed three distinct groups (A, B, and C; Figure 7B). In the clock mutant, the temperature response of CDF1 is significantly greater as compared to the response observed in WT, suggesting a repressive role of the clock on the heat stress response of CDF1. For TRFL3, a significant interaction between the effects of temperature, genotype, and the time of the day was determined (Temperature: Genotype: Time; Figure 7C). Tukey’s HSD tests showed that the temperature response of TRFL3 was significantly greater at night versus during the day in WT, and that the clock mutation significantly decreased the temperature response at night (Figure 7, B and C).
Figure 7.
Influence of the circadian clock on the heat stress response of selected TFs. A and B, Transcript levels of selected heat-responsive TFs measured by RNA-Seq (A) and RT-qPCR (B; means ± sd, n = 3). In (A), stars above the plots indicate a significant difference between measurements taken at 37°C and 22°C (FDR < 0.05 and log2 fold change > |1|). In (B), transcript levels were measured in the WT (Pro35S:HF-RPL18) and clock mutant (cca1 lhy/Pro35S:HF-RPL18). In (A) and (B), gray areas represent the subjective night. C, Multivariate analysis of variance of transcript abundance of selected TFs quantified in (B). Statistical significance (P < 0.05) is indicated in bold and green (see Supplemental Table S2 for the full table of results). Tukey’s HSD tests were performed to compare group levels for the significant sources of variation revealed by the multivariate analysis. To show the influence of the genotype on the temperature responses of the two selected TFs, letters were assigned to group levels significantly different based on Tukey’s HSD tests, for the interactions Temperature: Genotype (CDF1; A, B, C) and Temperature: Genotype: Time (TRFL3; a, b, c). For example, for CDF1, group levels for the significant interaction “Temperature: Genotype” were WT 22°C; WT 37°C; cca1 lhy 22°C and cca1 lhy 37°C. Tukey tests showed that cca1lhy 37°C group A was significantly higher than WT 37°C group B. These two group levels corresponding to heat stress data were significantly higher than group levels corresponding to data obtained at 22°C (WT 22°C and cca1 lhy 22°C, group C). Within each group level (e.g. WT 22°C), all times of the day and both mRNA populations were considered
Priorities for translation are quickly redefined following heat stress
Following heat stress, cells need to quickly readjust their ribosome pool (Merret et al., 2017; Zhang et al., 2017). To assess how priorities to access ribosomes change if a heat stress occurred in the middle of the day (ZT53 to ZT54), we analyzed the translatome after heat stress (ZT54, 0 h of recovery) and at 1, 3, and 6 h of recovery (ZT55, ZT57 and ZT60, respectively; Figure 8A;Supplemental Data Sets S8 and S9). Most (71%) circadian DRGs recovered within 1 h and showed no significant changes during the plant recovery, in both total and TRAP mRNAs (Figure 8, B and C; groups A and B). At the translatome level, 21 of the 29 (72%) TFs highlighted in Figure 6A recovered within 1 h, including DREB2A and DREB2B (Figure 8D). However, no clear connection can be made between the speed of recovery and the levels of priority to access ribosomes that were defined above. In addition, seven of the 29 (24%) selected TFs including CDF1 were downregulated at one or multiple times during the plant recovery (Figure 8D). This demonstrates that plants quickly redefined priorities for translation to survive rapid changes in their environmental conditions (Merret et al., 2017; Zhang et al., 2017; Crisp et al., 2017). This result also suggests that the vast majority of the 29 highly prioritized regulators play a role in the prime response to heat stress. Slower recovery profiles were also observed for groups C and D, which include protein folding genes and also TRFL3 that was still upregulated at 1 h of recovery (Figure 8, C and D;Supplemental Table S1). Together with DRGs exhibiting specific responses during the recovery but not during heat stress (groups G and H; Figure 8C), these genes may be involved in establishing heat stress memory and may be important for plant survival during growth resumption (group D; Figure 8, C and D; Supplemental Table S1; Forestan et al., 2020). Differential regulation of CCA1, LHY, and PRR9 during recovery also suggests readjustments of the clock and clock-controlled processes following heat stress (Supplemental Figure S13).
Figure 8.
Recovery of the circadian translatome following a heat stress. A, Experimental design of the recovery experiment. B, Numbers of DRGs after heat stress (0 h of recovery) and at 1, 3, and 6 h of recovery. C, Clustering analysis of the 2,824 TRAP circadian DRGs identified in the recovery experiment. The number of transcripts by group is indicated in parentheses. Clock genes and selected TFs are indicated in the corresponding group. On each cluster plot, the black dashed line represents a log2 fold change = 0 (i.e. no change between measurements taken at 37°C and at 22°C), the black solid line represents the mean and the gray shading represents the sd. D, Heatmap of selected TFs during recovery. The 29 selected TFs showed high priority to access ribosomes under heat stress (group 3, Figure 5A) and were identified as direct targets of clock proteins in published ChIP-Seq analyses (Figure 6A). These TFs were classified in groups B, D, or F in (C). Stars indicate significant differences between the recovery and control conditions. In (C) and (D), color scales represent log2 fold change values (Recovery versus Control)
Discussion
In planta protein detection of rhythmic TFs including clock genes is often challenged by their low abundance. Actively translating mRNAs can therefore provide a robust model to infer functional proteins that are indispensable to the heat stress response depending on the time of the day (Patole and Bindschedler, 2019). We have presented a robust and comprehensive analysis that reveals how the time of the day contributes to the heat stress response for the circadian translatome in plants. In addition to putative regulators, DRGs characterized through previous transcriptome or targeted approaches to be involved in the heat stress response pathway, controlled by the circadian clock, and exhibiting a gated response are reinforced at the translatome level.
Our transcriptome analysis revealed that ∼30% of the transcripts showed circadian oscillations when plants are transferred to free-running conditions, suggesting that the corresponding genes are regulated by the clock, and these observations correspond to previously published data sets (Mockler et al., 2007; Hsu and Harmer, 2012; Romanowski et al., 2020). However, the circadian translatome analysis showed that this proportion of transcripts is even higher at this level of regulation (∼40% of the translatome). Both the time of the day and the clock therefore highly influence the mRNA–ribosome associations (Supplemental Figure S14). Interesting similarities between our data and animal clocks were observed. We identified ∼36% of transcripts that are only rhythmic at the translatome level in our study, including a notable proportion even when comparing to several other publicly available data sets. This phenomenon is also conserved in mammalian and Neurospora clocks, where a significant proportion of the rhythmic translatome or proteome is thought to result from nonrhythmic mRNAs (Reddy et al., 2006; Robles et al., 2014; Mauvoisin et al., 2014; Hurley et al., 2018). Clock regulation of selected genes only at the translation level in both plant and animal systems, implies an intriguing possibility for the existence of a conserved circadian mechanism. However, experiments to dissect the functional relevance of this shared circadian discordance of gene regulation are needed. Furthermore, within the subset of circadian-regulated translatome-specific transcripts, cell cycle genes are overrepresented. In mammals, several important cell cycle genes are regulated in a circadian manner (Hunt and Sassone-Corsi, 2007; Sahar and Sassone-Corsi, 2009). These observations support the proposed hypothesis for the evolutionary conservation between the regulatory machinery that controls the clock and cell division, a feature that may be shared among eukaryotic clocks. We also found that the expression of several translation initiation factors is circadian regulated across all levels of gene regulation. Rhythmic expression of translation initiation factors has been documented in both Neurospora and mammals, further reinforcing that clock control of the translation regulatory mechanism is a conserved circadian property (Jouffe et al., 2013; Cao et al., 2013; Hurley et al., 2018; Karki et al., 2020).
Under heat stress, priorities of circadian and non-circadian mRNAs to bind ribosomes are highly perturbed (Figure 3). For circadian genes, the heat stress response was highly dependent on the time of the day, and this gating phenomenon was related to the transcript phase (Figure 5). Altering the clock also disturbed the temperature response of selected TFs (Figure 7). Together, we concluded that heat stress, daytime, the clock, and interactions between the different factors were therefore the main contributors to the changes in transcript–ribosome associations (Supplemental Figure S14). We postulate that this gating phenomenon is not restricted to circadian genes and would be observed for a high proportion of genes with diurnal or perhaps nonrhythmic expression patterns in natural conditions. The timing of recovery is also an important parameter to consider when trying to understand how the cell prioritizes mRNAs in cue to be translated. While priorities for translation rapidly recovered following heat stress, slower recovery profiles, and specific responses observed during the few hours succeeding the stress reflect another level in the hierarchy for translation (Figure 8). Analyzing how quickly (minutes or hours) the plants respond to heat stress, as previously performed at the transcriptome level, also provides important information to study the dynamics of the cellular response to stress (Li et al., 2019). In future experiments, simulating a heatwave happening during the day or night in natural conditions for Arabidopsis or other crops and analyzing the dynamics of the translatome response during and after the wave would be particularly useful to visualize how the timing and duration of translation of clock-controlled genes are adjusted (e.g. phase-shift phenomenon).
We identified highly responsive TFs, which to the best of our knowledge were not previously characterized to be involved in heat stress responses and linked these genes as key nodes in the regulation of plant thermotolerance (Figure 6). Similar to the well-described regulatory hubs DREBs and HSFs, the TFs identified here are also involved in other environmental stress responses, implicating these regulators as critical hubs to coordinate external signals with endogenous growth and developmental processes throughout the 24-h period (Ohama et al., 2017). For example, CDFs were initially characterized by their cycling expression and as important regulators of the photoperiod flowering pathway (Imaizumi et al., 2005; Fornara et al., 2009). Subsequently, CDFs have been shown to play a role in other growth, development, and metabolic processes, and abiotic stress responses in plants (Le Hir and Bellini, 2013; Fornara et al., 2015; Yanagisawa, 2016; Xu and Dai, 2016; Renau-Morata et al., 2017; Corrales et al., 2017). The Arabidopsis CDF3 is the best-characterized linking growth and plant thermotolerance in response to cold stress (Corrales et al., 2017). Of the six Arabidopsis CDFs (CDF1-6), CDF1, CDF2, and CDF5 are upregulated in response to heat stress at the transcriptome and translatome levels at all timepoints, CDF3 is upregulated only at night in both mRNA pools, while CDF4 and CDF6 are not differentially regulated. These observations suggest that while CDF3 is important for coordinating growth and cold responses, CDF1, CDF2, and CDF5 may be critical for the survival and tolerance to heat stress. Similarly, members of the MYB-like RVE TF subfamily, which encode RVE1-8 and are either central oscillator components or outputs of the clock, also show specific responses to heat stress (Gray et al., 2017). RVE4 and RVE8 have previously been shown to be important for plant heat tolerance and in a time-of-day-dependent context (Li et al., 2019). The rve4 rve8 double mutant shows decreased survival to heat stress when applied in the midmorning (Li et al., 2019). In our study, RVE1, RVE2, RVE4, RVE5, and RVE7 are significantly upregulated in total and TRAP mRNAs, with RVE7 showing the highest accumulation of transcripts at both levels (Supplemental Figure S11). For RVE8, we did not observe differential regulation or any significant change at the translatome level. We therefore speculate that the translation of other RVEs mRNAs is prioritized and may be more important for a longer duration (>30 min) of heat stress. Many of the TFs discussed in this study belong to large gene families. However, by analyzing the translational change for all members within a subfamily, we can detect subfamily-specific heat responses, as mentioned above for the CDF1-6 and eight RVE1-8 and observed for selected BBXs and Myb-like TRFLs (Karamysheva et al., 2004; Khanna et al., 2009). Furthermore, we propose that integrating a time dimension in regulatory stress networks may also be a powerful strategy to identify co-regulated and/or co-expressed genes acting in the same biological pathway.
In summary, the identification of orthologous TF nodes in crops, and characterization at the translatome and proteome levels in response to heat stress will help to comprehensively define the regulatory mechanisms of plant thermotolerance. Genetic manipulation of these TFs will help to guide studies aimed at balancing and improving beneficial growth-related traits in response to environmental stress.
Materials and methods
Plant materials and growth conditions
The previously reported Arabidopsis (Arabidopsis thaliana) Pro35S:HF-RPL18 line (Zanetti et al., 2005) was used as the wild-type (WT) in TRAP experiments. The cca1 lhy/Pro35S:HF-RPL18 was generated by crossing the cca1-1 lhy-20 double mutant into the Pro35S:HF-RPL18 background (Blair et al., 2019). Seeds were surface sterilized and were stratified at 4°C for 3 days in the dark. Seedlings were grown in square petri dishes on filter paper placed on Murashige and Skoog medium supplemented with 1.5% (w/v) sucrose. Plates were randomly arranged in the growth chamber and seedlings were grown for 10 days in 12-h white light (F17T8/TL841 Fluorescent Tube, ∼130 μE·m−2·s−1) and 12-h dark cycles at constant 22°C temperature. On day 11, plants were transferred to constant light and light intensity was reduced to 80–100 μE·m−2·s−1 to limit oxidative stress.
The time of the day is referred to as Zeitgeber time (ZT), i.e. ZT48 and ZT60, corresponding to the beginning of the light and subjective night periods on day 13, respectively. On day 13, a 1-h heat stress (37°C) that does not alter Arabidopsis development (Supplemental Figure S4) was applied to the seedlings (i.e. ZT47–ZT48), every 3 h (ZT48–ZT69) on a 24-h time course, on different sets of plants (Figure 3A). Control plants were maintained at 22°C (ZT48–ZT72).
To analyze the plant recovery, a 1 h heat stress was applied from ZT53 to ZT54 (Figure 8A). Stressed plants were transferred back to 22°C for recovery and control plants were maintained at 22°C. Seedlings were collected at ZT54, ZT55, ZT57, and ZT60, which correspond to 0 h, 1 h, 3 h, and 6 h of recovery, respectively. Three biological replicates were grown for each condition and time point. Replicates correspond to different plates, randomly arranged in the growth chamber. Control or stressed seedlings were snap frozen in liquid N2 and stored at −80°C until sample processing.
TRAP and mRNA isolation
To extract mRNAs associated with ribosomes, TRAP was performed as previously described (Reynoso et al., 2015). Briefly, one volume (∼1 mL) of frozen tissue powder was homogenized in five volumes of polysome extraction buffer (200-mM Tris–HCl, pH 9.0, 200-mM KCl, 25-mM EGTA–NaOH, pH 8.0, 35-mM MgCl2, 0.2% (w/v) polyoxyethylene(23)lauryl ether, 0.2% (v/v) Triton X-100, 0.2% (v/v) octylphenyl–polyethylene glycol, 0.2% (v/v) polyoxyethylene sorbitan monolaurate 20, 1% (v/v) polyoxyethylene (10) tridecyl ether, 1-mM DTT, 1-mM PMSF, 178-µM cycloheximide, 309-µM chloramphenicol) using a glass homogenizer. After centrifugation (16,000g for 15 min at 4°C), the supernatant was filtered through one layer of Miracloth. A small volume (200 µL) was conserved for the total mRNA extraction and the remaining extract was used for polysome immunoprecipitation. Dynabeads Protein G (50 µL, Thermo Fisher Scientific) were washed with a bead wash binding buffer (BB; 200-mM Tris–HCl, pH 9.0, 200-mM KCl, 25-mM EGTA–NaOH, pH 8.0, 35-mM MgCl2, 0.2% (v/v) polyoxyethylene sorbitan monolaurate 20) and incubated for 1 h with 5 µg of monoclonal ANTI-FLAG® M2 antibody (F1804, 1 mg·mL−1, MilliporeSigma). Dynabeads Protein G coupled with antibody were consecutively washed with BB and with wash buffer (WB; 200-mM Tris–HCl, pH 9.0, 200-mM KCl, 25-mM EGTA–NaOH, pH 8.0, 35-mM MgCl2, 1-mM DTT, 1-mM PMSF, 178-µM cycloheximide, 309-µM chloramphenicol) and then incubated with the filtered extract for 2 h at 4°C for polysome immunoprecipitation. Beads were washed six times with WB at 4°C. Each washing step consisted of the bead capture on a magnetic rack, the elimination of the supernatant and the bead incubation in the buffer. Immunoprecipitated polysomes coupled with protein G Dynabeads, and filtered extracts conserved for total mRNA isolation were stored at −80°C in 800 µL of lysis binding buffer (100-mM Tris–HCl, pH 8.0, 1 M LiCl, 10-mM EDTA–NaOH, pH 8.0, 1% (v/v) SDS, 5-mM DTT, 1.5% (v/v) Antifoam A) until mRNA isolation.
Total and TRAP mRNAs were isolated using biotinylated oligo(dT) and streptavidin magnetic beads (New England Biolabs) as previously described (Townsley et al., 2015). Purified mRNAs were either used for library preparation or RT-qPCR.
Library preparation and RNA-Seq
Libraries were prepared as previously described (Townsley et al., 2015) with the modifications previously detailed (Blair et al., 2019). In the final enrichment step, indexed adapter enrichment primers were used (Townsley et al., 2015) and 12 cycles were performed to amplify the libraries. The libraries were quantified using a Qubit 2.0 Fluorescence Reader (Thermo Fisher Scientific) and quality was verified using a Bioanalyzer 2100 (Agilent Genomics). Final libraries were multiplexed and sequenced on the NextSeq 500 (Illumina) at the University of California, Riverside (UCR) Institute for Integrated Genome Biology (IIGB) Genomics Core facility to obtain 75 nt single-end reads.
RNA-Seq data processing
RNA-Seq data analyses were performed on the UCR IIGB high-performance bioinformatics cluster (https://hpcc.ucr.edu/). Trimming of indexed adapter primers was performed on raw reads using cutadapt and quality reports of raw reads were generated with the FastQC package (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All other steps were conducted by following the SystemPipeR workflow (Backman and Girke, 2016). Trimmed reads were mapped to the TAIR10 Arabidopsis genome using the alignment program HISAT2 v2.2.0 (Kim et al., 2015). The number of sequenced and aligned reads per sample is provided in Supplemental Data Set S10. Read counting was realized with the summarizeOverlaps function from the GenomicRanges package (Lawrence et al., 2013) and using the Araport11 gff (201606). For visualization of expression data, rlog (regularized-logarithm) transformed expression values were generated with the DESeq2 package (Love et al., 2014). Expression data are provided in Supplemental Data Set S1 (time of the day data) and Supplemental Data Set S8 (recovery data).
RT-qPCR
Total and TRAP mRNAs were isolated from Pro35S:HF-RPL18 and cca1 lhy/Pro35S:HF-RPL18 seedlings using polyA mRNA extraction (Townsley et al., 2015). cDNA was obtained using iScript cDNA synthesis kit (Bio-Rad), and RT-qPCR was performed using SYBR Green Master Mix (Thermo Fisher Scientific) and the CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Primers used are provided in Supplemental Table S3. Protein Phosphatase 2A (PP2A) and isopentenyl-diphosphate delta-isomerase II (IPP2) were used as housekeeping genes. Two technical replicates and three biological replicates were analyzed. The mRNA expression levels relative to the housekeeping genes were calculated using the ΔΔCq method.
Analysis of circadian oscillations
Transcript levels (rlog normalized counts) quantified by RNA-Seq in total and TRAP mRNAs at 22°C and every 3 h during a 24 h time course were analyzed using Metacycle and the meta2d function (Wu et al., 2016). Only genes with a total number of raw counts >20 in the experiment were considered for this analysis (25,947 genes). Data are provided in Supplemental Data Set S2. A stringent cutoff (BH.Q < 0.01) has been applied to select transcripts exhibiting circadian oscillations (Supplemental Figure S1). However, to avoid the elimination of a high number of false negatives, the lists of transcripts with a 0.01 < BH.Q < 0.05 were compared to the lists of circadian transcripts identified in Hsu and Harmer (2012), Romanowski et al. (2020), and in DIURNAL data sets representing continuous light conditions (LL12_LDHH, LL_LLHC, LL_LDHC, LL23_LDHH; Supplemental Figure S1; Mockler et al., 2007). Genes overlapping with at least one of the mentioned data sets were selected (Supplemental Figure S1). Transcript phases calculated with the JTK_CYCLE program (Hughes et al., 2010) included in the Metacycle analysis were used for phase enrichment analyses.
Statistical analyses
All statistical analyses were performed with the statistical software program R v. 4.0.1 (R Core Team, 2020). For differential expression analysis, pairwise comparisons were performed to compare temperatures (37°C versus 22°C) at each time of the day, with the DESeq2 package (Love et al., 2014), using the SystemPipeR workflow (Backman and Girke, 2016). Results are provided in Supplemental Data Set S4 (time of the day data) and Supplemental Data Set S9 (recovery data). Transcripts with a P <0.05 after FDR correction using the Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995) and a log2 fold change (LFC, 37°C versus 22°C) > |1| were considered as differentially accumulated. To analyze interactions between the main effects of temperature and time of the day, and of temperature and mRNA population, a likelihood ratio test (LRT) was conducted using the DESeq2 package (Love et al., 2014). The LRT is conceptually similar to an analysis of variance (ANOVA) calculation in linear regression (Love et al., 2014). The full model was as follows: design = ∼ Temperature + mRNA + Time + Temperature: mRNA + Temperature: Time. Significant interaction effects were judged at FDR <0.05. Results are provided in Supplemental Data Set S4.
To compare the expression of CCA1 and LHY in control conditions in the Pro35S:HF-RPL18 and cca1 lhy/Pro35S:HF-RPL18 lines, unpaired Student’s t test were performed at each time of the day and statistical differences were considered to be significant at P < 0.05 (Supplemental Figure S12). To analyze the influence of the clock mutations on the temperature response of selected TFs in RT-qPCR data (Figure 7B), a multi-factor model including the effects of temperature, genotype, time of the day, mRNA population, and interactions between these factors was generated, and an ANOVA was performed. The model used was as follows: design= ∼ Temperature * Genotype * Time * mRNA. Significant effects were judged at P < 0.05 (Supplemental Table S2). Following this multivariate analysis, Tukey’s HSD tests were performed to compare group levels for the significant sources of variation. For example, for the significant interaction Temperature: Genotype revealed for CDF1, levels 22°C:WT, 22°C:cca1 lhy, 37°C:WT, and 37°C:cca1 lhy were compared and three significantly different groups were identified (A, B, and C; Figure 7, B and C).
To identify over- and underrepresented phases in the lists of circadian total and TRAP DRGs, proportions of phases in the lists of DRGs were compared to those in all circadian total and TRAP mRNAs, respectively (Figure 4;Supplemental Figure S8). χ2 tests were conducted and statistical differences were considered at P < 0.05. Results are provided in Supplemental Data Set S5. For gene ontology (GO) and TF family enrichment analyses, Fisher’s exact tests were used because of the small sample size for GO terms and TF families. GO enrichment analyses was performed using the interface of The Arabidopsis Information Resource for GO term enrichment for plants (https://www.arabidopsis.org/tools/go_term_enrichment.jsp), powered by Panther classification system (Mi et al., 2019). Biological processes significantly over- and underrepresented (Fisher’s exact test, P < 0.05 after FDR correction) in the lists of analyzed genes as compared to all Arabidopsis genes were selected (Supplemental Data Set S3). To identify overrepresented TF families in specific lists of DRGs (Supplemental Figure S9 and Supplemental Table S1), proportions or TF families in the lists of DRGs were compared to those in all Arabidopsis TFs previously described (Pruneda-Paz et al., 2014), and significant differences were judged at P < 0.05 (Supplemental Data Set S6).
Numbers of uORFs in circadian DRGs and groups of circadian DRGs having different responses at transcriptome and translatome levels were determined using the uORFlight database (Niu et al., 2020) and were compared using a Poisson linear regression and significant differences were considered at P < 0.05 (Supplemental Figure S6F).
Data mining and visualization
Heatmaps of circadian transcripts were generated using the R package “pheatmap” (Kolde, 2019). The Web server REVIGO (http://revigo.irb.hr/) was used to calculate similarity between enriched GO terms and to remove redundant terms (Supek et al., 2011). The selected enriched GO terms (Figures 1, F and 3, C) were then visualized as previously described (Bonnot et al., 2019). The bar plot depicting the overlapping transcripts between circadian data sets (Supplemental Figure S2C) was generated using the “UpSet” R package (Lex, 2014). A principal component analysis was performed on circadian transcripts differentially accumulated under heat stress (Figure 3D), using the multivariate data analysis R package “ade4” (Thioulouse et al., 1997). To define groups of circadian DRGs presented in Figure 8C, criteria used are described in Supplemental Table S4. Description of individual genes presented in the main and supplemental figures is provided in Supplemental Table S5.
For network analyses, TFs exhibiting circadian oscillations in TRAP mRNAs, differentially regulated under heat stress and identified as targets of proteins encoded by clock genes in published ChIP-Seq analyses were selected (Adams et al., 2018; Nagel et al., 2015; Liu et al., 2013, 2016; Kamioka et al., 2016; Ezer et al., 2017; Nakamichi et al., 2012; Huang et al., 2012). Networks were visualized using the CYTOSCAPE software v. 3.7.2 (Smoot et al., 2011), and nodes were manually sorted by TF family (Figure 6A) or based on their timing of peak of accumulation and their heat stress response (Figure 6B;Supplemental Data Set S11). All other plots were generated using the R package “ggplot2” (Wickham, 2016).
Data availability
All data reported in this manuscript are accessible from the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession no. GSE158444). R scripts and Cytoscape files are accessible from the following Github repository: https://github.com/Nagel-lab.
Accession numbers
Accession numbers of main discussed TFs are provided in Supplemental Data Set S11. Accession numbers and gene description of all circadian DRGs are provided in Supplemental Data Set S7.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Diagram showing the method of selection of circadian mRNAs.
Supplemental Figure S2. Comparison of Arabidopsis circadian transcriptomes and translatome.
Supplemental Figure S3. Overlap between the circadian transcriptome, translatome, a diel translatome and a rhythmic proteome.
Supplemental Figure S4. Effect of heat stress on Arabidopsis seedlings.
Supplemental Figure S5. Validation of clock gene transcript profiles by RT-qPCR.
Supplemental Figure S6. Differential heat stress response between the Arabidopsis transcriptome and translatome.
Supplemental Figure S7. Timing of response to heat of circadian transcripts relative to their peak of abundance.
Supplemental Figure S8. Timing effect of heat stress on the circadian transcriptome.
Supplemental Figure S9. Overrepresented TF families in circadian transcripts differentially accumulated at all times of the day.
Supplemental Figure S10. Time of the day gates the heat stress response of circadian TRAP DRGs especially during the day.
Supplemental Figure S11. Transcript profiles of selected circadian TFs with high priority to access ribosomes under heat.
Supplemental Figure S12. Alterations of CCA1 and LHY expressions in cca1 lhy/TRAP seedlings.
Supplemental Figure S13. Recovery of clock genes following heat stress.
Supplemental Figure S14 . Model of the main factors influencing mRNA-ribosome associations.
Supplemental Table S1. Enriched biological processes and transcription factor families in groups of circadian TRAP DRGs.
Supplemental Table S2. Multivariate analysis of variance of transcript abundance of selected circadian TFs.
Supplemental Table S3. Primer sequences used in RT-qPCR analyses.
Supplemental Table S4. Criteria used to group circadian TRAP DRGs during recovery.
Supplemental Table S5. Identification number and description of genes highlighted in main and supplemental figures.
The following Supplemental Data Sets were submitted to the Data Dryad Repository and are available at http://datadryad.org/resource/doi:10.6086/D1B69N
Supplemental Data Set S1. Arabidopsis transcript levels during the 24 h time course.
Supplemental Data Set S2. Metacycle results.
Supplemental Data Set S3. Gene ontology (GO) enrichment analysis.
Supplemental Data Set S4. Differential expression analysis.
Supplemental Data Set S5. Phase enrichment analysis.
Supplemental Data Set S6. Transcription factor (TF) family enrichment analysis.
Supplemental Data Set S7. Annotation and grouping of circadian DRGs.
Supplemental Data Set S8. Arabidopsis transcript levels during the plant heat stress recovery.
Supplemental Data Set S9. Differential expression analysis: pairwise comparisons during the plant recovery.
Supplemental Data Set S10. Number of sequenced and aligned reads per sample.
Supplemental Data Set S11. Interactions between clock proteins and circadian TFs translationally regulated by heat stress.
Supplementary Material
Acknowledgments
We thank Dr Morgane B. Gillard for helping with statistical analyses. We thank Dr Carolyn Rasmussen and Dr Xuemei Chen for critical reading of the manuscript. We thank Emily Blair for helping with data processing, and members of Dr Julia Bailey-Serres’ lab for providing the Pro35:HF-RPL18 line and for helpful and critical discussions.
Funding
This work was supported by NSF Early Career Award IOS 1942949 to D.H.N.
Conflict of interest statement: The authors declare no competing interests.
D.H.N. conceived the project, T.B. and D.H.N. designed and performed experiments, analyzed the data, and wrote the manuscript.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is Dawn H. Nagel (dawnn@ucr.edu).
References
- Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S, Carré IA (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol 220:893–907 [DOI] [PubMed] [Google Scholar]
- Backman TWH, Girke T (2016) systemPipeR: NGS workflow and report generation environment. BMC Bioinform 17:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Howell SH (2017) The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front Plant Sci 8:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300 [Google Scholar]
- Blair EJ, Bonnot T, Hummel M, Hay E, Marzolino JM, Quijada IA, Nagel DH (2019) Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci Rep 9:4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnot T, Gillard M, Nagel D (2019) A simple protocol for informative visualization of enriched gene ontology terms. Bio-101: e3429 [Google Scholar]
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, et al. (2013) Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron 79:712–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantarachot T, Bailey-Serres J (2018) Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol 176:254–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales A, Carrillo L, Lasierra P, Nebauer SG, Dominguez‐Figueroa J, Renau‐Morata B, Pollmann S, Granell A, Molina R, Vicente‐Carbajosa J, et al. (2017). Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ 40:748–764 [DOI] [PubMed] [Google Scholar]
- Crisp PA, Ganguly DR, Smith AB, Murray KD, Estavillo GM, Searle I, Ford E, Bogdanović O, Lister R, Borevitz JO, et al. (2017). Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29:1836–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309:630–633 [DOI] [PubMed] [Google Scholar]
- Ezer D, Jung J-H, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, et al. (2017). The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3:17087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Cumbie JS, Dharmawardhana P, Jaiswal P, Chang JH, Palusa SG, Reddy ASN, Megraw M, Mockler TC (2015) Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol Plant 8:207–227 [DOI] [PubMed] [Google Scholar]
- Forestan C, Farinati S, Zambelli F, Pavesi G, Rossi V, Varotto S (2020) Epigenetic signatures of stress adaptation and flowering regulation in response to extended drought and recovery in Zea mays. Plant Cell Environ 43:55–75 [DOI] [PubMed] [Google Scholar]
- Fornara F, Montaigu A, Sánchez‐Villarreal A, Takahashi Y, Ver Loren van Themaat E, Huettel B, Davis SJ, Coupland G (2015) The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81:695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17:75–86 [DOI] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137:961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil K-E, Kim W-Y, Lee H-J, Faisal M, Saquib Q, Alatar AA, Park C-M (2017) ZEITLUPE contributes to a thermoresponsive protein quality control system in Arabidopsis. Plant Cell 29:2882–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Shalit-Kaneh A, Chu DN, Hsu PY, Harmer SL (2017) The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol 173:2308–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AAR (2007) Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 30:333–349 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL (2012) Circadian phase has profound effects on differential expression analysis. PLoS One 7:e49853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336:75–79 [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25:372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Sassone-Corsi P (2007) Riding tandem: circadian clocks and the cell cycle. Cell 129:461–464 [DOI] [PubMed] [Google Scholar]
- Hurley JM, Jankowski MS, De Los Santos H, Crowell AM, Fordyce SB, Zucker JD, Kumar N, Purvine SO, Robinson EW, Shukla A, et al. (2018) Circadian proteomic analysis uncovers mechanisms of post-transcriptional regulation in metabolic pathways. Cell Syst 7:613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293–297 [DOI] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F (2013) The circadian clock coordinates ribosome biogenesis. PLoS Biol 11:e1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28:696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva ZN, Surovtseva YV, Vespa L, Shakirov EV, Shippen DE (2004) A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J Biol Chem 279:47799–47807 [DOI] [PubMed] [Google Scholar]
- Karki S, Castillo K, Ding Z, Kerr O, Lamb TM, Wu C, Sachs MS, Bell-Pedersen D (2020) Circadian clock control of eIF2α phosphorylation is necessary for rhythmic translation initiation. Proc Natl Acad Sci 117:10935–10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu S-H (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21:3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R (2019) pheatmap: pretty heatmaps.
- Krahmer J, Hindle M, Perby LK, Nielsen TH, VanOoijen G, Halliday KJ, Le Bihan T, Millar AJ (2019) Circadian protein regulation in the green lineage II. The clock gene circuit controls a phospho-dawn in Arabidopsis thaliana. bioRxiv 760892. [Google Scholar]
- Kwon Y-J, Park M-J, Kim S-G, Baldwin IT, Park C-M (2014) Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol 14:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir R, Bellini C (2013) The plant-specific Dof transcription factors family: new players involved in vascular system development and functioning in Arabidopsis. Front Plant Sci 4: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H (2014) UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 20: 1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gao Z, Liu X, Sun D, Tang W (2019) Transcriptional profiling reveals a time-of-day-specific role of REVEILLE 4/8 in regulating the first wave of heat shock-induced gene expression in Arabidopsis. Plant Cell 31:2353–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM (2013) Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J 76:101–114 [DOI] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu M-J, Shiu S-H, Farré EM (2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170:528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F (2014) Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci 111:167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R. (2006). Plant circadian rhythms. Plant Cell 18: 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merret R, Carpentier M-C, Favory J-J, Picart C, Descombin J, Bousquet-Antonelli C, Tillard P, Lejay L, Deragon J-M, Charng Y (2017) Heat shock protein HSP101 affects the release of ribosomal protein mRNAs for recovery after heat shock. Plant Physiol 174:1216–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merret R, Nagarajan VK, Carpentier M-C, Park S, Favory J-J, Descombin J, Picart C, Charng Y, Green PJ, Deragon J-M, et al. (2015) Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Res 43:4121–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A, Ernest B, Lohoff T, Jia Q, Satterlee J, Ke K, von Arnim AG (2015) The circadian clock modulates global daily cycles of mRNA ribosome loading. Plant Cell 27:2582–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The diurnal project: diurnal and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72:353–363 [DOI] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci 112:E4802–E4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Kay SA (2012) Complexity in the wiring and regulation of plant circadian networks. Curr Biol 22:R648–R657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109:17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu R, Zhou Y, Zhang Y, Mou R, Tang Z, Wang Z, Zhou G, Guo S, Yuan M, Xu G (2020) uORFlight: a vehicle toward uORF-mediated translational regulation mechanisms in eukaryotes. Database 2020:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22:53–65 [DOI] [PubMed] [Google Scholar]
- Patole C, Bindschedler LV (2019) Plant proteomics. In Meena SN and Naik MM, eds, Advances in Biological Science Research. Elsevier, New York, pp 45–67 [Google Scholar]
- Pruneda-Paz JL, Breton G, Nagel DH, Kang SE, Bonaldi K, Doherty CJ, Ravelo S, Galli M, Ecker JR, Kay SA (2014) A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep 8:622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020) R: a language and environment for statistical computing.
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GKY, Chesham J, Odell M, Lilley KS, et al. (2006) Circadian orchestration of the hepatic proteome. Curr Biol 16:1107–1115 [DOI] [PubMed] [Google Scholar]
- Renau-Morata B, Molina RV, Carrillo L, Cebolla-Cornejo J, Sánchez-Perales M, Pollmann S, Domínguez-Figueroa J, Corrales AR, Flexas J, Vicente-Carbajosa J, et al. (2017) Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front Plant Sci 8:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso MA, Juntawong P, Lancia M, Blanco FA, Bailey-Serres J, Zanetti ME (2015) Translating ribosome affinity purification (TRAP) followed by RNA sequencing technology (TRAP-SEQ) for quantitative assessment of plant translatomes. Methods Mol Biol 1284:185–207 [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40:253–266 [DOI] [PubMed] [Google Scholar]
- Robles MS, Cox J, Mann M (2014) In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet 10:e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski A, Schlaen RG, Perez‐Santangelo S, Mancini E, Yanovsky MJ (2020) Global transcriptome analysis reveals circadian control of splicing events in Arabidopsis thaliana. Plant J 103:889–902 [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P (2009) Metabolism and cancer: the circadian clock connection. Nat Rev Cancer 9:886–896 [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Miao C, Hanel M, Borthwick AGL, Duan Q, Ji D, Li H (2019) Global heat stress on health, wildfires, and agricultural crops under different levels of climate warming. Environ Int 128:125–136 [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD (2015) Regulation of mRNA translation during mitosis. eLife 4:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thioulouse J, Chessel D, Doledec S, Olivier JM (1997) ADE-4: a multivariate analysis and graphical display software. Stat Comput 7:75–83 [Google Scholar]
- Townsley BT, Covington MF, Ichihashi Y, Zumstein K, Sinha NR (2015) BrAD-seq: breath adapter directional sequencing: a streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front Plant Sci 6:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: elegant graphics for data analysis
- Wilkins O, Bräutigam K, Campbell MM (2010) Time of day shapes Arabidopsis drought transcriptomes. Plant J 63:715–727 [DOI] [PubMed] [Google Scholar]
- Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB (2016) MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32:3351–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Dai H (2016) Brassica napus Cycling Dof Factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul 80:315–322 [Google Scholar]
- Yanagisawa S (2016) Structure, function, and evolution of the Dof transcription factor family. In Gonzalez DH, ed, Plant Transcription Factors. Elsevier, New York, pp 183–197 [Google Scholar]
- Yángüez E, Castro-Sanz AB, Fernández-Bautista N, Oliveros JC, Castellano MM (2013) Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS One 8:e71425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Chang I-F, Gong F, Galbraith DW, Bailey-Serres J (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138:624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu X, Gaikwad K, Kou X, Wang F, Tian X, Xin M, Ni Z, Sun Q, Peng H, et al. (2017) Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana. Plant Cell 29:1952–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this manuscript are accessible from the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession no. GSE158444). R scripts and Cytoscape files are accessible from the following Github repository: https://github.com/Nagel-lab.