Abstract
Large numbers of mink have been infected with SARS-CoV2 containing the spike protein Y453F mutation in Europe, causing zoonosis concerns. To evaluate the genetic characteristics of the U.S. and Canadian mink–derived SARS-CoV2 sequences, we analyzed all animal-derived (977) and all Canadian (19,529) and U.S. (173,277) SARS-CoV2 sequences deposited in GISAID from December 2019 to March 12, 2021, and identified 2 dominant novel variants, the N501T-G142D variant and N501T-G142D-F486L variant, in the U.S. mink–derived SARS-CoV2 sequences. These variants were not found in mink from Canada or other countries. The Y453F mutation was not identified in the mink-derived sequences in the United States and Canada. The N501T mutation occurred 2 mo earlier in humans than in mink in the United States, and the novel N501T-G142D and N501T-G142D-F486L variants were found in humans prior to mink. Our results suggest that the novel SARS-CoV2 variants may have evolved during human infection and were then transmitted to mink populations in the United States.
Keywords: COVID-19, mink, N501T, N501Y, SARS-CoV2, spike protein, variant
Genomic surveillance of circulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) variants is critical for epidemiologic tracking and infection control. Rapid global spread of a variant harboring a mutation located on the SARS-CoV2 spike protein (S protein) has been reported.11,15 In the GISAID (Global Initiative on Sharing All Influenza Data, https://www.gisaid.org) database of SARS-CoV2 genomes as of March 17, 2021, 758,684 of 792,388 (95.7%) of the SARS-CoV2 genome had the D614G mutation in the S protein gene; and 178,186 of 792,388 (22%) had the N501Y mutation, which occurred in the more virulent U.K. variant lineage B.1.1.7, with transmissibility increased by 50% and potentially more virulence;2 a similar phenomenon was observed with the South African lineage B.1.35116 and Brazilian lineage P.1 (B.1.1.28.1) variants,7,17 both of which have the N501Y mutation. Large numbers of mutants with Y453F mutations were found in infected mink in Denmark, which may have spread from human to animal and back to humans.8,10 We analyzed the sequences of the SARS-CoV2 S protein gene collected from mink and identified 2 dominant novel variants with 2 and 3 mutations each in Michigan (MI) and Wisconsin (WI) mink–derived sequences.
From the GISAID, we downloaded the following SARS-CoV2 sequences collected from December 2019 to the date of writing (March 12, 2021): all animal-derived sequences (977), including 916 Neovison vison (mink)-derived sequences from WI (47 sequences), MI (54), Poland (14), Netherlands (333), Lithuania (7), Denmark (458), and France (1), plus all sequences collected in Canada (19,529) and the United States (173,277). The SARS-CoV2 isolate Wuhan-Hu-1 collected on December 19, 2019 (GenBank NC045512) was used as a reference for mutation analysis; all nucleotide position labeling in our study was based on alignment with this sequence. SARS-CoV2 full genome sequences were multiple aligned with Geneious v.11 (Biomatters) using “Geneious Multiple Alignment” and “Map to a Reference Assembly” functions. The aligned sequences were examined visually to confirm that they were aligned properly. The mutations were identified by the software automatically and verified by visual confirmation. Short fragments (30 nt) containing the novel mutations identified in our study were used as queries to BLAST search the local databases consisting of the downloaded sequences to determine the incidence of the novel mutations using the “BLAST” function of the Geneious software. The global incidence of “N501Y” was determined using the “search” function with substitution “N501Y” in GISAID. The global incidence of “N501T” was not determined because the query of substitution “N501T” was not available in GISAID.
The N501Y mutation that was common to U.K., South African, and Brazilian variants, and the Y453F mutation that widely prevailed in COVID-19–positive mink in Denmark and the Netherlands were absent in all U.S. and Canadian mink SARS-CoV2 sequences examined. Interestingly, an N501T mutation was found in almost all U.S. mink SARS-CoV2 sequences (100 of 101; 99%); furthermore, all N501T mutant sequences, except 2 of poor quality, had the G142D mutation (Suppl. Table 1). Based on GISAID records, all U.S. mink SARS-CoV2 sequences were collected from MI and WI in October 2020. We found that, except for 1 sequence with poor quality, all MI mink sequences had the F486L mutation and the Y144 deletion, in addition to the N501T and G142D mutations (Suppl. Table 1). Hereafter, we designated the SARS-CoV2 with N501T and G142D, but without F486L, as the N501T-G142D variant (WI mink variant), and the ones with all 3 novel mutations as the N501T-G142D-F486L variant (MI mink variant). In addition to the above 3 mutations and 1 deletion in S protein, G614D, the mutation that dominated the SARS-CoV2 sequences, existed in all U.S. and Canadian mink–derived sequences (Suppl. Tables 1, 2). No N501T-G142D or N501T-G142D-F486L variants were found in mink and other animals from Canada or other countries (Table 1).
Table 1.
Novel spike protein mutations identified in mink-derived and related human-derived sequences deposited in GISAID by March 12, 2021.
| Spike mutation variant | Incidence | Collection date |
|---|---|---|
| MI mink D614G-N501T-G142D-F486L (T23047G)-144delY | 53 of 54 | Oct 13–18, 2020 |
| WI mink D614G-N501T-G142D | 45 of 47 | Oct 13–15, 2020 |
| US (MI only) human D614G-N501T-G142D-F486L (T23047G) | 2 of 173,277 | Oct 6, 2020 |
| US (MI) human D614G-N501T-F486L (T23047G)-144-5delYY* | 4 of 8,369 | Nov 29–Dec 29, 2020 |
| US (WI only) human D614G-N501T-G142D | 3 of 173,277 | Oct 3, 2020 |
| US human D614G-N501T | 1,339 of 173,277 | Since Aug 4, 2020 |
| US OR mink D614G* | 4 of 4 | Nov 24, 2020 |
| Canada mink D614G | 4 of 4 | Dec 4, 2020 |
| NL mink D614G-F486L (T23045C) | 204 of 333 | Since Apr 2020 |
| NL and DK mink D614G-N501T | 7 of 791 | Since Apr 2020 |
DK = Denmark; MI = Michigan; NL = Netherlands; OR = Oregon; WI = Wisconsin.
Deposited in GISAID in April 2021.
Outside the United States, among 335 Netherlands mink–derived SARS-CoV2 S protein gene sequences, 5 collected from April–June 2020 had the N501T mutation without G142D or F486L mutations (Suppl. Table 3), which is consistent with a previous report on N501T in Netherlands mink–derived sequences.1,13 We also found the F486L mutation in 204 of 335 (60%) Netherlands mink–derived SARS-CoV2 S protein gene sequences, none of which were N501T-G142D variants. The F486L mutation was unique to Netherlands mink–derived sequences caused by nucleotide mutation T23045C, different from the F486L mutation found in the U.S. mink–derived sequences as a result of the nucleotide T23047G mutation. Among 454 Danish mink SARS-CoV2 sequences, 3 had N501T mutations as described earlier,14 none had G142D or F486L mutations (Table 1, Suppl. Table 3).
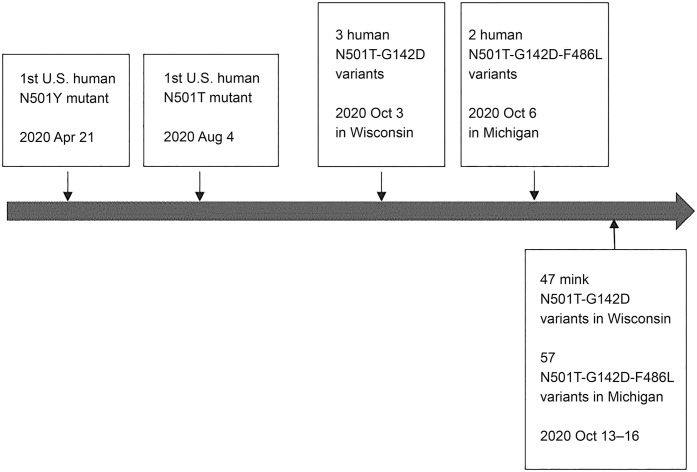
To determine the origin of the 2 novel variants, we analyzed all human-derived SARS-CoV2 sequences collected from the United States and Canada. Three WI N501T-G142D variant and 2 MI N501T-G142D-F486L variant sequences were identified only from the sequences collected from WI on October 3, 2020 and Michigan on October 6, and the sequences were identical to WI and MI mink variants, respectively, except that the MI human variants lacked the Y144 deletion in the S protein. Furthermore, we identified N501T mutants without G142D and F486L in U.S. human–derived sequences collected as early as August 2020, which was earlier than the mink-derived sequences collected in October 2020 (Suppl. Table 4, Fig. 1). In Canada, the N501T mutation only existed in human-derived sequences (Suppl. Table 2). The N501T mutation was described in 2 Italian human–derived sequences collected in August 2020.3 We therefore speculate that the N501T-G142D and N501T-F486L-G142D mutations may have evolved during human infection and were then transmitted to mink populations in the United States, and it is less likely that the mutations occurred in mink populations. The fact that the 2 novel variants were found in almost all infected mink in the United States within days also supports this speculation.
Figure 1.
Timeline of emergence of human-derived (above the timeline) and mink-derived (below the timeline) SARS-CoV2 spike protein N501T-G142D and N501T-G142D-F486L variants in the United States. Timeline scale is not proportional.
To evaluate the transmissibility of N501T mutants, the incidence of N501T and N501Y mutants was enumerated from U.S. and Canadian GISAID SARS-CoV2 sequences collected up to March 12, 2021: 4,097 of 173,277 (2.4%) and 1,339 of 173,277 (0.77%) of U.S. sequences had the N501Y and N501T maturation, respectively; and 315 of 19,529 (1.6%) and 12 of 19,529 (0.06%) of Canadian sequences had the N501Y and N501T mutation, respectively (Suppl. Table 5). Comparatively, the incidence of N501Y is much lower in the United States and Canada than globally. Our analysis suggested that the N501T mutants might have weaker transmissibility compared to the N501Y mutants, which include U.K. B.1.17, South Africa B.1.351, and Brazil P.1. However, it cannot be ruled out that the different time of introduction of N501Y and N501T (April 2020 vs. August 2020) may have affected the occurrences of the 2 mutants (Fig. 1). Further investigation of the transmissibility should be carried out with extensive epidemiologic studies or animal transmission studies.
It has been described that the mutations at S protein N501 of SARS-CoV2 can significantly increase the transmissibility, and likely the virulence; the U.K. variant N501Y is an example. It was predicted that a single N501T mutation might significantly enhance the binding affinity between 2019-nCoV (SARS-CoV2) receptor-binding domain (RBD) and human angiotensin-converting enzyme 2 (ACE2), and researchers were urged to closely monitor the emergence of the novel mutations at the 501 position.18 Multiple alignment of human and animal ACE2 and SARS-CoV2 protein sequences indicated that the hot spot amino acid 353 of ACE2, the binding site of SARS-CoV2 spike amino acid 501, is identical between humans and mink, and that the mink ACE2 G354H substitution could conserve the polar contacts between the mink ACE2 and SARS-CoV2 virus.4,5 In silico analysis indicated that T501 in SARS-CoV2 (T487 in SARS-CoV) provides extra support for hot spot 353 of human ACE2.18 It is likely that the N501T mutation in mink SARS-CoV2 provides the binding affinity to host ACE2 in a similar manner. It has been described that 486F of the SARS-CoV2 receptor binding motif (RBM) inserts into a hydrophobic pocket of M82 of ACE2 to further stabilize hotspot 31K,9 and the F486L mutation has the potential to change the RBD region protein structure.12 The actual effects of mink SARS-CoV2 spike protein mutations N501T, N501T-G142D, and N501T-G142D-F486L on virus-receptor interaction needs further investigation.
Our analysis revealed that the N501T and G142DL mutations occurred in 99% of mink-derived sequences collected in the United States; all, except one with poor quality, mink sequences collected from MI also contained the F486L mutation. The large number of new variants occurring in the mink population warrants further study of how these changes affected their interaction with the ACE2 receptor, and thereby the transmissibility, virulence, and immunogenicity in humans and mink. It is important to monitor the emerging new variants and determine their impact on human and animal health. During the revision of this manuscript, 4 more N501T-F486L-144-5del human–derived variant sequences collected in November 2020 from MI,6 and 4 D614G mink variant sequences (collected in November 2020 from Oregon) were deposited to GISAID in April 2021 (Table 1). These variants were not identical to the N501T-G142D and N501T-F486L-G142D variants revealed in our study.
Supplemental Material
Supplemental material, sj-pdf-1-jvd-10.1177_10406387211023481 for SARS-CoV2 spike protein gene variants with N501T and G142D mutation–dominated infections in mink in the United States by Hugh Y. Cai and Allison Cai in Journal of Veterinary Diagnostic Investigation
Supplemental material, sj-pdf-2-jvd-10.1177_10406387211023481 for SARS-CoV2 spike protein gene variants with N501T and G142D mutation–dominated infections in mink in the United States by Hugh Y. Cai and Allison Cai in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We gratefully acknowledge the authors and the originating and submitting laboratories of the sequences from the GISAID hCov-19 Database on which our research was based. See supplemental acknowledgment for details.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Hugh Y. Cai  https://orcid.org/0000-0002-1868-483X
https://orcid.org/0000-0002-1868-483X
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Hugh Y. Cai, Animal Health Laboratory, University of Guelph, Guelph, Ontario, Canada.
Allison Cai, Faculty of Science, University of British Colombia, BC, Canada.
References
- 1.Elaswad A, et al. Mutational spectra of SARS-CoV-2 isolated from animals. PeerJ 2020;8:e10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control. Risk assessment: risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA—first update, 2021Jan 21. https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update
- 3.Fiorentini S, et al. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. Lancet Infect Dis 2021;6:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi T, et al. Highly conserved binding region of ACE2 as a receptor for SARS-CoV-2 between humans and mammals. Vet Q 2020;40:243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan S, et al. SARS-CoV-2 infection in farmed minks, associated zoonotic concerns, and importance of the One Health approach during the ongoing COVID-19 pandemic. Vet Q 2021;41:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kravitz D, et al. CDC: Michigan taxidermist may have caught COVID-19 from infected mink. Detroit Free Press. 2021Apr29. https://www.freep.com/story/news/health/2021/04/29/mink-michigan-covid-coronavirus/7400913002/ [Google Scholar]
- 7.Kupferschmidt K.New mutations raise specter of ‘immune escape’. Science 2021;371:329–330. [DOI] [PubMed] [Google Scholar]
- 8.Larsen HD, et al. Preliminary report of an outbreak of SARS-CoV-2 in mink and mink farmers associated with community spread, Denmark, June to November 2020. Euro Surveill 2021;26:2100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc Natl Acad Sci U S A 2021;118:e2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munnink BBO, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021;371:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Na W, et al. A comprehensive review of SARS-CoV-2 genetic mutations and lessons from animal coronavirus recombination in one health perspective. J Microbiol 2021;59:332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen TT, et al. Genomic mutations and changes in protein secondary structure and solvent accessibility of SARS-CoV-2 (COVID-19 virus). Sci Rep 2021;11:3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oreshkova N, et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill 2020;25: 2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira F.SARS-CoV-2 variants lacking ORF8 occurred in farmed mink and pangolin. Gene 2021;784:145596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phan T.Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol 2020;81:104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tegally H, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. Preprint. doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 17.Voloch CM, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol 2021;95:JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan Y, et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 2020;94:e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jvd-10.1177_10406387211023481 for SARS-CoV2 spike protein gene variants with N501T and G142D mutation–dominated infections in mink in the United States by Hugh Y. Cai and Allison Cai in Journal of Veterinary Diagnostic Investigation
Supplemental material, sj-pdf-2-jvd-10.1177_10406387211023481 for SARS-CoV2 spike protein gene variants with N501T and G142D mutation–dominated infections in mink in the United States by Hugh Y. Cai and Allison Cai in Journal of Veterinary Diagnostic Investigation