Abstract
Felis catus gammaherpesvirus 1 (FcaGHV1) is a newly described virus that infects domestic cats. To identify FcaGHV1 antigens, we developed an immunofluorescent antibody assay by expressing FcaGHV1 open reading frames (ORFs) in feline cells and incubating fixed cells with sera from FcaGHV1-positive cats. Of the seven ORFs tested, ORF52 and ORF38 had the strongest, most consistent antibody responses. We used recombinant ORF52 and ORF38 proteins to develop two FcaGHV1 ELISAs. These assays were used to detect reactivity in cats previously tested by qPCR for FcaGHV1 in blood cell DNA. Results indicated 32% FcaGHV1 seroprevalence, compared to 15% qPCR-evaluated prevalence (n=133); all but one qPCR positive animal was seropositive. ELISA results confirmed infection risk factors previously identified by qPCR: geographic location, male sex, and adult age. These data suggest that FcaGHV1 is a common infection of domestic cats that has a seropositive but often qPCR negative state characteristic of herpesviral latency.
Keywords: ELISA, herpesvirus, antibody, antigen, virus, ORF52, ORF38, prevalence, risk factor, feline
Introduction
Viruses of the family Herpesviridae have DNA genomes, establish life-long persistent infection in their hosts, and are divided into three subfamilies: Alpha, Beta, and Gammaherpesvirinae (ICTV, 2014). Gammaherpesvirus (GHV) infection is typically characterized by a brief acute replication phase followed by an extended period of viral latency in which the viral genome persists within cells with little or no viral gene expression (Barton et al., 2011). In response to specific signals, viral gene expression can be reactivated resulting in production of infectious progeny virus (Speck and Ganem, 2010). Many animal species are infected with one or more GHVs and these viruses are in many cases associated with lymphoproliferative and other disease conditions (Ackermann, 2006; Cesarman, 2011). Felis catus gammaherpesvirus 1 (FcaGHV1) was recently identified in the domestic cat (Troyer et al., 2014) and likely has a worldwide distribution (Beatty et al., 2014; Ertl et al., 2015). We developed a real-time quantitative PCR (qPCR) assay that detected FcaGHV1 in 16% of US domestic cats (Troyer et al., 2014) and similar prevalence (10–20%) in Australia, Asia, and Europe (Beatty et al., 2014; Ertl et al., 2015). Results revealed several risk factors for FcaGHV1, including being male, adult, and being infected with other pathogens (Beatty et al., 2014; Troyer et al., 2014). Feline immunodeficiency virus (FIV) infection was a particularly strong risk factor for FcaGHV1; and FIV positive cats had significantly higher FcaGHV1 viral loads than FIV negative cats, suggesting potential reactivation of FcaGHV1 due to immune suppression (Beatty et al., 2014).
Host immunity plays an important role in control of GHV infections and GHVs limit this control through multiple mechanisms of immune evasion(Means et al., 2007). Despite this ability to evade immunity, GHV infections typically stimulate persistent antibody responses against a diverse repertoire of viral antigens (Bartley et al., 2014; Katano et al., 2000; Labo et al., 2014; Zheng et al., 2011). Serologic assays for detection of these responses are crucial for identifying infected individuals, particularly during latency when GHV nucleic acids may be at undetectably low levels (Morrison et al., 2015). Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) are human GHVs which pose important health risks in immunocompromised individuals (Cesarman, 2011; Ganem, 2006; Speck and Ganem, 2010) and their seroprevalence has been well characterized. EBV has a fairly uniform seroprevalence of about 95% (Jenson, 2011) while KSHV has a lower seroprevalence with regional geographic variation from 5–50% (Dow et al., 2014). Both KSHV and EBV trigger immune responses to multiple viral antigens with certain antigens being more immunogenic than others (Katano et al., 2000; Zheng et al., 2011). FcaGHV1 infects domestic cats worldwide (Beatty et al., 2014; Ertl et al., 2015), yet the immune response to this agent has not been characterized. Study of FcaGHV1 is relevant not only for the health of domestic cats, but also to provide additional details of GHV pathobiology.
We hypothesized that the measure of prevalence from our qPCR assay underestimates exposure to FcaGHV1 by not accounting for animals with low-level latent infection. We consequently designed an ELISA to assess humoral antibody status in order to indicate exposure in the absence of detectable viral DNA. We developed recombinant proteins to several FcaGHV1 antigens (Table 1) and screened them for seroreactivity to serum from cats with high qPCR loads. We selected two antigens to develop indirect ELISAs for screening sera from over 100 animals from three geographic regions with known FcaGHV1 qPCR status. Results were compared to estimate sensitivity and specificity of the ELISA and risk factors for seropositivity were assessed.
Table 1.
FcaGHV1 protein homologs are antigenic in other gammaherpesviruses.
| Gene | Type of protein | Antigen of | Possible function |
|---|---|---|---|
|
| |||
| ORF17.5 | Scaffold protein | OvHV2, AlHV11 | Involved in capsid assembly2 |
| ORF26 | Capsid | EBV (BDLF1)3 | Capsid protein4, late gene5 |
| ORF38 | Tegument | KSHV6 | Support virion maturation in the cytoplasm7 |
| ORF42 | Tegument | EBV (BBRF2)8 | May contribute to the regulation of mitochondrial function9 |
| ORF52 | Tegument | EBV (BLRF2)10,11 | Assist virion egress and secondary envelopment10 |
| ORF59 | Phosphoprotein | KSHV12 | Binds DNA polymerase and dsDNA to promote DNA synthesis by acting as a sliding clamp13 |
| ORF65 | Capsid | EBV (BFRF3)11, KSHV12, 14, OvHV2, AlHV11 | Small capsid protein4, late gene5 |
Ovine herpesvirus 2 (OvHV2) and Alcelaphine herpesvirus 1 (AlHV1) are malignant catarrhal fever-causing viruses. EBV has distinct gene nomenclature listed in parentheses.
Citations:
(Schulz, 2000).
RESULTS
Identification of FcaGHV1 antigens
To our knowledge, FcaGHV1 has not yet been propagated in culture despite several attempts (R. Troyer, unpublished data). Thus FcaGHV1 antibody detection required recombinant expression of viral antigens. We selected seven FcaGHV1 genes that code for potential antigens from the FcaGHV1 genome (Troyer et al., 2015) based on homology to antigens of KSHV, EBV, and malignant catarrhal fever-causing viruses: Ovine herpesvirus 2 (OvHV2), and Alcelaphine herpesvirus 1 (AlHV1) (Table 1). We cloned these genes into mammalian expression vector pKH3 with a 5’ (N-terminal) HA tag sequence. We also cloned the antigenic FIV capsid protein (FIVCA) into pKH3 for use as a positive control in conjunction with FIV-positive cat serum. To determine which FcaGHV1 proteins elicited IgG antibody during a natural infection, we developed an immunofluorescence assay to screen and evaluate the seven recombinant FcaGHV1 proteins expressed in feline CRFK cells. After incubation with anti-HA antibodies, we were able to visualize immunofluorescent cells indicative of recombinant protein expression for all seven FcaGHV1 proteins (Fig. 1). ORF59, a phosphoprotein that assists DNA polymerase (Massimelli et al., 2015), was the only protein that localized exclusively to the nucleus. All other proteins had visible fluorescence in the cytoplasm (Fig. 1). Serum from negative-control specific pathogen-free (SPF) cats was uniformly negative for immunofluorescence on all proteins tested (Fig. 1). Serum from FIV-positive cats consistently produced strong immunofluorescence on cells expressing FIV capsid.
Figure 1. IFA detects FcaGHV1 antibodies in infected cats.
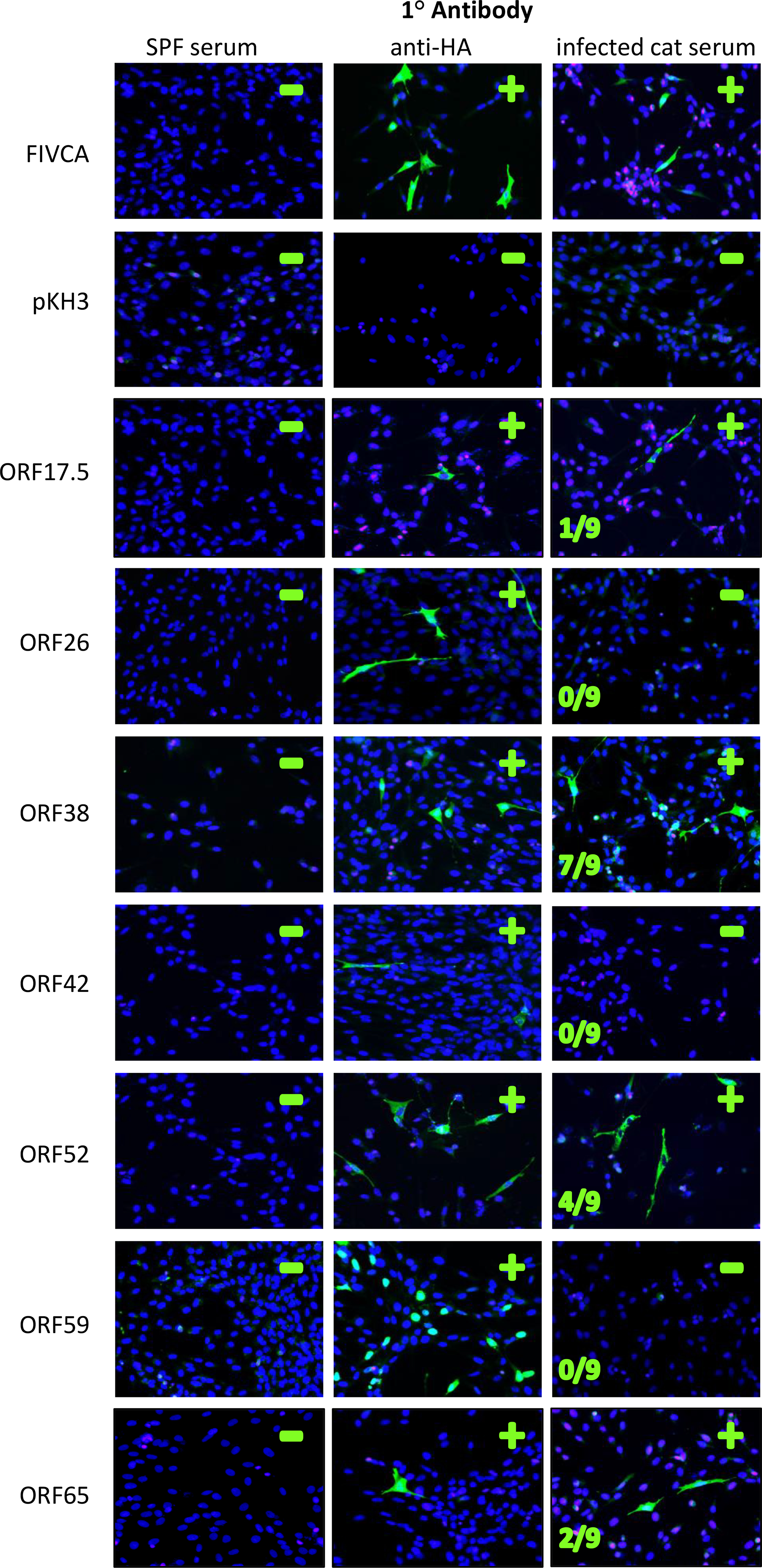
The left protein antigen that was transfected into each set of cells. SPF-naive cat serum was used as a Fluorescence in the second column (anti-HA) indicates cells expressing the protein of interest with an HA tag. Fluorescence in the third column indicates cells that bound cat serum antibody. Nuclei are stained with DAPI and appear blue. Serum from nine FcaGHV1-qPCR positive cats was used to screen each transfection reaction to determine if antibodies were present in cat sera for each antigen tested, number of cats positive is indicated. Representative results with serum from one FcaGHV1 qPCR-positive cat are shown here, see Table 2 for tabulation of these results. The FIV capsid (FIVCA) was used as a positive control for detection of viral antigen when exposed to sera from FIV+ cats. A vector-only negative control, pKH3, was also run with each transfection. FIVCA= FIV capsid protein.
To evaluate the ability of the IFA to detect anti-FcaGHV1 antibodies, we screened each of the seven FcaGHV1 antigens with serum from nine FcaGHV1 PCR-positive cats with high peripheral-DNA viral loads (Table 2, Fig. 1). ORF38, ORF65, ORF17.5, and ORF52 all had serum antibody reactivity against one or more of the FcaGHV1-positive cat sera (Table 2, Fig. 1). ORF38 and ORF52 antigens detected the greatest number of FcaGHV1 positive cats: 7/9 and 4/9, respectively (Table 2). No immunofluorescent antibody response was detected against ORF26, ORF59, or ORF42 (Table 2, Fig. 1).
Table 2.
ELISA assays increase detection of infection with FcaGHV1.
| ORF38 | ORF52 | ORF65 | ORF17.5 | ORF26 | ORF42 | ORF59 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| IFA | WB | ELISA | IFA | WB | ELISA | IFA | IFA | IFA | IFA | IFA | |
|
|
|||||||||||
| G1 (CA) | + | + | + | + | − | + | − | − | − | − | − |
| G2 (CO) | − | − | − | + | − | + | − | − | − | − | − |
| G3 (CA) | + | + | + | + | − | + | + | + | − | − | − |
| G4 (CA) | + | + | + | − | − | + | − | − | − | − | − |
| G5 (FL) | − | − | − | − | − | − | − | − | − | − | − |
| G6 (FL) | + | + | + | − | − | + | − | − | − | − | − |
| G7 (CA) | + | + | + | − | − | + | − | − | − | − | − |
| G8 (FL) | + | + | + | − | − | + | − | − | − | − | − |
| G9 (FL) | + | + | + | + | + | + | + | − | − | − | − |
|
|
|||||||||||
| Total + | 7 | 7 | 7 | 4 | 1 | 8 | 2 | 1 | 0 | 0 | 0 |
Nine cat samples positive for FcaGHV1 by qPCR were used for antigen identification screening. Table shows which cats displayed a serum antibody response to ORF52 and ORF38 proteins on each respective immunofluorescence assay (IFA), western blot (WB), and ELISA.
Western blot analysis
To further verify that the antibody responses detected by IFA were specific to the FcaGHV1 antigen of interest, we performed Western blot analysis. We immunoprecipitated recombinant ORF38 and ORF52 from transfected CRFK cell lysates and tested reactivity with serum from the same nine FcaGHV1-positive cats used for IFA (Fig. 2, Table 2). Sera from 3 SPF cats were consistently negative (Fig. 2). ORF38 blots showed bands of the expected size (~12kD) for 7/9 sera from FcaGHV1 positive cats (Fig. 2). ORF52 blots showed a band of the expected size (~19kD) for one individual (Fig. 2).
Figure 2. Western blot results confirm IFA.
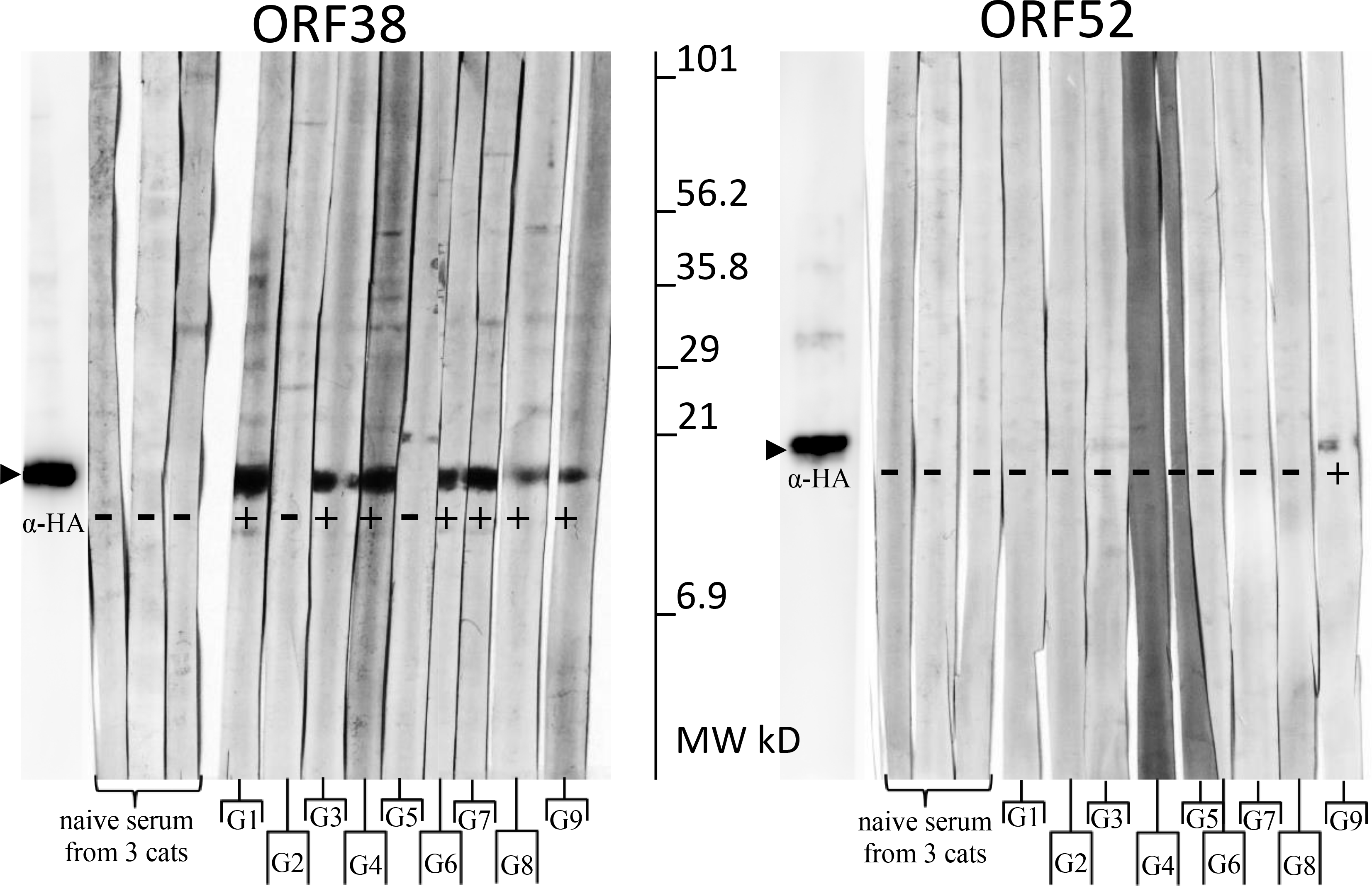
Immobilized protein indicated at top. Each strip was incubated with serum or anti-HA as primary antibody (listed below). Tabulated results are listed in Table 2. SPF cats (S1–3), GHV+ cats (G1-G9).
Determination of seroprevalence by ELISA and comparison to qPCR results
To enable rapid testing for FcaGHV1-specific antibodies in large numbers of domestic cat serum samples, we developed ELISAs as described in Methods utilizing recombinant ORF38 and ORF52 proteins as antigens. To compare serologic results with nucleic acid detection, we performed ELISAs on serum from 133 cats previously tested for FcaGHV1 prevalence via qPCR assay of peripheral whole-blood FcaGHV1 DNA (Beatty et al., 2014; Troyer et al., 2014). These cats represented animals free-roaming prior to intake by eight shelters in three states over a period of 2–3 years depending on the shelter.
This sample set included 15% (20/133) qPCR positive animals. Thirty two percent (43/133) overall seroprevalence was calculated after combining results of the two antigens for the ELISAs (Fig. 3). Of the 20 cats that tested positive on qPCR, 19 also tested positive by serology; this represented 44% of FcaGHV1 seropositive animals. Thus there were 22 cats testing negative on qPCR and positive by serology (Fig. 3).
Figure 3. FcaGHV1 ELISA results coincide with qPCR and show additional positive results.
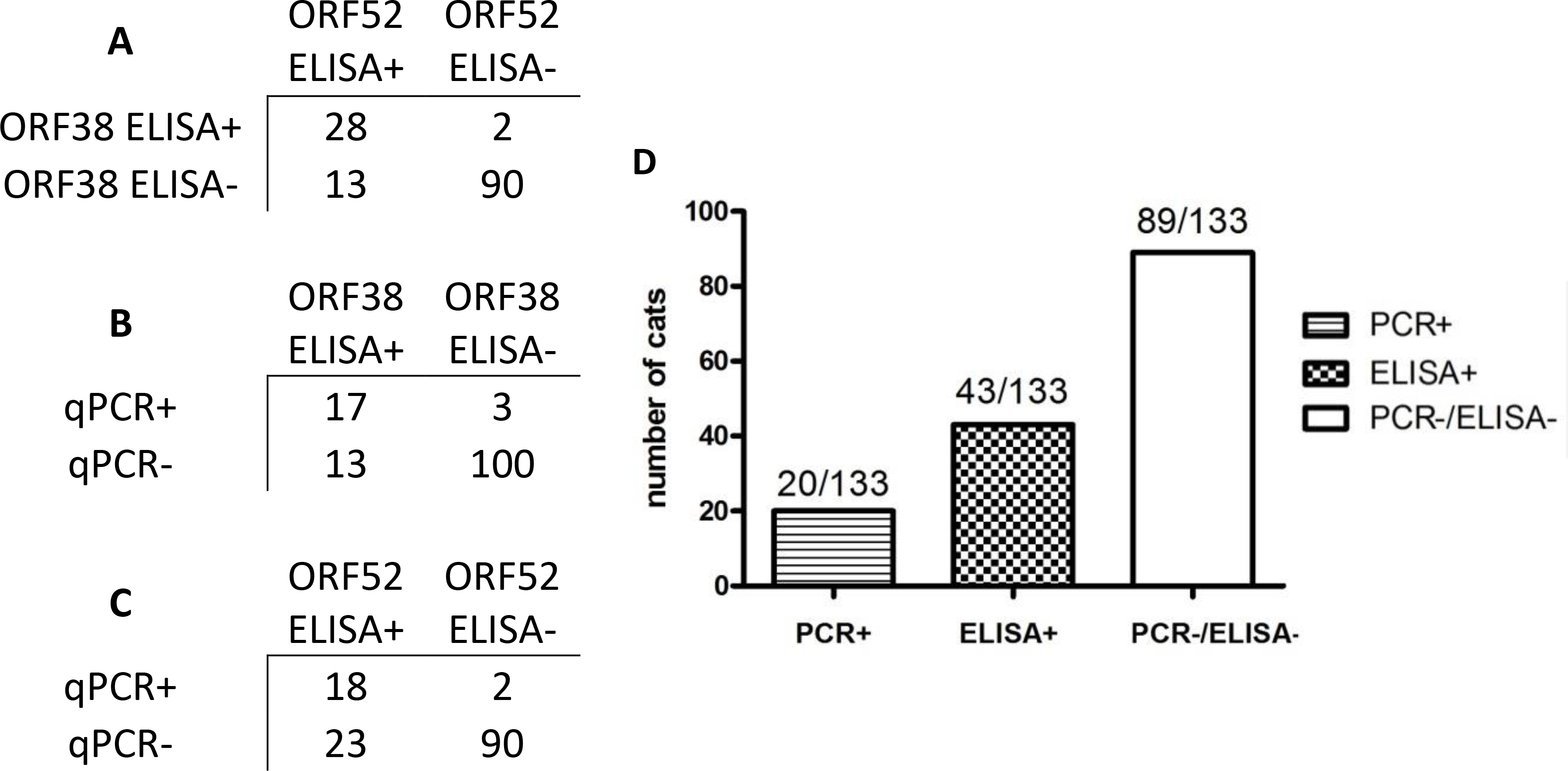
A.-C. Categorical data comparison of cats tested for all 3 assays: ORF38 ELISA, ORF52 ELISA, and FcaGHV1 qPCR. D. Comparison of FcaGHV1 serology to qPCR for 133 cats. One adult male cat from Florida was PCR+/ELISA- (data not shown). All other PCR+ animals were also ELISA+.
Figure 3 displays the categorical data results of each assay in comparison to the qPCR results from our previous publication (Beatty et al., 2014). ELISA results generally supported each other; only one cat was qPCR positive and ELISA negative, one cat was positive only with the ORF38 ELISA (ORF52 and qPCR negative) and 11 cats were only positive by ORF52 ELISA (ORF38 and qPCR negative). All other cats were confirmed by at least two assays.
ELISA sensitivity and specificity
Using a two stage Bayesian method (Liu et al., 2014), sensitivity of ORF52 ELISA was estimated as 74.3% (95% CI: 61.0 to 92.6), while specificity was estimated as 96.4% (95% CI: 90.7 to 99.8). ORF38 ELISA had a sensitivity of 57.9% (95% CI: 50.3 to 73.8) and specificity of 97.9% (95% CI: 93.5 to 99.9). Using this same Bayesian model, seroprevalence of FcaGHV1 was estimated at 30.6% (95% CI: 21.6 to 41.1).
FcaGHV1 risk factor analysis
For the rest of the analysis, we combined the ELISA results so that if a cat was positive for either antigen (ORF38, ORF52, or both) it was considered ELISA-positive for comparison of regional data and risk factors. We compared qPCR and ELISA assay results with demographic information including locations, sex, and age (Table 3). We noted a uniform increase from qPCR prevalence to ELISA prevalence by location, sex, and age categories reflective of the overall increase in prevalence with the ELISA assay (Figs. S1).
Table 3.
Risk factors identified by FcaGHV1 qPCR status are supported by FcaGHV1 ELISA result.
| y= ELISA | y=qPCR | |||||
|---|---|---|---|---|---|---|
| P (Wald) | OR | 95% CI | P (Wald) | OR | 95% CI | |
|
| ||||||
| Male | 0.0001 | 7.11 | 2.62 to 19.3 | 0.009 | 44.0 | 2.58 to 749 |
| Adult | 0.011 | 43.7 | 2.38 to 801 | 0.004 | 9.87 | 1.95 to ∞ |
|
| ||||||
| State | 0.0176 | - | - | 0.0520 | - | - |
| California vs Colorado | - | 4.65 | (1.60 to 13.5) | - | 4.92 | (1.29 to 18.8) |
| California vs Florida | - | 1.57 | (0.449 to 5.47) | - | 0.941 | (0.217 to 4.08) |
| Florida vs Colorado | - | 2.97 | (0.788 to 11.2) | - | 5.22 | (0.942 to 29.0) |
| Region | 0.0429 | - | - | 0.402 | - | - |
| Ventura vs San Diego | - | 8.61 | (1.26 to 59.0) | - | 2.19 | (0.320 to 15.0) |
| Ventura vs Corona | - | 1.94 | (0.336 to 11.2) | - | 0.70 | (0.135 to 3.62) |
P value from Wald testing and odds ratios (OR) show significance of binary logistic regression modeling with FcaGHV1 ELISA result or qPCR result as the response variable (y). State and regional analyses were performed using categorical logistic regression modeling with sex and age as co-independent variables and qPCR or ELISA result as the response variable (y). Odds ratios were calculated to compare regions of California: Animals taken in by Ventura Animal Services, San Diego Feral Cat Coalition, and Corona Animal Shelter. Odds ratios between Colorado and Florida regions were not significant (not shown).
We used logistic regression to find associations with the ELISA results for risk factors previously evaluated by qPCR. The risk factors of adult vs. young (p=0.011) and male vs. female (p=0.0001) were corroborated as risk factors previously identified with qPCR testing (Beatty et al., 2014; Troyer et al., 2014) (Table 3). Male cats were 7.1 (CI: 2.6, 19.3) times more likely to be seropositive and 44.0 (2.6, 749) times more likely to be qPCR-positive than females. Adult cats were 43.7 (2.4, 801) times more likely to be seropositive and 9.9 (2.0, ∞) times more likely to be qPCR-positive than young cats (Table 3). It should be noted that wide confidence intervals are a reflection of the statistical modeling limits. There were no young cats that were either seropositive or qPCR-positive for FcaGHV1 and no female cats that were qPCR-positive. The result of this is quasi-complete separation of the data set, which was accounted for using Firth’s penalized likelihood to allow calculation of odds ratios and p values, but with wide confidence intervals.
There was a significant difference in seroprevalence between capture locations by state, p=0.0176 (Table 3). Similar differences also exist in the qPCR data, p=0.052. Cats from California were 4.7 (CI: 1.6, 13.5) times more likely to be ELISA-positive and 4.9 (CI: 1.3, 18.8) times more likely to be qPCR-positive than cats from Colorado. There was not a significant difference in odds between California and Florida or between Florida and Colorado for qPCR or ELISA FcaGHV1 results (Table 3). In the qPCR analysis there was no significant difference between regions within the states (p=0.40). In contrast, there were significant differences among regions when comparing seroprevalence (p=0.043). When odds ratios were calculated to compare regions within states only (Table 3), there were no significant differences between regions of Colorado or Florida (data not shown). However in California, cats captured by Ventura Animal Services were 8.6 (CI: 1.3 to 59.0) times more likely to be seropositive when compared to cats captured by San Diego Feral Cat Coalition (Table 3). Although the locations of capture are within 250 miles of each other, cats located to Ventura Animal Services were found in areas of high human population density while the cats from San Diego were captured in rural areas of the Peninsular mountain range.
DISCUSSION
This study provides the first evidence of a cat immune response against FcaGHV1. As hypothesized, we detected a higher prevalence of FcaGHV1 in a population of 133 free-ranging cats via ELISA (32%) compared to qPCR-positive individuals (15%). Antibody response indicates exposure followed by seroconversion while qPCR-positivity indicates presence of the viral genome in blood cells. GHVs typically establish latent infection in a small number of lymphocytes, often within a particular lymphocyte subset, resulting in low levels of viral DNA present in the blood (Speck and Ganem, 2010). FcaGHV1 appears to follow a similar pattern of low level infection in blood cells - approximately half (44%) of FcaGHV1 seropositive cats had detectable FcaGHV1 DNA by qPCR. We infer that the animals diagnosed as FcaGHV1 seropositive and qPCR-negative are likely in a latent phase of GHV infection. It remains to be determined whether body fluids other than blood might also contain FcaGHV1 DNA. If virus were detectable by qPCR in a fluid such as saliva, this could potentially alter sensitivity of qPCR and interpretations of viral latency.
Our measured FcaGHV1 seroprevalence is much lower than seroprevalence of EBV in humans, which is greater than 95% (Jenson, 2011). Reported KSHV exposures range dramatically by region. North America, Asia, and Europe are considered low seroprevalence areas and most studies have indicated exposure rates of <5% (Dow et al., 2014). Higher KSHV seroprevalence (approximately 50%), is characteristic of endemic regions of Africa and the Brazilian Amazon (Dow et al., 2014). Thus in comparison to disease-inducing GHVs of humans, FcaGHV1 appears to have a distinct infection pattern with much lower seroprevalence than EBV, but higher seroprevalence than KSHV in North America. Future studies will determine whether the seroprevalence of FcaGHV1 varies in other locations relative to the level found in this study for North American cats.
In the absence of a gold standard for FcaGHV1 detection, ORF38 and ORF52 ELISA assays were evaluated using a Bayesian logistic regression model with qPCR data used as informative priors. From these calculations, the ELISAs had high specificity, while sensitivity was more moderate. The accuracy of these estimates is only as good as the probability distributions of the known parameters (Hadgu et al., 2005). A better method which could be used with future development of more assays (3 or more) would be to use latent class modeling, modified for sensitivity and specificity calculations (Alonzo and Pepe, 1999; Hadgu et al., 2005).
The results of ELISA data strongly corroborate adult age and male sex as risk factors for FcaGHV1 as identified previously (Beatty et al., 2014; Troyer et al., 2014). Being male and adult appears to be strongly associated both with qPCR-positive FcaGHV1 infection and ELISA-positive results (Table 3). Interestingly, no young cats (n=30) were seropositive for FcaGHV1, and while some female cats in this population were seropositive (7/54), none were qPCR-positive. This suggests that mother-to-kitten, vertical transmission, or vector borne disease transmission are unlikely modes of spread of FcaGHV1 in the US feral cat populations. Aggressive male contact is the most probable mode of transmission based upon infection data. Transmission via exposure to salivary excretions would be consistent with transmission of other GHVs such as EBV and KSHV (Baillargeon et al., 2002; Jenson, 2011; Mantina et al., 2001; Plancoulaine et al., 2000; Taylor et al., 2004).
Geographic variation was a risk factor for both FcaGHV1 seropositivity and FcaGHV1 qPCR-positive result. Cats captured in California were 4.9 (95% CI: 1.3, 18.8) times more likely to be qPCR positive and 4.7 (95% CI: 1.6, 13.5) times more likely to be ELISA positive than Colorado cats (Table 3). A closer analysis of the geography reveals that the majority of cats in California came from dense city populations while cats in Colorado were largely located in the rural Western Slope region of the state (Troyer et al., 2014). Moreover, a comparison by region revealed no significance in qPCR results; however, cats from Ventura County, California were 8.6 (95% CI: 1.3, 59.0) times more likely to be ELISA positive than cats captured in San Diego County (Table 3). The majority of cats captured in San Diego County were in mountainous areas outside of the city of San Diego with low human populations, whereas the Ventura County cats were captured in urban Ventura. Collectively, this information suggests that feline population density may play a role in FcaGHV1 transmission, resulting in higher virus prevalence in areas with high cat density. High density may also correspond with increased aggressive encounters further supporting this as a major mode of viral transmission.
Indirect ELISAs based upon FcaGHV1 tegument-associated antigens ORF52 and ORF38 provided an enhanced assay to detect viral exposure compared to FcaGHV1 qPCR on blood cells. We conclude that approximately half of seropositive cats have detectable FcaGHV1 viral genomes using qPCR. This suggests that FcaGHV1 maintains a latent state in which cats are seropositive but viral DNA is often undetectable in blood cells. A sensitive and specific serodiagnostic assay for FcaGHV1 is an important tool to inform further work to identify pathogenesis of this recently discovered agent and will allow comparisons with GHV induced human disease.
METHODS
Collection of samples
Domestic cat blood cells and serum were obtained from archived samples collected from free-ranging domestic cats upon admission to shelters in Florida, California, and Colorado as previously described (Troyer et al., 2014). Samples were collected in accordance with Colorado State University Animal Care and Use Committee protocol #11–2453A and demographic data were recorded for each cat (Bevins et al., 2012). FcaGHV1 qPCR assay was performed on DNA extracted from whole blood as previously described (Troyer et al., 2014)
Plasmids
Using the complete FcaGHV1 genome sequence (Troyer et al., 2015), we selected seven genes conserved among gammaherpesviruses to evaluate their encoded proteins for the potential to elicit humoral immunity in naturally-infected cats. These included ORF17.5, ORF26, ORF38, ORF42, ORF52, ORF59, and ORF65 that code for proteins homologous to antigenic virion-associated proteins of KSHV, EBV, and OvHV2 (Table 1). Specific primers that incorporated restriction sites to facilitate cloning were designed for each gene of interest (Table S1). All genes were PCR-amplified using High Fidelity Platinum Taq (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions from lymph node DNA, extracted from FcaGHV1 infected cat 31286. Amplicons were ligated into pKH3 mammalian expression vector, which contains a 5’ (N-terminal) HA tag.
Plasmid constructs were transformed in TOP10 cells (Life Technologies) and colonies were selected on LB agar with ampicillin. One clone with ORF sequence identical to the published FcaGHV1 genomic sequence (GenBank accession number KT595939) was selected for each gene and this plasmid was used for all cell transfections. The FIV Gag capsid (p27) gene sequence, used as a control for the immunofluorescence assay, was subcloned from pGEX2T (Wood et al., 2013) into pKH3 using BamHI and EcoRI enzymes (New England Biolabs).
Immunofluorescence assay
An immunofluorescence assay (IFA) was performed to detect serum antibodies specific to recombinant FcaGHV1 proteins expressed in feline CRFK cells. Positive-control serum was obtained from nine cats in CO, CA, or FL previously shown to have high peripheral FcaGHV1 DNA viral loads (Troyer et al., 2014). Each set of cells transfected to express recombinant FcaGHV1 protein was tested with serum from all nine FcaGHV1-positive cats. Transfected cells were also tested with negative control serum from three specific pathogen-free (SPF) cats from the Colorado State University Retrovirus Research cat colony. This colony has been extensively screened and shown to be FcaGHV1 negative by qPCR. Banked FIV-positive serum was used to detect recombinant FIV capsid p27 expression for IFA optimization and as an assay positive control.
CRFK cell cultures were maintained in DMEM media as previously described (Troyer et al., 2013). For the IFA, CRFK cells were plated at 5,000 cells per well in media without antibiotics on glass slides with twelve 0.4 mm wells and incubated for 2 days to allow slide adherence. Slides were incubated at 37°C in 5% CO2 in Petri dishes with sterile water to maintain humidity. Transfections with plasmid constructs were performed using 0.15 μL Lipofectamine 2000 (Life Technologies) and 60 ng plasmid per well. Cells were incubated for 21 hours post-transfection. After incubation, slides were gently washed with PBS to remove excess media. Cells were fixed using 2% paraformaldehyde and then 50:50 ethanol/methanol followed by air-drying. Cat serum diluted 1:20 in PBS with 2% bovine serum albumin (BSA) was spotted on the fixed cells and slides were incubated for 1 hour at 37°C. Alternatively, to test for protein expression by evaluating the presence of the N-terminal HA tag included in each construct, rabbit anti-HA antibody (Covance, Princeton, NJ) diluted 1:500 in BSA/PBS was used in place of serum. After incubation, slides were washed three times with PBS, rinsed with water, and air-dried. Secondary antibodies labeled with fluorescein isothiocyanate (FITC) were diluted in BSA/PBS and spotted on the slide, followed by incubation at 37°C in the dark for 1 hour. Anti-cat IgG FITC (Covance) diluted 1:50 was used to detect feline serum antibodies and goat anti-rabbit IgG FITC (Covance) diluted 1:500 was used for HA detection. After incubation, slides were washed three times with PBS and stained for 2 min with diamidino-2-phenylindole (DAPI) nuclear stain (Thermo Fisher Scientific) at 300 nM in PBS. Slides were washed twice with PBS, rinsed with water, and air-dried. Slide covers were sealed with ProLong Gold Antifade (Life Technologies) prior to light microscopic viewing with an Olympus BX60 (Tokyo, Japan) for immunofluorescence.
Positive results were recorded if there was at least one cell per well that had stronger immunofluorescence than the strongest background visible on the (negative) wells exposed to SPF/naïve serum. This immunofluorescence was also compared to surrounding negative cells within the well when evaluating positive versus negative. Immunofluorescence detections were repeated for confirmation and only samples which consistently had positive results were considered “positive” for publication purposes. Images were recorded with the microscope-connected Olympus DD71 digital capture system. Uniform adjustments were made with Photoshop CS5 12.1 (Adobe Systems, San Jose, California) to improve overall brightness of images for presentation.
Western blot
Immunoblot analysis was performed to confirm IFA observations on two candidate antigens (ORF38 and ORF52) demonstrating the most consistent immunofluorescence against qPCR positive FcaGHV1 cat sera. ORF38 and ORF52 plasmids were transfected into 293T cells in 6-well tissue culture plates using Lipofectamine 2000 according to the manufacturer protocol (Life Technologies). At 20 hours post-transfection, cells were lysed with Pierce magnetic anti-HA IP/co-IP kit lysis buffer and recombinant proteins were immunoprecipitated using the kit and following manufacturer instructions (Thermo Fisher Scientific). After immunoprecipitation, protein concentration was determined using the Pierce BCA kit (Thermo Fisher Scientific). Polyacrylamide gel electrophoresis was conducted at 160 V for 40 min using NuPage 1.5 mm, 4–12% Bis-Tris 10-well gels in a NuPage gel box with NuPage MES Running Buffer (1M Tris-Base, 1M MES, 2.0%SDS, 2mM EDTA) (Life Technologies). For each protein being evaluated, 1 μg of protein per well was used. The Bio-Rad Precision Plus Blue and Magic Mark protein standards were run for each gel (Hercules, CA).
Proteins were transferred to a PVDF membrane using NuPage transfer buffer (Life Technologies) with added 10% methanol in a Trans-Blot Turbo Transfer System (Bio-Rad) at 15 V for 30 min. The PVDF membrane was blocked for 1 hour with 5% non-fat dry milk protein in PBS, and then cut into strips for exposure to serum from 9 qPCR-positive cats and 3 SPF/naïve cats at 1:20 dilution in PBS for 1 hour. The strips were washed in PBS plus 2% tween (PBST). Strips were incubated with phosphatase labeled goat anti-cat IgG (KPL, Gaithersburg, MD) for 1 hour at 1:2000 in PBS. After an additional wash in PBST, strips were incubated with BCIP-NBT phosphatase substrate (Thermo Fisher Scientific) to visualize bands.
To ensure the correct protein was on the membrane, one strip with each protein was incubated with Covance HA.11 mouse anti-HA antibody at 1:5000 and secondary goat anti-mouse horseradish peroxidase (KPL, Gaithersburg, MD) at 1:10,000. Dilutions were in PBS and washes were performed as described earlier with serum-incubated strips. Equal amounts of peroxide solution and luminol (Immobilon Western HRP substrates, Millipore, Billerica, MA) were applied to the strip and then visualized using chemiluminescent detection imaging (ImageQuant LAS 4000, GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK).
ELISA
ORF38 and ORF52 were chosen as antigens for ELISA, requiring larger-scale production in 293T cells. 293T cells were maintained as previously described (Troyer et al., 2013) and eight million 293T cells were plated onto 100 mm tissue culture plates in antibiotic-free media and incubated overnight at 37°C. ORF38 and ORF52 plasmids were transfected the following day using 45 μL Lipofectamine 2000 (Life Technologies) with 18 μg plasmid per plate for ORF52 and 60 μL Lipofectamine 2000 with 15 μg plasmid for ORF38. OptiMEM serum-free media was used in the quantities recommended by Lipofectamine 2000 manufacturer protocols (Life Technologies). Transfection length was found to be optimal at 20 hours. Cells were collected and lysed and recombinant protein was immunoprecipitated using the Pierce anti-HA magnetic bead kit and included lysis buffer. Protein concentration was determined using the Pierce BCA kit (Thermo Fisher Scientific), and proteins were stored at 4°C.
ELISAs were performed in 96-well plates. Each protein was diluted in 50 mM carbonate buffer (pH 9.5), 40 ng ORF52 in 100 μL buffer per well and 100 ng ORF38 in 100 μL buffer per well. Plates were incubated overnight at 4°C. Contents were discarded and 300 μL of 2% BSA in imidazole-buffered saline (IBS; 2 mM imidazole, 160 mM NaCl, 0.5 mM EDTA) was used to block plates for 2 hours at room temperature. Contents were discarded and wells were incubated at room temperature for 2 hours with 100 μL of serum per well, diluted 1:100 in ELISA diluent (IBS with 2% BSA, 4% fetal bovine serum, and 0.5% Triton X-100). Plates were washed 5 times in a plate washer with IBS + 0.2% Tween (IBST). The secondary antibody incubation was 1 hour at room temperature with 100 μL of Cappel goat anti-cat IgG peroxidase conjugate (MP Biomedicals, Santa Ana, CA) diluted 1:5000 in ELISA diluent with 5% mouse serum (Sigma, St. Louis, MO). Wells were again washed in a plate washer 5 times with IBST. TMB-peroxidase detection solution (KPL, Gaithersburg, MD) was added to each well (100 μL) and incubated for 10 min at room temperature. The reaction was stopped using 50 μL per well of 2.5 N H2SO4 and absorbance was read at 450 nm. Serum from each cat and controls were run in triplicate. Every plate included controls: serum from 3 different SPF/naïve cats, no-antigen control wells, and diluent-only wells.
Optimization was performed to identify the most appropriate dilutions of feline serum, secondary antibody, and concentration of coated antigen. Feline serum from an FcaGHV1 qPCR-positive animal was used for the optimizations. A range of dilutions were initially attempted above and below the ones ultimately selected to identify the largest signal-to-background ratio. Prior to initiation of sample testing each ELISA was tested with serum from 10 SPF/naïve cats to ensure lack of non-specific reactions. Positive threshold was evaluated on a plate-by-plate basis and defined as the mean absorbance of the replicates of the 3 SPF/naïve cats with the addition of 3 standard deviations. Additionally, if this calculation yielded a number < 0.2, then 0.2 was considered the threshold for positive for that 96-well plate.
Statistical analysis
Logistic regression was used to make statistical analysis of geographic data and FcaGHV1 qPCR or ELISA result. Region and state were each modeled as categorical independent variables along with sex and age as binary co-variables. In each model, FcaGHV1 ELISA result or qPCR result was the dependent (response) variable. For sex and age, male was recorded as 1 and female as 0, similarly adult as 1 and young as 0. Odds ratios were calculated within the respective logistic models. Logistic regression modeling was performed in SAS University Edition (SAS Institute, Cary, NC).
Sensitivity and specificity was calculated using modeling techniques described by Liu et al (Liu et al., 2014). This Bayesian model requires a gold standard test used to evaluate only the positive responses from two dependent screening tests along with positive and negative results from those assays. This was adapted to our study by considering the two ELISAs (ORF38 and ORF52) to be the dependent assays. The qPCR assay was considered a gold standard only for the sake of specificity (100%). As with the model designed by Liu et al., two sensitivities were calculated for the ELISAs based on the association to qPCR specificity, only cats testing positive on qPCR were considered in this evaluation. This information was then used as informative priors for a second model to calculate specificity and sensitivity for each ELISA assay as well as prevalence. We used the open access program written by Liu et al. to make these calculations (Liu et al., 2014) and the freeware, WinBugs version 1.4.3 (MRC Biostatistics Unit, Cambridge Biomedical Campus).
Supplementary Material
Highlights.
FcaGHV1 is a newly described gammaherpesvirus that widely infects domestic cats
This study provides the first evidence of a host immune response against FcaGHV1
Cat antibody responses were detected against multiple recombinant FcaGHV1 antigens
Seroreactivity against FcaGHV1 ORF52 and ORF38 indicates 32% seroprevalence in USA
Approximately half of FcaGHV1 seropositive cats have detectable genome using qPCR
ACKNOWLEDGEMENTS
Anne Avery of the Colorado State University Clinical Immunology Lab generously provided DNA from FcaGHV1 infected cat 31286. We thank Michael Lappin and multiple humane societies and animal care centers for providing domestic cat samples. This work was supported by a Morris Animal Foundation award (D14FE-301) to RMT and an NIH T32 Fellowship (OD 12201) to KSR.
REFERENCES
- Ackermann M, 2006. Pathogenesis of gammaherpesvirus infections. Veterinary microbiology 113, 211–222. [DOI] [PubMed] [Google Scholar]
- Alonzo TA, Pepe MS, 1999. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Statistics in medicine 18, 2987–3003. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Loftus MS, Kedes DH, 2014. Maturation and Vesicle-Mediated Egress of Primate Gammaherpesvirus Rhesus Monkey Rhadinovirus Require Inner Tegument Protein ORF52. Journal of virology 88, 9111–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillargeon J, Leach CT, Deng JH, Gao SJ, Jenson HB, 2002. High prevalence of human herpesvirus 8 (HHV-8) infection in south Texas children. Journal of medical virology 67, 542–548. [DOI] [PubMed] [Google Scholar]
- Bartley K, Deane D, Percival A, Dry IR, Grant DM, Inglis NF, Mclean K, Manson ED, Imrie LH, Haig DM, 2014. Identification of immuno-reactive capsid proteins of malignant catarrhal fever viruses. Veterinary microbiology 173, 17–26. [DOI] [PubMed] [Google Scholar]
- Barton E, Mandal P, Speck SH, 2011. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annual review of immunology 29, 351–397. [DOI] [PubMed] [Google Scholar]
- Beatty JA, Troyer RM, Carver S, Barrs VR, Espinasse F, Conradi O, Stutzman-Rodriguez K, Chan CC, Tasker S, Lappin MR, 2014. Felis catus gammaherpesvirus 1; a widely endemic potential pathogen of domestic cats. Virology 460, 100–107. [DOI] [PubMed] [Google Scholar]
- Bevins SN, Carver S, Boydston EE, Lyren LM, Alldredge M, Logan KA, Riley SP, Fisher RN, Vickers TW, Boyce W, 2012. Three pathogens in sympatric populations of pumas, bobcats, and domestic cats: implications for infectious disease transmission. PLoS One 7, e31403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett 305, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS, 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869. [DOI] [PubMed] [Google Scholar]
- Dow DE, Cunningham CK, Buchanan AM, 2014. A Review of Human Herpesvirus 8, the Kaposi’s Sarcoma-Associated Herpesvirus, in the Pediatric Population. Journal of the Pediatric Infectious Diseases Society 3, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dry I, Haig DM, Inglis NF, Imrie L, Stewart JP, Russell GC, 2008. Proteomic analysis of pathogenic and attenuated alcelaphine herpesvirus 1. Journal of virology 82, 5390–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl R, Korb M, Langbein-Detsch I, Klein D, 2015. Prevalence and risk factors of gammaherpesvirus infection in domestic cats in Central Europe. Virology journal 12, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D, 2006. KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu. Rev. Pathol. Mech. Dis. 1, 273–296. [DOI] [PubMed] [Google Scholar]
- Hadgu A, Dendukuri N, Hilden J, 2005. Evaluation of nucleic acid amplification tests in the absence of a perfect gold-standard test: a review of the statistical and epidemiologic issues. Epidemiology 16, 604–612. [DOI] [PubMed] [Google Scholar]
- ICTV, 2014. Virus Taxonomy, July 2014 ed. International Committee on Taxonomy of Viruses (ICTV), Montreal Canada. [Google Scholar]
- Jenson HB, 2011. Epstein-Barr virus. Pediatrics in Review-Elk Grove 32, 375. [DOI] [PubMed] [Google Scholar]
- Katano H, Iwasaki T, Baba N, Terai M, Mori S, Iwamoto A, Kurata T, Sata T, 2000. Identification of antigenic proteins encoded by human herpesvirus 8 and seroprevalence in the general population and among patients with and without Kaposi’s sarcoma. Journal of virology 74, 3478–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ, 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus research 145, 173–186. [DOI] [PubMed] [Google Scholar]
- Kúdelová M, Belvončíková P, Halásová Z, Košovský J, Lapuníková B, Pančík P, Režuchová I, Supolíková M, Zelník V, 2013. Recombinant herpesviruses as tools for the study of herpesvirus biology. Acta virologica 57, 149. [DOI] [PubMed] [Google Scholar]
- Labo N, Miley W, Marshall V, Gillette W, Esposito D, Bess M, Turano A, Uldrick T, Polizzotto MN, Wyvill KM, 2014. Heterogeneity and breadth of host antibody response to KSHV infection demonstrated by systematic analysis of the KSHV proteome. PLoS pathogens 10, e1004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen F, Yu H, Zeng P, Liu L, 2014. A two-stage Bayesian method for estimating accuracy and disease prevalence for two dependent dichotomous screening tests when the status of individuals who are negative on both tests is unverified. BMC medical research methodology 14, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantina H, Kankasa C, Klaskala W, Brayfield B, Campbell J, Du Q, Bhat G, Kasolo F, Mitchell C, Wood C, 2001. Vertical transmission of Kaposi’s sarcoma-associated herpesvirus. International journal of cancer 94, 749–752. [DOI] [PubMed] [Google Scholar]
- Massimelli MJ, Majerciak V, Kang J-G, Liewehr DJ, Steinberg SM, Zheng Z-M, 2015. Multiple Regions of Kaposi’s Sarcoma-Associated Herpesvirus ORF59 RNA are Required for Its Expression Mediated by Viral ORF57 and Cellular RBM15. Viruses 7, 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means RE, Lang SM, Jung JU, 2007. Human gammaherpesvirus immune evasion strategies, in: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (Eds.), Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, Cambridge. [PubMed] [Google Scholar]
- Morrison BJ, Labo N, Miley WJ, Whitby D, 2015. Serodiagnosis for tumor viruses. Semin Oncol 42, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plancoulaine S, Abel L, van Beveren M, Tregouet DA, Joubert M, Tortevoye P, de The G, Gessain A, 2000. Human herpesvirus 8 transmission from mother to child and between siblings in an endemic population. Lancet 356, 1062–1065. [DOI] [PubMed] [Google Scholar]
- Schulz T, 2000. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8): epidemiology and pathogenesis. Journal of Antimicrobial Chemotherapy 45, 15–27. [DOI] [PubMed] [Google Scholar]
- Shedd D, Angeloni A, Niederman J, Miller G, 1995. Detection of human serum antibodies to the BFRF3 Epstein-Barr virus capsid component by means of a DNA-binding assay. Journal of Infectious Diseases 172, 1367–1370. [DOI] [PubMed] [Google Scholar]
- Shen S, Guo H, Deng H, 2014. Murine gammaherpesvirus-68 ORF38 encodes a tegument protein and is packaged into virions during secondary envelopment. Protein & cell 5, 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck SH, Ganem D, 2010. Viral latency and its regulation: lessons from the γ-herpesviruses. Cell host & microbe 8, 100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MM, Chohan B, Lavreys L, Hassan W, Huang ML, Corey L, Ashley Morrow R, Richardson BA, Mandaliya K, Ndinya-Achola J, Bwayo J, Kreiss J, 2004. Shedding of human herpesvirus 8 in oral and genital secretions from HIV-1-seropositive and -seronegative Kenyan women. The Journal of infectious diseases 190, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, Beatty JA, Stutzman-Rodriguez KR, Carver S, Lozano CC, Lee JS, Lappin MR, Riley SP, Serieys LE, Logan KA, 2014. Novel gammaherpesviruses in North American domestic cats, bobcats, and pumas: identification, prevalence, and risk factors. Journal of virology 88, 3914–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, Lee JS, Vuyisich M, Chain P, Lo C-C, Kronmiller B, Bracha S, Avery AC, VandeWoude S, 2015. First complete genome sequence of Felis catus gammaherpesvirus 1. Genome announcements 3, e01192–01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, Thompson J, Elder JH, VandeWoude S, 2013. Accessory genes confer a high replication rate to virulent feline immunodeficiency virus. Journal of virology 87, 7940–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Ho E, Wu T-T, Davis ZH, Zhang B, Huang J, Gong H, Deng H, Liu F, Glaunsinger B, Sun R, 2014. Unconventional sequence requirement for viral late gene core promoters of murine gammaherpesvirus 68. Journal of virology 88, 3411–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood BA, Carver S, Troyer RM, Elder JH, VandeWoude S, 2013. Domestic cat microsphere immunoassays: Detection of antibodies during feline immunodeficiency virus infection. Journal of immunological methods 396, 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Wan J, Cho YG, Wang L, Chiou C-J, Pai S, Woodard C, Zhu J, Liao G, Martinez-Maza O, 2011. Comparison of Humoral Immune Responses to Epstein-Barr Virus and Kaposi’s Sarcoma–Associated Herpesvirus Using a Viral Proteome Microarray. Journal of Infectious Diseases 204, 1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


