Abstract
Background:
In the Coronavirus Efficacy (COVE) trial, estimated mRNA-1273 vaccine efficacy against coronavirus disease-19 (COVID-19) was 94%. SARS-CoV-2 antibody measurements were assessed as correlates of COVID-19 risk and as correlates of protection.
Methods:
Through case-cohort sampling, participants were selected for measurement of four serum antibody markers at Day 1 (first dose), Day 29 (second dose), and Day 57: IgG binding antibodies (bAbs) to Spike, bAbs to Spike receptor-binding domain (RBD), and 50% and 80% inhibitory dilution pseudovirus neutralizing antibody titers calibrated to the WHO International Standard (cID50 and cID80). Participants with no evidence of previous SARS-CoV-2 infection were included. Cox regression assessed in vaccine recipients the association of each Day 29 or 57 serologic marker with COVID-19 through 126 or 100 days of follow-up, respectively, adjusting for risk factors.
Results:
Day 57 Spike IgG, RBD IgG, cID50, and cID80 neutralization levels were each inversely correlated with risk of COVID-19: hazard ratios 0.66 (95% CI 0.50, 0.88; p=0.005); 0.57 (0.40, 0.82; p=0.002); 0.42 (0.27, 0.65; p<0.001); 0.35 (0.20, 0.61; p<0.001) per 10-fold increase in marker level, respectively, multiplicity adjusted P-values 0.003–0.010. Results were similar for Day 29 markers (multiplicity adjusted P-values <0.001–0.003). For vaccine recipients with Day 57 reciprocal cID50 neutralization titers that were undetectable (<2.42), 100, or 1000, respectively, cumulative incidence of COVID-19 through 100 days post Day 57 was 0.030 (0.010, 0.093), 0.0056 (0.0039, 0.0080), and 0.0023 (0.0013, 0.0036). For vaccine recipients at these titer levels, respectively, vaccine efficacy was 50.8% (−51.2, 83.0%), 90.7% (86.7, 93.6%), and 96.1% (94.0, 97.8%). Causal mediation analysis estimated that the proportion of vaccine efficacy mediated through Day 29 cID50 titer was 68.5% (58.5, 78.4%).
Conclusions:
Binding and neutralizing antibodies correlated with COVID-19 risk and vaccine efficacy and likely have utility in predicting mRNA-1273 vaccine efficacy against COVID-19.
Trial registration number:
COVE ClinicalTrials.gov number, NCT04470427
Introduction
Multiple vaccines have demonstrated efficacy against coronavirus disease-19 (COVID-19) in phase 3 trials1 and have been authorized for emergency use.2,3 However, the manufacturing challenges posed by the global demand for doses, the need for affordable and widely accessible options that are safe and effective in diverse populations, the lack of efficacy data in important populations (e.g., pediatrics, pregnancy and immunocompromised patients), and the emergence of more transmissible viral variants, highlight the need for a large armamentarium of effective COVID-19 vaccines.4,5 The identification and validation of a correlate of protection6–8 could expedite the clinical evaluation and regulatory approval process for existing vaccines for new populations, for vaccine regimen modifications, and for new vaccines.
The Coronavirus Efficacy (COVE) phase 3 trial of the mRNA-1273 COVID-19 vaccine showed estimated vaccine efficacy against COVID-19 of 94.1%,9 leading to the US Food and Drug Administration’s Emergency Use Authorization of mRNA-1273 for prevention of COVID-19 in adults,10 and more than 130 million doses have been administered in the United States.11 The mRNA-1273 vaccine has been shown to be highly effective in the elderly and in essential and frontline workers, including healthcare workers,12 and to have non-inferior immunogenicity in adolescents vs. adults.13
Neutralizing antibodies (nAbs) or binding antibodies (bAbs) have been established as a correlate of protection/surrogate marker for vaccines against many viral diseases.7 The Spike protein14 and its receptor binding domain (RBD)15 are targets for nAbs produced by SARS-CoV-2 infection and by vaccination.16–18 The hypothesis that antibodies, whether elicited by natural infection or by Spike protein-based vaccines, are a correlate of protection against COVID-19 is supported by diverse lines of evidence,19–30 including evidence supporting a mechanistic correlate of protection that can be derived from passive transfer of antibodies in experimental challenge studies and from studies of human monoclonal antibodies.21,28–30 For the mRNA-1273 vaccine, multiple SARS-CoV-2 antibody markers (IgG bAbs to Spike, IgG bAbs to Spike RBD, 50% (ID50) inhibitory dilution nAb titer, and angiotensin-converting enzyme 2 (ACE2)-binding inhibition) each correlated with protection, defined as reduced SARS-CoV-2 replication after challenge, in vaccinated rhesus macaques.30 Here we assessed three of these same SARS-CoV-2 antibody markers (IgG bAbs to Spike, IgG bAbs to Spike RBD, ID50 nAb titer), as well as 80% inhibitory dilution (ID80) nAb titer, as correlates of risk of COVID-19 and as correlates of mRNA-1273 vaccine protection against COVID-19 in the COVE trial.
Methods
Trial design and COVID-19 endpoint evaluated for correlates.
From July 27, 2020 to October 23, 2020, 30,415 participants were randomized (1:1 ratio) to receive two injections of mRNA-1273 (100 μg) or placebo at Day 1 and at Day 29.9 Correlates were evaluated against the protocol-specified primary endpoint, referred to as “COVID-19”: first occurrence of acute symptomatic COVID-19 with virologically-confirmed SARS-CoV-2 infection in participants with no evidence of previous SARS-CoV-2 infection.9 Correlates analyses included COVID-19 endpoints diagnosed during two time periods: “intercurrent endpoints” starting 7 days post Day 29 visit through 6 days post Day 57 visit and “post Day 57 endpoints” starting 7 days post Day 57 visit and any time during follow-up while participants were still blinded to randomization assignment (Figure S1). The data cutoff date was March 26, 2021.
Laboratory methods.
A solid-phase electrochemiluminescence S-binding IgG immunoassay was developed for simultaneous quantitative detection of IgG antibodies to three distinct SARS-CoV-2 antigens in human serum. This validated assay uses custom plates manufactured by Meso Scale Discovery (Rockville, Maryland) provided as 4-plex, detecting SARS-CoV-2 prefusion stabilized Spike protein (S-2P), RBD, and Nucleocapsid, with a bovine serum albumin control ligand. The Spike and RBD markers were converted to International Units for assessment as immune correlates. Neutralizing antibodies were measured in a formally validated assay as a function of reductions in luciferase reporter gene expression after a single round of infection with SARS-CoV-2.D614G Spike-pseudotyped virus in 293T/ACE2 cells as described.31 ID50 and ID80 nAb titers were calculated based on a dose-response curve and were calibrated to the WHO anti-SARS-CoV-2 immunoglobulin International Standard32,33 and the resulting calibrated ID50 titer (cID50) and calibrated ID80 titer (cID80) assessed as immune correlates. Supplementary Text 1 and Table S1 provide additional details on both immunoassays.
Statistical methods.
All data analyses were pre-specified in the Statistical Analysis Plan (Supplementary Material), summarized here. A case-cohort sampling design was used for selecting participants for measurement of antibody markers at Day 1, 29, 57,34 comprising a random sample of all participants, stratified by cross-classified randomization arm, baseline SARS-CoV-2 positivity, protocol randomization strata (Age below 65 years At-Risk, Age below 65 years Not At-Risk, Age above 65 years), and underrepresented minority status, and all other participants who acquired an intercurrent or post Day 57 COVID-19 endpoint until database lock. At-Risk comprised six self-reported health/comorbidities.9 Analyses were based on the case-cohort set defined by the subset of selected participants with no evidence of previous SARS-CoV-2 infection and per-protocol received both doses without major protocol violations (Figure S2). Each participant had two inverse probability sampling (IPS) weights calculated, for Day 29 and Day 57 marker correlates analyses. Day 29 and Day 57 marker correlates analyses excluded participants with any evidence of SARS-CoV-2 infection before 7 days post Day 29 and Day 57 visit, respectively, because natural infection before the serology may have modulated the antibody response.
For each marker, values below the assay limit of detection (LOD) were set to LOD/2, and values above the upper limit of quantitation (ULOQ) were set to ULOQ. Positive responses for Spike IgG and RBD IgG were defined by IgG > 10.8424 IU/ml and 14.0858 IU/ml, respectively. Positive responses for cID50 and cID80 were defined by value > LOD (LOD = 2.42 and 15.02, respectively). All analyses were done reproducibly based on R scripts hosted at the Github code repository CoVPN/correlates_reporting, which are publicly available with application to a mock COVE trial data set.
The positive response rate for each marker and analyzed group was estimated by an IPS-weighted average, with 95% confidence interval (CI) calculated with an IPS-weighted Clopper-Pearson method. Differences in response rates with 95% CIs were calculated by the Wilson-Score method without continuity correction.35 Geometric mean concentration (GMC) or titer (GMT) and their ratios between cases vs. non-cases were estimated with 95% CIs, with hypothesis tests, based on the t-distribution with IPS weighting. Reverse cumulative distribution function curves were estimated by IPS-weighted nonparametric maximum likelihood.
Covariate-adjustment.
All correlates analyses adjusted for baseline variables prognostic for COVID-19: At-Risk status, membership in a community of color, and the logit of predicted COVID-19 risk score built from machine learning of placebo arm participants (Supplementary Text 2, Tables S2–S5, Figures S3–S4).
Correlates of risk in vaccine recipients.
Each antibody marker at Day 57 was assessed as a correlate of risk for COVID-19 starting 7 days post Day 57 visit; similarly each Day 29 marker was assessed as a correlate of risk for COVID-19 starting 7 days post Day 29 visit. For each marker, without replacement IPS-weighted Cox regression was applied to estimate the covariate-adjusted hazard ratio of COVID-19 across tertiles of the marker or per 10-fold increase in the quantitative marker level with 95% CIs and Wald-based p-values. The Cox model fits were used to estimate marker-conditional cumulative incidence of COVID-19 marginalizing over baseline variables, with 95% CIs computed using the bootstrap. The Cox models were fit using the survey R package.36 Additional analyses estimated the same quantity with a generalized additive model with degrees of freedom selected by generalized cross-validation,37 and by nonparametric threshold regression.38
Controlled vaccine efficacy and mediation.
Vaccine efficacy by marker level was estimated by a causal inference approach implemented using the IPS-weighted Cox regression described above for the vaccine arm and unweighted Cox regression for the placebo arm, marginalizing over the baseline variables for each randomization arm.39 A sensitivity analysis was conducted to assess robustness of findings to unmeasured confounding of the effect of the antibody marker on COVID-19 (Supplementary Text 3).39 For each antibody marker with at least 10% of vaccinated participants with value the same as for placebo recipients (i.e., negative or undetectable), the proportion of vaccine efficacy mediated by the antibody marker was estimated.40
Hypothesis testing.
All p-values were two-sided. For each set of hypothesis tests for Day 29 and Day 57 marker correlates of risk – which included eight Cox model Wald-tests for correlates of risk including both tertiles of markers and quantitative markers – Westfall-Young multiplicity adjustment41 was applied to obtain q-values and family-wise error rate p-values.
Software and data quality assurance.
The analysis was implemented in R version 4.0.3; the code used for analysis was verified using mock data.
Results
Participant demographics.
Tables 1, S6, and S7 summarize the case-cohort set data and demographics (N = 1010 vaccine, N = 137 placebo in the immunogenicity subcohort). Thirty-four percent of case-cohort participants were age 65 or over, 40% were At-Risk, 47% were female, 32% were Hispanic or Latino, 46% White and Non-Hispanic, and 54% communities of color, with 18% Black or African American.
Table 1:
Anti-Spike and anti-RBD IgG response rates and Geometric Mean Concentrations (GMCs) and pseudovirus calibrated neutralization titer cID50 and cID80 response rates and Geometric Mean Titers (GMTs) by COVID-19 outcome status. Analysis based on baseline negative per-protocol vaccine recipients in the case-cohort set.
| COVID-19 Cases* | Non-Cases | Comparison | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Visit for Marker | Marker | N | Response Rate | GMC or GMT | N | Response Rate | GMC or GMT | Response Rate Difference | Ratio of GM (Cases/Non-Cases) |
| Day 29 | Anti Spike IgG (IU/ml) | 46 | 97.8% (85.4%, 99.7%) | 183 (126, 266) | 1005 | 98.6% (97.4%, 99.2%) | 318 (292, 347) | −0.01 (−0.13, 0.01) | 0.57 (0.39, 0.84) |
| Day 29 | Anti RBD IgG (IU/ml) | 46 | 97.8% (85.4%, 99.7%) | 207 (147, 293) | 1005 | 98.4% (97.2%, 99.1%) | 327 (302, 354) | −0.01 (−0.13, 0.02) | 0.63 (0.44, 0.90) |
| Day 29 | Pseudovirus-nAb cID50 | 46 | 65.2% (50.1%, 77.8%) | 7.6 (5.4, 10.8) | 1005 | 81.7% (78.8%, 84.3%) | 13.0 (11.9, 14.1) | −0.17 (−0.32, −0.04) | 0.59 (0.41, 0.84) |
| Day 29 | Pseudovirus-nAb cID80 | 46 | 43.5% (29.7%, 58.4%) | 18.0 (13.3, 24.2) | 1005 | 63.9% (60.4%, 67.3%) | 29.0 (27.1, 31.0) | −0.2 (−0.35, −0.05) | 0.62 (0.46, 0.84) |
| Day 57 | Anti Spike IgG (IU/ml) | 36 | 100.0% (100.0%, 100.0%) | 1890 (1449, 2465) | 1005 | 99.4% (98.2%, 99.8%) | 2652 (2457, 2863) | 0.01 (0, 0.02) | 0.71 (0.54, 0.94) |
| Day 57 | Anti RBD IgG (IU/ml) | 36 | 100.0% (100.0%, 100.0%) | 2744 (2056, 3664) | 1005 | 99.4% (98.3%, 99.8%) | 3937 (3668, 4227) | 0.01 (0, 0.02) | 0.70 (0.52, 0.94) |
| Day 57 | Pseudovirus-nAb cID50 | 36 | 100.0% (100.0%, 100.0%) | 160 (117, 220) | 1005 | 98.7% (97.6%, 99.3%) | 247 (231, 264) | 0.01 (0.01, 0.02) | 0.65 (0.47, 0.90) |
| Day 57 | Pseudovirus-nAb cID80 | 36 | 97.2% (81.6%, 99.6%) | 332 (248, 444) | 1005 | 98.3% (97.1%, 99.1%) | 478 (450, 508) | −0.01 (−0.17, 0.02) | 0.69 (0.52, 0.93) |
Cases for Day 29 marker correlates analyses are baseline negative per-protocol vaccine recipients with the symptomatic infection COVID-19 primary endpoint diagnosed starting 7 days after the Day 29 study visit. Cases for Day 57 marker correlates analyses are baseline negative per-protocol vaccine recipients with the symptomatic infection COVID-19 primary endpoint diagnosed starting 7 days after the Day 57 study visit.
N is the number of participants with Day 1, 29, 57 antibody marker data included in data analyses (for Non-Cases and for COVID-19 Cases for Day 57 marker correlates analyses). For COVID-19 Cases for Day 29 marker correlates analyses, N is the number with Day 1, 29 antibody marker data included in analyses (Day 57 marker data are not used in Day 29 marker correlates analyses).
GM, geometric mean; GMC, geometric mean concentration; GMT, geometric mean titer.
cID50 and cID80: ID50 and ID80 neutralizing antibody (nAb) titers calibrated to the WHO International Standard.
COVID-19 endpoints.
For Day 57 marker correlates analyses, 47 vaccine recipients had a COVID-19 endpoint. For Day 29 marker correlates analyses, an additional 8 intercurrent COVID-19 endpoints occurred. Overall cumulative incidence from 7 to 126 days post Day 29 was 0.005 in vaccine recipients compared to 0.065 in placebo recipients. Average follow-up of vaccine recipients was 81 days (starting 7 days post Day 57 visit) for Day 57 marker correlates analyses and 109 days (starting 7 days post Day 29 visit) for Day 29 marker correlates analyses.
Antibody marker distributions.
At Day 57, almost 100% of vaccine recipients had positive/detectable antibody response by all four markers (Table 1). This was also true at Day 29 for Spike IgG and RBD IgG, whereas cID50 and cID80 titers were detectable in 82% and 64% of vaccine recipients, respectively. Each marker was moderately correlated between the Day 29 and Day 57 time points (Spearman rank r = 0.527 to 0.625, Figure S5). Median (interquartile range) days from dose one to Day 29 was 28 (28 to 30) and from Day 29 to Day 57 was 28 (28 to 30). The Spike IgG and RBD IgG markers were tightly correlated (Spearman rank r = 0.942, 0.969 at Day 29, 57; Figures S6, S7), as were the cID50 and cID80 markers (r = 0.968, 0.961 at Day 29, 57; Figures S6, S7). Accordingly, some results only show Spike IgG and cID50 for brevity; results were similar for RBD IgG and cID80 (Supplementary Appendix). Each binding antibody marker was correlated with each neutralization marker at each time point (r = 0.734 to 0.804).
Figures 1 and S8 show the Day 29 and Day 57 marker distributions by case/non-case status in vaccine recipients (Figure S9 shows analogous plots for placebo recipients), with Figures S10 and S11 showing marker values by participant age. For all eight markers the geometric mean was lower in cases, with 95% CIs about geometric mean ratios (cases/non-cases) below one (Table 1). Figures S12 and S13 show reverse cumulative distribution function curves of the eight markers, in the context of the overall vaccine efficacy estimates.42 Figure S14 shows the marker values of vaccine breakthrough cases by timing of COVID-19 endpoint diagnosis.
Figure 1:
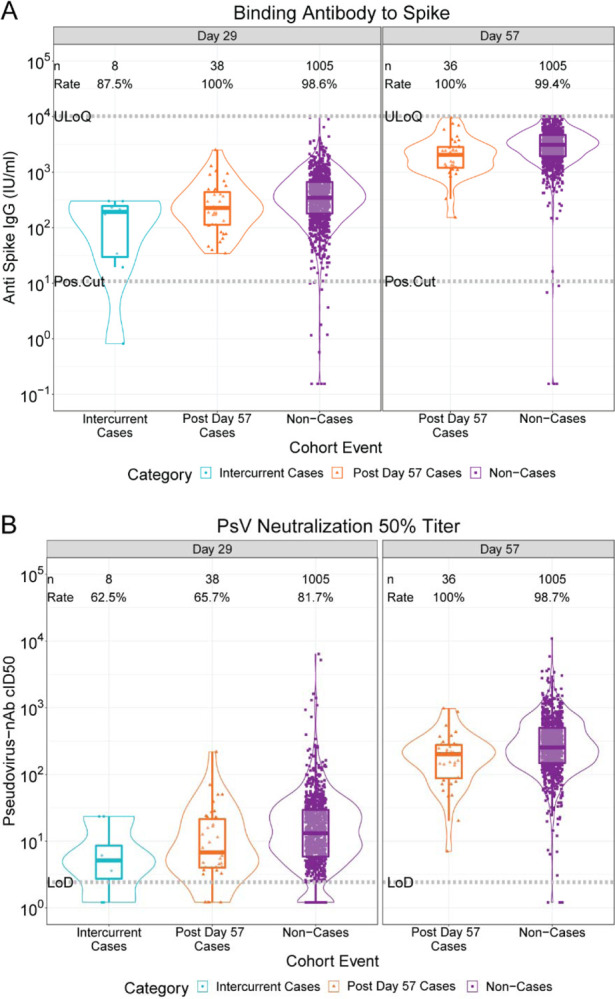
(A) Anti-Spike IgG concentration and (B) pseudovirus neutralization cID50 titer by COVID-19 outcome status. Data points are from baseline negative per-protocol vaccine recipients selected into the case-cohort set. The violin plots contain interior box plots with upper and lower horizontal edges the 25th and 75th percentiles of antibody level and middle line the 50th percentile. Each side shows a rotated probability density (estimated by a kernel density estimator with a default Gaussian kernel) of the data. Positive response rates were computed with inverse probability of sampling weighting. Pos.Cut, Positivity cut-off. LoD, limit of detection. ULoQ, upper limit of quantitation; ULOQ = 10,919 for cID50 (above all data points). Post Day 57 cases are COVID-19 endpoints starting 7 days post Day 57 visit; Intercurrent cases are COVID-19 endpoints starting 7 days post Day 29 visit through 6 days post Day 57 visit. cID50: ID50 nAb titer calibrated to the WHO International Standard.
Correlates of risk.
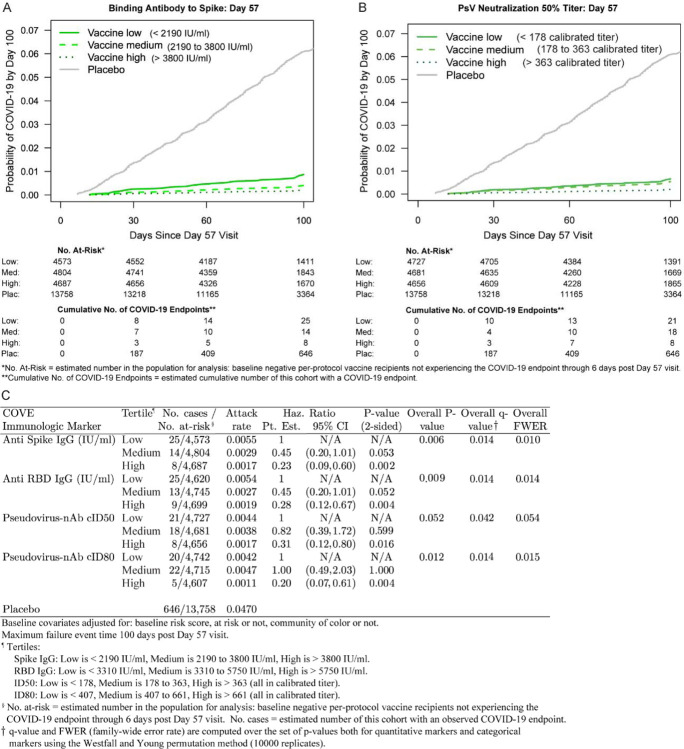
Figure 2 shows COVID-19 cumulative incidence curves for subgroups of vaccine recipients defined by tertile of Day 57 IgG Spike and cID50 (A, B) (results for IgG RBD and cID80 in Figure S15). A dose response was observed for each marker, with estimated hazard ratios for upper vs. lower tertiles between 0.20 and 0.31 and 95% CIs and multiplicity-adjusted p-values indicating significant inverse correlations with risk (Figure 2C). Results were similar for Day 29 markers, with smaller hazard ratios and smaller p-values indicating strengthened evidence for correlates of risk (Figures S16, S17).
Figure 2.
Covariate-adjusted cumulative incidence of COVID-19 by Low, Medium, High tertile of Day 57 IgG concentration or pseudovirus neutralization titer. (A) Anti-Spike IgG concentration; (B) cID50 titer; (C) IgG (Spike, RBD) and (cID50, cID80). The overall p-value is from a generalized Wald test of whether the hazard rate of COVID-19 differed across the Low, Medium, and High subgroups. cID50, cID80: ID50 or ID80 nAb titer calibrated to the WHO International Standard.
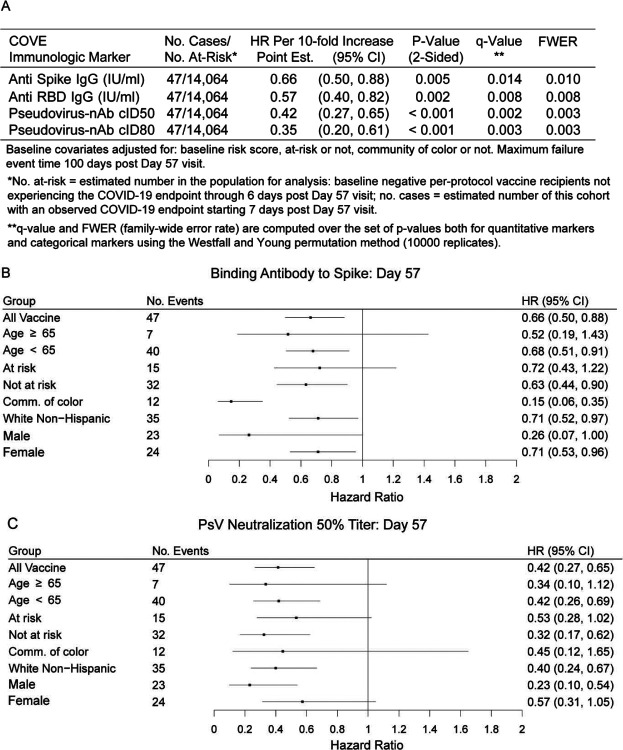
For quantitative Day 57 markers, the estimated hazard ratio per 10-fold increase in marker value ranged between 0.35 and 0.66 (Figure 3A), with 95% CIs below one and multiplicity-adjusted p-values indicating significant associations. Generally, similar results were obtained across pre-specified subgroups of vaccine recipients (Figure 3B and 3C, Figure S18).
Figure 3.
Covariate-adjusted hazard ratios of COVID-19 per 10-fold increase in each Day 57 antibody marker in baseline negative per-protocol vaccine recipients overall and in subgroups. (A) Inferences for IgG (Spike, RBD) and (cID50, cID80); (B) Forest plots for Spike IgG; (C) Forest plots for cID50. cID50, cID80: ID50 or ID80 neutralizing antibody titer calibrated to the WHO International Standard. Comm. of color = all participants other than White Non-Hispanic.
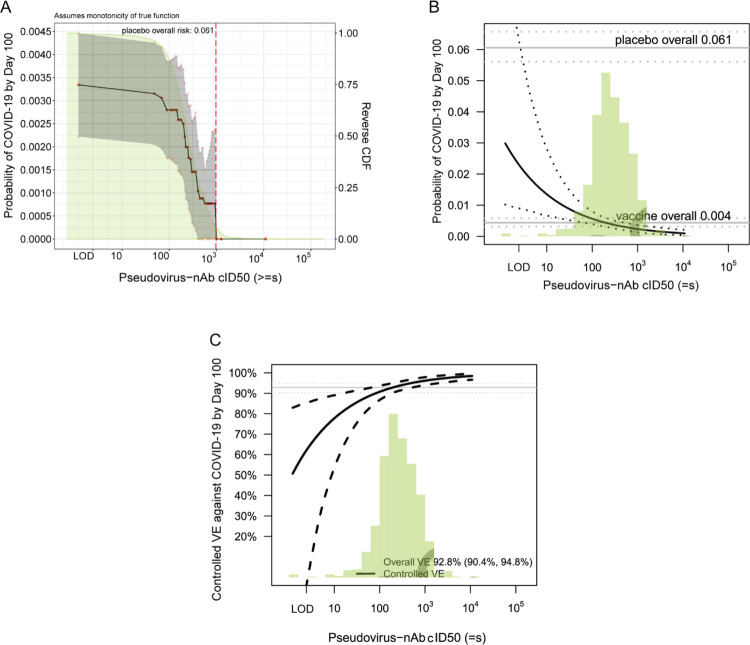
The estimated cumulative incidence of COVID-19 by end of follow-up (100 days post Day 57) for the entire vaccine group was 0.0033 (95% CI 0.0022, 0.0045). Based on nonparametric threshold regression this rate decreased across vaccinated subgroups with Day 57 cID50 titer above a given threshold, with estimates of 0.00088 (0.00, 0.0020) at cID50 titer above 500 and zero COVID-19 endpoints at cID50 titer above 1000 (Figure 4A, Figure 1B). In contrast, 5% of non-cases had cID50 titer above 1000 (Figure 1B). Based on the Cox model, the COVID-19 rate for vaccine recipients with undetectable Day 57 cID50 titer (value < LOD) was 0.030 (0.010, 0.093), and decreased to 0.0056 (0.0039, 0.0080) at titer of 100 and to 0.0023 (0.0013, 0.0036) at titer of 1000 (Figure 4B). Results using an alternative model (generalized additive) also supported inverse correlates of risk for all markers (Figures S19, S20).
Figure 4.
(A) Covariate-adjusted cumulative incidence of COVID-19 by 100 days post Day 57 by subgroups defined by Day 57 cID50 level above a threshold, with reverse cumulative distribution function of Day 57 cID50 level overlaid in green. The gray shaded area is pointwise 95% confidence intervals (CIs). The upper boundary of the green shaded area is the estimate of the reverse cumulative distribution function (CDF) of the marker in baseline SARS-CoV-2 negative per-protocol vaccine recipients. (B) Covariate-adjusted cumulative incidence of COVID-19 by 100 days post Day 57 by Day 57 cID50 level. The dotted lines indicate bootstrap point-wise 95% CIs. (C) Vaccine efficacy by Day 57 cID50 level, estimated using the method of Gilbert, Fong, and Carone.39 The dashed lines indicate bootstrap point-wise 95% CIs. In (B) and (C), covariate adjustment was based on an inverse probability samplingweighted Cox model; the green histograms are an estimate of the density of Day 57 cID50 level in baseline negative per-protocol vaccine recipients. LOD, limit of detection. cID50: ID50 nAb titer calibrated to the WHO International Standard.
Correlates of protection.
Vaccine efficacy estimates (obtained using the method of Gilbert, Fong, and Carone39) increased with Day 57 cID50 neutralization titer: At undetectable (<LOD) Day 57 cID50 titer, vaccine efficacy was 50.8% (−51.2, 83.0%). At Day 57 cID50 titer of 100, vaccine efficacy increased to 90.7% (86.7, 93.6%), and at Day 57 cID50 titer of 1000, vaccine efficacy further increased to 96.1% (94.0, 97.8%) (Figure 4C). Figures S21–S27 show these results for the other seven antibody markers. The sensitivity analysis allowing unmeasured confounding (Figure S28C) yielded estimates of vaccine efficacy of 89.8% (68.8, 96.5%) at undetectable Day 57 cID50 titer, 94.9% (92.6, 96.8%) at Day 57 cID50 titer of 500, and 95.8% (93.4, 97.6%) at Day 57 cID50 titer of 1000 (see Figures S28 and S29 for results for the other seven markers). The sensitivity analysis based on E-values43 of the vaccine recipient tertile subgroups supported the finding that vaccine efficacy was generally higher for the upper tertile subgroup, indicating some robustness to potential unmeasured confounding (Table S8).
Day 29 cID50 and cID80 titers could be assessed as mediators of VE by the Benkeser et al. method,40 given 18% and 36% of vaccine recipients had undetectable titer, respectively. An estimated 68.5% (58.5, 78.4%) of VE was mediated by Day 29 cID50 titer and 48.5% (34.5, 62.4%) by Day 29 cID80 titer (Table S9).
Discussion
For recipients of two doses of mRNA-1273 in the COVE trial, all four antibody markers at Day 29 and at Day 57 were inverse correlates of risk of COVID-19 occurrence through 3–4 months post second dose. Based on any of the antibody markers, COVID-19 risk was about 10 times lower for vaccine recipients with antibodies in the top 10% of values compared to those with negative/undetectable values (the same value as in baseline negative placebo recipients).
The correlates of protection analyses found that estimated vaccine efficacy against COVID-19 increased with Day 29 and with Day 57 antibody levels, where for the latter estimated vaccine efficacy was about 45–60% for vaccine recipients with negative/undetectable binding or neutralizing antibody response (Figure 4C; Figures S21C, S22C, S23C) and increased to above 98% at highest antibody levels. Positive vaccine efficacy for the vaccinated subgroup with negative/undetectable antibody level implies lack of full mediation of vaccine efficacy through the antibody marker,39 and formal causal analysis estimated that 68% of the overall vaccine efficacy was mediated through Day 29 cID50 titer. To interpret the mediation result: if neutralizing antibodies circulating on Day 29 could be removed but the other consequences of vaccination remained, overall vaccine efficacy would be expected to change from 92.3% to 56%, a 68% reduction (on the log scale) of overall vaccine efficacy. In comparison, hemagglutination inhibition titer against B/Victoria influenza virus mediated an estimated 57% of inactivated influenza vaccine efficacy against virologically confirmed B/Victoria influenza illness.44 As hemagglutination inhibition titer has been used to guide vaccine strain selection and approval, this defines a benchmark for a potential level of mediation relevant for supporting vaccine approval decisions.45 The very high overall vaccine efficacy and the small number of vaccine recipients with negative/undetectable antibody levels in COVE limited the ability to identify a high percentage of neutralization-mediated vaccine efficacy. Similar analyses of trials with lower vaccine efficacy and lower neutralization response rates may be able to detect a greater percentage of vaccine efficacy mediated through neutralizing antibody titers.
An interpretation also consistent with our results is that neutralization as a biological function did fully mediate the vaccine efficacy in COVE, but the cID50 and cID80 markers had inadequate sensitivity to quantify low-level neutralization below the detection limit that was partially protective. This hypothesis is supported by the finding that passive transfer of purified IgG from mRNA-1273-immunized rhesus macaques was sufficient to protect golden Syrian hamsters from disease after SARS-CoV-2 challenge.30 Yet, it is more likely that additional immune responses are needed to fully explain the observed vaccine efficacy, given that the vaccine provided more than 90% efficacy by Day 29,9 18% of vaccine recipients had undetectable neutralization response at Day 29 based on cID50 (Figure 1, Figure S8), and the vaccine elicited other immune responses by Day 29 in a high proportion of vaccine recipients.46 In addition to markers measuring distinct immunological functions, markers not measured fully in serum (e.g., mucosal markers) and/or anamnestic responses not fully represented by a single time point measurement may be needed to fully explain vaccine efficacy.
The immune correlates results of the AZD12222 trial25 for Spike IgG and RBD IgG can be quantitatively compared to the COVE results by virtue of the same MSD assay platform, conversion of IgG concentration to WHO International Units/ml, and the same antibody measurement time: 4 weeks post second dose. Estimated ChAdOx1 nCoV-19 vaccine efficacy was 70% and 90% at Spike IgG levels of 113 (95% CI < LOD = 0.31, 245) and 899 (369, NC) IU/ml, respectively, and at RBD IgG levels of 165 (< LOD = 1.59, 452) and 2360 (723, NC) IU/ml, respectively (NC = not calculated).25 In comparison, estimated mRNA-1273 vaccine efficacy was 70% and 90% at Day 57 Spike IgG levels of 1 (< LOD = 0.31, 170) and 298 (1, 1786) IU/ml, respectively, and at Day 57 RBD IgG levels of 8 (< LOD = 1.59, 243) and 775 (29, 2819) IU/ml, respectively. These results are suggestive that lower binding antibody levels in COVE are associated with the same level of vaccine efficacy.
Pseudovirus neutralization results can also be compared between the trials using calibrated ID50 titers, where estimated ChAdOx1 nCoV-19 vaccine efficacy was 70% and 90% at cID50 titer of 8 (< LOD = 2.42, 26) and 140 (43, NC), compared to COVE results at cID50 titer of 4 (< LOD = 2.42, 22) and 83 (16, 188). The neutralization assays employed for AZD12222 and COVE used the D614 and D614G pseudovirus, respectively, such that this comparison is affected by any difference in neutralization sensitivity of these two strains. As a sensitivity analysis, we multiplied the AZD12222 cID50 values by a second conversion factor, 2.19, which scales the values as if D614G had been used in the assay (2.19 was calculated from the validation study that used both strains, Supplementary Text 1). The cID50 titers at 70% and 90% ChAdOx1 nCoV-19 vaccine efficacy are 18 (< LOD = 2.42, 57) and 307 (95, NC), respectively.
With the caveats of different study endpoints/hosts, the COVE results are also consistent with results on Spike IgG and neutralizing antibody titers as correlates of protection against SARS-CoV-2 replication in mRNA-1273-vaccinated rhesus macaques. For instance, all macaques with Spike IgG > 336 IU/ml at 4 weeks post second dose were protected from >10,000 subgenomic RNA copies/ml in bronchoalveolar lavages,30 and in COVE, Day 57 Spike IgG of 336 IU/ml corresponded to 90% vaccine efficacy against COVID-19 (Figure S22).
Together with evidence from other studies, the current results support that neutralization titer is a potential surrogate marker for mRNA-1273 vaccination against COVID-19 that can be considered as a primary endpoint for basing certain accelerated approval decisions. The total body of evidence may support an immunogenicity non-inferiority approach to bridging vaccine efficacy of refined or novel mRNA vaccines, which has been proposed for adding vaccine spike variants and boosters.47 One open question challenging this approach is whether titers to different strains have different relationships with vaccine efficacy. A more challenging objective is bridging vaccine efficacy of new candidate vaccines in different platforms from mRNA. When correlates results are available from several phase 3 vaccine efficacy trials of different vaccine platforms, it will be possible to conduct validation analyses of how well an antibody marker can be used to predict vaccine efficacy across platforms.
We also note that the Day 29 markers correlated with COVID-19 at least as strongly as the Day 57 markers. If a Day 29 immune marker in recipients of two mRNA-1273 doses becomes established as a correlate of protection, it could be a more practical surrogate marker than a Day 57 marker by virtue of being measured earlier and because trials would not need participants to come back for a Day 57 study visit.
Limitations of this correlates study include the lack of data for assessing correlates against other outcomes besides COVID-19 (e.g., severe COVID-19, asymptomatic SARS-CoV-2 infection, infection regardless of symptomology, viral shedding), the lack of assessment of correlates for recipients of only one mRNA-1273 dose, and the fact that almost all COVID-19 cases resulted from infections with viruses bearing genetically and antigenically similar Spike as compared to the ancestral Spike in the vaccine, implying that it was not possible to assess robustness of correlates to SARS-CoV-2 variants of concern. However, the paucity of divergent circulating virus is also a strength in affording a clear interpretation as correlates against COVID-19 caused by variants genetically close to the vaccine. Another strength is the use of validated assays with all results reported in WHO International Units or calibrated to WHO International Standards, facilitating comparison of results from other studies and vaccine platforms. An additional strength is the racial and ethnic diversity of the trial participants and large number of diverse participants sampled for immunogenicity measurements.48
This study evaluated antibody markers measured at two specific time points (Day 29, Day 57) as immune correlates against subsequent COVID-19 occurrence over 3–4 months follow-up, but did not assess how the current level of antibody correlated with instantaneous risk of COVID-19. COVE follows participants for 2 years, enabling future analyses of when and how vaccine efficacy wanes as antibody levels wane and as new variants of SARS-CoV-2 emerge, informing decisions about the timing of a booster or the need to update vaccine composition, and potentially clarifying the mediation of protection. Future correlates analyses that study antibody markers over time in relation to the timing of breakthrough infections of variants with variable neutralization sensitivity might further clarify functional mediation of protection.
In conclusion, the result that all evaluated binding and neutralizing antibody markers strongly inversely correlated with COVID-19 risk, and directly correlated with vaccine efficacy, as well as the result that a substantial proportion of the overall vaccine efficacy was mediated through neutralizing antibody levels, adds evidence toward establishing an immune marker surrogate endpoint for COVID-19 vaccines.
Supplementary Material
Acknowledgments
We thank the volunteers who participated in the COVE trial. We also thank John Mascola, Matt Hepburn, Robert A. Johnson, Mary Marovich, and Merlin Robb.
Funding
Supported by Public Health Service Grant UM1 AI068635 to the HVTN Statistical Data and Management Center [SDMC], Fred Hutchinson Cancer Research Center [FHCRC] from the National Institute of Allergy and Infectious Diseases (NIAID) and by the Intramural Research Program of the NIAID. This work was also supported by Scientific Computing Infrastructure at Fred Hutch funded by ORIP grant S10OD028685. This project has been funded in whole or in part with Federal funds from the Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, under Contract No. 75A50120C00034, and Moderna, Inc.
References
- 1.Centers for Disease Control and Prevention. COVID-19 Vaccines and Vaccination. CDC COVID-19 Science Briefs. National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. Atlanta (GA). [Updated 2021 May 27]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK570435/. 2021. [PubMed] [Google Scholar]
- 2.McGill COVID19 Vaccine Tracker Team. COVID19 Vaccine Tracker. https://covid19.trackvaccines.org/ Access date 21 Jun, 2021. Last updated 19 Jun, 2021. 2021.
- 3.Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 Vaccines. JAMA 2021;325:1318–20. [DOI] [PubMed] [Google Scholar]
- 4.Weller C. Four reasons why we need multiple vaccines for Covid-19. Wellcome Opinion. 6 Jun 2021. Access date 21 Jun 2021. https://wellcome.org/news/four-reasons-why-we-need-multiplevaccines-covid-19. [Google Scholar]
- 5.Wouters OJ, Shadlen KC, Salcher-Konrad M, et al. Challenges in ensuring global access to COVID-19 vaccines: production, affordability, allocation, and deployment. Lancet 2021;397:1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plotkin SA, Gilbert PB. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis 2012;54:1615–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol 2010;17:1055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plotkin SA, Orenstein WA, Offit PA, Edwards KM. Plotkin’s Vaccines. Seventh Edition. 2018; Elsevier, Inc. Chapter 3: Correlates of Protection. Plotkin Stanley A. and Gilbert Peter. pp 35–40. [Google Scholar]
- 9.Baden LR, El Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med 2021;384:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US Food and Drug Administration. Moderna COVID-19 Vaccine. Updated Apr 1, 2021. Access date Jun 21, 2021. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease2019-covid-19/moderna-covid-19-vaccine.
- 11.Centers for Disease Control and Prevention. COVID Data Tracker. COVID-19 Vaccinations in the United States. Last updated 20 Jun 2021. Access date 21 Jun 2021. Available at https://covid.cdc.gov/covid-data-tracker/#vaccinations. [Google Scholar]
- 12.Thompson MG, Burgess JL, Naleway AL, et al. Interim Estimates of Vaccine Effectiveness of BNT162b2 and mRNA-1273 COVID-19 Vaccines in Preventing SARS-CoV-2 Infection Among Health Care Personnel, First Responders, and Other Essential and Frontline Workers - Eight U.S. Locations, December 2020-March 2021. MMWR Morb Mortal Wkly Rep 2021;70:495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moderna. Moderna Announces TeenCOVE Study of its COVID-19 Vaccine in Adolescents Meets Primary Endpoint and Plans to Submit Data to Regulators in Early June. Press Release. 25 May, 2021. Access date 21 Jun, 2021. Available at https://investors.modernatx.com/news-releases/news-releasedetails/moderna-announces-teencove-study-its-covid-19-vaccine.
- 14.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020;581:215–20. [DOI] [PubMed] [Google Scholar]
- 16.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020;5:562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 2020;117:11727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet 2020;396:1595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Tostanoski LH, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020;369:806–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He X, Chandrashekar A, Zahn R, et al. Low-dose Ad26.COV2.S protection against SARS-CoV-2 challenge in rhesus macaques. Cell 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021;590:630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021. [DOI] [PubMed] [Google Scholar]
- 23.Earle KA, Ambrosino DM, Fiore-Gartland A, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. [Preprint] Access date 22 Jun, 2021. 10.1101/2021.03.17.20200246 medRxiv 2021:2021.03.17.20200246. [DOI] [PMC free article] [PubMed]
- 24.Cromer D, Reynaldi A, Steain M, Triccas JA, Davenport MP, Khoury DS. Relating in vitro neutralisation level and protection in the CVnCoV (CUREVAC) trial. doi: 10.1101/2021.06.29.21259504 Access date 2 Jul, 2021. medRxiv 2021:2021.06.29.21259504. [DOI] [PMC free article] [PubMed]
- 25.Feng S, Phillips DJ, White T, et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. [Preprint] Access date 25 Jun, 2021 10.1101/2021.06.21.21258528 medRxiv 2021:2021.06.21.21258528. [DOI] [PMC free article] [PubMed]
- 26.Addetia A, Crawford KHD, Dingens A, et al. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J Clin Microbiol 2020;58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N Engl J Med 2021;384:1866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen MS, Nirula A, Mulligan MJ, et al. Effect of Bamlanivimab vs Placebo on Incidence of COVID-19 Among Residents and Staff of Skilled Nursing and Assisted Living Facilities: A Randomized Clinical Trial. JAMA 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regeneron. Phase 3 Prevention Trial Showed 81% Reduced Risk of Symptomatic SARS-CoV-2 Infections with Subcutaneous Administration of REGEN-COV™ (casirivimab with imdevimab). Press Release. 12 Apr, 2021. Access date 22 Jun, 2021. Available at https://investor.regeneron.com/newsreleases/news-release-details/phase-3-prevention-trial-showed-81-reduced-risk-symptomatic-sars.
- 30.Corbett KS, Nason MC, Flach B, et al. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science 2021:eabj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen X, Tang H, McDanal C, et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 2021;29:529–39 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristiansen PA, Page M, Bernasconi V, et al. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet 2021;397:1347–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Institute for Biological Standards and Control (NIBSC). Instructions for use of First WHO International Standard for anti-SARS-CoV-2 Immunoglobulin (Version 3.0, Dated 17/12/2020) NIBSC code: 20/136 https://www.nibsc.org/science_and_research/idd/cfar/covid-19_reagents.aspx Access date Jul 29, 2021.
- 34.Prentice RL. A Case-Cohort Design for Epidemiologic Cohort Studies and Disease Prevention Trials. Biometrika 1986;73:1–11. [Google Scholar]
- 35.Newcombe RG. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat Med 1998;17:857–72. [DOI] [PubMed] [Google Scholar]
- 36.Lumley T. Complex Surveys: A Guide to Analysis Using R. vol. 565, John Wiley and Sons. 2010. [Google Scholar]
- 37.Wood SN. Generalized Additive Models: An Introduction with R. Chapman and Hall/CRC Texts in Statistical Science, CRC Press, Boca Raton, FL. 10.1201/9781315370279 2017. [DOI] [Google Scholar]
- 38.van der Laan L, Zhang W, Gilbert PB. Efficient nonparametric estimation of the covariate-adjusted threshold-response function, a support-restricted stochastic intervention [Preprint] Access date 28 Jul, 2021. arXiv:2107.11459 [stat.ME]. 2021.
- 39.Gilbert PB, Fong Y, Carone M. Assessment of Immune Correlates of Protection via Controlled Risk and Controlled Vaccine Efficacy [Preprint] Access date 13 Jul, 2021. arXiv:2107.05734 [stat.ME]. 2021.
- 40.Benkeser D, Díaz I, Ran J. Inference for natural mediation effects under case-cohort sampling with applications in identifying COVID-19 vaccine correlates of protection [Preprint] Access date 13 Jul, 2021. arXiv:210302643 [statME] 2021.
- 41.Westfall PH, Young SS. Resampling-based multiple testing: Examples and methods for p-value adjustment: John Wiley & Sons; 1993. [Google Scholar]
- 42.Siber GR, Chang I, Baker S, et al. Estimating the protective concentration of anti-pneumococcal capsular polysaccharide antibodies. Vaccine 2007;25:3816–26. [DOI] [PubMed] [Google Scholar]
- 43.VanderWeele TJ, Ding P. Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann Intern Med 2017;167:268–74. [DOI] [PubMed] [Google Scholar]
- 44.Cowling BJ, Lim WW, Perera RAPM, et al. Influenza Hemagglutination-inhibition Antibody Titer as a Mediator of Vaccine-induced Protection for Influenza B. Clinical Infectious Diseases 2019;68:1713–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleming TR, Powers JH. Biomarkers and surrogate endpoints in clinical trials. Stat Med 2012;31:2973–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N Engl J Med 2020;383:1920–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.U.S. Department of Health and Human Services, Food and Drug Administration, and Center for Biologics Evaluation and Research. Emergency Use Authorization for Vaccines to Prevent COVID-19: Guidance for Industry. Appendix 2: pp 18–22. Issued on 25 May, 2021. https://www.fda.gov/regulatoryinformation/search-fda-guidance-documents/emergency-use-authorization-vaccines-prevent-covid-19.
- 48.Andrasik MP, Broder GB, Wallace SE, Chaturvedi R, Michael NL, et al. Increasing Black, Indigenous and People of Color participation in clinical trials through community engagement and recruitment goal establishment. PLOS One (in press) 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.