Abstract
Renal cell carcinomas (RCCs) are a diverse set of malignancies that have recently been shown to harbour mutations in a number of chromatin modifier genes — including PBRM1, SETD2, BAP1, KDM5C, KDM6A, and MLL2 — through high-throughput sequencing efforts. Current research focuses on understanding the biological activities that chromatin modifiers employ to suppress tumorigenesis and developing clinical approaches that take advantage of this knowledge. Unsurprisingly, several common themes unify the functions of these epigenetic modifiers, particularly regulation of histone post-translational modifications and nucleosome organization. Furthermore, chromatin modifiers also govern processes crucial for DNA repair and maintenance of genomic integrity, as well as the regulation of splicing and other key processes. Many chromatin modifiers have additional noncanonical roles in cytoskeletal regulation, which further contribute to genomic stability, expanding the repertoire of functions that might be essential in tumorigenesis. Our understanding of how mutations in chromatin modifiers contribute to tumorigenesis in RCC is improving, but remains an area of intense investigation. Importantly, elucidating the activities of chromatin modifiers offers intriguing opportunities for the development of new therapeutic interventions in RCC.
Introduction
Renal cell carcinoma (RCC) is among the top ten most commonly diagnosed malignancies in the United States, with an estimated 63,990 new cases having been diagnosed in 20181. RCC is a heterogeneous disease that includes several histological subtypes; the most common histology is clear cell renal cell carcinoma (ccRCC), which represents up to 75% of RCCs, whereas papillary RCC (pRCC), chromophobe renal cell carcinoma (chRCC) and several rare tumour types such as renal medullary carcinoma comprise the remaining ~25% (REF. 2). Major therapeutic advances in the treatment of metastatic RCC have been made over the past two decades, mainly owing to the advent of antiangiogenic targeted therapies and immunotherapies. However, only a fraction of patients show durable clinical responses and long-term remission3.In addition, the long-term prognosis for all patients with relapsed and metastatic disease remains poor4. Thus, an urgent clinical need exists for novel insights and therapeutic strategies.
The necessity for novel therapeutic strategies in RCC has motivated considerable research dedicated to dissecting the molecular biology underpinning this unique group of cancers. The discovery of mutations in the gene encoding von Hippel-Lindau disease tumour suppressor (VHL) in ccRCC was a major step forward5. Somatic and germline inactivating VHL mutations, as well as deletion of VHL, are present in a very large proportion of ccRCCs6–11. Loss of VHL function results in the inappropriate stabilization of hypoxia inducible factor (HIF), which triggers a pseudohypoxic response that includes a profound upregulation of angiogenesis12. This understanding inspired the clinical application of angiogenesis inhibitors in ccRCC, which now represent a clinical standard of care13. However, the field has struggled with the identification of co-drivers, given that VHL mutations are necessary, but not sufficient, for ccRCC tumorigenesis14,15.
In 2010, the targeted sequencing of 3,544 protein coding genes in 101 ccRCC samples uncovered mutations in four genes encoding histone modifying enzymes, namely: histone-lysine N-methyltransferase SETD2 (SETD2), a histone H3 lysine 36 methyltransferase; lysine-specific demethylase 5C (KDM5C; also known as JARID1C), a histone H3 lysine 4 demethylase; histone-lysine N-methyltransferase 2D (KMT2D; also known as MLL2), a histone H3 lysine 4 methyltransferase; and lysine-specific demethylase 6A (KDM6A; also known as UTX), a histone H3 lysine 27 demethylase16. The following year, mutations in protein polybromo-1 (PBRM1; also known as BAF180), ubiquitin carboxyl-terminal hydrolase BAP1 (BAP1), AT-rich interactive domain-containing protein 1A (ARID1A; also known as BAF250A) and ARID1B (also known as BAF250B) genes were identified in ccRCC17,18. BAP1 is a deubiquitinase that targets mono-ubiquitylation of lysine 119 on histone H2A (H2AK119ub1), whereas PBRM1, ARID1A and ARID1B are components of the switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex, which is involved in nucleosome repositioning19. Several large-scale sequencing studies have since confirmed these findings and have further characterized the repertoire of genetic alterations, not only in large numbers of ccRCCs, but also in pRCCs and chRCCs6,7,9,20,21. These enzymes all share a common theme — they participate in modifying chromatin structure.
In this Review, we focus on requisite co-drivers in RCC, which share common chromatin modifying activity and discuss how mutations in their encoding genes contribute to the development and progression of this disease. We cover key aspects of chromatin modifier biology and the repertoire of mutations in these genes in RCC, and discuss the canonical and noncanonical functions of chromatin modifiers and how their dysfunction contributes to this disease. Finally, we examine potential therapeutic opportunities created by loss-of-function of chromatin modifiers.
Chromatin modifiers in RCC
When eukaryotic cells are not dividing, their DNA is assembled around core histones (H2A, H2B, H3 and H4 family proteins), the basic structural units of the nucleosome, and packaged along with other protein complexes within the nucleus; the resulting complex macromolecule is called chromatin22. Linker histones attach to core histones at DNA entry and exit sites and function to mediate the arrangement of nucleosomes with respect to both each other and to their associated gene units. Access to nucleosomal DNA is further mediated by the action of chromatin remodeling factors, which are in turn aided by a complex coding system of post-translational modifications (PTMs) present on the histones23. These PTMs — including methylation, acetylation, phosphorylation, ubiquitylation, sumoylation, citrullination and ADP-ribosylation — occur in the long C-terminal tail domains of histones (which protrude from the nucleosome), altering their interaction with DNA and nuclear proteins. Histone PTMs and their associated functions are dynamically regulated by numerous enzymes that can be classified as ‘writers’ (enzymes that add PTMs), ‘erasers’ (enzymes that remove PTMs) and ‘readers’ (interacting proteins that recognize the histone PTM). This combination of nucleosome position and associated PTMs form a ‘histone code’ that can be interpreted by other proteins to elicit a variety of biological functions23. Thus, chromatin organization provides an additional layer of transcriptional regulation by controlling DNA accessibility. This remarkable architecture enables the dynamic genomic regulation that is necessary for a cell to orchestrate its regular functions.
Chromatin remodelers
When wrapped around histones, DNA is less accessible for transcription, as well as other processes; thus, DNA accessibility must be controlled by nucleosomal remodeling complexes, which interact with PTMs to guide localization and subsequently restructure chromatin by moving, destabilizing and ejecting nucleosomes24. To date, four families of chromatin remodelers have been identified — SWI/SNF, imitation switch (ISWI), chromodomain helicase DNA-binding (CHD) and INO8025.
Alterations in components of the SWI/SNF chromatin remodeling complex are frequently observed in RCC7,18,20,21, as well as in other cancers26–30. SWI/SNF complexes are assembled around a catalytic subunit — protein brahma homolog (BRM; encoded by SMARCA2) or transcription activator BRG1 (encoded by SMARCA4) — that provides energy for chromatin remodeling through ATP hydrolysis31 (FIG. 1). The BRM subunit is incorporated into a complex referred to as the BRM-associated factor (BAF) complex, whereas the BRG1 subunit can nucleate either BAF or polybromo-associate BAF (PBAF) complexes. The BAF and PBAF complexes differ in the composition of several subunits, including those involved in chromatin targeting, which are thought to mediate recruitment of the complex to specific chromatin regions. The ARID1A and ARID1B subunits are believed to participate in the targeting of BAF complexes32,33, whereas the AT-rich interactive domain-containing protein 2 (ARID2; also known as BAF200)34, bromodomain-containing protein 7 (BRD7)35 and PBRM136 subunits are thought to mediate PBAF complex targeting to DNA.
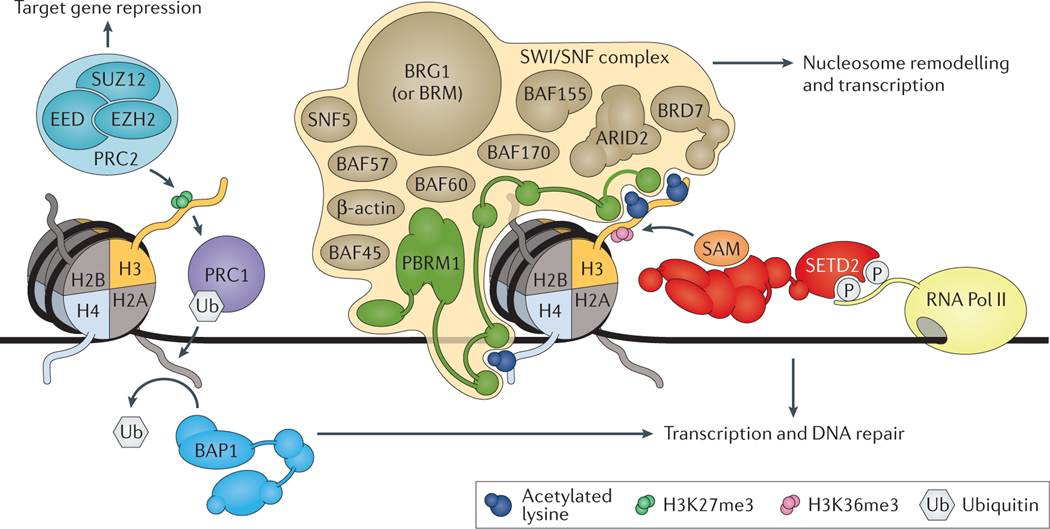
Figure 1 |. Chromatin modification by PBRM1, BAP1 and SETD2.
The switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex, shown here as polybromo-associate BAF (PBAF), is comprised of >11 subunits, including: a catalytic core subunit (protein brahma homolog (BRM) or transcription activator BRG1); accessory subunits (β-actin, SNF5, BAF45, BAF57, BAF60, BAF155, and BAF170); and DNA binding subunits (AT-rich interactive domain-containing protein 2 (ARID2), bromodomain-containing protein 7 (BRD7), and protein polybromo-1 (PBRM1); or alternatively, ARID1A and ARID1B). PBRM1 contains 6 bromodomains that mediate DNA targeting by binding acetylated histone residues. Histone-lysine N-methyltransferase SETD2 associates with hyperphosphorylated RNA polymerase II (RNA Pol II) and deposits the histone H3 lysine 36 trimethyl mark (H3K36me3) as it travels along with RNA Pol II during transcription; SETD2 uses S-Adenosyl methionine (SAM) as a one carbon donor during methylation. Consequently, SETD2 deposits the H3K36me3 mark primarily on exons of actively transcribed genes. Thus, SETD2 and its associated mark (H3K36me3) are involved in regulating transcription and mediating DNA damage repair. Polycomb repressive complex 1 (PRC1) and PRC2 are responsible for regulating cellular differentiation and lineage commitment and maintenance via transcriptional repression of target genes. PRC2, comprised of polycomb protein SUZ12, polycomb protein EED, and histone-lysine N-methyltransferases EZH1 or EZH2, catalyzes the addition of the trimethyl mark on lysine 27 of histone H3 (H3K27me3), a mark that is associated with gene silencing. Canonically, H3K27me3 subsequently recruits PRC1 to reinforce gene repression through mono-ubiquitylation of lysine 119 on histone H2A (H2AK119ub1). PRC1 activity can be reversed by ubiquitin carboxyl-terminal hydrolase BAP1, a protein deubiquitinase that catalyzes the deubiquitination of H2AK119ub1. BAP1-mediated deubiquitination of H2AK119ub1 has important roles in regulating transcription and mediating DNA damage repair.
PBRM1.
The PBRM1 subunit contains six tandem bromodomains, two bromo-adjacent homology (BAH) domains and a high-mobility group (HMG) domain (FIG. 2A). Bromodomains, composed of ~100 amino acids, recognize acetylated lysine residues on histone tails, a mark associated with active transcription37. Each bromodomain of PBRM1 has a distinct pattern of affinity for specific acetylated peptides on histones37. Although the overall affinity of each bromodomain for a peptide is low, PBRM1 is thought to bind cooperatively to a precise pattern of acetylated lysine residues in nucleosomes38. Thus, this process in part mediates PBAF complex recruitment to specific regions of DNA where nucleosome remodeling occurs.
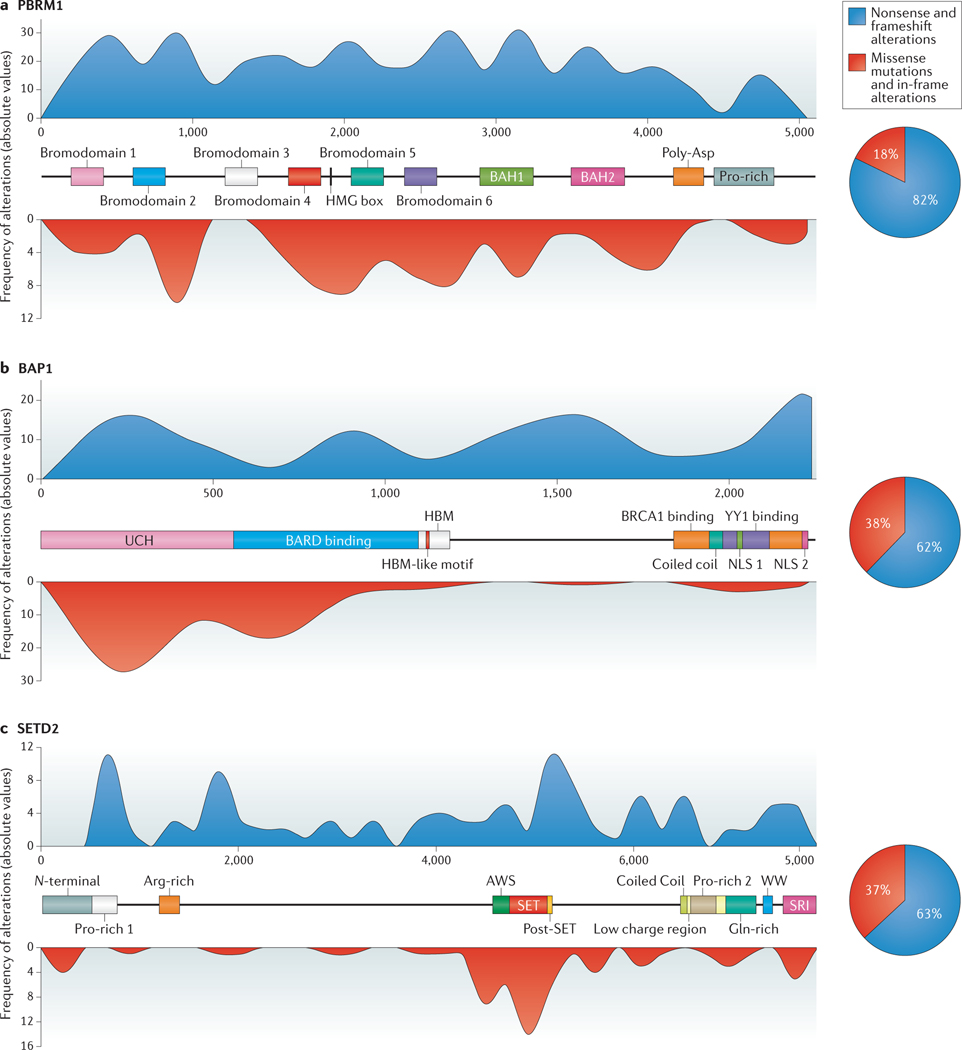
Figure 2 |. Structure and distribution of PBRM1, BAP1 and SETD2 mutations in RCC.
The known protein domains and motifs are indicated for protein polybromo-1 (PBRM1; also known as BAF180) (part a), ubiquitin carboxyl-terminal hydrolase BAP1 (part b) and histone-lysine N-methyltransferase SETD2 (part c). The overlaid blue (nonsense and frameshift alterations) and red (missense mutations and in-frame alterations) histograms represent the distribution of genetic alterations, previously reported in RCC, along each gene40,41. The X axis represents the position of cDNA bases and the Y axes for each respective histogram shows the frequency of alterations (absolute values). The associated pie charts show the proportion of nonsense and frameshift alterations (blue area) and missense and in-frame alterations (red area). Data from REFs 40,41.BAH, bromo-adjacent homology; HMG, high-mobility group; UCH, ubiquitin C-terminal hydrolase; HBM, HCF-C1 binding motif; SRI, SET2-Rpb1 interacting; NLS, nuclear localization signal.
The importance of BAH domains and bromodomains for tumour suppressive function is emphasized by the high frequency of PBRM1 missense mutations in ccRCC18,39 (FIG. 2A). Mutations in other SWI/SNF complex subunits have also been described in RCC, such as ARID1A6,7,20,21,40–42. Notably, ARID1A and ARID1B mutations have been observed in conjunction with PBRM1 mutations18, suggesting that dysfunction of these genes cooperate in tumorigenesis. The potential implications of BAF and PBAF complexes in RCC is also exemplified by the presence of mutations in the catalytic BRG1 subunit, which forms part of both complexes43.
The mechanisms by which mutations in PBRM1 or ARID1A and ARID1B contribute to tumorigenesis in RCC is incompletely understood. PBRM1-mutated RCCs have been found to have a characteristic gene expression signature44, suggesting that PBRM1 functions as a tumour suppressor as part of the targeting subunit of the PBAF nucleosome remodeling complex through its effects on DNA accessibility and gene expression. In this manner, PBRM1 and the SWI/SNF complex have been implicated in the regulation of diverse biological pathways, from hypoxia to interferon signaling45–49. For instance, loss-of-function of PBRM1 has been associated with favourable clinical outcomes with immune checkpoint therapy in RCC48 and melanoma50. How these pathways contribute to tumourigenesis and progression are just now beginning to be recognized.
BAP1
BAP1, a nuclear localized deubiquitinase enzyme and a tumour suppressor protein, is also frequently mutated in ccRCC39. BAP1 belongs to the ubiquitin C-terminal hydroxylase (UCH) family of deubiquitinases, which mediate the binding and cleavage of the ubiquitin isopeptide bond51. The PTM of proteins by covalent attachment of ubiquitin controls many essential cellular processes, such as targeting proteins for assembly into complexes, transport and degradation52. BAP1 is comprised of several functional domains, including the ubiquitin C-terminal hydrolase (UCH) region, the BARD1 binding domain, the HCF-C1 binding motif (HBM), the HBM-like motif, the BRCA1-binding domain and two putative nuclear localization signals (NLS1 and NLS2) (FIG. 2B). In addition, BAP1 contains a region of extreme acidity, multiple potential phosphorylation sites and N-linked glycosylation sites. BAP1 has been shown to interact with the INO80 family of chromatin remodelers, but its role in mediating nucleosome remodeling remains to be fully developed 53.
Classically, BAP1 is known to deubiquitinate H2AK119ub1, which is associated with polycomb-mediated gene repression54 (FIG. 1). Polycomb proteins form part of two complexes — the polycomb repressive complex 1 (PRC1) and PRC2. The PRC2 complex can trimethylate lysine 27 on histone H3 (H3K27me3) via the histone-lysine N-methyltransferases EZH1 or EZH255. The PRC1 complex mediates mono-ubiquitylation of histone H2A at lysine 119 (H2AK119ub1)56. The traditional view on polycomb-mediated gene silencing involves a consecutive model whereby PRC2 first trimethylates H3K27, followed by the binding of PRC1 to H3K27me3 and the subsequent addition of the H2AK119ub1 mark55,57. Noncanonical PRC1 complexes have been found to target chromatin independently of H3K27me358,59. For instance, noncanonical PRC1 targeting to chromatin has been shown to occur through the activity of lysine-specific demethylase 2B (KDM2B), which specifically targets nonmethylated CpG islands via its CxxC domain60. In addition, noncanonical PRC1 complexes can also induce H2AK119ub1 to initiate PRC2 recruitment and the subsequent placement of H3K27me361.
SETD2
Mutations in the histone H3 lysine 36 (H3K36) trimethyltransferase SETD2 have been reported in ccRCC16 (FIG. 2C). Several histone methyltransferases have been shown to mediate H3K36 monomethylation and dimethylation, whereas SETD2 is the only enzyme responsible for H3K36 trimethylation (H3K36me3) in mammalian cells62. The complexity of H3K36 regulation in higher eukaryotes suggests that this PTM has an important regulatory function.
The human SETD2 protein consists of several functional domains: the methyltransferase (SET), the SET2-Rpb1 interacting (SRI) and WW domains (FIG. 2C). The catalytic activity of the SET domain mediates histone methyltransferase activity63. The SRI domain binds to diphosphorylated serine 2 and serine 5 residues on the carboxyl-terminal domain (CTD) repeats of RNA polymerase II (RNA Pol II) 64, a domain with an important role in transcriptional regulation65. Unphosphorylated RNA Pol II is associated with assembly of the preinitiation complex; however, phosphorylation of RNA Pol II at serine 2 and serine 5 elicits promoter clearance and elongation, allowing other factors necessary for transcriptional elongation and termination to bind the CTD65. SETD2 preferentially binds to phosphorylated RBP1; thus, it travels along with RNA Pol II and is available to trimethylate H3K36 in actively transcribed genes.
KDM5C and KDM6A
Mutations in other chromatin modifier genes, such as KDM5C and KDM6A, have been identified in ccRCC16,18. Located on chromosome Xp11.22 and Xp11.3, KDM5C and KDM6A respectively code for demethylases that target trimethylated lysine 4 on histone H3 (H3K4me3) and H3K27me3. KDM5C has been described as a tumour suppressor in ccRCC. Overexpression of KDM5C, which is a HIF target gene, decreased the global H3K4me3 levels, suppressed global gene transcription and retarded in vivo tumour growth in xenograft models66. KDM5C also contributes to the maintenance of heterochromatin and directly binds to satellite repeats during heterochromatin duplication in S phase67. In the latter study, tumours harbouring KDM5C mutations from patients with ccRCC showed features of heterochromatin disruption and genomic rearrangement, and these patients had a poor prognosis67.
Repertoire of mutations in ccRCC
In ccRCC, mutations in PBRM1 or SETD2 and BAP1 have been described to be mutually exclusive, whereas mutations in PBRM1 and SETD2 have the tendency to co-occur11,39,68. Patients with RCC whose tumours harbour a BAP1 mutation have pathological features associated with aggressive disease, such as rhabdoid histology and tumour infiltration, and consistently have poorer survival outcomes than those with mutations in PBRM139,44,69,70. By contrast, evidence regarding the clinical significance of mutations in PBRM1 and SETD2 in RCC is inconsistent39,44,69,70. A study from 2017 reported that loss or low expression of SETD2, but not H3K36me3, by immunohistochemistry, was associated with a poor prognosis in patients with ccRCC71. Initially, PBRM1 mutation was reported to be associated with increased aggressiveness in ccRCC69. In addition, loss of PBRM1 expression by immunohistochemistry was associated with advanced stage, high Furman grade and poor overall survival72. However, other reports provide evidence suggesting that ccRCCs have similar rates of PBRM1 mutations regardless of stage and do not seem to influence patient survival outcomes44,70,73. The high prevalence of this mutation, and its inclusion as a truncal event, suggests that later events in tumour progression have a greater degree of influence on the ultimate behavior of the tumour.
Recently, the Tracking Renal Cell Cancer Evolution Through Therapy (TRACERx Renal) study examined the association between disease stage and clinical outcomes in a large cohort of primary ccRCC tumours11. Using multi-region genomic profiling to study tumour heterogeneity in ccRCC, the authors identified seven major evolutionary subtypes in ccRCC, namely: multiple clonal drivers; VHL wild-type; VHL monodriver; BAP1-driven (VHL→BAP1); VHL→PBRM1→SETD2; VHL→PBRM1→phosphoinositide 3-kinase (PI3K); and VHL→PBRM1→somatic copy number alteration (SCNA)11. Consistent with other reports74, the multiple clonal driver subtype, characterized by the clonal co-occurrence of drivers that are usually mutually exclusive (such as BAP1 and PBRM1, or BAP1 and SETD2), was associated with aggressive disease and comprised the largest tumours in the cohort11. The majority of tumours in the BAP1-driver (VHL→BAP1) mutational subtype had no other detectable mutational drivers, which confirmed the tendency of mutual exclusivity between BAP1 and PBRM1 mutations68, and suggested that BAP1 mutations drive strong clonal expansion11. Conversely, tumours in the the VHL→PBRM1→SETD2 subtype had extensive clonal branching and a preponderance for parallel evolution11. Although mutations in PBRM1 seem to be an early event, with evidence showing clonality of up to 74%, SETD2 mutations seem to occur later during RCC development, given that subclones with mutations in SETD2 showed limited expansion11. Interestingly, the spatial clustering of parallel SETD2 mutations suggested a potential role for SETD2 loss in niche-specific clone selection11. Importantly, these evidences suggest that the order in which driver events are acquired can have prognostic and therapeutic implications. These findings could in part explain the discrepancies regarding the clinical significance of mutations in SETD2 and PBRM1 in previous studies, which involved analysis of single tumour samples. This study11 and others75–78 indicate that multi-region sampling of RCCs might be necessary to account for tumour heterogeneity.
Co-drivers in RCC mouse models
In mice, loss of Vhl alone does not induce kidney cancer and additional mutations in co-drivers were, therefore, believed to be necessary for tumorigenesis79–81. Initial efforts to explore the potential roles of Pbrm1, Bap1 and Setd2 as co-drivers in mouse models were hampered because homozygous loss of these genes resulted in embryonic lethality36,82–84. It was not until Vhl and Bap1 were conditionally knocked out that mice developed kidney tumours85,86. Conditional knockout of Vhl and Pbrm1 was also shown to cause ccRCC, as well as large cysts, in mouse kidneys47,85. However, the conditional knockout of Setd2 and Vhl has not been reported to date. In mice, concurrent knockout of Vhl and Bap1 produced high-grade ccRCC tumours, whereas concurrent Vhl and Pbrm1 knockout was associated with the development of low-grade ccRCC85. These findings provide further support to the co-driver hypothesis, which posits that a VHL loss-of-function mutation alone is insufficient to promote tumorigenesis, but that a second mutation is required for the acquisition of malignant potential.
Mutations in other RCC histologies
Mutations in chromatin modifiers are not unique to ccRCC and have been detected in other RCC histologies. Although rare, type 1 pRCC tumours have been shown to harbour inactivating mutations in KDM6A and SMARCB1 (also known as BAF47, INI1 or SNF5; encoding SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1)7. Conversely, type 2 pRCC is associated with loss of PBRM1, BAP1, SETD2 and ARID2, although the frequencies are lower than those observed in ccRCC87,88. Interestingly, as observed in ccRCC, mutations in PBRM1 and BAP1 are mutually exclusive whereas mutations in PBRM1 and SETD2 tend to co-occur in pRCC7,68. Although loss of PBRM1, BAP1 and SETD2 is common to both ccRCC and type 2 pRCC, the path to tumorigenesis seems to proceed via different routes. Notably, loss of chromosome 3p (a consistently large deletion which encompasses VHL, PBRM1, BAP1 and SETD2) is rarely observed in pRCCs and inactivation of PBRM1, BAP1 and SETD2 occurs primarily through mutation10,11,76,77,87. Thus, pRCCs are often heterozygous for these genes, suggesting that PBRM1, BAP1 and SETD2 genes are haploinsufficient, such that a loss-of-function mutation in one gene copy cannot be fully compensated by the presence of the remaining wild-type allele. In chRCC and renal oncocytomas, mutations in PBRM1, BAP1 and SETD2 are rare88.
The involvement of these chromatin modifiers has also been described in sarcomatoid RCCs. Mutations in PBRM1, BAP1 and SETD2 were detected in sarcomatoid ccRCC, tumours characterized by an unusually high prevalence of TP53 mutations compared with conventional ccRCCs89,90. Similar to their ccRCC counterparts, sarcomatoid ccRCCs also harbour mutations in VHL, consistent with the observation. However, unlike conventional ccRCCs, sarcomatoid ccRCCs rarely have loss of chromosome 3 and are less likely to harbour PBRM1 mutations90. Furthermore, these findings also indicate that IHC might not be an appropriate method to screen for mutations in these proteins (which result in altered expression) in non-ccRCC tumours, and that genomic sequencing would be more suitable.
Canonical functions in RCC
Chromatin and chromatin modifiers mediate many important processes ranging from transcription and splicing to DNA repair. Furthermore, histone PTMs can regulate other epigenetic marks, for instance, via crosstalk with DNA methylation. Dysregulation of the canonical functions (DNA damage repair, transcriptional regulation, splicing and epigenetic crosstalk) of PBRM1, BAP1 and SETD2 contribute to the development and progression of RCC.
DNA damage repair
Cancer cells, including RCC, are known to have defects in the DNA mismatch repair (MMR) and DNA double-strand break repair (DSB repair) pathways91,92. Accordingly, defective MMR function and impaired DSB repair enables elevated mutation frequencies and can lead to certain cancers93. As DNA repair must occur in the context of chromatin, the processes of DNA repair and chromatin modification are intimately linked.
SETD2 and H3K36me3 in DNA repair.
Several studies have implicated SETD2 and its cognate PTM, H3K36me3, in both DNA MMR and the resolution of DSBs. In human cells, DNA mismatches are recognized by the MutSα (MSH2–MSH6 heterodimer) and MutSβ (MSH2–MSH3 heterodimer) complexes, triggering additional MMR components to repair the DNA mismatch94,95. Given that MMR (and most forms of DNA repair) is preferentially directed to actively transcribed genes, the H3K36me3 mark, which is associated with actively transcribed genes, might enable this preferential activity. Accordingly, MutS protein homolog 6 (MSH6) contains a PWWP domain that has been shown to bind to H3K36me3 and promote the recruitment of MutSα complexes to genomic regions that contain coding sequences94,96 (FIG. 3A). These discoveries have shed light on how the histone code contributes to the high fidelity of DNA replication in eukaryotic cells by enhancing MMR efficiency at coding regions, where it is the most crucial.
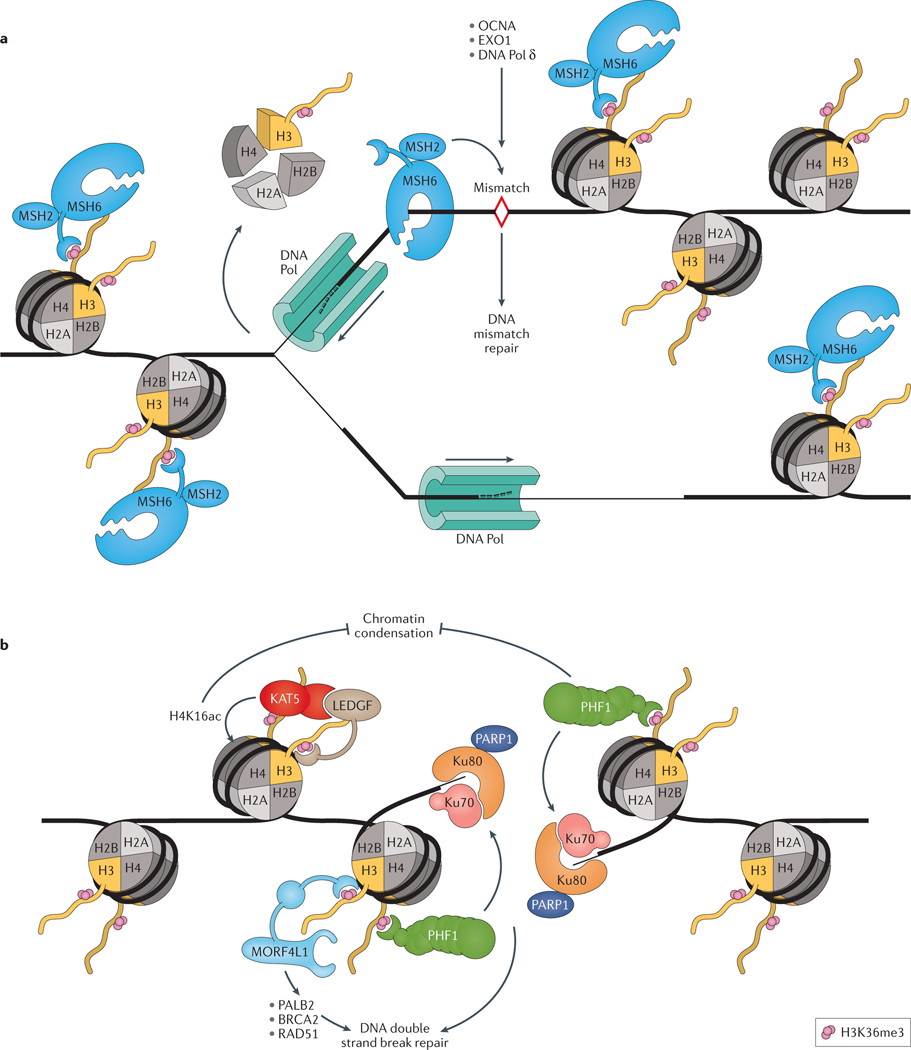
Figure 3 |. H3K36me3 facilitates DNA damage repair.
a | During DNA replication, misincorporated nucleotides and insertion–deletion mispairs are repaired through DNA mismatch repair (MMR). These replication errors can be recognized by the MutSα complexes (composed of a MSH2-MSH6 heterodimer), which can initiate MMR by recruiting proliferating cell nuclear antigen (PCNA), exonuclease 1 (EXO1) and DNA polymerase δ (DNA Pol δ) to sites of DNA damage. MutSα is recruited to genomic regions decorated with trimethylated histone H3 lysine 36 (H3K36me3), via the PWWP domain of MutS protein homolog 6 (MSH6). This nuance guarantees that MutSα is present and enriched for in coding sequences prior to DNA replication and ensures that MMR is concentrated at these regions. b | H3K36me3 also has a role in the repair of DNA double strand breaks (DSBs). When DSBs occur, H3K36me3 marks on nearby nucleosomes becomes exposed, leading to the recruitment of PHD finger protein 1 (PHF1) to the DSB site, where it locally recruits X-ray repair cross-complementing protein 6 (Ku70)–X-ray repair cross-complementing protein 5 (Ku80) complexes and PARP1 to promote the initiation of DSB repair. Mortality factor 4-like protein 1 (MORF4L1; also known as MRG15) also participates in DSB repair by recruiting partner and localizer of BRCA2 (PALB2), which then recruits breast cancer type 2 susceptibility protein (BRCA2) and DNA repair protein RAD51 homolog 1 (RAD51) to DSB sites. Maintaining an open chromatin state creates a favourable environment conducive to the resolution of DSBs. PHF1 also facilitates DSB repair by inhibiting the activity of factors involved in chromatin condensation, promoting a more relaxed, open chromatin state. Following DNA DSBs, MORF4L1 recruits lens epithelium-derived growth factor (LEDGF; also known as PSIP1), which subsequently recruits KAT5, a histone H4 lysine 16 (H4K16ac) acetyltransferase, and increases H4K16ac, which prevents chromatin condensation.
DNA DSBs are severe events in eukaryotic cells that can arise from genotoxic stress and that can be repaired through homologous recombination (HR) or non-homologous end joining (NHEJ)92. H3K36me3 can mediate DSB repair through its interaction with PHD finger protein 1 (PHF1) (FIG. 3B). The Tudor domain of PHF1 binds to H3K36me3 and stabilizes PHF1 at DNA DSBs, together with the X-ray repair cross-complementing protein 6 (Ku70)–X-ray repair cross-complementing protein 5 (Ku80) heterodimer and poly ADP-ribose polymerase 1 (PARP1), which constitute the DNA strand break recognition complex97. In addition to focusing this activity to actively transcribed genes, the binding of PHF1 to H3K36me3 promotes an open chromatin state (euchromatin ), which is also necessary for the successful resolution of DSBs.
The activity of two other H3K36me3 readers, lens epithelium-derived growth factor (LEDGF; also known as PSIP1) and mortality factor 4-like protein 1 (MORF4L1; also known as MRG15) were also shown to promote DSB repair (FIG. 3B). Following DNA damage, MORF4L1 interacts with partner and localizer of BRCA2 (PALB2) to activate the HR pathway98. LEDGF stimulates CtIP-mediated DNA end resection, a critical step during HR-mediated repair of DNA DSBs99,100, and also promotes DSB repair through the recruitment of histone acetyltransferase KAT5 to chromatin after DNA damage, which catalyzes the acetylation of lysine 16 on histone H4 (H4K16ac)101. This acetylation suppresses chromatin condensation and, therefore, promotes a favourable euchromatic environment for the resolution of DSBs102. The crosstalk between H3K36me3 and H4K16ac truly emphasizes the complexity and elegance of the histone code.
Interestingly, the differential modification of H3K36, including either methylation by Set2 (the yeast SETD2 homolog) or acetylation by histone acetyltransferase Gcn5 (the yeast KAT2A homolog), has been suggested to coordinate the choice of DSB repair pathway (that is, HR or NHEJ) in yeast103. In both yeast and mammals, modification of H3K36 is regulated during the cell cycle; H3K36me3 levels peak in G1 phase, during which NHEJ predominates, whereas H3K36 acetylation peaks in late S/G1 phase, during which HR occurs94,103. Cancer cells are known to compensate for defects in one strategy of DNA repair by using alterative repair pathways104. The differential preference for DNA repair pathways mediated by modification of H3K36 could in part explain why a drug synthetic lethality screen did not show the increased sensitivity of SETD2 mutant RCC cells to PARP inhibitors, DNA damaging drugs (such as etoposide) or microtubule toxins (such as taxanes and vinca alkaloids), compared with RCC cells harbouring wild-type SETD2105.
SETD2 loss and DNA damage repair in RCC.
Unsurprisingly, SETD2 loss has been reported to compromise DNA damage repair in RCC cell lines and in tumour samples. Following exposure to ionizing radiation, SETD2-depleted RCC cells showed delayed resolution of phosphorylated histone H2AX (γH2AX) foci, which marks ongoing DNA damage signalling and DSBs100,106. Furthermore, levels of H3K36me3 and SETD2 inversely correlated with γH2AX staining in human RCC tumour samples107. In addition, SETD2-depleted RCC cells showed reduced formation of DNA repair protein RAD51 homolog 1 (RAD51) foci following induction of DNA damage with ionizing radiation100,107. Chromatin immunoprecipitation sequencing (ChIP-seq) revealed that SETD2-deficient tumours from patients with RCC had an increased co-occurrence of chromosomal breaks and point mutations at sites marked by H3K36me3 compared with normal tissues107. These findings suggest that under normal physiological conditions, sites marked with H3K36me3 are less prone to having unrepaired DNA damage than other genomic locations that lack this modification, and that SETD2 loss contributes to disproportionate DNA damage in transcribed genes, which might be more relevant to the development of cancer.
Transcriptional regulation
Abnormal splicing patterns, although not well understood, are frequently associated with human disease, and are a hallmark of cancer108. Alternative splicing can occur through five modes, namely, exon skipping, mutually exclusive exons, alternative donor site, alternative acceptor site and intron retention, all of which predominantly occur co-transcriptionally. Chromatin modifiers and their associated histone PTMs are known to recruit other nuclear factors that mediate not only splicing, but also transcriptional elongation and the transcriptional response to stimuli.
H3K36me3 patterning in splicing.
The pattern of H3K36me3 distribution is recognized as an important factor in the governance of exon utilization. Indeed, lower levels of the H3K36me3 modification have been reported in alternative exons compared with constitutive exons109. Moreover, the presence of splicing factor 3B subunit 3 (SF3B3), a splicing factor that is enriched for at exon–intron junctions, was reported to influence the distribution of H3K36me3 marks109,110. On average, alternative exons assemble fewer spliceosomes than constitutive exons, which suggests that the activity of the spliceosome is influenced by the H3K36me3 modification111.
In addition to its roles in preventing cryptic transcription and promoting elongation, the H3K36me3 reader MORF4L1 has also been shown to influence splicing (FIG. 4A). MORF4L1 has been shown to cooperate with polypyrimidine tract-binding protein 1 (PTBP1) to promote exon inclusion112. In addition, MORF4L1 has also been reported to recruit lysine-specific demethylase 5B (KDM5B) — which targets H3K4me3, a modification associated with promoter regions of transcribed genes — to H3K36me3-marked intergenic regions to promote the removal of H3K4me3113. Thus, the interaction between H3K36me3, MORF4L1 and KDM5B ensures transcriptional fidelity by repressing cryptic intergenic transcription, whereas the MORF4L1–PTBP1 interaction mediates splicing through the regulation of alternative exon inclusion (FIG. 4A).
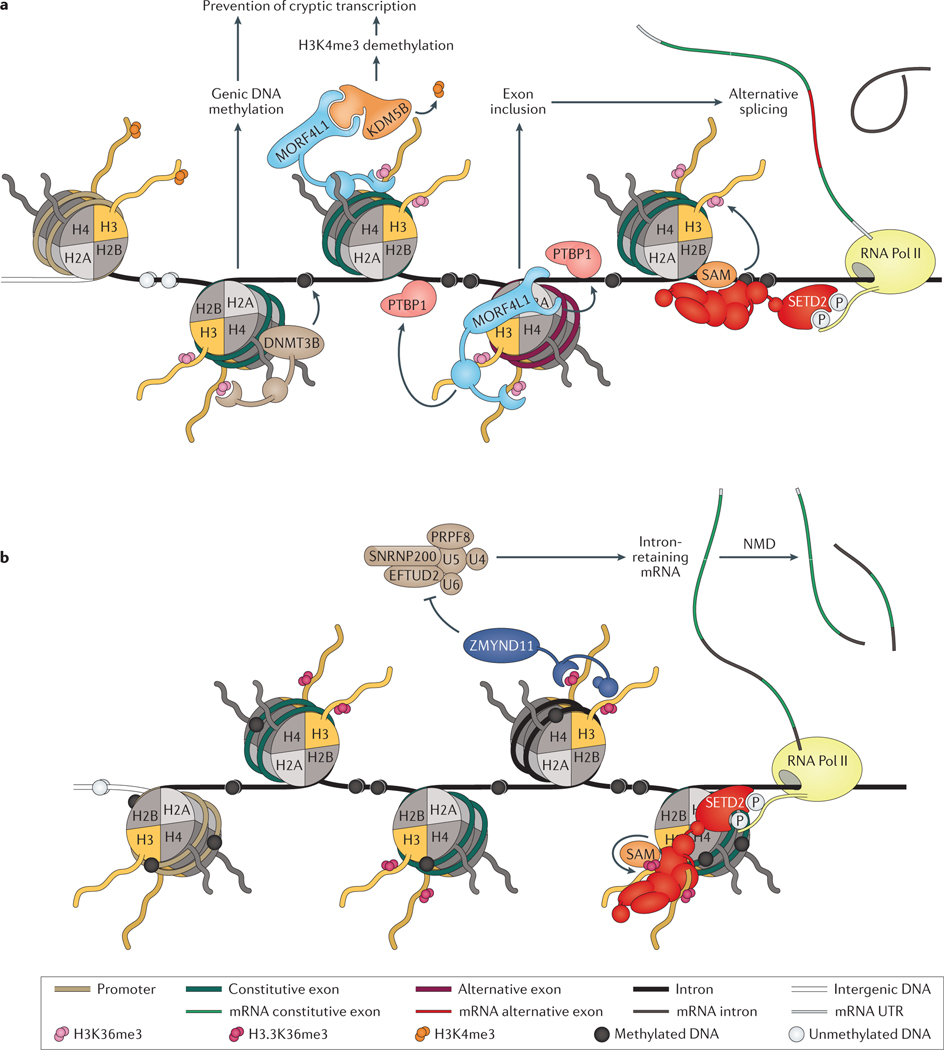
Figure 4 |. Maintenance of transcriptional fidelity.
a | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) mediates gene body DNA methylation through its interaction with trimethylated lysine 36 on histone H3 (H3K36me3) via its PWWP domain. DNMT3B can prevent spurious transcriptional initiation by promoting the DNA methylation of intragenic transcription factor binding motifs, which can influence the binding of some transcription factors. Mortality factor 4-like protein 1 (MORF4L1; also known as MRG15) also binds to H3K36me3 and can recruit lysine-specific demethylase 5B (KDM5B), the trimethylated lysine 4 on histone H3 (H3K4me3) lysine demethylase. The activity of KDM5B prevents spurious transcription initiation by removing H3K4me3, which is associated with active gene promoters. In addition, MORF4L1 can mediate gene splicing, whereby MORF4L1 recruits polypyrimidine tract-binding protein 1 (PTBP1) to DNA to promote exon inclusion in the mature mRNA transcript. b | A histone H3K36me3-specific reader, zinc finger MYND domain-containing protein 11 (ZMYND11; also known as BS69), mediates intron retention of certain target genes. ZMYND11 contains a bromo-PWWP domain that specifically binds trimethylated lysine 36 on the histone H3.3 variant (encoded by the H3F3A or H3F3B genes), rather than the canonical histone H3.1 or H3.2. Incorporation of the H3.3 variant histone into chromatin is independent of DNA synthesis, and it is deposited at distinct genomic regions, including gene bodies, telomeres, and pericentric chromatin. Histone H3.3 phosphorylation at serine 31 abrogates ZMYND11 binding to H3.3K36me3, suggesting that ZMYND11 recognition of H3.3K36me3 is regulated by signalling pathways that mediate histone H3.3 serine 31 phosphorylation. ZMYND11 inhibits the activity of the U5 small nuclear ribonucleoprotein particle protein complex — which comprises U5 small nuclear ribonucleoprotein 40 kDa protein (U5), 116 kDa U5 small nuclear ribonucleoprotein component (EFTUD2), pre-mRNA-processing-splicing factor 8 (PRPF8), U5 small nuclear ribonucleoprotein 200 kDa helicase (SNRNP200), U4 (RNU4–1), and U6 (RNU6–1) — to promote intron retention, which can trigger the nonsense mediated decay (NMD) pathway. The NMD surveillance pathway usually degrades erroneous transcripts, which contain premature a stop codon, and in this way, can reduce gene expression and could regulate transcriptional output.
Another H3K36me3 reader, zinc finger MYND domain-containing protein 11 (ZMYND11; also known as BS69), has been reported to regulate intron retention (FIG. 4B)114. Often, intron-retaining transcripts contain a premature stop codon, which triggers the nonsense-mediated decay pathway (NMD pathway), and therefore, is associated with decreased gene expression. In this context, ZMYND11 functions as a tumour suppressor by promoting intron retention in certain target genes, which are generally those with high expression and potential oncogenic functions.
Experimental evidence indicates that in the setting of SETD2 deficiency, these processes are in fact disrupted in ccRCC. Extensive genomic analysis has demonstrated that human ccRCC tumours harboring SETD2 mutations had more open chromatin, as measured by formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq) compared to either SETD2 wild-type tumours or matched normal tissue, and that regional chromatin opening was highly correlated with gene expression15. In addition, these same regions also displayed a higher frequency of alternate exon utilization as well as intron retention, suggesting an error-prone splicing process.
PBRM1 loss in hypoxia signalling.
Reports of the effects of PBRM1 loss on hypoxia signalling are inconsistent. In VHL-deficient RCC cells, PBRM1 loss has been described to both enhance and suppress hypoxia signalling46,115. The differential effect of PBRM1 loss on response to hypoxia seems to be cell-type specific and depends on the expression of expression VHL, HIF-1α and HIF-2α in RCC cells. In RCC, HIF-1α has long been thought to function as a tumour suppressor and is often deleted, whereas HIF-2α has been considered the primary oncogenic driver. Importantly, there is a consensus that PBRM1 functions as a tumour suppressor by facilitating the expression of HIF-1α target genes45,46,115. This hypothesis is in agreement with the observation that the SWI/SNF complex is required for expression of HIF-1α target genes and HIF-1α-induced cell cycle arrest in other cell types116. However, owing to discrepancies in these studies, whether PBRM1 is also required for expression of HIF-2α target genes is unclear. Nonetheless, it seems that PBRM1 loss contributes to ccRCC development and progression by downregulation of HIF-1α signalling.
Epigenetic crosstalk
Epigenetic modifications do not function alone, but rather work together in various combinations and can regulate one another, which diversifies their function. The result is a complex, but elegant, regulatory network. However, aberrations in chromatin modifiers, as well as loss or aberrant patterning of histone PTMs, frequently has far-reaching consequences that can further derange other epigenetic states.
Histone methylation.
As mentioned above, actively transcribed genes are marked by H3K36me3 in gene bodies and by H3K4me3 at promoters near the transcription start site. Repressed genes are characterized by H3K27me3, which is catalyzed by the PRC2 complex via EZH1 or EZH2. When present on the same histone tail, H3K4me3 and H3K36me3 are known to inhibit PRC2 complex activity and chromatin binding117. The switch from an actively transcribed to a repressive transcriptional state during differentiation is now known to be in part mediated by PHD finger protein 19 (PHF19), another H3K36me3 reader118,119. PHF19 has been shown to associate with the PRC2 complex to facilitate recruitment to H3K36me3 marked regions119,120. Interestingly, PHF19 also associates with the H3K36me3 demethylase NO66118. The opposing activities of SETD2 and NO66 mediate transitions between H3K36me3 and H3K27me3 modification118. Unsurprisingly, loss of SETD2 and H3K36me3 has been reported to result in increases in levels of H3K27me3 in gene bodies121.
DNA methylation.
DNA methylation, another important epigenetic regulatory mark, involves the covalent addition of a methyl group to the 5’-position of cytosine residues, and usually occurs in the context of CpG dinucleotides. Although the maintenance of DNA methylation through replication is ensured by DNA (cytosine-5)-methyltransferase 1 (DNMT1), the establishment of de novo DNA methylation is mediated by DNA (cytosine-5)-methyltransferase 3A (DNMT3A) and DNA (cytosine-5)-methyltransferase 3B (DNMT3B)122. DNA methylation can occur at promoters, as well as in intergenic regions, gene bodies and enhancers. Promoter DNA methylation is considered to be a mark of transcriptional repression, whereas the function of gene body DNA methylation has been more difficult to elucidate and is generally associated with a permissive transcriptional state. H3K36me3 has been shown to mediate gene body DNA methylation through interaction with DNMT3B123,124 (FIG. 4A).
Although both are capable of de novo DNA methylation, the genomic regions targeted by DNMT3A and DNMT3B differ. DNMT3A has been shown to preferentially methylate promoters and enhancers125, whereas DNMT3B-bound genomic loci were specifically enriched for features of active transcription. DNMT3B-mediated DNA methylation occurs predominately in the linker regions between nucleosomes because of occlusion from the core nucleosome (FIG. 4A). Interestingly, DNMT3B-mediated DNA methylation occurs at the introns of genes marked with H3K36me3, but not those lacking H3K36me3123. Moreover, loss of H3K36me3 resulted in diminished DNMT3B–nucleosome binding and reduced gene body DNA methylation. Genomic regions bound by DNMT3B showed a marked enrichment for several transcription factor binding motifs containing CpG sequences, including motifs related to transcription factor SP1 and members of the ETS family. Both of these transcription factors have been reported to recruit the transcription machinery in TATA-less promoters, and their binding to DNA has been reported to be negatively affected by CpG methylation126,127. Thus, gene body DNA methylation, in a H3K36me3-dependent manner, might reduce the occurrence of cryptic transcription by preventing the aberrant binding of transcription factors within the gene body, and/or facilitate gene expression in a dynamic environment.
Altered DNA methylation patterns in RCC.
Alterations in DNA methylation patterns has been described in SETD2-deficient RCC. However, discrepancies exist regarding whether SETD2 loss results in DNA hypomethylation or hypermethylation15,128–130. These discrepancies can be accounted for by differences in how the studies were conducted, which provides different clues into the complex crosstalk between epigenetic marks.
One study reported that SETD2 loss caused increased chromatin accessibility, characterized by nucleosome depletion, which substantially overlapped with gene bodies in RCC tumour specimens15. The authors subsequently assessed DNA methylation patterns in nucleosome-depleted regions associated with SETD2 loss using data from The Cancer Genome Atlas (TCGA) and predominantly observed DNA hypomethylation. These results suggest that SETD2 loss causes H3K36me3 depletion, and that those regions that lost H3K36me3 displayed increased chromatin accessibility and nucleosome depletion, and DNA hypomethylation. These findings are consistent with reports that H3K36me3 mediates DNMT3B-dependent DNA methylation123.
Other studies have reported that SETD2 loss is associated with DNA hypermethylation. A comparison of DNA methylation between SETD2 mutant and wild-type tumours from the ccRCC, pRCC, and chRCC TCGA datasets revealed that most changes involved increased global DNA methylation regardless of subtype128. The analysis was not restricted to any particular genomic location, and included promoters, gene bodies, enhancers or intragenic regions. Notably, the 450K DNA methylation assay (Illumina) that was used for the TCGA is enriched in promoter regions, thus ascertainment bias must be considered in interpreting these findings. Furthermore, SETD2 mutations were enriched in pRCC tumour samples that had a CpG island methylator phenotype (CIMP)128. CIMP tumours are characterized by global DNA hypermethylation, which tends to occur CpG islands and are generally associated with promoter regions. This CIMP phenotype could skew the differential DNA methylation pattern in the SETD2 mutant group towards DNA hypermethylation.
Another study noted that SETD2 loss caused increased DNA methylation in genes that are expressed at low levels, whereas highly expressed genes did not show changes in DNA methylation129. Other factors that govern the transcription of highly expressed genes, such as housekeeping genes, could possibly prevent aberrant DNA methylation from occurring. The authors also binned the probes on the 450K platform to identify differentially methylated regions (DMRs), which overlapped with regions that gained the H3K36me3 mark after SETD2 loss129; however, scaling presents a challenge in these studies in the absence of spike-in DNA references.
The examination of DNA methylation patterns using TCGA data identified three subclusters of RCC tumours that respectively showed no, low or high CIMP130. The CIMP-high group was associated with a poor prognosis and was enriched for BAP1 and SETD2 mutations. For the unsupervised clustering analysis that identified these three subclusters, the authors filtered the data to include only probes that displayed >20% methylation in any of the TCGA normal kidney samples (n=161)130. As noted by the authors, the resulting probes localized to promoter CpG islands. Thus, this analysis could be biased towards DNA hypermethylation, given that the probes included were selected so that they displayed hypomethylation in normal kidney samples (and presumably hypermethylation in tumours) and so that these probes were targeted to promoter CpG islands, which typically undergo DNA hypermethylation.
Noncanonical functions in RCC
Evidence indicates that chromatin modifiers have nonhistone substrates and, in some cases, even travel outside the nucleus. These findings suggest that these enzymes have noncanonical functions, including roles in metabolic control, cytoskeletal remodeling and interferon signalling, and reports suggest that these noncanonical functions, when lost, also contribute to RCC131–133.
Extranuclear roles of BAP1
Under normal growth conditions, BAP1 is predominantly localized in the nucleus. However, BAP1 can be ubiquitylated by ubiquitin-conjugating enzyme E2 O (UBE2O) at its NLS to promote nuclear export134. UBE2O-mediated ubiquitylation causes BAP1 to accumulate in the cytoplasm, whereas BAP1 autodeubiquitylation promotes nuclear retention. Although BAP1 localization and function is best characterized in the nucleus, it is conceivable that BAP1 could have extranuclear functions. Indeed, experimental evidence that BAP1 loss alters cellular metabolism — which primarily occurs in the cytoplasm and in mitochondria — supports this possibility.
BAP1+/− fibroblasts derived from patients with RCC with germline BAP1 mutations had an altered metabolism that was characteristic of the Warburg effect, as shown by increased glycolysis and reduced mitochondrial respiration135. In RCC cells, BAP1 inhibited glucose deprivation-induced apoptosis through interaction with the activating transcription factor 3 (ATF3) and DNA damage-inducible transcript 3 protein (DDIT3) gene promoters in a polycomb complex protein BMI-1-dependent manner136. Loss of BAP1 increased the production of reactive oxygen species (ROS) and activated endoplasmic reticium (ER) stress pathways. In addition, loss of BAP1 has also been described to deplete mitochondrial proteins in pancreas cells137. This observation is in contrast to a report in mesothelioma cells, in which BAP1 loss caused increased mitochondrial mass138. Notably, although BAP1 loss is associated with poor prognosis in patients with ccRCC, its loss is a favourable prognostic marker in mesothelioma139. Collectively, these evidences suggest that the metabolic effects of BAP1 loss could be cell-type specific.
Although the exact mechanism underpinning the metabolic alterations associated with BAP1 loss are currently unknown, a study from 2017 reported that BAP1 promotes calcium (Ca2+) release from the ER to the mitochondria135. This process was shown to be dependent on BAP1-mediated deubiquitylation of type 3 inositol-1,4,5-trisphosphate receptor (IP3R3), an ER channel that regulates Ca2+ release from the ER to the cytoplasm and mitochondria. Transient Ca2+ release leads to mitochondrial ATP production, whereas excessive or prolonged Ca2+ release promotes apoptosis via excessive mitochondrial Ca2+ levels and the consequent opening of the mitochondrial permeability transition pore140. The authors suggest that malignancies that are most frequently associated with the BAP1 cancer syndrome, defined by the germline loss of function of one allele of BAP1 (which predisposes to ccRCC, uveal and cutaneous melanoma, mesothelioma, and melanocytic BAP1-mutated atypical intradermal tumours), arise from cells or tissues in which Ca2+-induced apoptosis is crucial for preventing cellular transformation141.
SETD2 in microtubule midbody assembly
Historically, SETD2 was presumed to only act upon lysine 36 on histone H3. However, a study from 2016 reported that SETD2 could also trimethylate α-tubulin on lysine 40 (TubK40me3)131. The heterodimerization of α-tubulin with β tubulin gives rise to long, dynamic polymeric molecules called microtubules, and the acetylation of lysine residues on alpha tubulin is known to be involved in the polymerization process142. Alpha tubulin methylation and acetylation at lysine 40 were shown to be reciprocal marks that compete for the same lysine residue. Loss of both TubK40me3 and H3K36me3 was observed in mouse embryonic fibroblasts (MEFs) with conditional knockout of Setd2131. These SetD2-knockout MEFs displayed marked genomic instability and had mitotic and cytokinetic defects, including increased micronuclei, failure of chromosome congression and increased multinuclear spindles during prometaphase, lagging chromosomes during anaphase and chromosomal bridges during cytokinesis131. SETD2 loss was also shown to promote micronuclei formation in ccRCC133. This novel function of SETD2 in catalyzing TubK40me3 sheds light on how SETD2 loss contributes to cellular transformation and aggressiveness.
BAP1 in cytoskeleton remodeling
BAP1 has been reported to deubiquitylate γ-tubulin at lysine 48 and lysine 344143. In eukaryotes, the function of γ-tubulin is highly conserved and includes the regulation of microtubule nucleation and centrosome duplication144. Ubiquitination of γ-tubulin at lysine 48 and lysine 344 is mediated by the activity of the tumour suppressive complex breast cancer susceptibility gene 1 (BRCA1)–BRCA1-associated RING domain protein 1 (BARD1), a complex that includes BAP1145. Interaction between BAP1 and γ-tubulin occurs during metaphase, anaphase and telophase, but not during prophase, during which γ-tubulin is ubiquitylated. BAP1 was found to be necessary for proper mitotic spindle organization and prevention of chromosomal abnormalities, which include lagging chromosomes during metaphase, uncondensed mitotic chromosomes, anaphase and telophase bridges and tripolar mitotic spindles143.
In RCC cells, BAP1 has also been shown to have a role in mediating chromosome segregation by regulating the stability of microspherule protein 1 (MCRS1) — which is involved in various biological processes, such as transcription, proliferation, mitosis and senescence146 — through de-ubiquitylation, preventing its proteasomal degradation in RCC cells147. Knockout of MCRS1 resulted in chromosome instability and aneuploidy in mammary cells146. Similarly, BAP1 depletion induced multipolar spindle formation and multinucleation, which were partially rescued by the stable overexpression of MCRS1 in RCC cells147. These findings collectively suggest that BAP1 maintains chromosomal stability by modulating the ubiquitylation status of various substrates, including MCRS1 and γ-tubulin.
SETD2 and PBRM1 in interferon signalling
SETD2 has been reported to mediate signal transducer and activator of transcription 1 (STAT1) methylation on lysine 525132, which facilitates STAT1 phosphorylation by Janus kinase 1 (JAK1) and the consequent activation of STAT1 as a transcription factor to mediate transcriptional responses to interferon signals. SETD2 loss-of-function resulted in impaired STAT1 signalling,dampening the host response to Hepatitis C virus infection in human hepatocytes132. This finding further emphasizes the potential importance of the nonhistone substrates and noncanonical functions of SETD2. The impact of this activity of SETD2 in RCC remains to be determined.
The SWI/SNF complex has previously been described to have a role in regulating host defense against viral infection. Loss of SMARCB1 was shown to block cellular response to viral infection and impaired antiviral activity by inhibiting expression of virus-inducible and IFN-inducible genes in HeLa cells148. Similarly, PBRM1 knockdown in the ACHN RCC cell line altered the cytokine–cytokine receptor interaction pathway, characterized by upregulation of interleukin-6 receptor subunit beta (IL6ST) and C-C motif chemokine 2 (CCL2) and downregulation of IL-8, IL-6, and C-X-C motif chemokine 2 (CXCL2)149. These studies suggest that dysfunction of the SWI/SNF complex through loss-of-function of the PBRM1 and SMARCB1 subunits impairs cellular antiviral responses.
Conversely, other studies have shown that knockdown of PBRM1 results in unrestrained cellular antiviral responses and increased tumour immunogenicity. PBRM1 knockdown in colon cancer cells caused a robust upregulation of retinoic acid-inducible gene 1 protein (RIG-I)-like receptor signalling genes, such as viral RNA receptors (RIGI and MDA5) and their downstream transcriptional targets (IRF7, ISG15, ISG56, and IRF9)49. In Drosophila, PBRM1-deficient intestinal cells were susceptible to infection as a result of hyper-inflammation150. PBRM1 was shown to cooperate with EZH2 to directly suppress RIG-I and MDA5 transcription, and therefore, knockdown of PBRM1 resulted in unrestrained transcription of target genes49. In this regard, gene expression analyses in PBRM1-deficient and PBRM1-proficient RCC cells showed that PBRM1 deficiency induced expression of a number of genes related to immune response, such as those involved in IL-6–Janus kinase (JAK)–signal transducer and activator of transcription 3 (STAT3) signalling, TNF signalling via nuclear factor-κB (NF-κB), and IL-2–STAT5 signalling, as well as genes involved in hypoxia signalling48. These conflicting reports highlight the complexity of SWI/SNF complex-mediated regulation of viral defense genes and suggest that the effect that loss of SWI/SNF complex function has on host viral defense is dependent on which subunit is lost, as well as the cell type in which the subunit is lost. The relevance of PBRM1 loss in RCC and its effect on response to therapy is now beginning to be understood.
Therapeutic opportunities
In contrast to gain-of-function mutations in oncogenes, which are conceptually straightforward to target, loss-of-function mutations in tumour suppressor genes are more challenging to approach therapeutically. Synthetic lethality describes a relationship between two genes whereby loss of both genes is incompatible with cell survival, whereas loss of either gene does not induce cell death. Thus, by exploiting the phenomenon of synthetic lethality, loss-of-function mutations in chromatin modifiers could present unique therapeutic opportunities in RCC.
Loss of SETD2
Loss of the yeast SETD2 homolog Set2 was shown to be synthetically lethal with loss of mitosis inhibitor protein kinase Wee1 in Schizosaccharomyces pombe151. This synthetic lethality was also observed when the human RCC cell lines A498 and LB996 — which harbour naturally-occurring SETD2 mutations — were treated with MK1775, a selective inhibitor of the human Wee1 homolog Wee1-like protein kinase (WEE1)151. WEE1 is a serine/tyrosine protein kinase that has a crucial role in enabling DNA repair to occur before mitotic entry during the G2–M cell cycle checkpoint arrest. Normal cells repair damaged DNA during G1 arrest; however, cancer cells usually have a deficient G1–S checkpoint and, therefore, depend on a functional G2–M checkpoint to repair DNA damage152. Indeed, SETD2-deficient osteosarcoma cells treated with a WEE1 inhibitor showed signs of replicative stress, replication fork stalling and S phase arrest151. Ribonucleoside-diphosphate reductase subunit M2 (RRM2), a component of ribonucleotide reductase (RNR), was shown to mediate this synthetic lethality through two mechanisms in RCC as well as osteosarcoma cells. First, SETD2-mediated H3K36me3 was found to be necessary for RRM2 transcription, and loss of H3K36me3 reduced RRM2 transcription151. Second, WEE1 inhibition was found to mediate RRM2 degradation through activation of cyclin-dependent kinase 1 (CDK1) and/or CDK2151. In osteosarcoma cells, WEE1 inhibition has also been reported to cause aberrant origin firing through CDK activation, which, similar to RRM2 depletion, exhausts the deoxyribonucleotide triphosphate (dNTP) pool153. Aberrant origin firing, synergized with RRM2 depletion, was found to cause synthetic lethality in RCC cells151. These findings emphasize the importance of SETD2 in DNA repair and imply that exploiting this synthetic interaction could lead to effective therapies in at least selected cellular backgrounds.
Using the publicly available Genomics of Drug Sensitivity in Cancer (GDSC) database, TGX221, a selective phosphoinositide 3-kinase β (PI3Kβ) inhibitor, was identified as a potential drug candidate in ccRCC tumours with VHL and SETD2 mutations105. In vitro experiments demonstrated that ccRCC cells with mutations in both VHL and SETD2 were sensitive to TGX221. However, sensitivity to TGX221 was not observed in SETD2-competent ccRCC cells harbouring VHL-inactivating mutations. SETD2-deficient cells have also shown sensitivity to another PI3Kβ inhibitor, AZD6482154. Inhibition of PI3Kβ in SETD2-mutant ccRCC cells led to decreased phosphorylation of protein kinase B (AKT) at both Serine 473 and Threonine 308, which was not accompanied by a change in total AKT levels155. Interestingly, loss of MutL protein homolog 1 (MLH1) conferred resistance to PI3Kβ inhibitors in SETD2-mutant ccRCC cells, and analysis of the TCGA database revealed a high tendency of concurrent homozygous loss of SETD2 and MLH1155. Further investigation showed that SETD2 also mediates MMR via activation of AKT and repression of MLH1 transcription to regulate the expression of the mismatch repair endonuclease PMS2, an essential component of the MMR response. The authors speculate that SETD2 indirectly activates AKT via the JAK–STAT pathway. Indeed, the aforementioned report that SETD2 mediates STAT1 methylation on lysine 525 supports this theory132. These findings further emphasize the potential importance of the nonhistone substrates and noncanonical functions of SETD2 in normal cells, as well as in SETD2-deficient RCC cells.
Loss of BAP1
Loss of BAP1 has been shown to be synthetically lethal with inhibition of EZH2 or PRC2. In a mouse model, Bap1-knockout induced myeloid malignancy and resulted in increased H3K27me3 levels, elevated EZH2 protein expression and enhanced repression of PRC2 targets156. To further assess the role of PCR2-mediated H3K27me3 in cellular transformation mediated by Bap1 loss, the authors investigated the influence of Ezh2 loss on transformation in vivo. Ezh2 loss in Bap1-knockout mice resulted in decreased levels of H3K27me3 and abrogated the development of myeloid malignancy. Moreover, treatment of Bap1-knockout mice with EPZ011989, a small molecule inhibitor of EZH2, recapitulated the in vivo-effects observed with knockout of Ezh2156. In BAP1-deficient human mesothelioma cells, EZH2 inhibition also produced synthetic lethality156. However, this synthetic lethality between BAP1 and EZH2 was not observed in uveal melanoma cell lines157,158, suggesting cell-type specificity. Thus, studies are warranted to determine whether BAP1-deficient RCC cells display sensitivity to EZH2 inhibition.
Loss of SWI/SNF complex components
Synthetic lethality between some components of the SWI/SNF complex (PBRM1, SMARCB1, BRG1 and ARID1A) and EZH2 has also been reported in RCC159,160, as well as in other cancers159–161. This synthetically lethal relationship between SWI/SNF complex components and EZH2 was shown to be mitigated by mutations in members of the RAS–mitogen activated protein kinase (MAPK) pathway159. Preclinical studies have shown that PBRM1-deficient RCC cells and SMARCB1-deficient malignant rhabdoid tumour cells are sensitive to EZH2 inhibitors159,160. Interestingly, PBRM1 exhibited a synthetically lethal interaction with the H4K16ac acetyltansferase KAT5 in osteosarcoma cells162. ARID1A-deficient breast, ovarian, lung and colorectal cancer cells also showed sensitivity to PI3K–AKT pathway and PARP1 inhibitors163,164. However, monotherapy with PI3K–AKT–mechanistic target of rapamycin (mTOR) pathway inhibitors has shown limited clinical efficacy in RCC, suggesting that combinational therapies could be necessary163,165.
These evidences demonstrate that deciphering the downstream oncogenic sequelae of mutations in chromatin modifiers is crucial to the rational and hypothesis-driven development of novel therapeutics for RCC. In addition, the influence of other co-drivers is also important in determining response to therapy. Thus, the identification of mutations in chromatin modifer genes in RCC tumour samples could become important in the clinical management of patients with RCC.
Loss of PBRM1 and immunotherapy
Although immunotherapies have gained considerable attention, the mechanisms governing their efficacy remain poorly understood. The efficacy of immunotherapies is thought to lie in the ability of cytotoxic T cells to detect and eliminate transformed cells following recognition of peptide antigens displayed by major histocompatibility (MHC) class I proteins166. Interestingly, components of the PBAF complex, specifically Arid2, Brd7 and Pbrm1, were identified in a CRISPR–Cas9 functional genomic screen as tumour cell-intrinsic genes that conferred resistance to T cell-mediated killing of B16F1 mouse melanoma cells50. Genes involved in the NF-κB pathway, mTOR complex 1 (mTORC1) signalling, glycolysis, and nicotinate and nicotinamide metabolism, were also identified toconfer resistance to T cell-mediated B16F10 killing50. T cell-mediated killing of B16F10 melanoma cells was enhanced by loss of Arid2, Brd7 or Pbrm1. Pbrm1 loss in B16F10 cells conferred therapeutic benefit to anti-programmed cell death protein 1 (PD-1) and/or anti-cytotoxic T-lymphocyte protein 4 (CTLA-4) immune checkpoint blockade, whereas this treatment was ineffective in control B16F10 cells, which are resistant to these therapies50. In another study, patients with metastatic RCC harbouring truncating PBRM1 mutations derived more clinical benefit from immune checkpoint blockade than patients with PBRM1 wild-type tumours48. The authors suggested that the increased clinical benefit in patients with PBRM1 truncating mutations was due to the unique immune-related gene expression signature observed in PBRM1-deficient RCC cells48. These findings, together with evidence supporting the role of PBRM1 in regulating host antiviral response, suggest that PBRM1 loss promotes RCC immunogenicity through hyperactivation of interferon-responsive genes and causes RCC cells to be ‘visible’ to the immune system, which could in turn confer sensitivity to immune checkpoint blockade.
Conclusions
Loss-of-function of chromatin modifiers, including PBRM1, BAP1 and SETD2, among others, is prevalent in RCC, and these events have long been considered as important co-drivers in this disease. These chromatin modifiers have far-reaching, and sometimes unexpected, functions. Classically, chromatin modifiers are known for their canonical functions, which include roles in DNA repair and transcriptional regulation. In recent years, histone modifiers have been found have nonhistone substrates and engage in noncanonical activities, such as the regulation of cytoskeletal dynamics and interferon signalling. Although canonical functions are undoubtedly important, loss of these noncanonical functions more than likely also contributes to RCC tumorigenesis and progression.
Mounting evidence indicates that clinical behaviour and response to treatment modalities can be influenced by the particular repertoire of mutations in chromatin modifiers. Furthermore, loss-of-function mutations in these chromatin modifiers might create unique therapeutic opportunities, such as those afforded by exploiting synthetic lethal dependencies. Accordingly, there is renewed urgency to further understand their etiology and how loss-of-function of these chromatin modifiers contributes to RCC tumourigenesis and progression. This knowledge will not only be pivotal for the rational and hypothesis-driven development of novel therapeutic strategies in RCC, but will also be essential as we usher in the age of personalized medicine.
Key points.
Loss-of-function mutations in chromatin modifiers, common in renal cell carcinoma (RCC), can modify tumour biology and influence therapeutic responses; thus, understanding how these events contribute to RCC is paramount.
Chromatin modifiers classically regulate genomic architecture and, therefore, control DNA accessibility; these canonical functions are fundamental for essential cellular processes, such as gene expression programs and DNA damage repair.
Chromatin modifiers have non-histone substrates and participate in extranuclear processes; these noncanonical functions regulate important cellular processes such as cytoskeletal dynamics and immune responses.
Loss-of-function mutations in chromatin modifiers can be approached therapeutically by exploiting synthetically lethal dependencies between two genes; loss of both genes induces cell death, but loss of either is nonfatal.
Acknowledgements
The authors acknowledge the support of the NIH: grants R01CA198482 (W. K. R.), K24CA172355 (W. K. R.) and T32CA009582 (A. A. D.). This work was performed in part at the Aspen Center for Physics, which is supported by National Science Foundation grant PHY-1607611.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.ACS. Cancer Facts & Figures 2018. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf. (2018).
- 2.Miller DC et al. Contemporary clinical epidemiology of renal cell carcinoma: insight from a population based case-control study. J Urol 184, 2254–2258, doi: 10.1016/j.juro.2010.08.018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham J & Heng DY Real-world evidence in metastatic renal cell carcinoma. Tumori, 300891618761004, doi: 10.1177/0300891618761004 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Heng DY et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. J Clin Oncol 27, 5794–5799, doi: 10.1200/JCO.2008.21.4809 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Nabi S, Kessler ER, Bernard B, Flaig TW & Lam ET Renal cell carcinoma: a review of biology and pathophysiology. F1000Res 7, 307, doi: 10.12688/f1000research.13179.1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 45, 860–867, doi: 10.1038/ng.2699 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research, N. et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med 374, 135–145, doi: 10.1056/NEJMoa1505917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maher ER & Kaelin WG Jr. von Hippel-Lindau disease. Medicine (Baltimore) 76, 381–391 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Ricketts CJ et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep 23, 313–326e315, doi: 10.1016/j.celrep.2018.03.075 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell TJ et al. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell 173, 611–623e617, doi: 10.1016/j.cell.2018.02.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turajlic S. et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell 173, 595–610e511, doi: 10.1016/j.cell.2018.03.043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bratslavsky G, Sudarshan S, Neckers L & Linehan WM Pseudohypoxic pathways in renal cell carcinoma. Clin Cancer Res 13, 4667–4671, doi: 10.1158/1078-0432.CCR-06-2510 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Nielsen OH, Grimm D, Wehland M, Bauer J & Magnusson NE Anti-Angiogenic Drugs in the Treatment of Metastatic Renal Cell Carcinoma: Advances in Clinical Application. Curr Vasc Pharmacol 13, 381–391 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Kaelin WG Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 8, 865–873, doi: 10.1038/nrc2502 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Simon JM et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 24, 241–250, doi: 10.1101/gr.158253.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalgliesh GL et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363, doi: 10.1038/nature08672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo G. et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet 44, 17–19, doi: 10.1038/ng.1014 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Varela I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542, doi: 10.1038/nature09639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biegel JA, Busse TM & Weissman BE SWI/SNF chromatin remodeling complexes and cancer. Am J Med Genet C Semin Med Genet 166C, 350–366, doi: 10.1002/ajmg.c.31410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49, doi: 10.1038/nature12222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis CF et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330, doi: 10.1016/j.ccr.2014.07.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauer MH & Gasser SM Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev 31, 2204–2221, doi: 10.1101/gad.307702.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strahl BD & Allis CD The language of covalent histone modifications. Nature 403, 41–45, doi: 10.1038/47412 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Clapier CR & Cairns BR The biology of chromatin remodeling complexes. Annu Rev Biochem 78, 273–304, doi: 10.1146/annurev.biochem.77.062706.153223 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Conaway RC & Conaway JW The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci 34, 71–77, doi: 10.1016/j.tibs.2008.10.010 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Jones S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231, doi: 10.1126/science.1196333 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiegand KC et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 363, 1532–1543, doi: 10.1056/NEJMoa1008433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M. et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet 43, 828–829, doi: 10.1038/ng.903 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K. et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 43, 1219–1223, doi: 10.1038/ng.982 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Shain AH et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A 109, E252–259, doi: 10.1073/pnas.1114817109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reisman D, Glaros S & Thompson EA The SWI/SNF complex and cancer. Oncogene 28, 1653–1668, doi: 10.1038/onc.2009.4 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Nie Z. et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol 20, 8879–8888 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X. et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem J 383, 319–325, doi: 10.1042/BJ20040524 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan Z. et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev 19, 1662–1667, doi: 10.1101/gad.1323805 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaeser MD, Aslanian A, Dong MQ, Yates JR 3rd & Emerson BM BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem 283, 32254–32263, doi: 10.1074/jbc.M806061200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z. et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev 18, 3106–3116, doi: 10.1101/gad.1238104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brownlee PM, Chambers AL, Oliver AW & Downs JA Cancer and the bromodomains of BAF180. Biochem Soc Trans 40, 364–369, doi: 10.1042/BST20110754 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Thompson M. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie 91, 309–319, doi: 10.1016/j.biochi.2008.10.019 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pena-Llopis S. et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 44, 751–759, doi: 10.1038/ng.2323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forbes SA et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45, D777–D783, doi: 10.1093/nar/gkw1121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.COSMIC. Catalogue Of Somatic Mutations In Cancer, <http://cancer.sanger.ac.uk>. (2017).
- 42.Turajlic S, Larkin J & Swanton C. SnapShot: Renal Cell Carcinoma. Cell 163, 1556–1556 e1551, doi: 10.1016/j.cell.2015.11.026 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Rao Q. et al. Coexistent loss of INI1 and BRG1 expression in a rhabdoid renal cell carcinoma (RCC): implications for a possible role of SWI/SNF complex in the pathogenesis of RCC. Int J Clin Exp Pathol 7, 1782–1787 (2014). [PMC free article] [PubMed] [Google Scholar]
- 44.Kapur P. et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol 14, 159–167, doi: 10.1016/S1470-2045(12)70584-3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami A. et al. Context-dependent role for chromatin remodeling component PBRM1/BAF180 in clear cell renal cell carcinoma. Oncogenesis 6, e287, doi: 10.1038/oncsis.2016.89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao W, Li W, Xiao T, Liu XS & Kaelin WG Jr. Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL−/− clear cell renal carcinoma. Proc Natl Acad Sci U S A 114, 1027–1032, doi: 10.1073/pnas.1619726114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nargund AM et al. The SWI/SNF Protein PBRM1 Restrains VHL-Loss-Driven Clear Cell Renal Cell Carcinoma. Cell Rep 18, 2893–2906, doi: 10.1016/j.celrep.2017.02.074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao D. et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806, doi: 10.1126/science.aan5951 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shu XS et al. The epigenetic modifier PBRM1 restricts the basal activity of the innate immune system by repressing retinoic acid-inducible gene-I-like receptor signalling and is a potential prognostic biomarker for colon cancer. J Pathol 244, 36–48, doi: 10.1002/path.4986 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Pan D et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775, doi: 10.1126/science.aao1710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen DE et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16, 1097–1112 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Pickart CM Mechanisms underlying ubiquitination. Annu Rev Biochem 70, 503–533, doi: 10.1146/annurev.biochem.70.1.503 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Lee HS, Lee SA, Hur SK, Seo JW & Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun 5, 5128, doi: 10.1038/ncomms6128 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Scheuermann JC et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247, doi: 10.1038/nature08966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043, doi: 10.1126/science.1076997 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Wang H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878, doi: 10.1038/nature02985 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Bernstein E. et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26, 2560–2569, doi: 10.1128/MCB.26.7.2560-2569.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tavares L. et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678, doi: 10.1016/j.cell.2011.12.029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Boom V. et al. Non-canonical PRC1.1 Targets Active Genes Independent of H3K27me3 and Is Essential for Leukemogenesis. Cell Rep 14, 332–346, doi: 10.1016/j.celrep.2015.12.034 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Farcas AM et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 1, e00205, doi: 10.7554/eLife.00205 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blackledge NP et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459, doi: 10.1016/j.cell.2014.05.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edmunds JW, Mahadevan LC & Clayton AL Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27, 406–420, doi: 10.1038/sj.emboj.7601967 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strahl BD et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22, 1298–1306 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M. et al. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci U S A 102, 17636–17641, doi: 10.1073/pnas.0506350102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobor MS & Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim Biophys Acta 1577, 261–275 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Niu X. et al. The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene 31, 776–786, doi: 10.1038/onc.2011.266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rondinelli B. et al. Histone demethylase JARID1C inactivation triggers genomic instability in sporadic renal cancer. J Clin Invest 125, 4625–4637, doi: 10.1172/JCI81040 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pena-Llopis S, Christie A, Xie XJ & Brugarolas J. Cooperation and antagonism among cancer genes: the renal cancer paradigm. Cancer Res 73, 4173–4179, doi: 10.1158/0008-5472.CAN-13-0360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hakimi AA et al. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. Eur Urol 63, 848–854, doi: 10.1016/j.eururo.2012.09.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gossage L. et al. Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes Chromosomes Cancer 53, 38–51, doi: 10.1002/gcc.22116 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Liu L. et al. Loss of SETD2, but not H3K36me3, correlates with aggressive clinicopathological features of clear cell renal cell carcinoma patients. Biosci Trends 11, 214–220, doi: 10.5582/bst.2016.01228 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Pawlowski R. et al. Loss of PBRM1 expression is associated with renal cell carcinoma progression. Int J Cancer 132, E11–17, doi: 10.1002/ijc.27822 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Hakimi AA et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 19, 3259–3267, doi: 10.1158/1078-0432.CCR-12-3886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.da Costa WH et al. Prognostic impact of concomitant loss of PBRM1 and BAP1 protein expression in early stages of clear cell renal cell carcinoma. Urol Oncol 36, 243.e241–243 e248, doi: 10.1016/j.urolonc.2018.01.002 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Martinez P. et al. Parallel evolution of tumour subclones mimics diversity between tumours. J Pathol 230, 356–364, doi: 10.1002/path.4214 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Gerlinger M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 46, 225–233, doi: 10.1038/ng.2891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turajlic S. et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell 173, 581–594e512, doi: 10.1016/j.cell.2018.03.057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ricketts CJ & Linehan WM Multi-regional Sequencing Elucidates the Evolution of Clear Cell Renal Cell Carcinoma. Cell 173, 540–542, doi: 10.1016/j.cell.2018.03.077 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Mandriota SJ et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell 1, 459–468 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Frew IJ et al. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J 27, 1747–1757, doi: 10.1038/emboj.2008.96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rankin EB, Tomaszewski JE & Haase VH Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res 66, 2576–2583, doi: 10.1158/0008-5472.CAN-05-3241 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dey A. et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546, doi: 10.1126/science.1221711 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu M. et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc Natl Acad Sci U S A 107, 2956–2961, doi: 10.1073/pnas.0915033107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P & Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol 319, 258–266, doi: 10.1016/j.ydbio.2008.04.020 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Gu YF et al. Modeling Renal Cell Carcinoma in Mice: Bap1 and Pbrm1 Inactivation Drive Tumor Grade. Cancer Discov 7, 900–917, doi: 10.1158/2159-8290.CD-17-0292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang SS et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci U S A 111, 16538–16543, doi: 10.1073/pnas.1414789111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kovac M. et al. Recurrent chromosomal gains and heterogeneous driver mutations characterise papillary renal cancer evolution. Nat Commun 6, 6336, doi: 10.1038/ncomms7336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ho TH et al. Loss of PBRM1 and BAP1 expression is less common in non-clear cell renal cell carcinoma than in clear cell renal cell carcinoma. Urol Oncol 33, 23.e29–23 e14, doi: 10.1016/j.urolonc.2014.10.014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Malouf GG et al. Genomic Characterization of Renal Cell Carcinoma with Sarcomatoid Dedifferentiation Pinpoints Recurrent Genomic Alterations. Eur Urol 70, 348–357, doi: 10.1016/j.eururo.2016.01.051 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Wang Z. et al. Sarcomatoid Renal Cell Carcinoma Has a Distinct Molecular Pathogenesis, Driver Mutation Profile, and Transcriptional Landscape. Clin Cancer Res, doi: 10.1158/1078-0432.CCR-17-1057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dietlein F, Thelen L & Reinhardt HC Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet 30, 326–339, doi: 10.1016/j.tig.2014.06.003 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Chapman JR, Taylor MR & Boulton SJ Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47, 497–510, doi: 10.1016/j.molcel.2012.07.029 (2012). [DOI] [PubMed] [Google Scholar]
- 93.Risinger JI, Barrett JC, Watson P, Lynch HT & Boyd J. Molecular genetic evidence of the occurrence of breast cancer as an integral tumor in patients with the hereditary nonpolyposis colorectal carcinoma syndrome. Cancer 77, 1836–1843, doi: (1996). [DOI] [PubMed] [Google Scholar]
- 94.Li F. et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 153, 590–600, doi: 10.1016/j.cell.2013.03.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Acharya S. et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A 93, 13629–13634 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Laguri C. et al. Human mismatch repair protein MSH6 contains a PWWP domain that targets double stranded DNA. Biochemistry 47, 6199–6207, doi: 10.1021/bi7024639 (2008). [DOI] [PubMed] [Google Scholar]
- 97.Musselman CA et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19, 1266–1272, doi: 10.1038/nsmb.2435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hayakawa T. et al. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci 123, 1124–1130, doi: 10.1242/jcs.060178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Daugaard M. et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat Struct Mol Biol 19, 803–810, doi: 10.1038/nsmb.2314 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Pfister SX et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 7, 2006–2018, doi: 10.1016/j.celrep.2014.05.026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li L & Wang Y. Crosstalk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J Biol Chem, doi: 10.1074/jbc.M117.788224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shogren-Knaak M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847, doi: 10.1126/science.1124000 (2006). [DOI] [PubMed] [Google Scholar]
- 103.Pai CC et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun 5, 4091, doi: 10.1038/ncomms5091 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Palii SS, Cui Y, Innes CL & Paules RS Dissecting cellular responses to irradiation via targeted disruptions of the ATM-CHK1-PP2A circuit. Cell Cycle 12, 1105–1118, doi: 10.4161/cc.24127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feng C. et al. PI3Kbeta inhibitor TGX221 selectively inhibits renal cell carcinoma cells with both VHL and SETD2 mutations and links multiple pathways. Sci Rep 5, 9465, doi: 10.1038/srep09465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hacker KE et al. Structure/Function Analysis of Recurrent Mutations in SETD2 Protein Reveals a Critical and Conserved Role for a SET Domain Residue in Maintaining Protein Stability and Histone H3 Lys-36 Trimethylation. J Biol Chem 291, 21283–21295, doi: 10.1074/jbc.M116.739375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kanu N. et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene 34, 5699–5708, doi: 10.1038/onc.2015.24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA & Skotheim RI Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35, 2413–2427, doi: 10.1038/onc.2015.318 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Kolasinska-Zwierz P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41, 376–381, doi: 10.1038/ng.322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.de Almeida SF et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18, 977–983, doi: 10.1038/nsmb.2123 (2011). [DOI] [PubMed] [Google Scholar]
- 111.Convertini P. et al. Sudemycin E influences alternative splicing and changes chromatin modifications. Nucleic Acids Res 42, 4947–4961, doi: 10.1093/nar/gku151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sanidas I. et al. Phosphoproteomics screen reveals akt isoform-specific signals linking RNA processing to lung cancer. Mol Cell 53, 577–590, doi: 10.1016/j.molcel.2013.12.018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xie L. et al. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 30, 1473–1484, doi: 10.1038/emboj.2011.91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guo R et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell 56, 298–310, doi: 10.1016/j.molcel.2014.08.022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chowdhury B. et al. PBRM1 Regulates the Expression of Genes Involved in Metabolism and Cell Adhesion in Renal Clear Cell Carcinoma. PLoS One 11, e0153718, doi: 10.1371/journal.pone.0153718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kenneth NS, Mudie S, van Uden P & Rocha S. SWI/SNF regulates the cellular response to hypoxia. J Biol Chem 284, 4123–4131, doi: 10.1074/jbc.M808491200 (2009). [DOI] [PubMed] [Google Scholar]
- 117.Schmitges FW et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42, 330–341, doi: 10.1016/j.molcel.2011.03.025 (2011). [DOI] [PubMed] [Google Scholar]
- 118.Brien GL et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 19, 1273–1281, doi: 10.1038/nsmb.2449 (2012). [DOI] [PubMed] [Google Scholar]
- 119.Ballare C. et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19, 1257–1265, doi: 10.1038/nsmb.2434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cai L. et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 49, 571–582, doi: 10.1016/j.molcel.2012.11.026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ferrari KJ et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53, 49–62, doi: 10.1016/j.molcel.2013.10.030 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Goll MG & Bestor TH Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74, 481–514, doi: 10.1146/annurev.biochem.74.010904.153721 (2005). [DOI] [PubMed] [Google Scholar]
- 123.Baubec T. et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247, doi: 10.1038/nature14176 (2015). [DOI] [PubMed] [Google Scholar]
- 124.Hahn MA, Wu X, Li AX, Hahn T & Pfeifer GP Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS One 6, e18844, doi: 10.1371/journal.pone.0018844 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Manzo M. et al. Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J 36, 3421–3434, doi: 10.15252/embj.201797038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hogart A. et al. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res 22, 1407–1418, doi: 10.1101/gr.132878.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Clark SJ, Harrison J & Molloy PL Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene 195, 67–71 (1997). [DOI] [PubMed] [Google Scholar]
- 128.Chen F. et al. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell Rep 14, 2476–2489, doi: 10.1016/j.celrep.2016.02.024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tiedemann RL et al. Dynamic reprogramming of DNA methylation in SETD2-deregulated renal cell carcinoma. Oncotarget 7, 1927–1946, doi: 10.18632/oncotarget.6481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Su X. et al. NSD1 Inactivation and SETD2 Mutation Drive a Convergence toward Loss of Function of H3K36 Writers in Clear Cell Renal Cell Carcinomas. Cancer Res 77, 4835–4845, doi: 10.1158/0008-5472.CAN-17-0143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park IY et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 166, 950–962, doi: 10.1016/j.cell.2016.07.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen K. et al. Methyltransferase SETD2-Mediated Methylation of STAT1 Is Critical for Interferon Antiviral Activity. Cell 170, 492–506e414, doi: 10.1016/j.cell.2017.06.042 (2017). [DOI] [PubMed] [Google Scholar]
- 133.Chiang YC et al. SETD2 Haploinsufficiency for Microtubule Methylation is an Early Driver of Genomic Instability in Renal Cell Carcinoma. Cancer Res, doi: 10.1158/0008-5472.CAN-17-3460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mashtalir N. et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol Cell 54, 392–406, doi: 10.1016/j.molcel.2014.03.002 (2014). [DOI] [PubMed] [Google Scholar]
- 135.Bononi A. et al. Germline BAP1 mutations induce a Warburg effect. Cell Death Differ 24, 1694–1704, doi: 10.1038/cdd.2017.95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dai F. et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc Natl Acad Sci U S A 114, 3192–3197, doi: 10.1073/pnas.1619588114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baughman JM et al. NeuCode Proteomics Reveals Bap1 Regulation of Metabolism. Cell Rep 16, 583–595, doi: 10.1016/j.celrep.2016.05.096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hebert L. et al. Modulating BAP1 expression affects ROS homeostasis, cell motility and mitochondrial function. Oncotarget 8, 72513–72527, doi: 10.18632/oncotarget.19872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Luchini C. et al. Different prognostic roles of tumor suppressor gene BAP1 in cancer: A systematic review with meta-analysis. Genes Chromosomes Cancer 55, 741–749, doi: 10.1002/gcc.22381 (2016). [DOI] [PubMed] [Google Scholar]
- 140.Palmer AE, Jin C, Reed JC & Tsien RY Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci U S A 101, 17404–17409, doi: 10.1073/pnas.0408030101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bononi A. et al. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature 546, 549–553, doi: 10.1038/nature22798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Verhey KJ & Gaertig J The tubulin code. Cell Cycle 6, 2152–2160, doi: 10.4161/cc.6.17.4633 (2007). [DOI] [PubMed] [Google Scholar]
- 143.Zarrizi R, Menard JA, Belting M & Massoumi R. Deubiquitination of gamma-tubulin by BAP1 prevents chromosome instability in breast cancer cells. Cancer Res 74, 6499–6508, doi: 10.1158/0008-5472.CAN-14-0221 (2014). [DOI] [PubMed] [Google Scholar]
- 144.Kollman JM, Merdes A, Mourey L & Agard DA Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol 12, 709–721, doi: 10.1038/nrm3209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sankaran S, Starita LM, Groen AC, Ko MJ & Parvin JD Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol Cell Biol 25, 8656–8668, doi: 10.1128/MCB.25.19.8656-8668.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hsu CC et al. 58-kDa microspherule protein (MSP58) is novel Brahma-related gene 1 (BRG1)-associated protein that modulates p53/p21 senescence pathway. J Biol Chem 287, 22533–22548, doi: 10.1074/jbc.M111.335331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Peng J. et al. Stabilization of MCRS1 by BAP1 prevents chromosome instability in renal cell carcinoma. Cancer Lett 369, 167–174, doi: 10.1016/j.canlet.2015.08.013 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Cui K. et al. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol Cell Biol 24, 4476–4486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xiang W. et al. miR-106b-5p targets tumor suppressor gene SETD2 to inactive its function in clear cell renal cell carcinoma. Oncotarget 6, 4066–4079, doi: 10.18632/oncotarget.2926 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.He X. et al. Bap180/Baf180 is required to maintain homeostasis of intestinal innate immune response in Drosophila and mice. Nat Microbiol 2, 17056, doi: 10.1038/nmicrobiol.2017.56 (2017). [DOI] [PubMed] [Google Scholar]
- 151.Pfister SX et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell 28, 557–568, doi: 10.1016/j.ccell.2015.09.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bucher N & Britten CD G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer 98, 523–528, doi: 10.1038/sj.bjc.6604208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Beck H. et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol 32, 4226–4236, doi: 10.1128/MCB.00412-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang J. et al. High selectivity of PI3Kbeta inhibitors in SETD2-mutated renal clear cell carcinoma. J BUON 20, 1267–1275 (2015). [PubMed] [Google Scholar]
- 155.Feng C, Ding G, Jiang H, Ding Q & Wen H. Loss of MLH1 confers resistance to PI3Kbeta inhibitors in renal clear cell carcinoma with SETD2 mutation. Tumour Biol 36, 3457–3464, doi: 10.1007/s13277-014-2981-y (2015). [DOI] [PubMed] [Google Scholar]
- 156.LaFave LM et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med 21, 1344–1349, doi: 10.1038/nm.3947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schoumacher M. et al. Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status. Nat Med 22, 577–578, doi: 10.1038/nm.4098 (2016). [DOI] [PubMed] [Google Scholar]
- 158.LaFave LM et al. Reply to “Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status”. Nat Med 22, 578–579, doi: 10.1038/nm.4094 (2016). [DOI] [PubMed] [Google Scholar]
- 159.Kim KH et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 21, 1491–1496, doi: 10.1038/nm.3968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Knutson SK et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A 110, 7922–7927, doi: 10.1073/pnas.1303800110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bitler BG et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 21, 231–238, doi: 10.1038/nm.3799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hopkins SR, McGregor GA, Murray JM, Downs JA & Savic V. Novel synthetic lethality screening method identifies TIP60-dependent radiation sensitivity in the absence of BAF180. DNA Repair (Amst) 46, 47–54, doi: 10.1016/j.dnarep.2016.05.030 (2016). [DOI] [PubMed] [Google Scholar]
- 163.Samartzis EP et al. Loss of ARID1A expression sensitizes cancer cells to PI3K- and AKT-inhibition. Oncotarget 5, 5295–5303, doi: 10.18632/oncotarget.2092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shen J. et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov 5, 752–767, doi: 10.1158/2159-8290.CD-14-0849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Konings IR, Verweij J, Wiemer EA & Sleijfer S. The applicability of mTOR inhibition in solid tumors. Curr Cancer Drug Targets 9, 439–450 (2009). [DOI] [PubMed] [Google Scholar]
- 166.Zhang N & Bevan MJ CD8(+) T cells: foot soldiers of the immune system. Immunity 35, 161–168, doi: 10.1016/j.immuni.2011.07.010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]