Abstract
Recently, a rationally engineered SpCas9 variant (SpCas9-NG) that can recognize a minimal NG protospacer adjacent motif (PAM) was reported to expand the targeting scope in genome editing. However, increased genome-wide off-target mutations with this variant compared with SpCas9 were reported in previous studies. In addition, lower base editing frequencies and higher unintended off-target mutations were also found in Hoxc13-ablated rabbits generated by NG-BE4max in our study. Here, a high-fidelity base editor, NG-HiFi, in comparison to NG-BE4max, showed retention of on-target activity while exhibiting significantly decreased off-target activity in Hoxc13-ablated rabbits. Collectively, the improved specificity and reduced off-target effect of SpCas9-NG assisted in cytidine base editing with the NG-HiFi system, providing a promising tool to precisely model human diseases in rabbits.
Keywords: SpCas9-NG, NG-BE4max, Hoxc13, off-target, NG-HiFi, rabbit
Graphical abstract
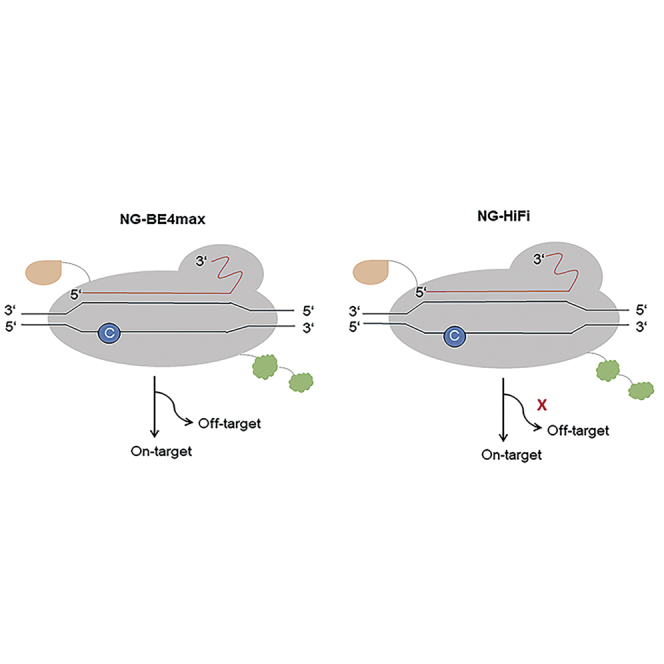
NG-BE4max that can recognize NG PAM was reported to expand the targeting space in genome editing but, however, with higher genome-wide off-target mutations compared with BE4max. Here, a high-fidelity base editor, NG-HiFi, reduces the off-target effect, which is comparable to NG-BE4max.
Introduction
CRISPR/Cas9-based cytosine base editors (CBEs) can mediate the direct conversion of C to T (or G to A), which does not induce double-stranded DNA breaks (DSBs) or require a donor template.1 CBEs enable C-to-T conversion at a target genomic locus with the requirement of a protospacer adjacent motif (PAM), such as NGG, possessing limited activity at noncanonical NGH (H = A, C, and T) PAM sites, which restricts the targetable genomic loci in applications.2
To address the PAM limitation, a rationally engineered SpCas9 variant (SpCas9-NG)3 that can recognize the more relaxed NG PAM, which broadens PAM compatibility and significantly expands the target scope, has been used in Arabidopsis4 and rice.5 In addition, the NG-BE4max and NG-ABEmax systems are highly efficient tools for targeted base editing and have been used to precisely mimic human pathogenic mutations in rabbits.6 However, a previous study showed that SpCas9-NG not only targeted the genome but also potentially increased off-target risk by generating new single-guide RNAs (sgRNAs).7 In addition, a higher frequency of off-target editing events was also observed in Hoxc13 mutant rabbits generated by the NG-ABEmax system in our previous study.6
In this study, significantly reduced off-target events and similar on-target effects in Hoxc13 mutant rabbits were generated by NG-HiFi, which greatly improved the base editing specificity and expanded the genome targeting scope of base editing in human disease modeling and gene therapy in the future.
Results
Significantly increased off-target effects in Hoxc13 (Q87Stop) rabbits generated by the NG-BE4max system
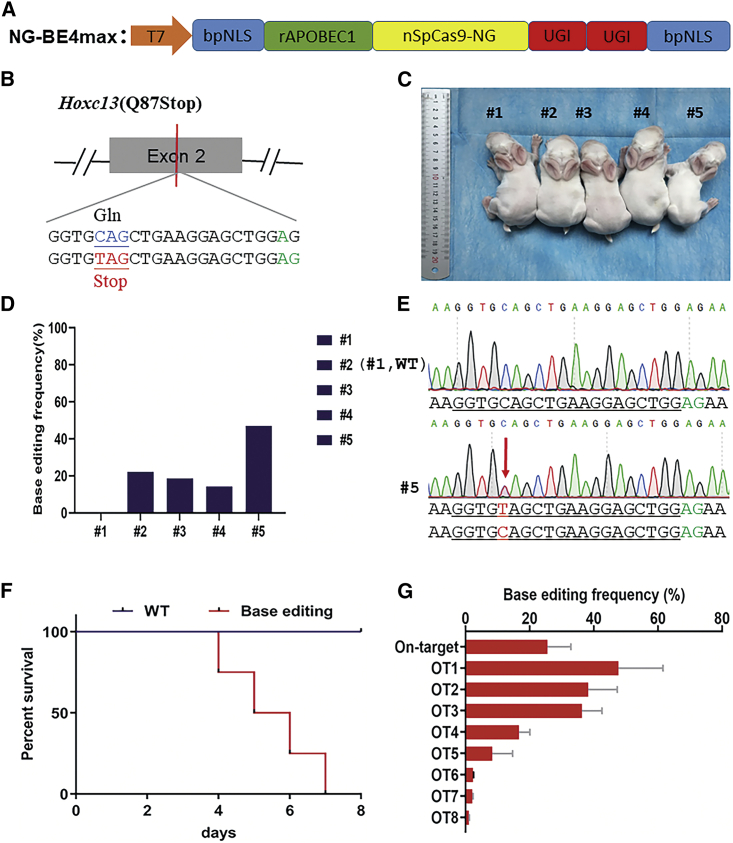
First, the more relaxed NG PAM in the NG-BE4max vector was constructed in our previous study6 (Figure 1A). An sgRNA targeting the second exon of rabbit Hoxc13 was designed (Figure 1B). Then, the base editing frequencies were evaluated by Sanger sequencing using EditR, a robust base editing quantification software.8 The results showed that four (#2, #3, #4, and #5) of five pups (80%) carried a desired nonsense mutation (Q87Stop) at the target site. However, there was no obvious phenotype (hairlessness) (Figure 1C) because of the low base editing frequencies for C-to-T conversion, ranging from 14.3% to 47% in these rabbits (Figures 1D and 1E; Table S1). In addition, the four base-edited rabbits died within a week (Figure 1F), which is not consistent with our previous study.9
Figure 1.
NG-BE4max-mediated C-T base editing of the Hoxc13 locus in rabbits
(A) Plasmid schematic of the NG-BE4max cytosine base editor used in this study. The D10A mutation inactivates the nuclease activity of the RuvC domain in SpCas9-NG. NLS, nuclear localization signal. (B) Schematic of sgRNA design at the rabbit Hoxc13 locus. (C) Photograph of F0 Hoxc13 (Q87Stop) rabbits generated by the NG-BE4max system. (D) On-target base editing frequency in F0 Hoxc13 (Q87Stop) rabbits generated by the NG-BE4max system. (E) Chromatograms of wild-type (WT) and mutant sequences showing C-T substitution in Hoxc13 (Q87Stop) by the NG-BE4max system. (F) Survival curves of F0 Hoxc13 (Q87Stop) rabbits generated by the NG-BE4max system. (G) On- and off-target base editing frequency in F0 Hoxc13 (Q87Stop) rabbits generated by the NG-BE4max system.
To confirm the increased off-target effect of SpCas9-NG, the sites of EMX1, DNMT1, HEK293 site4, FANCF site2, and VEGFA site2 were predicted according to Cas-OFFinder.10 The results showed significantly increased off-target effects in SpCas9-NG compared with wild-type SpCas9 (Figure S2). Consistent results were found in Hoxc13 (Q87Stop) rabbits, which showed 8.35%–47.68% off-target effects (Figure 1G; Figure S1). It was reported that OT1 (GALK1, p.Q259Stop) is a major enzyme for galactose metabolism11, defects in which are known to cause cataracts in infants and galactosemia type 2 (Table S2),12 while OT2 (SPTB, p.Q727Stop) plays an important role in the stability of the erythrocyte membrane,13 which is associated with hereditary spherocytosis (HS). Thus, we hypothesized that the high death rate of Hoxc13 (Q87Stop) rabbits may have been caused by the significantly increased off-target effect generated by the NG-BE4max system.
Improved specificity of NG-BE4max obtained by using NG-HiFi
CRISPR/Cas9 enables highly efficient genome editing in a variety of organisms but can also cause unwanted mutations at off-target sites that resemble the on-target sequence.14,15 To date, Cas9 variants have been used to reduce genome-wide off-target mutations, such as eSpCas9(1.1),16 SpCas9-HF1,17 Sniper-Cas9,18 HiFi-Cas9,19 and Opti-Cas9.20 Thus, to reduce the off-target base editing effect of the NG-BE4max system, five high-fidelity base editors were used to reduce off-target editing events in this study (Figure 2A).
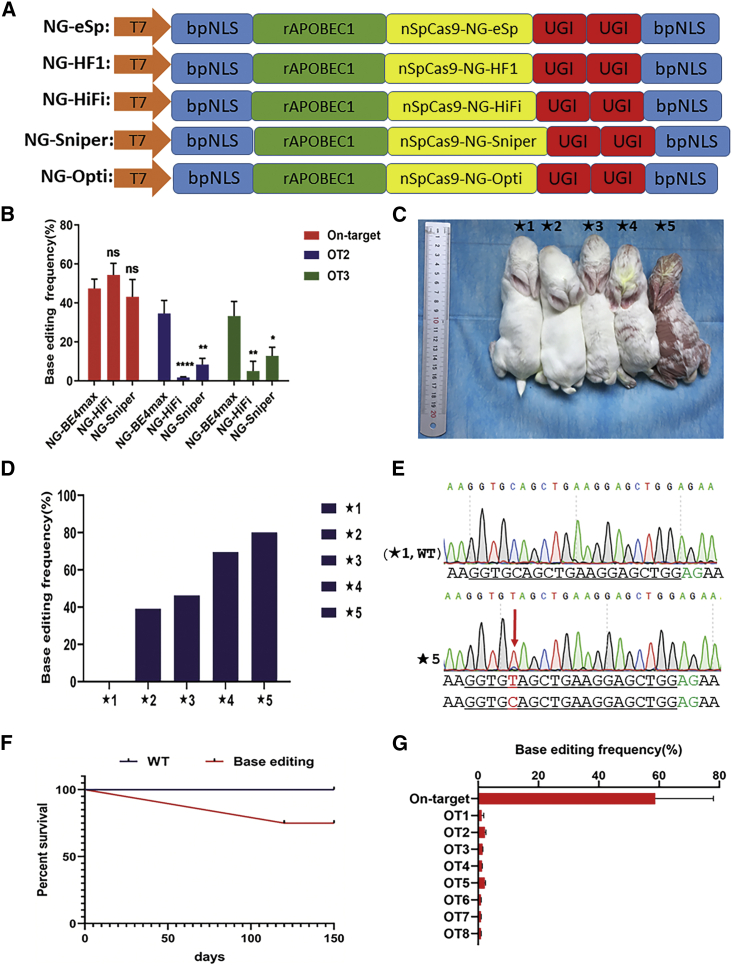
Figure 2.
Significantly reduced off-target effects in Hoxc13 (Q87Stop) rabbits using NG-HiFi
(A) Plasmid schematic of the five high-fidelity base editors NG-eSp, NG-HF1, NG-Sniper, NG-HiFi, and NG-Opti. (B) NG-HiFi induces efficient C-to-T base editing in rabbit blastocysts. (C) Photograph of F0 Hoxc13 (Q87Stop) rabbits generated by the NG-HiFi system. (D) On-target base editing frequency of F0 Hoxc13 (Q87Stop) rabbits generated by the NG-HiFi system. (E) Chromatograms of WT and mutant sequences showing C-T substitution in Hoxc13 (Q87Stop) rabbits by the NG-HiFi system. (F) Survival curves of F0 Hoxc13 (Q87Stop) rabbits generated by the NG-HiFi system. (G) On- and off-target base editing frequency of F0 Hoxc13 (Q87Stop) rabbits generated by the NG-HiFi system.
Then, five endogenous genomic loci were chosen to test the on-target (Figure S4) and off-target (Figure S5) effects of the high-fidelity base editors in 293T cells. The results showed significantly reduced off-target editing by using high-fidelity base editors compared with NG-BE4max, and the base-editing specificity of NG-Sniper and NG-HiFi was comparable to that of NG-BE4max (Figure S5). To test whether NG-Sniper and NG-HiFi could be used to generate Hoxc13 (Q87Stop) with reduced off-target effects, injected rabbit blastocysts were collected and subjected to analysis of the on-target effects and the off-target effects (on OT2 and OT3). The results showed that there was no significant difference in on-target effects and the off-target effects of NG-Sniper and NG-HiFi decreased significantly compared with those of NG-BE4max (Figure 2B). Moreover, NG-HiFi maintained on-target base editing while eliminating detectable off-target base editing. Therefore, NG-HiFi was used to generate Hoxc13 (Q87Stop) in the following study.
As shown in Figures 2D and 2E and Table S1, four (★2, ★3, ★4, and ★5) of five pups (80%) carried a desired nonsense mutation at the target site of Hoxc13 (Q87Stop), generated by using NG-HiFi. The base editing frequency was significantly increased compared with that of the NG-BE4max system (58.73% versus 25.55%) (Figure 2G). In addition, the desired hairless phenotype was detected in pups ★4 and ★5 (Figure 2C). Moreover, the normal survival rate and lack of off-target effects were observed in Hoxc13 (Q87Stop) rabbits generated by the NG-HiFi system (Figures 2F and 2G), and Hoxc13 (Q87Stop) could also be stably transmitted to the F1 offspring (Figure S6).
Discussion
In this study, we successfully generated a high-fidelity base editor, NG-HiFi, and demonstrated the significantly reduced off-target effect of NG-BE4max by using the NG-HiFi system. The desired hairless phenotype was obtained in Hoxc13 (Q87Stop) rabbits by using the NG-HiFi base editor, which greatly expanded the genome targeting scope and reduced the risk of off-target effects, providing a promising tool to precisely model human diseases in rabbits.
The possibility of creating off-target mutations with unknown consequences is a major concern associated with the CRISPR/Cas9 system. To date, numerous Cas9 homologs and variants have been used for cytidine base editing, such as SaCas9,21 ScCas9,22 Spy-macCas9,23 Nme2Cas9,24 St1Cas9,25 and Cas12a.26 In addition to CBEs, additional base editing tools, such as adenine base editors (ABEs),27 glycosylase base editors (GBEs),28 and prime editors (PEs),29 have been developed to increase versatility. Combining these tools with high-fidelity mutations may further improve the DNA specificity of these genome editors, as shown in this study.
In addition to Cas9-dependent DNA off-target mutations, it has been shown that CBEs may cause Cas9-independent off-target DNA and RNA mutations.30, 31, 32, 33 These unexpected off-target DNA and RNA mutations are mainly caused by deaminase domains rather than Cas9 domains. Additionally, off-target DNA and RNA editing could be eliminated by rational mutagenesis of the deaminase domain.34, 35, 36 Therefore, by further rationally engineering both Cas9 and deaminase domains, it is possible to produce a perfect base editor without off-target mutations in the future.
In summary, this study demonstrates the great value of a highly specific base editor in efficient C-to-T conversion at sites containing the broadened NG PAM, which greatly expands the genome targeting scope of base editing in human disease modeling and future gene therapy.
Materials and methods
Ethics statement
New Zealand white rabbits were obtained from the Laboratory Animal Center of Jilin University (Changchun, China). All animal studies were conducted according to experimental practices and standards approved by the Animal Welfare and Research Ethics Committee of Jilin University.
Plasmid construction
The BE4max plasmid was obtained from Addgene (#112093). Seven mutations (R1335A/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R) in CRISPR/Cas9 were introduced into BE4max to obtain NG-BE4max. Five high-fidelity CRISPR/Cas9 systems were introduced into NG-BE4max to obtain NG-eSp (K848A/K1003A/R1060A), NG-HF1 (N497A/R661A/Q695A/Q926A), NG-Sniper (F539S/M761I/K890N), NG-HiFi (R691A), and NG-Opti (R661A/K1003H). Site-directed mutagenesis of the plasmid was performed with the Fast Site-Directed Mutagenesis Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. All primers used for site-directed mutation are listed in Table S6.
mRNA and gRNA preparation
All plasmids were linearized with NotI and transcribed in vitro with the HiScribe T7 ARCA mRNA Kit (NEB). The RNeasy Mini Kit (QIAGEN) was used for mRNA purification according to the manufacturer’s instructions.
The sgRNA oligos were annealed into pUC57-sgRNA expression vectors containing a T7 promoter, transcribed in vitro with the MAXIscript T7 Kit (Ambion), and purified with the miRNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions.
Microinjection of rabbit zygotes and embryo transfer
The protocol used for microinjection of pronuclear-stage embryos has been described in detail in our previously published study.37 Briefly, a mixture of mRNA (200 ng/μL) and sgRNA (50 ng/μL) was co-injected into the cytoplasm of pronuclear-stage zygotes. The injected embryos were transferred into Earle's Balanced Salt Solution (EBSS) medium for short-term culture at 38.5°C, under 5% carbon dioxide and 100% humidity. Then, ~30–50 injected zygotes were transferred into the oviducts of recipient rabbits.
Single-embryo PCR amplification and rabbit genotyping
Single-embryo PCR amplification and rabbit genotyping were performed according to our previous study.38 The base editing frequencies were evaluated by EditR (baseEditR.com/). All primers used for genotyping are listed in Table S4.
Off-target detection
The potential off-target sites (POTs) were predicted according to an online design tool (http://www.rgenome.net/cas-offinder/). Selected POTs (Table S3 and Table S5) were amplified by PCR and Sanger sequencing. All primers used for the off-target assay are listed in Table S3 and Table S5. Mutations were detected with deep sequencing and Hi-TOM analysis according to a previous study.39
Cell culture and transfection
HEK293T cells were maintained in DMEM plus GlutaMax (Thermo Fisher) supplemented with 10% (v/v) fetal bovine serum at 37°C with 5% CO2. HEK293T cells were seeded on 6-well collagen-coated BioCoat plates (Corning) in an antibiotic-free medium and transfected at ~70% confluency. Then, BE and sgRNA plasmids were transfected with Lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s protocol.
Statistical analysis
All data are expressed as the mean ± SEM, with at least three individual determinations in all experiments. The data were analyzed with t tests using GraphPad Prism software 6.0. p < 0.05 was considered statistically significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Data availability
High-throughput sequencing reads were deposited in the NCBI Sequence Read Archive under PRJNA725675.
Acknowledgments
The authors thank Peiran Hu and Nannan Li for assistance at the Embryo Engineering Center for critical technical assistance. This study was financially supported by the National Key Research and Development Program of China Stem Cell and Translational Research (2017YFA0105101), The Program for Changjiang Scholars and Innovative Research Team in University (No. IRT_16R32), The Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030501 and XDA16030503), and Key Research & Development Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR110104004).
Author contributions
H.S., L.L., and Z. Li conceived and designed the experiments. H.S., Y.J., and Z. Liu performed the experiments. H.S., Y.J., and Z. Liu analyzed the data. M.C., Y.S., and T.S. contributed reagents/materials/analysis tools. H.S. and Z. Liu wrote the paper. All authors have read and approved the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.05.012.
Contributor Information
Liangxue Lai, Email: lai_liangxue@gibh.ac.cn.
Zhanjun Li, Email: lizj_1998@jlu.edu.cn.
Supplemental information
References
- 1.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eid A., Alshareef S., Mahfouz M.M. CRISPR base editors: genome editing without double-stranded breaks. Biochem. J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. 29891532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge Z., Zheng L., Zhao Y., Jiang J., Zhang E.J., Liu T., Gu H., Qu L.-J. Engineered xCas9 and SpCas9-NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol. J. 2019;17:1865–1867. doi: 10.1111/pbi.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong Z., Sretenovic S., Ren Q., Yang L., Bao Y., Qi C., Yuan M., He Y., Liu S., Liu X. Improving Plant Genome Editing with High-Fidelity xCas9 and Non-canonical PAM-Targeting Cas9-NG. Mol. Plant. 2019;12:1027–1036. doi: 10.1016/j.molp.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z., Shan H., Chen S., Chen M., Song Y., Lai L., Li Z. Highly efficient base editing with expanded targeting scope using SpCas9-NG in rabbits. FASEB J. 2020;34:588–596. doi: 10.1096/fj.201901587R. [DOI] [PubMed] [Google Scholar]
- 7.Qin R., Li J., Liu X., Xu R., Yang J., Wei P. SpCas9-NG self-targets the sgRNA sequence in plant genome editing. Nat. Plants. 2020;6:197–201. doi: 10.1038/s41477-020-0603-9. [DOI] [PubMed] [Google Scholar]
- 8.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: A Method to Quantify Base Editing from Sanger Sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng J., Chen M., Liu Z., Song Y., Sui T., Lai L., Li Z. The disrupted balance between hair follicles and sebaceous glands in Hoxc13-ablated rabbits. FASEB J. 2019;33:1226–1234. doi: 10.1096/fj.201800928RR. [DOI] [PubMed] [Google Scholar]
- 10.Bae S., Park J., Kim J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timson D.J., Reece R.J. Functional analysis of disease-causing mutations in human galactokinase. Eur. J. Biochem. 2003;270:1767–1774. doi: 10.1046/j.1432-1033.2003.03538.x. [DOI] [PubMed] [Google Scholar]
- 12.Sneha P., Ebrahimi E.A., Ghazala S.A., T K D., S R., Priya Doss C.,G., Zayed H. Structural analysis of missense mutations in galactokinase 1 (GALK1) leading to galactosemia type-2. J. Cell. Biochem. 2018;119:7585–7598. doi: 10.1002/jcb.27097. [DOI] [PubMed] [Google Scholar]
- 13.He B.J., Liao L., Deng Z.F., Tao Y.F., Xu Y.C., Lin F.Q. Molecular Genetic Mechanisms of Hereditary Spherocytosis: Current Perspectives. Acta Haematol. 2018;139:60–66. doi: 10.1159/000486229. [DOI] [PubMed] [Google Scholar]
- 14.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J.K., Jeong E., Lee J., Jung M., Shin E., Kim Y.-H., Lee K., Jung I., Kim D., Kim S., Kim J.S. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018;9:3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vakulskas C.A., Dever D.P., Rettig G.R., Turk R., Jacobi A.M., Collingwood M.A., Bode N.M., McNeill M.S., Yan S., Camarena J. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi G.C.G., Zhou P., Yuen C.T.L., Chan B.K.C., Xu F., Bao S., Chu H.Y., Thean D., Tan K., Wong K.H. Combinatorial mutagenesis en masse optimizes the genome editing activities of SpCas9. Nat. Methods. 2019;16:722–730. doi: 10.1038/s41592-019-0473-0. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee P., Jakimo N., Jacobson J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018;4:eaau0766. doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z., Shan H., Chen S., Chen M., Song Y., Lai L., Li Z. Efficient base editing with expanded targeting scope using an engineered Spy-mac Cas9 variant. Cell Discov. 2019;5:58. doi: 10.1038/s41421-019-0128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z., Chen S., Jia Y., Shan H., Chen M., Song Y., Lai L., Li Z. Efficient and high-fidelity base editor with expanded PAM compatibility for cytidine dinucleotide. Sci. China Life Sci. 2021 doi: 10.1007/s11427-020-1775-2. Published online January 6, 2021. [DOI] [PubMed] [Google Scholar]
- 25.Agudelo D., Carter S., Velimirovic M., Duringer A., Levesque S., Rivest J.-F., Loehr J., Mouchiroud M., Cyr D., Waters P.J. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. bioRxiv. 2019:321208. doi: 10.1101/gr.255414.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Wang Y., Liu Y., Yang B., Wang X., Wei J., Lu Z., Zhang Y., Wu J., Huang X. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 27.Matsoukas I.G. Commentary: Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Front. Genet. 2018;9:21. doi: 10.3389/fgene.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L., Park J.E., Paa P., Rajakumar P.D., Prekop H.-T., Chew Y.T., Manivannan S.N., Chew W.L. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 2021;12:1384. doi: 10.1038/s41467-021-21559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.-L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 31.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 32.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grünewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo E., Sun Y., Yuan T., He B., Zhou C., Ying W., Liu J., Wei W., Zeng R., Li Y., Yang H. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat. Methods. 2020;17:600–604. doi: 10.1038/s41592-020-0832-x. [DOI] [PubMed] [Google Scholar]
- 36.Yu Y., Leete T.C., Born D.A., Young L., Barrera L.A., Lee S.-J., Rees H.A., Ciaramella G., Gaudelli N.M. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 2020;11:2052. doi: 10.1038/s41467-020-15887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Y., Yuan L., Wang Y., Chen M., Deng J., Lv Q., Sui T., Li Z., Lai L. Efficient dual sgRNA-directed large gene deletion in rabbit with CRISPR/Cas9 system. Cell. Mol. Life Sci. 2016;73:2959–2968. doi: 10.1007/s00018-016-2143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S., Xie W., Liu Z., Shan H., Chen M., Song Y., Yu H., Lai L., Li Z. CRISPR Start-Loss: A Novel and Practical Alternative for Gene Silencing through Base-Editing-Induced Start Codon Mutations. Mol. Ther. Nucleic Acids. 2020;21:1062–1073. doi: 10.1016/j.omtn.2020.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q., Wang C., Jiao X., Zhang H., Song L., Li Y., Gao C., Wang K. Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019;62:1–7. doi: 10.1007/s11427-018-9402-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing reads were deposited in the NCBI Sequence Read Archive under PRJNA725675.