Abstract
Introduction:
Cytosolic sulfotransferases (SULTs)-mediated sulfation is critically involved in the metabolism of key endogenous compounds such as catecholamines and thyroid/steroid hormones, as well as a variety of drugs and other xenobiotics. Studies performed in the past three decades have yielded a good understanding about the enzymology of the SULTs and their structural biology, phylogenetic relationships, tissue/organ-specific/developmental expression, as well as the regulation of the SULT gene expression. An emerging area is related to the functional impact of the SULT genetic polymorphisms.
Areas covered:
The current review aims to summarize our current knowledge about the above-mentioned aspects of the SULT research. An emphasis is on the information concerning the effects of the polymorphisms of the SULT genes on the functional activity of the SULT allozymes and the associated physiological, pharmacological, and clinical implications.
Expert opinion:
Elucidation of how SULT SNPs may influence the drug-sulfating activity of SULT allozymes will help understand the differential drug metabolism and eventually aid in formulating personalized drug regimens. Moreover, the information concerning the differential sulfating activities of SULT allozymes toward endogenous compounds may allow for the development of strategies for mitigating anomalies in the metabolism of these endogenous compounds in individuals with certain SULT genotypes.
Keywords: Cytosolic sulfotransferase, SULT, sulfation, single-nucleotide polymorphism, SNP
1. Introduction
Biological sulfation in humans was first documented in 1876 with the detection of phenyl sulfate in the urine of a patient who was treated with phenol as an antiseptic [1]. This finding provided the initial clue that sulfation may play a role in the metabolism of xenobiotics. In line with this notion, many studies subsequently performed using experimental animals or human subjects have demonstrated the metabolism of drugs and other xenobiotics through sulfation [2,3]. It is generally accepted that sulfation may lead to the inactivation and/or facilitated excretion of xenobiotics. The enzymes responsible for catalyzing the sulfation reactions, now called the cytosolic sulfotransferases (SULTs) [4], however, were not identified until 1980 when two “phenol sulfotransferases” were isolated [5]. Interestingly, one of these two enzymes was found to display high affinity for dopamine and other catecholamines [6], indicating that sulfation may also play a role in the metabolism of endogenous compounds. It is now well documented that SULT-mediated sulfation is involved in the metabolism of not only catecholamines, but also thyroid and steroid hormones, as well as bile acids [7]. Sulfation of these endogenous compounds is believed to be involved in their homeostasis and/or transport in the body. Like other genes, genetic polymorphisms have been reported for the genes encoding SULTs [8–12]. In view of the physiological and pharmacological involvement of the SULTs, it is possible that the SULT genetic polymorphisms may predispose differential metabolism of catecholamines, steroid/thyroid hormones and other endogenous compounds, as well as xenobiotics including drugs, and therefore differential disease risks or clinical outcome for individuals with different SULT genotypes. This review aims to summarize our current knowledge about the SULTs, particularly the effects of the polymorphisms of the SULT genes on the functionality of the coded SULT allozymes and the possible resulting physiological/pharmacological implications. Figure 1 shows a typical SULT-mediated sulfation reaction. It is noted that during the reaction, the enzyme, SULT1A1, catalyzes the transfer of the sulfonate group from the co-substrate, 3’-phosphoadenosine 5’-phosphosulfate (PAPS), to the hydroxyl group of the substrate, acetaminophen.
Figure 1.
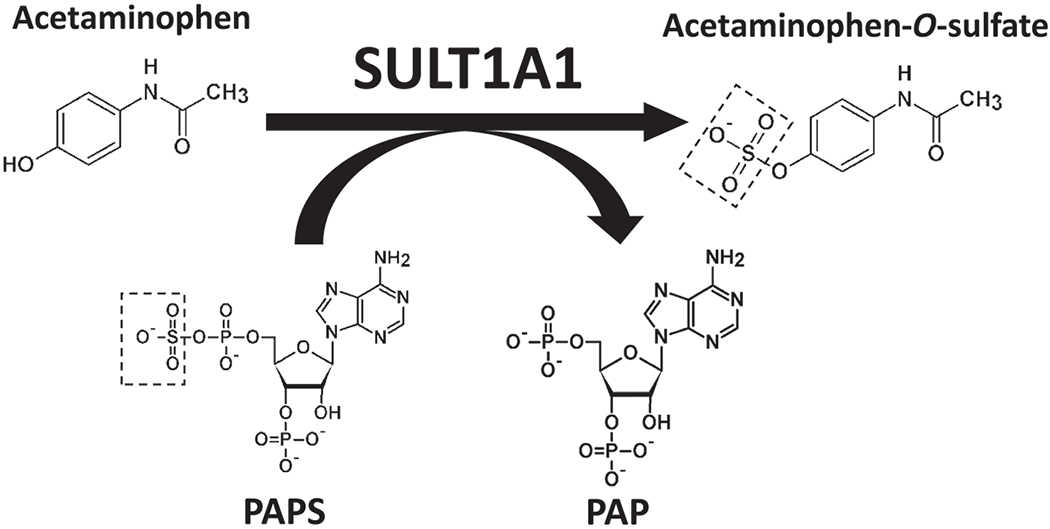
SULT-mediated sulfation of acetaminophen. SULT1A1 catalyzes the transfer of the sulfonate (-SO3−) group from 3-phosphoadenosine-5’-phosphosulfate (PAPS), a co-substrate, to the hydroxyl (-OH) group of acetaminophen.
2. Major classes of the SULTs and their roles in the metabolism of endogenous compounds and xenobiotics
Prior to the mid-1990s, the SULTs were mostly classified based on their substrate specificity and thus assigned names such as “monoamine”, “simple phenol”, or “estrogen” sulfotransferases [5,6,13]. Such a nomenclature system, however, suffered from the overlapping substrate specificity among SULT enzymes. With the advent of molecular cloning and nucleotide/amino acid sequence analysis, it has become clear that the all SULTs across vertebrate species constitute a gene superfamily [4]. Among vertebrates, six gene families have been classified within the SULT gene superfamily [4,14,15]. Members of the same SULT gene family share at least 45% amino acid sequence identity. Within each SULT gene family, members of each subfamily share greater than 60% identity in amino acid sequence [4]. In humans, eighteen SULT genes, including five pseudogenes, have been identified and classified into five gene families [15]. Of the eighteen human SULT genes, twelve belong to the SULT1 family and comprise five subfamilies: SULT1A, SULT1B, SULT1C, SULT1D and SULT1E. Three belong to the SULT2 gene family with two subfamilies: SULT2A1 and SULT2B1. In contrast to SULT1 and SULT2 families, SULT3A1P, SULT4A1 and SULT6B1 contain only sole members in their respective families. Figure 2 shows a diagram illustrating the possible phylogenetic relationship between the eighteen human SULT genes. It is noted that SULT1D1P, SULT1D2P, SULT1C5P, SULT2A2P, and SULT3A1P are pseudogenes, with no corresponding protein products [15].
Figure 2.

Diagram illustrating the possible phylogenetic relationship between human SULT genes and pseudogenes. Phylogenic tree was prepared with CDS nucleotide sequences of SULT1A1 (NM_001055), SULT1A2 (NM_001054), SULT1A3 (NM_177552), SULT1A4(NM_001017390), SULT1B1 (NM_014465), SULT1C2 (NM_001056), SULT1C3a (NM_001320878), SULT1C4 (NM_006588), SULT1C5P(AK056906), SULT1D1P (NG_002642), SULT1D2P (NC_000003.12), SULT1E1 (NM_005420), SULT2A1 (NM_003167), SULT2A2P (), SULT2B1b (NM_177973), SULT3A1P (NC_000014), SULT4A1 (NM_014351), SULT6A1 (NM_001367551). Hypothetical CDS nucleotide sequences of SULT1D1P were generated from the regions of exons 2 - 8 based on the NG_002642. For SULT1D2P, NC_000003.12, containing exons 2 - 8, was directly used as hypothetical CDS nucleotide sequences of SULT1D2P. For SULT1C5P, exons 2 - 7 were retrieved from AK056906 and NC_000002.12 using BLAST search as hypothetical CDS nucleotide sequences of SULT1C5P. For SULT2A2P, exons 2 - 3 were retrieved from NC_000019.10 using BLAST search as hypothetical CDS nucleotide sequences of SULT2A2P. For SULT3A1P, NC_000014, containing exons 2 - 7, was directly used as hypothetical CDS nucleotide sequences of SULT3A1P. Analysis of aligned sequence data was carried out in MEGA 11 ALPHA.
2.1. Human SULT1 subfamily
Human SULT1A subfamily consists of 4 SULT genes, encoding 3 SULT isoforms: SULT1A1, SULT1A2, and SULT1A3. Sequence analysis revealed that SULT1A3 and SULT1A4 genes encode identical protein products, designated SULT1A3 [16]. Of the three SULT1A isoforms, SULT1A1 is known to play a pivotal role in the sulfation of phenolic xenobiotics, including polyphenols and number of drug compounds (cf. Table 1) [17,18]. p-Nitrophenol, naphthol, acetaminophen, and minoxidil are typical substrates of SULT1A1. SULT1A3 is known to be involved in the sulfation of monoamines and structurally related compounds (Table 1) [17,18]. Dopamine, serotonin, and isoproterenol are typical substrates of SULT1A3. Sulfation of phenolic compounds as mediated by SULT1A members plays an important role in the metabolism, and usually, the inactivation and excretion of the substrate compounds. It is noted, however, that the biological activity of minoxidil is activated upon sulfation [19].
Table 1.
Human cytosolic sulfotransferases (SULTs) with their amino acid sequence length, gene location on chromosomes, substrate specificity and gene cloning timeline.
| SULT1 | No. of AA2 | Gene Location on Chromosome | Prototype Substrate | References |
|---|---|---|---|---|
| 1A1 | 295 | 16p12.1 | p-Nitrophenol | Wilborn et al., 1993; Dooley et al., 1994; |
| 1A2 | 295 | 16p12.1 | p-Nitrophenol | Zhu et al., 1996; Her et al., 1996 |
| 1A3 | 295 | 16p11.2 | Dopamine | Dooley et al., 1994; Aksoy and Weinshilboum, 1995 |
| 1B1 | 296 | 4q13.3 | Iodothyronines | Fujita et al., 1997 |
| 1C2 | 296 | 2q12.3 | p-Nitrophenol | Her et al., 1997; Sakakibara, 1998 |
| 1C3 | 304 | 2q12.3 | Bile acids; iodothyronines | Freimuth et al., 2000; Kurogi et al., 2017 |
| 1C4 | 302 | 2q12.3 | p-Nitrophenol | Freimuth et al., 2000; Sakakibara et al., 1998 |
| 1E1 | 294 | 4q13.1 | 17β-estradiol | Aksoy et al., 1994 |
| 2A1 | 285 | 19q13.3 | DHEA1 | Otterness et al., 1992; Kong et al., 1992 |
| 2B1a | 350 | 19q13.3 | Pregnenolone | Her et al., 1998 |
| 2B1b | 365 | 19q13.3 | Cholesterol | Her et al., 1998 |
| 4A1 | 284 | 22q13 | unknown | Walther et al., 1999; Falany et al., 2000 |
| 6B1 | 265 | 2p22.3 | unknown | Freimuth et al., 2004 |
Dehydroepiandrosterone
Human SULT1B1 is the sole member of SULT1B subfamily. The main physiological function of SULT1B1 has been proposed to be in the sulfation of thyroid hormones (Table 1) [20,21]. It is noted, however, that dopa and tyrosine have also been shown to be substrates for rat SULT1B isoform [22]. Sulfation of thyroid hormones is proposed to regulate the iodothyronine metabolism through iodothyronine deiodinase, since sulfated metabolites of thyroxine (T4) and triidothyronine (T3) undergo deiodination faster than unsulfated T4 and T3 [23].
Human SULT1C subfamily consists of 4 genes (including 1 pseudogene), encoding 4 SULT isoforms, SULT1C2, SULT1C3a, SULT1C3d, and SULT1C4. SULT1C5P gene is located between SULT1C2 and SULT1C4 and was originally designated SULT1C2P. In order to avoid redundant designation, the SULT1C pseudogene is designated SULT1C5P in this review. SULT1C5P contains some frameshifts, including start codon and the exons 2 - 5 and exons 6 - 7 [15]. SULT1C subfamily members have been proposed to catalyze the sulfation of some procarcinogenic compounds, e.g., N-hydroxy-2-acetylaminofluorene and hydroxybenzopyrene, leading to the activation of their carcinogenic activities [24–26]. Although the physiological substrates of SULT1C enzymes have not been fully elucidated, a number of endogenous and xenobiotic compounds have been reported to be sulfated by SULT1C enzymes (Table 1) [27]. Among these substrate compounds, thyroid hormones have been shown to be substrates for SULT1C2 [28]. SULT1C4 appeared to exhibit the broadest substrate specificity toward xenobiotic compounds including polyphenols and some drug compounds [27, 29–31]. Since the expression of human SULT1C subfamily members has been shown to be more prominent in fetal tissues, it has been suggested that SULT1C enzymes may play a role in the metabolism of xenobiotics and thyroid hormones during fetal development [24,27].
Human SULT1E1 is the sole member of SULT1E subfamily. The main physiological function of SULT1E1 is believed to be in the sulfation of estrogens, including estrone (E1) and 17β-estradiol (E2) [13,32] (Table 1). Xenobiotic estrogen-like compounds such as 17β-ethynylestradiol (EE2) and daidzein have also been shown to be substrates for SULT1E1 [32–34]. Since estrogen sulfates are the major circulatory estrogens and can be hydrolyzed by steroid sulfatase, sulfation of estrogens has been proposed to play a pivotal role in the steroid homeostasis, in conjunction with the sulfatase pathway [32,35]. Therefore, the balance of sulfation and desulfation may be critical in the adjustment of estrogenic activity in vivo.
2.2. Human SULT2 family
Human SULT2 family comprises three subfamilies, SULT2A and SULT2B. SULT2A1 is the sole member of the SULT2A subfamily. SULT2A1 and SULT2A2P are paralogous genes. Of the two, SULT2A2P is a pseudogene which contains some frameshifts, including start codon and reversed order of exons 4 - 6 [15]. SULT2A1 is known to play a fundamental role in the metabolism of androgens and bile acids (Table 1) [36,37]. Steroid drugs such as budesonide and tibolone have also been shown to be substrates for SULT2A1 [38,39]. For endogenous androgens, sulfation has been shown to act as a reversible metabolic pathway, as in the case of estrogen metabolism [35]. Therefore, sulfation of androgens plays an important role in the deactivation and storage of androgens. It is worthwhile noting that estrogens are generated from androgens under the action of aromatase (CYP19A1), implying that sulfation of androgens by SULT2A1 may also affect the homeostasis of estrogens.
While human SULT2B1 is the sole member of SULT2B subfamily, two N-terminal splice variants, SULT2B1a and SULT2B1b, have been reported [40,41]. SULT2B1a has been shown to play an important role in the sulfation of pregnenolone, while SULT2B1b is responsible for the sulfation of cholesterol. Similar to other steroid sulfates, pregnenolone sulfate and cholesterol sulfate are also hydrolyzed by steroid sulfatase, implying that the SULT2B1 isoforms play an important role in the homeostasis of steroid/sterol, in conjunction with steroid sulfatase [35]. Pregnenolone sulfate has been reported to act both as a neuromodulator and as a neurotransmitter via N-methyl-D-aspartate (NMDA) receptor and γ-aminobutyric acid type A (GABAA) receptors [42]. Sulfation of pregnenolone may be a key factor in the learning and memory, as well as other synaptic functions. Cholesterol sulfate has been reported to act as a second messenger that functions in keratinocyte differentiation [43]. Besides, recent studies using the mouse model have shown that cholesterol sulfate may exert immune regulatory functions [44,45]. Sulfation of cholesterol has been suggested to be an important reaction that converts cholesterol to a signaling molecule. Therefore, the two SULT2B1 enzymes, SULT2B1a and SULT2B1b, appear to play multiple physiological roles, not just the deactivation and metabolism of steroids/sterols.
2.3. Human SULT4 family
Human SULT4A1 is the sole member of the SULT4 family. Although a great many studies have been performed to investigate the enzymatic characteristics of SULT4A1, no substantial sulfating activity of SULT4A1 has been detected [27,46,47]. Therefore, SULT4A1 is now considered likely an orphan SULT, although it has been shown to be highly expressed in nervous system. In recent years, genetic analyses have suggested that mutation in the untranslated regions of SULT4A1 gene may contribute to the etiology of schizophrenia [48,49]. Nevertheless, no mutations (synonymous or nonsynonymous) in coding regions of the SULT4A1 gene have been found in schizophrenia patients [50]. More recently, SULT4A1 knockout mouse or zebrafish models, have been developed to investigate the physiological relevance of SULT4A1 [51,52]. These animal studies have suggested that SULT4A1 may play a role in physical activity and behavior.
2.4. Human SULT6 family
Human SULT6B1, the sole member of the SULT6 family, is likely an orphan SULT. No enzymatic activity of human SULT6B1 has been reported. In contrast, zebrafish, chicken, and mouse SULT6 enzymes have been shown to exhibit sulfating activities toward a variety of substrate compounds [53–55]. Zebrafish SULT6 showed strong sulfating activities toward simple phenols and polyphenols, while chicken SULT6 exhibited sulfating activities toward E2 and corticosterone [53,54]. Mouse SULT6B1, on the other hand, displayed weak sulfating activities toward thyroxine and bithionol [55]. It thus appears that physiological functions of SULT6 may not be conserved among vertebrates and further studies are needed in order to elucidate any possible functions of human SULT6B1.
Table 1 shows the thirteen human SULTs with their polypeptide length, the chromosomal location of their coding genes, and their prototype substrates.
3. SULT genetic polymorphisms and functional implications
Like many other genes, single nucleotide polymorphisms (SNPs) occur for genes encoding the SULTs [8–12], and, significantly, SULT SNPs have been shown to be ethnically distributed [56]. SULT SNPs were first reported for the gene encoding human SULT2A1 (dehydroepiandrosterone sulfotransferase) [8], and has since been demonstrated for the genes encoding SULT1A1 [8], SULT1A2 [9], SULT1A3 [10], SULT1C2 [11], and SULT1E1 [12]. SNPs among the SULTs may lead to individual differences in the metabolism of both endogenous compounds and xenobiotics, including drugs, through sulfation [9,11,56,57]. Such differences may correlate with the occurrence of certain pathophysiological conditions. For example, molecular epidemiology studies have demonstrated the correlations between certain SULT1A1 SNPs and risks for cancers [58]. In view of the involvement of SULT-mediated sulfation in the metabolism of certain drugs, it is possible that SULT SNPs may also influence the metabolism of drugs in individuals with different SULT genotypes [58]. To understand better the correlations between SULT SNPs and particular disease states/risks or differential metabolism and, therefore, efficacy of drugs in different individuals, it is important to clarify their functional impact on the resulting protein products, i.e., SULT allozymes. To date, a number of studies have been performed to investigate the differential sulfating activity of different SULTs. The results obtained from these studies, as summarized in Table 2, are described in detail below. Table 3 summarizes the clinical relevance of human SULT gene polymorphisms and chromosomal mutations in respective SULT genes.
Table 2.
Summary of differential sulfating activities of human SULT allozymes toward endogenous hormones and drugs.
| SULT | Allozymes | Effects on the activities with hormones and drugs | References |
|---|---|---|---|
| 1A1 | Arg37Gln | acetaminophen (↓)*, O-desmethylnaproxen (↓)2 | 78 |
| Pro47Ser | acetaminophen (↓), O-desmethylnaproxen (↓) | 78 | |
| Met77Ile | acetaminophen (↑)** | 78 | |
| His149Tyr | acetaminophen (↓), O-desmethylnaproxen (↓) | 78 | |
| Tyr169Asp | acetaminophen (↓), O-desmethylnaproxen (↓) | 78 | |
| Arg213His | thyroxine (↓), acetaminophen (↓), O-desmethylnaproxen (↓) | 71, 78 | |
| Thr227Pro | acetaminophen (↓), tapentadol (↓) | 78 | |
| Val243Asp | acetaminophen (↓), O-desmethylnaproxen (↓) | 78 | |
| Fhe247Leu | acetaminophen (↑), O-desmethylnaproxen (↑) | 78 | |
| 1A3 | Ser8Pro | O-desmethyltramadol (↓), salbutamol (↓) | 128, 129 |
| Arg9Cys | norepinephrine (↓), serotonine (↓), acetaminophen (↓), morphine (↓), O-desmethyltramadol (↓) | 127, 128 | |
| Pro101Leu | ritodorine (↓), serotonine (↓) | 57, 127 | |
| Pro101His | ritodorine (↓) | 57 | |
| Arg144Cys | ritodorine (↓), serotonine (↓), acetaminophen (↓), phenylephrine (↓), | 57, 128, 129 | |
| Lys234Asn | ritodorine (↓) | 57 | |
| Asn235Thr | dopamine (↓), epinephrine (↓), norepinephrine (↓), acetaminophen (↓), morphine (↓), tapentadol (↓), O-desmethyltramadol (↓), phenylephrine (↓), salbutamol (↓) | 127, 128, 129 | |
| 1E1 | Asp22Tyr | 17β-estradiol (↓) | 12 |
| Ala34Val | 17β-estradiol (↓) | 12 | |
| Ala43Asp | estrone (↑), estriol (↑), 17β-estradiol (↓) 4-HO-tamoxifen (↓), diethylstilbestrol (↓) | 156, 157 | |
| Arg186Leu | 17β-estradiol (↓), 4-HO-tamoxifen (↓) | 156 | |
| Pro214Thr | 17β-estradiol (↓), diethylstilbestrol (↓) | 156 | |
| Asp220Val | 17β-estradiol (↓), 4-HO-tamoxifen (↓), diethylstilbestrol (↓) | 156 | |
| 2A1 | |||
| Lys44Glu | 4-androstene-3, 17-dion (↓), progesterone (↓) tibolone (↓) | 172 | |
| Ala63Pro | DHEA (↓) | 164 | |
| Pro76Thr | DHEA (↓), pregnenolone (↓), 4-androstene-3, 17-dion (↓), progesterone (↓), tibolone (↓), budesonide (↓) | 171, 172 | |
| Glu147Lys | DHEA (↓), progesterone (↓), tibolone (↓) budesonide (↓) | 171, 172 | |
| Glu148Lys | DHEA (↓), pregnenolone (↓), 4-androstene-3, 17-dion (↓), progesterone (↓), tibolone (↓) budesonide (↓) | 171, 172 | |
| Leu159Val | DHEA (↑) | 170 | |
| Lys227Glu | DHEA (↓), tibolone (↓) | 164, 170 | |
| Leu246Pro | DHEA (↓), pregnenolone (↓), 4-androstene-3, 17-dion (↓), progesterone (↓), tibolone (↓), budesonide (↓) | 171, 172 | |
| Phe258Leu | DHEA (↓), pregnenolone (↓), 4-androstene-3, 17-dion (↓), progesterone (↓), tibolone (↓), budesonide (↓) | 171, 172 | |
| Gln262Glu | DHEA (↓) | 171 | |
| 2B1 | Pro69Ala | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 |
| Gly72Val | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 | |
| Thr73Met | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 | |
| Arg147His | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 | |
| Asp191Asn | DHEA (↓), cholesterol (↓) | 193, 194, 195 | |
| Arg230His | DHEA (↓), cholesterol (↓) | 193, 194, 195 | |
| Ser244Thr | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 | |
| Arg274Gln | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 | |
| Gly276Val | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 | |
| Pro345Leu | DHEA (↓), pregnenolone (↓), cholesterol (↓) | 194, 195 |
(↓) refers to decreased activity toward the corresponding compounds.
(↑) refers to increased activity toward the corresponding compounds.
Table 3.
Summary of potential diseases related to human SULT genetic polymorphisms.
| SULT | Polymorphism | Related diseases | References |
|---|---|---|---|
| 1A1 | Arg213His | brain tumor | 69 |
| breast cancer | 82, 84 | ||
| esophageal cancer | 83 | ||
| gastric cancer | 85 | ||
| lung cancer | 86, 87, 88 | ||
| colorectal cancer (reduced risk) | 82 | ||
| bladder cancer (reduced risk) | 94 | ||
| endometrial cancer | 148, 149 | ||
| Met223Val | endometrial cancer | 149 | |
| 3’-UTR variant (rs6839, rs1042157) | endometrial cancer | 148 | |
| 1A3 | low copy number | alzheimer’s disease | 120 |
| chromosomal mutation in SULT1A3/SULT1A4 gene area (16p11.2)* | autism | 115–119, 121, 122 | |
| obesity | 123 | ||
| schizophrenia | 124 | ||
| attention deficit hyperactivity disorder | 125 | ||
| 1C4 | Asp5Glu | higher post-treatment relapse rate in acute myeloblastic leukemia | 133 |
| 1E1 | His224Gln | breast cancer | 152 |
| 5’-UTR variant (rs3736599) | endometrial cancer | 148, 149 | |
| ischemic stroke in young adults | 150 | ||
| intron variant (rs3775778) | breast cancer | 103 | |
| reduced bone mineral density | 151 | ||
| intron variant (rs3775775) | breast cancer | 103 | |
| intron variant (rs1238574) | poor survival rate of colorectal cancer | 153 | |
| intron variant (rs3822172) | poor survival rate of colorectal cancer | 153 | |
| 2A1 | intron variant (rs182420) | prostate cancer, polycystic ovary syndrome | 168, 169 |
| intron variant (rs2547238) | prostate cancer | 168 | |
| 2B1 | Arg274Gln | autosomal recessive ichthyosis | 189 |
| Arg100Trp | congenital ichthyosiform erythroderma | 190 | |
| Glu78Lys | congenital ichthyosiform erythroderma | 190 | |
| intron variant (rs4149455) | esophageal squamous cell carcinoma | 191 | |
| intron variant (rs3760806) | colorectal cancer | 153 | |
| 4A1 | 5’-UTR variant (D22s1749e) | schizophrenia | 48 |
| intron variant (rs138060) | schizophrenia | 49 | |
| intron variant (rs138097) | schizophrenia | 49 |
These mutations (deletion and duplication) occur in the 16p11.2 at which SULT1A3/SULT1A4 genes are located in. No potential SULT allozymes have been identified in these studies.
SULT1A1 SNPs and SULT1A1 allozymes
As noted above, SULT1A1 is one of the major SULTs involved in xenobiotic metabolism and detoxification [59, 60]. Pertaining to this important function is the ubiquitous expression of SULT1A1 in human tissues/organs [59, 60]. It has been reported that SULT1A1 exhibits the highest hepatic expression level among all SULT1 enzymes. SULT1A1 is also expressed, albeit at lower levels, in almost every extrahepatic organ/tissue, including brain, breast, gastrointestinal tract (GIT), kidney, platelets, and skin [61–63]. Furthermore, from a developmental biology perspective, SULT1A1 has been reported to be widely expressed at fetal stages [64–66]. The extensive expression during fetal development implies an essential role of SULT1A1 in the chemical defense during fetal development, through neonatal/child development, onto adulthood [66].
SULT1A1 gene has long been recognized to have a polymorphic nature [8, 9, 67, 68]. Earlier studies revealed that SULT1A1 alleles encode allozymes with variations in not only the level of activity but also thermal stability [8, 60, 69]. Functional genomic studies have revealed that changes in nucleotide sequence could be translated into amino acid replacements with variations in enzyme function in the SULT gene family [8, 9, 70]. Distinct SULT1A1 allozymes were shown to be expressed as a result of single nucleotide polymorphisms (SNPs) in the SULT1A1 gene [71]. Studies have demonstrated an alteration in the processing of xenobiotics, including therapeutic drugs, due to SULT1A1 polymorphisms that influenced the SULT1A1 enzyme activity [72, 73].
Earlier studies have revealed a number of non-synonymous coding SNPs (cSNPs) of the SULT1A1 gene, including SULT1A1-R213H, SULT1A1*3 (M223V), SULT1A1*4 (R37Q), and SULT1A1*5 (F247L) [70, 74, 75]. Among these allelic variants, SULT1A1-R213H, which occurs in exon seven of the SULT1A1 gene, appeared to be the most prevalent SULT1A1 genetic polymorphism [60, 76]. The coded SULT1A1-R213H allozyme appeared to be less thermostable and displayed decreased binding affinity toward different substrate compounds, including some pro-mutagens, compared to the wild-type SULT1A1 [77]. A more recent study investigated the effect of nine missense SULT1A1 cSNPs, SULT1A1-R37Q, SULT1A1-P47S, SULT1A1-M77I, SULT1A1-H149Y, SULT1A1-Y169D, SULT1A1-R213H, SULT1A1-T227P, SULT1A1-V243D, and SULT1A1-F247L, on the sulfation of acetaminophen, O-desmethylnaproxen, and tapentadol [78]. Similar patterns of differential sulfating activities were found for the nine SULT1A1 allozymes towards all three drug compounds. Of the nine allozymes, SULT1A1-M77I and SULT1A1-F247L exhibited sulfating activities that were higher/comparable to that of the wild-type SULT1A1. The other seven SULT1A1 allozymes displayed lower specific activities than that of the wild-type enzyme, with SULT1A1-T227P showing the lowest activity [78]. It is noted that SULT1A1-R37Q and SULT1A1-R213H SNPs had also been identified in earlier polymorphic studies, which showed that the platelet samples corresponding to the SULT1A1-R213H genotype presented with a much lower sulfating activity than the platelet samples corresponding to wild-type SULT1A1 genotype [8, 9, 71].
Some human population studies have revealed that the enzymatic activity of SULT1A1 in platelet differs by gender [79, 80]. Distinct allelic frequencies of some SULT1A1 genotypes have been found in different ethnic groups [76, 81]. The allele occurrence of SULT1A1-R213H SNP in the population appeared the highest among a number of SULT1A1 genotypes studied. For instance, it occurred at a frequency of about 0.30 in the Caucasians and about 0.17 in the Japanese population [77]. Additionally, SULT1A1-R213H SNP was found to be common among African American subjects (about 29%) [76]. Chinese populace presented with allelic frequencies of about 0.08 and 0.006 for SULT1A1-R213H, SULT1A1*3 (M223V), respectively. Both of these two alleles presented with higher frequencies in African American subjects, 0.294 for SULT1A1-R213H and 0.229 for SULT1A1*3 (M223V) [74, 76]. These differences in SULT1A1 allele frequencies might contribute to the observed variability in drug disposition and metabolism among those different ethnic groups [72]. Due to the high frequency of SULT1A1-R213H SNP in the population and its potential effect on the bioconversion of different drugs and carcinogens, this variant has received much attention in genotyping studies [82].
A good number of molecular epidemiological studies has been carried out aiming to link SULT1A1-R213H polymorphism to disease susceptibility. An initial report suggested that the SULT1A1-R213H SNP is an independent risk factor for esophageal cancer in men [83]. Other studies suggested a positive correlation between SULT1A1-R213H allele and the risk of breast cancer [84], gastric cancer [85], as well as brain tumors [69]. Additionally, SULT1A1-R213H polymorphism has been shown to be a risk factor for lung cancer in different ethnicities/populations, including Caucasian [86], Turkish [87], and Bangladesh [88]. The exact mechanism underlying the risk for different cancers due to certain SULT1A1 genotypes remains unclear [8, 9, 83, 89]. Contradictory results, however, also exist that indicated a lack of association between SULT1A1 genotype and the risk for colorectal cancer [90] or prostate cancer [91] in Caucasian populations, as well as lung cancer [92] and urothelial epithelial cancers in a Japanese populace [93]. Moreover, in some other studies, SULT1A1-R213H SNP has been shown to be associated with a reduced incidence of bladder and colorectal cancers [82, 94]. It has been proposed that individual susceptibility to certain promutagenic and procarcinogenic compounds may ultimately be influenced by SULT1A1 genetic polymorphisms [95].
SULT1A3/SULT1A3 SNPs and SULT1A3 allozymes
As noted above, SULT1A3 and SULT1A4 genes encode identical protein products, collectively designated SULT1A3 [16]. SULT1A3 is the major enzyme responsible for the sulfation of the catecholamines such as dopamine, epinephrine, and norepinephrine, as well as a number of drug and dietary compounds [5]. Unlike other SULTs, SULT1A3 is expressed only in humans and closely related primates [113]. In adult humans, SULT1A3 is expressed predominantly in the upper gastrointestinal tract, constituting nearly one-third of the total amount of SULTs present in the small intestine. In contrast, it has a very low level of expression in the liver [62,114]. Substantial expression of SULT1A3 is also found in the brain, lung, and platelets [56]. From the developmental standpoint, hepatic expression of SULT1A3 was found to be high at early stages of fetal development, but decreased substantially in late fetal/early neonatal liver, and essentially absent in adult liver. In the lung, significant SULT1A3 activity was observed in the fetus, whereas neonatal levels were considerably lower. In brain, low and widespread activity was recognized for SULT1A3 in different regions other than choroid plexus [64].
Earlier studies revealed a link between the SULT1A locus and the predisposition to autism spectrum disorders [115,116,117]. More recently, a correlation between autism and mutations in chromosome 16 at position 16p11.2, where the SULT1A3 and SULT1A4 genes are located, was reported [118,119]. Furthermore, the sequences surrounding the SULT1A3 and SULT1A4 genes were found to be associated with several pathological disorders [120]. Individuals with microdeletions in these sequences were found to manifest high frequency of cognitive, developmental, and speech delay as well as behavior abnormalities [121] and autism spectrum disorders [117,122]. Moreover, duplications in the sequences surrounding the SULT1A3 and SULT1A4 genes have been found to be associated with obesity [123], schizophrenia [124] and attention deficit hyperactivity disorder [125]. Genetic polymorphisms of both SULT1A3 and SULT1A4 have been studied. Initial studies revealed ethnic-specific inherited differences in the capacity of catecholamine sulfation [10,126]. In an earlier study, eleven SULT1A3/SULT1A4 SNPs were identified and two of them, designated C302T, and C302A, were found to be cSNPs [16]. Corresponding SULT1A3 allozymes, expressed in COS-1 cells displayed lower enzymatic activity in comparison with the wild type enzyme, while without significant alterations in substrate kinetics [16]. In another study using DNA samples from 60 African-American (AA) and 60 Caucasian American (CA) subjects, eight single nucleotide polymorphisms (SNPs) were observed in AA and five in CA subjects, and a single amino acid change, Lys234Asn, has been shown to lead to accelerated SULT1A3 degradation [10]. In another study, four SULT1A3 allozymes (Lys234Asn, Pro101Leu, Pro101His and Arg144Cys) were found to display lower sulfating activity, compared with the wild-type enzyme, with ritodrine as a substrate [57].
More recently, the effects of SULT1A3/SULT1A4 cSNPs on the sulfating activity of SULT1A3 allozymes were systematically investigated [127,128,129]. Twelve known SULT1A3 allozymes (SULT1A3-T7P, SULT1A3-S8P, SULT1A3-R9C, SULT1A3-P10L, SULT1A3-V15M, SULT1A3-V18F, SULT1A3-P101L, SULT1A3-P101H, SULT1A3-R144C, SULT1A3-K234N, SULT1A3-N235T and SULT1A3-S290T), bacterially expressed and purified, showed differential sulfating activity toward catecholamines and serotonin as substrates. Interestingly, the variations in the sulfating activity of SULT1A3 allozymes toward dopamine were markedly smaller than those toward epinephrine, norepinephrine, and serotonin. Kinetic studies demonstrated differences in both substrate affinity and catalytic efficiency of the tested SULT1A3 allozymes. Of these allozymes, SULT1A3-N235T displayed the lowest sulfating activity and catalytic efficiency [127]. Several drugs including acetaminophen, morphine, tapentadol, O-desmethyltramadol, phenylephrine and salbutamol, were tested as substrates for these SULT1A3 allozymes [128,129]. The tested allozymes exhibited a similar pattern of differential sulfating activity toward these drugs, and kinetic studies showed further significant variations in their substrate-binding affinity and catalytic efficiency. The SULT1A3-N235T allozyme also exhibited the lowest sulfating activity toward the tested drugs. These findings may imply the differential pharmacokinetics and, consequently, efficacy and associated toxicity of these drugs when administered to individuals with distinct SULT1A3 and SULT1A4 genotypes [128,129].
SULT1B1 SNPs and SULT1B1 allozymes
Only a limited amount of information is available concerning the genetic polymorphism of SULT1B1. Studies have shown that certain SULT1B1 SNPs may influence the activity of thyroid hormones and the mutagenicity of polycyclic aromatic hydrocarbons [130, 131]. Three nonsynonymous cSNPs (Leu145Val, Glu186Gly, and Glu204Asp) have been identified in human populations [132–134]. Among them, Leu145Val, with a 17% frequency in African American, had been shown to display a higher affinity toward 1-hydroxypyrenes than the wild-type enzyme [134]. No significant correlation between the SULT1B1variants and pathologies (colorectal or prostate cancers) associated with the subjects studied, however, was observed.
SULT1C2/SULT1C4 SNPs and SULT1C2/SULT1C4 allozymes
SNPs of SULT1C2 and SULT1C4 genes have been reported in human population [11,133]. For SULT1C2, 4 nonsynonymous cSNPs (encoding Asp60Ala, Arg73Gln, Ser111Phe, and Ser193Ala) have been identified in Caucasians [11]. Among the corresponding SULT1C2 allozymes, Asp60Ala and Arg73Gln showed reduced sulfating activity (15% of wild-type) toward p-nitrophenol and Ser111Phe showed no detectable activity [11]. Arg73Gln showed a much higher Km with PAPS, while Asp60Ala showed no change in Km with PAPS. These observations suggested that substitution of Arg73 with Gln may affect the interaction with PAPS. Since Ser111 is located in catalytic region that includes also the catalytic residue His109, substitution of Ser111 with Ala may alter the interaction of the enzyme with the substrate, p-nitrophenol. On the other hand, Ser193Ala showed no change in the catalytic properties. For SULT1C4, a nonsynonymous cSNP (encoding Asp5Glu) has been reported [133]. Although no information on the catalytic properties of the corresponding allozyme is available, Asp5Glu has been reported to correlate with a higher post-treatment relapse rate in acute myeloblastic leukemia [135]. Therefore, the clinical significance of the SNP will need to be clarified. It should be pointed out that polymorphisms of SULT1C subfamily may be particularly important in the fetal physiology vs. that of the adult in view of their more prominent expression at fetal stages [24,27].
SULT1E1 SNPs and SULT1E1 allozymes
SULT1E1 is known to be the most efficient SULT enzyme in catalyzing the sulfation of endogenous estrogens (E1 and E2), while with a lower efficiency in sulfating other hydroxysteroids (e.g., DHEA, pregnenolone), as well as some xenobiotics, including a number of drug compounds [13,32,39, 136–143]. SULT1E1 has been shown to be expressed in adult human lung, liver, and small intestine [62]. SULT1E1 expression has also been detected in other organs including breast, endometrium, adrenal gland, placenta, jejunum, lung, skin, and testis [32, 144–147], as well as human fetal kidney, liver, lung, thyroid gland, and choroid plexus [66].
Genetic variations of the SULT1E1 gene have been found to be associated with the risk for certain cancers/disease etiologies, and responses to therapies [56,58]. In earlier studies, the SULT1E1-64G/A (rs3736599) genotype was found to be associated with a significant increase in the risk for developing endometrial tissue cancer [148], particularly in women receiving long-term hormone replacement therapy (HRT) [149]. A correlation between SULT1E1-64G/A (rs3736599) and catechol-O-methyl-transferase (COMT Val158Met), resulting in lower serum estradiol levels as well as increased risk for ischemic stroke, has also been reported [150]. Another study revealed that Korean women breast cancer patients with SULT1E1 *959G>A (rs3775778) and SULT1E1 IVS4-1653 T>C (rs3775775) base changes manifested a 4-fold increase in the risk for breast cancer [103]. Furthermore, a correlation between SULT1E1 *959 G>A (rs3775778) and reduced bone mineral density at the distal radius and calcaneus in healthy Korean women has been reported [151]. In another study, missense mutation (His224Gln) in SULT1E1 protein was implicated as a risk factor for breast cancer in Jewish women [152]. A more recent study revealed that two SULT1E1 SNPs (SULT1E1 rs1238574 and SULT1E1 rs3822172) were associated with poor survival rate of colorectal cancer patients [153]. In a clinical study, three variants of the SULT1E1 gene (SULT1E1 -9-899G>A, SULT1E1 -9-682A>G and SULT1E1 -9-469A>G) were found to result in inter-individual variations of plasma concentrations of tamoxifen metabolites in breast cancer patients treated with tamoxifen, indicating contribution of SULT1E1 to tamoxifen metabolism in vivo [154]. Another clinical study showed a significant correlation between genetic variations of SULT1E1 gene (SULT1E1 rs3775777, SULT1E1 rs4149534, SULT1E1 rs10009305, SULT1E1 rs3775770, SULT1E1 rs4149527, and SULT1E1 rs3775768) and the time to treatment failure (TTF) of abiraterone therapy in male patients with metastatic castration refractory prostate cancer [155]. The presence of one or more rare alleles among these six SNPs resulted in shorter TTF compared to those with wild-type alleles. These findings suggest that genetic variations of the SULT1E gene could potentially reduce the interval between abiraterone administration and its discontinuation due to different reasons including patient decision, cancer progression, adverse effects, and patient death.
Several studies on the effects of SULT1E1 cSNPs on the functional activity of SULT1E1 protein product have been reported [12,156,157]. In an earlier functional genomic study, COS-1 cells transfected with constructs containing two nonsynonymous SULT1E1 cSNPs (Ala32Val and Asp22Tyr) were found to exhibit reductions in both SULT1E1-sulfating activity toward E2 (40% and 90%, respectively) as well as enzyme expression level [12]. In a more recent study, five missense cSNPs of the SULT1E1 gene (SULT1E1-A43D, SULT1E1-A131P, SULT1E1-R186L, SULT1E1-P214T, and SULT1E1-D220V) were bacterially expressed, purified, and characterized in regard to their sulfating activity toward endogenous estrogens (E1, E2, estriol (E3)), EE2, 4-hydroxytamoxifen (4-OHT), and diethylstilbestrol (DES) [156,157]. Compared with the wild-type enzyme, all five SULT1E1 allozymes showed significant variations in the kinetics parameters (Vmax, Km, and Vmax/Km) toward E2, 4-OHT and DES, reflecting the effect of these cSNPs on the sulfoconjugation of E2, 4-OHT and DES by SULT1E1 allozymes [156]. With respect to the remaining substrates (E1, E3, and EE2), the sulfating activities of the five SULT1E1 allozymes were all significantly lower in comparison to the wild-type SULT1E1. The inhibitory effects of triclosan on E2 sulfation by these SULT1E1 allozymes has also been evaluated [157]. In the presence of triclosan (150 µM), SULT1E1-A43D, SULT1E1-A131P, SULT1E1-R186L, and SULT1E1-D220V displayed 80%, 9%, 22%, and 70% decrease in E2-sulfating activities, respectively, compared to the wild-type SULT1E1. These results demonstrated the significant inhibitory effect of triclosan on E2 sulfation by SULT1E1 allozymes, indicating that exposure to triclosan may impact the endogenous estrogens hemostasis as well as the bioavailability of the compounds that are metabolized by SULT1E1. These results provide support for the notion that individuals with different SULT1E1 genotypes may differ in biotransformation capacity in sulfating endogenous and exogenous estrogenic compounds as well as xenobiotics, suggesting inter-individual variations in susceptibility to certain diseases and responses to relevant therapies.
SULT2A1 SNPs and SULT2A1 allozymes
As noted above, SULT2A1 is known to display a strong activity toward DHEA and other hydroxysteroids, as well as a number of xenobiotic compounds including drugs [158–160]. Studies have demonstrated the expression of SULT2A1 at high levels in liver and intestine, as well as adrenal glands [161], and at low levels in the kidney and lung [161]. Being the primary enzyme responsible for the sulfation of DHEA, SULT2A1 plays an important role in the homeostasis of DHEA [162]. Effects of the variations of the SULT2A1 gene on the sulfating activity of SULT2A1 or DHEA homeostasis have been studied. In an earlier study, a 4.6-fold variation in SULT2A1 enzymatic activity level was detected among 94 human hepatic tissue samples analyzed [163]. In a later study sequencing the SULT2A1 gene in DNA samples from African-American and Caucasian-American individuals, a total of 15 SNPs were identified [164]. Three of these SULT2A1 SNPs were non-synonymous cSNPs, with corresponding amino acid changes: Ala63Pro, Lys227Glu and Ala261Thr [164]. Intriguingly, these SULT2A1 cSNPs were only detected in African-American individuals. The expression of SULT2A1 allozymes in COS-1 cells resulted in enzymes with varying levels of DHEA-sulfating activity when compared to wild-type SULT2A1 [164]. Another study conducted to analyze the SULT2A1 gene in DNA samples from normal African-American men revealed the presence of two cSNPs, Ala63Pro and Ala261Thr, similar to those reported earlier [165]. Interestingly, a significant increase in the DHEA:DHEA-sulfate ratio was observed in individuals with a heterozygous G187C/G781A genotype that codes for the amino acid change Ala63Pro [165]. In a genome-wide association study of 14,846 individuals, a SULT2A1 SNP (rs2637125) was identified as one of the eight common genetic variants linked to variations in serum DHEA-S levels [166]. In another study performed to investigate the association of SULT2A1 SNPs with plasma DHEA-S concentration, it was found that both SULT2A1 SNPs rs2637125 and rs182420 were associated with decreased levels of DHEA-S in 12-16 year old children [167]. In a more recent study, it was demonstrated that SULT2A1 SNP rs182420 was associated with prostate cancer risk in Caucasians, while SULT2A1 SNP rs2547238 was associated with prostate cancer risk in African-Americans [168]. Another epidemiological study revealed that SULT2A1 SNP rs182420 was associated with a variation in DHEA-S levels in women with polycystic ovary syndrome [169]. Collectively, these studies indicated that SULT2A1 SNPs have implications in the homeostasis of DHEA/DHEA-S as well as DHEA/sex hormone-associated diseases, including prostate cancer and polycystic ovary syndrome. In a study investigating the effects of SULT2A1 SNPs, nine SULT2A1 allozymes, in comparison with the wild-type enzyme, were shown to display differential sulfating activities toward tibolone [170]. More recently, the functional consequences of a set of seven SULT2A1 non-synonymous cSNPs were investigated [171,172]. The sulfating activities of these seven SULT2A1 allozymes were characterized with three kinds of substrates, those that carry hydroxyl group in their chemical structures, including DHEA, pregnenolone, tibolone, and budesonide, those that carry amine groups in their chemical structures, including ciprofloxacin and desipramine, and Δ4-3-ketosteroids, such as 4-androstene-3, 17-dione and progesterone, that do not carry hydroxyl or amine groups in their chemical structures. Results indicated that the seven SULT2A1 allozymes analyzed displayed differential sulfating activities toward all three types of substrates, compared with wild-type SULT2A1, reaffirming the impact of SULT2A1 SNPs on the functional activity of SULT2A1 protein products [172].
SULT2B1 SNPs and SULT2B1b allozymes
As noted above, two N-terminal variants, SULT2B1a and SULT2B1b, have been reported to be derived from the alternative splicing of the primary SULT2B1 transcript [40,41]. Of the two, SULT2B1b has been reported to display a strong activity toward cholesterol, and thus dubbed a cholesterol sulfotransferase [173]. Additionally, SULT2B1b has also been shown to display activity toward hydroxysteroids, particularly 3β-hydroxysteroids including dehydroepiandrosterone (DHEA), pregnenolone, and androstenediol [174, 43]. Moreover, SULT2B1b can also sulfate oxygenated derivatives of cholesterol, collectively called “oxysterols”, such as 7-ketocholesterol (7KC), 5α,6α-epoxycholesterol (5α,6α-EC), 5β,6β-epoxycholesterol (5β,6β -EC), 25-hydroxycholesterol (25HC), and 24-hydroxycholesterol (24HC) [175–177]. Besides the endogenous substrates, SUlT2B1b has been reported to display sulfating activity toward xenobiotics, including a number of drug compounds, e.g., 3-OH-tibolone, raloxifene, bisphenol A, 4-n-octylphenol, 4-n-nonylphenol, diethylstilbestrol, 17-α-ethynylestradiol, and p-nitrophenol [174, 140, 39]. On the other hand, several antiandrogens including galeterone, abiraterone, cyproterone, and danazol have been reported to inhibit DHEA sulfation by SULT2B1b [136].
The ability of SULT2B1b to sulfate some important steroids and hydroxysteroids and its expression in many tissues/organs (including prostate, placenta, breast, endometrium, uterus, ovary, small intestine, colon, lung, platelet, brain, and skin), suggest the critical involvement of SULT2B1b in some physiological and pathological conditions in the human body [178–183]. Indeed, reduction or elevation in SULT2B1b activity or expression level due to genetic variations have been linked to disease states like autosomal recessive ichthyosis and several types of cancers including prostate cancer, esophageal squamous cell carcinoma, hepatocellular carcinoma, gastric cancer, endometrial cancers, and colorectal cancer [184–192]. Several studies have been conducted to study the effect of SULT2B1 genetic polymorphisms on the sulfating activity of the expressed enzyme [193–195]. The first such study revealed that of the samples taken from 120 African American and Caucasian subjects, four non-synonymous cSNPs (SULT2B1b-Leu51Ser, SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, and SULT2B1b-Peo345Leu) were detected [193]. Corresponding SULT2B1b allozymes were transiently expressed in COS-1 cells and the sulfating activity was characterized using DHEA as a substrate [193]. Compared to wild-type SULT2B1b, the level of the tested allozymes in COS-1 cells varied from 79% to 112%, while their sulfating activity varied from 76% to 98% [193]. In a more recent study, the impact of ten SULT2B1 non-synonymous cSNPs on the functional activity of corresponding SULT2B1b allozymes was investigated using cholesterol as a substrate [194]. Amino acid changes in three of the ten allozymes, SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val, were found to result in a complete loss of the enzyme activity [194]. The other seven allozymes (SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, SULT2B1b-Ser244Thr, SULT2B1b-Arg274Gln, and SULT2B1b-Pro345Leu) all resulted in a dramatic decrease in the sulfating activity, substrate affinity, and catalytic efficiency [194]. Similar findings were later reported for these SUL2B1b allozymes using DHEA or pregnenolone as substrate [195]. Three allozymes (SULT2B1b-Gly72Val, SULT2B1b-Arg147His, and SULT2B1b-Gly276Val) exhibited no detectable sulfating activity toward DHEA or pregnenolone, whereas the other seven (SULT2B1b-Pro69Ala, SULT2B1b-Thr73Met, SULT2B1b-Asp191Asn, SULT2B1b-Arg230His, SULT2B1b-Ser244Thr, SULT2B1b-Arg274Gln, and SULT2B1b-Pro345Leu) displayed differential sulfating activity and affinity toward DHEA or pregnenolone [195]. Interestingly, only SULT2B1b-Pro69Ala and SULT2B1b-Arg274Gln showed a significant decrease in the co-substrate (PAPS) binding when tested using pregnenolone as the substrate [195]. Interestingly, among the ten SULT2B1 genotypes studied, SULT2B1-Arg274Gln heterozygous allele has been implicated in autosomal recessive ichthyosis (ARCI), previously reported to be associated with a reduced skin cholesterol-sulfating activity [189]. In a phenotype-genotype association study, two missense SULT2B1 cSNPs (p. Arg100Trp and p. Glu78Lys) have also been implicated in a specific subtype of ARCI called congenital ichthyosiform erythroderma for which the underlying molecular mechanisms is yet to be clarified [190].
4. Concluding remarks
Despite that the biological sulfation has been known for well over a century, the research on the responsible enzymes had been slow until the 1980s. The past three decades have witnessed significant progress made in the elucidation of the diversity of the SULT enzymes and their enzymatic characteristics, the phylogenetic relationships between the SULTs, the structural biology of the SULTs, the developmental expression of the SULTs, and the mechanisms underlying the regulation of the SULT gene expression, as well as the development of the zebrafish as a model for use in SULT research. While continued efforts need to be made in all these aspects of the SULT research, additional fronts, particularly the implications of the polymorphisms of the SULT genes and the systems biology regarding the physiological involvement of the SULTs - not only in the detoxification of xenobiotics but also in the homeostasis of key endogenous compounds such as thyroid/steroid hormones and catecholamine neurotransmitter/hormones - will need to be addressed.
5. Expert opinion
By virtue of their involvement in drug metabolism, drug-metabolizing enzymes play a crucial role in influencing the level of drugs in vivo [196]. With the advent of pharmacogenomics, mounting evidence has indicated that genetic variations of the genes coding for drug-metabolizing enzymes may lead to the differential metabolism and thus efficacy, as well as adverse drug reactions, of drugs in individuals with different genotypes. Variations in the genes coding for both Phase I enzymes, such as cytochrome P-450 2C9 [197] and 3A4 [198], and Phase II enzymes, such as COMT [199, 200] and N-acetyltransferase [201, 202], and UDP-glucuronosyltransferases [203, 204], have all been reported to significantly affect the efficacy and adverse effects of a variety of drugs. An important application of such information may lie in helping to formulate personalized drug regimens for individuals with unique drug-metabolizing enzyme genotypes. Compared with other Phase II enzymes, less is known concerning the impact of genetic variations of SULT genes on the metabolism of drugs that are metabolized, in part at least, by the coded SULT enzymes. A better understanding about how SULT SNPs may influence the drug-sulfating activity of SULT allozymes will similarly help understand the differential drug metabolism and aid in formulating personalized drug regimens. It should be reiterated that while nonsynonymous coding SULT SNPs have been shown to affect the drug-sulfating activities of resulting SULT allozymes, there is evidence that the stability and expression level of different SULT allozymes may also vary [8,10, 60,69]. More studies are warranted in order to fully elucidate the impact of SULT SNPs on the metabolism of relevant drugs.
In association with the role of SULT SNPs in influencing the efficacy of drugs are the associated adverse drug reactions. For example, doxorubicin, which has been reported to be metabolized by sulfation [205], is known to cause cardiotoxicity when used in the treatment of hematologic malignancies as well as solid and soft tumors [206,207]. Studies have shown that not all patients who received the same chemotherapeutic regimen developed cardiotoxicity, implying likely an underlying genetic predisposition [208,209]. The enzyme responsible for the sulfation of doxorubicin has been shown to be SULT1C4 [30]. It is an interesting question whether genetic polymorphisms of the SULT1C4 gene may influence the doxorubicin-sulfating activity of SULT1C4 allozymes, thereby influencing the susceptibility to the development of cardiotoxicity in patients with different SULT1C4 genotypes. Another example is the occurrence of idiosyncratic skin rash associated with the use of nevirapine (NVP) in the treatment of human immunodeficiency virus (HIV) infection [210,211]. Sulfation of 12-OH-NVP, a metabolite of NVP, has been proposed to be involved in this NVP-induced adverse drug reaction [212,213]. An interesting question is whether the genetic polymorphisms of the gene encoding SULT1A1, a major 12-OH-NVP-sulfating SULT [214] which is expressed in human skin cells [215,216], may influence the level of 12-OH-NVP generated, thereby dictating the development of skin rash in patients with different SULT1A1 genotypes.
Another area worthy of attention is the SULT-mediated sulfation of 3-nitrotyrosine and 3-chlorotyrosine as pertaining to intracellular oxidative/nitrative stress [217–219]. Studies have shown that 3-nitrotyrosine, generated via de novo nitration of tyrosine or degradation of tyrosine-nitrated proteins, can induce oxidative DNA damage [220] or trigger apoptosis in cultured cells [221], while 3-chlorotyrosine has been reported to increase free radical production and attenuate the intracellular NO synthase enzyme expression [222,223]. Studies have revealed SULT1A3 as the only SULT enzyme capable of sulfating 3-nitrotyrosine and 3-chlorotyrosine [217–219]. In view of the pathogenic effects of these latter compounds, it is an interesting question whether the genetic polymorphisms of the genes coding for SULT1A3, SULT1A3 and SULT1A4, may dictate the differential sulfating activity of coded SULT1A3 allozymes, thereby influencing the capacity in mitigating the adverse effects of 3-nitrotyrosine and 3-chlorotyrosine generated under oxidative/nitrative stress conditions.
Finally, the effects of SULT genetic polymorphisms on the sulfation of endogenous compounds should not be overlooked. Several human SULTs, including SULT1A3, SULT1B1, SULT1E1, SULT2A1, SULT2B1a, and SULT2B1b (cf. Table 1), have been shown to be involved in the metabolism of key endogenous compounds such as catecholamine neurotransmitters and thyroid/steroid hormones. Anomalies in the metabolism of these endogenous compounds in individuals with certain SULT genotypes have been shown to lead to increased risk for cancers and other pathological conditions as elaborated above.
Article highlights.
Cytosolic sulfotransferases (SULTs)-mediated sulfation is critically involved in the metabolism of key endogenous compounds such as catecholamines and steroid/thyroid hormones, as well as drugs and other xenobiotics.
In humans, eighteen SULT genes, including five pseudogenes, have been identified and classified into five gene families.
Like many other genes, single nucleotide polymorphisms (SNPs) occur for genes encoding the SULTs.
Studies have shown that certain SULT genotypes may predispose risks for diseases.
SULT allozymes, coded by distinct SULT genotypes, have been reported to display differential sulfating activities.
Elucidation of the functional relevance of SULT SNPs may eventually aid in formulating personalized drug regimens and help develop strategies for mitigating anomalies in the metabolism of key endogenous compounds.
Funding
This manuscript received NIH funding (grant number: R03HD071146).
Abbreviations:
- DHEA
dehydroepiandrosterone
- PAPS
3’-phosphoadenosine 5’-phosphosulfate
- SULT
cytosolic sulfotransferase
- SNP
single nucleotide polymorphism
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Bauman E [Ueber sulfosauren im harn,] Ber Dtsch Chem Ges. 1876;54–58. German. [Google Scholar]
- 2.Nelson SD, Gordon WP. Mammalian drug metabolism. J Nat Prod. 1983; 46:71–78. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Zhong D, Blume H. Stereoselective pharmacokinetics of propafenone and its major metabolites in healthy Chinese volunteers. Eur J Pharm Sci. 2000;10:11–16. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. [DOI] [PubMed] [Google Scholar]; * The paper proposed the systematic nomenclature for SULT genes.
- 5.Reiter C, Mwaluko G, Dunnette J, Van Loon J, Weinshilboum R. Thermolabile and thermostable human platelet phenol sulfotransferase. Substrate specificity and physical separation. Naunyn Schmiedebergs Arch Pharmacol. 1983;324(2):140–7. [DOI] [PubMed] [Google Scholar]
- 6.Young WF Jr, Okazaki H, Laws ER Jr, Weinshilboum RM. Human brain phenol sulfotransferase: biochemical properties and regional localization. J Neurochem. 1984;43(3):706–15. [DOI] [PubMed] [Google Scholar]
- 7.Falany CN. Enzymology of human cytosolic sulfotransferases, FASEB J. 1997;11:206–216. [DOI] [PubMed] [Google Scholar]
- 8.Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype, Biochem Biophys Res Commun. 1997;239:298–304. [DOI] [PubMed] [Google Scholar]
- 9.Raftogianis RB, Wood TC, Weinshilboum RM. Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations, Biochem Pharmacol. 1999;58:605–616. [DOI] [PubMed] [Google Scholar]
- 10.Thomae BA, Rifki OF, Theobald MA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catecholamine sulfotransferase (SULT1A3) pharmacogenetics: functional genetic polymorphism. J Neurochem. 2003;87:809–819. [DOI] [PubMed] [Google Scholar]
- 11.Freimuth RR, Eckloff B, Wieben ED, Weinshilboum RM. Human sulfotransferase SULT1C1 pharmacogenetics: gene resequencing and functional genomic studies, Pharmacogenetics. 2001;11:747–756. [DOI] [PubMed] [Google Scholar]
- 12.Adjei AA, Thomae BA, Prondzinski JL, Eckloff BW, Wieben ED, Weinshilboum RM. Human estrogen sulfotransferase (SULT1E1) pharmacogenomics: gene resequencing and functional genomics. Br J Pharmacol. 2003;139:373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aksoy IA, Wood TC, Weinshilboum R. Human liver estrogen sulfotransferase: identification by cDNA cloning and expression. Biochem Biophys Res Commun. 1994;16;200(3):1621–9. [DOI] [PubMed] [Google Scholar]
- 14.Liu TA, Bhuiyan S, Liu MY, et al. Zebrafish as a model for the study of the phase II cytosolic sulfotransferases. Curr Drug Metab. 2010;11:538–546. [DOI] [PubMed] [Google Scholar]
- 15.Freimuth RR, Wiepert M, Chute CG, et al. (2004) Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J. 2004;4:54–65. [DOI] [PubMed] [Google Scholar]; *The study identified novel and pseudogenes for human SULT genes.
- 16.Hildebrandt MA, Salavaggione OE, Martin YN, et al. Human SULT1A3 pharmacogenetics: gene duplication and functional genomic studies. Biochem Biophys Res Commun. 2004;321:870–878. [DOI] [PubMed] [Google Scholar]
- 17.Veronese ME, Burgess W, Zhu X, et al. Functional characterization of two human sulphotransferase cDNAs that encode monoamine- and phenol-sulphating forms of phenol sulphotransferase: substrate kinetics, thermal-stability and inhibitorsensitivity studies. Biochem J. 1994;302:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugahara T, Pai TG, Suiko M, et al. Differential roles of human monoamine (M)-form and simple phenol (P)-form phenol sulfotransferases in drug metabolism. J Biochem. 2003;133:259–262. [DOI] [PubMed] [Google Scholar]
- 19.Buhl AE, Waldon DJ, Baker CA, et al. Minoxidil sulfate is the active metabolite that stimulates hair follicles. J Invest Dermatol. 1990;95:553–557. [DOI] [PubMed] [Google Scholar]
- 20.Fujita KI, Nagata K, Yamazaki T, et al. Enzymatic characterization of human cytosolic sulfotransferases: identification of ST1B2 as a thyroid hormone sulfotransferase. Biol Pharm Bull. 1999;22:446–452. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Falany JL, Falany CN. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol Pharmacol. 1999;53:274–282. [DOI] [PubMed] [Google Scholar]
- 22.Sakakibara Y, Takami Y, Zwieb C, et al. Purification, characterization, and molecular cloning of a novel rat liver Dopa/tyrosine sulfotransferase. J Biol Chem. 1995;270:30470–30478. [DOI] [PubMed] [Google Scholar]
- 23.Wu SY, Green WL, Huang WS, et al. Alternate pathways of thyroid hormone metabolism. Thyroid. 2005;15:943–958. [DOI] [PubMed] [Google Scholar]
- 24.Sakakibara Y, Yanagisawa K, Katafuchi J, et al. Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem. 1998;273:33929–33935. [DOI] [PubMed] [Google Scholar]
- 25.Meinl W, Donath C, Schneider H, et al. SULT1C3, an orphan sequence of the human genome, encodes an enzyme activating various promutagens. Food Chem Toxicol. 2008;46:1249–1256. [DOI] [PubMed] [Google Scholar]
- 26.Meinl W, Ebert B, Glatt H, Lampen A. Sulfotransferase forms expressed in human intestinal Caco-2 and TC7 cells at varying stages of differentiation and role in benzo[a]pyrene metabolism. Drug Metab Dispos. 2008;36:276–283. [DOI] [PubMed] [Google Scholar]
- 27.Allali-Hassani A, Pan PW, Dombrovski L, et al. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Clemens DL, Anderson RJ. Sulfation of iodothyronines by human sulfotransferase 1C1 (SULT1C1)*. Biochem Pharmacol. 2000;60:1713–1716. [DOI] [PubMed] [Google Scholar]
- 29.Hui Y, Luo L, Zhang L, et al. Sulfation of afimoxifene, endoxifen, raloxifene, and fulvestrant by the human cytosolic sulfotransferases (SULTs): A systematic analysis. J Pharmacol Sci. 2015;128:144–149. [DOI] [PubMed] [Google Scholar]
- 30.Luo L, Zhou C, Hui Y, et al. Human cytosolic sulfotransferase SULT1C4 mediates the sulfation of doxorubicin and epirubicin. Drug Metab Pharmacokinet. 2016;31:163–166. [DOI] [PubMed] [Google Scholar]
- 31.Guidry AL, Tibbs ZE, Runge-Morris M, et al. Expression, purification and characterization of human cytosolic sulfotransferase (SULT) 1C4. Horm Mol Biol Clin Investig. 2017;29:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol. 1995;52:529–539. [DOI] [PubMed] [Google Scholar]
- 33.Barbosa ACS, Feng Y, Yu C, et al. Estrogen sulfotransferase in the metabolism of estrogenic drugs and in the pathogenesis of diseases. Expert Opin Drug Metab Toxicol. 2019;15: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano H, Ogura K, Takahashi E, et al. Regioselective monosulfation and disulfation of the phytoestrogens daidzein and genistein by human liver sulfotransferases. Drug Metab Pharmacokinet. 2004;19:216–226. [DOI] [PubMed] [Google Scholar]
- 35.Mueller JW, Gilligan LC, Idkowiak J, et al. The regulation of steroid action by sulfation and desulfation. Endocr Rev. 2015;36:526–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radominska A, Comer KA, Zimniak P, et al. Human liver steroid sulphotransferase sulphates bile acids. Biochem J. 1990;272:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forbes KJ, Hagen M, Glatt H, et al. Human fetal adrenal hydroxysteroid sulphotransferase: cDNA cloning, stable expression in V79 cells and functional characterisation of the expressed enzyme. Mol Cell Endocrinol. 1995;112:53–60. [DOI] [PubMed] [Google Scholar]
- 38.Meloche CA, Sharma V, Swedmark S, et al. Sulfation of budesonide by human cytosolic sulfotransferase, dehydroepiandrosterone-sulfotransferase (DHEA-ST). Drug Metab Dispos. 2002;30:582–585. [DOI] [PubMed] [Google Scholar]
- 39.Falany JL, Macrina N, Falany CN. Sulfation of tibolone and tibolone metabolites by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 2004;88:383–391. [DOI] [PubMed] [Google Scholar]
- 40.Her C, Wood TC, Eichler EE, et al. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53:284–295. [DOI] [PubMed] [Google Scholar]
- 41.Fuda H, Lee YC, Shimizu C, et al. Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. J Biol Chem. 2002;277:36161–36166. [DOI] [PubMed] [Google Scholar]
- 42.Smith CC, Gibbs TT, Farb DH. Pregnenolone sulfate as a modulator of synaptic plasticity. Psychopharmacology. 2014;231:3537–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strott CA, Higashi Y. Cholesterol sulfate in human physiology what’s it all about? J Lipid Res. 2003;44:1268–1278. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, Beck-García K, Zorzin C, et al. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat Immunol. 2016;17:844–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakurai T, Uruno T, Sugiura Y, et al. Cholesterol sulfate is a DOCK2 inhibitor that mediates tissue-specific immune evasion in the eye. Sci Signal. 2018;11:eaao4874. [DOI] [PubMed] [Google Scholar]
- 46.Falany CN, Xie X, Wang J, et al. Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochem J. 2000;346:857–864. [PMC free article] [PubMed] [Google Scholar]
- 47.Sakakibara Y, Suiko M, Pai TG, et al. Highly conserved mouse and human brain sulfotransferases: molecular cloning, expression, and functional characterization. Gene 2002;285:39–47. [DOI] [PubMed] [Google Scholar]
- 48.Brennan MD, Condra J. Transmission disequilibrium suggests a role for the sulfotransferase-4A1 gene in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2005;139:69–72. [DOI] [PubMed] [Google Scholar]
- 49.Meltzer HY, Brennan MD, Woodward ND, et al. Association of Sult4A1 SNPs with psychopathology and cognition in patients with schizophrenia or schizoaffective disorder. Schizophr Res. 2008;106:258–264. [DOI] [PubMed] [Google Scholar]
- 50.Lewis AG, Minchin RF. Lack of exonic sulfotransferase 4A1 mutations in controls and schizophrenia cases. Psychiatr Genet. 2009;19:53–55. [DOI] [PubMed] [Google Scholar]
- 51.Crittenden F, Thomas HR, Parant JM, Falany CN. Activity suppression behavior phenotype in SULT4A1 frameshift mutant zebrafish. Drug Metab Dispos. 2015;43:1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia PL, Hossain MI, Andrabi SA, Falany CN. Generation and characterization of SULT4A1 mutant mouse models. Drug Metab Dispos. 2018;46:41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugahara T, Liu CC, Pai TG, Liu MC. Molecular cloning, expression, and functional characterization of a novel zebrafish cytosolic sulfotransferase. Biochem Biophys Res Commun. 2003;300:725–730. [DOI] [PubMed] [Google Scholar]
- 54.Cao H, Agarwal SK, Burnside J. Cloning and expression of a novel chicken sulfotransferase cDNA regulated by GH. J Endocrinol. 1999;160:491. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi S, Sakakibara Y, Mishiro E, et al. Molecular cloning, expression and characterization of a novel mouse SULT6 cytosolic sulfotransferase. J Biochem. 2009;146:399–405. [DOI] [PubMed] [Google Scholar]
- 56.Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;13:25(11):1673–8. [DOI] [PubMed] [Google Scholar]; **The study demonstrated the ethnic variation of SULT1A1 and SULT2A1 polymorphism.
- 57.Hui Y, Liu MC. Sulfation of ritodrine by the human cytosolic sulfotransferases (SULTs): Effects of SULT1A3 genetic polymorphism, Eur J Pharmacol. 2015;761:125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daniels J, Kadlubar S. Sulfotransferase genetic variation: from cancer risk to treatment response. Drug Metab Rev. 2013;45(4):415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The study summarized the correlation between SULT1A1 polymorphism and cancer in human.
- 59.Gamage N, Barnett A, Hempel N, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90(1):5–22. [DOI] [PubMed] [Google Scholar]
- 60.Hempel N, Gamage N, Martin JL, et al. Human cytosolic sulfotransferase SULT1A1. Int J Biochem Cell Biol. 2007;39(4):685–689. [DOI] [PubMed] [Google Scholar]
- 61.Glatt H, Meinl W. Pharmacogenetics of soluble sulfotransferases (SULTs). Naunyn Schmiedebergs Arch Pharmacol. 2004;369(1):55–68. [DOI] [PubMed] [Google Scholar]
- 62.Riches Z, Stanley EL, Bloomer JC, et al. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab Dispos. 2009;37(11):2255–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salman ED, Kadlubar SA, Falany CN. Expression and localization of cytosolic sulfotransferase (SULT) 1A1 and SULT1A3 in normal human brain. Drug Metab Dispos. 2009;37(4):706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richard K, Hume R, Kaptein E, et al. Sulfation of thyroid hormone and dopamine during human development: ontogeny of phenol sulfotransferases and arylsulfatase in liver, lung, and brain. J Clin Endocrinol Metab. 2001;86(6):2734–2742. [DOI] [PubMed] [Google Scholar]
- 65.Vietri M, Pietrabissa A, Mosca F, et al. Human adult and foetal liver sulphotransferases: inhibition by mefenamic acid and salicylic acid. Xenobiotica. 2001;31(3):153–161. [DOI] [PubMed] [Google Scholar]
- 66.Stanley EL, Hume R, Coughtrie MW. Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol Cell Endocrinol. 2005;240(1–2):32–42. [DOI] [PubMed] [Google Scholar]
- 67.Price RA, Spielman RS, Lucena AL, et al. Genetic polymorphism for human platelet thermostable phenol sulfotransferase (TS PST) activity. Genetics. 1989;122(4):905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hebbring SJ, Adjei AA, Baer JL, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007;16(5):463–470. [DOI] [PubMed] [Google Scholar]
- 69.Bardakci F, Arslan S, Bardakci S, et al. Sulfotransferase 1A1 (SULT1A1) polymorphism and susceptibility to primary brain tumors. J Cancer Res Clin Oncol. 2008;134(1):109–114. [DOI] [PubMed] [Google Scholar]
- 70.Hildebrandt MA, Carrington DP, Thomae BA, et al. Genetic diversity and function in the human cytosolic sulfotransferases. Pharmacogenomics J. 2007;7(2):133–143. [DOI] [PubMed] [Google Scholar]
- 71.Li X, Clemens DL, Cole JR, et al. Characterization of human liver thermostable phenol sulfotransferase (SULT1A1) allozymes with 3,3’,5-triiodothyronine as the substrate. J Endocrinol. 2001;171(3):525–532. [DOI] [PubMed] [Google Scholar]
- 72.Nagar S, Walther S, Blanchard RL. Sulfotransferase (SULT) 1A1 polymorphic variants *1, *2, and *3 are associated with altered enzymatic activity, cellular phenotype, and protein degradation. Mol Pharmacol. 2006;69(6):2084–2092. [DOI] [PubMed] [Google Scholar]
- 73.Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19(1):56–61. [DOI] [PubMed] [Google Scholar]
- 74.Coughtrie MW. Sulfation through the looking glass--recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2(5):297–308. [DOI] [PubMed] [Google Scholar]
- 75.Lindsay J, Wang LL, Li Y, et al. Structure, function and polymorphism of human cytosolic sulfotransferases. Curr Drug Metab. 2008;9(2):99–105. [DOI] [PubMed] [Google Scholar]
- 76.Carlini EJ, Raftogianis RB, Wood TC, et al. Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African-American subjects. Pharmacogenetics. 2001;11(1):57–68. [DOI] [PubMed] [Google Scholar]
- 77.Ozawa S, Shimizu M, Katoh T, et al. Sulfating-activity and stability of cDNA-expressed allozymes of human phenol sulfotransferase, ST1A3*1 ((213)Arg) and ST1A3*2 ((213)His), both of which exist in Japanese as well as Caucasians. J Biochem. 1999;126(2):271–277. [DOI] [PubMed] [Google Scholar]
- 78.Rasool MI, Bairam AF, Gohal SA, et al. Effects of the human SULT1A1 polymorphisms on the sulfation of acetaminophen, O-desmethylnaproxen, and tapentadol. Pharmacol Rep. 2019;71(2):257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marazziti D, Palego L, Mazzanti C, et al. Human platelet sulfotransferase shows seasonal rhythms. Chronobiol Int. 1995;12(2):100–105. [DOI] [PubMed] [Google Scholar]
- 80.Marazziti D, Palego L, Rossi A, et al. Gender-related seasonality of human platelet phenolsulfotransferase activity. Neuropsychobiology. 1998;38(1):1–5. [DOI] [PubMed] [Google Scholar]
- 81.Coughtrie MW, Gilissen RA, Shek B, et al. Phenol sulphotransferase SULT1A1 polymorphism: molecular diagnosis and allele frequencies in Caucasian and African populations. Biochem J. 1999;337 (1):45–49. [PMC free article] [PubMed] [Google Scholar]
- 82.Dalhoff K, Buus Jensen K, Enghusen Poulsen H. Cancer and molecular biomarkers of phase 2. Methods Enzymol. 2005; 400:618–627. [DOI] [PubMed] [Google Scholar]
- 83.Wu MT, Wang YT, Ho CK, et al. SULT1A1 polymorphism and esophageal cancer in males. Int J Cancer. 2003;103(1):101–104. [DOI] [PubMed] [Google Scholar]
- 84.Han DF, Zhou X, Hu MB, et al. Sulfotransferase 1A1 (SULT1A1) polymorphism and breast cancer risk in Chinese women. Toxicol Lett. 2004;150(2):167–177. [DOI] [PubMed] [Google Scholar]
- 85.Boccia S, Persiani R, La Torre G, et al. Sulfotransferase 1A1 polymorphism and gastric cancer risk: a pilot case-control study. Cancer Lett. 2005;229(2):235–243. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Spitz MR, Tsou AM, et al. Sulfotransferase (SULT) 1A1 polymorphism as a predisposition factor for lung cancer: a case-control analysis. Lung Cancer. 2002;35(2):137–142. [DOI] [PubMed] [Google Scholar]
- 87.Arslan S, Silig Y, Pinarbasi H. An investigation of the relationship between SULT1A1 Arg(213)His polymorphism and lung cancer susceptibility in a Turkish population. Cell Biochem Funct. 2009;27(4):211–215. [DOI] [PubMed] [Google Scholar]
- 88.Tasnim T, Al-Mamun MMA, Nahid NA, et al. Genetic variants of SULT1A1 and XRCC1 genes and risk of lung cancer in Bangladeshi population. Tumour Biol. 2017;39(11):1010428317729270. [DOI] [PubMed] [Google Scholar]
- 89.Daniels J, Kadlubar S. Pharmacogenetics of SULT1A1. Pharmacogenomics. 2014;15(14):1823–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wong CF, Liyou N, Leggett B, et al. Association of the SULT1A1 R213H polymorphism with colorectal cancer. Clin Exp Pharmacol Physiol. 2002;29(9):754–758. [DOI] [PubMed] [Google Scholar]
- 91.Steiner M, Bastian M, Schulz WA, et al. Phenol sulphotransferase SULT1A1 polymorphism in prostate cancer: lack of association. Arch Toxicol. 2000;74(4–5):222–225. [DOI] [PubMed] [Google Scholar]
- 92.Tamaki Y, Arai T, Sugimura H, et al. Association between cancer risk and drug-metabolizing enzyme gene (CYP2A6, CYP2A13, CYP4B1, SULT1A1, GSTM1, and GSTT1) polymorphisms in cases of lung cancer in Japan. Drug Metab Pharmacokinet. 2011;26(5):516–522. [DOI] [PubMed] [Google Scholar]
- 93.Ozawa S, Katoh T, Inatomi H, et al. Association of genotypes of carcinogen-activating enzymes, phenol sulfotransferase SULT1A1 (ST1A3) and arylamine N-acetyltransferase NAT2, with urothelial cancer in a Japanese population. Int J Cancer. 2002;102(4):418–421. [DOI] [PubMed] [Google Scholar]
- 94.Zheng L, Wang Y, Schabath MB, et al. Sulfotransferase 1A1 (SULT1A1) polymorphism and bladder cancer risk: a case-control study. Cancer Lett. 2003;202(1):61–69. [DOI] [PubMed] [Google Scholar]
- 95.Lu J, Li H, Zhang J, et al. Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2. Biochem Biophys Res Commun. 2010;396(2):429–434. [DOI] [PubMed] [Google Scholar]
- 96.Zheng W, Xie D, Cerhan JR, et al. Sulfotransferase 1A1 polymorphism, endogenous estrogen exposure, well-done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10(2):89–94. [PubMed] [Google Scholar]
- 97.Tang D, Rundle A, Mooney L, et al. Sulfotransferase 1A1 (SULT1A1) polymorphism, PAH-DNA adduct levels in breast tissue and breast cancer risk in a case-control study. Breast Cancer Res Treat. 2003;78(2):217–222. [DOI] [PubMed] [Google Scholar]
- 98.Nowell S, Sweeney C, Winters M, et al. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94(21):1635–1640. [DOI] [PubMed] [Google Scholar]
- 99.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91(3):249–258. [DOI] [PubMed] [Google Scholar]
- 100.Lee H, Wang Q, Yang F, et al. SULT1A1 Arg213His polymorphism, smoked meat, and breast cancer risk: a case-control study and meta-analysis. DNA Cell Biol. 2012;31(5):688–699. [DOI] [PubMed] [Google Scholar]
- 101.Tengström M, Mannermaa A, Kosma VM, et al. SULT1A1 rs9282861 polymorphism-a potential modifier of efficacy of the systemic adjuvant therapy in breast cancer?. BMC Cancer. 2012; 12:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grabinski JL, Smith LS, Chisholm GB, et al. Genotypic and allelic frequencies of SULT1A1 polymorphisms in women receiving adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2006;95(1):13–16. [DOI] [PubMed] [Google Scholar]
- 103.Choi JY, Lee KM, Park SK, et al. Genetic polymorphisms of SULT1A1 and SULT1E1 and the risk and survival of breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1090–1095. [DOI] [PubMed] [Google Scholar]
- 104.Serrano D, Lazzeroni M, Zambon CF, et al. Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian Tamoxifen Prevention Trial. Pharmacogenomics J. 2011;11(2):100–107. [DOI] [PubMed] [Google Scholar]
- 105.Knechtel G, Hofmann G, Gerger A, et al. Analysis of common germline polymorphisms as prognostic factors in patients with lymph node-positive breast cancer. J Cancer Res Clin Oncol. 2010;136(12):1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tao P, Li H, Wang Q, et al. [A case-control study on association of SULT1A1 polymorphism, smoked meat intake with breast cancer risk.] Zhonghua Yu Fang Yi Xue Za Zhi. 2012;46(9):831–835. Chinese. [PubMed] [Google Scholar]
- 107.Li W, Gu M. SULT1A1 Arg213His polymorphism is associated with bladder cancer risk: a meta-analysis. Med Sci Monit. 2014; 20:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun XF, Ahmadi A, Arbman G, et al. Polymorphisms in sulfotransferase 1A1 and glutathione S-transferase P1 genes in relation to colorectal cancer risk and patients’ survival. World J Gastroenterol. 2005;11(43):6875–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lilla C, Risch A, Verla-Tebit E, et al. SULT1A1 genotype and susceptibility to colorectal cancer. Int J Cancer. 2007;120(1):201–206. [DOI] [PubMed] [Google Scholar]
- 110.Cotterchio M, Boucher BA, Manno M, et al. Red meat intake, doneness, polymorphisms in genes that encode carcinogen-metabolizing enzymes, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3098–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lopes BA, Emerenciano M, Gonçalves BA, et al. Polymorphisms in CYP1B1, CYP3A5, GSTT1, and SULT1A1 are associated with early age acute leukemia. PLoS One. 2015;10(5): e0127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nowell S, Ratnasinghe DL, Ambrosone CB, et al. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2004;13(2):270–276. [DOI] [PubMed] [Google Scholar]
- 113.Dajani R, Cleasby A, Neu M, et al. X-ray crystal structure of human dopamine sulfotransferase, SULT1A3. Molecular modeling and quantitative structure activity relationship analysis demonstrate a molecular basis for sulfotransferase substrate specificity. J Biol Chem. 1999;274(53):37862–37868. [DOI] [PubMed] [Google Scholar]
- 114.Rubin GL, Sharp S, Jones AL, et al. Design, production and characterization of antibodies discriminating between the phenol- and monoamine-sulphating forms of human phenol sulphotransferase. Xenobiotica. 1996;26(11):1113–1119. [DOI] [PubMed] [Google Scholar]
- 115.Dooley TP, Obermoeller RD, Leiter EH, et al. Mapping of the phenol sulfotransferase gene (STP) to human chromosome 16p12.1-p11.2. Genomics. 1993;18(2):440–443. [DOI] [PubMed] [Google Scholar]
- 116.Kumar RM, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17(4):628–638. [DOI] [PubMed] [Google Scholar]
- 117.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358(7):667–675. [DOI] [PubMed] [Google Scholar]
- 118.Steinman KJ, Spence SJ, Ramocki MB, et al. “16p11.2 deletion and duplication: Characterizing neurologic phenotypes in a large clinically ascertained cohort”. Am J Med Genet A. 2016;170(11):2943–2955. [DOI] [PubMed] [Google Scholar]
- 119.Angelakos CC, Watson AJ, O’Brien WT, et al. Hyperactivity and male-specific sleep deficits in the 16p11.2 deletion mouse model of autism. Autism Res. 2017;10(4):572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Butcher NJ, Horne MK, Mellick GD, et al. Sulfotransferase 1A3/4 copy number variation is associated with neurodegenerative disease. Pharmacogenomics J. 2018;18(2):209–214. [DOI] [PubMed] [Google Scholar]
- 121.Rosenfeld JA, Coppinger J, Bejjani BA, et al. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J Neurodev Disord. 2010;2(1):26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fernandez BA, Roberts W, Chung B, et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2010;47(3):195–203. [DOI] [PubMed] [Google Scholar]
- 123.Jacquemont S, Reymond A, Zufferey F, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478(7367):97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41(11):1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47(5):332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang L, Yee VC, Weinshilboum RM. Aggresome formation and pharmacogenetics: sulfotransferase 1A3 as a model system. Biochem Biophys Res Commun. 2004;325(2):426–433. [DOI] [PubMed] [Google Scholar]
- 127.Bairam AF, Rasool MI, Alherz FA, et al. Sulfation of catecholamines and serotonin by SULT1A3 allozymes. Biochem Pharmacol. 2018;151:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bairam AF, Rasool MI, Alherz FA, et al. Effects of human SULT1A3/SULT1A4 genetic polymorphisms on the sulfation of acetaminophen and opioid drugs by the cytosolic sulfotransferase SULT1A3. Arch Biochem Biophys. 2018;648:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bairam AF, Rasool MI, Alherz FA, et al. Impact of SULT1A3/SULT1A4 genetic polymorphisms on the sulfation of phenylephrine and salbutamol by human SULT1A3 allozymes. Pharmacogenet Genomics. 2019;29(5):99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Glatt H Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000;129(1–2):141–170. [DOI] [PubMed] [Google Scholar]
- 131.Meinl W, Tsoi C, Swedmark S, et al. Highly selective bioactivation of 1-and 2-hydroxy-3-methylcholanthrene to mutagens by individual human and other mammalian sulphotransferases expressed in Salmonella typhimurium. Mutagenesis. 2013;28(5):609–619. [DOI] [PubMed] [Google Scholar]
- 132.Fujita K, Nagata K, Ozawa S, et al. Molecular cloning and characterization of rat ST1B1 and human ST1B2 cDNAs, encoding thyroid hormone sulfotransferases. J Biochem. 1997;122(5):1052–1061. [DOI] [PubMed] [Google Scholar]
- 133.Iida A, Sekine A, Saito S, et al. Catalog of 320 single nucleotide polymorphisms (SNPs) in 20 quinone oxidoreductase and sulfotransferase genes. J Hum Genet. 2001;46(4):225–240. [DOI] [PubMed] [Google Scholar]
- 134.Tibbs ZE, Guidry AL, Falany JL, et al. A high frequency missense SULT1B1 allelic variant (L145V) selectively expressed in African descendants exhibits altered kinetic properties. Xenobiotica. 2018;48(1):79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Monzo M, Brunet S, Urbano-Ispizua A, et al. Genomic polymorphisms provide prognostic information in intermediate-risk acute myeloblastic leukemia. Blood. 2006;107(12):4871–4879. [DOI] [PubMed] [Google Scholar]
- 136.Yip CKY, Bansal S, Wong SY, et al. Identification of Galeterone and Abiraterone as Inhibitors of Dehydroepiandrosterone Sulfonation Catalyzed by Human Hepatic Cytosol, SULT2A1, SULT2B1b, and SULT1E1. Drug Metab Dispos. 2018;46(4):470–482. [DOI] [PubMed] [Google Scholar]
- 137.Zhang H, Varlamova O, Vargas FM, et al. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem. 1998;273(18):10888–10892. [DOI] [PubMed] [Google Scholar]
- 138.Petrotchenko EV, Doerflein ME, Kakuta Y, et al. Substrate gating confers steroid specificity to estrogen sulfotransferase. J Biol Chem. 1999;274(42):30019–30022. [DOI] [PubMed] [Google Scholar]
- 139.Schrag ML, Cui D, Rushmore TH, et al. Sulfotransferase 1E1 is a low km isoform mediating the 3-O-sulfation of ethinyl estradiol. Drug Metab Dispos. 2004;32(11):1299–1303. [DOI] [PubMed] [Google Scholar]
- 140.Falany JL, Pilloff DE, Leyh TS, et al. Sulfation of raloxifene and 4-hydroxytamoxifen by human cytosolic sulfotransferases. Drug Metab Dispos. 2006;34(3):361–368. [DOI] [PubMed] [Google Scholar]
- 141.Nishiyama T, Ogura K, Nakano H, et al. Reverse geometrical selectivity in glucuronidation and sulfation of cis- and trans-4-hydroxytamoxifens by human liver UDP-glucuronosyltransferases and sulfotransferases. Biochem Pharmacol. 2002;63(10):1817–1830. [DOI] [PubMed] [Google Scholar]
- 142.Suiko M, Sakakibara Y, Liu MC. Sulfation of environmental estrogen-like chemicals by human cytosolic sulfotransferases. Biochem Biophys Res Commun. 2000;267(1):80–84. [DOI] [PubMed] [Google Scholar]
- 143.Edavana VK, Yu X, Dhakal IB, et al. Sulfation of fulvestrant by human liver cytosols and recombinant SULT1A1 and SULT1E1. Pharmgenomics Pers Med. 2011;4:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res. 1996;56(7):1551–1555. [PubMed] [Google Scholar]
- 145.Falany JL, Falany CN. Regulation of estrogen sulfotransferase in human endometrial adenocarcinoma cells by progesterone. Endocrinology. 1996;137(4):1395–1401. [DOI] [PubMed] [Google Scholar]
- 146.Qian Y, Deng C, Song WC. Expression of estrogen sulfotransferase in MCF-7 cells by cDNA transfection suppresses the estrogen response: potential role of the enzyme in regulating estrogen-dependent growth of breast epithelial cells. J Pharmacol Exp Ther. 1998;286(1):555–560. [PubMed] [Google Scholar]
- 147.Song WC, Qian Y, Sun X, et al. Cellular localization and regulation of expression of testicular estrogen sulfotransferase. Endocrinology. 1997;138(11):5006–5012. [DOI] [PubMed] [Google Scholar]
- 148.Hirata H, Hinoda Y, Okayama N, et al. CYP1A1, SULT1A1, and SULT1E1 polymorphisms are risk factors for endometrial cancer susceptibility. Cancer. 2008;112(9):1964–1973. [DOI] [PubMed] [Google Scholar]
- 149.Rebbeck TR, Troxel AB, Wang Y, et al. Estrogen sulfation genes, hormone replacement therapy, and endometrial cancer risk. J Natl Cancer Inst. 2006;98(18):1311–1320. [DOI] [PubMed] [Google Scholar]
- 150.Hsieh YC, Jeng JS, Lin HJ, et al. Epistasis analysis for estrogen metabolic and signaling pathway genes on young ischemic stroke patients. PLoS One. 2012;7(10):e47773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee SA, Choi JY, Shin CS, et al. SULT1E1 genetic polymorphisms modified the association between phytoestrogen consumption and bone mineral density in healthy Korean women. Calcif Tissue Int. 2006;79(3):152–159. [DOI] [PubMed] [Google Scholar]
- 152.Cohen S, Laitman Y, Kaufman B, et al. SULT1E1 and ID2 genes as candidates for inherited predisposition to breast and ovarian cancer in Jewish women. Fam Cancer. 2009;8(2):135–144. [DOI] [PubMed] [Google Scholar]
- 153.Li S, Xie L, Du M, et al. Association study of genetic variants in estrogen metabolic pathway genes and colorectal cancer risk and survival. Arch Toxicol. 2018;92(6):1991–1999. [DOI] [PubMed] [Google Scholar]
- 154.Woo HI, Lee SK, Kim J, et al. Variations in plasma concentrations of tamoxifen metabolites and the effects of genetic polymorphisms on tamoxifen metabolism in Korean patients with breast cancer. Oncotarget. 2017;8(59):100296–100311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Agarwal N, Alex AB, Farnham JM, et al. Inherited Variants in SULT1E1 and Response to Abiraterone Acetate by Men with Metastatic Castration Refractory Prostate Cancer. J Urol. 2016;196(4):1112–1116. [DOI] [PubMed] [Google Scholar]
- 156.El Daibani AA, Alherz FA, Abunnaja MS, et al. Impact of Human SULT1E1 Polymorphisms on the Sulfation of 17β-Estradiol, 4-Hydroxytamoxifen, and Diethylstilbestrol by SULT1E1 Allozymes. Eur J Drug Metab Pharmacokinet. 2021;46(1):105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.El Daibani AA. Studies on the Functional Relevance of Genetic Polymorphisms of the Human Cytosolic Sulfotransferase 1E1 (SULT1E1) [dissertation]. University of Toledo; 2018. https://etd.ohiolink.edu/ [Google Scholar]
- 158.Comer KA, Falany CN. Immunological characterization of dehydroepiandrosterone sulfotransferase from human liver and adrenal. Mol Pharmacol. 1992;41(4):645–51. [PubMed] [Google Scholar]
- 159.Falany CN, Comer KA, Dooley TP, et al. Human dehydroepiandrosterone sulfotransferase. Purification, molecular cloning, and characterization. Ann N Y Acad Sci. 1995;774:59–72. [DOI] [PubMed] [Google Scholar]
- 160.Forbes KJ, Hagen M, Glatt H, et al. Human fetal adrenal hydroxysteroid sulphotransferase: cDNA cloning, stable expression in V79 cells and functional characterisation of the expressed enzyme. Mol Cell Endocrinol. 1995;112(1):53–60. [DOI] [PubMed] [Google Scholar]
- 161.Ekström L, Rane A. Genetic variation, expression and ontogeny of sulfotransferase SULT2A1 in humans. Pharmacogenomics J. 2015;15(4):293–7. [DOI] [PubMed] [Google Scholar]
- 162.Chang H, Shi R, Rehse P, et al. Identifying androsterone (ADT) as a cognate substrate for human dehydroepiandrosterone sulfotransferase (DHEA-ST) important for steroid homeostasis: structure of the enzyme-ADT complex. J Biol Chem. 2004;279(4):2689–96. [DOI] [PubMed] [Google Scholar]
- 163.Aksoy IA, Sochorová V, Weinshilboum RM. Human liver dehydroepiandrosterone sulfotransferase: nature and extent of individual variation. Clin Pharmacol Ther. 1993;54(5):498–506. [DOI] [PubMed] [Google Scholar]
- 164.Thomae BA, Eckloff BW, Freimuth RR, et al. Human sulfotransferase SULT2A1 pharmacogenetics: genotype-to-phenotype studies. Pharmacogenomics J. 2002;2(1):48–56. [DOI] [PubMed] [Google Scholar]
- 165.Wilborn TW, Lang NP, Smith M, et al. Association of SULT2A1 allelic variants with plasma adrenal androgens and prostate cancer in African American men. J Steroid Biochem Mol Biol. 2006;99(4–5):209–14. [DOI] [PubMed] [Google Scholar]
- 166.Zhai G, Teumer A, Stolk L, et al. Eight common genetic variants associated with serum DHEAS levels suggest a key role in ageing mechanisms. PLoS Genet. 2011;7(4):e1002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.García-Anguita A, Ortega L, Garcés C. Relationship of dehydroepiandrosterone sulfate with overweight and insulin sensitivity in 12–16-year-old Spanish children. Horm Metab Res. 2013;45(7):545–7. [DOI] [PubMed] [Google Scholar]
- 168.Kwon EM, Holt SK, Fu R, et al. Androgen metabolism and JAK/STAT pathway genes and prostate cancer risk. Cancer Epidemiol. 2012;36(4):347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Louwers YV, de Jong FH, van Herwaarden NAA, Stolk L, Fauser BCJM, Uitterlinden AG, Laven JSE. Variants in SULT2A1 affect the DHEA sulphate to DHEA ratio in patients with polycystic ovary syndrome but not the hyperandrogenic phenotype. J Clin Endocrinol Metab. 2013;98(9):3848–55. [DOI] [PubMed] [Google Scholar]
- 170.Miller E, Zalzala MH, Abunnaja MS, et al. Effects of Human Sulfotransferase 2A1 Genetic Polymorphisms 3 on the Sulfation of Tibolone. Eur J Drug Metab Pharmacokinet. 2018;43(4):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Abunnaja MS, Alherz FA, El Daibani AA, et al. Effects of genetic polymorphisms on the sulfation of dehydroepiandrosterone and pregnenolone by human cytosolic sulfotransferase SULT2A1. Biochem Cell Biol. 2018;96(5):655–662. [DOI] [PubMed] [Google Scholar]
- 172.Abunnaja MS. Investigation of the Genetic Polymorphisms of the Human Cytosolic Sulfotransferase SULT2A1: Potential Impact on the Metabolism of Hydroxysteroids and Drugs [dissertation]. University of Toledo. 2019. OhioLINK Electronic Theses and Dissertations Center. [Google Scholar]
- 173.He D, Falany CN. Characterization of proline-serine-rich carboxyl terminus in human sulfotransferase 2B1b: immunogenicity, subcellular localization, kinetic properties, and phosphorylation. Drug Metab Dispos. 2006;34(10):1749–55. [DOI] [PubMed] [Google Scholar]
- 174.Pai TG, Sugahara T, Suiko M, et al. Differential xenoestrogen-sulfating activities of the human cytosolic sulfotransferases: molecular cloning, expression, and purification of human SULT2B1a and SULT2B1b sulfotransferases. Biochim Biophys Acta. 2002;1573(2):165–70. [DOI] [PubMed] [Google Scholar]
- 175.Fuda H, Javitt NB, Mitamura K, et al. Oxysterols are substrates for cholesterol sulfotransferase. J Lipid Res. 2007;48(6):1343–52. [DOI] [PubMed] [Google Scholar]
- 176.Cook IT, Duniec-Dmuchowski Z, Kocarek TA, et al. 24-hydroxycholesterol sulfation by human cytosolic sulfotransferases: formation of monosulfates and disulfates, molecular modeling, sulfatase sensitivity, and inhibition of liver x receptor activation. Drug Metab Dispos. 2009;37(10):2069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ren S, Ning Y. Sulfation of 25-hydroxycholesterol regulates lipid metabolism, inflammatory responses, and cell proliferation. Am J Physiol Endocrinol Metab. 2014;306(2):E123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Geese WJ, Raftogianis RB. Biochemical characterization and tissue distribution of human SULT2B1. Biochem Biophys Res Commun. 2001;288(1):280–9. [DOI] [PubMed] [Google Scholar]
- 179.Higashi Y, Fuda H, Yanai H, et al. Expression of cholesterol sulfotransferase (SULT2B1b) in human skin and primary cultures of human epidermal keratinocytes. J Invest Dermatol. 2004;122(5):1207–13. [DOI] [PubMed] [Google Scholar]
- 180.Koizumi M, Momoeda M, Hiroi H, et al. Expression and regulation of cholesterol sulfotransferase (SULT2B1b) in human endometrium. Fertil Steril. 2010;93(5):1538–44. [DOI] [PubMed] [Google Scholar]
- 181.He D, Frost AR, Falany CN. Identification and immunohistochemical localization of Sulfotransferase 2B1b (SULT2B1b) in human lung. Biochim Biophys Acta. 2005;1724(1–2):119–26. [DOI] [PubMed] [Google Scholar]
- 182.Yanai H, Javitt NB, Higashi Y, et al. Expression of cholesterol sulfotransferase (SULT2B1b) in human platelets. Circulation. 2004;109(1):92–6. [DOI] [PubMed] [Google Scholar]
- 183.Salman ED, Faye-Petersen O, Falany CN. Hydroxysteroid sulfotransferase 2B1b expression and localization in normal human brain. Horm Mol Biol Clin Investig. 2011;8(1):445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.He D, Falany CN. Inhibition of SULT2B1b expression alters effects of 3beta-hydroxysteroids on cell proliferation and steroid hormone receptor expression in human LNCaP prostate cancer cells. Prostate. 2007;67(12):1318–29. [DOI] [PubMed] [Google Scholar]
- 185.Seo YK, Mirkheshti N, Song CS, et al. SULT2B1b sulfotransferase: induction by vitamin D receptor and reduced expression in prostate cancer. Mol Endocrinol. 2013;27(6):925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen W, Zhou H, Ye L, et al. Overexpression of SULT2B1b Promotes Angiogenesis in Human Gastric Cancer. Cell Physiol Biochem. 2016;38(3):1040–54. [DOI] [PubMed] [Google Scholar]
- 187.Hu L, Yang GZ, Zhang Y, et al. Overexpression of SULT2B1b is an independent prognostic indicator and promotes cell growth and invasion in colorectal carcinoma. Lab Invest. 2015;95(9):1005–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hong W, Guo F, Yang M, et al. Hydroxysteroid sulfotransferase 2B1 affects gastric epithelial function and carcinogenesis induced by a carcinogenic agent. Lipids Health Dis. 2019;18(1):203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Heinz L, Kim GJ, Marrakchi S, et al. Mutations in SULT2B1 Cause Autosomal-Recessive Congenital Ichthyosis in Humans. Am J Hum Genet. 2017;100(6):926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Youssefian L, Vahidnezhad H, Saeidian AH, et al. Autosomal recessive congenital ichthyosis: Genomic landscape and phenotypic spectrum in a cohort of 125 consanguineous families. Hum Mutat. 2019;40(3):288–298. [DOI] [PubMed] [Google Scholar]
- 191.Hyland PL, Freedman ND, Hu N, et al. Genetic variants in sex hormone metabolic pathway genes and risk of esophageal squamous cell carcinoma. Carcinogenesis. 2013;34(5):1062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hevir N, Sinkovec J, Rizner TL. Disturbed expression of phase I and phase II estrogen-metabolizing enzymes in endometrial cancer: lower levels of CYP1B1 and increased expression of S-COMT. Mol Cell Endocrinol. 2011;331(1):158–67. [DOI] [PubMed] [Google Scholar]
- 193.Ji Y, Moon I, Zlatkovic J, et al. Human hydroxysteroid sulfotransferase SULT2B1 pharmacogenomics: gene sequence variation and functional genomics. J Pharmacol Exp Ther. 2007;322(2):529–40. [DOI] [PubMed] [Google Scholar]
- 194.Alherz FA, Abunnaja MS, El Daibani AA, et al. On the role of genetic polymorphisms in the sulfation of cholesterol by human cytosolic sulphotransferase SULT2B1b. J Biochem. 2018;164(3):215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Alherz FA, El Daibani AA, Abunnaja MS, et al. Effect of SULT2B1 genetic polymorphisms on the sulfation of dehydroepiandrosterone and pregnenolone by SULT2B1b allozymes. Mol Cell Endocrinol. 2019;496:110535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–41. [DOI] [PubMed] [Google Scholar]
- 197.Hizbullah, Ahmed S, Mumtaz MN, et al. Genetic variations in drug-metabolizing enzyme CYP2C9 among major ethnic groups of Pakistani population. Gene. 2020;746:144659. [DOI] [PubMed] [Google Scholar]
- 198.Meaden CW, Mozeika A, Asri R, Santos CD. A review of the existing literature on buprenorphine pharmacogenomics. Pharmacogenomics J. 2020. doi: 10.1038/s41397-020-00198-1. [DOI] [PubMed] [Google Scholar]
- 199.Zhao Q, Liu B, Zhang J, et al. Association between a COMT polymorphism and clinical response to risperidone treatment: a pharmacogenetic study. Psychiatr Genet. 2012;22(6):298–9. [DOI] [PubMed] [Google Scholar]
- 200.Matsuoka H, Tsurutani J, Chiba Y, et al. Selection of opioids for cancer-related pain using a biomarker: a randomized, multi-institutional, open-label trial (RELIEF study). BMC Cancer. 2017;17(1):674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Semiz S, Dujic T, Ostanek B, et al. Association of NAT2 polymorphisms with type 2 diabetes in a population from Bosnia and Herzegovina. Arch Med Res. 2011;42(4):311–7. [DOI] [PubMed] [Google Scholar]
- 202.Selinski S, Blaszkewicz M, Ickstadt K, et al. Improvements in algorithms for phenotype inference: the NAT2 example. Curr Drug Metab. 2014;15(2):233–49. [DOI] [PubMed] [Google Scholar]
- 203.Lu Y, Fang Y, Wu X, et al. Effects of UGT1A9 genetic polymorphisms on monohydroxylated derivative of oxcarbazepine concentrations and oxcarbazepine monotherapeutic efficacy in Chinese patients with epilepsy. Eur J Clin Pharmacol. 2017;73(3):307–315. [DOI] [PubMed] [Google Scholar]
- 204.Li J, Peng P, Mei Q, et al. The impact of UGT2B7 C802T and CYP3A4*1G polymorphisms on pain relief in cancer patients receiving oxycontin. Support Care Cancer. 2018;26(8):2763–2767. [DOI] [PubMed] [Google Scholar]
- 205.Takanashi S, Bachur NR Adriamycin metabolism in man. Evidence from urinary metabolites. Drug Metab Dispos. 1976;4, 79–87. [PubMed] [Google Scholar]
- 206.Larsson R, Nygren P. Cytotoxic activity of topoisomerase II inhibitors in primary cultures of tumor cells from patients with human hematologic and solid tumors. Cancer 1994;74,2857–2862. [DOI] [PubMed] [Google Scholar]
- 207.Gianni L, Norton L, Wolmark N, et al. Role of anthracyclines in the treatment of early breast cancer. J Clin Oncol. 2009;27,4798–4808. [DOI] [PubMed] [Google Scholar]
- 208.Krischer JP, Epstein S, Cuthbertson DD, et al. Clinical cardiotoxicity following anthracycline treatment for childhood cancer: the Pediatric Oncology Group experience. J Clin Oncol. 1997;15,1544–1552. [DOI] [PubMed] [Google Scholar]
- 209.Deng S, Wojnowski L. Genotyping the risk of anthracycline-induced cardiotoxicity, Cardiovasc Toxicol. 2007;7,129–134. [DOI] [PubMed] [Google Scholar]
- 210.Montaner JS, Reiss P, Cooper D, et al. A randomized, double-blind trial comparing combinations of nevirapine, didanosine, and zidovudine for HIV-infected patients. J Am Med Assoc. 1998;279:930–937. [DOI] [PubMed] [Google Scholar]
- 211.Sweat MD, O’Reilly KR, Schmid GP, et al. Cost-effectiveness of nevirapine to prevent mother-to-child HIV transmission in eight African countries. AIDS. 2004;18:1661–1671. [DOI] [PubMed] [Google Scholar]
- 212.Sharma AM, Klarskov K, Uetrecht J. Nevirapine bioactivation and covalent binding in the skin. Chem Res Toxicol. 2013;26:410–421. [DOI] [PubMed] [Google Scholar]
- 213.Sharma AM, Novalen M, Tanino T, et al. 12-OH-nevirapine sulfate, formed in the skin, is responsible for nevirapine-induced skin rash. Chem Res Toxicol. 2013;26:817–827. [DOI] [PubMed] [Google Scholar]
- 214.Kranendonk M, Alves M, Antunes P, et al. Human sulfotransferase 1A1-dependent mutagenicity of 12-hydroxy-nevirapine: the missing link? Chem Res Toxicol. 2014;27: 1967–1971. [DOI] [PubMed] [Google Scholar]
- 215.Dooley TP, Haldeman-Cahill R, Joiner J, et al. Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem Biophys Res Commun. 2000;277:236–245. [DOI] [PubMed] [Google Scholar]
- 216.Luu-The V, Duche D, Ferraris C, et al. Expression profiles of phases 1 and 2 metabolizing enzymes in human skin and the reconstructed skin models Episkin™ and full thickness model from Episkin™. J Steroid Biochem Mol Biol. 2009;116:178–186. [DOI] [PubMed] [Google Scholar]
- 217.Yasuda S, Idell S, Liu MC. Generation and release of nitrotyrosine O-sulfate by HepG2 human hepatoma cells upon SIN-1 stimulation: identification of SULT1A3 as the enzyme responsible. Biochem J. 2007;401(2):497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Liu MC, Yasuda S, Idell S. Sulfation of nitrotyrosine: biochemistry and functional implications. IUBMB Life. 2007;59(10):622–7. [DOI] [PubMed] [Google Scholar]
- 219.Yasuda S, Yasuda T, Liu MY, et al. Sulfation of chlorotyrosine and nitrotyrosine by human lung endothelial and epithelial cells: role of the human SULT1A3. Toxicol Appl Pharmacol. 2011;251(2):104–9. [DOI] [PubMed] [Google Scholar]
- 220.Murata M, Kawanishi S. Oxidative DNA damage induced by nitrotyrosine, a biomarker of inflammation. Biochem Biophys Res Commun. 2004;316:123–128. [DOI] [PubMed] [Google Scholar]
- 221.Peluffo H, Shacka JJ, Ricart K, et al. Induction of motor neuron apoptosis by free 3-nitro-L-tyrosine. J Neurochem. 2004;89:602–612. [DOI] [PubMed] [Google Scholar]
- 222.Mohiuddin I, Chai H, Lin PH, et al. Nitrotyrosine and chlorotyrosine: clinical significance and biological functions in the vascular system. J Surg Res. 2006;133:143–149. [DOI] [PubMed] [Google Scholar]
- 223.Chai H, Mohiuddin I, Lin P, et al. Chlorotyrosine induces endothelial dysfunction in human coronary artery endothelial cells and porcine coronary arteries. J Surg Res. 2007;137:174. [Google Scholar]


