Abstract
Glioblastoma multiforme (GBM) is a highly invasive, central nervous system (CNS) cancer for which there is no cure. Invading tumor cells evade treatment, limiting the efficacy of the current standard of care regimen. Understanding the underlying invasive behaviors that support tumor growth may allow for generation of novel GBM therapies. Zebrafish (Danio rerio) are attractive for genetics and live imaging and have, in recent years, emerged as a model system suitable for cancer biology research. While other groups have studied CNS tumors using zebrafish, few have concentrated on the invasive behaviors supporting the development of these diseases. Previous studies demonstrated that one of the main mechanisms of GBM invasion is perivascular invasion, i.e., single tumor cell migration along blood vessels. Here, we characterize phenotypes, methodology, and potential therapeutic avenues for utilizing zebrafish to model perivascular GBM invasion. Using patient-derived xenolines or an adherent cell line, we demonstrate tumor expansion within the zebrafish brain. Within 24-h postintracranial injection, D54-MG-tdTomato glioma cells produce fingerlike projections along the zebrafish brain vasculature. As few as 25 GBM cells were sufficient to promote single cell vessel co-option. Of note, these tumor-vessel interactions are CNS specific and do not occur on pre-existing blood vessels when injected into the animal’s peripheral tissue. Tumor-vessel interactions increase over time and can be pharmacologically disrupted through inhibition of Wnt signaling. Therefore, zebrafish serve as a favorable model system to study perivascular glioma invasion, one of the deadly characteristics that make GBM so difficult to treat.
Keywords: glioblastoma multiforme, perivascular glioma invasion, zebrafish, tumor model, Wnt signaling
Introduction
Brain tumors are one of the most devastating forms of cancer, as they grow within an organ of limited regenerative capacity and can be highly invasive. Additionally, drug delivery to brain tumors is challenging due to the blood-brain barrier (BBB), a physiological structure that limits the open flow of foreign compounds into the brain tissue. Glioblastoma multiforme (GBM) is the most common, primary malignant brain tumor and is newly diagnosed in around 13,000 patients every year in the United States.1 The standard of care is a three-pronged approach of surgical resection, radiation, and chemotherapy.2 Even after over a decade of implementation, this multifaceted treatment regimen extends survival to only 14–19 months.3 These aggressive therapies mainly target the main tumor mass that is most easily identified by MRI for surgical removal or therapeutics that target actively dividing cells. Even in the era of personalized medicine, every GBM case presents with a different set of mutations, making it difficult to standardize a therapeutic regimen that effectively treats all patients. Research efforts have shown that the genetic heterogeneity of GBM tumors results in varied patient response to specific therapies, and individual mutations may not be present in the majority of primary GBM patients.4−7 Therefore, targeting how these brain tumor cells behave in the context of their mutations or environment may elucidate avenues for more promising therapies that have a wider reach within the GBM patient population.8
One of the main routes of GBM infiltration is cellular invasion within the tight spaces of the brain parenchyma. While the main neoplasm is targeted for surgical resection, migratory cells can remain undetected by MRI and form satellite tumors away from the original mass. The avenues by which these motile cells travel are white matter tracts and blood vessels, with the perivascular space presenting little physical resistance.9,10 Previous work using a mouse model intracranially injected with GBM cells demonstrated human tumor cell tropism along the mouse brain vasculature, and after 2–4 weeks of implantation, GBM cells caused microbreaches at the BBB.11 Furthermore, another GBM rodent model demonstrated the Wnt signaling cascade is necessary for glioma-vessel interactions.12 However, in vivo mouse models require the injection of thousands of cells, use of immune-compromised animals, and performing advanced surgical and imaging techniques. The emergence of an in vivo model that could more easily visualize and manipulate perivascular interactions important for tumor invasion would greatly benefit the field of GBM biology. In recent years, groups have established zebrafish (Danio rerio) specifically modeling the growth and invasion of brain tumors.13−15 These studies take different approaches, such as injecting brain tumor cells into the yolk of the animal or a large bolus into the brain to study invasion and tumor angiogenesis.14,16,17 While these zebrafish models provide exciting avenues to study human disease in a simpler vertebrate system, most have looked at the molecular features of tumors and not the mechanism of perivascular glioma invasion. Therefore, a more thorough characterization of the relationship between glioma cells with the zebrafish brain vasculature remains.
In the present study, we hypothesized that zebrafish could be utilized to specifically model perivascular glioma invasion. To test and visualize this hypothesis, we intracranially injected red fluorescently labeled human glioma cells into the translucent zebrafish vascular reporter line Tg(fli1a:eGFP)y1;casper, whereby all blood vessels are labeled with enhanced green fluorescent protein (eGFP). We injected both an adult glioma cell line (D54-MG) and a pediatric patient-derived xenoline (D2159MG) into Tg(fli1a:eGFP)y1;casper zebrafish and witnessed both of these glioma lines attach to the zebrafish vasculature. Gliomas retain salient disease features in the zebrafish brain such as attachment to secondary structures (blood vessels). Injected animals were imaged daily, and confocal microscopy revealed glioma cells survive and expand within the zebrafish brain. Perivascular glioma invasion was dependent on the number of tumor cells injected. When ≤25 cells were initially injected, tumors survived, grew, and had more contacts with the vasculature than if an animal was implanted with >25 cells. Tumor cell vessel co-option increased over time along the vessel and at vessel branch points. Glioma-vessel interactions were also dependent on the CNS microenvironment, as glioma cells injected into the animal’s peripheral tissue failed to interact with pre-existing vasculature. Furthermore, since prior studies have alluded to Wnt signaling being important in vessel co-option in a rodent glioma model, we examined whether requirement for Wnt signaling is conserved in zebrafish.12 We utilized the small molecule XAV939, an inhibitor of the Wnt signaling pathway, to pharmacologically disrupt glioma cell-vascular interactions in the zebrafish brain as it has been demonstrated in the mammalian brain. These proof-of-principle studies demonstrate valuable features of this established zebrafish model of perivascular glioma invasion, which could lead to the discovery of newly needed GBM therapeutics.
Methods
Cell Culture
D54-MG human glioma adherent cell lines (WHO IV, glioblastoma multiforme; gifted by Dr. D. Bigner, Duke University, Durham, NC) were genetically modified to express tdTomato or eGFP in a previous study.11 D54-MG cells were maintained in Dulbecco’s modified Eagle’s Media (DMEM) (Fisher #11320082), supplemented with 7% fetal bovine serum (Lot 105313, Aleken), and kept in a cell culture incubator with 10% CO2 at 37 °C
D2159MG and GBM22 patient-derived xenograft (PDX) lines (gifted by Darell D. Bigner, MD, Ph.D. and Stephen T. Keir, DrPH, MPH, Duke University, Tisch Brain Tumor Center and Dr. Yancey Gillespie, University of Alabama at Birmingham, Birmingham, AL, USA, respectively) were from primary brain tissue and maintained by serial passage in the flank of athymic nude mice, as previously described.11 PDX cells were grown in a DMEM complete media supplemented with B-27 (Fisher #12587010), amphotericin (1:100; Fisher #BP264550), sodium pyruvate (1:100; ThermoFisher #11360-070), EGF and FGF growth factors (10 ng/mL each, final; ThermoFisher #PHG0266 and #PHG0315), and gentamicin (1:1,000; Fisher #BW17 518Z). PDX cells were maintained in a cell culture incubator with 10% CO2 at 37 °C. The D2159MG pediatric glioma cells were genetically modified with a lentivirus (MGH Vector Core) to constitutively express mCherry. To visualize unlabeled GBM22 cells, cells were labeled overnight with 5 μM of the lipophilic DiL dye (Invitrogen #V22885).
For cell counting, D54-MG cells were dissociated using Trypsin-EDTA (ThermoFisher/LifeTech #25300054), and PDX cells were dissociated using Accutase (Sigma #A6964). Cells were counted on a hemocytometer and resuspended at 25,000 cells per μL in sterile PBS prior to microinjection.
Acute Mouse Brain Slice Invasion and Immunohistochemistry (IHC)
Experiments were performed in accordance with the University of Alabama, Birmingham Institutional Animal Care and Use Committee (IACUC). In situ tumor invasion was performed as previously described.18 For IHC, 300 μM brain sections were acutely sliced from 10-week-old NG2:dsRed mice and seeded with 80,000 D54-MG-eGFP tumor cells on multiwell filter chambers (Fisher #08-771). Tumor cells invaded brain slices for 4 h on filter chambers in a 10% CO2 cell culture incubator. Brain slices were fixed in 4% PFA and used for subsequent IHC. Slices were rinsed in PBST for 15 min, permeated with proteinase K (20 mg/mL stock, 1:800) for 15 min at room temperature, and then blocked in 10% goat serum for 1 h at room temperature. Sections were stained with a mouse-anti-Laminin primary antibody (Sigma L8271, 1:500) overnight at 4 °C. The next morning, slices were washed in PBST for 1.5 h, stained with donkey-antimouse-Alexa 555 secondary antibody (Invitrogen A31570,1:500) for 3 h at room temperature, washed in PBST for 1.5 h, and mounted with Fluoromount (VWR 100502-406) on glass slides. Z-stacks were acquired with an Olympus Fluoview FV1000 confocal microscope and a 60X(NA1.42) oil objective.
Zebrafish
Adult zebrafish were maintained according to Virginia Tech IACUC guidelines on a 14/10-h light/dark cycle in a Tecniplast system at 28.5 °C, pH 7.0, and 1000 conductivity. The Tg(fli1a:eGFP)y1 line was crossed onto the casper background19 to generate genetically translucent vascular reporter lines. These animals were specific-pathogen free (SPF) and came from the Sinnhuber Aquatic Research Laboratory at Oregon State University. SPF animals were used for these brain tumor studies to avoid confounding factors such as Pseudoloma neuropihila, a pathogen that resides in the zebrafish CNS and is common in zebrafish colonies. The Tg(glut1b:mCherry) line was obtained from Dr. Michael Taylor’s laboratory at the University of Wisconsin-Madison. For all experiments, embryos were collected from multiple pairwise crosses. Embryos were maintained in embryo water (1.5 g of Instant Ocean salt per 5 L of RO water) at 28.5 °C. For studies with the Tg(glut1b:mCherry) line, 24-h postfertilization embryos were treated with 200 μM N-phenylthiourea (Sigma # P7629) to prevent melanocyte formation. Embryos for tumor cell injections were gradually acclimated to develop at 32 °C, so as to accommodate a more desirable temperature for glioma cell growth.
Tumor Microinjections
Healthy animals (i.e., no developmental abnormalities) were screened before tumor implantation. Three-days postfertilization (dpf) Tg(fli1a:eGFP)y1;casper or Tg(glut1b:mCherry) larvae were anesthetized with 0.04% MS-222 (Sigma #A5040) prior to microinjection. During anesthesia, thin wall glass capillary needles with filament (World Precision Instruments #TW150F-4) were pulled with program 4 “Pro-Nuclear Injection” (Heat: 460, Pull:90, Vel: 70, Delay: 70, Pressure: 200, Ramp: 485) on a horizontal pipet puller (Sutter, Model P-1000). This program creates needles with a 0.7 μM tip and a taper of 6–7 mm. One microliter of Phenol Red (Sigma #P0290) was added to 9 μL of the 25,000 glioma cell/μL solution mixture. Capillaries were positioned in a round glass pipet holder and back loaded with 1 μL of the human glioma cell mixture. Each loaded needle was calibrated using a pneumatic pump and micrometer (Carolina Biological Supply #591430) to calculate the bolus and corresponding cell number per injection. Animals were positioned dorsal side up for intracranial injection or on their sides for trunk injection in a homemade agarose injection mold and then microinjected with the desired amount of 25 cells. While we attempted to only implant 25–50 cells (1 nL) per animal, microinjections are not trivial, and slight pressure differences between animals could lead to more or less cells implanted. After injections, zebrafish were tracked in individual wells of a 24-well plate. After tumor microinjection, animals recovered for at least 1 h, were anesthetized, and then embedded for live confocal microscopy to assess initial tumor volume.
Live Confocal Microscopy
Animals were removed from multiwell plates with a glass Pasteur pipet and anesthetized with 0.04% MS-222 in individual 35 mm Petri dishes containing a 14 mm glass coverslip (MatTek Corporation #P35G-1.5-14-C). A 1.2% low melting point agarose (ThermoFisher #16520050) solution was made in embryo water for embedding. MS-222 was removed from the animal, and a bolus of warm agarose the size of the coverslip was added to the animal/cover glass. A dissection probe was used to align animals either dorsal side down for brain imaging or sagittally for trunk imaging. After the agarose solidified, the Petri dish was filled with 0.04% MS-222 to keep the animal immobilized, and the dish was wrapped with parafilm along the edges. Live confocal microscopy was performed to take z-stacks at the optimal section thickness of animals with D54-MG and D2159MG cells with an Olympus Fluoview FV1000 confocal microscope and a 40X(NA0.75) objective. The Nyquist optimal section thickness value for z-stacks was calculated through an algorithm in the Olympus Fluoview software for all images. For whole zebrafish brain imaging after GBM22 injections, a Nikon A1R confocal microscope with a 10X(NA 0.45) objective and a 1.5 optical zoom was utilized. Because these microscopes are upright, the glass coverslip bottom dishes containing the animals were flipped upside down so that the glass side made contact with the objective. Supplemental Movie 1 was generated from a Maximum Intensity Projection Image in NIS-Elements software.
Small Molecule Treatment
After initial tumor implantation at 3dpf and subsequent imaging, tumors grew overnight before animals were unbiasedly sorted into treatment groups at 4dpf. The Wnt signaling inhibitor, XAV939, (Cayman Chemical #13596) was solubilized in dimethyl sulfoxide (DMSO) to generate 10 mM stock solutions and used at a final concentration of 30 μM in zebrafish egg water.
Glioma-Vessel Interaction Analysis
To assess the percentage of human glioma cells interacting with the vasculature, OIB z-stack files were analyzed in the Fluoview software. In brief, the total number of visible human glioma cells was initially counted. The orthogonal view option was used to detect whether the signal from a cell was associated with the signal from a blood vessel or blood vessel branch point as seen in both XZ and YZ planes. The total number of interacting cells on a blood vessel or cells specifically at a vessel branch point was counted, and then this value was divided by the total number of cells present at that point in time.
Tg(glut1b:mCherry) Volumetric Analysis
Fiji-ImageJ and NIS-Elements software were utilized to measure the volume and signal intensity of glut1 in tumor-associated and nontumor-associated vessels. In brief, confocal z-stacks were opened as split channels in FIJI, and tiff files were saved to import into NIS-Elements. After importing, channels were merged together, the document was calibrated, and files were saved as an ND2 file. Sections around vessels of interest were cropped out of the whole file, and a line was drawn along a vessel where it was ensheathed by a tumor cell body. The rotating rectangle feature was utilized, and this selected area underwent a binary threshold and 1X clean and smooth options. The same procedure and size line was drawn along a similarly placed vessel without a tumor cell in the contralateral brain as a paired control vessel, to measure the volume and intensity in a similar manner. Measurements were generated in the NIS Elements “3D Object Measurements” table.
Statistical Analysis
For analysis of perivascular glioma invasion at developmental time points 3–7dpf, a one-way ANOVA with multiple comparisons test was performed using GraphPad Prism software to determine p-values. For small molecule treatment experiments, a two-tailed, unpaired Student’s t-test was performed using GraphPad Prism software to determine p-values. For volumetric vessel and glut1 fluorescence analyses, a two-tailed, paired Student’s t-test was performed using GraphPad Prism software to determine p-values.
Results
Human Tumors Retain Salient Features in a Cell Number-Dependent Manner in the Zebrafish Brain
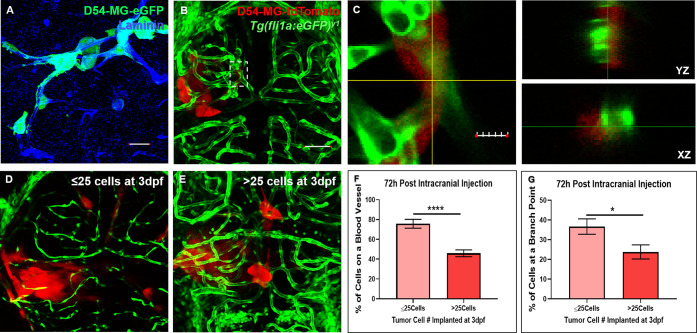
While traditional glioma models utilize rodents, studies have now demonstrated zebrafish are a suitable model to elucidate CNS cancer biology.16,17,20 As a means to ask whether glioma-vessel interactions may occur in the zebrafish brain, we opted to inject fewer cells than previously published. Animals from the optically translucent Tg(fli1a:eGFP)y1;casper vascular reporter line were injected with 25–50 human D54-MG-tdTomato cells at 3dpf, a time point when the BBB has started forming and sufficient CNS angiogenesis has occurred.21 Similar to the behavior of the D54-MG tumor line in acute mouse brain slices (Figure 1A), D54-MG glioma cells attached to the zebrafish brain vasculature within 24-h postintracranial injection (Figure 1B,C). We also performed microinjection into non-SPF animals and witnessed glioma-vascular interactions (Supplemental Figure 1), suggesting that while maintaining a SPF colony is the best practice for studying diseases of the CNS, it does not affect the growth of human brain cancer cells with regard to the zebrafish brain vasculature. This data suggests conserved signaling mechanisms exist to promote the interaction between glioma cells and the developing zebrafish vessel network.
Figure 1.
Glioma cells retain salient features in the zebrafish brain. (A) A maximum intensity projection of acute slice invasion with mouse brain seeded with 80,000 D54-MG-eGFP tumor cells. After 4 h of invasion, slices were fixed and stained for the vessel protein Laminin (blue) to visualize the close association of glioma cells (green) during perivascular glioma invasion in this acute ex vivo model. Scale bar = 25 μm. (B) The transgenic zebrafish vascular reporter line Tg(fli1a:eGFP)y1;casper (green) was intracranially injected with 25–50 D54-MG-tdTomato glioma cells (red) at the midbrain-hindbrain boundary at 3-days postfertilization (dpf). At 24-h postinjection, a maximum intensity projection of an injected animal reveals tumor-vessel associations as seen in the acute slice model. Scale bar = 50 μm. (C) An area (dotted white box in B) of a representative Z-plane from the confocal stack showing the corresponding orthogonal planes between the vessel (green) and glioma cell (red) signals. Scale bar 10 μm. (D,E) Representative confocal images of 6dpf animals initially injected with ≤25 cells (D) or >25 cells. (F,G) Representative live confocal images of Tg(fli1:eGFP)y1 larvae (red), 72-h postinjection with ≤25 cells (D) or >25 cells (E). (F,G) Quantification of tumor-vessel interactions after 72-h postinjection, comparing animals implanted on 3dpf with either ≤25 cells or >25 cells. Data shows tumor-vessel interactions along the vessel wall (F) or at a branch point. (G) A two-tailed, unpaired Student’s t test was performed. Error bars represent mean with standard error of the mean (**p = <0.01), *p = <0.05). (n = 16–21 animals).
To decipher whether glioma-vascular interactions depended on the volume of implanted cells, we intracranially injected a bolus of 25–50 and ≥50 glioma cells in 3dpf Tg(fli1a:eGFP)y1;casper animals. We observed that a larger bolus of ≥50 cells became more easily trapped within the ventricular space (data not shown), which is superficial to the developing zebrafish brain and therefore why we decided to perform the remaining experiments with lower injection volumes. These >50 cell compacted tumors were reminiscent of previously published zebrafish glioma studies that, due to cell number and size of the zebrafish brain versus human cells, could not examine the specific behaviors associated with perivascular glioma invasion.16 However, when 25–50 cells were injected into the midbrain-hindbrain boundary at 3dpf, we witnessed frequent extension of fingerlike processes as previously observed in GBM mouse models and human brain.11 While present, >25 cell injected animals had fewer tumor-vessels interactions, along the vessel wall or at a branchpoint, within a 72-h timespan compared to animals implanted with ≤25 cells at 3dpf (Figure 1D–G). While survival studies were not in the scope of this study, some animals would succumb to the disease 4-days postinjection (data not shown). Previous studies reported a median survival time of 10-days postinjection in zebrafish with glioma xenografts of the preferred cell number (∼25–50 cells) used in our study.14 Taken together, these experiments reveal that salient features of GBM and the process of perivascular invasion both occur in the developing brain of our zebrafish glioma model.
Glioma Cells Actively Invade the Brain along the Zebrafish Vasculature
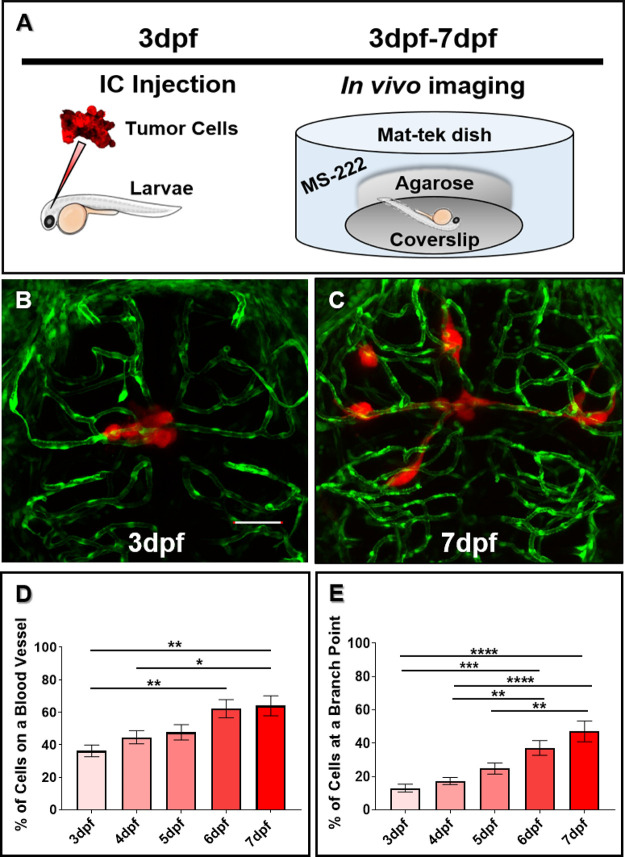
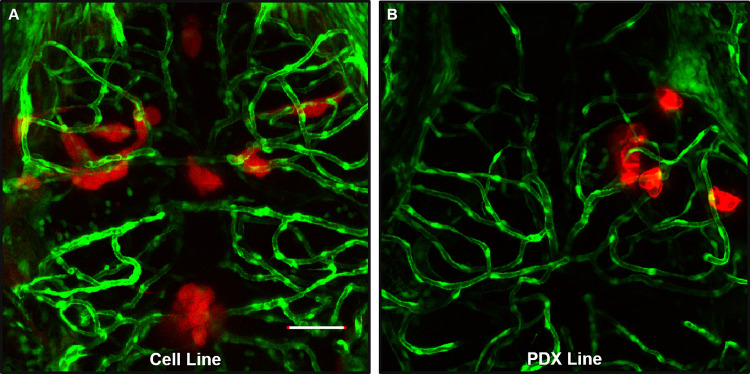
To analyze the dynamics of perivascular glioma invasion in the developing zebrafish brain, we microinjected D54-MG-tdTomato tumor cells into the midbrain-hindbrain boundary of 3dpf Tg(fli1a:eGFP)y1;casper larvae and monitored tumor behavior until 7dpf (Figure 2A). Because daily live imaging is easy to perform with the zebrafish model system, we wanted to assess tumor burden after the onset of injection and subsequent days. As shown by representative images in Figure 2, the few D54-MG-tdTomato glioma cells implanted at 3dpf (Figure 2B) not only survived in the zebrafish brain but also expanded and migrated along the vascular network by 7dpf (Figure 2C). These glioma-vessel interactions increased over time along the main vessel wall (Figure 2D) and at branch points (Figure 2E) as previously described in a GBM mouse model.11 Furthermore, these interactions occurred with both pediatric and adult glioma cells, as well as in adherent and PDX cell lines (Figure 3). With a variety of PDX lines at our disposal, we labeled the GBM22 PDX line with a lipophilic dye to visualize tumor cells postintracranial injection. Of note, lipophilic dye did not permeate the cell’s finer processes after injection and therefore does not retain the morphology that was visualized with D54-MG-tdTomato cells (Supplemental Figure 2). Therefore, it was determined that tumor cells that are transgenically labeled to express a fluorophore are more suitable for these imaging studies. These results demonstrate that even though glioma injections occurred in the developing brain, conserved signaling pathways support perivascular glioma invasion over time in our zebrafish model.
Figure 2.
Glioma cells actively associate with the developing zebrafish brain vasculature over time. (A) A cartoon schematic of intracranial (IC) injection and subsequent in vivo imaging of our zebrafish perivascular glioma model. Tg(fli1a:eGFP)y1;casper animals were injected with 25–50 glioma cells in the midbrain-hindbrain boundary at 3dpf. After recovering, animals were imaged every day until 7dpf to monitor tumor burden over time. (B,C) Representative maximum intensity projection confocal images of tumor cells implanted at 3dpf (B) and subsequent migration by 7dpf (C). Scale bar = 50 μm. (D,E) Quantification of tumor-vessel interactions in the developing brain. (D) Quantification of D54-MG-tdTomato cells attached to blood vessels over time (**p < 0.001, *p = <0.05). (E) Quantification of D54-MG-tdTomato cells at vessel branch points over time (****p = <0.0001, ***p = <0.001, **p = <0.01). n = 18–21 animals per time point. A one-way ANOVA with Tukey’s multiple comparisons test was performed. Error bars represent mean with standard error of the mean.
Figure 3.
Glioma-vascular interactions occur in the zebrafish brain with various GBM lines. An adult D54-MG-tdTomato cell line (A, red) and D2159MG pediatric xenoline (B, red) were intracranially implanted in 3dpf Tg(fli1a:eGFP)y1 larvae. Live confocal imaging of animals at 72-h postinjection reveals GBM growth and development in the larval zebrafish brain. Scale bar = 50 μm.
Tumor Microenvironment Affects Perivascular Glioma Invasion in Vivo
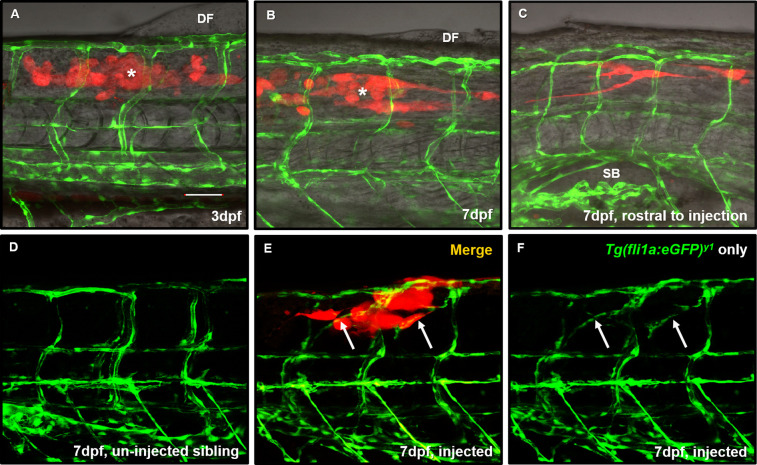
Cancer arises when normal cells within an organ become misprogrammed. Prior studies have injected non-CNS cancer cells into the periphery of the zebrafish to assess tumor associations with blood vessels.22,23 Therefore, it is logical to inject tumor cells in a tissue in which those cells naturally thrive. To validate the importance of the microenvironment in our perivascular GBM invasion model, we injected D54-MG-tdTomato cells in the trunk near the dorsal fin of 3dpf Tg(fli1a:eGFP)y1;casper animals. Surprisingly, these glioma cells not only survived but also radically moved rostrally away from the dorsal fin area and toward the brain by 7dpf (Figure 4A–C). These cells extended long projections during invasion (Supplemental Movie 1), and unlike when in the brain, these cells did not attach to nonlymphatic, pre-existing blood vessels. This data suggests there are unique trophic factors within the brain environment that attract glioma cells to the zebrafish vascular network. Interestingly, we observed instances of tumor-induced angiogenesis in 60% of injected animals (n = 5 animals), a biology witnessed in other zebrafish cancer models (Figure 4E,F).22,24 These experiments highlight the importance of the microenvironment when modeling and quantifying glioma-vessel interactions.
Figure 4.
Glioma cells do not migrate along pre-existing peripheral vessels but do evoke non-CNS angiogenesis. (A) Representative maximum intensity projection image of a 3dpf Tg(fli1a:eGFP)y1;casper animal (green) injected with 25–50 D54-MGtdTomato cells (red) into the trunk of the muscle (white asterisk) close to the dorsal fin (DF) to survey whether GBM would interact with non-CNS vasculature. (B) The same animal was imaged at 7dpf to survey tumor cell survival and movement from the point of injection (white asterisk). (C) Rostral from the point of injection in the same animal from (B). Tumor cells moved down the trunk toward the brain and appear near the swim bladder (SB). n = 9 animals. (D) A representative image of trunk intersegmental vessel patterning in a 7dpf uninjected Tg(fli1a:eGFP)y1;casper sibling. (E) We noted instances where tumor cells (red) reaching outward also ensheathed new blood vessels (E,F arrows) that were created after tumor implantation, not seen in uninjected sibling controls. Scale bar = 50 μm.
Gross Vessel Morphology and Glut1 Expression Is Not Altered after Tumor Invasion
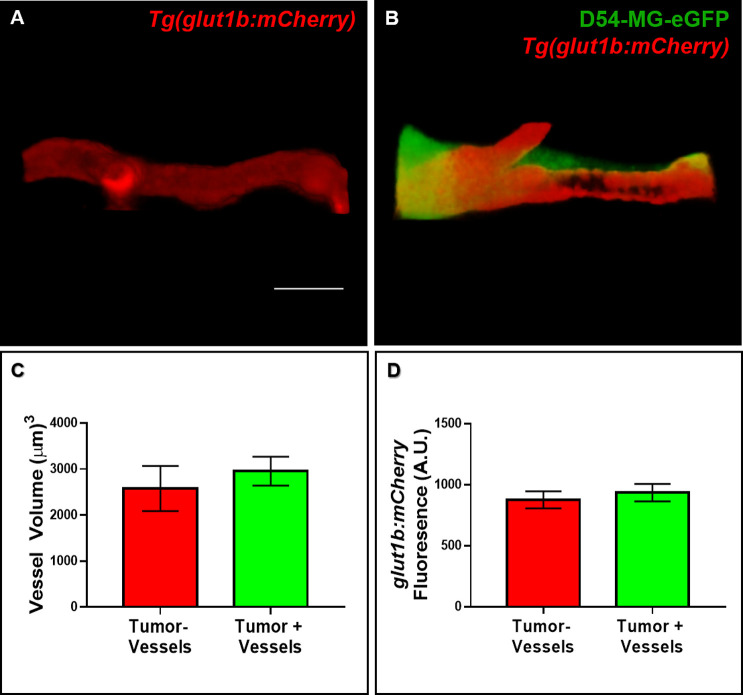
After determining that tumor cells failed to attach to pre-existing, non-CNS vasculature, we wanted to further investigate other parameters postinvasion. Due to the reports of vascular tone alterations and BBB disruption in both a GBM mouse model and in patient tissue, we assessed the zebrafish brain vascular morphology after 4 days of glioma invasion.11,25 We first asked whether vessel volume was changed in our zebrafish model of perivascular glioma invasion. We utilized volumetric analyses of vessels without tumor burden and vessels on the contralateral brain region contacted by D54-MG-eGFP tumor cells (Figure 5A,B). While tumor cells readily ensheathed vessels in the zebrafish brain, we did not find any difference in vessel volume 4-days postinvasion (Figure 5C). As the Tg(glut1b:mCherry) line expresses mCherry under the control of a reliable BBB marker, glut1, we next analyzed the mCherry fluorescence signal of tumor and nontumor associated vessels in 7dpf animals.21 Data from fluorescence intensity analysis revealed that glut1 expression did not differ between vessels co-opted by a tumor cell soma and vessels in a similar location on the contralateral side of the brain (Figure 5D). These live imaging results suggest that co-opted vessels are not visibly compromised, and the BBB specific marker glut1 was not altered after this 4-day time frame of tumor cell invasion.
Figure 5.
Vessel volume and glut1 expression are not altered after acute GBM invasion. (A,B) Representative 3D rendered volume views of a blood vessel from the contralateral brain (A) in a region similar to a vessel co-opted by a D54-MG-eGFP tumor cell in a 7dpf zebrafish brain (B). (C,D) The volume and signal intensity of glut1 in tumor-associated and nontumor associated vessels were measured in 3D rendered volume views. (C) Quantification of vessel volume. A two-tailed, paired Student’s t test was performed. (D) Quantification of glut1b:mCherry fluorescence. A two-tailed, paired Student’s t test was performed. Error bars represent mean with standard error of the mean. n = 8 animals total, 1–3 vessels per animal per group. Scale bar = 10 μm.
Glioma-Vessel Interactions Can Be Pharmacologically Inhibited in Vivo
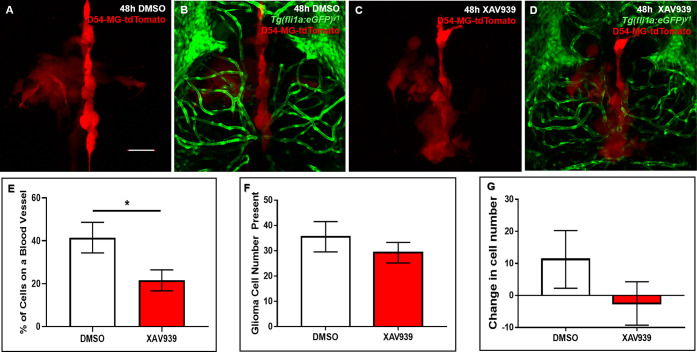
In order to demonstrate whether glioma-vessel interactions could be pharmacologically disrupted in vivo, we sought to perform proof-of-principle experiments targeting pathways important for glioma-vessel interactions in GBM mouse models, such as Wnt signaling.12 Therefore, we hypothesized that pathways important for mitigating tumor-vessel biology in rodent models would be conserved during perivascular glioma invasion in zebrafish brain.12 We hypothesized that adding a small molecule inhibitor of Wnt signaling (XAV939) to the water during tumor invasion would result in fewer cellular interactions in the zebrafish brain. To test this hypothesis, 4dpf animals were blindly sorted into the DMSO vehicle control or XAV939 treatment groups 24 h after tumor cell implantation. After 48 h of bath application, confocal images revealed significantly fewer glioma-vessel interactions (21.57% ± 4.908) in XAV939 treated animals compared to control treated animals (41.5% ± 7.106) (Figure 6E). Important to note, the total number of cells was not significantly different after 48 h of either treatment (Figure 6F). While there was a change in cell number after 48 h of XAV939 treatment (−2.5 ± 6.786, n = 6) compared to the DMSO vehicle control (11.25 ± 9.013, n = 4), this was not statistically significant (Figure 6G). These studies suggest that conserved signaling pathways in mammals attract glioma cells to the zebrafish brain vasculature, and this in vivo system is amenable to chemical perturbation.
Figure 6.
Pharmacological inhibition of Wnt signaling diminishes tumor-vessel interactions. (A–D) Representative maximum intensity projections of tumor burden (red) in the vehicle control (A,B) or 30 μM XAV939 treated (C,D) animals. Animals were injected with D54-MG-tdTomato cells at 3dpf, unbiasedly sorted, and treated between 4 and 6dpf. (E) Quantification of tumor-vessel interactions after 48 h of the vehicle control (DMSO) or 30 μM XAV939 treatment. A two-tailed, unpaired t test was performed (p = <0.05). n = 6 animals per group. Error bars represent mean with standard error of the mean. (F) Quantification of total glioma cells resulting after 48 h of the vehicle control (DMSO) or 30 μM XAV939 treatment. A two-tailed, unpaired t test was performed. n = 7–8 animals per group. Error bars represent mean with standard error of the mean. (G) Quantification of the change in tumor cell number after 48 h of the vehicle control (DMSO) or 30 μM XAV939 treatment. A two-tailed, unpaired t test was performed. n = 4–6 animals per group. Error bars represent mean with standard error of the mean. Scale bar = 50 μm.
Discussion
GBM is a fatal primary brain tumor studied in an extensive array of models. More recently, research groups have implemented xenograft studies to assess glioma invasion in the developing zebrafish. Zebrafish are genetically tractable, share a similar body plan and genetic conservation to mammals, and are optically transparent. The zebrafish does not develop a functional adaptive immune system until 4 weeks of age and, therefore, circumvents the need for requiring immunosuppressive chemicals or special animals for injection at early time points.26 The adult mouse brain contains roughly 700 times more neuronal cells than the larval zebrafish brain, thus fewer tumor cells need to be injected to visualize similar phenotypes.27,28 These features have presented zebrafish to cancer scientists as an attractive model for dissecting tumor biology and finding new therapeutics.16,29
As the zebrafish model has emerged as a suitable system for dissecting cancer biology, such studies have dissected the processes of tumor invasion and angiogenesis.15,23 However, tumor cells are commonly loaded with lipophilic dyes, which, compared to genetic expression of fluorescent proteins, are not as effective for thoroughly labeling cells and their finer processes for imaging studies.16,24 As it is now widely recognized in the field of GBM biology, gliomas are highly motile tumors known for rapid proliferation and migration. Establishing the biology of perivascular glioma invasion in the zebrafish model would open therapeutic screening possibilities toward a pathological tumor behavior commonly seen in a majority of GBM patients.
Our injection studies reveal the active process of perivascular glioma invasion along the zebrafish CNS vasculature in vivo. These observations are exciting for two reasons. First, we demonstrated that important glioma biology could be visualized in zebrafish with only 25–50 cells in 4 days. These experiments utilized significantly fewer cells in a fraction of the time compared to traditional mouse models.11,12 Second, a conserved biology must support the attraction of human cells to the developing zebrafish brain vascular network. One of the most critical elements in establishing a model of perivascular glioma invasion is cell number. Other research groups inject widely varied numbers of tumor cells, ranging from 50 to hundreds of tumor cells. We found that fewer cells injected allowed the tumors to migrate. While an individual glioma cell can shrink up to 30% of its volume to invade the mammalian brain, injecting fewer cells likely provides the glioma cells with more room to not only grow but also invade in a smaller space.18 By injecting only 25–50 cells, we observed glioma cells associating with the zebrafish brain vasculature within 24-h postimplantation. This denotes a salient feature of GBM biology: tumor cell attachment to secondary structures during invasion. We also witnessed brain necrosis in animals injected with >50 cells, as well as animals succumbing to their tumor burden (data not shown). Most interestingly, perivascular glioma invasion increased over time, from 3 to 7dpf. After only 4 days of growth, we witnessed a similar percentage of tumor-vessel interactions as previously demonstrated in a GBM mouse model that also utilized implantation of D54-MG cells.11 Furthermore, we observed tumor-vessel interactions with adherent cell lines and patient-derived xenolines. These experiments highlight the model’s accessibility due to what resources may be available to a research group, as PDX lines are time and resource intensive to maintain. Of note, our study is one of the few to utilize pediatric xenoline cells and demonstrate that tumor cell-vessel interactions arise in the context of pediatric CNS cancer in vivo.
The extracellular milieu plays a major role during tissue development and for cellular interactions. The unique composition of the brain space is also remodeled in the context of brain tumor progression.30 When assessing the possibility of perivascular glioma invasion in the periphery of the zebrafish, we did not observe glioma-vessel interactions with the pre-existing vasculature (Figure 4). While others have reported glioma-induced angiogenesis after tumor implantation in the zebrafish brain, little tumor-induced angiogenesis occurred after intracranial injections, possibly due to the lack of large tumors that require more nutrients or the presence of vascular endothelial growth factor (VEGF) already in the developing zebrafish brain during active CNS angiogenesis.16,31 Moreover, with distinct extracellular matrix features, such as the selective BBB and a functional lymphatic system, the brain creates an exclusive environment for cancer growth.32
While it is common for other cancers to metastasize into the brain, it is possible that specific tropism to the BBB and the surrounding microenvironment prevents CNS tumors from hematagenously spreading into the periphery. For example, studies have demonstrated how astrocytes produce connexin 43 mediated tumor-stromal interactions and how neuronal activity promotes cancer cell proliferation through neuroligin-3 upregulation.33,34 Glioma cells are known to release VEGF during tumor growth.35 Because the intersegmental vessels (ISVs) are already patterned by the time we injected cells, it is possible new vessels formed due to tumor cell secreted VEGF. Researchers have witnessed angiogenesis toward non-CNS tumor cells implanted in the zebrafish, a common process that provides a tumor mass with nutrients necessary to maintain uncontrolled cell growth.23 Furthermore, in the context of CNS tumors, the BBB vasculature is specialized under a different set of signals than the vasculature within the periphery in both rodents and zebrafish.36−39 Interestingly, recent studies injecting non-CNS tumor cells into circulation of the developing zebrafish demonstrated these cancers attached to ISVs unlike our brain tumor cells.40 Therefore, the process of angiogenesis (the formation of new blood vessels) and perivascular invasion (attraction of cells to pre-existing vessels for cell migration) underscores different biology in GBM. While these studies support the examination of brain tumors within their tissue of origin, the zebrafish could provide an interesting model to dissect brain tumor preferences for the BBB versus peripheral vasculature attachment. Additionally, such experiments would be easily achievable with the zebrafish model system.
As tumor cells secrete a myriad of destructive molecules during invasion, a theoretical side effect of perivascular glioma invasion is disruption of the vasculature’s integrity.41 GBM models and morphological analysis of patient tissue have demonstrated a change in vessel response and BBB breach via the loss of tight junction proteins.11,25 While the BBB was not a direct focus of this study, we looked at a few potential effects of perivascular glioma invasion on the zebrafish CNS vasculature. Zebrafish possess the chemical and physical barriers necessary to make a functional BBB early in development, also making it easy to survey in our model of perivascular glioma invasion.21,42 We found that vessel volume and BBB integrity via expression of glut1 were not altered in our zebrafish model of perivascular glioma invasion. Our glut1 data aligns with a study of xenograft models and patient tissue demonstrating normal expression of GLUT1 at vessels co-opted by tumor cells.43 While previous studies showed a decrease in tight junction protein expression, it would be an interesting focus to use this zebrafish model to investigate if changes in BBB proteins occur before or after BBB dysfunction during perivascular tumor invasion.
One of the ultimate goals of establishing a zebrafish model of perivascular glioma invasion was to demonstrate the feasibility of using small molecules that could target mechanisms necessary for glioma-vessel interactions. The zebrafish is an ideal organism for in vivo cancer drug discovery, as animals can be successfully xenografted with tumors in a short time span and are amenable to high-throughput chemical assays.44 In fact, zebrafish “avatars” are becoming successful platforms for drugs to make it into clinical trials.45 In the field of GBM biology, most studies assess overall tumor cell growth, but few have investigated means for the chemical disruption of glioma-vessel communication. Previous research has begun to dissect pathways involved in perivascular glioma invasion mouse models, specifically through Wnt and Bradykinin signaling.12,46 Zebrafish express homologous proteins for both the Wnt and Bradykinin pathways, but the function of these homologs in zebrafish cancer models remains unexplored.47,48 We demonstrate that tumor-vessel interactions in the zebrafish brain can be disrupted by the Wnt signaling antagonist XAV939, supporting the potential of BBB-specific cues that mediate perivascular glioma invasion.37 Wnt signaling inhibition was not novel in the context of glioma biology but was in regard to our zebrafish model of tumor invasion. As the Wnt/beta-catenin signaling cascade is targeted in clinical trials, the use of XAV939 in our zebrafish tumor xenograft model highlights the suitability of the zebrafish in the preclinical pipeline.49 These proof-of-principle small molecule experiments demonstrate the practicality of using this zebrafish model to identify compounds of interest that can target processes important for glioma invasion. Determining the underlying behavior that supports tumor growth would cultivate novel therapies, as migratory cell populations evade the current GBM standard of care. Therefore, this established zebrafish model of perivascular glioma invasion generates an additional platform for dissecting tumor cell biology and progressing cancer drug discovery.
Acknowledgments
The authors would like to acknowledge and thank Joelle Martin and AnnaLin Woo for constructive criticism of the manuscript, Tré Mills for assistance with the volumetric analysis protocol, Dr. Emily Thompson for transducing the D2159MG xenoline for mCherry expression, the Pan laboratory for use of their stereoscope and intellectual discussions, and Dr. Michael Taylor for use of the Tg(glut1b:mCherry) line. This research was supported by funding from the National Institutes of Health (R01CA227149 and R01NS082851) and Fralin Biomedical Research Institute at VTC operational funds (234859 and 175999) to H.W.S. A Virginia Tech ACC Creativity and Innovation Grant funded work for M.T.K.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00129.
Glioma invasion in a nonspecific pathogen free transgenic line, Tg(glut1b:mCherry), and lipophilic dye labeled glioma xenoline implanted into the Tg(fli1a:eGFP)y1 zebrafish brain (PDF)
3D-rendered movie of glioma cells in the periphery of a 10-day postfertilization Tg(fli1a:eGFP)y1 zebrafish larvae (MP4)
Author Contributions
R.A.U. and H.S. designed experiments, R.A.U., M.T.K., and C.P. analyzed data, R.A.U., M.T.K., and C.P. conducted experiments, and R.A.U. and H.S. wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Ostrom Q. T.; Gittleman H.; Liao P.; Vecchione-Koval T.; Wolinsky Y.; Kruchko C.; Barnholtz-Sloan J. S. (2017) CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol 19 (suppl_5), v1–v88. 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R.; Mason W. P.; van den Bent M. J.; Weller M.; Fisher B.; Taphoorn M. J.; Belanger K.; Brandes A. A.; Marosi C.; Bogdahn U.; Curschmann J.; Janzer R. C.; Ludwin S. K.; Gorlia T.; Allgeier A.; Lacombe D.; Cairncross J. G.; Eisenhauer E.; Mirimanoff R. O.; (2005) Radiotherapy plus concomitant and adjuvant Temozolomide for glioblastoma. N. Engl. J. Med. 352 (10), 987–996. 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Feng E.; Sui C.; Wang T.; Sun G. (2017) Temozolomide with or without Radiotherapy in Patients with Newly Diagnosed Glioblastoma Multiforme: A Meta-Analysis. Eur. Neurol. 77 (3–4), 201–210. 10.1159/000455842. [DOI] [PubMed] [Google Scholar]
- Wang X.; You C.; Mao Q. (2013) Mutant TP53 enhances the resistance of glioblastoma cells to Temozolomide by up-regulating O(6)-methylguanine DNA-methyltransferase. Neurol Sci. 34, 1421–1428. 10.1007/s10072-012-1257-9. [DOI] [PubMed] [Google Scholar]
- Hegi M. E.; Diserens A.-C.; Gorlia T.; Hamou M.-F.; de Tribolet N.; Weller M.; Kros J. M.; Hainfellner J. A.; Mason W.; Mariani L.; Bromberg J. E. C.; Hau P.; Mirimanoff R. O.; Cairncross J. G.; Janzer R. C.; Stupp R. (2005) MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 352 (10), 997–1003. 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Mellinghoff I. K.; Wang M. Y.; Vivanco I.; Haas-Kogan D. A.; Zhu S.; Dia E. Q.; Lu K. V.; Yoshimoto K.; Huang J. H. Y.; Chute D. J.; Riggs B. L.; Horvath S.; Liau L. M.; Cavenee W. K.; Rao P. N.; Beroukhim R.; Peck T. C.; Lee J. C.; Sellers W. R.; Stokoe D.; Prados M.; Cloughesy T. F.; Sawyers C. L.; Mischel P. S. (2005) Molecular Determinants of the Response of Glioblastomas to EGFR Kinase Inhibitors. N. Engl. J. Med. 353 (19), 2012–2024. 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- McNeill R. S.; Stroobant E. E.; Smithberger E.; Canoutas D. A.; Butler M. K.; Shelton A. K.; Patel S. D.; Limas J. C.; Skinner K. R.; Bash R. E.; Schmid R. S.; Miller C. R. (2018) PIK3CA missense mutations promote glioblastoma pathogenesis, but do not enhance targeted PI3K inhibition. PLoS One 13 (7), e0200014. 10.1371/journal.pone.0200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umans R. A.; Sontheimer H. (2018) Combating malignant astrocytes: Strategies mitigating tumor invasion. Neurosci. Res. 126, 22–30. 10.1016/j.neures.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah V. A.; Robel S.; Watkins S.; Sontheimer H. (2014) A neurocentric perspective on glioma invasion. Nat. Rev. Neurosci. 15 (7), 455–465. 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin A.; Suzuki S. O.; Weiker M.; Goldman J. E.; Bruce J. N.; Canoll P. (2006) Transplanted glioma cells migrate and proliferate on host brain vasculature: A dynamic analysis. Glia 53 (8), 799–808. 10.1002/glia.20334. [DOI] [PubMed] [Google Scholar]
- Watkins S.; Robel S.; Kimbrough I. F.; Robert S. M.; Ellis-Davies G.; Sontheimer H. (2014) Disruption of astrocyte–vascular coupling and the blood–brain barrier by invading glioma cells. Nat. Commun. 5 (1), 4196. 10.1038/ncomms5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griveau A.; Seano G.; Shelton S. J.; Kupp R.; Jahangiri A.; Obernier K.; Krishnan S.; Lindberg O. R.; Yuen T. J.; Tien A.-C.; Sabo J. K.; Wang N.; Chen I.; Kloepper J.; Larrouquere L.; Ghosh M.; Tirosh I.; Huillard E.; Alvarez-Buylla A.; Oldham M. C.; Persson A. I.; Weiss W. A.; Batchelor T. T.; Stemmer-Rachamimov A.; Suvà M. L.; Phillips J. J.; Aghi M. K.; Mehta S.; Jain R. K.; Rowitch D. H. (2018) A Glial Signature and Wnt7 Signaling Regulate Glioma-Vascular Interactions and Tumor Microenvironment. Cancer Cell 33 (5), 874–889.e7. 10.1016/j.ccell.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittori M.; Motaln H.; Turnšek T. L. (2015) The Study of Glioma by Xenotransplantation in Zebrafish Early Life Stages. J. Histochem. Cytochem. 63 (10), 749–761. 10.1369/0022155415595670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker A. M.; Jaros B. D.; Puduvalli V. K.; Imitola J.; Kaur B.; Beattie C. E. (2016) Standardized orthotopic xenografts in zebrafish reveal glioma cell-line-specific characteristics and tumor cell heterogeneity. Dis. Models & Mech. 9 (2), 199. 10.1242/dmm.022921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S.; La Du J.; Tanguay R. L.; Greenwood J. A. (2012) Calpain 2 is required for the invasion of glioblastoma cells in the zebrafish brain microenvironment. J. Neurosci. Res. 90 (4), 769–781. 10.1002/jnr.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A.; Ye T.; Cao D.; Huang X.; Yang Y.; Chen X.; Xie Y.; Yao S.; Zhao C. (2017) Identify a Blood-Brain Barrier Penetrating Drug-TNB using Zebrafish Orthotopic Glioblastoma Xenograft Model. Sci. Rep. 7 (1), 14372. 10.1038/s41598-017-14766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-j.; Cui W.; Gu A.; Xu C.; Yu S.-c.; Li T.-t.; Cui Y.-h.; Zhang X.; Bian X.-w. (2013) A Novel Zebrafish Xenotransplantation Model for Study of Glioma Stem Cell Invasion. PLoS One 8 (4), e61801. 10.1371/journal.pone.0061801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins S.; Sontheimer H. (2011) Hydrodynamic Cellular Volume Changes Enable Glioma Cell Invasion. J. Neurosci. 31 (47), 17250. 10.1523/JNEUROSCI.3938-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. M.; Sessa A.; Burke C.; Bowman T.; LeBlanc J.; Ceol C.; Bourque C.; Dovey M.; Goessling W.; Burns C. E.; Zon L. I. (2008) Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2 (2), 183–189. 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Patron L. A.; Agudelo-Dueñas N.; Madrid-Wolff J.; Venegas J. A.; González J. M.; Forero-Shelton M.; Akle V. (2019) Xenotransplantation of Human glioblastoma in Zebrafish larvae: in vivo imaging and proliferation assessment. Biol. Open 8 (5), bio043257. 10.1242/bio.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umans R. A.; Henson H. E.; Mu F.; Parupalli C.; Ju B.; Peters J. L.; Lanham K. A.; Plavicki J. S.; Taylor M. R. (2017) CNS angiogenesis and barriergenesis occur simultaneously. Dev. Biol. 425 (2), 101–108. 10.1016/j.ydbio.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.; Wang X.; Zhao Y.; Li Z.; Lin S.; Wei Y.; Yang H. (2011) A novel xenograft model in zebrafish for high-resolution investigating dynamics of neovascularization in tumors. PLoS One 6 (7), e21768–e21768. 10.1371/journal.pone.0021768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletov K.; Montel V.; Lester R. D.; Gonias S. L.; Klemke R. (2007) High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc. Natl. Acad. Sci. U. S. A. 104 (44), 17406–17411. 10.1073/pnas.0703446104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble J. T.; Reed-Harris Y.; Barton C. L.; La Du J.; Tanguay R.; Greenwood J. A. (2018) Quantification of glioblastoma progression in zebrafish xenografts: Adhesion to laminin alpha 5 promotes glioblastoma microtumor formation and inhibits cell invasion. Biochem. Biophys. Res. Commun. 506 (4), 833–839. 10.1016/j.bbrc.2018.10.076. [DOI] [PubMed] [Google Scholar]
- Liebner S.; Fischmann A.; Rascher G.; Duffner F.; Grote E. H.; Kalbacher H.; Wolburg H. (2000) Claudin-1 and claudin-5 expression and tight junction morphology are altered in blood vessels of human glioblastoma multiforme. Acta Neuropathol. 100 (3), 323–331. 10.1007/s004010000180. [DOI] [PubMed] [Google Scholar]
- Trede N. S.; Langenau D. M.; Traver D.; Look A. T.; Zon L. I. (2004) The Use of Zebrafish to Understand Immunity. Immunity 20 (4), 367–379. 10.1016/S1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- Friedrich R. W.; Genoud C.; Wanner A. A. (2013) Analyzing the structure and function of neuronal circuits in zebrafish. Front. Neural Circuits 7, 71. 10.3389/fncir.2013.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S.; Mota B.; Lent R. (2006) Cellular scaling rules for rodent brains. Proc. Natl. Acad. Sci. U. S. A. 103 (32), 12138–12143. 10.1073/pnas.0604911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hason M.; Bartunek P. (2019) Zebrafish Models of Cancer-New Insights on Modeling Human Cancer in a Non-Mammalian Vertebrate. Genes 10 (11), 935. 10.3390/genes10110935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini C.; Kozaki T.; Bruschi M.; Nguyen T. H. H.; Minard-Colin V.; Castel D.; Grill J.; Ginhoux F. (2020) Modeling the Interaction between the Microenvironment and Tumor Cells in Brain Tumors. Neuron 10.1016/j.neuron.2020.09.018. [DOI] [PubMed] [Google Scholar]
- Liang D.; Chang J. R.; Chin A. J.; Smith A.; Kelly C.; Weinberg E. S.; Ge R. (2001) The role of vascular endothelial growth factor (VEGF) in vasculogenesis, angiogenesis, and hematopoiesis in zebrafish development. Mech. Dev. 108 (1), 29–43. 10.1016/S0925-4773(01)00468-3. [DOI] [PubMed] [Google Scholar]
- Quail D. F.; Joyce J. A. (2017) The Microenvironmental Landscape of Brain Tumors. Cancer Cell 31 (3), 326–341. 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M.; Jung E.; Sahm F.; Solecki G.; Venkataramani V.; Blaes J.; Weil S.; Horstmann H.; Wiestler B.; Syed M.; Huang L.; Ratliff M.; Karimian Jazi K.; Kurz F. T.; Schmenger T.; Lemke D.; Gömmel M.; Pauli M.; Liao Y.; Häring P.; Pusch S.; Herl V.; Steinhäuser C.; Krunic D.; Jarahian M.; Miletic H.; Berghoff A. S.; Griesbeck O.; Kalamakis G.; Garaschuk O.; Preusser M.; Weiss S.; Liu H.; Heiland S.; Platten M.; Huber P. E.; Kuner T.; von Deimling A.; Wick W.; Winkler F. (2015) Brain tumour cells interconnect to a functional and resistant network. Nature 528 (7580), 93–8. 10.1038/nature16071. [DOI] [PubMed] [Google Scholar]
- Venkatesh H. S.; Johung; Tessa B.; Caretti V.; Noll A.; Tang Y.; Nagaraja S.; Gibson; Erin M.; Mount; Christopher W.; Polepalli J.; Mitra; Siddhartha S.; Woo P. J.; Malenka R. C.; Vogel H.; Bredel M.; Mallick P.; Monje M. (2015) Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell 161 (4), 803–816. 10.1016/j.cell.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate M. C.; Aghi M. K. (2009) Biology of angiogenesis and invasion in glioma. Neurotherapeutics 6 (3), 447–457. 10.1016/j.nurt.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B. W.; Gu C. (2015) The molecular constituents of the blood-brain barrier. Trends Neurosci. 38 (10), 598–608. 10.1016/j.tins.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R.; Agalliu D.; Zhou L.; Kuhnert F.; Kuo C. J.; Barres B. A. (2009) Wnt/β-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 106 (2), 641–646. 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreone B. J.; Chow B. W.; Tata A.; Lacoste B.; Ben-Zvi A.; Bullock K.; Deik A. A.; Ginty D. D.; Clish C. B.; Gu C. (2017) Blood-Brain Barrier Permeability Is Regulated by Lipid Transport-Dependent Suppression of Caveolae-Mediated Transcytosis. Neuron 94 (3), 581–594.e5. 10.1016/j.neuron.2017.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brown N. M.; Megason S. G.; Gu C. (2019) Suppression of transcytosis regulates zebrafish blood-brain barrier function. eLife 8, e47326. 10.7554/eLife.47326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan N.; Daetwyler S.; Bernas S. N.; Schmied C.; Vogler S.; Lambert K.; Wobus M.; Wermke M.; Kempermann G.; Huisken J.; Brand M.; Bornhäuser M. (2020) Long-term in vivo imaging reveals tumor-specific dissemination and captures host tumor interaction in zebrafish xenografts. Sci. Rep. 10 (1), 13254. 10.1038/s41598-020-69956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzinger D. B.; Demuth T.; Berens M. E. (2007) Autocrine Factors That Sustain Glioma Invasion and Paracrine Biology in the Brain Microenvironment. JNCI: Journal of the National Cancer Institute 99 (21), 1583–1593. 10.1093/jnci/djm187. [DOI] [PubMed] [Google Scholar]
- van Leeuwen L. M.; Evans R. J.; Jim K. K.; Verboom T.; Fang X.; Bojarczuk A.; Malicki J.; Johnston S. A.; van der Sar A. M. (2018) A transgenic zebrafish model for the < em > in vivo</em> study of the blood and choroid plexus brain barriers using < em > claudin 5</em>. Biol. Open 7 (2), bio030494. 10.1242/bio.030494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacioni S.; D’Alessandris Q. G.; Buccarelli M.; Boe A.; Martini M.; Larocca L. M.; Bolasco G.; Ricci-Vitiani L.; Falchetti M. L.; Pallini R. (2020) Brain Invasion along Perivascular Spaces by Glioma Cells: Relationship with Blood-Brain Barrier. Cancers 12 (1), 18. 10.3390/cancers12010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letrado P.; de Miguel I.; Lamberto I.; Díez-Martínez R.; Oyarzabal J. (2018) Zebrafish: Speeding Up the Cancer Drug Discovery Process. Cancer Res. 78 (21), 6048. 10.1158/0008-5472.CAN-18-1029. [DOI] [PubMed] [Google Scholar]
- Cully M. (2019) Zebrafish earn their drug discovery stripes. Nat. Rev. Drug Discovery 18 (11), 811–813. 10.1038/d41573-019-00165-x. [DOI] [PubMed] [Google Scholar]
- Montana V.; Sontheimer H. (2011) Bradykinin Promotes the Chemotactic Invasion of Primary Brain Tumors. J. Neurosci. 31 (13), 4858. 10.1523/JNEUROSCI.3825-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Lee J. E.; Dorsky R. I. (2009) Identification of Wnt-responsive cells in the zebrafish hypothalamus. Zebrafish 6 (1), 49–58. 10.1089/zeb.2008.0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromée T.; Kukkonen J. P.; Andersson P.; Conlon J. M.; Larhammar D. (2005) Pharmacological characterization of ligand-receptor interactions at the zebrafish bradykinin receptor. Br. J. Pharmacol. 144 (1), 11–16. 10.1038/sj.bjp.0706032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy N.; Kurzrock R. (2018) Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 62, 50–60. 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.