Although hematopoietic stem cells (HSCs) are the most thoroughly characterized type of adult stem cell, the intricate molecular machinery that regulates their self-renewal properties remains elusive. Here we showed that the E3 ubiquitin ligase Itch negatively regulated the development and functions of HSCs. Itch−/− mice had HSCs with enhanced frequency, competence and long-term repopulating activity. Itch-deficient HSCs showed accelerated proliferation rates and sustained progenitor properties, as well as more Notch1 signaling, due to more accumulation of activated Notch1. Knockdown of Notch1 in Itch-mutant HSCs resulted in reversion of the phenotype. Thus, we have identified Itch as a previously unknown negative regulator of HSC homeostasis and function.
Hematopoietic stem cells (HSCs) are a specialized subset of cells that give rise to the entire blood system throughout life1–3. Like any other stem cell, HSCs are able to self-renew and to differentiate into various lineages of the hematopoietic system. Self-renewal is a tightly controlled process through which stem cells divide and generate daughter stem cells with properties similar to those of the mother cell4. However, under certain conditions, HSCs differentiate into progenitor cells with less self-renewal properties. Since the discovery of stem cells, intense research aimed at understanding the genetic and molecular bases of self-renewal has identified candidates involved in HSC self-renewal. These include cell-intrinsic regulators, such as transcription factors, signal transducers, cell-cycle inhibitors and surface receptors, and cell-extrinsic regulators, such as the bone marrow niche and cytokines2,4–6. The Really Interesting New Gene (RING) finger–type E3 ubiquitin ligase c-Cbl has been reported to have a role in the self-renewal of HSCs7,8.
Ubiquitination is a post-translational modification of proteins by which polyubiquitin chains are added to lysine residues of target proteins9,10. The functional consequences of this modification include the targeting of proteins for proteasomal degradation, as well as other cellular functions, such as protein recycling, endocytic trafficking, DNA repair and transcriptional regulation11–13. Ubiquitination occurs through a three-step enzymatic cascade involving ubiquitin-activating (E1) enzymes, ubiquitin conjugating (E2) enzymes and ubiquitin ligase (E3) enzymes14. E3 ubiquitin ligases are considered crucial for the process, as they recognize, bind and recruit specific target proteins for ubiquitination. They are broadly classified into two main families based on their domains: the RING-finger domain, which promotes ubiquitination by simultaneously binding to the substrate and an E2 enzyme, and the Homologous to E6-Associated Protein (E6AP) C-Terminus (HECT) domain, that participates directly in catalysis by forming an obligate thiolester bond with ubiquitin during the ubiquitination reaction12. Because E3 ligases modify substrates at a specific time and place, comprehensive knowledge of their physiological roles is critical for understanding the events associated with post-translational modifications in vivo.
Itch is an E3 ubiquitin ligase that belongs to the HECT family15. It contains four amino-terminal domains containing two conserved tryptophan residues separated by 20–22 amino acids (the ‘WW’ domain), a C2 domain, and a HECT ligase domain. WW domains mediate protein-protein interactions, whereas the HECT domain recruits ubiquitin-loaded E2 ligases and transfers the ubiquitin to the substrate10. Biochemical studies, mainly in vitro, have identified more than 20 cellular targets of Itch proteins15. Mice deficient in Itch develop a skin-scratching phenotype (the ‘itchy’ phenotype) and severe immune dysregulation, including lymphadenopathy, splenomegaly and inflammation in the lungs and digestive tract16. On a C57BL/6 background, Itch-deficient mice die around ~6–8 months of age, most probably because of hypoxia associated with pulmonary chronic interstitial inflammation and alveolar proteinosis.
In correlation with the strong autoimmune disorder associated with Itch deficiency in mice, Itch has been shown to have important roles in T cells, such as controling AKT phosphorylation in thymic T cells17, ubiquitylating JunB in Th2 cells18, regulating the expression of Foxp3 in Tregs19 and restricting the development of pathogenic αβ & γδ T cells20. However, the role of Itch in other cell types remains largely unknown. Here we investigate the involvement of Itch in hematopoiesis. We found that Itch negatively regulated the development and function of HSCs. Itch deficiency in HSCs was associated with more Notch1 signaling. Our data suggest that the ubiquitination events mediated by Itch are critical for the development and function of HSCs.
RESULTS
Itch restricts the HSC pool in the bone marrow
In an attempt to identify the importance of ubiquitination events and E3 ligases in HSC development, we studied the expression of three well-characterized E3 ligases, c-Cbl, Cbl-B and Itch, in the lineage-negative (Lin−: CD11b−Gr-1−220−CD3ε−Ter119−) Sca-1+c-Kit+ (LSK) compartment of the bone marrow of C57BL/6 wild-type mice. Although real-time PCR analysis showed expression of all three candidate E3 ubiquitin ligases, Itch expression was much higher than that of c-Cbl and Cbl-B (Supplementary Fig. 1a). Direct comparison of LSK cells and CD4+ T cells showed that Itch mRNA expression was higher in bone marrow LSK cells (Supplementary Fig. 1b). Further real-time PCR analysis of Itch mRNA expression in sorted long-term HSCs (LT-HSCs; CD150+CD48− LSK cells), short-term HSCs (ST-HSCs; CD150+CD48+ LSK cells) and multipotent progenitors (MPPs; CD150−CD48+ LSK cells)21,22 from the bone marrow of wild-type mice showed that Itch mRNA was detectable at all these stages of HSC development. However, the LT-HSCs had much more Itch mRNA (Fig. 1a).
Figure 1.
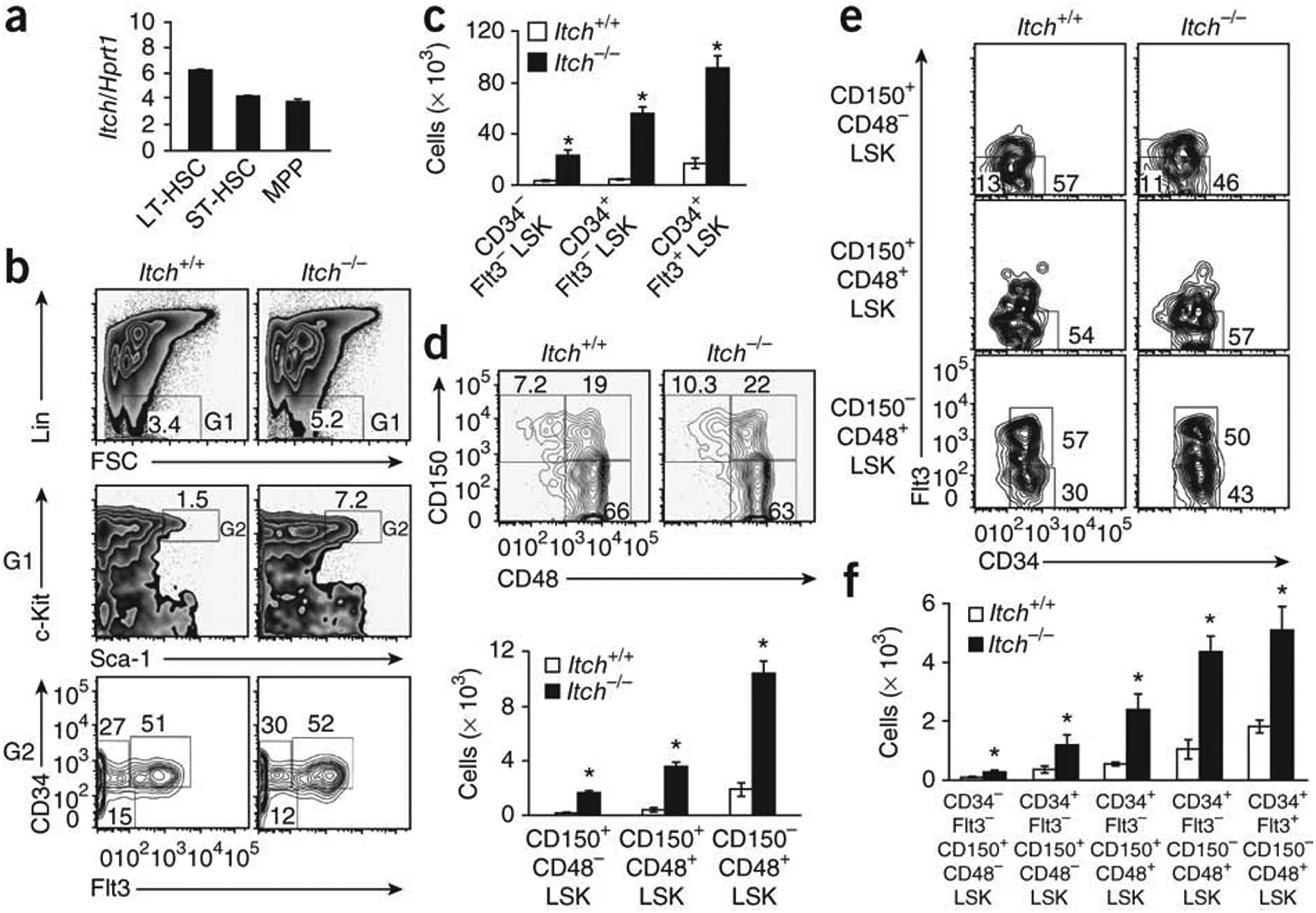
Itch deficiency results in a greater frequency of HSCs in the bone marrow. (a) Real-time PCR analysis of Itch mRNA expression in sorted LT-HSCs (LSK CD150+CD48−), ST-HSCs (LSK CD150+CD48+) and MPPs (LSK CD150−CD48+), presented relative to the expression of Hprt1 (encoding hypoxanthine guanine phosphoribosyl transferase). (b) Distribution of Lin− cells (top), LSK cells (middle) and LSK subsets defined on the basis of the expression of CD34 and Flt3 (bottom), among total wild-type and Itch−/− bone marrow cells. Numbers adjacent to outlined areas indicate percent cells in each throughout. (c) Absolute number of cells in HSC subsets in 4-week-old wild-type and Itch−/− mice (n = 5 per group) based on the gates presented in b. (d) Distribution (top) and absolute number (bottom) of LSK subsets in 4-week-old wild-type and Itch−/− mice (n = 5 per group), assessed on the basis of expression of CD150 and CD48. (e) Expression of Flt3 and CD34 on cells of wild-type and Itch−/− bone marrow LSK subsets identified as in d. (f) Absolute number of cells of LSK subsets from 4-week-old wild-type and Itch−/− mice (n = 5 per group), stained as in e. *P < 0.05 (Student’s t-test). Data are representative of two independent experiments (a; mean and s.e.m. of duplicates) or ten (b–e) or three (f) independent experiments (mean and s.e.m. in c,d,f).
To investigate whether Itch deficiency affects the bone marrow HSC compartment, we used previously characterized Itch−/−mice16. Our analysis showed that Itch-deficient mice had a much greater frequency of LSK cells in the bone marrow (Fig. 1b). Further analysis of the LSK compartment indicated Itch−/− mice had much greater absolute numbers of CD34−Flt3−LSK, CD34+Flt3−LSK and CD34+Flt3+LSK cells (Fig. 1c), although the frequencies of these subsets were similar to those of wild-type mice (Fig. 1b). We obtained similar results by CD150- and CD48-based immunophenotyping of LT-HSCs, ST-HSCs and MPPs (Fig. 1d). Of note, wild-type and Itch−/− mice had a similar absolute number of total bone marrow cells (Supplementary Fig. 1c).
According to a published study, LSK cells can be further subcategorized on the basis of their expression of CD150, CD48, CD34 and Flt3 into the following five subsets: a most primitive HSC subset (CD34−Flt3−CD150+CD48−LSK), and the increasingly differentiated MPP1 subset (CD34+Flt3−CD150+CD48−LSK), MPP2 subset (CD34+Flt3−CD150+CD48+LSK), MPP3 subset (CD34+Flt3−CD150−CD48+LSK) and MPP4 subset (CD34+Flt3+CD150−CD48+LSK)23. Using that developmental scheme, we found that none of those subsets was perturbed in the absence of Itch (Fig. 1e); nevertheless, the absolute number of the most primitive HSCs (CD34−Flt3−CD150+CD48−LSK) and their downstream progenitors (MPP1, MPP2, MPP3 and MPP4) was greater in Itch-deficient bone marrow (Fig. 1f). Analysis of cells of the myeloid, erythroid and lymphoid lineages indicated that Itch−/− and wild-type mice had similar frequencies of these (Supplementary Fig. 2a−d). Consistent with that, the total number of myeloid- and lymphoid-restricted progenitors was similar in wild-type and Itch−/− mice (Supplementary Fig. 2e,f). Together these data suggest that the E3 ligase Itch restricts the size of the HSC pool in the bone marrow.
Augmented repopulation ability of Itch−/− HSCs
During development of the mouse embryo, hematopoiesis first originates from the mesodermal precursors and involves various organs, including the intraembryonic aorta-gonad-mesonephros region, the extraembrionic yolk sac, the placenta, the fetal liver and the thymus24, 25. Starting from embryonic day 12 (E12) through birth, the most prominent site of hematopoiesis is the fetal liver, where HSCs expand their populations and differentiate into myeloid, erythroid and lymphoid lineages before migrating into the bone marrow25,26. To investigate whether the augmented HSC pool is detectable at early stages of development in Itch-deficient mice, we collected fetal livers from embryos (at E15) and neonatal pups (at the second day after birth) and the HSC pool was evaluated. The frequency of HSCs was greater both on E15 (Fig. 2a–c) and at the second day after birth (Supplementary Fig. 3a,b). In line with that observation, Itch mRNA was expressed in LSK cells obtained from the fetal liver of wild-type embryos at E15 (Supplementary Fig. 3c).
Figure 2.
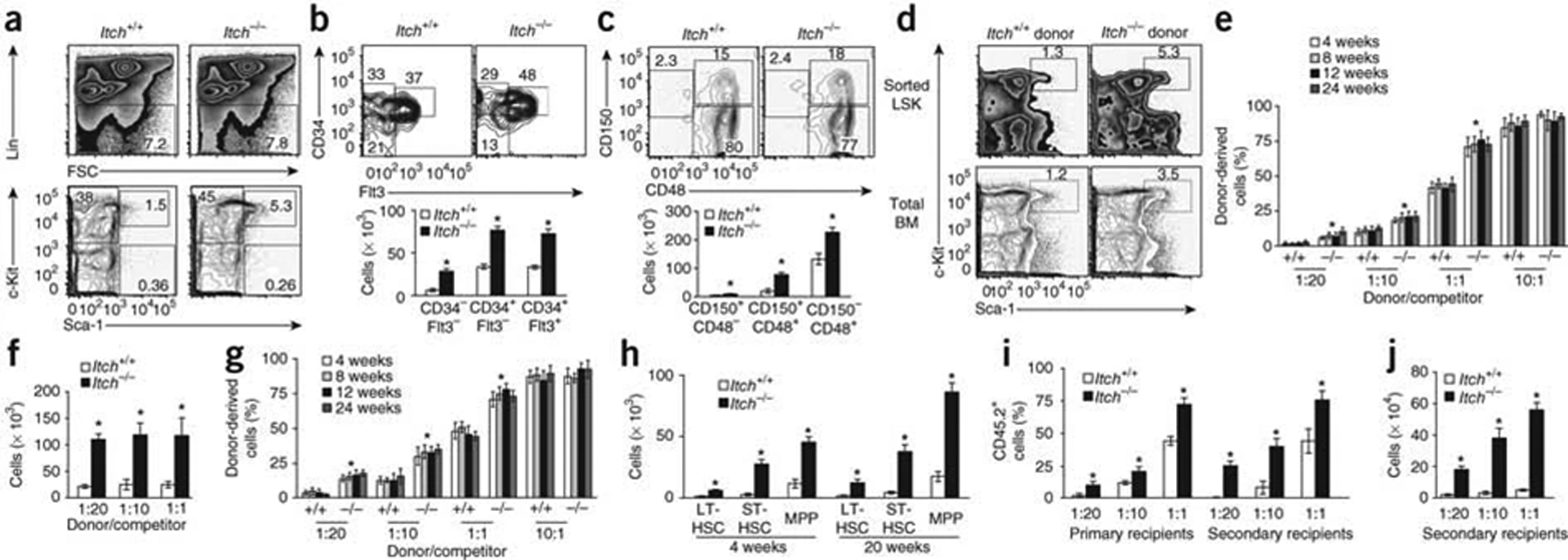
Cell-intrinsic defects and greater competence of Itch-mutant HSCs. (a) Distribution Lin− cells (top) and LSK cells (bottom) in wild-type and Itch−/− fetal livers (n = 5 mice per genotype). FSC, forward scatter. (b,c) Distribution (top) and absolute number (bottom) of cells of the LSK subsets in wild-type and Itch−/− fetal livers (n = 5 mice per genotype), assessed on the basis of the expression of CD34 and Flt3 (b) or CD150 and CD48 (c). (d) Distribution of donor-derived sorted LSK cells (top) or total bone marrow cells (bottom) from wild-type and Itch−/− (CD45.2+) mice in the total bone marrow of wild-type (CD45.1+) congenic recipient mice 18 weeks after transplantation. (e,f) Hematopoiesis of donor-derived (CD45.2+) cells in the peripheral blood (e) and absolute number of LSK cells in the bone marrow (f) of wild-type (CD45.1+) recipients (n = 10 per group) of donor wild-type or Itch−/− (CD45.2+) LSK cells mixed at various ratios (horizontal axes) with wild-type competitor (CD45.1+) cells. (g) Hematopoiesis of donor-derived (CD45.2+) cells in the peripheral blood of wild-type (CD45.1+) recipients (n = 10 per group) of total wild-type or Itch−/− (CD45.2+) bone marrow cells mixed at various ratios (horizontal axis) with wild-type competitor (CD45.1+) cells. (h) Absolute number of cells of various LSK subsets in 4- and 20-week-old wild-type and Itch−/− mice (n = 5 per genotype). (i,j) Hematopoiesis of either wild-type or Itch−/− donor-derived (CD45.2+) cells in primary and secondary wild-type recipients (i; n = 10 per group) or number of either wild-type or Itch−/− donor-derived LSK cells in secondary wild-type recipients (j). *P < 0.05 (Student’s t-test). Data are representative of three (a–c,h) or two (d–g,I,j) independent experiments (mean and s.e.m.).
Next we assessed whether the greater number of HSCs in Itch-deficient bone marrow was a cell-intrinsic phenomenon. We mixed LSK cells either from Itch−/− or wild-type animals (CD45.2) with bone marrow cells depleted of c-Kit+ (to provide efficient short-term hematopoiesis after irradiation) from wild-type (CD45.1+) mice and injected the mixture into lethally irradiated wild-type congenic (CD45.1+) recipients. At 18 weeks after transplantation, the bone marrow of recipients that received Itch−/− LSK cells had more LSK cells (Fig. 2d and Supplementary Fig. 4a). Consistent with the results reported above, transfer of total bone marrow from Itch−/− (CD45.2+) mice into lethally irradiated wild-type congenic recipients resulted in larger LSK pool (Fig. 2d and Supplementary Fig. 4b). Furthermore, we assessed the ability of Itch-deficient HSCs to repopulate the hematopoietic system under competitive settings. We mixed limiting dilutions of LSK cells derived either from Itch−/− or wild-type (CD45.2+) mice with defined ratios of wild-type (CD45.1+) LSK competitor cells and transplanted the mixtures into lethally irradiated wild-type (CD45.1+) recipients as described before7. We obtained blood from recipient mice at regular intervals (4, 8, 12 and 24 weeks after transplantation) and calculated the proportion of hematopoiesis derived from donor (CD45.2+) cells and competitor (CD45.1+) cells. Notably, at all time points analyzed, hematopoiesis derived from donor (CD45.2+) cells was greater in the recipients that received Itch−/− LSK cells than in those that received wild-type LSK cells (Fig. 2e). This was most probably due to the greater abundance LSK cells in the bone marrow (Fig. 2f), as the wild-type and Itch−/− groups had a similar abundance of donor-derived total bone marrow cells (Supplementary Fig. 5a). Although Itch−/− and wild-type donor cells had a similar multilineage-reconstitution capacity (analyzed by calculation of the frequency of CD45.2+ lymphoid and myeloid lineage cells in the peripheral blood) at all time points analyzed (Supplementary Fig. 5b–d), the frequency of hematopoietic cells derived from Itch-deficient cells was significantly greater than the frequency of cells derived from wild-type cells (Fig. 2e). Similarly, we did competition experiments in which we lysed red blood cells in total bone marrow from Itch−/− and wild-type mice and mixed the resultant cells with competitor Itch+/+ (CD45.1+) cells at various ratios and injected the resultant mixture into lethally irradiated wild-type recipients. We found that hematopoiesis derived from Itch−/−donor cells was greater in the peripheral blood of recipients at 4, 8, 12 and 24 weeks after transplantation (Fig. 2g).
We further strengthened our observations about the greater self-renewal properties of Itch−/− HSCs by radioprotection assays27 in which we injected limiting dilutions of bone marrow cells from wild-type and Itch−/− mice into lethally irradiated wild-type recipients. Itch-deficient bone marrow cells were better in providing radioprotection to recipients, even at very low frequencies (Table 1).
Table 1.
Radioprotective functions of young bone marrow cells
| Donor cells | Itch+/+ donor | Itch−/− donor |
|---|---|---|
| 0 | 0/10 | 0/10 |
| 1 × 102 | 0/10 | 0/10 |
| 1 × 103 | 0/10 | 0/10 |
| 5 × 103 | 0/10 | 4/10 |
| 1 × 104 | 2/10 | 8/10 |
| 5 × 104 | 3/10 | 9/10 |
| 1 × 105 | 9/10 | 10/10 |
Survival of lethally irradiated wild-type (CD45.1+) recipients (n = 10 per group) of limiting numbers of bone marrow cells from 4-week-old Itch+/+ or Itch−/− mice; results are presented as number of surviving mice/total mice. Data are representative of two independent experiments.
Next we assessed the size of the HSCs pool in Itch−/− mice at later stages of life. The enhancement of the HSC pool was even greater at 20 weeks than it was at 4 weeks (Fig. 2h). In radioprotection experiments, bone marrow cells from 20-week-old Itch−/− mice were able to ‘rescue’ lethally irradiated mice at very low frequencies (Table 2). This proved that Itch-deficient HSCs retained their self-renewal properties even at later stages of life.
Table 2.
Radioprotective functions of older Itch−/− bone marrow cells
| Donor cells | Itch+/+ donor | Itch−/− donor |
|---|---|---|
| 0 | 0/10 | 0/10 |
| 1 × 102 | 0/10 | 0/10 |
| 1 × 103 | 0/10 | 2/10 |
| 5 × 103 | 0/10 | 8/10 |
| 1 × 104 | 0/10 | 10/10 |
| 5 × 104 | 5/10 | 10/10 |
| 1 × 105 | 8/10 | 10/10 |
Survival of lethally irradiated wild-type (CD45.1+) recipients of limiting numbers of bone marrow cells from 20-week-old Itch+/+ and Itch−/− mice; results are presented as number of surviving mice/total mice. Data are representative of two independent experiments.
To investigate the long-term-repopulation abilities of Itch−/− HSCs, we did serial transplantation experiments under competitive settings7. We sacrificed primary recipients (wild-type mice that either received wild-type or Itch−/− BM cells) after 6 months of transplantation and injected their bone marrow cells into lethally irradiated wild-type secondary recipients. At 6 months after transplantation, we obtained blood from secondary recipient mice and analyzed hematopoiesis derived from donor (CD45.2+) cells. Our analysis indicated that hematopoiesis derived from Itch−/− donor cells was significantly greater in the secondary recipients than was hematopoiesis derived from wild-type donor cells (Fig. 2i), most probably because of the greater number of LSK cells in the bone marrow (Fig. 2j). Together these data document a cell-intrinsic effect of Itch deficiency and suggest that Itch−/− HSCs are more competent than wild-type HSCs and have a greater ability to repopulate the hematopoietic system.
Hyperproliferation of HSCs in the absence of Itch
We next used the thymidine analog BrdU to assess whether the larger HSC pool in the Itch-deficient mice was due to more proliferation. After a single intraperitoneal injection of BrdU, we provided mice with BrdU in the drinking water for 3 d. Analysis of LSK cells on the fourth day showed augmented proliferation of LSK cells in the Itch-deficient mice (Fig. 3a) with more proliferation of LT-HSCs, ST-HSCs and MPPs (Fig. 3a). Next, we directly assessed the cell-cycle status of the LSK compartment under homeostatic conditions. Pyronin Y is a dye that stains total cellular RNA, and the intensity of the staining is directly related to cell-cycle status28. Accordingly, quiescent cells are usually negative to low for pyronin Y, while actively proliferating cells are positive for pyronin Y. In line with the BrdU experimental data, pyronin Y staining showed that Itch-deficient HSCs were less quiescent (Fig. 3b). In addition, we stained sorted LT-HSCs from Itch−/− and wild-type mice with the cytosolic dye CFSE and cultured the cells in vitro in the presence of a cytokine ‘cocktail’ that promotes HSC proliferation in vitro (interleukin 3, stem cell factor, interleukin 6, the hematopoietic growth factor Flt3L, and thrombopoietin). Flow cytometry of hematopoietic stem and progenitor cells (HSPCs) on days 3 and 6 suggested augmented proliferation associated with Itch deficiency (Fig. 3c). Itch-deficient HSPCs showed augmented ex vivo population expansion rates in response to cytokines (Fig. 3d). Of note, we observed the greater proliferation of Itch−/− HSCs only in the presence of the complete HSC cytokine ‘cocktail’, as Itch−/− HSCs cultured in the presence of cytokines neither survived nor proliferated for 10 d of culture (Supplementary Fig. 6). Together these data suggest that Itch deficiency is associated with augmented proliferation and population expansion of the HSC compartment.
Figure 3.
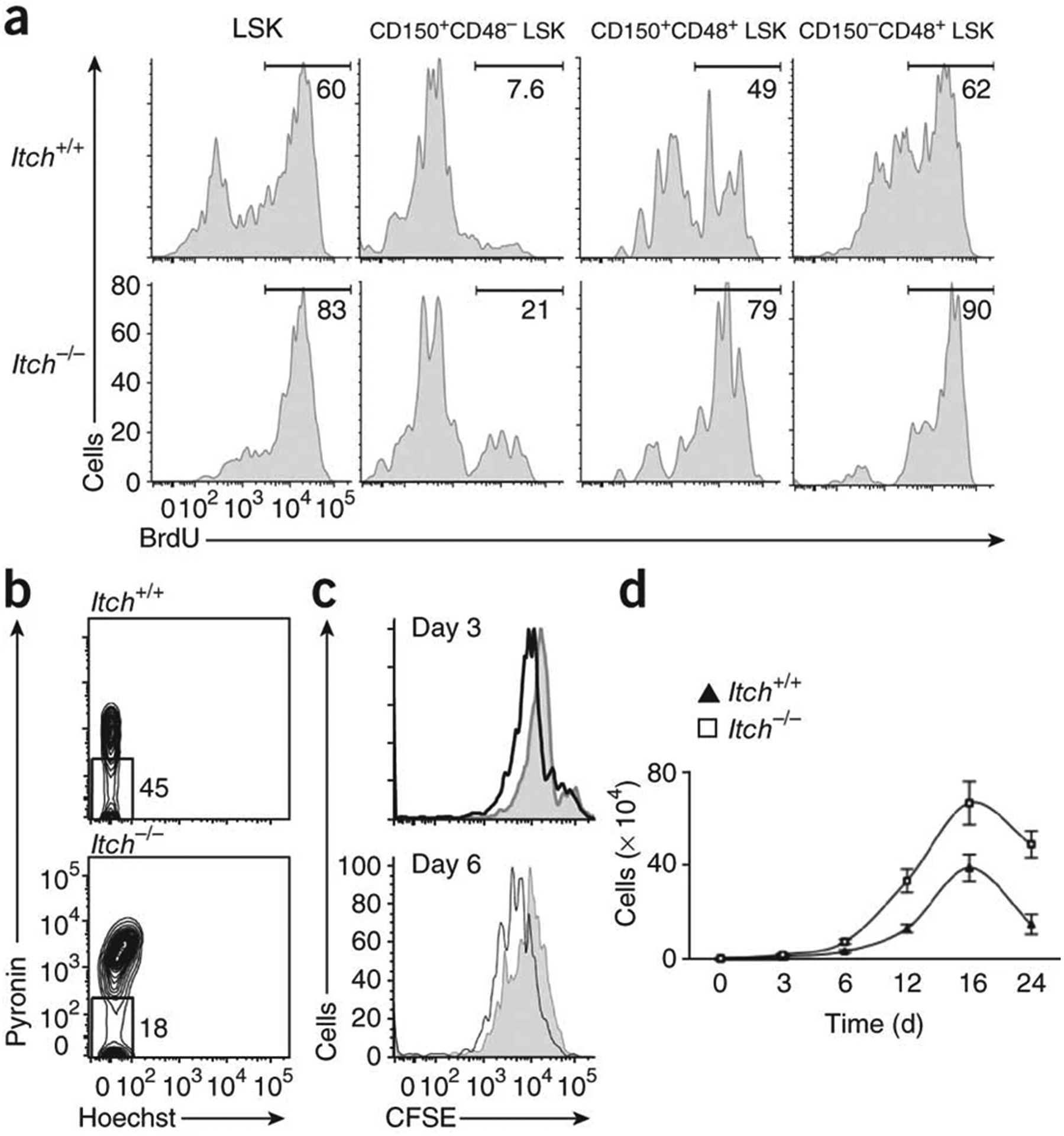
Accelerated proliferation of Itch-mutant HSCs. (a) In vivo proliferative potential of wild-type and Itch−/− LSK subsets, as assessed by BrdU incorporation. Numbers above bracketed lines indicate percentages of proliferating (BrdU+) cells. (b) Ex vivo cell cycle status of wild-type and Itch−/− LSK cells, as assessed by pyronin Y and Hoechst staining. (c) In vitro proliferative potential of wild-type (shaded histograms) and Itch−/− (black lines) CD150+CD48− LSK cells, as assessed by CFSE dilution. (d) Ex vivo population expansion of wild-type and Itch−/− CD150+CD48− LSK cells in response to a cytokine ‘cocktail’. Data are representative of two (a,b) or five (c,d) independent experiments (mean ± s.e.m. of duplicates in d).
Augmented recovery of the HSC pool in Itch−/− mice
To evaluate the in vivo consequences of Itch deficiency under physiological stimulation or stress, we analyzed hematopoietic recovery after hemoablation with 5-fluorouracil (5-FU). This reagent kills actively cycling cells and results in ablation of the progenitor pool but spares the quiescent stem cells responsible for long-term repopulation29. Analysis of the LSK compartment 2 d after injection of 5-FU showed that Itch−/− and wild-type mice had a similar proportion of LSK cells (Fig. 4a and Supplementary Fig. 7), which suggested that actively proliferating (5-FU-sensitive) LSK cells contributed to the larger HSC pool in the bone marrow of Itch−/− mice. At 8 days after 5-FU injection, there was augmented recovery of hematopoietic progenitors in the absence of Itch (Fig. 4a and Supplementary Fig. 7). We obtained blood from mice on days 0, 5 and 10 after 5-FU treatment and counted platelets, leukocytes and red blood cells and measured the hematocrit. Although we observed a similar drop in blood counts in both Itch−/− and wild-type mice on day 5, we noted augmented recovery of blood counts in Itch-deficient mice on day 10 (Fig. 4b).
Figure 4.
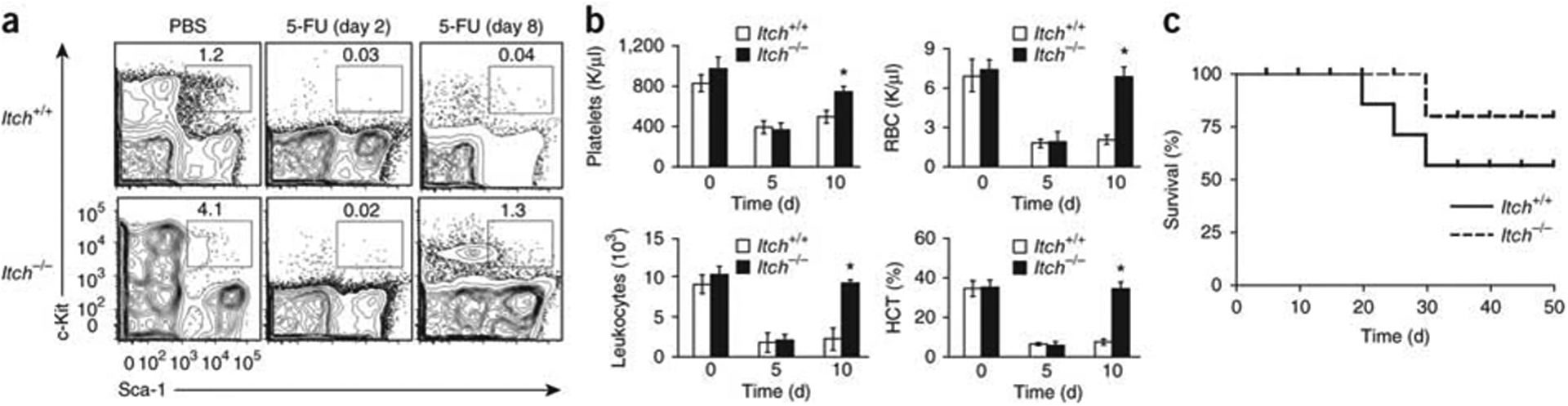
Augmented repopulation activity of Itch−/− HSCs show after myeloablation. (a) Distribution of LSK cells in wild-type and Itch−/− bone marrow (n = 5 mice per group) after injection of 5-FU. (b) Differential blood counts and hematocrit (HCT) of wild-type and Itch−/− mice (n = 5 per group) after injection of 5-FU. RBC, red blood cell. *P < 0.05 (Student’s t-test). (c) Survival of wild-type recipients (n = 10 per group) of wild-type or Itch−/− bone marrow cells, assessed with a log-rank nonparametric test after sequential 5-FU treatment and presented as a Kaplan-Meier survival curve. P = 0.0082 (Student’s t-test). Data are representative of two independent experiments (mean ± s.e.m. in b).
To investigate whether the enhanced hematopoietic recovery in Itch−/− mice was a cell-intrinsic phenomenon, we transferred wild-type or Itch−/− bone marrow cells into lethally irradiated wild-type recipients. At 8 weeks after transplantation, we administered 5-FU intraperitoneally to the recipient mice on a weekly basis and monitored their survival. Recipients of Itch−/− bone marrow cells had better survival rates than did mice that received wild-type bone marrow cells (Fig. 4c). These data indicate that Itch-deficient HSCs show enhanced recovery of hematopoiesis under stress conditions.
Sustained progenitor properties of Itch−/− HSPCs
To investigate the ability of Itch-deficient HSPCs to maintain progenitor properties under in vitro culture conditions, we sorted LT-HSCs and cultured then in the presence of the HSC cytokine ‘cocktail’. We obtained aliquots of cells on days 5, 10, 15 and 20 of culture and analyzed their expression of lineage markers (CD11b, Gr-1, B220, Ter119 and CD3ε). Starting from day 10, Itch-deficient HSPCs showed lower expression of these lineage-associated markers (Fig. 5a). The proportion of cells that maintained the progenitor immunophenotype (LSK) was much higher in Itch−/− cell cultures than in wild-type cell cultures (Fig. 5b). To assess whether these cells also maintained the functions of progenitor cells after in vitro culture, we sorted LSK cells after 20 d of in vitro culture and assessed colony-forming units (CFU). Although the wild-type cells generated modest number of colonies in the semisolid medium, Itch−/− cells generated an augmented number of colonies (Fig. 5c). We also assessed the ability of LT-HSCs cultured in vitro to repopulate the hematopoietic system. After 20 d of culture, we sorted wild-type or Itch−/− cells with an LSK phenotype and injected the cells into sublethally irradiated congenic (CD45.1+) recipient mice. As a control, we injected freshly isolated (naive) wild-type or Itch−/− LSKs into sublethally irradiated recipients. At 4 weeks after transplantation, we killed the recipient mice and calculated the frequency of CD45.2+ cells in the peripheral blood. Whereas only ~5% cells were CD45.2+ in recipients that received wild-type cells, ~36% cells were CD45.2+ in recipients that received Itch-deficient LSKs (Fig. 5d). Analysis of the multilineage differentiation of donor cells showed that wild-type cells cultured in vitro had an greater myeloid differentiation capacity and a lower lymphoid differentiation capacity than did naive wild-type LSKs. Itch−/− cells cultured in vitro showed multilineage-differentiation capacities similar to those of naive Itch-deficient LSKs (Fig. 5e). Overall, these experiments suggest that Itch-deficient HSPCs maintain their progenitor properties even after prolonged in vitro culture.
Figure 5.
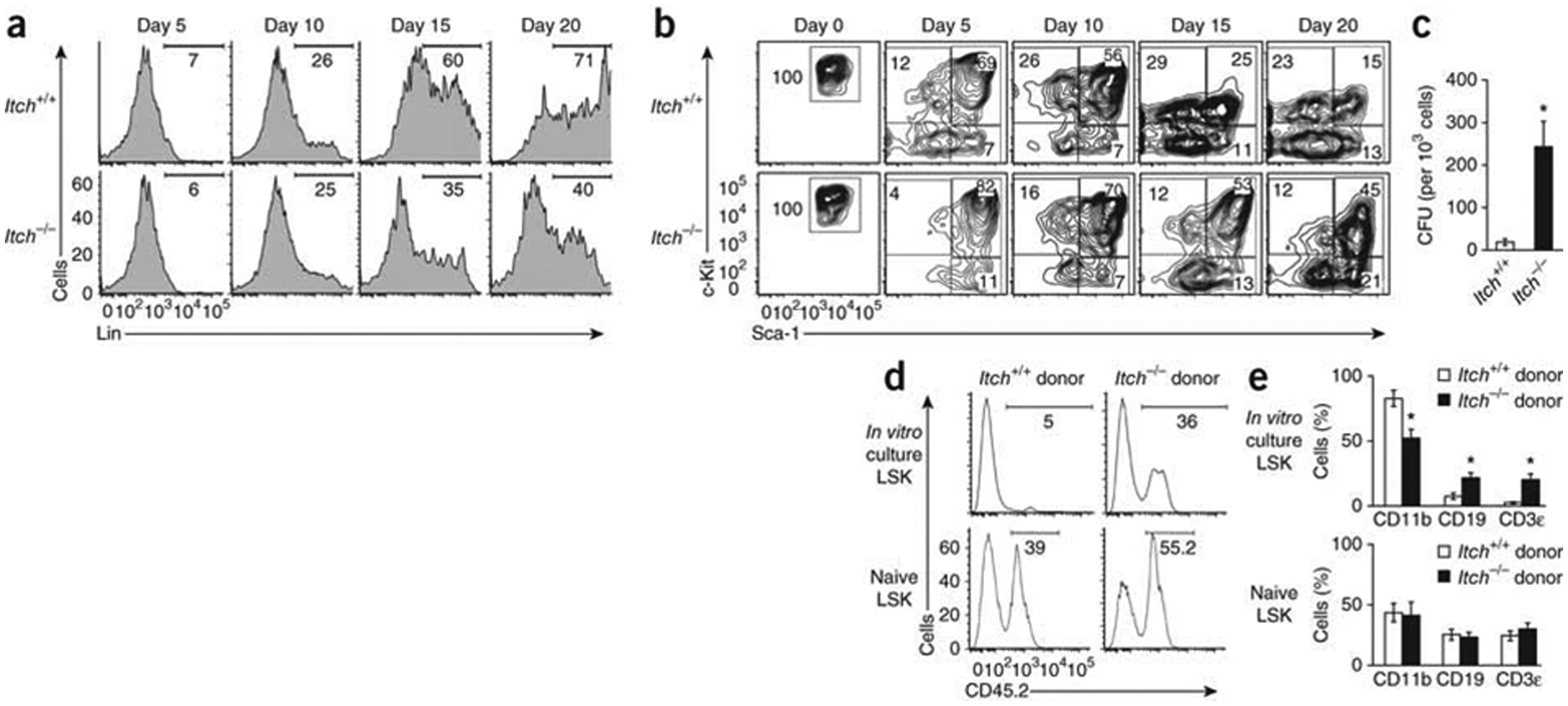
Less spontaneous differentiation by Itch-deficient HSCs in vitro. (a) Expression of lineage markers in wild-type and Itch−/− CD150+CD48− LSK cells after in vitro culture in the presence of a cytokine ‘cocktail’. Numbers below bracketed lines indicate percent Lin+ cells. (b) Expression of Sca-1 and c-Kit in wild-type and Itch−/− CD150+CD48− LSK cells after in vitro culture in the presence of a cytokine’ cocktail’; Lin− cells were pre-gated. (c) Colony-forming unit (CFU) potential of wild-type and Itch−/− CD150+CD48− LSK cells. (d) Hematopoiesis of donor-derived (CD45.2+) cells in the peripheral blood of wild-type (CD45.1+) recipients (n = 8 mice per group) that received LSK cells cultured in vitro (top) or freshly isolated LSK cells (Naive; bottom). Numbers below bracketed lines indicate percent CD45.2+ cells. (e) Multilineage reconstitution by donor-derived (CD45.2+) cells (myeloid, CD45.2+CD11b+; B lineage, CD45.2+ CD19+; T lineage, CD45.2+CD3ε+) in recipients (n = 8 mice per group) of cells as in d. *P < 0.05 (Student’s t-test). Data are representative of five (a,b) or two (c–e) independent experiments (mean and s.s.m. in c,e).
Augmented Notch1 signaling in Itch−/− HSPCs
Overexpression of the intracellular domain of the oncogenic transcription factor Notch1 (ICN1) in HSCs results in augmented proliferation, more competence and enhanced ex vivo expansion rates without the loss of self-renewal30,31. Our analysis here has identified many phenotypic similarities between Itch-deficient HSCs and Notch1-overexpressing HSCs. Moreover, Itch is directly involved in Notch1 ubiquitination17,32,33. On the basis of those observations, we hypothesized that Itch deficiency might result in defective ubiquitination of Notch1 and, thus, in augmented Notch1 signaling in HSCs. To test our hypothesis, we did biochemical analysis of Lin− hematopoietic progenitor cells (HPCs) from wild-type and Itch-deficient mice. First, with a Notch1-specific antibody, we detected more full-length Notch1 and a cleaved form consisting of the transmembrane and intracellular domain in HPCs (Fig. 6a). We also measured Notch1 protein in LT-HSCs sorted from Itch−/− and wild-type mice by flow cytometry. Itch-deficient LT-HSCs isolated from the bone marrow (Fig. 6b,c) and the fetal liver (Supplementary Fig. 8) had more Notch1. To further substantiate and independently document the augmented Notch1 signaling in Itch-deficient HSCs, we crossed Itch−/− mice with transgenic Notch reporter mice34. In these reporter mice, the expression of green fluorescent protein is directly proportional to the activity of Notch signaling in vivo34. Suggestive of the fact that Notch signaling is augmented in HSCs in the absence of Itch, HSCs from Itch−/− transgenic Notch reporter mice had higher expression of green fluorescent protein (Fig. 6d,e). Itch may also be involved in the regulation of various cellular proteins, such as c-Jun, JunB, PLC-γ1, Smad2 and p73 (ref. 15). Because of their critical roles in hematopoiesis and signal transduction, we analyzed the expression of these proteins in HPCs; however, we found similar expression of all in the absence of Itch (Supplementary Fig. 9).
Figure 6.
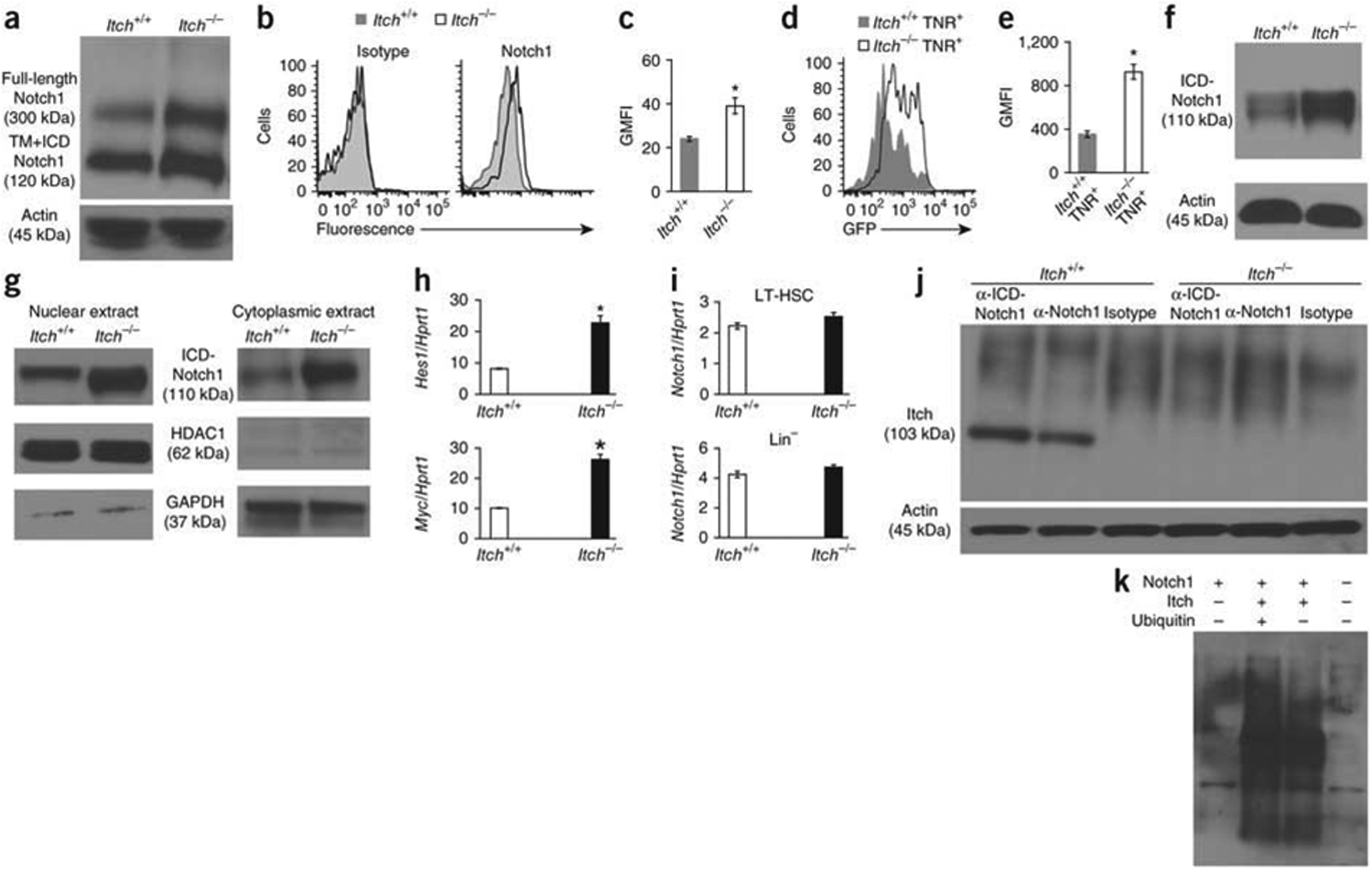
Itch deficiency results in more Notch1 protein and signaling in HPCs. (a) Full-length Notch1 and cleaved Notch1 (transmembrane and intracellular domains (TM+ICD)) in wild-type and Itch−/− Lin− bone marrow cells. Actin serves as a loading control throughout. (b) Intracellular Notch1 expression in wild-type and Itch−/− CD150+CD48− LSK cells; left, cells stained with isotype-matched control antibody (controls). (c) Geometric mean fluorescence intensity (GMFI) of the results in b. (d) Expression of green fluorescent protein (GFP) in CD150+CD48−LSK cells from Itch+/+ and Itch−/− transgenic Notch reporter (TNR+) mice. (e) Geometric mean fluorescence intensity of the results in d. (f) Cleaved Notch1 in wild-type and Itch−/− Lin−bone marrow cells. (g) cleaved Notch1 in the nucleus (left) and cytoplasm (right) of wild-type and Itch−/− Lin− bone marrow cells. HDAC1 (histone deacetylase) and GAPDH (glyceraldehyde phosphate dehydrogenase) serve as controls. (h,i) Real-time PCR analysis of Hes1 and Myc in wild-type and Itch−/− CD150+CD48− LSK cells (h) and of Notch1 in wild-type and Itch−/− CD150+CD48− LSK or Lin−cells (i); results are presented relative to Hrpt1 expression. (j) Immunoprecipitation analysis of the interaction between Itch and Notch1 in Lin− cells. (k) Ubiquitination assay of the involvement of Itch in the ubiquitination of Notch1 protein. kDa, kilodaltons. *P < 0.05 (Student’s t-test). Data are representative of five (a,f), three (b–e) or two (g–i) independent experiments (average and s.e.m. in c,e; mean and s.e.m. in h,i).
Notch1 signaling is initiated by engagement of the Notch ligand. Ligand binding leads to two successive proteolytic cleavage events that result in the separation of the intracellular domain from the transmembrane domain. The cleaved ICN1 translocates from the cell membrane to the cytoplasm and subsequently to the nucleus, where it interacts with the transcription factor CBF (RBP-jk) and activates the expression of target genes such as Hes1 and Myc35. Using antibodies specific for cleaved Notch1 (that recognize cleavage between Gly1743 and Val1744 of the intracellular domain), we detected more cleaved Notch1 in Itch−/− HPCs (Fig. 6f), which suggesting that more total Notch1 led to more release of ICN1. In addition, both the nuclear and cytoplasmic fractions of Itch-deficient HPCs had more ICN1 (Fig. 6g), which correlated with the augmented Hes1, Myc and Dtx1 mRNA in the LT-HSCs of Itch−/− mice detected by real-time PCR (Fig. 6h and Supplementary Fig. 10). Of note, Itch−/− and wild-type LT-HSC cells had similar amounts of Notch1 mRNA (Fig. 6i), which suggested that the greater abundance of Notch1 protein in Itch-deficient cells might have been a consequence of defective degradation.
To evaluate whether Itch interacts with Notch1 in HPCs, we transduced prestimulated HPCs with retrovirus overexpressing ICN1 and coimmunoprecipitated proteins from lysates with antibody to Notch1 (anti-Notch1). Our analysis showed interaction between Notch1 and Itch (Fig. 6j) and that Notch1 protein was ubiquitinated only in the presence of Itch (Fig. 6k). Finally, knockdown of Notch1 by small hairpin RNA in LT-HSCs in Itch-deficient mice resulted in a lower frequency of HSCs (Fig. 7a and Supplementary Fig. 11) and diminished radioprotection function (Table 3), which demonstrated that Notch1 activation was responsible for the phenotype of Itch−/− HSCs. Furthermore, knockdown of Notch1 in Itch−/− LSK cells led to modest proliferation in vivo (Fig. 7b) and in vitro (Fig. 7c) and lower expression of Hes1 mRNA and Myc mRNA (Fig. 7d,e). These data suggest that the phenotype of Itch−/− HSCs was at least in part due to augmented Notch1 signaling.
Figure 7.
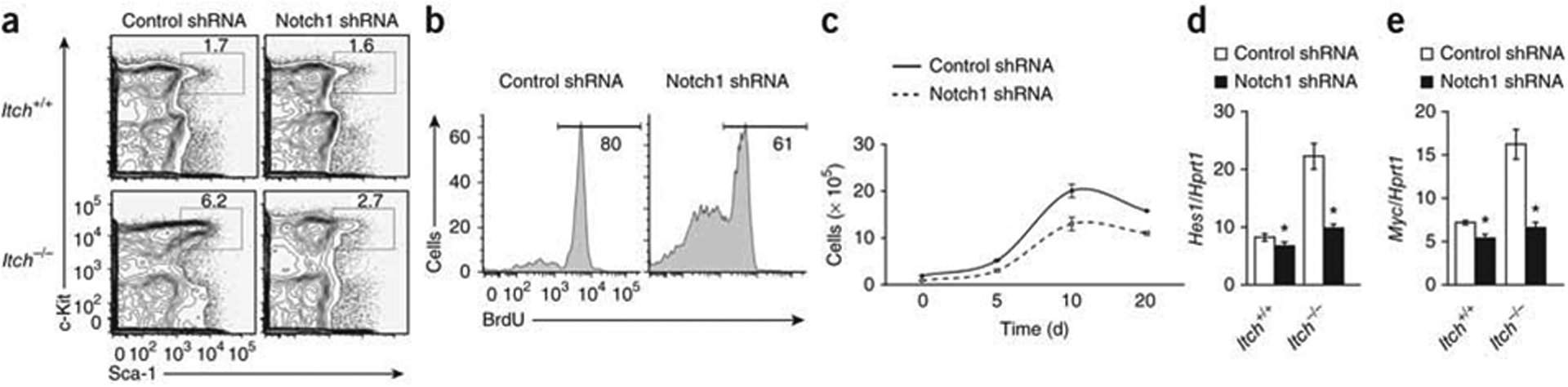
Knockdown of Notch1 in Itch-deficient HSCs results in reversion of the phenotype. (a) Distribution of donor-derived (CD45.2+) LSK cells in total bone marrow from wild-type (CD45.1+) congenic recipients 8 weeks after transplantation of wild-type and Itch−/− (CD45.2+) CD150+CD48− LSK cells transduced with control or Notch1-specific short hairpin RNA (shRNA). (b) Incorporation of BrdU in vivo by donor-derived (CD45.2+) LSK cells from wild-type (CD45.1+) congenic recipients 8 weeks after transplantation of cells as described in a. (c) Ex vivo population expansion of LSK cells obtained from wild-type (CD45.1+) congenic recipients 8 weeks after transplantation of cells as described in a, then cultured in vitro in the presence of an HSC cytokine ‘cocktail’. (d,e) Real-time PCR analysis of Hes1 (d) and Myc (e) in wild-type and Itch−/− CD150+CD48− LSK cells obtained from wild-type (CD45.1+) congenic recipients 8 weeks after transplantation of cells as described in a; results are presented relative to Hrpt1 expression. *P < 0.05 (Student’s t-test). Data are representative of two independent experiments (mean ± s.e.m. of duplicate samples in d,e).
Table 3.
Radioprotection by Itch−/− cells expressing Notch1-specific shRNA
| Donor cells | Control | Notch1 |
|---|---|---|
| 0 | 0/10 | 0/10 |
| 1 × 103 | 0/10 | 0/10 |
| 1 × 104 | 0/10 | 0/10 |
| 5 × 104 | 0/10 | 0/10 |
| 1 × 105 | 5/10 | 0/10 |
| 5 × 105 | 8/10 | 4/10 |
| 1 × 106 | 10/10 | 10/10 |
Survival of lethally irradiated wild-type (CD45.1+) recipients (n = 10 per group) of limiting numbers of bone marrow cells from wild-type recipients transplanted with CD150+CD48− LSK Itch−/− cells expressing control (middle) or Notch1-specific (left) shRNA; results are presented as the number of survivors/total mice. Data are repre- sentative of two independent experiments.
DISCUSSION
Here we have shown that Itch, a member of the HECT family of E3 ligases, is a negative regulator of HSC development and function. The self-renewal of stem cells is tightly controlled by various cell-intrinsic and cell-extrinsic factors1–6,36. Throughout their lifespan, HSCs respond to signals mediated by a spectrum of cytokines and growth factors that can promote their quiescence, proliferation, migration and differentiation. Under steady-state conditions, HSCs constantly replenish the entire hematopoietic system. During an immune insult, HSCs must proliferate and differentiate into specific cell types that accomplish the required effector functions37. Whenever HSCs switch from one physiological state to another, a tight balance between protein synthesis and protein degradation is probably vital for their proper function. Here we have shown that defective cellular pathways that resulted in deficient ubiquitination resulted in altered HSC self-renewal properties, and we have demonstrated an important role for Itch in this process. Other E3 ubiquitin ligases, such as the RING finger ligase c-Cbl, have been shown to control the development and function of HSCs7,8,38. Together, these studies highlight the pivotal roles of E3 ubiquitin ligases and the importance of post-translational modifications in the physiology of HSCs and the molecular control of HSC self-renewal.
The hallmark feature of adult stem cells is their relative proliferative quiescence2. Published studies suggest that two distinct HSC subsets—dormant HSCs and active HSCs—can exist in the HSC pool under steady-state conditions23,39. In the mouse, dormant HSCs are generally quiescent, with a division rate of approximately once every 150 d and are believed to divide only five times during the lifespan of the mouse, whereas active HSCs are thought to be more proliferative and to produce the progenitors and mature cells required for the maintenance of normal hematopoiesis23,39. Quiescence is critical for the maintenance, survival and self-renewal of HSCs. Published studies of mice deficient in p21, Gfi1, Pten, FoxO1, 3 and 4, Pbx1, Mi2β, TSC1, PML and Fbw7 have proven that unscheduled HSC proliferation results in loss of self-renewal or stem cell exhaustion2,6,39. We found that Itch-deficient mice had a larger HSC pool due to their hyper-proliferative properties but did not show the phenomenon of ‘stem cell exhaustion’. Instead, Itch-deficient HSCs were more competent and faster in repopulating the hematopoietic system under both steady-state and stress conditions, which indicatedthat in certain situations, self-renewal of HSCs is not compromised even after excessive rounds of proliferation. Similar observations have been obtained with c-Cbl-deficient mice7 and Egr1-deficient mice40, in which HSC functions are not compromised despite continuous and accelerated proliferation. Studies of these mice will be very useful in understanding the physiology and molecular mechanisms behind inexhaustible stem cell properties even after excessive rounds of proliferation.
In mammalian hematopoiesis, Notch signaling is essential for definitive hematopoiesis in the developing embryo, as well as for the development of T cells, marginal zone B cells and megakaryocytes41–43. Although the role of Notch in the differentiation of multiple hematopoietic lineages has become increasingly clear, its role in the development and maintenance of adult HSCs has remained controversial. The idea of Notch1 as an important modulator of adult HSCs has been provided by several gain-of-function studies through overexpression of the active form of Notch1 (ICN1) or its downstream target Hes1 in HSCs. ICN1 overexpression in HSCs results in ex vivo HSC population expansion without compromise of their self-renewal properties30,43,44. However, genetic studies of mice that are deficient in Notch1, Jagged1 or Rbp-jκ or express a dominant negative form of the mastermind-like protein DN-MAML, which functions as a total Notch inhibitor, have shown that canonical Notch signaling is dispensable for HSC homeostasis in the bone marrow42–44. A possible interpretation of these paradoxical findings might be that although Notch signaling might not be required for the homeostasis of adult HSCs, augmented Notch signaling may result in enhanced HSC self-renewal. In line with that idea, it has been proposed that Notch signaling is maintained at a minimal basal amount in HSCs42,43. As exaggerated Notch signaling results in altered self-renewal, pathways that negatively regulate Notch signaling should remain active in HSCs. Itch is known to directly participate in the degradation of Notch1 through ubiquitination15,17,32,33. Our analysis has provided direct evidence that the lack of Itch resulted in augmented Notch1 signaling in HSCs, leading to altered self-renewal.
LRF has also been identified as a negative regulator of Notch signaling in early hematopoiesis45. LRF-deficient mice have more HSCs, perturbed B cell development and ectopic T cell development in the bone marrow due to inappropriate activation of Notch signaling in the hematopoietic progenitor cells45. These observations are in line with the earlier idea that Notch signaling needs to be suppressed during the early stages of B cell development in the bone marrow46. We propose that Itch deficiency results in augmented Notch signaling, thus leading to enhanced self-renewal of HSCs. Although the phenotype of LRF-deficient and Itch-deficient HSCs was similar, we did not detect extrathymic T cell development or perturbed B cell development in the bone marrow of young (4-week-old) Itch-deficient mice. A possible explanation for the distinct phenotypes is that LRF deficiency might induce inappropriate Notch expression and activation in cells that usually lack Notch signaling, such as cells of the B lineage45. In contrast, Itch deficiency resulted in sustained activation of Notch signaling only in hematopoietic cells that usually express Notch.
Nonetheless, the phenotype of Itch-deficient HSCs should be interpreted with care, as it differs in several ways from the ICN1-overexpression phenotype47. Unlike HSCs with retrovirus-mediated, constitutive expression of ICN1, Itch−/− HSCs expressed physiological amounts of Notch1 mRNA and Notch1 protein, and Notch1 activation strictly required Notch engagement by its ligands expressed on neighboring cells in the niche. Those and other results may explain any differences that can be observed between Itch−/− HSCs and ICN1-overexpressing HSCs. Nevertheless, this is the first report to our knowledge describing a phenotype associated with augmented Notch signaling in HSCs under physiological settings.
Several human patients have been identified with mutations in ITCH that result in deficiency in Itch expression48. These patients have multiple defects, including multisystem autoimmunity, morphological and developmental abnormalities48. Although there is no information available yet on the HSC defects in these patients, our study warrants consideration of such a possibility. In essence, our study has identified a previously unknown function for Itch in the restriction of the HSC pool size and emphasizes the importance of post-translational modifications in the maintenance of HSC homeostasis. A better understanding of the roles of Itch in regulation of hematopoiesis might aid the development of new therapies for stem cell–based disorders.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
Note: Supplementary information is available on the Nature Immunology website.
ONLINE METHODS
Mice.
The Itch−/− mice have been described16. Transgenic Notch reporter mice (Jackson Laboratories) were crossed with Itch−/− mice to obtain Itch+/− (heterozygous) transgenic Notch reporter pups; those mice were interbred to generate Itch−/− and Itch+/+ transgenic Notch reporter mice. All mice were kept under specific pathogen–free conditions in the animal care facility at Yale University. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Yale University.
Competitive-repopulation and bone marrow–transplantation studies.
For competitive-repopulation experiments, various numbers of donor (CD45.2+) LSK cells (1 × 103, 2 × 103, 1 × 104, 1.8 × 104 or 2 × 104) were sorted from 8-week-old wild-type and Itch−/− mice and mixed with defined numbers of competitor LSK (CD45.1+) cells (2 × 104, 1.9 × 104, 1.8 × 104, 1.7 × 104 and 2 × 103) and the mixtures were transplanted intravenously into lethally irradiated (11 Gy) congenic (CD45.1+) recipients. For serial transplantation, 2 × 106 bone marrow cells from the primary transplants (6 months after transplantation) were injected into lethally irradiated congenic (CD45.1+) recipients. In some experiments, Itch+/+ and Itch−/− bone marrow was depleted of red blood cells and 1 × 106 total cells were transplanted into lethally irradiated (CD45.1+) recipient mice.
For radioprotection assays, red blood cells were lysed in Itch+/+ and Itch−/− donor bone marrow and limiting dilutions of the resulting bone marrow cells (1 × 102, 1 × 103, 5 × 103, 1 × 104, 5 × 104 and 1 × 105) were injected into lethally irradiated wild-type (CD45.1+) recipient mice. All mice survived at least 10 d, which suggested that fatalities were due to hematopoietic failure.
Cell culture.
Purified CD150+CD48− LSK cells or LSK cells were cultured in vitro in the presence all or some of the following recombinant cytokines: mouse interleukin 3 (10 ng/ml), mouse interleukin 6 (10 ng/ml), mouse stem cell factor (50 ng/ml), mouse thrombopoietin (10 ng/ml) and human Flt3L (50 ng/ml; all from Peprotech). Cells were cultured in Iscove’s modified Dulbecco medium supplemented with 10% FCS, 2 mM L-Glutamine, 1% Penicillin-Streptomycin and 1 mM non-essential amino acids.
Cell-proliferation studies.
In vivo proliferation experiments with BrdU and in vitro proliferation experiments with CFSE (carboxyfluorescein diacetate succinimidyl ester) were done as described7.
For assay of colony-forming units, cells cultured in vitro were sorted on day 20 on the basis of their LSK immunophenotype and were cultured in semisolid Methocult M3534 medium supplemented with cytokines to promote the proliferation of progenitor cells; colonies were detected on day 12 according to the manufacturer’s instructions (StemCell Technologies).
Analysis with 5-FU.
Mice were given a single intraperitoneal dose of 5-FU (150 mg per kg body weight) on day 0. Blood was obtained from mice or mice were killed on days 0, 2, 5 and 10 after injection, and their bone marrow LSK compartments were analyzed by flow cytometry. The Hemavet 950 LV system (DREW scientific) was used for differential blood count analysis.
In some experiments, wild-type CD45.1+ recipients were lethally irradiated and given transplantation of Itch+/+ or Itch−/− bone marrow cells. At 8 weeks after transplantation, 5-FU was administered intraperitoneally to recipients (150 mg per kg body weight on a weekly basis) and survival was monitored.
Pyronin and Hoechst staining.
The cell cycle status of the freshly isolated HSCs was determined with Hoechst 33342 (Molecular Probes) and pyronin Y staining (Sigma). Sorted LT-HSCs were resuspended in PBS containing 2% (vol/vol) FCS and 10 μM Hoechst 33342. Cells were then incubated for 30 min at 37 °C and then were washed and resuspended in PBS supplemented with 20 mM HEPES, pH 7.4, glucose (1 mg/ml), 10% (vol/vol) FCS, 10 μM Hoechst 33342 and pyronin Y (1 μg/ml). Cells were incubated for an additional 30 min at 37 °C, then were washed and analyzed by flow cytometry. Pyronin Y fluorescence was detected at 575 nm in the linear range.
Flow cytometry.
Single-cell suspensions were analyzed by flow cytometry with a FACSScan, FACSCalibur or LSR II (BD) and CellQuest software (BD Biosciences), FACSDiva software (BD Biosciences) or FlowJo software (Tree Star), respectively. Cells were sorted into defined subpopulations with a FACSAria (BD Biosciences). Cells incubated with biotinylated monoclonal antibodies were incubated with fluorochrome-conjugated streptavidin-phycoerythrin, streptavidin–peridinin chlorophyll protein, streptavidin-allophycocyanin and streptavidin-allophycocyanin-indotricarbocyanine (BD Pharmingen). The following monoclonal antibodies were used: CD3e (145-2C11; BD Biosciences), CD-4 (GK1.5; BD Biosciences), CD8 (53-6.7; BD Biosciences), CD11b (M1/70; BD Biosciences), CD11c (HL3; BD Biosciences), CD19 (1D3; BD Biosciences), CD34 (RAM34; BD Biosciences), CD45.1 (A20; BD Biosciences), CD45.2 (104; BD Biosciences), CD48 (HM48-1; BD Biosciences), CD117 (2B8; BD Biosciences), CD150 (TC15-12F12.2; Biolegend), B220 (RA3-6B2; BD Biosciences), Flt3 (A2F10.1; BD Biosciences), Gr-1 (RB6-8C5; BD Biosciences), Sca-1 (D7; BD Biosciences) and TER119 (TER119; BD Biosciences). Cells reacted with biotinylated monoclonal antibodies were incubated with fluorochrome-conjugated streptavidin-PE (#554061; BD Biosciences), streptavidin-PerCP-Cy5.5 (#551419; BD Biosciences), streptavidin-APC (#554067; BD Biosciences) and streptavidin-APC-Cy7 (#554063; BD Biosciences). All fluorescence intensity plots are presented in log scales.
Intracytoplasmic staining.
For detection of Notch1 by flow cytometry, CD150+CD48−LSK cells were first fixed and made permeable with a Phosflow kit (BD Pharmingen) and then were stained with phycoerythrin–anti-Notch1 (mN1A; BD Biosciences) according to the manufacturer’s instructions (BD Biosciences).
Immunoblot analysis and Immunoprecipitation.
Immunoblot analyses were done as described7. Primary antibodies were specific for c-Jun (60A8; Cell signaling), JunB (C37F9; Cell signaling), Fyn (# 4023; Cell signaling), SMAD2 (86F9; Cell signaling), p73 (#4662; Cell signaling), HDAC1 (#2062; Cell signaling), c-Myc (#9402; Cell signaling), ubiquitin (#3933; Cell signaling), Notch1 (full-length + transmembrane domain(C44H11; Cell signaling) and cleaved Notch1 (Val1744; Cell signaling), Hes1 (#Q14469; Millipore) and GAPDH (L18; Santa Cruz Biotechnologies) and actin (I-19; Santa Cruz Biotechnologies). Proteins were immunoprecipitated with antibody to cleaved Notch1(Val1744) with a commercially available kit according to the manufacturer’s instructions (Upstate).
RNA extraction and real-time PCR.
Total RNA was isolated with the RNeasy Mini kit (‘Qiagen), then cDNA was synthesized with oligo(dT) primer and Expand reverse transcriptase (Roche). PCR was done in duplicate with a 7500 RealTime PCR system and Power SYBR Green PCR Master Mix according to the manufacturer’s instructions (Applied Biosystems).
Retrovirology.
For short hairpin RNA (shRNA) studies, Notch1-specific shRNA was designed through VectorNTI (Invitrogen) software. Two shRNA constructs recognizing different regions of Notch1 mRNA (shRNA1 and shRNA2) were cloned into the pSicoR retroviral backbone (Addgene). Retroviruses were generated with PhoenixGP cell lines. Confirmation of knockdown of Notch1 mRNA with shRNA showed efficient downregulation with both shRNA1 and shRNA2; however, the efficiency of knockdown was better (~70%) with shRNA1 and thus only shRNA1 was used for the knockdown experiments. Cells transduced with control shRNA served as controls.
For immunoprecipitation studies, retrovirus overexpressing ICN1 was prepared as described30.
Ubiquitination studies.
For ubiquitination assays, human embryonic kidney 293T cells were cotransfected with plasmid that expressing c-Itch, Notch1 and ubiquitin, then, 48 h after transfection, proteins were immunoprecipitated with anti-Notch1 (C44H11; Cell signaling) as described above. Precipitates were analyzed by immunoblot, probed with anti-ubiquitin (#3933; Cell signaling).
Statistical analysis.
Statistical significance was assessed with a two-sided Student’s t-test. P values above 0.05 were considered not significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Manzo for help with manuscript submission, and the Yale Cell Sorter Facility for support. Supported by the Howard Hughes Medical Institute (R.A.F.).
We thank the National Institutes of Health Center for Biomedical Research Excellence (P20RR018757) administrative core at Roger Williams Medical Center.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Morrison SJ, Uchida N & Weissman IL The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol 11, 35–71 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Orford KW & Scadden DT Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet 9, 115–128 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Orkin SH & Zon LI Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He S, Nakada D & Morrison SJ Mechanisms of stem cell self-renewal. Annu. Rev. Cell Dev. Biol 25, 377–406 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Blank U, Karlsson G & Karlsson S Signaling pathways governing stem-cell fate. Blood 111, 492–503 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Wilson A, Laurenti E & Trumpp A Balancing dormant and self-renewing hematopoietic stem cells. Curr. Opin. Genet. Dev 19, 461–468 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Rathinam C, Thien CB, Langdon WY, Gu H & Flavell RA The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 22, 992–997 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokomizo T & Dzierzak E Fine-tuning of hematopoietic stem cell homeostasis: novel role for ubiquitin ligase. Genes Dev. 22, 960–963 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conaway RC, Brower CS & Conaway JW Emerging roles of ubiquitin in transcription regulation. Science 296, 1254–1258 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Hershko A & Ciechanover A The ubiquitin system. Annu. Rev. Biochem 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Haglund K, Di Fiore PP & Dikic I Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci 28, 598–603 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Hicke L & Dunn R Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol 19, 141–172 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Kodadek T, Sikder D & Nalley K Keeping transcriptional activators under control. Cell 127, 261–264 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Pickart CM Mechanisms underlying ubiquitination. Annu. Rev. Biochem 70, 503–533 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Bernassola F, Karin M, Ciechanover A & Melino G The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Perry WL et al. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet 18, 143–146 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Matesic LE, Haines DC, Copeland NG & Jenkins NA Itch genetically interacts with Notch1 in a mouse autoimmune disease model. Hum. Mol. Genet 15, 3485–3497 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Fang D et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol 3, 281–287 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Venuprasad K et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat. Immunol 9, 245–253 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parravicini V, Field AC, Tomlinson PD, Basson MA & Zamoyska R Itch−/− alphabeta and gammadelta T cells independently contribute to autoimmunity in Itchy mice. Blood 111, 4273–4282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C & Morrison SJ SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Yilmaz OH, Kiel MJ & Morrison SJ SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood 107, 924–930 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson A et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Dzierzak E & Speck NA Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol 9, 129–136 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X, Cho S & Spangrude GJ Hematopoietic stem cells: generation and self-renewal. Cell Death Differ. 14, 1851–1859 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Johnson GR & Moore MA Role of stem cell migration in initiation of mouse foetal liver haemopoiesis. Nature 258, 726–728 (1975). [DOI] [PubMed] [Google Scholar]
- 27.Spangrude GJ, Brooks DM & Tumas DB Long-term repopulation of irradiated mice with limiting numbers of purified hematopoietic stem cells: in vivo expansion of stem cell phenotype but not function. Blood 85, 1006–1016 (1995). [PubMed] [Google Scholar]
- 28.Gothot A, van der Loo JC, Clapp DW & Srour EF Cell cycle-related changes in repopulating capacity of human mobilized peripheral blood CD34+ cells in non-obese diabetic/severe combined immune-deficient mice. Blood 92, 2641–2649 (1998). [PubMed] [Google Scholar]
- 29.Lerner C & Harrison DE 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp. Hematol 18, 114–118 (1990). [PubMed] [Google Scholar]
- 30.Rathinam C et al. Generation and characterization of a novel hematopoietic progenitor cell line with DC differentiation potential. Leukemia 20, 870–876 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Varnum-Finney B et al. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med 6, 1278–1281 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Chastagner P, Israel A & Brou C AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE 3, e2735 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu L et al. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem 275, 35734–35737 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Duncan AW et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol 6, 314–322 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Allman D, Aster JC & Pear WS Notch signaling in hematopoiesis and early lymphocyte development. Immunol. Rev 187, 75–86 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Moore KA & Lemischka IR Stem cells and their niches. Science 311, 1880–1885 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Rathinam C & Flavell RA The hematopoiesis paradigm: clarity or ambiguity? Blood 112, 3534–3535 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrison BS & Rossi DJ Controlling stem cell fate one substrate at a time. Nat. Immunol 11, 193–194 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Trumpp A, Essers M & Wilson A Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol 10, 201–209 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Min IM et al. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell 2, 380–391 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Kopan R & Ilagan MX The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maillard I et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell 2, 356–366 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radtke F, Fasnacht N & Macdonald HR Notch signaling in the immune system. Immunity 32, 14–27 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Maillard I, Adler SH & Pear WS Notch and the immune system. Immunity 19, 781–791 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Maeda T et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science 316, 860–866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radtke F, Wilson A & MacDonald HR Notch signaling in T- and B-cell development. Curr. Opin. Immunol 16, 174–179 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Pui JC et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299–308 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Lohr NJ et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am. J. Hum. Genet 86, 447–453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


