ABSTRACT
Burkholderia cepacia complex (Bcc) and Burkholderia pseudomallei complex (Bpc) species include pathogens that are typically multidrug resistant. Dominant intrinsic and acquired multidrug resistance mechanisms are efflux mediated by pumps of the resistance-nodulation-cell division (RND) family. From comparative bioinformatic and, in many instances, functional studies, we infer that RND pump-based resistance mechanisms are conserved in Burkholderia. We propose to use these findings as a foundation for adoption of a uniform RND efflux pump nomenclature.
KEYWORDS: Burkholderia, multidrug resistance, RND efflux pumps
INTRODUCTION
The genus Burkholderia encompasses a diverse group of beneficial and pathogenic bacteria (1–3). The genus is separated into two main groups; the Burkholderia cepacia complex (Bcc; 22 members) (1, 4, 5) and the Burkholderia pseudomallei complex (Bpc; 6 members) (6). Both complexes contain potent opportunistic pathogens of plants, animals, and humans (1, 5, 7). In Bcc, Burkholderia cenocepacia and Burkholderia multivorans are pathogens that frequently afflict compromised individuals, most notably people with cystic fibrosis (7). In Bpc, B. pseudomallei and its close relative Burkholderia mallei are the etiologic agents of melioidosis and glanders, respectively (8, 9). A common hallmark of Burkholderia infections is that they are often recalcitrant to antibiotic therapy (10–12).
Efflux via pumps belonging to the resistance-nodulation-cell division (RND) family are the dominant intrinsic and acquired multidrug resistance mechanism in Burkholderia species (13). Despite the fact that Bpc species have been intensively studied for only a short time compared to Bcc species, the landscape of RND pumps in Bpc bacteria such as B. pseudomallei (14–20) and Burkholderia thailandensis (21, 22) has been well defined. RND pumps in Bcc have been studied to a significant extent in B. cenocepacia (12, 23–29) and more sporadically in other Bcc members, such as Burkholderia vietnamiensis (30) and Burkholderia ubonensis (31). We previously noted that the RND efflux pump nomenclature in Bcc species is nonuniform and confusing (13). Bioinformatic and, in many instances, functional analyses predict that RND efflux pump-based resistance mechanisms are conserved in Bcc and Bpc species. We propose to use these findings as a foundation for adoption of a more uniform RND efflux pump nomenclature.
(Part of this work was presented at the Multi-drug Efflux Gordon Research Conference, Barga [Lucca], Italy, 2019).
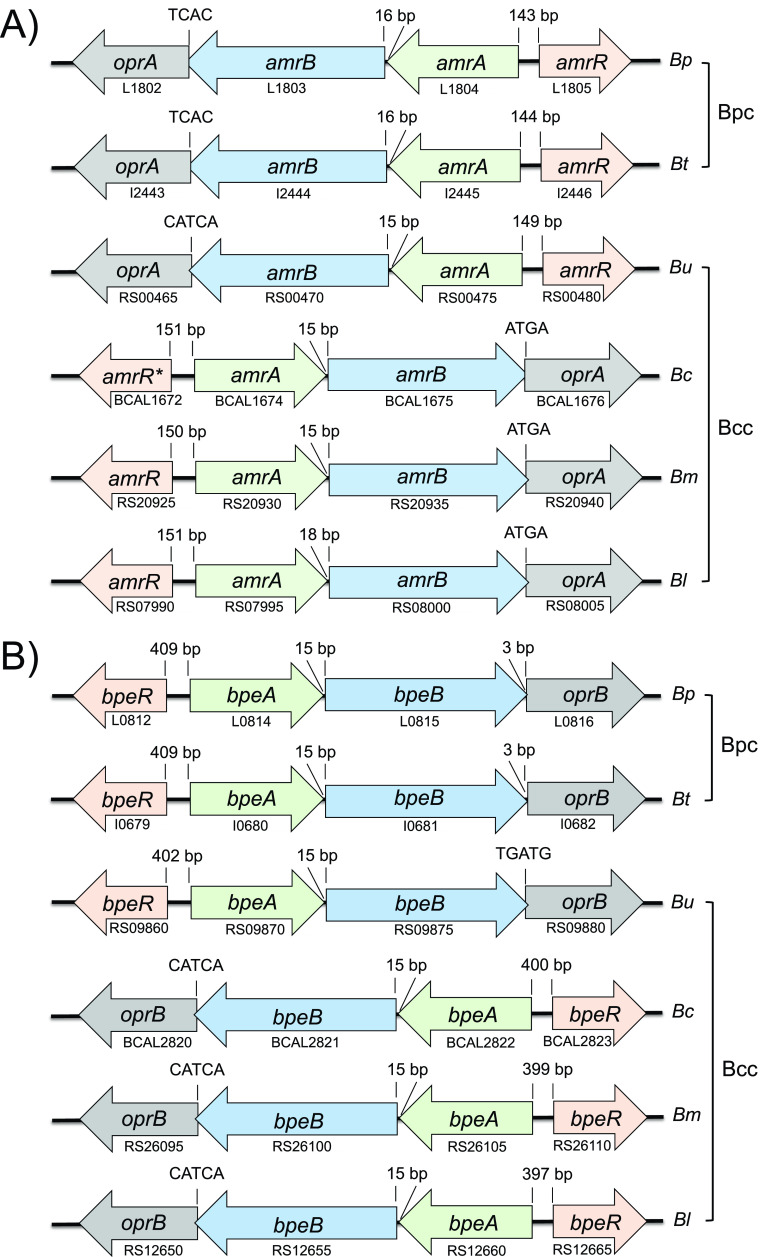
We focused on three Bcc and Bpc RND efflux pumps for which published and unpublished experimental data were available, as well as evidence of potential clinical significance with regard to the resistance profiles bestowed by these pumps (Table 1). Since these criteria are best defined in Bpc bacteria, especially B. pseudomallei (14–20) and the closely related B. thailandensis (21, 22), we chose these species’ well-characterized AmrAB-OprA, BpeAB-OprB, and BpeEF-OprC efflux pumps as models and queries for identification and naming of the corresponding efflux pumps in Bcc species. Lesser-known Bcc members were included either as an example of a potential environmental reservoir of novel antimicrobial resistance determinants (B. ubonensis) (31, 32) or as an emerging opportunistic pathogen whose biology and antimicrobial resistance potential are not yet well understood (Burkholderia latens) (33).
TABLE 1.
Established RND efflux pumps in Burkholderia speciesa
| Species | Efflux pump | Gene names | Gene annotation | Major antibiotic substrate(s) | Reference(s) |
|---|---|---|---|---|---|
| B. pseudomallei | AmrAB-OprA | amrAB-oprA | BPSL1804–1802 | AG, MAC, TET, CET | 14, 46 |
| BpeAB-OprB | bpeAB-oprB | BPSL0814–0816 | CHL, FQ, MAC, TETb | 15, 17 | |
| BpeEF-OprC | bpeEF-oprC | BPSS0292–0294 | CHL, FQ, TET, TMP, SMXc | 18, 19, 20 | |
| B. thailandensis | AmrAB-OprA | amrAB-oprA | BTH_I2445–I2443 | AG, MAC, TET | 22 |
| BpeAB-OprB | bpeAB-oprB | BTH_I0680–I0682 | TET | 22 | |
| BpeEF-OprC | bpeEF-oprC | BTH_II2106–II2104 | CHL, FQ, TET, TMP, SMX | 21, 22 | |
| B. cenocepacia | RND-3 | NA | BCAL1674–1676 | AG, FQ, NAL, TET | 25, 28 |
| RND-4 | NA | BCAL2822–2820 | ATM, CHL, FQ, TOB, TET | 26, 28 | |
| RND-10 | ceoAB-opcM | BCAM2551–2549 | CHL, FQ, TET, TMPc | 23, 47 |
For details, see Table S1. This is an updated and condensed version of a previously published table (13). Abbreviations: AG, aminoglycosides; ATM, azithromycin; CET, cethromycin; CHL, chloramphenicol; FQ, fluoroquinolones; MAC, macrolides; MEM, meropenem; NAL, nalidixic acid; SMX, sulfamethoxazole; TET, tetracycline(s); TMP, trimethoprim; TOB, tobramycin; NA, not applicable.
DIAMOND BLASTP (34) within the Burkholderia Genome Database (www.burkholderia.com) (35) was employed for identification of candidate efflux pump proteins in other Burkholderia species. These candidates were further scrutinized by using them in DIAMOND BLASTP, performing protein sequence alignments and similarity predictions using MUSCLE on the EMBL-EBI server (https://www.ebi.ac.uk/Tools/msa/muscle/) (36), and by genomic content analyses (e.g., chromosome localization and presence of predicted cognate transcriptional regulators). In addition to similarity predictions, transcriptional regulators were analyzed for conserved helix-turn-helix (HTH) DNA-binding domains by the Rhone-Alpes Bioinformatic Pole Gerland Site (https://npsa-prabi.ibcp.fr) (37). In four instances, DNA sequence alignments were used to reexamine and revise translational start sites (see Fig. S1 in the supplemental material).
By using these methods, we identified gene clusters encoding AmrAB-OprA, BpeAB-OprB, and BpeEF-OprC in the four Bcc species that we studied, namely, B. ubonensis, B. cenocepacia, B. multivorans, and B. latens (Fig. 1). In all instances, the architecture of the predicted structural and regulatory gene clusters was identical in Bcc and Bpc: (i) the genes encoding the AmrAB-OprA, BpeAB-OprB, and BpeEF-OprC efflux system components exhibit the same operon structure; (ii) the locations and transcriptional orientation of the operon-associated regulatory genes are the same; (iii) transcriptional regulator protein sequences and their predicted DNA-binding domains are highly conserved; and (iv) the gene clusters are located on the same chromosomes (amrAB-oprA and bpeAB-oprB on chromosome 1 and bpeEF-oprC on chromosome 2). These conclusions are supported by published data and new experimental findings presented below. Most previously published experimental data about RND pumps in Bcc were obtained from studies of B. cenocepacia. These and more recent studies lend credence to correct efflux gene assignments in B. cenocepacia and other Bcc species.
FIG 1.
Continued.
Studies with B. cenocepacia strain J2315 found that AmrAB-OprA (also known as RND-3) (see Table S1) plays a role in antimicrobial resistance (Table 1). Increased expression of AmrAB-OprA encoded by the genes BCAL1674 to BCAL1676 on chromosome 1 led to increased antimicrobial resistance (29). Conversely, deletion of AmrAB-OprA led to decreased antibiotic resistance (38). The amrAB-oprA-associated AmrR repressors are highly conserved in the examined Bcc and Bpc species (Fig. 1A), with the Bcc proteins exhibiting 72.1 to 72.7% identity to B. pseudomallei K96243 AmrR (Fig. S2A) and containing highly conserved predicted DNA-binding domains (Fig. S2B). In strain J2315, the amrR gene encoding the TetR family repressor AmrR is a frame-shifted pseudogene coding for a truncated protein (29). In its stead, the wild-type AmrR from K56-2 was therefore used for AmrR protein analyses (39). Several studies with environmental and clinical B. pseudomallei isolates showed that AmrAB-OprA is the major intrinsic aminoglycoside (AG) resistance determinant. AG-susceptible strains either lack a functional efflux pump due to deletion of the amrAB-oprA operon (16) or amrB point mutations (40, 41) or fail to express the efflux operon (16). Similar mutations are responsible for the AG susceptibility of at least some B. mallei strains (13). We recently confirmed that AmrAB-OprA is the intrinsic AG resistance determinant in B. ubonensis (32). A study with B. vietnamiensis showed that changes in AmrR-dependent expression of what was called AmrAB-OprM led to acquisition of AG resistance in vivo and in vitro (30). Studies with B. pseudomallei (42) and B. ubonensis (31) revealed that AmrAB-OprA also plays a role in resistance to tetracycline (TET) and its derivatives (e.g., doxycycline). Lastly, we show here that in B. latens AmrAB-OprA is induced in response to chloramphenicol (CHL) exposure (Table S2).
In B. cenocepacia J2315, the BpeAB-OprB pump (also known as RND-4 [Table S1]) encoded by the genes BCAL2822 to BCAL2820 has previously been implicated in antibiotic resistance (Table 1). Deletion of this pump leads to decreased antimicrobial resistance (43). Like its B. pseudomallei counterpart, BpeAB-OprB is expressed at only low levels in strain J2315 (17, 29). Although J2315 has two potential bpeAB-oprB gene clusters annotated, RND-4 on chromosome 1 and RND-2 (BCAS0764 to BCAS0766) on chromosome 3 (Fig. S3), only RND-4 (or BpeAB-OprB) was shown to play a role in antibiotic resistance. Two lines of evidence support the notion that there is only one BpeR-regulated bpeAB-oprB operon on chromosome 1 as indicated in Fig. 1B. The first is the absence of the conserved bpeR gene upstream of the RND-2 genes (Fig. S3). The bpeAB-oprB associated BpeR repressors (confirmed in B. pseudomallei [17]) are highly conserved in the examined Bcc and Bpc species, with the Bcc proteins exhibiting 80.1 to 82% identity to B. pseudomallei K96243 BpeR (Fig. S2A) and containing highly conserved predicted DNA-binding domains (Fig. S2B). The second is the location of RND-2 genes on the dispensable chromosome 3 (44). In contrast to B. pseudomallei where BpeAB-OprB seems to play a minor role in antibiotic resistance (17), this efflux system apparently plays a more dominant role in Bcc species, at least in B. cenocepacia (28).
The BpeEF-OprC pump (also known as RND-10 or CeoAB-OpcM [Table S1]) encoded by the genes BCAM2551 to BCAM2549 in strain J2315 was the first RND efflux pump characterized in B. cenocepacia (23, 24). Its substrates include CHL, TET,trimethoprim (TMP), and ciprofloxacin. Expression of CeoAB-OpcM was shown to be inducible by CHL and sulfamethoxazole/trimethoprim (SXT), likely involving a divergently transcribed gene, ceoR, which encodes an uncharacterized LysR-type transcriptional regulator (LTTR) (23, 47). In B. pseudomallei, BpeEF-OprC expression is transcriptionally regulated by two closely related LTTRs: (i) BpeT, encoded by the divergently from the llpE-bpeE-bpeF-oprC-transcribed bpeT gene on chromosome 2 (Fig. 1C), and (ii) BpeS, encoded by a distant gene on chromosome 1 (19, 20). BpeT and BpeS mutations are the main causes of acquired CHL, fluoroquinolone, TMP, and SXT resistance (19, 20). Bcc BpeS homologs are also found on chromosome 1 in all species examined, although in a different genomic context (Fig. S4).
BpeT and BpeS are highly conserved in the examined Bcc and Bpc species, with the Bcc proteins exhibiting 91.0 to 93.4% (BpeT) and 82 to 85.2% (BpeS) identity to the corresponding B. pseudomallei K96243 proteins (Fig. S2A) and preservation of conserved predicted DNA-binding domains (Fig. S2B). A unique hallmark of the ceoAB-opcM operon is that its three genes are cotranscribed with llpE, which encodes a putative lipase/esterase of unknown function. LlpE is not required for antibiotic efflux (20, 23), and its presence in an operon with bpeEF-oprC is conserved in all Bcc, all Bpc, and other Burkholderia species (e.g., Burkholderia gladioli [Fig. S5]) examined to date. Burkholderia cenocepacia CeoAB-OpcM shares many traits with B. pseudomallei BpeEF-OprC (e.g., efflux pump components, substrate spectrum, transcriptional regulation by two LTTRs, inducibility with select pump substrates, organization in an operon with the unique llpE, and location on chromosome 2), and these traits are conserved in other Bcc and Bpc species (Fig. 1C). For instance, we show in this study that in B. latens BpeEF-OprC is constitutively expressed in a strain whose multidrug resistance (MDR) profile matches that of B. pseudomallei and B. cenocepacia BpeEF-OprC (Fig. S6).
Comparative bioinformatic analyses and mounting evidence from functional studies in Bcc and Bpc species support the notion that RND efflux pump-based resistance mechanisms are largely conserved across Bcc and Bpc. Based on the work and findings reported here we propose a uniform RND efflux pump nomenclature as shown in Fig. 1. Its adoption by the research community would greatly facilitate following the literature and advancing the field.
ACKNOWLEDGMENTS
This work was funded by grant HDTRA1-17-1005-1 from the United States Defense Threat Reduction Agency (DTRA).
The contents are solely the responsibility of the authors and do not necessarily represent the official views of DTRA or the Department of Defense.
We acknowledge John J. LiPuma, Director, Burkholderia cepacia Research Laboratory & Repository, University of Michigan, for his insightful comments on the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 2.Suarez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonca-Previato L, James EK, Venturi V. 2012. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol 63:249–266. 10.1007/s00248-011-9929-1. [DOI] [PubMed] [Google Scholar]
- 3.Jones C, Webster G, Mullins AJ, Jenner M, Bull MJ, Dashti Y, Spilker T, Parkhill J, Connor TR, LiPuma JJ, Challis GL, Mahenthiralingam E. 2021. Kill and cure: genomic phylogeny and bioactivity of Burkholderia gladioli bacteria capable of pathogenic and beneficial lifestyles. Microb Genom 7:mgen000515. 10.1099/mgen.0.000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estrada-de los Santos P, Palmer M, Chávez-Ramírez B, Beukes C, Steenkamp E, Briscoe L, Khan N, Maluk M, Lafos M, Humm E, Arrabit M, Crook M, Gross E, Simon M, dos Reis Junior F, Whitman W, Shapiro N, Poole P, Hirsch A, Venter S, James E. 2018. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes (Basel) 9:389. 10.3390/genes9080389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallner A, King E, Ngonkeu ELM, Moulin L, Bena G. 2019. Genomic analyses of Burkholderia cenocepacia reveal multiple species with differential host-adaptation to plants and humans. BMC Genomics 20:803. 10.1186/s12864-019-6186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahl JW, Vazquez AJ, Hall CM, Busch JD, Tuanyok A, Mayo M, Schupp JM, Lummis M, Pearson T, Shippy K, Colman RE, Allender CJ, Theobald V, Sarovich DS, Price EP, Hutcheson A, Korlach J, LiPuma JJ, Ladner J, Lovett S, Koroleva G, Palacios G, Limmathurotsakul D, Wuthiekanun V, Wongsuwan G, Currie BJ, Keim P, Wagner DM. 2016. The effects of signal erosion and core genome reduction on the identification of diagnostic markers. mBio 7:e00846-16. 10.1128/mBio.00846-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipuma JJ. 2005. Update on the Burkholderia cepacia complex. Curr Opin Pulm Med 11:528–533. 10.1097/01.mcp.0000181475.85187.ed. [DOI] [PubMed] [Google Scholar]
- 8.Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, Dance DAB, Limmathurotsakul D. 2018. Melioidosis. Nat Rev Dis Primers 4:17107. 10.1038/nrdp.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitlock GC, Estes DM, Torres AG. 2007. Glanders: off to the races with Burkholderia mallei. FEMS Microbiol Lett 277:115–122. 10.1111/j.1574-6968.2007.00949.x. [DOI] [PubMed] [Google Scholar]
- 10.Schweizer HP. 2012. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol 7:1389–1399. 10.2217/fmb.12.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes KA, Schweizer HP. 2016. Antibiotic resistance in Burkholderia species. Drug Resist Updat 28:82–90. 10.1016/j.drup.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scoffone VC, Chiarelli LR, Trespidi G, Mentasti M, Riccardi G, Buroni S. 2017. Burkholderia cenocepacia infections in cystic fibrosis patients: drug resistance and therapeutic approaches. Front Microbiol 8:1592. 10.3389/fmicb.2017.01592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6:305. 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore RA, DeShazer D, Reckseidler S, Weissman A, Woods DE. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother 43:465–470. 10.1128/AAC.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan YY, Tan TMC, Ong YM, Chua KL. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob Agents Chemother 48:1128–1135. 10.1128/AAC.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trunck LA, Propst KL, Wuthiekanun V, Tuanyok A, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Peacock SJ, Keim P, Dow SW, Schweizer HP. 2009. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl Trop Dis 3:e0000519. 10.1371/journal.pntd.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mima T, Schweizer HP. 2010. The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob Agents Chemother 54:3113–3120. 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Podnecky NL, Wuthiekanun V, Peacock SJ, Schweizer HP. 2013. The BpeEF-OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei isolates. Antimicrob Agents Chemother 57:4381–4386. 10.1128/AAC.00660-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podnecky NL, Rhodes KA, Mima T, Drew HR, Chirakul S, Wuthiekanun V, Schupp JM, Sarovich DS, Currie BJ, Keim P, Schweizer HP. 2017. Mechanisms of resistance to folate pathway inhibitors in Burkholderia pseudomallei: deviation from the norm. mBio 8:e01357-17. 10.1128/mBio.01357-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes KA, Somprasong N, Podnecky NL, Mima T, Chirakul S, Schweizer HP. 2018. Molecular determinants of Burkholderia pseudomallei BpeEF-OprC efflux pump expression. Microbiology (Reading) 164:1156–1167. 10.1099/mic.0.000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biot FV, Valade E, Garnotel E, Chevalier J, Villard C, Thibault FM, Vidal DR, Pages JM. 2011. Involvement of the efflux pumps in chloramphenicol selected strains of Burkholderia thailandensis: proteomic and mechanistic evidence. PLoS One 6:e16892. 10.1371/journal.pone.0016892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biot FV, Lopez MM, Poyot T, Neulat-Ripoll F, Lignon S, Caclard A, Thibault FM, Peinnequin A, Pages JM, Valade E. 2013. Interplay between three RND efflux pumps in doxycycline-selected strains of Burkholderia thailandensis. PLoS One 8:e84068. 10.1371/journal.pone.0084068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair BM, CheungKJ, Jr, Griffith A, Burns JL. 2004. Salicylate induces an antibiotic efflux pump in Burkholderia cepacia complex genomovar III (B. cenocepacia). J Clin Invest 113:464–473. 10.1172/JCI200419710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair BM, Joachimiak LA, Chattopadhyay S, Montano I, Burns JL. 2005. Conservation of a novel protein associated with an antibiotic efflux operon in Burkholderia cenocepacia. FEMS Microbiol Lett 245:337–344. 10.1016/j.femsle.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Buroni S, Pasca MR, Flannagan RS, Bazzini S, Milano A, Bertani I, Venturi V, Valvano MA, Riccardi G. 2009. Assessment of three Resistance-Nodulation-Cell Division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol 9:200. 10.1186/1471-2180-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. 2011. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS One 6:e18902. 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mira NP, Madeira A, Moreira AS, Coutinho CP, Sa-Correia I. 2011. Genomic expression analysis reveals strategies of Burkholderia cenocepacia to adapt to cystic fibrosis patients' airways and antimicrobial therapy. PLoS One 6:e28831. 10.1371/journal.pone.0028831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buroni S, Matthijs N, Spadaro F, Van Acker H, Scoffone VC, Pasca MR, Riccardi G, Coenye T. 2014. Differential roles of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob Agents Chemother 58:7424–7429. 10.1128/AAC.03800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng SP, Tsai WC, Liang CY, Lin YS, Huang JW, Chang CY, Tyan YC, Lu PL. 2014. The contribution of antibiotic resistance mechanisms in clinical Burkholderia cepacia complex isolates: an emphasis on efflux pump activity. PLoS One 9:e104986. 10.1371/journal.pone.0104986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jassem AN, Forbes CM, Speert DP. 2014. Investigation of aminoglycoside resistance inducing conditions and a putative AmrAB-OprM efflux system in Burkholderia vietnamiensis. Ann Clin Microbiol Antimicrob 13:2. 10.1186/1476-0711-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somprasong N, Hall CM, Webb JR, Sahl JW, Wagner DM, Keim P, Currie BJ, Schweizer HP. 2021. Burkholderia ubonensis high-level tetracycline resistance is due to efflux pump synergy involving a novel TetA(64) resistance determinant. Antimicrob Agents Chemother 65:e01767-20. 10.1128/AAC.01767-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somprasong N, Hall CM, Webb JR, Sahl JW, Wagner DM, Keim P, Currie BJ, Schweizer HP. 2020. Burkholderia ubonensis meropenem resistance: insights into distinct properties of class A beta-lactamases in Burkholderia cepacia complex and Burkholderia pseudomallei complex bacteria. mBio 11:e00592-20. 10.1128/mBio.00592-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horsley A, Perry C, Martin K, Webb K, Turton J, Kenna D, Jones A. 2011. Burkholderia latens infection in cystic fibrosis. J Cyst Fibros 10:291–292. 10.1016/j.jcf.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 35.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FSL. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804. 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodd IB, Egan JB. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res 18:5019–5026. 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamad MA, Skeldon AM, Valvano MA. 2010. Construction of aminoglycoside-sensitive Burkholderia cenocepacia strains for use in studies of intracellular bacteria with the gentamicin protection assay. Appl Environ Microbiol 76:3170–3176. 10.1128/AEM.03024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varga JJ, Losada L, Zelazny AM, Kim M, McCorrison J, Brinkac L, Sampaio EP, Greenberg DE, Singh I, Heiner C, Ashby M, Nierman WC, Holland SM, Goldberg JB. 2013. Draft genome sequences of Burkholderia cenocepacia ET12 lineage strains K56-2 and BC7. Genome Announc 1:e00841-13. 10.1128/genomeA.00841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podin Y, Sarovich DS, Price EP, Kaestli M, Mayo M, Hii K, Ngian H, Wong S, Wong I, Wong J, Mohan A, Ooi M, Fam T, Wong J, Tuanyok A, Keim P, Giffard PM, Currie BJ. 2014. Burkholderia pseudomallei isolates from Sarawak, Malaysian Borneo, are predominantly susceptible to aminoglycosides and macrolides. Antimicrob Agents Chemother 58:162–166. 10.1128/AAC.01842-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bugrysheva JV, Sue D, Gee JE, Elrod MG, Hoffmaster AR, Randall LB, Chirakul S, Tuanyok A, Schweizer HP, Weigel LM. 2017. Antibiotic resistance markers in strain Bp1651 of Burkholderia pseudomallei identified by genome sequence analysis. Antimicrob Agents Chemother 61:e00010-17. 10.1128/AAC.00010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webb JR, Price EP, Currie BJ, Sarovich DS. 2017. Loss of methyltransferase function and increased efflux activity leads to doxycycline resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother 61:e00268-17. 10.1128/AAC.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubarry N, Du W, Lane D, Pasta F. 2010. Improved electrotransformation and decreased antibiotic resistance of the cystic fibrosis pathogen Burkholderia cenocepacia strain J2315. Appl Environ Microbiol 76:1095–1102. 10.1128/AEM.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agnoli K, Schwager S, Uehlinger S, Vergunst A, Viteri DF, Nguyen DT, Sokol PA, Carlier A, Eberl L. 2012. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol Microbiol 83:362–378. 10.1111/j.1365-2958.2011.07937.x. [DOI] [PubMed] [Google Scholar]
- 45.Kavanaugh LG, Harrison SK, Flanagan JN, Steck TR. 2021. Antibiotic cycling reverts extensive drug resistant Burkholderia multivorans. Antimicrob Agents Chemother 10.1128/AAC.00611-21. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mima T, Schweizer HP, Xu Z-Q. 2011. In vitro activity of cethromycin against Burkholderia pseudomallei and investigation of mechanism of resistance. J Antimicrob Chemother 66:73–78. 10.1093/jac/dkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sass A, Marchbank A, Tullis E, LiPuma JJ, Mahenthiralingam E. 2011. Spontaneous and evolutionary changes in the antibiotic resistance of Burkholderia cenocepacia observed by global gene expression analysis. BMC Genomics 12:373. 10.1186/1471-2164-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download AAC.00920-21-s0001.pdf, PDF file, 1.9 MB (1.9MB, pdf)