To the Editor:
Molecular lesions, either structural cytogenetic anomalies or gene mutations of known significance, occur in a step-wise fashion and are morphologic and prognostic markers in myeloid neoplasms (MNs). Current World Health Organization (WHO) classification categorizes patients with myelodysplastic syndrome (MDS) with ringed sideroblasts (RS) ≥15% or ≥5% RS with an SF3B1 mutation (SF3B1MT) as MDS-RS [1], a subtype in which the presence of SF3B1 mutations confers a favorable prognosis [2–6]. Although most patients with SF3B1MT have a classic phenotype, there is considerable heterogeneity within this subcategory including individual diversion of the originally favorable clinical phenotype. The disappearance of RS can be observed during the disease course of MNs, suggesting that new cellular shifts, due to the acquisition of additional lesions cooperating with/ or suppressing SF3B1MT might go along with evolution to acute myeloid leukemia (AML). Such courses are likely a result of the type and configuration of SF3B1MT, which potentially impact the disease course.
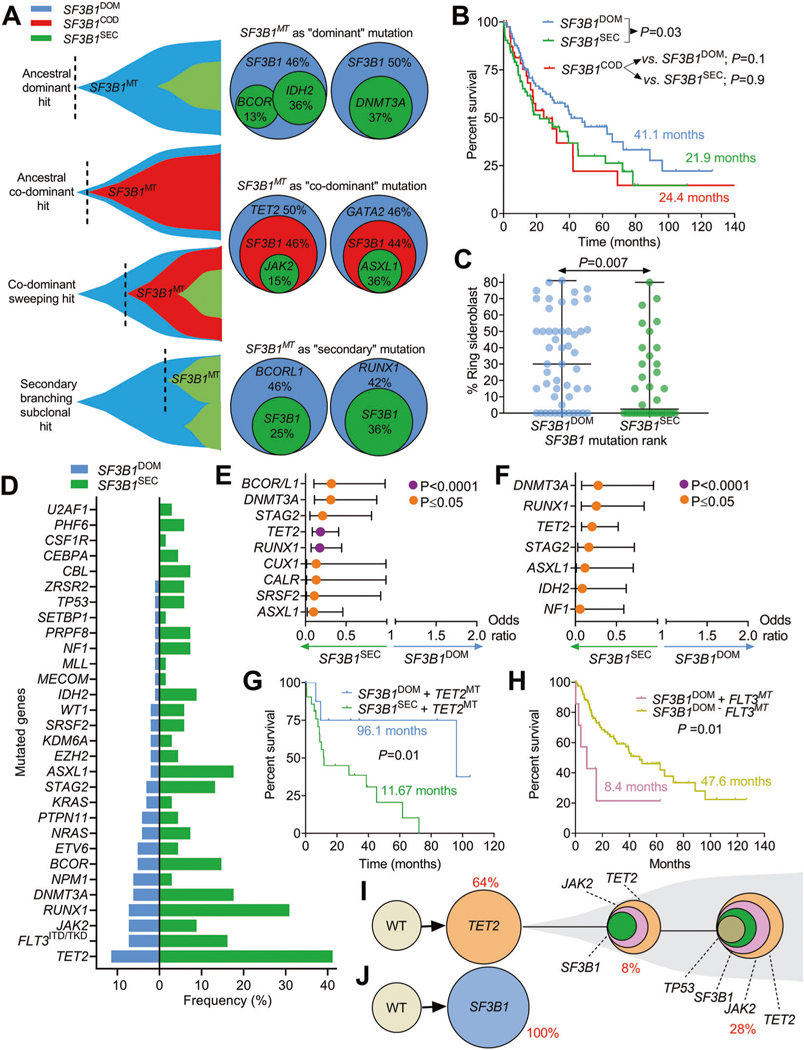
Inspired by a seminal work describing SF3B1MT as a disease-defining molecular lesion in terms of survival and independent risk of clonal evolution [6], we looked closer at our preliminary observations suggesting that important biological clues can be extrapolated from the molecular associations of SF3B1MT and their clonal architecture in the context of MNs [7]. To that end, we reviewed clinical and molecular annotations of patients with MNs with the intent of dissecting the SF3B1 mutatome and describing whether the clonal rank (ancestral/dominant vs. sub-clonal/secondary) might alter the phenotypic cell trajectories. We identified 209 SF3B1MT in about 6% of MN patients (209/3673). Clinical and molecular data of the cohort were collected at The Cleveland Clinic Foundation and retrieved from publicly available datasets (refer to Supplementary Material) for comparisons’ purposes. The demographic, clinical and cytogenetic characteristics are summarized in Table S1. Using variant allele frequencies (VAFs) estimation that was previously confirmed by PyClone method [7, 8], SF3B1MT were categorized as: (i) dominant (SF3B1DOM, n = 96, 46%), (ii) secondary (SF3B1SEC, n = 68, 33%) and as expected, due to the lack of resolution of the bioinformatics methods deployed, sub-categorization included transitional category of (iii) “co-dominant” (SF3B1COD, n = 45, 21%). Schematic representations of these configurations are described in Fig. 1A. Despite the different hierarchical assignments with respect to other concurrent mutations, SF3B1MT VAFs in the 3 groups did not statistically vary (P = 0.07; Fig. S1). In terms of survival, SF3B1COD had a similar median OS to that of SF3B1SEC (24.4 vs. 21.9 mo.) and a poorer outcome vs. SF3B1DOM (41.1 mo.; P = 0.03; Fig. 1B). In terms of disease phenotypes, SF3B1COD cases had no significant association with disease-type or cytogenetics vs. SF3B1DOM and SF3B1SEC (Table S2). The mutational profile of SF3B1COD resembled that of SF3B1SEC, while differences were found compared to SF3B1DOM with a significant association with ASXL1, DNMT3A, RUNX1 and TET2 mutations (Fig. S2A, B). Although some mutations (DNMT3A, 22%; TET2, 20%; RUNX1, 11%) were preferentially associated with SF3B1COD (Fig. S3A), they had no prognostic power (Fig. S3B–D). Therefore, we focused on non-ambivalent hierarchical SF3B1MT (mutations with >5% difference in VAFs; ΔVAF > 5%) as these showed higher degrees of distinction and a more stringent representation of the chronology of mutations defined as SF3B1DOM vs. SF3B1SEC.
Fig. 1. Characteristics, mutational associations and prognostic implications of clonal events in SF3B1 mutant myeloid neoplasms.
A Fish plots and pie charts depicting different examples of SF3B1 mutations clonal status and its secondary mutations in the case of SF3B1 dominant (SF3B1DOM), SF3B1 co-dominant (SF3B1COD) and SF3B1 secondary mutations (SF3B1SEC). B Kaplan–Meier curves showing the overall survival of cases with SF3B1 mutations per clonal status (dominant, secondary, co-dominant). The curves represent the percent survival of patients with dominant, co-dominant and secondary SF3B1 mutations. Comparisons of survival in dominant versus secondary SF3B1 and co-dominant versus dominant and secondary were done. Levels of statistical significance was calculated by P values. C Scatter plot describing the percentage of ringed sideroblasts in patients with SF3B1 dominant (blue) vs. secondary (green) mutations. Levels of statistical significance was assessed by P values using Mann–Whitney U test. D Bar graph showing the frequency (in percentage) of a panel of myeloid gene mutations co-occurring with SF3B1 mutations in dominant vs. secondary status. E Univariate analysis showing the odds ratio representing the strength of the association of SF3B1 mutations with other gene mutations. Levels of statistical significance is indicated in purple and orange colors (P < 0.0001 and P≤ 0.05) using Fisher’s Exact test. F Multivariate analysis showing the odds ratio representing the strength of the association of SF3B1 mutations with other gene mutations. Levels of statistical significance is indicated in purple and orange colors (P < 0.0001 and P≤ 0.05) using Fisher’s Exact test. G Kaplan–Meier curves showing the impact of TET2 mutations (TET2MT) on SF3B1 mutant cases per hierarchical SF3B1 mutational configuration: SF3B1 dominant (SF3B1DOM) vs. secondary (SF3B1SEC). Levels of statistical significance was calculated by P values. H Kaplan–Meier curves showing the impact of the presence of FLT3 mutations (FLT3MT) on SF3B1 dominant (SF3B1DOM) cases. Levels of statistical significance was calculated by P values. Single cell-DNA sequencing revealing the clonal trajectories of one sample with (J) ancestral/founder SF3B1 mutation and another sample with secondary/subclonal SF3B1 mutation (I). Different colors represent different mutations.
As compared to SF3B1SEC, SF3B1DOM were enriched in MDS (33 vs. 18%; P = 0.007), particularly with RS (27 vs. 9%; P = 0.006) while less present in sAML (12 vs. 23%; P = 0.03) (Table S3, Fig. S4). Eighty-percent of SF3B1DOM and 85% of SF3B1SEC occurred in the elderly (age ≥ 60 years). No differences in sex distribution and hematological parameters were reported except for bi-cytopenia (52 vs. 36%; P = 0.01), more dysplastic myeloid cells (53 vs. 29%; P = 0.01) and bilineage dysplasia (47 vs. 26%; P = 0.02, Table S3) in patients with SF3B1SEC vs. SF3B1DOM. In contrast, the bone marrow of SF3B1DOM patients was more often normocellular (46 vs. 29%; P = 0.02) and enriched with RS (Table S3); the latter also correlated with higher SF3B1MT VAFs (Fig. 1C, Fig. S5). Interestingly, although patients with SF3B1SEC had shorter OS than those with SF3B1DOM (21.9 vs. 41.1 mo.; P < 0.03; Fig. 1B), when dichotomized according to different SF3B1MT VAF ranges (>40%, 20–40%, <20%), no significant OS differences were observed (Fig. S6A) even when the SF3B1MT clonal ranks were considered (Fig. S6B–D).
Therefore, we investigated the prognostic importance of distinct molecular associations in SF3B1DOM and SF3B1SEC.The analysis of the mutational burden showed that patients with SF3B1DOM had fewer numbers of mutations per individual compared to those harboring SF3B1SEC [1.0 (95/96) vs. 2.6 (179/68)]. The frequencies of mutations in SF3B1DOM vs. SF3B1SEC are depicted in Fig. 1D. In total, 274 concurrent somatic mutations were detected, of which 4% included co-hits in other RNA-splicing factor genes (PRPF8, SRSF2, U2AF1, ZRSR2; Fig. S7A, B and S8A–E) as recently described [9]; and of which 75% had a normal karyotype (Fig. S7C). Targeted deep sequencing of the most recurrently mutated genes in MNs revealed that concurrent myeloid mutations were more associated with SF3B1SEC than SF3B1DOM, suggesting that this increased accumulation of concomitant mutations might reflect a more pronounced susceptibility to clonal expansion. Indeed, univariate analyses showed significantly higher odds of mutations in TET2 (41 vs. 11%; P < 0.0001), RUNX1 (31 vs. 7%; P < 0.0001), ASXL1 (18 vs. 2%; P = 0.0005), BCOR/BCORL1 (15 vs. 5%; P = 0.05), DNMT3A (18 vs. 6%; P = 0.02) and STAG2 (13 vs. 3%; P = 0.01) in individuals with SF3B1SEC vs. SF3B1DOM (Fig. 1E). We then applied multivariate analysis to confirm the independent relationships between SF3B1 clonal status and other distinct genomic associations and found that ASXL1, DNMT3A, IDH2, NF1, STAG2, TET2 and RUNX1 significantly shaped the molecular profile of SF3B1SEC (Fig. 1F). Moreover, mutations preceding SF3B1SEC mainly affected lineage-restricted genes associated with repression of erythroid programs (RUNX1, 12%), cohesin complex (STAG2, 9%), transcriptional corepressors (BCOR/ BCORL1, 7%), terminal monocytic differentiation (TET2, 7%), chromatin remodeling (ASXL1, 6%) and leukemogenesis (DNMT3A and FLT3, 4% each) (Fig. S9). Thereby, we investigated whether specific concurrent mutations can impact the survival outcomes of SF3B1MT MNs and further distinguish SF3B1DOM vs. SF3B1SEC subsets. Our analysis revealed markedly unfavorable outcomes in SF3B1SEC compared to SF3B1DOM when particular genes, e.g., TET2, co-existed (11.7 vs. 96.1 mo., P = 0.01; Fig. 1G) while the presence of FLT3MT contributed to dismal prognosis only in SF3B1DOM (8.4 vs. 47.6 mo., P = 0.01; Fig. 1H). The extent of prognostic variations between both SF3B1MT configurations suggests that new lesions might represent key contributors to the distinct clinical course of SF3B1MT MNs. The association between TET2 and SF3B1 mutations has been previously seen in MNs and in vivo functional studies described an exacerbated anemic phenotype when present [10]. The prognostic implication of FLT3 mutation concurrence with SF3B1 mutation might highlight a distinct subgroup of patients with more proliferative features.
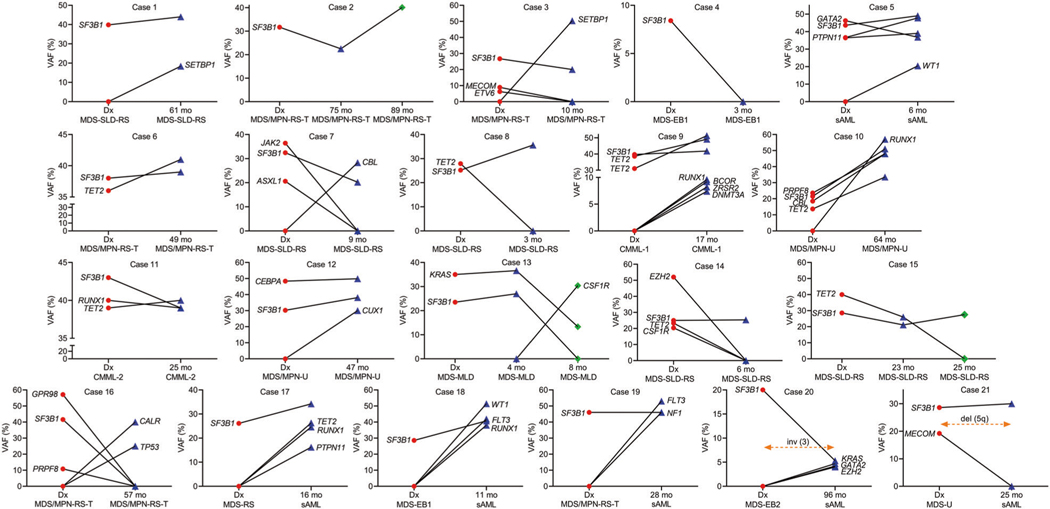
Given the fact that inferring clonal architecture from bulk sequencing might not accurately describe that SF3B1MT can exist in a secondary configuration and acquired during clonal evolution, we applied single-cell DNA sequencing to an MDS case with complex molecular architecture and dissected its clonal trajectory. We found a subclonal SF3B1MT (p.K700E) to an ancestral TET2MT (p.Y1618X), that coexisted in clones carrying TET2 and JAK2 (p.V617F) and TET2, JAK2 and TP53 (c.276+1G>A) (Fig. 1J). Additionally, to confidently assign the mutations we performed serial DNA sequencing on 21 patients’ specimens to deeply define the clonal trajectories of SF3B1MT in MNs. The study of the evolution of SF3B1MT VAFs (Fig. 2A–J) demonstrated that a founder SF3B1 clone (p.E622D) remained as such and increased in VAF (case #1, 2; the latter confirmed also by single-cell DNA sequencing; Fig. 1I), became secondary due to acquisition of other hits (cases #3) or codominant (case #11). Of note, we found that in one case, SF3B1DOM disappeared at the 2nd time point (case #4) possibly suggesting that the composition of the bone marrow at that time-point contained only a minimal fraction of cells carrying SF3B1MT. In addition, SF3B1COD clones remained co-dominant (cases #5, 6), became secondary (case #7) or dominant (case #8). The SF3B1SEC clone remained secondary (case #12), disappeared (case #13, 16) or became dominant due to the disappearance of other founder mutations (case #14, 15). The clonal evolution elucidated by serial samples as well as bone marrow reviews, unveiled that an evolving AML phenotype was accompanied by the acquisition of novel lesions (FLT3, PTPN11, RUNX1, TET2, WT1 in cases #17–19) or emerging cytogenetic abnormalities [inv(3) and del(5q) in cases #20, 21] driving transformation.
Fig. 2. The clonal trajectories of SF3B1 mutations.
Serial analysis of patients with myeloid neoplasia and SF3B1 mutations. DNA specimens from 21 samples with SF3B1 mutations were analyzed at 2 or 3 time points. Targeted sequencing panel for myeloid genes was applied to all samples. Line graphs indicate variant allele frequency in percentage for mutations considered somatic. Red, blue and green colors indicate mutations at first time point, second and third time point, respectively. Dx diagnosis, VAF variant allele frequency, mo months.
The mutational rank within clonal hierarchy is different from just assigning clonal burden and correlates with the outcomes of the mutations including SF3B1. Our study supports that specific disease phenotypes and clinical outcomes of patients with MNs are inferred by the clonal hierarchy of SF3B1MT, not only its VAF%, with respect to other founder or subclonal myeloid events. This distinction allows for an even higher level of precision in predicting clinical outcomes of patients with SF3B1 mutations. Both, methodological feasibility of VAF based clonal ranking and predictive value of SF3B1 clonal hierarchy assessment in MN will need further confirmation. Additionally, we deciphered the clonal trajectories of SF3B1MT and uncovered key events driving leukemic evolution in SF3B1MT low-risk MDS. Hence, SF3B1MT hierarchical configuration can potentially delineate the fate of SF3B1MT MNs and further define this newly proposed disease-entity.
Supplementary Material
Acknowledgements
We thank the following sources of funding: Aplastic Anemia and MDS International Foundation Research Grant (VV and JPM), Vera and Joseph Dresner Foundation-MDS Research Fund (VV), VeloSano Pilot Award (VV), NIH/NHLBI R35HL135795 (JPM), R01HL132071 (JPM), The Henry and Marilyn Taub Foundation (JPM). We thank the team of Mission Bio for technical expertise on single cell-DNA sequencing and The Cancer Genome Atlas (TCGA), The BEAT AML Master Trial and The German-Austrian Study Group for data accessibility.
Footnotes
Compliance with ethical standards
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41375-021-01176-7.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl J Med. 2011;365:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26: 542–5. [DOI] [PubMed] [Google Scholar]
- 5.Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jädersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015; 126:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malcovati L, Stevenson K, Papaemmanuil E, Neuberg D, Bejar R, Boultwood J, et al. SF3B1-mutant myelodysplastic syndrome as a distinct disease subtype - a Proposal of the International Working Group for the Prognosis of Myelodysplastic Syndromes (IWG-PM). Blood. 2020;136:157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagata Y, Makishima H, Kerr CM, Przychodzen BP, Aly M, Goyal A, et al. Invariant patterns of clonal succession determine specific clinical features of myelodysplastic syndromes. Nat Commun. 2019;10:5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awada H, Nagata Y, Goyal A, Asad MF, Patel B, Hirsch CM, et al. Invariant phenotype and molecular association of biallelic TET2 mutant myeloid neoplasia. Blood Adv. 2019;3:339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor J, Mi X, North K, Binder M, Penson A, Lasho T, et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood. 2020;136:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obeng E, Chappell R, Seiler M, Chen MC, Campagna DR, Schmidt PJ, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30: 404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.