Abstract
A missense variant in the cytoplasmic domain of the insulin receptor (INSR) was identified by exome sequencing in affected members of a four-generation family with fatty liver disease (FLD). The variant (rs766457461, c.4063T>C, p.Y1355H) results in the substitution of histidine for a tyrosine that undergoes autophosphorylation in response to insulin stimulation in vitro. Because insulin promotes lipogenesis in hepatocytes, we hypothesized that the variant was causally linked to FLD in the family. To test this hypothesis, we used CRISPR/Cas9 technology to replace the corresponding tyrosine in mouse INSR with histidine (Y1345H). No significant differences were found in hepatic insulin signaling, as assessed by phosphorylation of INSR or AKT levels or in activation of the insulin-responsive transcription factor SREBP-1c. Glucose tolerance and hepatic triglyceride (TG) content in Insr1345H/H mice fed a chow diet or diets rich in fat, sucrose or fructose did not differ significantly from WT littermates. Thus, our studies in mice failed to support the notion that INSR (Y1355H) is causally related to FLD in the family or that phosphorylation of this residue alters hepatic TG metabolism.
Keywords: Autophosphorylation, Hepatic triglyceride, Sterol regulatory element-binding protein 1c (SREBP-1c)
Highlights
-
•
We identified a missense variant in insulin receptor (INSR) (Y1355H) that segregates with fatty liver disease in family.
-
•
Mice expressing INSR(Y1355H) were established using CRISPR.
-
•
The hepatic TG content of INSR(Y1355H) mice did not differ from that of their littermate controls fed on a chow, high-fat, or high-sucrose diet.
1. Introduction
The metabolic and mitogenic effects of insulin are mediated through the insulin receptor (INSR). The INSR has two extracellular α subunits and two transmembrane β subunits interconnected by disulfide bonds. Autophosphorylation of the β subunits promotes both the catalysis of the INSR and phosphorylation of peptide substrates [1]. Six of the tyrosine residues in the cytoplasmic domain of the human INSR-β subunit undergo autophosphorylation upon insulin stimulation: one in the juxtamembrane domain (Tyr-999), three in the kinase domain (Tyr-1185, -1189, and -1190) and two in the C-terminal region (Tyr-1355 and -1361) [2]. The tyrosine residue in the juxtamembrane domain is involved in recruiting INSR substrates [3,4]. The three tyrosine residues in the kinase domain control insulin-induced metabolic and mitogenic effects [[4], [5], [6]]. The role of the two tyrosine residues in the C-terminal region of the INSR remains unclear [7,8]. Substitution of these two tyrosine residues (Tyr-1355 and Tyr-1361) with phenylalanine enhances mitogenic signaling in rat fibroblasts and Chinese Hamster Ovarian cells [9,10], suggesting the possibility that the post-translational modification of these two residues has functional consequences.
Here, we identified a family (Family L860548) in which a missense mutation (Y1355H, rs766457461) in INSR co-segregated with fatty liver disease (FLD). To determine whether this variant was causally related to FLD in this family, we generated mice expressing a mutant form of INSR (Y1345H), which corresponds to INSR (Y1355H) in humans. We found no indication in mice that substitution of histidine for tyrosine at this position of INSR alters hepatic glucose or TG metabolism.
2. Materials and methods
2.1. Study subjects
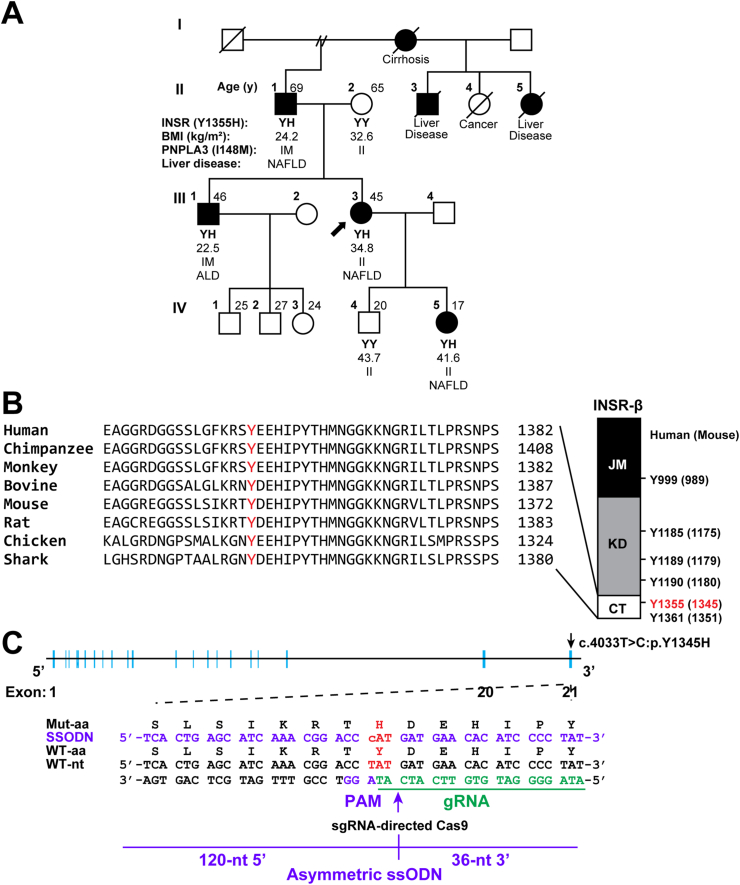
Blood samples and clinical information were obtained from six members (II.1, II.2, III.1, III.3, IV.4, and IV.5) of a four-generation European-American family with FLD (Family L860548) (Figure 1A). The proband was a 45-year-old woman (III.3) who had been diagnosed with nonalcoholic FLD (NAFLD) at the time of blood collection (Figure 1A). All participants signed a written consent form (in their native language) approved by the Institutional Review Board (IRB) of UT Southwestern Medical Center (UTSW) in accordance with the Helsinki Declaration of the World Medical Association. Blood samples were obtained after a 12-h fast and processed as described previously [11]. Genomic DNA was isolated from peripheral blood using the Easy-DNA kit (Invitrogen, Carlsbad, CA).
Figure 1.
Identification of INSR(Y1355H) in European-American family with fatty liver disease (FLD). A: A four-generation pedigree of a European-American family (Family L860548) in which several family members have FLD (non-alcoholic or alcoholic). The index case is indicated by an arrow. Family members with FLD are indicated by filled symbols. The genotypes for the PNPLA3(I148M) variant are provided. B: Conservation of Tyr1355 in the C-terminal domain (CT) of human INSR among different species (left) and a schematic of the cytoplasmic domain of INSR with the location of tyrosine residues (right). The corresponding residue number in mice is provided in parentheses. C: Schematic of genetic targeting strategy using CRISPR/Cas9. The thymine at position 4033 was replaced with cytosine (c.4033T>C), resulting in the substitution of histidine for tyrosine at residue 1345 (P. Y1345H). The gRNA sequence (bottom strand) is shown in green followed by the protospacer adjacent motif (PAM) sequence. The 156-nt asymmetric ssODN is designed to have two asymmetric homology arms (120-nt, 5′; 36-nt, 3′) flanking the predicted Cas9-cleavage site (indicated by an arrow). NFAFLD, nonalcoholic fatty liver disease; ALD, alcoholic liver disease; BMI, body mass index; INSR, insulin receptor; PNPLA3, patatin-like phospholipase domain containing protein; JM, juxtamembrane; KD, kinase domain, CT, C-terminal domain; ssODN, single-stranded donor oligonucleotides.
2.2. Whole exome sequencing
Whole exome sequencing was performed in six family members by using the SureSelect Human All Exon V4 kit on the Illumina platform. Sequencing read length was paired-end 2 × 100 basepairs. Sequences were aligned to the human reference genome b37. The mean coverage of the targeted regions for all samples was >100-fold, with >98% bases covered by >20 reads in all samples. Genetic variations were determined using the Genome Analysis Toolkit [12] and annotated using SnpEff [13]. Because FLD appears to be inherited in an autosomal dominant manner in the family, we focused on rare variants [minor allele frequency (MAF) < 0.001 in gnomAD (v2.1.1; https://gnomad.broadinstitute.org/)] that were shared in the four affected family members (II.1, III.1, III.3, IV.5) but absent in the two unaffected relatives (II.2, IV.4) (Figure 1). Variants with a genomic evolutionary rate profiling++ score [14] > 2.0 and a combined annotation-dependent depletion score [15] >15 were considered. Sanger sequencing of the candidate variants was performed to confirm the exome sequencing results.
2.3. Generation and maintenance of Insr1345Y/H knock-in (KI) mice
Insr1345Y/H KI mice (C57BL/6N) were generated in the UT Southwestern Transgenic Technology Center by using CRISPR/Cas9 technology [16]. The gRNA (crRNA: 5′-ATAGGGGATGTGTTCATCAT-3′) was synthesized by Integrated DNA Technologies and annealed to the trans-activating CRISPR RNA (tracrRNA). The annealed sgRNA, asymmetric single-stranded donor oligonucleotide (ssODN, 156-nt) and Cas9 protein (Integrated DNA Technologies) were microinjected into the cytoplasm of fertilized one-cell pronuclear eggs, and the surviving eggs were transferred into the oviducts of pseudo-pregnant ICR recipient mice (Envigo). DNA extracted from the mouse tails was PCR-amplified using two oligonucleotides (Forward: 5′-ACCTGATGCGCATGTGCTGGCAGTTCAACC 3′ and Reverse: 5′-AGAAGGCGCTGTTAGGAAGGGTTTGACC-3′), and the genotype was confirmed by Sanger sequencing.
Mice that were heterozygous for Insr1345H were backcrossed with wild-type (WT) C57BL/6N mice. Offspring of heterozygous mating were used for all experiments described in this manuscript. All animal experiments were approved and conducted under the guidance of the UT Southwestern Institutional Animal Care and Use Committee. Mice were fed a chow diet (Teklad Rodent Diet 2016, 16% protein) ad libitum with free access to water and housed under conventional conditions with a 12-h light/12-h dark cycle (lights on: 6:00 am to 6:00 pm). For some experiments, mice were fed diets containing high fat (no. D12451; 45% kcal from fat, Research Diets) for 16 weeks [17], high sucrose (no. 901683; 74% kcal from sucrose, MP Biomedicals) for 4 weeks [17], or high fructose (no. TD.89247; 66% kcal from fructose; Envigo) for 3 weeks. For all experiments, unless otherwise stated, mice were sacrificed 2 h after food was removed, and both blood and tissues were harvested. To assess whether feeding had an effect on the phenotypes of Insr1345H/H mice, the WT or Insr1345H/H mice were fasted for 18 h (F) or were fasted for 12 h and then re-fed for 6 h (R) with chow diet.
2.4. Biochemical measurements in mouse tissues
Lipids from liver (~100 mg) were extracted using the Folch and Lees method [18], and the levels were normalized to sample tissue weight. Blood was collected from the vena cava in heparin tubes, and plasma was isolated by centrifugation at 2000g for 10 min at 4 °C. TG, cholesterol, cholesteryl ester, free cholesterol, and phosphatidylcholine levels were measured using enzymatic assays (Infinity, Thermo Fisher Scientific). For fasting glucose and insulin measurement, blood was collected after a 16-h fast, and the levels were measured using Contour Blood Glucose Monitoring System (Bayer) and an ultrasensitive mouse insulin ELISA kit (no. 90080; Crystal Chem Inc.), respectively. Plasma levels of nonesterified fatty acids (NEFAs) were determined by the HR Series NEFA HR (Fujifilm Medical Systems). Plasma levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were determined using VITROS 250 Microslide Technology.
A glucose tolerance test was performed after the mice were fasted for 16 h. They were then intraperitoneally administered a bolus of 20% glucose (1–2 mg/kg body weight). Blood was collected from the tail vein at 0, 15, 30, 60, 90, and 120 min post-injection.
After a 6-h fast, mice were injected with saline or insulin (5 U/kg body weight) (no. 0002751001; Eli Lily) by intraperitoneal injection, and liver tissues were collected 5 min after insulin was injected.
2.5. Real-time PCR assay
Total RNA was extracted from the livers of mice, and real-time PCR was performed as described previously [19] using the primers listed in Supplemental Table 1.
2.6. Immunoblotting
An aliquot of liver (100 mg) was homogenized in RIPA buffer (1 ml) supplemented with protease inhibitors (no. 11836170001; Sigma). Proteins were extracted as described [20], and protein concentrations were measured using the BCA kit (Thermo Fisher Scientific). Proteins were adjusted to a concentration of 4 μg/μl in RIPA buffer plus 1/6 volume of 6× Laemmli SDS sample buffer with 9% beta-mercaptoethanol. Samples were incubated at 95 °C for 5 min before proteins were size-fractionated by SDS-PAGE (8%) at 100 V. After the proteins (40–80 μg) were transferred to a nitrocellulose membrane (GE Healthcare) using a wet transfer system (Bio-Rad), the membranes were incubated in TBST buffer (1 × TBS, 0.1% Tween-20, pH 7.4) plus 5% BSA for 60 min at room temperature. The indicated antibodies (Supplemental Table 2) diluted in TBST plus 5% BSA were added and incubated with the filter at 4 °C overnight with gentle shaking. The next morning, the membranes were washed with TBST buffer and incubated with anti-mouse or anti-rabbit IgG antibody (Supplemental Table 2). Intensities of bands were quantified using LI-COR Image Studio Lite version 5.2.5. Beta-actin was used as an internal control for quantification.
2.7. Statistical analysis
Differences between groups were analyzed using Student's unpaired two-tailed t-test as implemented in GraphPad Prism version 8.1.1 (GraphPad Software). Values are presented as mean ± SD (standard deviation). P < 0.05 was considered statistically significant.
3. Results
3.1. Co-segregation of INSR1355Y/H with FLD in family L860548
The proband (III.3) was a 45-year-old woman with a BMI of 34.8 kg/m2 who presented with elevated liver enzymes (AST, 60 U/L and ALT, 32 U/L) and hepatic steatosis (Figure 1A). She further developed cirrhosis and required liver transplantation. Her father (II.1) was diagnosed with NAFLD at the age of 59 years and subsequently died due to cirrhosis. Her elder brother (III.1), who has a BMI of 22.5 kg/m2, was diagnosed with alcoholic liver disease (ALD) and hepatic cirrhosis. Her daughter (III.5), who had a BMI of 41.6 kg/m2, developed NAFLD in her early teens and had some associated fibrosis on liver biopsy. A clinical description of the family members is provided in Supplemental Table 3.
Whole exome sequencing identified a heterozygous missense mutation in INSR (NC_000019: g.7117153A>G; NM_000208: c.4063T>C, NP_000199: p.Y1355H; rs766457461) that was shared by all affected (II1, III1, III3 and IV5) family members, but none of the unaffected (II.2 and IV.4) family members. The allele was present in gnomAD (v2.1.1) in two of 141,348 individuals (MAF 7.1 × 10−6). A similar very low frequency of the allele (MAF of 7.6 × 10−6) was seen in TOPMed (freeze 8).
Two other variants in ESCO2 (chr 8) and BCL7A (chr 12) co-segregated with FLD in the family (Supplemental Table 4). Because both genes are expressed at very low levels in the liver and adipose tissues (https://www.gtexportal.org/home/gene/ESCO2, https://www.gtexportal.org/home/gene/BCL7A) and neither gene has been previously linked to FLD, we eliminated these two variants from further consideration. None of the missense, nonsense, splicing, or frameshift variants found in the known FLD susceptibility genes (including PNPLA3, TM6SF2, GCKR, and MBOAT7), co-segregated with FLD in Family L860548.
The variant substitutes histidine for tyrosine in a residue in the C-terminal domain of INSR that has been completely conserved through vertebrate evolution: the residue corresponding to Y1355 is invariant in 250 jawed vertebrates (NCBI nr protein database) (Figure 1B), indicating that that this residue has been subject to strong purifying selection for over 400 million years.
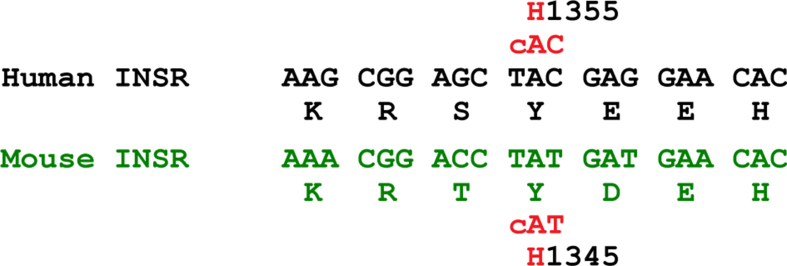
To further explore the relationship between the Y1355H and FLD, we introduced the mutation into the mouse Insr gene and determined its effects on hepatic glucose and TG metabolism. Tyr-1355 of human INSR corresponds to Tyr-1345 in mouse INSR (Figure 1B and Supplemental Figure 1). We generated Insr1345Y/H KI mice using CRISPR/Cas9 technology as summarized in Figure 1C. Both male and female KI mice were fertile. A total of 3 independent lines of mice were generated. The sex ratios were similar among WT, Insr1345Y/H, and Insr1345H/H offspring, which were born in the expected Mendelian ratios (Supplemental Table 5).
3.2. Insulin-stimulated phosphorylation of hepatic INSR(Y1345) and AKT in Insr1345H/H KI mice
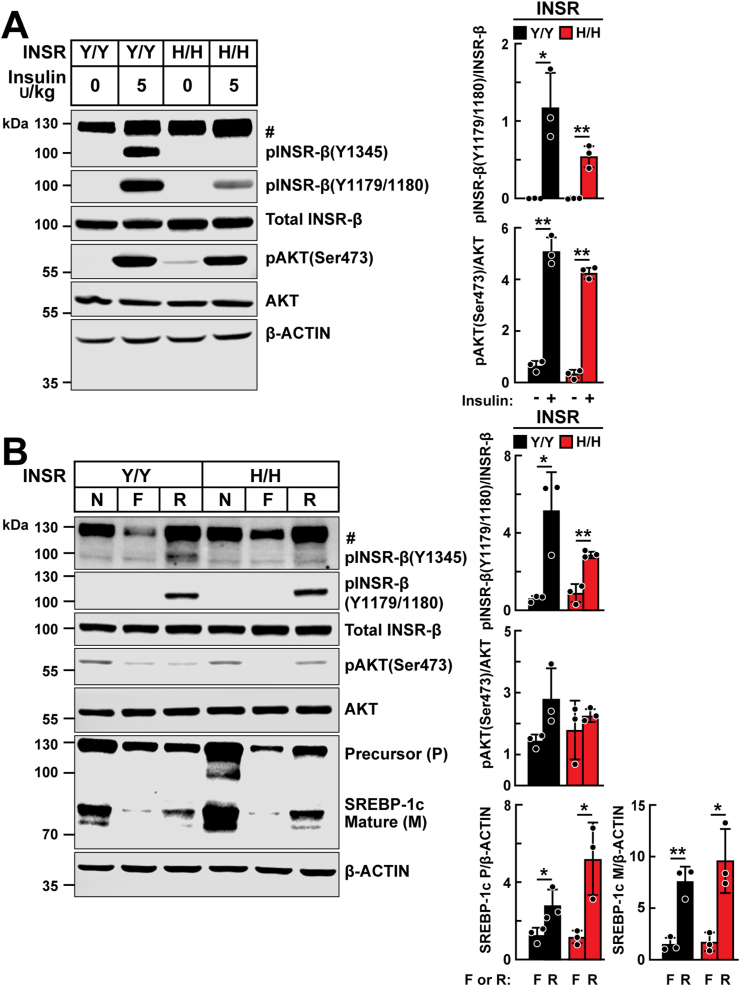
Mice were given insulin by intraperitoneal injection (5 U/kg body weight), and the phosphorylation of Tyr1345 was assessed in liver lysates from WT and Insr1345H/H mice using an antibody specific for this phosphotyrosine (Y1345) (Figure 2A). A strong signal was observed at the expected molecular weight (~100 kDa) in WT, but not in Insr1345H/H KI mice (Figure 2A). Phosphorylation of Y1179 and Y1180, a pair of residues required for insulin-activated kinase activity and uptake of 2-deoxyglucose [5], was not consistently different in the livers of Insr1345H/H mouse when compared with those of WT animals (Figure 2A). Phosphorylation of AKT, a key downstream of mediator of insulin signaling [21], was similar in WT and Insr1345H/H KI mice (Figure 2A).
Figure 2.
Hepatic insulin signaling in Insr1345H/H and Insr1345Y/Y littermates. A: Chow-fed 8-week old male Insr1345H/H and Insr1345Y/Y mice were fasted 6 h and then injected intraperitoneally with saline or insulin (5 U/kg). Livers were harvested after 5 min and immunoblotting of liver lysates was performed (40 μg protein) as described in the Methods. Band signals were quantitated using LI-COR Image Studio Lite version 5.2.5. The experiment was repeated twice, and the results were similar. B: WT and Insr1345H/H 7- to 9-week-old male mice were fasted for 18 h (F) or were fasted for 12 h and then re-fed a chow diet for 6 h (R). Mice in nonfasted (N) groups were fed a chow diet ad libitum. Mice were sacrificed and liver tissues were harvested, and immunoblotting and band quantification were performed as descried in Panel A. The experiments were repeated, and the results were similar. Values are expressed as mean ± SD. Difference between groups were analyzed using two-tailed Student's t test. ∗P < 0.05, ∗∗P < 0.01. #, non-specific band. pINSR-β, phosphorylated insulin receptor-β; pAKT, phosphorylated protein kinase B; SREBP-1c, sterol regulatory element-binding protein-1c; F, fasted; R, re-fed; N, nonfasted.
To examine the insulin signaling pathway during fasting and refeeding, WT and Insr1345H/H mice were fasted for 18 h (F) or were fasted for 12 h and then re-fed a chow diet for 6 h (R). Mice in nonfasted (N) groups were fed a chow diet ad libitum. Refeeding was associated with phosphorylation of Tyr-1345 in WT, but not Insr1345H/H KI mice (Figure 2B). No major differences were apparent between WT and Insr1345H/H mice in pINSR (Y1179/1180), or phosphorylated AKT during fasting or after refeeding (Figure 2B).
We also examined the levels and processing of sterol regulatory element-binding protein-1c (SREBP-1c), an insulin-responsive protein located in the endoplasmic reticulum that undergoes processing to release an active transcription factor that diffuses into the nucleus and upregulates fatty acid synthesis [22]. Expression of both the full-length precursor and mature forms of SREBP-1c in response to feeding were similar in the two genotype groups (Figure 2B).
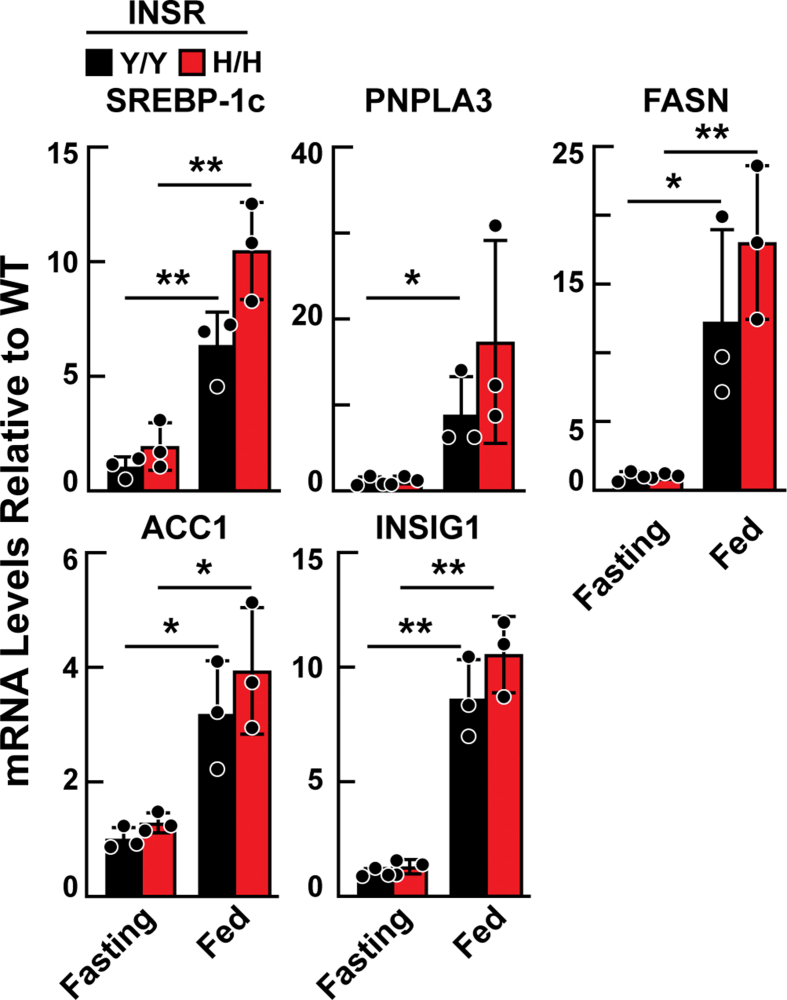
In addition, we compared the mRNA expression levels of SREBP-1c and selected genes involved in fatty acid synthesis in livers of Insr1345H/H and WT animals. As expected, refeeding induced mRNA expression of SREBP-1c, PNPLA3, FASN, ACC1 and INSIG1, but the increase was comparable to the WT and Insr1345H/H mice (Supplemental Figure 2).
3.3. Glucose and TG metabolism in Insr1345H/H KI and WT mice
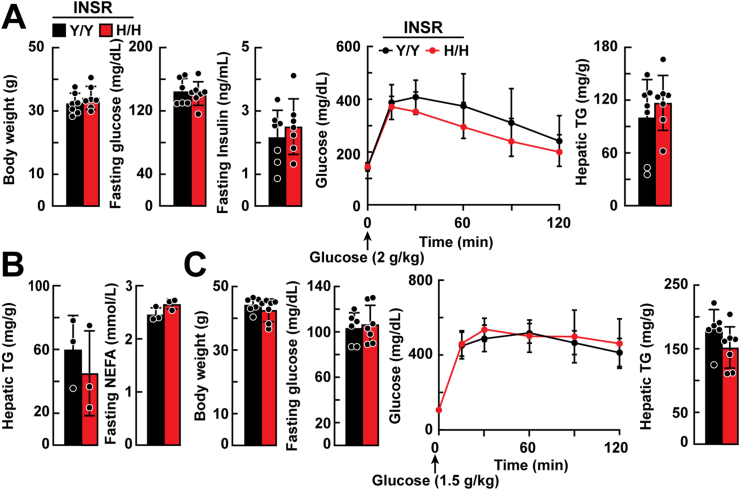
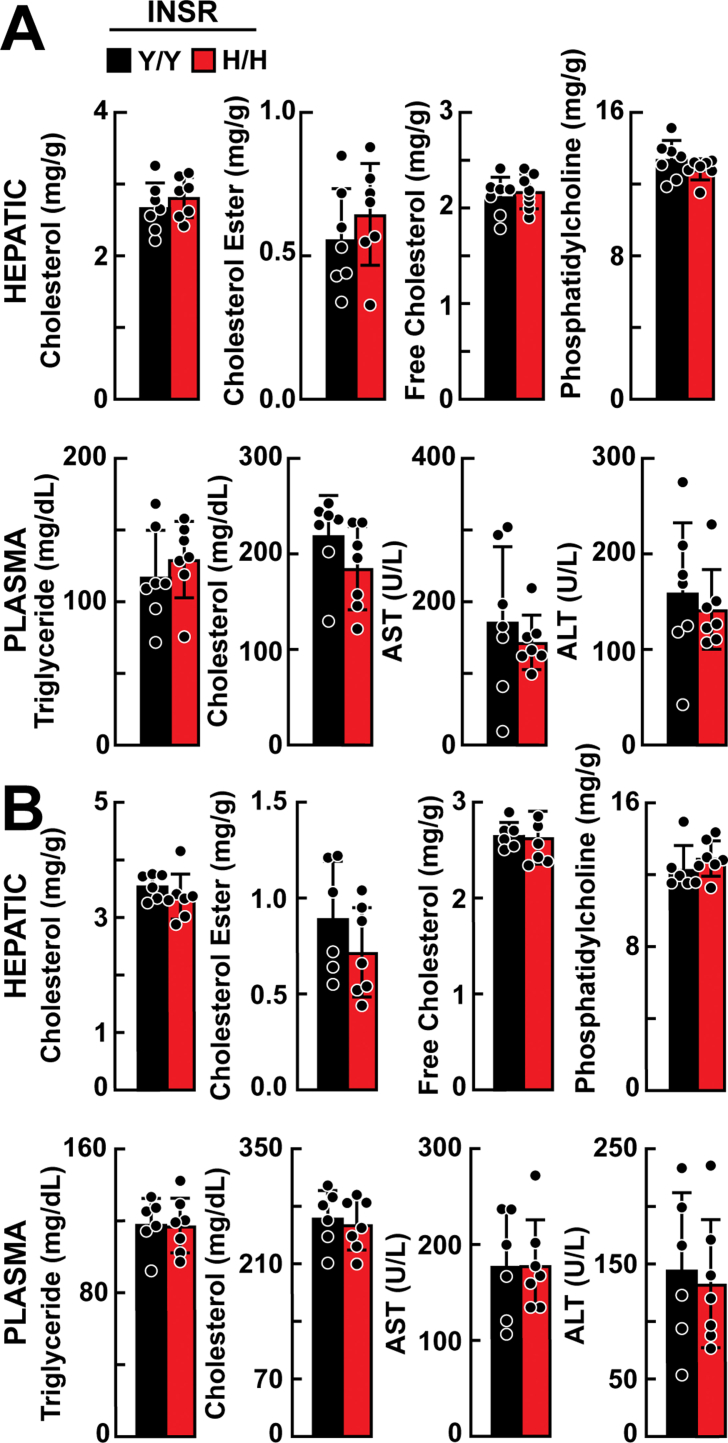
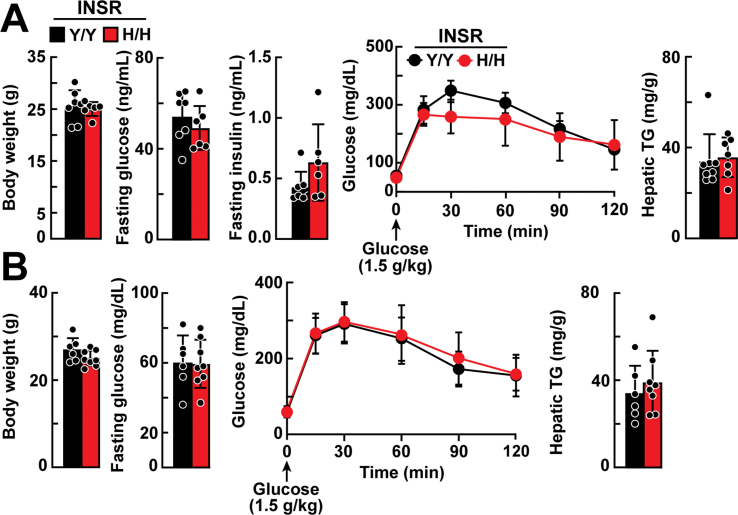
The mean body weights of chow-fed WT and Insr1345H/H KI mice did not differ significantly (Figure 3A). Fasting plasma levels of glucose and insulin as well as the response of the plasma glucose to a glucose challenge also did not differ significantly between the two groups of mice (Figure 3A). Hepatic TG measured in same mice at 49–51 weeks of age after a 2-h fast did not differ between the strains (Figure 3A). No significant differences were observed between the strains in either hepatic TG content or plasma NEFAs after an 18-hour fast (Figure 3B). Levels of selected other hepatic and plasma lipids as well as liver enzymes (AST, ALT) also did not differ between the two strains of mice (Supplemental Figure 3A).
Figure 3.
Hepatic glucose and TG metabolism in Insr1345H/H and Insr1345Y/Y littermates fed a chow or high-fat diet. A: Age-matched, chow-fed male mice (n = 7 mice/group, 15–17 weeks) were weighed and then fasted for 6 h before blood was sampled from the tail vein. Plasma was isolated and glucose and insulin levels were measured as described in the Methods. Glucose (2 g/kg) was administered by intraperitoneal injection, and blood was collected at the indicated times. Hepatic TG was measured in the same chow-fed mice when they reached 49–51 weeks after a 2-h fast. B: Age-matched, chow-fed male mice (n = 3 mice/group, 7–9 weeks) were fasted for 18 h before blood was sampled from the tail vein. Plasma nonesterified fatty acid (NEFA) and hepatic TG content were measured. C: Age-matched male mice (n = 6–7 mice/group, 7–8 weeks) were fed a high-fat (45%) diet for 16 weeks, and fasting glucose levels, glucose tolerance, and hepatic TG were measured as described in panel A. The experiment was repeated, and the results were similar. Values are expressed as mean ± SD. Difference between groups were analyzed using two-tailed Student's t test.
The same measurements were made in mice challenged with a high-fat diet (45% kcal from fat) for 16 weeks (Figure 3C and Supplemental Figure 3B) or with a high-sucrose diet for 4 weeks (Supplemental Figure 4A) or high-fructose diet for 3 weeks (Supplemental Figure 4B). No significant differences were found between the strains in body weight, hepatic TG content, fasting glucose or glucose tolerance on any of these diets. Hepatic TG levels did not differ between the strains.
4. Discussion
In this study, we identified a mutation in a highly conserved tyrosine residue in the cytoplasmic domain of INSR that co-segregated with FLD in a four-generation family. Previously, this tyrosine was shown to undergo phosphorylation in response to insulin in a cell-free system [4,23]. The strong evolutionary conservation, low population frequency (MAF < 7.6 × 10−6), and insulin-stimulated phosphorylation of this tyrosine in INSR are consistent with the notion that it is biologically important. To assess its potential contribution to the FLD in this family, we established mice expressing the mutant receptor. Careful phenotypic characterization of these mice failed to reveal any differences in glucose or TG metabolism, even after the mice were challenged with high-fat, high-sucrose, and high-fructose diets. Thus, we found no evidence that the variant that segregated with FLD in this family contributed to the liver disease.
Insulin resistance contributes to the development of NAFLD by increasing the flux of fatty acids to the liver and by stimulating hepatic de novo lipogenesis [24]. INSR(Y1345H) was not associated with hyperglycemia or an altered response to a glucose challenge in mice fed either chow or obesogenic diets (Figure 3 and Supplemental Figure 4), indicating the Y1345H mutation in mice did not have a major effect on glucose metabolism or insulin sensitivity. Activation of AKT by INSR plays a key role in regulating both glucose metabolism and SREBP-1c controlled TG metabolism [21,25]. The phosphorylation of AKT upon insulin challenge or refeeding was comparable in Insr1345H/H KI and WT (Figure 2).
The effects of insulin on TG metabolism in the liver are mediated primarily by SREBP-1c [22], and no differences between WT and Y1345 mice were apparent in the level of the transcription factor in the fasted or fed state (Figure 2B). Moreover, the expression of lipogenesis-associated genes (FASN, ACC1) did not differ between genotypes (Supplemental Figure 2). These data support the notion that Y1355H mutation in mice does not affect TG metabolism and are consistent with our finding that hepatic TG levels did not differ significantly between the Insr1345H/H KI and WT mice (Figure 3 and Supplemental Figure 4).
Based on our findings, we conclude that it is highly unlikely that INSR(Y1345H), which segregates with FLD in a family with FLD, is responsible for the liver disease in Family L860548. It remains possible that Tyr-1355 has effects on hepatic fuel homeostasis in humans that are not apparent in mice. It is also possible that mice have compensatory mechanisms for the effect of the mutation. More likely, another genetic (or possibly nongenetic) factor(s) causes the clustering of FLD in this family. If other families with FLD are identified in the future that have this same mutation, it would further implicate this sequence difference as a causative mechanism in this condition.
Author contributions
S.K.A. identified the proband and assisted with family collection. S.K.A., H.H.H., and J.C.C. conceived the project; H.H.H., J.C.C., and F.L. designed the experiments; F.L. performed the experimental work; S.L. and G.L. assisted with generation of the INSRY1345H mice. X.C. performed the analysis of the exome sequence data and H.H.H., J.C.C., F.L., and C.X. wrote the article. All authors read and approved the manuscript.
Acknowledgments
We acknowledge the technical assistance of Christina Zhao, Fang Xu, Tommy Hyatt, Mengle Shao, and Gabriela Perez-Garcia. We thanks Eriks Smagris, Yang Wang, Avash Das and Federico Oldoni for helpful discussions. This work was supported by a grant from the National Institute of Health-NIDDK (DK090066) and the Howard Hughes Medical Institute. Fei Luo was partially supported by the Chinese Scholarship Council.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101299.
Contributor Information
Fei Luo, Email: Fei.Luo@utsouthwestern.edu, LuoFei0058@csu.edu.cn.
Chao Xing, Email: Chao.Xing@utsouthwestern.edu.
Sumeet K. Asrani, Email: Sumeet.Asrani@BSWHealth.org.
Shili Li, Email: Shili.Li@utsouthwestern.edu.
Guosheng Liang, Email: Guosheng.Liang@utsouthwestern.edu.
Helen H. Hobbs, Email: Helen.Hobbs@utsouthwestern.edu.
Jonathan C. Cohen, Email: Jonathan.Cohen@utsouthwestern.edu.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Figure 1.
Alignment of human and mouse INSR sequence in the vicinity of Y1355H. Sequence of human and mouse INSR showing mutation identified in proband of Family L860548. INSR(Y1355H) in human INSR corresponds to INSR(Y1345H). INSR, insulin receptor.
Supplemental Figure 2.
Expression of selected genes involved in fatty acid synthesis in the livers of Insr1345Y/Y and Insr1345H/H mice. Age-matched female mice (n = 3/group, 7–9 weeks) were fasted 18 h or were fasted 12 h and then refed with chow diet for 6 h. Liver tissues were harvested and subjected to Real-time PCR assay using the olionucleotides shown in Supplemental Table 1. Values are expressed as mean ± SD. Difference between groups were analyzed using two-tailed Student's t test. ∗P < 0.05, ∗∗P < 0.01. SREBP-1c, sterol regulatory element-binding protein 1c; PNPLA3, patatin-like phospholipase domain-containing protein 3; FASN, fatty acid synthase; ACC1, acetyl-CoA carboxylase 1; INSIG1, insulin induced gene 1.
Supplementary Figure 3.
Levels of selected hepatic and plasma lipids of Insr1345Y/Y and Insr1345H/H mice. A: Chow-fed male mice (n = 7/group, 49–51 weeks) were killed after a 2-h fast and lipids were measured in liver lysates and plasma and liver enzymes were measure in plasma. B: Age-matched male mice (n = 6–7 mice/group, 7–8 weeks) were fed a high-fat (45%) diet for 16 weeks. Mice were sacrificed after a 2-h fast and lipids were measured in liver lysates and plasma, and liver enzymes were measure in plasma. The experiment was repeated, and the results were similar. Values are expressed as mean ± SD. Difference between groups were analyzed using two-tailed Student's t test.
Supplementary Figure 4.
Hepatic glucose and TG metabolism in Insr1345H/H KI and WT mice fed a high-sucrose or high-fructose diet. A: Age-matched male mice (n = 7–8 mice/group) were fed a high-sucrose diet for 4 weeks. Fasting glucose and insulin were measured, and glucose tolerance were performed after a 16-h fast. Hepatic triglyceride were measured after a 2-h fast. B: Age-matched male mice (n = 6–8 mice/group) were fed a high-fructose diet for 3 weeks. The mice were weighted prior to obtaining plasma glucose levels after a 16-h fast. A glucose tolerance was performed after a 16 h fast. Hepatic triglyceride was measured after a 2-h fast. Values are expressed as mean ± SD. Difference between groups were analyzed using two-tailed Student's t test.
References
- 1.Rosen O.M., Herrera R., Olowe Y., Petruzzelli L.M., Cobb M.H. Phosphorylation activates the insulin receptor tyrosine protein kinase. Proceedings of the National Academy of Sciences of the U S A. 1983;80(11):3237–3240. doi: 10.1073/pnas.80.11.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tornqvist H.E., Pierce M.W., Frackelton A.R., Nemenoff R.A., Avruch J. Identification of insulin receptor tyrosine residues autophosphorylated in vitro. Journal of Biological Chemistry. 1987;262(21):10212–10219. [PubMed] [Google Scholar]
- 3.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiological Reviews. 2018;98(4):2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murakami M.S., Rosen O.M. The role of insulin receptor autophosphorylation in signal transduction. Journal of Biological Chemistry. 1991;266(33):22653–22660. [PubMed] [Google Scholar]
- 5.Ellis L., Clauser E., Morgan D.O., Edery M., Roth R.A., Rutter W.J. Replacement of insulin receptor tyrosine residues 1162 and 1163 compromises insulin-stimulated kinase activity and uptake of 2-deoxyglucose. Cell. 1986;45(5):721–732. doi: 10.1016/0092-8674(86)90786-5. [DOI] [PubMed] [Google Scholar]
- 6.Wilden P.A., Backer J.M., Kahn C.R., Cahill D.A., Schroeder G.J., White M.F. The insulin receptor with phenylalanine replacing tyrosine-1146 provides evidence for separate signals regulating cellular metabolism and growth. Proceedings of the National Academy of Sciences of the U S A. 1990;87(9):3358–3362. doi: 10.1073/pnas.87.9.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers M.G., Backer J.M., Siddle K., White M.F. The insulin receptor functions normally in Chinese hamster ovary cells after truncation of the C terminus. Journal of Biological Chemistry. 1991;266(16):10616–10623. [PubMed] [Google Scholar]
- 8.Thies R.S., Ullrich A., McClain D.A. Augmented mitogenesis and impaired metabolic signaling mediated by a truncated insulin receptor. Journal of Biological Chemistry. 1989;264(22):12820–12825. [PubMed] [Google Scholar]
- 9.Takata Y., Webster N.J., Olefsky J.M. Mutation of the two carboxyl-terminal tyrosines results in an insulin receptor with normal metabolic signaling but enhanced mitogenic signaling properties. Journal of Biological Chemistry. 1991;266(14):9135–9139. [PubMed] [Google Scholar]
- 10.Ando A., Momomura K., Tobe K., Yamamoto-Honda R., Sakura H., Tamori Y. Enhanced insulin-induced mitogenesis and mitogen-activated protein kinase activities in mutant insulin receptors with substitution of two COOH-terminal tyrosine autophosphorylation sites by phenylalanine. Journal of Biological Chemistry. 1992;267(18):12788–12796. [PubMed] [Google Scholar]
- 11.Victor R.G., Haley R.W., Willett D.L., Peshock R.M., Vaeth P.C., Leonard D. The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. The American Journal of Cardiology. 2004;93(12):1473–1480. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 12.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Computational Biology. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang F., Shi X., Brown M.S., Goldstein J.L., Liang G. Growth hormone acts on liver to stimulate autophagy, support glucose production, and preserve blood glucose in chronically starved mice. Proceedings of the National Academy of Sciences of the U S A. 2019;116(15):7449–7454. doi: 10.1073/pnas.1901867116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smagris E., BasuRay S., Li J., Huang Y., Lai K.M., Gromada J. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61(1):108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 19.Oldoni F., Cheng H., Banfi S., Gusarova V., Cohen J.C., Hobbs H.H. ANGPTL8 has both endocrine and autocrine effects on substrate utilization. JCI Insight. 2020;5(17) doi: 10.1172/jci.insight.138777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smagris E., Gilyard S., BasuRay S., Cohen J.C., Hobbs H.H. Inactivation of Tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. Journal of Biological Chemistry. 2016;291(20):10659–10676. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu M., Wan M., Leavens K.F., Chu Q., Monks B.R., Fernandez S. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nature Medicine. 2012;18(3):388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown M.S., Goldstein J.L. Selective versus total insulin resistance: a pathogenic paradox. Cell Metabolism. 2008;7(2):95–96. doi: 10.1016/j.cmet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Tennagels N., Bergschneider E., Al-Hasani H., Klein H.W. Autophosphorylation of the two C-terminal tyrosine residues Tyr1316 and Tyr1322 modulates the activity of the insulin receptor kinase in vitro. FEBS Letters. 2000;479(1–2):67–71. doi: 10.1016/s0014-5793(00)01879-2. [DOI] [PubMed] [Google Scholar]
- 24.Buzzetti E., Pinzani M., Tsochatzis E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65(8):1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Yecies J.L., Zhang H.H., Menon S., Liu S., Yecies D., Lipovsky A.I. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metabolism. 2011;14(1):21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.