ABSTRACT
Accurate and reproducible antimicrobial susceptibility testing (AST) of polymyxin antibiotics is critical, as these drugs are last-line therapeutic options for the treatment of multidrug-resistant Gram-negative bacterial infections. However, polymyxin AST in the routine laboratory remains challenging. In this study, we evaluated the performance of an automated broth microdilution (BMD) system (Sensititre, ThermoFisher) compared to that of agar dilution (AD) for colistin and polymyxin B AST of 129 Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii complex clinical isolates. MICs derived from the Sensititre instrument based on two operator comparisons demonstrated overall categorical agreement (CA) of 86% and 89% compared to AD for colistin and 89% and 92% compared to AD for polymyxin B. However, error rates were higher than recommended by CLSI. Manual inspection of microdilution wells revealed microbial growth and skip wells which were erroneously interpreted by the Aris 2X instrument. Using manually interpreted BMD MICs read by two operators increased the overall categorical agreements to 88% and 95% compared to AD for colistin and 92% and 96% compared to AD for polymyxin B. Laboratories choosing to use the Sensititre platform for polymyxin AST should consider manual evaluation of wells as part of their algorithm.
KEYWORDS: assay, clinical methods, colistin, diagnostic, Gram-negative bacteria, polymyxin B, polymyxins, susceptibility
INTRODUCTION
For more than 2 decades, the polymyxins have been part of the armamentarium for the treatment of multidrug-resistant Gram-negative infections (1). The recent availability of newer combination agents that exhibit more favorable pharmacological profiles has eased reliance on polymyxins as a last line of intervention. Some studies have highlighted poor outcomes and/or high nephrotoxicity associated with colistin monotherapy or colistin combination therapy compared to those associated with alternative agents (2–5). However, polymyxins are still used for the treatment of serious infections caused by bacteria resistant to ceftazidime-avibactam, ceftolozane-tazobactam, meropenem-vaborbactam, imipenem-relebactam, or cefiderocol, multidrug-resistant Acinetobacter baumannii, and metallo-beta-lactamase-producing organisms, highlighting the important role of these drugs (6). The clinical significance of the differences in pharmacologic properties and toxicity of colistin versus those of polymyxin B has been discussed, and both polymyxins remain options for therapy, so antimicrobial susceptibility testing (AST) for both agents is relevant to current treatment strategies (7–11).
As contemporary use of polymyxins continues, the need for a reliable AST method for these compounds is obvious. Unfortunately, polymyxin AST is fraught with technical challenges that have prevented establishment of a unified testing modality. Poor diffusion of polymyxins into agar causes unacceptably variable results when Kirby-Bauer disk diffusion is used for AST (12). This has resulted in the removal of zone diameter interpretive criteria from AST guidance documents (12–14). Etests exhibit similar problems, demonstrating high error rates and an inability to reliably detect resistant isolates (15–17).
CLSI and EUCAST currently endorse the ISO 20776-1 manual broth microdilution (BMD) method as the reference method for colistin and polymyxin B AST (18–20), while broth disk elution and agar dilution methods are additional acceptable MIC methods for colistin for Enterobacterales and Pseudomonas aeruginosa (18). Despite this designation, colistin BMD remains prone to several technical challenges. Adsorption of the polymyxins to laboratory plastics used commonly in BMD assays (21), variability in AST media cation concentration (12), lack of consistency in the composition of commercial polymyxin powders (22, 23), and the presence of “skip wells” resulting in uninterpretable results (24) have all been reported as technical complications of polymyxin BMD. Additionally, manual BMD is labor intensive, requiring not only specialized reagents and procedures but also specialized training for individuals performing and interpreting the results of such assays. As such, polymyxin AST often necessitates reference laboratory utilization, which can result in longer turnaround times or empirical treatment of critically ill patients.
Agar dilution MIC methods are listed as acceptable alternatives to BMD for colistin, but not for polymyxin B, in the 2020 CLSI M100 (18). Investigations have revealed that AD MICs exhibit a high level of agreement with those derived from manual BMD for both colistin (15, 23, 25) and polymyxin B (25), and studies have demonstrated reproducible results with agar dilution (26). Similar to manual BMD, AD is highly laborious and requires significant technical skill, rendering it not easily adaptable into routine workflow of most clinical microbiology laboratories.
Automated methods would make polymyxin AST less cumbersome and facilitate integration into a routine workflow. Approved colistin breakpoints have been published (18); however, the lack of FDA-recognized polymyxin breakpoints for Gram-negative bacteria, except Pseudomonas aeruginosa, complicates manufacturer validation of automated device performance (27). Thus, some manufacturers have included polymyxins as part of specialized panels for research use only.
In this work, we investigated the performance of the Sensititre (ThermoFisher, Waltham, MA) automated BMD system for polymyxin AST testing of both Enterobacterales and non-Enterobacterales compared to that of manual AD, the method utilized by our reference laboratory. Recent evaluations of polymyxin AST using the Sensititre system reported good correlation with results obtained from BMD for members of the Enterobacterales; however, many of these studies examined only colistin (17, 28–33). The Sensititre platform allows for fully automated readings as well as manual plate viewing. There is no specific guideline for manual versus automated reads for polymyxins, so either method is acceptable per the manufacturer. This work expands on previous studies (17, 28–33) by performing Sensititre polymyxin B and colistin BMD in parallel via both fully automated and manual readings against a retrospective collection of clinical isolates representing multiple species and resistance phenotypes. The newly approved use of colistin breakpoints for the prediction of polymyxin B susceptibility by CLSI is also considered.
MATERIALS AND METHODS
Clinical isolates.
A total of 129 unique clinical isolates were used for this study. Species were restricted to those with either CLSI/EUCAST-established colistin breakpoints or epidemiologic cutoff values (ECVs). Isolates originated from patient specimens at the National Institutes of Health Clinical Center or were obtained from either the CDC/FDA Antibiotic Resistance Isolate Bank (34) or IHMA Inc. (IHMA Inc., Schaumburg, IL). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa 27853 were obtained from ATCC for the quality control of Sensititre panels and AD test plates.
Organism identification.
Prior to AST, bacterial identifications were confirmed by matrix-assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS). Briefly, banked isolates frozen at −70°C in Microbank cryovials (Pro-Lab Diagnostics, Round Rock, TX) were passaged twice on sheep blood agar (Remel, Lenexa, KS) prior to MALDI-TOF MS. Proteins for MALDI-TOF MS were extracted as described previously (35). MALDI-TOF MS spectra were analyzed using Bruker BioTyper software v.3.1 (Bruker Daltonics, Billerica, MA). Acceptable scores used to confirm organism identification were those of ≥2.0, with database entries for no other species within 10% of the obtained score.
Interpretive criteria for antimicrobial susceptibility testing.
2020 CLSI colistin and polymyxin B breakpoints were applied for all organisms: Enterobacterales (≥4 μg/ml resistant, ≤2 μg/ml intermediate), Acinetobacter baumannii complex (≥4 μg/ml resistant, ≤2 μg/ml intermediate), and Pseudomonas aeruginosa (≥4 μg/ml resistant, ≤2 μg/ml intermediate) (18).
Sensititre broth microdilution testing.
Frozen isolates were passaged twice on sheep blood agar (Remel) prior to AST. All isolates were tested using Sensititre GNX2F panels (ThermoFisher) containing wells for colistin and polymyxin B at concentrations of 0.25 μg/ml, 0.5 μg/ml, 1 μg/ml, 2 μg/ml, and 4 μg/ml. Sensititre cation-adjusted Mueller-Hinton broth (ThermoFisher) used for automated BMD studies did not contain Tween 80 in accordance with CLSI recommendations. All isolates were incubated on the Sensititre ARIS 2X instrument at 35°C. Susceptibility was determined after 18 h for all species with the exception of A. baumannii complex isolates, for which susceptibility was determined following 24 h of incubation per the manufacturer’s instructions. Automated reads were performed using the ARIS 2X system. Manual reads were performed by operator A or B, aided by Sensititre VIZION digital MIC viewing system. All isolates were tested using Sensititre GNX2F panels twice, by independent operators.
Manual Sensititre BMD readings.
Operator A repeated Sensititre AST for manual resulting on a subset (n = 17) of isolates. Operator B manually interpreted panels of all 129 isolates at initial AST, blinded from fully automated results. All panels were evaluated manually using CLSI interpretive standards (36). Isolates exhibiting skip wells to either polymyxin B or colistin were manually assigned an MIC of ≥4 μg/ml. Isolates for which growth was visible by eye in test wells that were not detected by the automated read were labeled as undetected microbial growth. All isolates had appropriate growth in control wells lacking antimicrobial agents.
Agar dilution.
Manual colistin and polymyxin B AD were performed for all isolates. A stock 1 mg/ml solution of polymyxin B sulfate (Sigma-Aldrich, Saint Louis, MO) was prepared in sterile water containing 0.002% Tween 80. No additional surfactant was added to further dilutions of the antibiotic or to the culture medium prior to pouring test plates. To verify that the small quantity of Tween 80 present in these plates did not affect the results, a subset of test isolates spanning all resistance phenotypes were inoculated to polymyxin B AD plates made from the antibiotic stock containing 0.002% Tween 80 or as second stock reconstituted in sterile water alone (Fig. S1). A 1 mg/ml stock solution of colistin (Sigma-Aldrich) was prepared in sterile water without Tween 80, and agar dilution assays were performed as follow-up experiments in accordance with CLSI guidelines. Mueller-Hinton AD test plates were prepared within 48 h of AST. Plates were poured at concentrations of 0 μg/ml, 0.5 μg/ml, 1 μg/ml, 2 μg/ml, 4 μg/ml, and 8 μg/ml polymyxin B or colistin by adding defined amounts of stock solution to molten agar prior to solidification. Test isolates were passaged twice from frozen stocks on sheep blood agar, resuspended to a concentration of 0.5 McFarland in sterile saline, and subsequently diluted 1:10. A 2-μl aliquot of this dilution was spotted to AD test plates, resulting in a final count of 1 × 104 CFU applied to the surface of AD plates (36). Spots were allowed to dry at room temperature, and the inoculated test plates were incubated at 35°C for 18 to 24 h prior to interpretation.
Data analysis.
Sensititre BMD was used as a comparative method in all analyses, with AD serving as a reference method. For the most comprehensive analysis of all data, MIC values and interpretations of two agar dilution (AD) reference sets were generated. AD references were determined by the mode value of the MIC when three or more replicates were available, and the modes are the same in both reference sets. When only two MIC values were available, and no mode could be determined, each MIC value was included in either reference set 1 or reference set 2. Breakpoint interpretations are identical between the two AD reference sets; see Table S2. Data from Sensititre were compared independently to each reference set, and then all comparison data were summarized. Percent categorical agreement (CA), essential agreement (EA), and minor Error (mE) were calculated according to established methods (37). The errors were then categorized into false resistance and false intermediate based on the direction the error occurred. False resistance is defined as an AD MIC of intermediate and a Sensititre BMD MIC of resistant. False intermediate is defined as an AD MIC of resistant and a Sensititre BMD MIC of intermediate. CA, EA, and mE were calculated using both automated and manual MIC calls for each independent operator. Percent EA was additionally calculated for each AD reference. Significance between changes in categorical agreement (CA) values for automated and manually interpretated reads was calculated by unpaired t test. P values of <0.05 were considered significant.
Data availability.
All primary data used in the analyses in this work are included in the supplemental material files: Table S2 (agar dilution reference data sets 1 and 2), Table S3 (polymyxin B agreements and error calculations between ThermoFisher Sensititre and AD reference 1), Table S4 (polymyxin B agreements and error calculations between ThermoFisher Sensititre and AD reference 2), Table S5 (colistin agreements and error calculations between ThermoFisher Sensititre and AD reference 1), and Table S6 (colistin agreements and error calculations between ThermoFisher Sensititre and AD reference 2).
RESULTS
Characterization of polymyxin resistance among the clinical isolates within this collection.
Of the 129 isolates selected for testing, 74 (57%) were Enterobacterales and 55 (43%) were non-Enterobacterales, comprised of 27 Pseudomonas aeruginosa and 28 Acinetobacter baumannii complex isolates. The number of organisms intermediate to colistin (n = 83) was similar to the number intermediate to polymyxin B (n = 82) as determined by AD, consistent with clinical isolates usually exhibiting overlapping resistance phenotypes to both polymyxins (Table 1).
TABLE 1.
Characterization of resistance to polymyxin antibiotics among Enterobacterales and non-Enterobacterales as determined by agar dilution
| Isolate | n (%) |
n (%) of organisms intermediate to: |
|
|---|---|---|---|
| Polymyxin B | Colistin | ||
| Enterobacter cloacae complex | 19 (26) | 5 (26) | 4 (21) |
| Escherichia coli | 20 (27) | 17 (85) | 18 (90) |
| Klebsiella aerogenes | 4 (5) | 4 (100) | 4 (100) |
| Klebsiella pneumoniae | 30 (41) | 6 (20) | 7 (23) |
| Raoultella ornithinolytica | 1 (1) | 0 | 0 |
| Total Enterobacterales | 74 (57) | 32 (43) | 33 (45) |
| Acinetobacter baumannii complex | 28 (51) | 23 (82) | 23 (82) |
| Pseudomonas aeruginosa | 27 (49) | 27 (100) | 27 (100) |
| Total non-Enterobacterales | 55 (43) | 50 (91) | 50 (91) |
| Total isolates | 129 | 82 (64) | 83 (64) |
Comparison of Sensititre BMD and AD for polymyxin AST.
Comparison of BMD MICs from Sensititre automated polymyxin B MIC calls to AD MICs revealed overall CA of 89% (n = 115) and 92% (n = 119) for operators A and B, respectively. CA of 81% (n = 60) and 88% (n = 65) were observed when analysis was restricted to Enterobacterales (Table 2). Up to 19% (n = 14) mEs were identified, the majority of which were false intermediate (Table 2). Evaluation of non-Enterobacterales isolates using the same parameters resulted in 100% (n = 55) and 98% (n = 54) CA, with only one mE (Table 2).
TABLE 2.
Evaluation of agreement between Sensititre-derived polymyxin B MICs via ARIS automated reads and MICs from agar dilutiona
| Isolate | n | Results for operator A |
Results for operator B |
||||
|---|---|---|---|---|---|---|---|
| n CA (%) |
n mE (%) |
n CA (%) |
n mE (%) |
||||
| False R | False I | False R | False I | ||||
| Enterobacter cloacae complex | 19 | 14 (74) | 0 | 5 (26) | 13 (68) | 0 | 6 (32) |
| Escherichia coli | 20 | 18 (90) | 0 | 2 (2) | 19 (95) | 0 | 1 (5) |
| Klebsiella aerogenes | 4 | 4 (100) | 0 | 0 | 3 (75) | 1 (25) | 0 |
| Klebsiella pneumoniae | 30 | 24 (80) | 0 | 6 (20) | 29 (97) | 1 (3) | 0 |
| Raoultella ornithinolytica | 1 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 |
| Total Enterobacterales | 74 | 60 (81) | 0 | 14 (19) | 65 (88) | 2 (3) | 7 (9) |
| Acinetobacter baumannii complex | 28 | 28 (100) | 0 | 0 | 28 (100) | 0 | 0 |
| Pseudomonas aeruginosa | 27 | 27 (100) | 0 | 0 | 26 (96) | 1 (4) | 0 |
| Total non-Enterobacterales | 55 | 55 (100) | 0 | 0 | 54 (98) | 1 (2) | 0 |
| Total isolates | 129 | 115 (89) | 0 | 14 (11) | 119 (92) | 3 (2) | 7 (5) |
CA, categorical agreement; mE, minor error; false R, false resistance, AD MIC intermediate and Sensititre BMD MIC resistant; false I, false intermediate, AD MIC resistant and Sensititre BMD MIC intermediate.
Colistin Sensititre automated MIC calls and AD-derived MICs were also compared (Table 3). The overall CA of 89% (n = 115) and 86% (n = 111), for operators A and B, were similar to the agreement values obtained for polymyxin B. Among the Enterobacterales, CA of 81% and 82% were observed, with up to 14% (n = 18) mEs, with both false resistance and false intermediate errors observed. In contrast, the non-Enterobacterales exhibited CA of 100% and 91% and a 9% mE rate consisting of only false resistance (Table 3).
TABLE 3.
Evaluation of agreement between Sensititre-derived colistin MICs via ARIS automated reads and MICs from agar dilutiona
| Isolate | n | Results for operator A |
Results for operator B |
||||
|---|---|---|---|---|---|---|---|
| n CA (%) |
n mE (%) |
n CA (%) |
n mE (%) |
||||
| False R | False I | False R | False I | ||||
| Enterobacter cloacae complex | 19 | 12 (63) | 0 | 7 (37) | 11 (58) | 0 | 8 (42) |
| Escherichia coli | 20 | 18 (90) | 1 (5) | 1 (5) | 18 (90) | 1 (5) | 1 (5) |
| Klebsiella aerogenes | 4 | 3 (75) | 1 (25) | 0 | 3 (75) | 1 (25) | 0 |
| Klebsiella pneumoniae | 30 | 27 (90) | 2 (7) | 1 (3) | 28 (93) | 2 (7) | 0 |
| Raoultella ornithinolytica | 1 | 0 | 0 | 1 (100) | 1 (100) | 0 | 0 |
| Total Enterobacterales | 74 | 60 (81) | 4 (5) | 10 (14) | 61 (82) | 4 (5) | 9 (12) |
| Acinetobacter baumannii complex | 28 | 28 (100) | 0 | 0 | 24 (86) | 4 (14) | 0 |
| Pseudomonas aeruginosa | 27 | 27 (100) | 0 | 0 | 26 (96) | 1 (4) | 0 |
| Total non-Enterobacterales | 55 | 55 (100) | 0 | 0 | 50 (91) | 5 (9) | 0 |
| Total isolates | 129 | 115 (89) | 4 (3) | 10 (8) | 111 (86) | 9 (7) | 9 (7) |
CA, categorical agreement; mE, minor error; false R, false resistance, AD MIC intermediate and Sensititre BMD MIC resistant; false I, false intermediate, AD MIC resistant and Sensititre BMD MIC intermediate.
Repeat Sensititre BMD testing with manual MIC call of categorically discrepant isolates.
A technical advantage of the Sensititre system is that operators can manually evaluate growth present in test wells following incubation of BMD panels. As polymyxin BMD is prone to technical issues, manual inspection of the colistin and polymyxin B test wells to derive polymyxin MICs of 17 categorically discrepant Enterobacterales isolates was undertaken during repeat testing by operator A.
Of the 17 isolates responsible for errors for either antibiotic compared to AD, 11 exhibited overlapping errors in both polymyxin B and colistin AST. All errors associated with polymyxin B AST were false intermediate, while both false resistance and false intermediate were encountered during colistin AST (Tables 2 and 3). In order to further examine these issues, repeat testing for manual interpretation of the MICs was undertaken for these categorically discrepant isolates. Manual MIC interpretations resolved 11 discrepancies. Of these, 5 isolates exhibited skip wells and 2 isolates had visible growth in BMD wells at concentrations of polymyxin that were not detected by the ARIS 2X instrument (Tables S3 to S6).
Evaluation of the impact of manual inspection of Sensititre BMD wells.
Due to discrepancies between automated reads and growth patterns observed upon manual inspection of BMD wells, the accuracy of manually interpreted Sensititre BMD MICs was determined. Percent CA and percent mE were calculated by comparing manually called BMD MICs to AD MICs (Tables 4 and 5), allowing for direct comparison of the accuracy of Sensititre automated MICs (Fig. 1). Two independent operators performed the manual calls. For operator A, the manual MICs for the 17 isolates from the previously described repeat testing were used in the analysis, whereas automated reads were used for isolates that did not have discrepant results between automated read and AD. For operator B, the isolates were tested subsequently and manual MICs were performed for all 129 isolates tested at time of initial Sensititre BMD AST so that we could better assess the impact of manual readings.
TABLE 4.
Evaluation of agreement between Sensititre-derived polymyxin B MICs via manual reads and MICs from agar dilutiona
| Isolate | n | Results for operator A (manual n = 17) |
Results for operator B (manual n = 129) |
||||
|---|---|---|---|---|---|---|---|
| n CA (%) |
n mE (%) |
n CA (%) |
n mE (%) |
||||
| False R | False I | False R | False I | ||||
| Enterobacter cloacae complex | 19 | 18 (95) | 0 | 1 (5) | 16 (84) | 0 | 3 (16) |
| Escherichia coli | 20 | 19 (95) | 0 | 1 (5) | 20 (100) | 0 | 0 |
| Klebsiella aerogenes | 4 | 3 (75) | 1 (25) | 0 | 3 (75) | 1 (25) | 0 |
| Klebsiella pneumoniae | 30 | 28 (93) | 1 (3) | 1 (3) | 29 (97) | 1 (3) | 0 |
| Raoultella ornithinolytica | 1 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| Total Enterobacterales | 74 | 69 (93) | 2 (3) | 3 (4) | 69 (93) | 2 (3) | 3 (4) |
| Acinetobacter baumannii complex | 28 | 28 (100) | 0 | 0 | 24 (86) | 4 (14) | 0 |
| Pseudomonas aeruginosa | 27 | 27 (100) | 0 | 0 | 26 (96) | 1 (4) | 0 |
| Total non-Enterobacterales | 55 | 55 (100) | 0 | 0 | 50 (91) | 5 (9) | 0 |
| Total isolates | 129 | 124 (96) | 2 (2) | 3 (2) | 119 (92) | 7 (5) | 3 (2) |
CA, categorical agreement; mE, minor error; false R, false resistance, AD MIC intermediate and Sensititre BMD MIC resistant; false I, false intermediate, AD MIC resistant and Sensititre BMD MIC intermediate. Operator A, manual reads for 17 isolates were used as MIC calls and the automated MIC calls were used for the remaining 112 isolates. Operator B, manual reads for all 129 isolates in this collection of clinical isolates were used as MIC calls for this data set.
TABLE 5.
Evaluation of agreement between Sensititre-derived colistin MICs via manual reads and MICs from agar dilutiona
| Isolate | n | Results for operator A (manual n = 17) |
Results for operator B (manual n = 129) |
||||
|---|---|---|---|---|---|---|---|
| n CA (%) |
n mE (%) |
n CA (%) |
n mE (%) |
||||
| False R | False I | False R | False I | ||||
| Enterobacter cloacae complex | 19 | 18 (95) | 0 | 1 (5) | 16 (84) | 0 | 3 (16) |
| Escherichia coli | 20 | 19 (95) | 1 (5) | 0 | 19 (95) | 1 (5) | 0 |
| Klebsiella aerogenes | 4 | 3 (75) | 1 (25) | 0 | 3 (75) | 1 (25) | 0 |
| Klebsiella pneumoniae | 30 | 28 (93) | 2 (7) | 0 | 28 (93) | 2 (7) | 0 |
| Raoultella ornithinolytica | 1 | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| Total Enterobacterales | 74 | 69 (93) | 4 (5) | 1 (1) | 67 (91) | 4 (5) | 3 (4) |
| Acinetobacter baumannii complex | 28 | 28 (100) | 0 | 0 | 21 (75) | 7 (7) | 0 |
| Pseudomonas aeruginosa | 27 | 27 (100) | 0 | 0 | 26 (96) | 1 (4) | 0 |
| Total non-Enterobacterales | 55 | 55 (100) | 0 | 0 | 47 (85) | 8 (15) | 0 |
| Total isolates | 129 | 124 (96) | 4 (3) | 1 (1) | 114 (88) | 12 (9) | 3 (2) |
CA, categorical agreement; mE, minor error; false R, false resistance, AD MIC intermediate and Sensititre BMD MIC resistant; false I, false intermediate, AD MIC resistant and Sensititre BMD MIC intermediate. Operator A, manual reads for 17 isolates were used as MIC calls and the automated MIC calls were used for the remaining 112 isolates. Operator B, manual reads for all 129 isolates in this collection of clinical isolates were used as MIC calls for this data set.
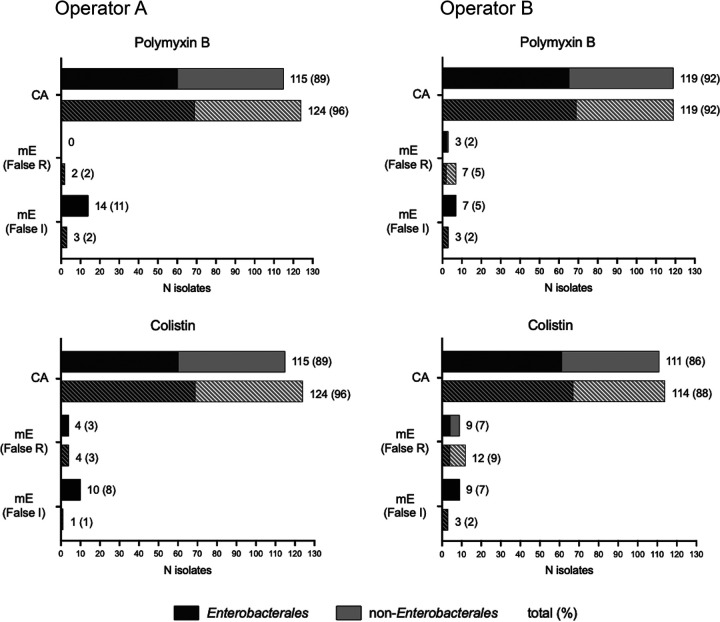
FIG 1.
Comparison of categorical agreements between automated and manually interpreted Sensititre-derived MICs and MICs from agar dilution. Values of and percent categorical agreement (CA) and minor error (mE) are shown for polymyxin B and colistin with either automated (solid) or manual (striped) MICs compared to agar dilution. False resistance is defined as AD MIC intermediate and Sensititre BMD MIC resistant. False intermediate is defined as AD MIC resistant and Sensititre BMD MIC intermediate. Proportions of Enterobacterales (black) and non-Enterobacterales (gray) are shown.
For polymyxin B, overall CA of 96% (n = 124) and 92% (n = 119) were observed for operators A and B, respectively. Additionally, the percent mE decreased to 4% (n = 5) and 7% (n = 10) (Table 4), indicating that the automatic reads incorrectly reported the MIC of a subset of isolates. While the number of false intermediate errors decreased, there was an increase in the percent false resistance. Specifically, a subset of Acinetobacter baumannii complex species were designated resistant when interpreted manually, disagreeing with the automatic MIC call.
Similar changes for colistin comparisons were observed (Table 5). Overall CA of 96% (n = 124) and 88% (n = 114) were observed for operators A and B, respectively. The lower CA percentage observed by operator B is due to an increase in the percentage of false resistant mEs in the Acinetobacter baumannii complex species (7%). If restricted to Enterobacterales, the overall CA is 91% for operator B, an increase from 82%. The percent false intermediate mEs observed was 1% and 2%, a decrease of more than half when error rates based on manual versus automated MIC calls are compared. These error rates are still higher than recommended by CLSI guidelines, so reference laboratory utilization is recommended in the discussion as the most conservative approach for adjudicating isolates with any automated/manual read discrepancy. Although the difference in CA between automated and manual reads did not reach statistical significance for this data set, it is important to note that automated readings repeatedly missed skipped wells, and statistical significance might be reached if a larger collection of isolates were tested.
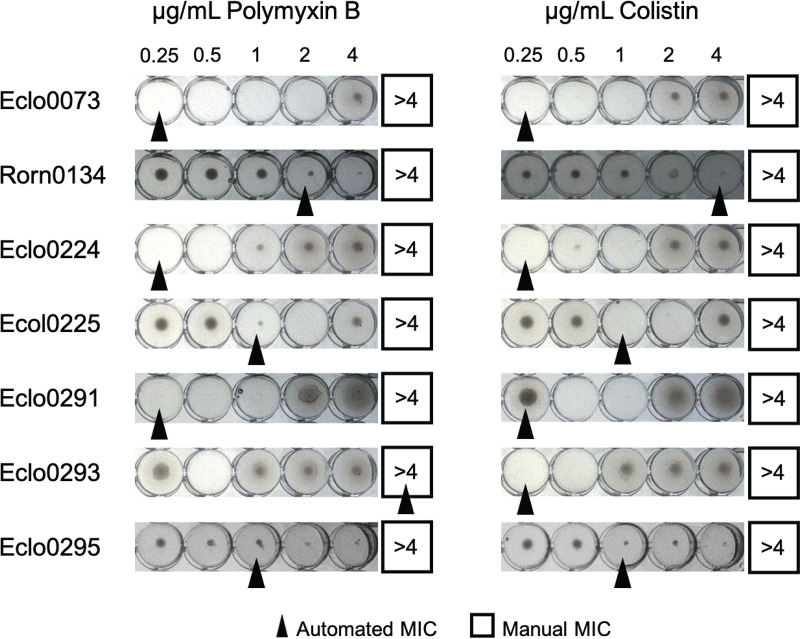
Most changes between the automated and manually interpreted BMD MICs were due to undetected microbial growth and skip wells. For isolates where the automated and manual polymyxin B MICs did not agree, half of the manual interpretations had visible undetected growth and the remaining half had observed skipped wells. For colistin, the majority of isolates had observed skipped wells, while only one had visible undetected growth. Out of 21 isolates that had disagreements between the automated and manual MICs, either for polymyxin B or for colistin, 6 isolates were observed to be discrepant for both polymyxins (Fig. 2). The manual MIC interpretation of these isolates resulted in the resolution of most discrepancies compared to AD MICs, as discussed above.
FIG 2.
Undetected microbial growth and skip wells observed via manual inspection of growth in Sensititre BMD wells. Automated calls are indicated by a black arrowhead. Manual calls are indicated with a black box.
Use of resistance to either polymyxin antibiotic as a predictor of polymyxin B or colistin susceptibility.
Interpretive guidance in the 2020 CLSI M100 document permits the use of colistin MICs to predict polymyxin B susceptibility (18). As both colistin and polymyxin B were tested in parallel on BMD panels, we sought to determine whether resistance to colistin determined by Sensititre BMD accurately predicts resistance to polymyxin B. Agreement between the two polymyxins within this isolate archive was observed to be 96% (n = 124) by AD and 98% and 95% by manual Sensititre BMD for operators A and B, respectively. Colistin and polymyxin B susceptibility did not agree for a total of 13 isolates across both methods, 9 of which were resistant to colistin and intermediate for polymyxin B. Only 3 of the 13 isolates in disagreement were within essential agreement. Additionally, a majority of isolates with disagreements via Sensititre BMD had growth patterns including skip wells and undetected microbial growth. These data indicate that although a high degree of agreement between the two polymyxins is observed, any laboratory testing both colistin and polymyxin B simultaneously will face potential discrepancies between the two results for specific isolates.
DISCUSSION
In this work, the performance of the Sensititre automated BMD platform was evaluated for polymyxin B and colistin AST of a broad collection of Enterobacterales and non-Enterobacterales clinical isolates. Previous investigations have reported that the Sensititre system performs adequately compared to manual BMD for the determination of polymyxin MICs (15, 17, 28–33). While a majority of these studies have focused on colistin AST, a proportionately smaller number have evaluated the Sensititre platform for polymyxin B (15, 28, 32, 38). Importantly, these studies also restricted test isolates to a specific organism group or species.
Automated MIC calls from the Sensititre ARIS 2X instrument exhibited rates of discrepancy that were high compared to those of colistin and polymyxin B AD (Tables 2 and 3 and Fig. 1). While delineation of the resistance mechanism was beyond the scope of this work, there were isolates for which the growth pattern was erroneously interpreted by the automated reads for both drugs (Fig. 2). No previous study to date has comprehensively evaluated bacterial growth within the wells used to derive automated MICs by the Sensititre ARIS 2X instrument against colistin and polymyxin B.
Skip wells are a technical challenge associated with polymyxin AST of Enterobacter cloacae complex isolates by BMD (24). It has been hypothesized that skip wells may be the result of heteroresistant subpopulations of bacterial cells which emerge in response to polymyxin exposure (39). Heteroresistance to both colistin (40) and polymyxin B (24) has been reported among members of the E. cloacae complex and recently has been shown to be underreported by BMD (41).
In this work, 7 of 19 E. cloacae complex isolates (37%) exhibited skip wells during colistin BMD, 4 of which also exhibited skip wells when tested against polymyxin B (Fig. 2). The ARIS 2X instrument interpreted the polymyxin B MICs of 3 of these isolates as intermediate over two independent assays. For one isolate with skip wells in both colistin and polymyxin B wells, Eclo0293, the automated MIC for polymyxin B, was interpreted as ≥4 μg/ml despite exhibiting skip wells (Fig. 2). Further investigation to examine more closely the mechanisms underlying the skip well phenomenon, its impact on automated susceptibility testing, and its clinical significance is warranted. As the ARIS 2X cannot consistently discern skip well patterns and CLSI directs noninterpretation of BMD assays exhibiting multiple skip wells (36), incorporation of a manual evaluation of BMD wells for polymyxin AST is critical.
Among test isolates, a strong correlation between polymyxin B and colistin MICs is observed and no currently described resistance mechanism affects only colistin or polymyxin B individually. In the 2020 CLSI M100 document, colistin and polymyxin B are listed as equivalent agents for Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii complex (18, 42, 43). In this work, there is a strong agreement between colistin and polymyxin B MICs for both AD and manually interpreted Sensititre BMD. However, the calls are not identical, so there is still a risk of a false call of intermediate when testing only one of the two drugs and referring the result. This is consistent with reported concerns about underreporting of resistance (41). Furthermore, no interpretive guidance is provided for a situation where both polymyxin antibiotics are tested simultaneously (i.e., in the context of a panel) and the results are discrepant. As CLSI directs that these drugs are equivalent, it may be confusing to clinicians and not consistent with current recommendations to report one as intermediate and one as resistant. Therefore, a laboratory might choose to call the attending physician to discuss the case and to ensure that the limitations of polymyxin AST are very clear and/or that repeat testing by an alternative reference method is performed.
Differences in polymyxin B and colistin MICs have been attributed to technical variation between AST systems during routine testing (12). Therefore, recommendations to perform multiple dilution tests when encountering skip wells have been made for colistin AST and E. cloacae complex organisms (24). Likewise, laboratories using panels that include both colistin and polymyxin B may detect more resistance or heteroresistance because the number of polymyxin test wells would be increased.
In this work, the majority of categorically discrepant isolates were members of the Enterobacterales. No non-Enterobacterales isolates exhibited false intermediate MICs between Sensititre automated and manual MIC calls and manual AD for either polymyxin antibiotic. However, non-Enterobacterales isolates had a high rate of false resistant mEs when MICs were determined by manual interpretation. This was due to a subset of Acinetobacter baumannii complex isolates that exhibited undetected growth and skipped wells. For clinical quality control purposes, 10 of these isolates were repeated and growth patterns including skip wells and undetected growth were observed for colistin and polymyxin B but not for other antibiotics on the panel (Table S7), indicating that there is high variability in the polymyxin resistance patterns of these Acinetobacter baumannii complex isolates. While only a small number of resistant non-Enterobacterales were included in the test battery, it is important to note how the instrument performed for the available set of isolates. Testing additional resistant non-Enterobacterales will provide important adjunctive information regarding the performance of the assay within this organism group.
There are still a number of unresolved questions surrounding optimal polymyxin AST modality. Broth disk elution and agar screening methods have been added as optional supplemental tests for determining colistin MICs for Enterobacterales and Pseudomonas species in the most recent CLSI M100 (18). The development of these assays was driven with the objective to identify AST methods with accuracy equal to but not greater than that of the current reference broth microdilution method. Recently, Humphries et al. tested these methods and reported that both exhibit strong agreement with the reference method (44); however, it should be noted that the exclusion criteria utilized by the authors for isolates which exhibit either inconsistent MICs or repeated skip wells bias the composition of the challenge set by eliminating problematic strains which could be clinically encountered. The authors urged careful interpretation of these assays for the presence of skipped dilutions over multiple replicates and noninterpretation of such isolates.
In conclusion, we evaluated the accuracy of the Sensititre system for automated polymyxin AST compared to that of AD for a varied collection of Enterobacterales and non-Enterobacterales. Agreement between the two methods was enhanced through the incorporation of a manual evaluation of bacterial growth within the polymyxin BMD test wells. Despite the technical challenges and the development of new manual assays, automated BMD is a viable option that is cost-effective, rapid, accessible, and adaptable to routine laboratories already utilizing the requisite instrumentation. Laboratories choosing to perform in-house polymyxin AST using the Sensititre system should consider adopting manual evaluation of bacterial growth in polymyxin BMD wells to improve performance. For the most conservative approach, if automated and manual reads do not agree, an alternative method or reference laboratory testing for relevant isolates is suggested.
ACKNOWLEDGMENTS
We thank Wendy Bishop, Laurie Flemming, and the technologists of the clinical microbiology laboratory at the National Institutes of Health Clinical Center for their assistance. This work was supported by the Intramural Research Programs of the National Institutes of Health Clinical Center and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, Bethesda, MD, USA. The findings and conclusions in this study are those of the authors and do not necessarily represent the official positions of the National Institutes of Health or the Department of Health and Human Services. We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Karen M. Frank, Email: karen.frank@nih.gov.
Nathan A. Ledeboer, Medical College of Wisconsin
REFERENCES
- 1.Perez F, El Chakhtoura NG, Yasmin M, Bonomo RA. 2019. Polymyxins: to combine or not to combine? Antibiotics (Basel) 8:38–51. doi: 10.3390/antibiotics8020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKinnell JA, Dwyer JP, Talbot GH, Connolly LE, Friedland I, Smith A, Jubb AM, Serio AW, Krause KM, Daikos GL, CARE Study Group. 2019. Plazomicin for infections caused by carbapenem-resistant Enterobacteriaceae. N Engl J Med 380:791–793. doi: 10.1056/NEJMc1807634. [DOI] [PubMed] [Google Scholar]
- 3.Motsch J, Murta de Oliveira C, Stus V, Koksal I, Lyulko O, Boucher HW, Kaye KS, File TM, Brown ML, Khan I, Du J, Joeng HK, Tipping RW, Aggrey A, Young K, Kartsonis NA, Butterton JR, Paschke A. 2020. RESTORE-IMI 1: a multicenter, randomized, double-blind trial comparing efficacy and safety of imipenem/relebactam vs colistin plus imipenem in patients with imipenem-nonsusceptible bacterial infections. Clin Infect Dis 70:1799–1808. doi: 10.1093/cid/ciz530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul M, Daikos GL, Durante-Mangoni E, Yahav D, Carmeli Y, Benattar YD, Skiada A, Andini R, Eliakim-Raz N, Nutman A, Zusman O, Antoniadou A, Pafundi PC, Adler A, Dickstein Y, Pavleas I, Zampino R, Daitch V, Bitterman R, Zayyad H, Koppel F, Levi I, Babich T, Friberg LE, Mouton JW, Theuretzbacher U, Leibovici L. 2018. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: an open-label, randomised controlled trial. Lancet Infect Dis 18:391–400. doi: 10.1016/S1473-3099(18)30099-9. [DOI] [PubMed] [Google Scholar]
- 5.van Duin D, Lok JJ, Earley M, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Doi Y, Kaye KS, Fowler VG, Paterson DL, Bonomo RA, Evans S, Antibacterial Resistance Leadership G. 2018. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 66:163–171. doi: 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenhard JR, Bulman ZP, Tsuji BT, Kaye KS. 2019. Shifting gears: the future of polymyxin antibiotics. Antibiotics (Basel) 8:42–55. doi: 10.3390/antibiotics8020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avedissian SN, Liu J, Rhodes NJ, Lee A, Pais GM, Hauser AR, Scheetz MH. 2019. A review of the clinical pharmacokinetics of polymyxin B. Antibiotics (Basel) 8:31–42. doi: 10.3390/antibiotics8010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Lee W, Kwa AL. 2015. Polymyxin B versus colistin: an update. Expert Rev Anti Infect Ther 13:1481–1497. doi: 10.1586/14787210.2015.1093933. [DOI] [PubMed] [Google Scholar]
- 9.Kassamali Z, Danziger L. 2015. To B or not to B, that is the question: is it time to replace colistin with polymyxin B? Pharmacotherapy 35:17–21. doi: 10.1002/phar.1510. [DOI] [PubMed] [Google Scholar]
- 10.Tran TB, Velkov T, Nation RL, Forrest A, Tsuji BT, Bergen PJ, Li J. 2016. Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet? Int J Antimicrob Agents 48:592–597. doi: 10.1016/j.ijantimicag.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zavascki AP, Nation RL. 2017. Nephrotoxicity of polymyxins: is there any difference between colistimethate and polymyxin B? Antimicrob Agents Chemother 61:e02319-16. doi: 10.1128/AAC.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jerke KH, Lee MJ, Humphries RM. 2016. Polymyxin susceptibility testing: a cold case reopened. Clin Microbiol Newsl 38:69–77. doi: 10.1016/j.clinmicnews.2016.04.003. [DOI] [Google Scholar]
- 13.Bakthavatchalam YD, Veeraraghavan B. 2017. Challenges, issues and warnings from CLSI and EUCAST working group on polymyxin susceptibility testing. J Clin Diagn Res 11:DL03–DL04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasoo S. 2017. Susceptibility testing for the polymyxins: two steps back, three steps forward? J Clin Microbiol 55:2573–2582. doi: 10.1128/JCM.00888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 51:1678–1684. doi: 10.1128/JCM.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulengowski B, Ribes JA, Burgess DS. 2019. Polymyxin B Etest((R)) compared with gold-standard broth microdilution in carbapenem-resistant Enterobacteriaceae exhibiting a wide range of polymyxin B MICs. Clin Microbiol Infect 25:92–95. doi: 10.1016/j.cmi.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Matuschek E, Ahman J, Webster C, Kahlmeter G. 2018. Antimicrobial susceptibility testing of colistin - evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect 24:865–870. doi: 10.1016/j.cmi.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 18.CLSI. 2020. Performance standards for antimicrobial susceptibility testing, 30th Edition. CLSI M100, CLSI, Wayne, PA. [Google Scholar]
- 19.EUCAST. 2016. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 20.Tsuji BT, Pogue JM, Zavascki AP, Paul M, Daikos GL, Forrest A, Giacobbe DR, Viscoli C, Giamarellou H, Karaiskos I, Kaye D, Mouton JW, Tam VH, Thamlikitkul V, Wunderink RG, Li J, Nation RL, Kaye KS. 2019. International consensus guidelines for the optimal use of the polymyxins: endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 39:10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karvanen M, Malmberg C, Lagerbäck P, Friberg LE, Cars O. 2017. Colistin is extensively lost during standard in vitro experimental conditions. Antimicrob Agents Chemother 61:e00857-17. doi: 10.1128/AAC.00857-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, Ledesma KR, Lam WY, Figueroa DA, Lim TP, Chow DS, Tam VH. 2010. Variability of polymyxin B major components in commercial formulations. Int J Antimicrob Agents 35:308–310. doi: 10.1016/j.ijantimicag.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Humphries RM. 2015. Susceptibility testing of the polymyxins: where are we now? Pharmacotherapy 35:22–27. doi: 10.1002/phar.1505. [DOI] [PubMed] [Google Scholar]
- 24.Landman D, Salamera J, Quale J. 2013. Irreproducible and uninterpretable Polymyxin B MICs for Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol 51:4106–4111. doi: 10.1128/JCM.02129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gales AC, Reis AO, Jones RN. 2001. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol 39:183–190. doi: 10.1128/JCM.39.1.183-190.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turlej-Rogacka A, Xavier BB, Janssens L, Lammens C, Zarkotou O, Pournaras S, Goossens H, Malhotra-Kumar S. 2018. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur J Clin Microbiol Infect Dis 37:345–353. doi: 10.1007/s10096-017-3140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.USFDA. Antibacterial susceptibility test interpretive criteria. https://www.fda.gov/drugs/development-resources/antibacterial-susceptibility-test-interpretive-criteria/.
- 28.Chew KL, La MV, Lin RTP, Teo JWP. 2017. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol 55:2609–2616. doi: 10.1128/JCM.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javed M, Ueltzhoeffer V, Heinrich M, Siegrist HJ, Wildermuth R, Lorenz FR, Neher RA, Willmann M. 2018. Colistin susceptibility test evaluation of multiple-resistance-level Pseudomonas aeruginosa isolates generated in a morbidostat device. J Antimicrob Chemother 73:3368–3374. [DOI] [PubMed] [Google Scholar]
- 30.Jayol A, Nordmann P, Andre C, Poirel L, Dubois V. 2018. Evaluation of three broth microdilution systems to determine colistin susceptibility of Gram-negative bacilli. J Antimicrob Chemother 73:1272–1278. doi: 10.1093/jac/dky012. [DOI] [PubMed] [Google Scholar]
- 31.Pfennigwerth N, Kaminski A, Korte-Berwanger M, Pfeifer Y, Simon M, Werner G, Jantsch J, Marlinghaus L, Gatermann SG. 2019. Evaluation of six commercial products for colistin susceptibility testing in Enterobacterales. Clin Microbiol Infect 25:1385–1389. doi: 10.1016/j.cmi.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 32.Richter SS, Karichu J, Otiso J, Van Heule H, Keller G, Cober E, Rojas LJ, Hujer AM, Hujer KM, Marshall S, Perez F, Rudin SD, Domitrovic TN, Kaye KS, Salata R, van Duin D, Bonomo RA. 2018. Evaluation of Sensititre broth microdilution plate for determining the susceptibility of carbapenem-resistant Klebsiella pneumoniae to polymyxins. Diagn Microbiol Infect Dis 91:89–92. doi: 10.1016/j.diagmicrobio.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yusuf E, van Westreenen M, Goessens W, Croughs P. 2020. The accuracy of four commercial broth microdilution tests in the determination of the minimum inhibitory concentration of colistin. Ann Clin Microbiol Antimicrob 19:42–50. doi: 10.1186/s12941-020-00383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CLSI. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI document M-100, CLSI, Wayne, PA. [Google Scholar]
- 35.Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn JH, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. 2014. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol 52:2804–2812. doi: 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CLSI. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. CLSI Document M-07, CLSI, Wayne, PA. [Google Scholar]
- 37.CLSI. 2015. Verification of commercial microbial identification and antimicrobial susceptibility testing systems, 1st ed. CLSI document M52, CLSI, Wayne, PA. [Google Scholar]
- 38.Sader HS, Rhomberg PR, Farrell DJ, Jones RN. 2015. Differences in potency and categorical agreement between colistin and polymyxin B when testing 15,377 clinical strains collected worldwide. Diagn Microbiol Infect Dis 83:379–381. doi: 10.1016/j.diagmicrobio.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 39.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Band VI, Satola SW, Smith RD, Hufnagel DA, Bower C, Conley AB, Rishishwar L, Dale SE, Hardy DJ, Vargas RL, Dumyati G, Kainer MA, Phipps EC, Pierce R, Wilson LE, Sorensen M, Nilsson E, Jordan IK, Burd EM, Farley MM, Jacob JT, Ernst RK, Weiss DS, Bonomo RA. 2021. Colistin heteroresistance is largely undetected among carbapenem-resistant Enterobacterales in the United States. mBio 12:e02881-20. doi: 10.1128/mBio.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogue JM, Jones RN, Bradley JS, Andes DR, Bhavnani SM, Drusano GL, Dudley MN, Flamm RK, Rodvold KA, Ambrose PG. 2020. Polymyxin susceptibility testing and interpretive breakpoints: recommendations from the United States committee on antimicrobial susceptibility testing (USCAST). Antimicrob Agents Chemother 64:e01495-19. doi: 10.1128/AAC.01495-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satlin MJ, Lewis JS, Weinstein MP, Patel J, Humphries RM, Kahlmeter G, Giske CG, Turnidge J. 2020. Clinical and Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility testing position statements on polymyxin B and colistin clinical breakpoints. Clin Infect Dis 71:e523–e529. doi: 10.1093/cid/ciaa121. [DOI] [PubMed] [Google Scholar]
- 44.Humphries RM, Green DA, Schuetz AN, Bergman Y, Lewis S, Yee R, Stump S, Lopez M, Macesic N, Uhlemann A, Kohner P, Cole N, Simner PJ. 2019. Multicenter evaluation of colistin broth disk elution and colistin agar test: a report from the Clinical and Laboratory Standards Institute. J Clin Microbiol 57:e01269-19. doi: 10.1128/JCM.01269-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1, and legends of Tables S2 to S7. Download JCM.00332-21-s0001.pdf, PDF file, 1.0 MB (978.7KB, pdf)
Table S2. Download JCM.00332-21-s0002.xlsx, XLSX file, 0.05 MB (55.5KB, xlsx)
Table S3. Download JCM.00332-21-s0003.xlsx, XLSX file, 0.03 MB (30.6KB, xlsx)
Table S4. Download JCM.00332-21-s0004.xlsx, XLSX file, 0.03 MB (30.7KB, xlsx)
Table S5. Download JCM.00332-21-s0005.xlsx, XLSX file, 0.03 MB (30.7KB, xlsx)
Table S6. Download JCM.00332-21-s0006.xlsx, XLSX file, 0.03 MB (30.7KB, xlsx)
Table S7. Download JCM.00332-21-s0007.xlsx, XLSX file, 0.01 MB (14.8KB, xlsx)
Data Availability Statement
All primary data used in the analyses in this work are included in the supplemental material files: Table S2 (agar dilution reference data sets 1 and 2), Table S3 (polymyxin B agreements and error calculations between ThermoFisher Sensititre and AD reference 1), Table S4 (polymyxin B agreements and error calculations between ThermoFisher Sensititre and AD reference 2), Table S5 (colistin agreements and error calculations between ThermoFisher Sensititre and AD reference 1), and Table S6 (colistin agreements and error calculations between ThermoFisher Sensititre and AD reference 2).