Abstract
The rapid adoption of electronic health records (EHRs) systems has made clinical data available in electronic format for research and for many downstream applications. Electronic screening of potentially eligible patients using these clinical databases for clinical trials is a critical need to improve trial recruitment efficiency. Nevertheless, manually translating free-text eligibility criteria into database queries is labor intensive and inefficient. To facilitate automated screening, free-text eligibility criteria must be structured and coded into a computable format using controlled vocabularies. Named entity recognition (NER) is thus an important first step. In this study, we evaluate 4 state-of-the-art transformer-based NER models on two publicly available annotated corpora of eligibility criteria released by Columbia University (i.e., the Chia data) and Facebook Research (i.e.the FRD data). Four transformer-based models (i.e., BERT, ALBERT, RoBERTa, and ELECTRA) pretrained with general English domain corpora vs. those pretrained with PubMed citations, clinical notes from the MIMIC-III dataset and eligibility criteria extracted from all the clinical trials on ClinicalTrials.gov were compared. Experimental results show that RoBERTa pretrained with MIMIC-III clinical notes and eligibility criteria yielded the highest strict and relaxed F-scores in both the Chia data (i.e., 0.658/0.798) and the FRD data (i.e., 0.785/0.916). With promising NER results, further investigations on building a reliable natural language processing (NLP)-assisted pipeline for automated electronic screening are needed.
CCS CONCEPTS: Computing methodologies → Information extraction
Keywords: Eligibility Criteria Parsing, Transformer-Based Model, Named Entity Recognition, Clinical Trial
1. INTRODUCTION
Clinical trials, which generate gold standard evidence for advancing science and developing effective treatments, often fail to recruit sufficient patients or suffer from delayed patient accrual, leading to (1) trial failures, and (2) potential low population representativeness [6, 16]. Since the Health Information Technology for Economic and Clinical Health (HITECH) Act was signed into law in 2009, electronic health record (EHR) systems have been rapidly adopted by more than 95% healthcare providers in the United States [1]. Identifying trial eligible patients from EHRs (i.e., electronic screening) for targeted recruitment has been shown to improve recruitment efficiency [13]. In clinical trials, eligibility criteria are used to characterize the patients who would be considered for the study; thus, the first step in electronic screening is to translate trial eligibility criteria into computable database queries against clinical databases. Nevertheless, eligibility criteria are free text and poorly standardized (not using controlled vocabularies), making development of corresponding database queries labor intensive and error prone. Natural language processing (NLP) is a key technology to help translate free-text eligibility criteria into EHR-based cohort identification queries for targeted recruitment.
To transform free-text eligibility criteria to a structured format, the first necessary step to perform named entity recognition (NER), and then map the named entities to concepts in controlled vocabularies based on their corresponding data domains in EHRs (e.g., conditions, medications, laboratory tests, and procedures). Even though a number of criteria parsers were developed before (e.g., [5, 17]), most of them are rule-based systems; it is not until recently that machine-learning-based NLP methods were employed (e.g., Criteria2Query, a system that parses trial criteria according to the widely used Observational Medical Outcomes Partnership (OMOP) common data model for EHRs [19]). Nonetheless, the performance of these existing systems is suboptimal, likely due to the limitations of using small training data and less powerful NER models. Recently, two large datasets with expert-annotated eligibility criteria were made publicly available: the Chia data (with 1000 trials) [9] and the Facebook Research data (FRD, with 3314 trials) [14]. Further, recent advancements in deep-learning based NLP models, especially the transformers, have shown remarkable state-of-the-art (SOTA) performance in biomedical NER tasks [4]. In this project, we benchmarked 4 SOTA transformer-based NER models pretrained with different corpora on the task of named entity recognition for eligibility criteria parsing using the two new annotated eligibility criteria datasets.
2. RELATED WORK
Previously, a number of NLP systems were developed to parse free-text clinical trial eligibility criteria. Earlier systems such as EliXR [17], EliXR-Time [17], ValX [5], and ERGO [15] are rule-based and/or ontology-based systems, which often have high precision but low recall, due to the limited coverage of the predefined rules and ontologies. Recent more robust systems including ELIIE [8] and Criteria2Query [19] use machine learning based NER models such as conditional random fields (CRF) and then combine with rule-based methods for negation detection, relationship extraction, and logic detection. No existing system explored recent SOTA transformer-based NER models for eligibility criteria parsing. Previously, these transformer-based models were benchmarked for clinical concept extraction from clinical narratives with SOTA performance [18].
3. METHODS
3.1. Problem Definition
Our goal is to perform NER of clinical trial eligibility criteria and recognize their entity types (e.g., condition, drug, procedure). Trial eligibility criteria are often partial sentences, e.g., “Diagnosed with COVID-19 pneumonia by RT-PCR” (from trial NCT04445272), where two named entities: “COVID-19” (Entity type: Condition) and “RT-PCR” (Entity type: Measurement) can be extracted. The NER task of this study is to recognize both the entities as well as their entity types in the criteria.
3.2. Model Description
We explored 4 widely-used SOTA transformer-based models including BERT, ALBERT, RoBERTa, and ELECTRA. BERT is a multilayer bidirectional transformer-based encoder model that can be pretrained with unlabelled data using masked language modeling and optimized by next sentence prediction [3]. ALBERT is a simplified version of BERT by reducing the token-embedding layer size and sharing parameters across all layers [10], and optimized using sentence-order prediction model. RoBERTa has the same architecture as BERT and is pretrained using a dynamic masked language modeling and optimized using strategies such as removing the next sentence prediction [11]. ELECTRA also has the same architecture as BERT but is pretrained using a strategy called replaced token detection [2].
We benchmarked these 4 models (1) pretrained with general English domain corpora, and (2) further pretrained with PubMed citations, clinical notes from the Medical Information Mart for Intensive Care III (MIMIC-III) database [7] and free-text eligibility criteria of clinical trials. The BERT, ALBERT, RoBERTa, and ELECTRA models are pretrained with general English domain corpora. The BERT-MIMIC, ALBERT-MIMIC, RoBERTa-MIMIC, and ELECTRA-MIMIC models are further pretrained with clinical notes (MIMIC-III) [7] based on the BERT, ALBERT, RoBERTa, and ELECTRA models. The BLUEBERT [12] model was a model further pretrained with PubMed abstracts and clinical notes (MIMIC-III) [7] based on the BERT model. Initial experiment showed the RoBERTa-MIMIC model achieved the best performance. In order to investigate performance of models further pretrained with eligibility criteria, we further pretrained the RoBERTa-MIMIC model with eligibility criteria extracted from more than 350,000 clinical trial summaries on ClinicalTrials.gov (i.e. the RoBERTa-MIMIC-Trial model). The Attention-based Bidirectional Long Short-Term Memory model with a CRF layer (Att-BiLSTM-CRF) was used for trial eligibility criteria parsing [14] with the FRD data; thus, we used Att-BiLSTM-CRF as the baseline in this study.
4. EXPERIMENTAL SETUP
4.1. Dataset Preparation
Two sets of annotated eligibility criteria data were used in our experiment: Chia and FRD.
Chia:
an annotated corpus of 12,409 eligibility criteria from 1,000 Phase IV trials in ClinicalTrials.gov [9]. It was annotated by two medical professionals and contains 41,487 distinctive entities of 15 entity types that are aligned with the data domains in the OMOP common data model. As transformer-based models and Att-BiLSTM-CRF require non-overlapping entity annotation, we retained the entity with largest text span when the original annotated entities overlap. We selected 11 major entity types in Chia and converted the annotations into the BIO format using SpaCy. We used 800 trials for training, 100 trials for validation, and 100 trials for testing. Table 1 shows the details of the Chia data.
Table 1:
Number of entities by type and by train/validation/test split in Chia
| Entity Type | Train | Val. | Test |
|---|---|---|---|
| Condition | 8,927 | 1,057 | 1,098 |
| Value | 2,990 | 373 | 327 |
| Drug | 2,892 | 345 | 311 |
| Procedure | 2,602 | 282 | 320 |
| Measurement | 2,532 | 316 | 280 |
| Temporal | 2,456 | 247 | 266 |
| Observation | 1,381 | 127 | 180 |
| Person | 1,239 | 152 | 140 |
| Mood | 467 | 47 | 49 |
| Device | 275 | 35 | 41 |
| Pregnancy_considerations | 160 | 12 | 18 |
| Total | 25,921 | 2,993 | 3,030 |
FRD:
an annotated corpus of about 50,000 eligibility criteria from 3,314 randomly selected trials in the United States [14]. It has 15 entity types as shown in Table 2. Unlike the Chia data, there were no overlapping entity annotations. Similarly, we used 80% (2,651 trials) for training, 10% (331 trials) for validation, and 10% (332 trials) for testing.
Table 2:
Number of entities by type and by train/validation/test split in FRD.
| Entity Type | Train | Val. | Test |
|---|---|---|---|
| treatment | 24,504 | 2,746 | 3,204 |
| chronic_disease | 20,999 | 2,371 | 2,614 |
| upper_bound | 11,136 | 1,228 | 1,371 |
| lower_bound | 10,734 | 1,179 | 1,380 |
| clinical_variable | 10,439 | 1,152 | 1,424 |
| cancer | 7,436 | 826 | 933 |
| gender | 2,830 | 323 | 358 |
| pregnancy | 2,174 | 248 | 315 |
| age | 2,095 | 223 | 262 |
| allergy_name | 1,504 | 183 | 194 |
| contraception_consent | 1,301 | 130 | 169 |
| language_fluency | 391 | 44 | 47 |
| bmi | 235 | 25 | 24 |
| technology_access | 108 | 13 | 11 |
| ethnicity | 64 | 7 | 8 |
| Total | 95,950 | 10,698 | 12,314 |
4.2. Model Implementations
For the transformer-based models for NER, we used the default implementation provided in [18]. For Att-BiLSTM-CRF, we used its implementation in [14] with their default setting, which uses word embeddings pretrained with FastText on trial description and eligibility criteria of over 300K trials from ClinicalTrials.gov as of May 2019. We evaluated model performance using precision, recall, and F1 based on both the strict matching criterion (i.e., exact match of both the entity type and entity with the gold-standard annotated entity) and relaxed criterion (i.e., only requires the predicted entity overlap with the gold-standard annotated entity).
5. EXPERIMENTAL RESULTS AND ANALYSES
5.1. Experimental Results
Table 3 and Table 4 show the performance of the transfomer-based NER models on the Chia and FRD datasets, respectively. All transformer-based models outperform the baseline Att-BiLSTM-CRF model on both datasets, even though Chia is much smaller than FRD. The RoBERTa-MIMIC-Trial model has the best strict-level F1-score and the best relax-level F1-score.
Table 3:
Performance of the transformer-based models vs. the baseline Att-BiLSTM-CRF model on Chia.
| Model | Strict Criterion | Relaxed Criterion | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| BERT | 0.6052 | 0.6653 | 0.6339 | 0.7646 | 0.8132 | 0.7882 |
| BERT-MIMIC | 0.5934 | 0.6749 | 0.6316 | 0.7559 | 0.8228 | 0.7879 |
| BLUEBERT | 0.6244 | 0.6634 | 0.6433 | 0.7819 | 0.8033 | 0.7925 |
| ALBERT | 0.6007 | 0.6488 | 0.6238 | 0.7715 | 0.8020 | 0.7864 |
| ALBERT-MIMIC | 0.6329 | 0.6475 | 0.6401 | 0.7871 | 0.7818 | 0.7845 |
| RoBERTa | 0.6312 | 0.6818 | 0.6556 | 0.7715 | 0.8155 | 0.7929 |
| RoBERTa-MIMIC | 0.6158 | 0.6766 | 0.6448 | 0.7711 | 0.8175 | 0.7936 |
| RoBERTa-MIMIC-Trial | 0.6209 | 0.6993 | 0.6578 | 0.7662 | 0.8333 | 0.7984 |
| ELECTRA | 0.5749 | 0.6498 | 0.6101 | 0.7369 | 0.8013 | 0.7678 |
| ELECTRA-MIMIC | 0.6086 | 0.6723 | 0.6389 | 0.7661 | 0.8149 | 0.7897 |
| Att-BiLSTM-CRF | 0.3586 | 0.3896 | 0.3735 | 0.7064 | 0.7344 | 0.7201 |
Table 4:
Performance of the transformer-based models vs. the baseline Att-BiLSTM-CRF model on FRD.
| Model | Strict Criterion | Relaxed Criterion | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| BERT | 0.7618 | 0.7919 | 0.7765 | 0.8972 | 0.9208 | 0.9089 |
| BERT-MIMIC | 0.7517 | 0.7949 | 0.7727 | 0.8895 | 0.9290 | 0.9088 |
| BLUEBERT | 0.7550 | 0.7948 | 0.7744 | 0.8933 | 0.9280 | 0.9103 |
| ALBERT | 0.7248 | 0.7671 | 0.7454 | 0.8644 | 0.9007 | 0.8822 |
| ALBERT-MIMIC | 0.7422 | 0.7682 | 0.7550 | 0.8820 | 0.8974 | 0.8896 |
| RoBERTa | 0.7610 | 0.8061 | 0.7829 | 0.8908 | 0.9333 | 0.9115 |
| RoBERTa-MIMIC | 0.7601 | 0.8026 | 0.7808 | 0.8918 | 0.9331 | 0.9120 |
| RoBERTa-MIMIC-Trial | 0.7680 | 0.8027 | 0.7849 | 0.9028 | 0.9290 | 0.9157 |
| ELECTRA | 0.7444 | 0.7910 | 0.7670 | 0.8840 | 0.9254 | 0.9042 |
| ELECTRA-MIMIC | 0.7683 | 0.7903 | 0.7792 | 0.9034 | 0.9185 | 0.9109 |
| Att-BiLSTM-CRF | 0.6799 | 0.7199 | 0.6993 | 0.8692 | 0.9039 | 0.8862 |
Table 5 and Table 6 shows the model performance by entity type on the best-performing RoBERTa-MIMIC-Trial model. In general, the entity types with more instances have better performance, while the entity types with the lowest F1-scores only have a handful of instances, i.e. “Pregnancy_considerations” in Chia and “technology_access” in FRD, respectively. When comparing the performance by entity type of the RoBERTa-MIMIC-Trial model to other models, we found that it significantly improved the performance on certain entity types such as Measurement, Procedure, Value in Chia and ethnicity, technology_access in FRD.
Table 5:
Performance of the best performing model (i.e., RoBERTa-MIMIC-Trial) by entity types on Chia.
| Model | Strict Criterion | Relaxed Criterion | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| Overall | 0.6209 | 0.6993 | 0.6578 | 0.7662 | 0.8333 | 0.7984 |
| Condition | 0.7324 | 0.7878 | 0.7591 | 0.8721 | 0.9144 | 0.8928 |
| Device | 0.4667 | 0.6829 | 0.5545 | 0.6167 | 0.8293 | 0.7073 |
| Drug | 0.6949 | 0.7910 | 0.7398 | 0.8418 | 0.9100 | 0.8746 |
| Measurement | 0.6127 | 0.6893 | 0.6487 | 0.7937 | 0.8464 | 0.8192 |
| Mood | 0.2727 | 0.2449 | 0.2581 | 0.3636 | 0.3265 | 0.3441 |
| Observation | 0.2933 | 0.2444 | 0.2667 | 0.4333 | 0.3556 | 0.3906 |
| Person | 0.6914 | 0.8643 | 0.7683 | 0.7257 | 0.8929 | 0.8007 |
| Pregnancy_considerations | 0.0000 | 0.0000 | 0.0000 | 0.3784 | 0.4444 | 0.4088 |
| Procedure | 0.5012 | 0.6375 | 0.5612 | 0.6560 | 0.8031 | 0.7222 |
| Temporal | 0.4800 | 0.6316 | 0.5455 | 0.6514 | 0.8008 | 0.7184 |
| Value | 0.7000 | 0.7278 | 0.7136 | 0.8324 | 0.8685 | 0.8500 |
Table 6:
Performance of the best performing model (i.e., RoBERTa-MIMIC-Trial) by entity types on FRD.
| Model | Strict Criterion | Relaxed Criterion | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| Overall | 0.7680 | 0.8027 | 0.7849 | 0.9028 | 0.9290 | 0.9157 |
| age | 0.9696 | 0.9733 | 0.9714 | 0.9696 | 0.9733 | 0.9714 |
| allergy_name | 0.8553 | 0.7010 | 0.7705 | 0.9434 | 0.7680 | 0.8467 |
| bmi | 0.9130 | 0.8750 | 0.8936 | 0.9565 | 0.9583 | 0.9574 |
| cancer | 0.7436 | 0.7835 | 0.7630 | 0.8983 | 0.9250 | 0.9114 |
| chronic_disease | 0.7297 | 0.7789 | 0.7535 | 0.8892 | 0.9304 | 0.9093 |
| clinical_variable | 0.7888 | 0.7633 | 0.7759 | 0.9151 | 0.8729 | 0.8935 |
| contraception_consent | 0.6473 | 0.7929 | 0.7128 | 0.8164 | 0.9349 | 0.8717 |
| ethnicity | 1.0000 | 0.7500 | 0.8571 | 1.0000 | 0.7500 | 0.8571 |
| gender | 0.9333 | 0.9385 | 0.9359 | 0.9861 | 0.9944 | 0.9902 |
| language_fluency | 0.8222 | 0.7872 | 0.8043 | 0.9778 | 0.9574 | 0.9675 |
| lower_bound | 0.8230 | 0.8623 | 0.8422 | 0.9163 | 0.9565 | 0.9360 |
| pregnancy | 0.8228 | 0.8698 | 0.8457 | 0.9489 | 0.9968 | 0.9723 |
| technology_access | 0.3846 | 0.4545 | 0.4167 | 1.0000 | 1.0000 | 1.0000 |
| treatment | 0.7178 | 0.7747 | 0.7451 | 0.8745 | 0.9223 | 0.8978 |
| upper_bound | 0.8208 | 0.8417 | 0.8311 | 0.9353 | 0.9555 | 0.9453 |
In fact, when comparing the performance between the two datasets, models trained with FRD have consistently better performance than models trained with Chia data, even in the Att-BiLSTM-CRF baseline models (i.e., an F1 of 0.8862 in FRD vs. an F1 of 0.7201 in Chia at relax-level). This may be attributed to the fact that FRD has much more data than Chia and deep learning based NER models require more data to achive optimal performance.
5.2. Error Analysis
Table 7 and Table 8 show the number of entities recognized in terms of strict match, relaxed match, and missed match by entity types in Chia and FRD for the best performing model (i.e. RoBERTa-MIMIC-Trial). Compared to FRD, the Chia data have considerably higher proportion of missed matched entities of all entity types, especially for Observation, Mood and Pregnancy_considerations. Further investigation was conducted to analyze the entities predicted in terms of relaxed match and missed match. Investigation of the incorrectly predicted entities revealed that most Observation and Mood entities in Chia are phrases that cannot be used to find patients in EHRs (i.e., uncomputable), while most Pregnancy_considerations entities contain long text that could be broken into multiple entities of different types. Most cases of miss-matched Observation and Mood entities were entities not recognized by the model, as illustrated in examples in Figure 1. Nonetheless, since these uncomputable entities missed by the model will not be used for developing database queries for finding potentially eligible patients, they have little impact on the utility of our approach for subsequent cohort identification. In other entity types, entities with long text spans or entities with a mixture of alphabets, numbers, and punctuations tend to be missed by the model, which is mainly due to the inherent limitation of NER models.
Table 7:
Number of entities recognized in terms of strict match, relaxed match and missed match by entity types in Chia for the best performing model (i.e. RoBERTa-MIMIC-Trial).
| Entity Type | Strict | Relaxed | Missed | Total |
|---|---|---|---|---|
| Condition | 865 | 139 | 94 | 1,098 |
| Value | 238 | 46 | 43 | 327 |
| Procedure | 204 | 53 | 63 | 320 |
| Drug | 246 | 37 | 28 | 311 |
| Measurement | 193 | 44 | 43 | 280 |
| Temporal | 168 | 45 | 53 | 266 |
| Observation | 44 | 20 | 116 | 180 |
| Person | 121 | 4 | 15 | 140 |
| Mood | 12 | 4 | 33 | 49 |
| Device | 28 | 6 | 7 | 41 |
| Pregnancy_considerations | 0 | 8 | 10 | 18 |
| Sum | 2,119 | 406 | 505 | 3,030 |
Table 8:
Number of entities recognized in terms of strict match, relaxed match and miss match predictions by entity types in FRD for the best performing model (i.e. RoBERTa-MIMIC-Trial).
| Entity Type | Strict | Relaxed | Missed | Total |
|---|---|---|---|---|
| treatment | 2,482 | 473 | 249 | 3,204 |
| chronic_disease | 2,036 | 396 | 182 | 2,614 |
| clinical_variable | 1,087 | 156 | 181 | 1,424 |
| lower_bound | 1,190 | 130 | 60 | 1,380 |
| upper_bound | 1,154 | 156 | 61 | 1,371 |
| cancer | 731 | 132 | 70 | 933 |
| gender | 336 | 20 | 2 | 358 |
| pregnancy | 274 | 40 | 1 | 315 |
| age | 255 | 0 | 7 | 262 |
| allergy_name | 136 | 13 | 45 | 194 |
| contraception_consent | 134 | 24 | 11 | 169 |
| language_fluency | 37 | 8 | 2 | 47 |
| bmi | 21 | 2 | 1 | 24 |
| technology_access | 5 | 6 | 0 | 11 |
| ethnicity | 6 | 0 | 2 | 8 |
| Sum | 9,884 | 1,556 | 874 | 12,314 |
Figure 1:
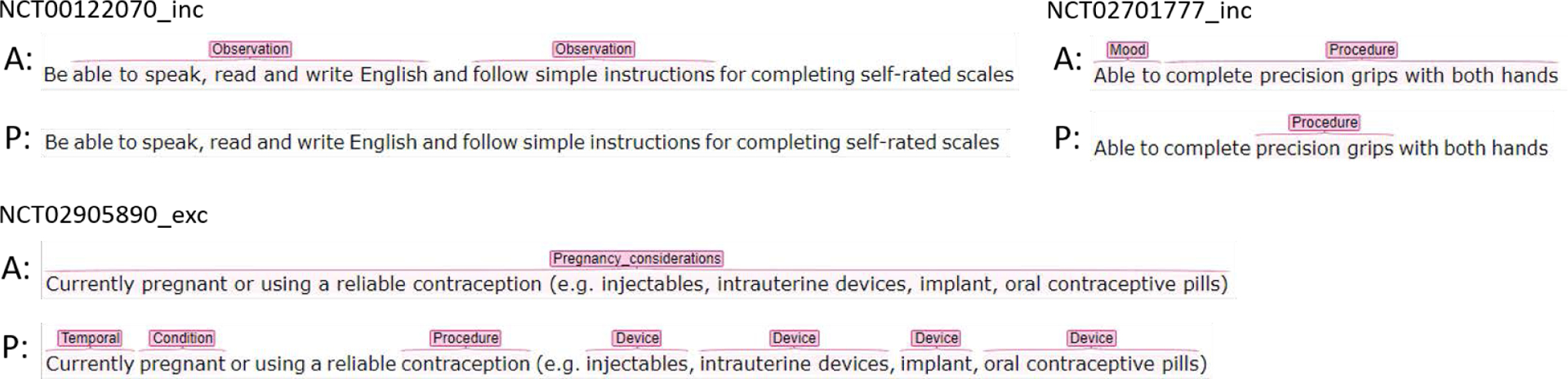
Examples of missed match predictions in Chia for the best performing model (i.e. RoBERTa-MIMIC-Trial). (A: Chia Annotation, P: Prediction)
When we looked at the entities recognized using relaxed match in Chia, there are 4 major types of relaxed match predictions, as illustrated by the examples in Figure 2. The most frequent type is that the model recognized part of the annotated span as an entity and missed the qualifiers. For example, in inclusion criterion of trial NCT00317148, the condition “hot flushes” was recognized but its qualifier “moderate to severe” was not. The second type of relaxed match is that the predicted entity span is longer than the span of the corresponding annotated entity, as shown in the exclusion criterion of trial NCT01098383. In the third type, the model split the annotated entity into 2 or more entities of the same entity type. In the exclusion criterion from trial NCT00317148, the annotated entity “Body mass index (BMI)” was recognized as two entities of Measurement. In this case, the annotated entities usually contain punctuations such as parentheses or slash. The fourth type of relaxed match is that the annotated entity is predicted as entities of multiple entity types which contains at least one entity of the same type as the annotated entity. This can be seen in the example of an inclusion criterion from trial NCT01483118. In this case, the annotated long entity was often recognized as multiple entities, possibly of different types. Other relaxed match types include cases where two or more consecutive entities of the same type were recognized as one entity, or a combination of two or more of the 4 aforementioned relaxed match types.
Figure 2:
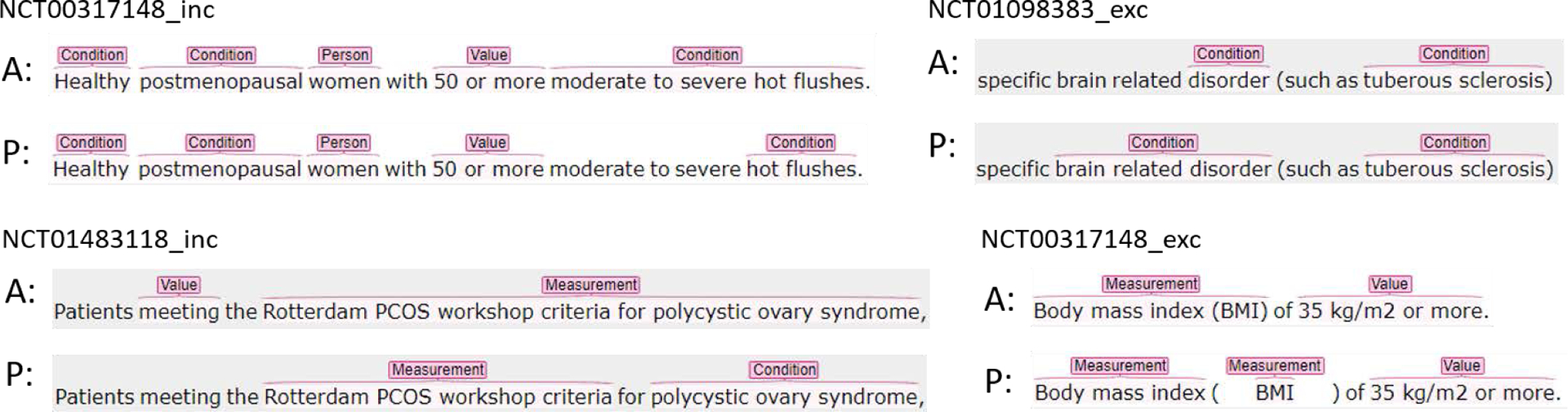
Examples of relaxed match predictions in Chia for the best performing model (i.e. RoBERTa-MIMIC-Trial). (A: Chia Annotation, P: Prediction)
6. DISCUSSION AND CONCLUSIONS
In this study, we evaluated 4 SOTA transformer-based NER models for parsing clinical trial eligibility criteria using two recent publicly available datasets, Chia and FRD. Our experiments showed that transformer-based models outperform the baseline Att-BiLSTM-CRF model (i.e., the current STOA in eligibility criteria parsing) in all evaluation metrics. Transformer-based models further pretrained with relevant clinical corpora such as clinical notes from MIMIC-III can improve their performance. Models further pretrained with eligibility criteria extracted from clinical trials can help to improve their performance further. These findings will advance the SOTA in the development of a reliable NLP-assisted pipeline for automated electronic screening. Further, a fully end-to-end solution that can automatically translate free-text eligibility criteria to database queries may not be operational currently, given the ambiguity in both the free-text eligibility criteria and the EHR databases. For example, translating the criterion “Diagnosed with COVID-19 pneumonia by RT-PCR” into database queries requires us to know (1) how COVID-19 information of a patient is recorded in EHRs (i.e., represented by a diagnostic code and/or a positive PCR test), and (2) the temporal constraints of the criterion (i.e., diagnosed before the time of screening), both of which cannot be extracted from parsing the criterion alone. Thus, merely mapping COVID-19 to “Condition” domain and PCR to “Measurement” domain in OMOP will unlikely yield an accurate database query. Future investigations should not merely focus on the performance of the NLP pipeline, but how to consider human-in-loop to build a realistic and useful workflow to facilitate clinical trial investigators. All the codes and detailed results of this study can be found in https://github.com/ctgatecci/Clinical-trial-eligibility-criteria-NER.
7. ACKNOWLEDGMENTS
This study was partially supported by the National Institute on Aging (NIA) of the National Institutes of Health (NIH) under Award Number R21AG061431; and in part by Florida State University-University of Florida Clinical and Translational Science Award funded by National Center for Advancing Translational Sciences under Award Number UL1TR001427.
Contributor Information
Jinfeng Zhang, Florida State University.
Jiang Bian, University of Florida.
Zhe He, Florida State University.
REFERENCES
- [1].Adler-Milstein Julia and Jha Ashish K. 2017. HITECH Act drove large gains in hospital electronic health record adoption. Health Affairs 36, 8 (2017), 1416–1422. [DOI] [PubMed] [Google Scholar]
- [2].Clark Kevin, Luong Minh-Thang, Le Quoc V, and Manning Christopher D. 2020. Electra: Pre-training text encoders as discriminators rather than generators. arXiv preprint arXiv:2003.10555 (2020). [Google Scholar]
- [3].Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. 2018. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 (2018). [Google Scholar]
- [4].Fu Sunyang, Chen David, He Huan, Liu Sijia, Moon Sungrim, Peterson Kevin J., Shen Feichen, Wang Liwei, Wang Yanshan, Wen Andrew, Zhao Yiqing, Sohn Sunghwan, and Liu Hongfang. 2020. Clinical concept extraction: A methodology review. Journal of Biomedical Informatics 109 (September. 2020), 103526. 10.1016/j.jbi.2020.103526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hao Tianyong, Liu Hongfang, and Weng Chunhua. 2016. Valx: a system for extracting and structuring numeric lab test comparison statements from text. Methods of information in medicine 55, 3 (2016), 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Heller Caren, Balls-Berry Joyce E, Nery Jill Dumbauld, Erwin Patricia J, Littleton Dawn, Kim Mimi, and Kuo Winston P. 2014. Strategies addressing barriers to clinical trial enrollment of underrepresented populations: a systematic review. Contemporary clinical trials 39, 2 (2014), 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Johnson Alistair EW, Pollard Tom J, Shen Lu, Li-Wei H Lehman, Feng Mengling, Ghassemi Mohammad, Moody Benjamin, Szolovits Peter, Anthony Celi Leo, and Mark Roger G. 2016. MIMIC-III, a freely accessible critical care database. Scientific data 3, 1 (2016), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kang Tian, Zhang Shaodian, Tang Youlan, Hruby Gregory W, Rusanov Alexander, Elhadad Noémie, and Weng Chunhua. 2017. EliIE: An open-source information extraction system for clinical trial eligibility criteria. Journal of the American Medical Informatics Association 24, 6 (2017), 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kury Fabrício, Butler Alex, Yuan Chi, Fu Li-heng, Sun Yingcheng, Liu Hao, Sim Ida, Carini Simona, and Weng Chunhua. 2020. Chia, a large annotated corpus of clinical trial eligibility criteria. Scientific data 7, 1 (2020), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lan Zhenzhong, Chen Mingda, Goodman Sebastian, Gimpel Kevin, Sharma Piyush, and Soricut Radu. 2019. Albert: A lite bert for self-supervised learning of language representations. arXiv preprint arXiv:1909.11942 (2019). [Google Scholar]
- [11].Liu Yinhan, Ott Myle, Goyal Naman, Du Jingfei, Joshi Mandar, Chen Danqi, Levy Omer, Lewis Mike, Zettlemoyer Luke, and Stoyanov Veselin. 2019. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692 (2019). [Google Scholar]
- [12].Peng Yifan, Yan Shankai, and Lu Zhiyong. 2019. Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. arXiv:1906.05474 [cs] (June 2019). http://arxiv.org/abs/1906.05474 arXiv: 1906.05474. [Google Scholar]
- [13].Thadani Samir R, Weng Chunhua, Thomas Bigger J, Ennever John F, and Wajngurt David. 2009. Electronic screening improves efficiency in clinical trial recruitment. Journal of the American Medical Informatics Association 16, 6 (2009), 869–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tseo Yitong, Salkola MI, Mohamed Ahmed, Kumar Anuj, and Abnousi Freddy. 2020. Information extraction of clinical trial eligibility criteria. arXiv preprint arXiv:2006.07296 (2020). [Google Scholar]
- [15].Tu Samson W, Peleg Mor, Carini Simona, Bobak Michael, Ross Jessica, Rubin Daniel, and Sim Ida. 2011. A practical method for transforming free-text eligibility criteria into computable criteria. Journal of biomedical informatics 44, 2 (2011), 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Unger Joseph M, Cook Elise, Tai Eric, and Bleyer Archie. 2016. The role of clinical trial participation in cancer research: barriers, evidence, and strategies. American Society of Clinical Oncology Educational Book 36 (2016), 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Weng Chunhua, Wu Xiaoying, Luo Zhihui, Boland Mary Regina, Theodoratos Dimitri, and Johnson Stephen B. 2011. EliXR: an approach to eligibility criteria extraction and representation. Journal of the American Medical Informatics Association 18, Supplement_1 (2011), i116–i124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang Xi, Bian Jiang, Hogan William R, and Wu Yonghui. 2020. Clinical concept extraction using transformers. Journal of the American Medical Informatics Association 27, 12 (October. 2020), 1935–1942. 10.1093/jamia/ocaa189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yuan Chi, Ryan Patrick B, Ta Casey, Guo Yixuan, Li Ziran, Hardin Jill, Makadia Rupa, Jin Peng, Shang Ning, Kang Tian, et al. 2019. Criteria2Query: a natural language interface to clinical databases for cohort definition. Journal of the American Medical Informatics Association 26, 4 (2019), 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]


